Abstract
Variants in the PLPP3 gene encoding for lipid phosphate phosphohydrolase 3 have been associated with susceptibility to atherosclerosis independently of classical risk factors. PLPP3 inactivates lysophosphatidic acid (LPA), a pro-inflammatory, pro-thrombotic product of phospholipase activity. Here we performed the first exploratory analysis of PLPP3, LPA, and LPA receptors (LPARs 1–6) in human atherosclerosis. PLPP3 transcript and protein were repressed when comparing plaques versus normal arteries and plaques from symptomatic versus asymptomatic patients, and they were negatively associated with risk of adverse cardiovascular events. PLPP3 localized to macrophages, smooth muscle, and endothelial cells (ECs) in plaques. LPAR 2, 5, and especially 6 showed increased expression in plaques, with LPAR6 localized in ECs and positively correlated to PLPP3. Utilizing in situ mass spectrometry imaging, LPA and its precursors were found in the plaque fibrous cap, co-localizing with PLPP3 and LPAR6. In vitro, PLPP3 silencing in ECs under LPA stimulation resulted in increased expression of adhesion molecules and cytokines. LPAR6 silencing inhibited LPA-induced cell activation, but not when PLPP3 was silenced simultaneously. Our results show that repression of PLPP3 plays a key role in atherosclerosis by promoting EC activation. Altogether, the PLPP3 pathway represents a suitable target for investigations into novel therapeutic approaches to ameliorate atherosclerosis.
Keywords: atherosclerosis, therapy, biobank profiling
Introduction
Therapies that can reduce the deposition of apolipoprotein-B-containing lipoproteins in lesion-susceptible arterial segments can slow down atherosclerotic cardiovascular disease (ACVD) progress.1 Efficient therapies for suppression of the inflammatory response, which accompanies lipoprotein deposition and contributes to plaque development and ACVD, may add further cardiovascular benefit, as shown recently with the CANTOS (Canakinumab Anti-inflammatory Thrombosis Outcomes Study) clinical trial.2
Several genome-wide association studies (GWASs), including a meta-analysis, have identified PLPP3, the gene encoding lipid phosphate phosphatase 3, as a candidate locus causally related to ACVD risk.3, 4 Furthermore, PLPP3 SNP rs17114036 was included in a genetic risk score (GRS) study showing that individuals with high GRSs appear to have a larger benefit in cardiovascular absolute risk reduction from statin treatment than those with low GRSs,5 thus suggesting that PLPP3 genetic data may be applied to enhance clinical benefit with already established therapy for ACVD. We identified PLPP3 and the associated lysophosphatidic acid (LPA) axis as possible anti-atherosclerotic drug candidates after applying a target selection workflow within the CarTarDis Consortium (http://cartardis.eu), aimed at identifying targets suitable for pharmaceutical drug development. In our approach, candidate targets were first assembled from published data related to human genetics of CVD, and subsequently they were prioritized using stringent target discovery filters, including genetic correlation to ACVD clinical phenotypes, novelty, feasibility to validate (in vitro and in vivo), and relation to known ACVD mechanisms.6
PLPP3 enzyme is an integral membrane protein that dephosphorylates and inactivates various lipid-phosphate mediators, such as LPA and sphingosine-1-phosphate, thus blocking the downstream signals of these bioactive lipids.7 Expression quantitative trait locus (eQTL) analyses linked the major ACVD risk-associated allele with lower expression of PLPP3 in human endothelial cells (ECs), but not in other tissues.8, 9 These results suggest that ACVD risk-associated SNPs may increase the disease susceptibility by regulating PLPP3 expression in an endothelial-specific manner. Elegant mouse in vitro and in vivo studies have shown that PLPP3 expression in the arterial wall is modulated by hemodynamic forces and that it is involved in arterial wall pathology by modulating vascular cell functions.9, 10, 11 Mechanistic studies with isolated arterial cells showed that lower expression of PLPP3 sensitizes the response of ECs to LPA.9, 12
Several studies have shown that LPA accumulates in human atheroma where it can exert pro-thrombotic effects.13, 14 LPA is an amphipatic phospholipid that can interact with cell membranes by its detergent properties but also by its association to LPA-specific receptors (LPARs 1–6).15 Experimental evidence supports a role for LPA and its G-protein-coupled receptors in promoting pro-atherogenic, pro-inflammatory, and pro-thrombotic processes.16, 17 LPA in lesions can be generated by enzymatic hydrolysis of lysophospholipids by extracellular lysophospholipase D, autotaxin, and by local deposition of modified apoB-lipoprotein carriers of LPA and its phospholipid precursors.7, 18, 19 For example, phosphatidylcholine (PC) (34:1) and PC (34:2) are two PC species connected to the lysophosphatidylcholine (LPC), which are the degradation products from PC hydrolysis by phospholipases and precursors for LPA production. LPA molecules can be inactivated by the enzymatic action of PLPP3, which removes the phosphate group generating the corresponding alcohols that are not agonists for LPA receptors (LPARs).20 Thus, PLPP3 expression levels in the arterial wall may have a relevant function in disease progression by affecting local LPAR-LPA-mediated signaling.
To explore the putative association of the PLPP3-LPAR(s) pathway with ACVD and its potential for drug development, we performed a comprehensive analysis on genetic, transcriptomic, and proteomic levels in human carotid plaques and normal arteries (NAs). In the same tissues, we also evaluated their possible co-localization with LPA and its phospholipid precursors. In-vitro-silencing experiments were conducted to characterize the role of PLPP3 and LPARs on endothelial activation. Our results from investigation of this human genetic association and the pathway biology as a marker of atherosclerosis provide insight for selection and identification of potential new therapeutic approaches to prevent ACVD and its complications.
Results
PLPP3 Is Repressed in Human Carotid Atherosclerotic Lesions
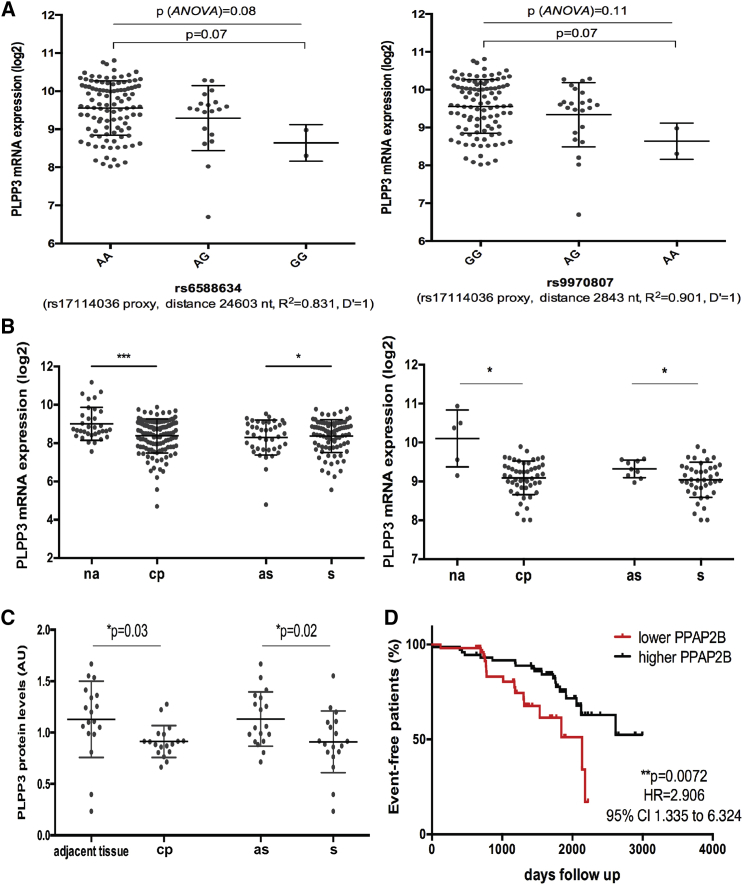
We first evaluated the genetic association of a previously reported rs17114036 variant with PLPP3 levels in human plaques by eQTL analyses. Two suitable proxies were identified (rs6588634 and rs9970807, both R2 > 0.8 and D’ = 1) that confirmed a marginal association with PLPP3 mRNA levels in plaques from n = 127 patients that were available for these analyses (p = 0.07; Figure 1A). Next, gene expression analysis of totally n = 177 lesions showed a significant downregulation of PLPP3 transcript in carotid plaques (CPs) compared to macroscopically NAs in two non-overlapping microarray datasets (n = 127 CP versus n = 33 NA from the larger discovery dataset and n = 50 CP plaque versus n = 5 NA from the smaller validation dataset; Figure 1B), with mean log2 difference ± SD = −0.6311 ± 0.1731 and mean log2 difference ± SD = −1.012 ± 0.3323, respectively. Notably, PLPP3 mRNA was also significantly downregulated in CPs from symptomatic patients compared with asymptomatic ones (mean difference ± SD = −0.2803 ± 0.1031). Importantly, these data were confirmed on the protein level by mass spectrometry analyses comparing plaques with adjacent control arterial tissue (n = 18 matched samples) and plaques from symptomatic versus asymptomatic patients (n = 9 in each group; Figure 1C), strongly suggesting a correlation between lower PLPP3 levels and a more severe clinical phenotype. Moreover, patients with below-median expression levels of PLPP3 in their plaques at surgery conferred a significantly higher risk of future adverse cardio- and cerebrovascular events during the follow-up period after carotid endoarterectomy (CEA) (p = 0.0072; Figure 1D).
