Significance
Immune evasion by the human pathogen Staphylococcus aureus includes the synthesis of cytotoxic deoxyadenosine (dAdo) from neutrophil extracellular traps to eliminate macrophages. The signaling pathway that triggers dAdo-mediated apoptosis in macrophages is not known. By using a genome-wide CRISPR-Cas9 knockout screen, we demonstrate that S. aureus-derived dAdo targets the purine salvage pathway to provoke caspase-3–dependent cell death in human macrophages, thereby antagonizing host immune responses and promoting the establishment of invasive disease. Interference with the staphylococcal dAdo signaling pathway may boost macrophage survival and phagocytic clearance of bacteria, providing new strategies for the treatment of S. aureus infections.
Keywords: adenosine synthase A, nuclease, deoxyadenosine, human equilibrative nucleoside transporter 1, adenosine kinase
Abstract
Staphylococcus aureus colonizes large segments of the human population and causes invasive infections due to its ability to escape phagocytic clearance. During infection, staphylococcal nuclease and adenosine synthase A convert neutrophil extracellular traps to deoxyadenosine (dAdo), which kills phagocytes. The mechanism whereby staphylococcal dAdo intoxicates phagocytes is not known. Here we used CRISPR-Cas9 mutagenesis to show that phagocyte intoxication involves uptake of dAdo via the human equilibrative nucleoside transporter 1, dAdo conversion to dAMP by deoxycytidine kinase and adenosine kinase, and signaling via subsequent dATP formation to activate caspase-3–induced cell death. Disruption of this signaling cascade confers resistance to dAdo-induced intoxication of phagocytes and may provide therapeutic opportunities for the treatment of infections caused by antibiotic-resistant S. aureus strains.
The pathogen Staphylococcus aureus persistently colonizes the nasopharynx of large segments of the human population and is also a frequent cause of soft tissue infections, pneumonia, osteomyelitis, septic arthritis, bacteremia, endocarditis, and sepsis (1–3). S. aureus colonization represents a key risk factor for invasive disease, which in hospital environments frequently manifests as surgical wound and medical device infections or ventilator-asscociated pneumonia (4, 5). Owing to the high incidence of hospital-acquired infection, antibiotics are used both for S. aureus decolonization and prophylaxis of nosocomial disease (6, 7). However, large-scale use of antibiotics selects for antibiotic-resistant strains, designated methicillin-resistant S. aureus (MRSA) (8). Due to limited efficacy of antibiotics to eradicate drug-resistant strains, MRSA infections, i.e., 22% of hospital-acquired S. aureus infections in the United States, are associated with increased morbidity and mortality, compared with infections caused by antibiotic-sensitive strains (1).
The hallmark of S. aureus disease is the formation of abscess lesions, where staphylococci replicate within fibrin-encapsulated communities and attract neutrophils and other phagocytes to implement the purulent destruction of host tissues (9). Activated neutrophils release extracellular traps (neutrophil extracellular traps, NETs), an extracellular matrix composed of nuclear and mitochondrial DNA armed with granular proteins, cell-specific proteases, and antimicrobial peptides (10, 11). NETs rapidly immobilize and kill many different bacterial pathogens (10); however S. aureus evolved escape strategies, using secreted proteases and nuclease (Nuc) to degrade antimicrobial peptides and DNA (12, 13). Interestingly, nuclease-mediated degradation of neutrophil NETs triggers the formation of deoxyadenosine monophosphate (dAMP), which is converted by S. aureus adenosine synthase A (AdsA) into deoxyadenosine (dAdo) (14). Macrophages and other immune cells are highly sensitive to dAdo intoxication, enabling staphylococci to block phagocyte infiltration of mouse abscess lesions and preventing bacterial clearance in vivo (14, 15). Recent work revealed further that other bacterial pathogens, including pathogenic bacilli and streptococcci, utilize secreted nucleases and 5′-nucleotidases (homologs of S. aureus AdsA) to escape phagocyte clearance, suggesting that dAdo production represents a general immune evasion mechanism for microbial pathogens (16–18). However, the mechanisms whereby S. aureus- or other pathogen-derived dAdo is able to kill phagocytes is not known.
Here, we used a genome-wide CRISPR-Cas9 knockout screen to identify genes required for dAdo intoxication. The results suggest that S. aureus targets the purine salvage pathway to eliminate host phagocytes. The identification of genes affecting phagocyte intoxication may aid in the development of therapeutic strategies that can improve the outcome of MRSA infections.
Results
A CRISPR-Cas9 Screen Identifies Genes Contributing to Deoxyadenosine Intoxication of Phagocytes.
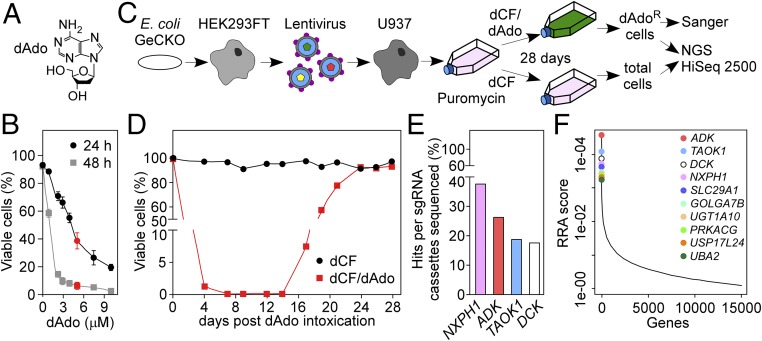
To conduct CRISPR-Cas9 knockout screens in the human U937 macrophage cell line, we determined the concentration of dAdo required to kill U937 cells within 24 and 48 h of incubation, respectively (Fig. 1 A and B). Human genome-scale CRISPR-Cas9 knockout (GeCKO) (19, 20) library-transduced U937 cells were treated with 5 μM dAdo or left untreated (nontreatment control) (Fig. 1 C and D). U937 cells with defective genes whose products otherwise contribute to intoxication are expected to proliferate in the presence of dAdo, leading to the enrichment of the corresponding small-guide RNAs (sgRNA) in treatment samples compared with control samples. This experimental scheme led to the isolation of dAdo-resistant U937 cell populations after 28 d of dAdo selection (Fig. 1D). Next, cells were subjected to a combined strategy of rapid Sanger sequencing of cloned sgRNAs and of next generation sequencing (NGS) of all sgRNAs in the samples (Fig. 1C). Sanger sequencing of 80 independent Escherichia coli-derived plasmids containing the sgRNA cassette amplified from dAdo-resistant cell populations identified sgRNAs targeting genes encoding for adenosine kinase (ADK), deoxycytidine kinase (DCK), TAO kinase 1 (TAOK1), and neurexophilin 1 (NXPH1) (Fig. 1E). NGS identified ADK, TAOK1, DCK, NXPH1, and SLC29A1 as the top hits of the CRISPR-Cas9 screen (Fig. 1F and Dataset S1). Deep sequencing revealed further that nontreatment control cells contained 96.13% of all sgRNAs present in the original GeCKO library, whereas dAdo treatment decreased sgRNAs below 2%. Two additional CRISPR-Cas9 screens for dAdo-resistant U937 cells generated results similar to those presented in Dataset S1. Together these results suggest that dAdo intoxication of U937 cells enriched for sgRNAs targeting five human genes (ADK, TAOK1, DCK, NXPH1, and SLC29A1) and that mutations in these genes may provide for phagocyte resistance to dAdo intoxication.
