Abstract
Alkaline proteases have applications in numerous industries. In this study, we have isolated and screened proteolytic bacteria from poultry wastes mixed soil and identified two bacterial isolates as Bacillus subtilis AKAL7 and Exiguobacterium indicum AKAL11 based on 16S rDNA sequencing. Maximum level of protease production was achieved after 24 h of fermentation in a basal medium. The optimal temperature, initial pH of the media and agitation for alkaline protease production by these two isolates were 30 °C, pH 9.0 and 120 rpm, respectively. The both bacterial isolates produced maximum level of protease with 3.0% organic municipal solid wastes (OMSW) as the sole source of carbon and nitrogen under previously optimized fermentation conditions. In comparison with the shake flask, protease production increased about 2.5-fold in the bioreactor with reduction in fermentation period. The partial purification of protease resulted in a final 45.67 and 34.86-fold purified protease with a specific activity of 8335.34 and 9918.91 U/mg protein and a typical yield of 9.75 and 9.41% from B. subtilis and E. indicum, respectively. The optimum temperature and pH of the partially purified protease from the both sources was 40 °C and pH 9.0, respectively. Protease from the both isolates was stable at pH 7.0–12.0 and at temperatures up to 50 °C. The effects of protease inhibitors indicated that the protease from B. subtilis might be serine and cysteine type and from E. indicum might be cysteine type. Mg2+, K+ and Ca2+ stimulated but Zn2+, Hg2+, Co2+ and Fe3+ strongly inhibited the protease activity. The partially purified protease from B. subtilis substantially dehaired cow skin and decomposed gelatinous compound from X-ray film. Our study revealed that OMSW can be used as raw material for production of bacterial extracellular protease and alkaline protease from B. subtilis might be potential for industrial and biotechnological applications.
Keywords: Biotechnology, Microbiology
1. Introduction
Proteases [EC 3.4.2124], which cause the cleavage of peptide bonds between the amino acid residues in other proteins, are one of the most important groups of industrial enzymes, accounting for more than 65% of the total worldwide sale of the enzymes (Shankar et al., 2011; Sundararajan et al., 2011; Annamalai et al., 2014). Based on the mode of action of proteases, they are classified into four categories such as alkaline, acid, thiol and metallo proteases (Rao et al., 1998). Among the different types of proteases, alkaline proteases have wide applications in different industries such as detergent, leather, pharmaceutical, protein processing, foods, diagnostic reagents, soy processing, peptide synthesis industries, extraction of silver from used X-ray film and wastes treatment (Shah et al., 2010; Kumar et al., 2014; Rathod and Pathak, 2016; Yildirim et al., 2017; Asha and Palaniswamy, 2018). Therefore, the demand for industrially important highly active alkaline proteases with high specificity and stability of pH, temperature, and organic solvents continues to enhance the search for new enzymes (Vijayaraghavan and Vincent, 2012). Currently, proteases are not produced commercially in Bangladesh and tons of proteases are imported every year to use in different industries (Azad et al., 2013). Bacterial proteases are the most significant when compared with plants, animal, and fungal proteases due to their extracellular nature, high yield of production, limited space and short period of time required for their cultivation and their feasibility to genetic manipulation (Breithaupt, 2001; Selvamohan and Sherin, 2010). Among all protease producing bacteria, Bacillus spp. are the most popular source of commercial alkaline proteases (Kumar and Takagi, 1999; Gupta et al., 2002).
Municipal solid wastes (MSW) management in Bangladesh involves collection and dumping of wastes in open field or throwing haphazardly resulting environmental pollution, climate change and public health hazards. About 16,000 tons of solid wastes are produced every day from the six divisional cities and other urban areas of Bangladesh (Bahauddin and Uddin, 2012). It is predicted that this amount will be up to 47,000 tons per day by 2025 due to increase in population and urbanization (Bahauddin and Uddin, 2012; Iqbal et al., 2018). Methane, the second most prevalent greenhouse gas having impact on climate change over 20 times greater than carbon dioxide is emitted from the rotten MSW (EPA, 2013). Almost 70–80% of the MSW is organic materials, which are mainly composed of cellulose, protein and fat (Alamgir and Ahsan, 2007; Hasan et al., 2017). The large amount of organic MSW (OMSW) can be used for production of commercially important enzymes as well as renewable biomass energy and thereby to mitigate the climate change, public health hazards and environmental pollution caused by unmanaged MSW (Azad et al., 2013, 2014; Iqbal et al., 2018).
In industries, the overall cost of enzyme production is high and the growth medium accounts for about 30–40% of the production cost (Joo et al., 2002). Reducing the cost of enzyme production by optimization of fermentation medium and process parameters is the major goal of basic research for industrial applications (Li et al., 2006). In our previous studies, the inexpensive cellulosic and proteinous materials of MSW were used as carbon and nitrogen sources in fermentation for neutral protease production by bacterial isolates (Azad et al., 2013; Iqbal et al., 2018). However, because the alkaline protease has wide range of industrial applications, our target is to produce alkaline protease from new bacterial isolates by using OMSW.
In the present study, we have isolated, screened and identified two bacteria producing alkaline protease. Further, the levels of protease production by these isolates using commercial carbon and nitrogen sources were compared to that with OMSW following optimization of some physicochemical parameters. Proteases produced from these bacterial isolates by using OMSW as a substrate in the bioreactor were partially purified and characterized, and applied for dehairing of cow skin, and decomposition of raw gelatinous compound from X-ray film.
2. Materials and methods
2.1. Isolation of protease producing bacterial isolates
Poultry waste mixed soil samples were randomly collected from near the poultry slaughtering house where residual biomass of chicken was discarded. One gram of the soil was suspended in 9 mL of sterile distilled water. After a serial dilution (101 to 106) of this suspension with sterile distilled water, 100 μL from each diluted suspension was spread on skim milk agar (SMA) plates (2.0% skim milk, 1.0% glucose, 1.5% agar and pH 9.0) and incubated at 37 °C for 24–48 h. Bacteria showing clear zones due to hydrolysis of casein in milk were selected as protease producers. Based on the ratio of clear zone (the ratio of diameter of clear zone to diameter of bacterial colonies), screening of protease producer was carried out and then pure cultures were obtained on SMA media.
2.2. Identification of bacterial isolates
The selected proteolytic bacterial isolates were initially identified on the basis of cultural, morphological and biochemical characteristics. Finally, the 16S rDNA gene sequencing was performed for molecular identification of the two isolates, which showed higher level of proteolytic activity on alkaline SMA media and produced higher level of alkaline protease in a basal media mentioned below.
Genomic DNA extraction and the polymerase chain reaction (PCR) for amplification of the 16S rDNA of the both bacterial isolates were done as described previously (Iqbal et al., 2018). PCR products were purified and sequenced as described by Iqbal et al. (2018). The 16S rDNA sequences were analyzed using a free computer program Chromas 2.6.2. The similarities of the sequence were searched with the BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) search program. The sequence was aligned with the similar sequences by using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) and a phylogenetic tree was constructed using Molecular Evolution Genetic Analysis (MEGA), version 5.0 (Tamura et al., 2011) as described previously (Azad et al., 2016).
2.3. Production of crude protease in the shake flask
A basal media (l.0% glucose, 0.5% peptone, 0.5% yeast extracts, 0.1% K2HPO4, 0.01% MgSO4 and 0.5% Na2CO3; pH 9.0) was used for alkaline protease production unless noted. For seed culture, 5 ml of sterile basal media was inoculated with an isolated colony and incubated at 37 °C and 120 rpm for 20 h. The seed culture was then transferred to 45 ml production media in a 250 ml conical flask and incubated under the conditions as indicated. The cell free supernatant was obtained by centrifugation at 8000 rpm for 20 min at 4 °C and used for determining the protease activity or further study.
2.4. Enzyme assay and protein estimation
Protease activity was determined by using azocasein (Sigma) as a substrate with some modifications of the method described by Kreger and Loekwood (1981). In brief, the reaction mixture was (500 μl of 1.0% azocasein in 50 mM Tris HCI, pH 9.0 and 1 ml of enzyme solution) incubated for 30 min at 40 °C or as indicated. The reaction was stopped by adding 2.8 ml of 5.0% (w/v) trichloroacetic acid (TCA), and was kept at 4 °C for 15 min. The precipitate was removed by centrifugation at 8,000 rpm for 20 min. A 2 ml of the supernatant was added to 2 ml of 1.0 M NaOH and the absorbance was measured at 440 nm using a UV-visible spectrophotometer (T80 UV/VIS Spectrometer, PG Instruments Ltd, UK). One unit of protease activity was defined as the amount of enzyme that produced an increase in absorbance of 0.01 under the above assay conditions. For negative control, 5.0% of the TCA solution was added before addition of enzyme preparation. Total protein concentration was determined with Bradford protein assay kit (1x dye, Bio-Rad, USA) using bovine serum albumin as a standard protein (Bradford, 1976).