Figure 1.
Genetic Association and Expression of PLPP3 in Advanced Human Atherosclerotic Plaques
(A) Genetic variant rs17114036 was found to be tentatively associated with PLPP3 mRNA expression in carotid plaques by expression quantitative train locus (eQTL) analysis of two suitable proxies in n = 127 patients. Plots show median ± SD. (B) Two microarray datasets (discovery set on the left and validation set on the right) showing the PLPP3 mRNA expression in carotid plaques (CPs) compared to normal arteries (NAs), along with the comparison between plaques from symptomatic (S) and asymptomatic (AS) patients. Discovery set contained n = 33 NAs and n = 127 CPs and validation set contained n = 5 NAs and n = 50 CPs. Values are expressed as log2 mean ± SD. *p < 0.05 and ***p < 0.001; ns, not significant. (C) Evaluation of PLPP3 protein levels by mass spectrometry comparing plaques (CPs) with adjacent control arterial tissue (n = 18 matched samples), and plaques from S versus AS patients (n = 9 per group). Values are expressed in a.u. as mean ± SD. (D) Survival curves illustrating MACCE-free survival of patients during the follow-up period after surgery, based on PLPP3 mRNA expression in BiKE plaques above (black) and below (red) the median values (x axis, days of event-free survival). Each mark along the lines indicates an event. Total number of events in the cohort was n = 58. MACCE, major adverse cerebro- and cardiovascular event.
To begin delineating the cellular processes associated with PLPP3 expression, correlation analyses were performed in plaques from the Biobank of Karolinska Endarterectomies (BiKE) discovery dataset between PLPP3 and various cellular markers (Table S1). These analyses showed positive correlation (Pearson r > 0.3, p < 0.0001) between PLPP3 expression and typical smooth muscle cell (SMC) markers; endothelial marker CD31; T lymphocyte marker CD45RA; pro-inflammatory markers nuclear factor κB (NF-κB) and BMP4; extracellular matrix degradation via SULF1; growth factors TGFB1, IGF1, PDGFB, and PDGFD; and chemokine CCR2. Weak positive correlations were found also with some macrophage markers.
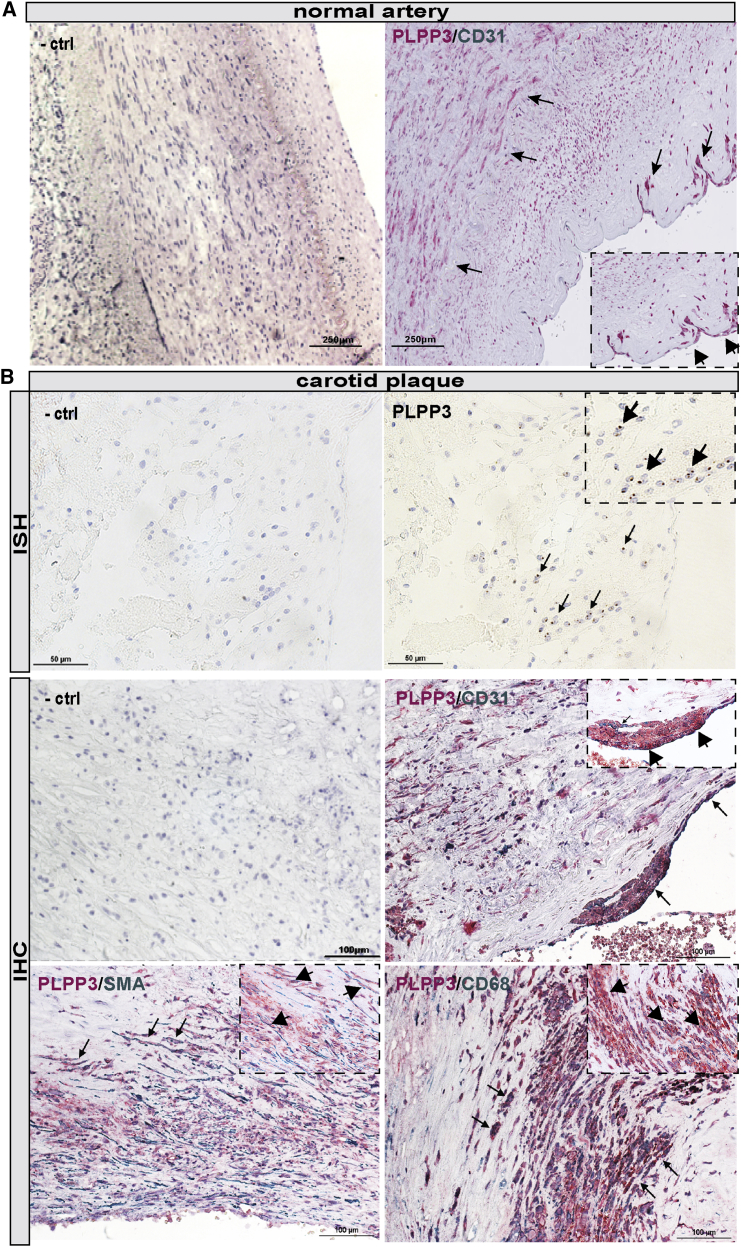
To evaluate these bioinformatic results, we further examined PLPP3 mRNA and protein expression levels and localization in various cell types in NAs and plaques in situ. Overall, PLPP3 protein signal was strong in NAs, particularly in CD31+ ECs, but it was also found in medial SMCs (Figure 2A). PLPP3 transcript and protein (Figure 2B) expression was also detected in CPs (n = 10 samples examined), particularly localized in the fibrous cap and in the areas lining the necrotic core. PLPP3 was absent from peripheral, subintimal plaque areas. Double immunostaining with specific cell markers showed PLPP3 protein localization in various cell types, such as SMCs (SMA+), ECs (CD31+), and macrophages (CD68+; Figure 2B), confirming the correlation analyses. These results confirmed overall lower levels of PLPP3 in advanced human plaques, but they indicated a wide expression pattern linked to several major plaque cell types.
Figure 2.
Localization of PLPP3 in Normal Arteries and Carotid Plaques
(A) Immunohistochemical staining shows the distribution of PLPP3 (red signal) in normal human artery, in medial smooth muscle cells, and CD31+ (green signal) luminal endothelial cells. (B) In situ hybridization detection of PLPP3 mRNA transcript in human lesions (top raw). Arrows indicate the RNA probe signals (also shown in the zoomed inset). Immunohistochemical stainings show the distribution of PLPP3 in human plaques. Double staining shows the co-localization of PLPP3 (red signal) with endothelial cell marker (CD31, green signal), smooth alpha actin (SMA, green signal), and macrophage cell marker (CD68, green signal). Arrows indicate double-positive cells and insets show higher magnification. Images were taken with the 40× objective.
LPAR6 Is the Most Abundantly Expressed LPAR in Human Plaques, Localized to ECs, and Positively Correlated with PLPP3
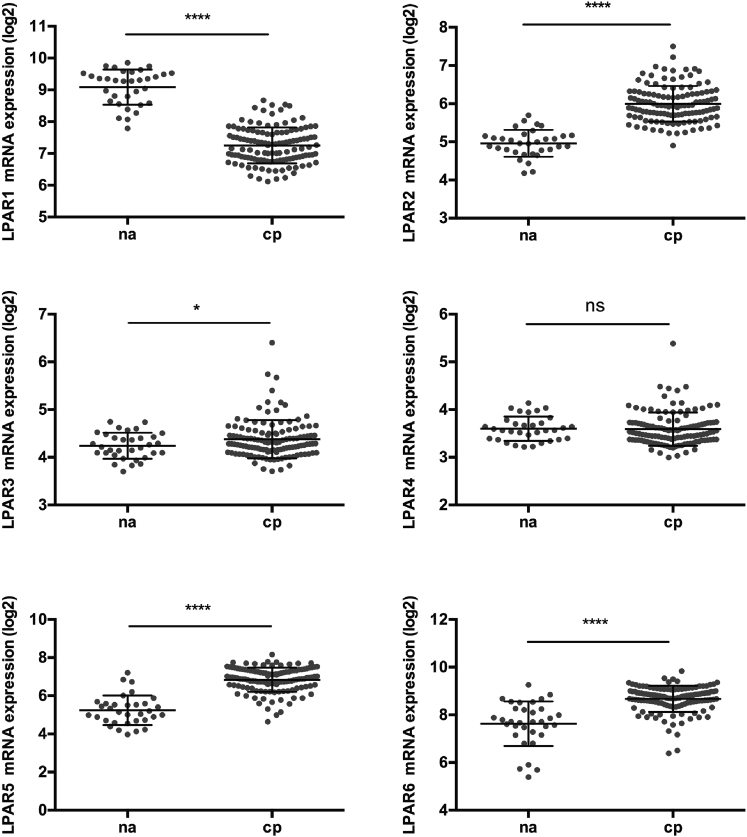
Expression of LPAR1–6 transcripts was then analyzed in microarrays from human CPs compared to NAs (discovery dataset) (Figure 3). LPAR1 was the only one downregulated in the CP versus NA comparison (mean log2 difference ± SD = −1.836 ± 0.1106), while LPAR4 did not show any significant difference and was detected at low levels, similar to LPAR3. LPARs 2, 5, and 6 were all significantly upregulated in this comparison and showed moderate-to-high expression levels in plaques, particularly LPAR6 (mean log 2 difference ± SD, LPAR2 = 1.037 ± 0.0876, LPAR5 = 1.604 ± 0.1306, and LPAR6 = 1.032 ± 0.1268).
