Abstract
Development and cancer share a variety of functional traits such as EMT, cell migration, angiogenesis, and tissue remodeling. In addition, many cellular signaling pathways are noted to coordinate developmental processes and facilitate aspects of tumor progression. The Forkhead box superfamily of transcription factors consists of a highly conserved DNA binding domain, which binds to specific DNA sequences and play significant roles during adult tissue homoeostasis and embryogenesis including development, differentiation, metabolism, proliferation, apoptosis, migration, and invasion. Interestingly, various studies have implicated the role of key Fox family members such as FOXP, FOXO, and FOXA during cancer initiation and metastases. FOXI3, a member of the Forkhead family affects embryogenesis, development, and bone remodeling; however, no studies have reported a role in cancer. In this review, we summarize the role of FOXI3 in embryogenesis and bone development and discuss its potential involvement in cancer progression with a focus on the bone metastasis. Moreover, we hypothesize possible mechanisms underlying the role of FOXI3 in the development of solid tumor bone metastasis.
Keywords: FOXI3, embryogenesis, bone, development, cancer, metastasis
Introduction
Embryogenesis, development, and solid tumor metastasis have many intersecting factors in regard to molecular events such as cell motility, changes in cellular differentiation, and changes in the interactions with cells of the local microenvironment (Kelleher et al., 2006; Heerboth et al., 2015). Indeed, these processes are tightly regulated and coordinated in part via gene regulation by transcription factors. Perhaps not surprisingly, a number of transcription factors have been shown to have overlapping roles in development, cancer progression, and cancer metastasis (Jackson et al., 1990; Schreiber et al., 2000; Hollern et al., 2014; Nowak et al., 2015; Zhang et al., 2016; To and Andrechek, 2018). Although some transcription factors have been well characterized for their roles in both development and in cancer; there remains many transcription factors with known significances in development that have not been examined for potential roles in carcinogenesis and metastatic progression. One such transcription factor with documented roles in development is FOXI3. Despite regulation of cancer-related processes such as epithelial to mesenchymal transition (Edlund et al., 2014; Shirokova et al., 2016); FOXI3 remains unexplored in tumorigenesis and metastasis. In this review, we summarize the role of FOXI3 in embryogenesis and bone development and examine its potential role in cancer progression using an informatics approach. Moreover, we discuss possible gene targets that might allow FOXI3 to facilitate solid tumor bone metastasis.
FOXI3 in embryogenesis and development
The transcription factors of the Forkhead box family of transcription factors consist of a highly conserved, 110-amino acid Forkhead or “winged helix” DNA binding domain (DBD) which usually binds to DNA through RYAAAYA motif (where R = purine and Y = pyrimidine) (Nakagawa et al., 2013; Chen et al., 2016). The FOX genes of this family play important roles in various biological processes including embryogenesis, development and metabolism (Lehmann et al., 2003; Coffer and Burgering, 2004; Tuteja and Kaestner, 2007a,b).
FOXI3 in early bone development
Similar to other FOX genes, FOXI3 plays a significant role in embryogenesis/development. In particular, a number of studies detail the FOXI3 transcription factor as a critical regulator of bone development (Jänicke et al., 2007; Janicke et al., 2010; Drogemuller et al., 2008; Khatri and Groves, 2013; Khatri et al., 2014). The formation of skeletal bone tissue takes place during development where bones are formed by intramembranous and endochondral ossification. The latter process is dependent upon mesenchymal progenitor cells; marked by local concentration and differentiation initially into cartilage-forming chondrocytes which generate an avascular template on which new bone is formed (Regard et al., 2012). Upon completion of the skeletal system, bone becomes a regenerative tissue and is maintained by continuous remodeling (Regard et al., 2012). Implicating FOXI3 in bone development, studies have shown that FOXI3 induces early bone and cartilage formation. For instance, proper pharyngeal arch, and otic placode development was shown to be dependent upon FOXI3 (Urness et al., 2010; Edlund et al., 2014; Khatri et al., 2014). Further, other work demonstrated FOXI3 expression during early craniofacial development and showed that FOXI3 maintains dental epithelial cells in an undifferentiated state (Jussila et al., 2015).
Indeed, additional studies point to wide ranging functions of FOXI3 in development as a number of phenotypes have been observed with loss of FOXI3 function. For example, mutation of FOXI3 in canines result in abnormalities such as dysplastic hair follicles, complete absence of the pinna, absence of middle-ear structures, and malformed teeth (Drogemuller et al., 2008; Wiener et al., 2013; Tassano et al., 2015). In addition, mutations of FOXI3 in humans result in Congenital Aural Atresia (CAA) (Tassano et al., 2015); a condition attributed to malformation of the outer ear. These findings demonstrate that FOXI3 is a critical regulator of early cartilage and bone development (Drogemuller et al., 2008; Wiener et al., 2013). While it is clear that FOXI3 is a key regulator of craniofacial development, there remains a gap in our knowledge of the mechanisms by which FOXI3 operates in these processes, which cell-types utilize FOXI3 to control development of cartilage and skeletal tissue and the genes the FOXI3 regulates to govern these processes.
