Abstract
Asthma is a heterogeneous group of conditions that typically begin in early life and result in recurrent, reversible bronchial obstruction. The role played by epigenetic mechanisms in the pathogenesis of childhood asthma is understood only in part. Here we discuss asthma epigenetics within a developmental perspective based on our recent demonstration that the epigenetic trajectory to childhood asthma begins at birth. We next discuss how this trajectory may be affected by prenatal environmental exposures. Finally, we examine in vitro studies that model the impact of asthma-associated exposures on the epigenome. All of these studies specifically surveyed human DNA methylation and involved a genome-wide component. In combination, their results broaden our understanding of asthma pathogenesis and the role the methylome plays in this process.
Keywords: : asthma trajectory, DNA methylation, epigenetics
Asthma is a heterogeneous group of conditions that result in recurrent, reversible bronchial obstruction [1]. It is also the most prevalent chronic disease of childhood, with an economic burden that exceeds that of tuberculosis and HIV/AIDS combined [2]. Epidemiological evidence shows that chronic symptoms may appear at any age but most often arise during the preschool years [3]. However, both the time at which the child's trajectory to asthma truly begins and the mechanisms underlying asthma inception remain largely unknown.
Subtle modifications of both innate and adaptive immune responses accompany and often precede a diagnosis of childhood asthma [4,5], consistent with the notion that immune and respiratory alterations at an early window of susceptibility converge to place the child on a trajectory to the disease. Genome-wide association studies (GWAS) have identified multiple genetic variants that influence asthma susceptibility [6] but have accounted for only a modest proportion of the total phenotypic variance, providing a compelling rationale for seeking additional asthma risk factors. In this context, epigenetic mechanisms appear especially worth investigating because environmental and developmental influences are essential for asthma pathogenesis [1], and epigenetic processes ensure the timed unfolding of developmental programs and plastic responses to environmental cues, including those delivered in utero by the maternal milieu [7].
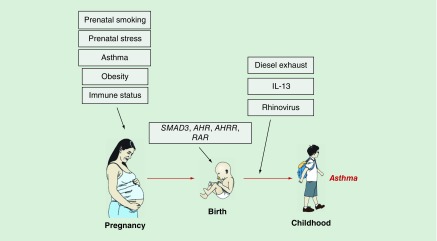
As noted in recent reviews [8–10], our understanding of the role of epigenetic mechanisms in asthma has evolved over the last few years, but only incrementally – in part because much of the work has relied on pre-existing cross-sectional studies that can only explore relationships with concurrent disease, in part because of limitations in the assessment technologies and sources of tissue (typically, unfractionated peripheral blood). However, experimental platforms have now improved and novel models are being proposed. The availability of more sophisticated algorithms to adjust for cellular heterogeneity in blood-based studies [11] (still the majority) also represents an important step forward. Thus, at least in our opinion, the field has now the potential to move to the next level and seek conceptual and experimental paradigm shifts. In an attempt to contribute to this evolving discourse, here we frame our discussion of asthma epigenetics within a developmental perspective that revolves around a fundamental question: is there an epigenetic trajectory to childhood asthma, and if so, when does it begin and which factors can influence it? Our novel findings that the trajectory to childhood asthma begins at birth [12] if not earlier [13], and is reflected in alterations of the neonatal methylome (Figure 1), provide a compelling rationale for this line of inquiry and outline a scenario in which the study of asthma epigenetics acts as a powerful searchlight to probe into the origins of a disease that remains mechanistically elusive.
Figure 1. . The epigenetic trajectory to asthma.
The figure shows key exposures that have been shown to influence the epigenetic trajectory to asthma.
Over the last few years, we have charted the rise of asthma epigenetics from different angles [9–10,14–16]. Here we review recent evidence that highlights the emerging role of epigenetic mechanisms as transducers of signals provided by pre- and/or perinatal exposures that influence the trajectory to childhood asthma and modify risk for this disease. We focused on genome-wide surveys of human DNA methylation because the role of other epigenetic modifications (e.g., post-translational histone modifications) has not yet been systematically addressed. Moreover, unlike candidate gene studies, genome-wide surveys provide a tool for discovering novel disease pathways – an urgent need, particularly in the childhood asthma field. We also discuss a novel, promising line of investigation that seeks to characterize the role of epigenetic events in asthma pathogenesis by relying on cellular in vitro models. Detailed information about the characteristics of the studies we reviewed (disease phenotype, sample size, age, exposure, sampled tissue and methylation platform) is presented in Table 1.
