Abstract
ZIC2 mutation is known to cause holoprosencephaly (HPE). A subset of ZIC2 HPE probands harbour cardiovascular and visceral anomalies suggestive of laterality defects. 3D-imaging of novel mouse Zic2 mutants uncovers, in addition to HPE, laterality defects in lungs, heart, vasculature and viscera. A strong bias towards right isomerism indicates a failure to establish left identity in the lateral plate mesoderm (LPM), a phenotype that cannot be explained simply by the defective ciliogenesis previously noted in Zic2 mutants. Gene expression analysis showed that the left-determining NODAL-dependent signalling cascade fails to be activated in the LPM, and that the expression of Nodal at the node, which normally triggers this event, is itself defective in these embryos. Analysis of ChiP-seq data, in vitro transcriptional assays and mutagenesis reveals a requirement for a low-affinity ZIC2 binding site for the activation of the Nodal enhancer HBE, which is normally active in node precursor cells. These data show that ZIC2 is required for correct Nodal expression at the node and suggest a model in which ZIC2 acts at different levels to establish LR asymmetry, promoting both the production of the signal that induces left side identity and the morphogenesis of the cilia that bias its distribution.
Introduction
ZIC2 mutation is one of four principal genetic risk factors for holoprosencephaly (HPE), the most common structural brain disease1–3. A similar HPE phenotype is seen in mice homozygous either for the ethylnitrosourea (ENU) mutant allele Zic2ku 4 or a hypomorphic allele, Zic2tm1Jaru 5. The midline organiser known as the node, which forms at the leading edge of the primitive streak at the end of gastrulation, plays a central role in the generation of mesendodermal structures required to pattern the neural tube, such as the pre-chordal plate (PCP) and the notochord. Zic2 is transiently expressed at the node from E7.0 to E7.56. Development of the PCP is impaired in the Zic2ku mutant at the late-streak stage4. Nodal, a member of the TGFβ family which signals via SMAD2, 3, is expressed in the epiblast giving rise to the primitive streak and in the primitive streak itself. It is required for primitive streak formation as well as for that of the node and of node-derived mesendoderm7. An epistatic genetic interaction exists between Nodal and Zic2 in forebrain development8.
Laterality of the heart and viscera is also determined by the NODAL pathway. Bilateral symmetry is broken through generation of a leftward fluid flow by cilia within the node9,10. Nodal is initially expressed bilaterally at the node within perinodal cells (at E7.5 in mouse), but in response to the nodal flow, this expression is reinforced in left perinodal cells, thus becoming asymmetric11,12. NODAL produced in perinodal cells is required to trigger its own expression in the left lateral plate mesoderm (LPM)13, where it induces the expression of Lefty214, a NODAL antagonist, and Pitx2c15,16. Pitx2c expression in the left LPM determines left-sided identity in mesoderm derivatives. In the absence of Pitx2c expression the default pattern is that of two morphologically right sides, a condition known as right-isomerism. Ectopic activation of Pitx2c in the right lateral mesoderm only results in situs inversus whereas bilateral expression of Pitx2c results in two morphologically left sides, known as left-isomerism. These phenotypes can be distinguished by examination of lung lobulation, atrial morphology and other characters17. The establishment of Left-Right (LR) polarity is essential for key aspects of cardiovascular, thoracic and abdominal development. It determines the lateralised identity of atria and lungs, influences looping morphogenesis of the linear heart tube and the gut, directs asymmetric remodelling of the vascular system and determines the positioning of visceral organs such as stomach and pancreas17–21. Mutations mapping to NODAL pathway genes are associated with heterotaxy22,23, a condition characterized by discordant LR arrangement of internal organs which accounts for approximately 3% of all congenital heart disease22.
Cardiac malformations have been noted amongst a number of extra-craniofacial anomalies reported in HPE patients with ZIC2 mutation, occurring in 9–14% of probands2,3, but have not previously been described in detail. We hypothesise these may result from an underlying laterality defect. The Zic2ku mutant shows randomised direction of heart tube looping during cardiac morphogenesis24, supporting this hypothesis. These embryos exhibit mid-gestation lethality, preventing a more detailed study of laterality defects24. Nodal cilia are shorter and morphologically abnormal in the Zic2ku mutant suggesting that Zic2 may function during cilia morphogenesis24. Expression of Nodal at the node and of downstream genes in the LPM is reduced in the Zic2ku mutant24, but ectopic right-sided or bilateral expression, which is present in iv embryos with nodal cilia defects12,25, is not observed.
Zic2 is also expressed earlier in development and is present in both embryonic (ESC) and epiblast (EpiSC) stem cells. ZIC2 is bound to the Nodal locus in both cell types26,27. EpiSCs have been shown to resemble cells from the anterior primitive streak28, from which the node is derived. ZIC2 appears to play a central role in the transition from the naive pluripotent state of ESCs to the primed pluripotent state of EpiSCs26,29 and has been proposed to act as a “pioneer factor” that functions to seed enhancers, recruiting additional transcription factors in order to prime loci for transcriptional activation at a later stage26. The MDB3-NURD chromatin remodelling complex is associated with ZIC2 at a subset of binding sites in ESCs27, this complex is known to play a role in fine-tuning dynamic expression of bivalent enhancers during development30, consistent with a role for ZIC2 in such a priming process.
Here, we examine clinical data and show that the cardiovascular, pulmonary and visceral phenotypes of ZIC2 HPE patients are consistent with an underlying laterality defect. We identify a novel Zic2 ENU mouse mutant in a screen for cardiovascular laterality phenotypes. We use 3D imaging to characterise in detail the phenotype of this mutant and that of a series of Zic2 alleles generated by TALEN gene editing31, revealing a complex set of cardiovascular, thoracic and abdominal malformations, in addition to the previously-described holoprosencephaly and heart tube looping phenotypes. Analysis of the phenotype reveals a strong bias towards right isomerism, indicative of defective left-sided identity specification during development. This is supported by gene expression data revealing weak or absent Nodal node expression and loss of downstream gene expression in the LPM. We use in vitro assays to demonstrate that ZIC2 can activate transcription from Nodal enhancer reporters. Our data suggest that ZIC2 acts upstream of Nodal expression at the node, possibly to prime the gene locus for the subsequent activation of its expression there. Together with a previous study that identified a role for ZIC2 in ciliogenesis, our results suggest a model in which ZIC2 acts at multiple levels during the establishment of laterality, upstream of genes and events that are critical for the process to take place.
Results
Cardiovascular and pulmonary malformations in human ZIC2 HPE cases suggest a laterality defect
Extra-craniofacial defects, including cardiovascular, visceral and urino-genital anomalies have previously been noted in patients with holoprosencephaly (HPE) carrying ZIC2 mutations2,3, but the details of these malformations have not previously been documented. We examined clinical reports derived from a previously published European series consisting of 645 HPE probands3, including both liveborns and medically terminated pregnancies. A total of 67 probands in this series have ZIC2 mutation, of which 8 (12%) exhibit cardiovascular or visceral anomalies suggesting a putative laterality defect (Table 1). The affected probands have mutations including three single amino acid substitutions (p.(His156Tyr); p.(Gln36Pro); p.(Phe314Cys); all affect highly conserved residues and are not found in 61,000 control exomes from the ExAC Project), alanine tract deletions and duplications, and larger chromosomal aberrations (Table 1; Fig. S1a,b).
Table 1.
Association of cardiovascular and situs defects with holoprosencephaly in human cases with ZIC2 mutation.
| Proband | Gender | Survival | Mutation | Isolated/Syndromic | Type of HPEc | Heart malformations | Other visceral anomalies | |
|---|---|---|---|---|---|---|---|---|
| DNAa | Protein | |||||||
| 1 | F | Fœtus MTPb | 107 A > C | p.(Gln36Pro) | I | Semilobar | N/A | Renal hypoplasia, Pulmonary hypoplasia, Adrenal hypoplasia, Spleen hypoplasia |
| 2 | F | Fœtus MTP | 466 C > T | p.(His156Tyr) | S | Semilobar | Hypoplastic ascendant aorta, VSD | Single umbilical artery |
| 3 | F | Fœtus MTP | 941 T > G | p.(Phe314Cys) | S | Pseudo-anencephaly | Hypoplastic ascendant aorta, Aortic stenosis, VSD | Diaphragmatic hernia, Unilateral renal agenesis, Unicorn uterus |
| 4 | F | Fœtus MTP | 1377–1406 del | Alanine tract deletion (aa456–465) | S | Lobar | N/A | Pulmonary hypoplasia Abnormal lung lobulation, Common mesentery, Single umbilical artery |
| 5 | F | Fœtus MTP | 1377–1406 dup | Alanine tract expansion (increased to 25 from 15) | I | Semilobar | Subaortic VSD | Single umbilical artery |
| 6 | M | Fœtus MTP | 1493 delG | p. (Gly498A fs*57)d | S | Semilobar | VSD | |
| 7 | M | Fœtus MTP | 13q deletion | N/A | S | Alobar | Duplicated superior vena cava Tetralogy of Fallot Ductus arteriosus agenesis |
Bilobulated lung Gallbladder agenesis |
| 8 | M | Born alive | Whole gene deletion | N/A | I | Syntelencephaly | Heart defect (not documented) | |
Notes
aDNA co-ordinates are shown for the cDNA sequence starting from the beginning of the coding sequence.
bMTP = medical termination of pregnancy.
cSeverity of HPE phenotype: Alobar > Semilobar > Lobar > Syntelencephaly.
dA single base deletion leads to a frameshift error resulting in translation of a false sequence with a stop codon after 57 amino acids.
Proband 7 has the most severe alobar form of HPE and also exhibits the most pronounced laterality defect (Table 1). This proband exhibits a loss of normal asymmetric thoracic anatomy, indicated by a duplicated superior vena cava (SVC) and a bilobulated lung. The right and left brachiocephalic veins are normally fused in man to form a single SVC which enters only the right atrium. Duplicated SVC therefore indicates bilateral connection to both atria, indicative of the laterality defect right atrial isomerism. Pulmonary morphology is also normally asymmetric in man such that, while the right lung has three lobes the left has only two. This patient has a symmetrical bilateral left-sided anatomy, suggesting left pulmonary isomerism. Thus, there is discordance between cardiovascular and pulmonary situs indicating situs ambiguus, a common phenomenon in heterotaxy.
Proband 4 also has abnormal lung lobulation indicating a laterality defect, but unfortunately the attending clinician did not record the details of this malformation and we are unable to assign situs. The same proband has additional features indicative of a laterality defect including a single umbilical artery (also seen in Probands 2 and 5) and a common mesentery. Proband 1 has spleen hypoplasia suggestive of right isomerism (which is also known as asplenia). This proband also has both adrenal and renal hypoplasia, features suggestive of abnormal abdominal situs. Pulmonary hypoplasia in this proband may indicate abnormal pulmonary situs.
Ventricular septal defect (VSD) is the most common cardiovascular anomaly observed (5 of 67 ZIC2 probands). This is associated with hypoplastic ascending aorta in two probands, while another exhibits Tetralogy of Fallot. VSD is frequent in mouse models with a laterality defect32 but is not diagnostic because it is also commonly associated with other genetic conditions, such as Chromosome 22 deletions33,34.
