Abstract
Background
Early life acute respiratory infection (ARI) with respiratory syncytial virus (RSV) has been strongly associated with the development of childhood wheezing illnesses, but the pathways underlying this association are poorly understood.
Objective
To examine the role of the nasopharyngeal microbiome in the development of childhood wheezing illnesses following RSV ARI in infancy.
Methods
We conducted a nested cohort study of 118 previously healthy, term infants with confirmed RSV ARI by RT-PCR. We used next-generation sequencing of the V4 region of the 16S ribosomal RNA gene to characterize the nasopharyngeal microbiome during RSVARI. Our main outcome of interest was 2-year subsequent wheeze.
Results
Of the 118 infants, 113 (95.8%) had 2-year outcome data. Of these, 46 (40.7%) had parental report of subsequent wheeze. There was no association between the overall taxonomic composition, diversity, and richness of the nasopharyngeal microbiome during RSV ARI with the development of subsequent wheeze. However, the nasopharyngeal detection and abundance of Lactobacillus was consistently higher in infants who did not develop this outcome. Lactobacillus also ranked first among the different genera in a model distinguishing infants with and without subsequent wheeze.
Conclusions
The nasopharyngeal detection and increased abundance of Lactobacillus during RSV ARI in infancy are associated with a reduced risk of childhood wheezing illnesses at age 2 years.
Keywords: Microbiome, Lactobacillus, Staphylococcus, nasopharynx, respiratory syncytial virus, asthma, wheezing, 16S ribosomal RNA sequencing, infants
Respiratory syncytial virus (RSV) is one of the most common causes of upper and lower acute respiratory infections (ARIs) in young children worldwide.1–3 In the United States, it is associated with ~132,000 to ~172,000 inpatient admissions among preschool-aged children and its frequency appears to be increasing.4,5 In addition to its short-term effects, early life RSVARI, particularly severe infection in the critical period of infancy (ie, between 0 and 12 months of age), has been strongly associated with the development of childhood wheezing illnesses including asthma, the most common chronic disease of childhood.6–8 However, the pathways underlying this association are poorly understood.
In recent years, we and others have demonstrated that the upper airway bacterial microbiome plays an important role in the pathogenesis and pulmonary sequelae of viral ARIs.9–17 For example, several studies in murine models have shown that priming of the nasal mucosa with certain taxa (such as Lactobacillus species) increases the resistance and beneficially modulates the immune response of mice against RSV, influenza, and pneumonia virus.9,10 In a recent study of young children with RSV ARI, higher nasopharyngeal abundances of Haemophilus influenzae and Streptococcus were associated with a more proinflammatory immune response and a higher risk of hospitalization.11 In the same context, we have shown increased nasal abundances of Haemophilus, Moraxella, and Streptococcus and lower nasal abundances of Lactobacillus, Staphylococcus, and Corynebacterium in infants with RSV ARI when compared with healthy infants.13 Taken together, these findings suggest that viral-bacterial interactions have a crucial role in the severity of the ARI, the host immune response, and possibly the development of childhood wheezing illnesses.
On the basis of these findings, we hypothesized that the taxonomic composition, diversity, richness, and abundance of certain bacterial taxa of the upper airway microbiome during RSV ARI in infancy are associated with the later development of childhood wheezing illnesses. To test this hypothesis, we examined the association of the nasopharyngeal microbiome with childhood wheezing illnesses in infants with at least 1 RSV ARI enrolled in the Infant Susceptibility to Pulmonary Infections and Asthma Following RSV Exposure (INSPIRE) study.
METHODS
Overview of INSPIRE
INSPIRE is a current population-based birth cohort of previously healthy, term infants born between June and December of 2012 to 2013, designed so that the first RSV ARI during infancy could be studied. Eligible infants were enrolled mainly during a well-child visit at a participating general pediatric practice throughout the middle Tennessee region. The recruitment area encompasses urban, suburban, and rural areas. At enrollment, 1 of the parents was administered an extensive questionnaire to obtain information on the infant’s sociodemographic characteristics, birth and family history, and respiratory health. In order to capture an infant’s first RSV ARI, biweekly respiratory illness surveillance was performed during the winter viral season (November to March) of each infant’s first year of life. Infants who met prespecified criteria for an ARI had an in-person visit, which included a nasal wash for viral identification and characterization of the nasopharyngeal microbiome, as well as a physical examination for assessment of the ARI severity using the respiratory severity score (RSS).18 Annual follow-up to assess the development of childhood wheezing illnesses is ongoing. The Institutional Review Board of Vanderbilt University approved this study. The detailed methods for INSPIRE have been previously reported and are presented in detail in this article’s Online Repository at www.jacionline.org.19
Study population
The current study included 118 infants with confirmed RSVARI enrolled in INSPIRE. A total of 125 nasal washes were included, as 5 infants had 2 RSV ARI episodes and 1 infant had 3. These 118 infants represent a nested cohort with nasopharyngeal microbiome assessment during a confirmed RSV ARI and available 2-year data (the most recent longitudinal follow-up visit available) at the time of the current study.
RSV detection
The detection of RSV was made by real-time RT-PCR. For this, TaqMan assays using RSV-specific primers and probes were run on the StepOnePlus platform (Applied Biosystems, Foster City, Calif) using the AgPath-ID OneStep RT-PCR Kit (Applied Biosystems) as per a previously described protocol.20,21
Characterization of the nasopharyngeal microbiome
We have previously described in detail the methods used to characterize the nasopharyngeal microbiome using nasal washes in infants enrolled in INSPIRE.12,13 In brief, following bacterial DNA extraction, the V4 region of the 16S ribosomal RNA (rRNA) gene was amplified using universal 515F/806R primers. The libraries were then sequenced on an Illumina MiSeq platform with 2 × 300 bp reads. Negative and positive controls (with known taxonomic composition) were amplified and sequenced concurrently for quality control. Further details on these steps are available in the Online Repository at www.jacionline.org.