Providing insight to these questions, recent work has uncovered some of the target genes by which FOXI3 controls development. For instance, several studies examined the transcriptional impact of FOXI3 loss using gene expression microarrays (Jussila et al., 2015; Shirokova et al., 2016). Jussila et al. performed expression profiling on FOXI3 knockout mice to identify Fgf15, Sfrp5 Dusp6, Etv5, Shh, and Ptch1 that were upregulated with FOXI3 loss (Jussila et al., 2015). Their work concluded that loss of FOXI3 led to abnormal upregulation of genes of Fgf, Shh and Bmp pathways, which in turn resulted in epithelial dysmorphogenesis during tooth development (Jussila et al., 2015). In a separate study, microarray analysis identified gene expression changes resulting from FOXI3 loss in hair follicles; key amongst their findings is that FOXI3 was essential for stem cell maintenance (Shirokova et al., 2016). Reanalyzing the gene expression data from these two studies (Jussila et al., 2015; Shirokova et al., 2016), using significant analysis of microarray (SAM), we highlight the genes significantly downregulated by FOXI3 loss (Figure 1). As shown in Table 1, many of these genes have critical roles in development. Examining other molecules, work by Edlund et al. demonstrated FGF8 as a key effector of FOXI3 signaling. In their work, Edlund et al. show that FOXI3 mutants lose expression of FGF8 and that loss of ectodermal FOXI3-Fgf8 signaling results in apoptosis of the neural crest cells (Edlund et al., 2014). Indeed, this work sheds light onto some of the details of FOXI3 regulation of craniofacial development and points to FOXI3 as a key regulator of neural crest cell survival. Together these studies point to FOXI3 as a critical regulator of developmental pathways tied to stem cell function. Perhaps related to the criticality of FOXI3 in maintenance of a de-differentiated state, recent work has implicated FOXI3 in the epithelial-mesenchymal transition (Khatri and Groves, 2013; Edlund et al., 2014; Khatri et al., 2014).
Figure 1.
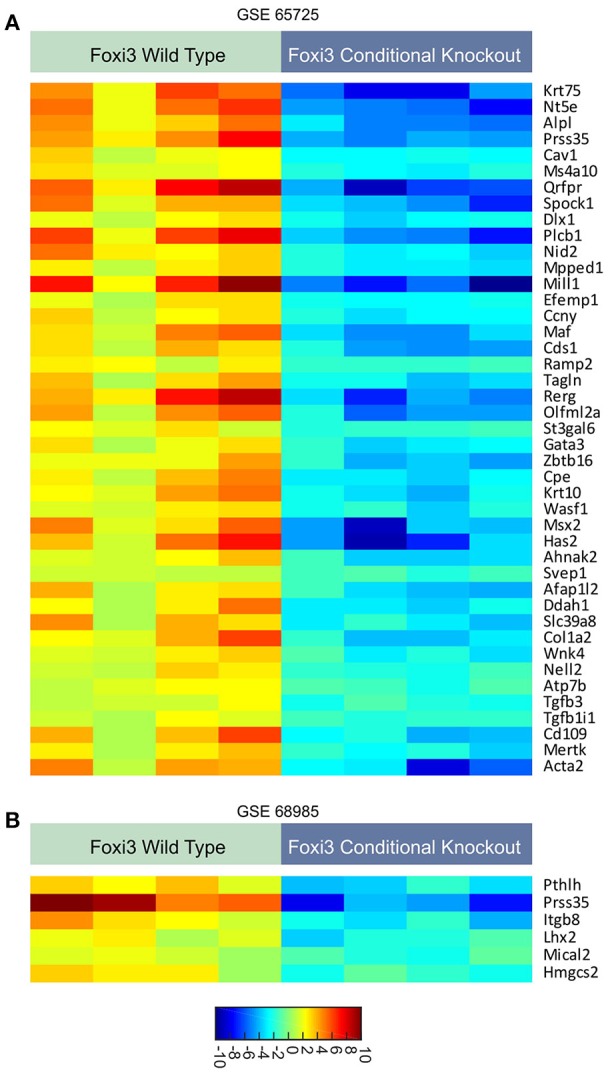
(A,B) Expression patterns of genes regulated with loss of FOXI3. Data from two published studies (Jussila et al., 2015; Shirokova et al., 2016) (accession numbers: GSE65725 and GSE68985) following download of series matrix files, data median centered, column standardized, and significant gene expression changes were identified using significant analysis of microarray (SAM) (Tusher et al., 2001). All depicted genes had a q-value < 0.0001. For both dataset, the FDR was minimized to 0.00%. The scale bar indicates the expression values corresponding to color.
Table 1.
Role of FOXI3 signature genes in early development.
| FOXI3 signature gene symbols | Negative fold change | Q-value | Accession number | Implications in development | References |
|---|---|---|---|---|---|
| Cav1 | 3.077057237 | 0 | GSE65725 | Required for thymocyte development, cochlear inner hair cell development and T cell homeostasis | A.Jha., et al. Science signaling, 2015 (PMID: 26486172) B.A. Brandt., et al. The journal of neuroscience, 2003 (PMID: 14645476) |
| Spock1 | 33.97310003 | 0 | GSE65725 | Implicated in murine development | S. Roll., et al. Matrix biology, 2006 (PMID: 16806869) F. Charbonnier., et al. Mechanisms of development, 2000 (PMID: 10640720) |
| Maf | 31.49087568 | 0 | GSE65725 | Essential for endochondral bone development | H. E. MacLean., et al. Developmental biology, 2003 (PMID: 14512017) |
| Ramp2 | 1.493731184 | 0 | GSE65725 | Plays a significant role in bone development, endocrine development, and angiogenesis | M. Kadmiel., et al. Molecular endocrinology, 2011 (PMID: 21566080) Y. Ichikawa-Shindo., et al. The journal of clinical investigation, 2008 (PMID: 18097473) |
| Gata3 | 3.117080473 | 0 | GSE65725 | Implicated in development and maintenance of type 2 innate lymphoid cells expressing IL-7Rα | T. Hoyler., et al. Immunity, 2012 (PMID: 23063333) R. Yagi., et al. Immunity, 2014 (PMID: 24631153) |
| Wasf1 | 2.190182385 | 0 | GSE65725 | Implicated in dendritic spine development and modulates oocyte transcription during embryogenesis | J. Y. Sung., et al. Proc Natl Acad Sci U S A, 2008 (PMID: 18287015) K. V., et al. Science, 2013 (PMID: 23990560) |
