Abstract
Strategies to fabricate biofunctionalized surfaces are essential for many biotechnological applications. Zwitterionic lipid bilayer coatings doped with lipids with chemically selective headgroups provide a robust platform for immobilization of biomolecules in an antifouling, protein resistant background. Herein, we assess the biological activity of two important components of the extracellular matrix (ECM), collagen type I (Col I) and fibronectin (FN), which are covalently attached to a supported lipid bilayer (SLB), and compare their activity with the same proteins, nonspecifically adsorbed onto a SiO2 surface. The characterization of protein coatings by quartz crystal microbalance with dissipation revealed that Col I and FN attached to SLB are less dense and have higher structural flexibility than when adsorbed onto SiO2. Cell adhesion, proliferation, and function, as well as Col I–FN interactions, were more efficient on the ECM-functionalized SLB, making it a promising platform for cell-based diagnostics, tissue engineering, medical implants, and biosensor development.
Introduction
Controlled functionalization of solid surfaces with proteins is a critical step in the development of biointeractive surfaces employed in advanced technologies such as cell-based diagnostics and therapies, tissue engineering, biomolecular therapies, and biosensors.1−6 Proteins usually physically adsorb to commonly used surfaces such as metal oxides, glass, polystyrene, and gold. The protein–surface interaction is a result of a complex interplay between many factors including (i) environmental conditions such as temperature,7 pH, and salinity,8 (ii) protein properties9 such as size, structural stability, and composition, as well as, (iii) surface properties such as polarity, charge, and roughness.10 However, depending on the interaction strength between proteins and the surface, proteins may undergo structural rearrangements or even denature upon adsorption.10−12 The biological activity and function of proteins, such as enzymatic activity or presentation of ligands for specific receptors, usually originate from the functional domains in their 3D structures that specifically interact with other biomolecules. Consequently, any surface-induced structural deformation may alter the protein function. Moreover, the part of the protein molecule which interacts with the surface is not available for other interactions. Thus, strategies to coat surfaces with proteins whilst preserving their biological activities are of importance for the successful development of tailored biomaterial surfaces. In this regard, supported lipid bilayers (SLBs)13 offer unique possibilities for controlled functionalization of solid surfaces.14,15
SLBs are biomembrane-mimetic coating materials that are biocompatible. Importantly, lipid membrane coatings composed of zwitterionic lipids that have both a positively and a negatively charged moiety within the same headgroup (such as phosphorylcholine) are nonfouling and resistant to nonspecific adsorption of proteins16 and cells.17 However, specific lipids containing reactive headgroups such as amine or free carboxylic acid,18 or specific chemical functionalities, such as biotin19 or DNA oligonucleotides,20 can be introduced into the membrane to act as linkers to specifically attach proteins or any biomolecule of interest to the membrane surface. The protein resistance but functionalizable characteristics of lipid membranes have led to many applications in biomedical research and biotechnology. The applications include the design of various innovative biointerfaces for optically or electrically active substrates in biosensors, passivation of micro and nanofluidics,21 and functionalization of nanoparticles for targeted drug delivery applications.22−24 In addition, SLBs functionalized with cell-binding peptides25−28 have been developed for studying cell behavior. In this regard, it is important to evaluate the biological activity of proteins that covalently attach to SLBs.
Collagen type I (Col I)29 and fibronectin (FN)30 belong to a collection of molecules called the extracellular matrix (ECM),31 which are secreted by cells and provide structural and biochemical support to the surrounding cells. They are of immense biomedical importance and have central roles in cell adhesion. This feature has been exploited to mediate adhesion of cells to synthetic surfaces coated with ECM for biomaterial and tissue-engineering applications.32−35 In addition, SLBs functionalized with Col I and FN have also been demonstrated to be an efficient platform for mammalian cell culture, and it has been shown that, for instance, smooth muscle cells retained normal growth behavior on such platforms.36,37
Herein, we provide a comparative assessment of the biological activity of Col I and FN when they are (i) attached to the SLB via covalent bonding and (ii) adsorbed nonspecifically to a SiO2 surface. To this end, protein adsorption to each surface was quantified by quartz crystal microbalance with dissipation (QCM-D). The amount of adsorbed protein to each platform was used as a framework for comparison of the protein biological activities including the cell attachment, proliferation, and function, as well as protein–protein interactions.
Results and Discussion
First, we demonstrate the preparation and characterization of ECM-functionalized bilayer platforms; then, we compare the biological activity of ECM proteins on SLB platforms with those prepared on SiO2 or glass. As mentioned before, the common strategy to functionalize lipid bilayers is to incorporate lipid-like molecules with chemically reactive headgroups into the bilayer structure.38 The functionalized lipids act as an anchor to immobilize specific biomolecules containing the corresponding binding moiety to the membrane surface. Importantly, the degree of biomolecule conjugation to the bilayer can be controlled by the fraction of functional lipids included in the bilayer.
