Abstract
Piriformospora indica, a root endophytic fungus, has been shown to enhance biomass production and confer tolerance to various abiotic and biotic stresses in many plant hosts. A growth chamber experiment of soybean (Glycine max) colonized by P. indica compared to uninoculated control plants showed that the fungus significantly increased shoot dry weight, nutrient content, and rhizobial biomass. RNA-Seq analyses of root tissue showed upregulation of 61 genes and downregulation of 238 genes in colonized plants. Gene Ontology (GO) enrichment analyses demonstrated that upregulated genes were most significantly enriched in GO categories related to lignin biosynthesis and regulation of iron transport and metabolism but also mapped to categories of nutrient acquisition, hormone signaling, and response to drought stress. Metabolic pathway analysis revealed upregulation of genes within the phenylpropanoid and derivative pathways such as biosynthesis of monolignol subunits, flavonoids and flavonols (luteolin and quercetin), and iron scavenging siderophores. Highly enriched downregulated GO categories included heat shock proteins involved in response to heat, high-light intensity, hydrogen peroxide, and several related to plant defense. Overall, these results suggest that soybean maintains an association with this root endosymbiotic fungus that improves plant growth and nutrient acquisition, modulates abiotic stress, and promotes synergistic interactions with rhizobia.
Introduction
Microbial symbionts, such as arbuscular mycorrhizal fungi (AMF), have the ability to promote the growth of plants through nutrient acquisition, growth promotion, and protection against biotic and abiotic stresses. Less well known are beneficial root associates belonging to the basidiomycete order Sebacinales, a ubiquitous and ecologically diverse group of fungi that universally form endophytic, and in some cases mycorrhizal-like associations, with a wide range of plants species1–3. They have been shown to increase nutrient acquisition4,5, while also improving resistance to a variety of biotic and abiotic stresses6.
Piriformospora indica, a root endophytic Sebacinaceae, was discovered in the Thar Desert of India7 and colonizes a wide range of plant species, forming intracellular pear shaped chlamydospores within roots8–10. It primarily colonizing the outer root cortex both inter- and intracellularly and generally requires host cell death for proliferation10. In barley, the fungus displays a biphasic lifestyle, initially colonizing living roots as a biotroph and subsequently initiating cell death that leads to proliferation and saprotrophic growth within dead root cells10,11. However, unlike traditional hemibiotrophs, cell death does not appear to harm the plant host and instead colonization by the fungus confers many benefits12, ranging from increased aboveground growth and biomass8,13–16 to increased flower and fruit production17,18 to resistance to various abiotic (e.g. salt, water, heat stress)19–22 and biotic (e.g. herbivores, and microbial pathogens) stresses13,23–25.
The mechanisms by which P. indica confers host benefits are not fully understood. The fungus has been shown to increase uptake of nutrients such as N and P as well as several micronutrients26–28 and impacts many other plant metabolic processes. The production and signaling of the phytohormones ethylene, jasmonic acid (JA), gibberellic acid (GA), salicylic acid (SA) and abscisic acid (ABA) have all been shown to be impacted by P. indica29–31. Changes in levels of these hormones likely mediate abiotic stress responses13,32 as well as regulate biotic responses such as induced and systemic plant defense responses, microbe-associated molecular patterns (MAMP) triggered immunity6,29,33, and autoregulation of mutualistic symbionts34–36. However, P. indica must maintain a complex communication with its host to both allow its own colonization while also enhancing defense responses against pathogens. Studies have also revealed that fungal colonization may increase plant production of defensive secondary metabolites like artemisinin37–39, bacoside40,41, forskolin18,42, volatile oils such as curcumin43, and lignans such as the anticancer podophyllotoxins that may derive from the phenylpropanoid pathway44–47. The phenylpropanoid pathway also provides precursors for many other plant secondary metabolite pathways, including synthesis of flavonoids and coumarins, among others48, and is often induced by plants in response to pathogens.
The beneficial effects of P. indica have been extensively researched using the model plant Arabidopsis thaliana and a number of important crop species including barley, wheat, and corn19,49–52, as well as angiosperms such as tomato17. While many studies have used molecular genetic approaches to investigate targeted sets of genes, genome scale transcriptional approaches to characterize host responses have only been conducted in Arabidopsis and barley29,53. Research investigating interactions between P. indica and legume species has been limited to a handful of studies on soybean25,54 and a few other legume hosts55,56. Soybean (Glycine max (L.) Merr.) is the most widely planted oil crop and is also a valuable source of protein in many parts of the world. Soybean is also known to form root symbioses with other microbes such as AMF and rhizobia, which also have been demonstrated to enhance tolerance against various abiotic and biotic stresses57,58. Here we report the first whole genome-scale transcriptional study of P. indica colonizing a legume host. Our results show many of the same plant growth responses observed in Arabidopsis and barley but also uncover responses unique to soybean, including potentially synergistic interactions with rhizobial symbionts. Our results also support findings that a mechanism of programmed cell death (PCD) distinct from the plant hypersensitivity response (HR) is involved in the symbiosis.
Results
Plant Growth Response and Nutrient Status
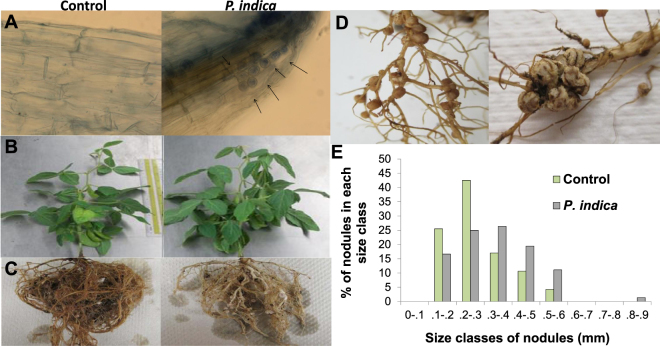
Soybean roots showed 41.47% colonization at 60 days after seeding in the P. indica inoculated plants, whereas the controls showed no colonization (Fig. 1A and Supplementary Table S1. Diagnostic pear-shaped spores of P. indica7 were observed under the microscope and chlamydospores were found as isolated spores, pairs, tetrads, long chains, or clusters (Fig. 1A). The shoot:root ratio and shoot dry weight both increased significantly as well as numbers of leaves (5.21%), flowers (27.23%), and pods (13.14%) at 60 days after seeding (Table 1). In contrast, the dry weights and volumes of roots decreased by 8.17% and 31.25%, respectively (Fig. 1C and Table 1). Thickening of both the primary and lateral roots was observed in the P. indica treatment (Fig. 1C). Colonization with P. indica also impacted nodule formation by rhizobia. While the number of nodules decreased slightly in the P. indica treatment (70.66 ± 12.42) compared to the control (96.33 ± 10.21), the ratio of dry weight nodules: dry weight roots (0.100 ± 0.013) was significantly larger than in the controls (0.043 ± 0.024 mm) (Table 1). The average diameter of nodules in the P. indica treatment was also observed to be slightly larger (treatment (0.29 ± 0.03 mm) than the control treatment (0.25 ± 0.04 mm), although this difference was not statistically significant (Fig. 1D, Table 1). A histogram of size classes of nodules showed that a larger proportion of nodules in the P. indica treatment fell into larger size classes (0.3 - 0.4, 0.4 - 0.5, and 0.5 - 0.6 mm) than those of the control (Fig. 1D and E).
Figure 1.
(A) Roots of control and P. indica treatments as seen under 40X magnification after staining with lactophenol cotton blue. Pear-shaped chlamydospores were observed in P. indica treatment (arrows) only while no chlamydospores was observed in the control. (B) Shoots of control and P. indica colonized plants. There was no significant difference in shoot lengths but the shoot dry weights increased in P. indica treatment. (C) Roots of control and P. indica colonized plants. The root dry weight decreased in P. indica colonized plants. (D) Nodules in control and P. indica treatments. Numbers of nodules decreased while average size increased in P. indica treatment.
Table 1.
Plant Growth and Nutrient Status.
| Control (C) | P. indica (Pi) | % increase of Pi/C | |
|---|---|---|---|
| Growth Parameters | |||
| Percent Colonization (%) | 0.00 ± 0.00 | 41.47 ± 7.12* | 41.47 |
| Dry shoot biomass(g) | 2.02 ± 0.45 | 2.28 ± 0.43* | 12.87 |
| Dry root biomass (g) | 0.90 ± 0.21 | 0.73 ± 0.17* | −18.89 |
| Root volume (mL) | 10.67 ± 1.15 | 7.33 ± 1.57* | −31.30 |
| Number of leaves | 31.00 ± 2.65 | 39.67 ± 3.06* | 27.96 |
| Number of branches | 11.00 ± 1.00 | 12.00 ± 2.65* | 9.09 |
| Number of flowers | 6.00 ± 2.64 | 9.33 ± 2.08* | 55.50 |
| Number of pods | 7.33 ± 2.08 | 8.00 ± 2.00 | 9.14 |
| Number of nodules | 96.33 ± 10.21 | 70.66 ± 12.42 | −26.65 |
| Dry nodule biomass (g) | 0.096 ± 0.038 | 0.147 ± 0.093 | 51.99 |
| Dry nodule biomass/dry root biomass (g) | 0.043 ± 0.024 | 0.101 ± 0.014* | 100.32 |
| Dry nodule biomass/dry shoot biomass (g) | 0.024 ± 0.009 | 0.050 ± 0.012* | 100.04 |
| Nutrient Status | |||
| Nitrogen (%) | 22.43 ± 0.13 | 23.71 ± 0.03* | 5.77 |
| Phosphorous (mg/kg) | 1493.33 ± 55.08 | 1663.33 ± 160.10* | 11.38 |
| Potassium (mg/kg) | 14728.00 ± 415.67 | 15730.33 ± 1615.09* | 6.81 |
The results show the mean and standard deviation calculated from three plant biological replicates. An asterisk (*) indicates significance at p = 0.05 using Dunnett’s multiple comparison tests between control and P. indica treatment.
The concentration of nitrogen (N), phosphorus (P), and potassium (K) increased significantly in shoots of P. indica colonized plants (Table 1). Micronutrients, including aluminum, manganese, zinc, and nickel as well as the beneficial nutrients including calcium, magnesium, sodium, and especially iron also increased significantly in colonized plants (Supplementary Table S2). The concentrations of boron, copper, cadmium, and chromium decreased, although not significantly (Supplementary Table S2).
RNA-Seq Analyses
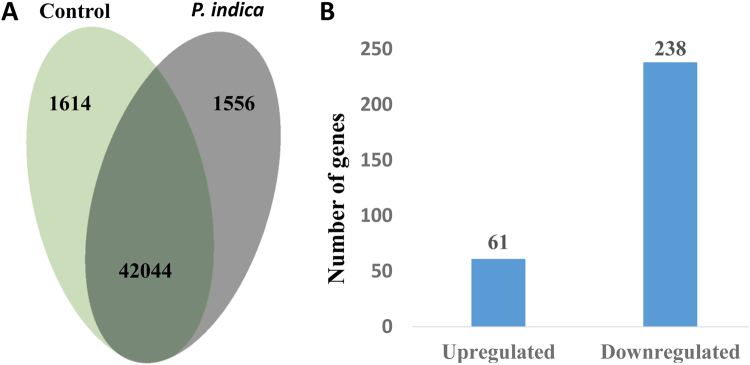
Over 90% of reads from each treatment aligned to the soybean Williams 82 genome a2.v1 and were evenly balanced across treatments with an average of 12 million reads per sample (Supplementary Table S3). We found that a large number of soybean genes, 42,044 out of a total of 56,044 models in Williams 82 a2.v1 annotations, or roughly 75% of annotated gene were expressed in both conditions, while 1556 were uniquely expressed in the P. indica treatment and 1614 were uniquely expressed in the control (Fig. 2). Differential expression of several genes was validated using qRT-PCR (Supplementary Table S4). In the differential expression analysis, 61 genes were significantly upregulated while 238 were downregulated (Fig. 2 and Supplementary Tables S5 and S6). Among the most highly upregulated genes, many belonged to functional categories known to be upregulated by P. indica in other crops, including hormone metabolism and nutrient acquisition (Table 2). Other categories of highly upregulated genes included lignin and cell-wall metabolism, amino acid metabolism, and sugar metabolism. (Table 2) Among downregulated genes, a majority were heat shock proteins with putative functions in abiotic stress response but also included genes involved in amino acid metabolism, cell-wall metabolism, and plant defense regulators of PCD and response to oxidative stress (Table 3).
Figure 2.
