Abstract
Intracellular symmetry breaking plays a key role in wide range of biological processes, both in single cells and in multicellular organisms. An important class of symmetry-breaking mechanisms relies on the cytoplasm/membrane redistribution of proteins that can autocatalytically promote their own recruitment to the plasma membrane. We present an analytical construction and a comprehensive parametric analysis of stable localized patterns in a reaction-diffusion model of such a mechanism in a spherical cell. The constructed patterns take the form of high-concentration patches localized into spherical caps, similar to the patterns observed in the studies of symmetry breaking in single cells and early embryos.
Main Text
Many important processes in cell and developmental biology are controlled by highly nonuniform intracellular distributions of protein concentrations and enzymatic activities (1, 2). Examples include polarized patterns of membrane localization observed in studies of mating responses in yeast (3, 4, 5, 6, 7) and early embryogenesis in Caenorhabditis elegans (8, 9, 10). In both cases, patterns take the form of a localized cap on a cell membrane and play an instructive role, leading to the formation of a budding protrusion in yeast and asymmetric localization of cell determinants in the C. elegans embryo, respectively.
The formation of such cap-like patterns can rely on autocatalytic membrane recruitment of a diffusible cytoplasmic protein. The molecular and cellular bases for this mechanism have been established in several cases, motivating an impressive number of mathematical models that are being used to explore the quantitative requirements for the formation of robust localized patterns (5, 8, 11, 12, 13, 14, 15, 16, 17, 18). The simplest of such models has a single chemical species that interconverts between membrane and cytoplasmic states (13, 14, 17). One critical aspect is that self-promoted membrane recruitment can give rise to bistability, whereby the membrane can be in two states with very different levels of accumulated protein. This is the model considered in this Letter (Fig. 1).
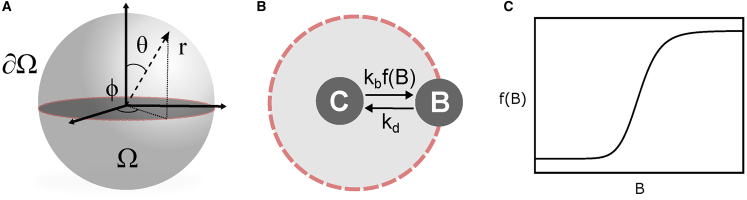
Figure 1.
(A) Schematic of a spherical cell. (B) The membrane association and dissociation processes are shown. (C) The function modeling the autocatalytic membrane recruitment is shown. To see this figure in color, go online.
In the system under consideration, the species with concentration C moves freely with diffusivity DC in the cell interior and reversibly binds to the cell membrane. The cell interior and the membrane are denoted by Ω and ∂Ω, respectively. The dissociation-rate constant is denoted by kd. The membrane-bound species, with surface density B, has diffusivity DB and promotes its own recruitment to the membrane. This is described by making the forward-binding rate a sigmoidal function of protein concentration at the membrane. The binding rate is modeled as a sum of two terms: the first term describes constitutive binding; the second term reflects autocatalytic recruitment, which follows a Hill-like dependence. These are characterized by the rate constant kb, constitutive rate β, threshold value Γ, and cooperativity parameter ν. The joint dynamics of the cytoplasmic and membrane species in a spherical cell of radius r is then governed by the following equations:
| (1) |
| (2) |
and
| (3) |
In these equations, is the outward unit normal to , is the spherical Laplacian in Ω, and is the Laplace-Beltrami operator on ∂Ω:
| (4) |
where θ ∈ [0, π] and ϕ ∈ [0, 2π] are the angular variables of the standard spherical coordinates.
This model and many of its relatives considered in the context of cell polarization belong to a broad class of reaction-diffusion systems with spatially segregated compartments of different dimensionality (15, 18, 19, 20, 21, 22). In combination with diffusion on the membrane, autocatalytic membrane recruitment can result in the formation and progressive expansion of a domain with high membrane concentration (11, 13, 14, 15, 16, 17, 18, 19, 20, 23, 24). However, it is counteracted by total mass conservation and fast cytoplasmic diffusion (12, 13, 14). For a proper choice of model parameters, localized accumulation on the membrane depletes concentration in the cytoplasm and can stabilize the expanding domain, resulting in a stable pattern.