Fig. 1.
Genome-wide CRISPR-Cas9 screen for mutations conferring resistance to cytotoxic deoxyadenosine in human macrophages. (A) Chemical structure of dAdo. (B) Survival of U937 cells treated with variable concentrations of dAdo (0–10 μM) in the presence of 50 μM dCF (2′ deoxycoformycin or pentostatin), an inhibitor of adenosine deaminase. Cell viability was analyzed after 24 h and 48 h of incubation; 5 μM dAdo (red) was used for selection of resistant variants. Data points represent the mean (±SD) of three independent determinations. (C) Experimental scheme for CRISPR-Cas9 knockout screen to identify genes conferring resistance to deoxyadenosine (dAdo) intoxication in U937 cells. (D) Analysis of cell viability for GeCKO library-transduced U937 cells intoxicated with 5 μM dAdo and 50 μM dCF for 28 d (red squares) or treated with 50 μM dCF alone (black circles). (E) Distribution of sgRNAs in dAdo-resistant U937 cells identified by Sanger sequencing of 80 plasmids containing cloned sgRNA cassettes. (F) Identification of the top candidate genes following dAdo treatment of U937 cells by next generation sequencing. Data were analyzed using the MaGeCK-based robust rank aggregation (RRA) score analysis.
hENT1 Mediates Uptake of Cytotoxic Deoxyadenosine into Macrophages.
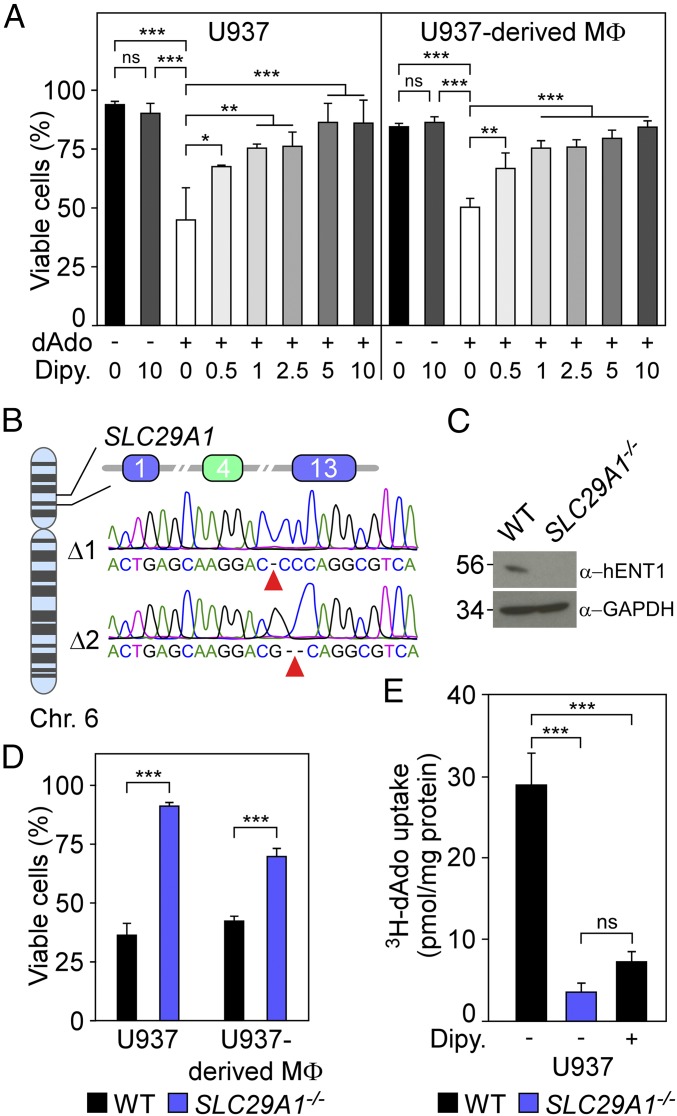
Secreted NXPH1 inhibits the proliferation of hematopoietic progenitor cells via neurexin 1α (NRXN1α) signaling (21). To test whether NXPH1 contributes to dAdo-mediated killing of phagocytes, U937 cells or U937-derived macrophages were incubated with human NXPH1, and cell viability was recorded. NXPH1 did not induce macrophage cell death (SI Appendix, Fig. S1). Conversely, inhibition of hENT1 (encoded by SLC29A1), a polytopic membrane protein, with dipyridamole (22) rendered U937 cells and U937-derived macrophages resistant to dAdo-induced cell death, suggesting that dipyridamole-mediated inhibition of hENT1 transport activity protects macrophages from dAdo intoxication (Fig. 2A). Targeted CRISPR-Cas9 mutagenesis with a SLC29A1-specific sgRNA was used to generate biallelic disruptions of SLC29A1, which were confirmed by sequencing exon 4 on human chromosome 6 (Fig. 2B). SLC29A1−/− cells were further analyzed by immunoblotting with hENT1-specific antibodies, which confirmed that hENT1 production had been abolished (Fig. 2C). Of note, U937 SLC29A1−/− cells as well as U937-derived SLC29A1−/− macrophages were resistant to dAdo intoxication (Fig. 2D). Together these data indicate that macrophage expression of hENT1 is essential for dAdo-mediated intoxication of these cells.
