Abstract
A tigecycline-susceptible (TGC-S) Sequence Type (ST) 5 clinical methicillin-resistant Staphylococcus aureus (MRSA) strain was cultured in escalating levels of tigecycline, yielding mutants eightfold more resistant. Their genomes were sequenced to identify genetic alterations, resulting in resistance. Alterations in rpsJ, commonly related to tigecycline resistance, were also investigated. Tigecycline resistance was mediated by loss-of-function mutations in the transcriptional repressor mepR, resulting in derepression of the efflux pump mepA. Increased levels of resistance were obtained by successive mutations in mepA itself. No alterations in RpsJ were observed in selected strains, but we observed a K57M substitution, previously correlated with resistance, among TGC-S clinical strains. Thus, the pathway to tigecycline resistance in CC5 MRSA in vitro appears to be derepression of mep operon as the result of mepR loss-of-function mutation, followed by alterations in MepA efflux pump. This shows that other evolutionary pathways, besides mutation of rpsJ, are available for evolving tigecycline resistance in CC5 MRSA.
Keywords: : tigecycline, MRSA, MepR, MepA
Introduction
Methicillin-resistant Staphylococcusaureus (MRSA) are a major public health threat worldwide as they are able to quickly develop antimicrobial resistance. Recently, the World Health Organization published a note of the bacteria for which new antibiotics are needed, and MRSA are among those of high priority.1 The antimicrobial agent tigecycline belongs to the tetracycline-derived class of glycylcyclines, and was designed to overcome known tetracycline-resistance mechanisms, such as ribosomal protection and drug efflux.2 Tigecycline is commonly used for the treatment of skin and intra-abdominal infections caused by both Gram-positive and Gram-negative bacteria.3 Since its introduction, tigecycline-resistant (TGC-R) S. aureus and related strains have arisen and genes so far associated with that resistance have been reported. McAleese et al.4 found that mutations in the repressor MepR of S. aureus led to overexpression of the efflux pump MepA and to an increase in the minimal inhibitory concentration (MIC) for tigecycline. Tigecycline resistance involving mutations in rpsJ that encodes ribosomal protein S10 has also been reported in S. aureus as well as in several other organisms.5–8 In Gram-positive Enterococcus faecium, the efflux pump protein TetL and the ribosomal protection protein TetM have been shown to contribute to tigecycline resistance.9 The tetX gene and its ortholog tetX1, encoding monooxygenases, have also been associated with tigecycline resistance in Enterococcus faecalis, and several Gram-negative microbes.10–12
Further, in selecting a hospital-derived CC5 S. aureus strain for resistance to tigecycline in the laboratory, we obtained mainly variants that lacked the previously described mutations known to confer tigecycline resistance. It was, therefore, of interest to characterize this new evolutionary trajectory, which appears available to the most common hospital clade of MRSA.
Materials and Methods
Bacterial strains and reagents
MRSA strain SA43 was isolated from an infection-derived specimen submitted to the diagnostic laboratory at the Risoleta Tolentino Neves Hospital (Belo Horizonte, Brazil), August 2009. It was reported to be susceptible to tigecycline, with an MIC = 0.125 mg/L. It was subsequently found to belong to Multilocus Sequence Type (ST) 5, and it was used in this study to select for the development of tigecycline resistance. For comparison, five additional ST105 (Single Locus Variant of ST5) MRSA clinical isolates (SA33, SA36, SA96, SA105, and SA107) from the same hospital and from the same pulsotype were used.
The media employed in this study were from BD (Franklin Lakes, NJ) or Neogen (Lansing, MI), and chemicals were purchased from Sigma-Aldrich (Saint Louis, MO), unless otherwise stated.
Antibiotic susceptibility testing and efflux pump inhibitor assay
Tigecycline MICs were determined by following the Clinical and Laboratory Standards Institute recommendations.13 The S. aureus breakpoint for tigecycline was assigned according to the European Committee on Antimicrobial Susceptibility Testing: MIC ≤0.5 mg/L for susceptibility,14 using 80% endpoint reading criteria. Tigecycline MICs were determined in the presence or absence of the efflux pump inhibitor verapamil (VER), with a decrease equal to or higher than fourfold the MIC in the presence of the inhibitor corresponding to tigecycline efflux.15 The concentration of verapamil tested was 200 mg/L.
In vitro selection of TGC-R strains
Three isolated colonies of strain SA43 were used in separate parallel experiments to select for tigecycline-resistant variants. An overnight culture of each colony, grown in cation-adjusted Mueller Hinton broth (MHB), was adjusted to an OD600 = 0.1, and 30 μl were inoculated into tubes containing 3 ml of MHB with graded concentrations of tigecycline: (1) 1/2 MIC; (2) 1× MIC; (3) 2× MIC; and (4) 4× MIC. All tubes were incubated at 37°C overnight without shaking and protected from light. The next day, the tube with visible growth at the highest tigecycline concentration was used as inoculum for the next series of tubes with increasing drug concentrations. This procedure was repeated for 15 days.
The strains recovered from each day of selection had their MIC and pulsotype determined, as described elsewhere.13,14,16 The samples were passaged for 3 days in MHB without tigecycline and had their MIC determined again to check the stability of the phenotype.17 Twelve strains from the three parallel in vitro TGC-R selection experiments were selected for further study: A2, A5, A7, A10, B2, B6, B7, B10, C2, C4, C6, and C7 (A, B, and C denote the three in vitro selection experiments, and the number is the corresponding experimental day).