(A) Total number of genes expressed in control only (light green), P. indica treatment only (gray), and genes expressed in both conditions (dark green). (B) Graph of the number of genes upregulated and down regulated in P. indica colonized roots compared to the control at FDR = 0.05.
Table 2.
Twenty most highly upregulated genes by fold-change in P. indica treatment.
| Soybean ID | Arabidopsis homolog | Log2 FoldChange | Description |
|---|---|---|---|
| Amino Acid Metabolism | |||
| Glyma.04G020200 | AT5G11880 | 2.38 | Pyridoxal-dependent decarboxylase family protein |
| Glyma.03G189800 | AT2G36570 | 3.01 | Leucine-rich repeat protein kinase family protein |
| Cell Wall Metabolism | |||
| Glyma.18G263700 | AT5G54160 | 2.41 | O-methyltransferase 1 |
| Glyma.06G286200 | AT4G35160 | 2.77 | O-methyltransferase family protein |
| Glyma.01G211000 | AT1G12240 | 2.11 | Glycosyl hydrolases family 32 protein |
| Hormone Metabolism | |||
| Glyma.18G250100 | AT2G04160 | 2.07 | Subtilisin-like serine endopeptidase family protein |
| Glyma.13G183600 | AT1G61120 | 2.09 | terpene synthase 04 |
| Glyma.03G168000 | AT1G15520 | 2.31 | pleiotropic drug resistance 12 |
| Glyma.17G092800 | AT5G14920 | 2.38 | Gibberellin-regulated family protein |
| Glyma.11G184800 | AT2G04160 | 2.89 | Subtilisin-like serine endopeptidase family protein |
| Nutrient Acquisition | |||
| Glyma.12G178500 | AT2G28160 | 2.01 | FER-like regulator of iron uptake |
| Glyma.10G139800 | AT3G62020 | 2.03 | germin-like protein 10 |
| Glyma.14G011800 | AT5G55970 | 2.10 | RING/U-box superfamily protein |
| Glyma.13G191400 | AT5G07010 | 2.31 | sulfotransferase 2 A |
| Glyma.07G014500 | AT4G25150 | 2.82 | HAD superfamily, subfamily IIIB acid phosphatase |
| Glyma.08G076000 | AT3G43660 | 2.71 | Vacuolar iron transporter (VIT) family protein |
| Lipid Metabolism | |||
| Glyma.17G139900 | AT4G12510 | 2.22 | Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein |
| Sugar Metabolism | |||
| Glyma.03G229800 | AT1G56600 | 2.82 | galactinol synthase 2 |
| Glyma.17G242400 | AT4G25000 | 3.20 | alpha-amylase-like |
| Unknown | |||
| Glyma.10G224300 | NA | 2.39 | N/A |
| Glyma.11G213500 | AT5G37478 | 2.23 | TPX2 (targeting protein for Xklp2) protein family |
| Glyma.15G014500 | AT1G79960 | 2.22 | ovate family protein 14 |
Table 3.
Twenty most highly downregulated genes by fold-change in P. indica treatment.
| Soybean ID | Arabidopsis homologue | Log2 Fold Change | Description |
|---|---|---|---|
| Abiotic Stress Response | |||
| Glyma.08G212000 | AT1G52560 | −6.29 | HSP20-like chaperones superfamily protein |
| Glyma.06G202200 | AT1G74310 | −5.95 | heat shock protein 101 |
| Glyma.04G229800 | AT4G27670 | −5.79 | heat shock protein 21 |
| Glyma.14G099900 | AT5G12020 | −4.92 | 17.6 kDa class II heat shock protein |
| Glyma.06G134900 | AT4G27670 | −4.91 | heat shock protein 21 |
| Glyma.10G029600 | AT3G22830 | −4.90 | heat shock transcription factor A6B |
| Glyma.16G206200 | AT4G10250 | −4.66 | HSP20-like chaperones superfamily protein |
| Glyma.16G012000 | AT1G54050 | −4.46 | HSP20-like chaperones superfamily protein |
| Glyma.02G205600 | AT1G16030 | −4.45 | heat shock protein 70B |
| Glyma.10G176400 | AT4G10250 | −4.78 | HSP20-like chaperones superfamily protein |
| Amino Acid Metabolism | |||
| Glyma.08G321200 | AT5G10770 | −6.32 | Eukaryotic aspartyl protease family protein |
| Glyma.08G321500 | AT5G10770 | −4.97 | Eukaryotic aspartyl protease family protein |
| Glyma.10G184700 | AT2G38870 | −4.73 | Serine protease inhibitor, potato inhibitor I-type family protein |
| Cell Wall Metabolism | |||
| Glyma.05G065700 | AT4G17030 | −6.77 | expansin-like B1 |
| Glyma.17G147400 | AT4G17030 | −6.30 | expansin-like B1 |
| Glyma.17G147500 | AT4G17030 | −4.51 | expansin-like B1 |
| Plant Defense | |||
| Glyma.09G284700 | AT4G11290 | −5.11 | Peroxidase superfamily protein |
| Glyma.07G061500 | AT2G46240 | −6.38 | BCL-2-associated athanogene 6 |
| Unknown | |||
| Glyma.16G069500 | AT5G01750 | −5.80 | Protein of unknown function (DUF567) |
| Glyma.01G036300 | NA | −4.37 | |
GO Term Analyses and Visualization with REVIGO
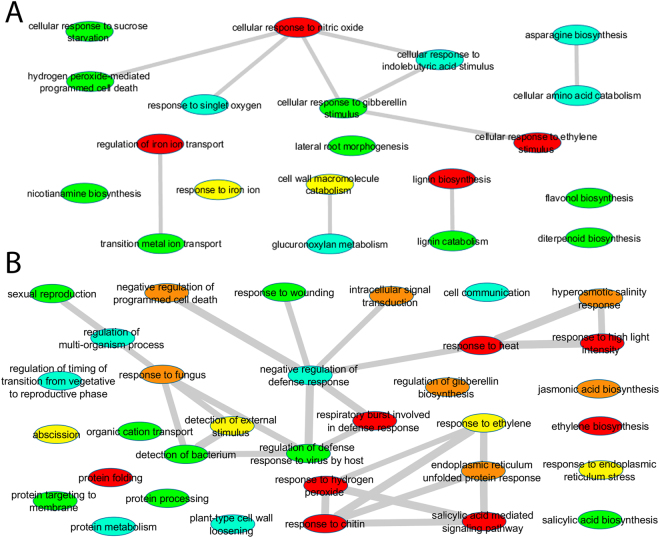
GO term enrichment analyses were performed using the Soybase (http://www.soybase.org) GO enrichment tool. GO categories were displayed as tree networks using ReviGO and were visualized in Cytoscape59–61. Upregulated genes showed the most significant (corrected p-value < 0.005) enrichment of the biological process categories of lignin biosynthesis (GO:0009809) (Fig. 3A and Supplementary Table S7). Other significantly (corrected p-value < 0.05) enriched categories included the regulation of iron transport (GO:0034756), cellular response to nitric oxide (GO:0071732) and ethylene stimulus (GO:0071369). GO categories that included upregulated genes but were not statistically enriched included cellular response to iron ion (GO:0071281), cell wall macromolecule catabolic process (GO:0016998), transition metal ion transport (GO:0000041), diterpenoid biosynthetic process (GO:0016102), flavonol biosynthesis (GO:0051555), response to sucrose starvation (GO:0043617), and hydrogen peroxide-mediated PCD (GO:0010421).
Figure 3.
Differentially expressed genes summarized as a tree network using REVIGO. (A) GO terms related to biological processes of upregulated genes in the comparison of control versus P. indica. (B) GO terms related to biological processes of downregulated genes in the comparison control versus P. indica. GO categories are represented by circles and are color coded according to corrected p-value for GO enrichment analysis. Red indicates categories that are significantly enriched at p < 0.05 while nonsignificant categories are color coded with corrected p < 0.5 = orange, p < 1 = yellow, p < 5 = green, and p < 10 = blue). Lines indicate connections between categories within the GO hierarchy and singletons have been grouped by similar function.
Downregulated genes that showed the most significant (corrected p-value < 0.0001) enrichment included biological processes related to response to heat (GO:0009408), response to high light intensity (GO:0009644), and response to hydrogen peroxide (GO:0042542) (Fig. 3B and Supplementary Table S8). Heat acclimation (GO:0010286), salicylic acid mediated signaling pathway (GO:0009863), protein folding (GO:0006457), and response to chitin (GO:0010200) were also highly statistically (corrected p-value < 0.003) enriched (Fig. 3B and Supplementary Table S8). Other significantly (corrected p-value < 0.05) enriched categories included several involved in plant defense, including respiratory burst involved in defense (GO:0002679), salicylic acid signaling (GO:0009863), and response to chitin (GO:0010200) as well as hormone metabolism including ethylene biosynthetic process (GO:0009693) and abscisic acid mediated signaling pathway GO:0009738). Downregulated GO categories that were not significantly enriched included others in plant defense or pathogen recognition, such as jasmonic acid (GO:0009867) mediated signaling, negative regulation of PCD (GO:0043069), regulation of plant-type hypersensitive response (GO:0010363), regulation of gibberellin biosynthesis (GO:0010370), response to endoplasmic reticulum stress (GO:0034976), response to fungus (GO:0009620), and response to bacterium (GO:0016045) (Fig. 3B and Supplementary Table S8). Other downregulated genes belonged to GO categories involved in abiotic stress responses including hyperosmotic salinity response (GO:0042538) (Fig. 3B and Supplementary Table S8).
Overall, these enriched GO categories suggest that, in response to P. indica, the soybean plant induces a growth response, increasing expression of genes in GO categories related to lignin biosynthesis, cell wall construction and remodeling, and iron uptake while downregulating genes involved in plant defense and heat shock proteins involved in response to several abiotic stresses (heat, high light, hydrogen peroxide, and salinity).
Metabolic Pathway Analysis
Differentially expressed genes and their associated log2 fold change values were mapped onto the SoyCyc metabolic pathway database. General categories of metabolism containing upregulated genes included anabolic processes involved in secondary metabolite biosynthesis, amino acid metabolism, fatty acid metabolism, hormone biosynthesis, amine and polyamine biosynthesis, and carbohydrate biosynthesis as well as catabolic processes of amino acid degradation and inorganic nutrient metabolism (Supplementary Table S9). Downregulated genes mapped to these same general categories as well as cell structure and aromatic compound biosynthesis and a number of catabolic processes such as carbohydrate, amino acid, fatty acid, secondary metabolite, amine/polyamine, and hormone degradation (Supplementary Table S9).
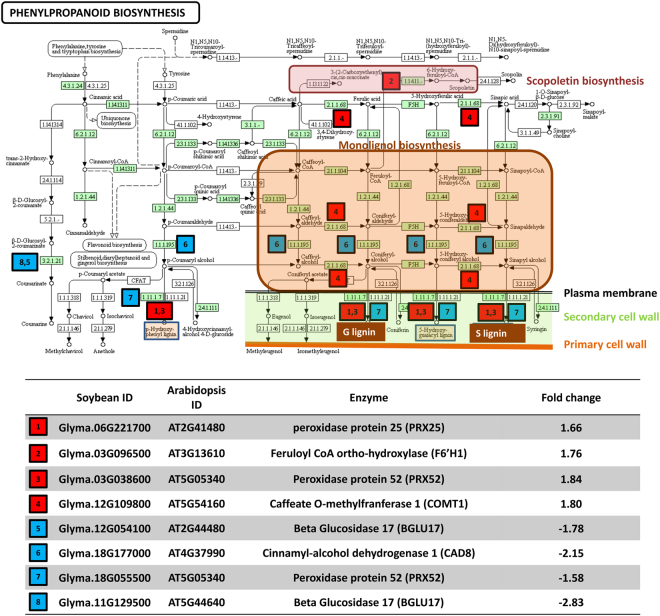
Among upregulated genes, several mapped to portions of the phenylpropanoid pathway responsible for production of monolignols and the iron scavenging compound scopoletin (Supplementary Table S9 and S10 and Fig. 4) as well as pathways derived from the phenylpropanoid pathway involved in biosynthesis of secondary metabolites such as flavonoids (luteolin, quercetin, leucodelphinidin, and leucocyanidin), resveratrol, phloridzin as well as furaneols and B series fagopyritols (Fig. 5 and Supplementary Tables S9 and S10). Upregulated genes also mapped to biosynthetic pathways of two terpenoid compounds, linalool and 1,3,5-trimethoxybenzene (Supplementary Table S9). Within primary metabolism, upregulated genes mapped to several fatty acid biosynthesis pathways (triacylglycerol and CDP-diacylglycerol), the carbohydrate biosynthetic pathway for stachyose, several amino acid pathways (citrulline, asparagine), and the biosynthesis of the amine/polyamine putrescine (Supplementary Tables S9 and S10). Other key pathways included inorganic nutrient metabolism, specifically phosphate acquisition through purple acid phosphatases62, and hormone metabolism (hydroxyjasmonate sulfate biosynthesis) (Supplementary Tables S9 and S10). Several catabolic pathways including betanidin, starch, chitin, glutamine, sucrose III, and superoxide radicals degradation also contained upregulated genes (Supplementary Tables S9 and S10).
