Abstract
Dysregulation of multiple long non-coding RNAs (lncRNAs) was reported to play major roles in breast cancer (BC). Here we aimed to collect most of the relevant literature to assess the prognostic value of lncRNAs in BC. To this end, we systematically searched PubMed, Embase, Web of Science, Chinese National Knowledge Infrastructure (CNKI), and Wanfang to identify published articles on the associations of lncRNAs with clinicopathology and/or survival of BC. Via this searching, we identified 70 articles involving 9,307 BC patients and regarding 48 lncRNAs. The expression of 41 lncRNAs was related to one or more clinicopathological parameters of BC, including tumor size; lymph node metastasis; histological grade; TNM stage; and estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER-2) statuses (p < 0.05). Dysregulation of 28 lncRNAs was associated with overall survival, and abnormal expression of 9 lncRNAs was linked to disease-free survival. Furthermore, the expression level of 3 lncRNAs was correlated with metastasis-free survival, 3 lncRNAs with relapse-free survival, and 3 lncRNAs with progression-free survival. Our analysis showed that multiple lncRNAs were significantly associated with BC clinicopathology and survival. A large-scale study is needed to verify the prognostic value of these lncRNAs in BC.
Keywords: long non-coding RNA, breast cancer, clinicopathology, survival, prognosis
Introduction
Breast cancer (BC) is the most common type of cancer among women and the main cause of female cancer death in the world.1 Although the survival rates of BC have been improved by early detection and progress in treatment, it remains to be a frequent malignancy with a poor prognosis, which seriously threatens the health of women.2, 3
Traditionally, we used clinicopathological features, including tumor size, lymph node status, TNM stage, histological grade, hormone receptor status, and human epidermal growth factor receptor 2 (HER-2) amplification, to predict the patient outcome.4 In addition, several biomarkers, such as tumor-associated macrophages (TAMs), microRNAs, matrix metalloproteinases (MMPs), retinoic acid receptor α (RARA), Ki-67, aromatase, osteopontin, etc., have also been identified.5, 6, 7, 8 In recent years, more and more BC studies have focused on long non-coding RNAs (lncRNAs) because of their key roles in human diseases, including cancer.9
lncRNAs are a class of RNA transcripts, with a length of >200 nt, that do not encode proteins. They were proven to be involved in diverse biological processes, such as chromosome remodeling, epigenetic modulation, and transcriptional and posttranscriptional modifications.10, 11 Studies have revealed that lncRNAs play an important role in cancer biology, and the expression of specific lncRNAs is implicated in the development and progression of cancer.12 For example, enforced expression of HOTAIR in epithelial cancer cells can induce genome-wide re-targeting of polycomb repressive complex 2 (PRC2), leading to altered histone H3 lysine 27 methylation and gene expression, and thus it promotes cancer invasiveness and metastasis in a manner dependent on PRC2.13 In BC, BCAR4 can bind to two transcription factors (SNIP1 and PNUTS) with extended regulatory consequences, and it relieves inhibition of RNA polymerase II (Pol II) via activation of the PP1 phosphatase. Thus, it activates a noncanonical Hedgehog/GLI2 pathway that promotes cell migration.14 Moreover, a large number of lncRNAs, such as MALAT1, MEG3, HOTAIR, CCAT2, H19, etc., are dysregulated in multiple tumors, including BC, hepatocellular carcinoma, and kidney cancer, possibly making them diagnostic or prognostic biomarkers or potential therapeutic targets for cancer.15, 16, 17
Many studies on the role of lncRNAs in BC revealed that the expression level of lncRNAs was associated with BC clinical features and outcome.17, 18, 19 By far, however, no study has evaluated these associations systematically. Therefore, we conducted this systematic review to clarify the present state of knowledge about the correlations between lncRNAs and BC clinicopathology and survival.
Results
Characteristics of Included Studies
A total of 991 articles was identified by mining databases and manual searching, and 732 articles were left after removing duplication. After screening titles and abstracts, 111 full-text articles remained for further assessment, and 41 articles were excluded according to the selection criteria. Finally, 70 articles involving 9,307 patients were included in the review. The main characteristics and quality score of studies included in the meta-analysis are presented in Table 1, and the information on the rest of the studies is shown in Table S1. Most of these articles were published within the last 3 years. Among all these articles, 63 articles involving 48 lncRNAs described the clinicopathological features of BC, and 48 articles involving 32 lncRNAs investigated the survival of BC.
Table 1.
Characteristics of Studies Included in the Meta-analysis
| lncRNAs | Reference | Country | Race | Number of Patients | Expression in Tumor | Method | Sample Type | Cutoff | Survival | Follow-up (Month) | Quality Score |
|---|---|---|---|---|---|---|---|---|---|---|---|
| MALAT1 | 26 | China | Asian | 43 | upregulated | qRT-PCR | tissue | median | OS | 60 | 7 |
| 25 | China | Asian | 118 | upregulated | qRT-PCR | tissue | NR | OS | 50 | 7 | |
| 20 | France | Caucasian | 446 | upregulated | qRT-PCR | tissue | 3.02-fold | NR | NR | 7 | |
| 24 | China | Asian | 139 | upregulated | qRT-PCR | tissue | median | OS | 55 | 7 | |
| 22 | China | Asian | 204 | upregulated | qRT-PCR | tissue | 75% expression | RFS | 65 | 8 | |
| 21 | China | Asian | 135 | downregulated | qRT-PCR | tissue | NR | NR | NR | 6 | |
| 23 | China | Asian | 78 | upregulated | qRT-PCR | tissue | median | DFS | 60 | 8 | |
| 27 | China | Asian | 86 | upregulated | qRT-PCR | tissue and serum | median | OS | NR | 5 | |
| CCAT2 | 31 | China | Asian | 120 | upregulated | qRT-PCR | tissue | NR | OS | 90 | 6 |
| 32 | Iran | Caucasian | 48 | normal | qRT-PCR | tissue | median | NR | NR | 6 | |
| 81 | Netherlands | Caucasian | 747 | upregulated | qRT-PCR | tissue | quartile | OS, MFS | >120 | 7 | |
| 80 | China | Asian | 67 | upregulated | qRT-PCR | tissue | 8-fold | OS | 60 | 7 | |
| 82 | Germany | Caucasian | 129 | NR | qRT-PCR | tissue | median | MFS | 120 | 8 | |
| HOTAIR | 13 | America | Caucasian | 132 | upregulated | qRT-PCR | tissue | 125-fold | OS, MFS | 180 | 8 |
| 83 | Denmark | Caucasian | 488 | NR | microarray | tissue | median | MFS | 217 | 7 | |
| 84 | Italy | Caucasian | 336 | NR | qRT-PCR | tissue | mean | OS, RFS | 86 | 8 | |
| 85 | China | Asian | 30 | NR | qRT-PCR | tissue | median | OS | 40 | 5 | |
| 44 | America | Caucasian | 94 | NR | ISH | tissue | median | NR | NR | 6 | |
| 43 | China | Asian | 112 | upregulated | qRT-PCR | serum | median | DFS | 48 | 7 | |
| MEG3 | 28 | China | Asian | 90 | downregulated | qRT-PCR | tissue | NR | OS, DFS | 80 | 6 |
| 29 | China | Asian | 207 | downregulated | qRT-PCR | tissue | median | OS, PFS | 60 | 8 | |
| 30 | China | Asian | 257 | downregulated | qRT-PCR | tissue | ΔCt = 8.065 | OS, RFS | 60 | 8 | |
| TUSC7 | 33 | China | Asian | 31 | downregulated | qRT-PCR | tissue | mean | NR | NR | 6 |
| 34 | China | Asian | 42 | downregulated | qRT-PCR | tissue | median | NR | NR | 6 | |
| BCAR4 | 35 | Germany | Caucasian | 96 | NR | qRT-PCR | tissue | NR | NR | NR | 6 |
| 36 | Netherlands | Caucasian | 786 | NR | qRT-PCR | tissue | detection limit | OS, PFS, MFS | 97 | 8 | |
| TP73-AS1 | 37 | China | Asian | 86 | upregulated | qRT-PCR | tissue | median | NR | NR | 8 |
| 38 | China | Asian | 36 | upregulated | qRT-PCR | tissue | median | OS | 48 | 8 | |
| NEAT1 | 40 | China | Asian | 118 | upregulated | qRT-PCR | tissue | NR | OS | 60 | 6 |
| 39 | China | Asian | 70 | upregulated | qRT-PCR | tissue | NR | OS | 60 | 6 | |
| 86 | China | Asian | 40 | upregulated | qRT-PCR | tissue | 2-fold | OS | 24 | 6 | |
| TUG1 | 41 | China | Asian | 100 | upregulated | qRT-PCR | tissue | mean | NR | NR | 7 |
| 42 | China | Asian | 58 | downregulated | qRT-PCR | tissue | mean | NR | NR | 6 | |
| CRNDE | 46 | China | Asian | 103 | upregulated | qRT-PCR | tissue | NR | OS | NR | 6 |
| 45 | China | Asian | 76 | upregulated | qRT-PCR | tissue& serum | 2-ΔΔCt = 1 | NR | NR | 6 |
OS, overall survival; DFS, disease-free survival; MFS, metastasis-free survival; RFS, relapse- free survival; PFS, progression-free survival; NR, not report; ISH, in situ hybridization.