Because the total amount of protein in the system is constant, the membrane and bulk protein concentrations are globally coupled by mass conservation. Although the general phenomenology associated with the formation of localized patterns in globally coupled reaction-diffusion problems is well understood (19, 20, 23), the number of results in the context of intracellular symmetry breaking is very limited (4, 16, 24). In fact, all existing analytical results are limited to one-dimensional “cells” and are based on ad hoc descriptions of nonlocal coupling of membrane dynamics by cytoplasmic diffusion (12, 13, 14, 17, 25). Here, we report exact analytical solutions for the spherical cap patterns constructed in the regime of large cytoplasmic diffusion that properly take into account the three-dimensional geometry of the cell.
We start by considering the conservation equation for the total amount of a protein, which is not synthesized or degraded but only redistributed between cytoplasm and membrane (5, 12, 13):
| (5) |
where C0 is the total amount of protein in the cell divided by cell volume. Because cytoplasmic diffusivity is typically much larger than membrane diffusivity, the cytoplasmic concentration may be assumed to be uniform throughout the cell (see Supporting Materials and Methods for further discussion):
| (6) |
Upon substitution of this expression into Eq. 1, our model is reduced to a single equation for the membrane species (26). This equation is rendered dimensionless by the following transformations: and . This leads to the following problem for the spatiotemporal dynamics of the dimensionless cell surface concentration, u = u (θ, ϕ, τ):
| (7) |
The rescaled problem has five dimensionless groups: ν and β are inherited directly from the original model; is the dimensionless threshold for the membrane recruitment nonlinearity; is the ratio of characteristic times for dissociation from the membrane and surface diffusion; and finally, may be interpreted as the ratio of the maximal fluxes to and from the cell membrane and quantifies the strength of the global coupling essential for the formation of localized patterns.
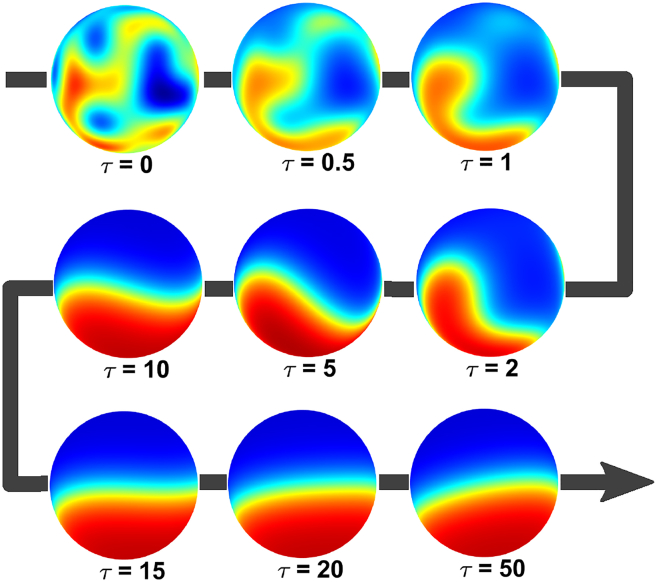
We started our analysis of this problem by solving it numerically using a spectral discretization implemented in the Chebfun package (27, 28). In all cases, the system converged to one of two types of behaviors: uniform steady state profiles and nonuniform axisymmetric cap-like patterns. Fig. 2 shows the snapshots from a representative numerical solution that eventually converges to such a pattern. Our numerical experiments indicate that such solitary caps, although placed at different locations on the surface, are the only spatially nonuniform attractors of Eq. 7.
Figure 2.
Snapshots of cell surface concentration from a representative numerical solution of Eq. 7 with α = 1, β = 0.1, γ = 0.3, δ2 = 0.05, and ν = 20. To see this figure in color, go online.
