Abstract
Homologous recombination enables the cell to access and copy intact DNA sequence information in trans, particularly to repair DNA damage affecting both strands of the double helix. Here, we discuss the DNA transactions and enzymatic activities required for this elegantly orchestrated process in the context of the repair of DNA double-strand breaks in somatic cells. This includes homology search, DNA strand invasion, repair DNA synthesis, and restoration of intact chromosomes. Aspects of DNA topology affecting individual steps are highlighted. Overall, recombination is a dynamic pathway with multiple metastable and reversible intermediates designed to achieve DNA repair with high fidelity.
Keywords: genomic instability, DNA recombination, DNA repair, DNA topology, DNA damage, DNA endonuclease, DNA helicase, DNA polymerase, DNA topoisomerase
Introduction
Homologous recombination (HR)2 is essential to access the redundancy of genetic information that exists in the form of sister chromatids or homologous chromosomes when both strands of the DNA double helix are compromised. Important biological contexts of HR are DNA replication support and the repair of DNA double-strand breaks (DSBs) in somatic cells and during meiosis (1). In this Minireview, we focus on the repair of DSBs by HR in somatic cells to illustrate the biochemistry of the enzymes involved in the individual steps.
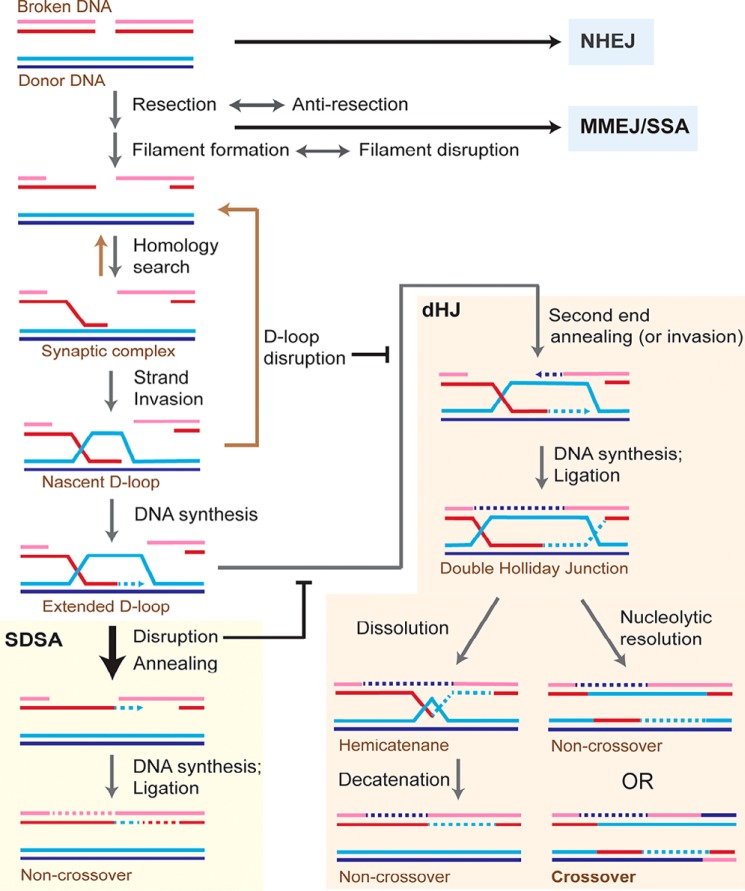
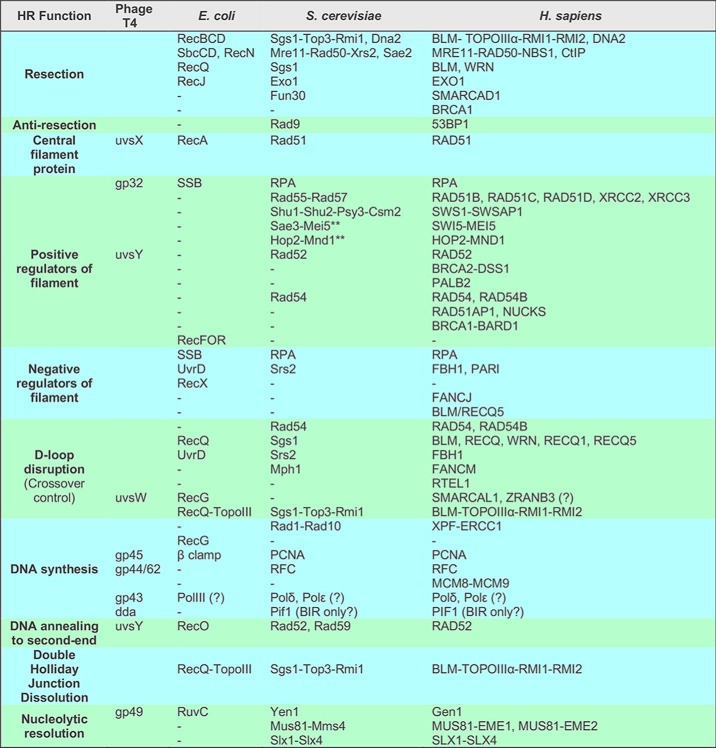
When a chromosome suffers a DSB, the DNA damage response coordinates cellular pathways to ensure genomic stability and survival, including the pathway choice of DSB repair (2). As summarized in Fig. 1, DSBs can be repaired by a number of different pathways that lead to the restoration of the chromosome. Other contributions in this Minireview series will discuss canonical nonhomologous end-joining (NHEJ) (3) and microhomology-mediated end-joining (MMEJ) (4). For HR, the DSB is resected 5′ to 3′ on one strand of the DSB ends producing terminal 3′-OH single-stranded DNA (ssDNA) tails. As control of DSB resection defines a mechanism of pathway choice between HR and end joining, we refer the reader to the contribution dedicated to this key step for a discussion of the nucleolytic processing of DSBs (5). Briefly, DSB resection is surprisingly complex and flexible (6). The nuclease Mre11-Rad50-Xrs2 and its cofactor Sae2 (Table 1 for a list of proteins)3 initiate resection with the capability of acting on DSB ends with nonstandard DNA chemistry or covalently attached proteins by delivering an endonucleolytic incision to release a terminal 5′-ending oligonucleotide. Bulk resection is accomplished by two pathways involving the 5′–3′ exonuclease Exo1 and the Sgs1–Top3–Rmi1 complex working in conjunction with the Dna2 nuclease. Both long-range resection pathways require the Fun30 chromatin–remodeling factor and are negatively regulated by the DNA damage–response protein Rad9 binding to specific histone marks (6).
Figure 1.
Model for repair of DNA double-strand breaks by homologous recombination in somatic cells. When a DNA double-strand break (DSB) occurs in a DNA molecule, repair can proceed by multiple pathways largely controlled by end resection. NHEJ is capable of repairing unresected or minimally resected DSBs in a template-independent fashion. MMEJ and single-strand annealing (SSA) rely on different extents of homology between the two DSB ends for repair independent of a donor molecule. Homologous recombination proceeds as shown in the figure using a homologous donor DNA. Most of the extended D-loops in somatic cells are disrupted and subsequently repaired by SDSA. The end result of the repair by SDSA is always a noncrossover outcome, thus avoiding loss of heterozygosity produced by somatic crossovers. SDSA occurs by disruption of the extended D-loop and annealing the newly synthesized DNA with the second end of the broken molecule. Alternatively, the newly synthesized strand may invade the second end as depicted in Fig. S1. The extended D-loop can also undergo second-end capture or invasion (Fig. S2) to form a double Holliday junction (dHJ). This may either lead to a crossover or a noncrossover outcome. Invasion by the second break end makes dHJ formation and hence crossover outcome more likely, as depicted in Fig. S2. See Fig. S3 for another model for crossover generation. dHJs can be dissolved into noncrossovers by the concerted action of the Sgs1–Top3–Rmi1 complex to migrate the two junctions toward each other and then decatenate the strands of the hemicatenane by the Top3 topoisomerase activity. Each colored line indicates a strand of DNA, and dotted lines represent DNA synthesis.
Table 1.
Homologous recombination: Proteins and their functions
** Meiosis-specific in S. cerevisiae.
The resected DSB end(s) must find, synapse with, and invade (intertwine with) a homologous donor locus to prime repair DNA synthesis. DSB repair by HR in somatic cells favors use of the sister chromosome over the homologous chromosome as a template donor (7) and primarily resolves interchromosomal joint molecules through the synthesis-dependent strand annealing pathway (SDSA) (Fig. 1 and Figs. S1 and S2) (8, 9). Both of these preferences serve to limit potential loss of heterozygosity through somatic crossover (10). In SDSA, DNA synthesis creates homology to the other broken DNA end, so that when the extended D-loop is unwound the two ends can anneal and achieve repair (Fig. 1 and Fig. S2). Alternatively, this step may involve a second DNA strand invasion event (Fig. S1). In the following, we will describe the individual steps of the pathway and discuss the reversibility of key intermediates that enhance the fidelity of HR and limit potential deleterious genomic rearrangements (11, 12).