Association of lncRNA Expression with Clinicopathological Features of BC
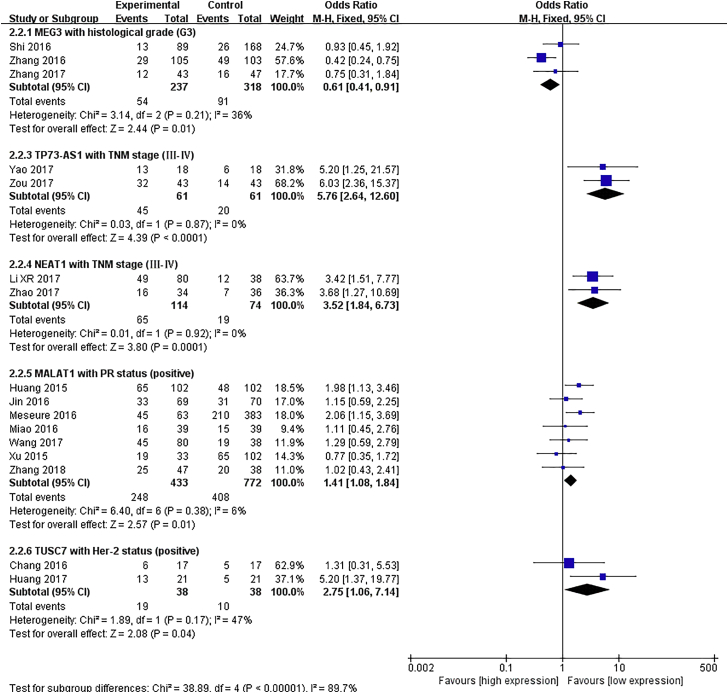
Ten lncRNAs, MALAT1, MEG3, CCAT2, BCAR, TUSC7, TP73-AS1, NEAT1, TUG1, HOTAIR, and CRNDE, were included in meta-analyses for clinicopathological features.20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42, 43, 44, 45, 46 Pooled results are presented in Table S2, and the significant associations are shown in Figure 1. We observed that an upregulated MALAT1 expression level was related to positive progesterone receptor (PR) status (odds ratio [OR] = 1.41, 95% confidence interval [CI]: 1.08–1.84, p = 0.01), and an elevated TUSC7 level was related to positive HER-2 status (OR = 2.75, 95% CI: 1.06–7.14, p = 0.04). The expression level of MEG3 was negatively correlated with tumor histological grade (OR = 0.61, 95% CI: 0.41–0.91, p = 0.01). Moreover, increased levels of NEAT1 and TP73-AS1 were associated with advanced TNM stage (OR = 3.52, 95% CI: 1.84–6.73 and OR = 5.76, 95% CI: 2.64–12.6, respectively; all p < 0.01).
Figure 1.
Forest Plots of the Significant Associations between the Expression of Five lncRNAs and Clinical Features of Breast Cancer
Each square indicates a study, and the area of squares is proportional to the weight of the study. The diamond represents the pooled OR and 95% CI.
Among the remaining 38 lncRNAs, 4 lncRNAs (MVIH, SOX2OT, PTPRG-AS1, and ANRIL) had no relationship with any clinicopathological features of BC.47, 48 The expression of the other 34 lncRNAs (HOTTIP, CCAT1, H19, HULC, etc.) was related to one or more clinical parameters of BC, including tumor size; lymph node metastasis; histological grade; TNM stage; and estrogen receptor (ER), PR, and HER-2 statuses (p < 0.05).48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75, 76, 77, 78, 79, 87 The p values of the correlations between these lncRNAs and BC clinicopathological features are shown in Table S3, and Table 2 summarizes all the lncRNAs that were related to clinicopathological features of BC.
Table 2.
Summary of lncRNAs Related to Clinicopathological Features of Breast Cancer
| Clinicopathological Feature | lncRNA |
|---|---|
| Tumor size | SNHG12, HOTTIP, H19, CRNDE, SPRY4-IT1, FGF14-AS2, APOC1P1-3, EGOT, 91H, HOXA-AS2, PVT1, CRALA, SNHG15, SUMO1P3, ARA |
| LN metastasis | NEAT1, SNHG12, HOTTIP, CCAT1, AFAP1-AS1, Z38, TUNAR, FGF14-AS2, HULC, EGOT, 91H, HIF1A-AS2, UCA1, linc-ROR, HOXA-AS2, GAS6-AS1, linc-ITGB1, DANCR, PVT1, OR3A4, CRALA, FENDRR, SNHG15, SUMO1P3 |
| Histological grade | MEG3, CCAT1, TUNAR, EPB41L4A-AS2, HULC, BC040587, GAS6-AS1, DANCR, OR3A4 |
| TNM stage | TP73-AS1, NEAT1, HOTTIP, CCAT1, AFAP1-AS1, ACTA2-AS1, Z38, SPRY4-IT1, FGF14-AS2, EPB41L4A-AS2, HULC, HOXA-AS2, linc-ITGB1, DANCR, PVT1, OR3A4, CRALA, HOXB-AS5, LINP1, SNHG15, SUMO1P3 |
| ER status | BCAR4, H19, LINC00978, EPB41L4A-AS2, CRALA |
| PR status | MALAT1, H19, EPB41L4A-AS2, CRALA, FENDRR |
| HER-2 status | TUSC7, 91H, ANRASSF1, OR3A4, FENDRR |
LN, lymph node; ER, estrogen receptor; PR, progesterone receptor; HER-2, human epidermal growth factor receptor 2.