Table 1. . Study characteristics of recent epigenome-wide studies in asthma.
| Methylation platform | Tissue | Sample size | Age | Exposure | Phenotype | Ref. |
|---|---|---|---|---|---|---|
| NimbleGen 2.1M Human Promoter Deluxe | CBMC | Genome-wide discovery phase: n = 36 (IIS) Targeted analysis phase: n = 60 (IIS) n = 30 (MAAS) n = 28 (COAST) |
Birth | Genome-wide discovery phase: NA Targeted analysis phase: maternal asthma |
Asthma (2–9 years) | [12] |
| Illumina HumanMethylation27 and HumanMethylation450 | Whole blood | n = 527 (CAMP) n = 526 (Asthma BRIDGE) n = 1062 (MoBa) |
Age 5–12 years | Prenatal smoking | Asthma (5–12 years) | [17] |
| Illumina HumanMethylation450 | Whole blood | n = 6685 newborns n = 3187 older children |
Birth and older children | Prenatal smoking | NA | [18] |
| Illumina HumanMethylation450 | Whole blood | n = 1062 (MoBa1) n = 685 (MoBa2) |
Birth | Prenatal smoking | NA | [19] |
| Illumina HumanMethylation27 | Whole blood | n = 40 | Age 12 months | Maternal asthma | NA | |
| Whole genome bisulfite-sequencing | Whole blood | n = 443 | Birth | Maternal prenatal stress | Persistent wheeze (5 years) | [20] |
| Illumina HumanMethylation450 | Whole blood | n = 1018 | Birth and older children | Maternal obesity | NA | [21] |
| Illumina HumanMethylation450 | Bronchial epithelial cells | n = 17 | Mean age: 28.4 years | Ex vivo diesel exhaust and allergen | Asthma (mean age: 28.4 years) | [22] |
| Illumina HumanMethylation450 | Airway epithelial cells | n = 58 primary AEC cultures n = 116 freshly isolated AEC |
Primary AEC cultures (mean age: 45 years) freshly isolated AEC (mean age: 38.41 years) |
In vitro IL-13 treatment | Asthma (mean age: 38.41 years) | [23] |
| Illumina HumanMethylation450 | Nasal epithelial cells | n = 29 | Mean age: 25 years | In vitro rhinovirus infection | Asthma (mean age: 25 years) | [24] |
AEC: Airway epithelial cell; Asthma BRIDGE: Asthma BioRepository for Integrative Genomic Exploration study; CAMP: Childhood Asthma Management Program; CBMC: Cord blood mononuclear cell; COAST: Childhood Origins of ASThma Study; IIS: Infant Immune Study; MAAS: Manchester Asthma and Allergy Study; MoBa: The Norwegian Mother and Child Cohort Study; NA: Not applicable; PBMC: Peripheral blood mononuclear cell.
The epigenetic trajectory to asthma begins at birth (if not earlier)
The impetus for our study, the first epigenome-wide search for signatures in the neonatal methylome that are associated with (and thus predict) childhood-onset asthma, was provided by the notion that the detection at birth of differentially methylated regions (DMRs) associated with asthma during childhood would both support a perinatal origin of the disease and highlight epigenetic mechanisms potentially contributing to asthma inception. To this purpose, we examined cord blood mononuclear cells from children enrolled in the Infant Immune Study (IIS), an unselected birth cohort in which the development of asthma and immune responses was carefully monitored from birth to age 9 [5,25]. This work showed that neonatal immune cells did indeed harbor 589 DMRs that distinguished IIS children who did and did not develop asthma by age 9. Network analysis revealed that these childhood asthma-associated DNA methylation signatures clustered in immunoregulatory and proinflammatory pathways; the transcription factor SMAD3, an asthma gene highly replicated in GWAS [26–28], was the most connected node within the network of asthma-associated DMRs. Of note, SMAD3 methylation was selectively increased in asthmatic children of asthmatic mothers and was associated with childhood asthma risk not only in IIS but also in two other comparable birth cohorts, the Manchester Asthma and Allergy Study and the Childhood Origins of ASThma study. A meta-analysis revealed that for each 10% increase in methylation at a representative CpG site in the SMAD3 DMR, there is nearly a twofold increased risk of childhood asthma (meta-analysis odds ratio [OR]: 1.95; 95% CI [1.23, 3.10]; p = 0.005). Moreover, SMAD3 methylation in IIS neonates with maternal asthma was strongly and positively associated with neonatal production of IL-1β, an innate inflammatory mediator that is increasingly recognized as a key asthma mediator in both children [29] and adults, especially in neutrophilic asthma [30,31].
These results strongly suggest that the trajectory to childhood asthma begins at birth and involves epigenetic modifications in immunoregulatory and proinflammatory pathways. As importantly, data from three birth cohorts showed that SMAD3 promoter hypermethylation was associated with childhood asthma selectively in neonates with a maternal history of asthma – one of the strongest risk factors for asthma in the child [32,33]. These findings are especially noteworthy in the context of the current dearth of characteristics measurable at birth that effectively predict asthma during childhood. To our knowledge, this is the first time that a neonatal epigenetic characteristic linked to asthma during childhood is robust enough to replicate across three independent birth cohorts.
