Abstract
Multidrug-resistant Plasmodium falciparum in the Greater Mekong Subregion of Southeast Asia is a major threat to malaria elimination and requires close surveillance. In this study, we collected 107 longitudinal clinical samples of P. falciparum in 2007–2012 from the malaria hypoendemic region of the China-Myanmar border and measured their in vitro susceptibilities to 10 antimalarial drugs. Overall, parasites had significantly different IC50 values to all the drugs tested as compared to the reference 3D7 strain. Parasites were also genotyped in seven genes that were associated with drug resistance including pfcrt, pfmdr1, pfmrp1, pfdhfr, pfdhps, pfnhe1, and PfK13 genes. Despite withdrawal of chloroquine and antifolates from treating P. falciparum, parasites remained highly resistant to these drugs and mutations in pfcrt, pfdhfr, and pfdhps genes were highly prevalent and almost reached fixation in the study parasite population. Except for pyronaridine, quinine and lumefantrine, all other tested drugs exhibited significant temporal variations at least between some years, but only chloroquine and piperaquine had a clear temporal trend of continuous increase of IC50s. For the pfmrp1 gene, several mutations were associated with altered sensitivity to a number of drugs tested including chloroquine, piperaquine, lumefantrine and dihydroartemisinin. The association of PfK13 mutations with resistance to multiple drugs suggests potential evolution of PfK13 mutations amid multidrug resistance genetic background. Furthermore, network analysis of drug resistance genes indicated that certain haplotypes associated multidrug resistance persisted in these years, albeit there were year-to-year fluctuations of the predominant haplotypes.
Keywords: Drug resistance, Plasmodium falciparum, Mutation, In vitro assay, China-Myanmar border
Graphical abstract

Highlights
-
•
P. falciparum from China-Myanmar border was followed for in vitro drug sensitivity.
-
•
Parasites displayed in vitro resistance to several antimalarial drugs.
-
•
Resistance to chloroquine and piperaquine escalated by years.
-
•
Genotyping genes associated with drug resistance confirmed in vitro assay results.
1. Introduction
Malaria, a life-threatening disease caused by the Plasmodium parasites, has claimed over 400 000 human lives globally in 2016 (WHO, 2017). In the tropical and subtropical areas of the Greater Mekong Subregion (GMS), recent achievements in malaria control have encouraged countries within this region to pursue malaria elimination, aiming to reach this goal by 2030. Chemotherapy is an essential tool for malaria management, but its effectiveness is compromised by the emergence and spread of drug-resistant Plasmodium falciparum strains. Chloroquine (CQ) was one of the most widely used antimalarial drugs. Only several years after its introduction, CQ-resistant cases emerged firstly in Southeast Asia, then appeared in Latin America, and spread to all other endemic areas (Wellems and Plowe, 2001). This also happened to the antifolates drug pyrimethamine (PY). The GMS is a breeding ground of antimalarial drug resistance, and P. falciparum has developed resistance to essentially all commonly used antimalarial drugs (Fairhurst and Dondorp, 2016). Multidrug-resistant (MDR) parasites have led to the deployment of artemisinin combination therapies (ACTs). However, artemisinin resistance in P. falciparum has also emerged in the same place of the GMS (Noedl et al., 2008; Dondorp et al., 2009), where CQ and PY resistance first emerged. Furthermore, resistance to the partner drug mefloquine (MQ) and recently piperaquine (PQ) have resulted in increased clinical failures of the artesunate (AS)-MQ and dihydroartemisinin (DHA)-PQ, respectively (Wongsrichanalai and Meshnick, 2008; Saunders et al., 2014; Spring et al., 2015). With the unfolding of malaria elimination campaign in the GMS, heightened surveillance of drug resistance in P. falciparum is required in order to monitor the situation, prevent the spread of resistant parasites, and make timely changes of the national drug treatment policies.
The identification of drug resistance mechanisms facilitates molecular surveillance of antimalarial drug resistance (Ekland and Fidock, 2007). The K76T mutation in the P. falciparum chloroquine resistance transporter, pfcrt, is a major determinant of CQ resistance (Fidock et al., 2000). Point mutations in dihydropteroate synthase (dhps) and dihydrofolate reductase (dhfr), two key enzymes in the folate biosynthesis pathway, mediate resistance to the antifolates sulfadoxine and PY, respectively (Gregson and Plowe, 2005). As its name indicates, point mutations in the multidrug resistance 1 (mdr1) gene confer resistance to a number of drugs, while mdr1 gene amplification is responsible for clinical resistance to MQ (Price et al., 2004), and in vitro resistance to other amino alcohol drugs (Sidhu et al., 2006). In recent years, the advancement of genomic tools allowed accelerated identification of resistance mechanisms to artemisinin and PQ. Through a combination of in vitro selection, genomics and population biology, artemisinin resistance was found to be associated with point mutations in the propeller domain of the PfK13 gene (Ariey et al., 2014), which were subsequently confirmed by genetic manipulations (Ghorbal et al., 2014; Straimer et al., 2015). Similarly, genome-wide association studies revealed that amplification of two protease genes plasmepsin 2/3 was associated with clinical resistance to PQ in Cambodia (Amato et al., 2017; Witkowski et al., 2017). In areas of low transmission where host immunity against the malaria parasites is low, molecular markers serve as proxies for the prediction of efficacies of antimalarial drugs and provide convenient assessment of the epidemiology of drug resistance in malaria parasites.
“Border malaria” – concentrated malaria transmission along international borders – brings extreme difficulties for surveillance, and malaria re-introduction by cross-border migratory human populations could plunge people of malaria eliminating countries into malaria resurgence (Delacollette et al., 2009; Cui et al., 2012). Since drug policies in neighboring countries may differ considerably, parasite populations at the border may experience divergent drug selection pressures, favoring the emergence of MDR parasites (Zeng et al., 2017). The China-Myanmar border used to be a malaria hyperendemic region with a distinct antimalarial drug use history. Since 1979, PQ has been used extensively as a replacement drug of CQ in China, which has led to clinical resistance to PQ (Gao et al., 1993; Yang et al., 1999). Also, artemisinin drugs had been deployed mostly as monotherapies prior to 2005. After 2005, the national antimalarial drug policy has changed to ACT, mostly DHA-PQ, as the frontline treatment for uncomplicated P. falciparum cases in this region. Clinical follow-ups in recent years showed that DHA–PQ remained highly efficacious for treating uncomplicated falciparum malaria (Liu et al., 2015; Wang et al., 2015a). Further studies of parasites from this region also showed that day-3 parasite positivity as well as delayed parasite clearance were associated with PfK13 mutations (Huang et al., 2015; Wang et al., 2015c). Moreover, consistent with extensive deployment of ACT, the proportions of parasites carrying the PfK13 mutations have been increasing (Wang et al., 2015b). Possibly reflecting the divergent antimalarial drug histories, parasites from this region showed a PfK13 mutation pattern that is distinct from that in Cambodia (Ariey et al., 2014; Tun et al., 2015; Wang et al., 2015b). Thus, continuous monitoring of antimalarial drug resistance in this region is warranted.
In the present study, we performed a longitudinal follow-up of in vitro sensitivities in P. falciparum clinical isolates collected during 2007–2012 to commonly-used antimalarial drugs and determined dynamic changes in drug sensitivities and polymorphisms of genes associated with drug resistance. These data combined allowed us to further detect and confirm associations between drug-resistant genes and in vitro sensitivities to several antimalarial drugs. Our results revealed the persistent circulation of MDR parasites and further highlighted the necessity of close drug-resistance monitoring in the GMS in order to use updated drug policy for a specific region and period.
2. Material and methods
2.1. Parasite sample collection
To longitudinally follow P. falciparum in vitro sensitivities to antimalarial drugs at the China-Myanmar border, we collected 107 clinical parasite samples from acute, uncomplicated P. falciparum infections from malaria clinics located near the Nabang township in west Yunnan Province, China, and the Laiza township, Kachin State, Myanmar, during 2007–2012. Malaria diagnosis was based on microscopy of Giemsa-stained blood smears, and 2–5 ml venous blood was drawn from patients with falciparum malaria. Blood samples were stored in liquid nitrogen and used for culture adaptation. All patients in this study signed informed consent forms voluntarily and the research project was approved by the institutional review board of Kunming Medical University.