Fig. 2.
Human equilibrative transporter 1 (hENT1) activity is required for dAdo-induced killing of human macrophages. (A) Survival of U937 cells and U937-derived macrophages (MΦ) treated with dAdo (+) and increasing concentrations of dipyridamole (Dipy) (0–10 μM), an inhibitor of hENT1. All samples received adenosine deaminase inhibitor (50 μM dCF). (B) Diagram illustrating the position of SLC29A1 on chromosome 6 and exons 1, 4, and 13 of SLC29A1 mRNA. Sequencing results for mutated exon 4 alleles (green box) cloned from SLC29A1−/− cells. (C) Immunoblotting of lysates from wild-type (WT) U937 and SLC29A1−/− variant cells with hENT1- and GAPDH-specific antibodies (α-hENT1, α-GAPDH). Numbers to the Left of blots indicate the migration of molecular weight markers in kilodaltons. (D) Survival of U937 cells or U937-derived macrophages (MΦ) (black bars) and their SLC29A1−/− (blue bars) variants after 24-h dAdo treatment. (E) Uptake of added [3H]dAdo into U937 cells or their SLC29A1−/− variants was quantified by liquid scintillation counting (LSC) of cell lysates. As control, hENT1-mediated import of [3H]dAdo into U937 cells was blocked with 10 μM Dipy. Data are the mean (±SD) of three independent determinations. Statistically significant differences were analyzed by one-way ANOVA with Bonferroni’s multiple comparison (A and E) or by Student’s t test (D). ns, not significant (P > 0.05); *P ≤ 0.05; **P < 0.01; ***P < 0.001.
As a member of the human equilibrative transporter family, hENT1 mediates uptake of nucleosides and of nucleoside derivatives used for the treatment of cancer and viral infections (23). Recent work demonstrated that hENT1 binds to dAdo in hENT1-overexpressing HEK293F cells (24). To test whether hENT1 transports dAdo into macrophages, we incubated U937 cells with [3H]dAdo and measured uptake via scintillation analysis of cell lysates. U937 cells imported extracellular [3H]dAdo into their cytoplasm (Fig. 2E). Mutations that abrogate hENT1 production abolished [3H]dAdo transport, as only small amounts of [3H]dAdo were detectable in the cytoplasm of SLC29A1−/− cells (Fig. 2E). Similarly, incubation with dipyridamole blocked [3H]dAdo transport into U937 cells, suggesting that hENT1 transports cytotoxic dAdo across the plasma membrane (Fig. 2E).
Purine Salvage Pathway Kinases Are Required for Deoxyadenosine Intoxication of Macrophages.
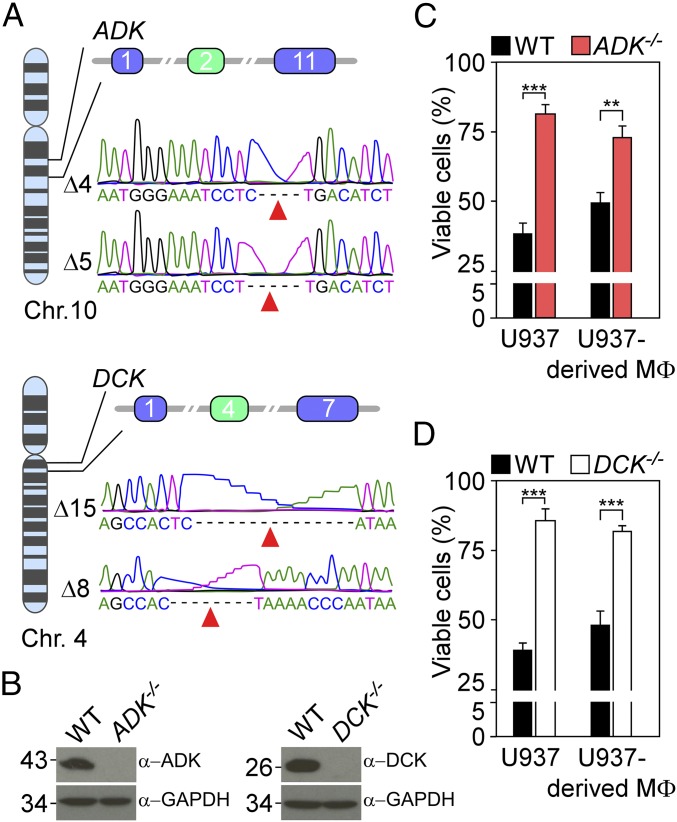
Earlier work reported that TAOK1 activates the p38 MAP kinase pathway (25). To test whether TAOK1 and the p38 MAP kinase pathway are involved in dAdo-mediated killing of macrophages, we incubated U937 cells and their derived macrophages with SB 203580, a p38-specific inhibitor. SB 203580 caused modest increases in the survival of dAdo-treated macrophages only at high concentrations, suggesting that the p38 MAP kinase pathway and TAOK1 do not play a major role in the killing of macrophages (SI Appendix, Fig. S2). To explore the role of ADK and DCK in dAdo-mediated cell death of macrophages, we first sought to inhibit the corresponding protein products. The substance 5′-amino-5′-deoxyadenosine is an inhibitor of ADK (26), whereas deoxycytidine is the physiological substrate and competitive inhibitor of DCK, but not of ADK. Addition of 5′-amino-5′-deoxyadenosine or deoxycytidine each reduced dAdo-mediated killing of U937 cells or U937-derived macrophages (SI Appendix, Fig. S3). To validate these observations, we used CRISPR-Cas9 mutagenesis and specific sgRNAs to generate U937 cells with mutations in the corresponding structural genes, which were confirmed by exon sequencing of the mutant alleles (Fig. 3A). Immunoblotting experiments with specific antibodies further confirmed the absence of ADK and DCK in ADK−/− and DCK−/− mutant cells, respectively (Fig. 3B). Both ADK−/− and DCK−/− cells and macrophages exhibited increased resistance to dAdo intoxication (Fig. 3 C and D). U937 cells and differentiated macrophages carrying mutations in the structural genes for both ADK and DCK exhibited increased resistance to dAdo-induced cell death (SI Appendix, Fig. S4). dAdo resistance in ADK−/−, DCK−/−, or DCK−/− ADK−/− cells was not associated with a defect in the uptake of [3H]dAdo, indicating that hENT1 is fully functional in cells with defects in the purine salvage pathway (Fig. 4A). Together these experiments suggest that purine salvage pathway kinases are required for dAdo-mediated intoxication of macrophages.
