Capsule Summary
The current studies demonstrate a critical role of IL-18 in transforming IL-5 generated naïve eosinophils into the distinct inflammatory CD101+CD274+ expressing mature and activated tissue eosinophils that promote disease pathogenesis.
Keywords: Eosinophil, CD101, CD274, IL-5, IL-18, allergic disease
Letter to Editor
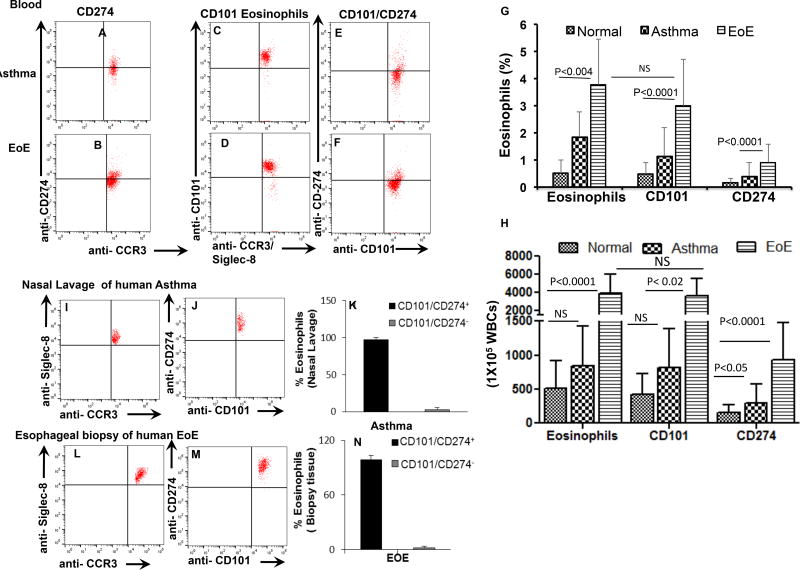
Eosinophils have very distinct roles, in maintaining the innate immunity in health and are also involved in disease pathogenesis for allergic, autoimmune diseases and allograft rejection.1, 2 Interleukin (IL)-5 is a well-established differentiation, growth and survival factor for eosinophils and earlier reports indicate that IL-5 and eotaxin(s) regulate baseline resident eosinophils.1, 3 Most recently, we and others reported a distinguishable subset of CD1014 and CD2745 expressing eosinophils, similar to the other immune cell subset populations that exhibit differences in their phenotypes and functions. Recently, CD101 eosinophils are described as IL-5 dependent species and lung-specific inflammatory eosinophils (iEOS) in HDM-induced mouse model of asthma, which is different from the IL-5 independent residential eosinophils (rEOS) detected in the healthy mouse lungs.4 Previously, we also reported CD101 and CD274 expressing eosinophils that are induced in the blood of eosinophilic esophagitis (EoE) patients, and their number increases with disease severity and decreases following the treatment.5 Herein, we validate our previous findings of CD101 and CD274 expressing eosinophils in EoE and further report that both these subsets are also present in the blood of other allergic diseases like asthma (Fig. 1 A–D). Interestingly, we observed that most CD101 expressed blood eosinophils are not CD274 expressing eosinophils (Fig. 1 E–F) and their percent and absolute number increases in the blood of allergic patients (Fig. 1, H, G). This observation indicates that both these eosinophil subsets are either not similar or in the different developmental stage. Most importantly, we further report that the tissue accumulated eosinophils in the nasal lavage of asthma patients and esophageal biopsies of EoE patients are CD101 and CD274 double positive (CD101+CD274+) eosinophils (Fig. 1 I–N). These observations confirm that CD101 and CD274 expressing eosinophils present in the blood are indeed intermediate stages eosinophils and following accumulation in tissue they mature into CD101+CD274+ double positive pathogenic eosinophils in allergic diseases. Earlier, CD101 expressed eosinophils are shown to be pathogenic by performing adaptive transfer experiments in the experimental mouse model of asthma.4
Figure 1. Human blood and tissue CD101 and CD274 expression on eosinophils in healthy and allergic patients.
