Abstract
Fidelity of histone gene regulation, and ultimately of histone protein biosynthesis, is obligatory for packaging of newly replicated DNA into chromatin. Control of histone gene expression within the 3-dimensional context of nuclear organization is reflected by two well documented observations. DNA replication-dependent histone mRNAs are synthesized at specialized subnuclear domains designated histone locus bodies (HLBs), in response to activation of the growth factor dependent Cyclin E/CDK2/HINFP/NPAT pathway at the G1/S transition in mammalian cells. Complete loss of the histone gene regulatory factors HINFP or NPAT disrupts HLB integrity that is necessary for coordinate control of DNA replication and histone gene transcription. Here we review the molecular histone-related requirements for G1/S-phase progression during the cell cycle. Recently developed experimental strategies, now enable us to explore mechanisms involved in dynamic control of histone gene expression in the context of the temporal (cell cycle) and spatial (HLBs) remodeling of the histone gene loci.
Keywords: higher order organization, HINFP, histones, NPAT
1 | INTRODUCTION
Regulatory mechanisms operative at the G1/S phase transition in normal and cancer cells (Baumbach, Stein, & Stein, 1987; Braastad, Hovhannisyan, van Wijnen, Stein, & Stein, 2004; Carozzi et al., 1984; Chrysogelos, Pauli, Stein, & Stein, 1989; Chrysogelos, Riley, Stein, & Stein, 1985; Green et al., 1984; Guo, Stein, van Wijnen, & Stein, 1997; Holmes et al., 2005; Hovhannisyan et al., 2003; Lichtler, Detke, Phillips, Stein, & Stein, 1980; Lichtler et al., 1982; Marashi et al., 1982; Miele et al., 2005; Mitra et al., 2003; Moreno, Chrysogelos, Stein, & Stein, 1986; Morris et al., 1986; Pauli, Chrysogelos, Stein, & Stein, 1988; Plumb, Stein, & Stein, 1983; Sierra et al., 1982; Sierra, Stein, & Stein, 1983; Stein, Park, Thrall, Mans, & Stein, 1975; Stein, Stein, & Marzluff, 1984; Stein, Stein, Van Wijnen, & Lian, 1996; van Wijnen et al., 1996; Vaughan et al., 1995) provide mechanistic and clinically relevant insight into biological control and the molecular pathology of the cell cycle. A critical parameter of cell proliferation is the relationship of histone gene expression with DNA replication. Compromised cell cycle control is catastrophic for the cell.
Advances in understanding histone gene organization, expression and regulation (Birnbaum et al., 1995; Vaughan et al., 1998) have been instrumental in elucidating the integration of physiological signals that mediate physiological control of histone gene expression at multiple levels including transcription, 3′ end mRNA processing and mRNA stability. Competency for genome replication depends on the availability of histone proteins during S-phase for immediate packaging of newly replicated DNA into chromatin. The initial rate-limiting step in the induction of histone protein synthesis at the G1/S-phase transition, however, is cell cycle-dependent modulation of histone gene transcription.
The regulatory machinery for transcription and processing of histone RNAs is focally organized at major and minor histone gene loci in specialized subnuclear domains, designated histone locus bodies (HLBs, Figure 1) (Bongiorno-Borbone et al., 2008; Dominski & Marzluff, 2007; Ghule et al., 2007, 2008; Ma et al., 2000; Miele et al., 2005; Ye et al., 2003; Zhao, Dynlacht, Imai, Hori, & Harlow, 1998; Zhao et al., 2000). Loss of the key HLB components HINFP and NPAT leads to disruption of HLB focal organization, loss of competency for histone synthesis, and perturbation of cell cycle control; these regulatory factors are unequivocally required for physiological control of cell proliferation (Bongiorno-Borbone et al., 2010; Dominski & Marzluff, 2007; Ghule et al., 2014; Salzler et al., 2013; Yang et al., 2014; Ye et al., 2003; Zhao et al., 2000).
FIGURE 1.
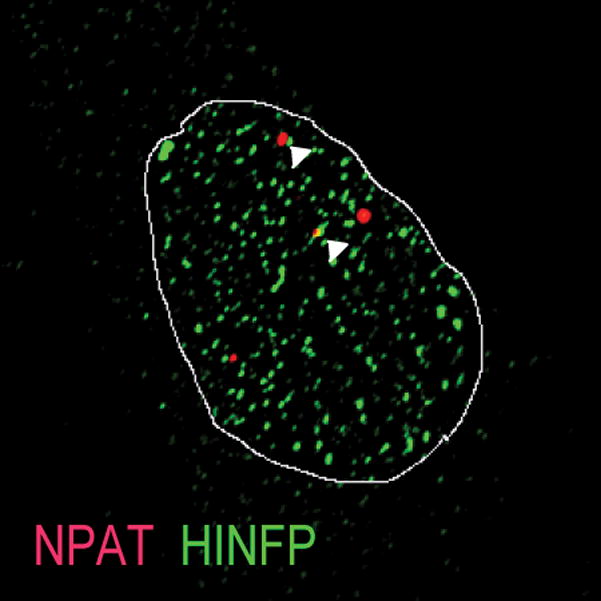
IF microscopy showing co-localization of histone gene transcription factors HINFP and NPAT at HLBs
An emerging dimension to transcriptional control is the contribution of higher-order nuclear organization and inter-/intra-chromosomal interactions in integrating activities of gene regulatory elements. Local interactions within the major histone gene locus (Fritz et al., 2018) have recently been identified. Understanding the higher-order genomic organization/chromatin architecture of histone genes within HLBs during cell cycle progression and the roles of HINFP and NPAT in regulating HLB structure and function is a compelling question.