Fig. 3.
Purine salvage pathway kinases are required for dAdo intoxication of macrophages. (A) Diagram illustrating the position of ADK on chromosome 10 and exons 1, 2, and 11 of ADK mRNA as well as the position of DCK on chromosome 4 and exons 1, 4, and 7 of DCK mRNA. Sequencing results for mutated exon 2 alleles cloned from ADK−/− cells and mutated exon 4 alleles cloned from DCK−/− cells (green boxes). (B) Immunoblotting of lysates from wild-type (WT) U937 cells and their ADK−/− and DCK−/− variants with ADK-, DCK-, and GAPDH-specific antibodies. Numbers to the Left of blots indicate the migration of molecular weight markers in kilodaltons. (C and D) Survival of U937 cells and U937-derived macrophages (MΦ) (black bars) and their ADK−/− (red bars) and their DCK−/− (white bars) variants after 24-h dAdo treatment. Data are the mean (±SD) of three independent determinations. Statistically significant differences were analyzed with the Student’s t test. ns, not significant (P > 0.05); **P < 0.01; ***P < 0.001.
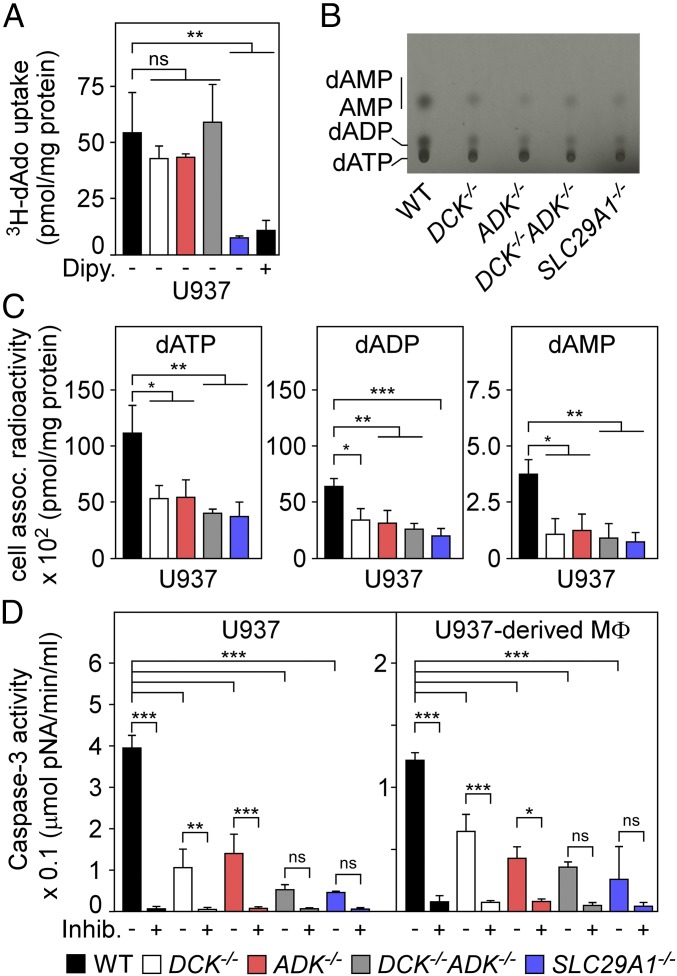
Fig. 4.
Mutations disrupting the purine salvage pathway of deoxyadenosine-intoxicated macrophages provide resistance to caspase-3–induced cell death. (A) Uptake of [3H]dAdo into wild-type U937 cells (WT, black) and their DCK−/− (white), ADK−/− (red), DCK−/− ADK−/− (gray), or SLC29A1−/− (blue) variants was quantified by LSC. As a control, dipyridamole (Dipy) (10 μM) inhibition of hENT1 blocked uptake of [3H]dAdo into WT U937 cells. (B) Analysis of [14C]dAdo-derived nucleotides from WT U937 cells and their DCK−/−, ADK−/−, DCK−/− ADK−/−, or SLC29A1−/− variants. An autoradiograph of the TLC plate is shown with migrational positions of dAMP, AMP, dADP, and dATP. (C) Nucleotides extracted from [14C]dAdo-treated cells were analyzed by TLC and LSC for the abundance of dATP, dADP, and dAMP. (D) WT U937 cells and U937-derived macrophages (MΦ) and their DCK−/−, ADK−/−, DCK−/− ADK−/−, or SLC29A1−/− variants were treated with dAdo and cell lysates were analyzed for caspase-3 activity using a colorimetric assay. As controls, lysates were treated with the caspase-3 inhibitor Ac-DEVD-CHO (+Inhib). All data are the mean (±SD) of three independent determinations. Statistically significant differences were analyzed with one-way ANOVA and Bonferroni’s multiple comparison test. ns, not significant (P > 0.05); *P ≤ 0.05; **P < 0.01; ***P < 0.001.
Disruption of the Purine Salvage Pathway Rescues Deoxyadenosine-Intoxicated Macrophages from Caspase-3–Induced Cell Death.
