Abstract
Human spermatogonial stem cells (SSCs) could have significant applications in reproductive medicine and regenerative medicine because of their great plasticity. The fate determinations of human SSCs are mediated by epigenetic factors. However, nothing is known about the regulation of non-coding RNA on human SSCs. Here we have explored for the first time the expression, function, and target of miR-663a in human SSCs. MiR-663a was upregulated in human spermatogonia compared with pachytene spermatocytes, as indicated by microRNA microarray and real-time PCR. CCK-8 and 5-Ethynyl-2′-deoxyuridine (EDU) assays revealed that miR-663a stimulated cell proliferation and DNA synthesis of human SSCs. Annexin V and propidium iodide (PI) staining and flow cytometry demonstrated that miR-663a inhibited early and late apoptosis of human SSCs. Furthermore, NFIX was predicted and verified as a direct target of miR-663a. NFIX silencing led to an enhancement of cell proliferation and DNA synthesis and a reduction of the early apoptosis of human SSCs. NFIX silencing neutralized the influence of miR-663a inhibitor on the proliferation and apoptosis of human SSCs. Finally, both miR-663a mimics and NFIX silencing upregulated the levels of cell cycle regulators, including Cyclin A2, Cyclin B1, and Cyclin E1, whereas miR-663a inhibitor had an adverse effect. Knockdown of Cyclin A2, Cyclin B1, and Cyclin E1 led to the decrease in the proliferation of human SSCs. Collectively, miR-663a has been identified as the first microRNA that promotes the proliferation and DNA synthesis and suppresses the early apoptosis of human SSCs by targeting NFIX via cell cycle regulators Cyclin A2, Cyclin B1, and Cyclin E1. This study thus provides novel insights into the molecular mechanisms underlying human spermatogenesis, and it could offer novel targets for treating male infertility and other human diseases.
Keywords: miRNA-663a, human spermatogonial stem cells, proliferation, apoptosis, NFIX, cell cycle proteins
Introduction
Spermatogonial stem cells (SSCs) are a subpopulation of type A spermatogonia in the testis. Significantly, SSCs could have great applications in reproductive medicine to treat male infertility and regenerative medicine for various kinds of human diseases, because SSCs can be induced to generate mature and functional spermatids1 and they acquire pluripotency to become embryonic stem-like (ES-like) cells that differentiate to mature and functional cells of three germ cell layers.2, 3, 4 In addition, SSCs are able to directly transdifferentiate to other cell lineages, including hepatocytes5, 6, 7 and neuron,8 and other tissues, e.g., prostatic, uterine, and skin epithelium.9 Therefore, it is essential to uncover the molecular mechanisms underlying the fate decisions of human SSCs, including the proliferation, differentiation, transdifferentiation, and apoptosis. In the seminiferous tubules, SSCs can divide to maintain the pool of stem cells, and they differentiate into spermatocytes and mature spermatozoa10, 11 and undergo apoptosis to remove extra cells. The fate determinations of SSCs are regulated tightly by epigenetic and genetic factors.
Emerging as a novel and crucial post-transcriptional regulator, microRNAs (miRNAs) are a class of endogenous small single-stranded RNA molecules (18–25 nt in length) that intricately participate in epigenetic regulation.12, 13 miRNAs negatively regulate gene expression via binding the complementary sequences in the 3′ UTRs of the targeting mRNAs, which results in the degradation of mRNA or translation inhibition.14 miRNAs were first found in mammalian animals in 2001,15 and increasing evidence has demonstrated that miRNAs play indispensable roles in the three stages of spermatogenesis in rodents.16, 17, 18 MiR-21 has been shown to promote the proliferation and maintain the survival of mouse SSCs through regulating transcription factor ETV5,19 while miR-221 and miR-222 retain the undifferentiated status of mouse spermatogonia via the suppression of KIT expression.20 We have demonstrated that miR-20 and miR-106a stimulate the self-renewal of mouse SSCs by targeting STAT3.21 MiR-34c has been found to be specifically expressed in mouse germ cells,22 and simultaneous inactivation of two miRNA clusters, namely miR-34b/c and miR-449, leads to severe testicular disruption and defective spermatogenesis.23 It has recently been reported that miR-7a2 regulates the level of follicle-stimulating hormone (FSH) and luteinizing hormone (LH) through pituitary prostaglandin and BMP4 signaling, and miR-7a2 ablation causes hypogonadism and eventual male infertility in mice.24 Moreover, miR-202 deficiency leads to the differentiation of mouse SSCs to the differentiating spermatogonia by suppressing Rbfox2, Cpeb1, and cell cycle regulators.25 These findings reflect the significant roles of miRNAs in determining the fate determinations of rodent SSCs. Nevertheless, previous studies were restricted to miRNA in mice. Notably, there are distinct cell types and biochemical phenotypes between rodent SSCs and human SSCs.26, 27 As a result, the molecular mechanisms in regulating SSCs in human and other species, including rodents, could be distinct. For example, the JAK/STAT pathway has been demonstrated to facilitate the self-renewal of Drosophila SSCs.28 Conversely, the STAT3 pathway has been shown to be required for the differentiation of mouse SSCs.29
Almost nothing is known about the function and mechanism of miRNAs on the regulation of human SSCs, due to the following factors, which impede a better understanding of the molecular mechanism of human SSCs. The number of human primary SSCs is very scarce, and it is rather difficult to obtain human testicular tissues. Additionally, long-term culture and expansion of human SSCs have not yet been available. We have established a human SSC line with an unlimited proliferation potential and high safety.30 Utilizing this stable human SSC line in the current study, we have demonstrated for the first time that miR-663a stimulates the proliferation and DNA synthesis and inhibits the apoptosis of human SSCs by targeting NFIX via cell cycle regulators, including Cyclin A2, Cyclin B1, and Cyclin E1. Significantly, this study offers novel insights into the epigenetic regulation of human SSCs, and it provides new targets for human SSCs in treating male infertility and other disorders.
Results
Isolation and Identification of Human Spermatogonia and Pachytene Spermatocytes from Testicular Tissues of OA Patients
A two-step enzymatic digestion followed by differential plating and STA-PUT sedimentation were employed to isolate the human spermatogonia and pachytene spermatocytes from testicular tissues of obstructive azoospermia (OA) patients. The seminiferous tubules were isolated after a first enzymatic digestion. Human germ cells, Sertoli cells, and myoid cells were then obtained after a second enzymatic digestion, and they were placed in a cell culture dish for differential plating. Due to different characteristics, human Sertoli cells and myoid cells attached to the culture plate, whereas male germ cells were suspended in medium. Human male germ cells were collected by centrifuging, and human spermatogonia and pachytene spermatocytes were further separated by STA-PUT velocity sedimentation.31
Freshly isolated human spermatogonia and pachytene spermatocytes were identified based on their morphological and phenotypic characteristics. Individual spherical spermatogonium could be observed under a phase-contrast microscope with large round or ovoid nuclei and a diameter of 9∼12 μm (Figure 1A). Notably, pachytene spermatocytes could be easily recognized because of their patchy condensed chromatin and diameter of 14∼16 μm (Figure 1B).
Figure 1.
Isolation, Identification, and MiR-663a Expression of Human Spermatogonia and Pachytene Spermatocytes
(A and B) Morphological characteristics of freshly isolated human spermatogonia (A) and pachytene spermatocytes (B) from testicular tissues of OA patients under phase-contrast microscope. (C) Real-time qPCR revealed the different expression levels of miR-663a in human spermatogonia and pachytene spermatocytes. *Statistically significant differences (p < 0.05) between human spermatogonia and pachytene spermatocytes. (D) RT-PCR revealed the expression of GPR125, GFRA1, UCHL1, and THY1 in human spermatogonia and testicular tissues of OA patients (positive control). (E) RT-PCR showed the transcripts of SYCP3, MLH1, and CREST in human pachytene spermatocytes and testicular tissues of OA patients (positive control). Samples without cDNA (no cDNA) but PCR with gene primers were employed as negative controls. GAPDH served as a loading control of total RNA. (F–I) Immunocytochemistry demonstrated the expression of GFRA1 (F), GPR125 (G), UCHL1 (H), and THY1 (I) proteins in freshly isolated human spermatogonia. Scale bars, 20 μm (F–I). (J) Meiotic spread assays displayed the co-expression of SYCP3 (red fluorescence), CREST (blue fluorescence), and MLH1 (green fluorescence) proteins in pachytene spermatocytes isolated from OA patients. Scale bar, 2 μm (J).
To further identify the phenotypic characteristics of human spermatogonia and pachytene spermatocytes, numerous markers, respectively, for these cells were used. RT-PCR showed that the transcripts of UCHL1, GPR125, GFRA1, and THY1 were detected in human spermatogonia (Figure 1D), and SYCP3, MLH1, and CREST mRNA were expressed in human pachytene spermatocytes (Figure 1E). RNA without RT (no cDNA) but with PCR of GAPDH primers served as negative controls (Figures 1D and 1E), and GAPDH was used as the loading control of total RNA (Figures 1D and 1E).
Immunocytochemistry further revealed that 90% of the freshly isolated human spermatogonia was positive for GFRA1 (Figure 1F), GPR125 (Figure 1G), UCHL1 (Figure 1H), and THY1 (Figure 1I), markers for human spermatogonia. Meiotic chromatin spread displayed that 92% of the freshly isolated human pachytene spermatocytes were co-expressing CREST, SYCP3, and MLH1 (Figure 1J), hallmarks for spermatocytes.32, 33, 34
Differential Expression of MiR-663a between Human Spermatogonia and Pachytene Spermatocytes
We have performed miRNA microarrays to compare global miRNA profiles among human spermatogonia, pachytene spermatocytes, and round spermatids.31 The representative miRNAs that are upregulated in human spermatogonia compared to pachytene spermatocytes were selected and are shown in Table 1. Among these differentially expressed miRNAs, miR-663a was expressed at a higher level in human spermatogonia than pachytene spermatocytes. Real-time qPCR revealed the higher level of miR-663a in human spermatogonia compared to pachytene spermatocytes (Figure 1C), which was consistent with miR-663a expression patterns by miRNA microarrays (Table 1). Therefore, we hypothesized that miR-663a might play a role in regulating the fate determinations of human spermatogonia and SSCs.
Table 1.
The Differentially Expressed miRNAs between Human Spermatogonia and Pachytene Spermatocytes
| miRNA Names | Normalized Intensity |
log2 (Ratio) |
p Value (Differentially Expressed) |
||
|---|---|---|---|---|---|
| S2 | S1 | S2/S1 | S2/S1 | S2/S1 | |
| hsa-miR-663a | 975.0828448 | 612.8868889 | 1.590967049 | 0.669903956 | 1.77408E−05 |
| hsa-miR-320c | 1,876.277424 | 1,206.225791 | 1.555494368 | 0.637373171 | 8.5358E−06 |
| hsa-miR-6126 | 21,918.16655 | 14,219.43578 | 1.541423085 | 0.624262902 | 3.33479E−05 |
| hsa-miR-4698 | 1,336.04519 | 873.9100104 | 1.528813235 | 0.612412173 | 0.000282942 |
| hsa-miR-150-3p | 794.6016723 | 524.3460063 | 1.51541475 | 0.599712696 | 1.32038E−05 |
| hsa-miR-3138 | 2,415.298376 | 1,605.234703 | 1.504638774 | 0.589417173 | 0.000384259 |
| hsa-miR-4730 | 266.4822681 | 400.1587943 | 0.665941301 | −0.586533077 | 0.001603233 |
| hsa-miR-99b-5p | 232.5663431 | 353.013649 | 0.658802694 | −0.602081639 | 3.9567E−05 |
| hsa-miR-6511a-3p | 9,508.57184 | 14,541.40262 | 0.65389647 | −0.61286586 | 0.00048655 |
| hsa-miR-34a-5p | 484.5132148 | 760.0717328 | 0.637457221 | −0.649599566 | 0.001450411 |
| hsa-miR-1281 | 5,075.275925 | 8,240.051493 | 0.615927695 | −0.699167094 | 0.000194081 |
| hsa-miR-107 | 300.3981932 | 501.3483744 | 0.599180547 | −0.738937309 | 0.001457314 |
S1, human pachytene spermatocytes; S2, human spermatogonia.