2 | CONTROL OF HISTONE GENE TRANSCRIPTION
The HINFP transcription factor mediates coordinate expression of multiple histone H4 genes at the G1/S transition, including the abbreviated human embryonic stem (hES) cell cycle (Becker, Stein, Lian, van Wijnen, & Stein, 2007; Ghule et al., 2008, 2014; Holmes et al., 2005; Miele et al., 2005; Mitra et al., 2003). Using in vivo mouse models, it has been determined that HINFP is essential for fidelity of histone H4 gene expression and required for early embryonic development (Ghule et al., 2014; Xie et al., 2009).
NPAT is an essential and prototypical HLB resident protein that regulates expression of all replication-dependent histone gene classes (H4, H3, H2A, H2B, H1). Unlike HINFP, NPAT exerts its effects without directly binding to DNA. Molecular and biochemical studies show that HINFP is the critical anchor that tethers NPAT to histone H4 genes (Medina et al., 2007; Medina, van Wijnen, Stein, & Stein, 2006). In response to activation of the cyclin E/CDK2 cell cycle signaling cascade at the onset of S phase, NPAT is phosphorylated to initiate histone gene transcription (Bongiorno-Borbone et al., 2008; Dominski & Marzluff, 2007; Ghule et al., 2007, 2008; Ma et al., 2000; Miele et al., 2005; Ye et al., 2003; Zhao et al., 1998, 2000). Additional key protein components of the histone mRNA processing machinery, as well as other proteins involved in both histone RNA transcription and processing, that include FLASH, also co-localize at HLBs (Bongiorno-Borbone et al., 2008; Burch et al., 2011; Dominski & Marzluff, 2007; Ghule et al., 2008, 2009).
These results support the requirements of HINFP and NPAT for subnuclear organization of HLBs to regulate transcription of histone genes. It can be anticiapated that dynamic regulatory interactions mediated by HINFP and NPAT at HLBs occur in association with specific higher-order chromatin conformation within histone gene clusters, and that these regulatory interactions remodel with, and are functionally related to, cell-cycle progression.
3 | HISTONE GENE EXPRESSION IN NUCLEAR MICROENVIRONMENTS
Deregulation of histone gene expression upon loss of HINFP or NPAT causes disruption of HLBs and has drastic consequences for cell division and cell survival in both normal and cancer cells (Di Fruscio et al., 1997; Ghule et al., 2014, 2015; Ma et al., 2000). The resulting changes in the organization of newly replicated chromatin create both genomic instability and chromosomal aberrations, that are frequently observed during tumorigenesis. These findings provide a compelling basis for characterizing the organization of HLBs in normal and early stage cancer cells.
A fundamental question is exactly how DNA is organized within the 3-dimensional context of the nucleus and rendered accessible to regulatory factors during the cell cycle. This question has been the focus of studies on subnuclear organization (Cremer & Cremer, 2006; Zaidi et al., 2014). Emerging evidence using genome-wide high-resolution chromosome conformation capture (Hi-C) technologies indicates that non-random higher-order organization of genes into structural domains supports regulatory activities involving non-contiguous sequences that influence transcription (Barutcu et al., 2016; Belton et al., 2012; Ching, Ahmed, Boutros, Penn, & Bazett-Jones, 2013; Dekker, Marti-Renom, & Mirny, 2013; Hamdi et al., 2016; Lieberman-Aiden et al., 2009; Martin et al., 2015; Matheson & Kaufman, 2016; Mifsud et al., 2015; Rowley & Corces, 2016; Tjong et al., 2016; Wang et al., 2016).
The human histone locus bodies are unique nuclear domains for dynamic regulation of histone genes. They include multiple protein factors and clustered genes and an array of intra-/inter-chromosomal interactions—analogous to organization of ribosomal gene clusters in nucleoli. Insights into 3D architecture of HLBs during the cell cycle, as well as the role of histone gene regulatory factors HINFP and NPAT in genomic organization and function of histone gene clusters can enhance understanding of cell cycle control that is necessary for development and tissue remodeling and is compromised in the onset and progression of cancer.
4 | HISTONE LOCUS BODY: A SUBNUCLEAR DOMAIN DEDICATED TO HISTONE BIOGENESIS
Insight into cell cycle control from regulatory and biological perspectives is provided by mechanisms that expedite G1 events that are required for the initiation of S phase. The subnuclear organization of the regulatory factors that control histone gene transcription: the cyclin E/CDK2-dependent NPAT/HINFP coactivation complex, histone mRNA-related processing factors, and histone locus bodies (HLBs) have been informative. It has been established that, although appearance of CDK2-phosphorylated NPAT in these domains occurs when cells enter S-phase, HLBs are formed ~6 hr before S-phase in human somatic cells. Furthermore, regulatory complexes that mediate transcriptional initiation of histone genes (e.g., NPAT, HINFP, FLASH) and processing of histone mRNA (e.g., Lsm11 and SLBP) co-localize at histone gene loci in HLBs that are distinct from Cajal bodies (Ghule et al., 2008, 2009).