The axial symmetry of the cap-like patterns allows us to reduce the dimensionality of the problem. Without loss of generality, we may place the cap center at the North Pole of the sphere and focus only on the ϕ-independent solutions. Furthermore, when the membrane-recruitment nonlinearity is sharp , it can be approximated by the Heaviside function. Using a common coordinate transformation, η = −cosθ ∈ [−1,1], we obtain the following ordinary differential equation for the axially symmetric steady state solution :
| (8) |
This equation is piecewise linear, which greatly simplifies the analysis of localized patterns. One can readily check that this equation may have at most two spatially uniform stable steady states, in which the membrane concentration is either above or below the threshold value:
In addition to these uniform steady states, we found patterned solutions that correspond to spherical caps. In constructing these solutions, we assumed that they decrease monotonically from η = −1 to η = 1, crossing the threshold at some value of the longitudinal coordinate: . In other words, the autocatalytic membrane recruitment is “on” for η < ηc and “off” for η > ηc. As shown in the Supporting Material, such steady states can be found analytically in terms of the Legendre function Pμ(η), where :
| (9) |
and
| (10) |
where
To evaluate these expressions, one must find the position ηc where . This position can be found from the dimensionless form of the conservation law for the total amount of protein, leading to the following implicit function that relates ηc to the dimensionless parameters of the problem:
| (11) |
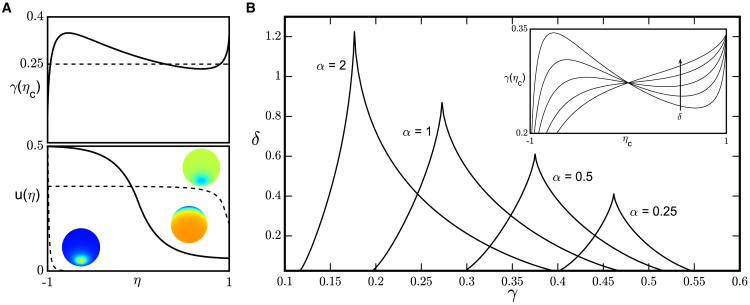
Graphical analysis of this equation reveals that it can have up to three roots, corresponding to three different values of ηc for which (Fig. 3). Each of these values gives rise to a different spatially nonuniform steady state. In particular, in the biologically significant regime of , there exists one solution in the form of a large spherical cap, for which
| (12) |
Figure 3.
(A) Construction of spatially nonuniform steady states. The graphical solution of Eq. 11, for α = 1, β = 0.1, γ = 0.25, and δ = 0.3, reveals three roots (top panel), giving rise to three different spatial profiles (bottom panel). (B) Two-dimensional cuts through the four-dimensional domain of model parameters that support the formation of stable localized patterns for β = 0.1 are shown. The boundaries of the cusps correspond to the extrema of the γ(ηc) function; this function becomes monotonic at the tip of the cusp (inset). Here, the inset shows γ(ηc) for β = 0.1, α = 1, and δ = {0.1, 0.25, 0.5, 1, 2}. To see this figure in color, go online.
and two small spherical cap solutions, for which whenever . Stability analysis of these solutions (see Supporting Materials and Methods) shows that for , a spherical cap solution is stable if and only if α >αc, where
| (13) |
According to this formula, for all δ sufficiently small and γ in the above range, the small-cap solutions are linearly unstable, whereas the large cap is linearly stable. We note that, as expected, the stable solutions constructed analytically above persist in the problem with ν finite, even for ν ≈ 5.
Equation 11 may be used to delineate the four-dimensional domain of parameters α, β, γ, and δ that can support states with broken symmetry. Fig. 3 shows a series of two-dimensional cuts through this domain, computed at a constant value of constitutive membrane recruitment and four different strengths of the global coupling parameter. For each of these cuts, the localized patterns exist in a cusp-like region that is characterized by an upper limit for dimensionless surface diffusivity δ. Beyond this value, the right-hand side of Eq. 11 becomes a single-valued function of γc (see the inset of Fig. 3 B), which corresponds to the disappearance of stable patterns. Our analysis also reveals how reducing the strength of global coupling moves the system outside of the domain corresponding to the symmetry-broken stable steady states.