Phenotypic Identification of the Human SSC Line
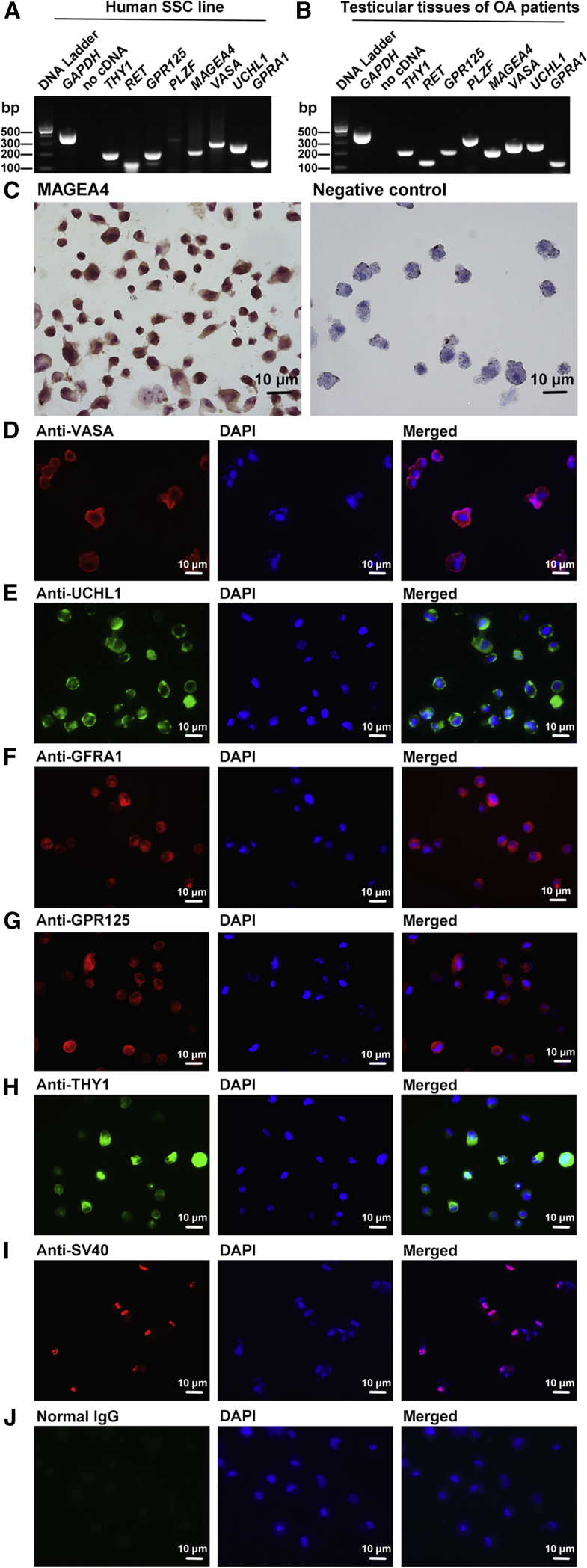
Since the human SSC line was employed to examine the role of miR-663a in determining the fate of human SSCs, we first identified the phenotypic identification of this cell line. RT-PCR showed that the human cell line expressed numerous genes for human germ cells and human spermatogonia, including VASA, MEGEA4, THY1, RET, GPR125, PLZF, UCHL1, and GFRA1 (Figure 2A). The expression of these genes in testicular tissues of OA patients was used as positive controls (Figure 2B), while RNA without RT (no cDNA) but with PCR of GAPDH primers served as negative controls (Figures 2A and 2B). Immunocytochemistry further revealed that the human cell line was positive for MAGEA4 (Figure 2C, left panel), VASA (Figure 2D), UCHL1 (Figure 2E), GFRA1 (Figure 2F), GPR125 (Figure 2G), THY1 (Figure 2H), and SV40 (Figure 2I). Replacement of primary antibodies with isotype immunoglobulin Gs (IgGs) was used as the negative control (Figure 2J), and no immunostaining was observed, thus verifying specific staining of the antibodies mentioned above in the cell line. Taken together, these results suggest that the human cell line is human SSCs phenotypically.
Figure 2.
Identification of the Human SSC Line
(A and B) RT-PCR showed the mRNA levels of VASA, MEGEA4, THY1, RET, GPR125, PLZF, UCHL1, and GFRA1 in the human SSC line (A) and testicular tissues of OA patients (B, positive control). Samples without cDNA (no cDNA) but PCR with gene primers were used as negative controls, and GAPDH served as a loading control of total RNA. (C) Immunocytochemistry of anti-MAGEA4 stained by diaminobenzine (DAB) showed the presence of MAGEA4 protein (left panel) in the human SSC line. Replacement of anti-MAGEA4 with isotype IgGs was used as a negative control (right panel). (D–I) Immunofluorescence demonstrated the expression of VASA (D), UCHL1 (E), GFRA1 (F), GPR125 (G), THY1 (H), and SV40 (I) in the human SSC line. (J) Normal IgG was substituted for primary antibodies as a negative control. Scale bars, 10 μm (C–J).
MiR-663a Promotes Proliferation and DNA Synthesis of Human SSCs
To explore the function of miR-663a in human SSCs, we employed miR-663a mimics and inhibitor. As shown by fluorescein amidite (FAM)-labeled miRNA oligonucleotides, transfection efficiency of miR-663a mimics and inhibitor in the human SSC line was over 80% (Figures 3A and 3B). miRNA mimics control, miR-663a mimics, miRNA inhibitor control, and miR-663a inhibitors were, respectively, transfected into the human SSC line. Real-time qPCR after transfection for 24 hr revealed that the expression level of miR-663a in these cells was significantly increased by miR-663a mimics when compared with miRNA mimics control (Figure 3C). In contrast, miR-663a level was statistically decreased by miR-663a inhibitor compared to miRNA inhibitor control (Figure 3C).
Figure 3.
Transfection Efficiency of MiR-663a Mimics and Inhibitor in the Human SSC Line and the Effects of Overexpression and Knockdown of MiR-663a on the Proliferation of Human SSCs
(A) Fluorescence microscope and phase-contrast microscope revealed transfection efficiency of miR-663a mimics and inhibitor using the FAM-labeled miRNA oligonucleotides. (B) Fluorescence microscope and phase-contrast microscope showed the control transfection of the cells treated by RNA oligonucleotides without FAM labeling. Scale bars, 20 μm (A and B). (C) Real-time qPCR revealed the relative levels of miR-663a in human SSCs after transfection of miR-663a mimics compared to miRNA mimics control and miR-663a inhibitor compared to miRNA inhibitor control. *Statistically significant differences (p < 0.05) between miRNA mimics- or inhibitor-treated groups and their control. (D and E) CCK-8 assays revealed the growth curve of the human SSC line treated with miR-663a mimics and miRNA mimics control for 5 days (D) or miR-663a inhibitor and miRNA inhibitor control for 5 days (E). *Statistically significant differences (p < 0.05) between miR-663a mimics- or inhibitor-treated groups and their controls.
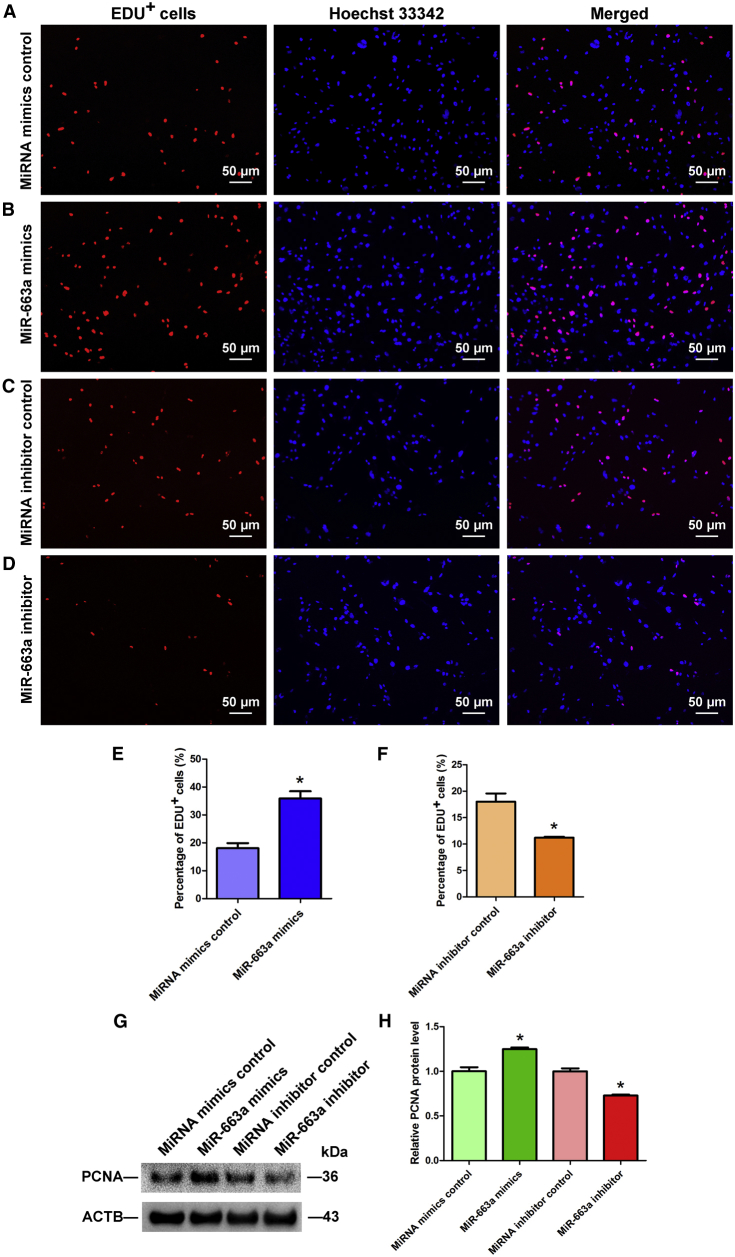
Proliferation ability of the human SSC line by miR-663a was determined by diverse assays. First, we conducted the cell proliferation assays from 24 to 120 hr after transfection of miR-663a mimics and miR-663a inhibitor; the growth of human SSCs was obviously enhanced by miR-663a mimics compared to miRNA mimics control (Figure 3D), whereas miR-663a inhibitor distinctly reduced the proliferation of the human SSC line compared to miRNA inhibitor control (Figure 3E). EDU (5-Ethynyl-2′-deoxyuridine) incorporation assay demonstrated that the percentage of EDU-positive cells was increased by miR-663a mimics and decreased by miR-663a inhibitor (Figures 4A–4F). A generally employed marker of cellular proliferation, PCNA (Proliferating cell nuclear antigen) was also detected. Western blots showed that protein level of PCNA was increased by miR-663a mimics versus the miRNA mimics control, whereas PCNA level was reduced by miR-663a inhibitor compared to the miRNA inhibitor control (Figures 4G and 4H). Collectively, these results indicate that miR-663a stimulates the proliferation and DNA synthesis of human SSCs.