Prognostic Value of lncRNA Expression for BC Survival
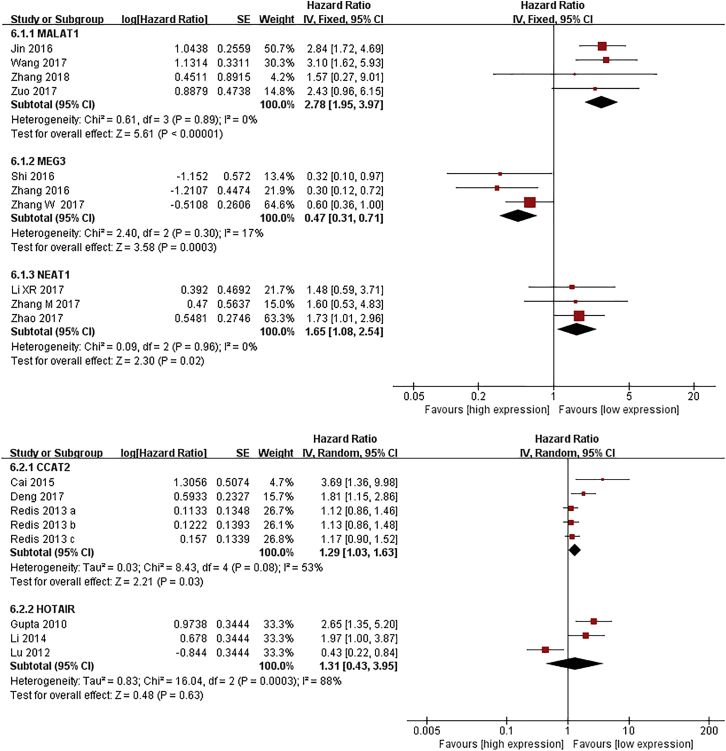
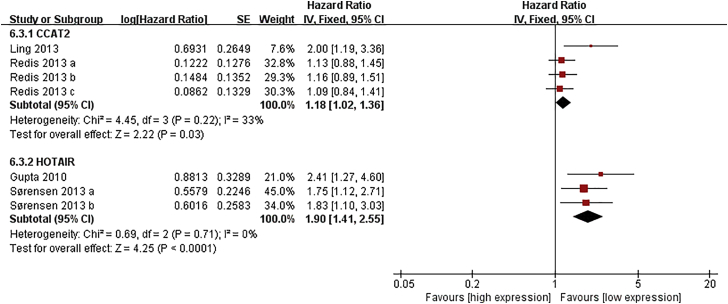
Five lncRNAs, including MALAT1, MEG3, CCAT2, HOTAIR, and NEAT1, were included in meta-analyses for survival.13, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 80, 81, 82, 83, 84, 85, 86 As shown in Figure 2, patients with high expression of CCAT2, MALAT1, or NEAT1 had shorter overall survival (OS) (hazard ratio [HR] = 1.29, 95% CI: 1.03–1.63, p = 0.03; HR = 2.78, 95% CI: 1.95–3.97, p < 0.01; HR = 1.65, 95% CI: 1.08–2.54, p = 0.02, respectively), while an increased level of MEG was associated with better OS (HR = 0.47, 95% CI: 0.37–0.71, p < 0.01). In addition, elevated expression of CCAT2 or HOTAIR was related to poor metastasis-free survival (MFS) (HR = 1.18, 95% CI: 1.02–1.36, p = 0.03; HR = 1.90, 95% CI: 1.41–2.55, p < 0.01, respectively) (Figure 3).
Figure 2.
Forest Plots of the Associations between the Expression of Five lncRNAs and Breast Cancer Overall Survival
Each square indicates a study and the area of squares is proportional to the weight of the study. The diamond represents the pooled HR and 95% CI.
Figure 3.
Forest Plots of the Associations between the Expression of Two lncRNAs and Breast Cancer Metastasis-free Survival
Each square indicates a study and the area of squares is proportional to the weight of the study. The diamond represents the pooled HR and 95% CI.
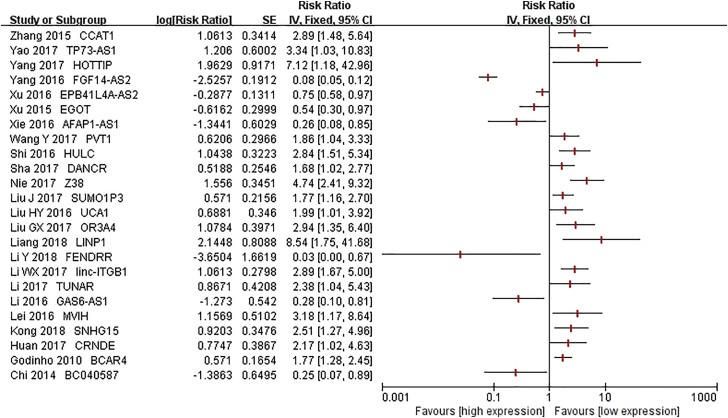
Another 24 lncRNAs were also correlated with OS of BC. Among them, the elevated expression of 7 lncRNAs (FGF14-AS2, AFAP1-AS1, EPB41L4A-AS2, BC040587, EGOT, GAS6-AS1, and FENDRR) related to a better survival,56, 59, 60, 61, 62, 66, 87 while increased expression of the 17 other lncRNAs (BCAR4, HOTTIP, CCAT1, Z38, TUNAR, CRNDE, HULC, MVIH, TP73-AS1, linc-ITGB1, PVT1, UCA1, OR3A4, DANCR, LINP1, SNHG15, and SUMO1P3) related to a worse survival36, 38, 46, 47, 51, 52, 53, 54, 55, 69, 71, 72, 73, 76, 77, 78, 88 (Figure 4). The expression of 9 lncRNAs (MALAT1, HOTTIP, MVIH, LINC00978, linc-ITGB1, MEG3, GAS6-AS1, HOTAIR, and LINP1) had an impact on disease-free survival (DFS) of BC. Furthermore, MALAT1, MEG3, and HOTAIR levels had a relationship with relapse-free survival (RFS); CCAT1, MEG3, and FENDRR levels were associated with progression-free survival (PFS); and the expression of BCAR4 was related to MFS. The detailed information is provided in Table 3.
Figure 4.
Forest Plots of the Associations between the Expression of lncRNAs and Breast Cancer Overall Survival in Single Studies
Each square indicates a study.
Table 3.
Summary of Other Significant Associations of lncRNAs with Breast Cancer Survival
| Survival | lncRNA | HR and 95% CI | Analysis | Reference |
|---|---|---|---|---|
| DFS | MALAT1 | 2.36 (1.04–5.38) | univariate | 23 |
| HOTTIP | 4.08 (1.13–14.71) | multivariate | 51 | |
| MVIH | 2.55 (1.06–6.12) | multivariate | 47 | |
| LINC00978 | 2.27 (1.24–4.16) | multivariate | 63 | |
| linc-ITGB1 | 3.13 (1.89–6.14) | multivariate | 69 | |
| MEG3 | 0.59 (0.36–0.96) | univariate | 28 | |
| GAS6-AS1 | 0.28 (0.13—0.60) | multivariate | 66 | |
| HOTAIR | 1.89 (1.15–3.11) | univariate | 43 | |
| LINP1 | 8.40 (1.72–41.06) | univariate | 76 | |
| RFS | MALAT1 | 2.02 (1.02–3.98) | multivariate | 22 |
| MEG3 | 0.37 (0.15–0.87) | multivariate | 30 | |
| HOTAIR | 0.47 (0.26–0.87) | multivariate | 84 | |
| PFS | CCAT1 | 3.59 (2.00–7.84) | multivariate | 52 |
| MEG3 | 0.37 (0.13–0.88) | multivariate | 29 | |
| FENDRR | 0.578 (0.454–0.735) | multivariate | 87 | |
| MFS | BCAR4 | 1.41 (1.03–1.94) | univariate | 36 |
DFS, disease-free survival; MFS, metastasis-free survival; RFS, relapse-free survival; PFS, progression-free survival.
Discussion
Increasing evidence has demonstrated that lncRNAs are involved in the initiation and progression of cancer and participate in multiple biological behaviors of cancer, including cell proliferation, apoptosis, migration, and metastasis.12, 89 Aberrant expression of lncRNAs has been observed in various types of cancer, including BC.17, 18 Previous reviews and meta-analyses have reported the prognostic values of lncRNAs in multiple cancers, such as colorectal cancer, ovarian cancer, prostate cancer, lung cancer, etc.90, 91, 92, 93 However, no one investigated BC specifically. Since many studies found that dysregulation of multiple lncRNAs may have an impact on the prognosis of BC, we conducted this systematic review to highlight the prognostic values of lncRNAs in BC. To our knowledge, this review is a thorough work that comprehensively clarifies the association of lncRNA expression with clinicopathological features and survival of BC.
