Abstract
The progesterone receptor (PGR) plays a central role in maintaining pregnancy and is significantly associated with medical conditions such as preterm birth that affects 12.6% of all the births in U.S. PGR has been evolving rapidly since the common ancestor of human and chimpanzee, and we herein investigated evolutionary dynamics of PGR during recent human migration and population differentiation. Our study revealed substantial population differentiation at the PGR locus driven by natural selection, where very recent positive selection in East Asians has substantially decreased its genetic diversity by nearly fixing evolutionarily novel alleles. On the contrary, in European populations, the PGR locus has been promoted to a highly polymorphic state likely due to balancing selection. Integrating transcriptome data across multiple tissue types together with large-scale genome-wide association data for preterm birth, our study demonstrated the consequence of the selection event in East Asians on remodeling PGR expression specifically in the ovary and determined a significant association of early spontaneous preterm birth with the evolutionarily selected variants. To reconstruct its evolutionary trajectory on the human lineage, we observed substantial differentiation between modern and archaic humans at the PGR locus, including fixation of a deleterious missense allele in the Neanderthal genome that was later introgressed in modern human populations. Taken together, our study revealed substantial evolutionary innovation in PGR even during very recent human evolution, and its different forms among human populations likely result in differential susceptibility to progesterone-associated disease conditions including preterm birth.
Keywords: preterm birth, prematurity, risk factors, archaic humans, pregnancy, genetics, population substructure
Introduction
In mammalian species, the steroid hormone progesterone plays a central role in the process of reproduction in females. Progesterone affects the estrus or menstrual cycle, ovulation, pregnancy, parturition, and lactation.1, 2, 3, 4, 5 These effects are mediated by the interaction of progesterone with the intracellular progesterone receptor (PR) that, in the human and most other species, exists as two major isoforms—the full-length PR-B and the truncated (by 164 N-terminal amino acids) PR-A—each encoded by single gene, PGR (MIM: 607311), controlled by two promoters.6, 7, 8 In humans and some higher primates, parturition is triggered by changes in PR isoform function such that the capacity for progesterone/PR signaling to maintain pregnancy (mainly promotion of uterine quiescence) is lost.9, 10, 11 This functional progesterone/PR withdrawal trigger for parturition appears to be unique to hominids, and appropriate PR signaling is essential for the establishment and maintenance of pregnancy and for successful and timely parturition. Therefore, mutations interfering with PGR function are associated with an elevated risk of preterm birth (PTB), as well as of breast (MIM: 114480) and ovarian (MIM: 167000) cancers.12, 13, 14, 15, 16, 17
PGR arose early in the vertebrate lineage, resulting from multiple rounds of expansions from an ancestral estrogen receptor.18 Its early origin indicates conserved mechanism(s) underlying female reproduction in vertebrates. However, comparative genomic studies also identified significant sequence divergence of PGR among species, indicating potential evolutionary innovations in specific species through altering PGR sequences. For example, comparisons across apes, Old World monkeys, New World monkeys, prosimian primates, and other mammalian species revealed that excessive amino acid replacements in PGR were pronounced only in humans and chimpanzees, a signature of adaptive evolution.19 These evolutionary changes likely explain the unique features of the PGR-mediated pathways involving pregnancy, gestation, and parturition shared between humans and chimpanzees. More importantly, diverging from the chimpanzee lineage ∼10 million years ago,20 human-specific evolution resulted in a smaller birth canal (by remodeling the pelvis during the emergence of bipedalism) and larger head (by expanding cranium associated with encephalization).10, 21 These structural changes are expected to be accompanied by modifications on the genetic basis of these processes. Indeed, the human PGR harbors unique sequence changes distinct from chimpanzees as well as other primates, and the human-specific amino acid replacements were clustered in the inhibitory region of PGR that prevents PGR from transcribing downstream target genes.19 This observation suggests a significant rewiring of the regulatory network to modulate human-specific progesterone-signaling pathways and may underlie the apparently unique form of PR-mediated functional progesterone withdrawal that is thought to trigger human parturition.
These observed sequence changes in PGR were fixed on the human lineage.19 However, what remains unknown is its evolutionary dynamics in modern humans, especially during human migration and population differentiation (∼75,000 years ago).22 Given the prime role of PGR in female reproduction, from an evolutionary perspective, answering this question is fundamental to identifying aspects of increased fitness for adaptation to challenging environments during early human migration and particularly in understanding the differentiation between modern and archaic humans (e.g., Neanderthals). From an etiologic perspective, given substantial disparity in female reproductive traits among today’s race/ethnic populations,23 e.g., the PTB rate for East Asians is 7%, compared with 15%–19% in Africans (WHO 2010 report), studying population diversity of this critical gene, PGR, may reveal a biologic basis for such disparities, help identify risk loci, and could inform personalized prevention and treatment.
In this study, we demonstrate substantial population differentiation of PGR, driven by natural selection, where very recent positive selection had nearly fixed beneficial alleles in East Asians. Conversely, we observed that this gene has experienced rapid haplotype decay in Europeans with an elevated state of polymorphisms likely due to balancing selection. Our integrated analysis further demonstrated the effect of positive selection on remodeling PGR expression specifically in the ovary among East Asians for local adaption. Comparisons between modern and archaic humans revealed substantial differentiation at the PGR locus, including fixation of a known deleterious missense mutation in Neanderthals associated with extant increased risk of preterm birth and ovarian cancer. So, our study has demonstrated substantial evolutionary innovation of the human PGR, giving rise to different “forms” of PGR in different human populations as well as in archaic humans. This observation allows us to reconstruct the evolutionary trajectory of this gene during very recent human evolution, and also contributes, at least in part, to the observed biologic disparities in aspects of human pregnancy and parturition.
Material and Methods
Population Genetic Analysis
This study used the human genome build hg19/GRCh37. The genomic coordinates of PGR were based on Ensembl annotation (ENSG00000082175) at chr11: 100,900,355–101,001,255. Population analyses were performed on CHB (Han Chinese in Beijing, China), YRI (Yoruba in Ibadan, Nigeria), and CEU (Utah Residents with Northern and Western European Ancestry) populations sequenced by the 1000 Genomes Project. Test statistics for positive selection were computed in 1000 Genomes Selection Browser,24 where we compiled XP-CLR, XP-EHH, derived allele frequencies (DAFs), population nucleotide diversity π, extended haplotype homozygosity (EHH), and Tajima’s D scores from the database. For the analysis of XP-CLR and XP-EHH, we empirically determined the statistical significance based on the upper 5% percentiles of the scores across the entire genome in each pairwise comparison among the three populations. XP-CLR, nucleotide diversity π, and Tajima’s D were computed in sequence windows; XP-EHH and DAFs were computed at each SNP locus. Fixation index (Fst) was also computed at each PGR-associated SNP site based on Weir and Cockerham’s definition.25 Ancestral allele states were obtained from Ensembl (GRCH37.p13) and were derived from multiple alignment for primate species. In the linkage disequilibrium (LD) analysis, we identified 150 common SNPs (minor allele frequency, MAF ≥ 0.2) within the PGR gene body, and then computed their pairwise LD (R2) in CEU YRI and CHB, respectively. In CHB, 11 SNPs have reached complete fixation, which were excluded from the LD analysis in CHB. LD calculation was based on LDlink.26 When necessary, SNP allele frequencies were also queried from ExAC27 and HGDP projects.28 To determine the enrichment of HD-SNPs (SNPs with high-frequency derived alleles) in the PGR locus in CHB, a random permutation test was used. Briefly, for a given population, we randomly sampled a genomic region of the same size as that of the PGR locus (encompassing the same number of contiguous SNPs) and computed the fraction of SNPs with high derived allele frequencies (DAF ≥ 0.7) in the random window. We repeated the randomized procedure 1,000 times and estimated the empirical p value for enrichment of SNPs with DAF ≥ 0.7 in the PGR locus, i.e., among 1,000 random permutations, how many times we could sample a region with the fraction of HD-SNPs (those with DAF ≥ 0.7) greater than or equal to what was observed from the PGR locus. The random permutation test was performed in YRI, CEU, and CHB, respectively. To determine the increase in Tajima’s D in CEU, we performed a random permutation test as follows: each time, we randomly sampled from the genome the same number of contiguous windows of the same size as those in the PGR locus and tested the null hypothesis that the windows covering the PGR locus had no difference in their Tajima’s D distribution from a random genomic locus of the same size (encompassing the same number of contiguous variants). Therefore, a p value could be derived from the comparison. We performed the same random sampling 1,000 times with p values corrected using q values based on the method described previously.29 Overall, from the random permutation tests in CEU, 90% of the tests were at the false discovery rate of q = 0.05, and the maximum false discovery rate across all simulations was less than q = 0.14.