In summary, this analysis indicates that HPE is seen in ZIC2 probands together with cardiovascular, pulmonary and visceral malformations suggestive of an underlying laterality defect.
Identification of the Zic2 iso mutant
We performed a recessive ENU mouse mutagenesis screen in which MRI screening was used to identify novel mutants exhibiting cardiovascular anomalies at E14.535,36. This resulted in isolation of the iso (isomeric) line. Mapping and sequencing revealed a stop-gain mutation (Y401X) in the Zic2 gene disrupting the fifth zinc finger domain of the protein (Fig. S2; Table S1). The phenotype was validated through generation of a series of additional alleles, each with mutation targeted to the fifth zinc finger domain using TALEN-based gene editing31. Line Zic2A8 encodes an identical protein to iso (Y401X), while Zic2A5, Zic2A10, Zic2A17 and Zic2A19 have short DNA deletions leading to a frameshift followed by premature termination (Zic2A5, Zic2A10, Zic2A17) or an internal deletion (Zic2A19). All mutations affect the fifth zinc finger domain (Fig. S2) and thus differ from the previously published Zic2ku mutant in which the fourth zinc finger is disrupted (C370S)37. iso fails to complement either Zic2A8 or Zic2A5 (Table 2).
Table 2.
Neural tube, visceral and cardiovascular defects identified in mouse Zic2 mutants.
| iso | TALEN | Trans-hets | ||||||
|---|---|---|---|---|---|---|---|---|
| iso/iso | A5/A5 | A10/A10 & A17/A17 | A19/A19 | iso/A5 | iso/A8 | |||
| Neural Tube Defects | Exencephaly | 17/17 | 10/10 | 3/3 | 3/3 | 2/2 | 2/2 | |
| Holoprosencephaly | Alobar | 13/16 | 5/7 | 3/3 | 2/3 | 2/2 | 2/2 | |
| Semilobar | 3/16 | 2/7 | 0/3 | 1/3 | 0/2 | 0/2 | ||
| Lobar | 0/16 | 0/7 | 0/3 | 0/3 | 0/2 | 0/2 | ||
| Eyes | Cyclopia | 8/17 | 3/7 | 1/3 | 2/3 | 1/2 | 2/2 | |
| Hypotelorism | 6/17 | 3/7 | 1/3 | 0/3 | 0/2 | 0/2 | ||
| Absent | 3/17 | 0/7 | 1/3 | 1/3 | 1/2 | 0/2 | ||
| Spina bifida | 17/17 | 10/10 | 3/3 | 3/3 | 2/2 | 2/2 | ||
| Curly tail | 17/17 | 10/10 | 3/3 | 3/3 | 2/2 | 2/2 | ||
| Viscera | Lungs | Right Isomerism | 13/17 | 6/10 | 2/3 | 3/3 | 1/2 | 2/2 |
| Left Isomerism | 0/17 | 1/10 | 0/3 | 0/3 | 0/2 | 0/2 | ||
| Reversed | 0/17 | 0/10 | 0/3 | 0/3 | 0/2 | 0/2 | ||
| Normal | 4/17 | 3/10 | 1/3 | 0/3 | 1/2 | 0/2 | ||
| Spleen | Reduced or absent | 1/6 | 4/7 | 0/2 | 0/0 | 0/1 | 0/0 | |
| Duplicated | 0/6 | 0/7 | 0/2 | 0/0 | 0/1 | 0/0 | ||
| Right sided | 2/6 | 0/7 | 0/2 | 0/0 | 0/1 | 0/0 | ||
| Heart | Dextrocardia | 4/16 | 4/10 | 0/3 | 2/3 | 0/2 | 1/2 | |
| Mesocardia | 7/16 | 2/10 | 0/3 | 1/3 | 2/2 | 1/2 | ||
| Stomach | Right sided | 7/17 | 2/10 | 0/3 | 0/3 | 0/2 | 2/2 | |
| Pancreas | Right sided | 5/14 | 2/7 | 0/3 | 0/2 | 0/2 | 0/0 | |
| Atria & Veins | Bilateral systemic venous sinus | 10/15 | 4/10 | 1/3 | 3/3 | 1/2 | 2/2 | |
| Left sided Inferior vena cava | 5/16 | 3/10 | 0/3 | 1/3 | 0/2 | 2/2 | ||
| Hepatic vein drainage direct to atria | 2/16 | 3/10 | 0/3 | 3/3 | 0/2 | 0/2 | ||
| Atrial septal defect | 8/11 | 5/10 | 1/3 | 2/2 | 1/2 | 2/2 | ||
| Ventricles & Arteries | Abnormal ventricular topology | 6/13 | 6/10 | 0/3 | 1/2 | 1/2 | 2/2 | |
| Great artery transposition | 0/13 | 0/10 | 1/3 | 0/3 | 0/2 | 0/2 | ||
| Double outlet right ventricle | 5/13 | 5/9 | 0/3 | 2/3 | 0/2 | 2/2 | ||
| Ventricular septal defect | 11/15 | 7/10 | 0/3 | 1/1 | 2/2 | 2/2 | ||
| Rightward looped aortic arch | 5/17 | 3/10 | 1/3 | 2/3 | 0/2 | 0/2 | ||
A summary of the phenotypes observed by MRI and µCT imaging in all Zic2 mutants examined including both the original iso line and TALEN-generated lines. All embryos are homozygous mutants, except for those labelled “trans-hets” which carry one iso allele and one TALEN allele as a test of complementarity. For each phenotype described, the first number indicates the number of embryos observed with that phenotype, while the second indicates the number examined. The latter number differs for different phenotypes because it was not possible to assess every phenotype in every embryo due to limitations of imaging.
Zic2 mutants exhibit holoprosencephaly
External examination revealed that all Zic2 mutant embryos (n = 37) had neural tube defects including exencephaly, spina bifida and curly tail (Table 2; Figs 1; 2a,b); spina bifida was also present in 2/27 heterozygotes, which also frequently show curly tail (Fig. S331), while wild-type embryos (n = 22) had no anomalies. We employed µCT and MRI imaging to analyse the phenotype more closely. All embryos examined had holoprosencephaly (Table 2; Fig. 1). A range of severity was seen, but the majority of embryos had the most severe alobar form in which the two hemispheres are completely fused (27 of 33; Fig. 1b2). The remaining 6 had the semilobar form, indicating partial hemisphere fusion. No embryos were observed to have the mild lobar form of HPE. Six of 33 embryos lacked eyes, cyclopia was seen in 17 of 33 embryos (Fig. 1b3). and hypotelorism in 10 of 33. These neural tube phenotypes are consistent with those of the previously described Zic2Ku mutant4,37, suggesting that Zic2iso may also carry a loss of function mutation.
Figure 1.
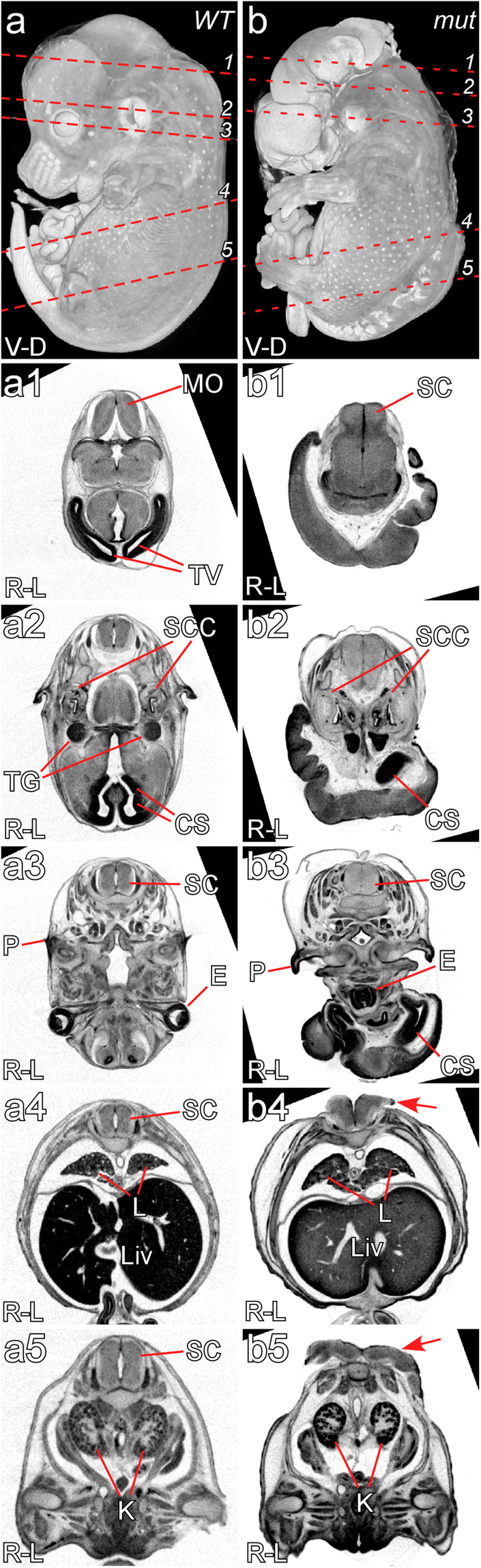
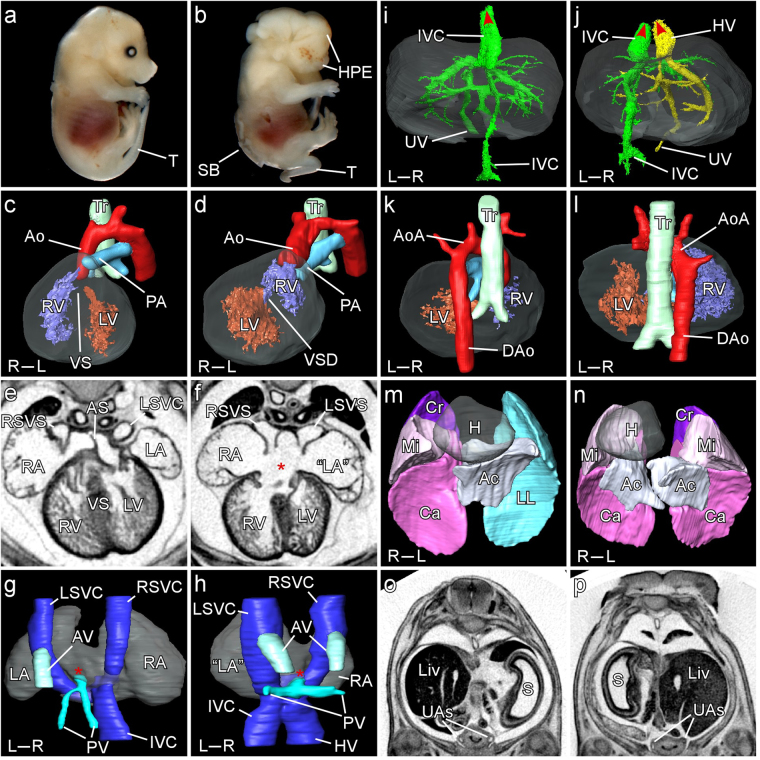
Holoprosencephaly, spina bifida and cyclopia in the Zic2 mouse mutant. Representative images illustrating a wildtype E14.5 embryo (a,a1–a5) and a Zic2A5/A5 embryo (b,b1–b5) at E14.5. Dashed red lines in a and b indicate the level of sections shown below. The mutant embryo exhibits alobar holoprosencephaly indicated by a lack of separation in forebrain structures (b1,b2), cyclopia (b3), spina bifida (b4,b5 arrows) and right pulmonary isomerism (b4). Abbreviations: V-D – ventral – dorsal; R-L – right – left; MO – medulla oblongata; SC – spinal cord; SCC – semi-circular canal; TG – trigeminal ganglion; TV – telencephalic vesicle; CS- corpus striatum; P – pinna; E – eye(s); L – lungs; Liv – liver; K – kidneys.