Figure 4.
Effects of Overexpression and Knockdown of MiR-663a on the DNA Synthesis of Human SSCs
(A–D) EDU incorporation assay showed the percentages of EDU-positive cells in the human SSC line affected by miRNA mimics control (A), miR-663a mimics (B), miRNA inhibitor control (C), and miR-663a inhibitors (D) in human SSCs. Scale bars, 50 μm (A–D). *Statistically significant differences (p < 0.05) between miR-663a mimics- or inhibitor-treated groups and their controls. (E and F) Qualification of EDU-positive cells in the human SSC line affected by miRNA mimics control, miR-663a mimics (E), miRNA inhibitor control, and miR-663a inhibitors (F) in human SSCs. (G) Western blots demonstrated PCNA expression in human SSCs at day 3 after transfection of miRNA mimic control, miR-663a mimics, miRNA inhibitor control, and miR-663a inhibitor. ACTB served as the loading control of protein. (H) The relative expression of PCNA in human SSCs at day 3 after transfection of miR-663a mimics to miRNA mimics control and miR-663a inhibitor to miRNA inhibitor control through normalization to the signals of their loading control. *Statistically significant differences (p < 0.05) between miR-663a mimic- or inhibitor-treated groups and their controls.
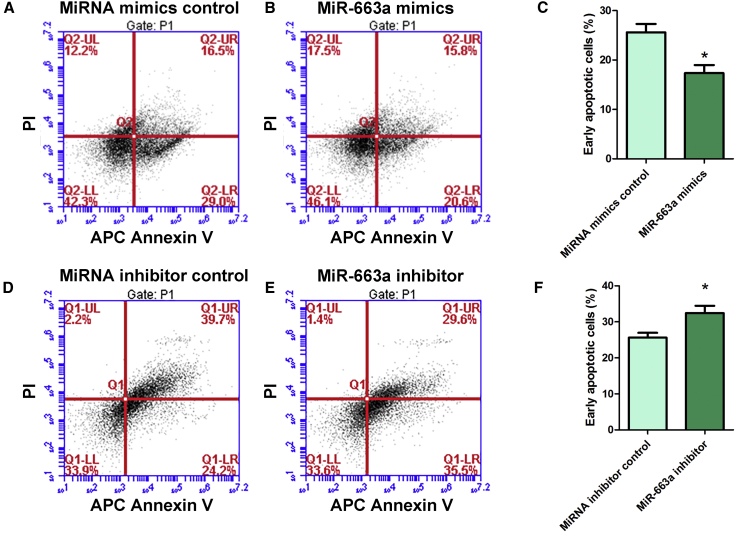
MiR-663a Inhibits the Early Apoptosis of Human SSCs
Annexin V and propidium iodide (PI) staining and flow cytometry assay indicated that miR-663a mimics could simultaneously suppress the early apoptosis of the human SSC line (Figures 5A–5C). Conversely, miR-663a inhibitor led to an increase of early apoptosis of the human SSC line when compared with miRNA inhibitor control (Figures 5D–5F). These data reflect that the miR-663a mimics suppresses the early apoptosis of human SSCs.
Figure 5.
Effect of Overexpression and Knockdown of MiR-663a on the Apoptosis of Human SSCs
(A–C) APC Annexin V and PI staining and flow cytometry depicted the percentages of early apoptosis in human SSCs transfected with miRNA mimics control (A and C) and miR-663a mimics (B and C). (D–F) APC Annexin V and PI staining and flow cytometry depicted the percentages of early apoptosis in human SSCs transfected with miRNA inhibitor control (D and F) and miR-663a inhibitor (E and F). *Statistically significant differences (p < 0.05) between miR-663a mimics- or inhibitor-treated groups and their respective controls (C and F).
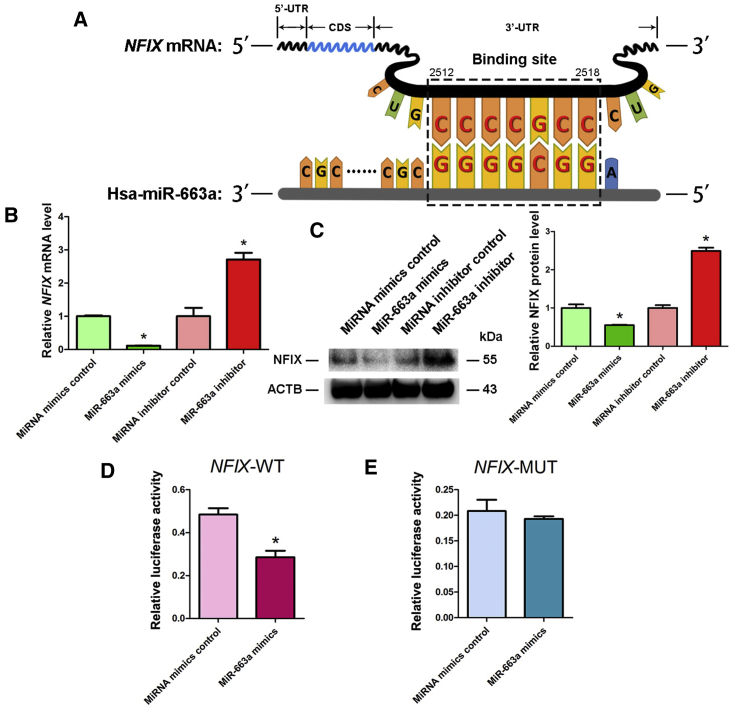
NFIX Was Directly Targeted by MiR-663a in the Human SSC Line
We further identified the targets of miR-663a since miRNAs act as epigenetic regulators by binding with the 3′ UTR of relative mRNAs. Using miRNA predict software, namely Targetscan and miRDB, we predicted that transcription factor NFIX was a potential binding target of miR-663a. As shown in Figure 6A, the seed region (the second to eighth nucleotides) of miR-663a could bind with the target 3′ UTR sequence of NFIX mRNA. Real-time qPCR showed that the expression levels of NFIX were downregulated by miR-663a mimics but increased by miR-663a inhibitor (Figure 6B). Simultaneously, Western blots demonstrated that the protein of NFIX was significantly repressed by miR-663a mimics but enhanced by miR-663a inhibitor (Figure 6C). Additionally, we conducted dual luciferase assays to further confirm the binding cite of NFIX mRNA. The predicted sequence in 3′ UTR of NFIX mRNA reduced luciferase activity of the fusion genes in response to the treatment of miR-663a mimics (Figure 6D). In contrast, mutated target sequences failed to exert any effect on the luciferase activity (Figure 6E). Considered together, these data implicate NFIX as a direct target of miR-663a in human SSCs.
Figure 6.
Identification and Verification of the Target NFIX of MiR-663a in Human SSCs
(A) Schematic diagram illustrated the binding site of miR-663a to NFIX mRNA. (B) Real-time qPCR demonstrated NFIX expression changes in human SSCs at day 2 after transfection of miRNA mimics control, miR-663a mimics, miRNA inhibitor control, and miR-663a inhibitor. (C) Western blots depicted level of NFIX protein in human SSCs at day 3 after transfection of miRNA mimics control, miR-663a mimics, miRNA inhibitor control, and miR-663a inhibitor (left panel). ACTB served as the control of loading proteins. The relative expression levels of NFIX protein in human SSCs at day 3 after transfection of miR-663a mimics to miRNA mimics control and miR-663a inhibitor to miRNA inhibitor control after normalization to the signals of their loading control (right panel). (D) Validation of the targeting of miR-663a to NFIX by dual luciferase reporter assays. (E) Validation of the binding of miR-663a to mutated NFIX by dual luciferase reporter assays.*Statistically significant differences (p < 0.05) between miR-663a mimics- or inhibitor-treated group and the corresponding control.
NFIX Silencing Stimulates the Proliferation and Suppresses the Early Apoptosis of Human SSCs
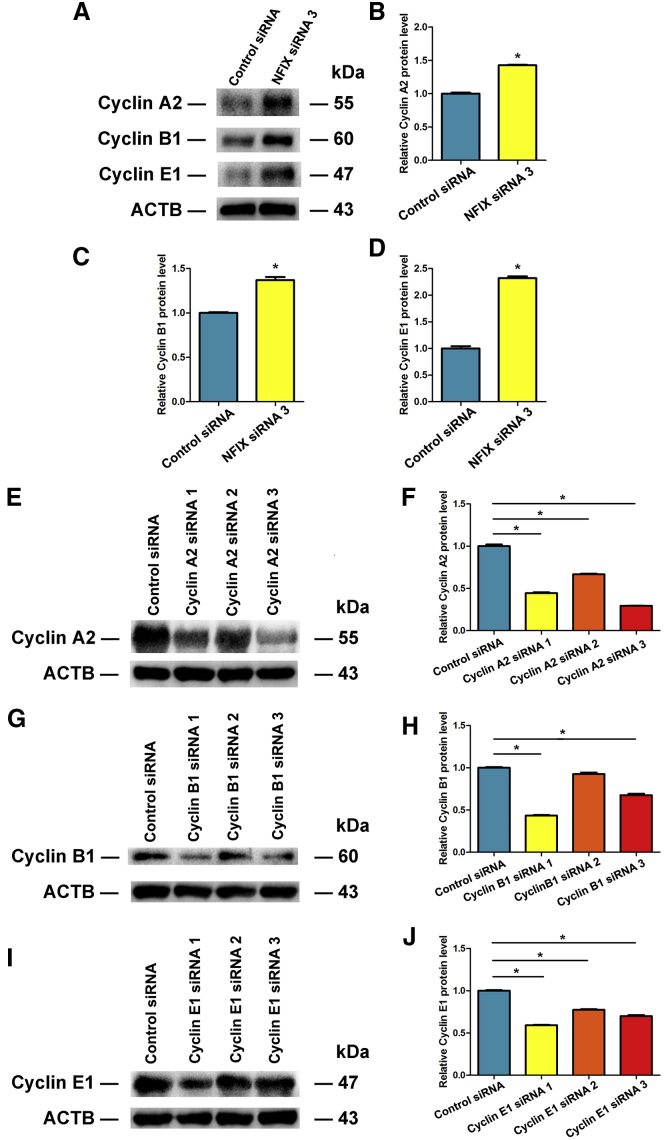
We next probed the influence of NFIX on the proliferation and apoptosis of the human SSC line. RNAi was used to silence NFIX in the human SSC line. Three pairs of NFIX small interfering RNAs (siRNAs) (i.e., NFIX siRNA 1, NFIX siRNA 2, and NFIX siRNA 3) with different binding sites were employed to obtain sequence-specific siRNAs of NFIX. The transfection efficiency of NFIX siRNAs in the human SSC line was over 80%, as evidenced by the transfection of FAM-labeled fluorescent oligo (Figures 3A and 3B). Expression of NFIX was examined by real-time qPCR and Western blots. All three NFIX siRNAs specifically knocked down the NFIX level, and NFIX siRNA 3 assumed the highest efficiency of NFIX silencing (Figure 7A). Western blots indicated that NFIX siRNAs 2 and 3 were more effective in reducing protein level of NFIX compared with NFIX siRNA 1 (Figure 7B).