A representative flow cytometer analysis was performed by selecting live CCR3 positive and anti-Siglec-8 double positive eosinophils for identifying CD101 and the CD274 expressing eosinophils in the blood of normal individuals, asthma and EoE patients, and EoE patients (A–D). The analysis shows that all blood CD101 expressing eosinophils do not express CD274 (G–H). The percent and absolute numbers of both CD101 and CD274 expressing CCR3+Siglec-8+ eosinophils in the blood of EoE and asthma patients compare to normal individuals are shown, respectively (H, G). Nasal lavage from asthma patients (I–K) and esophageal biopsies from EoE patients (L–N) detected only double positive CD101+CD274+ eosinophils. All data presented are expressed as the mean ± SD (normal, n=10 and asthma, n=14, EoE, n=16).
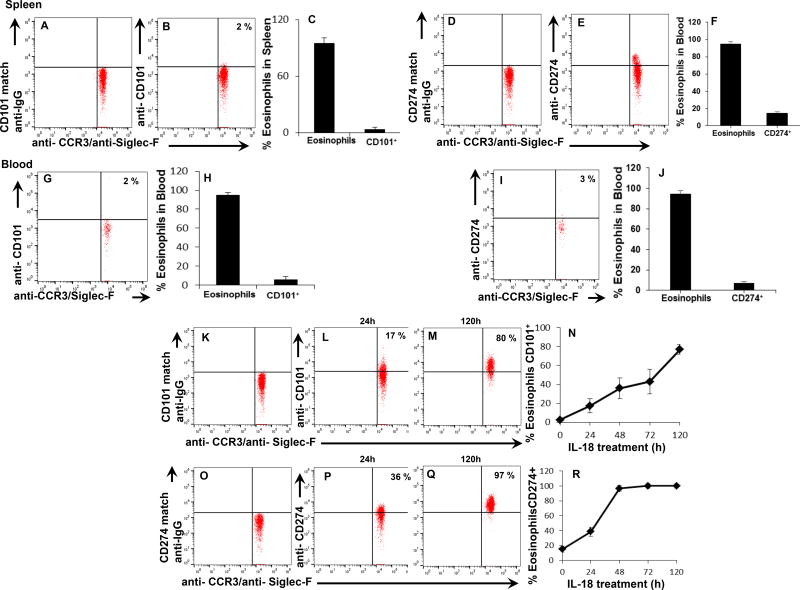
Mechanistically to understand the eosinophil subset development, we first analyzed IL-5 responsive eosinophils of CD2-IL-5 transgenic mice. The analysis showed that both spleen (Fig. 2 D–I) and blood (Fig. 2 J–O) most eosinophils do not express CD101 or CD274 in CD2-IL-5 transgenic mice. These observations are inconsistent with recently reported CD101 expressing eosinophils as IL-5 dependent;4 therefore, next we investigated the factors that develop the CD101 and CD274 expression on eosinophils. Since, several cytokines (IL-5, IL-13, IL-15, IL-18, IL-21 and IL-33) have been implicated in eosinophil associated allergic disease.6, 7 Thus, we investigated the role of IL-13, IL-15, IL-18, IL-21 and IL-33 in the eosinophil differentiation and transformation of both CD101 and CD274 subset of eosinophil. Herein, we show that CD2-IL-5 transgenic mice eosinophils become CD101 and CD274 expressing eosinophils kinetically only following ex vivo exposure to IL-18, no other cytokine was able to transform IL-5 generated eosinophils to CD101 or CD274 expressed eosinophils (Data not shown). Indicating that IL-18 is critical in transforming IL-5 responsive eosinophils to CD101 and CD274 expressing the subset of eosinophils (Fig. 2 K–R). Notably, IL-18 is reported in almost all allergic diseases including HDM-antigen induced mouse model of asthma.8 These mechanistic eosinophils transformation data indicates that due to the lack of understanding on the role of IL-18, the investigators of recently reported studies concluded that CD101 expressed eosinophils in the lungs are IL-5 dependent in asthmatic mice.4 Herein, we also show that IL-18 is capable of eosinophil differentiation apart from IL-5. The low-density CD34+ mouse bone marrow (LDEBM) precursors exposed to ex vivo IL-18 differentiate ~ 6–7% eosinophils despite the mouse genotypes, compared to rIL-5 (~ 45–52%) generated eosinophils (Fig. E1, D, E). This indicates that IL-5 independent eosinophils develop in response to IL-18; but still IL-5 is required for the growth and survival of IL-18 generated eosinophils (Fig. E1, F). No other cytokine (IL-13, IL-15, IL-18, IL-21, and IL-33) alone is able to differentiate eosinophils from LDEBM.