Our data also provide clues about potentially novel mechanisms of childhood asthma inception by emphasizing the functional connection between SMAD3, a regulator of both the asthma-protective T regulatory and the asthma-promoting Th17 cell differentiation programs [1,34], and IL-1β, a key asthma mediator in both children [29] and adults [30,31]. Indeed, in neonates who became asthmatic by age 9, SMAD3 promoter hypermethylation, an epigenetic configuration consistent with low SMAD3 expression, was strongly associated with high IL-1β production. This convergence may destabilize the T regulatory cell program, enhance inflammation and promote Th17 differentiation [35], ultimately favoring the development of asthma. While it is unclear whether these mechanisms operate pre- and/or perinatally, detection of the relationship between SMAD3 methylation and IL-1β production selectively among children of asthmatic mothers implies that the in utero environment is critical for directing the epigenetic trajectory toward childhood asthma. A scenario in which epigenetic modifications at an early window of susceptibility promote a long-term developmental trajectory to asthma is consistent with the emerging paradigm that chronic noncommunicable diseases have their origins in early life through an epigenetic calibration of set points for later responsiveness and function [36,37].
Impact of prenatal exposures on the epigenetic trajectory to asthma
The finding that the trajectory to childhood asthma begins at birth and involves epigenetic mechanisms [12] begs important questions – first and foremost, whether this epigenetic trajectory begins even before birth, under the influence of exposures that affect the pregnant mother and the in utero development of the child, thereby modifying the risk of developing asthma later in life. The recent studies reviewed below support this general notion by demonstrating that certain prenatal exposures associated with childhood asthma are also associated with DNA methylation changes in the child's peripheral blood cells (Figure 1).
Maternal smoking
Maternal smoking during pregnancy was shown to alter the child's methylome, implying that prenatal smoke exposure may lead to adverse outcomes in the fetus, including asthma. Indeed, a study conducted in 527 children showed that 5- to 12-year-old asthmatics differed in DNA methylation levels at 19 loci depending on in utero exposure to maternal smoking [17]. Findings for two of these loci (FRMD4A and C11orf52) were replicated in two independent cohorts, and four additional genes (XPNPEP1, PPEF2, SMPD3 and CRYGN) were nominally associated in at least one replication population. It is noteworthy that the expression of XPNPEP1 and PPEF2 was significantly decreased in subjects who had been exposed to prenatal smoking compared with those who had not been exposed. These results suggest that in utero smoke exposure can have lasting effects on both DNA methylation and gene expression during childhood.
A follow-up meta-analysis addressed these issues in a much larger population (n = 6685 neonates) [18]. Of the 3932 CpG sites that remained associated with prenatal smoke exposure after adjusting for cell type, the vast majority (2965 CpGs) had not been previously reported to be linked to maternal smoking during pregnancy [18]. When these novel CpGs were interrogated for their relationship with gene expression in adults, 254 unique CpGs were found to be significantly associated with the expression of a transcript located within 250 kb. In children (age 4 years), 35 CpGs were significantly associated with gene expression. Importantly, six genes (ENOSF1, HOXB2, IL32, NLRP2, PASK and TDRD9) exhibited significant CpG–transcript relationships in both adults and children. It is also worth emphasizing that the most significant hit of this study (cg05575921; AHRR) was also the top hit in other studies that have investigated the association between childhood DNA methylation and maternal smoking during pregnancy [18]. In combination, these data suggest that prenatal exposures may influence the epigenetic trajectory to asthma and elicit responses that persist into childhood.
The evaluation of individual CpG sites was more recently extended to gene-level and pathway-level analyses among 1062 participants in the Norwegian Mother and Child Cohort Study [19]. Maternal smoking in pregnancy was assessed using plasma cotinine as a biomarker. Novel bioinformatics tools (particularly, gene scores and the Sequence Kernel Association Test) were used to collapse epigenome-wide methylation data into gene- and pathway-level effects and test whether exposure to maternal smoking in utero results in differential methylation in genes enriched in biologic pathways. Fifteen genes were found to be associated with maternal plasma cotinine levels. Six of these 15 genes (GFI1, MYO1G, CYP1A1, RUNX1, LCTL and AHRR) contained individual CpGs that were differentially methylated relative to cotinine levels (p < 1.06 × 10−7). Replicated results from pathway analysis identified a cluster of pathways related to cancer, cell cycle, ER-α receptor signaling and angiogenesis, and a second cluster related to immune system function and T cell regulation. Thus, these data identify biological mechanisms that may underpin the adverse health effects of tobacco smoke exposure in utero.