Genome sequencing
Total DNA was purified from the parental strain SA43, and 12 strains selected in vitro, by using the DNeasy Blood & Tissue Kit (QIAGEN, Valencia, CA). DNA libraries were prepared from 1 ng by using the Nextera XT DNA Sample Preparation Kit (Illumina, San Diego, CA), with recommended modifications for 2 × 250 bp paired-end sequencing. Samples were multiplexed and sequenced on a MiSeq Sequencing System (Illumina) at the Ocular Genomics Institute at the Massachusetts Eye and Ear Infirmary. CLC Genomics Workbench v.8.5 (QIAGEN) was used for genome assembly and variant detection. Contigs were annotated through the NCBI Prokaryotic Genome Annotation Pipeline.
All genomes have been deposited at DDBJ/EMBL/GenBank under the accessions: JSBG00000000 (SA33), JSBH00000000 (SA36), JSBI00000000 (SA43), JSBL00000000 (SA96), JSBK00000000 (SA105), JSBJ00000000 (SA107), LELL00000000 (A2), LELM00000000 (A5), LELN00000000 (A7), LELO00000000 (A10), LELP00000000 (B2), LELQ00000000 (B6), LELR00000000 (B7), LELS00000000 (B10), LELT00000000 (C2), LELU00000000 (C4), LELV00000000 (C6), and LELW00000000 (C7). The versions described in this article are versions JSBG01000000, JSBH01000000, JSBI01000000, JSBL01000000, JSBK01000000, JSBJ01000000, LELL01000000, LELM01000000, LELN01000000, LELO01000000, LELP01000000, LELQ01000000, LELR01000000, LELS01000000, LELT01000000, LELU01000000, LELV01000000, and LELW01000000.
Variants were identified by mapping sequencing reads for each sample to the annotated reference genome from the starting culture of each selection. Variants at positions with a minimum coverage of 25, and occurring at frequencies higher than 95% were included.
Determination of the doubling time
Cultures were grown while shaking overnight in MHB at 37°C, and then each inoculum was adjusted to an OD600 = 0.05–0.1 by using a SpectraMax M5 Microplate Reader (Molecular Devices, Sunnyvale, CA) in a clear flat-bottom 96-well microplate filled with 200 μl of each culture.
Two hundred microliters of each adjusted inoculum were distributed in triplicate into the wells of a second sterile clear flat-bottom 96-well microplate, which was incubated for 20 hours at 37°C. The OD600 was determined every 5 minutes with 5 seconds of shaking before each read.18 Data were plotted for each sample with at least 20 points from the log phase of growth, and the doubling times were calculated by using a linear regression. One-way analysis of variance (ANOVA) test was applied to determine whether the differences in doubling times among the samples were significant. The significance level was set at p < 0.05.
Relative gene expression
All strains were cultivated overnight in Brain Heart Infusion (BHI) broth, at 37°C and shaking at 130 rpm. The next day, all cultures were diluted by inoculating 1 ml of the overnight pre-inoculum into 25 ml of fresh BHI, and cultures were grown until OD600 = 0.6–1.0. Cells were collected by centrifugation for 2 minutes at 14,000 × g at 4°C, supernatant was discarded, and cells were subjected to lysis by 10 mg/ml of lysostaphin.
RNA was extracted by using an SV Total RNA Isolation System (Promega Corporation, Madison, WI) according to the manufacturer's recommendations. The concentration and purity of total RNA was determined by using a NanoDrop 2000c Spectrophotometer (Thermo Fisher Scientific, Waltham, MA). For reverse transcription, a SuperScript™ III First-Strand kit (Invitrogen, Carlsbad, CA) was used according to the manufacturer's recommendations, using 50 ng of random hexamers and 1 μg of total RNA, generating 20 μl of complementary DNA (cDNA; 50 ng/μl).
For quantitative polymerase chain reaction (qPCR), the following housekeeping genes were selected as controls for normalizing the expression of mepA (Gene ID X998_0391): gmk (Gene ID BAB42304), tpi (Gene ID BAB41962), and pta (Gene ID BAB41777). Primers used for qPCR amplification were: mepA (F: TTATGGAAACTTCGCGATTGC, R: AACACCTTCACATAATCCCATGATAAT); gmk (F: ATCGTTTTATCGGGACCATC, R: CATTTGACGTGTTGTCATTG); tpi (F: TCGTTCATTCTGAACGTCGTGAA, R: CGTCTGTTTCACCAACACAAAT); and pta (F: GTTAAAATCGTATTACCTGAAGG, R: CCTAACACGATTGGTGTAACAT).
A cDNA pool generated from all the samples was used for optimization of annealing temperatures. This pool was also employed to create a standard curve for determination of primer efficiency, which was used to correct expression values.19 The same pool was used to evaluate the stability of the reference genes (gmk, tpi, and pta) with RefFinder.20 All three genes were considered stable for normalization of relative expression.