In the present study, we systematically reviewed all the published literature regarding the clinical and prognostic values of lncRNAs in BC. We identified a number of relevant lncRNAs, most of which have been studied only once. We found that the expression levels of these lncRNAs were most often linked to tumor size (n = 15), lymph node metastasis (n = 24), and TNM stage (n = 21), while fewer of them associated with histological grade (n = 9), hormone receptor status (n = 9), and HER-2 status (n = 6). Moreover, several lncRNAs were related to more than two clinical features of BC. However, all the lncRNA expression had no relationship with patient age. These results indicated the intrinsic role of lncRNAs in the pathogenesis and progression of BC, which suggested lncRNAs may be important biomarkers for BC. As for survival, most of the studies investigated the relationship between lncRNA expression and OS, and a majority of them (n = 28) had a statistically significant correlation with the OS of BC. Only a few studies evaluated the associations of lncRNA expression with other types of survival, including DFS, MFS, PFS, and RFS, and there was also a strong connection between them. These results revealed the significant prognostic value of lncRNAs in BC. Hence, these lncRNAs may be independent predictors of prognosis in BC.
The most frequently evaluated lncRNAs in BC included MALAT1, MEG3, CCAT2, and HOTAIR. All of them are statistically significant predictors of BC prognosis. The expression of MALAT1, CCAT2, and HOTAIR was increased in BC, and the upregulation was associated with shorter survival. The expression of MEG3 was downregulated in BC. Tumor with a lower MEG3 expression tended to be poorly differentiated, and the survival of patients was worse. This indicated the oncogenic role of MALAT1, CCAT2, and HOTAIR in BC, whereas MEG3 may be a tumor suppressor of BC. In terms of mechanism, MALAT1 was reported to mainly act as a competing endogenous RNA (ceRNA) to sponge microRNAs, thus regulating cell progression, invasion, and metastasis in BC through their targets.24, 25, 26 CCAT2 can promote BC tumor growth and metastasis by regulating Wnt- and transforming growth factor β (TGF-β)- signaling pathways.80, 94 HOTAIR was proven to promote BC metastasis through inducing or repressing critical genes in cell proliferation and migration as well as modulating the cancer epigenome.13, 95 As for MEG3, it can inhibit cell proliferation, invasion, and angiogenesis both by sponging microRNAs and through regulating signaling transduction, such as the AKT and TGF-β pathways.28, 96, 97
Overall, the results are comprehensive and credible because the quality of included articles is relatively high. However, there are still limitations in our analysis. First, heterogeneity exists between studies regarding the same lncRNA, and the heterogeneity is stubborn owing to the differences in methodology, such as sample selection, tissue preservation, determination of cutoff value, and statistical analysis. Second, almost all the studies in our review reported a statistically significant result. Although the Begg’s funnel plot suggested there is no publication bias on OS (Figure S1), we still suspect that selective reporting bias is prominent in the literature regarding lncRNA and BC prognosis. Third, about half of the included studies had a small sample size (<100), and small studies are considered associating with inflated estimates of effect size and higher heterogeneity.98, 99 Lastly, language bias may exist since only two languages were used in the literature review.
Our analysis demonstrated the prognostic value of lncRNAs in BC, and it highlighted the important biological function of lncRNAs in BC progression. These lncRNAs may exert their effects by directly binding to functional protein, modulation of DNA methylation, or post-transcriptional regulation of target genes.89, 100, 101 These genes and proteins include those that are involved in tumorgenesis and metastasis, such as Wnt, P53, PI3K, MYC, etc. Therefore, dysregulation of certain lncRNAs may have an effect on the development of BC, thus influencing the outcome of BC. Though the exact mechanisms are not yet fully clarified, we believe they will be better understood in the future with more studies in this field.
In conclusion, this systematic review identified a number of lncRNAs that were correlated with BC clinicopathological features and survival, and almost all the lncRNAs are statistically significant predictors of BC prognosis. The weightiness of these correlations is difficult to ascertain due to a lot of uncontrollable factors. Hence, a large-scale study with a standardized process of detection, analysis, and report is needed to further verify the prognostic value of these lncRNAs in BC.
Materials and Methods
This review has been performed based on preferred reporting items for systematic reviews and meta-analyses (PRISMA).102
Search Strategy
The databases of PubMed, Embase, Web of Science, as well as Chinese National Knowledge Infrastructure (CNKI) and Wanfang were systematically searched to identify all the eligible literature up to April 13, 2018. The following keywords and search terms were used: long noncoding RNA or long ncRNA or lncRNA or lincRNA or long intergenic non-coding RNA or long untranslated RNA, BC or breast carcinoma or breast tumor or breast neoplasm, and clinical or clinicopathological or clinicopathology or survival or odds ratio or OR or hazard ratio or HR. Additionally, references in relevant articles were also screened manually. The languages of the retrieved literature were confined to English and Chinese.
Inclusion and Exclusion Criteria
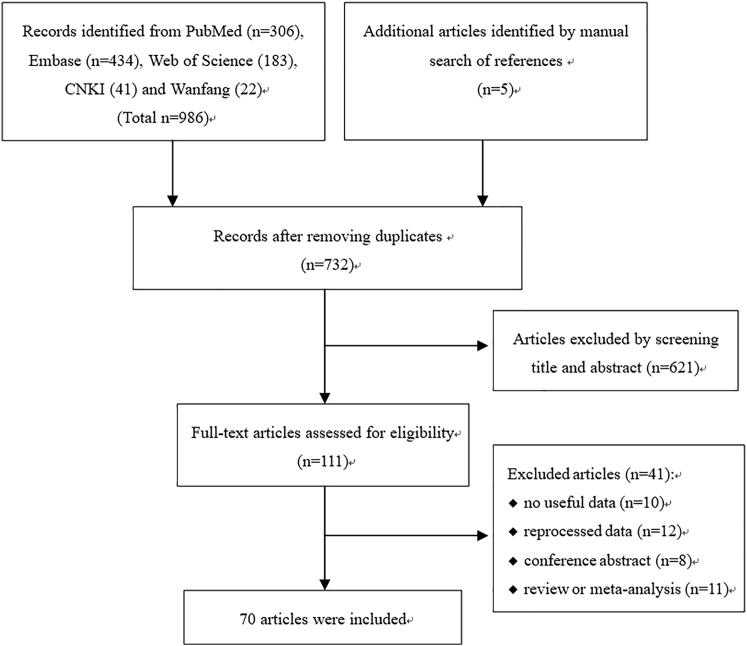
Studies were included if they fulfilled the following criteria: (1) original study focus on human beings, (2) investigated the relationship between lncRNA expression and clinicopathological features or survival of BC, (3) reported an OR or HR with 95% CI or there were sufficient data to calculate them, (4) full text was available. Exclusion criteria were as follows: (1) lacked key information, such as clinical parameters and survival curves, or lacked usable data; (2) reprocessed data from public databases; (3) HRs were for a combination of multiple lncRNAs; and (4) reviews, letters, single case reports, and conference abstracts. If multiple articles published by the same author reporting overlapped data, only the most complete one was included. The details about the selection process are shown in Figure 5.
Figure 5.
Flow Diagram of the Literature Review Process
A total of 991 articles were found by searching databases. After removing duplication, 732 articles were screened by title and abstract. Then, 111 articles were reviewed for full-text and 70 articles were finally included.
Quality Assessment and Data Extraction
Two authors (T.T. and M.W.) reviewed potentially eligible articles independently. The Newcastle-Ottawa Scale was used to assess the quality of each study.103 The following information was extracted from each included study: (1) basic information including first author’s name, publication year, country of origin, names of lncRNAs, sample size, expression levels of lncRNAs in BC, detection methods, sample type, outcome measurements, follow-up duration, cutoff value, and analysis method for survival; (2) p values of the correlation between lncRNA expression and clinicopathological features of BC and the original data for calculating ORs and their 95% CIs; and (3) HRs and their 95% CIs for survival analysis. If HRs were not directly accessible in the text, Kaplan-Meier survival curves were read using Engauge Digitizer (version 4.1) to obtain data. Different datasets for one lncRNA or one dataset concerning several lncRNAs in the same article was considered to be separate studies and the HR was extracted respectively; but, if multiple datasets were combined into a single dataset, we only extracted the pooled HR. Any discrepancy was discussed by all authors to reach a consensus.