| Col1a2 | 15.09160309 | 0 | GSE65725 | Regulates formation and activity of collagen during development and constitutes about 90% of the bone matrix | M. Ponticos., et al. Matrix biology, 2004 (PMID: 15062855) M. L. Sohaskey., et al. The journal of cell biology, 2010 (PMID: 20440000) |
| Tgfb3 | 1.005055429 | 0 | GSE65725 | Plays a pivotal role in differentiation of osteoblasts and overall bone development | M. Wu., G. Chen., and Y. P. Li. Bone research, 2016 (PMID: 27563484) G. Chen., C. Deng., and Y. P. Li. International journal of biological sciences, 2012 (PMID: 22298955) |
| Pthlh | 5.147615864 | 0 | GSE68985 | Implicated in murine embryogenesis, regulates endochondral bone development by inhibiting chondrocyte differentiation | L. Guo., et al. PLoS one, 2012 (PMID: 22808183) E. Minina., et al. Developmental cell, 2002 (PMID: 12361605) R. Flottmann., et al. European journal of human genetics, 2016 (PMID: 26733284) |
FOXI3 in EMT
Epithelial-mesenchymal transition (EMT) is a key process in embryogenesis, wound healing and cancer. In EMT, a variety of gene expression changes are observed that are typically associated with a de-differentiated cellular state, and cells become motile (Ye et al., 2015; Hollern et al., 2018). Certainly, this transition is a process key to ectodermal, mesodermal, endodermal, and neural crest development (Yang and Weinberg, 2008; Voulgari and Pintzas, 2009). Importantly, neural crest cells are of mesenchymal differentiation and express FOXI3 (Achilleos and Trainor, 2012; Khatri and Groves, 2013; Edlund et al., 2014; Niibe et al., 2017). During EMT, FOXI3 with other EMT genes plays a significant role in the development of the epibranchial placodes, inner ear, and pharyngeal arch derivatives (Khatri and Groves, 2013; Edlund et al., 2014; Khatri et al., 2014). Furthermore, during neurulation, neural crest cells undergo EMT and express many genes critical to ion channel formation; an integral part of neural cell differentiation (Takahashi and Okamura, 1998; Scarpa et al., 2015). Interestingly, FOXI3 is essential to ionocyte specification and differentiation in zebrafish (Hsiao et al., 2007; Jänicke et al., 2007). Like ion channels of human neural crest cells, ionocytes in zebrafish regulate ion transport, maintains homeostasis, and facilitates cell differentiation during EMT (Hsiao et al., 2007; Jänicke et al., 2007; Janicke et al., 2010; Thermes et al., 2010; Li and Xiong, 2011). During development, FOXI3 also functions as a molecular switch, which when turned on, converts cells to ionocytes, and when turned off, to non-keratinocytes (Jänicke et al., 2007). Therefore, FOXI3 has multimodal roles during development and the potent role of FOXI3 in cell differentiation insinuate us about its possible involvement in abnormal cell growth and migration.
FOXI3 in cancer progression
Given that many developmental pathways, transcription factors, and processes, are commonly implicated in cancer progression, we propose that FOXI3 may also play an important role in cancer. Indeed, with governance of developmental features such as stem cell maintenance and involvement in EMT, FOXI3 may enable resistance to chemotherapy or tumor metastasis (Anders and Carey, 2008; Chaffer et al., 2016). However, to date no studies have been published that demonstrate FOXI3's role in tumor progression. As such, we have utilized a variety of public data to examine FOXI3 across cancer types and test for association with key clinical measures of tumor progression.
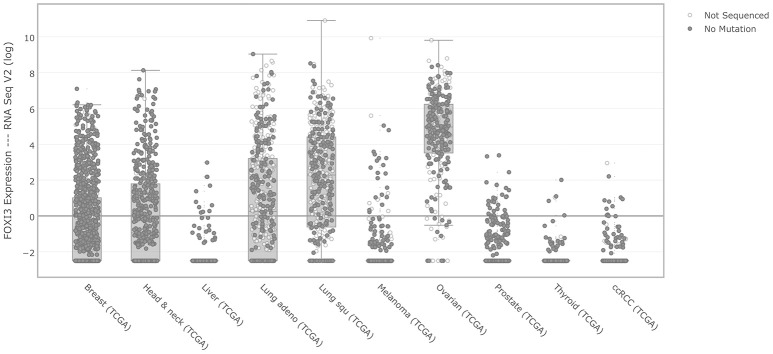
One of the largest coordinated efforts to document the genomic features of cancer has been the work of The Cancer Genome Atlas. Using the cBioPortal to access multiple cancer types (Cerami et al., 2012; Gao et al., 2013), we first interrogated FOXI3 expression across several major cancer types, (Figure 2). While relatively few liver and thyroid cancers show elevated FOXI3 expression; other cancers display a wide range of FOXI3 expression across patient tumors (breast, lung, prostate, ovarian, and head and neck cancer). With this, we questioned whether changes in FOXI3 expression associate with features of tumor progression in several of these cancer types.
Figure 2.
FOXI3 expression across foremost cancer types. RSEM normalized gene expression levels of FOXI3 as shown by cBioPortal (http://cbioportal.org) across TCGA cohorts.
