Abstract
The Xenopus laevis oocyte provides a unique model to study cell polarity. During oogenesis, numerous mRNAs and proteins become asymmetrically distributed in the oocyte cytoplasm and their analysis is important for understanding the establishment of developmental polarity. Microinjection of fluorescently labeled RNA can recapitulate the localization pattern of endogenous mRNA and can be combined with immunofluorescence analysis of proteins of interest. Such analyses can provide insight into the cytoplasmic interactions contributing to polarity and imaging by confocal microscopy allows a high-resolution view of RNA-protein interactions. Here, we present an updated method to image endogenous protein and microinjected RNA in immature X. laevis oocytes.
MATERIALS
It is essential that you consult the appropriate Material Safety Data Sheets and your institution’s Environmental Health and Safety Office for proper handling of equipment and hazardous materials used in this protocol.
Reagents
Ammonium acetate (7 M, RNase-free)
Collagenase solution <R>
Cy5-UTP or Cy3-UTP (GE Healthcare Life Sciences, PA55026/PA53026)
Ethanol (70%, RNase-free)
Ethanol (100%, RNase-free)
Glycogen (20 mg/mL; Thermo Scientific, R0561)
MBSH Buffer (10×) <R>
MEMFA <R>
Methanol (anhydrous, 100%)
mMessage machine SP6/T7/T3 transcription kit (Ambion, AM1340/AM1344/AM1348)
Murray’s clear <R>
Nuclease-free H2O (Thermo Fisher, AM9914G) or H2O treated with DEPC (Sigma D5758)
Oocyte culture medium (OCM) <R>
PBT for Xenopus oocytes/PBT-Plus <R>
Phenol/chloroform/isoamylalcohol (25:24:1, Fisher Scientific, AC327115000)
Plasmid containing sequence of interest downstream of an RNA polymerase promoter (T7, SP6, or T3)
Proteinase K buffer for Xenopus oocytes <R>
Primary antibody
Restriction enzyme appropriate for plasmid linearization
RNase-ZAP (Ambion, AM9780)
Secondary antibody, fluorescently-labeled
Equipment
Aluminum foil
Barrier pipette tips
Dissecting microscope
Fluorodishes (10 mm inside diameter; World Precision Instruments, FD3510-100)
Heat blocks (37°C and 72°C)
Ice bucket
Illustra ProbeQuant G-50 Micro Columns (GE Healthcare, 29-9034-08)
Inverted confocal microscope (such as Zeiss LSM 510 Meta Confocal Laser Scanning Microscope)
Microcentrifuges at room temperature and 4°C
Microcentrifuge tubes (1.5 mL, RNase-free)
Microcentrifuge tubes (1.5 mL, siliconized or low-retention)
Microinjection setup (such as Harvard Apparatus PLI-100 PicoInjector and Narishige MN-151 Joystick Micromanipulator)
NanoDrop spectrophotometer
Pasteur pipettes
Pulled glass pipette with a fine tip
Rocking platform
Surgical instruments for removing oocytes from Xenopus laevis females
Tissue culture plate (24-well)
Vortex mixer
Xenopus laevis females for oocyte preparation
METHOD
This protocol covers co-imaging of injected RNA and protein. Alternatively, RNA or protein can be visualized independently of each other. If imaging only endogenous protein, proceed to “Preparation of oocytes” (Step 16).
Preparation of fluorescently-labeled RNA
-
1
Linearize plasmid DNA containing the sequence of interest cloned downstream of a T7, SP6, or T3 promoter with an appropriate restriction enzyme.
-
2
Purify the plasmid DNA by phenol chloroform extraction followed by ethanol precipitation, and resuspend in nuclease-free H2O at 500–1000 ng/µL.
-
3Prepare an RNase-free working area by treating all surfaces and gloves with RNase-ZAP.For subsequent steps, up to Step 15, use only barrier pipette tips and RNase-free reagents.