Figure 4.
Effect of colonization with P. indica on the expression of genes within the phenylpropanoid biosynthetic pathway. Red colored squares indicate genes upregulated in the comparison of control versus P. indica; blue colored squares indicate down-regulated genes in this comparison. Homologs in Arabidopsis of all upregulated and downregulated soybean genes were mapped to the Arabidopsis KEGG pathway for phenylpropanoid biosynthesis144–146. Enzyme Commission (EC) codes colored in green boxes represent enzymatic steps present in soybean. Steps involved in biosynthesis of monolignol precursors sinapyl alcohol, coniferal alcohol, and 5-hydroxy-coniferal alcohol are shaded in the orange box. The final steps of production of G and S lignin (shaded brown box) are performed by peroxidases located within the plant cell wall. Biosynthesis of the iron-scavenging coumarin scopoletin is shaded in the pink box.
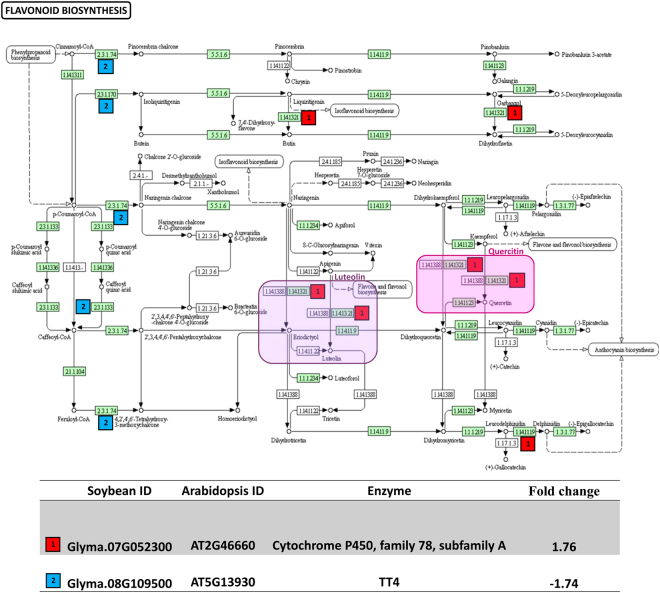
Figure 5.
Effect of colonization with P. indica on the expression of genes within the flavonoid biosynthetic pathway. Red colored squares indicate genes upregulated in the comparison of control versus P. indica; blue colored squares indicate down-regulated genes in this comparison. Homologs in Arabidopsis of all upregulated and downregulated soybean genes were mapped to the Arabidopsis KEGG pathway for flavonoid biosynthesis144–146. Enzyme Commission (EC) codes colored in green boxes represent enzymatic steps present in soybean. While several steps early in the pathway are downregulated, genes in steps leading to biosynthesis of luteolin (purple box) and quercetin (pink box) are upregulated.
Among downregulated genes, several also mapped to secondary metabolite pathways, including those involved in synthesis of kaemferol and dihydrokaemferol, the isoflavenoids pinkobanksin and dalochin, and a single step in the leucodelphinidin and leucocyanidin biosynthetic pathway (Fig. 5 and Supplementary Tables S9 and S10). Downregulated genes also mapped to primary metabolic anabolic pathways of fatty acid biosynthesis (very long chain fatty acids, sphingolipids, phosphatidamide, and triacylglycerol). Overall, more downregulated genes than upregulated genes mapped to catabolic pathways, including several carbohydrate (chitin, and sucrose III), amino acid (glutamine), fatty acid (triacylglycerol), secondary metabolite (betanidin, linamarin, and lotaustralin), and amine/polyamine degradation (spermidine) pathways.
Lignin and Cell Wall Biosynthesis
Lignin biosynthesis was one of the most highly enriched GO categories among upregulated genes in colonized plants and included genes involved in monolignol biosynthesis as well as several oxidative enzymes with potential roles in polymerization of monolignols to form lignin (Fig. 4 and Supplementary Table S7). The monomer subunits of lignin, or monolignols such as H (caffeic), C (coniferal), G (guaiacyl), and S (syringin) subunits, are produced through the phenylpropanoid pathway63 (Fig. 4). In our data, a number of genes involved in synthesis of monolignols, including a homolog of caffeate o-methyltransferase 1 (COMT; AT5G54160; Glyma.18G263700), involved in synthesis of precursors of G (coniferyl alcohol) and S (sinapyl alcohol) lignin, were upregulated (Fig. 4 and Supplementary Tables S5 and S7). During lignification of the cell wall, monolignols are transported across the plasma membrane where oxidative enzymes, including laccases and peroxidases, catalyze oxidative polymerization of the monolignol subunits63. The enriched GO category for lignin biosynthesis (GO:0009809) also included two peroxidases (Glyma.06G221700 and Glyma.03G038600) whose homologs in Arabidopsis (AT2G41480 and AT5G05340) are involved in plant cell wall biosynthesis (Fig. 4 and Supplementary Tables S5 and S7) and two chitinase-like proteins (ATG16920; Glyma.15G143600 and Glyma.09G038500) (Supplementary Table S7). Additionally, several members of the laccase-diphenol oxidase family (Glyma.14G062300 and Glyma.03G077900), with potential roles in polymerization of monolignols, were highly upregulated (Supplementary Tables S5 and S7). A mutant of the homolog of these laccases in Arabidopsis (LACCASE 4; AT2G38080) has shown defects in xylem lignin deposition64, specifically within protoxylem65.
In addition to lignin metabolism, upregulated genes mapped to several metabolic pathways involved in cell wall formation and remodeling. Genes involved in biosynthesis of suberin, a lipid compound found in the cell walls of root epidermal tissues that helps reduce water loss and entry of pathogenic fungi and bacteria66, were upregulated (Supplementary Tables S5 and S7). Synthesis of genes involved in biosynthesis of suberin precursors, such as octodecenedioate, and in biosynthesis of cutin, however, were downregulated (Supplementary Tables S6 and S8). Other upregulated genes with potential roles in cell wall remodeling included a xyloglucan endotransglucosylase/hydrolase 32 family gene (Supplementary Tables S5 and S7).
Flavonoid and Secondary Metabolite Biosynthesis
The phenylpropanoid pathway is also the starting point for the synthesis of many plant secondary compounds48. The first enzyme in the phenylpropanoid pathway, phenylalanine ammonium lyase, converts phenylalanine to p-coumaroyl CoA, a key branch-point leading to the synthesis of many different classes of plant secondary compounds including flavonoids, anthocyanins, tannins, coumarins, and lignans, among others48. Although a number of early steps in the flavonoid biosynthetic pathway and biosynthesis of some flavonoid compounds were downregulated, cytochrome P450 enzymes involved in production of the flavonoid compounds luteolin and quercetin were significantly upregulated (Fig. 5 and Supplementary Tables S5, S7, and S9). Upregulated genes also mapped to pathways involved in biosynthesis of aromatic and flavonoid related compounds derived from the phenylpropanoid pathway (1, 3, 5 trimethoxybenzene, furaneols) and the sugar-derived secondary metabolite B-type fagopyritols (Supplementary Table S9). Other classes of upregulated secondary metabolite genes included a terpene synthase (GES; AT1G61120; Glyma.13g183600), involved in biosynthesis of the volatile monoterpene alcohol linalool (Supplementary Tables S5 and S7).
Nutrient Acquisition
Colonization with P. indica increased shoot content of N, P, and K (Table 1). Upregulated genes mapped to the purple acid phosphatase (PAP) pathway62 (Supplementary Tables S7 and S9), known to be involved in phosphorus solubilization and acquisition67. The homolog of an Arabidopsis acid phosphatase gene (AT4G25150; Glyma.07G014500) that is induced by phosphate starvation and oxidative stress conditions68, was upregulated in colonized plants (Supplementary Tables S5 and S7). In uptake of N, GmSAT1 (AT4G37850; Glyma.15G061400), an ammonium transporter identified as being involved in transfer of ammonium from rhizobia to plants69, was not differentially expressed in our data (Supplementary Tables S5 and S6). In contrast, three high affinity ammonium transporters (AT5G60770, AT2G382901, and AT2G38290; Glyma.13G323800, Glyma.07G153800 and Glyma.01G123400, respectively) were downregulated (Supplementary Tables S6 and S8). Other sources of N include urea and amino acids. One urea transporter, an aquaporin called delta tonoplast integral protein (δ-TIP) (AtTIP2; 1; AT3G16240; Glyma.07g018000), was upregulated in the P. indica treatment70 (Supplementary Tables S5). No genes related to K uptake were differentially expressed in our dataset.
Iron is a limiting nutrient for growth of most organisms, including plants71,72, and also serves as a component of many enzymes involved in lignin biosynthesis73. The significantly enriched GO category of iron transport (GO:0034756) included two FER-like regulators of iron uptake (Glyma.13g322100 and Glyma.12g178500), both homologs of the FIT1 (AT2G28160) transcription factor that controls many genes involved in iron chelation and transport (Supplementary Table S7)74. A homolog of nicotianamine synthase (NAS1; AT5G04950; Glyma.15g251300)75, responsible for biosynthesis of the chelating agent nicotianamine which transports metal ions including iron76, was one of the most highly upregulated genes (Supplementary Tables S5 and S10). Nicotianamine is also an intermediate in the biosynthesis of the iron scavenging siderophore 2-hydroxymugineic acid67 (Fig. 4). Similarly, homologs of feruloyl-CoA 6-Hydroxylase 1 (F6′H1; AT3G13610; Glyma.03g096500 and Glyma.07g124400), were upregulated (Fig. 4 and Supplementary Table S5). This gene is induced under iron deficiency and is required for the biosynthesis of fluorescent coumarins and other Fe (III) mobilizing compounds such as scopoletin, which are released into the rhizosphere for iron scavenging77.
Hormone Metabolism
Upregulation of genes involved in abscisic acid, auxin, and gibberellin synthesis, signaling, or transport was observed. A membrane bound ATP binding cassette transporter that functions in the uptake of ABA into plant cells78,79 (ABCG40; AT1G15520; Glyma.03g168000) was upregulated (Supplementary Tables S5 and S7). Additionally, several ABA responsive genes with potential roles in drought tolerance, including a galactinol synthase (GolS2; AT1G56600; Glyma. 03g229800) and a homolog of a glycosyl hydrolase 32 (GH32) family protein sucrose vacuolar invertase (ATbetaFruct4; AT1G12240; Glyma.01g211000), were also upregulated (Supplementary Tables S5 and S7). An overexpression mutant of ATbetaFruct4 has been shown to be highly expressed in leaf stomatal guard cells and to contribute to increased drought tolerance in Arabidopsis80.
Although not significantly enriched, several upregulated genes belonged to GO categories cellular response to gibberellin stimulus (GO:0071370), gibberellic acid mediated signaling pathway (GO:0009740), and response to gibberellin stimulus (GO:0009739) (Fig. 3 and Supplementary Table S7). In addition to ATbetaFruct4 (AT1G12240; Glyma.01g211000) discussed above, these included homologs of the GA regulated gene GASA14 (AT5G14920; Glyma.17g092800) (Supplementary Tables S5 and S7). In contrast, a negative transcriptional regulator of GA biosynthesis, termed dwarf and delayed flowering phenotype (DDF1; AT1G12610; Glyma.01g216000 and Glyma.05g049900) was down-regulated (Supplementary Tables S5 and S7).
A number of auxin responsive genes with potential roles in shaping root development and architecture were also upregulated by P. indica, including two genes within the GO category lateral root morphogenesis (GO:0010102), Glyma.11g184800 and Glyma.18G250100, whose closest homolog in Arabidopsis is auxin-induced in root cultures 3 (AIR3; AT2G04160) (Supplementary Tables S5 and S7). AIR3 is a serine-peptidase that is localized to the cell wall and the extracellular region and is thought to have a role in remodeling of the cell wall to allow for lateral root morphogenesis81. An auxin transporter homologous to ABCG37 (AT3G53480; Glyma.17G034700), involved in distribution and homeostasis of auxin in roots and negative regulation of polar auxin transport inhibitors, was also upregulated (Supplementary Tables S5 and S7). ABCG37 shows broad substrate specificity for various auxin related compounds, including synthetic auxins and auxin transport inhibitors (IBA)82–84, and can transport caffeic acid and chlorogenic acid85 as well as iron scavenging compounds such as scopoletin85, and thus may also play a role in Fe uptake.