Deregulation of subnuclear organization and cell growth regulatory pathways are hallmarks of tumor cells (Stein, Davie, Knowlton, & Zaidi, 2008; Tai et al., 2014; Zaidi et al., 2007, 2014). It has been shown that HLB organization is disrupted in some cancer cell types in which histone gene transcription and histone mRNA processing factors are present in distinct compartments (i.e., HLBs and Cajal bodies, respectively) (Ghule et al., 2009).
Normal mammalian cells have two to four HLBs that encompass the two histone gene clusters at chromosomes 6p (major) and 1q (minor), as well as with factors involved in histone gene transcription (Figure 2) (Bongiorno-Borbone et al., 2008; Dominski & Marzluff, 2007; Ghule et al., 2007, 2008; Ma et al., 2000; Ye et al., 2003; Zhao et al., 1998, 2000). One key finding of our recent studies with Hinfp-depleted mouse embryonic fibroblasts is that loss of HINFP dramatically alters the number and/or focal organization of NPAT-containing bodies. Rather than forming a few distinct foci per nucleus as in wild-type cells, Hinfp-deficient cells exhibit diffuse NPAT staining. This result suggests that HINFP has an important architectural role in HLBs. Therefore, focal arrangement of NPAT and HINFP in HLBs is important for histone gene regulation (Figure 3) (Ghule et al., 2014).
FIGURE 2.
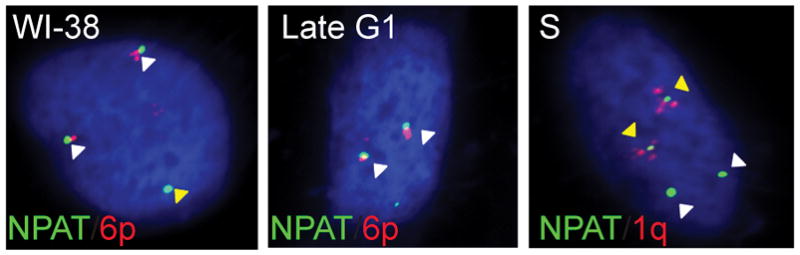
Immuno-FISH in normal diploid fibroblasts showing co-staining for HLB factor NPAT and the major Hist1 loci on 6p (white arrows) (left panel). Cell cycle stage-specific organization of the major HLB at 6p (Late G1) (middle panel) and both major (6p) and minor (1q) HLB (yellow arrows) (right panel) in S-phase
FIGURE 3.
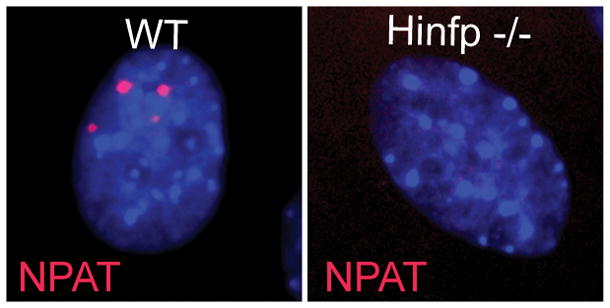
IF microscopy on WT and Hinp null cells showing loss of HLB factor HINFP causes deregulation of focal HLB organization
5 | CELL CYCLE CONTROL AND HISTONE GENE REGULATION IN HUMAN EMBRYONIC STEM CELLS
A fundamental mechanism that controls proliferation in pluripotent stem cells is a unique abbreviated cell cycle, with a shortened G1 phase and distinct differences in molecular parameters of cell cycle regulation (Becker et al., 2006, 2007; Ghule et al., 2007). hES cells are pre-mitotically committed to self-renewal and acquire a lengthened G1 phase, upon lineage programming (Becker, Stein, Lian, van Wijnen, & Stein, 2010). A unique chromatin architecture is associated with histone H4 genes in this abbreviated G1 phase, as reflected by major nuclease hypersensitive sites, atypical distribution of epigenetic histone marks, and a region devoid of histone octamers. Furthermore, cell-cycle progression in hES cells involves cell cycle stage-specific chromatin-remodeling events, and rapid spatial and temporal assembly of HLBs that activate histone gene transcription to accommodate nucleosomal packaging of newly replicated DNA (Ghule et al., 2008; Medina et al., 2012). These findings in hES cells underscore the crucial role of subnuclear and chromosomal structure in activation of histone gene expression.
6 | HINFP IS AN ESSENTIAL REGULATOR OF HISTONE GENE TRANSCRIPTION AND FIDELITY OF CELL CYCLE CONTROL
Because histones are required for packaging DNA into chromatin, competency for DNA replication is functionally coupled to the activation of histone gene expression at the onset of S phase. It has been established that the histone gene activator HINFP is a unique zinc finger transcription factor with a novel conserved auxiliary DNA-binding motif in the C-terminus. The CDK2/cyclin E/NPAT/HINFP/histone gene signaling pathway at the G1/S phase transition is an essential, cell cycle regulatory mechanism that is established early in embryogenesis (Xie et al., 2009). CDK inhibitors selectively diminish cell cycle-associated activation of the histone H4 gene promoter by NPAT and HINFP (Mitra et al., 2009).