-
4Modify the Ambion mMessage machine transcription kit by adding the reagents in the table below, in the order shown, to an RNase-free microfuge tube:
Reagent Volume Final
ConcentrationNotes 10× Transcription buffer 2.0 µL 1× 2× NTP/cap mixture 6.0 µL 3.3× The amount of NTP/cap mixture is altered from the manufacturer’s protocol to facilitate incorporation of fluorescent UTP Cy3-UTP or Cy5-UTP 1.5 µL 0.375 mM ChromaTide Alexa-Fluor UTPs (Thermo-Fisher) may also be used at a final concentration of 0.3 mM Linearized DNA (variable) 1 µg Less than 1 µg can be used per reaction but will reduce yield 10× enzyme mix 2.0 µL 1× Reactions using SP6 RNA polymerase require longer incubation times (4 hours) compared with those using T7 or T3 RNA polymerases (2 hours) for similar yields Nuclease-free H2O (variable) - Total 20 µl Reaction may be scaled up for increased RNA production. -
5
Mix reaction by gentle pipetting.
-
6
Incubate at 37°C for 2–4 hours. Protect tubes from light by covering with foil.
-
7
Pulse spin in a microcentrifuge. Add 1 µL Turbo DNase (included in mMessage machine kit).
-
8
Incubate at 37°C for 15 minutes with protection from light.
-
9
Add 179 µL 200 mM EDTA, pH 8.0, to stop the reaction. Mix gently by pipetting.
-
10
Add 200 µL phenol/chloroform/isoamylalchohol (1:1 ratio) to the reaction. Vortex for 15 seconds. Centrifuge at room temperature for 10 minutes at maximum speed.
-
11
Carefully remove the upper aqueous phase containing the RNA, and transfer to an RNAse-free tube. Avoid transferring any of the interphase.
-
12
To remove unincorporated nucleotides (including free fluorescent UTP) pass the RNA through an Illustra ProbeQuant G-50 Micro Column, following manufacturer’s instructions (GE Healthcare).
-
13Concentrate the RNA by ethanol precipitation.
- Add 500 µL 100% ethanol, (2.5× volume), 20 µL 7 M ammonium acetate (0.1× volume) and 1 µL 20 mg/mL glycogen. Mix well by vortexing, and incubate for 1 hour at −80°C.
- Centrifuge at maximum speed at 4°C for 20 minutes. Remove and discard the supernatant and add 1 mL 70% ethanol (stored at −20°C).
- Centrifuge at maximum speed at 4°C for 5 minutes. Remove and discard the supernatant, wash again with 70% ethanol. Centrifuge at maximum speed at 4°C for 5 minutes.
- Remove and discard the supernatant. Allow the RNA pellet to dry at room temperature for 10 minutes, or until most residual 70% ethanol has evaporated. Do not let the pellet dry completely.
- Resuspend RNA in 15 µL nuclease-free H2O.
-
14
Determine RNA concentration using a NanoDrop spectrophotometer, according to the manufacturer’s instructions. (The RNA should generate a curve with a single peak at 280 nM, and a 260/280 nM ratio of ~2.0.) Calculate RNA molarity, and adjust to a final concentration of 1 µM.
-
15
Store RNA at −80°C in 2 µL aliquots (for single use only). RNA will be stable for several months at −80°C.
Preparation of oocytes
For a detailed procedure see Protocol: Isolation of Xenopus oocytes<prot095851> (Newman et al. 2017)
-
16
Surgically remove oocytes from Xenopus laevis females. For imaging oocytes beyond stage II, albino frogs must be used.
-
17
Defolliculate oocytes using collagenase solution.
-
18
Wash oocytes 3–5 times in 1× MBSH buffer, until the wash buffer is clear.
-
19
Sort oocytes to obtain stages of interest and place in OCM.
-
20
Allow oocytes to recover in OCM at 18°C for at least 1 hour. Overnight recovery is preferable.
Microinjection
For a detailed microinjection protocol, see Protocol: Microinjection of Xenopus Oocytes<prot096974> (Aguero et al. 2017). If imaging only endogenous protein, go to “Fixation and immunofluorescence”, Step 28.
-
21
Thaw an aliquot of fluorescently-labeled RNA (from Step 15) on ice. Dilute the RNA to a final concentration of 125–500 nM using nuclease-free H2O.
-
22
Denature RNA at 72°C for 5 minutes. Immediately place on ice for 3 minutes.
-
23
Centrifuge RNA at maximum speed for 10 minutes at 4°C to remove any particulates. Carefully pipette RNA into a fresh microcentrifuge tube and place on ice.