Abiotic Stress Responses
Although our experiment did not expose plants to high salinity or osmotic stress, several genes induced by these stresses were observed to be upregulated in response to colonization by P. indica. In addition to homologs of the two ABA-inducible drought response genes discussed above (GolS2 and ATbetaFruct4), a homolog of the SNF1-related protein kinase (SnRK2.4; AT1G10940; Glyma.08g127000), whose overexpression showed enhanced survival rates under low water potential potentially due to their stronger ability to retain water86, was also upregulated (Supplementary Table S5).
Among the most highly downregulated and enriched GO categories were response to heat (GO:0009408), response to high light intensity (GO:0009644), and response to hydrogen peroxide (GO:0042542) (Fig. 3B and Supplementary Table S8). All of these categories included a large and diverse number of heat-shock family proteins and molecular chaperones as well as trypsin and other protease inhibitors (Supplementary Table S8). Several proteins involving in sensing and signaling high light or high temperature were also downregulated, including a homolog of thermosensitive gametophytic male sterility TMS1 (AT3G08970; Glyma.07g152900), which contributes to normal pollen tube development at high-temperatures (Supplementary Tables 6 and 8).
Biotic Stress Response
A number of genes involved in the plant immune response were downregulated in response to P. indica, including several MAMP responsive genes, a homolog of AtWRKY33 (AT2G38470; Glyma.01G128100) as well as two other WRKY transcription factors, WRKY40 (AT1G80840; Glyma.13G370100) and WRKY 41 (AT4G11070; Glyma.08G021900) (Supplementary Tables S6 and S8). WRKY 40, in particular, belongs to the GO category response to fungus (GO:0009620) and is also known to be involved in response to the hemibiotrophic bacterial pathogen Pseudomonas syringae87. Additionally, RIPK (AT2G05940; Glyma.09g069900), a positive regulator of pattern triggered immunity (PTI)88–90, as well as genes in the GO category response to chitin (GO:0010200), were downregulated (Supplementary Tables S6 and S8).
Genes belonging to several GO categories involved in the typical response of plants against pathogens were downregulated, including respiratory burst involved in defense response (GO:0002679), and defense response to fungus, incompatible interaction (GO:0009817) (Fig. 3B and Supplementary Tables S6 and S8). Similarly, homologs of the MLO family of seven membrane transcription factors (AT2G39200; Glyma.12G119300, Glyma.06G286800, Glyma.16G145500) involved in incompatible responses to pathogens (e.g. MLO12 for barley mildew resistance locus)91,92, and homologs of 2-oxoglutarate (2OG; AT3G13610; Glyma.03g096500), an Fe(II)-dependent oxygenase involved in hydrogen peroxide mediated PCD, were downregulated (Supplementary Tables S6 and S8). However, several genes in the GO category negative regulation of PCD (GO:0043069) were downregulated. Interestingly, downregulated genes also fell into several GO categories for response to endoplasmic reticulum stress (GO:0034976), endoplasmic reticulum unfolded protein response (GO:0030968), and protein folding (GO:0006457) (Fig. 3B and Supplementary Table S8).
Discussion
Overall, soybean showed growth responses to P. indica colonization similar to those observed in other hosts, including increased shoot biomass, decreased root biomass, and increased number of flowers and pods (Table 1). The fungus also improved nutrient uptake of macronutrients N, P, and K as well as a number of micronutrients and beneficial nutrients, especially Fe (Table 1 and Supplementary Table S2). Interesting, potential synergistic interactions with rhizobia were observed with P. indica, with a greater ratio of dry weight of nodules: roots (Table 1). Although differences in average size of nodules was not significantly different, the data showed a trend of fewer but slightly larger nodules in the P. indica treatment compared to the control. Previous studies have found a strong correlation between between average nodule size and the symbiotic efficiency of nitrogen fixation and accumulation in plant tissues93,94.
Lignin biosynthesis, whose monolignol precursors are produced through the phenylpropanoid pathway, was one of the most highly enriched upregulated GO categories. Several other recent studies have shown that colonization with P. indica increases activity of genes within the phenylpropanoid pathway. A joint metabolomics and transcriptomics study of P. indica colonizing Arabidopsis detected enrichment of transcripts in the phenylpropanoid pathway as well as increased levels of oligolignol metabolites within roots95. However, another metabolomics study in Arabidopsis detected significantly lower levels of oligolignols in root exudates53. Together, these studies suggest that oligolignols are likely utilized within roots, potentially through transformation into lignin. Lignification and changes in cell wall metabolism are often a component of the plant defense response to strengthen cell walls against invading pathogens. The phenylpropanoid pathway has been shown to be upregulated in response to various types of pathogens including plant parasitic nematodes96. Interestingly, increased lignification in leaves was also observed in tropical plants colonized by horizontally transmitted endophytes97. We hypothesize that while endophytes may be able to evade systemic plant defenses, they may still trigger localized responses and increased lignification may occur in response to both pathogenic and endophytic fungi. Alternatively, increased lignin production could serve other specialized functions such as strengthening of xylem vessels, supported by the upregulation of a homolog of Arabidopsis LACCASE 4 (AT2G38080) (Supplementary Table S5), potentially contributing to the increased water use efficiency and drought tolerance observed for plants colonized by P. indica.
Previous studies have also demonstrated increased production of other secondary metabolites, including phenolic compounds derived from either the phenylpropanoid or flavonoid biosynthetic pathways. Tashackori et al.46 documented the accumulation of the toxic and anticancer lignan podophyllotoxin, as well as other phenolic defense compounds (cinnamic, coumaric, caffeic and ferulic acid), in hairy roots of Linum album exposed to P. indica. Volatile terpenoids have also been shown to be induced by colonization with P. indica18,37,39,40,98,99. In our data, upregulation of genes involved in biosynthesis of the flavonoids luteolin and quercitin (Fig. 4), as well as terpenoids, was observed (Fig. 5 and Supplementary Tables S5 and S10). Flavonoids are important secondary compounds in soybean and may play roles ranging from promoting colonization by rhizobia to protecting plants from both abiotic (UV light) and biotic stresses100. Luteolin, in particular, has been shown to play a key role in signaling of nodulation genes101 and may be one factor contributing to the larger dry-weight of nodules per dry-weight or roots observed in P. indica colonized roots (Fig. 1 and Table 1). The bioflavonoid quercetin is a powerful anti-oxidant that provides protection from oxidative stress102,103.
The mechanisms by which P. indica and other endophytic fungi uptake macronutrients and beneficial nutrients are poorly understood and remain controversial104,105. Although some studies have shown that P. indica enhances P uptake and accumulation in some hosts, others have failed to show this effect106,107. Our results showed significant increases in N, P, and K in plant shoot tissue, demonstrating that in soybean, the fungus improves macronutrient uptake (Table 1). Potential mechanisms of uptake and transfer could involve either fungal or plant genes or both. A high-affinity phosphate transporter (PiPT) from P. indica was previously shown to be involved in uptake and transfer of P by P. indica to Zea mays27. Other studies have shown that P. indica is able to solubilize inorganic P by lowering the pH through secretion of organic acids108,109. Four different fungal high-affinity phosphate transporters, as well as two acid phosphatases in P. indica, were shown to be differentially expressed in response to different concentrations of inorganic P105. Studies of plant responses to P. indica have shown increased expression of plant genes such as PHOSPHATE 1 (PHO1; AT3G23430.1), involved in transfer of P from root cortical cells to xylem in Arabidopsis110. Homologs of PHOSPHATE 1 were not upregulated in our data, but we show increased expression of other plant genes involved in P acquisition, including a PAP gene (Supplementary Tables S5 and S7). PAPs are upregulated in plants in response to phosphorus deficiency67 and during phosphorus acquisition in interactions with other symbionts including rhizobia111 and AMF112. Thus, increased solubility of plant-unavailable P in the rhizosphere through activity of both fungal and plant PAPs is likely an important factor contributing to improved P nutrition in P. indica colonized plants.
The effects of P. indica on N uptake have been less well characterized. The genome of P. indica contains a lower number of nitrate reductases and nitrate transporters than most fungi but contains transporters for other nitrogen sources, including a urea permease (DUR3), high-affinity ammonium transporters, and amino acid-transporters11. However, the fungus has been shown to induce nitrate assimilation genes in plant hosts, including an NADH-dependent nitrate reductase (AT1G37130)26. We did not find a homolog of this gene differentially expressed in soybean (Supplementary Tables S5 and S6). We speculate that P. indica may either utilize its own, to date uncharacterized, ammonium, urea, or amino acid transporter for uptake and transfer of N to the plant or may stimulate rhizobial symbionts. In the plant, we observed downregulation of several plant high-affinity ammonium transporters (Table 1 and Supplementary Table S6), but upregulation of one urea transporter (AtTIP2; 1; AT3G16240; Glyma. 07g018000) by P. indica, suggesting that urea may be important in this symbiosis. Alternatively, P. indica may improve N uptake indirectly by stimulating rhizobial symbionts, as suggested by the increased rhizobial biomass:root biomass (Table 1) and upregulation of genes involved in biosynthesis of the flavonoid luteolin (Supplementary Table S5), which has been reported to regulate nodulation113, in P. indica colonized plants. However, we did not observe upregulation of ammonium transporters known to be involved in transfer of N from rhizobia to plants (Supplementary Table S5). P. indica also significantly improved shoot content of the beneficial nutrient iron, potentially through upregulation of two FER-like regulators of iron uptake (Glyma.13g322100 and Glyma.12g178500) and production of iron-scavenging compounds like scopoletin (Fig. 4 and Supplementary Table S5).
Induction of plant hormones by P. indica has been previously documented and is thought to play a role in its strong growth promoting and stress modulating effects23,32. Gibberellins, naturally occurring plant hormones that promote cell growth and stem elongation as well as aspects of flowering and fruiting114–116, have also been shown to have roles in primary root development, cell elongation117,118, and regulation of whole plant carbon metabolism119. While GA has been shown to increase root growth in some plants, it may also preferentially direct growth to the shoot, as observed in this study (Table 1). Our data showed upregulation of GASA14, (Supplementary Tables S5 and S7) a GA responsive gene shown to improve salt and ABA tolerance of transgenic Arabidopsis plants120. Additionally, a negative transcriptional regulator of GA biosynthesis (DDF1) was downregulated (Supplementary Table S5). Overexpression mutants of this gene (ddf1) showed decreased GA production, reduced height, and delayed flowering in Arabidopsis. The increased shoot biomass growth, faster flowering, and shorter time to seed production observed in our study could be due in part to downregulation of this or other negative regulators of GA.
The phytohormone ABA plays a crucial role in adaptive responses to environmental stresses, as higher ABA expression in response to drought and high salinity induces massive changes in gene expression resulting in stomatal closure31,121. An overexpression mutant of a homolog of an ABA responsive gene that was upregulated in our data (Supplementary Tables S5 and S7), ATbetaFruct4, showed higher levels in leaf stomatal guard cells and led to increased drought tolerance in Arabidopsis80. Transgenic Arabidopsis overexpressing GolS2, a homolog of another upregulated ABA responsive and drought-inducible gene involved in synthesis of galactinol (Supplementary Tables S5 and S7), showed greater drought tolerance due to higher intracellular osmolarity from galactinol and raffinose122. Other upregulated genes mapped to the pathways involved in biosynthesis of B-series fagopyritols and stachyose, compounds documented in both soybean and grasses123 to enhance seed desiccation tolerance during seed maturation (Supplementary Table S9). While these compounds have not previously been implicated in desiccation tolerance in vegetative parts of the plant, further research is needed to investigate their potential role in controlling osmotic stress.