For qPCR amplification, the fluorescent DNA-binding dye PowerUP™ SYBR® Green PCR Master Mix (Thermo Fisher Scientific) was used, according to manufacturer's recommendations, with 5 ng cDNA per reaction. A CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA) was used for amplification, and cycling conditions included: one hold at 50°C for 2 minutes, followed by one hold at 95°C for 2 minutes, then 40 cycles of 15 seconds at 95°C, and 1 minute at optimal annealing temperature (varying from 56°C to 60°C, depending on the primers). A melting curve was performed after each run (raising 0.5°C per second, from 65°C to 95°C), to verify amplicon specificity in each reaction. The qPCR reaction was carried out in technical triplicates, and it was repeated in two independent experiments. Relative expression was calculated by using the ratios of the three reference genes, using the comparative ΔΔCT method with PCR efficiency correction.21
Comparison of rpsJ, mepR, and mepA gene sequences from tigecycline-susceptible and TGC-R MRSA strains
To determine whether single-nucleotide polymorphisms (SNPs) found in the genomes of in vitro generated variants also occurred in clinical isolates, we examined the gene sequences of five tigecycline-susceptible hospital strains: SA33, SA36, SA96, SA105, and SA107. This was done for mepR and mepA genes, as well as the rpsJ gene, which codes for the ribosomal protein S10. This was accomplished by using the alignment tool of CLC Genomics Workbench v.8.5 (QIAGEN).
Results
To identify pathways available to MRSA of the common hospital lineage CC5, in evolving to tigecycline resistance, tigecycline-susceptible MRSA strain SA43 was exposed to graded levels of the drug, starting at 0.125 mg/L. From these experiments, nine variants (three from each experiment) were selected for genome sequencing, to identify changes that correlate with acquisition of tigecycline resistance. Genome sequence and assembly statistics are provided in Supplementary Table S1 (Supplementary Data are available online at www.liebertpub.com/mdr). To control for possible confounding spontaneous irrelevant SNPs occurring in the routine course of cultivation, an isolated colony from day 2 passage of each experiment was sequenced (Table 1). All the in vitro selected strains remained belonging to the same pulsotype (data not shown).
Table 1.
Strains from the In Vitro Tigecycline-Resistant Selection Experiment Selected for This Study and Their Tigecycline Minimal Inhibitory Concentrations
| Experiment A | Experiment B | Experiment C | ||||||
|---|---|---|---|---|---|---|---|---|
| Sample | Day | Tigecycline MIC (mg/L) | Sample | Day | Tigecycline MIC (mg/L) | Sample | Day | Tigecycline MIC (mg/L) |
| A2 | 2 | 0.125 | B2 | 2 | 0.125 | C2 | 2 | 0.125 |
| A5 | 5 | 0.25 | B6 | 6 | 0.25 | C4 | 4 | 0.25 |
| A7 | 7 | 0.5 | B7 | 7 | 0.25 | C6 | 6 | 0.25 |
| A10 | 10 | 1.0 | B10 | 10 | 1.0 | C7 | 7 | 1.0 |
MIC, minimal inhibitory concentration.
Efflux of the drug is the mechanism of tigecycline resistance in “in vitro” selected MRSA strains
All three independent experiments followed the same evolutionary trajectory in achieving tigecycline resistance, although the specific bases mutated in each gene varied (Table 2). Within 4–6 days of passage, all three experiments yielded strains with mutations in mepR (A5, B6, C4). The mutation in A5 generated a frameshift, and in B6 introduced a nonsense codon, resulting in premature translation termination of the MepR repressor in each case. C4 possessed a missense mutation that resulted in replacement of a glycine hydrogen with a large, strongly positively charged arginine side chain. Substitutions of this specific amino acid (Gly-97) previously have been shown to result in MepR loss of function.22 Introduction of a positive charge at position 97 in the known structure of MepR23 would be predicted to alter the charge environment affecting allosteric interactions, resulting in reduced ability of MepR to repress mepA.22,24
Table 2.
Mutations in the Genes mepR and mepA in the In Vitro Selected Tigecycline-Resistant Strains from This Study
| Gene | Strain | Mutationa | Result |
|---|---|---|---|
| mepR | A5/A7/A10 | AB877_02550: 58delG | Premature stop codon |
| B6/B7/B10 | AB881_09200: 76C>T | Premature stop codon | |
| C4/C6/C7 | AB885_00385: 289G>A | G97R | |
| mepAb | A7 | AB877_02555: 509T>A | I170N |
| A10 | AB877_02555: 536A>G | N179S | |
| B10 | AB881_09205: 1288G>A | A430T | |
| C6 | AB885_00380: 149A>T | H50L | |
| C7 | AB885_00380: 1244T>C | V415A |
The locus refers to the strain used for comparison: A2, in the case of Experiment A strains; B2, in the case of Experiment B strains; and C2, in the case of Experiment C strains.
All retained the respective prior mutation in mepR.
All subsequent mutations in the TGC-R strains occurred in the efflux pump-encoding gene, mepA, resulting in a further increase in tigecycline MIC. These mutations occurred in the transmembrane and exterior domains only.25 No mutations occurred in the cytoplasmic portion of the pump. None of the mutations we detected were identical to those found by Schindler et al. to increase or decrease efflux activity,25 although some are in close proximity.