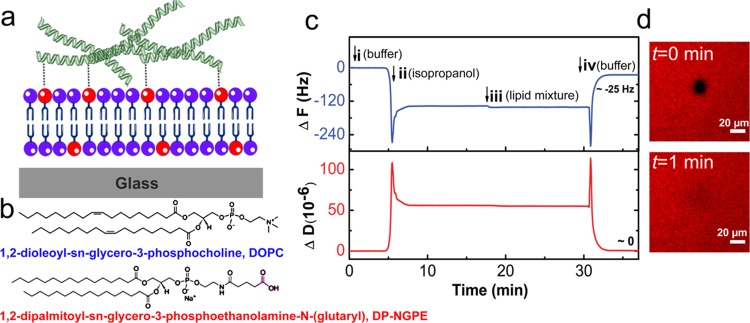
Figure 1a illustrates the schematics of a solid surface coated with a lipid bilayer that is functionalized with ECM proteins. In the current work, we prepared bilayers composed of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) doped with 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(glutaryl) (DP-NGPE). DOPC (Figure 1b) is a neutral, zwitterionic lipid that provides resistance to protein adhesion and gives the bilayer its antifouling capabilities. DP-NGPE serves as a functionalized lipid whose headgroup bears a free carboxylic acid (Figure 1b). After activation, DP-NGPE can react covalently with amine-containing biomolecules (e.g., proteins).
Figure 1.
ECM-functionalized SLB platform. (a) Schematic illustration of model membranes functionalized with ECM molecules via covalent bonding to lipids with reactive headgroups. (b) Structures of the lipids used in this study, together with their chemical names. (c) QCM-D frequency shift (ΔF, top panel) and dissipation (ΔD, bottom panel) for the third overtone were measured as a function of time during the formation of the planar bilayer on silicon dioxide. SLBs were formed on SiO2 via a solvent-assisted lipid bilayer (SALB) formation method using an isopropanol solution of a zwitterionic lipid (DOPC) and a lipid bearing a free carboxylic acid for amine coupling (DP-NGPE). Arrows indicate the injection of (i) tris buffer (10 mM tris, 150 mM NaCl, pH 7.5), (ii) isopropanol, (iii) lipid mixture [0.5 mg/mL of DOPC/DP-NGPE (5:1) in isopropanol], and (iv) tris buffer solution leading to a final ΔF and ΔD of −25 ± 1 Hz and 0.4 ± 0.2 × 10–6, respectively, which correspond to a planar bilayer. (d) Fluorescence recovery after photobleaching (FRAP) snapshots of DP-NGPE doped bilayer containing 0.5 wt % Rho-PE lipid, immediately (top) and 1 min (below) after photobleaching, showing almost complete recovery, indicating the fluidity of the SLB.
Previously, Huang et al.36,37 used the vesicle fusion method to form a SLB containing various fractions of DP-NGPE. Vesicle fusion, which involves the spontaneous adsorption and rupture of phospholipid vesicles, is the most commonly used method to form SLBs. However, under physiological conditions, vesicle fusion is limited to only a subset of lipid compositions and solid supports. In this work, the surface was coated with phospholipid bilayer using the so-called SALB formation method.39 The SALB method is based on the reverse-phase evaporation phenomenon and involves lipid deposition onto a hydrophilic solid surface in an alcohol, for example, isopropanol, followed by the slow exchange of the alcohol with water, resulting in the formation of a lipid bilayer on the surface.40 The SALB method has been applied successfully to a wide variety of surfaces including SiO2, and those which are intractable to the conventional vesicle fusion method, such as Au,39 Al2O3,41 and graphene.42 In addition, the SALB method enables formation of bilayers composed of lipids that cannot be formed by the vesicle fusion method, for example, those containing large fractions of cholesterol.43,44
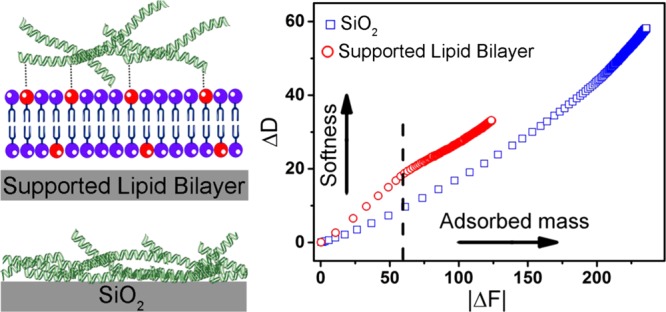
The bilayer formation on a solid surface and subsequent protein conjugation were monitored by the QCM-D monitoring measurement technique.36,37 QCM-D allows real-time monitoring of adsorption processes involving lipid membranes and proteins.45 The method records the frequency change (ΔF), which depends on the total effective mass of the adlayer, which consists of the mass of the coating material plus entrapped and coupled water. A negative change in ΔF indicates an increase in the mass of the adlayer. In addition, QCM-D measures energy dissipation (ΔD), which reflects the softness/rigidness of the adlayer. A perfectly rigid film, such as SLB, has a near zero energy dissipation, whereas softer adlayers, such as proteins, exhibit greater energy dissipation.