Figure 7.
Influence of NFIX Silencing on the Proliferation, DNA Synthesis, and Apoptosis of Human SSCs
(A) Real-time qPCR showed changes of NFIX mRNA by NFIX siRNA 1, NFIX siRNA 2, and NFIX siRNA 3 in the human SSC line. (B) Western blots revealed changes of NFIX protein by NFIX siRNA 1, NFIX siRNA 2, and NFIX siRNA 3 in human SSCs. *Statistically significant differences (p < 0.05) of NFIX siRNA 1-, 2-, and 3-treated cells compared with the control siRNA. (C–E) EDU incorporation assay demonstrated the percentages of EDU-positive cells affected by control siRNA (C and E) and NFIX siRNA 3 (D and E) in human SSCs. Scale bars, 50 μm (C and D). *Statistically significant differences (p < 0.05) between NFIX siRNA 3-treated cells and the control siRNA. (F) CCK-8 assays showed the growth curve of human SSCs treated with control siRNA and NFIX siRNA 3 for 5 days. *Statistically significant differences (p < 0.05) between control siRNA and NFIX siRNA 3. (G and H) Western blots illustrated the changes of PCNA protein in human SSCs at day 3 after transfection of control siRNA, NFIX siRNA 1, NFIX siRNA 2, and NFIX siRNA 3. ACTB served as the control of the loading proteins. The relative protein level of PCNA in human SSCs at day 3 after transfection of NFIX siRNA 1, NFIX siRNA 2, and NFIX siRNA 3 to control siRNA through normalization to the signals of their loading control was shown. *Statistically significant differences (p < 0.05) between NFIX siRNA-treated groups and the control siRNA. (I–K) APC Annexin V and PI staining and flow cytometry depicted the percentages of early apoptosis in human SSCs transfected with control siRNA (I and K) and NFIX siRNA 3 (J and K). *Statistically significant differences (p < 0.05) between control siRNA and NFIX siRNA 3.
Treatment of NFIX siRNA 3 from 24 to 120 hr led to enhancement in the proliferation of the human SSC line (Figure 7F). As shown in Figures 7D and 7E, the percentage of EDU-positive cells in S phase with DNA replication was obviously increased by NFIX siRNA 3 compared with control siRNA. The level of PCNA protein was enhanced by three NFIX siRNAs, particularly by NFIX siRNA 3 in the human SSC line (Figures 7G and 7H).
Annexin V and PI staining and flow cytometry assay illustrated that knockdown of NFIX reduced the early apoptosis of the human SSC line (Figures 7I–7K). Considered together, NFIX silencing significantly promotes the division and inhibits early apoptosis of human SSCs, which is consistent with the effect of miR-663a mimics.
The Synergetic Effect of MiR-663a and NFIX on DNA Synthesis, Proliferation, and Apoptosis of Human SSCs
We checked whether NFIX had a synergetic effect with miR-663a on human SSCs. As shown in Figures 8A–8D, the decline in the percentages of EDU+ cells by miR-663a inhibitor was counteracted by NFIX siRNA 3 in the human SSC line. Similarly, the decrease in the proliferation of the human SSC line caused by miR-663a inhibitor was neutralized by NFIX siRNA 3 at day 4 and day 5 (Figure 8E).
Figure 8.
The Effects of MiR-663a Inhibitor and NFIX Silencing on DNA Synthesis and Proliferation of Human SSCs
(A–D) EDU incorporation assay showed the percentages of EDU+ cells in the human SSC line treated with miRNA inhibitor control (A and D), miR-663a inhibitor (B and D), as well as miR-663a inhibitor and NFIX siRNA 3 (C and D). Scale bars, 50 μm (A–C). (E) CCK-8 assay displayed the proliferation of human SSC line treated with miRNA inhibitor control, miR-663a inhibitor, and miR-663a inhibitor and NFIX siRNA 3 for 5 days. *Statistically significant differences (p < 0.05).
Furthermore, the percentage of terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate-biotin nick end-labeling (TUNEL)+ cells was decreased in human SSCs by miR-663a mimics compared to miRNA mimics control (Figures 9A, 9B, and 9F). In contrast, miR-663a inhibitor led to an enhancement of TUNEL+ cells in human SSCs compared with miRNA inhibitor control (Figures 9C, 9D, and 9G). NFIX siRNA 3 resulted in the reduction in the TUNEL+ cells of human SSCs (Figures 9H and 9I). Notably, the increase in TUNEL+ cells by miR-663a inhibitor was also neutralized by NFIX siRNA 3 (Figures 9C–9E and 9G). Taken together, these results indicate that NFIX is a target of miR-663a in regulating human SSCs.
Figure 9.
The Effects of MiR-663a and NFIX Silencing on Apoptosis of Human SSCs
(A–I) TUNEL assay revealed the percentages of TUNEL+ cells in the human SSC line treated with miRNA mimics control (A and F), miR-663a mimics (B and F), miRNA inhibitor control (C and G), miR-663a inhibitor (D and G), as well as miR-663a inhibitor and NFIX siRNA 3 (E and G), control siRNA (H and I), and NFIX siRNA 3 (H and I). Scale bars, 50 μm (A–E and H). (E) *Statistically significant differences (p < 0.05).
MiR-663a Upregulates the Levels of Cell Cycle Proteins Cyclin A2, Cyclin B1, and Cyclin E1, but Not CDK2
We asked whether miR-663a changed the levels of cell cycle proteins in fate determination of the human SSC line. The progression of cell cycle is modulated by diverse cyclins and cyclin-dependent kinases (CDKs).35 Here we examined several cell cycle regulators, including Cyclin A2, Cyclin B1, Cyclin E1, and CDK2, in the human SSC line after transfection of miR-663a mimic and inhibitor. Western blots demonstrated that miR-663a mimics significantly elevated the expression levels of Cyclin A2 (Figures 10A and 10B), Cyclin B1 (Figures 10A and 10C), and Cyclin E1 (Figures 10A and 10D), whereas miR-663a inhibitor decreased the protein levels of the three cell cycle proteins (Figures 10A–10D). Additionally, no statistically significant change of CDK2 was observed in the human SSC line with treatment of miR-663a mimics, inhibitor, or control. Therefore, miR-663a upregulates the levels of Cyclin A2, Cyclin B1, and Cyclin E1 rather than CDK2.
Figure 10.
The Effect of MiR-663a on Levels of Cell Cycle Regulators Cyclin A2, Cyclin B1, Cyclin E1, and CDK2 in Human SSCs
(A) Western blots showed the changes of Cyclin A2, Cyclin B1, Cyclin E1, and CDK2 proteins in human SSCs at day 3 after transfection of miRNA mimics control, miR-663a mimics, miRNA inhibitor control, and miR-663a inhibitor. ACTB served as the control of the loading proteins. (B–E) The relative levels of Cyclin A2 (B), Cyclin B1 (C), Cyclin E1 (D), and CDK2 (E) proteins in human SSCs at day 3 after transfection of miR-663a mimics to miRNA mimics control and miR-663a inhibitor to miRNA inhibitor control through normalization to the signals of their loading control. *Statistically significant differences (p < 0.05) between miR-663a mimics- or inhibitor-treated group and the corresponding control.
NFIX Silencing Enhances the Levels of Cyclin A2, Cyclin B1, and Cyclin E1, and siRNAs against These Cell Cycle Genes Effectively Decrease Their Expression in Human SSCs
We also checked if NFIX silencing modified the levels of cell cycle proteins of the human SSC line. Western blots showed that NFIX siRNA 3 led to the enhancement in the expression levels of Cyclin A2 (Figures 11A and 11B), Cyclin B1 (Figures 11A and 11C), and Cyclin E1 (Figures 11A and 11D).
Figure 11.
The Influence of NFIX and Cell Cycle Genes’ Silencing on the Levels of Cyclin A2, Cyclin B1, and Cyclin E1 in Human SSCs
(A–D) Western blots illustrate the changes of Cyclin A2 (A and B), Cyclin B1 (A and C), and Cyclin E1 (A and D) in the human SSC line treated with control siRNA and NFIX siRNA. (E–J) Western blots show the modification of Cyclin A2 (E and F), Cyclin B1 (G and H), and Cyclin E1 (I and J) in the human SSC line treated with control siRNA and the siRNAs against cell cycle genes. *Statistically significant differences (p < 0.05).
We designed three siRNAs targeting different sequences of genes, namely Cyclin A2, Cyclin B1, and Cyclin E1. Western blots revealed that the levels of Cyclin A2 (Figures 11E and 11F), Cyclin B1 (Figures 11G and 11H), and Cyclin E1 (Figures 11I and 11J) proteins were significantly reduced by siRNAs against these genes.
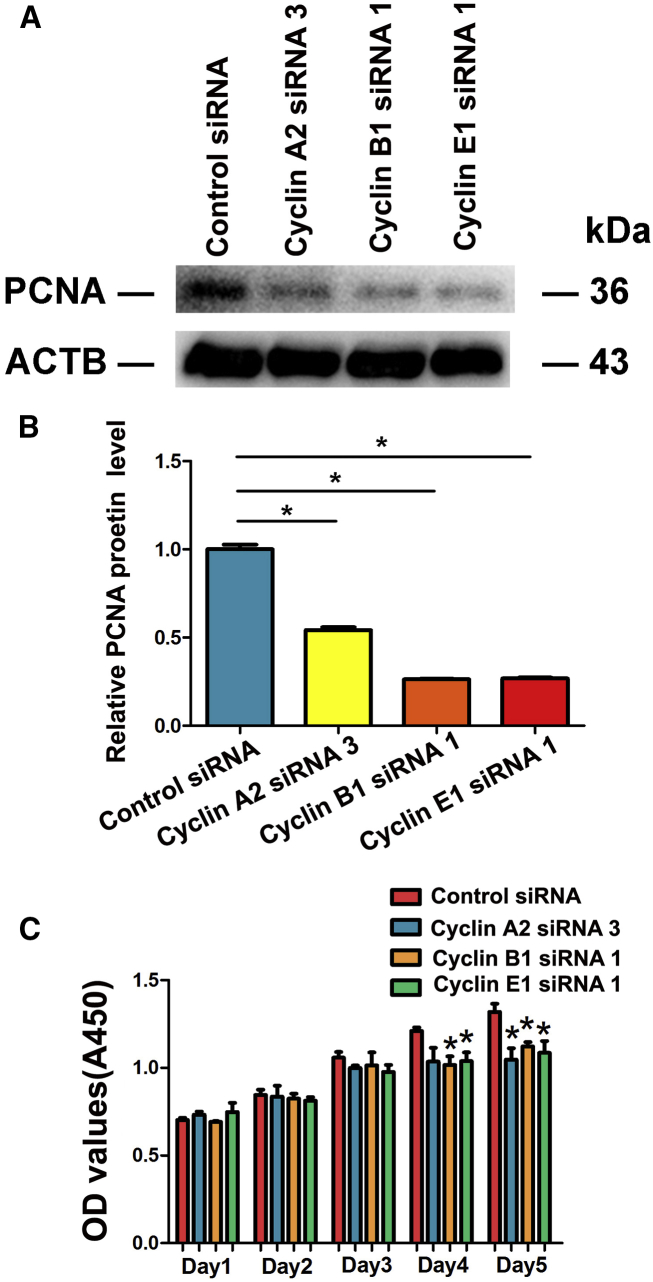
Cyclin A2, Cyclin B1, and Cyclin E1 Knockdown Results in the Decrease in Proliferation of Human SSCs
We finally explored the influence of Cyclin A2, Cyclin B1, and Cyclin E1 silencing on the proliferation of human SSCs. Western blots revealed that Cyclin A2, Cyclin B1, and Cyclin E1 siRNAs led to a significant decrease in the levels of PCNA, a hallmark for cell proliferation, in the human SSC line (Figures 12A and 12B). CCK-8 assay further demonstrated that cell growth was significantly decreased by Cyclin B1 and Cyclin E1 siRNAs in the human SSC line at day 4 and day 5 (Figure 12C).