Assembly and maintenance of the nucleoprotein filament scaffold on ssDNA
With a representative in every organism, Rad51 family proteins are central to HR, forming right-handed helical filaments on ssDNA that act as nucleoprotein scaffolds to direct their own and interacting protein activities (13–15). Nucleation of the Rad51 filament is challenged by competition with the ssDNA-binding protein RPA (16). Once nucleated, cooperative interactions between Rad51 protomers dominate, and filament growth ensues. The term mediator is used to describe proteins enabling Rad51 to overcome the inhibition of RPA by acting as nucleation platforms bridging Rad51 to ssDNA to displace RPA (13, 15). The RecFOR complex in bacteria (17) and Brh2 protein in the fungus Ustilago maydis (18) recognize a specific DNA structure to begin filament growth, the junction formed by ssDNA transitioning to dsDNA at the resection boundary. Other mediators, such as Rad52 and BRCA2, appear to lack a clear DNA junction preference (19). In yeast, Rad52 mediates the replacement of RPA with Rad51 in a mechanism where ssDNA wraps around Rad52, destabilizing the RPA–ssDNA interaction while promoting Rad51 binding through physical interaction between Rad51 and Rad52 (20, 21). Yeast Rad52 defines its epistasis group of HR proteins, being necessary for all Rad51 filament formation in vivo (22, 23). Interestingly, although RAD52 is conserved as a protein, its dominant mediator function is not. In vertebrates, genetic and biochemical evidence suggests that the BRCA2–DSS1 complex is critical to RAD51 filament nucleation (19, 24–28). BRCA2 appears to act in concert with the human RAD51 paralogs, as suggested by epistasis analysis (29). However, it is not yet fully clear why such a large protein with a complex interactome and nine RAD51-binding sites is required to accomplish what in other species is achieved by much simpler proteins with only one to two binding sites for the filament protein (e.g. RecFOR and Brh2-Dss1) (30). Interestingly, in BRCA2-deficient human cells, RAD52 becomes essential, and it was suggested that RAD52 exerts a mediator function under these conditions (31).
Rad51 family ATPases display complex behavior regulated by bound nucleotide cofactor (13–15, 32). First, it controls Rad51 DNA affinity (ATP = high affinity and ADP = low affinity) with the ATP-bound state favored for filament nucleation and growth. Second, Rad51 ATPase controls the extent of filament elongation through transitions in filament pitch. As it binds ssDNA, Rad51-ATP stretches it to 150% of B-form length, but in a nonuniform manner. In the RecA filament–ssDNA crystal structure trapped in its most extended form with ADP-AlF4 (33), each RecA monomer binds three nucleotides of DNA and holds them with near B-DNA rise values (3.8–4.2 Å per nucleotide; B-DNA is 3.4 Å). Most of the extension is achieved by the stretched (7.1–7.8 Å rise) base steps between nucleotide triplets and the next monomer, as RecA inserts hydrophobic residues of its L2 loop into the intertriplet base stacks. The arrangement of triplet bases is likely conserved among all family members and appears critical for homology search (34, 35).
The fully extended Rad51 filament is referred to as the active form of Rad51; however, this may be an oversimplification as the filament is not a static structure in the presence of ATP. RAD51 bound to a nonhydrolyzable ATP analog extends filaments the most (99 Å pitch), whereas RAD51-ADP, in contrast, forms compressed, low pitch filaments (76 Å) (36). Under conditions that permit ATP hydrolysis, intermediate RAD51 filament extension is observed (37). Although the ADP-bound state has low DNA affinity, Rad51 does not necessarily dissociate from ssDNA upon ATP hydrolysis. Dissociation of RAD51-ADP is concentrated at filament ends (38), whereas internal monomers, stabilized by interaction with two adjacent monomers, exchange ADP back for ATP without dissociating (39, 40). This general behavior has also been observed with bacterial RecA protein, which hydrolyzes ATP ∼100 times faster than eukaryotic Rad51 (Fig. 2A). In single molecule experiments, switching between a buffer with and without ATP, RecA filaments elongate and shorten, respectively (40). The local changes in filament pitch that occur with ATP hydrolysis promote nucleoprotein filament movement (41). Third, the RecA ATPase enhances heteroduplex DNA extension (see below), and the ATPase becomes necessary when mismatches are present in the path of the exchanging strands (42, 43).
Figure 2.
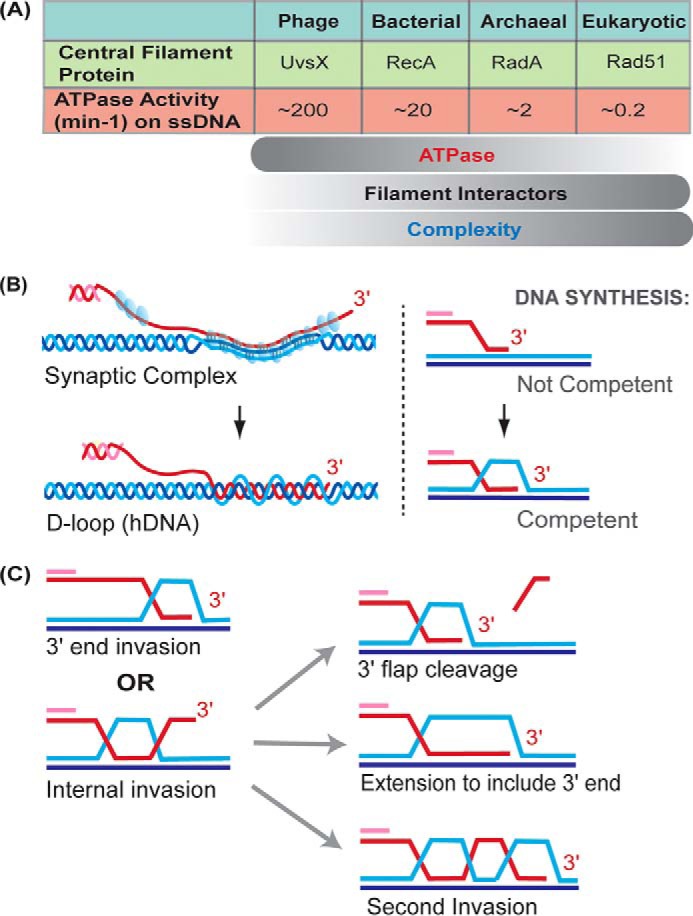
Homology search and DNA strand invasion. A, ATPase activity of central filament proteins decreases with organism complexity while filament interactors increase in number. From T4 phage UvsX to Rad51, the ssDNA-dependent ATPase activity decreases roughly four orders of magnitude. B, hypothetical cartoon of the synaptic complex intermediate. The arrangement of DNA strands within the Rad51 filament (blue spheres) is not known, but there is no net intertwining of DNA strands. Bases are shown paired in triplets based on the RecA crystal structure (33). Watson-Crick base pairing occurs between the invading strand and its complement and may be partially retained between the two complementary strands of the donor molecule. Intertwining of DNA strands leads to D-loop formation, containing heteroduplex DNA (hDNA). C, two types of invasion are 3′ end invasion, which is favored (81), and internal invasion away from the end. In the latter case, the D-loop must be processed to create a primer–template junction containing the 3′ end of the invading DNA strand for extension by a DNA polymerase, and different possibilities are depicted.
Accompanying ascending organism complexity, the central HR protein becomes less and less autonomous, its ATPase slows, and the list of interacting proteins grows larger (Fig. 2A; Table 1). At the upper extreme, vertebrates have evolved five different RAD51 paralogs, each required for normal RAD51 foci formation and each mutant displaying DNA damage sensitivity, suggesting that each paralog has a unique function (13, 15, 44). The functions of the Rad51 paralogs appear to be accomplished through their integration into the Rad51 filament (45, 46), although their arrangement of contacts and frequency within or capping filament segments is unknown. Rad51 paralog-dependent changes in nucleoprotein–ssDNA nuclease sensitivity suggest their interaction with Rad51 filaments leads to changes in its conformation (46). In Saccharomyces cerevisiae, the Srs2 helicase disrupts Rad51 filaments, preventing hyper-recombination (47–49). The Rad51 paralogs Rad55 and Rad57 form a heterodimer that opposes the action of Srs2, acting as a roadblock on ssDNA in the path of the helicase (45). Many facets and interspecies variations by which Rad51 filaments are modulated by its paralogs are still awaiting elucidation.
In eukaryotes, the Sae3–Mei5 heterodimer is important for mitotic and/or meiotic recombination, depending on the organism (50). For example, S. cerevisiae Sae3–Mei5 is only required for HR during meiosis, whereas in vertebrates the homologous complex SWI5–MEI5 functions during HR in somatic cells (Table 1). Some species utilize the heterodimer for both programs (e.g. Schizosaccharomyces pombe), and plants appear to lack homologs (50). Swi5–Sfr1, the fission yeast homologs, display an elongated structure that binds in the groove of the extended Rad51 filament (51). It stabilizes Rad51 filaments on ssDNA and promotes Rad51-dependent DNA strand exchange reactions (50). Mouse SWI5–SFR1 was shown to enhance the RAD51 ATPase by promoting ADP-ATP exchange (52). The mechanisms and evolutionary driving forces to explain why the Rad51 filament should become dependent on Sae3–Mei5 only in certain contexts await elucidation.