2.5. Bacterial growth analysis
Bacterial growth was analyzed by total viable cell count. During alkaline protease production, samples taken from the culture were diluted 104 to 108-fold. An aliquot of 100 μl from each dilution was spread on nutrient agar medium. After 24 h of incubation at 37 °C, the total viable cell count was performed manually as colony forming unit (CFU). At least three replicas were done for each dilution of a specific sample.
2.6. Optimization of fermentation conditions for protease production
2.6.1. Cultivation period
To determine the optimum cultivation period for maximum alkaline protease production, the seed culture was inoculated in the basal media. The fermentation was carried out at shake flask level at 37 °C and 120 rpm. The initial pH of the production media was adjusted to 9.0. After 18 h of cultivation, samples of the culture were withdrawn aseptically with 6 h intervals to investigate the proteolytic activity and the bacterial growth.
2.6.2. Optimum temperature and pH
To investigate the effects of temperature on alkaline protease production, cultures were incubated at different temperatures ranging 25–45 °C with agitation at 120 rpm. The initial pH of the media was adjusted to 9.0. In order to observe the effects of different initial pH on the production of protease, the basal media was adjusted to pH 7.0 to 10.0. The culture was carried out at 30 °C and 120 rpm.
2.6.3. Carbon and nitrogen sources
The basal media was supplemented with l.0% each of glucose, sucrose, maltose, lactose and starch to investigate the effects of different carbon sources (Azad and Hoq, 2000). To observe the effects of various inorganic nitrogen sources on protease production, peptone and yeast extract of the basal media were replaced by 1.0% of ammonium sulfate, ammonium nitrate and ammonium chloride and urea. Fermentation at the shake flask was carried out for optimum period under optimum temperature and pH at 120 rpm.
2.6.4. Agitation
To study the effects of agitation on the production of protease, fermentation was done at optimum temperature and pH under different agitation rates ranging 110.0–140.0 rpm.
2.7. Protease production in the shake flask and bioreactor with OMSW
Protease production in the shake flask with OMSW was done as described previously (Azad et al., 2013). Fermentation of OMSW for protease production was carried out in a bioreactor (Fermac 360, Electrolab, UK). The inoculum size of the seed culture was 10% of the total fermentation broth. A 10 ml seed culture was produced in the basal media at 30 °C as described above. This culture was inoculated to 190 ml of OMSW media (2–4% of proteinous (chicken flesh, skin, feather etc) and cellulosic (peels of potato, pumpkin and banana) OMSW, 0.1 % K2HPO4, and 0.01% MgSO4, pH 9.0) and incubated in a shaker incubator at 30 °C and 120 rpm for 20 h. For OMSW media preparation, the proteinous and cellulosic kitchen garbage were collected from dustbins situated in the Sylhet City Corporation of Bangladesh. Two hundred ml of this seed culture was aseptically transferred to the bioreactor containing 1.8 L OMSW media. Fermentation was carried out at pH 9.0 and 30 °C for 30 h by B. subtilis and at pH 9.0 and 30 °C for 36 h by E. indicum. During fermentation, the aeration was 1 vvm and the agitation was 120 rpm. After fermentation, cells were separated by centrifugation at 8000 rpm for 15 min at 4 °C, and the supernatant was used as a source of protease.
2.8. Partial purification of protease
The partial purification of protease was done with ammonium sulphate fractionation and anion exchange chromatography by the method as described previously (Iqbal et al., 2018).
2.9. Effects of temperature and pH on protease activity and stability
The effects of temperature and pH on the activity and stability of the partially purified protease was investigated as described previously (Phadatare et al., 1993).
2.10. Effects of metal ions and protease inhibitors on the partially purified protease
The effects of metal ions on the activities of the partially purified protease were investigated by treating protease with 10 mM metal ion prepared in 50 mM Tris-HCl buffer, pH 9.0 for 1 h at room temperature. The metal salts tested were CaCl2, FeCl3, HgCl2, MgCl2, NaCl, KCl, CoCl2 and ZnCl2. The protease activity was then assayed as described above.
To investigate the type of protease, the partially purified proteases were treated for 1 h with 1 mM PMSF, ethylene diamine tetra-acetic acid (EDTA), 1, 10 phenanthroline, mercuric chloride, 25 μM pepstatin and leupeptin. PMSF, pepstatin and 1, 10 phenanthroline were solubilized in 50% (w/v) ethanol in 50 mM Tris-HCl buffer (pH 9.0) and rests of the inhibitors were dissolved in 50 mM Tris-HCl buffer (pH 9.0).
2.11. Dehairing of cow skin
Cow skin was selected and washed with water to remove salt and other debris. It was cut into the small pieces (5 × 5 cm2) and treated with partially purified enzyme (∼1000 U) and 7% sodium sulphide and lime for 24 hour under shaking at 120 rpm. Pieces of cow skin that were considered as negative controls were treated only with distilled water. The pH and temperature for all treatments were adjusted to 9.0 and 40 °C, respectively. After the treatment, the process of depilation was evaluated by examining the features such as colour, scud removal and general appearance of the leather.
2.12. Recovery of silver from waste X-ray photographic film
The pieces of used X-ray films (2 × 2 cm2) were washed with distilled water and wiped with cotton impregnated with ethanol. The washed film was dried in an oven at 40 °C for 30 min and then treated with 15 ml of partially purified alkaline protease (1000 U/ml) at 40 °C, pH 9.0 with continuous shaking. The film was checked for decomposition of gelatinous coating after different incubation periods. For a negative control, a similar piece of used X-ray film was treated with 15 ml distilled water instead of enzyme solution.
2.13. Statistical analysis
For statistical analysis, Student's ‘t’ test was used. A p value of <0.05 was considered to be statistically significant. Data were presented as the means ± standard errors of the means (SEM) of at least three independent experiments, or as noted in the figure legends.
3. Results and discussion
3.1. Isolation, screening and identification of proteolytic bacterial isolates
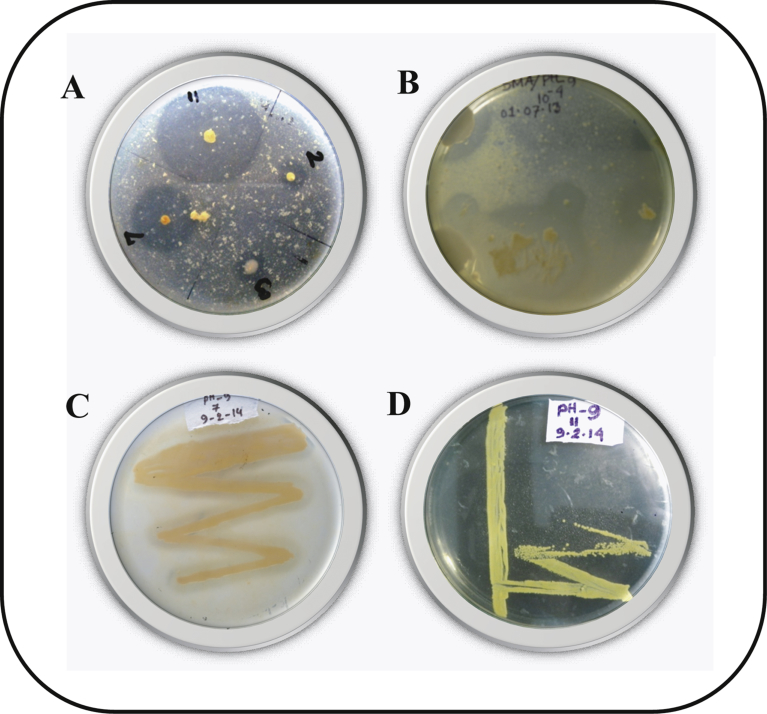
From a comprehensive screening on the SMA plate, twelve isolates showed clear zone around the colonies due to hydrolysis of milk protein casein, indicating that these bacterial isolates had proteolytic activity. All these twelve isolates were further sub-cultured on SMA media. Based on the cultural and morphological characteristics as well as the intensity of clear zones produced following subculture on the SMA plates (Fig. 1), two distinguished isolates (isolates 7 and 11) were obtained as pure culture.
Fig. 1.
Isolation of proteolytic bacteria from poultry wastes mixed soil. Representative plates of screening for proteolytic bacteria on SMA media is shown in A and B. Clear zones due to proteolytic activity of pure culture of Bacillus sp. (C) and Exiguobacterium sp. (D) were observed on SMA media after incubation at 37 °C for 24 h.