Given P. indica’s documented ability to confer adaptation to abiotic stresses such as drought20,124,125 and salt stress22,126,127, the downregulation of heat shock and other proteins involved in heat acclimation was surprising. However, another recent whole transcriptome study of the response of Arabidopsis to both P. indica and the related sebacinoid fungus Sebacina vermifera, showed that several classes of heat shock proteins were consistently downregulated by both fungi128, suggesting this may represent a conserved response in plants to colonization by sebacinoid fungi. It is possible that P. indica may induce other mechanisms to increase tolerance to heat, high light, and hydrogen peroxide. We hypothesize that downregulation of expression of heat shock proteins in response to these abiotic stressors in P. indica plants compared to control plants experiencing the same conditions may indicate that the colonized plants are reducing the negative effects of abiotic stresses using other mechanisms and thus do not need to upregulate heat shock proteins. For example, the upregulation of homologs of a galactinol synthase (GolS2; AT1G56600; Glyma. 03g229800) and a sucrose vacuolar invertase (ATbetaFruct4; AT1G12240; Glyma.01g211000), which increase drought tolerance in Arabidopsis through increased intracellular osmolarity and regulation of stomatal opening, respectively, are possible mechanisms that may mitigate cellular stress and ROS production under drought stress and thus decrease expression of proteins involved in response to ROS.
During the biotrophic phase of colonization, P. indica has been shown to suppress root innate immunity in order to colonize host root tissue33, potentially through modulation of GA and SA mediated signaling29. In addition to GA, ABA also controls growth and proliferation of the fungus through suppression of plant innate immunity in barley29. Similarly, external exposure of Arabidopsis seedlings to ABA increased levels of colonization by P. indica29. ABA exposure also decreased expression of two MAMP responsive genes whose homologs in soybean were also downregulated in our data (Supplementary Tables S6 and S8)31. Recent work also suggests that effectors produced by P. indica mediate suppression of innate immunity to allow for colonization129.
In barley, after the initial biotrophic phase, P. indica induces PCD10,11, which is required for root colonization by this symbiont in barley. Our data suggest that PCD may also be required for colonization of soybean roots by P. indica. PCD induced by P. indica, however, has different characteristics from the typical hypersensitive response (HR) of plants to pathogens in that neither typical defense markers such as pathogenesis related proteins130 nor accumulation of ROS29 have been detected at high concentrations in colonized roots. Instead, transmission electron microscopy suggests that ultrastructural changes associated with endoplasmic reticulum (ER) stress, as well as a simultaneous suppression of the plant’s adaptive ER stress response (e.g. unfolded protein response pathway), lead to cell death131. In this study, genes belonging to several GO categories involved in the respiratory burst response and the typical incompatible HR of plants against pathogens were downregulated (Fig. 3B and Supplementary Table S8), while those involved in ER stress response and protein folding were upregulated (Fig. 3A and Supplementary Table S7). These results provide support for the hypothesis that P. indica induces PCD through an ER mediated mechanism distinct from HR.
Conclusions
Our results demonstrate that colonization of soybean with P. indica induced shoot growth and biomass accumulation, leading to higher shoot: root ratios as well as earlier flowering and pod formation. In this study, the fungus also increased nutrient acquisition of key macronutrients, including N, P, and K, as well as of several micro and beneficial nutrients, especially Fe. Colonization by P. indica resulted in larger dry biomass of rhizobial nodules per root dry weight in colonized plants, suggesting a potential synergistic interaction of this fungal symbiont with rhizobia.
Transcriptomic analyses revealed that among the most highly upregulated genes and GO categories were those involved in lignin and cell wall synthesis, cell wall remodeling, and iron acquisition. In particular, genes (e.g. COMT) in the phenypropanoid pathway involved in monolignol biosynthesis, as well as several classes of oxidative enzymes such as laccases and peroxidases involved in polymerization of monolignol subunits in plant cell walls, were upregulated. Several genes involved in synthesis of plant secondary compounds, including aromatic compounds (1, 3, 5 trimethoxybenzene) and flavonoids (luteolin and, quercetin), both derived from the phenylpropanoid pathway, as well as furaneols and the volatile terpene linalool, were also upregulated. The upregulation of genes involved in biosynthesis of the flavonoid luteolin, which has been reported to regulate nodulation113, suggests a possible mechanism for the increased rhizobial biomass observed in the P. indica treatment. Synergistic interactions with rhizobia may be a potential mechanism for increased N uptake observed in P. indica colonized plants. We also demonstrate that P. indica induces increased expression of a plant PAP gene that is involved in solubilization and uptake of P. Other upregulated GO categories included those involved in hormone signaling. Our results support previous findings in other plant hosts that growth and abiotic stress responses induced during colonization by P. indica may be modulated by plant hormones. In particular, homologs of a number of Arabidopsis ABA responsive genes (GolS2 and ATbetaFruct4) with roles in response to drought stress were upregulated. Increased expression of genes involved in synthesis of other potential osmoregulatory compounds, B-series fagopyritols and stachyose, which have been shown to increase desiccation tolerance in seeds, was also observed.
Many downregulated genes belonged to GO categories involved in the plant defense response to fungal colonization. As is previous studies in Arabidopsis33, MAMP and PTI innate immunity pathways in soybean were suppressed in soybean roots colonized with P. indica. Similarly, suppression of the respiratory burst and some gene involved in HR in our data also support generalized suppression of the plant defense response to allow for colonization of roots by this symbiont. However, our results suggest that like in barley, a form of PCD is important for colonization of soybean roots. Results showed upregulation of genes in GO categories involved in response to ER stress and ER unfolded protein response, supporting microscopic findings that PCD involved in colonization by P. indica may not involve a typical HR response but is mediated by a distinct mechanism that involves ER stress. One of the more surprising results of this study was the pervasive downregulation of heat-shock proteins belonging to GO categories involved in response to heat, high light, hydrogen peroxide, and salinity stress. We hypothesize that P. indica may use other mechanisms to increase plant tolerance and decrease negative effects of abiotic stresses. Thus, the downregulation of heat shock proteins may reflect the ability of P. indica colonized plants to better tolerate or ameliorate these stresses without inducing oxidative stress to the plant.
Overall, this research sheds light on molecular mechanisms contributing to the increased growth, improved nutrient status, and stress tolerance of soybean colonized by P. indica and provides a foundation for exploring the tri-partite interactions between soybean, fungal, and rhizobial symbionts.
Materials and Methods
Growth of Plants and Fungus
Seeds of G. max Williams 82 were surface-sterilized and seeds of equal size were selected and pre-germinated. P. indica strain ATCC (204458) was propagated in potato dextrose broth as described previously25. Soil mix (Sunshine Professional Growing Mix) was autoclaved two times for 90 minutes with a 24-hour interval to sterilize. For inoculation of plants with P. indica, 2 g of mycelium were thoroughly mixed with 100 g of soil in peat pots before sowing. The control treatment contained no inoculum. Plants were grown in a Conviron growth chamber with temperatures of 22 °C at night and 24 °C during the day and a 16:8 hour light: dark cycle. Two different treatments including an uninoculated control and P. indica inoculated treatment were employed. Each treatment consisted of three biological replicate plants grown in separate peat pots. All the treatments were inoculated after 15 days of sowing with 10 mL of Optimize® LCO Promoter Technology® for soybean, Novozymes, according to manufacturer’s protocol. The inoculum contained 5 × 106 spores/mL of Bradyrhizobium japonicum. Plants were harvested 60 days after sowing. Shoot length, fresh weight of aerial parts, and the number of pods were counted at harvest. The roots were thoroughly washed and fresh root weight was determined. The root volume was determined by water displacement using a measuring cylinder.
Staining of Roots and Percent Colonization
Washed roots were cleaned by soaking in 10% KOH for 4 days, acidified with 1 N HCl for 5 minutes, and then stained with lacto-phenol cotton blue. The roots were observed under 630X with an optical light microscope (Olympus BH2, Leeds Precision Instrument, Inc.). Estimation of percent colonization was done using the grid line-intersect method132. Chlamydospores within roots were counted under the microscope and percent root colonization was calculated with the formula: % colonization = number of colonized roots × 100/number of roots observed133,134.
Estimation of Macro, Micro and Beneficial Elements
The macroelements (N, P, K), microelements (Al, Mn, Ni, Zn, B, Cd, Cr, Cu) and other elements (Ca, Fe, Mg, Na) were estimated using Dumas method for total N135,136 and the ICP-dry ash method for other elements137 by nutrient analysis of shoot tissue. Plant nutrient testing was outsourced to the Research Analytical Laboratory, University of Minnesota.
Statistical Analyses of Plant Growth Parameters and Nutrient Content
For analyses of plant growth parameters and nutrient content, the mean and standard deviation of three biological replicates, consisting of three individual plants grown in separate peat pots, were calculated. Statistical analyses of differences in plant growth responses and nutrient content across treatments were analyzed using a Dunnett’s multiple comparisons test. A p-value less than 0.05 was considered statistically significant. GraphPad Prism v6.04 was used for the analysis (GraphPad Software, La Jolla California USA, www.graphpad.com).
RNA Isolation and Purification
Roots from the two biological replicate plants per treatment were randomly selected for RNA sequencing. For each biological replicate, three root sections of approximately 5 cm in length were randomly collected from three locations on each plant root system and were pooled for RNA extraction. Soybean root tissue samples were ground in liquid nitrogen using a mortar and pestle immediately after harvest and directly mixed with TRIzolTM (Invitrogen) solution for extraction. Total RNA was isolated from the roots using the standard TRIzolTM (Invitrogen) protocol including a chloroform extraction followed by precipitation in 0.1% 3 M NaAcetate pH 5 and 0.8 volumes cold isopropanol. Contaminating DNA was removed by treatment with DNAase A138 for 15 minutes at 37 °C. RNA was further purified and concentrated using the RNA cleanup method on a Plant RNeasy column (Qiagen). RNA quality was evaluated by gel electrophoresis, spectrophotometer, and Agilent 2100. Only samples with an RNA integrity number (RIN) >7.5 were used for sequencing. Total RNA was sent to the University of Minnesota Genomics Center for library prep using the Illumina TruSeq 2 RNA kit and adaptors. Samples were barcoded and sequenced using 100 bp single end reads on the Illumina Hi-Seq 2000.
RNA-Seq Data Analysis
Data analysis packages within the Galaxy platform (www.galaxy.umn.edu) were used to analyze and perform quality control of the Illumina data, align sequenced reads to the G. max (Williams 82 a2.v1 downloaded from Phytozyme 10) reference genome and perform differential expression analyses. The Fastx Toolkit139 was used to assess the quality of the data. Trimmomatic140 was then used to remove any remaining Illumina adapters and to trim off 15 bases from the beginning of the reads and 10 bases at the end of the reads of poorer quality sequence to give a total read length of 75 bp. The average quality score for all reads was above 30. The reads were then aligned to the reference genome of Williams 82 using TopHat2141 and differential expression was performed with CuffDiff142 using the Williams 82 a2.v1 annotations. We used the geometric method for library normalization and a false discovery rate of 0.05. Genes with an FDR corrected p-value (q-value) less than 0.05 were considered differentially expressed. Functional annotation and Arabidopsis homologs were identified using SoyBase annotations.
Validation of Gene Expression by qRT-PCR
To validate the expression of upregulated genes of G. max, cDNA was synthesized using approximately 0.3 μg of total RNA isolated from all three biological replicates of colonized and non-colonized roots samples. The cDNA was synthesized using a cDNA synthesis kit (Two-Step RT-PCR Kit from Affymetrix). Gene specific primers were designed with IDT-primer quest (http://www.idtdna.com/primerquest/home/index). Primer pairs used for gene expression analysis were designed using published cDNA sequences (https://www.soybase.org/) for G. max for selected genes (Glyma.13G191400, Glyma.05G188900, Glyma.11G213500, Glyma.08G076000, and Glyma.13G191400) (Supplementary Table S11). Primers previously tested for Elongation factor1b (Ef1b) gene of G. max was used as the housekeeping gene143. The relative expression of genes was quantified using qRT- PCR (Applied Biosystems 7500 Real-Time PCR System). To prepare the reaction mixture, 1.9 μl of template of a 1:5 dilution of prepared cDNA sample), 6.25 μl iTaq™ Universal SYBR® Green Supermix, and 0.5 μl of each gene specific primer pair were mixed with 3.35 μl of nuclease-free water to make a total volume of 12.5 μl. Thermal cycle conditions included hot start at 95 °C for 3 min followed by 40 cycles at 95 °C (30 s) and 58 °C (30 s). The melt curve was done at 95 °C (15 s), 58 °C (1 min), 95 °C (15 s) and 58 °C (15 s). All reactions were run with three technical triplicates for each of the two biological replicates. The ΔΔCT method was used to determine relative fold change in gene expression levels.
Gene Ontology Analyses
A Gene Ontology (GO) enrichment analysis was performed on the SoyBase website (http://soybase.org/) to identify GO categories to which upregulated and downregulated genes belonged and to identify those that were over- or under-represented. In order to provide a broad overview of up and down regulated GO categories, both genes that were statistically enriched at a corrected p-value of 0.05 as well as those that were not significantly enriched but had a corrected p-value < 10 were displayed using the web server for REVIGO (http://revigo.irb.hr/) using an intermediate similarity cutoff of 0.759. The resulting clusters were exported as a tree network and visualized in Cytoscape61. Clusters were color coded in Cytoscape according to their corrected p-value.