Curiously, vertebrate RAD51, unlike yeast Rad51, prokaryotic or phage proteins, does not form stable RAD51–ssDNA filaments on its own in vitro. Calcium enables RAD51 filament formation by inhibiting its ATPase activity (53, 54). Furthermore, calcium ions appear to change the orientation of bases within RAD51 filaments, relative to magnesium ions (55). It is unclear whether calcium is a physiological cofactor or a substitute for a protein cofactor such as SWI5–MEI5, as the fission homolog Swi5–Sfr1 was found to stimulate the same step as calcium but by a different mechanism (56, 57).
A key difference between prokaryotic and eukaryotic recombination machineries is the evolution of a tightly coupled dsDNA-dependent motor activity complementing the Rad51 filament, the Rad54 protein. Although its primary role requires its ATPase activity (see below), Rad54 associates with Rad51–ssDNA filaments and stabilizes them, independent of its ATPase activity (58, 59).
Rad51 regulation by the nucleotide-binding/hydrolysis cycle, associated proteins, and potentially other cofactors is critical in its assembly and function. Once the nucleoprotein scaffold is assembled, other Rad51 filament-interacting proteins enhance the homology search and DNA strand invasion activities of this dynamic entity.
The homology search
The broken DSB end, now resected to ssDNA and assembled with the Rad51 filament and cofactors, must find a homology donor from which to initiate DNA synthesis. The nature of the search entity as a filament allows the entire sequence of ssDNA within to be exploited. The filament must interact with genomic dsDNA as it scans for sequence complementarity. In addition to the primary DNA-binding site for ssDNA, RecA has a secondary dsDNA-binding site that promotes lateral contact with duplex DNA (60). In a poorly understood fashion, one strand of duplex DNA is destabilized to open the helix and to allow bases to be sampled for complementarity to those within the filament, termed base-flipping (61, 62). In the case of eukaryotic Rad51, additional proteins may be involved in this step, and a role of bridging RAD51 filaments to dsDNA has been proposed for vertebrate RAD51AP1, PALB2, and HOP2-MND1 (63–66). In genome-wide anti-Rad51 CHIP assays, S. cerevisiae Rad54 or its paralog Rdh54 are required to measure significant Rad51 association outside the region of the DSB, suggesting homology search does not occur in their absence (67). Besides the well-established role of BRCA1 in DSB resection control (5), human BRCA1–BARD1 was identified to play an unanticipated role in RAD51-mediated homologous pairing that correlated with the interaction between BRCA1 and RAD51 (68). Thus, extrinsic factors may enhance dsDNA probing by Rad51 in eukaryotes.
For efficient homology search, dsDNA must be transiently bound and turned over rapidly when the sequence is incorrect. Microhomologies of as few as eight nucleotides promote extended lifetimes of Rad51–ssDNA–dsDNA complexes (69, 70). A clear role of filament ATPase activity in the homology search was lacking until recently when RecA ATPase was shown to be involved in the release of dsDNA bound by the filament (71). In contrast to RecA, Rad51 from yeast and human has ATPases of roughly two orders of magnitude lower than RecA (Fig. 2A), and therefore it is not clear whether the ATPase of Rad51 plays a role in dsDNA release during homology search or whether additional protein cofactors now assume this role in eukaryotes.
The synaptic complex, also known as a paranemic joint (72, 73), forms upon identification of homology in a donor dsDNA (Fig. 2B). By EM, all three strands are seen passing into the filament along the paired region until they exit again (74). The arrangement of base pairing in this three-stranded intermediate is poorly understood (Fig. 2B), but there is no net intertwining of the invading strand with its complement in the donor, such that removal of proteins results in total disruption of the structure. However, synaptic complexes are more stable than filament interactions observed with nonhomologous dsDNA (75). The structure, size, and lifetimes of synaptic complexes in vivo are not known; however, it was recently proposed that they exist for an extended period of time before being triggered to proceed to form the D-loop (76).
The RecA–ssDNA structure suggests that for greater than three nucleotides in dsDNA to be consecutively base-paired with nucleotides in the extended filament, the dsDNA too must extend (untwist) in the same pattern of triplet stacked bases intervened by unstacked and stretched intertriplet gaps. This arrangement may facilitate the homology search by preventing cooperative base stacking interactions in the absence of homology, to favor the stabilization of pairing through Watson-Crick base pairing (35). It also helps explain why negatively supercoiled DNA donors are highly favored for the formation of metastable synaptic complexes (72). As the dsDNA is stretched, the negative supercoils are relaxed like a coiled spring to provide energy to drive expansion of the paired region. RecA synaptic complexes formed with donor plasmids of defined supercoil content and observed by EM have paired regions where the length corresponds to that needed to dissipate all negative supercoils (74).
Formation of heteroduplex DNA in the D-loop and the ATP-dependent role of Rad54
After formation of the synaptic complex, the 3′ end of the invading strand must be intertwined with its complement in the donor to form a primer–template junction competent for DNA synthesis (Fig. 2B). This key intermediate is known as the displacement or D-loop, because the original base-pairing interactions in the donor dsDNA were disrupted and replaced by heteroduplex DNA (hDNA), a term that describes the region within the D-loop where the invading strand has intertwined with the complementary strand in the donor. The DNA-strand invasion reaction is fundamentally different from DNA strand annealing, because it requires DNA strand displacement and the original base pair to be broken.
Rad54 activity is tightly coupled to the activity of Rad51, to the extent that yeast Rad54 is required for D-loop formation in vitro and in vivo, dependent on its ATPase activity (77). Human RAD54 highly stimulates the calcium-supported RAD51 reaction (76, 78). Rad51 stimulates Rad54 ATPase activity manyfold, and this activity is coupled to the removal of Rad51 filaments from dsDNA (79). At least two nonexclusive models have been proposed to explain Rad54 motor activity in promoting D-loop formation. In one, Rad54 translocation on donor dsDNA induces supercoiling, and this in turn leads to transient separation of dsDNA to allow the Rad51 filament access to the complementary strand (80). Another model posits that Rad54 is an hDNA pump that threads in the invading strand and donor dsDNA and threads out hDNA and the displaced strand. Establishing and migrating an hDNA branch point (junction), the motor domain translocates on the newly uniting strands of the hDNA, while the displaced strand is guided out of the donor duplex through the N-terminal ssDNA-binding domain identified in Rad54 (81). In this model, Rad51 is removed during the process of forming hDNA, with Rad54 acting essentially as a three-way zipper (81).
Why have eukaryotes parsed out functions of the central DNA strand exchange protein to other protein factors, such as Rad54? First, it may provide greater function or versatility than the filament protein alone. For example, the Rad51/Rad54 pair can invade a linear donor, whereas RecA requires negatively supercoiled donor DNA (81). In eukaryotes, the supercoiling density is not uniform throughout the genome (82), and therefore more topological versatility may be required. Rad51 and Rad54 have also been reported to be competent to form D-loops in donors with bound nucleosomes, a reaction also not supported by RecA (83, 84). Second, less autonomy in the DNA strand-exchange protein translates to greater opportunity for regulation. Human RAD54 phosphorylation by NEK1 kinase is necessary for RAD51 foci to be turned over in the G2 phase of the cell cycle (76). One possible interpretation is that synaptic complexes, marked by RAD51 foci, are only triggered to form hDNA when RAD54 is phosphorylated. However, the purified phosphorylation site S572A mutant formed D-loops, bound DNA and RAD51-like WT protein (76), suggesting that the RAD54 phosphorylation-dependent triggering of RAD51 foci removal cannot be explained by a simple model of direct enhancement of RAD54 activity.