Within the TCGA prostate cancer cohort (Cerami et al., 2012; Gao et al., 2013; Cancer Genome Atlas Research Network, 2015), we find that FOXI3 expression increases with tumor stage (Figure 3A). While T4 tumors were infrequent in this cohort, many T3 stage tumors as compared to T2 stage have elevated FOXI3 expression. The progression from T2 to T3 stage is classified in part by the presence of tumor cell invasion to the surrounding tissues of the prostate gland (T3) (Buyyounouski et al., 2017). This marks the first signs of metastatic potential of FOXI3. Indeed, given the bounty of research demonstrating EMT as critical to the invasive potential of prostate cancer, future research examining whether FOXI3 expression functions in EMT, invasion, and metastasis could bring together these clinical associations to key processes of tumor progression controlled by FOXI3. Within the TCGA breast cancer cohort (Cancer Genome Atlas Research Network, 2012), some of the primary tumors were annotated for the site where distant metastasis was observed. Examining FOXI3 expression in this data showed that breast cancers that metastasized to the bone often had high FOXI3 expression (Figure 3B). While the number of cases are limited, additional experiments may illuminate the role of in facilitating bone metastasis. In support of this hypothesis, we noted that many of the genes shown to be downregulated with FOXI3 loss (Figure 1), also have demonstrated roles in tumor progression and metastasis (Table 2), including bone metastasis.
Figure 3.
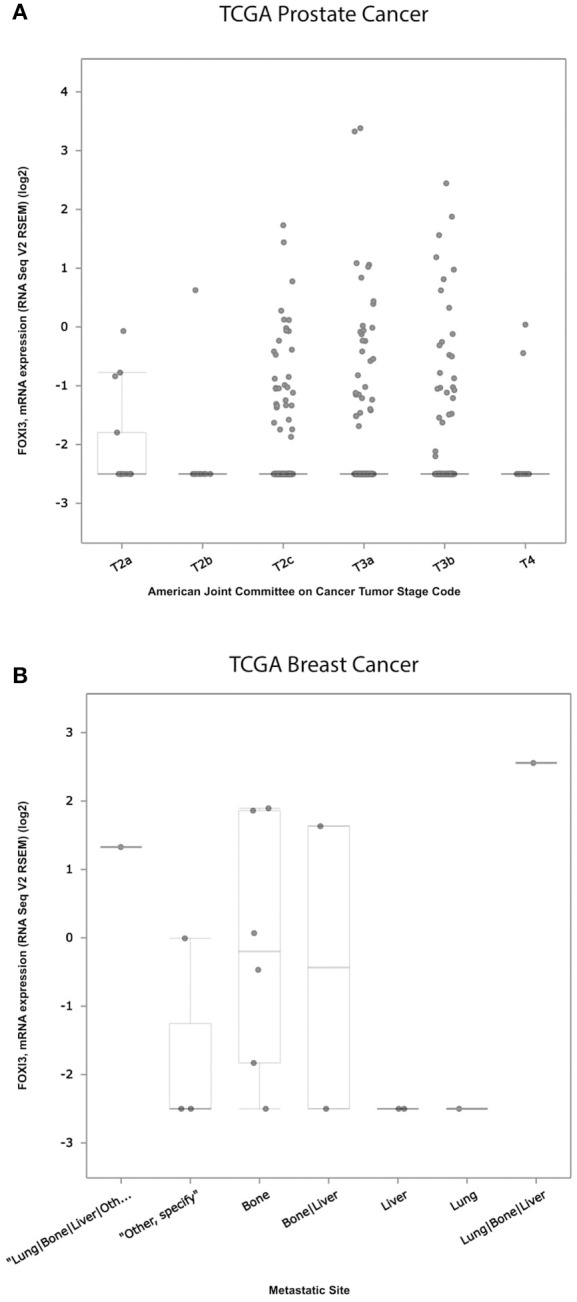
Expression of FOXI3 in prostate and breast cancers. (A) RSEM normalized expression values of the TCGA prostate cancer cohort with tumor stage (Cancer Genome Atlas Research Network, 2015). (B) RSEM normalized expression values for TCGA breast tumors annotated according to site of distant metastasis (Anders and Carey, 2008). Both panels: Data was interrogated using cBioPortal (http://cbioportal.org).
Table 2.
Role of FOXI3 signature genes in cancer metastasis.
| FOXI3 signature gene symbols | Negative fold change | Q-value | Accession number | Implications in cancer metastasis | References |
|---|---|---|---|---|---|
| Cav1 | 3.077057237 | 0 | GSE65725 | Favors pancreatic cancer progression including invasion and migration to distant sites | M. Chatterjee., et al. Scientific reports, 2015 (PMID: 26065715). |
| Spock1 | 33.97310003 | 0 | GSE65725 | Promotes prostate cancer metastasis | Q. Chen., et al. Drug design, development and therapy, 2016 (PMID: 27486308) |
| Maf | 31.49087568 | 0 | GSE65725 | Mediates breast cancer bone metastasis | M. Pavlovic., et al. Journal of the cancer institute, 2015 (PMID: 26376684) |
| Ramp2 | 1.493731184 | 0 | GSE65725 | Significantly promotes bone metastasis of breast cancer and is upregulated in prostate cancer | A. Cappariello., et al. Bone abstracts, 2013. M. Logan., et al. The American journal of pathology, 2013 (PMID: 23867798) |
| Gata3 | 3.117080473 | 0 | GSE65725 | Implicated in luminal breast cancer subtype | B. C. McCleskey., et al. American journal of clinical pathology, 2015 (PMID: 26486740) |
| Wasf1 | 2.190182385 | 0 | GSE65725 | Implicated in prostate cancer progression | H. S. Fernando., et al. The journal of urology, 2008 (PMID: 18710763) |