Outcome definitions and assessment
Our outcome of interest was subsequent wheeze, defined as parental report of any wheeze since the last birthday. To test the robustness of our results, we also conducted sensitivity analyses using the outcome of recurrent wheeze, defined as parental report of ≥2 episodes of wheeze since the last birthday. Both of these outcomes were assessed at age 2 years (the most recent longitudinal follow-up visit available) using the International Study of Asthma and Allergy in Children questionnaire.22
Data processing and statistical analyses
A mothur-based automated annotation pipeline,23 YAP,24 was used to perform initial processing of the 16S rRNA gene sequencing datasets. Low-quality sequences, chimeras, and nonbacterial sequences are discarded as part of this pipeline. Samples with ≤1000 final reads (n = 1) were discarded prior to statistical analysis. Statistical analyses were performed with the open source MGSAT package in R.25,26 The MGSAT pipeline calls on a number of R tests and packages to compare the taxonomic composition, diversity, richness, and abundance of taxa between groups, including the permutation-based ANOVA test, Shannon index, inverse Simpson index, Chao1 estimator, observed taxa counts, DESeq2,27 stabsel,28,29 and GeneSelector.30 For analyses using operational taxonomic units (OTUs), OTUs were clustered at 97% sequence identity.
Our main method to test for differential abundance of taxa in association with childhood wheezing illnesses was DESeq2.27 DESeq2 models raw absolute counts of each taxon with a negative binomial distribution and uses the estimated depth of sequencing of each sample to scale the (unknown) relative abundance that is the parameter of the negative binomial distribution. Compared with using either simple proportion-based normalization or rarefaction for controlling for differential sequencing depth, the DESeq2 approach provides improved sensitivity and specificity.31 Reported Q values are the result of a Wald test with Benjamini and Hochberg correction for multiple comparisons.32 To build alternative rankings of taxa in regard to their importance in predicting the same childhood wheezing phenotype, we also used stabsel and GeneSelector. The stabsel stability selection approach aims to build the relative ranking of the predictor variables (taxa in our case) according to their importance for predicting the outcome.28 It does so by building multiple “base” models on random subsamples of the data (n = 400 in our study). We have used the elastic net model from the R package glmnet as the base feature selection method to be wrapped by the stability protocol.33 The ranking of taxa and their probability of being selected into the model were reported, as well as the probability cutoff corresponding to the per-family error rate that is controlled by this method. The GeneSelector package was used as a stability feature ranking method that is based on a nonparametric univariate test.30 In brief, the same ranking method (package function RankingWilcoxon) was applied to multiple random subsamples of the full set of observations (400 replicates, sampling 50% of observations without replacement). RankingWilcoxon ranks features in each replicate according to the test statistic from Wilcoxon rank-sum test with regard to the outcome group (eg, subsequent wheeze vs no subsequent wheeze). Consensus ranking between replicates was then found with a Monte Carlo procedure (package function AggregateMC) and the features were reported in the order of that consensus. To account for different sequencing depth, the absolute abundance counts were normalized to simple proportions within each observation. For each feature, we also obtained several types of the effect size, such as common language effect size and rank biserial correlation.34 If a taxon received similar ranking in all 3 statistical analyses (ie, DESeq2, stabsel, and GeneSelector), then findings for that particular taxon were considered robust and unlikely to occur due to chance.
For the DESeq2 analyses, we built both unadjusted and adjusted models. Because of their well-established association with the nasopharyngeal microbiome or childhood wheezing illnesses, our initial multivariable models include the following a priori selected covariates: infant’s age, sex, maternal asthma, and early life exposure to antibiotics (either in utero or after birth). Other models were then constructed by adding additional covariates, such as RSS or mode of delivery.
Based on our initial results, we also conducted several exploratory analyses to better understand the relation between early life nasopharyngeal colonization with Lactobacillus during RSV ARI and childhood wheezing illnesses. First, we examined the association of Lactobacillus detection (based on absolute counts) and relative abundance (simple proportions) with other infants’ baseline sociodemographic and clinical characteristics (ie, age, sex, race or ethnicity, gestational age, birth weight, mode of delivery, early life exposure to antibiotics, any breastfeeding, maternal smoking, maternal asthma, RSS, and type of insurance [a marker of socioeconomic status]). To this end, we used univariable logistic regression (for Lactobacillus detection) and proportional odds models (for Lactobacillus abundance) with robust sandwich standard error estimation (Huber-White method) to account for subjects’ cluster. Next, to obtain an estimate of the magnitude of the association and further evaluate the possibility of confounding, we used multivariable logistic regression to assess the association between Lactobacillus detection with the subsequent and recurrent wheeze outcome definitions while adjusting for age, sex, maternal asthma, and early-life exposure for antibiotics. Other models were then constructed by adding additional covariates, such as RSS or mode of delivery. The Huber-White method was also used to account for subjects’ cluster.
Statistical significance was defined as P <.05 after controlling for multiple comparisons when appropriate. Further details on the statistical analyses are available in the Online Repository at www.jacionline.org.
RESULTS
Baseline characteristics of the study population
The baseline characteristics of the 118 infants with RSV ARI included in this study (as a whole and according to the outcomes of interest) are presented in Table I and in Table E1 in this article’s Online Repository at www.jacionline.org. The median age at the time of the RSV ARI was 21.8 (interquartile range [IQR]: 12.1–27.1) weeks. A total of 113 (95.8%) of the 118 infants had 2-year outcome data. Of these, 46 (40.7%) and 36 (31.9%) had parental report of subsequent and recurrent wheeze, respectively. Infants with subsequent and recurrent wheeze were more likely to be male, to be born by cesarean section, to have been exposed to antibiotics in early life, to have a nonsmoking mother, and to have a maternal history of asthma when compared with those without these outcomes, although none of these differences was statistically significant (P > .05 for all estimates). Infants with recurrent wheeze had a significantly higher RSS when compared with those without (P = .03).
TABLE I.