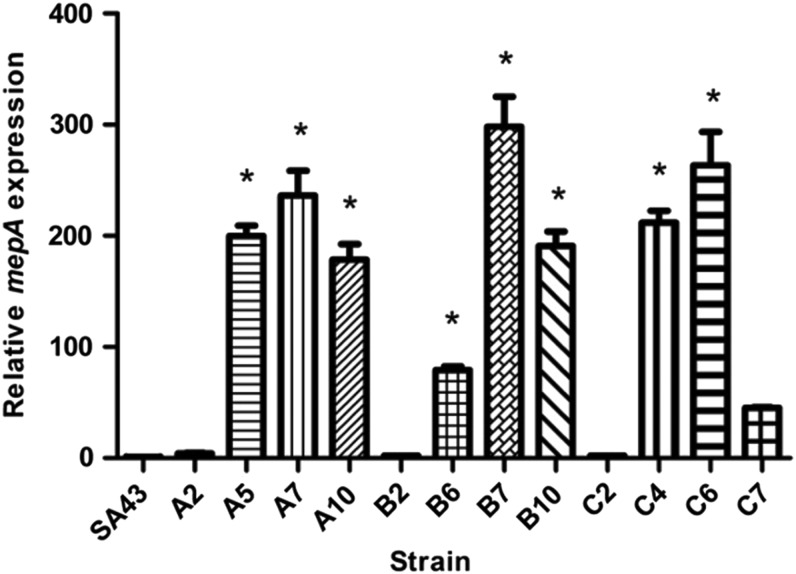
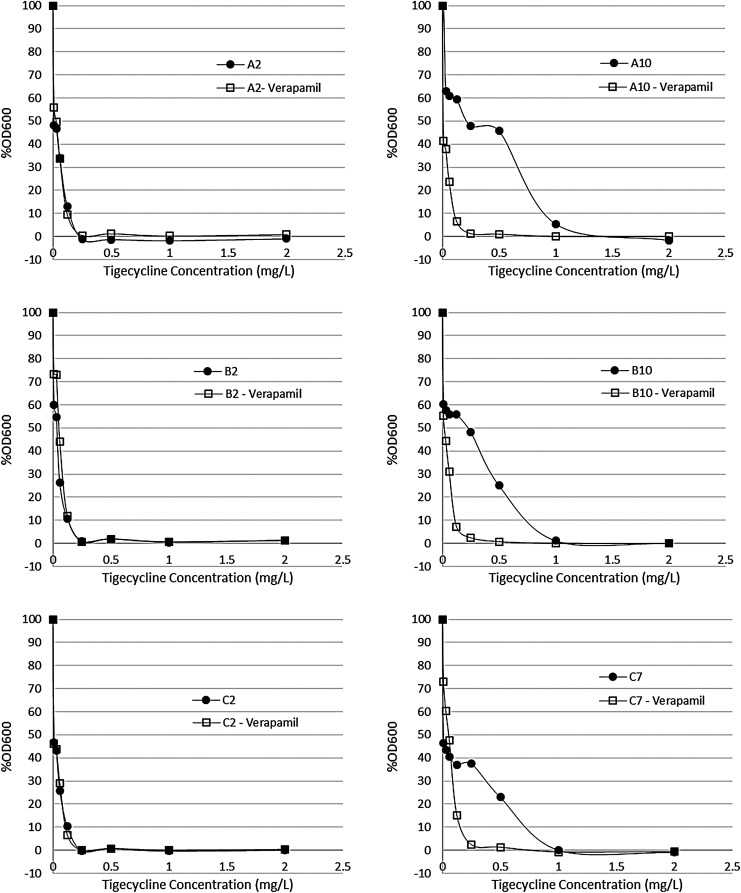
To test whether the mutations in mepR resulted in loss of function, levels of expression of mepA occurring in each mutant lineage were compared with parental S. aureus SA43 (Fig. 1 and Supplementary Table S2). Premature termination resulted in loss of mepR function as predicted, as did introduction of a strong positive charge in the fifth α helix, near the C-terminal domain,23 which occurred in strains C4, C6, and C7. Constitutive expression of mepA resulted in an average 200-fold increase in mepA messenger RNA (mRNA) abundance, and this correlated in each case with increases in tigecycline resistance. Since MepA is known to be affected by efflux pump inhibitors,4,26,27 to prove that increased MepA expression accounted for the resistance we observed (as opposed to another idiosyncratic SNP in one or the other mutant), the ability of verapamil to reverse tigecycline resistance28 was examined. Although not affecting the wild type, where little expression of mepA was observed, verapamil resulted in up to 8-fold reduction in tigecycline MIC in the de-repressed mutants (Fig. 2).
FIG. 1.
Relative expression of mepA for in vitro selected strains. Expression of mepA is plotted relative to the reference genes gmk, tpi, and pta. Relative mepA expression was normalized to SA43 (p > 0.05, as determined by ANOVA), and statistically significant differences are marked with *. Levels of transcript, standard deviation, and statistical significance are listed in Supplementary Table S2. ANOVA, analysis of variance.
FIG. 2.
Effect of the presence of the efflux pump inhibitor verapamil on the tigecycline MIC of in vitro selected TGC-R MRSA strains. Each graph shows bacterial growth after 24 hours in increasing concentrations of tigecycline, plotted as a percentage of the growth in no drug. Filled circles are dose–response curves for tigecycline only, whereas open squares show curves for tigecycline+verapamil. MIC, minimal inhibitory concentration; MRSA, methicillin-resistant Staphylococcus aureus; TGC-R, tigecycline-resistant.
To check whether the detected mepR and mepA mutations were exclusive to our TGC-R in vitro generated MRSA strains, we decide to look for these genes in the genomes of some clinical tigecycline-susceptible (TGC-S) MRSA strains we had available (SA33, SA36, SA96, SA105, and SA107), as well as in all S. aureus genomes available in NCBI. None of the mutations were observed, reinforcing the importance of such mutations to tigecycline resistance in MRSA.