Figure 1c represents the time course of ΔF (top panel) and ΔD (bottom panels) shift during SALB formation on silicon dioxide. A typical SALB formation experiment involved baseline recording in the aqueous buffer solution (i), the addition of isopropanol (ii), the addition of the lipid mixture [0. 5 m/mL, DOPC/DP-NGPE (8:2)] in isopropanol (iii), and solvent exchange back into the initial buffer solution (iv). The final frequency and dissipation values after complete solvent replacement were −25 ± 1 Hz and 0.4 ± 0.2, respectively, corresponding to formation of a planar bilayer. The formation of the bilayer was further confirmed by FRAP analysis showing lateral mobility of lipids in the two-dimensional plane of the membrane with a diffusion coefficient of 2 ± 0.3 μm2/s (Figure 1d), which is comparable to previously reported values.39
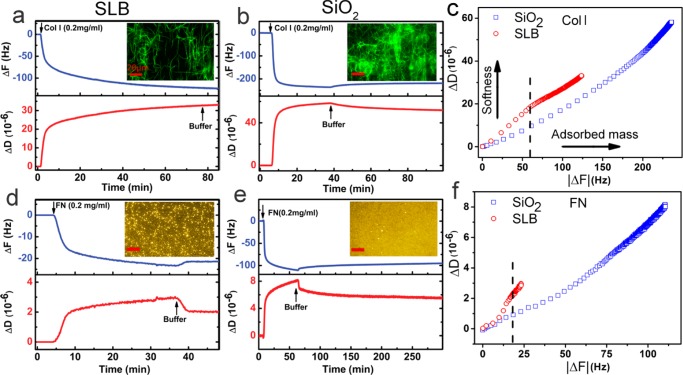
Next, we studied the adsorption of Col I and FN to the SLB and compared the results with their adsorption to SiO2. All steps were monitored by QCM-D. To this end, after the SLB formed, it was incubated with 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide (EDC)/N-hydroxysuccinimide (NHS) to activate the carboxylic groups of DP-NGPE, followed by injection of the protein solution. Figure 2a shows the changes in ΔF and ΔD during Col I conjugation. In this and other QCM-D graphs, the bilayer formation and EDC/NHS incubation were not shown and the frequency shift was normalized with ΔF = 0 Hz corresponding to the planar lipid bilayer in each experiment. During the adsorption of Col I to the SLB, ΔF decreased and saturated at ΔF = −122 ± 5 Hz. At the same time, ΔD increased and stabilized at ΔD = 32.7 ± 0.6 × 10–6. After the protein adsorption approached the saturation level, the pure buffer was injected, which resulted in almost no change in ΔF and ΔD, indicating that Col I had irreversibly adsorbed to the surface. The high energy dissipation indicates the softness and viscoelastic nature of the Col I film, suggesting that Col I fibers did not rigidly attach to the underlying bilayer. In addition, the high dissipation indicates the presence of a high amount of water molecules coupled to the Col I fibers resulting in a hydrated layer with high mass and dissipation. The immunofluorescence microscopy of the Col I-coated SLBs, prepared using the same protocol in a microfluidic chamber for microscopy, also showed attachment of Col I fibers randomly to the SLB surface (Figure 2a, inset).
Figure 2.
QCM-D analysis of ECM adsorption to SLB and SiO2 platforms. QCM-D monitoring of (a) Col I and (d) FN attachment to bilayer via covalent bonding. The SLB on silicon dioxide was initially formed by the SALB method, and then the measurement time was normalized. Frequency shift (ΔF, blue) and energy dissipation shift (ΔD, red) for the third overtone (n = 3) were measured as a function of time. Protein solution (0.2 mg/mL) was introduced to the surface after treatment of SLBs with EDC/NHS. The insets show the fluorescence immunostaining micrograph of SLB functionalized with Col I fibers (a) and FN (d). Proteins were stained using fluorescently labeled anti-Col I or anti-FN antibodies. QCM-D monitoring of (b) Col I and (e) FN adsorption on SiO2 substrate with no SLB coating. Dissipation–frequency curves (ΔD vs ΔF) upon (c) Col I and (f) FN adsorption to the SLB and SiO2 surface. The dashed line shows that at any given frequency, the corresponding dissipation was bigger for protein layer on the SLB. Scale bars are 20 μm.