Baseline characteristics of infants with RSV ARI included in this study (n = 118)
| Age (wk) | 21.8 (12.1–27.1) |
| Female sex | 50 (42.4) |
| Race or ethnicity | |
| Black non-Hispanic | 20 (17.0) |
| White non-Hispanic | 74 (62.7) |
| Hispanic | 11 (9.3) |
| Other* | 13 (11.0) |
| Gestational age (wk) | 39.0 (38.5–40.0) |
| Birth weight (g) | 3377 (2894–3859) |
| Birth by cesarean section | 42 (35.6) |
| Exposure to antibiotics in utero or after birth | 62 (52.5) |
| Any breastfeeding | 86 (72.9) |
| Maternal smoking at enrollment | 26 (22.0) |
| Maternal asthma | 20 (17.0) |
| Respiratory severity score | 3.0 (2.0–5.0) |
| Insurance type | |
| Medicaid | 59 (50.0) |
| Private | 56 (47.5) |
| Other | 3 (2.5) |
Data are presented as median (interquartile range) for continuous variables or n (%) for binary variables. Percentages calculated for children with complete data.
Other includes mixed race and unknown.
Overall characteristics of the nasopharyngeal microbiome
The total high-quality sequence count was 2,138,976 with a median sequence count per sample of 18,130 (IQR: 13,240–25,970). These sequences represented a total of 357 bacterial genera, with a median of 19 (IQR: 13–31) estimated observed genera per sample. The overall taxonomic composition of the nasopharyngeal microbiome during RSV ARI in infancy was characterized by high relative abundances of Moraxella (37.6%), Streptococcus (19.7%), Haemophilus (13.5%), Corynebacterium (10.0%), and Dolosigranulum (4.7%), and low relative abundances of the remaining bacterial genera (with a combined relative abundance of 14.5%).
Main analyses
The overall taxonomic composition, diversity, and richness of the nasopharyngeal microbiome during RSV ARI in infancy were not associated with the development of subsequent wheeze
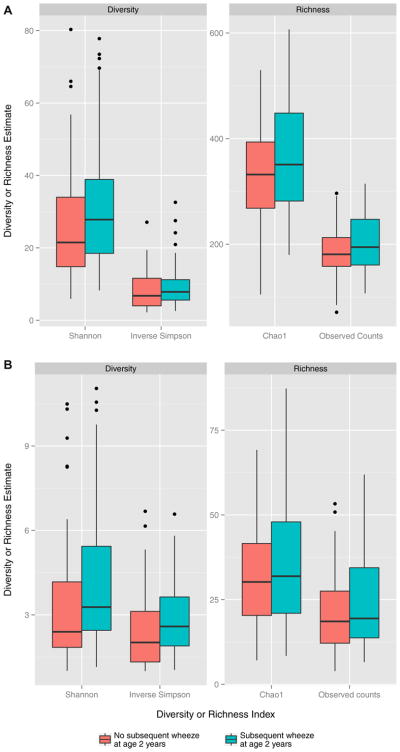
The overall taxonomic composition of the nasopharyngeal microbiome between infants with and without subsequent wheeze did not differ at the OTU (P = .9) or genus level (P = .7) in the permutation-based ANOVA test of pairwise Bray-Curtis dissimilarities. There was a trend toward higher diversity and richness in infants with subsequent wheeze using the Shannon index, inverse Simpson index, Chao1 estimator, and observed taxa counts at both the OTU (Fig 1, A) and genus (Fig 1, B) levels, although these did not reach statistical significance (P > .05 for all estimates).
FIG. 1.
Boxplots of nasopharyngeal diversity and richness in infants with RSV ARI at the OTU (A) or genus (B) level by 2-year subsequent wheeze. After rarefaction to the lowest library size, α diversity and richness estimates were calculated per each sample. This process was repeated 400 times and results were averaged. The Shannon and inverse Simpson indices were calculated to estimate abundance-based OTU or genus diversity, while the Chao1 estimator and observed taxa counts were calculated to estimate abundance-based OTU or genus richness. Both OTU and genus richness and diversity were lower in infants without 2-year subsequent wheeze, although this was not significant in any case.
Nasopharyngeal Lactobacillus was associated with reduced risk of subsequent wheeze following RSV ARI in infancy
In the initial DESeq2 analyses, the absolute counts of 3 genera were detected to be significantly different according to the development of subsequent wheeze: the absolute counts of Lactobacillus (DESeq2 test base mean = 4.93; log2 fold change = −5.21; Q = 4.69e-08) and Staphylococcus (DESeq2 test base mean = 59.29; log2 fold change = −1.47; Q = 2.08e-02) were lower in infants with subsequent wheeze compared with those without, whereas the absolute counts of Pseudomonas (DE-Seq2 test base mean = 5.84; log2 fold change = 2.22; Q = 4.95e-04) were higher. To control for potential confounders of these associations, we built a set of alternative DESeq2 models while adjusting for different relevant covariates as described in the Methods section. Only the changes in Lactobacillus and Staphylococcus absolute counts remained statistically significant in all models (Table II).
TABLE II.
Differences in the abundance of nasopharyngeal bacterial genera in infants with RSV ARI by the development of 2-year subsequent wheeze
| Genus | Model 1*
|
Model 2*
|
Model 3*
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Base mean | Log2-fold change (SE)† | Q value | Base mean | Log2-fold change (SE)† | Q value | Base mean | Log2-fold change (SE)† | Q value | |
| Lactobacillus | 8.24 | −4.45 (0.73) | 3.25e-08 | 8.42 | −4.56 (0.74) | 3.22e-08 | 8.24 | −4.57 (0.74) | 2.02e-08 |
|
| |||||||||
| Staphylococcus | 901.28 | −2.59 (0.57) | 1.00e-04 | 947.73 | −2.94 (0.58) | 6.75e-06 | 901.28 | −2.84 (0.57) | 1.36e-05 |
|
| |||||||||
| Pseudomonas | 53.98 | 2.29 (0.60) | 1.39e-03 | 56.34 | 2.39 (0.61) | 9.17e-04 | ‡ | ‡ | ‡ |
Data presented are the results of the DESeq2 test. Base means are calculated after normalizing read counts for each sample to account for differences in sequencing depth. Reported Q values are the result of a Wald test with Benjamini and Hochberg correction for multiple comparisons. Genera are ordered from lowest to highest Q value. Only genera with Q values below the statistical significance threshold of .05 in the initial DESeq2 test are presented (see main text for details).