The autonomous biological role of Hinfp has been established using conditional-null Hinfp-mouse embryonic fibroblasts. Recently it was demonstrated that Hinfp-mediated deregulation of histone H4 results in cellular and molecular defects that lead to genomic instability. Functional evidence has been provided that the tight coupling between DNA replication and histone synthesis is reciprocal (Ghule et al., 2014) Furthermore, simultaneous loss of Hinfp and p53 exacerbates cellular defects (Ghule et al., 2015). We observed that siRNA-depletion of HINFP-regulated H4 mRNAs causes genomic instability (unpublished data). This finding is consistent with the biological effects of genetic deletion of HINFP. Using a Hinfp-null mouse model, it has been observed that oocytes contain sufficient maternally-derived Hinfp-gene transcripts and/or protein for the earliest stages of embryogenesis, and that expression of zygote-derived Hinfp commences at the 4-cell stage of development (Ghule et al., 2016).
These results establish a linear causality between cyclin E/CDK2 mediated stimulation of the NPAT/HINFP coactivation complex, temporal and functional fidelity of H4 gene expression, chromatin integrity and genomic stability. These studies defined HLBs as crucial regulatory hubs that organize histone genes into specialized subnuclear domains in which transcriptional and processing pathways are juxtaposed. It was demonstrated that HLB organization is disrupted in at least some cancer cells. There is a necessity to understand functional changes in the higher-order genomic architecture of HLBs during the cell cycles of normal and cancer cells, and the functions of HINFP and NPAT in structuring the genomic organization of HLBs.
7 | HIGHER ORDER CHROMATIN ORGANIZATION OF THE MAJOR HISTONE LOCUS
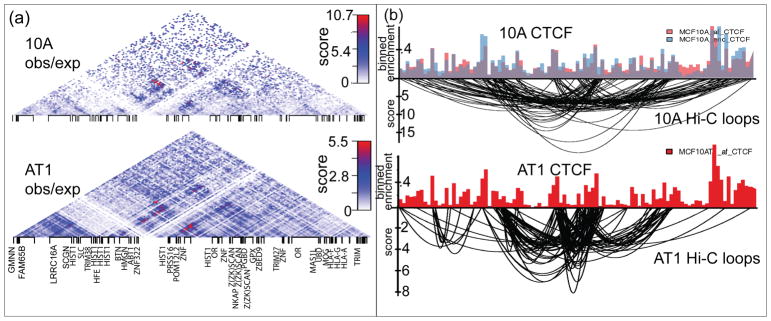
Alterations in nuclear morphology are common in cancer progression. However, the degree to which gross morphological abnormalities translate into compromised higher-order chromatin organization is poorly understood. Functional links between gene expression and chromatin structure in breast cancer, have been established by global gene expression profiling on the MCF10 basal breast cancer progression model cells. Positional gene enrichment identified the major histone gene cluster at chromosome 6p22 (Hist1) as one of the most significantly upregulated (and not amplified) clusters of genes from the normal-like MCF10A to premalignant MCF10AT1 and metastatic MCF10CA1a cells. The Hist1 cluster is subdivided into three sub-clusters of histone genes that are organized into hierarchical topologically associating domains (TADs). Interestingly, the sub-clusters of histone genes are located at TAD boundaries and interact more frequently with each other than the regions in-between them, suggesting that the histone subclusters form an active chromatin hub. The anchor sites of loops within this hub are occupied by CTCF, a known chromatin organizer (Figure 4). While the overall chromatin structure of the major histone gene locus is maintained across breast cancer progression, the role of HLB genomic organization in regulating histone gene expression needs further validation. Importantly, breast tumor specimens also exhibit a coordinate pattern of upregulation across the Hist1 locus (Fritz et al., 2018). Our results provide a novel insight into the higher-order chromatin organization of the major histone gene locus during breast cancer progression that may be clinically informative.
FIGURE 4.
(Adapted from Figures 3 and 4 published in Fritz et al. (2018)) The higher-order chromatin organization of the major histone gene locus. (a) Pairwise DNA-DNA interactions were analyzed via Hi-C within the major histone gene cluster in the MCF10 cells. The structure of the region is organized into hierarchical topologically associating domains. Interactions that are enriched above expected linear sequence-interactions are shown. (b) CTCF is a structural component of the major histone gene locus. CTCF ChIP-seq showing CTCF is coincident with looping of chromatin at the major histone gene cluster
8 | CONCLUSION AND PERSPECTIVE
A compelling question and novel dimension to cell cycle control at the G1/S-phase transition is how higher-order organization and genomic context at HLBs change during the cell cycle to regulate histone RNA transcription. A “working hypothesis” is that higher order genomic organization of histone genes and dynamic recruitment of HINFP and NPAT at histone locus bodies are functionally linked to competency and fidelity of histone gene expression during the cell cycle. Compromised cell cycle control is a hallmark of cancer development/progression. Pursuing nuclear structure and gene expression relationships will elucidate new dimensions for control of the cell cycle and cell proliferation. Chromosome conformation capture approaches can identify higher-order genomic organization of, and regulation at, HLBs during the cell cycle. These strategies will define the dynamic function roles of two principal cell-cycle regulators, including HINFP and NPAT, in higher-order genomic organization of HLBs, which coordinate histone gene expression with DNA replication for competency to package DNA as chromatin and for genome integrity. We expect new insights into fundamental relationships of higher order nuclear structure with cell cycle regulation of biological control and perturbations in cancer.