Statistical Analysis
ORs and their 95% CIs were used to estimate the association of lncRNAs with clinical features of BC. Patients were divided into two groups for comparison (for instance, histological grade III versus I and II, TNM stages III and IV versus I and II, and ER/PR status positive versus negative). As for survival rates, HRs with corresponding 95% CIs were used. All the ORs and HRs were calculated for high expression of lncRNAs. When two or more different studies investigated the same lncRNA, a meta-analysis was carried out to combine the effect size. The Z test was used to determine the significance of ORs or HRs. Heterogeneity between studies was tested using Q statistic and I2 test. When I2 value was more than 50%, which indicated a significant heterogeneity, the random-effects model was utilized. Otherwise, the fixed-effects model was used. All statistical analyses were done with the software Review Manager 5.3 (Cochrane Collaboration, London, UK). A p value less than 0.05 was considered statistically significant.
Author Contributions
T.T. and Zhijun Dai conceived and designed the study. T.T. and M.W. searched and reviewed literature. S.L., Y.G., Zhiming Dai, K.L., and C.D. contributed to data collection, analysis, and interpretation. P.Y., Y. Zhu, Y. Zheng, and P.X. prepared tables and figures. T.T. drafted the manuscript. Zhijun Dai and W.Z. revised the manuscript. All authors approved the final manuscript.
Conflicts of Interest
The authors declare that they have no competing interest.
Acknowledgments
This work was supported by National Natural Science Foundation, People’s Republic of China (81471670); the International Cooperative Project of Shaanxi province, China (2016KW-008); and the Key Research and Development Plan, Shaanxi Province, People’s Republic of China (2017ZDXM-SF-066). The sponsors had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Footnotes
Supplemental Information includes one figure and three tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.05.018.
Contributor Information
Wenge Zhu, Email: wz6812@gwu.edu.
Zhijun Dai, Email: dzj0911@xjtu.edu.cn.
Supplemental Information
References
- 1.Torre L.A., Bray F., Siegel R.L., Ferlay J., Lortet-Tieulent J., Jemal A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Smith R.A., Andrews K., Brooks D., DeSantis C.E., Fedewa S.A., Lortet-Tieulent J., Manassaram-Baptiste D., Brawley O.W., Wender R.C. Cancer screening in the United States, 2016: A review of current American Cancer Society guidelines and current issues in cancer screening. CA Cancer J. Clin. 2016;66:96–114. doi: 10.3322/caac.21336. [DOI] [PubMed] [Google Scholar]
- 3.Shi J.F., Huang H.Y., Guo L.W., Shi D., Gu X.Y., Liang H., Wang L., Ren J.S., Bai Y.N., Mao A.Y. Quality-of-life and health utility scores for common cancers in China: a multicentre cross-sectional survey. Lancet. 2016;388(Suppl 1):S29. [Google Scholar]
- 4.Hayes D.F. Prognostic and predictive factors revisited. Breast. 2005;14:493–499. doi: 10.1016/j.breast.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 5.Tang X. Tumor-associated macrophages as potential diagnostic and prognostic biomarkers in breast cancer. Cancer Lett. 2013;332:3–10. doi: 10.1016/j.canlet.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 6.Bahrami A., Aledavood A., Anvari K., Hassanian S.M., Maftouh M., Yaghobzade A., Salarzaee O., ShahidSales S., Avan A. The prognostic and therapeutic application of microRNAs in breast cancer: Tissue and circulating microRNAs. J. Cell. Physiol. 2018;233:774–786. doi: 10.1002/jcp.25813. [DOI] [PubMed] [Google Scholar]
- 7.Ren F., Tang R., Zhang X., Madushi W.M., Luo D., Dang Y., Li Z., Wei K., Chen G. Overexpression of MMP Family Members Functions as Prognostic Biomarker for Breast Cancer Patients: A Systematic Review and Meta-Analysis. PLoS ONE. 2015;10:e0135544. doi: 10.1371/journal.pone.0135544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kutomi G., Mizuguchi T., Satomi F., Maeda H., Shima H., Kimura Y., Hirata K. Current status of the prognostic molecular biomarkers in breast cancer: A systematic review. Oncol. Lett. 2017;13:1491–1498. doi: 10.3892/ol.2017.5609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wapinski O., Chang H.Y. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–361. doi: 10.1016/j.tcb.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 11.Beermann J., Piccoli M.T., Viereck J., Thum T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 12.Prensner J.R., Chinnaiyan A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta R.A., Shah N., Wang K.C., Kim J., Horlings H.M., Wong D.J., Tsai M.C., Hung T., Argani P., Rinn J.L. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xing Z., Lin A., Li C., Liang K., Wang S., Liu Y., Park P.K., Qin L., Wei Y., Hawke D.H. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159:1110–1125. doi: 10.1016/j.cell.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klingenberg M., Matsuda A., Diederichs S., Patel T. Non-coding RNA in hepatocellular carcinoma: Mechanisms, biomarkers and therapeutic targets. J. Hepatol. 2017;67:603–618. doi: 10.1016/j.jhep.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 16.Martens-Uzunova E.S., Böttcher R., Croce C.M., Jenster G., Visakorpi T., Calin G.A. Long noncoding RNA in prostate, bladder, and kidney cancer. Eur. Urol. 2014;65:1140–1151. doi: 10.1016/j.eururo.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 17.Wang J., Ye C., Xiong H., Shen Y., Lu Y., Zhou J., Wang L. Dysregulation of long non-coding RNA in breast cancer: an overview of mechanism and clinical implication. Oncotarget. 2017;8:5508–5522. doi: 10.18632/oncotarget.12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerk S., Schwarzenbacher D., Adiprasito J.B., Stotz M., Hutterer G.C., Gerger A., Ling H., Calin G.A., Pichler M. Current Status of Long Non-Coding RNAs in Human Breast Cancer. Int. J. Mol. Sci. 2016;17:E1485. doi: 10.3390/ijms17091485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serghiou S., Kyriakopoulou A., Ioannidis J.P. Long noncoding RNAs as novel predictors of survival in human cancer: a systematic review and meta-analysis. Mol. Cancer. 2016;15:50. doi: 10.1186/s12943-016-0535-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meseure D., Vacher S., Lallemand F., Alsibai K.D., Hatem R., Chemlali W., Nicolas A., De Koning L., Pasmant E., Callens C. Prognostic value of a newly identified MALAT1 alternatively spliced transcript in breast cancer. Br. J. Cancer. 2016;114:1395–1404. doi: 10.1038/bjc.2016.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu S., Sui S., Zhang J., Bai N., Shi Q., Zhang G., Gao S., You Z., Zhan C., Liu F., Pang D. Downregulation of long noncoding RNA MALAT1 induces epithelial-to-mesenchymal transition via the PI3K-AKT pathway in breast cancer. Int. J. Clin. Exp. Pathol. 2015;8:4881–4891. [PMC free article] [PubMed] [Google Scholar]