Model 1 includes age, sex, maternal asthma, and exposure to antibiotics in utero or after birth as covariates. Model 2 includes the same covariates as model 1 plus respiratory severity score. Model 3 includes the same covariates as model 1 plus mode of delivery.
A log2-fold change of >0 indicates that the abundance of that particular genus was detected to be higher in infants with RSV ARI who later developed subsequent wheeze when compared with those who did not developed this outcome, while a log2-fold change <0 indicates the opposite.
Indicates Q value was not <.05 for the genus in the specified model.
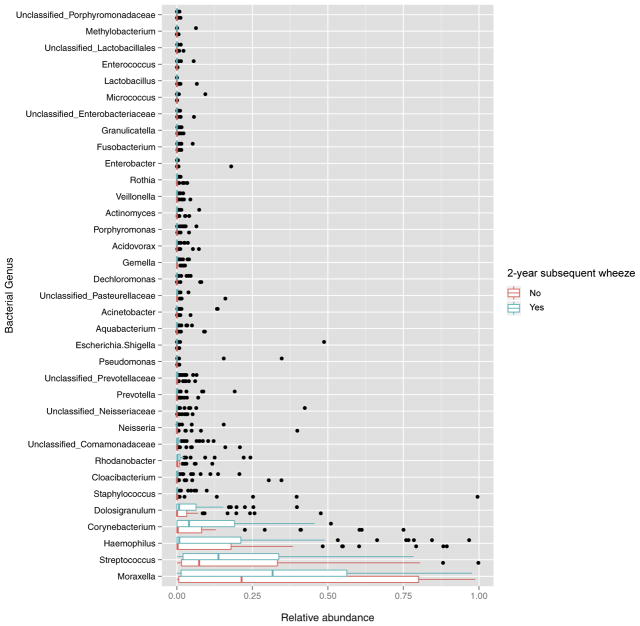
The relative abundances of the 35 most common genera expressed as simple proportions and split according to the development of subsequent wheeze are shown in Fig 2. Lactobacillus was the 31st most abundant genus, while Staphylococcus was the 15th. The mean ± SD relative abundances of Lactobacillus and Staphylococcus over all samples where subsequent wheeze data was available were 9.39e-04 ± 6.53e-03 and 0.02 ± 0.11, respectively. In spite of its low relative abundance, the mean ± SD relative abundance of Lactobacillus was ~2 orders of magnitude lower in infants with subsequent wheeze compared with those without (2.47e-05 ± 5.51e-05 vs 1.94e-03 ± 1.94e-03; P = .04 using a Mann-Whitney U-test). In contrast, there was no significant difference in the mean ± SD relative abundance of Staphylococcus between infants with and without subsequent wheeze (6.89e-03 ± 1.84e-02 vs 3.67e-02 ± 1.52e-01; P =.9 using a Mann-Whitney U-test).
FIG. 2.
Boxplots of relative abundance of nasopharyngeal bacterial genera in infants with RSV ARI by 2-year subsequent wheeze. Within each sample, counts were normalized to simple proportions. The relative abundance of the 35 most abundant genera is shown; all other genera are not shown in this figure. The median (middle bar), third quartile (right-most bar), and first quartile (left-most bar) abundances are shown. Outliers are represented as dots.
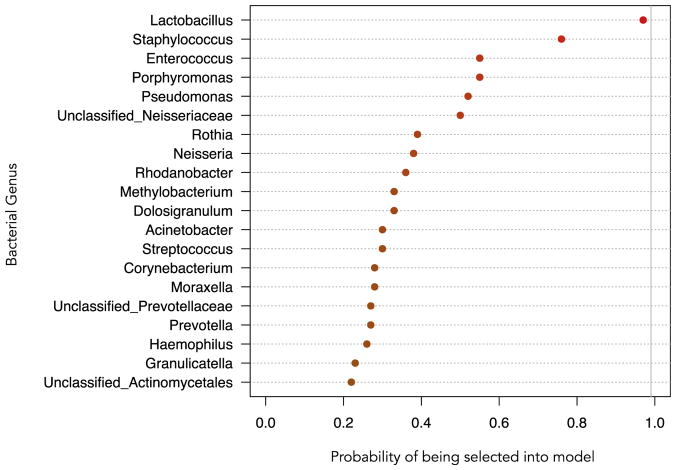
In a stabsel stability selection protocol distinguishing infants with and without subsequent wheeze using elastic net regression, Lactobacillus ranked first among the different genera, with a probability of being included into the model of 0.97, while Staphylococcus ranked second, with a probability of being included into the model of 0.76 (Fig 3). The mean relative abundance and standard deviation of the top 20 ranked genera in the stability selection model according to subsequent wheeze are shown in Table E2 in this article’s Online Repository at www.jacionline.org.
FIG. 3.
Probability of a nasopharyngeal bacterial genus being selected into a stability selection model distinguishing infants with RSV ARI with and without 2-year subsequent wheeze. The probability is plotted along the x-axis. The genera are indicated along the y-axis. The top 20 ranked genera are shown in this figure. Lactobacillus ranks highest among taxa selected into the model with a probability of being selected of 0.97.
In a GeneSelector stability ranking procedure that wraps a nonparametric Wilcoxon rank-sum test, Lactobacillus was again ranked first among all genera (rank-biserial correlation effect size of subsequent wheeze relative to the group without subsequent wheeze = −0.23), while Staphylococcus was ranked 32nd (rank-biserial correlation effect size of subsequent wheeze relative to the group without subsequent wheeze = 0.01).