Figure 3.
LPAR Expression in Comparison between Normal Arteries and Plaques
mRNA levels of LPA receptors (LPARs 1–6) in normal arteries (n = 33) and in human plaques (n = 127) were interrogated in the BiKE discovery microarray dataset. Values are expressed as log2 mean ± SD. *p < 0.05, and ****p < 0.0001; ns, not significant.
Expression correlations of LPARs with markers of cell types and processes in plaques showed an association between LPAR1 and typical SMC markers, whereas LPAR2 was negatively correlated with SMCs and showed moderate positive correlation with inflammatory cells (T lymphocytes and macrophages). Correlation analyses for LPAR3 and LPAR4 were inconclusive, likely due to low expression levels of these receptors in the tissue. LPAR5 and LPAR6 correlated to endothelial and inflammatory cell markers (dendritic cells, T lymphocytes, and macrophages) (Table S2).
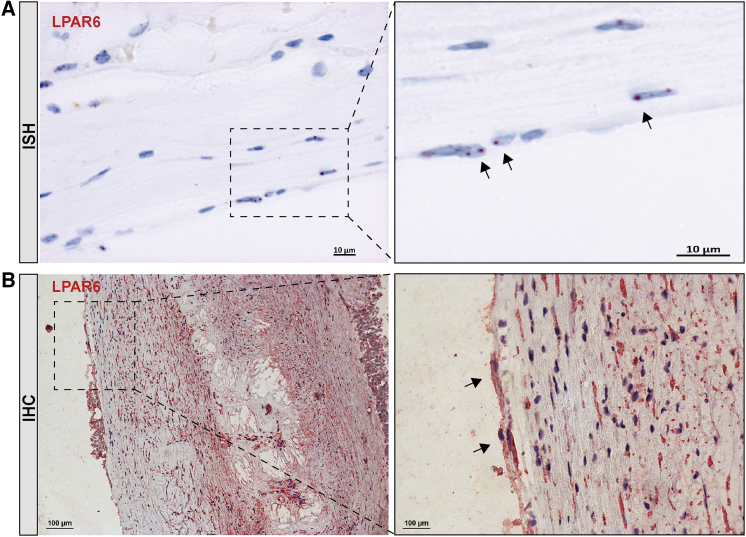
These analyses were followed by in situ hybridization (ISH) and immunohistochemistry (IHC) of the LPARs in consecutive sections of human lesions, which generally confirmed the differential expression data and correlation pattern of the receptors (Figure S1). LPAR1 was detected at low levels while LPARs 3 and 4, in line with the low expression levels from gene arrays, could not be detected in plaques (data not shown). LPAR2 localized exclusively in inflammatory cells, as determined by double staining with CD3, CD8, CD68, and CD163 (Figure S2). LPAR6 showed the most abundant expression of all receptors, both by ISH (Figure 4A) and IHC (Figure 4B), with localization in CD34+ ECs and in the fibrous cap smooth muscle-like cells. Together our analyses on transcript and protein levels showed the differential expression of LPARs in human plaques, and they revealed a strong enrichment of primarily LPAR6, but also LPAR2 and LPAR5 to a lesser extent.
Figure 4.
Localization of LPAR6 in Plaques
(A) In situ hybridization detection of LPAR6 mRNA transcript in human lesions. Arrows indicate the RNA probe signals (red). (B) Immunohistochemistry staining of LPAR6 in plaque (red signal) depicts its strong overall expression and localization in cells lining the luminal wall of the fibrous cap (arrows) as well as in the underlying cells with elongated nuclei. Nuclei (purple) are stained with hematoxylin. Images were taken with 20×, 40×, and 63× objectives.
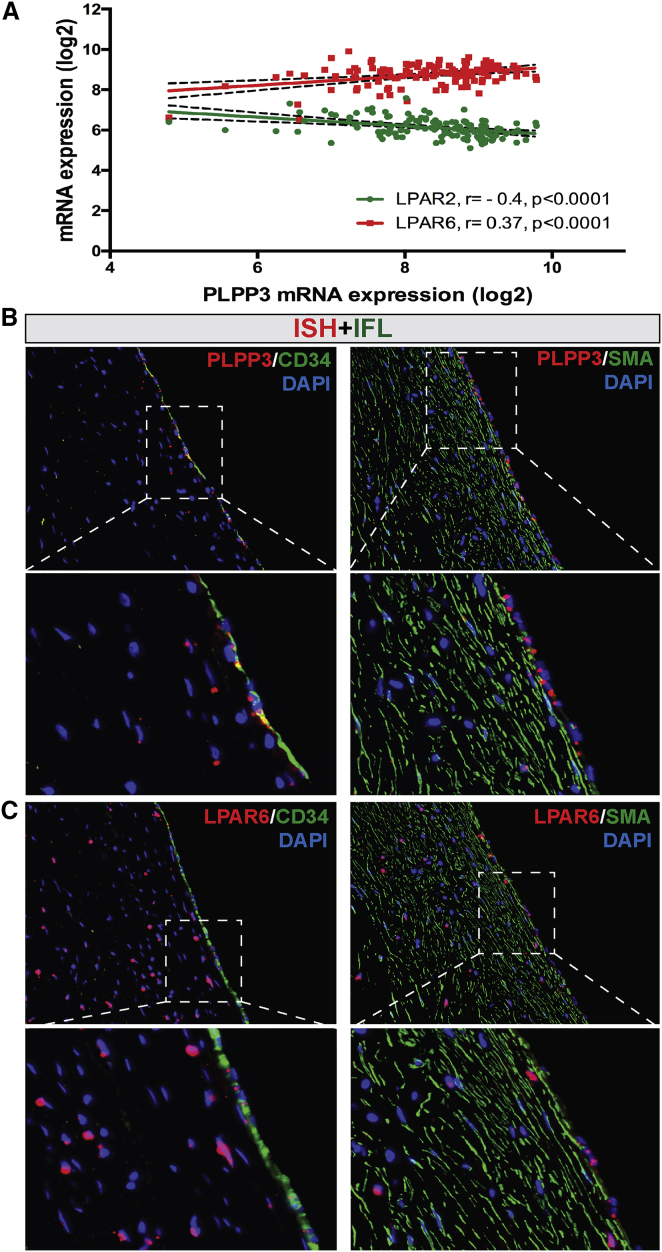
Next, the relationship between PLPP3 and LPARs in plaques was assessed by expression correlations from microarrays (Table S3). These analyses indicated a significantly positive association of PLPP3 with LPAR6 and negative association with LPAR2 (Figure 5A). Non-significant correlations were found between PLPP3 and other LPARs. A combined ISH/IFL staining was thus performed for PLPP3 and LPAR6 in human early fibroatheroma lesions, including markers of ECs and SMCs. These experiments convincingly showed that both PLPP3 and LPAR6 mRNA localize in CD34+ ECs on the luminal side of the fibrous cap, and localization in SMCs could also be confirmed (Figures 5B and 5C).
Figure 5.
Association between PLPP3 and LPAR6 in Human Lesions
(A) Correlation plot showing significantly positive association of PLPP3 with LPAR6 mRNA expression levels in plaques, while inverse with LPAR2. Pearson correlation calculated based on the discovery dataset (n = 127 samples). (B and C) Localization of (B) PLPP3 and (C) LPAR6 mRNA in CD34+ endothelial and SMA+ smooth muscle cells in the fibrous cap, shown by combined in situ hybridization (red signal) and immunofluorescence (green signal) in human early fibroatheroma lesions. Nuclei are stained with DAPI. Images were taken with 20× objective. IFL, immunofluorescence.