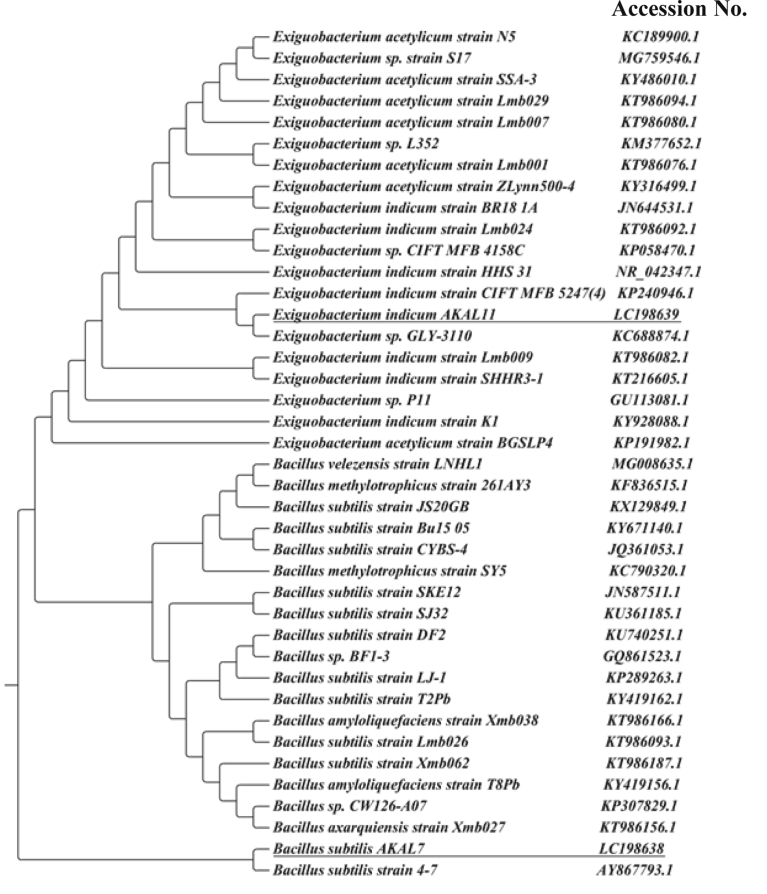
Based on the results of cultural, morphological and biochemical characteristics, isolates 7 and 11 were identified as Bacillus sp. and Exiguobacterium sp., respectively. The 16S rDNA gene sequencing was performed for molecular identification of the species of these two bacterial isolates. The PCR amplicons from the genomic DNA of Bacillus and Exiguobacterium isolates were approximately 1516 and 1449 base pair, respectively. BLAST similarity search with the DNA sequences of the PCR amplicons obtained from Bacillus (DDBJ, EMBL and Gene Bank accession no. LC198638) and Exiguobacterium (DDBJ, EMBL and Gene Bank accession no. LC198639) revealed that these isolates were similar to Bacillus and Exiguobacterium spp. Phylogenetic tree constructed with the similar sequences showed that the Bacillus and Exiguobacterium isolates were closely related to Bacillus subtilis and Exiguobacterium indicum, respectively (Fig. 2). We named them as Bacillus subtilis AKAL7 and Exiguobacterium indicum AKAL11.
Fig. 2.
Phylogenetic tree showing the relationships of Bacillus subtilis AKAL7 and Exiguobacterium indicum AKAL11 with other Bacillus and Exiguobacterium species, respectively.
3.2. Optimization of fermentation parameters for protease production
3.2.1. Cultivation period
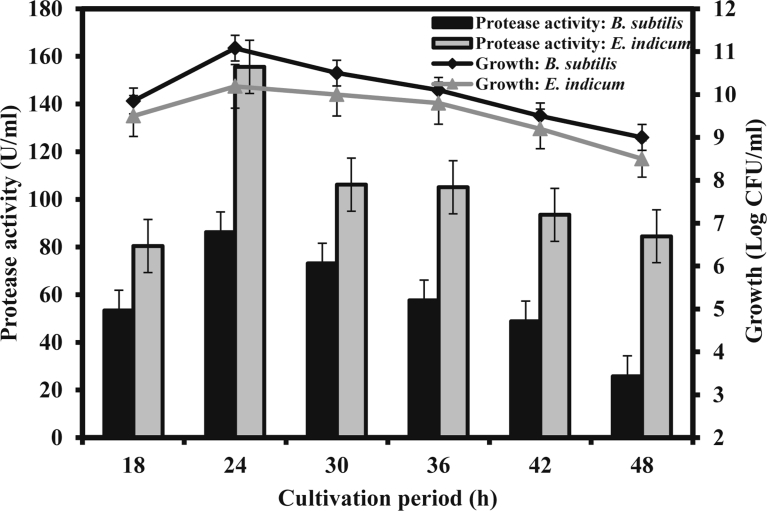
Cultivation period plays an important role in the extracellular alkaline protease production (Olajuyigbe and Ajele, 2005, 2011; Rahman et al., 2005). Time course study in the shake flask fermentation with the basal media revealed that the cultivation period for production of the maximum amount of protease was 24 h for the both bacterial isolates, B. subtilis and E. indicum (Fig. 3). However, E. indicum produced almost 2-fold more protease compared to B. subtilis. Protease level gradually decreased after 24 h of fermentation possibly due to denaturation, degradation and autolysis of protease (Chu et al., 1992). Growth kinetics during the time course showed that the protease production was almost growth associated. It was observed that the rate of protease production by these two bacterial isolates was low during early exponential growth phase, but a considerably increased rate was observed after the mid exponential growth and the highest levels of protease appeared at the end of the exponential growth phase. The residual glucose concentration was negatively correlated with the bacterial growth and production of protease was directly correlated with the production of extracellular protein (data not shown). These results suggested that the protein secretion continued from the cells along the growth phase.
Fig. 3.
Time course of protease production and growth in the basal media. The fermentation at shake flask was carried out at 37 °C and 120 rpm with the initial pH of the media 9.0.
3.2.2. Optimum temperature and pH
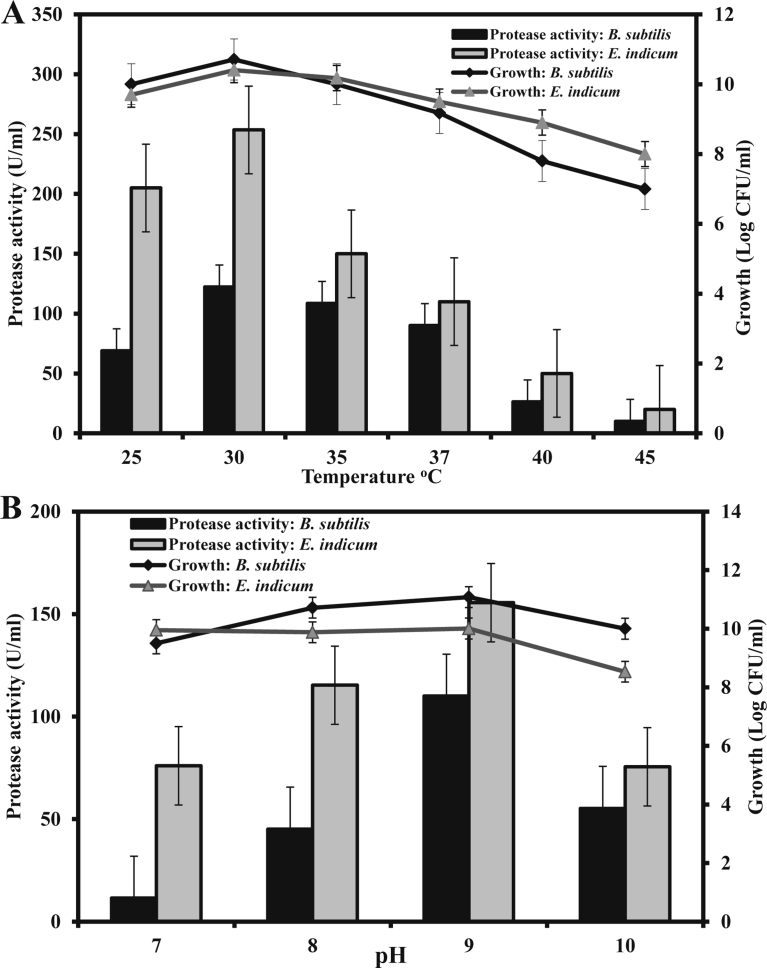
In bioprocess, specific temperature requirement and its regulation is one of the most critical parameters for the growth and production of metabolites by bacteria (Banerjee and Bhattacharyya, 1992; Singh et al., 2011). The optimum temperature for maximum production of protease by B. subtilis and E. indicum was 30 °C although considerable amount of protease was produced in the temperature range at 25–37 °C (Fig. 4A). This optimum temperature for protease production was in consistent with Bacillus sp. MIG (Gouda, 2006) and E. profundum BK-P23 (Anbu et al., 2013). The protease production was decreased considerably when the temperature increased above 37 °C. However, the optimum temperature for protease production varies from species to species, and it was reported that B. licheniformis and Bacillus sp. SMIA-2 produced maximum level of protease at 50 and 60 °C, respectively (Al-Shehri et al., 2004; Nascimento and Martins, 2004). The pH of the culture strongly influences many enzymatic processes and the transport of various components across the cell membranes (Ellaiah et al., 2002). We investigated the effects of initial pH of the media on the production of protease by the two bacterial isolates. The optimum pH for protease production by B. subtilis and E. indicum isolates was 9.0 (Fig. 4B). However, a significant level of protease was produced by the both isolates over a broad pH range. This result, in agreement with other reports (Joshi et al., 2008), indicated that the highest amount of protease was secreted by the both bacterial isolates at alkaline pH. It is also reported that the maximum level of alkaline protease is produced by B. licheniformis MP1 at pH 8.0 (Jellouli et al., 2011) and by B. circulans and B. infantis SKS1 at pH 10.0 (Prakasham et al., 2005; Rao et al., 2009; Saggu and Mishra, 2017).