2.2. Parasite culture and in vitro drug assay
Culture-adapted parasite isolates were assayed for their in vitro sensitivities to 10 antimalarial drugs. CQ, MQ, quinine (QN), and PY were purchased from Sigma (St. Louis, MO, USA). PQ was from Chongqing Kangle Pharmaceutical Co. (Chongqing, China), pyronaridine (PND) was obtained from the China Institute of Pharmaceutical and Biological Products (Beijing, China), while naphthoquine (NQ), lumefantrine (LMF), AS, and DHA were from Kunming Pharmaceutical Co. (Kunming, Yunnan, China). Stock solutions of CQ, NQ, PND, and PQ were prepared in distilled water, MQ, QN, LMF, AS and DHA in ethanol, and PY in 1% acetic acid. Only monoclonal isolates were used for drug assays (Meng et al., 2010; Yuan et al., 2013). Parasite culture, synchronization and drug assay using the SYBR Green I-based method were performed as described (Smilkstein et al., 2004; Wang et al., 2016). Drugs were added to each well of a 96-well microplate at an initial concentration of 3.75 μM for CQ and PY, 256 nM for NQ and MQ, 1.5 μM for AS and DHA, 160 nM for PND, 320 nM for PQ, 10.24 μM for QN, and 800 nM for LMF, which were serially diluted. Each parasite strain was assayed with three technical repeats and two biological replications, and the 3D7 strain was included in all assays as an internal reference.
2.3. Sequencing analysis of drug resistance genes
Parasite genomic DNA was extracted from cultured parasites using a QiaAmp DNA minikit (Qiagen). Polymorphisms in drug resistance genes were determined by PCR and sequencing as previously reported (Yang et al., 2011; Gupta et al., 2014; Wang et al., 2015b). These include two pfcrt fragments covering codons 72–76 and 220, two pfmdr1 fragments including codons 86, 184, 1042 and 1246, a pfdhfr fragments containing codons 51, 59, 108 and 164, two pfdhps fragments containing codons 436, 437, 540, and 581, a pfnhe1 fragment containing the ms4760 minisatellite, and two pfmrp1 gene fragments containing codons 191, 325, 437, 785, 876, 1007, 1390 and the complete sequence of PfK13 gene.
2.4. Statistical analyses
The geometric mean of the half-maximal inhibitory concentration (IC50) was calculated by fitting the drug response data to a sigmoid curve. Median and interquartile range (IQR) were used since the data were not normally distributed. IQR, mean and standard deviation (SD) were determined using GraphPad Prism 6.0 for Windows. IC50 values of parasite isolates among the years as well as between the field isolates and 3D7 were compared by Mann-Whitney U test. Correlations between IC50s of drugs were determined using Spearman's test in the R package and MATLAB R2013a. Associations between IC50s and mutations were investigated by multiple t-tests. The relationship among the haplotypes was analyzed by using the neighbor-joining algorithm in MEGA version 7 (Kumar et al., 2016) and the haplotype network program pegas in the R package (https://cran.r-project.org/web/packages/pegas/index.html).
3. Results
3.1. In vitro susceptibilities of parasite isolates to antimalarial drugs
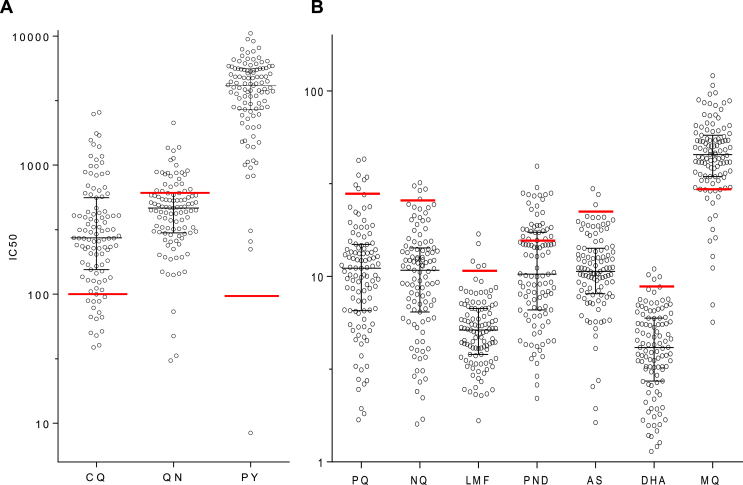
A total of 107 clinical P. falciparum samples from the China-Myanmar border in 2007 (22 isolates), 2008 (41 isolates), 2009 (22 isolates), 2010 (10 isolates), and 2012 (13 isolates) were culture-adapted and assayed for in vitro sensitivities to 10 antimalarial drugs (Table 1, Fig. 1). Overall, the field parasite isolates had significantly higher IC50 values to all drugs than 3D7 (P < 0.0001, Mann-Whitney U test) (Table 1). For CQ, 14.0% and 86.0% parasite isolates were considered moderately resistant (25 nM ≤ IC50 < 100 nM) and highly resistant (IC50 ≥ 100 nM) based on criterion described earlier (Table 1) (Ringwald et al., 1996; Nkhoma et al., 2007; Chaijaroenkul et al., 2010). The other two 4-aminoquinoline drugs PQ and NQ remained highly active against parasites in culture with median IC50s being in the lower nanomolar range, at 11.0 and 10.8 nM, respectively. Since the in vitro resistance thresholds for PQ and NQ were not defined, we arbitrarily estimated the cutoff values for resistance using the tradition of calculating the mean plus 2 SDs of the IC50s of the 107 field isolates (Pascual et al., 2015), which were both close to 30 nM (Table 1). If these cutoff values were used, 6.5% and 4.7% parasite isolates were above these cutoffs for PQ and NQ, respectively (Table 1, Fig. 1). For both drugs, the IC50s had relatively wide ranges, and the IC50s between the least and most sensitive parasite isolates differed by 25–30 folds, suggesting the presence of parasite strains with much decreased sensitivity to these drugs (Hao et al., 2013).
Table 1.
In vitro IC50 values (nM) of P. falciparum field isolates from the China-Myanmar border to 10 antimalarial drugs.
| Drugs | Median (IQR) | Range | 3D7 (Mean ± SD) |
P* | Cutoff (nM) | # (%) of isolates above cutoff |
|---|---|---|---|---|---|---|
| Chloroquine | 273.4 (154.9–559.2) | 38.7–2563.0 | 17.8 ± 8.1 | <0.0001 | 100# | 92 (86.0%) |
| Piperaquine | 11.0 (6.5–14.9) | 1.7–43.0 | 5.1 ± 2.0 | <0.0001 | 29.0 | 7 (6.5%) |
| Naphthoquine | 10.8 (6.4–14.3) | 1.6–32.0 | 8.5 ± 5.0 | <0.0001 | 25.4 | 5 (4.7%) |
| Mefloquine | 45.4 (34.5–57.7) | 5.7–121.1 | 18.1 ± 7.6 | <0.0001 | 30#; 90.6 | 89 (83.2%); 4 (3.7%) |
| Lumefantrine | 5.1 (3.8–6.7) | 1.7–17.0 | 4.8 ± 2.9 | <0.0001 | 10.6 | 5 (4.7%) |
| Quinine | 464.8 (298.7–605.5) | 30.6–2123.0 | 83.3 ± 41.5 | <0.0001 | 600# | 27 (25.2%) |
| Pyrimethamine | 4129.0 (2698.0–5588.0) | 8.4–10519.0 | 62.5 ± 44.9 | <0.0001 | 100# | 106 (99.1%) |
| Pyronaridine | 10.3 (6.6–17.3) | 2.2–39.3 | 5.6 ± 6.2 | <0.0001 | 15§; 27.4 | 34 (31.8%); 7 (6.5%) |
| Artesunate | 10.7 (8.1–14.2) | 1.6–29.7 | 7.4 ± 4.4 | <0.0001 | 22.2 | 3 (2.8%) |
| Dihydroartemisinin | 4.1 (2.7–6.0) | 1.1–11.0 | 3.0 ± 2.5 | <0.0001 | 8.9 | 4 (3.7%) |
IQR, interquartile range; SD, standard deviation; ND, not defined.
*P values are from Mann-Whitney U test for comparison between field isolates and 3D7.
Cutoffs for resistance are based on earlier report in Ringwald et al. (1996)# and Pradines et al. (1998)§. The rest of the cutoff values were based on calculated based on mean + 2 × SD of IC50s from the field isolates. Note for mefloquine and pyronaridine, the second cutoff value was based on mean + 2 × SD of IC50s from the field isolates in this study.
Fig. 1.
In vitro susceptibilities of parasite isolates from the China-Myanmar border to 10 antimalarial drugs. Graph A shows the dot plot of IC50s of the 107 parasite isolates to chloroquine (CQ), quinine (QN) and pyrimethamine (PY), while graph B shows their IC50s to piperaquine (PQ), naphthoquine (NQ), lumefantrine (LMF), pyronaridine (PND), artesunate (AS), dihydroartemisinin (DHA) and mefloquine (MQ). Red bars indicate the cutoffs for resistance as defined in Table 1. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Among the aryl aminoalcohol drugs, LMF was highly active against the parasites with a median IC50 in the lower nanomolar concentration (5.1 nM) and a relatively narrow range (∼13 folds between the least and most sensitive strains), but five (4.7%) parasite isolates had IC50s of >10.6 nM. Though MQ has not been used in here, the median IC50 (45.4 nM) was greater than the 30 nM threshold used to define resistance in an earlier study (Ringwald et al., 1996), and 83.2% (89/107) parasite isolates had IC50 values above this cutoff. The IC50s of three parasite isolates were close to 100 nM, about 25 times of that in the most sensitive isolates. QN has been extensively used to treat falciparum malaria in the study area. Although the median IC50 of 464.8 nM was below 600 nM, the arbitrarily defined threshold for resistance (Ringwald et al., 1996), 25.2% (27/107) parasites displayed IC50 values above this threshold. Moreover, five samples had IC50s exceeding 1000 nM and one isolate had IC50 of >2000 nM, suggesting potential QN resistance in the study parasite population, a result that is consistent with our previous report (Meng et al., 2010).