| Col1a2 | 15.09160309 | 0 | GSE65725 | Upregulated in gastric cancer and implicated in gastric cancer metastasis | C. Zhuo., et al. Cell physiology and biochemistry, 2016 (PMID: 27997896). N. Oue., et al. Cancer research, 2004 (PMID: 15059891) |
| Tgfb3 | 1.005055429 | 0 | GSE65725 | Associated with poor breast cancer prognosis. Implicated in prostate cancer metastasis. In bone metastasis, Tgfb triggers metastatic cancer cells to secrete factors in the bone microenvironment that in turn favors osteolytic bone destruction | A. Ghellal., et al. Anticancer research, 2000 (PMID: 11205281) X. Zhang., et al. Experimental and therapeutic medicine, 2016 (PMID: 27446297) J. T. Buijs., et al. Bonekey reports, 2012 (PMID: 23951484) |
| Pthlh | 5.147615864 | 0 | GSE68985 | Promotes bone metastasis of breast cancer and nuclear PTHrP contributes to prostate cancer metastasis | K. Boras-Granic., and J. J. Wysolmerski. Breast cancer research, 2012 (PMID: 22546075) S. I. Park., and L. K. McCauley. Endocrine-related cancer, 2012 (PMID:22291434) |
These putative targets of FOXI3 (Figure 1) also provide an additional tool for informatics inquiries. Initially, one of the limitations in our examination was the omission of probes for FOXI3 on the microarrays of a variety of clinically rich gene expression datasets. However, the use of FOXI3 putative target genes as a signature of FOXI3 activity bypasses this limitation and allows for additional analyses. One dataset of particular interest was the published gene expression dataset across distant prostate cancer metastatic sites (Haider et al., 2016). As shown in Figure 4A, FOXI3 signature levels were high in bone, lung, and lymph node metastases. FOXI3 target genes also showed an association with metastasis in breast cancer. Probing expression data from primary breast tumors annotated for time to distant metastasis, we observed that tumors with the highest expression of the FOXI3 signature genes present distant metastasis earlier than those with low expression (Figure 4B). Taken together with the observations in Figure 3A, FOXI3 may mark and function in tumor cell invasion and metastasis. Given the clues from developmental studies, it might be that this function occurs by EMT and is an area that should be investigated. Additionally, data shown in Figures 3B, 4A, suggests that FOXI3 may facilitate colonization of tumor cells to the bone. Indeed, a number of the putative targets of FOXI3 have been implicated in bone metastasis and supports the need for future studies to examine FOXI3 specifically in the bone metastasis setting.
Figure 4.
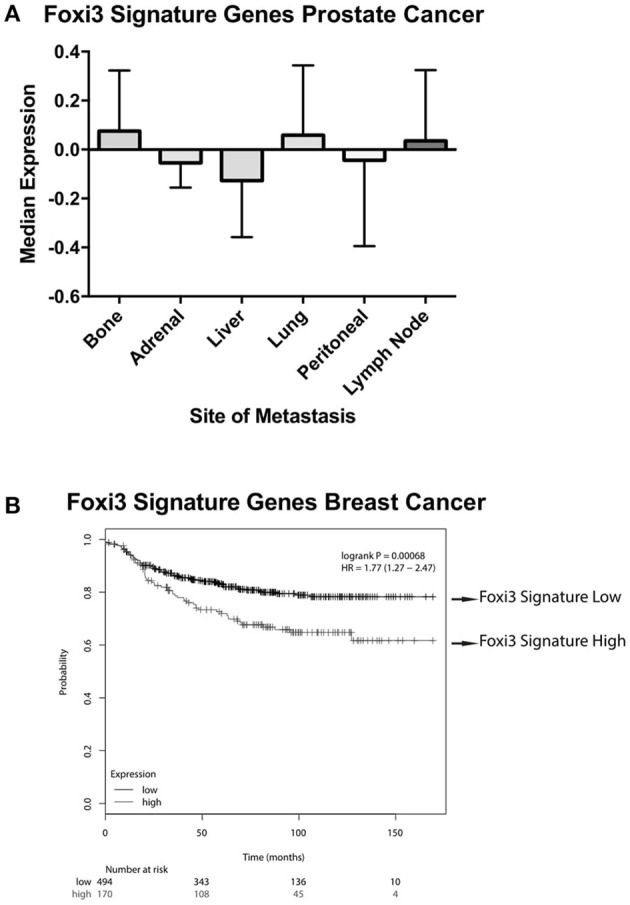
FOXI3 signature genes in prostate and breast cancer metastasis. (A) Publicly available data (Haider et al., 2016) (accession number: GSE74685) downloaded from the series matrix file. Data was median centered and column standardized. Next, tumors were scored for median expression of FOXI3 signature genes depicted in Figure 1. The boxplots show median expression of these genes for tumors at distant sites of metastasis (B) Distant metastasis free survival is shown for breast cancer patient tumors scored for average expression of the FOXI3 signature genes [p-value = 0.00068 and hazard ratio (HR) = 1.77 (1.27–2.47)]. Patient tumors were split into high and low expression groups using the autoselect threshold option on http://kmplot.com/analysis/ (Györffy et al., 2010).