- 22.Huang N.S., Chi Y.Y., Xue J.Y., Liu M.Y., Huang S., Mo M., Zhou S.L., Wu J. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 (MALAT1) interacts with estrogen receptor and predicted poor survival in breast cancer. Oncotarget. 2016;7:37957–37965. doi: 10.18632/oncotarget.9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miao Y., Fan R., Chen L., Qian H. Clinical Significance of Long Non-coding RNA MALAT1 Expression in Tissue and Serum of Breast Cancer. Ann. Clin. Lab. Sci. 2016;46:418–424. [PubMed] [Google Scholar]
- 24.Jin C., Yan B., Lu Q., Lin Y., Ma L. Reciprocal regulation of Hsa-miR-1 and long noncoding RNA MALAT1 promotes triple-negative breast cancer development. Tumour Biol. 2016;37:7383–7394. doi: 10.1007/s13277-015-4605-6. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y., Zhou Y., Yang Z., Chen B., Huang W., Liu Y., Zhang Y. MiR-204/ZEB2 axis functions as key mediator for MALAT1-induced epithelial-mesenchymal transition in breast cancer. Tumour Biol. 2017;39 doi: 10.1177/1010428317690998. 1010428317690998. [DOI] [PubMed] [Google Scholar]
- 26.Zuo Y., Li Y., Zhou Z., Ma M., Fu K. Long non-coding RNA MALAT1 promotes proliferation and invasion via targeting miR-129-5p in triple-negative breast cancer. Biomed. Pharmacother. 2017;95:922–928. doi: 10.1016/j.biopha.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 27.Zhang P., Zhou H., Lu K., Lu Y., Wang Y., Feng T. Exosome-mediated delivery of MALAT1 induces cell proliferation in breast cancer. OncoTargets Ther. 2018;11:291–299. doi: 10.2147/OTT.S155134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang W., Shi S., Jiang J., Li X., Lu H., Ren F. LncRNA MEG3 inhibits cell epithelial-mesenchymal transition by sponging miR-421 targeting E-cadherin in breast cancer. Biomed. Pharmacother. 2017;91:312–319. doi: 10.1016/j.biopha.2017.04.085. [DOI] [PubMed] [Google Scholar]
- 29.Zhang J.J., Guo S.H., Jia B.Q. Down-regulation of long non-coding RNA MEG3 serves as an unfavorable risk factor for survival of patients with breast cancer. Eur. Rev. Med. Pharmacol. Sci. 2016;20:5143–5147. [PubMed] [Google Scholar]
- 30.Shi W., Xia S., Yin Y., Qi X., Xing C. Decreased expression of lncRNA MEG3 in breast cancer is associated with poor prognosis. Int. J. Clin. Exp. Pathol. 2016;9:5327–5333. [Google Scholar]
- 31.Deng X., Zhao Y., Wu X., Song G. Upregulation of CCAT2 promotes cell proliferation by repressing the P15 in breast cancer. Biomed. Pharmacother. 2017;91:1160–1166. doi: 10.1016/j.biopha.2017.05.030. [DOI] [PubMed] [Google Scholar]
- 32.Sarrafzadeh Sh., Geranpayeh L., Tasharrofi B., Soudyab M., Nikpayam E., Iranpour M., Mirfakhraie R., Gharesouran J., Ghafouri-Fard S., Ghafouri-Fard S. Expression Study and Clinical Correlations of MYC and CCAT2 in Breast Cancer Patients. Iran. Biomed. J. 2017;21:303–311. doi: 10.18869/acadpub.ibj.21.5.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang L.J. Inner Mongolia Medical University; 2016. The Expression of Long Non-coding RNA-Loc285194 in Breast Cancer and Its Association with Clinicopathological Characteristics. [Google Scholar]
- 34.Huang Q.X., Chen G., Feng Z.B., Wei K.L., Chen H. Expression and clinical significance of long chain non-coding RNA LOC285194 in human breast cancer tissue. Chongqing Med. 2017;46:1223–1225. [Google Scholar]
- 35.van Agthoven T., Dorssers L.C., Lehmann U., Kreipe H., Looijenga L.H., Christgen M. Breast Cancer Anti-Estrogen Resistance 4 (BCAR4) Drives Proliferation of IPH-926 lobular Carcinoma Cells. PLoS ONE. 2015;10:e0136845. doi: 10.1371/journal.pone.0136845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Godinho M.F., Sieuwerts A.M., Look M.P., Meijer D., Foekens J.A., Dorssers L.C., van Agthoven T. Relevance of BCAR4 in tamoxifen resistance and tumour aggressiveness of human breast cancer. Br. J. Cancer. 2010;103:1284–1291. doi: 10.1038/sj.bjc.6605884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zou Q., Zhou E., Xu F., Zhang D., Yi W., Yao J. A TP73-AS1/miR-200a/ZEB1 regulating loop promotes breast cancer cell invasion and migration. J. Cell. Biochem. 2018;119:2189–2199. doi: 10.1002/jcb.26380. [DOI] [PubMed] [Google Scholar]
- 38.Yao J., Xu F., Zhang D., Yi W., Chen X., Chen G., Zhou E. TP73-AS1 promotes breast cancer cell proliferation through miR-200a-mediated TFAM inhibition. J. Cell. Biochem. 2018;119:680–690. doi: 10.1002/jcb.26231. [DOI] [PubMed] [Google Scholar]
- 39.Zhao D., Zhang Y., Wang N., Yu N. NEAT1 negatively regulates miR-218 expression and promotes breast cancer progression. Cancer Biomark. 2017;20:247–254. doi: 10.3233/CBM-170027. [DOI] [PubMed] [Google Scholar]
- 40.Li X., Wang S., Li Z., Long X., Guo Z., Zhang G., Zu J., Chen Y., Wen L. The lncRNA NEAT1 facilitates cell growth and invasion via the miR-211/HMGA2 axis in breast cancer. Int. J. Biol. Macromol. 2017;105:346–353. doi: 10.1016/j.ijbiomac.2017.07.053. [DOI] [PubMed] [Google Scholar]
- 41.Li T., Liu Y., Xiao H., Xu G. Long non-coding RNA TUG1 promotes cell proliferation and metastasis in human breast cancer. Breast Cancer. 2017;24:535–543. doi: 10.1007/s12282-016-0736-x. [DOI] [PubMed] [Google Scholar]
- 42.Fan S., Yang Z., Ke Z., Huang K., Liu N., Fang X., Wang K. Downregulation of the long non-coding RNA TUG1 is associated with cell proliferation, migration, and invasion in breast cancer. Biomed. Pharmacother. 2017;95:1636–1643. doi: 10.1016/j.biopha.2017.09.076. [DOI] [PubMed] [Google Scholar]
- 43.Lv R., Zhang J., Zhang W., Huang Y., Wang N., Zhang Q., Qu S. Circulating HOTAIR expression predicts the clinical response to neoadjuvant chemotherapy in patients with breast cancer. Cancer Biomark. 2018 doi: 10.3233/CBM-170874. https://doi.org/10.3233/CBM-170874 Published online March 30, 2018. [DOI] [PubMed] [Google Scholar]
- 44.Gökmen-Polar Y., Vladislav I.T., Neelamraju Y., Janga S.C., Badve S. Prognostic impact of HOTAIR expression is restricted to ER-negative breast cancers. Sci. Rep. 2015;5:8765. doi: 10.1038/srep08765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Z.B., Zhou J., Zhou L., Yang F.H. Value of detection of long non-coding RNA CRNDE in the diagnosis of breast cancer. Pract. Prev. Med. 2018;25:276–279. [Google Scholar]
- 46.Huan J., Xing L., Lin Q., Xui H., Qin X. Long noncoding RNA CRNDE activates Wnt/β-catenin signaling pathway through acting as a molecular sponge of microRNA-136 in human breast cancer. Am. J. Transl. Res. 2017;9:1977–1989. [PMC free article] [PubMed] [Google Scholar]
- 47.Lei B., Xu S.P., Liang X.S., Li Y.W., Zhang J.F., Zhang G.Q., Pang D. Long non-coding RNA MVIH is associated with poor prognosis and malignant biological behavior in breast cancer. Tumour Biol. 2016;37:5257–5264. doi: 10.1007/s13277-015-4360-8. [DOI] [PubMed] [Google Scholar]
- 48.Iranpour M., Soudyab M., Geranpayeh L., Mirfakhraie R., Azargashb E., Movafagh A., Ghafouri-Fard S. Expression analysis of four long noncoding RNAs in breast cancer. Tumour Biol. 2016;37:2933–2940. doi: 10.1007/s13277-015-4135-2. [DOI] [PubMed] [Google Scholar]
- 49.Wang O., Yang F., Liu Y., Lv L., Ma R., Chen C., Wang J., Tan Q., Cheng Y., Xia E. C-MYC-induced upregulation of lncRNA SNHG12 regulates cell proliferation, apoptosis and migration in triple-negative breast cancer. Am. J. Transl. Res. 2017;9:533–545. [PMC free article] [PubMed] [Google Scholar]