PLPP3, LPAR6, and LPA Pathway-Related Lipids Co-localize in Plaques
To examine the spatial relationship among PLPP3, LPAR6, and lipid species that constitute PLPP3 substrates in human plaques, a combination of immune- and lipid-imaging in situ was employed (n = 4 CPs tested). First, ten lipid species between 400 and 800 Da were detected in plaques (Table S4). Replicates of MALDI-mass spectrometry imaging (MSI) in detection mode at 30 μm showed the presence of cholesteryl ester (CE) stearate (18:0); oleate (18:1), and linoleate (18:2), localized particularly in the necrotic core and upper border of the fibrous cap. PC, the main phospholipid type, was detected in the necrotic core and in the middle area of the fibrous cap. LPC stearic acid (18:0), oleic acid (18:1), and linoleic acid (18:2) were mainly detected in the fibrous cap and shoulder region lining the necrotic core (Figure S3).
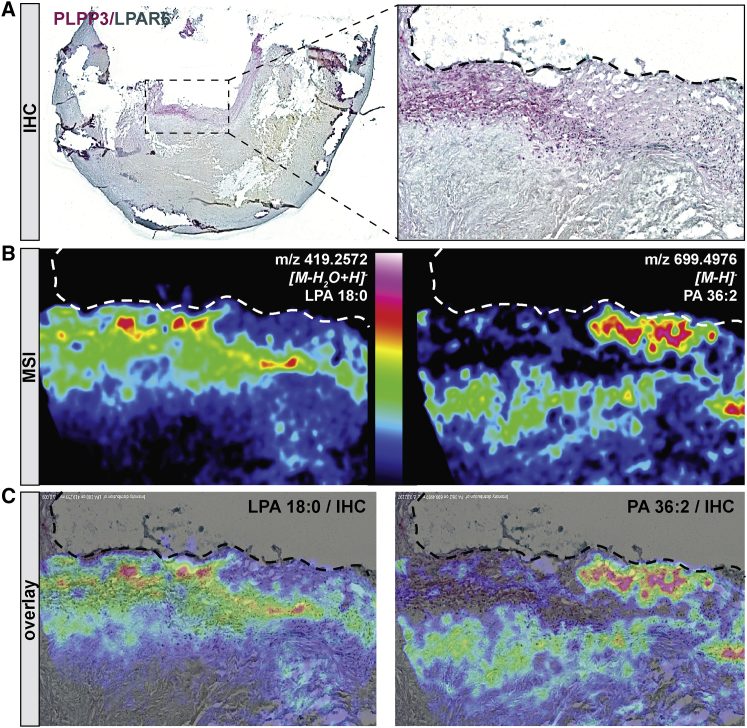
PLPP3 is an enzyme that de-phosphorylates LPA into monoacylglycerol (MAG); therefore, we next sought to examine the spatial relationship among PLPP3, LPAR6, and LPA in plaques. We could show that positive PLPP3 immunodetection (Figure 6A) co-localized with a high-intensity signal for LPA, a PLPP3 substrate, but not with PA signal, an LPA precursor (Figure 6B and overlay in Figure 6C). Similarly, co-localization was observed between LPAR6 and LPA, an LPAR ligand, but not with PA. Moreover, LPA and PA appeared to be mutually anti-localized based on the low spatial correlation factor of r = 0.2.
Figure 6.
Relationship of PLPP3, LPAR6, and LPA Pathway-Related Lipids in Atherosclerotic Plaques
(A) Double immunostaining of PLPP3 (red) and LPAR6 (green) in atherosclerotic plaques, showing their co-localization and accumulation of positive signal in the fibrous cap and shoulder region. (B) Distribution of lysophosphatidic acid (LPA 18:0, to the left) and PA 36:2 (to the right) on consecutive plaque sections by mass spectrometry imaging (MSI). Red signal indicates the highest amount of detected lipid (color scale reported in the middle). (C) Overlay of the PLPP3 and LPAR6 staining by IHC with MSI for LPA (left panel) or PA (right panel), showing an increase of LPA and a decrease of PA in PLPP3/LPAR6 double-positive regions. Images were taken with 2× objective, representative of n = 4 independent experiments.
Silencing of LPAR6 Diminishes LPA-Induced Endothelial Activation In Vitro, but Not after PLPP3 Silencing
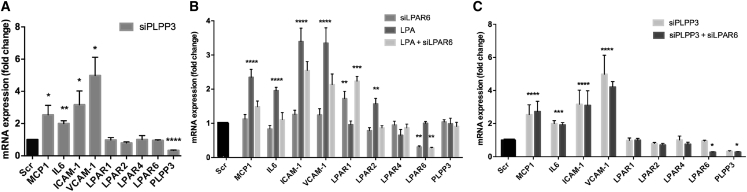
The possible effect of a reduced PLPP3 expression in ECs, which would resemble that observed in human plaques, was assessed using small interfering RNA (siRNA)-mediated gene silencing. Silencing of PLPP3 in human umbilical vein ECs (HUVECs) (by 47% as compared with a non-targeting scramble siRNA) resulted in a significant increase in mRNA expression of pro-inflammatory cytokines MCP-1 and interleukin-6 (IL-6) as well as the adhesion molecules VCAM-1 and ICAM-1 (Figure 7A). Importantly, a similar effect of PLPP3 silencing was previously reported in human aortic ECs.9, 12 Together, these results supported the anti-inflammatory function of PLPP3 in ECs.
Figure 7.
Effect of PLPP3 and LPAR6 Silencing on Endothelial Cell Activation In Vitro
(A) Silencing of PLPP3 in cultured endothelial cells leads to induced mRNA expression of the inflammatory cytokines MCP1 and IL-6 and adhesion factors VCAM-1 and ICAM-1 compared to control cells (scramble). No significant change in expression of LPARs 1–6 could be observed upon PLPP3 silencing. (B) Inhibition of LPAR6 alone had no prominent effect on MCP1, IL-6, VCAM-1, or ICAM-1, but the upregulation of these genes caused in response to LPA stimulation (10 μM, 2-hr treatment) could be significantly attenuated by simultaneous silencing of LPAR6. PLPP3 expression was not affected by LPA stimulation or the LPAR6 silencing. (C) Combined silencing of both LPAR6 and PLPP3 did not result in any diminishing effects on endothelial activation compared to those already observed after silencing PLPP3 alone. Note also that neither inhibition of PLPP3 nor LPA stimulation had any effect on the LPAR1 expression, but its induction was observed when LPAR6 was silenced. The expression of LPAR2 was strongly enhanced after LPA stimulation alone, but not in combination with the inhibition of LPAR6 or PLPP3. Expression of LPAR4 was not affected in any of these experiments. All experiments were repeated 4 times, n = 4 samples per group in all analyses. Significance indicated above bars calculated in comparison to scramble/control cells (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001).
To further explore this finding, we next stimulated the HUVECs with LPA, which led to the increased expression of cytokines IL-6 and MCP-1 as well as ICAM-1 and VCAM-1 (Figure 7B). These effects were comparable to when PLPP3 was silenced, which again suggested that lower PLPP3 levels may cause endothelial activation through enhanced LPA signaling. We further hypothesized that the effect of LPA would be mediated via LPAR6, shown to be the most prominent LPAR expressed in ECs in human plaques from our cohort. Interestingly, silencing of LPAR6 in HUVECs (by 57% on an mRNA level and confirmed also on a protein level; Figure S4) inhibited LPA-induced endothelial activation, observed by the reduced expression of ICAM-1, VCAM-1, IL-6, and MCP-1 (Figure 7B).
Surprisingly, simultaneous silencing of both LPAR6 and PLPP3 did not result in any diminishing effects on endothelial activation compared to those observed after silencing PLPP3 alone (Figure 7C). In addition, expression of LPAR1 was enhanced after silencing of LPAR6 and LPA stimulation, but not when in combination with the silencing of PLPP3. The mRNA expression of other LPARs (LPAR2, 4, and 6) was not significantly affected by the knockdown of PLPP3, and LPAR3 and LPAR5 were undetectable in ECs (data not shown). LPAR2 was only upregulated after the stimulation with LPA, and this effect could be inhibited by the silencing of LPAR6.