Implications for the future
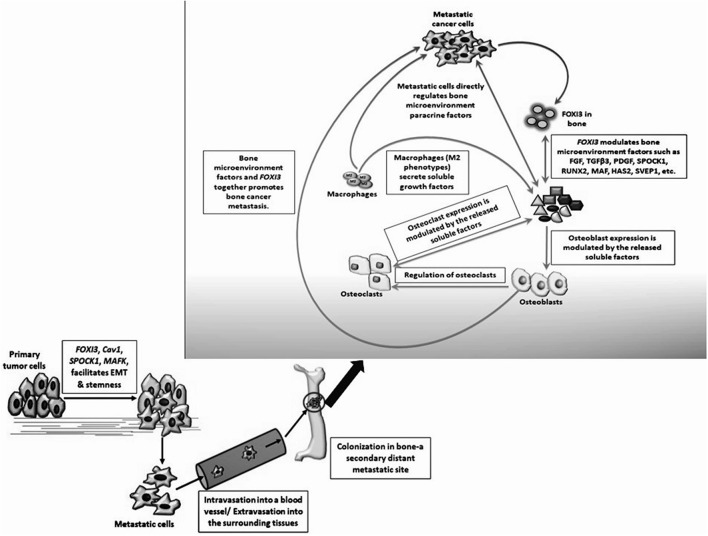
In this review, we note that FOXI3 a regulator of early bone development may play a potential role in cancer progression (Table 3). We further note that several of the genes that are downregulated with the loss of FOXI3, such as SPOCK1, Dlx, MAF, PthrP, HAS2, SVEP1, and TGFβ3 (Jussila et al., 2015; Shirokova et al., 2016) have been shown to play important roles in distant metastasis to other sites including bone (Table 2). Based on our analysis of publicly available clinical data regarding FOXI3 expression in different cancer types and stages, we speculate that FOXI3 may have multiple roles throughout metastatic progression. At the primary tumor, FOXI3 may allow for stem cell maintenance or EMT; the latter in turn may implicate FOXI3 in tumor cell invasion into the local tissues and also into circulation. Further, in advanced cancer stage, during colonization of distant sites, FOXI3 may facilitate reconfiguring of the tumor microenvironment by expression of paracrine factors. These mechanisms may come into play to facilitate bone metastasis as evidenced by several of the above listed potential target genes having previously demonstrated roles in bone metastasis (Table 2). Thus, we propose that when FOXI3 in tumor cells modulate the secretion of other bone factors including SPOCK1, Dlx, MAF, PthrP, HAS2, SVEP1, TGFβ3, and FGF (Jussila et al., 2015; Meng et al., 2016; Shirokova et al., 2016) which further primes the microenvironment, creates a crosstalk to help tumors grow in the bone (Figure 5). Understanding the intersect of the regulatory behavior of FOXI3 on factors prominent in hallmark processes of tumor progression to bone is essential to elucidating its role when these processes are hijacked and go awry. As a whole, this review highlights promising directions for research of FOXI3 and its potential roles in cancer progression. Given no such studies exist to date, there is much to be discovered for this key developmental factor in the cancer setting and is a direction our lab plans to pursue.
Table 3.
Summary describing the role of FOXI3 in development and its hypothesized role in cancer progression.
| Processes | Role of FOXI3 in embryogenesis | Hypothesized role of FOXI3 in cancer |
|---|---|---|
| Epithelial mesenchymal transition (EMT) | Early cartilage differentiation including development of epibranchial placodes, inner ear, and pharyngeal arch derivatives (Jussila et al., 2015) | May promote EMT, inducing metastasis |
| Bone development |
1. Promotes murine craniofacial development (Jussila et al., 2015) 2. Induces early bone and cartilage formation, i.e., pharyngeal and otic placode development in mice and chickens (Coffer and Burgering, 2004; Voulgari and Pintzas, 2009) 3. Regulates ectodermal development in Chinese crested dogs and Mexican and Peruvian hairless dogs (Tuteja and Kaestner, 2007b; Shirokova et al., 2016). |
Regulates bone matrix proteins, promoting osteomimicry of cancer cells, promoting bone metastasis |
| Bone microenvironment |
Fibroblast growth factor 1. Regulates FGF in vertebrate bone development (Khatri and Groves, 2013) 2. Osteoclast/blast differentiation (Khatri and Groves, 2013) Bone morphogenetic protein Distal limb morphogenesis, mesoderm formation, and cell differentiation (Wiener et al., 2013; Tassano et al., 2015) |
FGFs, SPOCK1, Dlx, RUNX2, MAF, PthrP, HAS2, SVEP1, TGFβ3, and other factors in bone matrix may modulate FOXI3 expression, promoting solid tumor bone metastasis |
| Ionocyte and ion channels |
1. Regulates ionocyte differentiation in zebrafish embryos (Gao et al., 2013; Chen et al., 2016) 2. Functions as a molecular switch converting cells to ionocytes (Chen et al., 2016) |
Mutation of sub-cellular FOXI3A/B interferes with ion channel function, disrupting cell homeostasis and promoting tumor metastasis |
| Notch signaling |
Jagged-2 Regulates skin ionocyte formation and differentiation in zebrafish embryos by regulating Jagged-2 in the Notch Signaling Pathway (Gao et al., 2013; Chen et al., 2016) |
FOXI3 overexpression may modulate Jagged-2, dysregulating the Notch pathway, promoting cancer invasion |
Figure 5.
Hypothesized mechanisms for FOXI3 role in cancer metastasis. FOXI3 may facilitate primary tumor cells to undergo EMT. Once the tumor cells invade the basement membrane, they enter the nearby blood or lymphatic vessel (intravasation) and tumors cells invade local microenvironment of distant sites via extravasation. Once these invasive migratory tumor cells colonize a fertile secondary tumor site such as bone, FOXI3 may regulate the expression of bone matrix factors such as SPOCK1, TGFβ3, FGFs, and others to prepare the microenvironment in a way that aggravates cancer cell progression.
Author contributions
JJ and AM devised the concept with CY. AM, JJ, and OW developed the theory. DH, AM and OW developed and analyzed the tables. AM and DH, developed and interpreted the figures. AM, JJ, DH, OW, TR, WB, and CY drafted the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer ADU and handling Editor declared their shared affiliation.
Acknowledgments
The authors wish to thank Glen Cohen and Honghe Wang for their valuable critiques and guidance for this manuscript.
Footnotes
Funding. This research was supported by grants JDJTU092015 (OFD/Troy University) [JJ], Tri-Beta Research Award [JJ, WB]; G12 RR03059-21A1 (NIH/RCMI) [CY], U54 CA118623 (NIH/NCI) [CY], (NIH/NCI) 1 R21 CA188799-01 [CY]; and the Department of Defense Grant, PC120913, W81XWH-10-1-0543.