It is important to note that other mutations did occur during the in vitro selection experiment (data not shown), but only mutations in genes mepR and mepA were found common to the triplicate, reinforcing their role in the development of resistance to tigecycline.
Amino acid substitution at position 57 of RpsJ is not the only determinant of tigecycline resistance in MRSA
Since alterations in ribosomal protein S10 had been related to tigecycline resistance in many organisms, we examined rpsJ genes from all our in vitro generated strains (A2, A5, A7, A10, B2, B6, B7, B10, C2, C4, C6, and C7), the parental MRSA strain (SA43), clinical TGC-S MRSA strains we had sequenced (SA33, SA36, SA96, SA105, and SA107), and from the strain MRSA131.5 In strains SA33, SA36, SA96, SA105, and SA107, we observed a lysine-to-methionine substitution at amino acid 57 (Supplementary Fig. S1), but this did not correlate with a detectably increased MIC for tigecycline. Evidently, amino acid substitution in position 57 of RpsJ protein does not always result in a TGC-R phenotype.
Resistance to tigecycline does not present a fitness cost for MRSA
To assess whether the development of tigecycline resistance strains was accompanied by an associated fitness cost, we measured doubling times for all strains derived in this study (Supplementary Fig. S2), since changes in the growth rate are usually observed in cells that have become resistant. When compared with the parent, no significant differences in the doubling times (p > 0.05, as determined by ANOVA) were noted.
Discussion
In this study, we observed that tigecycline resistance in a clinical MRSA strain subjected to in vitro selection was caused by increased efflux of the antimicrobial due to mutations in the transcriptional regulator MepR and in the efflux pump MepA.
Increased efflux of antimicrobials from bacterial cells appears to be the most common mechanism of resistance to tigecycline for several species.4,29–32 He et al. showed that tigecycline MICs for Klebsiella pneumoniae isolates decreased 4- to 16-fold in the presence of the efflux pump inhibitor 1-(1-naphthylmethyl)-piperazine,33 and a similar effect was observed with carbonyl cyanide m-chlorophenyl hydrazone (CCCP) by Zhong et al.34 In the case of Acinetobacter baumannii, Peleg et al. observed a fourfold decrease in the tigecycline MIC in the presence of another efflux pump inhibitor, phenyl-arginine-β-naphthylamide.35
Both mepR and mepA occur in the operon mepRAB.26 mepR encodes a transcriptional regulator of the multiple antibiotic resistance regulator (MarR) family that functions as an autorepressor, and a repressor of expression of efflux pump encoding mepA.36 MepA has been previously reported as being overexpressed in TGC-R S. aureus.4 The third gene on the mepRAB operon, mepB, is a gene of unknown function.4
The mutations we observed in the efflux pump MepA and its repressor MepR in the TGC-R MRSA strains selected in vitro lead us to hypothesize that tigecycline resistance is due to increased efflux of the drug. As would be predicted for this mechanism, the efflux pump inhibitor, verapamil, reversed most of the resistance phenotype (Fig. 2).
As predicted for loss-of-function mutations in mepR, mepA expression was increased in all strains that contained mutations in mepR (Fig. 1). Derepression, as occurred in strains A5, B6, and C4, was the first mutational event in the trajectory toward tigecycline resistance. An interesting fact that remains to be elucidated is the diminished expression level of mepA in strains A10, B10, and C7 in relation to A7, B7, and C6, besides the general trend to increase. The crystal structure of MepR was solved by Kumaraswami et al. in 2009,23 and since that time much has been learned about this repressor. Birukou et al. solved the crystal structure of MepR variants from multidrug-resistant S. aureus strains, and verified that none of the observed mutations were located on the DNA-binding domain, suggesting that allosteric mechanisms affected MepR function.37 Another study from the same year revealed markedly reduced repressor activity in the presence of Q18P, F27L, G97E, and A103V substitutions, all of which led to mepA overexpression.24 The structure of the mepR operator was also solved, which led to the observation that mutating Asp-85 and Arg-87, both conserved throughout the MarR family, markedly affect MepR repressor activity.38,39 Conversely, as observed in our study, mutations in mepR affect its ability to regulate expression of mepA. The fact that strains with mutations only in mepR (A5, B6, B7, and C4) showed overexpression of mepA supports the role of MepR in the mechanism of tigecycline resistance in our in vitro TGC-R MRSA strains.