Discussion
To the best of our knowledge, this is the first study to systematically characterize the expression of the PLPP3 pathway, including all six LPARs as well as substrate LPA and its precursors phosphatidic acid (PA) and LPC, in advanced human atherosclerotic plaques. A reduced PLPP3 mRNA and protein expression was demonstrated in human plaques compared with normal disease-free artery specimens, suggesting more permissive conditions for signaling through LPA and its receptors in late-stage human atherosclerosis. This assumption was further supported by indications of even lower levels of PLPP3 in plaques from symptomatic patients than in those of asymptomatic ones, and an inverse relationship between plaque PLPP3 levels and future adverse cardio- and cerebrovascular events. Together with our eQTL studies confirming the genetic link between PLPP3 and carotid atherosclerosis, these findings suggest that suppression of PLPP3 is associated with more severe disease and confirm a potential athero-protective role for the enzyme, as previously proposed.9
This study is the first to definitively localize LPA and PA molecular species in situ in the fibrous cap of human plaques using MSI and overlay with PLPP3 and LPAR6 by immunodetection. Our analyses showed that PLPP3 and LPAR6 protein in the fibrous cap co-localize with LPA, the substrate of PLPP3 and the ligand for LPAR6, but not with PA that is a potential precursor of LPA generated by A-type phospholipase activity. Lipoprotein associated phospholipase A2 (Lp-PLA2), a platelet-activating factor (PAF) acetyl-hydrolase or group VII phospholipase A-secreted enzyme that catalyzes the degradation of PAF-producing LPA and acetate, could be a source of LPA. However, in a large-scale human genetic study, none of a series of Lp-PLA2-lowering alleles, including a loss-of-function mutation leading to full Lp-PLA2 deficiency, was related to coronary heart disease risk, suggesting that Lp-PLA2 is unlikely to be a causal risk factor for ACVD.21 This conclusion is also supported by the lack of cardiovascular benefit of the anti-PAFAH/Lp-PLA2 drug darapladib in large phase three clinical trials.22 Thus, we suggest that other extra- and intracellular enzymatic pathways may be involved in the local production of LPA independent of Lp-PLA2 activity.23 We identified saturated LPC (LPC18:0) as the main LPC species present in the advanced atherosclerotic lesions, confirming the previous findings that showed the presence and the enzymatic activity of secretory phospholipase A2 in human lesions.24 LPC associated with plasma lipoproteins accumulating in lesions can be another source of local LPC species.25 LPA has been reported by others to be significantly higher in coronary than in peripheral systemic arterial blood.14 Thus, together with our results, these findings support the hypothesis that higher concentrations of LPA might be associated with atherogenesis and its clinical consequence, ACVD.
LPA is a potent lipid mediator with broad cellular effects that are relevant to athero-thrombosis.13, 26, 27 PLPP3 deactivates LPA, and it has been suggested that the enhancement of its expression and activity might serve for pharmacological therapeutic purposes; however, considering that its control mechanisms are not yet understood, this could be a challenging task. An alternative approach to reduce LPA-mediated pro-inflammatory activation could be to target LPA-mediated signaling through modulation of its different receptors (LPARs). However, this requires a detailed investigation of the expression and cellular distribution of LPARs and other components of the pathway in human atherosclerosis, and our results should be considered as an initial step in this area.
Our investigation suggests that the repression of PLPP3 in atherosclerotic arterial tissue could contribute to the elevated local accumulation of LPA and increase its biological effects in lesions. It should be noted that PLPP3 fulfills also non-catalytic functions, such as its interaction with integrins that can promote EC-to-cell adhesion.28 Although PLPP3 expression levels are lower in plaques, we showed that the residual protein localizes to ECs, SMCs, and CD68+ cells. However, further studies will be needed to establish whether differential expression of PLPP3 in normal and diseased tissue is translated into differences in the hydrolytic activity required for deactivation of LPA. Similarly, we do not know if increased expression of the enzyme affects its non-enzymatic roles in endothelium and foam cells.29
When it comes to the LPARs, our results indicated that LPAR2, LPAR5, and especially LPAR6 were the most prominent receptors expressed in lesions. LPAR2 and LPAR6 were the only receptors also showing significant correlation with PLPP3 expression at the mRNA level. We found that LPAR6 expression was mostly associated with luminal ECs and with SMCs, but less frequently with inflammatory cells. Interestingly, LPAR6 has previously been involved in LPA-induced actin stress formation in ECs.30 LPAR2 was expressed in a specific subset of inflammatory cells, thus, it is possibly limited to certain types of plaques that express less PLPP3 combined with increased leukocyte content. These results are in line with previous data indicating that LPA can stimulate T cell migration and secretion of matrix metalloproteinases when overexpressing LPAR2.31 Together, this suggests that LPAR2 might contribute to LPA-mediated atheroma progression and may be a promising LPAR for pharmacological intervention to reduce atherogenic inflammation. With respect to the other LPARs, we found that the expression of LPARs 3 and 4 was either absent or too low to be detected both in plaques and specifically in ECs, indicating that these receptors may be of limited importance in the regulation of LPA activation in atherosclerotic lesions.
Our in vitro experiments were targeted to ECs as a plausible cell type to facilitate the future therapeutic application of the PLPP3 pathway. We show that silencing of PLPP3 did not affect the expression levels of LPARs 1, 2, 4, and 6 in ECs, although there was a significantly positive correlation between PLPP3 and LPAR6 in the whole-plaque tissue. This suggests a mechanism by which the loss of PLPP3 expression, and the potential subsequent increase in LPA, does not lead to a compensatory increase in the expression of LPARs in ECs. Knockdown of LPAR6 appeared to inhibit the upregulation of LPAR2 after LPA stimulation, which indicates that the LPA-LPAR6 interaction may be involved not only in endothelial activation but also in the regulation of LPAR2. Furthermore, the inhibition of LPAR6 induced the expression of LPAR1, suggesting a compensatory mechanism between these two receptors in ECs. Based on our in vitro data, it is possible to speculate that, although silencing of PLPP3 and LPA stimulation induce similar phenotypes in ECs, they may not share the same LPA-LPAR6 signaling. Our results also indicate a complexity and flexibility in signaling patterns among LPARs that can be dictated by repression of PLPP3 in combination with some yet unknown factors in the atherosclerotic tissue environment, considering that silencing of LPAR6 did not have a diminishing effect on the endothelial activation already observed after silencing of PLPP3 alone. Further experiments are needed to better understand the interaction among PLPP3, LPA, and the different LPARs.
Limitations
Although the BiKE cohort is one of the world’s largest when it comes to the molecular profiles of human carotid atherosclerotic plaques, an even larger number of subjects may be necessary to validate our findings in independent biobanks and generalize the observations to other vascular beds. Replication is warranted in heterogeneous cohorts that will permit adjustment for traditional cardiovascular risk factors and evaluation within subgroups of interest, such as defined by age, gender, symptoms, and comorbidities. This is also important in the context of genetic investigations of PLPP3 and LPARs. Consensus is lacking in the field regarding the selection of appropriate control tissues, and it is worth noting that arteries of various embryonic origins have been used for this purpose as well as adjacent macroscopically intact parts of lesions. In addition, more detailed in vitro studies were precluded due to the lack of suitable PLPP3 antibodies that would allow assessment of the protein levels. Finally, molecular mechanisms behind the interactions among PLPP3, LPA, and LPARs that could modulate atherosclerosis need to be explored beyond in vitro findings, using atherosclerotic animal models.
Conclusions
We found that PLPP3 is repressed in the advanced stages of human atherosclerosis compared with NAs. The enzyme co-localizes in lesions mainly with LPARs 5 and 6. Furthermore, expression of LPARs is high in the atheroma, and LPA co-localizes within plaque regions where the cells expressing PLPP3 and LPAR6 are also found. Collectively, our findings indicate that LPA-mediated inflammatory cascades are relevant in the pathophysiology of human atherosclerosis and that the PLPP3 enzyme and LPARs can modulate the potential atherogenicity of LPA signaling.
Clinical Impact
Altogether, our results support the hypothesis that PLPP3 serves to suppress inflammatory pathways in atherosclerotic plaques and that diminished PLPP3 activity may carry higher risk for developing complications of atherosclerosis. In human lesions, the LPARs LPAR2, LPAR5, and LPAR6 are expressed in ECs and regulate the response to LPA-induced EC activation in advanced plaques. Low PLPP3 expression levels in plaques appear to increase the local accumulation of the specific LPA molecular species (18:0). Our proposal is that this phospholipid should be investigated as the plasma biomarker for risk associated with low activity of LPP3 and, consequently, of LPA-related receptor activation. Moreover, our findings open the possibilities for investigations into the anti-atherosclerotic effects that may be achieved by therapies aimed to (1) increase the levels of PLPP3, thus reducing the levels of pro-inflammatory products; or (2) modulate the activity of the LPA membrane receptor(s). The first option would require the development of therapeutic approaches for increasing the PLPP3 gene product expression or the PLPP3 enzyme activity (i.e., localized gene therapy, modified RNA, or recombinant protein in stable liposomes).32 However, increasing PLPP3 expression is hazardous as this enzyme hydrolyzes also S1P, which is critical for lymphocyte egress and blood vessel integrity. A high-affinity S1P receptor agonist is currently in clinical development as an immunomodulator for transplantation and autoimmunity.33 Alternatively, modulation of LPAR signaling, a target of known drug-treatable class,15 would imply the development of more frequently used therapeutic strategies, such as small molecules or biologicals.