Genetic Association Study
The association study was based on Boston Birth Cohort, including 1,733 African American women, among which 698 were of preterm birth (PTB) and 1,035 term birth.30 In addition to splitting the samples into PTB case and control subjects, we further categorized the case samples into 461 with spontaneous preterm birth (sPTB) and the remaining 237 with medically indicated preterm birth (mPTB). From the sPTB group, we identified 115 case subjects that had less than 32 gestational weeks (early sPTB). The GWAS model and calculation followed the standard procedures in the SNPTEST package for dichotomous phenotypes (with both Bayesian and Frequentists tests).31 We used the table output directly from the software, including allele frequencies, genotype count, missing data fraction, minor allele frequencies, risk allele odds ratios, and risk genotype odds ratios.30 Odds ratios were computed at each SNP locus for the alternative allele; to standardize the odds ratios for derived alleles, the ancestral states of alternative alleles were determined; in case an alternative allele is not the derived allele, the reciprocal of its original odds ratio will be used. Detailed sample description and information for the genotyping platform can be found in the original study.30
Functional Genomic Analysis
The 86 HD-SNPs were examined for their role in regulating PGR expression in multiple human GTEx tissues. Specifically, we examined association between all PGR-associated SNPs with PGR expression in different tissues, and the significance of associations was multiple-hypothesis corrected for the 86 HD-SNPs queried. Loci with false discovery rate (FDR) ≤ 0.1 were considered significant. SNP effect sizes on PGR expression were also examined, where we considered their absolute values in our analysis. PGR expression was also queried from GTEx. Happloinsufficiency scores were obtained from DECIPHER (GRCH37), based on the method described by Huang et al.32 The ovary ATAC-seq data were recently generated as part of the ENCODE project, and the data have been deposited in ENCODE Data Portal (ENCFF181UOS). Two-fold increase in ATAC-seq peak intensity over control was considered significant.
Results
Differentiation of PGR among Human Populations
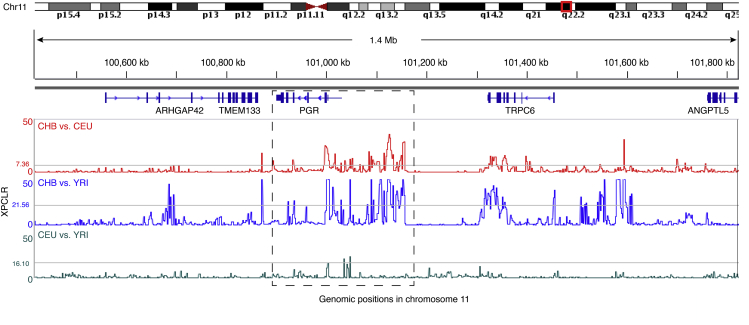
In the human genome, locus 11q22.2 encodes PGR (Figure 1). We analyzed populations in the 1000 Genomes Project,33 including CEU (Utah Residents with Northern and Western European Ancestry), YRI (Yoruba in Ibadan, Nigeria), and CHB (Han Chinese in Beijing, China), and determined the degree of population differentiation at the PGR locus referenced with that of the genome background. Extreme population differentiation at a particular genomic locus is usually indicative of positive selection resulting from population-specific local adaptation.34, 35 Since the conventional measure of population differentiation, the fixation index (Fst) at a single locus, is of low signal-to-noise ratio for detecting natural selection,36 we employed an improved approach (the XP-CLR test: the cross-population composite likelihood ratio test) to characterize population differentiation based on differences of multi-locus allele frequencies between populations.37 This multiple-locus composite likelihood ratio method has been shown to be robust against many factors including ascertainment bias, recombination rate heterogeneity, as well as variation in underlying population dynamics and substructure.37 We scanned the entire human genome with XP-CLR for pairwise comparisons involving CHB versus YRI, CEU versus YRI, and CHB versus CEU, and set YRI as the reference population given its ancestral state to non-African genomes.38, 39
Figure 1.
XP-CLR Scan to Detect Positive Selection in the PGR Locus
The peaks indicate signatures of positive selection that has differentiated the multi-locus allele frequency spectrum in one population from the other. Comparisons were performed for CHB-CEU, CHB-YRI, and CEU-YRI. Significance was empirically determined by the upper 5% XP-CLR scores across the entire genome for each comparison (the gray horizontal line). XP-CLR scan was performed with a maximum window of 0.1 cM.
Referenced with the genome background, we observed major XP-CLR peaks in the PGR locus (above the upper 5% of the genome background, the gray horizontal lines in Figure 1) from the CHB-YRI and CHB-CEU comparisons, but the signal was not apparent from the CEU-YRI comparison (Figure 1). Importantly, these XP-CLR peaks were distant from the neighboring genes surrounding PGR (ARHGAP42 [MIM: 615936], TMEM133 [MIM: NA], and TRPC6 [MIM: 603652]), precluding the possibility of a hitchhiking effect from other proximal genomic loci. Thus, these extreme XP-CLR peaks indicate differential allele usage in PGR between populations. To confirm the observed population differentiation at the PGR locus, we employed a complementary approach, XP-EHH (cross-population extended haplotype homozygosity), to scan the human genome, which is designed to identify genomic regions with extended haplotype homozygosity in a given population relative to a reference population.40 Because natural selection acts on haplotype blocks, loci with extended linkage disequilibrium (LD) in one population, but not the other, indicate recent positive selective sweeps due to population-specific local adaptation.40 Similar to the XP-CLR scan (Figure 1), the XP-EHH scan recapitulated the positive selection signals in the PGR locus, with extreme XP-EHH peaks (above the upper 95% of the genome background) in CHB-CEU and CHB-YRI comparisons, but not in the CEU-YRI comparison (Figure S1). Moreover, studying XP-EHH peaks surrounding the PGR locus excluded the explanation of a hitchhiking effect from its neighboring regions (Figure S1). Because XP-CLR and XP-EHH signals in the PGR locus were identified as extreme outliers from genome-wide scans (the upper 5% of the entire genome, Figures 1 and S1), the observations were less likely to be simply explained by population demographics (such as population bottlenecks) which presumably would affect the entire genome. Using the fully sequenced 1000 Genomes data minimized the potential impact from ascertainment bias, and, as demonstrated in previous studies, the test methods we employed are robust against ascertainment bias, heterogeneity in recombination rate, and population demography.37, 40 Thus, with these cross-population comparisons, we established that positive natural selection for local adaptation has differentiated the human PGR among populations, showing substantial genetic differences in the CHB-CEU and CHB-YRI comparisons, but not significant in the CEU-YRI comparison.