-
24
Calibrate a beveled micropipette with nuclease-free H2O to deliver 2 nL per injection.
-
25
Place oocytes into an injection dish containing 1× MBSH.
-
26
Load RNA into the calibrated micropipette and inject each oocyte with 2 nL of RNA.
-
27
Culture oocytes at 18°C for 8–48 hrs in a 24-well dish containing 500 µL OCM containing antibiotics.
Fixation and immunofluorescence
-
28
Examine cultured oocytes for survival.
If more than 25% of oocytes have lysed, consider repeating microinjections with new oocytes.
-
29
Place cultured oocytes in a siliconized microfuge tube.
If visualizing only fluorescent RNA and not co-imaging protein, proceed to Step 33.
-
30
Remove OCM and wash oocytes with proteinase K buffer (without enzyme added).
-
31
Add 500 µl proteinase K solution. Gently rock oocytes for 5 minutes at room temperature.
-
32
Immediately remove proteinase K solution.
-
33
To fix oocytes, add 500 µL MEMFA. Incubate for 1 hr., with rocking, in the dark.
-
34
Remove MEMFA and add 500 µL PBT. Wash oocytes for 15 minutes in the dark with rocking.
-
35
Wash twice more in PBT, for a total of three washes.
If only visualizing fluorescent RNA, proceed to Step 41.
-
36
Add 500 µL PBT-Plus blocking solution. Rock oocytes in the dark for at least 2 hours.
-
37
Remove PBT-Plus. Incubate oocytes with primary antibody diluted in 500 µL PBT-Plus, overnight at 4°C with rocking. If antibody is limiting, oocytes may be incubated in a 250 µL dilution of primary antibody.
-
38
Replace primary antibody with 500 µL of PBT and rock in the dark at room temperature for 1.5 – 2 hrs. Repeat wash twice for a total of three washes.
-
39
Add 500 µL of PBT-Plus containing an appropriate fluorescently-labeled secondary antibody. Incubate overnight at 4°C with rocking.
Xenopus oocytes exhibit strong autofluorescence at 488 nm, limiting the choice of secondary antibodies. When possible, use secondary antibodies with emission at higher wavelengths.
-
40
Wash oocytes three times in PBT for 1.5–2 hrs at room temperature to remove secondary antibody. After the final wash, resuspend oocytes in 1 mL PBT.
-
41
Dehydrate oocytes stepwise into 100% anhydrous methanol:
Oocytes must be completely dehydrated in anhydrous methanol to be compatible with Murray’s clear for imaging.- Remove 100 µL of PBT solution and replace with 100 µL anhydrous methanol. Rock oocytes for 5–10 minutes at room temperature. Repeat twice.
- Remove 200 µL and replace with 200 µL anhydrous methanol. Rock oocytes for 5–10 minutes at room temperature. Repeat twice.
- Remove 300 µL and replace with 300 µL anhydrous methanol. Rock for 5–10 minutes at room temperature. Repeat twice.
- Remove 400 µL and replace with 400 µL anhydrous methanol. Rock for 5–10 minutes at room temperature. Repeat twice.
- Remove 500 µL and replace with 500 µL anhydrous methanol. Rock for 5–10 minutes at room temperature. Repeat twice.
- Remove 750 µL and replace with 750 µL anhydrous methanol. Rock for 5–10 minutes at room temperature. Repeat twice.
- Remove all buffer, and perform three washes with 1 ml 100% anhydrous methanol. At the end of dehydration, the solution should be clear. If solution is cloudy or Schlieren lines are visible, continue washing with anhydrous methanol.
-
42
Store oocytes in 100% anhydrous methanol at −20°C until ready for imaging. Oocytes may be stored for 2–3 weeks in methanol.
Confocal Imaging
-
43
Use a Pasteur pipette to transfer oocytes to a fluorodish. Try to keep oocytes in the center of the dish and transfer as little methanol as possible.
-
44
Using a pulled glass pipette with a fine tip, remove any methanol transferred to the fluorodish.
-
45
With a Pasteur pipette, add Murray’s clear dropwise to the fluorodish, until oocytes are covered completely.