For the two artemisinin derivatives tested, both AS and DHA were highly active against these isolates with median IC50s being in the sub-nanomolar range (Table 1). Using mean + 2 SDs to arbitrarily define the cutoffs for reduced sensitivity, 2.8% and 3.7% of parasites had IC50s above the cutoffs for AS and DHA, respectively (Table 1). For the Mannich base drug PND, the median IC50 was 10.3 nM, below the 15 nM threshold value for resistance defined earlier (Pradines et al., 1998). Despite this, 31.8% of the tested parasites had IC50s above this cutoff value. If we arbitrarily defined the cutoff value by using the mean + 2 SDs, 6.5% parasites showed reduced sensitivity to this drug (Table 1).
Antifolates have been deployed extensively for the treatment of P. falciparum malaria as well as for malaria prophylaxis in the past. In vitro resistance to PY was very high, and only one parasite isolate from the 2007 samples was considered sensitive based on the threshold of 100 nM for resistance (Table 1, Fig. 1) (Ringwald et al., 1996; Aubouy et al., 2003). Moreover, 80.2% parasite isolates had PY IC50 values exceeding 2000 nM, a value to define high PY resistance.
3.2. Temporal trends of in vitro sensitivities
Except for PND, QN and LMF, all other tested drugs exhibited significant temporal variations at least between some years (Fig. S1, Table S1). Consistent with an earlier observation (Hao et al., 2013), in vitro IC50s to CQ displayed a clear trend of continuous annual increase from a median value of 145.0 nM in 2007 to 692.3 nM in 2012 (Fig. S1). In addition, IC50s for PQ and PY also increased annually during the study period except in 2012 for PY. Despite extensive use of artemisinin drugs in this region, there were no significant annual increases in IC50s to AS and DHA.
3.3. Relationships between in vitro sensitivities to different drugs
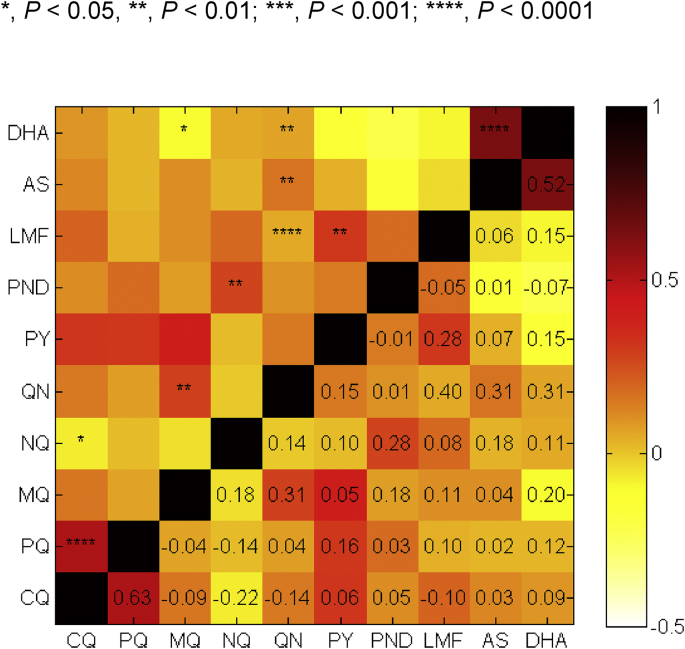
Pairwise comparison showed that there were highly significant, positive correlations between sensitivities to AS and DHA (Fig. 2, P < 0.0001, Spearman's test). In addition, as we have reported earlier (Hao et al., 2013), susceptibilities to the two 4-aminoquinoline drugs CQ and PQ were significantly correlated (P < 0.0001). Also, sensitivities to aminoalcohol drugs and artemisinin derivatives were correlated. Specifically, the sensitivity to MQ was correlated with those to QN and DHA (P < 0.05). Similarly, QN showed significant correlations with DHA and AS (P < 0.01) as well as LMF (P < 0.0001). Sensitivities to NQ and PND were significantly correlated (P < 0.01). In addition, PY and LMF also showed significant correlation (P < 0.01).
Fig. 2.
Correlation between IC50values of parasite isolates to 10 antimalarial drugs. Correlations between IC50 values were analyzed by Spearman's test and the degree of correlation between two drugs was colored coded. The coefficients are shown below the diagonal, while their levels of significance are shown as asterisks above the diagonal. Abbreviations of the drugs are the same as in Fig. 1.
To determine whether the CQ resistance background affects the in vitro susceptibilities to other antimalarial drugs, parasite isolates were divided into moderately CQ-resistant (15 parasites) and highly CQ-resistant (92 parasites) groups (Table S2). Among the drugs tested, only PQ and DHA were less active against highly CQ-resistant parasite strains, whereas other drugs were similarly active against parasites in these two groups.
3.4. Polymorphisms in drug resistance genes
We genotyped seven genes to determine the prevalence of mutations associated with drug resistance. For pfcrt, M74I, N75E, K76T and A220S all reached fixation in the study parasite population. For pfmdr1, the N86Y mutation was rare in the parasite population with a prevalence of 0.9%, whereas the Y184F mutation reached 30.8% and appeared to have been decreasing through the years. Of the other two mutations N1042D and D1246Y, the former was detected in some years with an overall prevalence of 3.7%, whereas the latter was not detected in the study population (Table 2).
Table 2.
The prevalence of mutations in genes associated with drug resistance in different years.a
| Gene | Residue position | 2007 (n = 21) | 2008 (n = 41) | 2009 (n = 22) | 2010 (n = 10) | 2012 (n = 13) | Total (n = 107) |
|---|---|---|---|---|---|---|---|
| Pfcrtc | C72S | – | – | – | – | – | – |
| M74I | 100 | 100 | 100 | 100 | 100 | 100 | |
| N75E | 100 | 100 | 100 | 100 | 100 | 100 | |
| K76T | 100 | 100 | 100 | 100 | 100 | 100 | |
| A220S | 100 | 100 | 100 | 100 | 100 | 100 | |
| Pfmdr1 | N86Y | 4.8 | – | – | – | – | 0.9 |
| Y184F | 28.6 | 43.9 | 31.8 | 10 | 7.7 | 30.8 | |
| N1042D | 4.8 | – | 13.6 | – | – | 3.7 | |
| Pfdhfr | N51I | 61.9 | 65.9 | 63.6 | 70 | 76.9 | 66.4 |
| C59R | 95.2 | 100 | 100 | 100 | 100 | 99.1 | |
| S108N | 95.2 | 100 | 100 | 100 | 100 | 99.1 | |
| I164L | 71.4 | 85.4 | 77.2 | 80 | 61.5 | 77.5 | |
| Pfdhps | S436A | 61.9 | 51.2 | 68.2 | 40 | 30.8 | 53.3 |
| A437G | 95.2 | 95.1 | 95.4 | 100 | 100 | 96.3 | |
| K540E/N | 90.5 | 85.4 | 90.9 | 100 | 61.5 | 86.0 | |
| A581G | 23.8 | 46.3 | 36.4 | 40 | 69.2 | 42.1 | |
| Pfmrp1 | H191Y | 66.7 | 75.6 | 63.6 | 30 | 84.6 | 68.2 |
| N325S | 4.8 | 12.2 | 9.1 | 10 | 15.4 | 10.3 | |
| S437A | 66.7 | 70.7 | 68.2 | 30 | 69.2 | 65.4 | |
| H785N | 19.0 | 9.8 | 4.8 | 10 | 38.5 | 14.0 | |
| I876V | 57.1 | 61 | 68.2 | 50 | 53.8 | 59.8 | |
| T1007M | 19.0 | 22 | 31.8 | 20 | 46.2 | 26.2 | |
| F1390I | 9.5 | 14.6 | 10 | – | 7.7 | 10.3 | |
| PfK13c | NNb | 42.9 | 61.5 | 68.2 | 80 | 69.2 | 60.7 |
| K189T | – | – | – | 10 | – | 1.0 | |
| E252Q | 4.8 | 5.1 | – | – | – | 2.9 | |
| P441L | 4.8 | – | – | – | – | 1.0 | |
| F446I | 19.0 | 30.8 | 18.1 | 10 | 46.2 | 25.7 | |
| R539T | – | 7.7 | – | – | 7.7 | 3.8 | |
| P574L | 9.5 | 2.6 | – | – | – | 2.9 | |
| C580Y | – | 2.6 | – | – | – | 1.0 | |
| A676D | – | 5.1 | – | – | – | 2.9 | |
| H719N | – | – | 18.1 | – | – | 3.8 |
ND, not done.