Materials and Methods
Human Material
Atherosclerotic plaques were obtained from patients undergoing surgery for high grade (>50% North American Symptomatic Carotid Endarterectomy Trial [NASCET])34 carotid stenosis at the Department of Vascular Surgery, Karolinska University Hospital, Stockholm, Sweden, and clinical data were recorded upon admission. Symptomatic patients (S) were defined based on the following symptoms: transitory ischemic attack (TIA), minor stroke, and amaurosis fugax (AF). Patients without qualifying symptoms within 6 months prior to surgery were categorized as asymptomatic (AS) and indication for CEA based on results from the Asymptomatic Carotid Surgery Trial (ACST). A mandatory examination of the carotid arteries (by duplex ultrasound [US] and/or computed tomography [CT] angiography) was performed prior to surgery. Patients with severe disability after major stroke were excluded since the remaining cost-benefit of stroke prevention was limited and did not outweigh the risk of surgery. In the study group, patients with atrial fibrillation were also excluded in order to minimize analysis of carotid lesions in patients with symptoms from cardioembolic rather than atheroembolic origin.
As previously described,35 human carotid endarterectomy samples (CPs) and blood were collected at surgery and retained within the BiKE. NA controls were obtained from nine macroscopically disease-free iliac arteries and one aorta from organ donors without history of cardiovascular disease. The BiKE study is approved by the Ethical Committee of Northern Stockholm with the following ethical permits: EPN DNr 95-276/277; 02-146; 02-147, 2005/83-31; 2009/512-31/2; 2009/295-31/2; 2011/950-32; 2012/619-32; and 213/2137-32. The project was performed under the Swedish biobank regulations, and prospective sampling was approved with informed consent procedure (DNr 2009/512-31/2). BiKE is registered at Socialstyrelsen (The National Board of Health and Welfare) and Biobank of Karolinska and approved by the Swedish Data Inspection Agency (approval date/number 2002-09-30 DNr 916-2002). The BiKE database was merged with the Swedish Hospital Discharge Register and the Swedish Cause of Death Register for follow-up of major adverse cardiovascular, cerebrovascular, and vascular events (MACCEs). All samples were collected with informed consent from patients or organ donor guardians.
This study involved 3 non-overlapping subsets of BIKE patients, where 2 sets of plaques were analyzed by Affymetrix microarrays (larger set with n = 127 plaques of which 87 were from S + 40 from AS patients and smaller set with n = 50 plaques where 40 from S + 10 from AS patients). DNA genotyping by Illumina chips was carried out on patients from the larger subset (n = 127) and used for eQTL analyses. The third subset of n = 18 BiKE plaques (n = 9 from S + 9 from AS patients, matched for gender, age, and statin medication) was analyzed using liquid chromatography-tandem mass spectrometry (LC-MS/MS) as previously described.35 For proteomic analyses, a central portion of the plaque corresponding to the maximum stenosis was separated from the respective downstream peripheral end (adjacent tissue) of the plaque and used in comparisons. The BiKE study cohort demographics, details of sample collection, processing, and analyses were as previously described.36
Additional control vascular tissues (n = 23) and early fibroateroma lesions were obtained from the SOCRATES biobank (Leiden University Medical Center, the Netherlands). Details of this biobank have been described previously.37 Briefly, this biobank contains aortic wall patches obtained during kidney transplantation with grafts derived from cadaveric donors. All patches were from grafts that were eligible for transplantation (i.e., all donors met the criteria set by The Eurotransplant Foundation). Sample collection and handling were performed in accordance with the guidelines of the Medical and Ethical Committee in Leiden, the Netherlands, and the code of conduct of the Dutch Federation of Biomedical Scientific Societies (https://www.federa.org/?s=1&m=82&p=0&v=4#827).
Altogether, n = 33 NAs of different embryonic origins were used as controls in this study. No additional demographic information is available for these samples obtained from organ donors.
ISH
Chromogenic ISH for the detection of the LPAR mRNAs was performed in a Ventana Discovery ULTRA instrument (Ventana Medical Systems, AZ, USA) using the ACD RNAscope 2.5 Red Kit (Advanced Cell Diagnostics, Newark, CA, USA) and the mRNA Discovery ULTRA RED 4.0 procedure. RNAscope 2.5 VS Probes for Hs-LPAR1 (483889), Hs-LPAR2 (428809), Hs-LPAR3 (428819), Hs-LPAR4 (458859), Hs-LPAR5 (#456369), and Hs-LPAR6 (409359) were designed by the probe manufacturer (Advanced Cell Diagnostics). Formalin fixed paraffin embedded (FFPE) sections (5 μm) were applied to Superfrost Plus (Thermo Fisher Scientific) slides, and all operations, including baking, deparaffinization, conditioning, pretreatment, ISH, and counterstaining using hematoxylin, were performed in a Ventana Discovery ULTRA instrument. Following the ISH procedure in the Ventana instrument, slides were washed in lukewarm tap water with detergent until oil from the slides was fully removed. Slides were finally washed in demineralized water and allowed to air dry before mounting in EcoMount mounting medium (Advanced Cell Diagnostics). Slides were subsequently inspected in bright-field microscopy using an AxioImager.Z1 microscope (Zeiss, Oberkochen, Germany), and digital images of selected regions of interest were acquired using 40× and 63× objectives.
Double Staining by ISH Combined with Immunofluorescence
Non-decalcified FFPE sections (5 μm) from early fibroatheroma lesion were used for co-staining of mRNA and protein markers. First, as described in the previous section, ISH analyses were performed for LPAR6 and PLPP3 mRNA using the Ventana Discovery ULTRA instrument (Ventana Medical Systems, AZ, USA), the ACD RNAscope 2.5 Red Kit (Advanced Cell Diagnostics, Newark, CA, USA), and the mRNA Discovery ULTRA RED 4.0 procedure. Hs-LPAR6 (409359) and Hs-PLPP3 (456371) for detection of mRNA were designed by the probe manufacturer (Advanced Cell Diagnostics). Second, following ISH and washes as previously described, two slides each from LPAR6 and PPAP2B mRNA detection were stained for CD34 and SMA, respectively. Briefly, slides were washed for 5 min in 1×PBS, followed by blocking in 0.1 M Tris-HCl, 0.15 M NaCl, 10% fetal bovine serum, and 0.01% Tween-20 for 15 min. Anti-CD34 primary antibody (mAb mouse anti human CD34 Class II clone QBEnd10, Dako, Glostrup, Denmark) diluted 1:100 and anti-SMA primary antibody (rabbit mAb anti human SMA, EPR5368, Abcam, Cambridge, UK) diluted 1:2,000, respectively, were added and incubated 30 min at room temperature (RT). Slides were washed three times in 1×PBS, and then fluorophore-labeled secondary antibodies were added. For detection of CD34, Alexa Flour 488 Goat anti Mouse IgG(H+L) (Jackson ImmunoResearch Laboratories, West Grove, PA, USA) (1:50) was added for 30 min at RT. For detection of SMA, Alexa Flour 488 Goat anti Rabbit IgG (H+L) (Jackson ImmunoResearch Laboratories, West Grove, PA, USA) (1:50) was added for 30 min at RT. Slides were washed 2 times in 1×PBS and mounted with DAPI for epifluorescence microscopy inspection and imaging.
Lipid MSI In Situ
Snap-frozen human carotid plaque tissues were sectioned (12-μm thickness) using cryostat CM3050S (Leica, Germany) and thaw mounted on SuperFrost glass slide to enable multiple staining modalities or on indium tin oxide (ITO) conductive glass slide for MSI experiments and then dried in a vacuum chamber.38 Optimal cutting temperature (OCT) compound was purchased from Fisher Scientific (Illkirch, France). For the MALDI imaging experiment, two MALDI matrices were used to enable the detection of lipid species, the 2,5-Dihydroxybenzoic acid (2,5-DHB) and the 9-aminoacridine (9AA, Sigma-Aldrich, saint-quentin Fallavier, France) for positive and negative detection mode, respectively. 2,5-DHB powder (150 mg) was used and vaporized on tissue sample using a home-built sublimation apparatus (150°C, 8 min, 2.10–3 mbar). 9AA solution was prepared at 10 mg/mL in methanol (MeOH)/water (7:3). The 9AA matrix solution was sprayed onto tissue sections using the SunCollect automatic sprayer (SunChrom, Friedrichsdorf, Germany).