Fig. 4.
Effects of temperature (A) and pH (B) on protease production by the bacterial isolates and their growth. (A) The initial pH of the media was 9.0 at temperatures as indicated. (B) The fermentation at shake flask level was carried out at 37 °C. Fermentation in (A) and (B) was done at 120 rpm for the optimum period for protease production.
Agitation plays an important role in transfer rate of nutrients, solubility of oxygen, cell dispersion and thereby enhanced aerobic metabolism of microbes (Patil and Chaudhari, 2013). In the shake flask fermentation, 120 rpm was observed as the optimum agitation for the highest level of protease production by the both bacterial isolates (data not shown). The amount of protease produced under optimum temperature, pH, agitation and fermentation period was approximately 250 U/ml.
3.2.3. Carbon and nitrogen sources
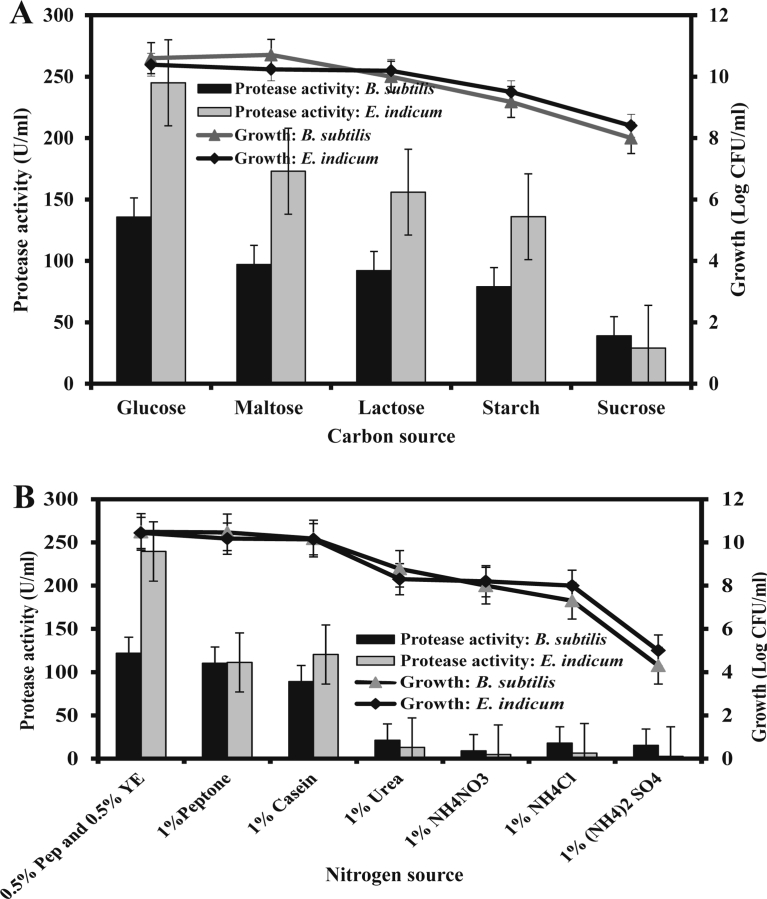
Production of alkaline protease is highly influenced by the nature of the carbon and nitrogen sources used in the media (Sen et al., 2009; Sharma et al., 2017). The result showed that the media containing 1.0% glucose as a carbon source supported highest alkaline protease production compared to the other carbon sources (Fig. 5A). We further observed that the substrate inhibition was occurred above l.0% of glucose, and below this concentration, insufficient substrate caused lower growth as well as lower production of alkaline protease (Data not shown). The similar results were reported previously with other bacterial strains (Ellaiah et al., 2002; Vijayaraghavan et al., 2012).
Fig. 5.
Effects of carbon (A) and nitrogen (B) sources on protease production. Shake flask fermentation carried out at optimum temperature, pH and agitations were 30 °C, 9.0 and 120.0 rpm, respectively for protease production by the both isolates.
Among the different nitrogen sources investigated, 0.5% peptone with 0.5% yeast extract supported the highest level of alkaline protease production (Fig. 5B). This result indicated that the organic nitrogen compounds favored higher amount of alkaline protease production than the inorganic ones. It is also reported that the presence of yeast extract and peptone in the production medium had shown substantially enhanced protease production by Bacillus sp MA6 (Azad and Hoq, 2000), B. acidophilus subsp. Halodurans (Takii et al., 1990), B. subtilis (Vanitha et al., 2014; Badhe et al., 2016).
3.3. Production of protease in the bioreactor using OMSW
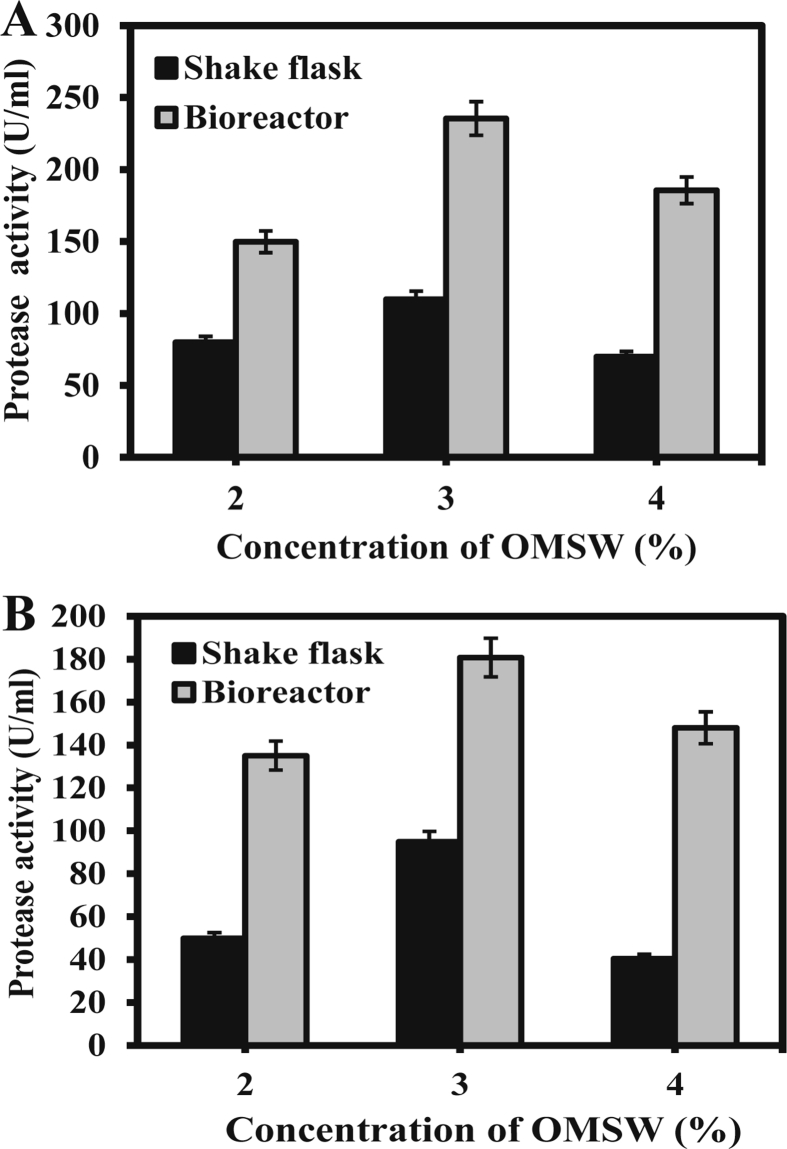
In order to produce alkaline protease using OMSW, the carbon and nitrogen (glucose, peptone and yeast extract) sources of the basal media were replaced by 2, 3 and 4% each of cellulosic and proteinous OMSW. The highest level of alkaline protease was produced with 3% OMSW (Fig. 6). Almost similar level of protease was produced when the commercial carbon and nitrogen sources of the basal media were substituted by 3% of cellulosic and proteinous OMSW, indicating that OMSW might be adequate carbon and nitrogen source for protease production. Protease production by the both bacterial isolates with OMSW was optimized in shake flask and bioreactor. The optimum temperature, pH and agitation for fermentation of OMSW for maximum production of protease by both bacterial isolates at shake flask and bioreactor was 30 °C, 9.0 and 120 rpm, respectively. Optimum fermentation period for protease production from OMSW by B. subtilis and E. indicum at shake flask was 48 h and 60 h, respectively (data not shown). However, optimum fermentation period for protease production in the bioreactor by the both isolates was reduced by 18 h compared to shake flask. In our previous study, the fermentation period for protease production in the bioreactor by Serratia marcescens and Pseudomonas putida was reduced by 6 h compared to that in the shake flask (Iqbal et al., 2018).