Figure 2. IL-18 is critical for the development of CD101 and CD274 eosinophil subsets.
A representative flow cytometer analysis on eosinophil surface expression on 7AAD−/CCR3+ leukocytes selected for the detection of CD101 and Cd274 on anti-CCR3 and anti-Siglec-F double positive eosinophils of CD2-IL-5 transgenic mouse splenocytes based on CD101 and CD274 isotype-matched anti-IgG. Very few (> 2%) CD101 (G, H) and CD274 (I, J) expressed eosinophils (CCR3+Siglec-F+) was detected in the blood. CD2-IL-5 transgenic mouse splenocytes exposed to IL-18 ex vivo showed a time-dependent kinetic increase in the expression of CD101 on CCR3+Siglec-F+ eosinophils (K–N) and CD274 on CCR3+Siglec-F+ eosinophils (O–R). Data are expressed as the mean ± SD, n=3 individual experiments.
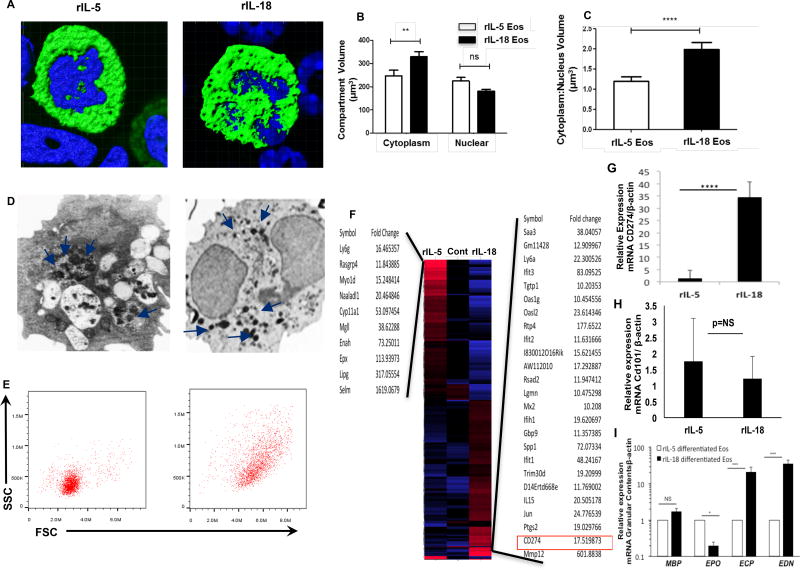
These observations prompted us to further characterize both IL-5 and IL-18 differentiated ex vivo eosinophils including their size, nuclear shape, the volume of cytoplasmic:nuclear ratio, granular contents and the expression of differentially regulated genes. The confocal microscopic images of anti-MBP stained IL-5 and IL-18 differentiated eosinophils from LDEBM indicated differences in their nuclear shape, the volume of cytoplasm: nuclear ratio (Fig. 3 A–C). Further, electron microscopic analysis shows intact eosinophil granules in IL-5 generated eosinophils compare to degranulated eosinophils in IL-18 generated eosinophils (Fig. 3D). Finally, the differences in eosinophils size and granularity of IL-18 and IL-5 generated eosinophils was established by performing flow cytometer analysis, which indicated that IL-18 derived eosinophils are more granular and bigger in size (Fig. 3 E). A similar electron microscopic characteristics was recently reported for CD101 expressing lung eosinophils of asthmatic mice, similar to the observed in IL-18 differentiated eosinophils.4 Interestingly, microarray analysis also identified highly induced CD274 transcript expression in IL-18 differentiated eosinophils (Fig. 3F) compared to IL-5 differentiated eosinophils that were further confirmed by performing qPCR analysis (Fig. 3G). Notably, CD101 expression was not detected by microarray analysis and a very low and comparable level of CD101 expression in IL-5 and IL-18 generated eosinophils (Fig. 1H). A detailed gene expression list of IL-5 and IL-18 differentiated transcript analysis is provided in (Sup, T2). These data further confirm that IL-18 generated eosinophils are indeed distinct subset of eosinophils. Despite high expression of CD274 transcript analysis, a distinct hierarchical clustering characteristic also revealed that several pro-inflammatory genes such as CLEC4D, PTGS2, IL-15, PF4, NT5C3b, Saa3, CXCL2, Spp1, and Rsad2 are also found highly induced in IL-18 derived eosinophils compared to IL-5 derived eosinophils (Fig. 3 F). The role of IL-18 in eosinophil maturation is further supported by the detection of a multiple-fold increase in eosinophil granules (ECP and EDN) in IL-18 differentiated eosinophils as compared to IL-5 generated eosinophils (Fig. 3 I). The enhanced level of MBP, EDN and ECP granules was reported earlier in a number of allergic diseases including asthma, EoE, and colitis.9 The details of the methods used to identify and characterize these subsets of eosinophils are reported in Online Repository at www.jacionline.org.