MS images were obtained using a SolariX MALDI-FTICR 7.0T (Bruker Daltonics, Bremen, Germany) equipped with a Smartbeam II laser used at a repetition rate of 1 kHz. Mass spectra were acquired in the 100–1,200 m/z range in positive and negative detection mode depending on MALDI matrix. The mass spectrum was obtained by mass spectra average of 300 consecutive laser shots on the same location, with a time domain of 1MWord, subsequent single zero filling, and sine wave apodization. An image curve reduction (ICR) noise threshold was fixed at 0.97 for imaging acquisition. An image raster size of 30 μm was selected for human plaque tissues analysis. FTMSControl 3.0 and FlexImaging 4.2 software packages (Bruker Daltonics, Bremen, Germany) were used to control the mass and set imaging parameters. The visualization and statistical analysis of imaging data were performed using MultiImaging 1.1 software (ImaBiotech, Lille, France). To combine and compare tissue staining with the lipid distribution, MALDI imaging acquisition was performed on adjacent carotid tissue sections and scanned using a 40× magnification objective on an Olympus IX18 microscope (Olympus, Germany). Scans were then integrated in the MultiImaging software for a co-registration with imaging data to accurately correlate the distribution of molecular species with human plaque histological regions. Assignment of lipid species was performed by searching the accurate m/z values against the Lipidmap database using a mass tolerance of 3 ppm.39
Bionformatics and Statistical Analyses
RNA extracted from endarterectomy (CP) and control NA specimens was analyzed by Affymetrix microarrays. Robust multiarray average (RMA) normalization and correction for batch effect was performed, and processed gene expression data were returned in log2 scale. The microarray dataset is available with the following accession number from Gene Expression Omnibus: GSE21545. Target gene and protein analyses were performed with GraphPad Prism 6 using a two-sided Student’s t test assuming non-equal deviation. Pearson correlations were calculated to determine the association between mRNA expression levels from microarrays, with Bonferroni correction for multiple comparisons. Survival analysis was done using the Cox regression model40 with event-free survival as the response variable and log2-transformed gene expression levels as the explanatory variable. The covariates age and gender were tested and had no effect on results. Results from qPCR were evaluated by multiple t tests assuming non-equal deviation. In comparisons with more than two groups, ANOVA was used as appropriate. In all analyses, p value < 0.05 was considered to indicate statistical significance.
Additional methods are described in the Supplemental Information.
Author Contributions
S.A., L.P.M., G.H., D.v.K., D.T., K.H., A.S., B.S.N., V.E., R.A.-B., and M.L. performed experiments and analyzed data. G.P.-B., J.S., P.E., and J.H.N.L. contributed human material, datasets, and method development. All authors were involved in designing the study, joint discussions, and writing of the manuscript and comments. A.J.G., U.H., and E.H.-C. supervised the study.
Conflicts of Interest
G.H., J.S., and R.A.-B. are employees of ImaBiotech, France. D.v.K. and D.T. are employees of Quorics, the Netherlands. K.H. and B.S.N. are employed by Bioneer A/S, Denmark. A.J.G. is employed by TNO, the Netherlands. A.S. and E.H.-C. are employees of AstraZeneca, Sweden.
Acknowledgments
We thank Germán Camejo for carefully reading the manuscript and valuable comments. The research leading to these results has received major funding from the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement 602936 (CarTarDis project). This work was conducted with support from the Swedish Heart and Lung Foundation, the Swedish Research Council (K2009-65X-2233-01-3, K2013-65X-06816-30-4, and 349-2007-8703), Uppdrag Besegra Stroke (P581/2011-123), the Strategic Cardiovascular Programs of Karolinska Institutet and Stockholm County Council, the Stockholm County Council (ALF2011-0260 and ALF-2011-0279), the Foundation for Strategic Research, and the European Commission (CarTarDis, AtheroRemo, VIA, and AtheroFlux projects). L.P.M. is the recipient of fellowships from the Swedish Society for Medical Research (SSMF) and the Heart and Lung Foundation (HLF, Sweden), and L.P.M. acknowledges research grants from Tore Nilsson, Magnus Bergvall, and Karolinska Institutet Foundations of Sweden.
Footnotes
Supplemental Information includes Supplemental Materials and Methods, four figures, and four tables and can be found with this article online at https://doi.org/10.1016/j.omtm.2018.05.003.
Supplemental Information
References
- 1.Borén J., Williams K.J. The central role of arterial retention of cholesterol-rich apolipoprotein-B-containing lipoproteins in the pathogenesis of atherosclerosis: a triumph of simplicity. Curr. Opin. Lipidol. 2016;27:473–483. doi: 10.1097/MOL.0000000000000330. [DOI] [PubMed] [Google Scholar]
- 2.Ridker P.M., MacFadyen J.G., Thuren T., Everett B.M., Libby P., Glynn R.J., CANTOS Trial Group Effect of interleukin-1β inhibition with canakinumab on incident lung cancer in patients with atherosclerosis: exploratory results from a randomised, double-blind, placebo-controlled trial. Lancet. 2017;390:1833–1842. doi: 10.1016/S0140-6736(17)32247-X. [DOI] [PubMed] [Google Scholar]
- 3.Schunkert H., König I.R., Kathiresan S., Reilly M.P., Assimes T.L., Holm H., Preuss M., Stewart A.F., Barbalic M., Gieger C., Cardiogenics. CARDIoGRAM Consortium Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat. Genet. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson A.D., Handsaker R.E., Pulit S.L., Nizzari M.M., O’Donnell C.J., de Bakker P.I. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–2939. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mega J.L., Stitziel N.O., Smith J.G., Chasman D.I., Caulfield M., Devlin J.J., Nordio F., Hyde C., Cannon C.P., Sacks F. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet. 2015;385:2264–2271. doi: 10.1016/S0140-6736(14)61730-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cook D., Brown D., Alexander R., March R., Morgan P., Satterthwaite G., Pangalos M.N. Lessons learned from the fate of AstraZeneca’s drug pipeline: a five-dimensional framework. Nat. Rev. Drug Discov. 2014;13:419–431. doi: 10.1038/nrd4309. [DOI] [PubMed] [Google Scholar]
- 7.Ren H., Panchatcharam M., Mueller P., Escalante-Alcalde D., Morris A.J., Smyth S.S. Lipid phosphate phosphatase (LPP3) and vascular development. Biochim. Biophys. Acta. 2013;1831:126–132. doi: 10.1016/j.bbalip.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erbilgin A., Civelek M., Romanoski C.E., Pan C., Hagopian R., Berliner J.A., Lusis A.J. Identification of CAD candidate genes in GWAS loci and their expression in vascular cells. J. Lipid Res. 2013;54:1894–1905. doi: 10.1194/jlr.M037085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu C., Huang R.T., Kuo C.H., Kumar S., Kim C.W., Lin Y.C., Chen Y.J., Birukova A., Birukov K.G., Dulin N.O. Mechanosensitive PPAP2B Regulates Endothelial Responses to Atherorelevant Hemodynamic Forces. Circ. Res. 2015;117:e41–e53. doi: 10.1161/CIRCRESAHA.117.306457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Humtsoe J.O., Liu M., Malik A.B., Wary K.K. Lipid phosphate phosphatase 3 stabilization of beta-catenin induces endothelial cell migration and formation of branching point structures. Mol. Cell. Biol. 2010;30:1593–1606. doi: 10.1128/MCB.00038-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Panchatcharam M., Miriyala S., Salous A., Wheeler J., Dong A., Mueller P., Sunkara M., Escalante-Alcalde D., Morris A.J., Smyth S.S. Lipid phosphate phosphatase 3 negatively regulates smooth muscle cell phenotypic modulation to limit intimal hyperplasia. Arterioscler. Thromb. Vasc. Biol. 2013;33:52–59. doi: 10.1161/ATVBAHA.112.300527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Touat-Hamici Z., Weidmann H., Blum Y., Proust C., Durand H., Iannacci F., Codoni V., Gaignard P., Thérond P., Civelek M. Role of lipid phosphate phosphatase 3 in human aortic endothelial cell function. Cardiovasc. Res. 2016;112:702–713. doi: 10.1093/cvr/cvw217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Siess W., Zangl K.J., Essler M., Bauer M., Brandl R., Corrinth C., Bittman R., Tigyi G., Aepfelbacher M. Lysophosphatidic acid mediates the rapid activation of platelets and endothelial cells by mildly oxidized low density lipoprotein and accumulates in human atherosclerotic lesions. Proc. Natl. Acad. Sci. USA. 1999;96:6931–6936. doi: 10.1073/pnas.96.12.6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dohi T., Iida O., Soga Y., Hirano K., Suzuki K., Takahara M., Uematsu M., Nanto S. Incidence, predictors, and prognosis of in-stent occlusion after endovascular treatment with nitinol stents for femoropopliteal lesions. J. Vasc. Surg. 2014;59:1009–1015.e1. doi: 10.1016/j.jvs.2013.10.050. [DOI] [PubMed] [Google Scholar]