Our results provide insights into the parametric dependence and robustness of the localized patterns. For instance, changes in the cell radius affect two dimensionless groups: the dimensionless-cell-surface diffusivity, δ ∼ 1/r, and the strength of global coupling, α ∼ 1/r. Because the existence of patterned states is promoted by small values of surface diffusivity and strong global coupling, patterned states are realized for an intermediate range of cell sizes (see Fig. S2).
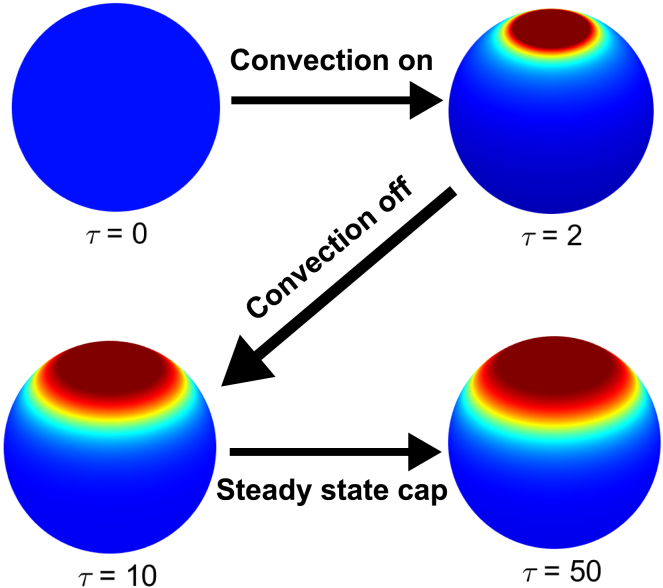
Further research is required to understand how cells can not only form the localized patterns but also direct them to specific positions. Studies in the early C. elegans embryo suggest that such control may be provided by cortical flows that transiently localize the diffusible species at a specific location on the cell membrane (1, 8, 10). Our model can be readily modified to account for such flows. As an illustration, Fig. 4 shows the results of a numerical experiment in which a model cell starts from a steady state in which most of the protein is cytoplasmic. A transient unidirectional flow then creates a region of high membrane concentration, triggering the formation of a self-sustained localized pattern.
Figure 4.
Directed control of pattern formation by a transient unidirectional flow. The system starts from a uniform “off” steady state. The flow is from the South to North Pole. α = 1, β = 0.1, γ = 0.35, and δ2 = 0.05. See the Supporting Materials and Methods for details. To see this figure in color, go online.
An additional layer of control may be provided by variations in the curvature of the cell surface. Our preliminary simulations in spheroid geometries indicate that large spherical cap solutions persist under perturbations of spherical symmetry but at the same time are guided to specific locations by variations of curvature. Analyzing the effects of such variations, which can be either static or dynamic, should provide additional insights into the mechanisms of intracellular symmetry breaking (1, 2).
Author Contributions
S.Y.S. and C.B.M. designed the research. C.B.M. and R.D. performed analytical and numerical studies. H.M. provided software and numerical analysis.
Acknowledgments
The authors thank Dr. Mahim Misra for his contributions during the early stages of this work.
This work was supported by funding from the National Science Foundation Science and Technology Center for Emergent Behaviors of Integrated Cellular Systems (CBET-0939511) and National Institutes of Health (R01GM107103). This research was partially supported by the Allen Discovery Center program through The Paul G. Allen Frontiers Group.
Editor: Anatoly Kolomeisky.
Footnotes
Supporting Materials and Methods and three figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(18)30672-6.
Contributor Information
Cyrill B. Muratov, Email: muratov@njit.edu.