Figure 3. Confocal, electron microscopic, flow cytometry and microarray analyses of eosinophils derived from LDEBM precursors in response to rIL-5 and rIL-18.
Representative confocal photomicrographs of anti-MBP and anti-IgG-FITC (green) stained and DAPI stained (blue) rIL-5 and rIL-18 differentiated eosinophils (A). A total computerized quantitative area of the compartmentalized nucleus, cytoplasm and their ratio analyzed by IMARIS software is presented (mean ± SD, n=30) from rIL-5 and rIL-18 differentiated eosinophils (B and C). Electron photomicrographs of rIL-5 differentiated eosinophils (original magnifications ×24000) and rIL-18 differentiated eosinophils (original magnifications ×1800, n=15 eosinophil analyzed) (D). A representative flow cytometer analysis (n=5) of eosinophil granularity and size analysis on side-scatter (SSC) and forward-scatter (FSC) plots for rIL-5 and rIL-18 treated eosinophils (E). The microarray analysis of CD34+ LDEBM precursors treated with FLT3L for 4 days (control), rIL-5-derived eosinophils, and rIL-18 differentiated FACS purified eosinophils was performed. Overexpressed genes are represented in red and down-regulated genes in blue. Each column represents combined experiment datasets analyzed using multiple independently generated eosinophil RNA and the gene list of more than 10-fold change in response to IL-18 and IL-5 are presented (F). Highly expressed CD274 (PDL1) mRNA relative expression along with CD101 was also examined by real-time PCR (G, H). A comparative real-time PCR analysis data of the transcript levels (fold change) in IL-5 and IL-18 generated eosinophils granules (MBP, EPO, ECP, and EDN) are presented and demonstrate more granular content in IL-18 differentiated eosinophils compared to IL-5 generated eosinophils, except EPO (I). Arrows demonstrate eosinophils granules in the eosinophils. Data are expressed as mean ± SD, n=3, *p<0.04, **p<0.01, *** p<0.0001, **** p<0.00001.
Furthermore, the significance of IL-18 in the development of both CD101 and CD274 expression on eosinophils in vivo was also examined in the mouse model of aspergillus–induced experimental asthma. We show that asthmatic mice also show CD101 and CD274 expressed double positive eosinophils (CD101+CD274+) in wild type mice that were not detected in the absence of endogenous IL-18 in mice (Fig. E2, A–M). This further confirms that IL-18 has a critical role of in the development of CD101+CD274+ pathogenic eosinophils in the lungs of allergic mice that are also present in the tissues of human allergic diseases. However, some ~8% eosinophils in IL-18−/− mice that seems to weak positive for CD274 this may be due to the influence of allergen-induced IFN-γ. The IFN-γ induction in the lungs of aspergillus challenged asthmatic mice is recently reported (Online report, doi: 10.1016/j.jaci.2017.05.025) and it has been reported that CD274 expression on immune cells is influenced by IFN-γ; therefore, possibility exist that in the absence of in vivo endogenous IL-18, allergen-induced IFN-γ influences a week CD274 expression on lung eosinophils. Notably, even though few eosinophils of IL-18−/− mice show weak expression of CD274 but they are not double positive eosinophils (CD101+CD274+) as observed in IL-5−/− and WT mice (Figure E2, M–O). This further confirms that IL-18 has a critical role of in the development of CD101+CD274+ pathogenic eosinophils in the lungs of allergic mice that are also present in the tissues of human allergic diseases.