Figure 2.
Cardiovascular, pulmonary and visceral phenotypes in the Zic2 mouse mutant. Representative images illustrating range of phenotypes observed in wildtype (a,c,e,g,i,k,m,o) and homozygous mutants (iso and TALEN; b,d,f,h,j,l,n,p) at E14.5. (a,b) Holoprosencephaly (HPE), spina bifida (SB) and curly tail (T). (c,d) Reversed ventricular topology. The left ventricle (LV) is dextral to the right (RV), which gives rise to both Ao and PA (double-outlet right ventricle). (e,f) Right atrial isomerism indicated by bilateral systemic venous sinuses (SVS) and a large atrio-ventricular septal defect (asterisk). (g,h) Systemic and pulmonary veins (dorsal view). In the control embryo, the great veins (Inferior vena cava (IVC), left superior vena cava (LSVC), right superior vena cava (RSVC)) converge to form the systemic venous sinus which drains into the right atrium (RA) while the pulmonary veins (PV) drain into the left atrium (red asterisk). The azygos vein (AV) drains into the LSVC just above the level of the atria. In the mutant embryo, the IVC is left-sided while the hepatic vein (HV) and PV also drain into sinus venosus (red asterisk) which opens bilaterally into the atria. The AV is duplicated. (i,j) Hepatic venous anatomy (ventral view). In the wildtype, the IVC passes through the liver (grey shading) to drain into the right atrium (red arrowhead) and is connected to the umbilical vein (UV). In the mutant embryo, the IVC drains into the left atrium. The UV connects to the right hepatic vein (HV) which drains directly into the right atrium (red arrowheads). (k,l) Right-sided aortic arch (AoA). The descending aorta (Dao) may be seen to be ectopically located to the right of the trachea (Tr) in the mutant. (m,n) Right pulmonary isomerism. H – heart; LL – left lung; Cr – cranial lobe; Mi – middle lobe; Ca – caudal lobe; Ac – accessory lobe. (o,p) Ectopic right sided stomach (S). Liv – liver; S – stomach; UAs – umbilical arteries.
Zic2iso mutants have extensive cardiovascular, thoracic and abdominal malformations
CT imaging also revealed extensive defects within the cardiovascular system, thorax and abdomen of Zic2iso and TALEN mutants (Fig. 2; Table 2), many of which have not been previously described for Zic2.
We observed severe cardiovascular malformations in the majority of embryos. Abnormal ventricular topology was seen in half of all embryos examined (16 of 32; Table 2) such that the morphologically right ventricle was positioned on the left side of the embryo (Fig. 2c,d), while it was normal in the other 16. This 50–50 split indicates that the direction of heart tube looping was randomly assigned in these embryos, consistent with the phenotype previously described in younger Zic2Ku embryos24. The majority of embryos had an ostium primum atrial or atrio-ventricular septal defect (19 of 30; Fig. 2e,f asterisk). 21 of 35 embryos exhibited bilateral systemic venous sinuses (Fig. 2e–h), often with bilateral right-sided atrial appendages. Double outlet right ventricle was seen in 14 of 32 embryos examined (Fig. 2c,d) and ventricular septal defect was present in 23 of 33 embryos (Fig. 2c,d). Defects were observed in the vascular system at reduced penetrance relative to the cardiac defects, and were generally present in about a third of embryos. The inferior vena cava (IVC) was found to be aberrantly left-sided in 11 of 36 embryos (Fig. 2g–j), while 8 of 36 exhibited hepatic vein drainage directly into the atrium, bypassing the IVC (Fig. 2g,h). A right-sided aortic arch was observed in 11 of 37 embryos (Fig. 2k,l).
In mouse, the right lung is divided into four distinct lobes while the left consists of a single lobe (Fig. 2m). We observed bilateral multilobed lungs in 27 of 37 mutant embryos (Figs 1b4; 2n), while a single embryo was observed with a 1:1 lobed arrangement (Table 2). All control and heterozygous embryos showed a normal 4:1 lobed arrangement. The stomach and pancreas are normally located on the left side of the body but were observed to be ectopically right sided in 11 of 37 and 7 of 28 mutant embryos respectively (Fig. 2p; Table 2). The spleen was reduced or absent in 5 of 16 embryos and right-sided in another two (Table 2).
Zic2iso mutants have right isomerism
A random distribution in the direction of looping of the heart tube and gut among mutant embryos is indicative of a generic laterality defect, but does not provide information on the specific situs of affected individuals. This may be assigned based on careful examination of the anatomy of the lungs and atria, the positioning of visceral organs and the organisation of the vascular system, all of which show precise phenotypes that may be classified as right-isomerism, left-isomerism or situs inversus17,38.
Pulmonary anatomy is a useful indicator of thoracic situs because the lungs have a lateralised morphology determined by Pitx2c expression. Bilateral multi-lobed lungs are indicative of a lack of Pitx2c expression (right isomerism) while bilateral unilobar lungs indicate bilateral Pitx2c expression (left isomerism). Situs inversus, in contrast (right-sided Pitx2c expression), would be expected to result in a four-lobed lung on the left side and a single lobed lung on the right. Our data indicate that 73% of Zic2 embryos (27/37) exhibit right pulmonary isomerism, 3% (1/37) exhibit left isomerism, 24% (9/37) have the normal anatomy (situs solitus) and no embryos have reversed situs. This distribution is significantly different from a random distribution of the four phenotypes, and reveals a strong bias towards the phenotype of right isomerism (Chi squared test, p = 5.7 × 10−11). Similarly, bilateral systemic venous sinus and bilateral right atrial appendage is indicative of right atrial isomerism. We observe bilateral systemic venous sinus (right atrial isomerism) in 60% of embryos (21/35), with no evidence for left isomerism or reversed situs. This distribution is also significantly different from a random distribution (Chi squared test, p = 3.1 × 10−8). Double outlet right ventricle (44%; 14/32) is associated with right but not with left isomerism32,39. Abnormal right-sided aortic arch looping is not seen in left isomerism40 but is present in right isomerism32,41 and in situs inversus. We did not observe vascular phenotypes associated with left isomerism such as interruption of the inferior vena cava and partial anomalous pulmonary return17. Asplenia is commonly associated with right isomerism42, and indeed the human disease is sometimes known as Asplenia. The spleen is reduced in mice lacking the NODAL receptor Acvr2b, although asplenia seems to be observed only in Cfc1 (Cryptic) mutants41,43. We observe reduced or right-sided spleen in 44% of embryos (7/16) while none exhibited polysplenia, a feature of left isomerism.
These data thus indicate a strong bias towards right isomerism. Only one embryo exhibits any feature of left isomerism (bilateral unilobed lungs) and this embryo does not show atrial left-isomerism nor any vascular phenotype associated with left-isomerism.
The NODAL pathway is downregulated in Zic2 mutants
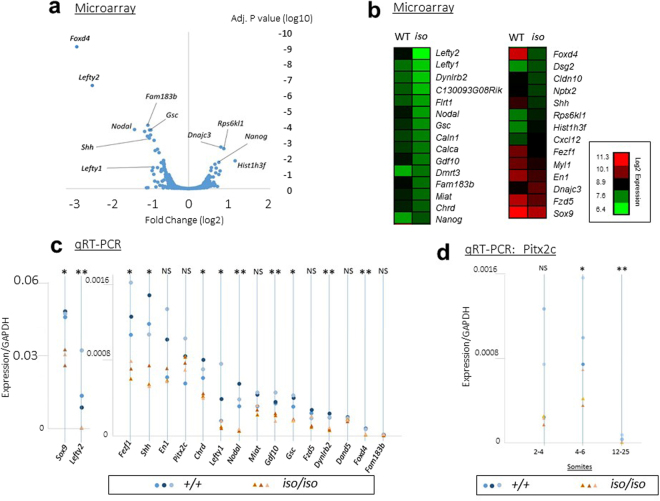
We performed a microarray experiment in mice as an unbiased screen to identify putative Zic2 transcriptional targets. Global gene expression was assayed in whole Zic2iso/iso embryos harvested at E8.0 -E8.5 (0–4 somites) relative to wild-type. Only 29 genes showed a significant change in their expression level (with a false discovery rate of 5%) and the majority (23/29) were downregulated (Fig. 3a,b; Table S2). Some of these, such as Dmrt3 and Fzd5, are specifically expressed in the developing head. Other downregulated genes have an established role in left-right patterning including Lefty2 (−5.1), Nodal (−2.5), Lefty1 (−1.8) and Shh (−2.0). Downregulated genes also include Gdf10 (−2.01) and Chordin (−1.53), which are, like Nodal and Lefty, associated with TGFβ signalling, and Sox9 which has not so far been associated with LR establishment in the mouse although it is known to be involved in sea urchin44. Many of the changed genes, including Nodal, Shh, Fam183b, Foxd4 and Dynlrb2 are known to be expressed within the node, while others including Gsc and Lefty1 are expressed in node-derived cells at the midline. Upregulated genes included the pluripotency factor Nanog (+1.67). To validate these changes we performed quantitative real-time polymerase chain reaction (qPCR) assays for changed genes, as well as for the NODAL pathway genes Pitx2c and Dand5 (also known as Cerl2; Fig. 3c). This analysis confirmed the downregulation of Nodal, Lefty2, Lefty1 and Shh. Surprisingly, Pitx2c was not changed. We hypothesised that this might be because the assay was performed at an age before Pitx2c is upregulated in the LPM, and therefore repeated the analysis using older embryos. In older embryos (aged 4–6 somites), we observed a significant reduction in expression, which persists in embryos of 12–25 somites (Fig. 3d), indicating that Pitx2c is downregulated in the iso embryo during the time at which it is normally expressed in the LPM. Thus, these data suggest downregulation of the NODAL pathway in the iso embryo.
Figure 3.