Significant differences in mutation prevalence are highlighted in bold (χ2 test): pfdhps S436A between 2009 and 2012 (P = 0.0358) and between 2010 and 2012 (P = 0.0266); pfdhps A581G 2007 and 2012 (P = 0.0089); pfmrp1 H191Y between 2008 and 2010 (P = 0.0061) and between 2010 and 2012 (P = 0.0078); pfmrp1 S437A between 2008 and 2010 (P = 0.0169) and between 2009 and 2010 (P = 0.0436); pfmrp1 H785N between 2008 and 2012 (P = 0.0155) and between 2009 and 2012 (P = 0.0101); PfK13 H719N between 2008 and 2009 (P = 0.0140).
NN insertion between amino acids 136 and 137.
39 samples were genotyped in 2008, giving a total of 105 samples genotyped for the pfk13 gene.
Major mutations mediating resistance to PY in pfdhfr were all highly prevalent in the study parasites (Table 2). In particular, C59R and S108N almost reached fixation with only one parasite isolate from 2007 remaining as the wild type. The other two major mutations N51I and I164L also reached high prevalence (66.4% and 77.5%, respectively). Among the mutations in pfdhps that confer resistance to sulfadoxine, the A437G mutation was approaching fixation at 96.3%. The K540E/N mutation was also highly prevalent at 86.0%.
Pfmrp1 gene had seven mutations with 10.3–68.2% frequencies, among which two mutations H191Y and S437A exceeded 60% in the parasite population. The minisatellite in pfnhe1 was associated with altered sensitivity to QN in some parasite populations (Ferdig et al., 2004; Bennett et al., 2007; Henry et al., 2009; Meng et al., 2010). In this study, a total of 11 pfnhe1 ms4760 alleles were found; two (MS-6 and MS-7) reached substantial levels (13.1% and 58.9%, respectively) in the parasite population.
Genotyping PfK13 gene detected the NN insertion between amino acids 136 and 137 in 60.7% parasites and nine point mutations in 45% of parasites. Among these point mutations, seven were located in the propeller domain (>440 amino acids). Consistent with our earlier findings (Wang et al., 2015b, 2015c), F446I was the predominant mutation with a prevalence of 25.7%. The other six were rare mutations P441L (1.0%), R539T (3.8%), P574L (2.9%), C580Y (1.0%), A676D (2.9%) and H719N (3.8%) (Table 2, Table 3).
Table 3.
Prevalence of major haplotypes of known drug resistance genes in parasites from different years.
| Gene | H | Haplotypea | 2007 (n = 21) | 2008 (n = 41) | 2009 (n = 22) | 2010 (n = 10) | 2012 (n = 13) | Total (n = 107) |
|---|---|---|---|---|---|---|---|---|
|
Pfcrt |
1 |
CIETS |
100 |
100 |
100 |
100 |
100 |
100 |
|
Pfmdr1 |
5 |
NYN | 66.7 | 56.1 | 54.5 | 90.0 | 92.3 | 65.4 |
| NFN |
23.8 |
43.9 |
31.8 |
10.0 |
7.7 |
29.9 |
||
|
Pfdhfr |
4 |
IRNI | 19.0 | 12.2 | 22.7 | 20.0 | 23.1 | 17.8 |
| NRNL | 28.6 | 31.7 | 36.4 | 30.0 | 7.7 | 29.0 | ||
|
IRNL |
42.9 |
53.7 |
40.9 |
50.0 |
53.8 |
48.6 |
||
|
Pfdhps |
12 |
SGEA | 9.5 | 7.3 | – | 20.0 | – | 6.5 |
| AGEA | 47.6 | 31.7 | 45.5 | 40.0 | 23.1 | 37.4 | ||
| SGEG | 19 | 31.7 | 22.7 | 30.0 | 38.5 | 28.0 | ||
|
AGEG |
– |
9.8 |
9.1 |
– |
– |
5.6 |
||
|
Pfmrp1 |
22 |
HNSHITF | 28.6 | 17.1 | 22.7 | 50.0 | 15.4 | 23.4 |
| YNAHVTF | 33.3 | 24.4 | 18.2 | – | – | 19.6 | ||
| YSAHITF | – | 9.8 | 4.5 | – | 7.7 | 5.6 | ||
| YNAHVTI | 4.8 | 12.2 | 9.1 | – | 7.7 | 8.4 | ||
| YNAHVMF | 4.8 | 9.8 | 27.3 | 10.0 | – | 11.2 | ||
|
YNANVMF |
4.8 |
9.8 |
– |
10.0 |
30.8 |
9.3 |
||
| Pfnhe1 | 11 | MS-1 (2) | 14.3 | 9.8 | 9.1 | 10.0 | 7.7 | 10.3 |
| MS-5 (4) | – | 4.9 | 9.1 | 20.0 | – | 5.6 | ||
| MS-6 (1) | 19.0 | 14.6 | 18.2 | – | – | 13.1 | ||
| MS-7 (3) | 52.4 | 58.5 | 50.0 | 50.0 | 84.6 | 58.9 |
H, number of haplotypes. Only haplotypes with prevalence ≥5% were included.
Significant differences in the annual prevalence of haplotypes are highlighted in bold (χ2 test): Pfmdr1 NYN between 2008 and 2010 (P = 0.0468), 2008 and 2012 (P = 0.0172) and between 2009 and 2012 (P = 0.0201); pfmdr1 NFN between 2008 and 2010 (P = 0.0468) and 2008 and 2012 (P = 0.0172); pfmrp1 HNSHITF between 2008 and 2010 (P = 0.0277), and pfnhe1 MS-7 between 2008 and 2012 (P = 0.0067) and between 2009 and 2012 (P = 0.0406).
Haplotypes were based on amino acids at the positions pfcrt (72, 74, 75, 76, 220); pfmdr1(86, 184,1042); pfdhfr(51, 59, 108, 164), pfdhps(436, 437, 540, 581); andpfmrp1(191, 325, 437, 785, 876, 1007, 1390). Mutant residues are in bold. For the pfnhe1 haplotypes, the number in parenthesis indicates the copy number of DNNND repeats.
3.5. Association of gene polymorphisms with in vitro drug sensitivities
We compared the IC50s between parasites carrying the wild-type alleles and those with the mutant alleles. Fixation of the major CQ-resistant alleles (K76T and A220S) was consistent with the in vitro assay result. For the pfdhfr gene, mutations in the codon N51I was significantly associated with increased in vitro resistance to PY, whereas parasites carrying I164 and 164L had similar PY sensitivities (Fig. S2A). Mutations in pfmdr1 and pfmrp1 may affect parasite's sensitivities to multiple drugs. In Africa, the N86Y mutation is associated decreased sensitivity to aminoquinolines but increased sensitivity to arylamino alcohol drugs such as MQ, LMF and halofantrine (Dokomajilar et al., 2006; Mwai et al., 2009). In our sample set, this mutation was found in only one parasite isolate. For the rest of pfmdr1 mutations, we identified that parasites with the mutation N1042D showed significantly increased sensitivity to PND (Fig. S2B). For the pfmrp1 gene, several mutations were associated with altered sensitivity to a number of drugs tested. The prevalent T1007M was correlated with elevated IC50 values to two 4-aminoquinoline drugs CQ and PQ (Fig. S2C, D). In addition, H785N was linked to decreased sensitivity to PQ and LMF, whereas F1390I was associated with increased sensitivity to PY and DHA (Fig. S2D-G).
Consistent with our previous finding, parasites with the pfnhe1 ms4760 haplotype MS-7 (with three type 1 repeats) exhibited significantly reduced sensitivity to QN compared to those carrying MS-1 (with two type 1 repeats) and MS-5 allele (with four type 1 repeats). In addition, parasites with MS-6 (with one type 1 repeat) expressed significantly increased sensitivity to MQ compared to those carrying the MS-1, MS-5 and MS-7 allele (Fig. S2H, I).
Artemisinin resistance shown as delayed clearance is associated with mutations in the propeller domain of the PfK13 gene. Whereas none of the propeller mutations identified in this study were linked to increased IC50s to AS and DHA, F446I was linked to increased resistance to both CQ and PQ (Fig. S2J, K). In addition, the N-terminal insertion was also found to be associated with altered in vitro sensitivity to CQ and MQ. The H719N and the P574L were associated with reduced sensitivity to QN and increased sensitivity to PY, respectively (Fig. S2J, L).