Different Forms of PGR Have Been Created by Recent Natural Selection in Different Populations
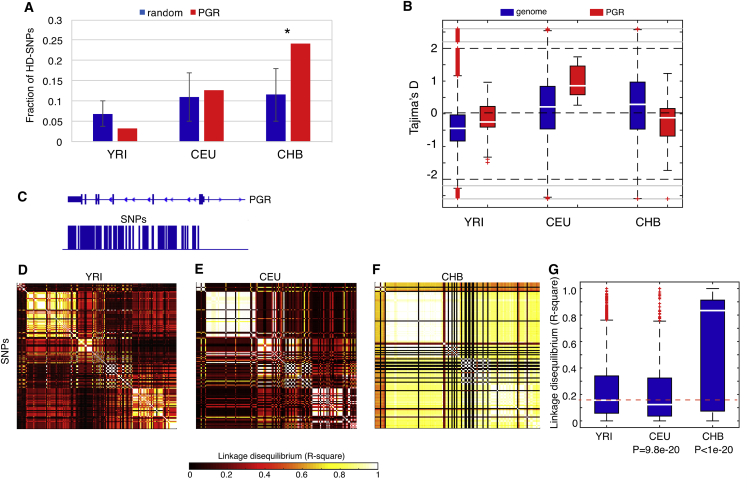
A parsimonious explanation for these observations is local adaptation acting on the PGR locus specifically in CHB, i.e., one that drove the pattern of its genetic variation away from that of other populations. Because both XP-CLR and XP-EHH detect selections that have (nearly) fixed derived beneficial alleles (e.g., the newly emerged alleles during human evolution, as opposed to the ancestral alleles also seen in other primate species) in a population,34 we asked whether in CHB, SNPs with high-frequency derived alleles (HD-SNPs) were enriched in the PGR locus, which is an expected signal from a recent selective sweep. Across the human genome, the ancestral state for each of the variants was identified from multiple primate species alignment (see Material and Methods), and derived allele frequencies (DAFs) were computed as one minus ancestral allele frequencies in CHB, CEU, and YRI (see Material and Methods). For each population, we considered HD-SNPs as those with DAF ≥ 0.7 (in the upper ∼10% across the genomes in three populations, Table S2) and observed that 24.2% of variants in PGR have DAFs ≥ 0.7 in CHB. We performed a random permutation test (see Material and Methods) and observed that the fraction was significantly higher than the CHB genome background at 12% (p = 0.037, Figure 2A). With the same procedure, the enrichment was not observed in YRI and CEU (Figure 2A), confirming the CHB-specific selection acting on this PGR locus.
Figure 2.
Intra-population Analysis Identified Differential Evolutionary Dynamics in the PGR Locus
(A) Enrichment of SNPs with high-frequency derived alleles (frequencies ≥ 0.7, HD-SNPs) in CHB but not in other populations (YRI and CEU). For each comparison, HD-SNP distribution was compared between the PGR locus and the genome background estimated from random permutation tests. Error bars indicate standard deviation, and the asterisk indicates statistical significance p < 0.05.
(B) Tajima’s D analysis for the PGR locus. This locus received a negative D score less than the genome background in CHB (a signature of positive selection), but the opposite trend in CEU (suggesting balancing selection)..
(C) The spatial distribution of the selected common SNPs for haplotype analysis.
(D–F) Linkage disequilibrium (LD, R2) analysis in the PGR locus for YRI (D), CEU (E), and CHB (F) based on the selected common SNPs.
(G) Distribution of R2 in each population, where CHB displayed increased LD, indicating long-range haplotypes, where the reduced R2 in CEU indicates rapid LD breakdown in CEU. Comparisons were referenced with YRI.
To further these observations, we examined nucleotide diversity (π) across genomes of the different populations, which is a statistical measure to quantify the degree of polymorphism within a population.41 As expected, the PGR locus exhibited a significantly reduced π among CHB relative to the genome background (p = 0.03, Wilcoxon rank-sum test, Figure S2), a signature of selective sweep that had eliminated genetic diversity as a consequence of promoting the frequencies of derived beneficial alleles. YRI exhibited a similar level of π at the PGR locus, compared with the genome background (p = 0.43, Wilcoxon rank-sum test, Figure S2), whereas a substantial increase of π in PGR was significant in CEU (p = 3.19e-4, Wilcoxon rank-sum test, Figure S2), indicating an excess of highly polymorphic sites at the PGR locus. To test whether the increased genetic diversity was potentially driven by selection forces (e.g., heterozygote advantage), we examined Tajima’s D scores.42 Consistent with the signature from positive selection, the PGR locus in CHB displayed a negative Tajima’s D, showing a substantial reduction from the genome background (Figure 2B). The same locus in CEU displayed the opposite trend with positive Tajima’s D values (Figure 2B). To confirm this observation, we performed a random permutation test, where the PGR locus was compared against 1,000 sets of randomly sampled genomic regions encompassing the same number of contiguous SNPs, and the PGR site consistently displayed increased Tajima’s D (Material and Methods). This observation, together with the increased nucleotide diversity, revealed that the PGR locus has been promoted to a highly polymorphic state during evolution in CEU. Although a significant positive Tajima’s D could be explained by either balancing selection or a substantial change in population size, the PGR locus is likely attributable to balancing selection, given its increased Tajima’s D compared with the genome background (population demography influences the entire genome). The PGR locus in YRI has remained negative (Figure 2B, close to zero). Taken together, natural positive selection has reshaped the PGR locus in East Asians (represented by CHB) by eliminating its genetic diversity to fix beneficial derived alleles for local adaption. In contrast, the PGR locus in Europeans (represented by CEU) has been promoted to a highly polymorphic state, likely resulting from balancing selection.
As a direct consequence of these differential selection forces, the PGR locus is expected to display differential haplotype structures among populations. For example, haplotypes with extended length (long-range association with other alleles), without having been broken down by recombination, are a typical signature of very recent positive selection, whereas a region with promoted sequence polymorphism is expected to exhibit rapid haplotype decay. To reveal haplotype structures, we identified 150 loci in PGR with common SNPs (minor allele frequency, MAF ≥ 0.2, Figure 2C, also see Material and Methods, Table S2), and computed their pairwise linkage disequilibrium (LD, R2) in YRI, CEU, and CHB (Figures 2D–2F). Note that these SNPs cover the major gene body of PGR (Figure 2C), and thus a complete genetic linkage map in the PGR locus could be revealed. Referenced with YRI and CEU, the PGR locus in CHB exhibited exceptionally strong LD even between distant loci (p < 1e−20, Wilcoxon rank-sum test, Figures 2F and 2G), demonstrating a strong effect from positive selection. Such long-range haplotype associations indicate that the selection event should be very recent (having not been significantly interfered with by recombination). However, the distribution of pairwise LD (R2) among loci in CEU showed a significant reduction relative to YRI (Figures 2E and 2G, p = 9.8e−20, Wilcoxon rank-sum test), reflecting a rapid LD decay in CEU due to increased polymorphism in PGR. We additionally performed the EHH (extended haplotype homozygosity) test43 and confirmed that the long haplotypes in the PGR locus indeed resulted from positive selection in CHB but not in CEU and YRI (Figure S3). Taken together, our LD analyses demonstrate differential haplotype structures of PGR in different human populations, resulting from differential evolutionary forces.