Loss of Zic2 leads to downregulation of the NODAL pathway. (a,b) Microarray analysis of gene expression changes in the Zic2iso/iso embryo compared to wildtype at E8.5. (a) Volcano plot. The data are plotted as log fold change (relative to wildtype) against adjusted p-value. (b) Heat map showing wildtype (WT; left panel) and Zic2iso/iso (KO; right panel) embryos. Data show mean expression for three independent biological replicates of pooled embryos and are expressed as log2 values. (c) SYBR Green based qRT-PCR analysis of genes identified in the microarray screen, and known NODAL pathway genes. Expression values shown are normalised to Gapdh. Three independent biological replicates of pooled embryos (0–4 somites) were performed for each condition and these are plotted as individual data points (blue circles indicate wildtype; orange triangles indicate Zic2iso/iso). Asterisks above each column indicate result of a one tailed t-test of samples with unequal variance testing the null hypothesis that loss of Zic2 has no effect on gene expression: ***p < 0.0005; **p < 0.005; *p < 0.05, NS = not significant. (d) Taqman qRT-PCR analysis of Pitx2c expression at three different ages (2–4 somites, 4–6 somites and 12–25 somites). Three independent biological replicates of pooled embryos were performed for each condition. Labels as per panel c.
Zic2 is required for Nodal expression in perinodal crown cells
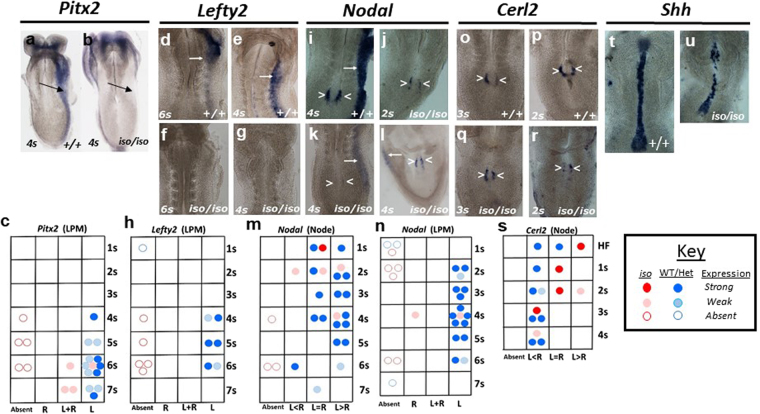
We used in situ hybridisation to further analyse changes in the core NODAL pathway genes. Pitx2 expression was investigated using a probe which recognises all three Pitx2 isoforms45 and was found to be expressed in the head folds and in the LPM in the wildtype embryo (Fig. 4a). In Zic2iso/iso embryos Pitx2 expression was seen in the head folds, but was weak or, in many cases, absent from the LPM (Fig. 4b, arrow). Expression appeared to be delayed from the 4 to the 6-somite stage, it was weaker, and its laterality was perturbed, often bilateral (Fig. 4c). Lefty2 was expressed exclusively in the left LPM of wildtype embryos aged between 4 to 6 somites (Fig. 4d,e). All Zic2iso/iso embryos failed to express Lefty2 (Fig. 4f–h). Nodal was expressed in the node of all wildtype embryos examined (Fig. 4i, arrowhead) and was either bilaterally expressed or was enriched on the left side of the node (Fig. 4m). In the LPM, Nodal was expressed in all wildtype embryos from 2 to 6 somites and was restricted to the left side (Fig. 4i, arrow; Fig. 4n). Nodal expression at the node of Zic2iso/iso embryos was in most cases (7 of 8) weak or absent (Fig. 4j–m). Expression was only detectable in early somite stages (1 to 4 somites) mutant embryos (5 of 8), and its abnormal laterality in one of them suggested a defective nodal flow, which would be consistent with the requirement for Zic2 in node cilia development or function as previously proposed24. Most Zic2iso/iso embryos (6 of 8) failed to express Nodal in the left LPM (Fig. 4j,n). In the two mutant embryos that did express Nodal in the LPM, it was weak and was observed in both left (Fig. 4k) and right LPM (Fig. 4l). Cerl2 (Dand5), known to be the earliest asymmetrically biased gene expressed at the node, (L < R)46 was expressed exclusively within the node of wildtype embryos (Fig. 4o,p) and was observed to be enriched on the right side of the node in 8 of 9 embryos (Fig. 4s). Cerl2 appeared to be maintained at wildtype levels in Zic2iso/iso embryos (Fig. 4q–s), but the onset of its laterality appeared to be delayed and slightly perturbed (Fig. 4s), an observation again consistent with the possibility that the mutation of Zic2 initially results in defective nodal flow and randomisation of gene expression at the node. We also studied the expression of Shh, which was detected along the embryonic midline in wildtype embryos in a solid band (Fig. 4t). In Zic2iso/iso embryos, expression levels were maintained but there seemed to be fewer positive cells and the band of expression appeared disrupted (Fig. 4u) suggestive of impaired development of Shh-expressing cells in these embryos.
Figure 4.
Disrupted left-side specification in the Zic2 mutant. (a,b; d–g; i–l; o–r; t,u) in situ hybridisation. Genotype and age of embryos (expressed as somite number) are indicated. All images are oriented with the left side on the right of the photo and show the ventral surface of the embryo. Arrows indicate LPM, arrowheads: node. t and u are composite images. (c,h,m,n,s) Charts summarise the results of in situ experiments illustrating the relationship of expression pattern and expression level to embryo age and genotype. Each dot represents an individual embryo, blue dots are control embryos (+/+ and +/iso), red are mutants (iso/iso). A dark colour indicates strong expression while a lighter colour indicates weak or absent expression. Data are plotted as expression pattern (one of 4 categories shown on the x axis) against embryo age (expressed as somite number: 1 s, 2 s etc., y axis). HF = headfold stage. L = left, R = right.
In summary, we find some anomalies suggestive of an occasional randomisation of LPM identity (bilateral Pitx2c expression, right-sided expression of Nodal). These anomalies match the laterality defects we characterized in the expression of Nodal and Cerl2 at the node, defects which are consistent with the Zic2 mutation leading to nodal flow perturbations, most likely as a result from the node cilia defect described in Zic2Ku24. However, our data show that the predominant phenotype is the absence of expression in the LPM. This failure to activate NODAL-dependent gene expression (Nodal, Pitx2c, Lefty2) in the LPM is consistent with the later bias towards right-isomerism revealed by our analysis of the Zic2iso mutant phenotype. Nodal expression within the node, a prerequisite for downstream gene expression in the LPM13,46, is reduced or absent in most embryos. These data suggest that ZIC2 may be required for Nodal expression at the node.
ZIC2 binds to the Nodal locus and can activate transcription
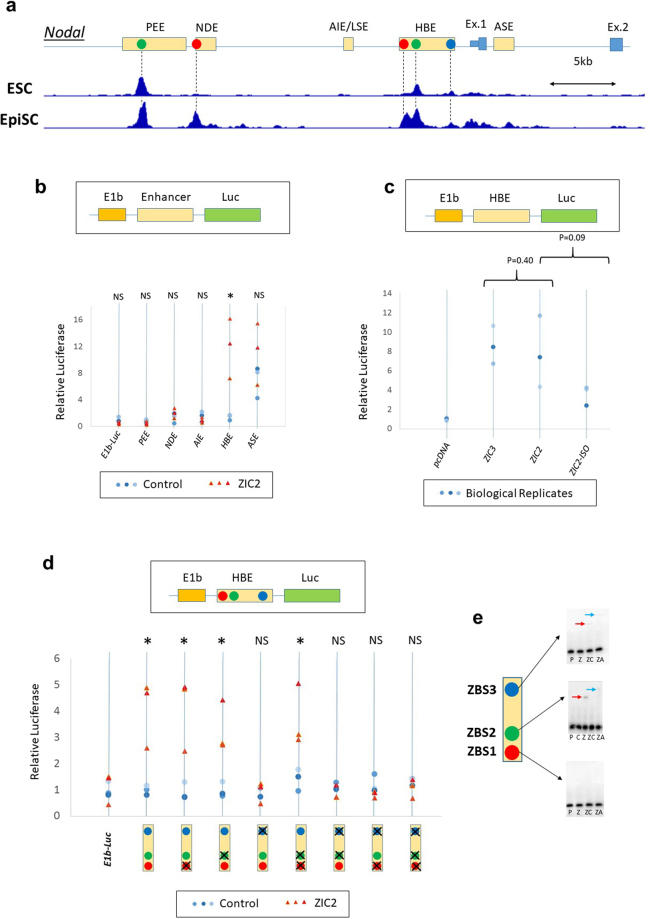
The expression of Zic2 in the epiblast overlaps with that of Nodal from the blastocyst stage to the late gastrula stage, up to and including formation of the node6,7. The dynamic expression of Nodal during development is regulated by five enhancers47–49. Analysis of previously published ChIP-seq datasets26,27 shows that ZIC2 is bound to the Nodal locus in both ESCs and in EpiSCs (Fig. 5a). In ESCs, prominent binding peaks were mapped to the Proximal Epiblast Enhancer (PEE)50 and to the Highly Bound Element (HBE)49 (Fig. 5a, green dots). These binding sites are also occupied in EpiSCs, but two additional high-affinity binding peaks are seen within the Node Dependent Enhancer (NDE)51 and in HBE (Fig. 5a, red dots) indicating that ZIC2 binding is more widespread in these cells. Further low-affinity peaks map to the Asymmetric Enhancer (ASE)48,51, to Exon 1 and to HBE (blue dot).
Figure 5.
ZIC2 binds to the Nodal locus and promotes transcriptional activation. (a) ChIP-seq data to show ZIC2 binding at the Nodal locus in embryonic stem cells (ESC)27 and in epiblast stem cells (EpiSC)26. The five previously-characterised enhancer elements are indicated by yellow boxes in the cartoon above, while exons are indicated by blue boxes. Coloured dots in the cartoon indicate putative ZIC2 binding sites. In ESCs (upper trace), two high-affinity binding sites are seen, located within the PEE and HBE enhancers (green dots). In EpiSCs (lower trace), four high-affinity sites are observed which includes the two bound in ESCs (green dots) as well as two additional sites located within the NDE and HBE enhancers (red dots). Several lower-affinity binding sites map to ASE, HBE and exon 1, the site located within HBE is indicated by a blue dot. (b) Luciferase assays performed using reporter constructs consisting of each of the known Nodal enhancers linked to a viral E1b promoter and luciferase, as shown in the cartoon. Blue circles indicate control transfections, orange triangles ZIC2 transfections. Luciferase activity is plotted relative to control. The data show that the HBE reporter is activated by ZIC2 above background. (c) Luciferase assays using the native HBE reporter, performed as in b. Cells were transfected with the reporter and an expression plasmid for ZIC3, ZIC2 or ZIC2-ISO, or a control empty vector (pcDNA). (d) Luciferase assays using modified HBE reporters, performed as in b. HBE contains 3 putative ZIC2 binding sites, indicated by the coloured dots in the cartoon, deleted binding sites are indicated by an “X”. Deletion of sites ZBS1 and ZBS2 (red and green dots in the cartoon) has no effect on the ability of ZIC2 to activate the reporter, while deletion of site ZBS3 (blue dot) eliminates this activity. (e) Gel shift (EMSA) assays to study the binding of ZIC2 to HBE. P = probe only, Z = probe + HA-ZIC2, ZC = probe + HA-ZIC2 + unlabelled competitor, ZA = probe + HA-ZIC2 + αHA antibody. Red arrow = gel shift, blue arrow = supershift. Images show cropped gel pictures, uncropped images are shown in the supplemental data.