3.6. Haplotype diversity of drug resistance genes
For pfcrt, CIETS at positions 72–76 occurred at 100%, consistent with this being the most prevalent CQ-resistant haplotype in Southeast Asia. For antifolate resistance, 95% parasites carried triple and quadruple mutations in pfdhfr, while 77.5% parasites carried triple and quadruple mutations in pfdhps, confirming that the parasite population was highly resistant to sulfadoxine-PY. However, more than 65% parasites carried the wild-type pfmdr1, and it reached 90% or higher in samples after 2010, albeit the sample size was small. For pfmrp1, seven mutations were detected, resulting in 22 haplotypes with six exceeding 5%. For the pfnhe1 gene, 11 haplotypes were detected and MS-7 occurred in 58.9% of the parasites. This haplotype has been associated with reduced in vitro IC50 to quinine. For the PfK13 gene, nine point mutations and 11 haplotypes were identified. All parasites contained single mutations in the propeller domain, whereas only one parasite carried the E252Q and F446I double mutations.
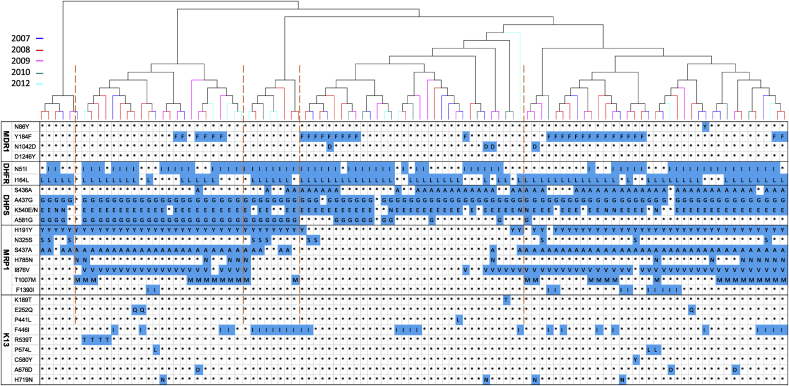
When all sequenced drug resistance-related genes with the exclusion of pfnhe1 were considered, 96 haplotypes were identified (Fig. 3). These haplotypes include those with mutations in all sequenced genes, indicating MDR phenotypes. Phylogenetic analysis revealed five major groups, with each of the haplotype groups containing isolates from different years (Fig. 3). Similarly, haplotype network analysis did not identify substantial clustering of the haplotypes by years (Fig. S3). Although there were haplotypes that persisted through these years, the predominant haplotypes showed changes in each year.
Fig. 3.
Phylogenetic clustering of the haplotypes based on the mutations in five genes associated with drug resistance in P. falciparum. The tree was constructed using the neighbor-joining method implemented in the MEGA program. Parasites were color-coded by the years of collection. Haplotypes are shown with mutations in each gene highlighted in blue color and wild-type residues shown as asterisks. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
4. Discussion
Chemotherapy remains a cornerstone in malaria treatment, but the P. falciparum parasite is highly adept at developing resistance to antimalarial drugs. Especially, parasite strains from the GMS have developed resistance to essentially all antimalarials deployed so far. This arms race between the use of new drugs and evolution of resistance has forced malaria-endemic nations to frequently change their drug policies to maintain relatively high efficacies of the frontline antimalarials. This demands close surveillance of drug efficacy and resistance development and spread. In vitro assays of clinical parasite isolates for sensitivity to antimalarials and monitoring known molecular markers associated resistance offer complementary ways of resistance surveillance. In this study, we used these two approaches to monitor the longitudinal trends of in vitro susceptibilities of P. falciparum using an archived collection of parasite isolates from the China-Myanmar border region. All drugs except MQ have been used with varied extents in this region. Consistently, these clinical parasite isolates exhibited significant differences in their in vitro susceptibilities to all drugs compared to 3D7. However, except for CQ and PQ, sensitivities to other drugs did not show a consistent temporal trend of continuous decrease over the years.
CQ has been withdrawn from treating falciparum malaria in this region in the 1970s, but parasites remained highly resistant to CQ. Interestingly, we even observed continued escalation of in vitro CQ IC50 values during longitudinal monitoring of the field parasite isolates. This is in contrast to reports from other malaria endemic areas where cessation of CQ use is accompanied with a gradual decline of parasites carrying the pfcrt K76T mutation and concomitant return of CQ sensitivity (Kublin et al., 2003; Laufer et al., 2010). The reasons for the persistent and continuous evolution of CQ resistance in our study site could include 1) continued selection pressure due to the use of CQ for treating sympatric P. vivax cases, and 2) an almost complete lack of competing wild type parasites as the major pfcrt mutant alleles are fixed. It is also possible that the extensive use of PQ as the ACT partner drug has exerted selective pressure on CQ resistance given their structural similarity, which is also reflected in the significant correlation in sensitivities to CQ and PQ. Since we only determined the sequence polymorphisms around the K76 and A220 position of the pfcrt gene, it is possible that the parasites, like those identified in Cambodia, may evolve new pfcrt mutations, which could confer CQ resistance without incurring fitness cost (Gabryszewski et al., 2016). Future studies will need to elucidate the evolution of the pfcrt locus and its pleotropic effects on resistance development to other antimalarial drugs.
As a replacement drug for CQ, extensive deployment of PQ as a monotherapy has led to the development of clinical resistance to PQ in China (Gao et al., 1993; Yang et al., 1999). Though clinical resistance to DHA-PQ has recently emerged in Cambodia (Amato et al., 2017; Witkowski et al., 2017), this ACT remained highly efficacious for treating falciparum cases in the China-Myanmar border region (Liu et al., 2015; Wang et al., 2015a). In Cambodia, the DHA-PQ failure involves resistance to both DHA and PQ; the latter apparently evolved in the background of artemisinin resistance and was associated with amplification of the aspartic protease genes plasmepsin 2/3 (Amato et al., 2017; Witkowski et al., 2017). The decrease of in vitro PQ sensitivity observed in some parasite isolates used in this study may involve different mechanisms. Two mutations in the pfmrp1 gene were associated with reduced susceptibility to PQ. In addition, the F446I mutation in PfK13 was also associated with reduced susceptibility to PQ and CQ. Though PfK13 mutations may not be mediating PQ resistance per se, it is possible that artemisinin resistance might have evolved in the background of PQ resistance or vice versa, like what was observed in Cambodia (Duru et al., 2015; Spring et al., 2015). The other 4-aminoquinoline drug NQ, an antimalarial drug developed in China, has not been applied widely in this region and parasites were relatively sensitive to this drug. Interestingly, sensitivities to NQ and CQ appear to be negatively correlated, suggesting pfcrt mutations do not confer cross resistance to NQ.
The antifolate drugs have also been discontinued for malaria treatment, but resistance to PY remained very high, and mutations in dhfr and dhps conferring resistance to PY and sulfadoxine, respectively, were highly prevalent in the parasite population. We did not notice a consistently declining trend of dhfr and dhps mutant alleles through the years of monitoring, which is different from what has been observed in other malaria regions such as Ethiopia after withdrawal of sulfadoxine -PY (Tessema et al., 2015). It is noteworthy that intermittent preventive treatment based on sulfadoxine-PY has not been carried out in the study area. It is curious whether selective pressure maintaining the dhfr and dhps mutations is due to the use of antifolate drugs for treating bacterial infections, a common practice in the study area.
Artemisinins have been used for more than three decades in the China-Myanmar border area, mostly as monotherapies prior to 2005. There are indications that artemisinin resistance also has emerged in this area, and clinical cases remaining parasitemic three days after treatment with ACT were associated with increased in vitro ring-stage survival and mutations in the propeller domain of PfK13 (Wang et al., 2015c). Unlike other parts of the GMS, parasites from this border area have high prevalence of the F446I mutation (Huang et al., 2015; Wang et al., 2015b; Ye et al., 2016). Yet, it is noteworthy that the conventional IC50 values of the parasite strains could not predict artemisinin resistance that is restricted to the early ring stage because this method subjects the parasites to continuous exposure to antimalarials through most of the developmental cycle (Dondorp et al., 2009; Witkowski et al., 2013). Nonetheless, our genotyping result was consistent with previous findings, showing F446I as the predominant mutation in this region. Importantly, we also identified the presence of some mutations such as R539T and C580Y, which were highly prevalent in the Cambodian parasite populations and were confirmed genetically to confer artemisinin resistance in vitro (Straimer et al., 2015). Thus, despite excellent ACT efficacy at the China-Myanmar border, strenuous monitoring is needed.