Fig. 6.
Production of protease from B. subtilis (A) and E. indicum (B) by using OMSW. Black and gray bars indicated the protease level produced during fermentation at shake flask and bioreactor, respectively. The fermentation with the isolates was carried out under temperature, pH, and agitation at 30 °C, 9.0 and 120 rpm, respectively. The aeration in the bioreactor was maintained at 1 vmm. Results are mean ± SEM of repeated experiments (n = 3).
The protease production by B. subtilis and E. indicum in the bioreactor increased about 2–2.5 fold compared to that at the shake flask (Fig. 6). This result is consistent with our previous study in which protease production by S. marcescens and P. putida in the bioreactor increased about 2–2.5 fold compared to that at shake flask (Iqbal et al., 2018). However, the bacterial isolates in the present study produced ∼2-fold more alkaline protease compared to our previous bacterial isolates. Interestingly, the B. subtilis isolate was the better alkaline protease producer using OMSW as the raw material may be due to its different enzymatic system compared to E. indicum. Growth kinetics during the time course study showed that the protease production was growth-associated (data not shown).
3.4. Characterization of partially purified protease
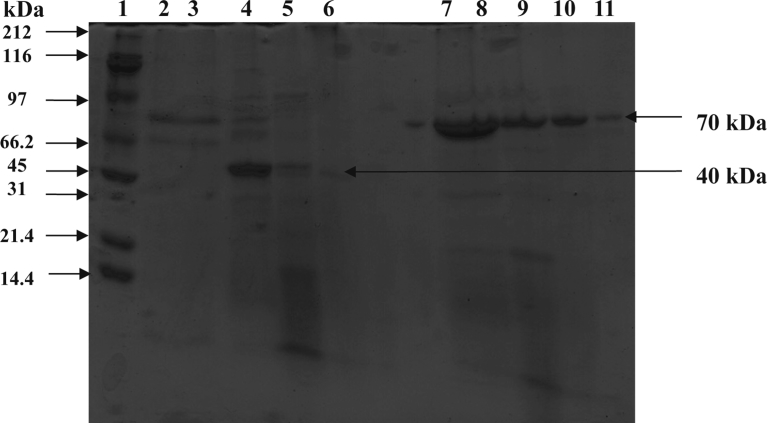
Partial purification of protease obtained from B. subtilis resulted in a final 45-fold purified protease with specific activity of 8335 U/mg protein and a typical yield of about 10% (Table 1), and that of protease from E. indicum resulted in a final 35-fold purified protease with specific activity of ∼9920 U/mg protein and a typical yield of 9% (Table 2). The predicted molecular mass of the partially purified protease from B. subtilis and E. indicum were 40 and 70 kDa, respectively (Fig. 7). The partially purified protease from the both sources was used for partial characterization as follows.
Table 1.
Summary of partial purification of crude protease from B. subtilis isolate.
| Steps | Volume (ml) | Protein (mg/ml) | Total protein (mg) | Activity (U/ml) | Total activity (U) | Specific activity (U/mg) | Purification fold | Yield (%) |
|---|---|---|---|---|---|---|---|---|
| Crude | 150 | 0.777 | 116.55 | 141.8 | 21270 | 182.5 | 1 | 100 |
| 30% (NH4)2SO4 ppt. | 2 | 1.614 | 3.228 | 722.5 | 1445 | 447.64 | 2.45 | 6.8 |
| 60% (NH4)2SO4 ppt. | 2 | 2.284 | 4.568 | 5362.5 | 10725 | 2347.85 | 12.86 | 50.42 |
| 90% (NH4)2SO4 ppt. | 2 | 1.812 | 3.624 | 252.5 | 505 | 278.7 | 1.52 | 2.37 |
| DEAE- Cellulose | 1 | 0.249 | 0.249 | 2075.5 | 2075.5 | 8335.34 | 45.67 | 9.75 |
Table 2.
Summary of partial purification of crude protease from E. indicum isolate.
| Steps | Volume (ml) | Protein (mg/ml) | Total protein (mg) | Activity (U/ml) | Total activity (U) | Specific activity (U/mg) | Purification fold | Yield (%) |
|---|---|---|---|---|---|---|---|---|
| Crude | 150 | 0.457 | 68.55 | 130 | 19500 | 284.46 | 1 | 100 |
| 30% (NH4)2SO4 ppt. | 2 | 1.115 | 2.230 | 520.5 | 1041 | 466.81 | 1.46 | 5.33 |
| 60% (NH4)2SO4 ppt. | 2 | 1.884 | 3.768 | 3172.5 | 6345 | 1683.92 | 5.91 | 32.53 |
| 90% (NH4)2SO4 ppt. | 2 | 1.513 | 3.026 | 882.5 | 1765 | 583.27 | 2.05 | 9.05 |
| DEAE- Cellulose | 1 | 0.185 | 0.185 | 1835 | 1835 | 9918.91 | 34.86 | 9.41 |
Fig. 7.
SDS-PAGE of the partially purified protease from B. subtilis and E. indicum. Molecular masses of the standard proteins are shown in lane 1. Lanes 2, 3, 4, 5 and 6 indicated the crude, 30% ppt, 60% ppt, 90% ppt and partially purified protease by DEAE cellulose from B. subtilis, respectively. Lane 7, 8, 9, 10 and 11 indicated crude, 30% ppt, 60% ppt, 90% ppt and partially purified protease by DEAE cellulose from E. indicum, respectively.
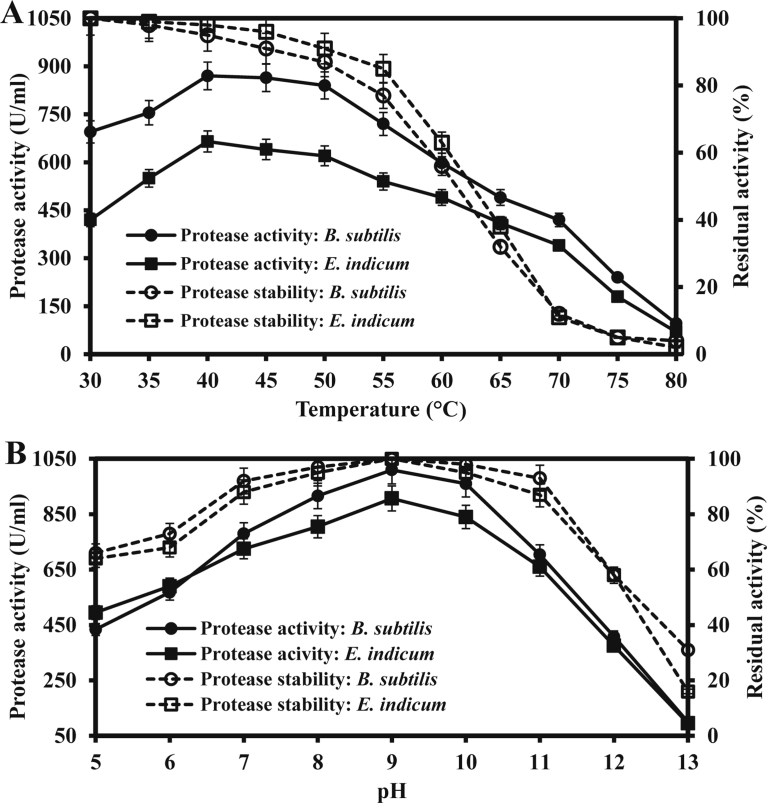
3.4.1. Effects of temperature and pH on protease activity and stability
The partially purified protease from the both sources was active within a wide range of temperature with the optimum temperature at 40 °C (Fig. 8A). However, almost similar protease activity was observed at 40–50 °C. The protease was stable from 30 to 50 °C. However, approximately 15–25% of protease activity was lost when the enzyme was treated at 55 °C. These results indicated that the protease from the both sources can be applied at a wide range of temperature above the room temperature. The temperature optima and thermostability of protease vary based on the bacterial species (Iqbal et al., 2018). It is a well-known fact that protein conformation changes at higher temperatures, and hence, causes a decrease in the protease activity (Johnvesly and Naik, 2001). Nevertheless, the protease from halo tolerant B. subtilis retains its full activity after 30 min of incubation in the temperature ranging from 37 to 55 °C (Huang et al., 2003; Karbalaei-Heidari et al., 2009; Abusham et al., 2009).
Fig. 8.
Effects of temperature (A) and pH (B) on activity and stability of protease from B. subtilis and E. indicum. Filled and open circles indicate the activity and stability of protease from B. subtilis, respectively and filled and open rectangles indicate those of protease from E. indicum, correspondingly.