Acknowledgments
Funding information
American Cancer Society, Grant number: 14-196-01; The Charlotte Perelman Fund for Cancer Research; National Cancer Institute, Grant numbers: P01 CA082834, R01 CA139322, 1F32CA220935
This work was supported by National Institutes of Health grants R01 CA139322 and The Charlotte Perelman Fund for Cancer Research to GSS, Institutional Research Grant 14-196-01 from the American Cancer Society (project# 033807) to PNG and DJS, NIH/NCI P01 CA082834 (project 1) to JLS and GSS, NIH/NCIP01 CA082834 (project 3) to JBL, NIH/NCI U01 CA196383 to JLS, and NIH/NCI 1F32CA220935 fellowship to AJF. The authors thank the members of Stein-Lian laboratories for helpful discussions and suggestions. The authors also thank Caitlin Maloney for assistance with manuscript preparation. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
References
- Barutcu AR, Hong D, Lajoie BR, McCord RP, van Wijnen AJ, Lian JB, … Stein GS. RUNX1 contributes to higher-order chromatin organization and gene regulation in breast cancer cells. Biochimica et Biophysica Acta. 2016;1859(11):1389–1397. doi: 10.1016/j.bbagrm.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumbach LL, Stein GS, Stein JL. Regulation of human histone gene expression: Transcriptional and posttranscriptional control in the coupling of histone messenger RNA stability with DNA replication. Biochemistry. 1987;26(19):6178–6187. doi: 10.1021/bi00393a034. [DOI] [PubMed] [Google Scholar]
- Becker KA, Ghule PN, Therrien JA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. Journal of Cellular Physiology. 2006;209(3):883–893. doi: 10.1002/jcp.20776. [DOI] [PubMed] [Google Scholar]
- Becker KA, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Establishment of histone gene regulation and cell cycle checkpoint control in human embryonic stem cells. Journal of Cellular Physiology. 2007;210(2):517–526. doi: 10.1002/jcp.20903. [DOI] [PubMed] [Google Scholar]
- Becker KA, Stein JL, Lian JB, van Wijnen AJ, Stein GS. Human embryonic stem cells are pre-mitotically committed to self-renewal and acquire a lengthened G1 phase upon lineage programming. Journal of Cellular Physiology. 2010;222(1):103–110. doi: 10.1002/jcp.21925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belton JM, McCord RP, Gibcus JH, Naumova N, Zhan Y, Dekker JH-C. A comprehensive technique to capture the conformation of genomes. Methods. 2012;58(3):268–276. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum MJ, Wright KL, van Wijnen AJ, Ramsey-Ewing AL, Bourke MT, Last TJ, Aziz F, Frenkel B, Rao BR, Aronin N, et al. Functional role for Sp1 in the transcriptional amplification of a cell cycle regulated histone H4 gene. Biochemistry. 1995;34(23):7648–7658. doi: 10.1021/bi00023a011. [DOI] [PubMed] [Google Scholar]
- Bongiorno-Borbone L, De Cola A, Vernole P, Finos L, Barcaroli D, Knight RA, … De Laurenzi V. FLASH and NPAT positive but not coilin positive cajal bodies correlate with cell ploidy. Cell Cycle. 2008;7(15):2357–2367. doi: 10.4161/cc.6344. [DOI] [PubMed] [Google Scholar]
- Bongiorno-Borbone L, De Cola A, Barcaroli D, Knight RA, Di Ilio C, Melino G, De Laurenzi V. FLASH degradation in response to UV-C results in histone locus bodies disruption and cell-cycle arrest. Oncogene. 2010;29(6):802–810. doi: 10.1038/onc.2009.388. [DOI] [PubMed] [Google Scholar]
- Braastad CD, Hovhannisyan H, van Wijnen AJ, Stein JL, Stein GS. Functional characterization of a human histone gene cluster duplication. Gene. 2004;342(1):35–40. doi: 10.1016/j.gene.2004.07.036. [DOI] [PubMed] [Google Scholar]
- Burch BD, Godfrey AC, Gasdaska PY, Salzler HR, Duronio RJ, Marzluff WF, Dominski Z. Interaction between FLASH and Lsm11 is essential for histone pre-mRNA processing in vivo in Drosophila. RNA. 2011;17(6):1132–1147. doi: 10.1261/rna.2566811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carozzi N, Marashi F, Plumb M, Zimmerman S, Zimmerman A, Coles LS, … Stein J. Clustering of human H1 and core histone genes. Science. 1984;224(4653):1115–1117. doi: 10.1126/science.6719136. [DOI] [PubMed] [Google Scholar]
- Ching RW, Ahmed K, Boutros PC, Penn LZ, Bazett-Jones DP. Identifying gene locus associations with promyelocytic leukemia nuclear bodies using immuno-TRAP. The Journal of Cell Biology. 2013;201(2):325–335. doi: 10.1083/jcb.201211097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrysogelos S, Riley DE, Stein G, Stein J. A human histone H4 gene exhibits cell cycle-dependent changes in chromatin structure that correlate with its expression. Proceedings of the National Academy of Sciences of the United States of America. 1985;82(22):7535–7539. doi: 10.1073/pnas.82.22.7535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrysogelos S, Pauli U, Stein G, Stein J. Fine mapping of the chromatin structure of a cell cycle-regulated human H4 histone gene. The Journal of Biological Chemistry. 1989;264(2):1232–1237. [PubMed] [Google Scholar]