Cytoplasmic dAdo can be detoxified by adenosine deaminase (ADA), which catalyzes the irreversible conversion of dAdo to deoxyinosine (27). Alternatively, ADK and DCK can convert dAdo into dAMP for subsequent synthesis of dADP and dATP via adenylate kinase and nucleoside-diphosphate kinase, respectively. If so, S. aureus-mediated synthesis of dAdo may trigger not only import of dAdo into macrophages but also intracellular synthesis of dAMP and the accumulation of dADP and dATP. To test this, we extracted nucleotides from [14C]dAdo-treated U937 cells and analyzed the abundance of dAMP, dADP, and dATP via TLC. The experiments revealed the accumulation of [14C]dATP, [14C]dADP, [14C]dAMP, and [14C]AMP following incubation of U937 cells with [14C]dAdo (Fig. 4B). Compared with their U937 parent, ADK−/−, DCK−/−, DCK−/− ADK−/−, and SLC29A1−/− cells exhibited reduced abundance of intracellular [14C]dATP, [14C]dADP, and [14C]AMP (Fig. 4B). Quantification of [14C]dAdo-derived nucleotides using liquid scintillation counting (LSC) confirmed that [14C]dAdo exposure caused accumulation of dATP and its precursors (dADP/dAMP), which was diminished in ADK−/−, DCK−/−, DCK−/− ADK−/−, and SLC29A1−/− cells (Fig. 4C). Thus, mutations that disrupt the purine salvage pathway prevent the accumulation of dATP following dAdo exposure, suggesting that dATP may represent a key signaling molecule triggering cell death in macrophages. Earlier work suggested that intracellular dATP activates the apoptotic protease-activating factor 1 (APAF-1)/caspase-9 complex that further activates caspase-3 (28). To test whether dAdo-treatment increases caspase-3 activity in a manner requiring factors identified via CRISPR-Cas9 screening, U937 macrophages and their ADK−/−, DCK−/−, DCK−/− ADK−/−, and SLC29A1−/− variants were treated with dAdo, and caspase-3 activity in cell lysates was measured via the hydrolysis of the peptide substrate (Ac-DEVD-pNA) by caspase-3, resulting in the release of p-nitroaniline (pNA). Consistent with earlier studies, dAdo intoxication of U937 cells increased caspase-3 activity (Fig. 4D) (14, 29). In contrast, dAdo treatment of ADK−/−, DCK−/−, DCK−/− ADK−/−, or SLC29A1−/− mutant cells led to diminished caspase-3 activation (Fig. 4D). Similar results were observed with U937 macrophages and their ADK−/−, DCK−/−, DCK−/− ADK−/−, or SLC29A1−/− variants (Fig. 4D). Thus, dAdo import via hENT1 as well as ADK- and DCK-mediated synthesis of dAMP and accumulation of dADP and dATP activates the caspase-3 pathway to trigger apoptotic cell death in macrophages.
S. aureus-Derived Deoxyadenosine Targets the Purine Salvage Pathway to Kill Human Macrophages.
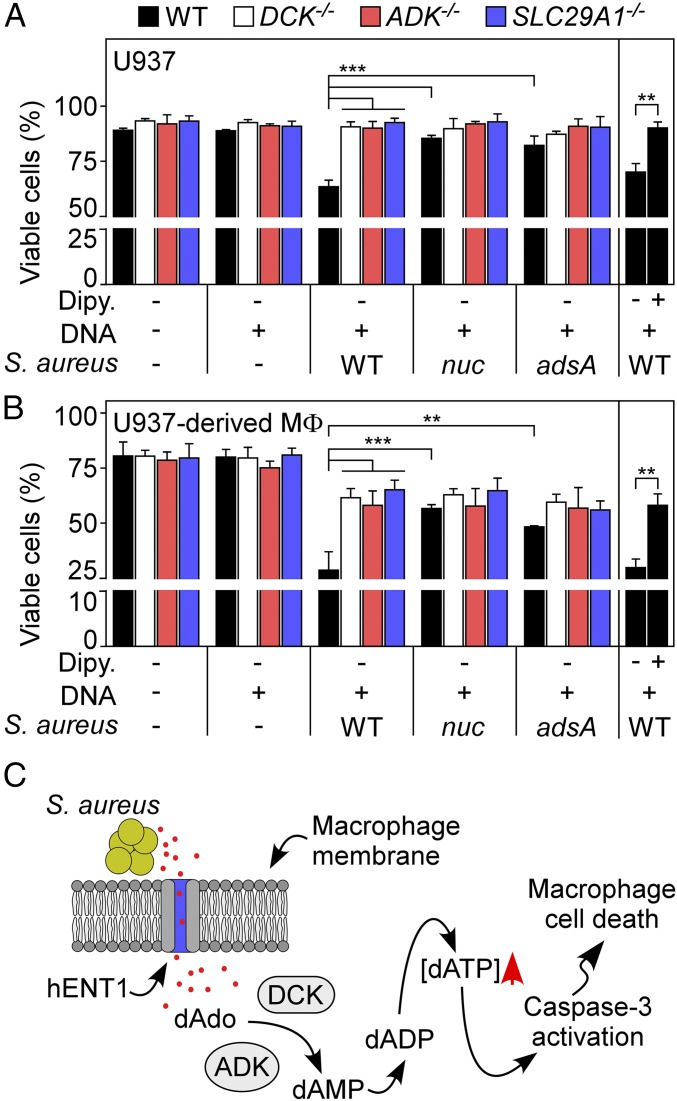
Finally, we asked whether U937 cells or macrophages lacking ADK, DCK, or hENT1 are resistant to S. aureus-derived dAdo. To test this, wild-type S. aureus Newman or its nuclease (nuc) and adenosine synthase (adsA) mutants were grown in chemically defined medium supplemented with host DNA. Samples were then centrifuged and bacteria-free, filter-sterilized conditioned media were then added to U937 cells or their genetic variants. As reported earlier, S. aureus and host DNA conditioned culture medium intoxicated U937 cells in a manner requiring staphylococcal nuclease and AdsA (Fig. 5A) (14). However, S. aureus-derived dAdo did not kill mutant cells lacking ADK, DCK, or hENT1 (Fig. 5A). Further, differentiated U937 macrophages lacking ADK, DCK, or hENT1 were also resistant to S. aureus-mediated dAdo intoxication (Fig. 5B). Last, treatment of U937 cells or macrophages with dipyridamole abrogated S. aureus-derived dAdo intoxication, indicating that hENT1-mediated transport of bacterial dAdo is essential for macrophage killing (Fig. 5 A and B). Together these data indicate that the S. aureus escapes macrophage-mediated phagocytosis and clearance from host tissues via secretion of Nuc and AdsA to produce dAdo from host DNA, targeting the purine salvage pathway for apoptotic destruction of phagocytes (Fig. 5C).