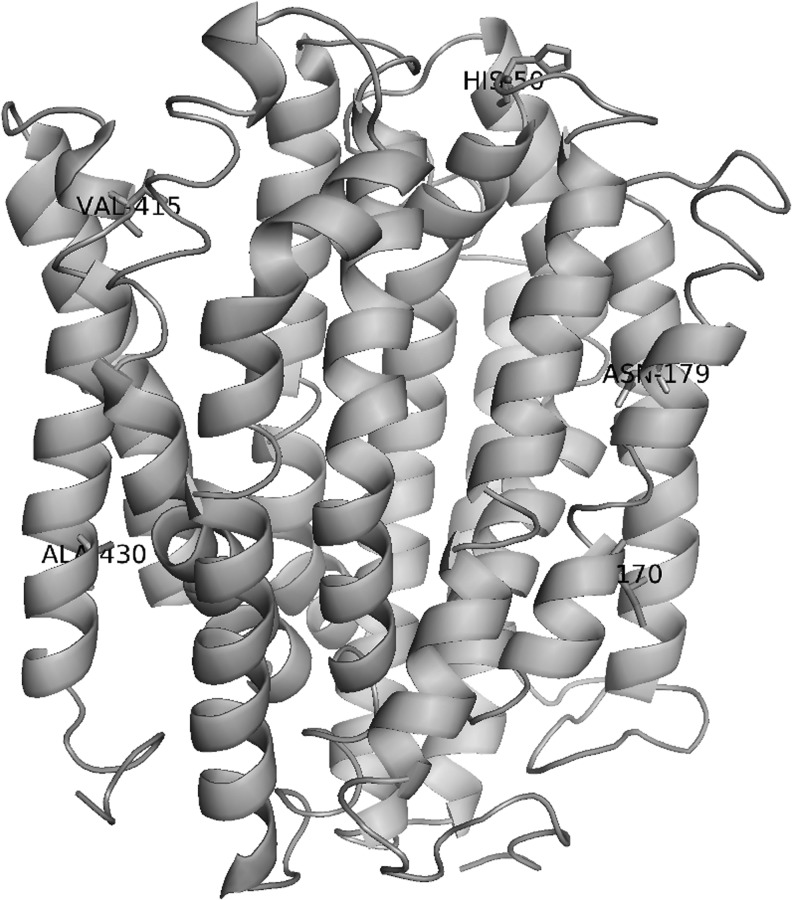
None of the mutations in MepA that we identified have been previously described to affect efflux activity,25 or drug resistance. The mutations we identified in MepA all reside either in the transmembrane portion (A7, A10, and B10) or in the exterior portion (C6 and C7) of the protein, with no mutations occurring in the cytoplasmic region of the pump (Fig. 3). The transmembrane portion of MepA would be predicted to form the interior of the efflux pump channel, and the changes identified in mutants A7 (I170N) and A10 (N179S) introduce polar residues that would be predicted to alter the hydrogen-bonding environment of the channel. Mutant B10 also possesses a mutation in the pump channel with the introduction of a polar residue (A430T), but in the C-terminal extremity of MepA. Finally, the mutations occurring in the exterior region of MepA (C6: H50L and C7: V415A) do not seem to significantly affect the charge of the protein. Therefore, we speculate that the mutations we observe in MepA confer increased tigecycline resistance to these strains via enhanced efflux activity.
FIG. 3.
Structural model of MepA. Sites of amino acid substitutions are highlighted: His-50 (Leu in C6 strain), Ile-170 (Asn in A7 strain), Asn-179 (Ser in A10 strain), Val-415 (Ala in C7 strain), and Ala-430 (Thr in B10 strain).
Antibiotic resistance is not always accompanied by a fitness cost, which depends on a variety of factors, including the mechanism of resistance itself.40 Others have found that the development of tigecycline resistance in Escherichia coli and K. pneumoniae was accompanied by little or no fitness cost.6,7 This is consistent with our finding that the TGC-R MRSA strains derived in vitro here exhibited no measurable change in doubling time compared with sensitive progenitors. It is important to have in mind that growth rate is only one way to measure the fitness cost, which involves many other manifestations.
Analysis of the rpsJ gene sequence from several clinical and in vitro selected MRSA strains showed that some had a point mutation encoding a Lys57Met change in the S10 protein (Supplementary Fig. S1). This is one of the mutations that Beabout et al. observed when they evolved S. aureus strain MRSA131, from an initial TGC MIC of 0.5 mg/L, to 11.2 mg/L TGC.5 They observed RpsJ amino acid 57 to be a common site of change in Gram-positive and Gram-negative pathogens after TGC exposure, and they suggest that the S10 protein is a general target for reduced TGC susceptibility. Importantly, this study did not investigate other possible resistance mechanisms, such as drug efflux. We did not find rpsJ mutated in the TGC-resistant variants selected here. Further, clinical strains SA33, SA36, SA96, SA105, and SA107 were found to have a K57M substitution, but they are not resistant to TGC (MIC = 0.25 mg/L).
Thus, ribosomal protein S10 may be involved with reduced susceptibility to TGC in some organisms or lineages, but this does not appear to be generalizable.
Overall, we have shown that in vitro, the most common trajectory to tigecycline resistance for an MRSA strain of the common hospital CC5 lineage involves an initial derepression of the efflux pump MepA, caused by changes in the repressor MepR, followed by alterations in the pump itself, leading to an increased level of non-susceptibility. Precisely what dictates this trajectory, and whether it is determined by genetic background or environment in which selection occurs, are points of considerable interest as the spread of antibiotic resistance forces increased reliance on tigecycline and other recently introduced drugs.
Supplementary Material
Acknowledgments
The authors wish to thank José T. Saavedra for assistance in preparing the Illumina sequencing libraries. This work was supported by grants from the Sao Paulo Research Foundation (grant Nos. FAPESP 2010/02619-0 to I.L.B.C.C., and FAPESP 2013/24952-0 to A.N.G.D.), Coordination for the Improvement of Higher Education Personnel (grant Nos. CAPES/PVE 88881.030524/2013-01 to I.L.B.C.C., and CAPES 88887.114824/2015-00 to J.S.A.C.), and the National Institutes of Health (DHHS/NIH/NIAID grant numbers AI072360 and AI083214 to M.S.G., and grant number AI109855 to D.V.T.).