Maternal asthma
Maternal smoking in pregnancy is not the only prenatal exposure that is associated with asthma risk during the child's subsequent life. Infants exposed in utero to the biological milieu associated with maternal asthma, the strongest and most replicated predictor of asthma during childhood [32], had distinct DNA methylation profiles in their peripheral blood cells at 12 months of age [38]. Of the 70 loci identified as being differentially methylated depending on maternal asthma status, 12 showed differences >10%, with increased methylation at FAM181A, MRI1, PIWIL1, CHFR, DEFA1, MRPL28 and RKA, and decreased methylation at NALP1L5, MAP8KIP3, ACAT2 and PM20D1, in children with maternal asthma. Moreover, MAP8KIP3 methylation was also significantly associated with maternal blood eosinophils and serum total IgE, and PM20D1 methylation was significantly lower in infants born to asthmatic mothers treated with inhaled corticosteroids. These findings raise the possibility that maternal asthma modifies childhood asthma risk at least in part through epigenetic mechanisms.
Maternal stress during pregnancy
Because prenatal maternal stress is known to influence the risk for persistent wheeze [39] and childhood asthma [40], a recent study investigated whether and how this exposure affects DNA methylation at birth. Prenatal maternal stress was found to be associated with cord blood DNA methylation at over 2000 regions. KEGG pathway analysis showed that maternal stress-induced differential methylation was enriched for neuroactive G-protein-coupled receptors also associated with the calcium and Wnt signaling pathways. In particular, methylation of NMUR1 was significantly associated with neonatal production of IL-4, IL-5 and IL-6 selectively in children who developed persistent wheeze later in life [20]. Neonatal GNA11 methylation was significantly higher in children exposed to high maternal stress, and was associated with decreased gene expression. Additionally, higher GNA11 methylation was observed in 4-year-old children with persistent wheeze. This study further emphasizes the long lasting effects of prenatal exposures on a child's trajectory to asthma and related phenotypes.
Maternal obesity
The possibility that maternal obesity promotes asthma in the child by leveraging both inflammatory and metabolic pathways has been gaining momentum over the last few years [41]. A meta-analysis of 14 studies and 108,321 mother–child pairs showed that maternal overweight or obesity in pregnancy was associated with increased risks of childhood asthma or wheeze ever (OR: 1.31; 95% CI [1.16, 0.49]) and current asthma or wheeze (OR: 1.21; 95% CI [1.07, 0.37]), independent of offspring BMI [42]. The studies that explored the links between maternal obesity and DNA methylation in the offspring are somewhat inconsistent in their results but point to an association between maternal BMI and neonatal DNA methylation at a limited number of CpG sites. Associations of maternal obesity with offspring methylation were stronger than associations of paternal obesity, supporting an intrauterine mechanism [21].
Maternal immune milieu during pregnancy
Although, as discussed, various maternal characteristics during pregnancy have been shown to relate to childhood asthma, little is known about the relationship between maternal immune status during pregnancy and asthma in the child. Our group has recently provided the first evidence of maternal immune status indicators during pregnancy that relate to risk for childhood asthma. Indeed, a higher ratio of IFN-γ:IL-13 secreted by maternal mitogen-stimulated peripheral blood mononuclear cells isolated during the third trimester of pregnancy was associated with decreased risk of childhood asthma (OR: 0.33; 95% CI [0.17, 0.65]; p = 0.001). Interestingly, this relation was evident for non-asthmatic mothers (OR: 0.18; 95% CI [0.08, 0.41]; p = 0.00004) but not for asthmatic mothers (p = 0.18), and was apparent in maternal prenatal but not postnatal immune cells. Paternal cytokine ratios did not relate to childhood asthma [43]. Interestingly, preliminary analyses suggest that the relationship between the maternal prenatal immune profile and childhood asthma involves the neonatal epigenome [13]. In children of non-asthmatic mothers, methylation at 553 CpG sites was significantly associated with the prenatal maternal IFN-γ:IL-13 ratio (false discovery rate [FDR] <0.15). In marked contrast, no CpG sites were associated with the IFN-γ:IL-13 ratio in children of asthmatic mothers. IFN-γ:IL-13 ratio associated CpG sites clustered primarily in the RAR and AHR pathways, and had TGFB1 as a prominent upstream regulator. These results suggest that the maternal prenatal immune milieu influences DNA methylation selectively in neonates born to non-asthmatic mothers, primarily affecting genes important for the induction of functional T regulatory cells (RAR: [44]) and the establishment of microbial communities (AHR: [45]). More generally, these results are consistent with the hypothesis that the epigenetic trajectory to childhood asthma begins prenatally [13].
In combination, the findings discussed above support the notion that prenatal exposures can alter the trajectory to asthma and allergic disease through epigenetic mechanisms. The fact that some of these exposures have effects on DNA methylation that persist in early life and throughout childhood points to the unique properties of the epigenome as a sensor of exposures at critical windows of development. It should also be noted that in general, these epigenetic hits do not map to genes associated with asthma in GWAS. Therefore, epigenetic and genetic studies might provide complementary information.
