Abstract
Trimethyllysine (Kme3) reader proteins are targets for inhibition due to their role in mediating gene expression. Although all such reader proteins bind Kme3 in an aromatic cage, the driving force for binding may differ; some readers exhibit evidence for cation–π interactions whereas others do not. We report a general unnatural amino acid mutagenesis approach to quantify the contribution of individual tyrosines to cation binding using the HP1 chromodomain as a model system. We demonstrate that two tyrosines (Y24 and Y48) bind to a Kme3-histone tail peptide via cation–π interactions, but linear free energy trends suggest they do not contribute equally to binding. X-ray structures and computational analysis suggest that the distance and degree of contact between Tyr residues and Kme3 plays an important role in tuning cation–π-mediated Kme3 recognition. Although cation–π interactions have been studied in a number of proteins, this work is the first to utilize direct binding assays, X-ray crystallography, and modeling, to pinpoint factors that influence the magnitude of the individual cation–π interactions.
Lysine methylation plays a critical role in the regulation of gene expression by recruiting proteins involved in chromatin remodeling.1,2 Methyllysine reader proteins discriminate both the degree (Kmen, n = 0–3) and position of methylation on histone tails, triggering downstream processes that control gene function. Dysregulation of Kmen formation and recognition is associated with varied disease states including cancer.3–5Accordingly, Kmen reader proteins are emerging as important therapeutic targets.6
Reader proteins that recognize trimethyllysine (Kme3) contain an “aromatic cage” consisting of 2–4 aromatic residues that bind the cationic ammonium.2 Although only a few studies have probed the mechanism of Kme3 recognition, cation–π interactions have been shown to play a key role in some Kme3 readers.7,8 Due to the importance of Kme3 recognition in regulating gene expression, as well as efforts to develop inhibitors of this interaction,6,9 there is a need for methods that provide detailed insight into the mechanisms by which reader proteins bind and discriminate Kmen adducts. We report a general, unnatural amino acid (UAA)-based approach to examine the contribution of individual Tyr residues toward the recognition of Kme3-containing histone tail peptides. Coupled with computational studies, we show that two Tyr residues in a model reader protein contribute differentially to cation–π-mediated binding of Kme3.
Initial investigations into cation–π interactions in Kme3 reader proteins were performed using the chromodomain of heterochromatin protein 1 (HP1), a model reader from Drosophila melanogaster that binds to K9me3 on the tail of histone H3, leading to epigenetic silencing.11 The binding pocket is composed of two Tyr residues (Y24 and Y48, Figure 1) and one Trp residue (W45).12 It has been shown that mutation of any of these aromatic residues to Ala significantly reduces binding to Kme3.12 Furthermore, the importance of the cation has been demonstrated by a reduced affinity of >2 kcal/mol for a histone peptide containing a neutral Kme3 isostere.7 As HP1 has been extensively characterized, it provides an excellent model system for detailed mechanistic studies of cation–π interactions in methyllysine reader proteins.
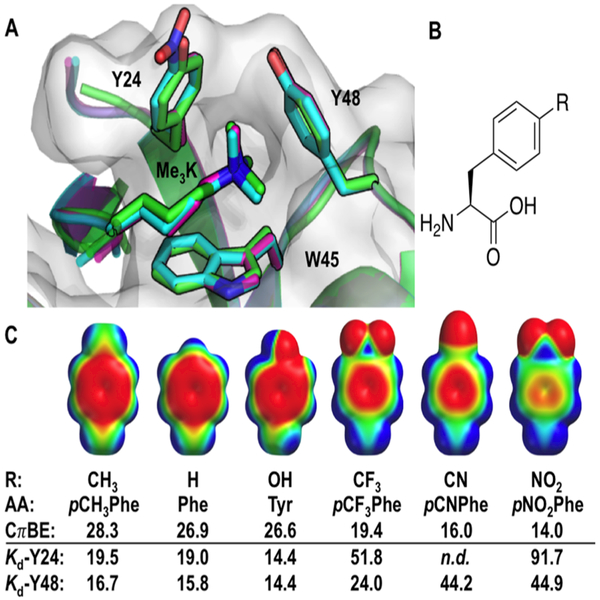
Figure 1.