Metabolic Pathway Analyses
The normalized expression levels of significantly up and down regulated genes for the control versus P. indica were mapped to the SoyCyc metabolic database in Soybase in order to identify metabolic pathways affected. All of the significantly upregulated or downregulated genes were displayed with a cutoff threshold of 0.05. The SoyCyc pathway database does not perform statistical analyses but provides a color coded scale of relative fold change of expression levels ranging from red (most upregulated) to blue (most down-regulated). Arabidopsis homologs of up and down regulated genes were also mapped to selected pathways in the KEGG pathway database to display up- or downregulated genes144–146.
Data availability
Transcriptome raw Illumina reads are deposited in the short read archive at NCBI under submission number SUB3229927.
Electronic supplementary material
Acknowledgements
We would like to thank the the University of Minnesota Genomics Center (UMGC) for preparation of libraries and sequencing of RNA samples, the Minnesota Supercomputing Institute for computational programs and resources and Startup funding from the University of Minnesota to KEB for sequencing costs.
Author Contributions
R.B. and K.E.B. conceived the research. R.B. and Y.H. conducted plant growth experiments and isolated RNA. S.B., B.M., H.B., and R.B. performed RNA-Seq quality control and data analysis. R.B. and K.E.B. wrote the manuscript with contributions and editing from R.P. and A.V.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-26809-3.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Weiss M, et al. Sebacinales Everywhere: Previously Overlooked Ubiquitous Fungal Endophytes. Plos One. 2011;6:7. doi: 10.1371/journal.pone.0016793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weiss M, Selosse MA, Rexer KH, Urban A, Oberwinkler F. Sebacinales: a hitherto overlooked cosm of heterobasidiomycetes with a broad mycorrhizal potential. Mycological Research. 2004;108:1003–1010. doi: 10.1017/S0953756204000772. [DOI] [PubMed] [Google Scholar]
- 3.Selosse MA, Weiss M, Jany JL, Tillier A. Communities and populations of sebacinoid basidiomycetes associated with the achlorophyllous orchid Neottia nidus-avis (L.) LCM Rich. and neighbouring tree ectomycorrhizae. Molecular Ecology. 2002;11:1831–1844. doi: 10.1046/j.1365-294X.2002.01553.x. [DOI] [PubMed] [Google Scholar]
- 4.Barazani, O., Benderoth, M., Groten, K., Kuhlemeier, C. & Baldwin, I.T. Piriformospora indica and Sebacina vermifera increase growth performance at the expense of herbivore resistance in Nicotiana attenuate. Oecologia146 (2005). [DOI] [PubMed]
- 5.Weiß M, Waller F, Zuccaro A, Selosse M-A. Sebacinales – one thousand and one interactions with land plants. New Phytologist. 2016;211:20–40. doi: 10.1111/nph.13977. [DOI] [PubMed] [Google Scholar]
- 6.Waller F, et al. Systemic and local modulation of plant responses by Piriformospora indica and related Sebacinales species. Journal of Plant Physiology. 2008;165:60–70. doi: 10.1016/j.jplph.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 7.Verma S, et al. Piriformospora indica, gen. et sp. nov., a new root-colonizing fungus. Mycologia. 1998;90:896–903. doi: 10.2307/3761331. [DOI] [Google Scholar]
- 8.Varma A, et al. Piriformospora indica, a cultivable plant-growth-promoting root endophyte. Applied and Environmental Microbiology. 1999;65:2741–2744. doi: 10.1128/aem.65.6.2741-2744.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bagheri AA, Saadatmand S, Niknam V, Nejadsatari T, Babaeizad V. Effect of endophytic fungus, Piriformospora indica, on growth and activity of antioxidant enzymes of rice (Oryza Sativa L.) under salinity stress. International Journal of Advanced Biological and Biomedical Research. 2013;1:1337–1350. [Google Scholar]
- 10.Deshmukh S, et al. The root endophytic fungus Piriformospora indica requires host cell death for proliferation during mutualistic symbiosis with barley. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18450–18457. doi: 10.1073/pnas.0605697103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuccaro, A. et al. Endophytic life strategies decoded by genome and transcriptome analyses of the mutualistic root symbiont Piriformospora indica. Plos Pathogens7 (2011). [DOI] [PMC free article] [PubMed]
- 12.Jogawat A, et al. Piriformospora indica rescues growth diminution of rice seedlings during high salt stress. Plant Signaling & Behavior. 2013;8:e26891. doi: 10.4161/psb.26891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waller F, et al. The endophytic fungus Piriformospora indica reprograms barley to salt-stress tolerance, disease resistance, and higher yield. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:13386–13391. doi: 10.1073/pnas.0504423102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee Y-C, et al. Growth promotion of chinese cabbage and Arabidopsis by Piriformospora indica is not stimulated by mycelium-synthesized auxin. Molecular Plant-Microbe Interactions. 2011;24:421–431. doi: 10.1094/MPMI-05-10-0110. [DOI] [PubMed] [Google Scholar]
- 15.Varma A, et al. Functions of novel symbiotic fungus- Piriformospora indica. Proceedings of Indian National Science Academy. 2014;80:429–441. doi: 10.16943/ptinsa/2014/v80i2/55119. [DOI] [Google Scholar]
- 16.Peškan-Berghöfer, T. et al. Association of Piriformospora indica with Arabidopsis thaliana roots represents a novel system to study beneficial plant-microbe interactions and involves early plant protein modifications in the endoplasmic reticulum and at the plasma membrane. Physiol Plantarum122 (2004).
- 17.Fakhro A, et al. Impact of Piriformospora indica on tomato growth and on interaction with fungal and viral pathogens. Mycorrhiza. 2010;20:191–200. doi: 10.1007/s00572-009-0279-5. [DOI] [PubMed] [Google Scholar]
- 18.Das A, et al. The root endophyte fungus Piriformospora indica leads to early flowering, higher biomass and altered secondary metabolites of the medicinal plant, Coleus forskohlii. Plant Signaling & Behavior. 2012;7:103–112. doi: 10.4161/psb.7.1.18472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zarea MJ, et al. Effect of Piriformospora indica and Azospirillum strains from saline or non-saline soil on mitigation of the effects of NaCl. Soil Biology & Biochemistry. 2012;45:139–146. doi: 10.1016/j.soilbio.2011.11.006. [DOI] [Google Scholar]
- 20.Sun CA, et al. Piriformospora indica confers drought tolerance in Chinese cabbage leaves by stimulating antioxidant enzymes, the expression of drought-related genes and the plastid-localized CAS protein. Journal of Plant Physiology. 2010;167:1009–1017. doi: 10.1016/j.jplph.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 21.Husaini AM, et al. Modifying strawberry for better adaptability to adverse impact of climate change. Current Science. 2012;102:1660–1673. [Google Scholar]
- 22.Baltruschat H, et al. Salt tolerance of barley induced by the root endophyte Piriformospora indica is associated with a strong increase in antioxidants. New Phytologist. 2008;180:501–510. doi: 10.1111/j.1469-8137.2008.02583.x. [DOI] [PubMed] [Google Scholar]
- 23.Serfling A, Wirsel SGR, Lind V, Deising HB. Performance of the biocontrol fungus Piriformospora indica on wheat under greenhouse and field conditions. Phytopathology. 2007;97:523–531. doi: 10.1094/PHYTO-97-4-0523. [DOI] [PubMed] [Google Scholar]
- 24.Dolatabadi HK, et al. Biocontrol potential of root endophytic fungi and Trichoderma species against Fusarium wilt of lentil under in vitro and greenhouse conditions. Journal of Agricultural Science and Technology. 2012;14:407–420. [Google Scholar]
- 25.Bajaj R, et al. The beneficial root endophyte Piriformospora indica reduces egg density of the soybean cyst nematode. Biological Control. 2015;90:193–199. doi: 10.1016/j.biocontrol.2015.05.021. [DOI] [Google Scholar]
- 26.Sherameti I, et al. The endophytic fungus Piriformospora indica stimulates the expression of nitrate reductase and the starch-degrading enzyme glucan-water dikinase in tobacco and Arabidopsis roots through a homeodomain transcription factor that binds to a conserved motif in their promoters. Journal of Biological Chemistry. 2005;280:26241–26247. doi: 10.1074/jbc.M500447200. [DOI] [PubMed] [Google Scholar]
- 27.Yadav V, et al. A phosphate transporter from the root dndophytic fungus Piriformospora indica plays a role in phosphate transport to the host plant. Journal of Biological Chemistry. 2010;285:26532–26544. doi: 10.1074/jbc.M110.111021. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Gosal, S. K., Karlupia, A., Gosal, S. S., Chhibba, I. M. & Varma, A. Biotization with Piriformospora indica and Pseudomonas fluorescens improves survival rate, nutrient acquisition, field performance and saponin content of micropropagated Chlorophytum sp. Indian Journal of Biotechnology9 (2010).
- 29.Schafer P, et al. Manipulation of plant innate immunity and gibberellin as factor of compatibility in the mutualistic association of barley roots with Piriformospora indica. Plant Journal. 2009;59:461–474. doi: 10.1111/j.1365-313X.2009.03887.x. [DOI] [PubMed] [Google Scholar]
- 30.Khatabi B, et al. Ethylene supports colonization of plant roots by the mutualistic fungus Piriformospora indica. Plos One. 2012;7:8. doi: 10.1371/journal.pone.0035502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peskan-Berghofer T, et al. Sustained exposure to abscisic acid enhances the colonization potential of the mutualist fungus Piriformospora indica on Arabidopsis thaliana roots. New Phytologist. 2015;208:873–886. doi: 10.1111/nph.13504. [DOI] [PubMed] [Google Scholar]
- 32.Gill SS, et al. Piriformospora indica: Potential and significance in plant stress tolerance. Frontiers in Microbiology. 2016;7:20. doi: 10.3389/fmicb.2016.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jacobs S, et al. Broad-Spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by the fungus Piriformospora indica. Plant Physiology. 2011;156:726–740. doi: 10.1104/pp.111.176446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hause B, Schaarschmidt S. The role of jasmonates in mutualistic symbioses between plants and soil-born microorganisms. Phytochemistry. 2009;70:1589–1599. doi: 10.1016/j.phytochem.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 35.Ding YL, et al. Abscisic ccid coordinates Nod factor and cytokinin Ssgnaling during the regulation of nodulation in Medicago truncatula. Plant Cell. 2008;20:2681–2695. doi: 10.1105/tpc.108.061739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Noorden GE, Ross JJ, Reid JB, Rolfe BG, Mathesius U. Defective long-distance auxin transport regulation in the Medicago truncatula super numeric nodules mutant. Plant Physiology. 2006;140:1494–1506. doi: 10.1104/pp.105.075879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arora M, Saxena P, Choudhary DK, Abdin MZ, Varma A. Dual symbiosis between Piriformospora indica and Azotobacter chroococcum enhances the artemisinin content in Artemisia annua L. World Journal of Microbiology and Biotechnology. 2016;32:19. doi: 10.1007/s11274-015-1972-5. [DOI] [PubMed] [Google Scholar]
- 38.Ahlawat S, Saxena P, Alam P, Wajid S, Abdin MZ. Modulation of artemisinin biosynthesis by elicitors, inhibitor, and precursor in hairy root cultures of Artemisia annua L. Journal of Plant Interactions. 2014;9:811–824. doi: 10.1080/17429145.2014.949885. [DOI] [Google Scholar]
- 39.Sharma, G. & Agrawal, V. Marked enhancement in the artemisinin content and biomass productivity in Artemisia annua L. shoots co-cultivated with Piriformosporaindica. World Journal of Microbiology and Biotechnology29, 1133–1138 (2013). [DOI] [PubMed]
- 40.Prasad R, Kamal S, Sharma PK, Oelmuller R, Varma A. Root endophyte Piriformospora indica DSM 11827 alters plant morphology, enhances biomass and antioxidant activity of medicinal plant Bacopa monniera. Journal of Basic Microbiology. 2013;53:1016–1024. doi: 10.1002/jobm.201200367. [DOI] [PubMed] [Google Scholar]
- 41.Prasad, R. et al. In Mycorrhiza: State of the art, genetics and molecular biology, cco-function, biotechnology, eco-physiology, structure and systematics (ed. Varma, A.) 655–678 (Springer Berlin Heidelberg, Berlin, Heidelberg 2008).