Disclosure Statement
No competing financial interests exist.
References
- 1.Willyard C. 2017. The drug-resistant bacteria that pose the greatest health threats. Nature 543:15. [DOI] [PubMed] [Google Scholar]
- 2.Petersen P.J., Jacobus N.V., Weiss W.J., Sum P.E., and Testa R.T. 1999. In vitro and in vivo antibacterial activities of a novel glycylcycline, the 9-t-butylglycylamido derivative of minocycline (GAR-936). Antimicrob. Agents Chemother. 43:738–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shen F., Han Q., Xie D., Fang M., Zeng H., and Deng Y. 2015. Efficacy and safety of tigecycline for the treatment of severe infectious diseases: an updated meta-analysis of RCTs. Int. J. Infect. Dis. 39:25–33 [DOI] [PubMed] [Google Scholar]
- 4.McAleese F., Petersen P., Ruzin A., Dunman P.M., Murphy E., Projan S.J., and Bradford P.A. 2005. A novel MATE family efflux pump contributes to the reduced susceptibility of laboratory-derived Staphylococcus aureus mutants to tigecycline. Antimicrob. Agents Chemother. 49:1865–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beabout K., Hammerstrom T.G., Perez A.M., Magalhaes B.F., Prater A.G., Clements T.P., Arias C.A., Saxer G., and Shamoo Y. 2015. The ribosomal S10 protein is a general target for decreased tigecycline susceptibility. Antimicrob. Agents Chemother. 59:5561–5566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang L., Chen Q., Shi K., Li X., Shi Q., He F., Zhou J., Yu Y., and Hua X. 2016. Step-wise increase in tigecycline resistance in Klebsiella pneumoniae associated with mutations in ramR, lon and rpsJ. PLoS One 11:e0165019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X., Mu X., Yang Y., Hua X., Yang Q., Wang N., Du X., Ruan Z., Shen X., and Yu Y. 2016. Rapid emergence of high-level tigecycline resistance in Escherichia coli strains harbouring blaNDM-5 in vivo. Int. J. Antimicrob. Agents 47:324–327 [DOI] [PubMed] [Google Scholar]
- 8.Cattoir V., Isnard C., Cosquer T., Odhiambo A., Bucquet F., Guerin F., and Giard J.C. 2015. Genomic analysis of reduced susceptibility to tigecycline in Enterococcus faecium. Antimicrob. Agents Chemother. 59:239–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiedler S., Bender J.K., Klare I., Halbedel S., Grohmann E., Szewzyk U., and Werner G. 2016. Tigecycline resistance in clinical isolates of Enterococcus faecium is mediated by an upregulation of plasmid-encoded tetracycline determinants tet(L) and tet(M). J. Antimicrob. Chemother. 71:871–881 [DOI] [PubMed] [Google Scholar]
- 10.Bartha N.A., Soki J., Urban E., and Nagy E. 2011. Investigation of the prevalence of tetQ, tetX and tetX1 genes in Bacteroides strains with elevated tigecycline minimum inhibitory concentrations. Int. J. Antimicrob. Agents 38:522–525 [DOI] [PubMed] [Google Scholar]
- 11.Costello S.E., Gales A.C., Morfin-Otero R., Jones R.N., and Castanheira M. 2016. Mechanisms of resistance, clonal expansion, and increasing prevalence of Acinetobacter baumannii strains displaying elevated tigecycline mic values in Latin America. Microb. Drug Resist. 22:253–258 [DOI] [PubMed] [Google Scholar]
- 12.Werner G., Gfrorer S., Fleige C., Witte W., and Klare I. 2008. Tigecycline-resistant Enterococcus faecalis strain isolated from a German intensive care unit patient. J. Antimicrob. Chemother. 61:1182–1183 [DOI] [PubMed] [Google Scholar]
- 13.Clinical and Laboratory Standards Institute. 2017. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Seventh Informational Supplement M100-S27. CLSI, Wayne,PA [Google Scholar]
- 14.European Committee on Antimicrobial Susceptibility Testing. 2017. Breakpoint tables for interpretation of MICs and zone diameters, on breakpoint tables for interpretation of MICs and zone diameters. Available at www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_7.1_Breakpoint_Tables.pdf (accessed May8, 2017)
- 15.Costa S.S., Falcao C., Viveiros M., Machado D., Martins M., Melo-Cristino J., Amaral L., and Couto I. 2011. Exploring the contribution of efflux on the resistance to fluoroquinolones in clinical isolates of Staphylococcus aureus. BMC Microbiol. 11:241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tenover F.C., Arbeit R.D., Goering R.V., Mickelsen P.A., Murray B.E., Persing D.H., and Swaminathan B. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mascio C.Chesnel T.M., L., Thorne G., and Silverman J.A. 2014. Surotomycin demonstrates low in vitro frequency of resistance and rapid bactericidal activity in Clostridium difficile, Enterococcus faecalis, and Enterococcus faecium. Antimicrob. Agents Chemother. 58:3976–3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall B.G., Acar H., Nandipati A., and Barlow M. 2014. Growth rates made easy. Mol. Biol. Evol. 31:232–238 [DOI] [PubMed] [Google Scholar]
- 19.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., and Wittwer C.T. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55:611–622 [DOI] [PubMed] [Google Scholar]
- 20.Xie F., Xiao P., Chen D., Xu L., and Zhang B. 2012. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 80:75–84 [DOI] [PubMed] [Google Scholar]