(A) Aromatic residues (Y24, Y48, and W45) in the HP1 Kme3-binding pocket complexed with Kme3-containing peptide. Wild type HP1 (green, PDB: 1KNE)11 has been overlaid with Y24pNO2Phe (cyan, PDB: 6AT0) and Y24F (purple, PDB: 6ASZ). (B) General structure of p-substituted phenylalanines. (C) The effect of R-groups on electrostatic surface potential (colored maps) and calculated cation–π binding energy (CπBE, kcal/mol) for substituted benzenes and Na+.10 Measured Kd values (μM) for HP1-UAA variants at positions 24 and 48, against a H3K9me3 peptide, are also provided. AA = amino acid abbreviations used in the text. n.d. = not determined.
Pioneering studies by Dougherty and co-workers on cationic-ligand gated ion channels elucidated the contribution of “aromatic cage” residues to cation–π interactions via replacement with fluorinated aromatic amino acid isosteres. Residues involved in cation–π interactions showed attenuated ion channel activity with increasing fluorination and a linear free energy relationship (LFER) between the log of activity and calculated cation–π strength.13 This approach, however, makes use of stoichiometric UAA-suppressor tRNAs, which restrict UAA-protein yields.14,15 Consequently, such investigations have been limited to systems that can be studied via single channel or single cell activity assays.13–15 Using this method, direct measurements of protein–ligand interactions (e.g., Ka and structural investigations have not been shown.
Genetic encoding via orthogonal tRNA/tRNA synthetase pairs permits the expression and purification of UAA-proteins in high yield, a feature that we envisioned would enable direct measurements of reader protein-peptide binding interactions and structure determination efforts. For example, Roberts and co-workers have confirmed the presence of multiple tyrosine-mediated cation–π interactions between phosphatidylcholine and amphitrophic phosphatidylinositol-specific phospholipase C (PI-PLC) via the genetic incorporation of pentafluorophyenyalanine or 3,5-difluorotyrosine.16 Moreover, this work permitted structural characterization of an intraprotein cationπ interaction between a Tyr residue and a positively charged histidine. Nevertheless, isosteric fluorinated amino acids are difficult targets for genetic incorporation due to their similar size and shape with respect to canonical amino acids. Accordingly, only a limited number of highly fluorinated isosteres have been shown to be selectively incorporated into proteins, precluding more detailed LFER investigations.17–20 As an alternative approach, a large number of p-substituted Phe derivatives bearing electron-withdrawing or -donating groups have been incorporated into proteins using this methodology (Figure 1B,C).21–25 In light of the recognized role of substituent effects in tuning cation–π interactions, we hypothesized that UAA-Phes might provide a convenient alternative for measuring cation–π interactions via LFER analysis.
We chose to replace each tyrosine residue in the HP1 aromatic cage (Tyr24 and Tyr48) with p-substituted Phe derivatives because the p-hydroxyl projects into solvent and substitutions at this position were least likely to interfere with the binding of Kme3 peptides (Figure 1). Accordingly, we chose a series of six amino acids possessing substituents that span a large range of electrostatic potential (R = CH3, H, OH, CF3, CN, NO2, Figure 1C and Table S1), for incorporation into HP1. Although tRNA/tRNA synthetase pairs have been evolved for each UAA,21,23,26–28 we exploited a single tRNA synthetase (pCNPheRS) that was evolved for the recognition of pCNPhe (Figure 1C), but possesses a particularly broad substrate scope.26
To incorporate UAAs into HP1, mutants containing a TAG stop codon in place of tyrosine codons at positions 24 or 48 were cloned into a pET expression system and paired with a single accessory plasmid (pUltra-pCNPheRS) that contains the orthogonal tRNA/tRNA synthetase pair.30 Expressions in the absence of amino acid yielded little to no HP1; however, addition of 5–20 mM pCNPhe, pNO2Phe, pCF3Phe, or pCH3Phe led to high levels of UAA containing protein (Figure S1). Although pCNPheRS has been shown to incorporate CN- and NO2-substituted Phe,26 to our knowledge this is the first demonstration of promiscuous pCNPheRS-mediated incorporation of pCF3Phe, and pCH3Phe. UAA-incorporation was confirmed by ESI-LCMS, and canonical amino acid contamination was not detected (Tables S2–S5, Figures S2–S15). Circular dichroism showed UAA-mutations did not affect folding of HP1 (Figure S16). We note that Y24pCNPhe was poorly behaved at the high concentrations necessary for ITC, and thus we were unable to obtain reliable binding data for this mutant.