We tested the ability of ZIC2 to regulate the transcriptional activity of each of the five Nodal enhancers using luciferase reporter constructs in which the enhancer is linked to the minimal E1b promoter49 (Fig. 5b). Each construct was co-transfected in U2-OS cells together with an expression plasmid encoding ZIC2 or an empty vector (pcDNA).
The ASE enhancer showed a strong basal activity in control cells suggesting activation by endogenous factors present in U2-OS cells (Fig. 5b). For this reason, we were not able to accurately test the ability of ZIC2 to activate this enhancer. Low basal activity was observed for the remaining enhancers. We found no evidence for ZIC2-mediated transcriptional activation from the PEE, NDE or AIE enhancers. HBE, in contrast, shows a robust ZIC2-mediated transcriptional activation. HBE has been shown to be transcriptionally active in primitive streak-like cells (node precursors)49.
Zic3 has a phenotype closely resembling that of Zic2, including laterality-related malformations and HPE52,53, and also exhibits reduced or absent expression of Nodal52. ZIC3 has previously been shown to be able to activate an NDE enhancer54. We therefore asked whether ZIC3 can also activate expression from the HBE enhancer. Luciferase assays demonstrate that ZIC3 activates HBE at a level similar to that of ZIC2 (Fig. 5c), lending further support to the hypothesis that this enhancer may be important in LR patterning.
We next asked whether the iso mutation reduces the ability of ZIC2 to activate the HBE enhancer, and thus whether a failure to activate Nodal might explain the phenotype. The Zic2 kumba allele has been shown to evade nonsense mediated decay (NMD) but to produce a protein which cannot activate the ApoE promoter in luciferase assays55. While the kumba mutation disrupts a cysteine residue in the fourth zinc finger37, the iso mouse carries a Y401X mutation which disrupts the fifth zinc finger of ZIC2 (Fig. S1). Two lines of evidence indicate that iso, like kumba, may evade NMD. Firstly, Zic2 mRNA is not significantly reduced in iso embryos by microarray analysis (Table S2). Secondly, qRT-PCR analysis indicates that the mRNA can be detected in mutant embryos (Fig. S4). We used site-directed mutagenesis to generate a Y401X mutation within the ZIC2 expression construct (named ZIC2-ISO) and tested the response of the HBE luciferase reporter to this. ZIC2-ISO shows a reduced ability to activate HBE (Fig. 5c), just above the threshold of significance (p = 0.09), but still maintains some transcriptional activity. This, together with potential redundancy with ZIC3, may explain the partial penetrance of the Zic2iso phenotype.
To gain further insights into the regulation of HBE by ZIC2, we investigated ZIC2 binding sites within HBE. ChIP-seq data suggests the presence of three putative binding sites, and we found that each of these contains sequence matching the consensus ZIC2 binding motif. ZIC2 Binding Site 1 (ZBS1) is located at position 99–109 bp within HBE and has the sequence CACCTCCTGGG (Fig. 5a,d,e red dot), ZBS2 at position 615–625 bp with sequence CCCCTGGGGTG (Fig. 5a,d,e green dot), and ZBS3 at 1845–1855 bp with sequence GCCCTCCTGGG (Fig. 5a,d,e blue dot). We used site-directed mutagenesis to delete each of these sites within the HBE-luciferase reporter construct. Luciferase assays indicated that while deletion of sites ZBS1 and ZBS2 has no effect on ZIC2-mediated transcriptional activation, reporters lacking ZBS3 completely lost their responsiveness to ZIC2, indicating that an intact site ZBS3 is an absolute requirement (Fig. 5d).
An electrophoretic mobility shift assay (EMSA) confirmed binding of ZIC2 to ZBS2 (occupied in both ESCs and EpiSCs) and ZBS3 (required for activation but demonstrating only low-affinity binding in ESCs and EpiSCs) but not to ZBS1 (occupied in EpiSCs but not in ESCs) in vitro (Fig. 5e).
Thus, these data demonstrate that the HBE enhancer, which is bound by ZIC2, both in vivo in node precursors (Fig. 5a) and in vitro (Fig. 5e), can mediate ZIC2-dependent and ZIC3-dependent transcriptional activation in vitro (Fig. 5b,c). An identified ZIC2 site within HBE, bound at low-affinity in precursor cells (Fig. 5a) is required for this activity in vitro (Fig. 5d) and a ZIC2 mutation reproducing the iso mouse mutant, which has impaired Nodal node expression (Fig. 4j–m), can reduce the ability of ZIC2 to activate HBE (Fig. 5c).
Discussion
In this work, we reveal that ZIC2/Zic2 loss in both man and mouse is associated with a complex set of previously unappreciated malformations affecting the cardiovascular, pulmonary and digestive systems, in addition to the better known neural tube defects. The mouse phenotype reveals a laterality defect which shows a strong bias towards right-isomerism, indicative of a lack of left-sided identity in the lateral plate mesoderm during morphogenesis of these organ systems. This is supported by molecular data indicating that the left-determining NODAL signalling cascade fails to be activated in the LPM. Nodal expression at the node is also impaired, implying that ZIC2 acts upstream of this event. Binding of ZIC2 to the Nodal locus in EpiSCs, together with in vitro data revealing ZIC2-mediated transcriptional activation of regulatory sequences, suggests that a direct interaction between ZIC2 and Nodal is critical for the establishment of left-sided mesoderm identity.
The association of Zic2 with cardiovascular laterality defects was first demonstrated in the Zic2Ku mouse, in which the direction of heart tube looping was shown to be randomly assigned24. Node cilia were found to be reduced in length in these mutants from 4 µm to 2.5 µm, and this, together with the earlier observation that Zic2 expression in the node is turned off just before Nodal expression in perinodal cells is initiated6, led to the conclusion that the laterality defect (interpreted as random cardiac situs) resulted from impaired nodal cilia development24. A randomised distribution in the direction of heart tube looping within a population of mutants does not in itself indicate randomised cardiac situs. Such a phenotype is also associated with specific, non-random laterality defects such as isomerism32,56. This is because, in the absence of laterality, heart looping lacks directionality and may turn in either direction. The Zic2Ku mutant exhibits mid-gestation lethality, preventing further investigation. In contrast, most Zic2iso and TALEN mutants survive until E15.5, which has allowed us to perform a more detailed analysis of the laterality phenotype. We confirmed the heart tube looping phenotype but revealed that this results not from randomisation of situs, but from right isomerism. Zic2iso and TALEN mutants show a strong bias towards right isomerism over other laterality phenotypes. Pulmonary situs was assessed in a total of 37 Zic2 mutant embryos, of these 73% showed right pulmonary isomerism, a single embryo exhibited left isomerism and none had situs inversus. The same phenotype was seen in multiple independent Zic2 alleles. Laterality of the cardiovascular system matched that of the thorax, albeit at reduced penetrance, as demonstrated, for example, in right atrial isomerism, which was seen in 60% of embryos. Many other features indicate right isomerism and not left isomerism or situs inversus.
This conclusion is consistent with the observed molecular phenotype. Both Zic2Ku and Zic2iso mutants fail to activate NODAL signalling in the LPM. Zic2Ku mutants lack Nodal and Lefty2 LPM expression, and have reduced Pitx2 expression24. Our analysis reveals a similar molecular phenotype in the Zic2iso mutant. Mutations which impair the motility of node cilia such as the iv mutant of the cilia dynein Dnah112, or mutations in transcription factors required for cilia development such as Noto57, result in the stochastic activation of NODAL signalling in the LPM. This is visualised via detection of the expression of Nodal/Pitx2c/Lefty2 with the same frequency on the left, on the right, on both sides or in none, and leads at later stages to equal proportions of embryos with situs solitus, situs inversus, right isomerism or left isomerism. Only 4 out of 20 E8.5 Zic2iso mutant embryos examined showed ectopic expression of these genes in the LPM. 15 failed entirely to express them in this tissue, an observation consistent with the fact that 73% of those examined at E14.5 exhibited right isomerism. These numbers do not support the hypothesis that a defect in cilia motility is the major cause of the laterality defect.
Although there is ample evidence that cilia motility conditions the asymmetry of Nodal expression in perinodal cells, this asymmetry in mRNA expression is not itself required to induce Nodal expression in the left LPM47,58,59. What appears to matter is the amount of NODAL protein produced. Pioneering studies relying on the removal of specific Nodal enhancers have established that Nodal expression at the node is required to induce Nodal expression in the LPM13,58,59 thus providing an explanation for the right isomerism of Zic2 mutant embryos that fail to express Nodal at the node. However these studies also showed that residual Nodal expression at the node could be sufficient to induce correct expression in the LPM, and therefore call into question whether the low level of Nodal we detect at the node in some of our Zic2 mutants is the only reason for their failure to induce NODAL downstream targets in the LPM. Assessing the exact contribution of the loss of Nodal expression at the node to the observed phenotype would require a rescue experiment in which a transgenic construct is used to drive Nodal expression at the node in a Zic2iso mutant background.
The situation is arguably complex in these mutants, because Nodal may not be the only gene in the pathway regulated by ZIC2. The level of Nodal expression in the node required to induce its own expression in the LPM may thus be different, and perhaps higher, than that in wildtype embryos, to compensate for a possible concomitant down-regulation of partners or agonists. Gdf1, encoding a co-ligand of NODAL, is likewise expressed in perinodal cells where it is required to ensure the adequate transfer of NODAL to the LPM60, and could be one such partner as its absence from the node also leads to right isomerism. No ZIC2-binding peaks are present at the Gdf1 locus26,27 and our microarray analyses of Zic2iso mutant embryos detected no alteration of Gdf1 expression (Table S2), but, given that Gdf1 is also bilaterally expressed in lateral plate mesoderm at this stage, this experiment may not be sensitive enough to detect its specific misregulation in perinodal cells. Interestingly, the expression of Gdf1 in these cells, like that of Nodal and Cerl2, is known to be dependent on NOTCH signalling61. Our observation that in most Zic2iso mutant embryos Cerl2 shows normal levels of expression in the node indicates that NOTCH signalling is likely to be intact in these mutants. This, together with a previous report that absence of Nodal expression in the node does not affect the expression of Gdf1 there58 make it quite possible that its expression is similarly unaffected in the node of Zic2iso mutant embryos. However, further investigations are necessary to make certain that this is case, not just for Gdf1 but also for all the genes expressed at the node that contribute to the production and propagation of the left identity-inducing signal.