Figure 12.
The Effect of Cyclin A2, Cyclin B1, and Cyclin E1 Knockdown on the Proliferation of Human SSCs
(A and B) Western blots show the level of PCNA in the human SSC line with treatment of Cyclin A2, Cyclin B1, and Cyclin E1 siRNAs. (C) CCK-8 assay displayed the cell number of the human SSC line affected by Cyclin A2, Cyclin B1, and Cyclin E1 siRNAs in the human SSC line for 5 days. *Statistically significant differences (p < 0.05).
Discussion
Mammalian SSCs share the potential of differentiating into various kinds of male germ cells, and a sufficient population maintained by the proliferation of SSCs determines the capacity of mammalian fertility. Therefore, the studies on the proliferation and mechanisms of SSCs are of great significance for preservation and reestablishment of male reproduction. Although much progress has been achieved in uncovering the mechanisms underlying the spermatogenesis in rodents, epigenetic and genetic regulation of male germ cells in humans remains largely unknown. We have previously revealed a large scale of differentially expressed miRNAs between human spermatogonia and pachytene spermatocytes.31 Among the differentially expressed miRNAs, we found that miR-663a was expressed at a higher level in human spermatogonia compared to pachytene spermatocytes, as shown by miRNA microarray and real-time qPCR. It has been reported that miR-663a suppresses the proliferation and invasion by targeting JunD in non-small-cell lung cancer cells,36 and miR-663a may be involved in chordoma development.37 However, it remains unclear about the role and molecular mechanism of miR-663a in regulating human SSCs.
We have recently established a human SSC line with an unlimited proliferation potential and high safety.30 Notably, this cell line assumes the phenotype of human primary SSCs, since they express numerous markers of SSCs and male germ cells, including THY1, RET, GPR125, PLZF, MAGEA4, VASA, UCHL1, and GFRA1, which has been verified in this study. This human SSC line possesses stronger proliferation ability than primary human SSCs, and it can colonize and proliferate in the recipient mice, a characteristic of primary human SSCs in vivo. Due to scarce human testicular tissues, this SSC line emerges as an excellent and important cell resource for basic research of SSCs. In this study, cell proliferation and apoptosis assays were conducted by transfecting miRNA mimics and inhibitor oligonucleotides into the human SSC line with a transfection efficiency of over 80%. We have demonstrated that miR-663a stimulates the proliferation and DNA synthesis of human SSCs, as indicated by CCK-8 assay, EDU incorporation assay, and PCNA expression. We have also revealed that miR-663a inhibits the early apoptosis of human SSCs, as evidenced by Annexin V and PI staining and flow cytometry as well as by TUNEL assay.
NFIX is a member of the nuclear factor I (NFI) family of transcription factors. The NFI genes are dual-directional factors that act as both positive and negative transcriptional regulators of cellular gene expression. There are four NFI members, namely NFIA, NFIB, NFIC, and NFIX, and they are expressed in various kinds of organs and exert different functions in a cell type-dependent manner.38 The NFI genes are highly evolutionarily conserved in higher eukaryotes, especially in vertebrates.39 Like other mRNAs, NFIX comprises a short 5′ UTR, which codes sequence (CDS), and a huge 3′ UTR that is the target of different miRNAs. The level of NFIX is obviously upregulated in neural stem cells (NSCs) at the transition from proliferation to quiescence, and it is most abundant in quiescent NSCs.40 It has been reported that Nfix plays a role in inducing the quiescence of the NSCs in vitro and controls the cell adhesion properties of NSCs in vivo.40 In addition, NFIX has been found to regulate the metastasis in lung cancer by transcribing the genes, e.g., IL6ST, TIMP1, and ITGB1, and it promotes the inflammation, proliferation, migration, and invasion of cancer cells.41 It has been suggested that NFIX is related to overgrowth syndrome and Marshall-Smith syndrome, and it promotes the commitment of radial glia into intermediate progenitor cells in neuron generation during forebrain development.42, 43, 44
Predicted by Targetscan and miRDB software, a panel of potential miR-663a targets was selected and classified by functional annotation clustering. Fortunately, we identified NFIX as a potential target of miR-663a. Subsequently, our real-time qPCR and western blots validated the inverse level of NFIX in response to miR-663a mimics and inhibitor, indicating that NFIX is a direct target of miR-663a in human SSCs. Dual luciferase reporter assays further confirmed the targeting of miR-663a to NFIX. Notably, NFIX silencing in the human SSC line stimulated the propagation and suppressed the early apoptosis of the human SSC line, which is consistent with the effect of miR-663a overexpression. Significantly, we found that NFIX silencing could neutralize the influence of the miR-663a inhibitor on DNA synthesis, proliferation, and apoptosis of human SSCs. Considered together, NFIX has been identified as a direct target of miR-663a in human SSCs.
We finally checked the level changes of cell cycle regulators, including Cyclin A2, Cyclin B1, Cyclin E1, and CDK2, for a better understanding of the molecular mechanism of miR-663a in human SSCs. Cyclin A2 is expressed in the male germline in mitotic spermatogonia and preleptotene spermatocytes, and it drives the progression through S and G2/M phases of cell cycle.45 Cyclin B1 is expressed in both mitotic and meiotic cells, and it plays an indispensable role in the G2/M phase.45, 46 It has been reported that E-type cyclins (Cyclin E1 and E2) are functional redundancy regulators. Cyclin E is involved in G1 progression and G1/S transition, and the absence of Cyclin E has been shown to be substituted by other cyclins in the regulation of cell proliferation.47, 48 Additionally, Cdk2 is dispensable for the mitotic cell cycle, while Cdk1 and Cdk4 can replace the Cdk2.49, 50 Utilizing miR-663a mimics and inhibitor, we found that the levels of Cyclin A2, Cyclin B1, and Cyclin E1 were upregulated by miR-663a mimics in the human SSC line, whereas their levels were reduced in response to miR-663a inhibitor. There was no statistically significant difference in the level of CDK2 among miR-663a mimics, miR-663a inhibitor, and the control in the human SSC line. Taken together, these results implicate that miR-663a regulates Cyclin A2, Cyclin B1, and Cyclin E1 in human SSCs. Furthermore, we revealed that NFIX negatively controlled Cyclin A2, Cyclin B1, and Cyclin E1 in human SSCs and that knockdown of Cyclin A2, Cyclin B1, and Cyclin E1 led to the decrease in proliferation of human SSCs.
In summary, we have demonstrated for the first time that miR-663a promotes the proliferation and DNA synthesis and suppresses the early apoptosis of human SSCs. We have also identified NFIX as a direct target of miR-663a in human SSCs. Furthermore, we have observed that miR-663a mediates cell cycle proteins Cyclin A2, Cyclin B1, and Cyclin E1 rather than CDK2 in human SSCs. Collectively, miR-663a facilitates the division and inhibits the early apoptosis of human SSCs via targeting NFIX and controlling cell cycle proteins Cyclin A2, Cyclin B1, and Cyclin E1 in human SSCs. This study thus sheds novel insights into epigenetic regulation of human SSCs, and it might provide new molecular targets for treating male infertility and other human diseases.
Materials and Methods
Procurement of Testicular Tissues from OA Patients with Normal Spermatogenesis
Testicular tissues were obtained from OA patients who underwent microdissection of testicular biopsy and testicular sperm extraction at Ren Ji Hospital affiliated with Shanghai Jiao Tong University School of Medicine. These OA cases were caused by inflammation and vasoligation, and normal spermatogenesis with sperm production was observed in their testes. This study was approved by The Institutional Ethical Review Committee of Ren Ji Hospital (license number of ethics statement: 2012-01), Shanghai Jiao Tong University School of Medicine. Informed consents for testicular biopsies were obtained from the donors for research only.
Isolation of Human Spermatogonia and Pachytene Spermatocytes by STA-PUT Velocity Sedimentation
Testicular biopsies from OA patients were pooled and washed three times aseptically in DMEM containing antibiotics with penicillin and streptomycin (Gibco). Human male germ cells were separated from testicular tissues of OA patients via a two-step enzymatic digestion and followed by differential plating, as previously described.27 Seminiferous tubules were isolated from human testis tissues by the first enzymatic digestion comprising 2 mg/mL collagenase IV (Gibco) and 1 mg/mL DNase I (Gibco) in 34°C water bath for 15 min. After sedimentation, human male germ cells, Sertoli cells, and myoid cells were obtained from seminiferous tubules using a second enzymatic digestion with 4 mg/mL collagenase IV, 2.5 mg/mL hyaluronidase (Sigma), 2 mg/mL trypsin (Sigma), and 1 mg/mL DNase I. Differential plating of the mixed cells was performed in a 10-cm-diameter cell culture dish in DMEM/F12 supplemented with 10% fetal bovine serum (FBS) (Gibco) at 34°C in 5% CO2 for 1 day. Sertoli cells and myoid cells attached to the culture plates after incubating, whereas male germ cells remained in suspension and were collected by centrifuging at 1,000 rpm for 5 min.
A linear BSA gradient and velocity sedimentation was employed using the STA-PUT method to further separate human spermatogonia and pachytene spermatocytes from male germ cells in terms of their sizes, mass, and gravities. Approximately 3 × 106 male germ cells were re-suspended in 15 mL 0.5% sterile BSA solution, followed by filtering through a 70-μm mesh to remove cell aggregates. The STA-PUT apparatus included two gradient glass chambers, namely, cell loading chamber and standard sedimentation chamber, plastic tubing, and baffles (ProScience Glass Shop Division, Scarborough, ON, Canada). In total, 15 mL cell suspension was loaded into the loading chamber, and the stirrer under the tube started to agitate and allow the cells to move into the sedimentation chamber (∼5 min). The stirrer under the 2% BSA solution started to work, and artery clips were removed so that both 2% and 4% BSA solutions were introduced into the chamber (∼10 min). A gradient of BSA was formed after 2.5 hr of sedimentation in the standard cell sedimentation chamber. Centrifuge tubes of 15 mL were used to collect cell fractions and designated from 1 to 40, respectively. After centrifuging at 1,000 rpm for 5 min, cells in each tube were re-suspended in 1 mL cold PBS. Each fraction was observed carefully under a phase-contrast microscope to assess cellular integrity and identify cell types. Fractions of similar cell size and morphology were pooled together and centrifuged at 1,000 rpm for 5 min for subsequent analyses by real-time qPCR, RT-PCR, and immunocytochemistry.