DNA topology and structure affect hDNA formation and stability
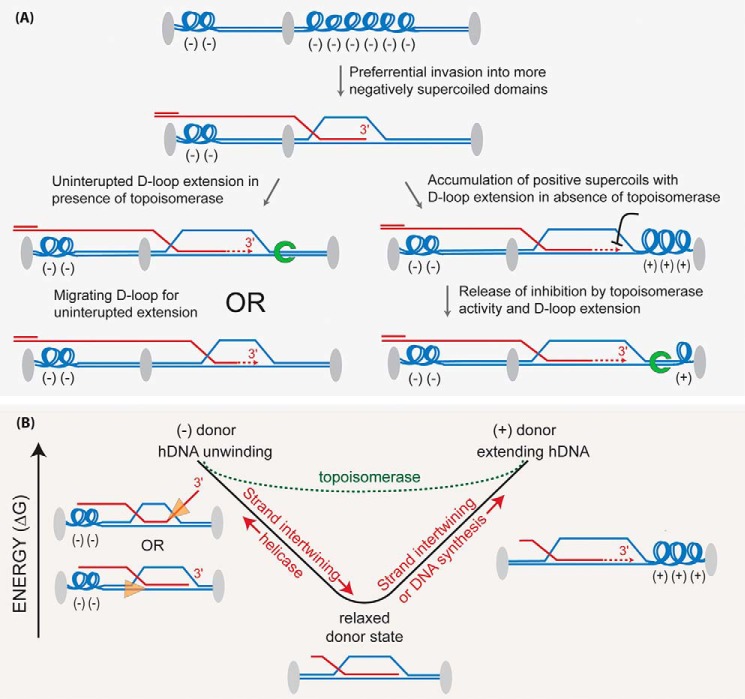
Where the initial invasion occurs along a long resected DSB end depends on multiple factors, including where the initial homology is encountered. The synaptic complex will likely spread to the extent allowed by supercoiling (see Fig. 2 legend); then, the invading strand will intertwine with the complementary donor strand forming hDNA, beginning at some point relative to the 3′ end to form a D-loop. Homologous ssDNA ends (physiologically the 3′ end) are favored over internal homologous sequence, yet hDNA may form anywhere along the homologous sequence (81). Supercoiling determines the extent of the hDNA tract that can be accommodated, with about 10.5 bp per negative supercoil that can be intertwined before there is a topological block encountered. Highly negatively supercoiled domains in the donor may favor the initial synaptic complex formation and subsequent invasion (Fig. 3A). The stored energy within the natural negative superhelicity of chromatin, analogous to a compressed spring, creates an energy well encouraging homologous strands to intertwine as favored by the energy release (Fig. 3B). Superhelicity also affects the DNA synthesis step as well as the unwinding of hDNA by helicases necessary to dissociate the D-loop (Fig. 3B). In the latter case, the donor is unwound toward a relaxed state as it intertwines with the invading strand, and so it must be rewound to eject the D-loop (reinduce negative supercoiling). In vivo, the supercoiling status is neither random nor static (82, 85). Besides ATP-dependent motor proteins, topoisomerases also have the potential to lower energetic barriers to extending hDNA, either by branch migration or DNA synthesis, and to accomplish hDNA disruption (Fig. 3B). The Sgs1–Top3–Rmi1 complex is composed of a potent 3′–5′ DNA helicase, a type 1A topoisomerase, and its co-factor, respectively. Specifically in the context of a reconstituted D-loop reaction, but not with protein-free DNA, Top3–Rmi1 relaxes the plasmid donor and promotes the dissociation of the D-loop (86). Mutants in the complex have a slow growth phenotype that is rescued by deletions that impair HR (87). Hence, Top3 is a good candidate for an HR-associated topoisomerase activity. Other type I or type II topoisomerases have the potential to affect HR outcome as well, even if indirectly by changing the topological landscape of the domain being invaded (Fig. 3). These topological aspects of hDNA dynamics underpin the energy landscape of DNA strand exchange reactions and may rationalize the need for multiple motor proteins driving reactions in defined directions.
Figure 3.
Topological considerations in DNA strand invasion. A, supercoiling density in various chromatin domains may influence the propensity to invade (invasion is a term inclusive of synaptic complex formation and hDNA formation through interwining of the invading strand) a particular domain in the donor. Synaptic complex formation initially relaxes the donor when the extended Rad51–ssDNA filament aligns with homologous bases present in the complementary donor strand, causing them to extend as well. Invading strand intertwining to produce hDNA is a separate consumption of the negative supercoils in the donor, and unlike synaptic complexes, hDNA is stable even in the absence of the bound proteins. The topological status of the donor will also influence the DNA synthesis step, as indicated in the figure and discussed in the text. Gray ovals indicate a specific DNA-bound protein(s), which creates discrete topological domains within linear chromosomes. Green horseshoe, topoisomerase. B, negative supercoiling creates an energy well that influences DNA strand intertwining or unwinding. Invasion into a supercoiled donor will favor stand intertwining as negative supercoils release their stored energy as they are dissipated, with a relaxed donor defining the lowest energy state. Once all negative supercoils are relaxed, extending hDNA through further intertwining, or DNA synthesis, will induce positive supercoils. Unwinding the D-loop in a relaxed donor by a DNA helicase will require they rewind negative supercoils in the donor. The dotted green line represents the altered energy landscape made possible by the action of topoisomerases, in effect making the energy well more shallow. Orange triangles depict DNA helicases and their translocation orientation to disrupt D-loops.
At a minimum, only the very 3′ end of the invading strand needs be in hDNA to form a primer–template junction for DNA synthesis. However, the hDNA region may be hundreds of bases pairs. It is becoming increasingly clear that the sequence away from the 3′ end can readily form hDNA, as evident by robust Rad51/Rad54-dependent D-loop formation using substrates with terminal heterologies (81). In addition, multi-invasion species, where the invading strand forms hDNA in more than one dsDNA donor molecule, occur in vitro and in vivo (11, 81). Therefore, it seems unlikely that as the DNA intertwines internally, the rest of the ssDNA molecule passes through the strands of the donor helix. Rather, the hDNA region may intertwine while dissipating the torsion created through rotation in the ssDNA. The prevalence of internal invasions brings up new questions as to how they would be processed. These D-loops could be disrupted to try end-invasion again, the hDNA region could be extended or migrated toward the 3′ end by as yet unknown enzymes, or the unincorporated strand extremity might be cleaved by a nuclease such as Rad1–Rad10 (Fig. 2C; Table 1). Alternatively, by anchoring the DSB end, the internal invasion may facilitate a secondary invasion to occur at the 3′ end (Fig. 2C). Internal or end invasion creates unique hDNA branch point structures that have the potential to influence HR protein activities, as found to be the case for the Rad54 and Srs2 motor proteins (81, 88).
Clearly, much is unknown about the formation of hDNA in vivo for the lack of assays to physically measure D-loops and their extent in somatic cells. For example, there is little information on the role of topoisomerases on HR other than Top3. The length of the hDNA tract can influence biological outcome because longer hDNA tracts favor dHJ formation thereby promoting crossovers (CO) (89, 90). COs, in turn, risk potential loss of heterozygosity of critical genes such as tumor suppressors (10). This evolutionary pressure is evidently intense enough on organisms that it has selected for pathways of somatic CO avoidance to favor SDSA.
D-loop reversal activities ensure high-fidelity DSB repair and CO avoidance
In SDSA, the disruption of the extended D-loop is ultimately required for chromosome resolution, but this is not the only need for D-loop disruption (Fig. 1 and Fig. S2). First, it may help reject hDNA formed with homeologous (partial homology) donors to aid the homology search. Second, D-loop disruption lowers the probability that the two ends of the DSB will simultaneously invade the donor, and DNA synthesis allow maturation into a dHJ with potential CO outcome (Fig. S2) (8, 9, 91). Third, it prevents the preponderance of multi-invasions (one ssDNA end invading two donors), which can lead to translocations of the two donors and a cascade of further rearrangements (11).
In S. cerevisiae, at least three enzymes/complexes have been implicated in D-loop disruption: Sgs1–Top3–Rmi1 (86), and two 3′–5′ helicases, Srs2 (88) and Mph1 (93). In vertebrates, the number of helicases increases and includes the five RecQ family helicases (Table 1). Likely, each helicase is specialized to recognize subtly different hDNA intermediates, whether they are demarcated by the topological status, DNA junction structure, and/or interaction with other bound proteins. Often these factors have roles in support of DNA replication in addition to DSB repair. D-loop disruption activities maintain a dynamic balance with hDNA formation and extension, enhancing overall HR fidelity and promoting CO avoidance.
DNA synthesis at the D-loop
Once the 3′ end has been incorporated into hDNA in the D-loop or a 3′ end has been generated by cleavage, the stage is set for DNA synthesis (Fig. 2C) (94). In vitro, this reaction has been reconstituted with a short oligonucleotide ssDNA paired with a supercoiled plasmid donor, including Rad51, Rad54, and RPA to make the D-loop and RFC1–5, PCNA, and DNA polymerase δ as minimal components for D-loop extension (95–97). On its own, Pol δ is unable to extend D-loops, yet the inclusion of PCNA and its loader RFC1–5 transforms the polymerase into a robust hDNA extender (95). A genetic system in yeast revealed that both Pol δ and ϵ are required for HR (98), but the precise role of Pol ϵ remains to be determined as it cannot perform displacement synthesis (99).
When a short ssDNA is employed, the donor is only partially relaxed by its intertwining in the D-loop, and DNA synthesis can proceed until a topological block is encountered, as D-loop extension by DNA synthesis consumes one negative supercoil every helical turn (∼10.5 bp) (Fig. 3). In vivo the ssDNA substrate is relatively long and might be expected to fully relax the negative supercoils in the donor, as is observed in vitro with long ssDNA substrates (74, 81). This means the D-loop will start DNA synthesis in a relaxed donor, and its extension will induce positive supercoiling, which topologically blocks DNA synthesis (Fig. 3). One way to overcome this block is to peel off the D-loop from the 5′ side (relative to invading strand) of the hDNA to establish a migrating bubble to prevent topological stalling (100). In yeast, Pif1 helicase enables long-range DNA synthesis during break-induced replication (101), and BLM helicase is required for long SDSA events in Drosophila (102). Both may act in a similar fashion to dda helicase, which carries out bubble migration in the phage T4 (100). Alternatively, topoisomerases might relax the donor ahead of the extending D-loop, as they do for normal replicative DNA synthesis. However, topoisomerases cannot deal with roadblocks in the template itself, such as RNA–DNA hybrids (R-loops), transcription machinery, and bound proteins. Interestingly, two homologs of the replicative helicase (MCM2–7), MCM8 and MCM9, have been proposed as a recombination-specific DNA helicase (103). MCM8–9 and RAD51 are important for replication fork-associated DNA synthesis after degradation of the replicative helicase by MCM2 depletion, and their knockout decreases DSB-induced HR measured in genetic assays in human cells (103). However, the details of their interactions with the recombination machinery have not been established by their reconstitution into D-loop extension assays.