Gene amplification such as pfmdr1 and plasmepsins 2/3 are associated with drug resistance in P. falciparum. Especially in some GMS countries where MQ has been extensively deployed, pfmdr1 amplification is highly prevalent and is associated with increased risk of recrudescence in patients treated with MQ and LUM (Price et al., 2004; Alker et al., 2007; Lim et al., 2009). Whereas increased pfmdr1 copies are associated with in vitro MQ resistance (Price et al., 1999), in vitro selection for PQ resistance found deamplification of the pfmdr1 gene in PQ-resistant parasites (Eastman et al., 2011). Consistent with this in vitro finding, the recently emerged resistance in Cambodia to DHA-PQ was also associated with single-copy pfmdr1 (Amato et al., 2017; Witkowski et al., 2017), suggesting that PQ selects against pfmdr1 amplification. Our earlier studies of pfmdr1 copy numbers in parasites from the China-Myanmar border region did not identify pfmdr1 amplification (Meng et al., 2010; Wang et al., 2012), which is consistent with no MQ but extensive DHA-PQ use in this area. Nevertheless, analysis of amplification of these drug resistance-associated genes in future parasite isolates from this region needs to be vigorously followed up.
Molecular surveillance of the genetic markers associated with drug resistance agreed well with the in vitro drug assay results for well-characterized genes such as pfcrt, dhfr and dhps. The high prevalence of resistance-conferring mutations in these genes is consistent with MDR phenotypes of the parasites. However, given that the sample sizes in this study are relatively small, especially in year 2010, caution is needed to make conclusions on drug resistance. Furthermore, the lack of well-defined cutoffs for in vitro drug resistance for most of the drugs assayed also warns against dichotomous division of drug resistance based solely on in vitro assay data and arbitrary cutoffs. Whereas the in vitro data clearly confirmed drug resistance to CQ and PY, the IC50 data for other drugs can only serve as references to guide future work, which should certainly involve clinical efficacy studies. Network analysis in this study showed that certain haplotypes associated MDR have been collected in multiple years, indicating relative persistence of these MDR parasites in this region. In addition, there have been year-to-year fluctuations of the predominant haplotypes, which could be due to the introduction of parasite genotypes as the result of migration of internally displaced people to the border region (Lo et al., 2015).
Acknowledgements
The authors wish to thank the field team for their assistance in sample collection. We are grateful to the local communities and hospitals for their participation in this research. This project was funded by the National Natural Science Foundation of China (31260508 to ZY, 81761128017 to YC), and the National Institutes of Health (U19 AI089672 and R01 AI128940 to LC). YW and CL were supported by grant 2014FB005 and 2017FE468-185 from Yunnan Province. HL was supported by the Ph.D. Starting Fund from Kunming Medical University (#1).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ijpddr.2018.05.003.
Contributor Information
Zhaoqing Yang, Email: zhaoqingy92@hotmail.com.
Liwang Cui, Email: luc2@psu.edu.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
Figure S1. In vitro sensitivities of parasite isolates collected in different years from the China-Myanmar border area to 10 antimalarial drugs. (A) chloroquine, (B) piperaquine, (C) naphthoquine, (D) mefloquine, (E) pyronaridine, (F) pyrimethamine, (G) quinine, (H) lumefantrine, (I) artesunate, and (J) dihydroartemisinin. Significant differences in drug sensitivity between two years are indicated (*, P < 0.05; **, P < 0.01; ***P < 0.001; Mann Whitney U test).
Figure S2. Association of SNPs in pfdhfr (A), pfmdr1 (B), pfmrp1 (C-G), minisatellites in pfnhe1 (H, I), and PfK13 (J-L) with sensitivities to different antimalarials (shown in Y axis). *, **, and **** indicate significant difference of in vitro sensitivities between parasites carrying the two alleles at P < 0.05, P < 0.01, and P < 0.0001, respectively (Mann-Whitney U test).
Figure S3. Median-joining haplotype network of P. falciparum isolates harboring mutations in five drug resistance-associated genes. The size of each circle corresponds to the number of samples sharing the same haplotype. Samples were colored coded by the years of collection.
References
- Alker A.P., Lim P., Sem R., Shah N.K., Yi P., Bouth D.M., Tsuyuoka R., Maguire J.D., Fandeur T., Ariey F., Wongsrichanalai C., Meshnick S.R. Pfmdr1 and in vivo resistance to artesunate-mefloquine in falciparum malaria on the Cambodian-Thai border. Am. J. Trop. Med. Hyg. 2007;76:641–647. [PubMed] [Google Scholar]
- Amato R., Lim P., Miotto O., Amaratunga C., Dek D., Pearson R.D., Almagro-Garcia J., Neal A.T., Sreng S., Suon S., Drury E., Jyothi D., Stalker J., Kwiatkowski D.P., Fairhurst R.M. Genetic markers associated with dihydroartemisinin-piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype-phenotype association study. Lancet Infect. Dis. 2017;17:164–173. doi: 10.1016/S1473-3099(16)30409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariey F., Witkowski B., Amaratunga C., Beghain J., Langlois A.C., Khim N., Kim S., Duru V., Bouchier C., Ma L., Lim P., Leang R., Duong S., Sreng S., Suon S., Chuor C.M., Bout D.M., Menard S., Rogers W.O., Genton B., Fandeur T., Miotto O., Ringwald P., Le Bras J., Berry A., Barale J.C., Fairhurst R.M., Benoit-Vical F., Mercereau-Puijalon O., Menard D. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubouy A., Jafari S., Huart V., Migot-Nabias F., Mayombo J., Durand R., Bakary M., Le Bras J., Deloron P. DHFR and DHPS genotypes of Plasmodium falciparum isolates from Gabon correlate with in vitro activity of pyrimethamine and cycloguanil, but not with sulfadoxine-pyrimethamine treatment efficacy. J. Antimicrob. Chemother. 2003;52:43–49. doi: 10.1093/jac/dkg294. [DOI] [PubMed] [Google Scholar]
- Bennett T.N., Patel J., Ferdig M.T., Roepe P.D. Plasmodium falciparum Na+/H+ exchanger activity and quinine resistance. Mol. Biochem. Parasitol. 2007;153:48–58. doi: 10.1016/j.molbiopara.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaijaroenkul W., Wisedpanichkij R., Na-Bangchang K. Monitoring of in vitro susceptibilities and molecular markers of resistance of Plasmodium falciparum isolates from Thai-Myanmar border to chloroquine, quinine, mefloquine and artesunate. Acta Trop. 2010;113:190–194. doi: 10.1016/j.actatropica.2009.10.016. [DOI] [PubMed] [Google Scholar]
- Cui L., Yan G., Sattabongkot J., Cao Y., Chen B., Chen X., Fan Q., Fang Q., Jongwutiwes S., Parker D., Sirichaisinthop J., Kyaw M.P., Su X.Z., Yang H., Yang Z., Wang B., Xu J., Zheng B., Zhong D., Zhou G. Malaria in the greater Mekong subregion: heterogeneity and complexity. Acta Trop. 2012;121:227–239. doi: 10.1016/j.actatropica.2011.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacollette C., D'Souza C., Christophel E., Thimasarn K., Abdur R., Bell D., Dai T.C., Gopinath D., Lu S., Mendoza R., Ortega L., Rastogi R., Tantinimitkul C., Ehrenberg J. Malaria trends and challenges in the greater Mekong subregion. Southeast Asian J. Trop Med. Publ. Health. 2009;40:674–691. [PubMed] [Google Scholar]
- Dokomajilar C., Nsobya S.L., Greenhouse B., Rosenthal P.J., Dorsey G. Selection of Plasmodium falciparum pfmdr1 alleles following therapy with artemether-lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrob. Agents Chemother. 2006;50:1893–1895. doi: 10.1128/AAC.50.5.1893-1895.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dondorp A.M., Nosten F., Yi P., Das D., Phyo A.P., Tarning J., Lwin K.M., Ariey F., Hanpithakpong W., Lee S.J., Ringwald P., Silamut K., Imwong M., Chotivanich K., Lim P., Herdman T., An S.S., Yeung S., Singhasivanon P., Day N.P., Lindegardh N., Socheat D., White N.J. Artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duru V., Khim N., Leang R., Kim S., Domergue A., Kloeung N., Ke S., Chy S., Eam R., Khean C., Loch K., Ken M., Lek D., Beghain J., Ariey F., Guerin P.J., Huy R., Mercereau-Puijalon O., Witkowski B., Menard D. Plasmodium falciparum dihydroartemisinin-piperaquine failures in Cambodia are associated with mutant K13 parasites presenting high survival rates in novel piperaquine in vitro assays: retrospective and prospective investigations. BMC Med. 2015;13:305. doi: 10.1186/s12916-015-0539-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman R.T., Dharia N.V., Winzeler E.A., Fidock D.A. Piperaquine resistance is associated with a copy number variation on chromosome 5 in drug-pressured Plasmodium falciparum parasites. Antimicrob. Agents Chemother. 2011;55:3908–3916. doi: 10.1128/AAC.01793-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekland E.H., Fidock D.A. Advances in understanding the genetic basis of antimalarial drug resistance. Curr. Opin. Microbiol. 2007;10:363–370. doi: 10.1016/j.mib.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairhurst R.M., Dondorp A.M. Artemisinin-resistant plasmodium falciparum malaria. Microbiol. Spectr. 2016;4 doi: 10.1128/microbiolspec.EI10-0013-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdig M.T., Cooper R.A., Mu J., Deng B., Joy D.A., Su X.Z., Wellems T.E. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol. Microbiol. 2004;52:985–997. doi: 10.1111/j.1365-2958.2004.04035.x. [DOI] [PubMed] [Google Scholar]