Stanislav Y. Shvartsman, Email: stas@princeton.edu.
Supporting Citations
References (29, 30, 31, 32, 33, 34, 35, 36) appear in the Supporting Material.
Supporting Material
References
- 1.Mogilner A., Allard J., Wollman R. Cell polarity: quantitative modeling as a tool in cell biology. Science. 2012;336:175–179. doi: 10.1126/science.1216380. [DOI] [PubMed] [Google Scholar]
- 2.Kim, E. J. Y., E. Korotkevich, and T. Hiiragi. Coordination of cell polarity, mechanics and fate in tissue self-organization. Trends Cell Biol. Published online March 28, 2018. https://doi.org/10.1016/j.tcb.2018.02.008. [DOI] [PubMed]
- 3.Chiou J.G., Balasubramanian M.K., Lew D.J. Cell polarity in yeast. Annu. Rev. Cell Dev. Biol. 2017;33:77–101. doi: 10.1146/annurev-cellbio-100616-060856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giese W., Eigel M., Klipp E. Influence of cell shape, inhomogeneities and diffusion barriers in cell polarization models. Phys. Biol. 2015;12:066014. doi: 10.1088/1478-3975/12/6/066014. [DOI] [PubMed] [Google Scholar]
- 5.Goryachev A.B., Pokhilko A.V. Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS Lett. 2008;582:1437–1443. doi: 10.1016/j.febslet.2008.03.029. [DOI] [PubMed] [Google Scholar]
- 6.Goryachev A.B., Leda M. Many roads to symmetry breaking: molecular mechanisms and theoretical models of yeast cell polarity. Mol. Biol. Cell. 2017;28:370–380. doi: 10.1091/mbc.E16-10-0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klünder B., Freisinger T., Frey E. GDI-mediated cell polarization in yeast provides precise spatial and temporal control of Cdc42 signaling. PLoS Comput. Biol. 2013;9:e1003396. doi: 10.1371/journal.pcbi.1003396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goehring N.W., Trong P.K., Grill S.W. Polarization of PAR proteins by advective triggering of a pattern-forming system. Science. 2011;334:1137–1141. doi: 10.1126/science.1208619. [DOI] [PubMed] [Google Scholar]
- 9.Lang C.F., Munro E. The PAR proteins: from molecular circuits to dynamic self-stabilizing cell polarity. Development. 2017;144:3405–3416. doi: 10.1242/dev.139063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mittasch M., Gross P., Kreysing M. Non-invasive perturbations of intracellular flow reveal physical principles of cell organization. Nat. Cell Biol. 2018;20:344–351. doi: 10.1038/s41556-017-0032-9. [DOI] [PubMed] [Google Scholar]
- 11.Alonso S., Bär M. Phase separation and bistability in a three-dimensional model for protein domain formation at biomembranes. Phys. Biol. 2010;7:046012. doi: 10.1088/1478-3975/7/4/046012. [DOI] [PubMed] [Google Scholar]
- 12.Otsuji M., Ishihara S., Kuroda S. A mass conserved reaction-diffusion system captures properties of cell polarity. PLoS Comput. Biol. 2007;3:e108. doi: 10.1371/journal.pcbi.0030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mori Y., Jilkine A., Edelstein-Keshet L. Wave-pinning and cell polarity from a bistable reaction-diffusion system. Biophys. J. 2008;94:3684–3697. doi: 10.1529/biophysj.107.120824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mori Y., Jilkine A., Edelstein-Keshet L. Asymptotic and bifurcation analysis of wave-pinning in a reaction-diffusion model for cell polarization. SIAM J. Appl. Math. 2011;71:1401–1427. doi: 10.1137/10079118X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rätz A., Röger M. Turing instabilities in a mathematical model for signaling networks. J. Math. Biol. 2012;65:1215–1244. doi: 10.1007/s00285-011-0495-4. [DOI] [PubMed] [Google Scholar]