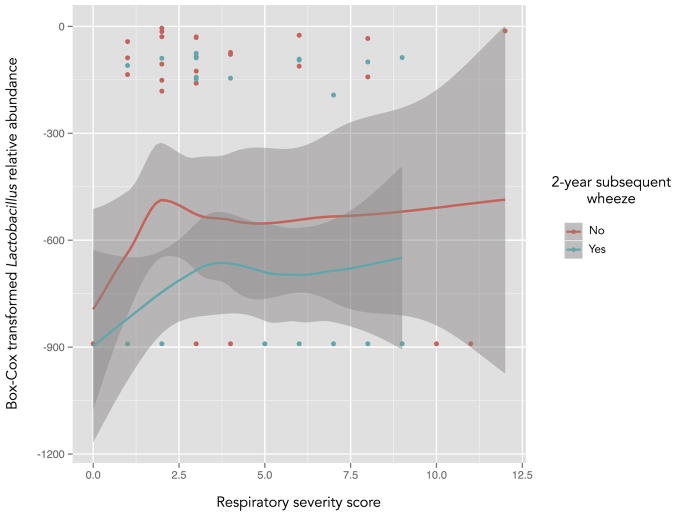
To further evaluate the possibility of confounding by RSVARI severity, we then examined the abundance of Lactobacillus and Staphylococcus in each subsequent wheeze group over the different values of the RSS. The relative abundance of Lactobacillus was generally lower among infants with subsequent wheeze over all values of the RSS (Fig 4). In contrast, there was no longer a clear trend for Staphylococcus, which suggests a potential confounding effect of RSVARI severity for this particular genus (see Fig E1 in this article’s Online Repository at www.jacionline.org).
FIG. 4.
Box-Cox transformed nasopharyngeal relative abundance of Lactobacillus in infants with RSV ARI with (blue line) and without (red line) 2-year subsequent wheeze, plotted by RSS. Lines are local regression (LOESS) smoothed curves and gray areas are the 95% CIs. For the y-axis, values closer to 0 indicate a higher abundance. Not all individual data points are shown; a single data point is displayed for infants who had the same RSS and Lactobacillus abundance. In general, the relative abundance of Lactobacillus was lower among infants with subsequent wheeze over all values of the RSS.
We performed several additional analyses to examine the reliability of the association for Lactobacillus and Staphylococcus with subsequent wheeze, which can be found in the Online Repository at www.jacionline.org. In these analyses, none of the infants with a relative abundance of Lactobacillus greater than ~0.001 developed subsequent wheeze when using either unrarefied or rarified datasets (see Figs E2 and E3 in this article’s Online Repository at www.jacionline.org).
Sensitivity analyses
In spite of the smaller sample size of infants with recurrent wheeze, we found similar results when using this outcome instead of subsequent wheeze. The overall taxonomic composition, diversity, and richness of the nasopharyngeal microbiome did not differ at the genus level in infants with RSVARI according to the development of recurrent wheeze (P > .05 for all estimates); however, infants with recurrent wheeze also had significantly lower Lactobacillus abundance than did those without in DESeq2 analyses (DESeq2 test base mean = 2.81; log2 fold change = −3.58; Q = 1.47e-03), and the relative abundance of Lactobacillus was also generally lower among infants with recurrent wheeze over all values of the RSS (see Fig E4 in this article’s Online Repository at www.jacionline.org).
Exploratory analyses
In exploratory analyses, there was no association between Lactobacillus detection or abundance and any of the covariates shown in Table I (P > .05 for all estimates). The genus Lactobacillus was detected in 39 of the 125 samples (31.2%) and the proportion of samples with this genus was lower in infants with subsequent wheeze than in those without (26% vs 49%). In a multivariable model adjusting for age, sex, maternal asthma, and early life exposure for antibiotics, the detection of Lactobacillus decreased the odds of subsequent wheeze by ~70% (odds ratio [OR]: 0.34; 95% CI:0.14–0.83; P = .02) and of recurrent wheeze by ~80% (OR: 0.21; 95% CI: 0.07–0.65; P = .006). Including other potential confounders in these multivariable models, such as RSS or mode of delivery, or conducting the exploratory analyses after rarefication to the lowest library size found among all samples with the nonzero initial count of Lactobacillus did not significantly change our results (data not shown).
DISCUSSION
In our study, we found that the nasopharyngeal detection and increased abundance of Lactobacillus during RSVARI in infancy are associated with a reduced risk childhood wheezing illnesses at age 2 years. Despite its low abundance compared with other taxa, Lactobacillus was still identified as being the strongest, most consistent discriminating taxon between infants with and without childhood wheezing illnesses by age 2 years across several different statistical methods. Furthermore, our results suggest that the detection of Lactobacillus in the nasopharynx of RSV-infected infants could be used as a biomarker for the later development of childhood wheezing illnesses. There was also a trend toward higher diversity and richness of the nasopharyngeal microbiome during RSV ARI in infants who developed childhood wheezing illnesses at age 2 years, although this was not statistically significant. We have previously shown a higher nasopharyngeal microbial diversity and richness in infants born via cesarean section compared with infants born via vaginal delivery.35 A higher diversity and richness of the gut microbiome has been associated with better outcomes, whereas a higher microbial diversity and richness of the vaginal microbiome has been associated with worse outcomes.36,37 Thus, the relationship of diversity and richness with health appears to be specific to each human body habitat.37
One other study has examined the association of the early life respiratory microbiome and childhood wheezing illnesses using culture-independent, next-generation sequencing techniques.14 In this study, the investigators found that a high nasopharyngeal abundance of Streptococcus in infancy was a strong predictor of current wheeze at age 5 years but not at age 10 years. Unlike ours, this study assessed the upper airway microbiome during health and included only children born to atopic parents. In addition, the nasopharyngeal detection and abundance of Lactobacillus were not reported and the statistical analyses used focused only on the most abundant genera, which could explain the contradictory results.
To our knowledge, this is the first study to show a protective effect of upper airway colonization with Lactobacillus on asthma-related outcomes in children. In biologic support of our findings, Tomosada et al9 previously showed that nasal administration of L rhamnosus improves the antiviral immune response, enhances viral clearance, and reduces the lung injury associated with RSV ARI in mice. In humans, 1 study found a low abundance of L sakei in the maxillary sinus in adults with chronic rhinosinusitis,38 while another found a low abundance of the genus Lactobacillus in the trachea of premature newborns who later developed bronchopulmonary dysplasia.39 In the same context, we have previously shown a higher nasopharyngeal abundance of Lactobacillus in healthy infants when compared with infants with RSV ARI.13 Lactobacilli are also the primary bacteria of the vaginal flora of healthy women, and birth by cesarean section (which likely reduces upper airway colonization with Lactobacillus species) has been repeatedly associated with an increased risk of childhood wheezing illnesses.35,40–42 Taken together, these findings strongly suggest a protective role of upper airway colonization with Lactobacillus in childhood respiratory outcomes.