Fig. 5.
Staphylococcus aureus-generated deoxyadenosine kills macrophages by targeting the purine salvage pathway. Survival of U937 cells (A) and U937-derived macrophages (B) and their ADK−/−, DCK−/−, or SLC29A1−/− variants following treatment with culture medium (RPMI) that had been conditioned by incubation with either wild-type S. aureus Newman or its nuc or adsA mutants in the presence of host DNA or dipyridamole (Dipy) (10 μM) as indicated with + and − signs. Data are the mean (±SD) of three independent determinations. Statistically significant differences were analyzed with one-way ANOVA and Bonferroni’s multiple comparison test and with the Student’s t test for the ±Dipy experiment. ns, not significant (P > 0.05); **P < 0.01; ***P < 0.001. (C) Diagram illustrating S. aureus killing of macrophages via hENT1-mediated uptake of AdsA/Nuc-derived dAdo, ADK/DCK-catalyzed conversion of dAdo to dAMP, as well as subsequent conversions to dADP and dATP, triggering caspase-3 activation and macrophage apoptosis.
Discussion
Histopathological features of S. aureus invasive disease include infiltration of large numbers of neutrophils, a process that is dependent on staphylococcal lipoproteins as inducers of TLR2 signaling by host immune cells (30). Neutrophils discharge their degradative enzymes and NETs at the site of infection, triggering liquefaction necrosis and purulent drainage of abscess lesions (9, 31). Unless treated with antibiotics (and surgical drainage), staphylococci replicating in deep-seated abscess lesions cannot be cleared by the immune system, subsequently causing disseminated abscess lesions and lethal bacteremia (9, 31). In the United States, S. aureus causes each year 3.4 million community-acquired diseases and 460,000 hospital-acquired infections. Antibiotic-resistant MRSA infections (7% of community- and 22% of hospital-acquired staphylococcal disease) are associated with poor clinical outcomes and represent a frequent cause of mortality (1).
Development of new therapeutic strategies to combat MRSA infections requires deeper understanding of the immune evasive attributes that enable staphylococci to survive in host tissues. Unlike neutrophils, macrophages are not found in close proximity to staphylococcal abscess communities (14, 31). Exclusion of these immune cells is at odds with physiological clearance mechanisms, which rely on macrophages for the removal of neutrophil remnants and the killing of bacteria. Earlier work showed that S. aureus secretes nuclease, an enzyme that degrades neutrophil NETs into its monophosphate deoxynucleosides (12). Staphylococcal AdsA, a cell wall-anchored surface protein, catalyzes the conversion of adenosine nucleotides (released by damaged host tissues) to adenosine and of deoxyadenosine nucleotides (derived by nuclease digestion of neutrophils NETs) to deoxyadenosine (14, 18, 32). Increases in adenosine concentration dampen inflammatory and pathogen-specific immune responses, whereas increases in deoxyadenosine trigger macrophage apoptosis in the vicinity of staphylococcal abscess communities formed during the infection of mice (14, 18). Unlike neutrophils, macrophages and other myeloid cells retain the ability to synthesize DNA and divide. Due to reduced levels of de novo nucleotide synthesis, these cells rely on the import of nucleosides for their DNA synthesis and cell division.
To identify genetic determinants associated with S. aureus deoxyadenosine intoxication, we used CRISPR-Cas9 mutagenesis of human U937 macrophages and identified SLC29A1, ADK, and DCK as top hits. CRISPR-Cas9–specific sgRNAs were used to generate U937 macrophage variants unable to produce hENT1, ADK, or DCK, which resulted in resistance against dAdo-mediated killing. S. aureus intoxication involves hENT1 import of dAdo into macrophages, ADK/DCK catalyzed conversion of dAdo to dAMP, subsequent adenylate kinase- and nucleoside-diphosphate kinase-dependent accumulation of dADP and dATP, as well as caspase-3–induced apoptosis. Thus, S. aureus intoxication targets the purine salvage pathway of macrophages with rapid increases in intracellular dATP, likely activating caspase-3–induced apoptosis. Other signaling pathways may also contribute to macrophage cell death, including p53 activation via DNA damage. Nevertheless, adenylate kinase (AK), nucleoside-diphosphate kinase (NDPK), and caspase-3 (CASP3) were not identified as top hits in the CRISPR-Cas9 screen. Various isoenzymes have been described for AK and NDPK (AK1–9; NDPK-A–D), and accordingly, only deletion of all variants in a single cell may provide for dAdo resistance (33, 34). U937 CASP3−/− cells presumably remain sensitive to dAdo intoxication, as earlier work reported that CASP3−/− colon cancer cells are more sensitive to DNA damaging agents than their parent (35). If so, dAdo may suppress proliferation of CASP3−/− cells while selecting SLC29A1−/−, DCK−/−, and ADK−/− cells during the CRISPR-Cas9 screen. While ADK is essential for viability and DCK is required for the development of B and T lymphocytes (36–38), the immune system of mice lacking SLC29A1 is not impaired (39). SLC29A1−/− mice exhibit accelerated bone synthesis with periarticular calcifications due to increases in blood adenosine and adenosine receptor signaling (40). In humans, SLC29A1 is the genetic determinant of the Augustine (Ata) blood type (41). The single nucleotide polymorphism rs45458701 (c.1171G > A in SLC29A1) gives rise to a missense mutation and identifies individuals with Ata− blood type, which occurs predominantly in individuals with African ancestry (41). Ata− individuals expressing variant hENT1 (p.Glu391Lys) may produce allotype antibodies recognizing wild-type ENT1 on erythrocytes, which trigger severe hemolytic reactions following transfusion with Ata blood. SLC29A1 null mutations have been identified in the genomes of individuals with European ancestry where they are associated with periarticular calcifications (41). Some of the identified SNPs in human SLC29A1, DCK, or ADK may provide for resistance against bacterial pathogens, which could explain their abundance in human populations. In this regard, we note that homologs of staphylococcal nuclease and adsA are found in other Firmicutes causing invasive disease in mammals, including members of the genera Bacillus, Streptococcus, and Staphylococcus (18). As analyzed in Streptococcus pyogenes, Streptococcus suis, and Streptococcus equi, products of homologous genes function also as 5′-nucleotidases, similar to S. aureus AdsA (16, 17, 42). The compelling findings of our CRISPR-Cas9 screen suggest that inhibitors of AdsA, adenosine signaling, hENT1, as well as ADK or DCK, may disrupt the immune evasive signaling cascades associated with AdsA 5′-nucleotidase and Nuc nuclease activities. If so, knockout mutation in the mouse gene for ENT1 (SLC29A1) as well as pharmacological inhibitors of AdsA, ENT1, ADK, or DCK may exhibit improved outcomes for MRSA infections in animal models for staphylococcal diseases.