For comparison, Col I adsorption on the SiO2 substrate with no SLB coating was examined by QCM-D, as shown in Figure 2b. Adsorption of Col I to the surface was revealed by a negative frequency shift and a simultaneous increase in the dissipation reaching a saturation value of ΔFCol I = −186 ± 60 Hz and ΔDCol I = 42 ± 19 × 10–6. A buffer wash step resulted in a small change in ΔF (1.75 ± 1 Hz) and ΔD (1.25 ± 0.5), indicating the majority of Col I irreversibly adsorbed to the surface. Comparing the ΔFCol I for SLB and SiO2 suggests that more Col I adsorbed to SiO2 than to SLB. The frequency shift for Col I adsorption to SiO2 was almost 1.5 times greater than that of SLB. The higher frequency shift indicates that more Col I adsorption to the bare SiO2 surface. In agreement with QCM-D results, the fluorescent images also revealed that a greater amount of Col I fibers adsorbed to the bare glass (Figure 2b, inset).
The proteins adsorbed nonspecifically to the glass surface. In contrast, zwitterionic lipid bilayer has previously been shown to be quite efficient in preventing protein adsorption at interfaces. The exact mechanism for the nonfouling behavior of zwitterionic bilayers is not known; however, the cooperative contribution of the following factors has been proposed to be important:16 (i) the charged neutrality of zwitterionic lipids in a wide range of pH weakens the electrostatic interaction between bilayer and proteins, (ii) the presence of a hydration layer which strongly associates with lipid headgroups acts as a barrier for protein adsorption, and (iii) the high lateral mobility of lipids in the membrane plane. Because of the nonfouling feature of the bilayer, Col I fibers attach to the bilayer surface exclusively via the anchoring lipids (DP-NGPE). The activated carboxylic acid of DP-NGPE covalently binds to lysine residues which contain the primary amine side chain. Therefore, the Col I fibers attached to the bilayer surface via a limited number of lysine residues that are dispersed along the fibers. As a result, fewer fibers attach to the membrane surface in comparison to those on the SiO2 substrate. To gain more information about the structure of the Col I film on SLB and SiO2, the correlation between energy dissipation and frequency change was studied. Figure 2c shows ΔD versus ΔF during Col I adsorption on SLB and SiO2. At any given frequency, the corresponding energy dissipation is higher for Col I on the bilayer. In other words, the Col I film on SLB has a higher structural flexibility than that on SiO2.This is expected as, due to the limited number of anchoring points along the fiber, the Col I film on SLB is less dense than that on SiO2.
The same series of experiments were performed to prepare and characterize the FN-coated bilayer and SiO2, as shown in Figure 2d–f. The QCM-D results show a frequency and dissipation shift of −21 ± 2.1 Hz and 1.4 ± 0.4 × 10–6 for the FN-coated bilayer, respectively. The fluorescence micrograph of FN on SLB shows the presence of some unevenly distributed FN aggregates which appear as particularly brighter spots. The interaction of FN with the supported bilayer has been previously studied and the formation of FN aggregates at the membrane interface has been shown by atomic force microscopy (AFM).46 The frequency and dissipation shifts for FN adsorption to SiO2 were 101.3 ± 6 Hz and 6.9 ± 1.3 × 10–6, respectively (Figure 2d–e), which are similar to the Col I samples, and are higher than those of FN adsorption to SLB. In contrast to FN on SLB, the fluorescence micrograph of FN on SiO2 revealed a uniform, highly packed layer (Figure 2e, inset). FN adsorbed to the bilayer only through the covalent binding to DP-NGPE, but nonspecifically, entirely covering the SiO2 surface. Similar to the trend observed for the Col I samples (Figure 2c), the FN film on SLB was softer than that on SiO2 (Figure 2f). In addition, the frequency and dissipation corresponding to the adsorption of FN on SLB and SiO2 were lower than those of Col I, due to its lower molecular weight and compact globular structure.47