-
46
Wait at least 5 minutes for oocytes to optically clear. Larger oocytes (stages III–VI) will take longer to clear than smaller oocytes.
-
47Image the oocytes using an inverted confocal microscope.
- Locate individual oocytes using a 488 nm laser and a 10× objective. Oocytes can be easily located due to autofluorescence at 488 nm.
- Once located, image oocytes at the desired magnification. It may be necessary to open the pinhole >1 airy unit to detect an RNA fluorescence signal.
- To decrease cross-excitation between channels, image with each laser separately (not simultaneously).
TROUBLESHOOTING
- Problem (Step 14): Low RNA yield
- Solution: If using Cy3-UTP, switch to the brighter Cy5-UTP. Increase concentration of RNA injected, or standardize injection concentration based on fluorescence intensity (e.g., 5,000 RFU, which can be measured with a NanoDrop 3300 Fluorospectrometer).
- Problem (Step 47): RNA Fluorescence signal is dim
- Solution: If using Cy3-UTP, switch to the brighter Cy5-UTP. Increase concentration of RNA injected, or standardize injection concentration based on fluorescence intensity (e.g., 5,000 RFU, which can be measured with a NanoDrop 3300 Fluorospectrometer).
- Problem (Step 47): Antibody does not penetrate oocyte interior
- Solution: Increase incubation with proteinase K. A time-series may be necessary to determine optimal digestion time for a particular antibody. Increase incubation period with primary antibody to 48 hours.
DISCUSSION
The X. laevis oocyte provides an important model to study cell polarity. Oogenesis in X. laevis proceeds through six distinct stages, during which the cytoplasm becomes increasingly polarized (Dumont 1972). Both mRNAs and proteins can be asymmetrically distributed in the oocyte cytoplasm, in many cases acting to specify embryonic polarity (reviewed in Houston 2013). Thus, analysis of protein and mRNA distributions in vertebrate oocytes is important for understanding establishment of developmental polarity. The X. laevis oocyte, in particular, offers many technical advantages for studying polarity. A single X. laevis ovary provides thousands of oocytes in a mixture of different maturity levels (Dumont 1972). Due to their large size, even the youngest oocytes can be manually microinjected (Yisraeli and Melton 1988; Chang et al. 2004). Microinjection of fluorescently labeled RNA can recapitulate the localization pattern of endogenous mRNAs and can be combined with immunofluorescence analysis of proteins of interest (Yoon and Mowry 2004). Furthermore, imaging by confocal microscopy allows a high-resolution view of RNA-protein interactions. Such analyses can provide insight into the cytoplasmic interactions contributing to polarity. The protocol presented here provides an updated method for imaging endogenous protein and microinjected RNA in immature X. laevis oocytes.
RECIPES
Collagenase Solution (3 mg/mL)
0.1 M KPO3+ (pH 7.4)
3 mg/mL collagenase I (Sigma C0130)
Prepare fresh. Do not store.
MBSH Buffer (10×)
880 mM NaCl
10 mM KCl
24 mM NaHCO3
8.2 mM MgSO4·7H2O
3.3 mM Ca(NO3)2·4H2O
4.1 mM CaCl2·6H2O
100 mM HEPES (pH 7.6)
Store as a sterilized 10× stock solution and prepare fresh 1× MBSH prior to use.
Oocyte Culture Media (OCM)
50% Leibovitz’s L-15 Medium (Thermo Fisher Scientific, 11415064)
15 mM HEPES, pH 7.6
1 mg/mL insulin
Sterilize though a 0.22 µm filter. OCM without antibiotics can be stored at 4°C for up to two months.
Add the following antibiotics to OCM just prior to culture. Do not store. Do not reuse.
50 U/mL nystatin (Sigma-Aldrich, N1638)
100 U/mL penicillin/streptomycin (Life Technologies, 15070063)
0.1 mg/mL gentamicin (Thermo Fisher Scientific, 15750060)
PBT for Xenopus oocytes/PBT-Plus
137 mM NaCl
2.7 mM KCl
10 mM Na2HPO4
1.8 mM KH2PO4
0.2% (w/v) BSA (Sigma-Aldrich, A2153)
0.1% (v/v) Triton X-100 in 1×PBS.