- 15.Llona-Minguez S., Ghassemian A., Helleday T. Lysophosphatidic acid receptor (LPAR) modulators: The current pharmacological toolbox. Prog. Lipid Res. 2015;58:51–75. doi: 10.1016/j.plipres.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 16.Panchatcharam M., Miriyala S., Yang F., Rojas M., End C., Vallant C., Dong A., Lynch K., Chun J., Morris A.J., Smyth S.S. Lysophosphatidic acid receptors 1 and 2 play roles in regulation of vascular injury responses but not blood pressure. Circ. Res. 2008;103:662–670. doi: 10.1161/CIRCRESAHA.108.180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bot M., Bot I., Lopez-Vales R., van de Lest C.H., Saulnier-Blache J.S., Helms J.B., David S., van Berkel T.J., Biessen E.A. Atherosclerotic lesion progression changes lysophosphatidic acid homeostasis to favor its accumulation. Am. J. Pathol. 2010;176:3073–3084. doi: 10.2353/ajpath.2010.090009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouchareb R., Mahmut A., Nsaibia M.J., Boulanger M.C., Dahou A., Lépine J.L., Laflamme M.H., Hadji F., Couture C., Trahan S. Autotaxin Derived From Lipoprotein(a) and Valve Interstitial Cells Promotes Inflammation and Mineralization of the Aortic Valve. Circulation. 2015;132:677–690. doi: 10.1161/CIRCULATIONAHA.115.016757. [DOI] [PubMed] [Google Scholar]
- 19.Rosengren B., Peilot H., Umaerus M., Jönsson-Rylander A.C., Mattsson-Hultén L., Hallberg C., Cronet P., Rodriguez-Lee M., Hurt-Camejo E. Secretory phospholipase A2 group V: lesion distribution, activation by arterial proteoglycans, and induction in aorta by a Western diet. Arterioscler. Thromb. Vasc. Biol. 2006;26:1579–1585. doi: 10.1161/01.ATV.0000221231.56617.67. [DOI] [PubMed] [Google Scholar]
- 20.Lin M.E., Herr D.R., Chun J. Lysophosphatidic acid (LPA) receptors: signaling properties and disease relevance. Prostaglandins Other Lipid Mediat. 2010;91:130–138. doi: 10.1016/j.prostaglandins.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gregson J.M., Freitag D.F., Surendran P., Stitziel N.O., Chowdhury R., Burgess S., Kaptoge S., Gao P., Staley J.R., Willeit P., CKDGen consortium. International Consortium for Blood Pressure. CHARGE inflammation working group. MICAD Exome consortium. EPIC-CVD consortium and the CHD Exome+ consortium Genetic invalidation of Lp-PLA2 as a therapeutic target: Large-scale study of five functional Lp-PLA2-lowering alleles. Eur. J. Prev. Cardiol. 2017;24:492–504. doi: 10.1177/2047487316682186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wallentin L., Held C., Armstrong P.W., Cannon C.P., Davies R.Y., Granger C.B., Hagström E., Harrington R.A., Hochman J.S., Koenig W., STABILITY Investigators Lipoprotein-Associated Phospholipase A2 Activity Is a Marker of Risk But Not a Useful Target for Treatment in Patients With Stable Coronary Heart Disease. J. Am. Heart Assoc. 2016;5:e003407. doi: 10.1161/JAHA.116.003407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gora S., Lambeau G., Bollinger J.G., Gelb M., Ninio E., Karabina S.A. The proinflammatory mediator Platelet Activating Factor is an effective substrate for human group X secreted phospholipase A2. Biochim. Biophys. Acta. 2006;1761:1093–1099. doi: 10.1016/j.bbalip.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Giordanetto F., Pettersen D., Starke I., Nordberg P., Dahlström M., Knerr L., Selmi N., Rosengren B., Larsson L.O., Sandmark J. Discovery of AZD2716: A Novel Secreted Phospholipase A2 (sPLA2) Inhibitor for the Treatment of Coronary Artery Disease. ACS Med. Chem. Lett. 2016;7:884–889. doi: 10.1021/acsmedchemlett.6b00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stübiger G., Aldover-Macasaet E., Bicker W., Sobal G., Willfort-Ehringer A., Pock K., Bochkov V., Widhalm K., Belgacem O. Targeted profiling of atherogenic phospholipids in human plasma and lipoproteins of hyperlipidemic patients using MALDI-QIT-TOF-MS/MS. Atherosclerosis. 2012;224:177–186. doi: 10.1016/j.atherosclerosis.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 26.Kurano M., Suzuki A., Inoue A., Tokuhara Y., Kano K., Matsumoto H., Igarashi K., Ohkawa R., Nakamura K., Dohi T. Possible involvement of minor lysophospholipids in the increase in plasma lysophosphatidic acid in acute coronary syndrome. Arterioscler. Thromb. Vasc. Biol. 2015;35:463–470. doi: 10.1161/ATVBAHA.114.304748. [DOI] [PubMed] [Google Scholar]
- 27.Zhou Z., Subramanian P., Sevilmis G., Globke B., Soehnlein O., Karshovska E., Megens R., Heyll K., Chun J., Saulnier-Blache J.S. Lipoprotein-derived lysophosphatidic acid promotes atherosclerosis by releasing CXCL1 from the endothelium. Cell Metab. 2011;13:592–600. doi: 10.1016/j.cmet.2011.02.016. [DOI] [PubMed] [Google Scholar]
- 28.Tang X., Benesch M.G., Brindley D.N. Lipid phosphate phosphatases and their roles in mammalian physiology and pathology. J. Lipid Res. 2015;56:2048–2060. doi: 10.1194/jlr.R058362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reschen M.E., Gaulton K.J., Lin D., Soilleux E.J., Morris A.J., Smyth S.S., O’Callaghan C.A. Lipid-induced epigenomic changes in human macrophages identify a coronary artery disease-associated variant that regulates PPAP2B Expression through Altered C/EBP-beta binding. PLoS Genet. 2015;11:e1005061. doi: 10.1371/journal.pgen.1005061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yukiura H., Kano K., Kise R., Inoue A., Aoki J. LPP3 localizes LPA6 signalling to non-contact sites in endothelial cells. J. Cell Sci. 2015;128:3871–3877. doi: 10.1242/jcs.172098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zheng Y., Kong Y., Goetzl E.J. Lysophosphatidic acid receptor-selective effects on Jurkat T cell migration through a Matrigel model basement membrane. J. Immunol. 2001;166:2317–2322. doi: 10.4049/jimmunol.166.4.2317. [DOI] [PubMed] [Google Scholar]
- 32.Valeur E., Guéret S.M., Adihou H., Gopalakrishnan R., Lemurell M., Waldmann H., Grossmann T.N., Plowright A.T. New Modalities for Challenging Targets in Drug Discovery. Angew. Chem. Int. Ed. Engl. 2017;56:10294–10323. doi: 10.1002/anie.201611914. [DOI] [PubMed] [Google Scholar]
- 33.Brinkmann V., Cyster J.G., Hla T. FTY720: sphingosine 1-phosphate receptor-1 in the control of lymphocyte egress and endothelial barrier function. Am. J. Transplant. 2004;4:1019–1025. doi: 10.1111/j.1600-6143.2004.00476.x. [DOI] [PubMed] [Google Scholar]
- 34.Naylor A.R., Rothwell P.M., Bell P.R. Overview of the principal results and secondary analyses from the European and North American randomised trials of endarterectomy for symptomatic carotid stenosis. Eur. J. Vasc. Endovasc. Surg. 2003;26:115–129. doi: 10.1053/ejvs.2002.1946. [DOI] [PubMed] [Google Scholar]
- 35.Perisic Matic L., Rykaczewska U., Razuvaev A., Sabater-Lleal M., Lengquist M., Miller C.L., Ericsson I., Röhl S., Kronqvist M., Aldi S. Phenotypic Modulation of Smooth Muscle Cells in Atherosclerosis Is Associated With Downregulation of LMOD1, SYNPO2, PDLIM7, PLN, and SYNM. Arterioscler. Thromb. Vasc. Biol. 2016;36:1947–1961. doi: 10.1161/ATVBAHA.116.307893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perisic L., Aldi S., Sun Y., Folkersen L., Razuvaev A., Roy J., Lengquist M., Åkesson S., Wheelock C.E., Maegdefessel L. Gene expression signatures, pathways and networks in carotid atherosclerosis. J. Intern. Med. 2016;279:293–308. doi: 10.1111/joim.12448. [DOI] [PubMed] [Google Scholar]
- 37.van Dijk R.A., Virmani R., von der Thüsen J.H., Schaapherder A.F., Lindeman J.H. The natural history of aortic atherosclerosis: a systematic histopathological evaluation of the peri-renal region. Atherosclerosis. 2010;210:100–106. doi: 10.1016/j.atherosclerosis.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 38.Stegemann C., Drozdov I., Shalhoub J., Humphries J., Ladroue C., Didangelos A., Baumert M., Allen M., Davies A.H., Monaco C. Comparative lipidomics profiling of human atherosclerotic plaques. Circ Cardiovasc Genet. 2011;4:232–242. doi: 10.1161/CIRCGENETICS.110.959098. [DOI] [PubMed] [Google Scholar]
- 39.Caprioli R.M., Farmer T.B., Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 1997;69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- 40.Folkersen L., Persson J., Ekstrand J., Agardh H.E., Hansson G.K., Gabrielsen A., Hedin U., Paulsson-Berne G. Prediction of ischemic events on the basis of transcriptomic and genomic profiling in patients undergoing carotid endarterectomy. Mol. Med. 2012;18:669–675. doi: 10.2119/molmed.2011.00479. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.