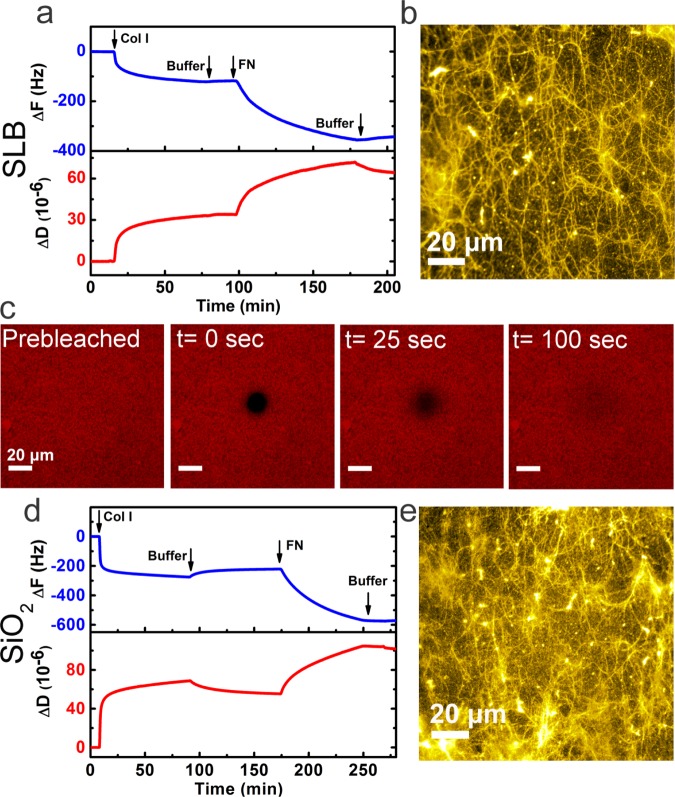
Next, we examined the biological activity of Col I and FN adsorbed sequentially to SLB and SiO2. We began by studying the Col I–FN interaction on the SLB and SiO2 platforms. The interaction of Col I and FN is essential for cell adhesion and migration. FN molecules can bind to a specific domain sequence within the Col I fiber.48 To study the Col I–FN interaction, FN was added to the Col I-coated platforms, and its binding was monitored by QCM-D and visualized by fluorescence microscopy. Figure 3a shows the QCM-D results for the adsorption of FN to Col I-coated SLB. After bilayer formation and subsequent Col I conjugation, FN was injected. Due to the organization of Col I in a fiber structure, there are many FN binding sites available on Col I fibers. The significant changes of resonance frequency (ΔFCol I+FN = −420.7 ± 70.5 Hz) and energy dissipation (ΔDCol I+FN = 84.7 ± 19.2 × 10–6) indicate the tendency of FN to bind to Col I on SLB as shown in Figure 3a. As a control, BSA was added to the Col I-decorated bilayer (see Figure S1). No change in ΔF and ΔD was observed after BSA injection, indicating that BSA did not adsorb to the Col I-decorated SLB. This observation suggests that FN adsorption to this platform was the result of specific interactions between Col I and FN.
Figure 3.
Binding of FN to Col I fibers on SLB and SiO2 platforms. (a) QCM-D curves of frequency and dissipation shifts for the adsorption of FN to Col I-functionalized SLB. (b) Fluorescence micrograph of Col I-coated SLB after incubation with FN, stained by fluorescently labeled anti-FN antibodies. (c) FRAP snapshots of Col I–FN-coated bilayer showing almost complete recovery, indicating the fluidity of the SLB. (d) QCM-D measurement of FN adsorption to Col I-coated SiO2. (e) Fluorescence micrograph of Col I-coated glass, after incubation with FN, stained by fluorescently labeled anti-FN antibodies. The initial baseline values in (a) and (d) are the normalized frequency and dissipation shifts for the bilayer (defined as ΔFSLB = 0). Immunostaining of FN revealed its co-localization with Col I fibers. All scale bars are 20 μm.
Figure 3b shows the fluorescence micrograph of Col I-coated SLB after addition of FN. In this figure, only FN was stained using a specific antibody. Fluorescence staining of FN in this sample revealed the co-localization with Col I fibers due to Col I–FN binding. Also, we checked if the underlying bilayer retains its fluidity after conjugation with Col I and FN (Figure 3c). FRAP analysis indicated that the bilayer maintains its mobility after functionalization with multiple protein systems (Col I–FN) with a long-range lateral mobility of 0.7 ± 0.2 μm2 s–1. The high lateral mobility of the underlying membrane suggests that protein conjugation was mainly mediated via the anchoring lipids, and the rest of the lipids were free to diffuse. The mobility of the bilayer may assist the lipid anchor in displacing and thus helping the fibers to assume a conformation with less strain.