While the effect of upper airway colonization with Lactobacillus on asthma-related outcomes in both children and adults has been understudied, numerous studies have focused on the effect of gastrointestinal supplementation with Lactobacillus species on childhood wheezing illnesses, yielding conflicting results.43 Because the nasopharyngeal route can induce local and systemic immune responses superior to those obtained using the oral route,9,10 respiratory supplementation with Lactobacillus in early life could be a potential strategy to prevent childhood wheezing illnesses.
In addition to Lactobacillus, we also found an association of upper airway colonization with Staphylococcus on childhood wheezing illnesses following RSV ARI in infancy. However, unlike Lactobacillus, this effect was not consistent across all statistical analyses and this lack of consistency (described in greater detail in the Online Repository at www.jacionline.org) prevents us from confidently concluding that Staphylococcus is protective against childhood wheezing illnesses within this cohort. Staphylococcus is one of the most common colonizers in the nasopharynx of healthy infants and its abundance appears to decrease over time.14,16,44,45 We have previously shown a higher nasopharyngeal abundance of Staphylococcus in healthy infants when compared with infants with RSV ARI.13 In the only other study examining the association of the early life respiratory microbiome and childhood wheezing illnesses using next-generation sequencing, no association was noted between nasopharyngeal colonization with Staphylococcus in infancy and current wheeze at age 5 or 10 years.14
The gut microbiome has been shown to play an important role in preserving host health.46,47 More recently, the manipulation of the gut microbiome has emerged as a strategy to prevent and treat certain diseases.48,49 In contrast to the gut microbiome, the upper airway microbiome has been relatively understudied. However, the upper airway is of germane interest in viral ARIs and childhood allergic diseases, such as asthma, as it is the first portal of entry of respiratory pathogens and aeroallergens. Our findings of nasopharyngeal bacterial genera independently associated with childhood wheezing illnesses are meaningful, as the implications are that targeted modifications of the infant upper airway microbiome could be a strategy to decrease morbidity from both RSV ARIs and childhood asthma, the most common acute and chronic diseases of infancy and childhood, respectively.
Our study has considerable strengths, including the population-based longitudinal design of the parent study, the inclusion of infants with different degrees of RSV ARI severity, the use of predefined criteria for ARIs, and the close surveillance during the first winter viral season to capture each infant’s initial RSV ARI. We also acknowledge several limitations. First, due to the inherent limits of 16S rRNA gene sequencing-based analyses, we were unable to identify bacterial taxa below the genus level and thus cannot identify whether particular Lactobacillus strains or species, or a consortium of members of the Lactobacillus genus, protect against childhood wheezing illnesses. The V4 region of the 16S rRNA gene is not sufficient to identify Lactobacillus species and our OTU-based analyses were also limited due to the low abundance of Lactobacillus in the nasopharynx of infants with RSV ARI. Unlike the genus Lactobacillus, which contains species generally regarded as safe and beneficial for human health, the genus Staphylococcus contains species that can be potentially detrimental and, thus, translation of our results into preventive strategies for this particular genus should be done with caution. Second, the nasopharyngeal abundance of Lactobacillus was generally low, which may have affected our estimates. Third, due to the observational design, there could also be residual confounding by variables not measured in our study (such as prior infection or coinfection with other respiratory viruses). Fourth, because childhood wheezing illnesses are a heterogeneous group of disorders,50 the effect of upper airway colonization with Lactobacillus on later asthma phenotypes will need to be examined. Last, our results might not be generalizable to infants without RSV ARI. However, RSV is a ubiquitous infection and a major cause of infant morbidity worldwide, thus our findings are relevant to a group at a high risk of developing childhood wheezing illnesses.
In summary, we found that the nasopharyngeal detection and increased abundance of Lactobacillus during RSVARI in infancy are associated with a reduced risk of childhood wheezing illnesses at age 2 years. While these preliminary findings merit replication in larger longitudinal cohorts, they provide novel data that could support the development of new prognostic or preventive strategies for childhood wheezing illnesses.
Supplementary Material
Key messages.
The nasopharyngeal detection and increased abundance of Lactobacillus during ARI with RSV in infancy are associated with a reduced risk of childhood wheezing illnesses at age 2 years.
Our results suggest that the detection of Lactobacillus in the nasopharynx of RSV-infected infants could be used as a biomarker for the later development of childhood wheezing illnesses.
Furthermore, providing Lactobacillus during RSV ARI in infancy could be a potential primary prevention intervention strategy for the development of childhood wheezing illnesses.
Acknowledgments
This work was supported in whole or in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under award numbers U19AI095227, K24AI77930, HHSN272200900007 C, and U19AI110819; the Vanderbilt Institute for Clinical and Translational Research grant support (National Center for Advancing Translational Sciences, National Institutes of Health, Department of Health and Human Services, under award numbers UL1 TR000445 and U54RR24975); the Vanderbilt Faculty Research Scholars Program; and the Parker B. Francis Fellowship Program.
Abbreviations used
- ARI
Acute respiratory infection
- INSPIRE
Infant Susceptibility to Pulmonary Infections and Asthma Following RSV Exposure
- IQR
Interquartile range
- OTU
Operational taxonomic unit
- rRNA
Ribosomal RNA
- RSS
Respiratory severity score
- RSV
Respiratory syncytial virus
Footnotes
Disclosure of potential conflict of interest: C. Rosas-Salazar has received a grant from the Francis Family Foundation. M. Shilts has received a grant and travel support from the National Institutes of Health. A. Tovchigrechko has received grants from J. Craig Venter Institute and the National Institutes of Health and is employed by and receives stock/stock options from MedImmune LLC. S. Schobel has received a grant from the J. Craig Venter Institute and the National Institutes of Health and is employed by the Henry M. Jackson Foundation. J. Chappell has received a grant from the National Institutes of Health. M. Moore has received a grant from the National Institutes of Health and is the founder of Meissa Vaccines. R. Peebles has received a grant from the National Institutes of Health. S. Das has received grants from the National Institutes of Health, Emergent Biosolution, Cargill, Abviro, and the Centers for Disease Control and is employed by Vanderbilt University Medical Center. T. Hartert has received grants from the National Institutes of Health and the Agency for Healthcare Research and Quality and is Associate Editor for the American Journal of Respiratory and Critical Care Medicine. The rest of the authors declare that they have no relevant conflicts of interest.