The partially purified protease of the both bacterial isolates was significantly active over a broad pH range from 7.0 to 12.0 having the maximum activity at pH 9.0 (Fig. 8B). These results were in agreement with those of Bacillus sp. (El Hadj-Ali et al., 2007). Generally, the commercial microbial proteases have pH optima in the alkaline ranges from 8.0 to 12.0 (Rao et al., 1998). However, pH optima may differ based on different substrates (Malathi and Chakraborty, 1991; Takami et al., 1992). The purified protease from the isolates was stable at pH 7.0–11.0. However, protease activity was significantly lost when the enzyme solution was treated under acidic conditions and pH 12.0 and above. Protease from B. pumilus CBS and Aeribacillus pallidus C10 showed similar pH stability (Jaouadi et al., 2008; Yildirim et al., 2017).
3.4.2. Effects of metal ions and protease inhibitors on protease activity
Mg2+, Ca2+ and K+ significantly increased the activities of protease from the both isolates (Table 3). In the presence of Mg2+, Ca2+ and K+, the activity of protease from B. subtilis increased 20, 25 and 23%, respectively and that from E. indicum increased 32, 12 and 5%, respectively. In contrast, protease from the both isolates was strongly inhibited by Fe3+, Zn2+, Hg+ and Co2+. It is reported that the activity of alkaline protease from Bacillus sp. and Bacillus megaterium RRM2 is increased by Ca2+ (Gupta et al., 2005; Rajkumar et al., 2011) and decreased by Hg2+, Zn2+ and Co2+ (Venugopal and Saramma, 2007; Olajuyigbe and Falade, 2014).
Table 3.
Effects of metal ions on the activity of the partially purified protease of B. subtilis and E. indicum.
| Metal ions (10 mM) | Relative residual activity (%) of partially purified protease from |
|
|---|---|---|
| B. subtilis | E. indicum | |
| None (control)a | 100 | 100 |
| K+ | 123 | 105 |
| Mn2+ | 100 | 95 |
| Fe3+ | 43.2 | 57 |
| Mg2+ | 120 | 132.4 |
| Hg+ | 30 | 15.5 |
| Zn2+ | 25 | 18 |
| Na+ | 96 | 85 |
| Ca2+ | 125 | 112 |
| Co2+ | 59 | 65 |
The activity of protease without treatment with any metal ion (control) was considered as 100%.
The protease of B. subtilis and E. indicum isolates was classified on the basis of their response to different protease inhibitors. The protease obtained from B. subtilis was strongly inhibited by PMSF and HgCl2 and that of E. indicum inhibited by HgCl2 (Table 4). However, other inhibitors showed almost no inhibitory effects. This result indicated that the protease from B. subtilis was serine and cysteine type and from E. indicum was cysteine type (Azad et al., 2013).
Table 4.
Effects of protease inhibitors on the activity of the partially purified protease from B. subtilis and E. indicum.
| Inhibitors | Concentrations | Relative residual activity (%) of purified protease from |
|
|---|---|---|---|
| B. subtilis | E. indicum | ||
| Controla | None | 100 | 100 |
| PMSF | 1 mM | 23 | 47 |
| 1, 10 Phenanthroline | 1 mM | 95.5 | 91.4 |
| HgCl2 | 1 mM | 30 | 9.6 |
| EDTA | 1 mM | 94.4 | 84.5 |
| Leupeptin | 25 μM | 86 | 92.5 |
| Pepstatin | 25 μM | 90.5 | 88.3 |
The activity of protease treated without any protease inhibitor (control) was considered as 100%.
3.4.3. Dehairing of hide by partially purified protease
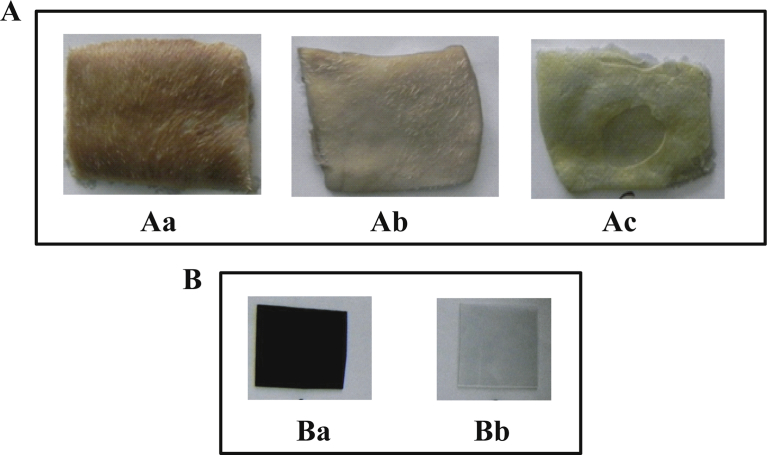
Complete dehairing of the skin treated with protease was achieved after 12 h, whereas the skin treated with only distilled water remained unchanged (Fig. 9A). It was reported by Nilegaonkar et al. (2007) that 1.0 % crude protease from B. cereus MCM B-326 dehaired buffalo hide after 21 h of incubation at 28 °C and pH 7.0. It was also observed that the sodium sulphide treatment dehaired the cow skin. Sodium sulphide removes the hair above the epidermis for which the leather doesn't become silky, however, the proteases attack the hair bellow the dermis and improve the silkier quality of the final leather preparations (Malathi and Dhar, 1987). Nevertheless, protease showed better dehairing capability in comparison with the conventional chemical dehairing method. This result suggests that the alkaline serine protease in the present study might be potential for dehairing of hide in the leather industries. Enzymatic dehairing process is gaining attraction as an alternative chemical methodology for reduction of toxicity in addition to improvement of leather quality (Azad et al., 2002; Sivasubramanian et al., 2008).
Fig. 9.
Application of partially purified protease from B. subtilis. (A) Dehairing of cow skin by protease. Aa, Ab, Ac were treated with distilled water (control), partially purified protease and chemicals (Lime sulfide), respectively. (B) Decomposition of gelatinous compound from X-ray film. Ba and Bb were treated with distilled water (control) and partially purified protease, respectively.
3.4.4. Decomposition of gelatinous compound from X-ray film
The crude and partially purified protease from B. subtilis isolate completely decomposed the gelatinous coating on the X-ray film with incubation for 24 h (Fig. 9B). This result indicated that the alkaline serine protease preparation from the B. subtilis isolate might be potential for recovery of silver from the wastes X-ray film. It has been shown that alkaline protease is useful for recovery of silver from used X-ray photographic film (Al-Abdalall and Al-Khaldi, 2016; Lakshmi and Hemalatha, 2016).
4. Conclusions
Two proteolytic bacterial strains were isolated and identified as B. subtilis and E. indicum based on the 16S rDNA sequence. Some physicochemical parameters were optimized for production of protease by the both bacterial isolates by using a basal media. The OMSW was successfully used in the bioreactor as raw material instead of commercial carbon and nitrogen source for the production of industrially important alkaline protease. Proteases produced from the two bacterial isolates by fermentation of OMSW were partially purified by ammonium sulphate fractionation followed by DEAE-cellulose chromatography. The partially purified protease from B. subtilis was serine and cysteine type and that from E. indicum was cysteine type. Removal of hair from raw cow skin and decomposition of gelatinous compound from X-ray film collectively revealed that the alkaline protease from B. subtilis might be potential for biotechnological applications.
Declarations
Author contribution statement
Al Hakim: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Farhana Rumzum Bhuiyan, Asif Iqbal, Tanvir Hossain Emon, Jahed Ahmed: Performed the experiments.
Abul Kalam Azad: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This work was partially supported by grants in aid from The World Academy of Sciences (TWAS) for the Advancement of Sciences in Developing Countries (No. 12-150 RG/BIO/AS_G), the Ministry of Science and Technology, Bangladesh (No. 39.009.006.01.00.042.2012–13/BS-135/518), Ministry of Education, Bangladesh (No. 37.20.0000.004.033.020.2016.7725), University Grant Commission of Bangladesh and Research Centre, Shahjalal University of Science and Technology, Sylhet, Bangladesh.
Competing interest statement
The authors declare no conflict of interest.
Additional information
Data associated with this study (nucleotide sequences) has been deposited at the DDBJ/EMBL/GenBank nucleotide sequence databases under the accession number LC198638 and LC198639 for Bacillus sp. and Exiguobacterium sp., respectively.
Acknowledgements
The research was done in the ‘Microbiology, Fermentation and Environmental Biotechnology Laboratory’ and ‘TWAS Laboratory’ of the Department of Genetic Engineering and Biotechnology, Shahjalal University of Science and Technology, Sylhet-3114, Bangladesh.