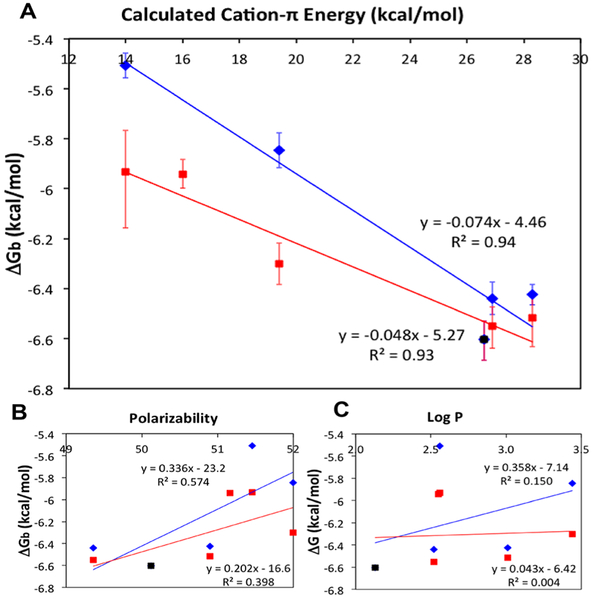
The binding constants between each HP1-UAA variant and an H3K9me3 peptide (amino acids 1–15) were measured using ITC (Table S16, Figure S17). When the free energy of binding (ΔGb) is plotted against calculated cation–π binding energies based on gas phase interactions of substituted benzene with Na+,10 a LFER is observed (Figure 2A), revealing the presence of a tunable cation–π interaction at each position. High correlations were also observed when plotted against other methods that have been used to calculate cation–π energies (Figure S18). Intriguingly, comparison of the relationship between ΔGb and calculated cation–π binding energies (Figure 2A) suggests a difference in magnitude of the effect at the two Tyr positions in which Tyr24 participates in a stronger cation–π interaction with Kme3 than Tyr48.
Figure 2.
(A) Relationship between ΔGb of H3K9me3/HP1-UAA variant interactions and calculated gas-phase cation–π binding energies between C6H5R and Na+.10 Y24 mutants (blue): slope = −0.074, R2 = 0.94; Y48 mutants (red): slope = −0.048, R2 = 0.93. (B) Relationship between ΔGb and calculated polarizability.29 (C) Relationship between ΔGb and hydrophobicity parameter (log P). In each panel, the two data sets share the wild type (WT) point shown in black.
To support our observation that changes in binding affinity result from modification of the cation—π interaction, the data were also plotted against polarizability29 (Figure 2B) and log P (Figure 2C, see Supporting information) to observe contributions of polarizability (van der Waals (VDW) interactions) and hydrophobicity to binding, respectively. A weak negative correlation between binding affinity and polarizability was observed. Increased polarizability of a substituent leads to a greater ability to participate in VDW interactions; if VDW interactions were contributing significantly, binding affinities would be expected to increase as this parameter increases, contrary to the relationship observed here. Log P is based on octanol/water partition coefficients and is used as a measure of hydrophobicity. There is no significant correlation between ΔGb and log P. These results indicate that VDW interactions and the hydrophobic effect are not strong drivers of the observed substituent effects.