Mix and sterilize through 0.22 µm filter. Store at 4°C.
For PBT-Plus, supplement PBT with:
2% (v/v) goat serum
2% (w/v) BSA (for a total of 2.2% BSA).
Mix and sterilize through a 0.22 µm filter. Store at 4°C.
Proteinase K Buffer for Xenepus oocytes
10 mM EDTA, pH 8.0 100 mM Tris-HCl, pH 7.5
For Proteinase K solution:
Add proteinase K to a final concentration of 50 µg/mL. Prepare fresh, do not re-use.
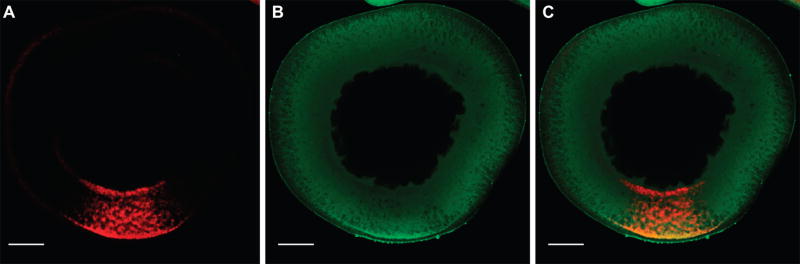
Figure 1.
Co-localization of endogenous Staufen protein and injected VLE RNA. Stage II oocytes were injected with 500 nM Cy5-UTP-labeled VLE RNA (Mowry and Melton, 1992) and cultured for 18 hr to allow for vegetal localization. Immunofluorescence was performed using an anti-XStau antibody (Yoon and Mowry 2004) at 1:250 dilution and an Alexa-546-secondary antibody (Thermo-Fisher) at 1:500 dilution. Oocytes were imaged on a Zeiss LSM 510 Meta Confocal Laser Scanning Microscope using a 20× objective. Shown is a confocal section with (A) VLE RNA in red, (B) Staufen protein in green, and (C) merged red and green channels, showing Staufen protein and VLE RNA colocalization. The oocyte is oriented with the vegetal pole at the bottom and the scale bars = 50 µm.
Acknowledgments
Development of this method was supported by NIH grant GM071049 to KLM.
References
- Aguero T, Newman K, King ML. Microinjection of Xenopus Oocytes. Cold Spring Protoc. 2017 doi: 10.1101/pdb.prot096974. [DOI] [PubMed] [Google Scholar]
- Chang P, Torres J, Lewis RA, Mowry KL, Houliston E, King ML. Localization of RNAs to the mitochondrial cloud in Xenopus oocytes through entrapment and association with endoplasmic reticulum. Mol Biol Cell. 2004;15:4669–4681. doi: 10.1091/mbc.E04-03-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont JN. Oogenesis in Xenopus laevis (Daudin). I. Stages of oocyte development in laboratory maintained animals. J Morph. 1972;136:153–179. doi: 10.1002/jmor.1051360203. [DOI] [PubMed] [Google Scholar]
- Houston DW. Regulation of cell polarity and RNA localization in vertebrate oocytes. Int Rev Cell Mol Biol. 2013;306:127–185. doi: 10.1016/B978-0-12-407694-5.00004-3. [DOI] [PubMed] [Google Scholar]
- Mowry KL, Melton DA. Vegetal messenger RNA localization directed by a 340-nt RNA sequence element in Xenopus oocytes. Science. 1992;255:991–994. doi: 10.1126/science.1546297. [DOI] [PubMed] [Google Scholar]
- Newman K, Aguero T, King ML. Isolation of Xenopus Oocytes. Cold Spring Protoc. 2017 doi: 10.1101/pdb.prot095851. [DOI] [PubMed] [Google Scholar]
- Yisraeli JK, Melton DA. The material mRNA Vg1 is correctly localized following injection into Xenopus oocytes. Nature. 1988;336:592–595. doi: 10.1038/336592a0. [DOI] [PubMed] [Google Scholar]
- Yoon YJ, Mowry KL. Xenopus Staufen is a component of a ribonucleoprotein complex containing Vg1 RNA and kinesin. Development. 2004;131:3035–3045. doi: 10.1242/dev.01170. [DOI] [PubMed] [Google Scholar]