Next, for comparison, we examined the sequential adsorption of Col I and FN to the SiO2 or glass surfaces. Figure 3d shows the time course of frequency and dissipation shifts upon subsequent addition of Col I and FN to the SiO2 substrate. The adsorption of FN to the Col I-coated SiO2 surface resulted in frequency and dissipation shifts of ΔFCol I+FN = −465 ± 144.2 Hz and ΔDCol I+FN = 72.5 ± 37.5 × 10–6, respectively. These results suggest that in both platforms, FN could specifically bind to the Col I fibers. However, the ratio of FN to Col (ΔFFN/ΔFCol I) was greater for SLB (2.4 ± 0.4) than that for SiO2 (1.5 ± 0.1). In other words, the amount of FN molecules per Col I fiber was greater (p ≤ 0.01) for SLB than that for SiO2. The data suggest that the FN binding sites on Col I fibers that adsorbed to SLB were more accessible than the ones on SiO2. This is consistent with QCM-D results showing that Col I fibers on SLB have a higher structural flexibility.
Next, we studied the cell-adhering activity of FN and Col I adsorbed to SLB and glass. The various ECM components including Col49,50 and FN51 interact specifically with different cells through integrins as specialized cell surface receptors. Integrins bind to specific domains within the ECM proteins and mediate adhesion of cells to the matrix proteins. In addition, integrin receptor interaction with ECM proteins also initiates an intracellular signaling cascade that directs cellular processes such as cell survival,52 proliferation,53 differentiation,54 and migration.55
We plated the same number of cells on the Col I- or FN-coated SLB and glass surfaces and measured the number and area of adhered cells. The aim was to determine the effect of protein coverage on the extent of cell adhesion and spreading. As a model, we used Huh 7.5 cells, which are a human hepatocyte cell line. The hepatic ECM consists mostly of FN and some Col I, which makes Huh 7.5 cells a suitable model for our study.56
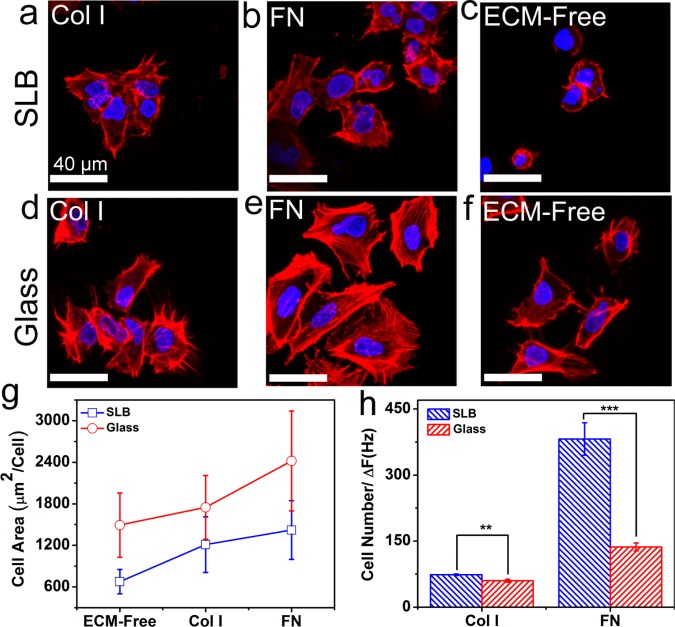
To assess cell attachment and spreading, cells were stained and analyzed with an epifluorescence microscope. Figure 4a,b presents the fluorescence images for actin-stained Huh 7.5 cells attached on Col I- and FN-coated SLB after 24 h and shows cells attached and spread well on these platforms. However, in agreement with previous reports,17,25,36,37 when cells were seeded on ECM-free SLB, only a limited number of cells attached and remained rounded and did not appreciably spread over 24 h. As cells attach to surfaces via their surface proteins, the limited cell attachment to the ECM-free bilayer can be related to the SLB’s resistance to the adsorption of cell adhesive proteins.
Figure 4.
Quantification of cell spreading on ECM-functionalized SLBs and glass. Representative confocal images of one day plated Huh 7.5 cells on (a) Col I- and (b) FN-functionalized SLB and (d) Col I- and (e) FN-functionalized glass. Representative confocal images of one day plated Huh 7.5 cells on ECM-free (c) SLB and (f) glass. F-actin was stained by Alexa Flour 555-labeled phalloidin (red), and nuclei were stained by DAPI (blue). All scale bars are 40 μm. (g) The average projected area of Huh 7.5 cells plated on different platforms after 24 h. (h) Number of cells per absolute values of the maximum QCM-D frequency shifts due to the amount of adsorbed proteins. The images were taken by optical microscope and analyzed with ImageJ. The observation field was 0.5 mm2 (n = 3, mean ± SD, ***P ≤ 0.001).