- Cremer T, Cremer C. Rise, fall and resurrection of chromosome territories: A historical perspective. Part II. Fall and resurrection of chromosome territories during the1950s to 1980s. Part III. Chromosome territories and the functional nuclear architecture: Experiments and models from the 1990s to the present. European Journal of Histochemistry. 2006;50(4):223–272. [PubMed] [Google Scholar]
- Dekker J, Marti-Renom MA, Mirny LA. Exploring the three-dimensional organization of genomes: Interpreting chromatin interaction data. Nature Reviews Genetics. 2013;14(6):390–403. doi: 10.1038/nrg3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fruscio M, Weiher H, Vanderhyden BC, Imai T, Shiomi T, Hori TA, … Gray DA. Proviral inactivation of the Npat gene of Mpv 20 mice results in early embryonic arrest. Molecular and Cellular Biology. 1997;17(7):4080–4086. doi: 10.1128/mcb.17.7.4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA: Getting closer to the end. Gene. 2007;396(2):373–390. doi: 10.1016/j.gene.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz AJ, Ghule PN, Boyd JR, Tye CE, Page NA, Hong D, … Stein GS. Intranuclear and higher-order chromatin organization of the major histone gene cluster in breast cancer. Journal of Cellular Physiology. 2018;233(2):1278–1290. doi: 10.1002/jcp.25996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghule PN, Becker KA, Harper JW, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Cell cycle dependent phosphorylation and subnuclear organization of the histone gene regulator p220(NPAT) in human embryonic stem cells. Journal of Cellular Physiology. 2007;213(1):9–17. doi: 10.1002/jcp.21119. [DOI] [PubMed] [Google Scholar]
- Ghule PN, Dominski Z, Lian JB, Stein JL, van Wijnen AJ, Stein GS. The subnuclear organization of histone gene regulatory proteins and 3′ end processing factors of normal somatic and embryonic stem cells is compromised in selected human cancer cell types. Journal of Cellular Physiology. 2009;220(1):129–135. doi: 10.1002/jcp.21740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghule PN, Dominski Z, Yang XC, Marzluff WF, Becker KA, Harper JW, … Stein GS. Staged assembly of histone gene expression machinery at subnuclear foci in the abbreviated cell cycle of human embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(44):16964–16969. doi: 10.1073/pnas.0809273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghule PN, Xie RL, Colby JL, Jones SN, Lian JB, Wijnen AJ, … Stein GS. P53 checkpoint ablation exacerbates the phenotype of Hinfp dependent histone H4 deficiency. Cell Cycle. 2015;14(15):2501–2508. doi: 10.1080/15384101.2015.1049783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghule PN, Xie RL, Colby JL, Rivera-Perez JA, Jones SN, Lian JB, … Stein GS. Maternal expression and early induction of histone gene transcription factor Hinfp sustains development in pre-implantation embryos. Developments in Biologicals. 2016;419(2):311–320. doi: 10.1016/j.ydbio.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghule PN, Xie RL, Medina R, Colby JL, Jones SN, Lian JB, … Stein GS. Fidelity of histone gene regulation is obligatory for genome replication and stability. Molecular and Cellular Biology. 2014;34(14):2650–2659. doi: 10.1128/MCB.01567-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green L, Van Antwerpen R, Stein J, Stein G, Tripputi P, Emanuel B, … Croce C. A major human histone gene cluster on the long arm of chromosome 1. Science. 1984;226(4676):838–840. doi: 10.1126/science.6494913. [DOI] [PubMed] [Google Scholar]
- Guo B, Stein JL, van Wijnen AJ, Stein GS. ATF1 and CREB trans-activate a cell cycle regulated histone H4 gene at a distal nuclear matrix associated promoter element. Biochemistry. 1997;36(47):14447–14455. doi: 10.1021/bi971781s. [DOI] [PubMed] [Google Scholar]
- Hamdi Y, Soucy P, Kuchenbaeker KB, Pastinen T, Droit A, Lemacon A, … Simard J. Association of breast cancer risk in BRCA1 and BRCA2 mutation carriers with genetic variants showing differential allelic expression: Identification of a modifier of breast cancer risk at locus 11q22.3. Breast Cancer Research and Treatment. 2016;161(1):117–134. doi: 10.1007/s10549-016-4018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes WF, Braastad CD, Mitra P, Hampe C, Doenecke D, Albig W, … Stein GS. Coordinate control and selective expression of the full complement of replication-dependent histone H4 genes in normal and cancer cells. The Journal of Biological Chemistry. 2005;280(45):37400–37407. doi: 10.1074/jbc.M506995200. [DOI] [PubMed] [Google Scholar]