Many questions still shroud the control of recombination-associated DNA synthesis. Besides Pol δ, which DNA polymerases are involved, and what is their function? How is the 3′ end generated for extension (Fig. 2C)? What is the influence of topoisomerases (and which) on DNA synthesis? Is a migrating bubble established, and which protein(s) are involved? How is the extent of DNA synthesis balanced by extended D-loop disruption activities to achieve SDSA?
Extended D-loop disruption, second-end annealing, and crossover avoidance
Pol δ DNA synthesis must extend the invading strand enough so that when the D-loop is disrupted, there is sufficient sequence homology to anneal to the second resected end of the DSB. Does the machinery sense when enough DNA synthesis has been accomplished? There may be no need, as genetic studies support a model where DNA synthesis is achieved through successive cycles of invasion, short DNA synthesis, and D-loop disruption (104, 105). Srs2 and Mph1 have been implicated genetically and demonstrated biochemically to disrupt extended D-loops (88, 97). These proteins might promote a stochastic disruption of the extended D-loop. However, in the case of Srs2, there is evidence that cell signaling can tip the balance in favor of disruption upon D-loop extension. PCNA-SUMO was shown to give a modest preference to Srs2, which has a SUMO-interacting motif, for disruption of extending D-loops. Structurally, preference was shown for D-loops that contained hDNA proximal to the 3′ end (88). As Rad54, Srs2 is another example of a complex DNA translocase whose motor and accessory regulatory domains receive inputs from cell signaling, DNA substrate structure, and interaction with partner proteins (107).
Srs2, Sgs1 (RecQ family), and Mph1 are 3′ to 5′ ssDNA translocases with respect to the DNA strand the motors translocate on. To disrupt the hDNA, they must first load onto a strand in the D-loop substrate. Continuing the Srs2 example, preference for the modified PCNA and 3′ proximal hDNA substrate leads to a model where it translocates on the invading strand starting at the 3′ end to disrupt hDNA (Fig. 3B). However, another possible mode of disruption would be for a helicase to load at the 5′ hDNA branch point on the donor template strand and translocate toward the 3′ side of the hDNA tract (Fig. 3B). In either case, helicase ssDNA translocation to unwind the heteroduplex and to eject the invading strand returns it to ssDNA, which will quickly be bound by RPA.
Now disengaged from D-loops, the two DSB ends have the chance to anneal in the region of homology created from DNA synthesis steps within the D-loops. In yeast, Rad52 is intermediary for the annealing step between the RPA-coated ssDNAs (108, 109). Likewise, RAD52 is the only human protein known to be capable of annealing RPA-coated ssDNA (110). However, the phenotype of RAD52 mutants or knockdowns in vertebrate cells is very subtle and inconsistent with a critical role during HR-mediated DSB repair (31, 111). Although the primary mediator function of Rad52 has been usurped by BRCA2, BRCA2 does not anneal RPA-coated ssDNA (19). Because SDSA is also the predominant HR-mode of DSB repair in somatic human cells (112), it is unclear which protein catalyzes the second-end annealing step. It is possible that an unidentified protein is involved, as the subtle phenotype of RAD52-deficient human cells is not consistent with a critical role in this step. Alternatively, we suggest a modified SDSA model, which replaces a second-end annealing step with a second-end DNA strand invasion step, obviating the need for an annealing protein altogether (Fig. S1).
Regardless of whether second-end rejoining proceeds by annealing or invasion, DNA synthesis primed by the second end is required to restore the integrity of the chromosome. The identity of the DNA polymerase involved in second-end DNA synthesis remains to be determined (94). Ligation of any remaining nicks restores the integrity of the chromosome, and CO was successfully avoided (Fig. 1).
Double Holliday junction processing and the possibility of a crossover
Although SDSA is the preferred DSB repair pathway in somatic cells, a fraction of repair events proceed through formation of a dHJ, as supported by their physical detection in somatic cells (113). En route to dHJ formation, one of two things can happen: the second resected DSB end anneals to the displaced strand of the D-loop (Fig. 1), or both DSB ends simultaneously invade the donor and extend through DNA synthesis (Fig. S2). Ligation of the resultant nicked duplexes then creates a dHJ, where each HJ is a four-way branched DNA joint molecule (Fig. 1 and Fig. S2). These dHJs are processed by either dissolution, giving strictly noncrossover (NCO), or endonucleolytic resolution that generates CO or NCO products.
In Escherichia coli, RuvC has set the paradigm for the resolution of single HJs (114). The RuvC dimer symmetrically cleaves two phosphodiester bonds across the HJ (Fig. S3), resulting in nicked duplexes capable of being ligated without further processing. If the cleavages of the dHJ across each of the two single HJs are in different planes, a CO product is formed, although incisions in the same plane result in an NCO outcome. This gives a 50% chance of CO formation, if the cleavages are random (Fig. S3). Thus, dHJ resolution has the potential for CO formation and is largely avoided in somatic cells.
The RuvC paradigm encouraged identification of similar resolvases in eukaryotes and led to the recognition of Mus81–Mms4, Slx1–Slx4, and Yen1. These nucleases have been shown to act on a variety of joint DNA molecules with different specificities as reviewed in Refs. 114, 115. For example, Yen1 cleaves HJs symmetrically to produce nicked duplex products, but it also cleaves other joint molecules such as 5′-flaps and model replication forks. Mus81–Mms4 cleaves intact HJs poorly, displaying a strong preference for joint DNA molecules containing a nick at the branch point. Human MUS81–EME1 has been proposed to cooperate with SLX1–SLX4 such that SLX1–SLX4 introduces the initial rate-limiting cut to form a nicked HJ, which is further processed by MUS81–EME1 (116). However, the SLX–MUS complex, unlike GEN1, often cleaves asymmetrically, leaving products with gaps and flaps that require further processing. Mus81–Mms4 may also act on unligated nicked HJs before they are ligated. Whether HJs are always ligated before cleavage by resolvases is unknown. Moreover, the relaxed substrate selectivity among the eukaryotic endonucleases opens the possibility that cuts in other junctions like the D-loop or half-junctions (Fig. S3) may also lead to crossover formation (92, 115). These alternative models do not necessarily involve dHJ formation, yet still produce COs. Thus, HJ resolution in eukaryotes and CO formation are still poorly understood.
Even after dHJ formation, as a last measure of CO avoidance, the cell has the opportunity for dHJ dissolution carried out by the Sgs1–Top3–Rmi1 complex leading to a strict NCO outcome (Fig. 1) (106). The Sgs1 helicase promotes branch migration of the two HJs toward each other to converge into a hemicatenane. Top3 is required to resolve topological constraints during branch migration and to dissolve the topological connection by passing the two DNA strands of the hemicatenane to result in NCO products. Whether migration of HJs is random or directed is unknown. Dissolution is important to avoid excessive inter-sister COs and to prevent loss of heterozygosity in somatic cells. This is exemplified in Bloom's syndrome, in which the human Sgs1 homolog BLM is mutated, resulting in excessive sister chromatid exchanges and genome instability (106).
Concluding remarks
HR enables cells to achieve high-fidelity repair of DSBs and other complex DNA damage. When properly regulated, the chances of deleterious rearrangements or loss of heterozygosity through CO are low. Because the complexity of HR increases with that of the organism, this makes studies in simpler systems valuable for establishing paradigms that can help deconvolute the complexity in humans. Continuing effort will be important to paint the full picture of this elegant DNA repair modus.
Supplementary Material
Acknowledgments
We are grateful to past and present members of the Heyer laboratory, especially Aurele Piazza, as well as Neil Hunter and Steve Kowalczykowski and their laboratory members for stimulating discussions. We thank Damon Meyer, Eliana Tavares, Steven Gore, and Hang Le for helpful comments on the manuscript.
This work was supported by National Institutes of Health Grants GM58015 and CA92276 (to the W. D. H. laboratory). This is the third article in the Thematic Minireview series “DNA double-strand break repair and pathway choice.” The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S3.
Throughout the paper, S. cerevisiae names and nomenclature are used when referring to protein families discussed in general. For the names of homologs in other species, see Table 1.
- HR
- homologous recombination
- NHEJ
- nonhomologous end-joining
- MMEJ
- microhomology-mediated end-joining
- DSB
- double-strand break
- ssDNA
- single-stranded DNA
- dsDNA
- double-stranded DNA
- SDSA
- synthesis-dependent strand-annealing
- NCO
- noncrossover
- CO
- crossover
- HJ
- Holliday junction
- dHJ
- double Holliday junction
- RPA
- replication protein A
- hDNA
- heteroduplex DNA
- PCNA
- proliferating cell nuclear antigen
- SUMO
- small ubiquitin-like modifier.