- 16.Rätz A., Röger M. Symmetry breaking in a bulk-surface reaction-diffusion model for signaling networks. Nonlinearity. 2013;27:1805–1827. [Google Scholar]
- 17.Trong P.K., Nicola E.M., Grill S.W. Parameter-space topology of models for cell polarity. New J. Phys. 2014;16:065009. [Google Scholar]
- 18.Anguige K., Röger M. Global existence for a bulk/surface model for active-transport-induced polarisation in biological cells. J. Math. Anal. Appl. 2017;448:213–244. [Google Scholar]
- 19.Middya U., Luss D., Sheintuch M. Impact of global interaction and symmetry on pattern selection and bifurcation. J. Chem. Phys. 1994;101:4688–4696. [Google Scholar]
- 20.Pismen L.M. Turing patterns and solitary structures under global control. J. Chem. Phys. 1994;101:3135–3146. [Google Scholar]
- 21.Cònsul N. On equilibrium solutions of diffusion equations with nonlinear boundary conditions. Z. Angew. Math. Phys. 1996;47:194–209. [Google Scholar]
- 22.Přibyl M., Muratov C.B., Shvartsman S.Y. Long-range signal transmission in autocrine relays. Biophys. J. 2003;84:883–896. doi: 10.1016/S0006-3495(03)74906-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Battogtokh D., Hildebrand M., Mikhailov A. Nucleation kinetics and global coupling in reaction-diffusion systems. Phys. Rep. 1997;288:435–456. [Google Scholar]
- 24.Madzvamuse A., Chung A.H.W. The bulk-surface finite element method for reaction-diffusion systems on stationary volumes. Finite Elem. Anal. Des. 2016;108:9–21. [Google Scholar]
- 25.Rubinstein B., Slaughter B.D., Li R. Weakly nonlinear analysis of symmetry breaking in cell polarity models. Phys. Biol. 2012;9:045006. doi: 10.1088/1478-3975/9/4/045006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hale J.K., Sakamoto K. Shadow systems and attractors in reaction-diffusion equations. Appl. Anal. 1989;32:287–303. [Google Scholar]
- 27.Driscoll, T. A., N, Hale, and L. N., Trefethen, eds. 2014. Chebfun Guide: Pafnuty Publications, Oxford, UK.
- 28.Wright G.B., Javed M., Trefethen L.N. Extension of Chebfun to periodic functions. SIAM J. Sci. Comput. 2015;37:C554–C573. [Google Scholar]
- 29.Abramowitz M., Stegun I.A. National Bureau of Standards; Dover, New York: 1964. Handbook of Mathematical Functions with Formulas, Graphs, and Mathematical Tables. [Google Scholar]
- 30.Kerner B.S., Osipov V.V. Springer-Science + Business Media; Dordrecht, the Netherlands: 1994. Autosolitons. [Google Scholar]
- 31.Kerner B.S., Osipov V.V. Long-range signal transmission in autocrine relays. Sov. Phys. JETP. 1978;47:874–885. [Google Scholar]
- 32.Kerner B.S., Osipov V.V. Pulsating “heterophase” regions in nonequilibrium systems. Sov. Phys. JETP. 1982;56:1275–1282. [Google Scholar]
- 33.Muratov C.B., Osipov V.V. General theory of instabilities for patterns with sharp interfaces in reaction-diffusion systems. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 1996;53:3101–3116. doi: 10.1103/physreve.53.3101. [DOI] [PubMed] [Google Scholar]
- 34.Bose A., Kriegsmann G.A. Stability of localized structures in non-local reaction-diffusion equations. Meth. Appl. Anal. 1998;5:351–366. [Google Scholar]
- 35.Muratov C.B., Osipov V.V. Stability of static spike autosolitons in the Gray-Scott model. SIAM J. Appl. Math. 2002;62:1463–1487. [Google Scholar]
- 36.Montanelli H., Nakatsukasa Y. Fourth-order time-stepping for stiff PDEs on the sphere. SIAM J. Sci. Comput. 2018;40:A421–A451. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.