Materials and Methods
Tissue Culture.
U937 cells were grown in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco) supplemented with 10% heat-inactivated FBS. HEK293FT cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% FBS, 0.1 mM MEM nonessential amino acids, 6 mM l-glutamine, 1 mM sodium pyruvate, and 500 μg/mL geneticin. Cells were grown at 37 °C under 5% CO2.
CRISPR-Cas9 Screen and Mutagenesis.
The human CRISPR-Cas9 GeCKO v2 library was a gift from Feng Zhang (Massachusetts Institute of Technology, Cambridge, MA) and mutagenesis was performed as described (19, 20). sgRNA library-transduced U937 were intoxicated with dAdo and dCF, or dCF alone for 28 d. The genomic DNA was isolated to prepare a sgRNA library by a two-step PCR (19, 20), which was sequenced with HiSeq2500 (Illumina). Analysis of essential sgRNAs and genes was performed using MAGeCK (43). CRISPR-Cas9–mediated mutagenesis was performed using lentiCRISPR v2 plasmids expressing specific sgRNAs as described before (19). Details are described in SI Appendix.
Cytotoxicity Assays.
dAdo-mediated cytotoxicity was analyzed as described earlier (14). Cytotoxicity mediated by S. aureus-derived dAdo was analyzed by incubating S. aureus strains in the presence of thymus DNA to generate dAdo-containing culture supernatants. Cell viability was analyzed after a 24-h incubation by Trypan Blue staining and microscopy (SI Appendix).
Biochemical Assays.
Caspase-3 activity was determined with the colorimetric caspase-3 detection kit (Sigma). dAdo uptake was analyzed by incubating cells with 1 μCi [3H]dAdo (specific activity: 38.1 Ci/mmol; Moravek Biochemicals). Radioactivity in washed cells was quantified by LSC and dAdo uptake reported as pmol/mg of total protein concentration in extracts. dAdo-derived nucleotides were quantified by treating cells with 0.5 μCi radiolabeled [14C]dAdo (specific activity: 48.8 mCi/mmol; American Radiolabeled Chemicals, Inc.) and 50 μM dCF. Cellular nucleotides were extracted using ice-cold 60% methanol (44) and separated by TLC (SIL G plates, Macherey-Nagel) using a water/isopropanol/ammonium bicarbonate mixture (25%:75%:0.2 M). Chromatograms were developed by autoradiography. Spots were excised and radioactivity was quantified by LSC to calculate nucleotide as picomoles per milligrams of protein (SI Appendix).
Supplementary Material
Acknowledgments
This work was supported by Grants AI038897 and AI052474 from the National Institute of Allergy and Infectious Diseases (to O.S.). V.W. acknowledges fellowship support (WI 4582/1-1) from the Deutsche Forschungsgemeinschaft.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805622115/-/DCSupplemental.
References
- 1.Klevens RM, et al. Active Bacterial Core surveillance (ABCs) MRSA Investigators Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 2.Kuehnert MJ, et al. Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001-2002. J Infect Dis. 2006;193:172–179. doi: 10.1086/499632. [DOI] [PubMed] [Google Scholar]
- 3.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 4.Kluytmans J, van Belkum A, Verbrugh H. Nasal carriage of Staphylococcus aureus: Epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev. 1997;10:505–520. doi: 10.1128/cmr.10.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wertheim HF, et al. Risk and outcome of nosocomial Staphylococcus aureus bacteraemia in nasal carriers versus non-carriers. Lancet. 2004;364:703–705. doi: 10.1016/S0140-6736(04)16897-9. [DOI] [PubMed] [Google Scholar]
- 6.Ammerlaan HS, Kluytmans JA, Wertheim HF, Nouwen JL, Bonten MJ. Eradication of methicillin-resistant Staphylococcus aureus carriage: A systematic review. Clin Infect Dis. 2009;48:922–930. doi: 10.1086/597291. [DOI] [PubMed] [Google Scholar]
- 7.Simor AE, et al. Randomized controlled trial of chlorhexidine gluconate for washing, intranasal mupirocin, and rifampin and doxycycline versus no treatment for the eradication of methicillin-resistant Staphylococcus aureus colonization. Clin Infect Dis. 2007;44:178–185. doi: 10.1086/510392. [DOI] [PubMed] [Google Scholar]
- 8.Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng AG, DeDent AC, Schneewind O, Missiakas D. A play in four acts: Staphylococcus aureus abscess formation. Trends Microbiol. 2011;19:225–232. doi: 10.1016/j.tim.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brinkmann V, et al. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 11.Papayannopoulos V. Neutrophil extracellular traps in immunity and disease. Nat Rev Immunol. 2018;18:134–147. doi: 10.1038/nri.2017.105. [DOI] [PubMed] [Google Scholar]
- 12.Berends ET, et al. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J Innate Immun. 2010;2:576–586. doi: 10.1159/000319909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Joo HS, Otto M. Mechanisms of resistance to antimicrobial peptides in staphylococci. Biochim Biophys Acta. 2015;1848:3055–3061. doi: 10.1016/j.bbamem.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thammavongsa V, Missiakas DM, Schneewind O. Staphylococcus aureus degrades neutrophil extracellular traps to promote immune cell death. Science. 2013;342:863–866. doi: 10.1126/science.1242255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carson DA, Kaye J, Matsumoto S, Seegmiller JE, Thompson L. Biochemical basis for the enhanced toxicity of deoxyribonucleosides toward malignant human T cell lines. Proc Natl Acad Sci USA. 1979;76:2430–2433. doi: 10.1073/pnas.76.5.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma F, Guo X, Fan H. Extracellular Nucleases of Streptococcus equi subsp. zooepidemicus degrade neutrophil extracellular traps and impair macrophage activity of the host. Appl Environ Microbiol. 