References
- 1. Kowalczykowski S. C., Hunter N., and Heyer W.-D. (eds) (2016) DNA Recombination, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 2. Hustedt N., and Durocher D. (2016) The control of DNA repair by the cell cycle. Nat. Cell Biol. 19, 1–9 10.1038/ncb3452 [DOI] [PubMed] [Google Scholar]
- 3. Pannunzio N. R., Watanabe G., and Lieber M. R. (2018) Nonhomologous DNA end-joining for repair of DNA double-strand breaks. J. Biol. Chem. 293, 10512–10523 10.1074/jbc.TM117.000374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sallmyr A., and Tomkinson A. (2018) Repair of DNA double-strand breaks by mammalian alternative end-joining pathways. J. Biol. Chem. 293, 10536–10546 10.1074/jbc.TM117.000375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Her J., and Bunting S. F. (2018) How cells ensure correct repair of DNA double-strand breaks. J. Biol. Chem. 293, 10502–10511 10.1074/jbc.TM118.000371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Symington L. S. (2014) End resection at double-strand breaks: mechanism and regulation. Cold Spring Harb. Perspect. Biol. 6, a016436 10.1101/cshperspect.a016436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kadyk L. C., and Hartwell L. H. (1992) Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics 132, 387–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nassif N., Penney J., Pal S., Engels W. R., and Gloor G. B. (1994) Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 14, 1613–1625 10.1128/MCB.14.3.1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pâques F., and Haber J. E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moynahan M. E., and Jasin M. (2010) Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Biol. 11, 196–207 10.1038/nrm2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Piazza A., Wright W. D., and Heyer W. D. (2017) Multi-invasions are recombination by-products that induce chromosomal rearrangements. Cell 170, 760–773 10.1016/j.cell.2017.06.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heyer W. D., Ehmsen K. T., and Liu J. (2010) Regulation of homologous recombination in eukaryotes. Annu. Rev. Genet. 44, 113–139 10.1146/annurev-genet-051710-150955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sung P., Krejci L., Van Komen S., and Sehorn M. G. (2003) Rad51 recombinase and recombination mediators. J. Biol. Chem. 278, 42729–42732 10.1074/jbc.R300027200 [DOI] [PubMed] [Google Scholar]
- 14. Yu X., Jacobs S. A., West S. C., Ogawa T., and Egelman E. H. (2001) Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proc. Natl. Acad. Sci. U.S.A. 98, 8419–8424 10.1073/pnas.111005398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu J., Ehmsen K. T., Heyer W. D., and Morrical S. W. (2011) Presynaptic filament dynamics in homologous recombination and DNA repair. Crit. Rev. Biochem. Mol. Biol. 46, 240–270 10.3109/10409238.2011.576007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bell J. C., and Kowalczykowski S. C. (2016) Mechanics and single-molecule interrogation of DNA recombination. Annu. Rev. Biochem. 85, 193–226 10.1146/annurev-biochem-060614-034352 [DOI] [PubMed] [Google Scholar]
- 17. Morimatsu K., and Kowalczykowski S. C. (2003) RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol. Cell 11, 1337–1347 10.1016/S1097-2765(03)00188-6 [DOI] [PubMed] [Google Scholar]
- 18. Yang H., Li Q., Fan J., Holloman W. K., and Pavletich N. P. (2005) The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature 433, 653–657 10.1038/nature03234 [DOI] [PubMed] [Google Scholar]
- 19. Jensen R. B., Carreira A., and Kowalczykowski S. C. (2010) Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467, 678–683 10.1038/nature09399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. New J. H., Sugiyama T., Zaitseva E., and Kowalczykowski S. C. (1998) Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature 391, 407–410 10.1038/34950 [DOI] [PubMed] [Google Scholar]
- 21. Song B., and Sung P. (2000) Functional interactions among yeast Rad51 recombinase, Rad52 mediator, and replication protein A in DNA strand exchange. J. Biol. Chem. 275, 15895–15904 10.1074/jbc.M910244199 [DOI] [PubMed] [Google Scholar]
- 22. McKee R. H., and Lawrence C. W. (1980) Genetic analysis of gamma-ray mutagenesis in yeast. III. Double-mutant strains. Mutat. Res. 70, 37–48 10.1016/0027-5107(80)90056-1 [DOI] [PubMed] [Google Scholar]
- 23. Symington L. S., Rothstein R., and Lisby M. (2014) Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics 198, 795–835 10.1534/genetics.114.166140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu J., Doty T., Gibson B., and Heyer W. D. (2010) Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 17, 1260–1262 10.1038/nsmb.1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thorslund T., McIlwraith M. J., Compton S. A., Lekomtsev S., Petronczki M., Griffith J. D., and West S. C. (2010) The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat. Struct. Mol. Biol. 17, 1263–1265 10.1038/nsmb.1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sharan S. K., Morimatsu M., Albrecht U., Lim D. S., Regel E., Dinh C., Sands A., Eichele G., Hasty P., and Bradley A. (1997) Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature 386, 804–810 10.1038/386804a0 [DOI] [PubMed] [Google Scholar]
- 27. Yuan S. S., Lee S. Y., Chen G., Song M., Tomlinson G. E., and Lee E. Y. (1999) BRCA2 is required for ionizing radiation-induced assembly of rad51 complex in vivo. Cancer Res. 59, 3547–3551 [PubMed] [Google Scholar]
- 28. Moynahan M. E., Pierce A. J., and Jasin M. (2001) BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell 7, 263–272 10.1016/S1097-2765(01)00174-5 [DOI] [PubMed] [Google Scholar]
- 29. Jensen R. B., Ozes A., Kim T., Estep A., and Kowalczykowski S. C. (2013) BRCA2 is epistatic to the RAD51 paralogs in response to DNA damage. DNA Repair 12, 306–311 10.1016/j.dnarep.2012.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Prakash R., Zhang Y., Feng W., and Jasin M. (2015) Homologous recombination and human health: the roles of BRCA1, BRCA2, and associated proteins. Cold Spring Harb. Perspect. Biol. 7, a016600 10.1101/cshperspect.a016600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Feng Z., Scott S. P., Bussen W., Sharma G. G., Guo G., Pandita T. K., and Powell S. N. (2011) Rad52 inactivation is synthetically lethal with BRCA2 deficiency. Proc. Natl. Acad. Sci. U.S.A. 108, 686–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Spirek M., Mlcouskova J., Belan O., Gyimesi M., Harami G. M., Molnar E., Novacek J., Kovacs M., and Krejci L. (2018) Human RAD51 rapidly forms intrinsically dynamic nucleoprotein filaments modulated by nucleotide binding state. Nucleic Acids Res. 2018 10.1093/nar/gky111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen Z., Yang H., and Pavletich N. P. (2008) Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 453, 489–494 10.1038/nature06971 [DOI] [PubMed] [Google Scholar]
- 34. Lee J. Y., Terakawa T., Qi Z., Steinfeld J. B., Redding S., Kwon Y., Gaines W. A., Zhao W., Sung P., and Greene E. C. (2015) DNARECOMBINATION. Base triplet stepping by the Rad51/RecA family of recombinases. Science 349, 977–981 10.1126/science.aab2666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kowalczykowski S. C. (2008) Structural biology: snapshots of DNA repair. Nature 453, 463–466 10.1038/453463a [DOI] [PubMed] [Google Scholar]
- 36. Yu X., Jacobs S. A., West S. C., Ogawa T., and Egelman E. H. (2001) Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proc. Natl. Acad. Sci. U.S.A. 98, 8419–8424 10.1073/pnas.111005398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hilario J., Amitani I., Baskin R. J., and Kowalczykowski S. C. (2009) Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc. Natl. Acad. Sci. U.S.A. 106, 361–368 10.1073/pnas.0811965106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. van Mameren J., Modesti M., Kanaar R., Wyman C., Peterman E. J., and Wuite G. J. (2009) Counting RAD51 proteins disassembling from nucleoprotein filaments under tension. Nature 457, 745–748 10.1038/nature07581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Robertson R. B., Moses D. N., Kwon Y., Chan P., Chi P., Klein H., Sung P., and Greene E. C. (2009) Structural transitions within human Rad51 nucleoprotein filaments. Proc. Natl. Acad. Sci. U.S.A. 106, 12688–12693 10.1073/pnas.0811465106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nishinaka T., Doi Y., Hara R., and Yashima E. (2007) Elastic behavior of RecA-DNA helical filaments. J. Mol. Biol. 370, 837–845 10.1016/j.jmb.2007.05.044 [DOI] [PubMed] [Google Scholar]
- 41. Ramreddy T., Sen S., Rao B. J., and Krishnamoorthy G. (2003) DNA dynamics in RecA-DNA filaments: ATP hydrolysis-related flexibility in DNA. Biochemistry 42, 12085–12094 10.1021/bi034667k [DOI] [PubMed] [Google Scholar]
- 42. Kim J. I., Cox M. M., and Inman R. B. (1992) On the role of ATP hydrolysis in RecA protein-mediated DNA strand exchange. I. Bypassing a short heterologous insert in one DNA substrate. J. Biol. Chem. 267, 16438–16443 [PubMed] [Google Scholar]
- 43. Bedale W. A., and Cox M. (1996) Evidence for the coupling of ATP hydrolysis to the final (extension) phase of RecA protein-mediated DNA strand exchange. J. Biol. Chem. 271, 5725–5732 10.1074/jbc.271.10.5725 [DOI] [PubMed] [Google Scholar]