The occurrence of right isomerism has been described in embryos carrying point mutations in either Pkd1l1 or Pkd2, which are believed to affect the detection of nodal flow by immotile cilia62, rather than affecting motile cilia function (node morphology and cilia motility is unaffected in these mutants). Crucially, not only does the NODAL signalling cascade fail to be activated in the LPM of these mutants, both Nodal and Cerl2 expression at the node remain symmetrical, unlike in Zic2 mutants. Pkd1l1 and Pkd2 expression is restricted to the node at E7.0–E7.75. Our microarray data shows no evidence for a downregulation of their transcripts in the Zic2iso mutants, but in situ data suggests they may be reduced in Zic2Ku24 mutants. The anomalies we detected in the laterality of Nodal and Cerl2 expression around the node of Zic2iso mutant embryos confirmed that their nodal flow is perturbed, and suggest that motile cilia at the node present defects similar to those characterized in Zic2Ku mutants24. However, the emergence of these anomalies, and their occasional consistency with corresponding anomalies in the expression of Nodal or Pitx2 in the LPM, suggest immotile cilia are functional and argue against this mutant version of ZIC2 having a major impact downstream of the nodal flow.
The expression of Cerl2 in perinodal cells of Zic2iso mutant embryos indicates NOTCH signalling is intact. This suggests that the Nodal locus in this mutant is either unresponsive to NOTCH signalling or unable to maintain its own expression after it is induced. The dynamic expression of Nodal is regulated by five distinct enhancers, three of which (NDE, PEE and HBE) were found to harbour significant ZIC2-binding peaks in ESCs and/or in EpiSCs (Fig. 5a). NDE is active in perinodal cells and its deletion eliminates most, but not all, of Nodal expression at the node. Its transcriptional activity has been shown to be dependent on NOTCH signalling63 and a previous study using Xenopus cells, has shown that it can be activated by ZIC354, a transcription factor closely related to ZIC2. NDE therefore would appears as an ideal candidate to mediate the influence of ZIC2 on Nodal expression at the node, however we did not detect an effect of ZIC2 on its transcriptional activity in our luciferase assay and further analysis will be necessary to find out whether it does mediate this influence in vivo.
PEE transcriptional activity is detected in the proximal epiblast and in the anterior primitive streak, but not in the node50. Deletion of PEE has been shown to result in a range of defects, including heart abnormalities50, which are reminiscent of those of Zic2 mutants. However, like for NDE, we did not detect an effect of ZIC2 on the transcriptional activity of PEE in our luciferase assay. These data leave open the possibility that this enhancer mediates the influence of ZIC2 on a domain of Nodal expression that is critical for anterior patterning and the establishment of laterality, but again further analysis will be necessary to confirm this is the case.
HBE was the only Nodal enhancer showing a ZIC2-mediated transcriptional activation response. Mutagenesis analysis indicated both that a defined ZIC2-binding site within HBE, ZBS3, is required for this response, and that a protein carrying the same mutation as the iso mouse shows a reduced ability to activate HBE. While the transcriptional activity of HBE is highest at preimplantation stages it is still detectable in the post-implantation epiblast, in the primitive streak and in the early node49. The impact on embryonic development of its deletion at these stages is not yet known, but these data suggest that an interaction between ZIC2 and HBE may be required for Nodal to be correctly expressed at the node. Testing this hypothesis calls for an investigation of the impact of a targeted mutation of ZBS3 on Nodal expression and the establishment of laterality.
Our analyses indicate that ZBS3 is a low-affinity ZIC2-binding site, and that higher affinity ZIC2-binding sites in HBE are dispensable for its transcriptional activity. This is consistent with current models of how transcription factors regulate gene expression, which place greater emphasis on the critical role played by low-affinity binding sites64. This result led us to consider the possibility that another Nodal enhancer, ASE, in which a low-affinity ZIC2-binding site similar to ZBS3 was identified in EpiSCs, might also mediate the influence of ZIC2 on Nodal expression at the node. ASE is an auto-regulatory enhancer, known to be dependent on Activin/Nodal signalling. Mouse embryos deleted for ASE show very weak Nodal expression in the left LPM, which leads to partial right isomerism later on47. Crucially, the level of Nodal expression at the node at E7.5 and E8.5 in these mutants is similar to that of wildtype embryos, except that it remains symmetrical. This observation appears to rule out the possibility that ASE plays a critical role in mediating the influence of ZIC2 on Nodal node expression.
Thus, a review of the evidence relating to NDE, PEE and HBE leaves open the possibility that they may all be, directly or indirectly, involved in promoting the influence of ZIC2 on the expression of Nodal during the time window in which ZIC2 function at the midline is critical for the establishment of a left identity in the LPM, which broadly extends from E6.5 to E7.5. The expression of Zic2 and the transcriptional activity of NDE only overlap briefly in the young node, which seems to preclude a prolonged interaction. In contrast, the overlap with the transcriptional activities of HBE and PEE in the epiblast and in the primitive streak, ahead of node formation, is extensive. This may be important as ZIC2 has been shown to act as a pioneer factor in ESCs, binding to target genes before they are transcriptionally active, seeding the locus to facilitate binding of additional factors later in more differentiated cell types26,29. Furthermore, in the case of Nodal, the presence of one of the ZIC2-bound enhancers, HBE, has been shown to condition the later activation of at least one of the other Nodal enhancers49. These observations may explain how the impact of ZIC2 absence on Nodal node expression could be delayed until a time when the transcription factor is no longer expressed. ZIC2 associates at a subset of loci in ESCs with the NURD chromatin remodelling complex27, a complex known to regulate bivalent enhancers. ZIC2 is bound to the Nodal locus in epiblast stem cells, precursors of the node, and is able to activate expression in in vitro assays from one such bound enhancer (HBE), yet there is no obvious phenotype at this stage of development in Zic2 mutants. All available data is therefore compatible with the possibility that ZIC2 interaction with regulatory sequences at the Nodal locus in node cell precursors in the primitive streak primes it for later activation, a pre-requisite for the correct establishment of left-sided identity. Further work will be required to test this hypothesis and to elucidate the precise molecular mechanisms by which ZIC2 may regulate Nodal expression.
Our analysis lends support to the hypothesis that cardiovascular and other visceral defects seen in ZIC2 HPE patients may result from an underlying laterality defect. However, it would appear that the co-morbidity of cardiovascular laterality defects with HPE is a relatively rare event in reported ZIC2 HPE cases, occurring in only between 9 and 14% of cases2,3. Cardiovascular malformations of this kind are embryonic lethal in mice and thus many pregnancies may be lost before term, indeed only one of the probands we describe was born alive. Thus, many cases may go unreported. It should also be noted that most patients have heterozygous ZIC2 mutation; heterozygosity in mice is associated with mild neural tube defects but is not associated with cardiovascular malformations. Thus, milder forms of HPE reported in living patients would not be expected to be associated with cardiovascular anomalies. Situs abnormalities do not always manifest in right isomerism in human cases as in the mouse model, and we find evidence for discordance between cardiac and pulmonary situs suggesting heterotaxy in one patient. This is likely to be for a number of reasons. The patient with heterotaxy (Proband 7) carries a large chromosomal deletion encompassing many genes in addition to ZIC2 and therefore genetic interactions are likely to be a factor. Genetic background is known to affect the penetrance of cardiovascular laterality phenotypes32, indicating that genetic interactions at multiple loci influence the phenotype. This may also reflect the influence of environmental factors, which are known to impact upon the expression of cardiovascular phenotypes65. Finally, in many cases it is not possible to make a definitive diagnosis based on the information provided.
Our data nevertheless explain the co-morbidity of holoprosencephaly with congenital heart disease and suggest that ZIC2 should be considered as a candidate for screening for the latter disease.
Methods
Human genetics
All protocols were approved by the local ethics committee of Rennes Hospital. All work was carried out in accordance with this protocol. All samples were obtained and analysed with informed consent according to the protocols approved by the local ethics committee (Rennes Hospital).
Mouse genetics
All animal procedures were approved by the Committee for Animal Care and Ethical Review at the University of Oxford, and all the experiments conformed to the UK Animals (Scientific Procedures) Act, 1986, incorporating Directive 2010/63/EU of the European Parliament. All animal procedures were performed in accordance with UK Home Office regulations (PPL 30/3174). The iso allele was isolated in a random recessive ENU mutagenesis screen in which cardiovascular anomalies were identified by MRI screening of E14.5 embryos35,36. Male C57Bl6/J males were mutagenized then crossed to C3H/HeH females. G3 progeny were screened for a phenotype. TALEN alleles of Zic2 have been previously described31. All Zic2 alleles were subsequently backcrossed for several generations and then maintained on a C3H/HeH background (mice obtained from MRC Harwell, Oxfordshire, UK) and embryos of both sexes were used in experiments. Heterozygous animals were crossed and pregnant dams were sacrificed by cervical dislocation before the embryos were dissected and processed for further analyses. Genotyping was performed using allele-specific Taqman probes on DNA obtained from ear biopsies. In all experiments, mutant embryos were compared to littermate controls.
Genetic mapping and sequencing
Mutations were mapped using a panel of SNP markers that differentiate between C57BL6/J and C3H/HeH mouse strains, as described previously35, identifying a minimal homozygous segregating interval lying on chromosome 14 between SNP rs13482392 (118.9 Mb) and the end of the chromosome at 124 Mb. The identified chromosomal interval was then exome sequenced using SureSelectXT mouse all exon kit (Agilent, as recommended by the manufacturer). Captured libraries were sequenced on the Illumina platform as paired-end 76-bp reads. Study accession number ERP000530. The only exonic mutation identified within this interval was a C to A substitution at position 122877892 indicating a stop-gain mutation (Y401X) in the Zic2 gene disrupting the fifth zinc finger domain (Fig. S1; Table S1). This was confirmed by capillary sequencing. No other exonic mutations were identified within the mapped interval.
Phenotyping by MRI/µCT scanning
Embryos were harvested at either E14.5 or E15.5, exsanguinated in warm HANKS saline (SIGMA, H4641), cooled down in ice cold PBS and then fixed in 4% paraformaldehyde. Imaging was performed using either MRI (Iso, A8, A10/17, A19) or by µCT (A5). MRI was performed as previously reported66. µCT was performed using SkyScan 1172 scanner (Kontich, Belgium). Prior to scanning, embryos were incubated in 0.025 N Lugol’s solution for 4 days to achieve soft tissue contrast and embedded in a tube with 1% agarose. Generated datasets were analysed and 3D reconstructions generated using Amira 5.3.3 software (FEI Visualization Sciences Group, Merignac, France).
Microarray analysis
E8.5 wildtype and Zic2iso/iso whole embryos were collected and snap frozen in liquid nitrogen. Eight stage-matched embryos were pooled per sample and three replicates of each genotype were performed. RNA was prepared using the RNeasy Plus Micro kit (Qiagen) and hybridised to Illumina Mouse WG6 v2 arrays at Oxford Genomics Centre (following manufacturers’ protocol). Raw data were imported into R statistical software for processing and analysis (http://www.R-project.org). Pre-processing and normalisation steps were performed with the BioConductor67 package ‘Variance Stabilisation and Normalisation’ (VSN)68. Quality control analyses showed the data were high quality with no outlier samples. Statistical analysis was performed on the full dataset (approximately 45,000 probes) with the Linear Models for Microarray Analysis (LIMMA) package69. Raw p-values were corrected for multiple testing using the false discovery rate (FDR) controlling procedure70. At 5% FDR, this resulted in 30 significantly changed genes. A heatmap was generated using the Expander programme71. Microarray data has been submitted to the Gene Expression Omnibus (accession number will be provided following acceptance).
qPCR
Total RNA was harvested from whole embryos as above. RNA was reverse transcribed using the Quantitect reverse transcription kit with genomic DNA eliminator (Qiagen), and analysed by SYBR Green based qPCR (Fig. 2c; Supplementary Table 3 lists primers used) or using Taqman probes (Figs 2d, S4). Graphs show the mean +/− standard deviation for three biological replicates, each consisting of a pool of embryos. A one tailed t-test was performed on linear expression values normalised to GAPDH to test the hypothesis that expression of selected genes is reduced in the mutant relative to wildtype.