- 42.Das A, Tripathi S, Varma A. In vitro plant development and root colonization of Coleus forskohlii by Piriformospora indica. World Journal of Microbiology and Biotechnology. 2014;30:1075–1084. doi: 10.1007/s11274-013-1526-7. [DOI] [PubMed] [Google Scholar]
- 43.Bajaj R, et al. Co-cultivation of Curcuma longa with Piriformospora indica enhances the yield and active ingredients. American Journal of Current Microbiology. 2014;2:12. [Google Scholar]
- 44.Baldi A, et al. Enhanced production of podophyllotoxins by co-culture of transformed Linum album cells with plant growth-promoting fungi. Pure and Applied Chemistry. 2010;82:227–241. doi: 10.1351/PAC-CON-09-02-09. [DOI] [Google Scholar]
- 45.Tahsili J, Sharifi M, Safaie N, Esmaeilzadeh-Bahabadi S, Behmanesh M. Induction of lignans and phenolic compounds in cell culture of Linum album by culture filtrate of Fusarium graminearum. Journal of Plant Interactions. 2014;9:412–417. doi: 10.1080/17429145.2013.846419. [DOI] [Google Scholar]
- 46.Hannaneh Tashackori MS, Chashmi NA, Safaie N, Behmanesh M. Induced-differential changes on lignan and phenolic acid compounds in Linum album hairy roots by fungal extract of Piriformospora indica. Plant Cell Tiss Organ Cult. 2016;127:187–194. doi: 10.1007/s11240-016-1041-2. [DOI] [Google Scholar]
- 47.Baldi A, Jain A, Gupta N, Srivastava AK, Bisaria VS. Co-culture of arbuscular mycorrhiza-like fungi (Piriformospora indica and Sebacina vermifera) with plant cells of Linum album for enhanced production of podophyllotoxins: a first report. Biotechnology Letters. 2008;30:1671. doi: 10.1007/s10529-008-9736-z. [DOI] [PubMed] [Google Scholar]
- 48.Vogt T. Phenylpropanoid biosynthesis. Molecular Plant. 2010;3:2–20. doi: 10.1093/mp/ssp106. [DOI] [PubMed] [Google Scholar]
- 49.Murphy BR, Doohan FM, Hodkinson TR. Yield increase induced by the fungal root endophyte Piriformospora indica in barley grown at low temperature is nutrient limited. Symbiosis. 2014;62:29–39. doi: 10.1007/s13199-014-0268-0. [DOI] [Google Scholar]
- 50.Kumar M, Yadav V, Tuteja N, Johri AK. Antioxidant enzyme activities in maize plants colonized with Piriformospora indica. Microbiology-Sgm. 2009;155:780–790. doi: 10.1099/mic.0.019869-0. [DOI] [PubMed] [Google Scholar]
- 51.Shahabivand S, Maivan HZ, Goltapeh EM, Sharifi M, Aliloo AA. The effects of root endophyte and arbuscular mycorrhizal fungi on growth and cadmium accumulation in wheat under cadmium toxicity. Plant Physiology and Biochemistry. 2012;60:53–58. doi: 10.1016/j.plaphy.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 52.Siddhanta S, Paidi SK, Bushley K, Prasad R, Barman I. Exploring morphological and biochemical linkages in fungal growth with label-free light sheet microscopy and raman spectroscopy. ChemPhysChem. 2017;18(1):72–78. doi: 10.1002/cphc.201601062. [DOI] [PubMed] [Google Scholar]
- 53.Lahrmann U, et al. Mutualistic root endophytism is not associated with the reduction of saprotrophic traits and requires a noncompromised plant innate immunity. New Phytologist. 2015;207:841–857. doi: 10.1111/nph.13411. [DOI] [PubMed] [Google Scholar]
- 54.Rathod DP, Brestic M, Shao HB. Chlorophyll a fluorescence determines the drought resistance capabilities in two varieties of mycorrhized and non-mycorrhized Glycine max Linn. African. Journal of Microbiology Research. 2011;5:4197–4206. [Google Scholar]
- 55.Mansotra P, Sharma P, Sharma S. Bioaugmentation of Mesorhizobium cicer, Pseudomonas spp. and Piriformospora indica for sustainable chickpea production. Physiology and Molecular Biology of Plants. 2015;21:385–393. doi: 10.1007/s12298-015-0296-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kumar V, et al. Effect of formulated root endophytic fungus Piriformospora indica and plant growth promoting rhizobacteria fluorescent pseudomonads R62 and R81 on Vigna mungo. World Journal of Microbiology and Biotechnology. 2012;28:595–603. doi: 10.1007/s11274-011-0852-x. [DOI] [PubMed] [Google Scholar]
- 57.Grumberg BC, Urcelay C, Shroeder MA, Vargas-Gil S, Luna CM. The role of inoculum identity in drought stress mitigation by arbuscular mycorrhizal fungi in soybean. Biology and Fertility of Soils. 2015;51:1–10. doi: 10.1007/s00374-014-0942-7. [DOI] [Google Scholar]
- 58.Spagnoletti FN, Balestrasse K, Lavado RS, Giacometti R. Arbuscular mycorrhiza detoxifying response against arsenic and pathogenic fungus in soybean. Ecotoxicology and Environmental Safety. 2016;133:47–56. doi: 10.1016/j.ecoenv.2016.06.012. [DOI] [PubMed] [Google Scholar]
- 59.Supek F, Bosnjak M, Skunca N, Smuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. Plos One. 2011;6:9. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Supek, F. & Škunca, N. In The gene ontology handbook (eds. Dessimoz, C. & Škunca, N.) 207–220 (Springer New York, New York, NY 2017).
- 61.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ueda, M. et al. The HALTED ROOT gene encoding the 26S proteasome subunit RPT2a is essential for the maintenance of Arabidopsis meristems. Development131 (2004). [DOI] [PubMed]
- 63.Barros J, Serk H, Granlund I, Pesquet E. The cell biology of lignification in higher plants. Annals of Botany. 2015;115:1053–1074. doi: 10.1093/aob/mcv046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berthet S, et al. Disruption of LACCASE4 and 17 results in tissue-specific alterations to lignification of Arabidopsis thaliana stems. Plant Cell. 2011;23:1124–1137. doi: 10.1105/tpc.110.082792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schuetz M, et al. Laccases direct lignification in the discrete secondary cell wall domains of protoxylem. Plant Physiology. 2014;166:798–U489. doi: 10.1104/pp.114.245597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schönherr, J. In Physiological plant ecology II: Water relations and carbon assimilation (eds Lange, O. L., Nobel, P. S., Osmond, C. B. & Ziegler, H.) 153–179 (Springer Berlin Heidelberg, Berlin, Heidelberg 1982).
- 67.Li CC, et al. Identification of soybean purple acid phosphatase genes and their expression responses to phosphorus availability and symbiosis. Annals of Botany. 2012;109:275–285. doi: 10.1093/aob/mcr246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Del Pozo JC, et al. A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. The Plant Journal. 1999;19:579–589. doi: 10.1046/j.1365-313X.1999.00562.x. [DOI] [PubMed] [Google Scholar]
- 69.Kaiser BN, et al. Characterization of an ammonium transport protein from the peribacteroid membrane of soybean nodules. Science. 1998;281:1202–1206. doi: 10.1126/science.281.5380.1202. [DOI] [PubMed] [Google Scholar]
- 70.Liu LH, Ludewig U, Gassert B, Frommer WB, von Wiren N. Urea transport by nitrogen-regulated tonoplast intrinsic proteins in Arabidopsis. Plant Physiology. 2003;133:1220–1228. doi: 10.1104/pp.103.027409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lemanceau, P., Expert, D., Gaymard, F., Bakker, P. & Briat, J. F. In Plant innate immunity (ed. VanLoon, L. C.) 491–549 (Academic Press Ltd-Elsevier Science Ltd, London, 2009).
- 72.Lemanceau, P., Expert, D., Gaymard, F., Bakker, P. A. H. M. & Briat, J. F. In Advances in botanical research 491–549 (Academic Press 2009).
- 73.Boerjan W, Ralph J, Baucher M. Lignin biosynthesis. Annual Review of Plant Biology. 2003;54:519–546. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 74.Lemanceau P, Bauer P, Kraemer S, Briat JF. Iron dynamics in the rhizosphere as a case study for analyzing interactions between soils, plants and microbes. Plant and Soil. 2009;321:513–535. doi: 10.1007/s11104-009-0039-5. [DOI] [Google Scholar]
- 75.Klatte M, et al. The analysis of Arabidopsis nicotianamine synthase mutants reveals functions for nicotianamine in seed iron loading and iron deficiency responses. Plant Physiology. 2009;150:257–271. doi: 10.1104/pp.109.136374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Takahashi M, et al. Role of nicotianamine in the intracellular delivery of metals and plant reproductive development. Plant Cell. 2003;15:1263–1280. doi: 10.1105/tpc.010256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schmid NB, et al. Feruloyl-CoA 6′-hydroxylase1-dependent coumarins mediate iron acquisition from alkaline substrates in Arabidopsis. Plant Physiology. 2014;164:160–172. doi: 10.1104/pp.113.228544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kamboj, A., Ziemann, M. & Bhave, M. Identification of salt-tolerant barley varieties by a consolidated physiological and molecular approach. Acta Physiologiae Plantarum37 (2015).
- 79.Umezawa T, et al. Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant and Cell Physiology. 2010;51:1821–1839. doi: 10.1093/pcp/pcq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen SF, et al. Ectopic expression of a tobacco vacuolar invertase inhibitor in guard cells confers drought tolerance in Arabidopsis. Journal of Enzyme Inhibition and Medicinal Chemistry. 2016;31:1381–1385. doi: 10.3109/14756366.2016.1142981. [DOI] [PubMed] [Google Scholar]
- 81.Swarup K, et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nature Cell Biology. 2008;10:946–954. doi: 10.1038/ncb1754. [DOI] [PubMed] [Google Scholar]
- 82.Strader LC, Bartel B. The Arabidopsis PLEIOTROPIC DRUG RESISTANCE8/ABCG36 ATP binding cassette transporter modulates sensitivity to the auxin precursor indole-3-butyric acid. The Plant Cell. 2009;21:1992–2007. doi: 10.1105/tpc.109.065821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Strader, L. C., Monroe-Augustus, M., Rogers, K. C., Lin, G. L. & Bartel, B. Arabidopsis ibaresponse5 suppressors separate responses to various hormones. Genetics180, 2019–2031 (2008). [DOI] [PMC free article] [PubMed]
- 84.Ruzicka K, et al. Arabidopsis PIS1 encodes the ABCG37 transporter of auxinic compounds including the auxin precursor indole-3-butyric acid. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:10749–10753. doi: 10.1073/pnas.1005878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fourcroy P, et al. Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New Phytologist. 2014;201:155–167. doi: 10.1111/nph.12471. [DOI] [PubMed] [Google Scholar]
- 86.Mao X, Zhang H, Tian S, Chang X, Jing R. TaSnRK2.4, an SNF1-type serine/threonine protein kinase of wheat (Triticum aestivum L.), confers enhanced multistress tolerance in Arabidopsis. Journal of Experimental Botany. 2010;61:683–696. doi: 10.1093/jxb/erp331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schon M, et al. Analyses of wrky18 wrky40 plants reveal critical roles of SA/EDS1 signaling and indole-glucosinolate biosynthesis for Golovinomyces orontii resistance and a loss-of resistance towards Pseudomonas syringae pv. tomato AvrRPS4. Molecular Plant-Microbe Interactions. 2013;26:758–767. doi: 10.1094/MPMI-11-12-0265-R. [DOI] [PubMed] [Google Scholar]
- 88.Zhang J, et al. Receptor-like Cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host & Microbe. 2010;7:290–301. doi: 10.1016/j.chom.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 89.Lu DP, et al. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:496–501. doi: 10.1073/pnas.0909705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu J, Elmore JM, Lin Z-JD, Coaker G. A receptor-like cytoplasmic kinase phosphorylates the host target RIN4, leading to the activation of a plant innate immune receptor. Cell Host & Microbe. 2011;9:137–146. doi: 10.1016/j.chom.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Devoto A, et al. Molecular phylogeny and evolution of the plant-specific seven-transmembrane MLO family. Journal of Molecular Evolution. 2003;56:77–88. doi: 10.1007/s00239-002-2382-5. [DOI] [PubMed] [Google Scholar]
- 92.Lorek J, Griebel T, Jones AM, Kuhn H, Panstruga R. The role of Arabidopsis heterotrimeric G-protein subunits in MLO2 function and MAMP-triggered immunity. Molecular Plant-Microbe Interactions. 2013;26:991–1003. doi: 10.1094/MPMI-03-13-0077-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.de Araujo ASF, Lopes ACD, Teran J, Palkovic A, Gepts P. Nodulation ability in different genotypes of Phaseolus lunatus by rhizobia from California agricultural soils. Symbiosis. 2017;73:7–14. doi: 10.1007/s13199-016-0465-0. [DOI] [Google Scholar]
- 94.Hungria M, Bohrer TRJ. Variability of nodulation and dinitrogen fixation capacity among soybean cultivars. Biology and Fertility of Soils. 2000;31:45–52. doi: 10.1007/s003740050622. [DOI] [Google Scholar]
- 95.Strehmel N, et al. Piriformospora indica stimulates root metabolism of Arabidopsis thaliana. International Journal of Molecular Sciences. 2016;17:19. doi: 10.3390/ijms17071091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kong LA, et al. Large-scale identification of wheat genes resistant to cereal cyst nematode Heterodera avenae using comparative transcriptomic analysis. BMC Genomics. 2015;16:18. doi: 10.1186/s12864-015-2037-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mejia LC, et al. Pervasive effects of a dominant foliar endophytic fungus on host genetic and phenotypic expression in a tropical tree. Frontiers in Microbiology. 2014;5:16. doi: 10.3389/fmicb.2014.00479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bagde, U. S., Prasad, R. & Varma, A. Interaction of mycobiont: Piriformosporaindica with medicinal plants and plants of economic importance. African Journal of Biotechnology9, 9214–9226 (2010).