Functional Implication of Positive Selection on PGR in CHB
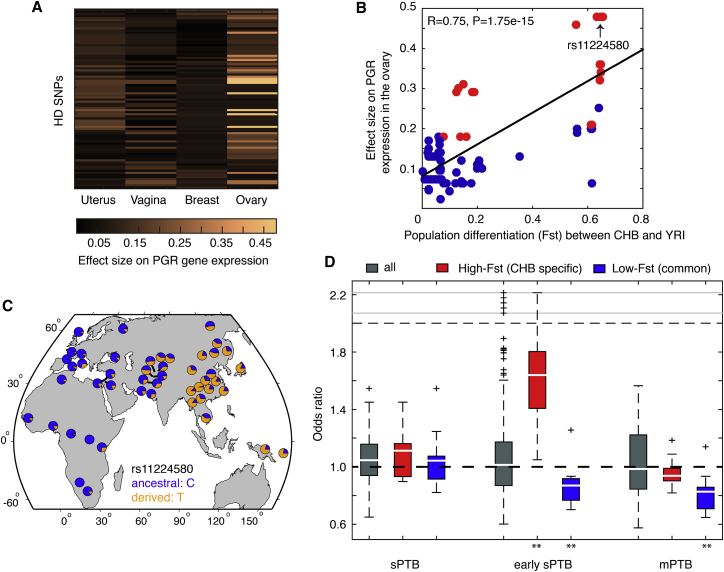
Positive selection implies a significant increase in organismal fitness by rapidly fixing evolutionarily novel alleles (derived alleles) in a population. We investigated the functional role(s) of the derived alleles that had risen to high frequency in CHB. In total, 86 loci in PGR have derived allele frequencies above 0.7 in CHB (the HD-SNPs, Figure 2A), constituting the signature of positive selection (Figure 2A). These SNPs (Table S1) are localized in the intronic and 3′ untranslated regions (3′ UTRs) of PGR, and thus we hypothesized their regulatory role in regulating PGR expression. We retrieved data from the Genotype-Tissue Expression project (GTEx, v7 release),44 which identifies genomic variants (by whole-genome sequencing) that influence gene expression (by RNA-seq) in 53 different human tissue types sampled from 714 donors (11,688 tissue samples in total). A regression model was built by the GTEx consortium to correlate genotypes of each SNP site with mRNA expression of its associated genes (the cis-effect, expression quantitative trait loci), taking into account many other co-variates, such as sex, age, tissue quality, and experimental platforms.44 For each SNP locus, the degree of contribution to gene expression was quantified by its effect size, where greater effect size indicates stronger expression alterations by varying genotypes of this SNP across individuals in the GTEx cohort. The comprehensive transcriptome data provided a global view of PGR expression across multiple human tissues, and its strong tissue-specific gene expression pattern was immediately revealed. PGR was most pronounced in the cervix, fallopian tube, ovary, uterus, and vagina, moderate in the artery and breast mammary tissue, and depleted in all others (e.g., the brain, whole blood, etc., Figure S4). We analyzed the effect sizes of the HD-SNPs on PGR expression in the uterus, vagina, ovary, as well as in the breast mammary tissue (data from the cervix and fallopian tube were unavailable in the current GTEx release). Interestingly, 23 among the 86 HD-SNPs (27%, Table S3) exhibited statistically significant effect size on modulating PGR expression in the ovary (FDR ≤ 0.1, Figure 3A), whereas none showed significance in all the other tissue types, including the uterus, vagina, and breast mammary tissue (FDR > 0.1, Figure 3A).
Figure 3.
Functional Analysis of the Loci Affected by Positive Selection in CHB
(A) The effect size of the 86 HD-SNPs on modulating PGR expression in the uterus, breast mammary tissue, vagina, and ovary, where 23 loci showed statistical significance (FDR ≤ 0.1) only in the ovary. Multiple-hypothesis correction was performed with the Benjamini-Hochberg procedure across the query SNPs.
(B) A significant correlation between the effect size and population differentiation (Fst, CHB versus YRI) for the HD-SNPs, where rs11224580 represents an exemplar site with a strong effect on modulating PGR in the ovary, and its high derived allele frequency is specific in CHB but not in CEU and YRI.
(C) Allelic distribution of rs11224580 across the world populations. Allele frequency data were obtained by the Human Genome Diversity Project (HGDP).28 The ancestral and derived alleles were also labeled.
(D) Distribution of odds ratios for spontaneous preterm birth (sPTB), early spontaneous preterm birth (≤32 gestational weeks, early sPTB), and medically indicated preterm birth (mPTB) for all the variants in the PGR locus, as well for the subsets of HD-SNPs with high derived allele frequencies specific in CHB, or shared among all populations. Odds ratios were all standardized on the derived alleles. Statistical significance was determined by Wilcoxon rank-sum test, where the asterisks indicate p values less than 0.01.
We then computed the degree of population differentiation (Fst) for each of the HD-SNPs (CHB versus CEU and CHB versus YRI): the high-Fst sites are those with high derived allele frequencies only in CHB, but low in others, whereas low-Fst sites indicate that the status of high derived allele frequencies is shared among human populations (compared with ancestral alleles in other primate species). Interestingly, for these HD-SNPs, we observed a strong positive correlation between Fst and the effect size on modulating PGR expression in the ovary (R = 0.75, p = 1.75e−15, Figures 3B and S5), indicating that, for these HD-SNPs, if their status of high derived allele frequencies is more specific in CHB, they tend to have stronger effects on PGR expression in the ovary. Therefore, positive selection in CHB on PGR was to remodel PGR expression specifically in the ovary. One extreme example is the SNP rs11224580 (in the 3rd intron, upstream of the 4th exon) with a strong effect size on PGR ovary expression (FDR = 0.02, Figure 3B) and substantial population differentiation (Fst > 0.6 for both CHB-CEU and CHB-YRI comparisons, Figure 3B). Examining its derived allele frequencies across worldwide indigenous populations,28 we observed the strong population differentiation of rs11224580 as illustrated in Figure 3C, where East Asia and the neighboring regions preferentially use the derived allele T, while the ancestral form, C, is dominant in African and European populations. This integrative analysis determined the regulatory role of the evolutionarily novel alleles in PGR that had been promoted to high frequencies in East Asians. The detected positive selection signal thus suggested regulatory innovations for local adaptation.
To determine the physiological importance of altering PGR expression, we investigated dosage sensitivity data across all human genes (haploinsufficiency scores)32 and observed extreme dosage sensitivity of PGR, ranking at the top 3% of the human genome (Figure S6). This observation confirmed the functional consequences of altering PGR expression. Therefore, remodeling PGR expression in East Asians by rapidly fixing the regulatory derived alleles substantially contributed to selective advantages for local adaptation. This observation predicts that, because of PGR dosage sensitivity, the remodeled PGR expression is only advantageous in East Asians, not compatible with other populations. In other words, the regulatory alleles specifically expanded in East Asians (the high-Fst HD-SNPs) would be expected to be deleterious in other populations.
We tested this prediction at the phenotypic level, allowing us to associate phenotypic traits with these evolutionarily selected alleles. Given the pivotal role of PGR in maintaining pregnancy, we examined subjects enrolled in the Boston Birth Cohort, including 1,733 African American women, 461 of whom had spontaneous PTB (sPTB), 237 had medically indicated PTB (mPTB), and 1,035 had term births.30 From the sPTB group, we identified 115 that occurred at less than 32 gestational weeks (early sPTB). Genome-wide association studies (GWASs) were then performed by comparing sPTB, mPTB, and early sPTB groups against the term control group. SNPs with low allele frequencies in this cohort (minor allele frequency less than 10%, estimated from the term control group) were excluded from analysis due to their numerical instability in estimating risk odds ratios, and GWAS odds ratios were standardized on derived alleles. Referenced with the African population (YRI), among the 86 HD-SNPs, we identified 20 with derived allele frequencies specifically high in East Asians (CHB, Fst ≥ 0.5, see the distribution in Figure S7), and 42 SNPs with high derived allele frequencies common in both CHB and YRI (Fst ≤ 0.1, Figure S7). African American women carrying these CHB-specific derived alleles exhibited a significant increase in the odds ratio for early sPTB, relative to all the derived alleles in the PGR locus (p = 4.01e−8, Wilcoxon rank-sum test, Figure 3D). This observation supports our prediction that remodeling PGR expression by expanding the derived regulatory alleles is advantageous only in East Asians for local adaptation (given positive selection signals), but is deleterious in other populations. Note that the risk increase was only significant in the early sPTB group (Figure 3D), suggesting a strong fitness consequence associated with early sPTB. In the meantime, we also examined variants with derived alleles at high frequencies across all human populations. These high-frequency derived alleles exhibited significantly reduced risk for early sPTB (p = 1.71e−6, Wilcoxon rank-sum test, Figure 3D) and mPTB (p = 2.26e−8, Wilcoxon rank-sum test, Figure 3D). Because the derived alleles were evolutionarily novel on the human lineage, this observation suggests evolutionary innovations in the PGR locus, prior to population differentiation, that had increased organismal fitness by reducing early sPTB and mPTB risk.