In situ hybridisation
Embryos were harvested at E8.5 and fixed in 4% paraformaldehyde, 0.2% glutaraldehyde for 3 hours at 4 °C before being dehydrated and stored in methanol at −20 °C until use. Prior to hybridising, embryos were bleached in 6% H2O2 and digested for 6 minutes in 10 µg/ml proteinase K. Hybridisation was performed with digoxigenin-labelled RNA probes (2 µg/ml) at 70 °C following standard protocols72. Probes used are listed in Supplementary Table 4.
Chip-Seq analysis
ChIP-seq data used in this paper are from published studies26,27; GEO accession numbers GSE61188, GSE74636. Binding peaks were visualised using the Integrated Genome Viewer software73.
Luciferase assays
All assays were performed with U2-OS cells (obtained from ATCC) grown in DMEM with 10% FBS. Cells were tested and confirmed free of mycoplasma contamination. Luciferase reporters carrying Nodal enhancers linked to a viral E1b promoter have been previously described49. Generation of Y401X ZIC2 and deletion of ZIC2 binding sites within HBE was performed by site directed mutagenesis (NEB) using primers listed in Supplemental Table 5. Cells were transfected using Fugene reagent (Promega) with either HA-ZIC2 (Zic2), HA-ZIC3 (Zic3), Y401X ZIC2 (Zic2-iso) or an empty vector (pcDNA) along with a 5× excess of reporter plasmid. A Renilla luciferase reporter was co-transfected to control for transfection efficiency. Cells were harvested after 48–72 hrs and assayed using the Dual Luciferase Reporter assay system (Promega). Each data point represents the mean of three technical replicates performed during the same experiment, and each experiment was repeated three times, as shown on the graphs. Asterisks indicate results of a one tailed t-test of samples with unequal variance testing the null hypothesis either that transfection with ZIC2 does not result in a change in activation of a given reporter over transfection with pcDNA (Fig. 5b,d) or that there is no difference between the two indicated conditions (Fig. 5c). *p < 0.05, NS = not significant.
EMSA assays
Crude protein extracts were derived from HEK293T cells (obtained from ATCC, tested for mycoplasma contamination) transfected with a Zic2-HA expression plasmid or a control pcDNA plasmid. Cells were lysed in a solution of 10 mM HEPES pH7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT and nuclei pelleted by centrifugation. Nuclei were lysed in 20 mM HEPES pH7.9, 25% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT. Short 31 bp double-stranded biotin labelled probes (listed in Supplemental Table 6) were synthesised and EMSA assays performed using the Lightshift Chemiluminescent EMSA kit (Life Technologies). Unlabelled oligos were used as competitors and a supershift was performed with a monoclonal αHA antibody (Covance #MMS-101R).
Data availability
The microarray datasets generated in the current study are available in the GEO repository, accession number GSE106350. Luciferase constructs and mouse alleles described in this work are available on request.
Electronic supplementary material
Acknowledgements
We thank Dominic Norris, Hiroshi Hamada, and Jose Belo for sharing in situ probes, Jun Aruga for sharing expression plasmids and Aude Vernet and Victoria Thornton for MRI/µCT assistance. This work was supported by the British Heart Foundation [RE/13/1/30181 to I.M.D, RG/10/17/28553 to S.B., CH/09/003/26631 to S.B., RM/13/3/30159 to S.B., RE/08/004 to J.E.S., FS/11/50/29038 to J.E.S]; the Wellcome Trust [083228 to S.B., 090532/Z/09/Z to S.B., 077012/Z/05/Z to D.J.A.] and the Agence Nationale de la Recherche [ANR-11-BSV2-0028 to J.C.].
Author Contributions
I.M.D.: designed experiments, performed experiments, analysed data, wrote paper. D.S.: performed experiments, analysed data. L.K.: performed experiments, maintained mouse colony. R.P.: maintained mouse colony. C.C.: performed experiments. C.P.: contributed luciferase reporter constructs. H.L.: performed statistical analysis of microarray data. C.D.: shared human genetics data (Table 1). V.D.: shared human genetics data (Table 1). J.S.: developed CT imaging technology used in Figures 1,2. T.M.K.: performed exome capture of iso allele (Sup. Table 1). D.J.A.: designed exome capture experiment (Sup. Table 1). S.D.M.B.: designed ENU mutagenesis screen. S.M.: shared human genetics data (Table 1). S.O.: shared human genetics data (Table 1). J.C.: wrote paper, contributed luciferase reporter constructs. S.B.: designed experiments, obtained funding for study, wrote paper.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-28714-1.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Iain M. Dykes, Email: I.M.Dykes@ljmu.ac.uk
Shoumo Bhattacharya, Email: sbhattac@well.ox.ac.uk.
References
- 1.Brown SA, et al. Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nat Genet. 1998;20:180–183. doi: 10.1038/2484. [DOI] [PubMed] [Google Scholar]
- 2.Solomon BD, et al. Mutations in ZIC2 in human holoprosencephaly: description of a novel ZIC2 specific phenotype and comprehensive analysis of 157 individuals. J Med Genet. 2010;47:513–524. doi: 10.1136/jmg.2009.073049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mercier S, et al. New findings for phenotype-genotype correlations in a large European series of holoprosencephaly cases. J Med Genet. 2011;48:752–760. doi: 10.1136/jmedgenet-2011-100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warr N, et al. Zic2-associated holoprosencephaly is caused by a transient defect in the organizer region during gastrulation. Hum Mol Genet. 2008;17:2986–2996. doi: 10.1093/hmg/ddn197. [DOI] [PubMed] [Google Scholar]
- 5.Nagai T, et al. Zic2 regulates the kinetics of neurulation. Proc Natl Acad Sci USA. 2000;97:1618–1623. doi: 10.1073/pnas.97.4.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elms P, et al. Overlapping and distinct expression domains of Zic2 and Zic3 during mouse gastrulation. Gene Expr Patterns. 2004;4:505–511. doi: 10.1016/j.modgep.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Conlon FL, et al. A primary requirement for nodal in the formation and maintenance of the primitive streak in the mouse. Development. 1994;120:1919–1928. doi: 10.1242/dev.120.7.1919. [DOI] [PubMed] [Google Scholar]
- 8.Houtmeyers, R. et al. Zic2 mutation causes holoprosencephaly via disruption of NODAL signalling. Hum Mol Genet, 10.1093/hmg/ddw235 (2016). [DOI] [PubMed]
- 9.Norris DP. Cilia, calcium and the basis of left-right asymmetry. BMC Biol. 2012;10:102. doi: 10.1186/1741-7007-10-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nonaka S, Shiratori H, Saijoh Y, Hamada H. Determination of left-right patterning of the mouse embryo by artificial nodal flow. Nature. 2002;418:96–99. doi: 10.1038/nature00849. [DOI] [PubMed] [Google Scholar]
- 11.Collignon J, Varlet I, Robertson EJ. Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature. 1996;381:155–158. doi: 10.1038/381155a0. [DOI] [PubMed] [Google Scholar]
- 12.Lowe LA, et al. Conserved left-right asymmetry of nodal expression and alterations in murine situs inversus. Nature. 1996;381:158–161. doi: 10.1038/381158a0. [DOI] [PubMed] [Google Scholar]
- 13.Brennan J, Norris DP, Robertson EJ. Nodal activity in the node governs left-right asymmetry. Genes Dev. 2002;16:2339–2344. doi: 10.1101/gad.1016202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saijoh Y, et al. Distinct transcriptional regulatory mechanisms underlie left-right asymmetric expression of lefty-1 and lefty-2. Genes Dev. 1999;13:259–269. doi: 10.1101/gad.13.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ammirabile G, et al. Pitx2 confers left morphological, molecular, and functional identity to the sinus venosus myocardium. Cardiovasc Res. 2012;93:291–301. doi: 10.1093/cvr/cvr314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen CM, Norris D, Bhattacharya S. Transcriptional control of left-right patterning in cardiac development. Pediatr Cardiol. 2010;31:371–377. doi: 10.1007/s00246-009-9610-3. [DOI] [PubMed] [Google Scholar]
- 17.Dykes IM. Left Right Patterning, Evolution and Cardiac Development. J. Cardiovasc. Dev. Dis. 2014;1:52–72. doi: 10.3390/jcdd1010052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramsdell AF. Left-right asymmetry and congenital cardiac defects: getting to the heart of the matter in vertebrate left-right axis determination. Dev Biol. 2005;288:1–20. doi: 10.1016/j.ydbio.2005.07.038. [DOI] [PubMed] [Google Scholar]
- 19.Nakamura T, Hamada H. Left-right patterning: conserved and divergent mechanisms. Development. 2012;139:3257–3262. doi: 10.1242/dev.061606. [DOI] [PubMed] [Google Scholar]
- 20.Shiratori H, Hamada H. The left-right axis in the mouse: from origin to morphology. Development. 2006;133:2095–2104. doi: 10.1242/dev.02384. [DOI] [PubMed] [Google Scholar]
- 21.Campione, M. & Franco, D. Current Perspectives in CardiacLaterality. J Cardiovasc Dev Dis 310.3390/jcdd3040034 (2016). [DOI] [PMC free article] [PubMed]
- 22.Sutherland MJ, Ware SM. Disorders of left-right asymmetry: heterotaxy and situs inversus. Am J Med Genet C Semin Med Genet. 2009;151C:307–317. doi: 10.1002/ajmg.c.30228. [DOI] [PubMed] [Google Scholar]
- 23.Mohapatra B, et al. Identification and functional characterization of NODAL rare variants in heterotaxy and isolated cardiovascular malformations. Hum Mol Genet. 2009;18:861–871. doi: 10.1093/hmg/ddn411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barratt KS, Glanville-Jones HC, Arkell RM. The Zic2 gene directs the formation and function of node cilia to control cardiac situs. Genesis. 2014;52:626–635. doi: 10.1002/dvg.22767. [DOI] [PubMed] [Google Scholar]
- 25.Campione M, et al. Pitx2 expression defines a left cardiac lineage of cells: evidence for atrial and ventricular molecular isomerism in the iv/iv mice. Dev Biol. 2001;231:252–264. doi: 10.1006/dbio.2000.0133. [DOI] [PubMed] [Google Scholar]
- 26.Matsuda, K. et al. ChIP-seq analysis of genomic binding regions of five major transcription factors in mouse epiblast stem cells that highlights a central role for ZIC2. Development, 10.1242/dev.143479 (2017). [DOI] [PMC free article] [PubMed]