To ensure that changes in binding were not due to structural perturbations induced by the UAAs, X-ray crystal structures of two variants were determined. Because pNO2Phe and Phe have the largest and smallest p-substituents, and represent opposite extremes of calculated cation–π binding energies, these two structures should provide evidence as to whether changes in ligand binding result from unexpected conformational changes. Because mutations to Tyr24 show a more pronounced effect on binding, the Tyr24Phe and Tyr24pNO2Phe variants were crystallized, and their structures were determined to 1.52 and 1.28 Å resolution, respectively (Table S7 and Figure S19). Changes in binding affinity do not appear to be the result of changes in protein structure as wild type, Tyr24Phe, and Tyr24pNO2Phe crystal structures overlay with an RMSD of less than 0.3 Å (Figure 1A, Figures S20 and S21), and the distances between Kme3 atoms and each phenyl ring do not significantly change (Figure S22).
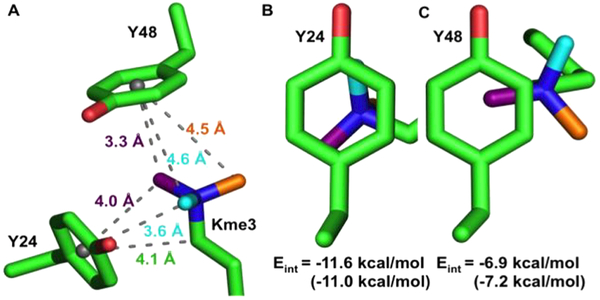
Inspection of the X-ray structures provides a qualitative assessment of the differences in measured binding affinity upon UAA mutagenesis at Tyr24 and Tyr48 (Figure 3). Two methyl groups and the methylene of Kme3 make van der Waals contact with Tyr24 (<4.5 Å, Figure 3A,B), whereas only a single methyl group makes close contact with Tyr48 (Figure 3C). Computational studies by Dougherty have predicted that a 3-point contact of NMe4 with benzene is about 1.67-fold stronger when compared to a single methyl group in gas-phase calculations.30 These calculations are consistent with our data in Figure 2, in which the slopes for Y24 and Y48 differ by a factor of 1.6.
Figure 3.
(A) Measured distances between the center of Y24 and Y48 with respect to atoms on Kme3 (PDB: 1KNE). Distances for Y24pNO2Phe and Y24F are provided in Figure S22. (B and C) Contact surface of Kme3 with Y24 (B) and Y48 (C) viewed normal to the plane of the ring. Kme3 atoms are colored as in panel A. Interaction energies (Eint) for Kme3 and each tyrosine are shown and were calculated by M06/6-31G(d,p) or (M06/6-311+G(d,p)).
To further support this observation, interaction energies (Eint) between Kme3 and Tyr24 or Tyr48 were calculated using geometries obtained from the wild type Kme3-bound HP1 crystal structure (1KNE). Eint values were calculated at the M06/6-31G(d,p) level of theory (see SI), which has previously been shown to predict cation–π strength in good agreement with experimental gas-phase measurements for tetramethylammonium interactions with benzene.30 Calculated Eint values predict a stronger interaction with Kme3 for Tyr24 (Eint = −11.6 kcal/mol) when compared to Tyr48 (Eint = −6.9 kcal/mol; Figure 3B,C). Furthermore, calculations at the M06 level performed using the larger 6-311+G(d,p) basis set provided similar results (Tyr24-Eint = −11.0 kcal/mol; Tyr48-Eint = −7.2 kcal/mol). Our experimental results are consistent with both levels of theory, which predict that the magnitude of the substituent effect differs by a factor of 1.5 and 1.7, respectively.
In summary, we have developed a method for detailed mechanistic and structural investigations of cation–π interactions in proteins, which we have applied here to a methyllysine reader protein. This work provides a rare direct measurement of the electronic tunability of discrete cation–π binding interactions in aqueous solutions.31,32 Interestingly, though our data demonstrate that both Tyr24 and Tyr48 of HP1 contribute to Kme3 binding via a cation–π interaction, our combined experimental and computational results indicate that these positions do not participate to the same degree, with Y24 exhibiting a greater influence on Kme3 binding. ITC binding analyses and X-ray crystal structures provide the first experimental data demonstrating that the distance and degree of contact influence the magnitude of the cation–π interaction, as had been predicted computationally.30 Few examples exist of different magnitudes of cation–π interactions within the same binding pocket, and these studies lack structural insight into the molecular basis of such differences.