The Neanderthal Form of PGR
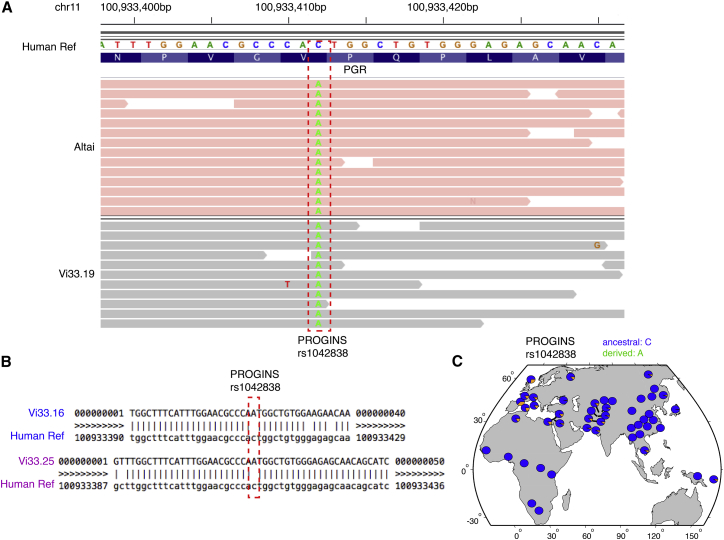
Our population and functional genomic analyses have revealed substantial evolutionary plasticity of PGR in modern human populations. To reconstruct its evolutionary trajectory on the human lineage, it is necessary to examine PGR in archaic humans, represented by Neanderthals that diverged from anatomical modern humans 520,000–630,000 years ago.45 We examined the PGR sequences in two female Neanderthals, whose genomes were sequenced with high coverage and quality. One Neanderthal, identified in the Altai Mountains in southern Siberia, lived ∼122,000 years ago (the Altai Neanderthal, 50× coverage),46 and the other was found in Vindija Cave in Croatia and lived ∼52,000 years ago (the Vindija 33.19 Neanderthal, 30× coverage).45
The Neanderthal PGR locus (spanning 100.9 kb) displayed extended sequence homozygosity. For example, the Vindija 33.19 genome even had no heterozygous sites, in contrast with hundreds of highly polymorphic sites in the same region in modern humans (Figure 2C). This observation is consistent with substantial inbreeding and small population size in Neanderthals45, 46 and suggests that deleterious mutations likely had been fixed in the Neanderthal population. Among many variants identified in PGR, the PROGINs alleles are the most extensively studied, characterized by one missense mutation, V660L (rs1042838, exon 4), one synonymous mutation, H770H (rs1042839, exon 5), and one 320-bp Alu insertion between exon 7 and 8.47, 48 Specifically, for the missense variant (rs1042838), its minor allele (A) has been associated with ovarian cancer49, 50 and preterm birth.51 This risk allele reduces the responsiveness of PGR to progesterone,52 consistent with the predicted deleteriousness of this missense mutation at the top 1% of the human genome by CADD.53
By analyzing the diploid genomes of the two female Neanderthals, we observed a homozygous state of this risk allele (A) in both Neanderthal individuals (Figure 4A). To confirm this observation, we further examined the low-coverage genome sequences of three additional female Neanderthals in an earlier study (Vi33.16, Vi33.25, and Vi33.26 from Vindija Cave in Croatia),54 where only Vi33.16 and Vi33.25 had sequenced reads covering this locus. Again, the risk allele A was observed in these Neanderthals, with the absence of the reference allele C at this locus (Figure 4B). Given the geographical distance between the Altai Mountains and Vindija Cave, as well as the temporal distance (∼130,000−145,000 years ago) between the Altai and Vindija Neanderthals,45 it is likely that the risk allele of the missense PROGINs site had been fixed in Neanderthals. Therefore, fixation of the risk allele in this pregnancy-associated gene in Neanderthals likely posed a significant selective disadvantage.
Figure 4.
Genetic Differentiation between Modern Humans and Neanderthals at the Missense Site (rs1042838) of the Well-Known PROGINS Alleles in the PGR Locus
(A) Sequenced reads surrounding the missense site in Altai and Vindija (Vi33.19) Neanderthals (both females), where the missense mutation, A, were in a homozygous state in the Neanderthals.
(B) Validation on two additional Neanderthal genomes (Vi33.16 and Vi33.25, both females, with low sequencing coverage) confirmed the prevalence of the missense mutated base, A, in Neanderthals.
(C) Allelic distribution of this missense SNP (rs1042838) in modern human populations. Allele frequency data were obtained by the Human Genome Diversity Project (HGDP).28 Note that the mutated base, A, is a derived allele.
To trace the evolutionary origin of this risk allele, we first determined the derived status of this allele by referencing with other primate species. In modern humans, this derived risk allele is underrepresented (Figure 4C, data from Human Genome Diversity Project28), with increased frequencies in European (18%, the 1000 Genomes estimate) and South Asian (7%, the 1000 Genomes estimate) populations, compared with its absence in African populations (Figure 4C). Given the fixed status of this allele in Neanderthals, this allele frequency pattern in human populations suggests a potential Neanderthal origin of this risk allele that was later introgressed in modern human’s genome. This notion is confirmed by the Neanderthal introgression map55 as well as by a more recent study using rigorous criteria to identify alleles of Neanderthal origin in modern human’s genome.56 We further investigated the sequence of the high-coverage Denisovan genome,57 an extinct archaic human who lived 72,000 years ago,45 but did not observe the risk allele from Denisovans, confirming its specific Neanderthal origin. Given its biochemical effect on progesterone responsiveness52 and its phenotypic consequences on preterm birth51 and ovarian cancer,49, 50 introgression of this archaic Neanderthal allele in modern humans likely contributes to phenotypic variances underlying these conditions.
Discussion
We performed population and functional genomic analyses for the human PGR, a gene critical for female pregnancy and reproduction. Our study revealed substantial evolutionary influence on the PGR locus during human migration and population differentiation, where rapid fixation of derived alleles by positive selection was evident in East Asians. On the contrary, a substantial increase in sequence polymorphisms was identified in European populations, likely driven by balancing selection. Therefore, different “forms” of PGR have been shaped by natural selection for local adaption, and thus its functional diversity is anticipated among modern human populations, which may constitute the biologic basis of many disparities associated with the progesterone-mediated pathways.
We performed cross-population comparisons and identified a positive selection signature acting on the PGR locus in East Asians (CHB). The selection signals in the PGR locus were identified as outliers across the genome: for example, the strongest XP-CLR signal at PGR for CHB-CEU comparison was ranked at the 9,453th position across the ∼1.2 million XPCLR windows examined, representing the top 0.79% across the genome. Notably, the XP-EHH test for CHB-CEU comparison indicated that the signal at PGR was ranked at the 42th position across ∼15 million SNPs examined, representing an extreme outlier of the genome (the 0.00028% upper percentile). In addition to these cross-population analysis, we independently confirmed the signals with complementary test statistics in robust intra-population analyses. Most notably, LD analysis revealed a long haplotype range of this locus in CHB, suggesting that the selection force was recent in human history. Given the different patterns observed from the African and European populations (Figures 1 and 2), the positive selection likely occurred after the population split between Europeans and Asians. Interestingly, examining sequences of a 40,000-year-old individual from Tianyuan Cave, China,58 several high-frequency derived alleles observed only in CHB were also identified in the Tianyuan individual, dating the selection event back to at least 40,000 years ago. Because East Asians are genetically related to American Indians,59, 60 it is interesting to further test the selection signature in native Americans. We examined the admixed American populations in the 1000 Genomes database (data not shown), and indeed observed similar allele usage at the PGR locus in PEL (Peruvians from Lima, Peru, although the signal is much attenuated), but not in PUR (Puerto Ricans from Puerto Rico) and CLM (Colombians from Medellin, Colombia). However, the observations could also be attributed to recent population admixture since the 19th century. Therefore, these observations await further confirmation when more indigenous American Indian genomes are available.
To investigate the physiological significance of the selection event in East Asians, we specifically examined the evolutionarily novel alleles (i.e., derived alleles) rising to high frequencies in CHB (i.e., the HD-SNPs), particularly the ones whose high derived allele frequencies were specific in CHB, an indicator of targets of positive selection. We leveraged the GTEx dataset and determined the molecular function of these selected alleles in remodeling PGR expression in the ovary. With the Boston Birth Cohort, we were able to associate the early sPTB risk with these evolutionarily novel alleles. Because the identified variants were primarily localized in intronic regions, their regulatory role is presumably achieved by perturbing regulatory elements in the PGR introns. We recently generated ATAC-seq data in the ovary (as part of the ENCODE project61), which mark all potential regulatory elements in the ovary genome. Indeed, we identified abundant regulatory elements in PGR introns (Figure S8), which will help identify the causal variants driving the observed adaptive selection in East Asians.