- Fidock D.A., Nomura T., Talley A.K., Cooper R.A., Dzekunov S.M., Ferdig M.T., Ursos L.M., Sidhu A.B., Naude B., Deitsch K.W., Su X.Z., Wootton J.C., Roepe P.D., Wellems T.E. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabryszewski S.J., Dhingra S.K., Combrinck J.M., Lewis I.A., Callaghan P.S., Hassett M.R., Siriwardana A., Henrich P.P., Lee A.H., Gnadig N.F., Musset L., Llinas M., Egan T.J., Roepe P.D., Fidock D.A. Evolution of Fitness Cost-Neutral Mutant PfCRT Conferring P. falciparum 4-aminoquinoline drug resistance is accompanied by altered parasite metabolism and digestive vacuole physiology. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005976. e1005976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Fu L., Fu Y., Qian B., Li G. Randomised comparison on the treatment of falciparum malaria with dihydroartemisinin and piperaquine. Natl. Med. J. Chin. 1993;73:602–604. [PubMed] [Google Scholar]
- Ghorbal M., Gorman M., Macpherson C.R., Martins R.M., Scherf A., Lopez-Rubio J.J. Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat. Biotechnol. 2014;32:819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- Gregson A., Plowe C.V. Mechanisms of resistance of malaria parasites to antifolates. Pharmacol. Rev. 2005;57:117–145. doi: 10.1124/pr.57.1.4. [DOI] [PubMed] [Google Scholar]
- Gupta B., Xu S., Wang Z., Sun L., Miao J., Cui L., Yang Z. Plasmodium falciparum multidrug resistance protein 1 (pfmrp1) gene and its association with in vitro drug susceptibility of parasite isolates from north-east Myanmar. J. Antimicrob. Chemother. 2014;69:2110–2117. doi: 10.1093/jac/dku125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao M., Jia D., Li Q., He Y., Yuan L., Xu S., Chen K., Wu J., Shen L., Sun L., Zhao H., Yang Z., Cui L. In vitro sensitivities of Plasmodium falciparum isolates from the China-Myanmar border to piperaquine and association with polymorphisms in candidate genes. Antimicrob. Agents Chemother. 2013;57:1723–1729. doi: 10.1128/AAC.02306-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry M., Briolant S., Zettor A., Pelleau S., Baragatti M., Baret E., Mosnier J., Amalvict R., Fusai T., Rogier C., Pradines B. Plasmodium falciparum Na+/H+ exchanger 1 transporter is involved in reduced susceptibility to quinine. Antimicrob. Agents Chemother. 2009;53:1926–1930. doi: 10.1128/AAC.01243-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F., Takala-Harrison S., Jacob C.G., Liu H., Sun X., Yang H., Nyunt M.M., Adams M., Zhou S., Xia Z., Ringwald P., Bustos M.D., Tang L., Plowe C.V. A single mutation in K13 Predominates in southern China and is associated with delayed clearance of plasmodium falciparum following artemisinin treatment. J. Infect. Dis. 2015;212:1629–1635. doi: 10.1093/infdis/jiv249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kublin J.G., Cortese J.F., Njunju E.M., Mukadam R.A., Wirima J.J., Kazembe P.N., Djimde A.A., Kouriba B., Taylor T.E., Plowe C.V. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J. Infect. Dis. 2003;187:1870–1875. doi: 10.1086/375419. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer M.K., Takala-Harrison S., Dzinjalamala F.K., Stine O.C., Taylor T.E., Plowe C.V. Return of chloroquine-susceptible falciparum malaria in Malawi was a reexpansion of diverse susceptible parasites. J. Infect. Dis. 2010;202:801–808. doi: 10.1086/655659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim P., Alker A.P., Khim N., Shah N.K., Incardona S., Doung S., Yi P., Bouth D.M., Bouchier C., Puijalon O.M., Meshnick S.R., Wongsrichanalai C., Fandeur T., Le Bras J., Ringwald P., Ariey F. Pfmdr1 copy number and arteminisin derivatives combination therapy failure in falciparum malaria in Cambodia. Malar. J. 2009;8:11. doi: 10.1186/1475-2875-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Yang H.L., Tang L.H., Li X.L., Huang F., Wang J.Z., Li C.F., Wang H.Y., Nie R.H., Guo X.R., Lin Y.X., Li M., Wang J., Xu J.W. In vivo monitoring of dihydroartemisinin-piperaquine sensitivity in Plasmodium falciparum along the China-Myanmar border of Yunnan Province, China from 2007 to 2013. Malar. J. 2015;14:47. doi: 10.1186/s12936-015-0584-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo E., Zhou G., Oo W., Lee M.C., Baum E., Felgner P.L., Yang Z., Cui L., Yan G. Molecular inference of sources and spreading patterns of Plasmodium falciparum malaria parasites in internally displaced persons settlements in Myanmar-China border area. Infect. Genet. Evol. 2015;33:189–196. doi: 10.1016/j.meegid.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H., Zhang R., Yang H., Fan Q., Su X., Miao J., Cui L., Yang Z. In vitro sensitivity of Plasmodium falciparum clinical isolates from the China-Myanmar border area to quinine and association with polymorphism in the Na+/H+ exchanger. Antimicrob. Agents Chemother. 2010;54:4306–4313. doi: 10.1128/AAC.00321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwai L., Kiara S.M., Abdirahman A., Pole L., Rippert A., Diriye A., Bull P., Marsh K., Borrmann S., Nzila A. In vitro activities of piperaquine, lumefantrine, and dihydroartemisinin in Kenyan Plasmodium falciparum isolates and polymorphisms in pfcrt and pfmdr1. Antimicrob. Agents Chemother. 2009;53:5069–5073. doi: 10.1128/AAC.00638-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nkhoma S., Molyneux M., Ward S. In vitro antimalarial susceptibility profile and prcrt/pfmdr-1 genotypes of Plasmodium falciparum field isolates from Malawi. Am. J. Trop. Med. Hyg. 2007;76:1107–1112. [PubMed] [Google Scholar]
- Noedl H., Se Y., Schaecher K., Smith B.L., Socheat D., Fukuda M.M. Evidence of artemisinin-resistant malaria in western Cambodia. N. Engl. J. Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- Pascual A., Madamet M., Briolant S., Gaillard T., Amalvict R., Benoit N., Travers D., Pradines B. Multinormal in vitro distribution of Plasmodium falciparum susceptibility to piperaquine and pyronaridine. Malar. J. 2015;14:49. doi: 10.1186/s12936-015-0586-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradines B., Tall A., Parzy D., Spiegel A., Fusai T., Hienne R., Trape J.F., Doury J.C. In-vitro activity of pyronaridine and amodiaquine against African isolates (Senegal) of Plasmodium falciparum in comparison with standard antimalarial agents. J. Antimicrob. Chemother. 1998;42:333–339. doi: 10.1093/jac/42.3.333. [DOI] [PubMed] [Google Scholar]
- Price R.N., Cassar C., Brockman A., Duraisingh M., van Vugt M., White N.J., Nosten F., Krishna S. The pfmdr1 gene is associated with a multidrug-resistant phenotype in Plasmodium falciparum from the western border of Thailand. Antimicrob. Agents Chemother. 1999;43:2943–2949. doi: 10.1128/aac.43.12.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price R.N., Uhlemann A.C., Brockman A., McGready R., Ashley E., Phaipun L., Patel R., Laing K., Looareesuwan S., White N.J., Nosten F., Krishna S. Mefloquine resistance in Plasmodium falciparum and increased pfmdr1 gene copy number. Lancet. 2004;364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringwald P., Bickii J., Basco L.K. In vitro activity of antimalarials against clinical isolates of Plasmodium falciparum in Yaounde, Cameroon. Am. J. Trop. Med. Hyg. 1996;55:254–258. doi: 10.4269/ajtmh.1996.55.254. [DOI] [PubMed] [Google Scholar]