Impact of asthma-associated exposures on the methylome: in vitro studies
In parallel with the in vivo investigation of early life and concurrent exposures that may alter the epigenetic trajectory to asthma and/or the pathogenesis of the disease, ex vivo and in vitro studies centered on bronchial epithelial cells seek to elucidate mechanisms of disease by exploring epigenetic (methylome) responses to individual asthma-associated exposures [46]. Below we discuss three recent papers that exemplify this interesting emerging trend. In view of the ethical hurdles involved in obtaining sufficient amounts of viable airway cells from children, especially very young ones, these studies were performed using cells from adults, a choice that drastically limits their ability to illuminate mechanisms of asthma inception. However, as cell number requirements are scaled down and sampling techniques improve, the ability of this approach to effectively explore asthma development will also substantially improve.
Diesel exhaust
Air pollution, and more specifically diesel exhaust particles, are known to be associated with both risk for asthma and allergy [47], and changes in DNA methylation profiles [48–50]. Recent work assessed how in vivo exposure to diesel exhaust (through a dedicated apparatus) and allergen (through challenge within the lung) affects DNA methylation in human epithelial cells obtained by bronchoscopy. A single exposure to allergen, diesel exhaust or both was sufficient to induce significant methylation changes at seven CpG sites within 48 h However, significant changes in more than 500 CpGs were observed if the same individual was exposed to allergen and diesel exhaust 4 weeks apart [22]. In individuals who had been previously exposed to diesel exhaust, 70 CpGs showed changes ≥10% and were significantly associated with allergen exposure after correcting for multiple testing. On the other hand, in individuals who had been previously exposed to allergen, 548 CpGs showed changes ≥10% and were significantly associated with diesel exhaust exposure after correcting for multiple testing. These findings suggest that initial exposures can prime DNA methylation at a subset of CpG sites that are uniquely sensitive to subsequent exposures.
Interleukin-13
Because IL-13 is a key mediator of asthma and allergic diseases, another recent study examined IL-13 driven epigenetic responses in airway epithelial cells and also assessed whether IL-13 induced epigenetic changes are recapitulated in asthmatics [23]. Of 6522 CpG sites that were differentially methylated after IL-13 treatment, 1590 were located near genes that were also differentially expressed in response to IL-13. These IL-13 responsive CpG gene pairs were also enriched for correlations between methylation and expression levels, pointing to an epigenetic response of airway epithelial cells to IL-13. Interestingly, IL-13 responsive CpG-gene pairs were also significantly enriched for asthma genes, and this epigenetic link between IL-13 and asthma was confirmed in vivo. Indeed, the epigenetic response profile seen in IL-13 stimulated cultured airway epithelial cells was largely recapitulated in airway epithelial cells freshly isolated from asthmatics. Over 2000 of the original 6522 IL-13 responsive CpGs were replicated in vivo. It is noteworthy that according to network analysis, CpGs found to be differentially methylated in patients clustered into two comethylation modules, one that correlated with asthma severity and one that correlated with blood eosinophilia. These modules included networks centered around ERK1/2 (module 1) and IFN-γ and NF-κB (module 2). This result highlights pathways that may coordinate IL-13 mediated DNA methylation changes to promote asthma pathophysiology.
Human rhinovirus
Viral infections of the respiratory tract play are known to play a major role in asthma exacerbations [1], but the underlying mechanisms remain incompletely understood. Recent work compared and contrasted the impact of human rhinovirus (HRV) infection on nasal epithelial cells isolated from asthmatics and healthy controls. Over 300 CpG sites were found to be differentially methylated by disease or HRV infection status. SNORA12 was differentially methylated in response to in vitro HRV infection in both asthmatics and healthy controls, but the effects on DNA methylation went in opposite directions in the two groups [24]. Interestingly, SNORA12 methylation was associated with gene expression changes that were in turn associated with subsequent changes in lung function. These results demonstrate that the DNA methylation changes induced by HRV in nasal epithelial cells depend on asthma status. These differences might contribute to linking respiratory viral infections and asthma exacerbations.
Future perspective
In previous surveys of asthma epigenetics [9–10,14–15] we expressed some skepticism about the achievements of this field. Some of our concerns remain. For instance, the regions where disease-associated differential methylation was detected are spread throughout the genome, generally with no obvious functional links to asthma and/or allergy-related pathways and limited replication across studies. Even when statistically significant, disease-associated differences in DNA methylation are typically modest, on the order of a few percent, raising questions about their biological significance. In many studies, sample sizes are limited and environmental exposures are left uncharacterized. Common approaches to DNA methylation assessment do not distinguish between 5-methylcytosine and other cytosine modifications. Finally, gene or protein expression data can provide information critical to complement the interpretation of epigenetic studies and their biological relevance, but samples for such analyses are rarely available.