As computational modeling has become a tool for more efficient drug design, this study also highlights the importance of accurately modeling cation–π interactions in therapeutic targets. The combined binding and structural information from this work provides an experimental benchmark for validating computational methods. Furthermore, as many methyllysine reader proteins share an aromatic cage motif in their binding pocket,2,33 this work suggests that differences in degree of contacts among reader proteins may be exploited to enhance selective inhibition. By understanding how a protein recognizes its natural substrate, we provide a new framework for the study and design of probes with the necessary affinity and selectivity for therapeutic use.
Supplementary Material
ACKNOWLEDGMENTS
A.L.K. was supported by a fellowship from the NSF (DGE-1144081). This work was supported in part by an NSF Career Award (CHE-1552718) E.M.B., an NSF grant (CHE-1608333) to M.L.W., and an NIH grant (GM118499) to E.M.B. and M.L.W.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI:10.1021/jacs.7b09223.
Experimental procedures for UAAM, binding measurements, and X-ray crystallography (PDF)
Notes
The authors declare no competing financial interest.
REFERENCES
- (1).Bannister AJ; Kouzarides T Cell Res. 2011, 21, 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Taverna SD; Li H; Ruthenburg AJ; Allis CD; Patel DJ Nat. Struct. Mol. Biol 2007, 14, 1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Khuong-Quang DA; Buczkowicz P; Rakopoulos P; Liu XY; Fontebasso AM; Bouffet E; Bartels U; Albrecht S; Schwartzentruber J; Letourneau L; Bourgey M; Bourque G; Montpetit A; Bourret G; Lepage P; Fleming A; Lichter P; Kool M; von Deimling A; Sturm D; Korshunov A; Faury D; Jones DT; Majewski J; Pfister SM; Jabado N; Hawkins C Acta Neuropathol. 2012, 124, 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).McGrath J; Trojer P Pharmacol. Ther 2015, 150, 1. [DOI] [PubMed] [Google Scholar]
- (5).Schwartzentruber J; Korshunov A; Liu XY; Jones DT; Pfaff E; Jacob K; Sturm D; Fontebasso AM; Quang DA; Tonjes M; Hovestadt V; Albrecht S; Kool M; Nantel A; Konermann C; Lindroth A; Jager N; Rausch T; Ryzhova M; Korbel JO; Hielscher T; Hauser P; Garami M; Klekner A; Bognar L; Ebinger M; Schuhmann MU; Scheurlen W; Pekrun A; Fruhwald MC; Roggendorf W; Kramm C; Durken M; Atkinson J; Lepage P; Montpetit A; Zakrzewska M; Zakrzewski K; Liberski PP; Dong Z; Siegel P; Kulozik AE; Zapatka M; Guha A; Malkin D; Felsberg J; Reifenberger G; von Deimling A; Ichimura K; Collins VP; Witt H; Milde T; Witt O; Zhang C; Castelo-Branco P; Lichter P; Faury D; Tabori U; Plass C; Majewski J; Pfister SM; Jabado N Nature 2012, 482, 226. [DOI] [PubMed] [Google Scholar]
- (6).Liu Y; Liu K; Qin S; Xu C; Min J Pharmacol. Ther 2014, 143, 275. [DOI] [PubMed] [Google Scholar]
- (7).Hughes RM; Wiggins KR; Khorasanizadeh S; Waters ML Proc. Natl. Acad. Sci. U. S. A 2007, 104, 11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Kamps JJ; Huang J; Poater J; Xu C; Pieters BJ; Dong A; Min J; Sherman W; Beuming T; Matthias Bickelhaupt F; Li H; Mecinovic J Nat. Commun 2015, 6, 8911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).James LI; Frye SV Curr. Opin. Chem. Biol 2016, 33, 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Wheeler SE; Houk KN J. Am. Chem. Soc 2009, 131, 3126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Jacobs SA; Taverna SD; Zhang Y; Briggs SD; Li J; Eissenberg JC; Allis CD; Khorasanizadeh S EMBO J. 2001, 20, 5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Jacobs SA; Khorasanizadeh S Science 2002, 295, 2080. [DOI] [PubMed] [Google Scholar]