RNA Extraction and RT-PCR
Total RNA was extracted from human spermatogonia, pachytene spermatocytes, testicular tissues of OA patients, and the human SSC line, respectively, using the RNAiso Plus reagent (Takara, Kusatsu, Japan). The concentrations of total RNA were determined by Nanodrop (Thermo Scientific, MA, USA). RNA with the ratio of A260/A280 = 1.9–2.0 was utilized for RT-PCR. RT was performed using the First Strand cDNA Synthesis Kit (Thermo Scientific, USA), and PCR of the cDNA was carried out according to the protocol described previously.1 The primers of the chosen genes, including GPR125 (G protein-coupled receptor 125), GFRA1 (GDNF family receptor alpha 1), UCHL1 (Ubiquitin C-terminal hydrolase L1), THY1, SYCP3 (Synaptonemal complex protein 3), MLH1, CREST, RET, PLZF, MEGEA4, VASA, and GAPDH (Glyceraldehyde-3-phosphate dehydrogenase), were designed and listed in Table 2. The PCR reaction started at 95°C for 5 min and was performed as follows: denaturation at 95°C for 30 s, annealing at 52°C–60°C as indicated in Table 2 for 30 s, elongation at 72°C for 45 s, for 35 cycles. The samples were incubated for an additional 5 min at 72°C. PCR products were separated by electrophoresis on 2% agarose gels, which were stained with Safer Ethidium Bromide Alternatives-GelGreen (Biotium, USA). Images were recorded by Image Analyzer ChemiDoc XRS+ (Bio-Rad). Samples without cDNA (no cDNA) but with PCR of gene primers served as negative controls.
Table 2.
The Sequences of Gene Primers Used for RT-PCR
| Genes | Primer Sequences | Product Size (bp) | Tm (°C) | |
|---|---|---|---|---|
| GPR125 | forward | CGGGATACCAGGTGTGTTTATC | 557 | 60 |
| reverse | TTTCTCTCATCCTGTGGGTTTC | |||
| forward | GCGTCATTACGGTCTTTGGAA | 199 | 61 | |
| reverse | ACGGCAATTCAAGCGGAGG | |||
| GFRA1 | forward | GGACTCCTGCAAGACGAATTA | 543 | 62 |
| reverse | GAAGCACCGAGACCTTCTTT | |||
| forward | GACTCCTGCAAGACGAATTACA | 87 | 61 | |
| reverse | GCTGCTGACAGACCTTGACT | |||
| UCHL1 | forward | AGCTGAAGGGACAAGAAGTTAG | 265 | 62 |
| reverse | TTGTCATCTACCCGACATTGG | |||
| THY1 | forward | CAGAAGGTGACCAGCCTAAC | 233 | 62 |
| reverse | TTGCTAGTGAAGGCGGATAAG | |||
| SYCP3 | forward | TGCAGAAAGCTGAGGAACAA | 247 | 58 |
| reverse | TGCTGCTGAGTTTCCATCAT | |||
| MLH1 | forward | TGAGGAAGGGAACCTGATTG | 245 | 60 |
| reverse | TCCAGGAGTTTGGAATGGAG | |||
| CREST | forward | CGCAGCAGCAGACGTACTC | 204 | 60 |
| reverse | GGCCCTGTTCATAGCCGTAG | |||
| RET | forward | CTCGTTCATCGGGACTTG | 126 | 60 |
| reverse | ACCCTGGCTCCTCTTCAC | |||
| PLZF | forward | CGGTTCCTGGATAGTTTGC | 317 | 60 |
| reverse | GGGTGGTCGCCTGTATGT | |||
| MAGEA4 | forward | CTTACCCACTACCATCAGCTTC | 212 | 62 |
| reverse | TGATGACTCTCTCCAGCATTTC | |||
| VASA | forward | ACTGGTCGTTGTGGGAATAC | 255 | 62 |
| reverse | GGGAGCTCGTGAAGAAGAAA | |||
| GAPDH | forward | AATCCCATCACCATCTTCC | 382 | 55 |
| reverse | CATCACGCCACAGTTTCC | |||
Real-Time qPCR
RNA was extracted from human spermatogonia; pachytene spermatocytes; and human SSCs without or with treatment of miR-663a mimics, miR-663a inhibitor, miRNA mimic control, miRNA inhibitor control, NFIX siRNAs, or siRNA control using the RNAiso Plus reagent (Takara, Kusatsu, Japan). The concentrations of total RNA were measured by Nanodrop (Thermo Scientific, MA, USA), and the ratios of A260/A280 of total RNA were set as 1.9–2.0 to ensure good quality. For miRNA real-time PCR, RT reaction was performed using TransScript miRNA First-Strand cDNA Synthesis SuperMix Kit (Transgene). Each RT reaction was composed of 100 ng RNA, 1 μL miRNA RT Enzyme Mix, 10 μL 2× TS miRNA Reaction Mix, and RNase-free water in a total volume of 20 μL. Reactions were performed in a Veriti 96-Well Thermal Cycler (Applied Biosystems) for 60 min at 37°C and followed by heat inactivation of RT for 5 s at 85°C. RT reaction mix was diluted by 5 times in nuclease-free water and held at −20°C. Primer sequences of miRNAs used for real-time PCR are listed in Table 3. Real-time PCR was performed in triplicate using Power SYBR Green PCR Master Mix (Applied Biosystems, Woolston Warrington, UK) and a 7500 Fast Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA), according to the protocol described previously.51 Melting curve analysis was performed to validate the specific generation of the expected PCR products. The expression levels of miRNAs were normalized to U6 and calculated using the 2−ΔΔCt method.51
Table 3.
The Sequences of Gene and miRNA Primers Used for Real-Time PCR
| Genes | Primer Sequences (5′ to 3′) | Tm (°C) | |
|---|---|---|---|
| Has-miR-663a | AGGCGGGGCGCCGCGGGACCGC | 60 | |
| U6 | CGCTTCGGCAGCACATATAC | 60 | |
| NFIX | forward | CGGCTCTACAAGTCGCCTC | 60 |
| reverse | GCAGTGGTTTGATGTCCGC | ||
| GAPDH | forward | CAGGAGGCATTGCTGATGAT | 60 |
| reverse | GAAGGCTGGGGCTCATTT | ||
Real-time PCR was also carried out to evaluate the expression of NFIX in freshly isolated human spermatogonia and pachytene spermatocytes from OA patients or human SSCs with treatment of miR-663a mimics, miR-663a inhibitor, miRNA mimic control, miRNA inhibitor control, NFIX-siRNAs, or siRNA control pursuant to the method described previously.51 The primers of these genes were designed and are listed in Table 3, and their expression levels were normalized to GAPDH and calculated using the 2−ΔΔCt method.51
Immunocytochemistry
For immunocytochemical staining, human spermatogonia, pachytene spermatocytes, and human SSCs were fixed with 4% paraformaldehyde (PFA) for 30 min, washed three times with cold PBS (Medicago, Uppsala, Sweden), and permeabilized in 0.5% Triton X-100 (Sigma) for 15 min. After washing with PBS, the cells were blocked in 2% BSA for 60 min, followed by incubation with primary antibodies (the detailed information is described in Table 4) overnight at 4°C. Replacement of primary antibodies with isotype IgGs served as negative controls. After extensive washes with PBS for 30 min, the cells were incubated with the secondary antibody IgG (Sigma) conjugated with fluorescein isothiocyanate (FITC) or rhodamine at a 1:200 dilution for 1 hr at room temperature. DAPI was used to label the nuclei, and images were captured with a Nikon fluorescence microscope (Tokyo, Japan).
Table 4.
Information on Primary Antibodies Used for Immunocytochemistry and Western Blots
| Antibodies | Vendors | Sources | Working Dilutions |
|---|---|---|---|
| GFRA1 | R&D Systems | mouse | 1:500 |
| GPR125 | Abcam | rabbit | 1:500 |
| UCHL1 | Bio-Rad | mouse | 1:500 |
| THY1 | Abcam | rabbit | 1:500 |
| SYCP3 | Abcam | rabbit | 1:100 |
| MLH1 | Abcam | mouse | 1:50 |
| CREST | Immunovision | human | 1:80 |
| VASA | Santa Cruz Biotechnology | rabbit | 1:200 |
| SV40 | Santa Cruz Biotechnology | mouse | 1:200 |
| NFIX | Novus | rabbit | 1:500 |
| PCNA | CST | rabbit | 1:1,000 |
| Cyclin A2 | Santa Cruz Biotechnology | rabbit | 1:200 |
| Cyclin B1 | Santa Cruz Biotechnology | rabbit | 1:200 |
| Cyclin E1 | Santa Cruz Biotechnology | mouse | 1:200 |
| CDK2 | Santa Cruz Biotechnology | rabbit | 1:200 |
| ACTB | Proteintech | mouse | 1:2,000 |
For the detection of MAGEA4, peroxidase-conjugated goat anti-mouse IgG (Envision detection kit, Dako) was used as the secondary antibody, and the immunostaining was examined under a light microscope.
Meiotic Spread Assays
Meiotic spread assays were performed to determine the identity of pachytene spermatocytes isolated from OA patients according to a method described previously.31 Cells were lysed by a hypotonic solution and spread evenly over slides. The cells were fixed with 1% PFA and permeabilized in 0.15% Triton X-100. Slides were dried for 24 hr at room temperature in a humid chamber. The cells were treated with 0.04% photoflo for 5 min and blocked with 4% goat serum. Triple immunostaining was performed in the cells by incubation with primary antibodies, including SYCP3 (Abcam, ab15093, 1:100), CREST (Immunovision, HCT-0100, 1:100), and MLH1 (Abcam, ab14206, 1: 50), overnight at 37°C in a humid chamber. Goat anti-rabbit Alexa Fluor 555 (Invitrogen), AMCA-AffiniPure donkey anti-human IgG, and goat anti-mouse Alexa 488 secondary antibodies (Jackson ImmunoResearch Laboratories) were used as the secondary antibodies at 1:1,000 dilution and incubated for 90 min at 37°C. Cells were washed three times with PBS, and images were captured with a fluorescence microscope (Nikon, Tokyo, Japan).
Transfection of MiR-663a Mimics, MiR-663a Inhibitors, and NFIX siRNAs into the Human SSC Line
The miR-663a mimics and inhibitors were purchased from GenePharma (Shanghai, China). The oligonucleotides of miRNA mimics and inhibitors are shown in Table 5. Human SSCs were seeded at 1 × 105/cm2 density and cultured in DMEM/F12 supplemented with 10% FBS overnight.
Table 5.