On the other hand, there are unmistakable, if still timid, signs suggesting that the conceptual foundations on which epigenetic studies of asthma rely may be maturing. The increasing focus on the prenatal origins of the disease and relevant environmental exposures is perhaps the most explicit and encouraging among those signs. Despite its current limitations, the rise of cellular in vitro models designed to dissect the epigenetic effects of asthma-associated exposures is another one. Needless to say, moving in these directions will not be easy – these studies need to be performed in carefully phenotyped mother/child birth cohorts, only a few of which already exist, and require ad hoc collected samples, only a few of which are currently available. Despite these hurdles, if these approaches provide results that fundamentally reshape our understanding of the asthma trajectory and its epigenetic components, it is reasonable to hope that appropriately designed population studies will be launched to fill the current gaps in our knowledge.
Executive summary.
The field of asthma epigenetics is currently driven by a fundamental question: is there an epigenetic trajectory to asthma, and if so, when does it begin and which factors can influence it?
In a proportion of children with childhood-onset asthma, a distinctive methylome is in place already at birth, particularly within innate immunoregulatory and proinflammatory pathways, and promotes a trajectory that may ultimately leads to clinical disease.
Prenatal exposures (e.g., smoking, asthma, stress, obesity and immune status in the mother) influence the epigenetic trajectory to asthma.
The rise of cellular in vitro models designed to dissect the epigenetic effects of asthma-associated exposures promises to advance our knowledge of the asthma trajectory.
Footnotes
Financial & competing interests disclosure
This work was supported by grant RC1HL100800 (to D Vercelli). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: •• of considerable interest
- 1.Martinez FD, Vercelli D. Asthma. Lancet. 2013;382(9901):1360–1372. doi: 10.1016/S0140-6736(13)61536-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.W.H.O. Bronchial asthma - Fact sheet n. 206. 2015. www.who.int/mediacentre/factsheets/fs206/en/
- 3.Stern DA, Morgan WJ, Halonen M, Wright AL, Martinez FD. Wheezing and bronchial hyper-responsiveness in early childhood as predictors of newly diagnosed asthma in early adulthood: a longitudinal birth-cohort study. Lancet. 2008;372(9643):1058–1064. doi: 10.1016/S0140-6736(08)61447-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Halonen M, Lohman IC, Stern DA, Ellis WL, Rothers J, Wright AL. Perinatal TNF-alpha production influenced by maternal pregnancy weight gain, predicts childhood asthma. Am. J. Respir. Crit. Care Med. 2013;188(1):35–41. doi: 10.1164/rccm.201207-1265OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rothers J, Halonen M, Stern DA, et al. Adaptive cytokine production in early life differentially predicts total IgE levels and asthma through age 5 years. J. Allergy Clin. Immunol. 2011;128(2):397–402. doi: 10.1016/j.jaci.2011.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meyers DA, Bleecker ER, Holloway JW, Holgate ST. Asthma genetics and personalised medicine. Lancet Respir. Med. 2014;2(5):405–415. doi: 10.1016/S2213-2600(14)70012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barua S, Junaid MA. Lifestyle, pregnancy and epigenetic effects. Epigenomics. 2015;7(1):85–102. doi: 10.2217/epi.14.71. [DOI] [PubMed] [Google Scholar]
- 8.Yang IV, Schwartz DA. Epigenetic mechanisms and the development of asthma. J. Allergy Clin. Immunol. 2012;130(6):1243–1255. doi: 10.1016/j.jaci.2012.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeVries A, Vercelli D. Epigenetics in allergic diseases. Curr. Opin. Pediatr. 2015;27(6):719–723. doi: 10.1097/MOP.0000000000000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeVries A, Vercelli D. Early predictors of asthma and allergy in children: the role of epigenetics. Curr. Opin. Allergy. Clin. Immunol. 2015;15(5):435–439. doi: 10.1097/ACI.0000000000000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bakulski KM, Feinberg JI, Andrews SV, et al. DNA methylation of cord blood cell types: applications for mixed cell birth studies. Epigenetics. 2016;11(5):354–362. doi: 10.1080/15592294.2016.1161875. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Presents a novel method for using DNA methylation data to infer the proportion of immune cell populations in cord blood mononuclear cell samples.
- 12.DeVries A, Wlasiuk G, Miller SJ, et al. Epigenome-wide analysis links SMAD3 methylation at birth to asthma in children of asthmatic mothers. J. Allergy Clin. Immunol. 2016 doi: 10.1016/j.jaci.2016.10.041. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Identifies DNA methylation signatures at birth that are associated with asthma during childhood, thereby pointing to a perinatal origin of the asthma trajectory.