On glass, the cells attached and spread on the ECM-coated (Figure 4d,e), as well as bare, surface (Figure 4f). A quantitative analysis of the images (see Figure S2) indicated that the projected area of cells adhered to glass platforms was larger than that of their SLB counterparts (Figure 4g). This is because Col I and FN on SLBs are less resistant to displacement when experiencing force from the cell. The reason is that the functional lipids, which anchor the proteins to the membrane, are free to move in the plane of the fluid underlying the membrane. In addition, for both SLB and glass, the same trend for cell area was observed; the cell area was larger on the FN-coated surfaces as compared to that on the Col I-coated and uncoated surfaces.37
To determine how the cell-adhering activity of Col I and FN was influenced by adsorption to SLB and glass, the number of attached cells on each platform was divided by protein surface coverage. The protein coverage can be estimated from the QCM-D frequency change. Although the Sauerbrey model57 can be used to convert the frequency shift to mass for rigid films, mass determination for soft layers is not trivial. However, as the exact value for protein mass is not required for our analysis, we use frequency shifts as an indicator for the adsorbed mass and apply them for comparative analysis. Figure 4h shows the number of cells normalized as cells/ΔFCol I or FN for glass and SLB.
For both Col I and FN, the normalized cell number was higher for SLB than that for glass. This result suggests that even though the protein coverage on the glass surface was greater than that on SLB, the biological activity of Col I and FN was greater on SLB. The cell attachment is mediated by integrin interaction with specific domains in Col I and FN. RGD is the principal integrin-binding domain present within ECM proteins such as Col I and FN.58 As Col I and FN interact strongly with the glass surface, they form a relatively dense layer upon surface adsorption and possibly undergo surface-induced conformational changes. In addition, there is a possibility of denaturing when the interaction between a passively adsorbed protein and the surface is too strong.59,60 As a result, the cell-binding sites (RGD) of a substantial population of adsorbed protein molecules may not be accessible to the cells in the solution phase. However, on SLB, the proteins retain their structural flexibility. Also, the local chemical environment of the phospholipid bilayer mimics that of the natural cell membrane and helps the immobilized protein molecules to maintain their activity and accessibility.
To further investigate the biological activity of Col I and FN on SLB and glass, we compared the viability and function of the cells plated on the Col I, FN, and Col I–FN platforms (Figure 5). The physical contact between cell and ECM through integrin and nonintegrin cell surface proteins can trigger chemical pathways leading to cell growth and division. Therefore, we measured Huh 7.5 proliferation on ECM-coated platforms as a function of protein coverage.
Figure 5.
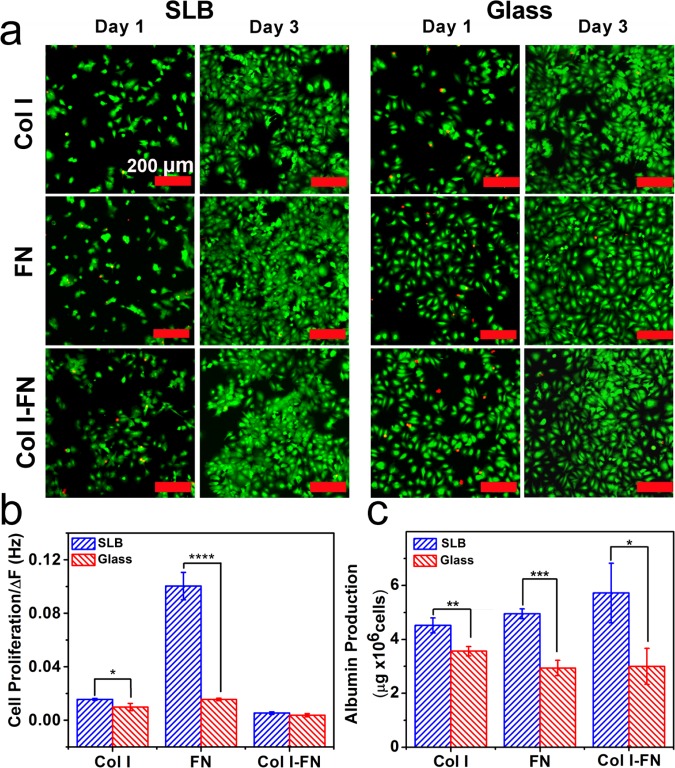
Cell viability and metabolic functions on ECM-coated platforms. (a) Representative fluorescence micrograph images of detecting live (green) and dead (red) Huh 7.5 cells cultured on SLB and glass coated with Col I and FN and Col I–FN at 1 and 3 days post seeding. Scale bar is 200 μm. (b) Proliferation of Huh 7.5 cells was measured over a 3 day period. The values were normalized to the absolute frequency shifts due to the amount of adsorbed proteins. (c) Albumin production by Huh 7.5 cells plated on different platforms measured after 24 h of cell growth. The values were normalized to the total number of cells (n = 3, mean ± SD, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001).