- Hovhannisyan H, Cho B, Mitra P, Montecino M, Stein GS, Van Wijnen AJ, Stein JL. Maintenance of open chromatin and selective genomic occupancy at the cell cycle-regulated histone H4 promoter during differentiation of HL-60 promyelocytic leukemia cells. Molecular and Cellular Biology. 2003;23(4):1460–1469. doi: 10.1128/MCB.23.4.1460-1469.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtler AC, Detke S, Phillips IR, Stein GS, Stein JL. Multiple forms of H4 histone mRNA in human cells. Proceedings of the National Academy of Sciences of the United States of America. 1980;77(4):1942–1946. doi: 10.1073/pnas.77.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtler AC, Sierra F, Clark S, Wells JR, Stein JL, Stein GS. Multiple H4 histone mRNAs of HeLa cells are encoded in different genes. Nature. 1982;298(5870):195–198. doi: 10.1038/298195a0. [DOI] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, … Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Van Tine BA, Wei Y, Garrett MD, Nelson D, Adams PD, … Harper JW. Cell cycle-regulated phosphorylation of p220 (NPAT) by cyclin E/Cdk2 in cajal bodies promotes histone gene transcription. Genes & Development. 2000;14(18):2298–2313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marashi F, Baumbach L, Rickles R, Sierra F, Stein JL, Stein GS. Histone proteins in HeLa S3 cells are synthesized in a cell cycle stage specific manner. Science. 1982;215(4533):683–685. doi: 10.1126/science.7058333. [DOI] [PubMed] [Google Scholar]
- Martin P, McGovern A, Orozco G, Duffus K, Yarwood A, Schoenfelder S, … Eyre S. Capture Hi-C reveals novel candidate genes and complex long-range interactions with related autoimmune risk loci. Nature Communications. 2015;6:10069. doi: 10.1038/ncomms10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheson TD, Kaufman PD. Grabbing the genome by the NADs. Chromosoma. 2016;125(3):361–371. doi: 10.1007/s00412-015-0527-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina R, Ghule PN, Cruzat F, Barutcu AR, Montecino M, Stein JL, … Stein GS. Epigenetic control of cell cycle-dependent histone gene expression is a principal component of the abbreviated pluripotent cell cycle. Molecular and Cellular Biology. 2012;32(19):3860–3871. doi: 10.1128/MCB.00736-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina R, van der Deen M, Miele-Chamberland A, Xie RL, van Wijnen AJ, Stein JL, Stein GS. The HiNF-P/p220NPAT cell cycle signaling pathway controls nonhistone target genes. Cancer Research. 2007;67(21):10334–10342. doi: 10.1158/0008-5472.CAN-07-1560. [DOI] [PubMed] [Google Scholar]
- Medina R, van Wijnen AJ, Stein GS, Stein JL. The histone gene transcription factor HiNF-P stabilizes its cell cycle regulatory co-activator p220NPAT. Biochemistry. 2006;45(51):15915–15920. doi: 10.1021/bi061425m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele A, Braastad CD, Holmes WF, Mitra P, Medina R, Xie R, … Stein GS. HiNF-P directly links the cyclin E/CDK2/p220NPAT pathway to histone H4 gene regulation at the G1/S phase cell cycle transition. Molecular and Cellular Biology. 2005;25(14):6140–6153. doi: 10.1128/MCB.25.14.6140-6153.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mifsud B, Tavares-Cadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, … Osborne CS. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nature Genetics. 2015;47(6):598–606. doi: 10.1038/ng.3286. [DOI] [PubMed] [Google Scholar]
- Mitra P, Ghule PN, van der Deen M, Medina R, Xie RL, Holmes WF, … van Wijnen AJ. CDK inhibitors selectively diminish cell cycle controlled activation of the histone H4 gene promoter by p220NPAT and HiNF-P. Journal of Cellular Physiology. 2009;219(2):438–448. doi: 10.1002/jcp.21687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra P, Xie RL, Medina R, Hovhannisyan H, Zaidi SK, Wei Y, … Stein GS. Identification of HiNF-P, a key activator of cell cycle-controlled histone H4 genes at the onset of S phase. Molecular and Cellular Biology. 2003;23(22):8110–8123. doi: 10.1128/MCB.23.22.8110-8123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno ML, Chrysogelos SA, Stein GS, Stein JL. Reversible changes in the nucleosomal organization of a human H4 histone gene during the cell cycle. Biochemistry. 1986;25(19):5364–5370. doi: 10.1021/bi00367a003. [DOI] [PubMed] [Google Scholar]
- Morris T, Marashi F, Weber L, Hickey E, Greenspan D, Bonner J, … Stein G. Involvement of the 5′-leader sequence in coupling the stability of a human H3 histone mRNA with DNA replication. Proceedings of the National Academy of Sciences of the United States of America. 1986;83(4):981–985. doi: 10.1073/pnas.83.4.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli U, Chrysogelos S, Stein J, Stein G. Native genomic blotting: High-resolution mapping of DNase I-hypersensitive sites and protein-DNA interactions. Proceedings of the National Academy of Sciences of the United States of America. 1988;85(1):16–20. doi: 10.1073/pnas.85.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumb M, Stein J, Stein G. Coordinate regulation of multiple histone mRNAs during the cell cycle in HeLa cells. Nucleic Acids Research. 1983;11(8):2391–2410. doi: 10.1093/nar/11.8.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley MJ, Corces VG. The three-dimensional genome: Principles and roles of long-distance interactions. Current Opinion in Cell Biology. 2016;40:8–14. doi: 10.1016/j.ceb.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzler HR, Tatomer DC, Malek PY, McDaniel SL, Orlando AN, Marzluff WF, Duronio RJ. A sequence in the Drosophila H3-H4 promoter triggers histone locus body assembly and biosynthesis of replication-coupled histone mRNAs. Developmental Cell. 2013;24(6):623–634. doi: 10.1016/j.devcel.