- Saunders D.L., Vanachayangkul P., Lon C. Dihydroartemisinin-piperaquine failure in Cambodia. N. Engl. J. Med. 2014;371:484–485. doi: 10.1056/NEJMc1403007. [DOI] [PubMed] [Google Scholar]
- Sidhu A.B., Uhlemann A.C., Valderramos S.G., Valderramos J.C., Krishna S., Fidock D.A. Decreasing pfmdr1 copy number in plasmodium falciparum malaria heightens susceptibility to mefloquine, lumefantrine, halofantrine, quinine, and artemisinin. J. Infect. Dis. 2006;194:528–535. doi: 10.1086/507115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smilkstein M., Sriwilaijaroen N., Kelly J.X., Wilairat P., Riscoe M. Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob. Agents Chemother. 2004;48:1803–1806. doi: 10.1128/AAC.48.5.1803-1806.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spring M.D., Lin J.T., Manning J.E., Vanachayangkul P., Somethy S., Bun R., Se Y., Chann S., Ittiverakul M., Sia-ngam P., Kuntawunginn W., Arsanok M., Buathong N., Chaorattanakawee S., Gosi P., Ta-aksorn W., Chanarat N., Sundrakes S., Kong N., Heng T.K., Nou S., Teja-isavadharm P., Pichyangkul S., Phann S.T., Balasubramanian S., Juliano J.J., Meshnick S.R., Chour C.M., Prom S., Lanteri C.A., Lon C., Saunders D.L. Dihydroartemisinin-piperaquine failure associated with a triple mutant including kelch13 C580Y in Cambodia: an observational cohort study. Lancet Infect. Dis. 2015;15:683–691. doi: 10.1016/S1473-3099(15)70049-6. [DOI] [PubMed] [Google Scholar]
- Straimer J., Gnadig N.F., Witkowski B., Amaratunga C., Duru V., Ramadani A.P., Dacheux M., Khim N., Zhang L., Lam S., Gregory P.D., Urnov F.D., Mercereau-Puijalon O., Benoit-Vical F., Fairhurst R.M., Menard D., Fidock D.A. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science. 2015;347:428–431. doi: 10.1126/science.1260867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessema S.K., Kassa M., Kebede A., Mohammed H., Leta G.T., Woyessa A., Guma G.T., Petros B. Declining trend of Plasmodium falciparum dihydrofolate reductase (dhfr) and dihydropteroate synthase (dhps) mutant alleles after the withdrawal of Sulfadoxine-Pyrimethamine in North Western Ethiopia. PLoS One. 2015;10:e0126943. doi: 10.1371/journal.pone.0126943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun K.M., Imwong M., Lwin K.M., Win A.A., Hlaing T.M., Hlaing T., Lin K., Kyaw M.P., Plewes K., Faiz M.A., Dhorda M., Cheah P.Y., Pukrittayakamee S., Ashley E.A., Anderson T.J., Nair S., McDew-White M., Flegg J.A., Grist E.P., Guerin P., Maude R.J., Smithuis F., Dondorp A.M., Day N.P., Nosten F., White N.J., Woodrow C.J. Spread of artemisinin-resistant Plasmodium falciparum in Myanmar: a cross-sectional survey of the K13 molecular marker. Lancet Infect. Dis. 2015;15:415–421. doi: 10.1016/S1473-3099(15)70032-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Yang Z., Yuan L., Zhou G., Parker D., Lee M.C., Yan G., Fan Q., Xiao Y., Cao Y., Cui L. Clinical efficacy of dihydroartemisinin-piperaquine for the treatment of uncomplicated Plasmodium falciparum malaria at the China-Myanmar border. Am. J. Trop. Med. Hyg. 2015;93:577–583. doi: 10.4269/ajtmh.15-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Cabrera M., Yang J., Yuan L., Gupta B., Liang X., Kemirembe K., Shrestha S., Brashear A., Li X., Porcella S.F., Miao J., Yang Z., Su X.Z., Cui L. Genome-wide association analysis identifies genetic loci associated with resistance to multiple antimalarials in Plasmodium falciparum from China-Myanmar border. Sci. Rep. 2016;6 doi: 10.1038/srep33891. 33891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Parker D., Meng H., Wu L., Li J., Zhao Z., Zhang R., Miao M., Fan Q., Wang H., Cui L., Yang Z. In vitro sensitivity of Plasmodium falciparum from China-Myanmar border area to major ACT drugs and polymorphisms in potential target genes. PLoS One. 2012;7:e30927. doi: 10.1371/journal.pone.0030927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Shrestha S., Li X., Miao J., Yuan L., Cabrera M., Grube C., Yang Z., Cui L. Prevalence of K13-propeller polymorphisms in Plasmodium falciparum from China-Myanmar border in 2007-2012. Malar. J. 2015;14:168. doi: 10.1186/s12936-015-0672-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wang Y., Cabrera M., Zhang Y., Gupta B., Wu Y., Kemirembe K., Hu Y., Liang X., Brashear A., Shrestha S., Li X., Miao J., Sun X., Yang Z., Cui L. Artemisinin resistance at the China-Myanmar border and association with mutations in the K13 propeller gene. Antimicrob. Agents Chemother. 2015;59:6952–6959. doi: 10.1128/AAC.01255-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellems T.E., Plowe C.V. Chloroquine-resistant malaria. J. Infect. Dis. 2001;184:770–776. doi: 10.1086/322858. [DOI] [PubMed] [Google Scholar]
- WHO . 2017. World Malaria Report 2017. [Google Scholar]
- Witkowski B., Amaratunga C., Khim N., Sreng S., Chim P., Kim S., Lim P., Mao S., Sopha C., Sam B., Anderson J.M., Duong S., Chuor C.M., Taylor W.R., Suon S., Mercereau-Puijalon O., Fairhurst R.M., Menard D. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect. Dis. 2013;13:1043–1049. doi: 10.1016/S1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkowski B., Duru V., Khim N., Ross L.S., Saintpierre B., Beghain J., Chy S., Kim S., Ke S., Kloeung N., Eam R., Khean C., Ken M., Loch K., Bouillon A., Domergue A., Ma L., Bouchier C., Leang R., Huy R., Nuel G., Barale J.C., Legrand E., Ringwald P., Fidock D.A., Mercereau-Puijalon O., Ariey F., Menard D. A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype-genotype association study. Lancet Infect. Dis. 2017;17:174–183. doi: 10.1016/S1473-3099(16)30415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongsrichanalai C., Meshnick S.R. Declining artesunate-mefloquine efficacy against falciparum malaria on the Cambodia-Thailand border. Emerg. Infect. Dis. 2008;14:716–719. doi: 10.3201/eid1405.071601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H., Liu D., Huang K., Yang Y., Yang P., Liao M., Zhang C. Assay of sensitivity of Plasmodium falciparum to chloroquine, amodiaquine, piperaquine, mefloquine and quinine in Yunnan province. Chin. J. Parasitol. Parasitic. Dis. 1999;17:43–45. [PubMed] [Google Scholar]
- Yang Z., Li C., Miao M., Zhang Z., Sun X., Meng H., Li J., Fan Q., Cui L. Multidrug-resistant genotypes of Plasmodium falciparum. Myanmar. Emerg. Infect. Dis. 2011;17:498–501. doi: 10.3201/eid1703.100870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R., Hu D., Zhang Y., Huang Y., Sun X., Wang J., Chen X., Zhou H., Zhang D., Mungthin M., Pan W. Distinctive origin of artemisinin-resistant Plasmodium falciparum on the China-Myanmar border. Sci. Rep. 2016;6:20100. doi: 10.1038/srep20100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L., Zhao H., Wu L., Li X., Parker D., Xu S., Zhao Y., Feng G., Wang Y., Yan G., Fan Q., Yang Z., Cui L. Plasmodium falciparum populations from northeastern Myanmar display high levels of genetic diversity at multiple antigenic loci. Acta Trop. 2013;125:53–59. doi: 10.1016/j.actatropica.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W., Bai Y., Wang M., Wang Z., Deng S., Ruan Y., Feng S., Yang Z., Cui L. Significant divergence in sensitivity to antimalarial drugs between neighboring Plasmodium falciparum populations along the eastern border of Myanmar. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.01689-16. e01689–01616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. In vitro sensitivities of parasite isolates collected in different years from the China-Myanmar border area to 10 antimalarial drugs. (A) chloroquine, (B) piperaquine, (C) naphthoquine, (D) mefloquine, (E) pyronaridine, (F) pyrimethamine, (G) quinine, (H) lumefantrine, (I) artesunate, and (J) dihydroartemisinin. Significant differences in drug sensitivity between two years are indicated (*, P < 0.05; **, P < 0.01; ***P < 0.001; Mann Whitney U test).
Figure S2. Association of SNPs in pfdhfr (A), pfmdr1 (B), pfmrp1 (C-G), minisatellites in pfnhe1 (H, I), and PfK13 (J-L) with sensitivities to different antimalarials (shown in Y axis). *, **, and **** indicate significant difference of in vitro sensitivities between parasites carrying the two alleles at P < 0.05, P < 0.01, and P < 0.0001, respectively (Mann-Whitney U test).
Figure S3. Median-joining haplotype network of P. falciparum isolates harboring mutations in five drug resistance-associated genes. The size of each circle corresponds to the number of samples sharing the same haplotype. Samples were colored coded by the years of collection.