References
- Achilleos A., Trainor P. (2012). Neural crest stem cells: discovery, properties and potential for therapy. Cell Res. 22, 288–304. 10.1038/cr.2012.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders C., Carey L. A. (2008). Understanding and treating triple-negative breast cancer. Oncology 22, 1233–1243. [PMC free article] [PubMed] [Google Scholar]
- Buyyounouski M. K., Choyke P. L., McKenney J. K., Sartor O., Sandler H. M., Amin M. B., et al. (2017). Prostate cancer - major changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J. Clin. 67, 245–253. 10.3322/caac.21391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2015). The molecular taxonomy of primary prostate cancer. Cell 163, 1011–1025. 10.1016/j.cell.2015.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2012). Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70. 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffer C. L., San Juan B. P., Lim E., Weinberg R. A. (2016). EMT, cell plasticity and metastasis. Cancer Metast. Rev. 35, 645–654. 10.1007/s10555-016-9648-7 [DOI] [PubMed] [Google Scholar]
- Chen X., Ji Z., Webber A., Sharrocks A. D. (2016). Genome-wide binding studies reveal DNA binding specificity mechanisms and functional interplay amongst Forkhead transcription factors. Nucleic Acids Res. 44, 1566–1578. 10.1093/nar/gkv1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffer P. J., Burgering B. M. (2004). Forkhead-box transcription factors and their role in the immune system. Nat. Rev. Immunol. 4, 889–899. 10.1038/nri1488 [DOI] [PubMed] [Google Scholar]
- Drogemuller C., Karlsson E. K., Hytönen M. K., Perloski M., Dolf G., Sainio K., et al. (2008). A mutation in hairless dogs implicates FOXI3 in ectodermal development. Science 321:1462 10.1126/science.1162525 [DOI] [PubMed] [Google Scholar]
- Edlund R. K., Ohyama T., Kantarci H., Riley B. B., Groves A. K. (2014). Foxi transcription factors promote pharyngeal arch development by regulating formation of FGF signaling centers. Dev. Biol. 390, 1–13. 10.1016/j.ydbio.2014.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 6:pl1 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Györffy B., Lanczky A., Eklund A. C., Denkert C., Budczies J., Li Q., et al. (2010). An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 123, 725–731. 10.1007/s10549-009-0674-9 [DOI] [PubMed] [Google Scholar]
- Haider M., Zhang X., Coleman I., Ericson N., True L. D., Lam H. M., et al. (2016). Epithelial mesenchymal-like transition occurs in a subset of cells in castration resistant prostate cancer bone metastases. Clin. Exp. Metastasis 33, 239–248. 10.1007/s10585-015-9773-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heerboth S., Housman G., Leary M., Longacre M., Byler S., Lapinska K., et al. (2015). EMT and tumor metastasis. Clin. Transl. Med. 4:6. 10.1186/s40169-015-0048-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollern D. P., Honeysett J., Cardiff R. D., Andrechek E. R. (2014). The E2F transcription factors regulate tumor development and metastasis in a mouse model of metastatic breast cancer. Mol. Cell Biol. 34, 3229–3243. 10.1128/MCB.00737-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollern D. P., Swiatnicki M. R., Andrechek E. R. (2018). Histological subtypes of mouse mammary tumors reveal conserved relationships to human cancers. PLoS Genet. 14:e1007135. 10.1371/journal.pgen.1007135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao C. D., You M. S., Guh Y. J., Ma M., Jiang Y. J., Hwang P. P. (2007). A positive regulatory loop between FOXI3a and FOXI3b is essential for specification and differentiation of zebrafish epidermal ionocytes. PLoS ONE 2:e302. 10.1371/journal.pone.0000302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson T., Allard M. F., Sreenan C. M., Doss L. K., Bishop S. P., Swain J. L. (1990). The c-myc proto-oncogene regulates cardiac development in transgenic mice. Mol. Cell Biol. 10, 3709–3716. 10.1128/MCB.10.7.3709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jänicke M., Carney T. J., Hammerschmidt M. (2007). FOXI3 transcription factors and Notch signaling control the formation of skin ionocytes from epidermal precursors of the zebrafish embryo. Dev. Biol. 307, 258–271. 10.1016/j.ydbio.2007.04.044 [DOI] [PubMed] [Google Scholar]
- Janicke M., Renisch B., Hammerschmidt M. (2010). Zebrafish grainyhead-like1 is a common marker of different non-keratinocyte epidermal cell lineages, which segregate from each other in a FOXI3-dependent manner. Int. J. Dev. Biol. 54, 837–850. 10.1387/ijdb.092877mj [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jussila M., Aalto A. J., Sanz Navarro M., Shirokova V., Balic A., Kallonen A., et al. (2015). Suppression of epithelial differentiation by FOXI3 is essential for molar crown patterning. Development 142, 3954–3963. 10.1242/dev.124172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher F. C., Fennelly D., Rafferty M. (2006). Common critical pathways in embryogenesis and cancer. Acta Oncol. 45, 375–388. 10.1080/02841860600602946 [DOI] [PubMed] [Google Scholar]
- Khatri S. B., Edlund R. K., Groves A. K. (2014). FOXI3 is necessary for the induction of the chick otic placode in response to FGF signaling. Dev. Biol. 391, 158–169. 10.1016/j.ydbio.2014.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri S. B., Groves A. K. (2013). Expression of the Foxi2 and Foxi3 transcription factors during development of chicken sensory placodes and pharyngeal arches. Gene Expr. Patterns 13, 38–42. 10.1016/j.gep.2012.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann O. J., Sowden J. C., Carlsson P., Jordan T., Bhattacharya S. S. (2003). Fox's in development and disease. Trends Genet. 19, 339–344. 10.1016/S0168-9525(03)00111-2 [DOI] [PubMed] [Google Scholar]