Taken together, we provide the significance of IL-18 in eosinophil differentiation, maturation, activation and transformation from naïve eosinophils to pathogenic eosinophils. These provided data authenticate that both CD101 and CD274 expressing eosinophil subsets are not IL-5 dependent as reported earlier.4 These investigations also indicate that IL-5 independent hidden eosinophils identified in earlier report may developing stage residential eosinophils in normal lung that mature following HDM challenge in asthmatic in mice.4 The mature and activated pathogenic eosinophils accumulate in the tissues of allergic diseases are CD101+CD274+. These double positive pathogenic eosinophils are observed in the tissue of both mice and human in allergic condition. Moreover, the study also showed some discrepancy on the expression levels of CD101 in the blood of normal human and mice that may be due to the well-controlled age, environment, and food provided to the mice. In contrast, analyzed normal human have some limitations like analyzed age group (14–48 years), different food habit and living environmental conditions. The range of CD101 expression levels in the blood eosinophils of normal individuals clearly denote the possibility and may be the reason for this observed discrepancy. This CD101 expression discrepancy also explains the wisdom of earlier reported CD101 expressed eosinophils in allergic mice, but not in allergic human compare to the respective normal.4 In conclusion, presented data first time authenticates a critical role of IL-18 in the development and transformation of distinct pathogenic eosinophils in allergic diseases.
Supplementary Material
Acknowledgments
This work was supported by NIH R01 AI080581 (AM) grants. We also appreciate the help of Bruce Aronow, PhD, for microarray data analysis, and Matthew Kofron, PhD for confocal microscopy data analysis at CCHMC, Cincinnati, OH. We also thank Ms. Christine Glynn, RN, Tulane University, New Orleans, for obtaining patient’s consent, collecting blood, nasal lavage and tissue specimens and organizing data are pertaining to patient clinical characteristics. The anti-MBP antibody was purchased from the laboratory of Dr. Lee’s laboratory at Mayo Clinic, Scottsdale, AZ.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of all sources of funding: This author discloses the following: Dr. Anil Mishra has served as a consultant for Axcan Pharma, Aptalis, Elite Biosciences, Calypso Biotech SA, and Enumeral Biomedical. The remaining authors disclose no conflicts of interest.
References
- 1.Asosingh K, Hanson JD, Cheng G, Aronica MA, Erzurum SC. Allergen-induced, eotaxin-rich, proangiogenic bone marrow progenitors: a blood-borne cellular envoy for lung eosinophilia. J Allergy Clin Immunol. 2010;125:918–25. doi: 10.1016/j.jaci.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mishra A, Hogan SP, Brandt EB, Rothenberg ME. An etiological role for aeroallergens and eosinophils in experimental esophagitis. J Clin Invest. 2001;107:83–90. doi: 10.1172/JCI10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mishra A, Hogan SP, Lee JJ, Foster PS, Rothenberg ME. Fundamental signals that regulate eosinophil homing to the gastrointestinal tract. J Clin Invest. 1999;103:1719–27. doi: 10.1172/JCI6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mesnil C, Raulier S, Paulissen G, Xiao X, Birrell MA, Pirottin D, et al. Lung-resident eosinophils represent a distinct regulatory eosinophil subset. J Clin Invest. 2016;126:3279–95. doi: 10.1172/JCI85664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venkateshaiah SU, Manohar M, Verma AK, Blecker U, Mishra A. Possible Noninvasive Biomarker of Eosinophilic Esophagitis: Clinical and Experimental Evidence. Case Rep Gastroenterol. 2016;10:685–692. doi: 10.1159/000452654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhiguang X, Wei C, Steven R, Wei D, Wei Z, Rong M, et al. Over-expression of IL-33 leads to spontaneous pulmonary inflammation in mIL-33 transgenic mice. Immunol Lett. 2010;131:159–65. doi: 10.1016/j.imlet.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Strengell M, Matikainen S, Siren J, Lehtonen A, Foster D, Julkunen I, et al. IL-21 in synergy with IL-15 or IL-18 enhances IFN-gamma production in human NK and T cells. J Immunol. 2003;170:5464–9. doi: 10.4049/jimmunol.170.11.5464. [DOI] [PubMed] [Google Scholar]
- 8.Lin LH, Zheng P, Yuen JW, Wang J, Zhou J, Kong CQ, et al. Prevention and treatment of allergic inflammation by an Fcgamma-Der f2 fusion protein in a murine model of dust mite-induced asthma. Immunol Res. 2012;52:276–83. doi: 10.1007/s12026-012-8339-x. [DOI] [PubMed] [Google Scholar]
- 9.Inamura H, Tomita M, Okano A, Kurosawa M. Serial blood and urine levels of EDN and ECP in eosinophilic colitis. Allergy. 2003;58:959–60. doi: 10.1034/j.1398-9995.2003.00135.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.