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra F, Lichtler A, Marashi F, Rickles R, Van Dyke T, Clark S, … Stein J. Organization of human histone genes. Proceedings of the National Academy of Sciences of the United States of America. 1982;79(6):1795–1799. doi: 10.1073/pnas.79.6.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierra F, Stein G, Stein J. Structure and in vitro transcription of a human H4 histone gene. Nucleic Acids Research. 1983;11(20):7069–7086. doi: 10.1093/nar/11.20.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein G, Park W, Thrall C, Mans R, Stein J. Regulation of cell cycle stage-specific transcription of histone genes from chromatin by non-histone chromosomal proteins. Nature. 1975;257(5529):764–767. doi: 10.1038/257764a0. [DOI] [PubMed] [Google Scholar]
- Stein GS, Stein JL, Marzluff WF. Histone Genes: Structure, Organization, and Regulation. New York, NY: John Wiley & Sons; 1984. p. 510. [Google Scholar]
- Stein GS, Davie JR, Knowlton JR, Zaidi SK. Nuclear microenvironments and cancer. Journal of Cellular Biochemistry. 2008;104(6):1949–1952. doi: 10.1002/jcb.21846. [DOI] [PubMed] [Google Scholar]
- Stein GS, Stein JL, Van Wijnen AJ, Lian JB. Transcriptional control of cell cycle progression: The histone gene is a paradigm for the G1/S phase and proliferation/differentiation transitions. Cell Biology International. 1996;20(1):41–49. doi: 10.1006/cbir.1996.0007. [DOI] [PubMed] [Google Scholar]
- Tai PW, Zaidi SK, Wu H, Grandy RA, Montecino M, van Wijnen AJ, … Stein JL. The dynamic architectural and epigenetic nuclear landscape: Developing the genomic almanac of biology and disease. Journal of Cellular Physiology. 2014;229(6):711–727. doi: 10.1002/jcp.24508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjong H, Li W, Kalhor R, Dai C, Hao S, Gong K, … Alber F. Population-based 3D genome structure analysis reveals driving forces in spatial genome organization. Proceedings of the National Academy of Sciences of the United States of America. 2016;113(12):E1663–E1672. doi: 10.1073/pnas.1512577113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijnen AJ, van Gurp MF, de Ridder MC, Tufarelli C, Last TJ, Birnbaum M, … Stein GS. CDP/cut is the DNA-binding subunit of histone gene transcription factor HiNF-D: A mechanism for gene regulation at the G1/S phase cell cycle transition point independent of transcription factor E2F. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(21):11516–11521. doi: 10.1073/pnas.93.21.11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan PS, Aziz F, van Wijnen AJ, Wu S, Harada H, Taniguchi T, … Stein GS. Activation of a cell-cycle-regulated histone gene by the oncogenic transcription factor IRF-2. Nature. 1995;377(6547):362–365. doi: 10.1038/377362a0. [DOI] [PubMed] [Google Scholar]
- Vaughan PS, van der Meijden CM, Aziz F, Harada H, Taniguchi T, van Wijnen AJ, … Stein GS. Cell cycle regulation of histone H4 gene transcription requires the oncogenic factor IRF-2. The Journal of Biological Chemistry. 1998;273(1):194–199. doi: 10.1074/jbc.273.1.194. [DOI] [PubMed] [Google Scholar]
- Wang Q, Sawyer IA, Sung MH, Sturgill D, Shevtsov SP, Pegoraro G, … Dundr M. Cajal bodies are linked to genome conformation. Nature Communications. 2016;7:10966. doi: 10.1038/ncomms10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie R, Medina R, Zhang Y, Hussain S, Colby J, Ghule P, … Stein GS. The histone gene activator HINFP is a nonredundant cyclin E/CDK2 effector during early embryonic cell cycles. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(30):12359–12364. doi: 10.1073/pnas.0905651106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XC, Sabath I, Kunduru L, van Wijnen AJ, Marzluff WF, Dominski Z. A conserved interaction that is essential for the biogenesis of histone locus bodies. The Journal of Biological Chemistry. 2014;289(49):33767–33782. doi: 10.1074/jbc.M114.616466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Franco AA, Santos H, Nelson DM, Kaufman PD, Adams PD. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Molecular Cell. 2003;11(2):341–351. doi: 10.1016/s1097-2765(03)00037-6. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Javed A, Pratap J, Montecino M, van Wijnen A, … Stein GS. Nuclear microenvironments in biological control and cancer. Nature Reviews Cancer. 2007;7(6):454–463. doi: 10.1038/nrc2149. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Grandy RA, Lopez-Camacho C, Montecino M, van Wijnen AJ, Lian JB, … Stein GS. Bookmarking target genes in mitosis: A shared epigenetic trait of phenotypic transcription factors and oncogenes? Cancer Research. 2014;74(2):420–425. doi: 10.1158/0008-5472.CAN-13-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Dynlacht B, Imai T, Hori T, Harlow E. Expression of NPAT, a novel substrate of cyclin E-CDK2, promotes S-phase entry. Genes & Development. 1998;12(4):456–461. doi: 10.1101/gad.12.4.456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, Harlow E. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes & Development. 2000;14(18):2283–2297. [PMC free article] [PubMed] [Google Scholar]