- 27.Luo Z, et al. Zic2 is an enhancer-binding factor required for embryonic stem cell specification. Mol Cell. 2015;57:685–694. doi: 10.1016/j.molcel.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kojima Y, et al. The transcriptional and functional properties of mouse epiblast stem cells resemble the anterior primitive streak. Cell Stem Cell. 2014;14:107–120. doi: 10.1016/j.stem.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 29.Buecker C, et al. Reorganization of enhancer patterns in transition from naive to primed pluripotency. Cell Stem Cell. 2014;14:838–853. doi: 10.1016/j.stem.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu G, Wade PA. NuRD and pluripotency: a complex balancing act. Cell Stem Cell. 2012;10:497–503. doi: 10.1016/j.stem.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies B, et al. Site specific mutation of the Zic2 locus by microinjection of TALEN mRNA in mouse CD1, C3H and C57BL/6J oocytes. PLoS One. 2013;8:e60216. doi: 10.1371/journal.pone.0060216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bamforth SD, et al. Cited2 controls left-right patterning and heart development through a Nodal-Pitx2c pathway. Nat Genet. 2004;36:1189–1196. doi: 10.1038/ng1446. [DOI] [PubMed] [Google Scholar]
- 33.Scambler PJ. 22q11 deletion syndrome: a role for TBX1 in pharyngeal and cardiovascular development. Pediatric cardiology. 2010;31:378–390. doi: 10.1007/s00246-009-9613-0. [DOI] [PubMed] [Google Scholar]
- 34.Dykes IM, et al. HIC2 is a novel dosage-dependent regulator of cardiac development located within the distal 22q11 deletion syndrome region. Circ Res. 2014;115:23–31. doi: 10.1161/CIRCRESAHA.115.303300. [DOI] [PubMed] [Google Scholar]
- 35.Szumska D, et al. VACTERL/caudal regression/Currarino syndrome-like malformations in mice with mutation in the proprotein convertase Pcsk5. Genes Dev. 2008;22:1465–1477. doi: 10.1101/gad.479408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider JE, et al. Identification of cardiac malformations in mice lacking Ptdsr using a novel high-throughput magnetic resonance imaging technique. BMC Dev Biol. 2004;4:16. doi: 10.1186/1471-213X-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Elms P, Siggers P, Napper D, Greenfield A, Arkell R. Zic2 is required for neural crest formation and hindbrain patterning during mouse development. Dev Biol. 2003;264:391–406. doi: 10.1016/j.ydbio.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 38.Seo JW, Brown NA, Ho SY, Anderson RH. Abnormal laterality and congenital cardiac anomalies. Relations of visceral and cardiac morphologies in the iv/iv mouse. Circulation. 1992;86:642–650. doi: 10.1161/01.CIR.86.2.642. [DOI] [PubMed] [Google Scholar]
- 39.Hildreth V, et al. Left cardiac isomerism in the Sonic hedgehog null mouse. J Anat. 2009;214:894–904. doi: 10.1111/j.1469-7580.2009.01087.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meno C, et al. lefty-1 is required for left-right determination as a regulator of lefty-2 and nodal. Cell. 1998;94:287–297. doi: 10.1016/S0092-8674(00)81472-5. [DOI] [PubMed] [Google Scholar]
- 41.Yan YT, et al. Conserved requirement for EGF-CFC genes in vertebrate left-right axis formation. Genes Dev. 1999;13:2527–2537. doi: 10.1101/gad.13.19.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartram U, Wirbelauer J, Speer CP. Heterotaxy syndrome–asplenia and polysplenia as indicators of visceral malposition and complex congenital heart disease. Biol Neonate. 2005;88:278–290. doi: 10.1159/000087625. [DOI] [PubMed] [Google Scholar]
- 43.Oh SP, Li E. The signaling pathway mediated by the type IIB activin receptor controls axial patterning and lateral asymmetry in the mouse. Genes Dev. 1997;11:1812–1826. doi: 10.1101/gad.11.14.1812. [DOI] [PubMed] [Google Scholar]
- 44.Duboc V, Rottinger E, Lapraz F, Besnardeau L, Lepage T. Left-right asymmetry in the sea urchin embryo is regulated by nodal signaling on the right side. Dev Cell. 2005;9:147–158. doi: 10.1016/j.devcel.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 45.Ryan AK, et al. Pitx2 determines left-right asymmetry of internal organs in vertebrates. Nature. 1998;394:545–551. doi: 10.1038/29004. [DOI] [PubMed] [Google Scholar]
- 46.Kawasumi A, et al. Left-right asymmetry in the level of active Nodal protein produced in the node is translated into left-right asymmetry in the lateral plate of mouse embryos. Dev Biol. 2011;353:321–330. doi: 10.1016/j.ydbio.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Norris DP, Brennan J, Bikoff EK, Robertson EJ. The Foxh1-dependent autoregulatory enhancer controls the level of Nodal signals in the mouse embryo. Development. 2002;129:3455–3468. doi: 10.1242/dev.129.14.3455. [DOI] [PubMed] [Google Scholar]
- 48.Adachi H, et al. Determination of left/right asymmetric expression of nodal by a left side-specific enhancer with sequence similarity to a lefty-2 enhancer. Genes Dev. 1999;13:1589–1600. doi: 10.1101/gad.13.12.1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papanayotou C, et al. A novel nodal enhancer dependent on pluripotency factors and smad2/3 signaling conditions a regulatory switch during epiblast maturation. PLoS Biol. 2014;12:e1001890. doi: 10.1371/journal.pbio.1001890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vincent SD, Dunn NR, Hayashi S, Norris DP, Robertson EJ. Cell fate decisions within the mouse organizer are governed by graded Nodal signals. Genes Dev. 2003;17:1646–1662. doi: 10.1101/gad.1100503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Norris DP, Robertson EJ. Asymmetric and node-specific nodal expression patterns are controlled by two distinct cis-acting regulatory elements. Genes Dev. 1999;13:1575–1588. doi: 10.1101/gad.13.12.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Purandare SM, et al. A complex syndrome of left-right axis, central nervous system and axial skeleton defects in Zic3 mutant mice. Development. 2002;129:2293–2302. doi: 10.1242/dev.129.9.2293. [DOI] [PubMed] [Google Scholar]
- 53.Jiang Z, et al. Zic3 is required in the extra-cardiac perinodal region of the lateral plate mesoderm for left-right patterning and heart development. Hum Mol Genet. 2013;22:879–889. doi: 10.1093/hmg/dds494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ware SM, Harutyunyan KG, Belmont JW. Heart defects in X-linked heterotaxy: evidence for a genetic interaction of Zic3 with the nodal signaling pathway. Dev Dyn. 2006;235:1631–1637. doi: 10.1002/dvdy.20719. [DOI] [PubMed] [Google Scholar]
- 55.Brown L, Paraso M, Arkell R, Brown S. In vitro analysis of partial loss-of-function ZIC2 mutations in holoprosencephaly: alanine tract expansion modulates DNA binding and transactivation. Hum Mol Genet. 2005;14:411–420. doi: 10.1093/hmg/ddi037. [DOI] [PubMed] [Google Scholar]
- 56.Lowe LA, Yamada S, Kuehn MR. Genetic dissection of nodal function in patterning the mouse embryo. Development. 2001;128:1831–1843. doi: 10.1242/dev.128.10.1831. [DOI] [PubMed] [Google Scholar]
- 57.Beckers A, Alten L, Viebahn C, Andre P, Gossler A. The mouse homeobox gene Noto regulates node morphogenesis, notochordal ciliogenesis, and left right patterning. Proc Natl Acad Sci USA. 2007;104:15765–15770. doi: 10.1073/pnas.0704344104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saijoh Y, Oki S, Ohishi S, Hamada H. Left-right patterning of the mouse lateral plate requires nodal produced in the node. Dev Biol. 2003;256:160–172. doi: 10.1016/S0012-1606(02)00121-5. [DOI] [PubMed] [Google Scholar]
- 59.Shiratori H, Hamada H. TGFbeta signaling in establishing left-right asymmetry. Semin Cell Dev Biol. 2014;32:80–84. doi: 10.1016/j.semcdb.2014.03.029. [DOI] [PubMed] [Google Scholar]
- 60.Tanaka C, Sakuma R, Nakamura T, Hamada H, Saijoh Y. Long-range action of Nodal requires interaction with GDF1. Genes Dev. 2007;21:3272–3282. doi: 10.1101/gad.1623907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kitajima K, Oki S, Ohkawa Y, Sumi T, Meno C. Wnt signaling regulates left-right axis formation in the node of mouse embryos. Dev Biol. 2013;380:222–32. doi: 10.1016/j.ydbio.2013.05.011. [DOI] [PubMed] [Google Scholar]
- 62.Field S, et al. Pkd1l1 establishes left-right asymmetry and physically interacts with Pkd2. Development. 2011;138:1131–1142. doi: 10.1242/dev.058149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Raya A, et al. Notch activity induces Nodal expression and mediates the establishment of left-right asymmetry in vertebrate embryos. Genes Dev. 2003;17:1213–1218. doi: 10.1101/gad.1084403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Crocker J, Noon EP, Stern DL. The Soft Touch: Low-Affinity Transcription Factor Binding Sites in Development and Evolution. Curr Top Dev Biol. 2016;117:455–469. doi: 10.1016/bs.ctdb.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 65.Bentham J, et al. Maternal high-fat diet interacts with embryonic Cited2 genotype to reduce Pitx2c expression and enhance penetrance of left-right patterning defects. Hum Mol Genet. 2010;19:3394–3401. doi: 10.1093/hmg/ddq251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schneider JE, et al. Rapid identification and 3D reconstruction of complex cardiac malformations in transgenic mouse embryos using fast gradient echo sequence magnetic resonance imaging. J Mol Cell Cardiol. 2003;35:217–222. doi: 10.1016/S0022-2828(02)00291-2. [DOI] [PubMed] [Google Scholar]
- 67.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–104. doi: 10.1093/bioinformatics/18.suppl_1.S96. [DOI] [PubMed] [Google Scholar]
- 69.Smyth, G. K. In Bioinformatics and Computational Biology Solutions using R and Bioconductor (ed. R. C. Gentleman) 397–420 (Springer-Verlag, 2005).
- 70.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- 71.Ulitsky I, et al. Expander: from expression microarrays to networks and functions. Nat Protoc. 2010;5:303–322. doi: 10.1038/nprot.2009.230. [DOI] [PubMed] [Google Scholar]
- 72.Correia KM, Conlon RA. Whole-mount in situ hybridization to mouse embryos. Methods. 2001;23:335–338. doi: 10.1006/meth.2000.1145. [DOI] [PubMed] [Google Scholar]
- 73.Robinson JT, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The microarray datasets generated in the current study are available in the GEO repository, accession number GSE106350. Luciferase constructs and mouse alleles described in this work are available on request.