- 13.De Vries A, Stern DA, Wright AL, Halonen M, Vercelli D. ATS International Conference. DC, USA: 19–24 May 2017. Neonatal DNA methylation profiles are associated with the maternal prenatal immune milieu selectively in children with non-asthmatic mothers. Presented at. [Google Scholar]
- 14.DeVries A, Vercelli D. The epigenetics of human asthma and allergy: promises to keep. Asian Pac. J. Allergy Immunol. 2013;31(3):183–189. [PubMed] [Google Scholar]
- 15.DeVries A, Vercelli D. Epigenetic mechanisms in asthma. Ann. Am. Thorac. Soc. 2016;13(Suppl. 1):S48–S50. doi: 10.1513/AnnalsATS.201507-420MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vercelli D. Does epigenetics play a role in human asthma? Allergol. Int. 2016;65(2):123–126. doi: 10.1016/j.alit.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Breton CV, Siegmund KD, Joubert BR, et al. Prenatal tobacco smoke exposure is associated with childhood DNA CpG methylation. PLoS ONE. 2014;9(6):e99716. doi: 10.1371/journal.pone.0099716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joubert BR, Felix JF, Yousefi P, et al. DNA methylation in newborns and maternal smoking in pregnancy: genome-wide consortium meta-analysis. Am. J. Hum. Genet. 2016;98(4):680–696. doi: 10.1016/j.ajhg.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Results from this meta-analysis suggest that prenatal exposure to smoking may influence the epigenetic trajectory to asthma and elicit DNA methylation responses that persist into childhood.
- 19.Rotroff DM, Joubert BR, Marvel SW, et al. Maternal smoking impacts key biological pathways in newborns through epigenetic modification in utero . BMC Genomics. 2016;17(1):976. doi: 10.1186/s12864-016-3310-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trump S, Bieg M, Gu Z, et al. Prenatal maternal stress and wheeze in children: novel insights into epigenetic regulation. Sci. Rep. 2016;6:28616. doi: 10.1038/srep28616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharp GC, Lawlor DA, Richmond RC, et al. Maternal pre-pregnancy BMI and gestational weight gain, offspring DNA methylation and later offspring adiposity: findings from the Avon Longitudinal Study of Parents and Children. Int. J. Epidemiol. 2015;44(4):1288–1304. doi: 10.1093/ije/dyv042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clifford RL, Jones MJ, Macisaac JL, et al. Inhalation of diesel exhaust and allergen alters human bronchial epithelium DNA methylation. J. Allergy Clin. Immunol. 2016;139(1):112–121. doi: 10.1016/j.jaci.2016.03.046. [DOI] [PubMed] [Google Scholar]
- 23.Nicodemus-Johnson J, Naughton KA, Sudi J, et al. Genome-wide methylation study identifies an IL-13-induced epigenetic signature in asthmatic airways. Am. J. Respir. Crit. Care Med. 2016;193(4):376–385. doi: 10.1164/rccm.201506-1243OC. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Examines whether in vitro IL-13 responsive DNA methylation profiles in airway epithelial cells were recapitulated in asthmatic patients.
- 24.McErlean P, Favoreto S, Jr, Costa FF, et al. Human rhinovirus infection causes different DNA methylation changes in nasal epithelial cells from healthy and asthmatic subjects. BMC Med. Genomics. 2014;7:37. doi: 10.1186/1755-8794-7-37. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• These investigators demonstrated that rhinovirus-induced changes in DNA methylation depend on asthma status, possibly linking respiratory viral infections and asthma.
- 25.Oddy WH, Halonen M, Martinez FD, et al. TGF-β in human milk is associated with wheeze in infancy. J. Allergy Clin. Immunol. 2003;112(4):723–728. doi: 10.1016/s0091-6749(03)01941-9. [DOI] [PubMed] [Google Scholar]
- 26.Moffatt MF, Gut IG, Demenais F, et al. A large-scale, consortium-based genomewide association study of asthma. N. Engl. J. Med. 2010;363(13):1211–1221. doi: 10.1056/NEJMoa0906312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferreira MA, Matheson MC, Tang CS, et al. Genome-wide association analysis identifies 11 risk variants associated with the asthma with hay fever phenotype. J. Allergy Clin. Immunol. 2014;133(6):1564–1571. doi: 10.1016/j.jaci.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hinds DA, Mcmahon G, Kiefer AK, et al. A genome-wide association meta-analysis of self-reported allergy identifies shared and allergy-specific susceptibility loci. Nat. Genet. 2013;45(8):907–911. doi: 10.1038/ng.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raedler D, Ballenberger N, Klucker E, et al. Identification of novel immune phenotypes for allergic and nonallergic childhood asthma. J. Allergy Clin. Immunol. 2015;135(1):81–91. doi: 10.1016/j.jaci.2014.07.046. [DOI] [PubMed] [Google Scholar]
- 30.Simpson JL, Phipps S, Baines KJ, Oreo KM, Gunawardhana L, Gibson PG. Elevated expression of the NLRP3 inflammasome in neutrophilic asthma. Eur. Respir. J. 2014;43(4):1067–1076. doi: 10.1183/09031936.00105013. [DOI] [PubMed] [Google Scholar]