- (13).Zhong W; Gallivan JP; Zhang Y; Li L; Lester HA; Dougherty DA Proc. Natl. Acad. Sci. U. S. A 1998, 95, 12088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Saks ME; Sampson JR; Nowak MW; Kearney PC; Du F; Abelson JN; Lester HA; Dougherty DA J. Biol. Chem 1996, 271, 23169. [DOI] [PubMed] [Google Scholar]
- (15).Xiu X; Puskar NL; Shanata JA; Lester HA; Dougherty DA Nature 2009, 458, 534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).He T; Gershenson A; Eyles SJ; Lee YJ; Liu WR; Wang J; Gao J; Roberts MF J. Biol. Chem 2015, 290, 19334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Tang Y; Ghirlanda G; Vaidehi N; Kua J; Mainz DT; Goddard IW; DeGrado WF; Tirrell DA Biochemistry 2001, 40, 2790. [DOI] [PubMed] [Google Scholar]
- (18).Bacher JM; Ellington AD J. Bacteriol 2001, 183, 5414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Fluorinated tyrosines (Fn-Ys) have been incorporated by exploiting the decreased pKa of the Fn-Y phenol. These amino acids are anionic at physiological pH however, which may also complicate cation–π investigations. Minnihan EC; Young DD; Schultz PG; Stubbe J. J. Am. Chem. Soc 2011, 133, 15942.21913683 [Google Scholar]
- (20).Fluorinated Phe isosteres have been incorporated using orthogonal tRNA synthetases, but this technology is restricted to Phe variants with high degrees of fluorination. Contamination with wild type Phe is also observed in these proteins. This system was used to measure Kme3 binding in a methyllysine reader protein; however, weak binding could not be determined with accuracy. Lee Y-J; Schmidt MJ; Tharp JM; Weber A; Koenig AL; Zheng H; Gao J; Waters ML; Summerer D; Liu WR Chem. Commun 2016, 52, 12606. [Google Scholar]
- (21).Wang L; Brock A; Herberich B; Schultz PG Science 2001, 292, 498. [DOI] [PubMed] [Google Scholar]
- (22).Brustad E; Bushey ML; Lee JW; Groff D; Liu W; Schultz PG Angew. Chem., Int. Ed 2008, 47, 8220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Grunewald J; Tsao ML; Perera R; Dong L; Niessen F; Wen BG; Kubitz DM; Smider VV; Ruf W; Nasoff M; Lerner RA; Schultz PG Proc. Natl. Acad. Sci. U. S. A 2008, 105, 11276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Wan W; Tharp JM; Liu WR Biochim. Biophys. Acta, Proteins Proteomics 2014, 1844, 1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Liu CC; Schultz PG Annu. Rev. Biochem 2010, 79, 413. [DOI] [PubMed] [Google Scholar]
- (26).Young DD; Young TS; Jahnz M; Ahmad I; Spraggon G; Schultz PG Biochemistry 2011, 50, 1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Miyake-Stoner SJ; Refakis CA; Hammill JT; Lusic H; Hazen JL; Deiters A; Mehl RA Biochemistry 2010, 49, 1667. [DOI] [PubMed] [Google Scholar]
- (28).Schultz KC; Supekova L; Ryu Y; Xie J; Perera R; Schultz PG J. Am. Chem. Soc 2006, 128, 13984. [DOI] [PubMed] [Google Scholar]
- (29).Hwang J; Dial BE; Li P; Kozik ME; Smith MD; Shimizu KD Chemical Science 2015, 6, 4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Davis MR; Dougherty DA Phys. Chem. Chem. Phys 2015, 17, 29262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Lee Y-J; Schmidt MJ; Tharp JM; Weber A; Koenig AL; Zheng H; Gao J; Waters ML; Summerer D; Liu WR Chem. Commun 2016, 52, 12606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Mahadevi AS; Sastry GN Chem. Rev 2013, 113, 2100. [DOI] [PubMed] [Google Scholar]
- (33).Adams-Cioaba MA; Min J Biochem. Cell Biol 2009, 87, 93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.