References
- Abusham R.A., Rahman R.N.Z.R., Salleh A.B., Basri M. Optimization of physical factors affecting the production of thermo-stable organic solvent-tolerant protease from a newly isolated halo tolerant Bacillus subtilis strain Rand. Microb. Cell Fact. 2009;8(1):1. doi: 10.1186/1475-2859-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Abdalall A.H., Al-Khaldi E.M. Recovery of silver from used X-ray film using alkaline protease from Bacillus subtilis sub sp. subtilis. Afr. J. Biotechnol. 2016;15(26):1413–1416. [Google Scholar]
- Al-Shehri L., Abdul-Rahman M., Yassar S. Production and some properties of protease produced by Bacillus licheniformis isolated from Tihamet Aseer, Saudi Arabia. Pak. J. Biol. Sci. 2004;7:1631–1635. [Google Scholar]
- Alamgir M., Ahsan A. Municipal solid waste and recovery potential: Bangladesh perspective. J. Environ. Health Sci. Eng. 2007;4(2):67–76. [Google Scholar]
- Anbu P., Annadurai G., Hur B. Production of alkaline protease from a newly isolated Exiguobacterium profundum BK-P23 evaluated using the response surface methodology. Biologia. 2013;68(2):186–193. [Google Scholar]
- Annamalai N., Rajeswari M.V., Balasubramanian T. Extraction, purification and application of thermostable and halostable alkaline protease from Bacillus alveayuensis CAS 5 using marine wastes. Food Bioprod. Process. 2014;92(4):335–342. [Google Scholar]
- Asha B., Palaniswamy M. Optimization of alkaline protease production by Bacillus cereus FT 1 isolated from soil. J. App. Pharm. Sci. 2018;8(02):119–127. [Google Scholar]
- Azad A.K., Nahar A., Hasan M., Islam K., Azim M., Hossain M., Rahman M., Ojha R., Mahmud G., Kayes R. Fermentation of municipal solid wastes by bacterial isolates for production of raw protein degrading proteases. Asian J. Microbiol. Biotechnol. Environ. Sci. 2013;15:365–374. [Google Scholar]
- Azad A.K., Ahmed J., Alum M.A., Hasan M.M., Ishikawa T., Sawa Y., Katsuhara M. Genome-wide characterization of major intrinsic proteins in four grass plants and their non-aqua transport selectivity profiles with comparative perspective. PLoS One. 2016;11(6):e0157735. doi: 10.1371/journal.pone.0157735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad A.K., Hoq M.M. Production of alkaline serine protease by Bacillus sp. MA6. Bangladesh J. Microbiol. 2000;11(2):143–149. [Google Scholar]
- Azad A.K., Yousuf A., Ferdoush A., Hasan M.M., Karim M.R., Jahan A. Production of microbial lipids from rice straw hydrolysates by lipomyces starkeyi for biodiesel synthesis. J. Microb. Biochem. Technol. 2014;8(2) [Google Scholar]
- Azad A.K., Shibata H., Hoq M.M. Hide processing with alkaline protease from Bacillus sp. MA6. Bangladesh J. Microbiol. 2002;19(1):67–71. [Google Scholar]
- Badhe P., Joshi M., Adivarekar R. Optimized production of extracellular proteases by Bacillus subtilis from degraded abattoir waste. J. BioSci. Biotechnol. 2016;5(1) [Google Scholar]
- Bahauddin K.M., Uddin M.H. Prospect of solid waste situation and an approach of Environmental Management Measure (EMM) model for sustainable solid waste management: case study of Dhaka city. J. Environ. Sci. Nat. Res. 2012;5(1):99–111. [Google Scholar]
- Banerjee R., Bhattacharyya B. Extracellular alkaline protease of newly isolated Rhizopus oryzae. Biotechnol. Lett. 1992;14(4):301–304. [Google Scholar]
- Bradford M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Breithaupt H. The hunt for living gold. EMBO Rep. 2001;2(11):968–971. doi: 10.1093/embo-reports/kve238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu I.-M., Lee C., Li T.-S. Production and degradation of alkaline protease in batch cultures of Bacillus subtilis ATCC 14416. Enzym. Microb. Technol. 1992;14(9):755–761. [Google Scholar]
- El Hadj-Ali N., Agrebi R., Ghorbel-Frikha B., Sellami-Kamoun A., Kanoun S., Nasri M. Biochemical and molecular characterization of a detergent stable alkaline serine-protease from a newly isolated Bacillus licheniformis NH1. Enzym. Microb. Technol. 2007;40(4):515–523. [Google Scholar]
- Ellaiah P., Srinivasulu B., Adinarayana K. A review on microbial alkaline proteases. J. Sci. Ind. Res. 2002;61:690–704. [Google Scholar]
- EPA . 2013. Inventory of U.S. Greenhouse Gas Emissions and Sinks: 1990–2011.https://www.epa.gov/ghgemissions/inventory-us-greenhouse-gas-emissions-and-sinks-1990-2011 [Google Scholar]
- Gouda M.K. Optimization and purification of alkaline proteases produced by marine Bacillus sp. MIG newly isolated from Eastern Harbour of Alexandria. Pol. J. Microbiol. 2006;55(2):119–126. [PubMed] [Google Scholar]
- Gupta A., Roy I., Patel R., Singh S., Khare S., Gupta M. One-step purification and characterization of an alkaline protease from haloalkaliphilic Bacillus sp. J. Chromatogr. A. 2005;1075(1):103–108. doi: 10.1016/j.chroma.2005.03.127. [DOI] [PubMed] [Google Scholar]
- Gupta R., Beg Q., Lorenz P. Bacterial alkaline proteases: molecular approaches and industrial applications. Appl. Microbiol. Biotechnol. 2002;59(1):15–32. doi: 10.1007/s00253-002-0975-y. [DOI] [PubMed] [Google Scholar]
- Hasan M.M., Marzan L.W., Hosna A., Hakim A., Azad A.K. Optimization of some fermentation conditions for the production of extracellular amylases by using Chryseobacterium and Bacillus isolates from organic kitchen wastes. J. Genet. Eng. Biotechnol. 2017;15(1):59–68. doi: 10.1016/j.jgeb.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Peng Y., Li X., Wang H., Zhang Y. Purification and characterization of an extracellular alkaline serine protease with dehairing function from Bacillus pumilus. Curr. Microbiol. 2003;46(3):0169–0173. doi: 10.1007/s00284-002-3850-2. [DOI] [PubMed] [Google Scholar]
- Iqbal A., Hakim A., Hossain M.S., Rahman M.R., Islam K., Azim M.F., Ahmed J., Assaduzzaman M., Hoq M.M., Azad A.K. Partial purification and characterization of serine protease produced through fermentation of organic municipal solid wastes by Serratia marcescens A3 and Pseudomonas putida A2. J. Genet. Eng. Biotechnol. 2018;16(2018):29–37. doi: 10.1016/j.jgeb.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaouadi B., Ellouz-Chaabouni S., Rhimi M., Bejar S. Biochemical and molecular characterization of a detergent-stable serine alkaline protease from Bacillus pumilus CBS with high catalytic efficiency. Biochimie. 2008;90(9):1291–1305. doi: 10.1016/j.biochi.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Jellouli K., Ghorbel-Bellaaj O., Ayed H.B., Manni L., Agrebi R., Nasri M. Alkaline-protease from Bacillus licheniformis MP1: purification, characterization and potential application as a detergent additive and for shrimp waste deproteinization. Process Biochem. 2011;46(6):1248–1256. [Google Scholar]
- Johnvesly B., Naik G. Studies on production of thermostable alkaline protease from thermophilic and alkaliphilic Bacillus sp. JB-99 in a chemically defined medium. Process Biochem. 2001;37(2):139–144. [Google Scholar]
- Joo H.-S., Kumar C.G., Park G.-C., Kim K.T., Paik S.R., Chang C.-S. Optimization of the production of an extracellular alkaline protease from Bacillus horikoshii. Process Biochem. 2002;38(2):155–159. [Google Scholar]
- Joshi R., Dodia M., Singh S. Production and optimization of a commercially viable alkaline protease from a haloalkaliphilic bacterium. Biotechnol. Bioprocess Eng. 2008;13(5):552–559. [Google Scholar]
- Karbalaei-Heidari H.R., Amoozegar M.A., Hajighasemi M., Ziaee A.-A., Ventosa A. Production, optimization and purification of a novel extracellular protease from the moderately halophilic bacterium Halobacillus karajensis. J. Ind. Microbiol. Biotechnol. 2009;36(1):21–27. doi: 10.1007/s10295-008-0466-y. [DOI] [PubMed] [Google Scholar]
- Kreger A., Lockwood D. Detection of extracellular toxin (s) produced by Vibrio vulnificus. Infect. Immun. 1981;33(2):583–590. doi: 10.1128/iai.33.2.583-590.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar C.G., Takagi H. Microbial alkaline proteases: from a bioindustrial viewpoint. Biotechnol. Adv. 1999;17(7):561–594. doi: 10.1016/s0734-9750(99)00027-0. [DOI] [PubMed] [Google Scholar]
- Kumar R.S., Ananthan G., Prabhu A.S. Optimization of medium composition for alkaline protease production by Marinobacter sp. GA CAS9 using response surface methodology–a statistical approach. Biocat. Agric. Biotechnol. 2014;3(2):191–197. [Google Scholar]