Because of strong dosage sensitivity, we showed that remodeling PGR ovary expression was advantageous only in East Asians for local adaptation but was deleterious in other populations (Figure 3D). We particularly note that although East-Asian-specific alleles are at low frequencies in other populations, such as the derived allele of SNP rs11224580 (Figure 3C), they likely constitute the biologic basis for disparate PTB risk in a population. These observations revealed a significant association between PGR expression in the ovary and the clinical outcome of preterm birth. Therefore, future study is warranted to identify the molecular mechanisms for the regulation of gestational timing and preterm birth risk by the ovary-specific PGR expression. Further, studying derived alleles in the PGR locus with high frequencies common in human populations, they displayed strong effects in reducing early sPTB and mPTB risk, strongly suggesting evolutionary innovations on the human lineage to confer additional selective advantage since the divergence from chimpanzees. On the other hand, it also indicates strong effects of early sPTB and mPTB on organismal fitness, but not late PTBs (>32 gestational weeks), which is consistent with the clinical outcomes of early sPTB and mPTB. For example, without appropriate medical care, early sPTBs would be associated with a high infant mortality rate. The majority of mPTB in modern times is a consequence of preeclampsia.62 Induced delivery, often preterm, is the only medical measure that can fully ameliorate such pregnancy-associated hypertension. Short of medically induced preterm delivery, the mother and baby will die. Thus, in historical periods, such a pregnancy complication was synonymous with maternal and fetal mortality. Taken together, during early human population differentiation, reducing early sPTB and mPTB risk would be expected to have a significant impact on enhancing organismal fitness.
In the European population, we observed an opposite trend, where the PGR locus had been highly diversified, likely driven by balancing selection given its positive Tajima’s D substantially increased from the genome background (Figure 2B). An increase in polymorphisms is usually required by the immune system,63, 64 which is also an important component to the parturition trigger mechanism,65 and PGR itself also modulates IFN-γ in CD4+ T cells.66 As such, susceptibility to inflammation-induced PTB might be affected by PGR, and its diversification might very well reflect a need for local adaptation when migrating to the European continent.
We also examined Neanderthal genomes and identified the fixation of the risk allele at the missense PROGINS site (rs1042838) in the Neanderthal population. This risk allele interferes with the responsiveness of PGR to progesterone,52 and thus likely posed a significant selective disadvantage to Neanderthals, perhaps contributing to their extinction 38,000 years ago.67 Intriguingly, we examined genome-wide data for alleles of Neanderthal origin55, 56 and confirmed the introgressed status of this risk allele in modern human populations from the Neanderthal genome. Therefore, our observation provides new evidence for the contribution to human diseases from Neanderthal alleles.68, 69
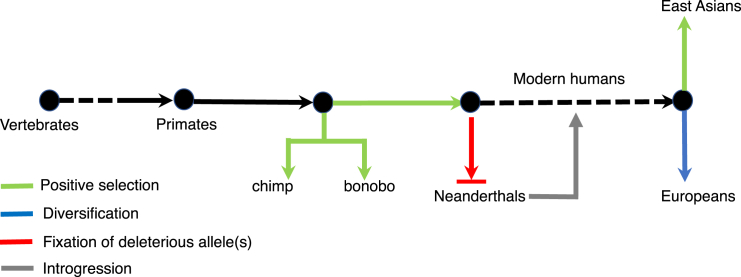
Our study allowed us to reconstruct a more complete evolutionary trajectory of PGR (Figure 5). This gene evolved from an ancestral estrogen receptor after multiple rounds of duplication on the vertebrate lineage.18 In primate species, positive selection had accelerated its evolution since the common ancestor of human and chimpanzee19 and fixed human-specific amino acid replacements specifically in the IF domain of the protein product of this gene.19 These alterations likely established human-specific traits by rewiring the PGR-mediated regulatory network. During early human evolution, substantial inbreeding and population bottleneck likely fixed novel deleterious alleles in archaic humans, exemplified by the PROGINs-derived allele that were specific in Neanderthals (Figure 4), and this allele was subsequently introgressed in the modern human genome. In modern humans, prior to population differentiation, many derived alleles in PGR had risen to high frequencies, which substantially reduced the rate of early sPTB and mPTB (Figure 3D), thereby contributing to organismal fitness. After population migration and differentiation, positive selection had acted on PGR in East Asians for local adaptation, whereas the PGR locus had been diversified in European populations (Figure 5).
Figure 5.
Reconstruction of the Evolutionary Dynamics of PGR
Arrows are colored by evolutionary events, and the dashed lines indicate relatively long-time window with no sufficient number of species sampled. The length of the arrows is not scaled by evolutionary time.
Overall, in this study, we presented our evolutionary analysis for a critical human reproductive gene, PGR, during human population differentiation. Even within such a short time window, natural selection has significantly differentiated this gene among human populations for local adaptation, highlighting the unique role of this gene in increasing human fitness during migration to and settlement in new challenging environments. We have shown that PGR has presented different forms among different populations with presumably differential molecular functions, likely resulting in differential disease susceptibility associated with progesterone, such as preterm birth and ovarian cancer.
Acknowledgments
The authors thank the anonymous reviewers for their constructive comments. This study was funded by March of Dimes Prematurity Research Center at Stanford University School of Medicine (22-FY18-808) and NIH/NHLBI (grant number RC2 HL101748). M.S. acknowledges NIH (grant 5P50HG00773502) and CIRM (grant GC1R-06673-A). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: June 21, 2018
Footnotes
Supplemental Data include eight figures and three tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.05.009.
Contributor Information
David K. Stevenson, Email: dstevenson@stanford.edu.
Gary M. Shaw, Email: gmshaw@stanford.edu.