- Li M., Xiong Z. (2011). Ion channels as targets for cancer therapy. Int. J. Physiol. Pathophysiol. Pharmacol. 3, 156–166. [PMC free article] [PubMed] [Google Scholar]
- Meng X., Vander Ark A., Lee P., Hostetter G., Bhowmick N. A., Matrisian L. M., et al. (2016). Myeloid-specific TGF-β signaling in bone promotes basic-FGF and breast cancer bone metastasis. Oncogene 35, 2370–2378. 10.1038/onc.2015.297 [DOI] [PubMed] [Google Scholar]
- Nakagawa S., Gisselbrecht S. S., Rogers J. M., Hartl D. L., Bulyk M. L. (2013). DNA-binding specificity changes in the evolution of forkhead transcription factors. Proc. Natl. Acad. Sci. U.S.A. 110, 12349–12354. 10.1073/pnas.1310430110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niibe K., Zhang M., Nakazawa K., Morikawa S., Nakagawa T., Matsuzaki Y., et al. (2017). The potential of enriched mesenchymal stem cells with neural crest cell phenotypes as a cell source for regenerative dentistry. Jpn. Dent. Sci. Rev. 53, 25–33. 10.1016/j.jdsr.2016.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak D. G., Cho H., Herzka T., Watrud K., DeMarco D. V., Wang V. M., et al. (2015). MYC drives Pten/Trp53-deficient proliferation and metastasis due to IL6 secretion and AKT suppression via PHLPP2. Cancer Discov. 5, 636–651. 10.1158/2159-8290.CD-14-1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regard J. B., Zhong Z., Williams B. O., Yang Y. (2012). Wnt signaling in bone development and disease: making stronger bone with Wnts. Cold Spring Harb. Perspect. Biol. 4:a007997. 10.1101/cshperspect.a007997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpa E., Szabó A., Bibonne A., Theveneau E., Parsons M., Mayor R. (2015). Cadherin Switch during EMT in neural crest cells leads to contact inhibition of locomotion via repolarization of forces. Dev. Cell 34, 421–434. 10.1016/j.devcel.2015.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber M., Wang Z. Q., Jochum W., Fetka I., Elliott C., Wagner E. F. (2000). Placental vascularisation requires the AP-1 component fra1. Development 127, 4937–4948. Available online at: http://dev.biologists.org/content/127/22/4937 [DOI] [PubMed]
- Shirokova V., Biggs L. C., Jussila M., Ohyama T., Groves A. K., Mikkola M. L. (2016). Foxi3 deficiency compromises hair follicle stem cell specification and activation. Stem Cells. 34, 1896–1908. 10.1002/stem.2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Okamura Y. (1998). Ion channels and early development of neural cells. Physiol. Rev. 78, 307–337. 10.1152/physrev.1998.78.2.307 [DOI] [PubMed] [Google Scholar]
- Tassano E., Jagannathan V., Drögemüller C., Leoni M., Hytönen M. K., Severino M., et al. (2015). Congenital aural atresia associated with agenesis of internal carotid artery in a girl with a FOXI3 deletion. Am. J. Med. Genet. A 167A, 537–544. 10.1002/ajmg.a.36895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermes V., Lin C. C., Hwang P. P. (2010). Expression of Ol-FOXI3 and Na(+)/K(+)-ATPase in ionocytes during the development of euryhaline medaka (Oryzias latipes) embryos. Gene Expr. Patterns 10, 185–192. 10.1016/j.gep.2010.04.001 [DOI] [PubMed] [Google Scholar]
- To B., Andrechek E. R. (2018). Transcription factor compensation during mammary gland development in E2F knockout mice. PLoS ONE 13:e0194937. 10.1371/journal.pone.0194937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher V. G., Tibshirani R., Chu G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 98, 5116–5121. 10.1073/pnas.091062498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja G., Kaestner K. H. (2007a). Forkhead transcription factors II. Cell 131:192 10.1016/j.cell.2007.09.016 [DOI] [PubMed] [Google Scholar]
- Tuteja G., Kaestner K. H. (2007b). SnapShot: forkhead transcription factors I. Cell 130:1160 10.1016/j.cell.2007.09.005 [DOI] [PubMed] [Google Scholar]
- Urness L. D., Paxton C. N., Wang X., Schoenwolf G. C., Mansour S. L. (2010). FGF signaling regulates otic placode induction and refinement by controlling both ectodermal target genes and hindbrain Wnt8a. Dev. Biol. 340, 595–604. 10.1016/j.ydbio.2010.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voulgari A., Pintzas A. (2009). Epithelial-mesenchymal transition in cancer metastasis: mechanisms, markers and strategies to overcome drug resistance in the clinic. Biochim. Biophys. Acta 1796, 75–90. 10.1016/j.bbcan.2009.03.002 [DOI] [PubMed] [Google Scholar]
- Wiener D. J., Gurtner C., Panakova L., Mausberg T. B., Müller E. J., Drögemüller C., et al. (2013). Clinical and histological characterization of hair coat and glandular tissue of Chinese crested dogs. Vet. Dermatol. 24, 274–e62. 10.1111/vde.12008 [DOI] [PubMed] [Google Scholar]
- Yang J., Weinberg R. A. (2008). Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell. 14, 818–829. 10.1016/j.devcel.2008.05.009 [DOI] [PubMed] [Google Scholar]
- Ye X., Tam W. L., Shibue T., Kaygusuz Y., Reinhardt F., Ng Eaton E., et al. (2015). Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature 525, 256–260. 10.1038/nature14897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wu J., Luo S., Lechler T., Zhang J. Y. (2016). FRA1 promotes squamous cell carcinoma growth and metastasis through distinct AKT and c-Jun dependent mechanisms. Oncotarget 7, 34371–34383. 10.18632/oncotarget.9110 [DOI] [PMC free article] [PubMed] [Google Scholar]