- Lakshmi B., Hemalatha K. Eco friendly recovery of silver from used X-ray films by alkaline protease of Bacillus cereus strain S8. Front. Environ. Microbiol. 2016;2(6):45–48. [Google Scholar]
- Li Y., Lin J., Meng D., Lu J., Gu G., Mao Z. Effect of pH, cultivation time and substrate concentration on the endoxylanase production by Aspergillus awamori ZH-26 under submerged fermentation using central composite rotary design. Food Technol. Biotechnol. 2006;44(4):473–477. [Google Scholar]
- Malathi S., Chakraborty R. Production of alkaline protease by a new Aspergillus flavus isolate under solid-substrate fermentation conditions for use as a depilation agent. Appl. Environ. Microbiol. 1991;57(3):712–716. doi: 10.1128/aem.57.3.712-716.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malathi S., Dhar S. Production of extracellular protease by an Aspergillus flavus isolate and its application in the depilation of skins. Leather Sci. 1987;34:67–76. [Google Scholar]
- Nascimento W.C.A.D., Martins M.L.L. Production and properties of an extracellular protease from thermophilic Bacillus sp. Braz. J. Microbiol. 2004;35(1–2):91–96. [Google Scholar]
- Nilegaonkar S., Zambare V., Kanekar P., Dhakephalkar P., Sarnaik S. Production and partial characterization of dehairing protease from Bacillus cereus MCM B-326. Bioresour. Technol. 2007;98(6):1238–1245. doi: 10.1016/j.biortech.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Olajuyigbe F., Ajele J.O. Thermostable alkaline protease from Bacillus licheniformis lbbl-11 isolated from traditionally fermented African locust bean (Parkia biglobosa) J. Food Biochem. 2011;35(1):1–10. [Google Scholar]
- Olajuyigbe F.M., Ajele J.O. Production dynamics of extracellular protease from Bacillus species. Afr. J. Biotechnol. 2005;4(8):776. [Google Scholar]
- Olajuyigbe F.M., Falade A.M. Purification and partial characterization of serine alkaline metalloprotease from Bacillus brevis MWB-01. Bioresour. Bioprocess. 2014;1(1):8. [Google Scholar]
- Patil U., Chaudhari A. Production of alkaline protease by solvent-tolerant alkaliphilic Bacillus circulans MTCC 7942 isolated from hydrocarbon contaminated habitat: process parameters optimization. ISRN Biochem. 2013 doi: 10.1155/2013/942590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadatare S.U., Deshpande V.V., Srinivasan M.C. High activity alkaline protease from Conidiobolus coronatus (NCL 86.8. 20): enzyme production and compatibility with commercial detergents. Enzym. Microb. Technol. 1993;15(1):72–76. [Google Scholar]
- Prakasham R., Rao C., Rao R.S., Sarma P. Alkaline protease production by an isolated Bacillus circulans under solid-state fermentation using agroindustrial waste: process parameters optimization. Biotechnol. Prog. 2005;21(5):1380–1388. doi: 10.1021/bp050095e. [DOI] [PubMed] [Google Scholar]
- Rahman R.N.Z.A., Geok L.P., Basri M., Salleh A.B. Physical factors affecting the production of organic solvent-tolerant protease by Pseudomonas aeruginosa strain K. Bioresour. Technol. 2005;96(4):429–436. doi: 10.1016/j.biortech.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Rajkumar R., Kothilmozhian J., Ramasamy R. Production and characterization of a novel protease from Bacillus sp. RRM1 under solid state fermentation. J. Microbiol. Biotechnol. 2011;21(6):627–636. [PubMed] [Google Scholar]
- Rao C.S., Sathish T., Ravichandra P., Prakasham R. Characterization of thermo-and detergent stable serine protease from isolated Bacillus circulans and evaluation of eco-friendly applications. Process Biochem. 2009;44(3):262–268. [Google Scholar]
- Rao M.B., Tanksale A.M., Ghatge M.S., Deshpande V.V. Molecular and biotechnological aspects of microbial proteases. Microbiol. Mol. Biol. Rev. 1998;62(3):597–635. doi: 10.1128/mmbr.62.3.597-635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathod M.G., Pathak A.P. Optimized production, characterization and application of alkaline proteases from taxonomically assessed microbial isolates from Lonar soda lake, India. Biocatal. Agric. Biotechnol. 2016;7:164–173. [Google Scholar]
- Saggu S.K., Mishra P.C. Characterization of thermostable alkaline proteases from Bacillus infantis SKS1 isolated from garden soil. PLoS One. 2017;12(11):e0188724. doi: 10.1371/journal.pone.0188724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvamohan T., Sherin S. Optimization of protease production from Bacillus cereus using different substrates. Plant Arch. 2010;10(2):651–656. [Google Scholar]
- Sen S., Veeranki V.D., Mandal B. Effect of physical parameters, carbon and nitrogen sources on the production of alkaline protease from a newly isolated Bacillus pseudofirmus SVB1. Ann. Microbiol. 2009;59(3):531–538. [Google Scholar]
- Shah K., Mody K., Keshri J., Jha B. Purification and characterization of a solvent, detergent and oxidizing agent tolerant protease from Bacillus cereus isolated from the Gulf of Khambhat. J. Mol. Catal. B Enzym. 2010;67(1):85–91. [Google Scholar]
- Shankar S., Rao M., Laxman R.S. Purification and characterization of an alkaline protease by a new strain of Beauveria sp. Process Biochem. 2011;46(2):579–585. [Google Scholar]
- Sharma K.M., Kumar R., Panwar S., Kumar A. Microbial alkaline proteases: optimization of production parameters and their properties. J. Genet. Eng. Biotechnol. 2017 doi: 10.1016/j.jgeb.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.K., Singh S.K., Tripathi V.R., Khare S.K., Garg S.K. Comparative one-factor-at-a-time, response surface (statistical) and bench-scale bioreactor level optimization of thermoalkaline protease production from a psychrotrophic Pseudomonas putida SKG-1 isolate. Microb. Cell Fact. 2011;10(1):114. doi: 10.1186/1475-2859-10-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasubramanian S., Manohar B.M., Rajaram A., Puvanakrishnan R. Ecofriendly lime and sulfide free enzymatic dehairing of skins and hides using a bacterial alkaline protease. Chemosphere. 2008;70(6):1015–1024. doi: 10.1016/j.chemosphere.2007.09.036. [DOI] [PubMed] [Google Scholar]
- Sundararajan S., Kannan C.N., Chittibabu S. Alkaline protease from Bacillus cereus VITSN04: potential application as a dehairing agent. J. Biosci. Bioeng. 2011;111(2):128–133. doi: 10.1016/j.jbiosc.2010.09.009. [DOI] [PubMed] [Google Scholar]
- Takami H., Nakamura S., Aono R., Horikoshi K. Degradation of human hair by a thermostable alkaline protease from alkaliphilic Bacillus sp. no. AH-101. Biosci. Biotechnol. Biochem. 1992;56(10):1667–1669. [Google Scholar]
- Takii Y., Kuriyama N., Suzuki Y. Alkaline serine protease produced from citric acid by Bacillus alcalophilus subsp. halodurans KP 1239. Appl. Microbiol. Biotechnol. 1990;34(1):57–62. doi: 10.1007/BF00170924. [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanitha N., Rajan S., Murugesan A. Optimization and production of alkaline protease enzyme from Bacillus subtilis 168 isolated from food industry waste. Int. J. Curr. Microbiol. App. Sci. 2014;3(6):36–44. [Google Scholar]
- Venugopal M., Saramma A. An alkaline protease from Bacillus circulans BM15, newly isolated from a mangrove station: characterization and application in laundry detergent formulations. Indian J. Microbiol. 2007;47(4):298–303. doi: 10.1007/s12088-007-0055-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayaraghavan P., Vijayan A., Arun A., Jenisha J.K., Vincent S.G.P. Cow dung: a potential biomass substrate for the production of detergent-stable dehairing protease by alkaliphilic Bacillus subtilis strain VV. SpringerPlus. 2012;1(1):76. doi: 10.1186/2193-1801-1-76. eCollection 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayaraghavan P., Vincent S.G.P. Cow dung as a novel, inexpensive substrate for the production of a halo-tolerant alkaline protease by Halomonas sp. PV1 for eco-friendly applications. Biochem. Eng. J. 2012;69:57–60. [Google Scholar]
- Yildirim V., Baltaci M.O., Ozgencli I., Sisecioglu M., Adiguzel A., Adiguzel G. Purification and biochemical characterization of a novel thermostable serine alkaline protease from Aeribacillus pallidus C10: a potential additive for detergents. J. Enzym. Inhib. Med. Chem. 2017;32(1):468–477. doi: 10.1080/14756366.2016.1261131. [DOI] [PMC free article] [PubMed] [Google Scholar]