Web Resources
1000 Genomes, http://www.internationalgenome.org/
1000 Genomes Selection Browser, http://hsb.upf.edu
Boston Birth Cohort, https://www.jhsph.edu/departments/population-family-and-reproductive-health/center-on-early-life-origins-of-disease/projects.html
DECIPHER (for PGR), https://decipher.sanger.ac.uk/gene/PGR#overview/clinical-info
ENCODE Portal, https://www.encodeproject.org
Ensembl, http://grch37.ensembl.org/index.html
GTEx Portal, https://www.gtexportal.org/home/
Human Genome Diversity Project (HGDP), http://www.hagsc.org/hgdp/
SNPTEST, https://mathgen.stats.ox.ac.uk/genetics_software/snptest/old/snptest_v2.3.0.html
Supplemental Data
Reference
- 1.Csapo A.I. The onset of labour. Lancet. 1961;2:277–280. doi: 10.1016/s0140-6736(61)90576-1. [DOI] [PubMed] [Google Scholar]
- 2.Henson M.C. Pregnancy maintenance and the regulation of placental progesterone biosynthesis in the baboon. Hum. Reprod. Update. 1998;4:389–405. doi: 10.1093/humupd/4.4.389. [DOI] [PubMed] [Google Scholar]
- 3.Mesiano S. Roles of estrogen and progesterone in human parturition. Front. Horm. Res. 2001;27:86–104. doi: 10.1159/000061038. [DOI] [PubMed] [Google Scholar]
- 4.Zalányi S. Progesterone and ovulation. Eur. J. Obstet. Gynecol. Reprod. Biol. 2001;98:152–159. doi: 10.1016/s0301-2115(01)00361-x. [DOI] [PubMed] [Google Scholar]
- 5.Barbieri R.L. The endocrinology of the menstrual cycle. Methods Mol. Biol. 2014;1154:145–169. doi: 10.1007/978-1-4939-0659-8_7. [DOI] [PubMed] [Google Scholar]
- 6.Giangrande P.H., McDonnell D.P. The A and B isoforms of the human progesterone receptor: two functionally different transcription factors encoded by a single gene. Recent Prog. Horm. Res. 1999;54:291–313, discussion 313–314. [PubMed] [Google Scholar]
- 7.Li X., O’Malley B.W. Unfolding the action of progesterone receptors. J. Biol. Chem. 2003;278:39261–39264. doi: 10.1074/jbc.R300024200. [DOI] [PubMed] [Google Scholar]
- 8.Wen D.X., Xu Y.F., Mais D.E., Goldman M.E., McDonnell D.P. The A and B isoforms of the human progesterone receptor operate through distinct signaling pathways within target cells. Mol. Cell. Biol. 1994;14:8356–8364. doi: 10.1128/mcb.14.12.8356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mesiano S., Chan E.C., Fitter J.T., Kwek K., Yeo G., Smith R. Progesterone withdrawal and estrogen activation in human parturition are coordinated by progesterone receptor A expression in the myometrium. J. Clin. Endocrinol. Metab. 2002;87:2924–2930. doi: 10.1210/jcem.87.6.8609. [DOI] [PubMed] [Google Scholar]
- 10.Smith R. Parturition. N. Engl. J. Med. 2007;356:271–283. doi: 10.1056/NEJMra061360. [DOI] [PubMed] [Google Scholar]
- 11.Oh S.Y., Kim C.J., Park I., Romero R., Sohn Y.K., Moon K.C., Yoon B.H. Progesterone receptor isoform (A/B) ratio of human fetal membranes increases during term parturition. Am. J. Obstet. Gynecol. 2005;193:1156–1160. doi: 10.1016/j.ajog.2005.05.071. [DOI] [PubMed] [Google Scholar]
- 12.Ehn N.L., Cooper M.E., Orr K., Shi M., Johnson M.K., Caprau D., Dagle J., Steffen K., Johnson K., Marazita M.L. Evaluation of fetal and maternal genetic variation in the progesterone receptor gene for contributions to preterm birth. Pediatr. Res. 2007;62:630–635. doi: 10.1203/PDR.0b013e3181567bfc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manuck T.A., Lai Y., Meis P.J., Dombrowski M.P., Sibai B., Spong C.Y., Rouse D.J., Durnwald C.P., Caritis S.N., Wapner R.J. Progesterone receptor polymorphisms and clinical response to 17-alpha-hydroxyprogesterone caproate. Am. J. Obstet. Gynecol. 2011;205:135.e1–135.e9. doi: 10.1016/j.ajog.2011.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manuck T.A., Major H.D., Varner M.W., Chettier R., Nelson L., Esplin M.S. Progesterone receptor genotype, family history, and spontaneous preterm birth. Obstet. Gynecol. 2010;115:765–770. doi: 10.1097/AOG.0b013e3181d53b83. [DOI] [PubMed] [Google Scholar]
- 15.Diep C.H., Daniel A.R., Mauro L.J., Knutson T.P., Lange C.A. Progesterone action in breast, uterine, and ovarian cancers. J. Mol. Endocrinol. 2015;54:R31–R53. doi: 10.1530/JME-14-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langmia I.M., Apalasamy Y.D., Omar S.Z., Mohamed Z. Progesterone Receptor (PGR) gene polymorphism is associated with susceptibility to preterm birth. BMC Med. Genet. 2015;16:63. doi: 10.1186/s12881-015-0202-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohammed H., Russell I.A., Stark R., Rueda O.M., Hickey T.E., Tarulli G.A., Serandour A.A., Birrell S.N., Bruna A., Saadi A. Progesterone receptor modulates ERα action in breast cancer. Nature. 2015;523:313–317. doi: 10.1038/nature14583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thornton J.W. Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions. Proc. Natl. Acad. Sci. USA. 2001;98:5671–5676. doi: 10.1073/pnas.091553298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C., Opazo J.C., Erez O., Uddin M., Santolaya-Forgas J., Goodman M., Grossman L.I., Romero R., Wildman D.E. The human progesterone receptor shows evidence of adaptive evolution associated with its ability to act as a transcription factor. Mol. Phylogenet. Evol. 2008;47:637–649. doi: 10.1016/j.ympev.2007.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moorjani P., Amorim C.E., Arndt P.F., Przeworski M. Variation in the molecular clock of primates. Proc. Natl. Acad. Sci. USA. 2016;113:10607–10612. doi: 10.1073/pnas.1600374113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenberg K., Trevathan W. Birth, obstetrics and human evolution. BJOG. 2002;109:1199–1206. doi: 10.1046/j.1471-0528.2002.00010.x. [DOI] [PubMed] [Google Scholar]
- 22.Rasmussen M., Guo X., Wang Y., Lohmueller K.E., Rasmussen S., Albrechtsen A., Skotte L., Lindgreen S., Metspalu M., Jombart T. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science. 2011;334:94–98. doi: 10.1126/science.1211177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Owen C.M., Goldstein E.H., Clayton J.A., Segars J.H. Racial and ethnic health disparities in reproductive medicine: an evidence-based overview. Semin. Reprod. Med. 2013;31:317–324. doi: 10.1055/s-0033-1348889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pybus M., Dall’Olio G.M., Luisi P., Uzkudun M., Carreño-Torres A., Pavlidis P., Laayouni H., Bertranpetit J., Engelken J. 1000 Genomes Selection Browser 1.0: a genome browser dedicated to signatures of natural selection in modern humans. Nucleic Acids Res. 2014;42:D903–D909. doi: 10.1093/nar/gkt1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weir B.S., Cockerham C.C. Estimating F-Statistics for the Analysis of Population Structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 26.Machiela M.J., Chanock S.J. LDlink: a web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2015;31:3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lek M., Karczewski K.J., Minikel E.V., Samocha K.E., Banks E., Fennell T., O’Donnell-Luria A.H., Ware J.S., Hill A.J., Cummings B.B., Exome Aggregation Consortium Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cavalli-Sforza L.L. The Human Genome Diversity Project: past, present and future. Nat. Rev. Genet. 2005;6:333–340. doi: 10.1038/nrg1596. [DOI] [PubMed] [Google Scholar]
- 29.Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hong X., Hao K., Ji H., Peng S., Sherwood B., Di Narzo A., Tsai H.J., Liu X., Burd I., Wang G. Genome-wide approach identifies a novel gene-maternal pre-pregnancy BMI interaction on preterm birth. Nat. Commun. 2017;8:15608. doi: 10.1038/ncomms15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wellcome Trust Case Control C., Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang N., Lee I., Marcotte E.M., Hurles M.E. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6:e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sabeti P.C., Schaffner S.F., Fry B., Lohmueller J., Varilly P., Shamovsky O., Palma A., Mikkelsen T.S., Altshuler D., Lander E.S. Positive natural selection in the human lineage. Science. 2006;312:1614–1620. doi: 10.1126/science.1124309. [DOI] [PubMed] [Google Scholar]
- 35.Barreiro L.B., Laval G., Quach H., Patin E., Quintana-Murci L. Natural selection has driven population differentiation in modern humans. Nat. Genet. 2008;40:340–345. doi: 10.1038/ng.78. [DOI] [PubMed] [Google Scholar]