- 31.Hastie AT, Moore WC, Meyers DA, et al. Analyses of asthma severity phenotypes and inflammatory proteins in subjects stratified by sputum granulocytes. J. Allergy Clin. Immunol. 2010;125(5):1028–1036. doi: 10.1016/j.jaci.2010.02.008. e1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lim RH, Kobzik L, Dahl M. Risk for asthma in offspring of asthmatic mothers versus fathers: a meta-analysis. PLoS ONE. 2010;5(4):e10134. doi: 10.1371/journal.pone.0010134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paaso EM, Jaakkola MS, Rantala AK, Hugg TT, Jaakkola JJ. Allergic diseases and asthma in the family predict the persistence and onset-age of asthma: a prospective cohort study. Respir. Res. 2014;15:152. doi: 10.1186/s12931-014-0152-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat. Med. 2012;18(5):716–725. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
- 35.Basu R, Whitley SK, Bhaumik S, et al. IL-1 signaling modulates activation of STAT transcription factors to antagonize retinoic acid signaling and control the Th17 cell-iTreg cell balance. Nat. Immunol. 2015;16(3):286–295. doi: 10.1038/ni.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang G, Walker SO, Hong X, Bartell TR, Wang X. Epigenetics and early life origins of chronic noncommunicable diseases. J. Adolesc. Health. 2013;52(2 Suppl. 2):S14–S21. doi: 10.1016/j.jadohealth.2012.04.019. [DOI] [PubMed] [Google Scholar]
- 37.Mcdade TW. Early environments and the ecology of inflammation. Proc. Natl Acad. Sci. USA. 2012;109(Suppl. 2):17281–17288. doi: 10.1073/pnas.1202244109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gunawardhana LP, Baines KJ, Mattes J, Murphy VE, Simpson JL, Gibson PG. Differential DNA methylation profiles of infants exposed to maternal asthma during pregnancy. Pediatr. Pulmonol. 2014;49(9):852–862. doi: 10.1002/ppul.22930. [DOI] [PubMed] [Google Scholar]
- 39.Chiu YH, Coull BA, Cohen S, Wooley A, Wright RJ. Prenatal and postnatal maternal stress and wheeze in urban children: effect of maternal sensitization. Am. J. Respir. Crit. Care Med. 2012;186(2):147–154. doi: 10.1164/rccm.201201-0162OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Marco R, Pesce G, Girardi P, et al. Foetal exposure to maternal stressful events increases the risk of having asthma and atopic diseases in childhood. Pediatr. Allergy Immunol. 2012;23(8):724–729. doi: 10.1111/j.1399-3038.2012.01346.x. [DOI] [PubMed] [Google Scholar]
- 41.Godfrey KM, Reynolds RM, Prescott SL, et al. Influence of maternal obesity on the long-term health of offspring. Lancet Diabetes Endocrinol. 2017;5(1):53–64. doi: 10.1016/S2213-8587(16)30107-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forno E, Young OM, Kumar R, Simhan H, Celedon JC. Maternal obesity in pregnancy, gestational weight gain, and risk of childhood asthma. Pediatrics. 2014;134(2):e535–e546. doi: 10.1542/peds.2014-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rothers J, Stern DA, Lohman IC, et al. Prenatal maternal cytokine production relates to asthma prevalence differentially in children of asthmatic vs non-asthmatic mothers. Submitted for publication.
- 44.Coombes JL, Siddiqui KR, Arancibia-Carcamo CV, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J. Exp. Med. 2007;204(8):1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murray IA, Nichols RG, Zhang L, Patterson AD, Perdew GH. Expression of the aryl hydrocarbon receptor contributes to the establishment of intestinal microbial community structure in mice. Sci. Rep. 2016;6:33969. doi: 10.1038/srep33969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ober C. Asthma genetics in the post-GWAS era. Ann. Am. Thorac. Soc. 2016;13(Suppl. 1):S85–S90. doi: 10.1513/AnnalsATS.201507-459MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peden DB, Bush RK. Advances in environmental and occupational disorders in 2013. J. Allergy Clin. Immunol. 2014;133(5):1265–1269. doi: 10.1016/j.jaci.2014.02.027. [DOI] [PubMed] [Google Scholar]
- 48.Tang WY, Levin L, Talaska G, et al. Maternal exposure to polycyclic aromatic hydrocarbons and 5′-CpG methylation of interferon-gamma in cord white blood cells. Environ. Health Perspect. 2012;120(8):1195–1200. doi: 10.1289/ehp.1103744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Breton CV, Salam MT, Wang X, Byun HM, Siegmund KD, Gilliland FD. Particulate matter, DNA methylation in nitric oxide synthase, and childhood respiratory disease. Environ. Health Perspect. 2012;120(9):1320–1326. doi: 10.1289/ehp.1104439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rossnerova A, Tulupova E, Tabashidze N, et al. Factors affecting the 27K DNA methylation pattern in asthmatic and healthy children from locations with various environments. Mutat. Res. 2013;741–742:18–26. doi: 10.1016/j.mrfmmm.2013.02.003. [DOI] [PubMed] [Google Scholar]