References
- 1.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, et al. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet. 2010;375:1545–55. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hall CB, Weinberg GA, Iwane MK, Blumkin AK, Edwards KM, Staat MA, et al. The burden of respiratory syncytial virus infection in young children. N Engl J Med. 2009;360:588–98. doi: 10.1056/NEJMoa0804877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall CB, Weinberg GA, Blumkin AK, Edwards KM, Staat MA, Schultz AF, et al. Respiratory syncytial virus-associated hospitalizations among children less than 24 months of age. Pediatrics. 2013;132:e341–8. doi: 10.1542/peds.2013-0303. [DOI] [PubMed] [Google Scholar]
- 4.Shay DK, Holman RC, Newman RD, Liu LL, Stout JW, Anderson LJ. Bronchiolitis-associated hospitalizations among US children, 1980–1996. JAMA. 1999;282:1440–6. doi: 10.1001/jama.282.15.1440. [DOI] [PubMed] [Google Scholar]
- 5.Stockman LJ, Curns AT, Anderson LJ, Fischer-Langley G. Respiratory syncytial virus-associated hospitalizations among infants and young children in the United States, 1997–2006. Pediatr Infect Dis J. 2012;31:5–9. doi: 10.1097/INF.0b013e31822e68e6. [DOI] [PubMed] [Google Scholar]
- 6.Regnier SA, Huels J. Association between respiratory syncytial virus hospitalizations in infants and respiratory sequelae: systematic review and meta-analysis. Pediatr Infect Dis J. 2013;32:820–6. doi: 10.1097/INF.0b013e31829061e8. [DOI] [PubMed] [Google Scholar]
- 7.Feldman AS, He Y, Moore ML, Hershenson MB, Hartert TV. Toward primary prevention of asthma: reviewing the evidence for early-life respiratory viral infections as modifiable risk factors to prevent childhood asthma. Am J Respir Crit Care Med. 2015;191:34–44. doi: 10.1164/rccm.201405-0901PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malveaux FJ. The state of childhood asthma: introduction. Pediatrics. 2009;123(suppl 3):S129–30. doi: 10.1542/peds.2008-2233B. [DOI] [PubMed] [Google Scholar]
- 9.Tomosada Y, Chiba E, Zelaya H, Takahashi T, Tsukida K, Kitazawa H, et al. Nasally administered Lactobacillus rhamnosus strains differentially modulate respiratory antiviral immune responses and induce protection against respiratory syncytial virus infection. BMC Immunol. 2013;14:40. doi: 10.1186/1471-2172-14-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitazawa H, Villena J. Modulation of respiratory TLR3-anti-viral response by probiotic microorganisms: lessons learned from Lactobacillus rhamnosus CRL1505. Front Immunol. 2014;5:201. doi: 10.3389/fimmu.2014.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Steenhuijsen Piters WA, Heinonen S, Hasrat R, Bunsow E, Smith B, Suarez-Arrabal MC, et al. Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am J Respir Crit Care Med. 2016;194:1104–15. doi: 10.1164/rccm.201602-0220OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosas-Salazar C, Shilts MH, Tovchigrechko A, Schobel S, Chappell JD, Larkin EK, et al. Differences in the nasopharyngeal microbiome during acute respiratory tract infection with human rhinovirus and respiratory syncytial virus in infancy. J Infect Dis. 2016;214:1924–8. doi: 10.1093/infdis/jiw456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosas-Salazar C, Shilts MH, Tovchigrechko A, Chappell JD, Larkin EK, Nelson KE, et al. Nasopharyngeal microbiome in respiratory syncytial virus resembles profile associated with increased childhood asthma risk. Am J Respir Crit Care Med. 2016;193:1180–3. doi: 10.1164/rccm.201512-2350LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teo SM, Mok D, Pham K, Kusel M, Serralha M, Troy N, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015;17:704–15. doi: 10.1016/j.chom.2015.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyde ER, Petrosino JF, Piedra PA, Camargo CA, Jr, Espinola JA, Mansbach JM. Nasopharyngeal proteobacteria are associated with viral etiology and acute wheezing in children with severe bronchiolitis. J Allergy Clin Immunol. 2014;133:1220–2. doi: 10.1016/j.jaci.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biesbroek G, Tsivtsivadze E, Sanders EA, Montijn R, Veenhoven RH, Keijser BJ, et al. Early respiratory microbiota composition determines bacterial succession patterns and respiratory health in children. Am J Respir Crit Care Med. 2014;190:1283–92. doi: 10.1164/rccm.201407-1240OC. [DOI] [PubMed] [Google Scholar]
- 17.Chonmaitree T, Jennings K, Golovko G, Khanipov K, Pimenova M, Patel JA, et al. Nasopharyngeal microbiota in infants and changes during viral upper respiratory tract infection and acute otitis media. PLoS One. 2017;12:e0180630. doi: 10.1371/journal.pone.0180630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodriguez H, Hartert TV, Gebretsadik T, Carroll KN, Larkin EK. A simple respiratory severity score that may be used in evaluation of acute respiratory infection. BMC Res Notes. 2016;9:85. doi: 10.1186/s13104-016-1899-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larkin EK, Gebretsadik T, Moore ML, Anderson LJ, Dupont WD, Chappell JD, et al. Objectives, design and enrollment results from the Infant Susceptibility to Pulmonary Infections and Asthma Following RSV Exposure Study (INSPIRE) BMC Pulm Med. 2015;15:45. doi: 10.1186/s12890-015-0040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kodani M, Yang G, Conklin LM, Travis TC, Whitney CG, Anderson LJ, et al. Application of TaqMan low-density arrays for simultaneous detection of multiple respiratory pathogens. J Clin Microbiol. 2011;49:2175–82. doi: 10.1128/JCM.02270-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emery SL, Erdman DD, Bowen MD, Newton BR, Winchell JM, Meyer RF, et al. Real-time reverse transcription-polymerase chain reaction assay for SARS-associated coronavirus. Emerg Infect Dis. 2004;10:311–6. doi: 10.3201/eid1002.030759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asher MI, Montefort S, Bjorksten B, Lai CK, Strachan DP, Weiland SK, et al. Worldwide time trends in the prevalence of symptoms of asthma, allergic rhino-conjunctivitis, and eczema in childhood: ISAAC phases one and three repeat multicountry cross-sectional surveys. Lancet. 2006;368:733–43. doi: 10.1016/S0140-6736(06)69283-0. [DOI] [PubMed] [Google Scholar]
- 23.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tovchigrechko A. [Accessed September 10, 2017];Yet Another Pipeline (YAP) 2015 Available at: https://github.com/andreyto/YAP.