- 21.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., and Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:Research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huet A.A., Raygada J.L., Mendiratta K., Seo S.M., and Kaatz G.W. 2008. Multidrug efflux pump overexpression in Staphylococcus aureus after single and multiple in vitro exposures to biocides and dyes. Microbiology 154:3144–3153 [DOI] [PubMed] [Google Scholar]
- 23.Kumaraswami M., Schuman J.T., Seo S.M., Kaatz G.W., and Brennan R.G. 2009. Structural and biochemical characterization of MepR, a multidrug binding transcription regulator of the Staphylococcus aureus multidrug efflux pump MepA. Nucleic Acids Res. 37:1211–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schindler B.D., Seo S.M., Jacinto P.L., Kumaraswami M., Birukou I., Brennan R.G., and Kaatz G.W. 2013. Functional consequences of substitution mutations in MepR, a repressor of the Staphylococcus aureus mepA multidrug efflux pump gene. J. Bacteriol. 195:3651–3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schindler B.D., Patel D., Seo S.M., and Kaatz G.W. 2013. Mutagenesis and modeling to predict structural and functional characteristics of the Staphylococcus aureus MepA multidrug efflux pump. J. Bacteriol. 195:523–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaatz G.W., McAleese F., and Seo S.M. 2005. Multidrug resistance in Staphylococcus aureus due to overexpression of a novel multidrug and toxin extrusion (MATE) transport protein. Antimicrob. Agents Chemother. 49:1857–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schindler B.D., Jacinto P., and Kaatz G.W. 2013. Inhibition of drug efflux pumps in Staphylococcus aureus: current status of potentiating existing antibiotics. Future Microbiol. 8:491–507 [DOI] [PubMed] [Google Scholar]
- 28.DeMarco C.E., Cushing L.A., Frempong-Manso E., Seo S.M., Jaravaza T.A., and Kaatz G.W. 2007. Efflux-related resistance to norfloxacin, dyes, and biocides in bloodstream isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 51:3235–3239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dean C.R., Visalli M.A., Projan S.J., Sum P.E., and Bradford P.A. 2003. Efflux-mediated resistance to tigecycline (GAR-936) in Pseudomonas aeruginosa PAO1. Antimicrob. Agents Chemother. 47:972–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirata T., Saito A., Nishino K., Tamura N., and Yamaguchi A. 2004. Effects of efflux transporter genes on susceptibility of Escherichia coli to tigecycline (GAR-936). Antimicrob. Agents Chemother. 48:2179–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hornsey M., Ellington M.J., Doumith M., Hudson S., Livermore D.M., and Woodford N. 2010. Tigecycline resistance in Serratia marcescens associated with up-regulation of the SdeXY-HasF efflux system also active against ciprofloxacin and cefpirome. J. Antimicrob. Chemother. 65:479–482 [DOI] [PubMed] [Google Scholar]
- 32.Hentschke M., Christner M., Sobottka I., Aepfelbacher M., and Rohde H. 2010. Combined ramR mutation and presence of a Tn1721-associated tet(A) variant in a clinical isolate of Salmonella enterica serovar Hadar resistant to tigecycline. Antimicrob. Agents Chemother. 54:1319–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He F., Fu Y., Chen Q., Ruan Z., Hua X., Zhou H., and Yu Y. 2015. Tigecycline susceptibility and the role of efflux pumps in tigecycline resistance in KPC-producing Klebsiella pneumoniae. PLoS One 10:e0119064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong X., Xu H., Chen D., Zhou H., Hu X., and Cheng G. 2014. First emergence of acrAB and oqxAB mediated tigecycline resistance in clinical isolates of Klebsiella pneumoniae pre-dating the use of tigecycline in a Chinese hospital. PLoS One 9:e115185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peleg A.Y., Adams J., and Paterson D.L. 2007. Tigecycline efflux as a mechanism for nonsusceptibility in Acinetobacter baumannii. Antimicrob. Agents Chemother. 51:2065–2069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaatz G.W., DeMarco C.E., and Seo S.M. 2006. MepR, a repressor of the Staphylococcus aureus MATE family multidrug efflux pump MepA, is a substrate-responsive regulatory protein. Antimicrob. Agents Chemother. 50:1276–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Birukou I., Tonthat N.K., Seo S.M., Schindler B.D., Kaatz G.W., and Brennan R.G. 2013. The molecular mechanisms of allosteric mutations impairing MepR repressor function in multidrug-resistant strains of Staphylococcus aureus. MBio. 4:e00528–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Birukou I., Seo S.M., Schindler B.D., Kaatz G.W., and Brennan R.G. 2014. Structural mechanism of transcription regulation of the Staphylococcus aureus multidrug efflux operon mepRA by the MarR family repressor MepR. Nucleic Acids Res. 42:2774–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schindler B.D., Seo S.M., Birukou I., Brennan R.G., and Kaatz G.W. 2015. Mutations within the mepA operator affect binding of the MepR regulatory protein and its induction by MepA substrates in Staphylococcus aureus. J. Bacteriol. 197:1104–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vogwill T., and MacLean R.C. 2015. The genetic basis of the fitness costs of antimicrobial resistance: a meta-analysis approach. Evol. Appl. 8:284–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.