The Sequences of MiR-663a Mimics, MiR-663a Inhibitor, NFIX siRNA Oligonucleotides, and siRNAs against Cell Cycle Genes
| miRNAs and siRNAs | Sequence (5′ to 3′) | |
|---|---|---|
| Hsa-miR-663a mimics | sense | AGGCGGGGCGCCGCGGGACCGC |
| antisense | GGUCCCGCGGCGCCCCGCCUUU | |
| miRNA mimics control | sense | UUCUCCGAACGUGUCACGUTT |
| antisense | ACGUGACACGUUCGGAGAATT | |
| Hsa-miR-663a inhibitor | GCGGUCCCGCGGCGCCCCGCCU | |
| miRNA inhibitor control | CAGUACUUUUGUGUAGUACAA | |
| NFIX siRNA 1 | sense | GGAAUCCGGACAAUCAGAUTT |
| antisense | AUCUGAUUGUCCGGAUUCCTT | |
| NFIX siRNA 2 | sense | CUCUAAAGAAGUCAGGAAATT |
| antisense | UUUCCUGACUUCUUUAGAGTT | |
| NFIX siRNA 3 | sense | CCAUCACCUUCAUUCGCAATT |
| antisense | UUGCGAAUGAAGGUGAUGGTT | |
| Cyclin A2 siRNA 1 | sense | CACCAUUCAUGUGGAUGAATT |
| antisense | UUCAUCCACAUGAAUGGUGTT | |
| Cyclin A2 siRNA 2 | sense | GGAGGUUAAAUGUAAACCUTT |
| antisense | AGGUUUACAUUUAACCUCCTT | |
| Cyclin A2 siRNA 3 | sense | GUAGCAGAGUUUGUGUACATT |
| antisense | UGUACACAAACUCUGCUACTT | |
| Cyclin B1 siRNA 1 | sense | CCAAGCCCAAUGGAAACAUTT |
| antisense | AUGUUUCCAUUGGGCUUGGTT | |
| Cyclin B1 siRNA 2 | sense | CUGGCUAGUACAGGUUCAATT |
| antisense | UUGAACCUGUACUAGCCAGTT | |
| Cyclin B1 siRNA 3 | sense | GCCAUGUUUAUUGCAAGCATT |
| antisense | UGCUUGCAAUAAACAUGGCTT | |
| Cyclin E1 siRNA 1 | sense | CCGGUAUAUGGCGACACAATT |
| antisense | UUGUGUCGCCAUAUACCGGTT | |
| Cyclin E1 siRNA 2 | sense | GCUUGUUCAGGAGAUGAAATT |
| antisense | UUUCAUCUCCUGAACAAGCTT | |
| Cyclin E1 siRNA 3 | sense | CCUGGAUGUUGACUGCCUUTT |
| antisense | AAGGCAGUCAACAUCCAGGTT | |
| Control siRNA | sense | UUCUCCGAACGUGUCACGUTT |
| antisense | ACGUGACACGUUCGGAGAATT | |
| FAM-siRNA | sense | UUCUCCGAACGUGUCACGUTT |
| antisense | ACGUGACACGUUCGGAGAATT | |
Human SSCs were classified into four groups in terms of transfecting different miRNAs: (1) miRNA mimics control, (2) miR-663a mimics, (3) miRNA inhibitor control, and (4) miR-663a inhibitor. For RNAi assays, human SSCs were classified into four groups based on transfecting different siRNAs: (1) control siRNA, (2) NFIX siRNA 1, (3) NFIX siRNA 2, and (4) NFIX siRNA 3. Transfection of miRNA mimics or inhibitor and siRNAs were conducted using lipofectamine 3000 transfection agent (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s protocol. After 48 hr of culture, cells were harvested for examining the expression changes of various genes and proteins accordingly.
CCK-8 Assay
Human SSCs were seeded at a density of 2,000 cells/well in 96-well microtiter plates in DMEM/F12 supplemented with 1% FBS, and they were transfected with miRNA mimics control; miR-663a mimics; miRNA inhibitor control; miR-663a inhibitor; miR-663a inhibitor and NFIX siRNA 3; NFIX siRNA 3; Cyclin A2, Cyclin B1, and Cyclin E1 siRNAs; or control siRNA. After 5 days of culture, the proliferation potential of human SSCs was examined by CCK-8 assay (Dojin Laboratories, Kumamoto, Japan), according to the manufacturer’s instruction.
Western Blots
Human SSCs with treatment of miR-663a mimics; miRNA mimics control; miR-663a inhibitor; miRNA inhibitor control; NFIX siRNAs; Cyclin A2, Cyclin B1, and Cyclin E1 siRNAs; or siRNA control were lysed with RIPA buffer (Santa Cruz Biotechnology). After 30 min of lysis on ice, cell lysates were cleared by centrifugation at 12,000 × g for 20 min, and the concentrations of proteins were measured by BCA kit (Dingguo, China). Cell lysate (30 μg) from each sample was used for SDS-PAGE (Bio-Rad), and Western blots were conducted according to the protocol described previously.52 The detailed information of chosen antibodies was listed in Table 4. After extensive washes with tris-buffered saline and Tween 20 (TBST), the blots were detected by chemiluminescence (Chemi-Doc XRS, Bio-Rad, Hercules, CA, USA).
EDU Incorporation Assay
The human SSC line was seeded at a density of 5,000 cells/well in a 96-well plate containing DMEM/F12 medium with 50 μM EDU (RiboBio, Guangzhou, China). Human SSCs were treated with miRNA mimics control, miR-663a mimics, miRNA inhibitor control, miR-663a inhibitor, miR-663a inhibitor and NFIX siRNA 3, control siRNA, or NFIX siRNA 3. After 12 hr of culture, the cells were washed with DMEM and fixed with 4% PFA. Cells were neutralized with 2 mg/mL glycine and permeabilized in 0.5% Triton X-100 for 10 min at room temperature. EDU immunostaining was performed with Apollo staining reaction buffer. The nuclei of cells were stained with Hoechst 33342, and the EDU-positive cells were counted from at least 500 cells under fluorescence microscopy (Nikon, Tokyo, Japan).
Annexin V and PI Staining and Flow Cytometry
The percentages of early and late apoptotic cells in human SSCs transfected without or with treatment of miR-663a mimics, miR-663a inhibitor, miRNA mimics control, miRNA inhibitor control, NFIX siRNAs, or control siRNA were measured using the APC Annexin V and PI apoptosis detection kit and flow cytometry, according to the manufacturer’s instruction. Human SSC cells were seeded at a density of 5 × 104 cells/well in 12-well plates. Cells were collected by centrifuging at 1,000 rpm for 5 min and washed twice with PBS at day 3 after transfection. The cells were simultaneously stained with Annexin V-FITC (green fluorescence) and the non-vital dye PI (red fluorescence), which allowed the identification of intact cells (FITC−PI−), early apoptotic cells (FITC+PI−), and late apoptotic cells (FITC+PI+).
TUNEL Assay
Further analysis of apoptotic cells was conducted by TUNEL Apoptosis Detection Kit (Yeasen, Shanghai, China). Human SSCs were treated with miRNA mimics control, miR-663a mimics, miRNA inhibitor control, miR-663a inhibitor, miR-663a inhibitor and NFIX siRNA 3, control siRNA, or NFIX siRNA 3, according to the methods mentioned above. The cells were fixed with 4% PFA for 25 min at 4°C. After several washes, the cells were incubated with Proteinase K (20 μg/mL) and 1× DNaseI Buffer for 5 min at room temperature. Cells were then treated with 10 U/mL DNaseI for 10 min at room temperature and followed by washes in deionized water. These cells were incubated with 1× Equilibration Buffer for 30 min at room temperature and labeled by Alexa Fluor 647 in buffer premixed with TdT Enzyme for 60 min at 37°C. After being washed with PBS, the cells were finally stained with DAPI and analyzed under a fluorescence microscope (Nikon, Tokyo, Japan).
Dual Luciferase Assay
The human SSC line was seeded in a 48-well culture plate. Approximately 24 hr later, miRNA mimics were first transfected to the cells using lipofectamine 3000 transfection agent (Life Technologies, Carlsbad, CA, USA), according to the manufacturer’s protocol. Then 10 hr later, 500 ng plasmids with the binding sequence in 3′ UTRs of NFIX, Firefly Luciferase (reporter), and Renilla Luciferase (internal control) (pMIR-GIO, Genecreate, Wuhan, China) were transfected to human SSCs by lipofectamine 3000 reagent (Life Technologies, Carlsbad, CA, USA). Cells were lysed after 48 hr of transfection, and luciferase activity was measured using the tube luminometer (Berthold, Germany), according to the manufacturer’s protocol. Data were normalized to miRNA mimic control-transfected cells.
Statistical Analysis
All data were presented as mean ± SEM from at least three independent experiments and analyzed by t test using Prism (version 5, GraphPad), and p < 0.05 was considered statistically significant.
Author Contributions
F.Z., Q.Y., W.Z., M.N., H.F., Q.Q., G.M., Hong Wang, and L.W. performed the laboratory experiments and collected the data. Hongxiang Wang, M.L., and Z.L. collected the testis tissues. Z.H. and F.Z. analyzed the data and wrote the manuscript. Z.H. conceived the project, designed the experiments, and supervised the entire study.
Conflicts of Interest
The authors declare no competing financial interests.
Acknowledgments
We thank Professor Giulio C. Spagnoli, University Hospital of Basel, Switzerland, for providing an antibody to MAGEA4. This work was supported by grants from the National Nature Science Foundation of China (31671550 and 31230048), the Chinese Ministry of Science and Technology (2016YFC1000606 and 2014CB943101), the Program for Professor of Special Appointment (Eastern Scholar) at Shanghai Institutions of Higher Learning (2012.53), Shanghai Municipal Education Commission-Gaofeng Clinical Medicine Grant Support (20152511), and Shanghai Hospital Development Center (SHDC12015122).