- 44. Takata M., Sasaki M. S., Tachiiri S., Fukushima T., Sonoda E., Schild D., Thompson L. H., and Takeda S. (2001) Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol. 21, 2858–2866 10.1128/MCB.21.8.2858-2866.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Liu J., Renault L., Veaute X., Fabre F., Stahlberg H., and Heyer W. D. (2011) Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature 479, 245–248 10.1038/nature10522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Taylor M. R. G., Špírek M., Chaurasiya K. R., Ward J. D., Carzaniga R., Yu X., Egelman E. H., Collinson L. M., Rueda D., Krejci L., and Boulton S. J. (2015) Rad51 paralogs remodel pre-synaptic Rad51 filaments to stimulate homologous recombination. Cell 162, 271–286 10.1016/j.cell.2015.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rong L., Palladino F., Aguilera A., and Klein H. L. (1991) The hyper-gene conversion hpr-1 mutation of Saccharomyces cerevisiae is an allele of the SRS2/RADH gene. Genetics 127, 75–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Krejci L., Van Komen S., Li Y., Villemain J., Reddy M. S., Klein H., Ellenberger T., and Sung P. (2003) DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423, 305–309 10.1038/nature01577 [DOI] [PubMed] [Google Scholar]
- 49. Veaute X., Jeusset J., Soustelle C., Kowalczykowski S. C., Le Cam E., and Fabre F. (2003) The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423, 309–312 10.1038/nature01585 [DOI] [PubMed] [Google Scholar]
- 50. Argunhan B., Murayama Y., and Iwasaki H. (2017) The differentiated and conserved roles of Swi5–Sfr1 in homologous recombination. FEBS Lett. 591, 2035–2047 10.1002/1873-3468.12656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kuwabara N., Murayama Y., Hashimoto H., Kokabu Y., Ikeguchi M., Sato M., Mayanagi K., Tsutsui Y., Iwasaki H., and Shimizu T. (2012) Mechanistic insights into the activation of Rad51-mediated strand exchange from the structure of a recombination activator, the Swi5–Sfr1 complex. Structure 20, 440–449 10.1016/j.str.2012.01.005 [DOI] [PubMed] [Google Scholar]
- 52. Su G. C., Chung C. I., Liao C. Y., Lin S. W., Tsai C. T., Huang T., Li H. W., and Chi P. (2014) Enhancement of ADP release from the RAD51 presynaptic filament by the SWI5–SFR1 complex. Nucleic Acids Res. 42, 349–358 10.1093/nar/gkt879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ristic D., Modesti M., van der Heijden T., van Noort J., Dekker C., Kanaar R., and Wyman C. (2005) Human Rad51 filaments on double- and single-stranded DNA: correlating regular and irregular forms with recombination function. Nucleic Acids Res. 33, 3292–3302 10.1093/nar/gki640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bugreev D. V., and Mazin A. V. (2004) Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc. Natl. Acad. Sci. U.S.A. 101, 9988–9993 10.1073/pnas.0402105101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fornander L. H., Frykholm K., Reymer A., Renodon-Cornière A., Takahashi M., and Nordén B. (2012) Ca2+ improves organization of single-stranded DNA bases in human Rad51 filament, explaining stimulatory effect on gene recombination. Nucleic Acids Res. 40, 4904–4913 10.1093/nar/gks140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ito K., Murayama Y., Takahashi M., and Iwasaki H. (2018) Two three-strand intermediates are processed during Rad51-driven DNA strand exchange. Nat. Struct. Mol. Biol. 25, 29–36 10.1038/s41594-017-0002-8 [DOI] [PubMed] [Google Scholar]
- 57. Fornander L. H., Renodon-Cornière A., Kuwabara N., Ito K., Tsutsui Y., Shimizu T., Iwasaki H., Nordén B., and Takahashi M. (2014) Swi5–Sfr1 protein stimulates Rad51-mediated DNA strand exchange reaction through organization of DNA bases in the presynaptic filament. Nucleic Acids Res. 42, 2358–2365 10.1093/nar/gkt1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mazin A. V., Alexeev A. A., and Kowalczykowski S. C. (2003) A novel function of Rad54 protein. Stabilization of the Rad51 nucleoprotein filament. J. Biol. Chem. 278, 14029–14036 10.1074/jbc.M212779200 [DOI] [PubMed] [Google Scholar]
- 59. Sanchez H., Kertokalio A., van Rossum-Fikkert S., Kanaar R., and Wyman C. (2013) Combined optical and topographic imaging reveals different arrangements of human RAD54 with presynaptic and postsynaptic RAD51-DNA filaments. Proc. Natl. Acad. Sci. U.S.A. 110, 11385–11390 10.1073/pnas.1306467110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Aihara H., Ito Y., Kurumizaka H., Terada T., Yokoyama S., and Shibata T. (1997) An interaction between a specified surface of the C-terminal domain of RecA protein and double-stranded DNA for homologous pairing. J. Mol. Biol. 274, 213–221 10.1006/jmbi.1997.1403 [DOI] [PubMed] [Google Scholar]
- 61. Gupta R. C., Folta-Stogniew E., O'Malley S., Takahashi M., and Radding C. M. (1999) Rapid exchange of A:T base pairs is essential for recognition of DNA homology by human Rad51 recombination protein. Mol. Cell 4, 705–714 10.1016/S1097-2765(00)80381-0 [DOI] [PubMed] [Google Scholar]
- 62. Folta-Stogniew E., O'Malley S., Gupta R., Anderson K. S., and Radding C. M. (2004) Exchange of DNA base pairs that coincides with recognition of homology promoted by E. coli RecA protein. Mol. Cell 15, 965–975 10.1016/j.molcel.2004.08.017 [DOI] [PubMed] [Google Scholar]
- 63. Dray E., Etchin J., Wiese C., Saro D., Williams G. J., Hammel M., Yu X., Galkin V. E., Liu D., Tsai M. S., Sy S. M., Schild D., Egelman E., Chen J., and Sung P. (2010) Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat. Struct. Mol. Biol. 17, 1255–1259 10.1038/nsmb.1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wiese C., Dray E., Groesser T., San Filippo J., Shi I., Collins D. W., Tsai M. S., Williams G. J., Rydberg B., Sung P., and Schild D. (2007) Promotion of homologous recombination and genomic stability by RAD51AP1 via RAD51 recombinase enhancement. Mol. Cell 28, 482–490 10.1016/j.molcel.2007.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Chi P., San Filippo J., Sehorn M. G., Petukhova G. V., and Sung P. (2007) Bipartite stimulatory action of the Hop2-Mnd1 complex on the Rad51 recombinase. Genes Dev. 21, 1747–1757 10.1101/gad.1563007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Modesti M., Budzowska M., Baldeyron C., Demmers J. A., Ghirlando R., and Kanaar R. (2007) RAD51AP1 is a structure-specific DNA binding protein that stimulates joint molecule formation during RAD51-mediated homologous recombination. Mol. Cell 28, 468–481 10.1016/j.molcel.2007.08.025 [DOI] [PubMed] [Google Scholar]
- 67. Renkawitz J., Lademann C. A., Kalocsay M., and Jentsch S. (2013) Monitoring homology search during DNA double-strand break repair in vivo. Mol. Cell 50, 261–272 10.1016/j.molcel.2013.02.020 [DOI] [PubMed] [Google Scholar]
- 68. Zhao W. X., Steinfeld J. B., Liang F., Chen X., Maranon D. G., Jian Ma C., Kwon Y., Rao T., Wang W., Sheng C., Song X., Deng Y., Jimenez-Sainz J., Lu L., Jensen R. B., et al. (2017) BRCA1–BARD1 promotes RAD51-mediated homologous DNA pairing. Nature 550, 360–365 10.1038/nature24060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Qi Z., Redding S., Lee J. Y., Gibb B., Kwon Y., Niu H., Gaines W. A., Sung P., and Greene E. C. (2015) DNA sequence alignment by microhomology sampling during homologous recombination. Cell 160, 856–869 10.1016/j.cell.2015.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Greene E. C. (2016) DNA sequence alignment during homologous recombination. J. Biol. Chem. 291, 11572–11580 10.1074/jbc.R116.724807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lee J. Y., Qi Z., and Greene E. C. (2016) ATP hydrolysis promotes duplex DNA release by the RecA presynaptic complex. J. Biol. Chem. 291, 22218–22230 10.1074/jbc.M116.740563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wong B. C., Chiu S. K., and Chow S. A. (1998) The role of negative superhelicity and length of homology in the formation of paranemic joints promoted by RecA protein. J. Biol. Chem. 273, 12120–12127 10.1074/jbc.273.20.12120 [DOI] [PubMed] [Google Scholar]
- 73. Bianchi M., DasGupta C., and Radding C. M. (1983) Synapsis and the formation of paranemic joints by E. coli RecA protein. Cell 34, 931–939 10.1016/0092-8674(83)90550-0 [DOI] [PubMed] [Google Scholar]
- 74. Christiansen G., and Griffith J. (1986) Visualization of the paranemic joining of homologous DNA molecules catalyzed by the RecA protein of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 83, 2066–2070 10.1073/pnas.83.7.2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ristic D., Kanaar R., and Wyman C. (2011) Visualizing RAD51-mediated joint molecules: implications for recombination mechanism and the effect of sequence heterology. Nucleic Acids Res. 39, 155–167 10.1093/nar/gkq766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Spies J., Waizenegger A., Barton O., Sürder M., Wright W. D., Heyer W. D., and Löbrich M. (2016) Nek1 regulates Rad54 to orchestrate homologous recombination and replication fork stability. Mol. Cell 62, 903–917 10.1016/j.molcel.2016.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Petukhova G., Van Komen S., Vergano S., Klein H., and Sung P. (1999) Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J. Biol. Chem. 274, 29453–29462 10.1074/jbc.274.41.29453 [DOI] [PubMed] [Google Scholar]