- 99.Bajaj R, et al. The beneficial root endophyte Piriformospora indica reduces of the soybean cyst nematode. Biological Control. 2015;90:193–199. doi: 10.1016/j.biocontrol.2015.05.021. [DOI] [Google Scholar]
- 100.Mierziak J, Kostyn K, Kulma A. Flavonoids as important molecules of plant interactions with the environment. Molecules. 2014;19:16240. doi: 10.3390/molecules191016240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Peters NK, Frost JW, Long SR. A plant flavone, luteolin, induces expression of Rhizobium-meliloti nodulation genes. Science. 1986;233:977–980. doi: 10.1126/science.3738520. [DOI] [PubMed] [Google Scholar]
- 102.Pourcel L, Routaboul J-M, Cheynier V, Lepiniec L, Debeaujon I. Flavonoid oxidation in plants: from biochemical properties to physiological functions. Trends in Plant Science. 2007;12:29–36. doi: 10.1016/j.tplants.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 103.Desentis-Mendoza RM, et al. Enzymatic polymerization of phenolic compounds using laccase and tyrosinase from Ustilago maydis. Biomacromolecules. 2006;7:1845–1854. doi: 10.1021/bm060159p. [DOI] [PubMed] [Google Scholar]
- 104.Behie SW, Bidochka MJ. Nutrient transfer in plant-fungal symbioses. Trends in Plant Science. 2014;19:734–740. doi: 10.1016/j.tplants.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 105.Ngwene B, Boukail S, Sollner L, Franken P, Andrade-Linares DR. Phosphate utilization by the fungal root endophyte Piriformospora indica. Plant and Soil. 2016;405:231–241. doi: 10.1007/s11104-015-2779-8. [DOI] [Google Scholar]
- 106.Karandashov V, Nagy R, Wegmuller S, Amrhein N, Bucher M. Evolutionary conservation of a phosphate transporter in the arbuscular mycorrhizal symbiosis. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:6285–6290. doi: 10.1073/pnas.0306074101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Achatz B, et al. Root colonization by Piriformospora indica enhances grain yield in barley under diverse nutrient regimes by accelerating plant development. Plant and Soil. 2010;333:59–70. doi: 10.1007/s11104-010-0319-0. [DOI] [Google Scholar]
- 108.Swetha S, Padmavathi T. Study of acid phosphatase in solubilization of inorganic phosphates by Piriformospora indica. Polish Journal of Microbiology. 2016;65:407–412. doi: 10.5604/17331331.1227666. [DOI] [PubMed] [Google Scholar]
- 109.Malla R, et al. Phosphorus solubilizing symbiotic fungus: Piriformospora indica. Endocytobiosis Cell Res. 2004;15:579–600. [Google Scholar]
- 110.Bakshi M, et al. WRKY6 restricts Piriformospora indica-stimulated and phosphate-induced root development in Arabidopsis. Bmc Plant Biology. 2015;15:19. doi: 10.1186/s12870-015-0673-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Araujo AP, Plassard C, Drevon JJ. Phosphatase and phytase activities in nodules of common bean genotypes at different levels of phosphorus supply. Plant and Soil. 2008;312:129–138. doi: 10.1007/s11104-008-9595-3. [DOI] [Google Scholar]
- 112.Ezawa T, Hayatsu M, Saito M. A new hypothesis on the strategy for acquisition of phosphorus in arbuscular mycorrhiza: Up-regulation of secreted acid phosphatase gene in the host plant. Molecular Plant-Microbe Interactions. 2005;18:1046–1053. doi: 10.1094/MPMI-18-1046. [DOI] [PubMed] [Google Scholar]
- 113.Maj D, Wielbo J, Marek-Kozaczuk M, Skorupska A. Response to flavonoids as a factor influencing competitiveness and symbiotic activity of Rhizobium leguminosarum. Microbiological Research. 2010;165:50–60. doi: 10.1016/j.micres.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 114.Paspatis EA. Effects of external application of gibberellic acid (GA3) on yield and nitrate content of spinach. Zizaniology. 1990;2:161–166. [Google Scholar]
- 115.Edwards PB, Wanjura WJ, Brown WV. Selective herbivory by Christmas beetles in response to intraspecific variation in Eucalyptus terpenoids. Oecologia. 1993;95:551–557. doi: 10.1007/BF00317440. [DOI] [PubMed] [Google Scholar]
- 116.Yildirim AN, San B, Koyuncu F, Yildirim F. Variability of phenolics, alpha-tocopherol and amygdalin contents of selected almond (Prunus amygdalus Batsch.) genotypes. Journal of Food Agriculture & Environment. 2010;8:76–79. [Google Scholar]
- 117.Wang L, et al. Evidence that high activity of vacuolar invertase is required for cotton fiber and Arabidopsis root elongation through osmotic dependent and independent pathways, respectively. Plant Physiology. 2010;154:744–756. doi: 10.1104/pp.110.162487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gonzalez MC, Roitsch T, Cejudo FJ. Circadian and developmental regulation of vacuolar invertase expression in petioles of sugar beet plants. Planta. 2005;222:386–395. doi: 10.1007/s00425-005-1542-4. [DOI] [PubMed] [Google Scholar]
- 119.Nägele T, et al. Mathematical modeling of the central carbohydrate metabolism in Arabidopsis reveals a substantial regulatory influence of vacuolar invertase on whole plant carbon metabolism. Plant Physiology. 2010;153:260–272. doi: 10.1104/pp.110.154443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sun SL, et al. GASA14 regulates leaf expansion and abiotic stress resistance by modulating reactive oxygen species accumulation. Journal of Experimental Botany. 2013;64:1637–1647. doi: 10.1093/jxb/ert021. [DOI] [PubMed] [Google Scholar]
- 121.Zhang JH, Jia WS, Yang JC, Ismail AM. Role of ABA in integrating plant responses to drought and salt stresses. Field Crops Research. 2006;97:111–119. doi: 10.1016/j.fcr.2005.08.018. [DOI] [Google Scholar]
- 122.Taji T, et al. Important roles of drought- and cold-inducible genes for galactinol synthase in stress tolerance in Arabidopsis thaliana. The Plant Journal. 2002;29:417–426. doi: 10.1046/j.0960-7412.2001.01227.x. [DOI] [PubMed] [Google Scholar]
- 123.Obendorf RL, Horbowicz M, Ueda T, Steadman KJ. Fagopyritols: Occurrence, biosynthesis, analyses, and possible roles. The European Journal of Plant Science and Biotechnology. 2012;6:27–36. [Google Scholar]
- 124.Sherameti I, Tripathi S, Varma A, Oelmuller R. The root-colonizing endophyte Pirifomospora indica confers drought tolerance in Arabidopsis by stimulating the expression of drought stress-related genes in leaves. Molecular Plant-Microbe Interactions. 2008;21:799–807. doi: 10.1094/MPMI-21-6-0799. [DOI] [PubMed] [Google Scholar]
- 125.Ghabooli M, et al. Proteomics study reveals the molecular mechanisms underlying water stress tolerance induced by Piriformospora indica in barley. Journal of Proteomics. 2013;94:289–301. doi: 10.1016/j.jprot.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 126.Alikhani M, et al. A proteomics approach to study the molecular basis of enhanced salt tolerance in barley (Hordeum vulgare L.) conferred by the root mutualistic fungus Piriformospora indica. Molecular BioSystems. 2013;9:1498–1510. doi: 10.1039/c3mb70069k. [DOI] [PubMed] [Google Scholar]
- 127.Trivedi DK, et al. Structure of RNA-interacting Cyclophilin A-like protein from Piriformospora indica that provides salinity-stress tolerance in plants. Scientific Reports. 2013;3:3001. doi: 10.1038/srep03001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lahrmann U, et al. Host-related metabolic cues affect colonization strategies of a root endophyte. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:13965–13970. doi: 10.1073/pnas.1301653110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Akum FN, Steinbrenner J, Biedenkopf D, Imani J, Kogel KH. The Piriformospora indica effector PIIN_08944 promotes the mutualistic Sebacinalean symbiosis. Frontiers in Plant Science. 2015;6:12. doi: 10.3389/fpls.2015.00906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Deshmukh SD, Kogel KH. Piriformospora indica protects barley from root rot caused by Fusarium graminearum. Journal of Plant Diseases and Protection. 2007;114:263–268. doi: 10.1007/BF03356227. [DOI] [Google Scholar]
- 131.Qiang XY, Zechmann B, Reitz MU, Kogel KH, Schafer P. The mutualistic fungus Piriformospora indica colonizes Arabidopsis roots by inducing an endoplasmic reticulum stress-triggered caspase-dependent cell death. Plant Cell. 2012;24:794–809. doi: 10.1105/tpc.111.093260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.McGonigle TP, Miller MH, Evans DG, Fairchild GL, Swan JA. A new method which gives an objective measure of colonization of roots by vesicular—arbuscular mycorrhizal fungi. New Phytologist. 1990;115:495–501. doi: 10.1111/j.1469-8137.1990.tb00476.x. [DOI] [PubMed] [Google Scholar]
- 133.Phillips JM, Hayman DS. Improved procedures for clearing roots and staining parasitic and vesicular-arbuscular mycorrhizal fungi for rapid assessment of infection. Transactions of the British Mycological Society. 1970;55:158–IN18. doi: 10.1016/S0007-1536(70)80110-3. [DOI] [Google Scholar]
- 134.Giovannetti M, Mosse B. An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytologist. 1980;84:489–500. doi: 10.1111/j.1469-8137.1980.tb04556.x. [DOI] [Google Scholar]
- 135.Simonne EH, Mills HA, Jones JB, Smittle DA, Hussey CG. A comparison of analytical methos for nitrogen analysis in plant-tissues. Communications in Soil Science and Plant Analysis. 1994;25:943–954. doi: 10.1080/00103629409369090. [DOI] [Google Scholar]
- 136.Matejovic I. Total nitrogen in plant material determinated by means of dry combustion: A possible alternative to determination by Kjeldahl digestion. Communications in Soil Science and Plant Analysis. 1995;26:2217–2229. doi: 10.1080/00103629509369441. [DOI] [Google Scholar]
- 137.Munter RC, Halverson TL, Anderson RD. Quality assurance for plant tissue analysis by ICP‐AES. Communications in Soil Science and Plant Analysis. 1984;15:1285–1322. doi: 10.1080/00103628409367559. [DOI] [Google Scholar]
- 138.Schiestl FP, et al. Evolution of ‘pollinator’ - attracting signals in fungi. Biology Letters. 2006;2:401–404. doi: 10.1098/rsbl.2006.0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Patel RK, Jain M. NGS QC Toolkit: A Toolkit for quality control of next generation sequencing data. Plos One. 2012;7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Jian B, et al. Validation of internal control for gene expression study in soybean by quantitative real-time PCR. Bmc Molecular Biology. 2008;9:14. doi: 10.1186/1471-2199-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Research. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Research. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Research. 2016;44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Transcriptome raw Illumina reads are deposited in the short read archive at NCBI under submission number SUB3229927.