References
- 1.Yang S., Ping P., Ma M., Li P., Tian R., Yang H., Liu Y., Gong Y., Zhang Z., Li Z., He Z. Generation of haploid spermatids with fertilization and development capacity from human spermatogonial stem cells of cryptorchid patients. Stem Cell Reports. 2014;3:663–675. doi: 10.1016/j.stemcr.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanatsu-Shinohara M., Inoue K., Lee J., Yoshimoto M., Ogonuki N., Miki H., Baba S., Kato T., Kazuki Y., Toyokuni S. Generation of pluripotent stem cells from neonatal mouse testis. Cell. 2004;119:1001–1012. doi: 10.1016/j.cell.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 3.Seandel M., James D., Shmelkov S.V., Falciatori I., Kim J., Chavala S., Scherr D.S., Zhang F., Torres R., Gale N.W. Generation of functional multipotent adult stem cells from GPR125+ germline progenitors. Nature. 2007;449:346–350. doi: 10.1038/nature06129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kossack N., Meneses J., Shefi S., Nguyen H.N., Chavez S., Nicholas C., Gromoll J., Turek P.J., Reijo-Pera R.A. Isolation and characterization of pluripotent human spermatogonial stem cell-derived cells. Stem Cells. 2009;27:138–149. doi: 10.1634/stemcells.2008-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Z., Gong Y., Guo Y., Hai Y., Yang H., Yang S., Liu Y., Ma M., Liu L., Li Z. Direct transdifferentiation of spermatogonial stem cells to morphological, phenotypic and functional hepatocyte-like cells via the ERK1/2 and Smad2/3 signaling pathways and the inactivation of cyclin A, cyclin B and cyclin E. Cell Commun. Signal. 2013;11:67. doi: 10.1186/1478-811X-11-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z., Sun M., Yuan Q., Niu M., Yao C., Hou J., Wang H., Wen L., Liu Y., Li Z., He Z. Generation of functional hepatocytes from human spermatogonial stem cells. Oncotarget. 2016;7:8879–8895. doi: 10.18632/oncotarget.7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Z., Niu M., Sun M., Yuan Q., Yao C., Hou J., Wang H., Wen L., Fu H., Zhou F. Transdifferentiation of human male germline stem cells to hepatocytes in vivo via the transplantation under renal capsules. Oncotarget. 2017;8:14576–14592. doi: 10.18632/oncotarget.14713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang H., Liu Y., Hai Y., Guo Y., Yang S., Li Z., Gao W.Q., He Z. Efficient Conversion of Spermatogonial Stem Cells to Phenotypic and Functional Dopaminergic Neurons via the PI3K/Akt and P21/Smurf2/Nolz1 Pathway. Mol. Neurobiol. 2015;52:1654–1669. doi: 10.1007/s12035-014-8966-4. [DOI] [PubMed] [Google Scholar]
- 9.Simon L., Ekman G.C., Kostereva N., Zhang Z., Hess R.A., Hofmann M.C., Cooke P.S. Direct transdifferentiation of stem/progenitor spermatogonia into reproductive and nonreproductive tissues of all germ layers. Stem Cells. 2009;27:1666–1675. doi: 10.1002/stem.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanatsu-Shinohara M., Shinohara T. Spermatogonial stem cell self-renewal and development. Annu. Rev. Cell Dev. Biol. 2013;29:163–187. doi: 10.1146/annurev-cellbio-101512-122353. [DOI] [PubMed] [Google Scholar]
- 11.van den Driesche S., Sharpe R.M., Saunders P.T., Mitchell R.T. Regulation of the germ stem cell niche as the foundation for adult spermatogenesis: a role for miRNAs? Semin. Cell Dev. Biol. 2014;29:76–83. doi: 10.1016/j.semcdb.2014.04.006. [DOI] [PubMed] [Google Scholar]
- 12.Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 13.Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 14.Kawasaki H., Taira K. Retraction: Hes1 is a target of microRNA-23 during retinoic-acid-induced neuronal differentiation of NT2 cells. Nature. 2003;426:100. doi: 10.1038/nature02141. [DOI] [PubMed] [Google Scholar]
- 15.Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 16.Tang F., Kaneda M., O’Carroll D., Hajkova P., Barton S.C., Sun Y.A., Lee C., Tarakhovsky A., Lao K., Surani M.A. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi K., Chuva de Sousa Lopes S.M., Kaneda M., Tang F., Hajkova P., Lao K., O’Carroll D., Das P.P., Tarakhovsky A., Miska E.A., Surani M.A. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS ONE. 2008;3:e1738. doi: 10.1371/journal.pone.0001738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang R., Peng Y., Wang W., Su B. Rapid evolution of an X-linked microRNA cluster in primates. Genome Res. 2007;17:612–617. doi: 10.1101/gr.6146507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niu Z., Goodyear S.M., Rao S., Wu X., Tobias J.W., Avarbock M.R., Brinster R.L. MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. USA. 2011;108:12740–12745. doi: 10.1073/pnas.1109987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Q.E., Racicot K.E., Kaucher A.V., Oatley M.J., Oatley J.M. MicroRNAs 221 and 222 regulate the undifferentiated state in mammalian male germ cells. Development. 2013;140:280–290. doi: 10.1242/dev.087403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He Z., Jiang J., Kokkinaki M., Tang L., Zeng W., Gallicano I., Dobrinski I., Dym M. MiRNA-20 and mirna-106a regulate spermatogonial stem cell renewal at the post-transcriptional level via targeting STAT3 and Ccnd1. Stem Cells. 2013;31:2205–2217. doi: 10.1002/stem.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bouhallier F., Allioli N., Lavial F., Chalmel F., Perrard M.H., Durand P., Samarut J., Pain B., Rouault J.P. Role of miR-34c microRNA in the late steps of spermatogenesis. RNA. 2010;16:720–731. doi: 10.1261/rna.1963810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J., Bao J., Kim M., Yuan S., Tang C., Zheng H., Mastick G.S., Xu C., Yan W. Two miRNA clusters, miR-34b/c and miR-449, are essential for normal brain development, motile ciliogenesis, and spermatogenesis. Proc. Natl. Acad. Sci. USA. 2014;111:E2851–E2857. doi: 10.1073/pnas.1407777111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ahmed K., LaPierre M.P., Gasser E., Denzler R., Yang Y., Rülicke T., Kero J., Latreille M., Stoffel M. Loss of microRNA-7a2 induces hypogonadotropic hypogonadism and infertility. J. Clin. Invest. 2017;127:1061–1074. doi: 10.1172/JCI90031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J., Cai T., Zheng C., Lin X., Wang G., Liao S., Wang X., Gan H., Zhang D., Hu X. MicroRNA-202 maintains spermatogonial stem cells by inhibiting cell cycle regulators and RNA binding proteins. Nucleic Acids Res. 2017;45:4142–4157. doi: 10.1093/nar/gkw1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clermont Y. The cycle of the seminiferous epithelium in man. Am. J. Anat. 1963;112:35–51. doi: 10.1002/aja.1001120103. [DOI] [PubMed] [Google Scholar]
- 27.He Z., Kokkinaki M., Jiang J., Dobrinski I., Dym M. Isolation, characterization, and culture of human spermatogonia. Biol. Reprod. 2010;82:363–372. doi: 10.1095/biolreprod.109.078550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiger A.A., Jones D.L., Schulz C., Rogers M.B., Fuller M.T. Stem cell self-renewal specified by JAK-STAT activation in response to a support cell cue. Science. 2001;294:2542–2545. doi: 10.1126/science.1066707. [DOI] [PubMed] [Google Scholar]
- 29.Oatley J.M., Kaucher A.V., Avarbock M.R., Brinster R.L. Regulation of mouse spermatogonial stem cell differentiation by STAT3 signaling. Biol. Reprod. 2010;83:427–433. doi: 10.1095/biolreprod.109.083352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hou J., Niu M., Liu L., Zhu Z., Wang X., Sun M., Yuan Q., Yang S., Zeng W., Liu Y. Establishment and Characterization of Human Germline Stem Cell Line with Unlimited Proliferation Potentials and no Tumor Formation. Sci. Rep. 2015;5:16922. doi: 10.1038/srep16922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y., Niu M., Yao C., Hai Y., Yuan Q., Liu Y., Guo Y., Li Z., He Z. Fractionation of human spermatogenic cells using STA-PUT gravity sedimentation and their miRNA profiling. Sci. Rep. 2015;5:8084. doi: 10.1038/srep08084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panula S., Medrano J.V., Kee K., Bergström R., Nguyen H.N., Byers B., Wilson K.D., Wu J.C., Simon C., Hovatta O., Reijo Pera R.A. Human germ cell differentiation from fetal- and adult-derived induced pluripotent stem cells. Hum. Mol. Genet. 2011;20:752–762. doi: 10.1093/hmg/ddq520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holloway J.K., Booth J., Edelmann W., McGowan C.H., Cohen P.E. MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet. 2008;4:e1000186. doi: 10.1371/journal.pgen.1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kee K., Angeles V.T., Flores M., Nguyen H.N., Reijo Pera R.A. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature. 2009;462:222–225. doi: 10.1038/nature08562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lim S., Kaldis P. Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development. 2013;140:3079–3093. doi: 10.1242/dev.091744. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y., Xu X., Zhang M., Wang X., Bai X., Li H., Kan L., Zhou Y., Niu H., He P. MicroRNA-663a is downregulated in non-small cell lung cancer and inhibits proliferation and invasion by targeting JunD. BMC Cancer. 2016;16:315. doi: 10.1186/s12885-016-2350-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Long C., Jiang L., Wei F., Ma C., Zhou H., Yang S., Liu X., Liu Z. Integrated miRNA-mRNA analysis revealing the potential roles of miRNAs in chordomas. PLoS ONE. 2013;8:e66676. doi: 10.1371/journal.pone.0066676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gronostajski R.M. Roles of the NFI/CTF gene family in transcription and development. Gene. 2000;249:31–45. doi: 10.1016/s0378-1119(00)00140-2. [DOI] [PubMed] [Google Scholar]
- 39.O’Connor C., Campos J., Osinski J.M., Gronostajski R.M., Michie A.M., Keeshan K. Nfix expression critically modulates early B lymphopoiesis and myelopoiesis. PLoS ONE. 2015;10:e0120102. doi: 10.1371/journal.pone.0120102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martynoga B., Mateo J.L., Zhou B., Andersen J., Achimastou A., Urbán N., van den Berg D., Georgopoulou D., Hadjur S., Wittbrodt J. Epigenomic enhancer annotation reveals a key role for NFIX in neural stem cell quiescence. Genes Dev. 2013;27:1769–1786. doi: 10.1101/gad.216804.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rahman N.I.A., Abdul Murad N.A., Mollah M.M., Jamal R., Harun R. NFIX as a Master Regulator for Lung Cancer Progression. Front. Pharmacol. 2017;8:540. doi: 10.3389/fphar.2017.00540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gurrieri F., Cavaliere M.L., Wischmeijer A., Mammì C., Neri G., Pisanti M.A., Rodella G., Laganà C., Priolo M. NFIX mutations affecting the DNA-binding domain cause a peculiar overgrowth syndrome (Malan syndrome): a new patients series. Eur. J. Med. Genet. 2015;58:488–491. doi: 10.1016/j.ejmg.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 43.Oshima T., Hara H., Takeda N., Hasumi E., Kuroda Y., Taniguchi G., Inuzuka R., Nawata K., Morita H., Komuro I. A novel mutation of NFIX causes Sotos-like syndrome (Malan syndrome) complicated with thoracic aortic aneurysm and dissection. Hum. Genome Var. 2017;4:17022. doi: 10.1038/hgv.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris L., Zalucki O., Gobius I., McDonald H., Osinki J., Harvey T.J., Essebier A., Vidovic D., Gladwyn-Ng I., Burne T.H. Transcriptional regulation of intermediate progenitor cell generation during hippocampal development. Development. 2016;143:4620–4630. doi: 10.1242/dev.140681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Satyanarayana A., Kaldis P. Mammalian cell-cycle regulation: several Cdks, numerous cyclins and diverse compensatory mechanisms. Oncogene. 2009;28:2925–2939. doi: 10.1038/onc.2009.170. [DOI] [PubMed] [Google Scholar]
- 46.Soh Y.Q.S., Mikedis M.M., Kojima M., Godfrey A.K., de Rooij D.G., Page D.C. Meioc maintains an extended meiotic prophase I in mice. PLoS Genet. 2017;13:e1006704. doi: 10.1371/journal.pgen.1006704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Geng Y., Yu Q., Sicinska E., Das M., Schneider J.E., Bhattacharya S., Rideout W.M., Bronson R.T., Gardner H., Sicinski P. Cyclin E ablation in the mouse. Cell. 2003;114:431–443. doi: 10.1016/s0092-8674(03)00645-7. [DOI] [PubMed] [Google Scholar]
- 48.Ohtsubo M., Theodoras A.M., Schumacher J., Roberts J.M., Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol. Cell. Biol. 1995;15:2612–2624. doi: 10.1128/mcb.15.5.2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jablonska B., Aguirre A., Vandenbosch R., Belachew S., Berthet C., Kaldis P., Gallo V. Cdk2 is critical for proliferation and self-renewal of neural progenitor cells in the adult subventricular zone. J. Cell Biol. 2007;179:1231–1245. doi: 10.1083/jcb.200702031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aleem E., Kiyokawa H., Kaldis P. Cdc2-cyclin E complexes regulate the G1/S phase transition. Nat. Cell Biol. 2005;7:831–836. doi: 10.1038/ncb1284. [DOI] [PubMed] [Google Scholar]
- 51.Wang H., Yuan Q., Sun M., Niu M., Wen L., Fu H., Zhou F., Chen Z., Yao C., Hou J. BMP6 Regulates Proliferation and Apoptosis of Human Sertoli Cells Via Smad2/3 and Cyclin D1 Pathway and DACH1 and TFAP2A Activation. Sci. Rep. 2017;7:45298. doi: 10.1038/srep45298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He Z., Jiang J., Hofmann M.C., Dym M. Gfra1 silencing in mouse spermatogonial stem cells results in their differentiation via the inactivation of RET tyrosine kinase. Biol. Reprod. 2007;77:723–733. doi: 10.1095/biolreprod.107.062513. [DOI] [PMC free article] [PubMed] [Google Scholar]