- 78. Mazina O. M., and Mazin A. V. (2004) Human Rad54 protein stimulates DNA strand exchange activity of hRad51 protein in the presence of Ca2+. J. Biol. Chem. 279, 52042–52051 10.1074/jbc.M410244200 [DOI] [PubMed] [Google Scholar]
- 79. Solinger J. A., Kiianitsa K., and Heyer W. D. (2002) Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol. Cell 10, 1175–1188 10.1016/S1097-2765(02)00743-8 [DOI] [PubMed] [Google Scholar]
- 80. Van Komen S., Petukhova G., Sigurdsson S., Stratton S., and Sung P. (2000) Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol. Cell 6, 563–572 10.1016/S1097-2765(00)00055-1 [DOI] [PubMed] [Google Scholar]
- 81. Wright W. D., and Heyer W. D. (2014) Rad54 functions as a heteroduplex DNA pump modulated by its DNA substrates and Rad51 during D loop formation. Mol. Cell 53, 420–432 10.1016/j.molcel.2013.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Kouzine F., Gupta A., Baranello L., Wojtowicz D., Ben-Aissa K., Liu J., Przytycka T. M., and Levens D. (2013) Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat. Struct. Mol. Biol. 20, 396–403 10.1038/nsmb.2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jaskelioff M., Van Komen S., Krebs J. E., Sung P., and Peterson C. L. (2003) Rad54p is a chromatin remodeling enzyme required for heteroduplex DNA joint formation with chromatin. J. Biol. Chem. 278, 9212–9218 10.1074/jbc.M211545200 [DOI] [PubMed] [Google Scholar]
- 84. Alexiadis V., Lusser A., and Kadonaga J. T. (2004) A conserved N-terminal motif in Rad54 is important for chromatin remodeling and homologous strand pairing. J. Biol. Chem. 279, 27824–27829 10.1074/jbc.M402648200 [DOI] [PubMed] [Google Scholar]
- 85. Baranello L., Levens D., Gupta A., and Kouzine F. (2012) The importance of being supercoiled: how DNA mechanics regulate dynamic processes. Biochim. Biophys. Acta 1819, 632–638 10.1016/j.bbagrm.2011.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Fasching C. L., Cejka P., Kowalczykowski S. C., and Heyer W. D. (2015) Top3–Rmi1 dissolve Rad51-mediated D loops by a topoisomerase-based mechanism. Mol. Cell 57, 595–606 10.1016/j.molcel.2015.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Shor E., Gangloff S., Wagner M., Weinstein J., Price G., and Rothstein R. (2002) Mutations in homologous recombination genes rescue top3 slow growth in Saccharomyces cerevisiae. Genetics 162, 647–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Liu J., Ede C., Wright W. D., Gore S. K., Jenkins S. S., Freudenthal B. D., Todd Washington M., Veaute X., and Heyer W. D. (2017) Srs2 promotes synthesis-dependent strand annealing by disrupting DNA polymerase delta-extending D-loops. eLife 6, e22195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Neuwirth E. A., Honma M., and Grosovsky A. J. (2007) Interchromosomal crossover in human cells is associated with long gene conversion tracts. Mol. Cell. Biol. 27, 5261–5274 10.1128/MCB.01852-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Aguilera A., and Klein H. L. (1989) Yeast intrachromosomal recombination: long gene conversion tracts are preferentially associated with reciprocal exchange and require the RAD1 and RAD3 gene products. Genetics 123, 683–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Szostak J. W., Orr-Weaver T. L., Rothstein R. J., and Stahl F. W. (1983) The double-strand-break repair model for recombination. Cell 33, 25–35 10.1016/0092-8674(83)90331-8 [DOI] [PubMed] [Google Scholar]
- 92. Osman F., Dixon J., Doe C. L., and Whitby M. C. (2003) Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell 12, 761–774 10.1016/S1097-2765(03)00343-5 [DOI] [PubMed] [Google Scholar]
- 93. Prakash R., Satory D., Dray E., Papusha A., Scheller J., Kramer W., Krejci L., Klein H., Haber J. E., Sung P., and Ira G. (2009) Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 23, 67–79 10.1101/gad.1737809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. McVey M., Khodaverdian V. Y., Meyer D., Cerqueira P. G., and Heyer W.-D. (2016) Eukaryotic DNA polymerases in homologous recombination. Annu. Rev. Genet. 50, 393–421 10.1146/annurev-genet-120215-035243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Li X., Stith C. M., Burgers P. M., and Heyer W. D. (2009) PCNA is required for initiation of recombination-associated DNA synthesis by DNA polymerase δ. Mol. Cell 36, 704–713 10.1016/j.molcel.2009.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sneeden J. L., Grossi S. M., Tappin I., Hurwitz J., and Heyer W. D. (2013) Reconstitution of recombination-associated DNA synthesis with human proteins. Nucleic Acids Res. 41, 4913–4925 10.1093/nar/gkt192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Sebesta M., Burkovics P., Haracska L., and Krejci L. (2011) Reconstitution of DNA repair synthesis in vitro and the role of polymerase and helicase activities. DNA Repair 10, 567–576 10.1016/j.dnarep.2011.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wang X., Ira G., Tercero J. A., Holmes A. M., Diffley J. F., and Haber J. E. (2004) Role of DNA replication proteins in double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 24, 6891–6899 10.1128/MCB.24.16.6891-6899.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Ganai R. A., Zhang X. P., Heyer W. D., and Johansson E. (2016) Strand displacement synthesis by yeast DNA polymerase ϵ. Nucleic Acids Res. 44, 8229–8240 10.1093/nar/gkw556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Formosa T., and Alberts B. M. (1986) DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell 47, 793–806 10.1016/0092-8674(86)90522-2 [DOI] [PubMed] [Google Scholar]
- 101. Wilson M. A., Kwon Y., Xu Y., Chung W. H., Chi P., Niu H., Mayle R., Chen X., Malkova A., Sung P., and Ira G. (2013) Pif1 helicase and Pol δ promote recombination-coupled DNA synthesis via bubble migration. Nature 502, 393–396 10.1038/nature12585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Adams M. D., McVey M., and Sekelsky J. J. (2003) Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science 299, 265–267 10.1126/science.1077198 [DOI] [PubMed] [Google Scholar]
- 103. Natsume T., Nishimura K., Minocherhomji S., Bhowmick R., Hickson I. D., and Kanemaki M. T. (2017) Acute inactivation of the replicative helicase in human cells triggers MCM8–9-dependent DNA synthesis. Genes Dev. 31, 816–829 10.1101/gad.297663.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. McVey M., Adams M., Staeva-Vieira E., and Sekelsky J. J. (2004) Evidence for multiple cycles of strand invasion during repair of double-strand gaps in Drosophila. Genetics 167, 699–705 10.1534/genetics.103.025411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Smith C. E., Llorente B., and Symington L. S. (2007) Template switching during break-induced replication. Nature 447, 102–105 10.1038/nature05723 [DOI] [PubMed] [Google Scholar]
- 106. Bizard A. H., and Hickson I. D. (2014) The dissolution of double Holliday junctions. Cold Spring Harb. Perspect. Biol. 6, a016477 10.1101/cshperspect.a016477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Niu H. Y., and Klein H. L. (2017) Multifunctional roles of Saccharomyces cerevisiae Srs2 protein in replication, recombination and repair. FEMS Yeast Res. 17, 10.1093/femsyr/fow111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Lao J. P., Oh S. D., Shinohara M., Shinohara A., and Hunter N. (2008) Rad52 promotes postinvasion steps of meiotic double-strand-break repair. Mol. Cell 29, 517–524 10.1016/j.molcel.2007.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Sugiyama T., New J. H., and Kowalczykowski S. C. (1998) DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl. Acad. Sci. U.S.A. 95, 6049–6054 10.1073/pnas.95.11.6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Rothenberg E., Grimme J. M., Spies M., and Ha T. (2008) Human Rad52-mediated homology search and annealing occurs by continuous interactions between overlapping nucleoprotein complexes. Proc. Natl. Acad. Sci. U.S.A. 105, 20274–20279 10.1073/pnas.0810317106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Rijkers T., Van Den Ouweland J., Morolli B., Rolink A. G., Baarends W. M., Van Sloun P. P., Lohman P. H., and Pastink A. (1998) Targeted inactivation of mouse RAD52 reduces homologous recombination but not resistance to ionizing radiation. Mol. Cell. Biol. 18, 6423–6429 10.1128/MCB.18.11.6423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Zapotoczny G., and Sekelsky J. (2017) Human cell assays for synthesis-dependent strand annealing and crossing over during double-strand break repair. G3 7, 1191–1199 10.1534/g3.116.037390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Bzymek M., Thayer N. H., Oh S. D., Kleckner N., and Hunter N. (2010) Double Holliday junctions are intermediates of DNA break repair. Nature 464, 937–941 10.1038/nature08868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Wyatt H. D., and West S. C. (2014) Holliday junction resolvases. Cold Spring Harb. Perspect. Biol. 6, a023192 10.1101/cshperspect.a023192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Schwartz E. K., and Heyer W. D. (2011) Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma 120, 109–127 10.1007/s00412-010-0304-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Wyatt H. D., Laister R. C., Martin S. R., Arrowsmith C. H., and West S. C. (2017) The SMX DNA repair tri-nuclease. Mol. Cell 65, 848–860 10.1016/j.molcel.2017.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.