- 50.Adriaenssens E., Dumont L., Lottin S., Bolle D., Leprêtre A., Delobelle A., Bouali F., Dugimont T., Coll J., Curgy J.J. H19 overexpression in breast adenocarcinoma stromal cells is associated with tumor values and steroid receptor status but independent of p53 and Ki-67 expression. Am. J. Pathol. 1998;153:1597–1607. doi: 10.1016/S0002-9440(10)65748-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Y., Qian J., Xiang Y., Chen Y., Qu J. The prognostic value of long noncoding RNA HOTTIP on clinical outcomes in breast cancer. Oncotarget. 2017;8:6833–6844. doi: 10.18632/oncotarget.14304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang X.F., Liu T., Li Y., Li S. Overexpression of long non-coding RNA CCAT1 is a novel biomarker of poor prognosis in patients with breast cancer. Int. J. Clin. Exp. Pathol. 2015;8:9440–9445. [PMC free article] [PubMed] [Google Scholar]
- 53.Nie Z.L., Wang Y.S., Mei Y.P., Lin X., Zhang G.X., Sun H.L., Wang Y.L., Xia Y.X., Wang S.K. Prognostic significance of long noncoding RNA Z38 as a candidate biomarker in breast cancer. J. Clin. Lab. Anal. 2018;32 doi: 10.1002/jcla.22193. https://doi.org/10.1002/jcla.22193 Published online March 1, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H., Zhu L., Xu L., Qin K., Liu C., Yu Y., Su D., Wu K., Sheng Y. Long noncoding RNA linc00617 exhibits oncogenic activity in breast cancer. Mol. Carcinog. 2017;56:3–17. doi: 10.1002/mc.22338. [DOI] [PubMed] [Google Scholar]
- 55.Shi F., Xiao F., Ding P., Qin H., Huang R. Long Noncoding RNA Highly Up-regulated in Liver Cancer Predicts Unfavorable Outcome and Regulates Metastasis by MMPs in Triple-negative Breast Cancer. Arch. Med. Res. 2016;47:446–453. doi: 10.1016/j.arcmed.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 56.Xie B. The expression and clinical significance of a long non-coding RNA AFAP1-AS1 in breast cancer. Mod. Oncol. 2016;24:3739–3742. [Google Scholar]
- 57.Huang Q.X., Chen G., Feng Z.B., Wei K.L., Chen H. The expression and significance of long non-coding RNA UC001kfo in human breast cancer. J. Clin. Pathol. Res. 2015;35:1992–1998. [Google Scholar]
- 58.Shi Y., Li J., Liu Y., Ding J., Fan Y., Tian Y., Wang L., Lian Y., Wang K., Shu Y. The long noncoding RNA SPRY4-IT1 increases the proliferation of human breast cancer cells by upregulating ZNF703 expression. Mol. Cancer. 2015;14:51. doi: 10.1186/s12943-015-0318-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang F., Liu Y.H., Dong S.Y., Ma R.M., Bhandari A., Zhang X.H., Wang O.C. A novel long non-coding RNA FGF14-AS2 is correlated with progression and prognosis in breast cancer. Biochem. Biophys. Res. Commun. 2016;470:479–483. doi: 10.1016/j.bbrc.2016.01.147. [DOI] [PubMed] [Google Scholar]
- 60.Xu S., Wang P., You Z., Meng H., Mu G., Bai X., Zhang G., Zhang J., Pang D. The long non-coding RNA EPB41L4A-AS2 inhibits tumor proliferation and is associated with favorable prognoses in breast cancer and other solid tumors. Oncotarget. 2016;7:20704–20717. doi: 10.18632/oncotarget.8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chi Y., Huang S., Yuan L., Liu M., Huang N., Zhou S., Zhou B., Wu J. Role of BC040587 as a predictor of poor outcome in breast cancer. Cancer Cell Int. 2014;14:123. doi: 10.1186/s12935-014-0123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu S.P., Zhang J.F., Sui S.Y., Bai N.X., Gao S., Zhang G.W., Shi Q.Y., You Z.L., Zhan C., Pang D. Downregulation of the long noncoding RNA EGOT correlates with malignant status and poor prognosis in breast cancer. Tumour Biol. 2015;36:9807–9812. doi: 10.1007/s13277-015-3746-y. [DOI] [PubMed] [Google Scholar]
- 63.Deng L.L., Chi Y.Y., Liu L., Huang N.S., Wang L., Wu J. LINC00978 predicts poor prognosis in breast cancer patients. Sci. Rep. 2016;6:37936. doi: 10.1038/srep37936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liao X.H., Wang J.G., Li L.Y., Zhou D.M., Ren K.H., Jin Y.T., Lv L., Yu J.G., Yang J.Y., Lu Q. Long intergenic non-coding RNA APOC1P1-3 inhibits apoptosis by decreasing α-tubulin acetylation in breast cancer. Cell Death Dis. 2016;7:e2236. doi: 10.1038/cddis.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vennin C., Spruyt N., Robin Y.M., Chassat T., Le Bourhis X., Adriaenssens E. The long non-coding RNA 91H increases aggressive phenotype of breast cancer cells and up-regulates H19/IGF2 expression through epigenetic modifications. Cancer Lett. 2017;385:198–206. doi: 10.1016/j.canlet.2016.10.023. [DOI] [PubMed] [Google Scholar]
- 66.Li X., Zhang R., Liu Z., Li C., Xu H. Low expression of long noncoding RNA GAS6-AS1 as a novel biomarker of poor prognosis for breast cancer. Int. J. Clin. Exp. Med. 2016;9:15820–15827. [Google Scholar]
- 67.Liu M., Xing L.Q., Liu Y.J. A three-long noncoding RNA signature as a diagnostic biomarker for differentiating between triple-negative and non-triple-negative breast cancers. Medicine (Baltimore) 2017;96:e6222. doi: 10.1097/MD.0000000000006222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen Y.M., Liu Y., Wei H.Y., Lv K.Z., Fu P. Linc-ROR induces epithelial-mesenchymal transition and contributes to drug resistance and invasion of breast cancer cells. Tumour Biol. 2016;37:10861–10870. doi: 10.1007/s13277-016-4909-1. [DOI] [PubMed] [Google Scholar]
- 69.Li W.X., Sha R.L., Bao J.Q., Luan W., Su R.L., Sun S.R. Expression of long non-coding RNA linc-ITGB1 in breast cancer and its influence on prognosis and survival. Eur. Rev. Med. Pharmacol. Sci. 2017;21:3397–3401. [PubMed] [Google Scholar]
- 70.Fang Y., Wang J., Wu F., Song Y., Zhao S., Zhang Q. Long non-coding RNA HOXA-AS2 promotes proliferation and invasion of breast cancer by acting as a miR-520c-3p sponge. Oncotarget. 2017;8:46090–46103. doi: 10.18632/oncotarget.17552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sha S., Yuan D., Liu Y., Han B., Zhong N. Targeting long non-coding RNA DANCR inhibits triple negative breast cancer progression. Biol. Open. 2017;6:1310–1316. doi: 10.1242/bio.023135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang Y., Zhou J., Wang Z., Wang P., Li S. Upregulation of SOX2 activated lncRNA PVT1 expression promotes breast cancer cell growth and invasion. Biochem. Biophys. Res. Commun. 2017;493:429–436. doi: 10.1016/j.bbrc.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 73.Liu G., Hu X., Zhou G. Long non-coding RNA OR3A4 promotes proliferation and migration in breast cancer. Biomed. Pharmacother. 2017;96:426–433. doi: 10.1016/j.biopha.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 74.Li Y., Wang B., Lai H., Li S., You Q., Fang Y., Li Q., Liu Y. Long non-coding RNA CRALA is associated with poor response to chemotherapy in primary breast cancer. Thorac. Cancer. 2017;8:582–591. doi: 10.1111/1759-7714.12487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rui J., Chunming Z., Binbin G., Na S., Shengxi W., Wei S. IL-22 promotes the progression of breast cancer through regulating HOXB-AS5. Oncotarget. 2017;8:103601–103612. doi: 10.18632/oncotarget.22063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liang Y., Li Y., Song X., Zhang N., Sang Y., Zhang H., Liu Y., Chen B., Zhao W., Wang L. Long noncoding RNA LINP1 acts as an oncogene and promotes chemoresistance in breast cancer. Cancer Biol. Ther. 2018;19:120–131. doi: 10.1080/15384047.2017.1394543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kong Q., Qiu M. Long noncoding RNA SNHG15 promotes human breast cancer proliferation, migration and invasion by sponging miR-211-3p. Biochem. Biophys. Res. Commun. 2018;495:1594–1600. doi: 10.1016/j.bbrc.2017.12.013. [DOI] [PubMed] [Google Scholar]