2016;83:e02468-16. doi: 10.1128/AEM.02468-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng L, et al. Streptococcal 5′-nucleotidase A (S5nA), a novel Streptococcus pyogenes virulence factor that facilitates immune evasion. J Biol Chem. 2015;290:31126–31137. doi: 10.1074/jbc.M115.677443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thammavongsa V, Kern JW, Missiakas DM, Schneewind O. Staphylococcus aureus synthesizes adenosine to escape host immune responses. J Exp Med. 2009;206:2417–2427. doi: 10.1084/jem.20090097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kinzfogl J, Hangoc G, Broxmeyer HE. Neurexophilin 1 suppresses the proliferation of hematopoietic progenitor cells. Blood. 2011;118:565–575. doi: 10.1182/blood-2010-12-325381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Visser F, et al. Mutation of residue 33 of human equilibrative nucleoside transporters 1 and 2 alters sensitivity to inhibition of transport by dilazep and dipyridamole. J Biol Chem. 2002;277:395–401. doi: 10.1074/jbc.M105324200. [DOI] [PubMed] [Google Scholar]
- 23.Young JD, Yao SY, Baldwin JM, Cass CE, Baldwin SA. The human concentrative and equilibrative nucleoside transporter families, SLC28 and SLC29. Mol Aspects Med. 2013;34:529–547. doi: 10.1016/j.mam.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 24.Huang W, Zeng X, Shi Y, Liu M. Functional characterization of human equilibrative nucleoside transporter 1. Protein Cell. 2017;8:284–295. doi: 10.1007/s13238-016-0350-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raman M, Earnest S, Zhang K, Zhao Y, Cobb MH. TAO kinases mediate activation of p38 in response to DNA damage. EMBO J. 2007;26:2005–2014. doi: 10.1038/sj.emboj.7601668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller RL, et al. Adenosine kinase from rabbit liver. II. Substrate and inhibitor specificity. J Biol Chem. 1979;254:2346–2352. [PubMed] [Google Scholar]
- 27.Whitmore KV, Gaspar HB. Adenosine deaminase deficiency–More than just an immunodeficiency. Front Immunol. 2016;7:314. doi: 10.3389/fimmu.2016.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li P, et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 29.Niitsu N, Yamaguchi Y, Umeda M, Honma Y. Human monocytoid leukemia cells are highly sensitive to apoptosis induced by 2′-deoxycoformycin and 2′-deoxyadenosine: Association with dATP-dependent activation of caspase-3. Blood. 1998;92:3368–3375. [PubMed] [Google Scholar]
- 30.Bubeck Wardenburg J, Williams WA, Missiakas D. Host defenses against Staphylococcus aureus infection require recognition of bacterial lipoproteins. Proc Natl Acad Sci USA. 2006;103:13831–13836. doi: 10.1073/pnas.0603072103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheng AG, et al. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J. 2009;23:3393–3404. doi: 10.1096/fj.09-135467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thammavongsa V, Schneewind O, Missiakas DM. Enzymatic properties of Staphylococcus aureus adenosine synthase (AdsA) BMC Biochem. 2011;12:56. doi: 10.1186/1471-2091-12-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boissan M, et al. The mammalian Nm23/NDPK family: From metastasis control to cilia movement. Mol Cell Biochem. 2009;329:51–62. doi: 10.1007/s11010-009-0120-7. [DOI] [PubMed] [Google Scholar]
- 34.Panayiotou C, Solaroli N, Karlsson A. The many isoforms of human adenylate kinases. Int J Biochem Cell Biol. 2014;49:75–83. doi: 10.1016/j.biocel.2014.01.014. [DOI] [PubMed] [Google Scholar]
- 35.Brown MF, et al. Loss of caspase-3 sensitizes colon cancer cells to genotoxic stress via RIP1-dependent necrosis. Cell Death Dis. 2015;6:e1729. doi: 10.1038/cddis.2015.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Austin WR, et al. Nucleoside salvage pathway kinases regulate hematopoiesis by linking nucleotide metabolism with replication stress. J Exp Med. 2012;209:2215–2228. doi: 10.1084/jem.20121061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toy G, et al. Requirement for deoxycytidine kinase in T and B lymphocyte development. Proc Natl Acad Sci USA. 2010;107:5551–5556. doi: 10.1073/pnas.0913900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boison D, et al. Neonatal hepatic steatosis by disruption of the adenosine kinase gene. Proc Natl Acad Sci USA. 2002;99:6985–6990. doi: 10.1073/pnas.092642899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi DS, et al. The type 1 equilibrative nucleoside transporter regulates ethanol intoxication and preference. Nat Neurosci. 2004;7:855–861. doi: 10.1038/nn1288. [DOI] [PubMed] [Google Scholar]
- 40.Warraich S, et al. Loss of equilibrative nucleoside transporter 1 in mice leads to progressive ectopic mineralization of spinal tissues resembling diffuse idiopathic skeletal hyperostosis in humans. J Bone Miner Res. 2013;28:1135–1149. doi: 10.1002/jbmr.1826. [DOI] [PubMed] [Google Scholar]
- 41.Daniels G, et al. Lack of the nucleoside transporter ENT1 results in the Augustine-null blood type and ectopic mineralization. Blood. 2015;125:3651–3654. doi: 10.1182/blood-2015-03-631598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu P, et al. Streptococcus suis adenosine synthase functions as an effector in evasion of PMN-mediated innate immunit. J Infect Dis. 2014;210:35–45. doi: 10.1093/infdis/jiu050. [DOI] [PubMed] [Google Scholar]
- 43.Li W, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson PM, et al. A novel fluorescence-based assay for the rapid detection and quantification of cellular deoxyribonucleoside triphosphates. Nucleic Acids Res. 2011;39:e112. doi: 10.1093/nar/gkr350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.