- 36.Holsinger K.E., Weir B.S. Genetics in geographically structured populations: defining, estimating and interpreting F(ST) Nat. Rev. Genet. 2009;10:639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen H., Patterson N., Reich D. Population differentiation as a test for selective sweeps. Genome Res. 2010;20:393–402. doi: 10.1101/gr.100545.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tishkoff S.A., Dietzsch E., Speed W., Pakstis A.J., Kidd J.R., Cheung K., Bonné-Tamir B., Santachiara-Benerecetti A.S., Moral P., Krings M. Global patterns of linkage disequilibrium at the CD4 locus and modern human origins. Science. 1996;271:1380–1387. doi: 10.1126/science.271.5254.1380. [DOI] [PubMed] [Google Scholar]
- 39.Zietkiewicz E., Yotova V., Jarnik M., Korab-Laskowska M., Kidd K.K., Modiano D., Scozzari R., Stoneking M., Tishkoff S., Batzer M., Labuda D. Genetic structure of the ancestral population of modern humans. J. Mol. Evol. 1998;47:146–155. doi: 10.1007/pl00006371. [DOI] [PubMed] [Google Scholar]
- 40.Sabeti P.C., Varilly P., Fry B., Lohmueller J., Hostetter E., Cotsapas C., Xie X., Byrne E.H., McCarroll S.A., Gaudet R., International HapMap Consortium Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–918. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nei M., Li W.H. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA. 1979;76:5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sabeti P.C., Reich D.E., Higgins J.M., Levine H.Z., Richter D.J., Schaffner S.F., Gabriel S.B., Platko J.V., Patterson N.J., McDonald G.J. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 44.Battle A., Brown C.D., Engelhardt B.E., Montgomery S.B., GTEx Consortium. Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group. Statistical Methods groups—Analysis Working Group. Enhancing GTEx (eGTEx) groups. NIH Common Fund. NIH/NCI. NIH/NHGRI. NIH/NIMH. NIH/NIDA. Biospecimen Collection Source Site—NDRI. Biospecimen Collection Source Site—RPCI. Biospecimen Core Resource—VARI. Brain Bank Repository—University of Miami Brain Endowment Bank. Leidos Biomedical—Project Management. ELSI Study. Genome Browser Data Integration &Visualization—EBI. Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz. Lead analysts. Laboratory, Data Analysis &Coordinating Center (LDACC) NIH program management. Biospecimen collection. Pathology. eQTL manuscript working group Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. [Google Scholar]
- 45.Prüfer K., de Filippo C., Grote S., Mafessoni F., Korlević P., Hajdinjak M., Vernot B., Skov L., Hsieh P., Peyrégne S. A high-coverage Neandertal genome from Vindija Cave in Croatia. Science. 2017;358:655–658. doi: 10.1126/science.aao1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prüfer K., Racimo F., Patterson N., Jay F., Sankararaman S., Sawyer S., Heinze A., Renaud G., Sudmant P.H., de Filippo C. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 2014;505:43–49. doi: 10.1038/nature12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rowe S.M., Coughlan S.J., McKenna N.J., Garrett E., Kieback D.G., Carney D.N., Headon D.R. Ovarian carcinoma-associated TaqI restriction fragment length polymorphism in intron G of the progesterone receptor gene is due to an Alu sequence insertion. Cancer Res. 1995;55:2743–2745. [PubMed] [Google Scholar]
- 48.Agoulnik I.U., Tong X.W., Fischer D.C., Körner K., Atkinson N.E., Edwards D.P., Headon D.R., Weigel N.L., Kieback D.G. A germline variation in the progesterone receptor gene increases transcriptional activity and may modify ovarian cancer risk. J. Clin. Endocrinol. Metab. 2004;89:6340–6347. doi: 10.1210/jc.2004-0114. [DOI] [PubMed] [Google Scholar]
- 49.Terry K.L., De Vivo I., Titus-Ernstoff L., Sluss P.M., Cramer D.W. Genetic variation in the progesterone receptor gene and ovarian cancer risk. Am. J. Epidemiol. 2005;161:442–451. doi: 10.1093/aje/kwi064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu T., Chen L., Sun X., Wang Y., Li S., Yin X., Wang X., Ding C., Li H., Di W. Progesterone receptor PROGINS and +331G/A polymorphisms confer susceptibility to ovarian cancer: a meta-analysis based on 17 studies. Tumour Biol. 2014;35:2427–2436. doi: 10.1007/s13277-013-1322-x. [DOI] [PubMed] [Google Scholar]
- 51.Tiwari D., Bose P.D., Das S., Das C.R., Datta R., Bose S. MTHFR (C677T) polymorphism and PR (PROGINS) mutation as genetic factors for preterm delivery, fetal death and low birth weight: A Northeast Indian population based study. Meta Gene. 2015;3:31–42. doi: 10.1016/j.mgene.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Romano A., Delvoux B., Fischer D.C., Groothuis P. The PROGINS polymorphism of the human progesterone receptor diminishes the response to progesterone. J. Mol. Endocrinol. 2007;38:331–350. doi: 10.1677/jme.1.02170. [DOI] [PubMed] [Google Scholar]
- 53.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Green R.E., Krause J., Briggs A.W., Maricic T., Stenzel U., Kircher M., Patterson N., Li H., Zhai W., Fritz M.H. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sankararaman S., Mallick S., Dannemann M., Prüfer K., Kelso J., Pääbo S., Patterson N., Reich D. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507:354–357. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dannemann M., Kelso J. The contribution of Neanderthals to phenotypic variation in modern humans. Am. J. Hum. Genet. 2017;101:578–589. doi: 10.1016/j.ajhg.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Meyer M., Kircher M., Gansauge M.T., Li H., Racimo F., Mallick S., Schraiber J.G., Jay F., Prüfer K., de Filippo C. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–226. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang M.A., Gao X., Theunert C., Tong H., Aximu-Petri A., Nickel B., Slatkin M., Meyer M., Pääbo S., Kelso J., Fu Q. 40,000-year-old individual from Asia provides insight into early population structure in Eurasia. Curr. Biol. 2017;27:3202–3208.e9. doi: 10.1016/j.cub.2017.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rasmussen M., Anzick S.L., Waters M.R., Skoglund P., DeGiorgio M., Stafford T.W., Jr., Rasmussen S., Moltke I., Albrechtsen A., Doyle S.M. The genome of a Late Pleistocene human from a Clovis burial site in western Montana. Nature. 2014;506:225–229. doi: 10.1038/nature13025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moreno-Mayar J.V., Potter B.A., Vinner L., Steinrücken M., Rasmussen S., Terhorst J., Kamm J.A., Albrechtsen A., Malaspinas A.S., Sikora M. Terminal Pleistocene Alaskan genome reveals first founding population of Native Americans. Nature. 2018;553:203–207. doi: 10.1038/nature25173. [DOI] [PubMed] [Google Scholar]
- 61.Consortium E.P., ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ananth C.V., Vintzileos A.M. Medically indicated preterm birth: recognizing the importance of the problem. Clin. Perinatol. 2008;35:53–67, viii. doi: 10.1016/j.clp.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 63.Fijarczyk A., Babik W. Detecting balancing selection in genomes: limits and prospects. Mol. Ecol. 2015;24:3529–3545. doi: 10.1111/mec.13226. [DOI] [PubMed] [Google Scholar]
- 64.Croze M., Živković D., Stephan W., Hutter S. Balancing selection on immunity genes: review of the current literature and new analysis in Drosophila melanogaster. Zoology (Jena) 2016;119:322–329. doi: 10.1016/j.zool.2016.03.004. [DOI] [PubMed] [Google Scholar]
- 65.Gomez-Lopez N., StLouis D., Lehr M.A., Sanchez-Rodriguez E.N., Arenas-Hernandez M. Immune cells in term and preterm labor. Cell. Mol. Immunol. 2014;11:571–581. doi: 10.1038/cmi.2014.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hughes G.C., Clark E.A., Wong A.H. The intracellular progesterone receptor regulates CD4+ T cells and T cell-dependent antibody responses. J. Leukoc. Biol. 2013;93:369–375. doi: 10.1189/jlb.1012491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Higham T., Douka K., Wood R., Ramsey C.B., Brock F., Basell L., Camps M., Arrizabalaga A., Baena J., Barroso-Ruíz C. The timing and spatiotemporal patterning of Neanderthal disappearance. Nature. 2014;512:306–309. doi: 10.1038/nature13621. [DOI] [PubMed] [Google Scholar]
- 68.McCoy R.C., Wakefield J., Akey J.M. Impacts of Neanderthal-introgressed sequences on the landscape of human gene expression. Cell. 2017;168:916–927.e12. doi: 10.1016/j.cell.2017.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Williams A.L., Jacobs S.B., Moreno-Macías H., Huerta-Chagoya A., Churchhouse C., Márquez-Luna C., García-Ortíz H., Gómez-Vázquez M.J., Burtt N.P., Aguilar-Salinas C.A., SIGMA Type 2 Diabetes Consortium Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature. 2014;506:97–101. doi: 10.1038/nature12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.