- 78.Liu J., Song Z., Feng C., Lu Y., Zhou Y., Lin Y., Dong C. The long non-coding RNA SUMO1P3 facilitates breast cancer progression by negatively regulating miR-320a. Am. J. Transl. Res. 2017;9:5594–5602. [PMC free article] [PubMed] [Google Scholar]
- 79.Jiang M., Guan H.G. Long noncoding RNA adriamycin resistance associated is overexpressed in triple negative breast cancer and contributes to shorter survival. Chin. J. Exp. Surg. 2017;34:1757–1759. [Google Scholar]
- 80.Cai Y., He J., Zhang D. Long noncoding RNA CCAT2 promotes breast tumor growth by regulating the Wnt signaling pathway. OncoTargets Ther. 2015;8:2657–2664. doi: 10.2147/OTT.S90485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Redis R.S., Sieuwerts A.M., Look M.P., Tudoran O., Ivan C., Spizzo R., Zhang X., de Weerd V., Shimizu M., Ling H. CCAT2, a novel long non-coding RNA in breast cancer: expression study and clinical correlations. Oncotarget. 2013;4:1748–1762. doi: 10.18632/oncotarget.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ling H., Spizzo R., Atlasi Y., Nicoloso M., Shimizu M., Redis R.S., Nishida N., Gafà R., Song J., Guo Z. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013;23:1446–1461. doi: 10.1101/gr.152942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sørensen K.P., Thomassen M., Tan Q., Bak M., Cold S., Burton M., Larsen M.J., Kruse T.A. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Res. Treat. 2013;142:529–536. doi: 10.1007/s10549-013-2776-7. [DOI] [PubMed] [Google Scholar]
- 84.Lu L., Zhu G., Zhang C., Deng Q., Katsaros D., Mayne S.T., Risch H.A., Mu L., Canuto E.M., Gregori G. Association of large noncoding RNA HOTAIR expression and its downstream intergenic CpG island methylation with survival in breast cancer. Breast Cancer Res. Treat. 2012;136:875–883. doi: 10.1007/s10549-012-2314-z. [DOI] [PubMed] [Google Scholar]
- 85.Li J.T., Wang L.F., Zhao Y.L., Yang T., Li W., Zhao J., Yu F., Wang L., Meng Y.L., Liu N.N. Nuclear factor of activated T cells 5 maintained by Hotair suppression of miR-568 upregulates S100 calcium binding protein A4 to promote breast cancer metastasis. Breast Cancer Res. 2014;16:454. doi: 10.1186/s13058-014-0454-2. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 86.Zhang M., Wu W.B., Wang Z.W., Wang X.H. lncRNA NEAT1 is closely related with progression of breast cancer via promoting proliferation and EMT. Eur. Rev. Med. Pharmacol. Sci. 2017;21:1020–1026. [PubMed] [Google Scholar]
- 87.Li Y., Zhang W., Liu P., Xu Y., Tang L., Chen W., Guan X. Long non-coding RNA FENDRR inhibits cell proliferation and is associated with good prognosis in breast cancer. OncoTargets Ther. 2018;11:1403–1412. doi: 10.2147/OTT.S149511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu H., Wang G., Yang L., Qu J., Yang Z., Zhou X. Knockdown of Long Non-Coding RNA UCA1 Increases the Tamoxifen Sensitivity of Breast Cancer Cells through Inhibition of Wnt/β-Catenin Pathway. PLoS ONE. 2016;11:e0168406. doi: 10.1371/journal.pone.0168406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tehrani S.S., Karimian A., Parsian H., Majidinia M., Yousefi B. Multiple Functions of Long Non-Coding RNAs in Oxidative Stress, DNA Damage Response and Cancer Progression. J. Cell. Biochem. 2018;119:223–236. doi: 10.1002/jcb.26217. [DOI] [PubMed] [Google Scholar]
- 90.Wang J., Du S., Wang J., Fan W., Wang P., Zhang Z., Xu P., Tang S., Deng Q., Yang W., Yu M. The prognostic value of abnormally expressed lncRNAs in colorectal cancer: A meta-analysis. PLoS ONE. 2017;12:e0179670. doi: 10.1371/journal.pone.0179670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Luo P., Liu X.F., Wang Y.C., Li N.D., Liao S.J., Yu M.X., Liang C.Z., Tu J.C. Prognostic value of abnormally expressed lncRNAs in ovarian carcinoma: a systematic review and meta-analysis. Oncotarget. 2017;8:23927–23936. doi: 10.18632/oncotarget.14760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ma W., Chen X., Ding L., Ma J., Jing W., Lan T., Sattar H., Wei Y., Zhou F., Yuan Y. The prognostic value of long noncoding RNAs in prostate cancer: a systematic review and meta-analysis. Oncotarget. 2017;8:57755–57765. doi: 10.18632/oncotarget.17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jing W., Li N., Wang Y., Liu X., Liao S., Chai H., Tu J. The prognostic significance of long noncoding RNAs in non-small cell lung cancer: a meta-analysis. Oncotarget. 2017;8:3957–3968. doi: 10.18632/oncotarget.13956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wu Z.J., Li Y., Wu Y.Z., Wang Y., Nian W.Q., Wang L.L., Li L.C., Luo H.L., Wang D.L. Long non-coding RNA CCAT2 promotes the breast cancer growth and metastasis by regulating TGF-β signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2017;21:706–714. [PubMed] [Google Scholar]
- 95.Yu X., Li Z. Long non-coding RNA HOTAIR: A novel oncogene (Review) Mol. Med. Rep. 2015;12:5611–5618. doi: 10.3892/mmr.2015.4161. [DOI] [PubMed] [Google Scholar]
- 96.Zhang C.Y., Yu M.S., Li X., Zhang Z., Han C.R., Yan B. Overexpression of long non-coding RNA MEG3 suppresses breast cancer cell proliferation, invasion, and angiogenesis through AKT pathway. Tumour Biol. 2017;39 doi: 10.1177/1010428317701311. 1010428317701311. [DOI] [PubMed] [Google Scholar]
- 97.Mondal T., Subhash S., Vaid R., Enroth S., Uday S., Reinius B., Mitra S., Mohammed A., James A.R., Hoberg E. MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat. Commun. 2015;6:7743. doi: 10.1038/ncomms8743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ioannidis J.P. Why most discovered true associations are inflated. Epidemiology. 2008;19:640–648. doi: 10.1097/EDE.0b013e31818131e7. [DOI] [PubMed] [Google Scholar]
- 99.IntHout J., Ioannidis J.P., Borm G.F., Goeman J.J. Small studies are more heterogeneous than large ones: a meta-meta-analysis. J. Clin. Epidemiol. 2015;68:860–869. doi: 10.1016/j.jclinepi.2015.03.017. [DOI] [PubMed] [Google Scholar]
- 100.Nagano T., Fraser P. No-nonsense functions for long noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 101.Forrest M.E., Khalil A.M. Review: Regulation of the cancer epigenome by long non-coding RNAs. Cancer Lett. 2017;407:106–112. doi: 10.1016/j.canlet.2017.03.040. [DOI] [PubMed] [Google Scholar]
- 102.Moher D., Liberati A., Tetzlaff J., Altman D.G., PRISMA Group Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int. J. Surg. 2010;8:336–341. doi: 10.1016/j.ijsu.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 103.Zeng X.T., Liu H., Chen X., Leng W.D. Meta Analysis series four: quality assessment tools for observational studies. Chin. J. Evid. Based. Cardiovasc Med. 2012;4:297–299. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.