- 25.R Development Core Team. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2006. [Google Scholar]
- 26.Tovchigrechko A. [Accessed September 10, 2017];MGSAT: statistical analysis of microbiome and proteome abundance matrices with automated report generation. 2015 Available at: https://bitbucket.org/andreyto/mgsat.
- 27.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hofner B, Hothorn T. [Accessed August 23, 2017];Stabs: stability selection with error control. 2014 Available at: http://CRAN.R-project.org/package=stabs.
- 29.Hofner B, Boccuto L, Goker M. Controlling false discoveries in high-dimensional situations: boosting with stability selection. BMC Bioinformatics. 2015;16:144. doi: 10.1186/s12859-015-0575-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boulesteix A-L, Slawski M. Stability and aggregation of ranked gene lists. Brief Bioinform. 2009;10:556–68. doi: 10.1093/bib/bbp034. [DOI] [PubMed] [Google Scholar]
- 31.McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol. 2014;10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 33.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 34.Grissom RJ, Kim JJ. Effect sizes for research: univariate and multivariate applications. 2. London: Routledge; 2012. [Google Scholar]
- 35.Shilts MH, Rosas-Salazar C, Tovchigrechko A, Larkin EK, Torralba M, Akopov A, et al. Minimally invasive sampling method identifies differences in taxonomic richness of nasal microbiomes in young infants associated with mode of delivery. Microb Ecol. 2016;71:233–42. doi: 10.1007/s00248-015-0663-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fredricks DN, Fiedler TL, Marrazzo JM. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med. 2005;353:1899–911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 37.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abreu NA, Nagalingam NA, Song Y, Roediger FC, Pletcher SD, Goldberg AN, et al. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci Transl Med. 2012;4:151ra24. doi: 10.1126/scitranslmed.3003783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lal CV, Travers C, Aghai ZH, Eipers P, Jilling T, Halloran B, et al. The airway microbiome at birth. Sci Rep. 2016;6:31023. doi: 10.1038/srep31023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thavagnanam S, Fleming J, Bromley A, Shields MD, Cardwell CR. A meta-analysis of the association between caesarean section and childhood asthma. Clin Exp Allergy. 2008;38:629–33. doi: 10.1111/j.1365-2222.2007.02780.x. [DOI] [PubMed] [Google Scholar]
- 41.Bager P, Wohlfahrt J, Westergaard T. Caesarean delivery and risk of atopy and allergic disease: meta-analyses. Clin Exp Allergy. 2008;38:634–42. doi: 10.1111/j.1365-2222.2008.02939.x. [DOI] [PubMed] [Google Scholar]
- 42.Wu P, Feldman AS, Rosas-Salazar C, James K, Escobar G, Gebretsadik T, et al. Relative importance and additive effects of maternal and infant risk factors on childhood asthma. PLoS One. 2016;11:e0151705. doi: 10.1371/journal.pone.0151705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Elazab N, Mendy A, Gasana J, Vieira ER, Quizon A, Forno E. Probiotic administration in early life, atopy, and asthma: a meta-analysis of clinical trials. Pediatrics. 2013;132:e666–76. doi: 10.1542/peds.2013-0246. [DOI] [PubMed] [Google Scholar]
- 44.Tsai MH, Chiu CY, Shih HJ, Liao SL, Hua MC, Huang SH, et al. Longitudinal investigation of nasopharyngeal methicillin-resistant Staphylococcus aureus colonization in early infancy: the PATCH birth cohort study. Clin Microbiol Infect. 2017;23:121e1–7. doi: 10.1016/j.cmi.2016.10.020. [DOI] [PubMed] [Google Scholar]
- 45.Biesbroek G, Bosch AA, Wang X, Keijser BJ, Veenhoven RH, Sanders EA, et al. The impact of breastfeeding on nasopharyngeal microbial communities in infants. Am J Respir Crit Care Med. 2014;190:298–308. doi: 10.1164/rccm.201401-0073OC. [DOI] [PubMed] [Google Scholar]
- 46.Lynch SV, Pedersen O. The human intestinal microbiome in health and disease. N Engl J Med. 2016;375:2369–79. doi: 10.1056/NEJMra1600266. [DOI] [PubMed] [Google Scholar]
- 47.Marchesi JR, Adams DH, Fava F, Hermes GD, Hirschfield GM, Hold G, et al. The gut microbiota and host health: a new clinical frontier. Gut. 2016;65:330–9. doi: 10.1136/gutjnl-2015-309990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scott KP, Antoine JM, Midtvedt T, van Hemert S. Manipulating the gut microbiota to maintain health and treat disease. Microb Ecol Health Dis. 2015;26:25877. doi: 10.3402/mehd.v26.25877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young VB. Therapeutic manipulation of the microbiota: past, present, and considerations for the future. Clin Microbiol Infect. 2016;22:905–9. doi: 10.1016/j.cmi.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reddy MB, Covar RA. Asthma phenotypes in childhood. Curr Opin Allergy Clin Immunol. 2016;16:127–34. doi: 10.1097/ACI.0000000000000252. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.