The viability of cells on the ECM-coated SLB and glass surfaces were quantified using a live/dead assay combined with a fluorescence microscope at day 1 and 3. As shown in Figure 5a over 90% of Huh 7.5 cells were viable on all platforms within three days. Next, cell proliferation was measured using the CCK-8 assay61,62 by dividing the number of cells at day 3 by those at day 1. The obtained proliferation value for each surface was then normalized to its corresponding QCM-D frequency shift upon protein adsorption (Figure 5b). For all cases, the normalized proliferation was higher for the SLB platforms. In particular, the cell proliferation on FN-coated SLB was significantly greater than that of glass.
The interaction of cells with ECM can also alter the cell functions by stimulating the transcription of a subset of genes.63 In hepatocytes, it has been shown that the interaction with ECM can promote albumin production. Therefore, we selected albumin secretion as one of the major functions of Huh 7.5 and measured its production on cells plated on the SLB and glass platforms. Figures 5c shows the average amount of albumin produced by each cell after 24 h. On average, cells grown on the SLB platforms produced more albumin than that of those grown on glass-based surfaces.
Conclusions
The adsorption behavior and biological activity of Col I and FN attached to SLB via covalent bonding were compared with those of Col I and FN adsorbed nonspecifically to SiO2 surfaces. The characterization of protein adsorption by QCM-D revealed that Col I and FN attached to SLB have a higher structural flexibility than those adsorbed onto SiO2. The QCM-D study also showed that the interaction of FN with Col I is more efficient on SLB than on SiO2, suggesting that the Col I–FN interaction on the SiO2 surface may be inhibited by limited accessible binding sites on the surface-adsorbed Col I. In addition, the ratio of the number of cells attached to protein-coated SLB and SiO2 to the amount of adsorbed protein revealed that the interaction of cells with the anchorage sites for cell adhesion on FN and Col I is more efficient on SLB than on SiO2. The same trend was observed for cell proliferation and albumin production by cells attached to the functionalized SLB and SiO2. Taken together, our results show that Col I and FN attached to SLB via covalent bonding retain their biological activities better than when attached on glass. Thus, SLB functionalization can serve as an effective strategy to interface biological molecules to inorganic surfaces. The benefit of this platform is expected to be more pronounced when smaller protein/peptide motifs are needed to be used for functionalization.
Materials and Methods
Lipid Bilayer Formation
The lipid reagents DOPC and DP-NGPE were purchased in a lyophilized powder form from Avanti Polar Lipid (Alabaster, AL). Isopropanol and ethanol were used to dissolve DOPC and DP-NGPE to a concentration of 10 and 5 mg/mL, respectively. The DP-NGPE solution was gently vortexed and heated to 50 °C for 3 min, to enhance the solubility. All of the stock solutions were diluted and mixed in isopropanol to prepare a lipid solution containing 80% DOPC and 20% DP-NGPE to a final concentration of 0.5 mg/mL. Lipid bilayers were formed using the SALB method, whereby an isopropanol mixture of DOPC and DP-NGPE was introduced to a hydrophilic SiO2/glass substrate and was incubated for 10 min. A buffer solution (10 mM Tris, 100 mM NaCl, pH 7.2) was carefully added and replaced with the isopropanol mixture of lipids.
Activating Carboxyl Groups of DP-NGPE Lipid Using EDC/NHS Chemistry
N-Hydroxysulfosuccinimide sodium (sNHS) and EDC hydrochloride were received from VWR. Immediately before the experiment, EDC/NHS powders were dissolved in 10 mM HEPES buffer and 100 mM NaCl (pH 5.5) at a final concentration of 10 mg/mL. The solution was incubated with SLBs for 1 h at room temperature. After carboxylic activation, the SLBs were washed three times with a HEPES solution.
Functionalization of SLB and Glass/SiO2 with ECM Proteins
The proteins, Col I (Sigma-Aldrich) and FN (Calbiochem, U.K.), were diluted in PBS (pH 7.5) at a concentration of 0.2 mg/mL. The protein solutions were added to an activated SLB and glass for 1.5 h using a peristaltic pump (Ismatec Reglo Digital) with a flow rate of 50 μL/min. The surface was then rinsed thoroughly with PBS to remove unbound proteins.
QCM-D
The formation of a lipid bilayer and binding of proteins to the lipid interface was monitored and characterized by a Q-Sense E4 instrument (Q-sense AB, Gothenburg, Sweden). SiO2-coated AT-cut piezoelectric quartz crystals were cleaned with 1% sodium dodecyl sulfate and rinsed with Milli-Q water (Millipore). The crystals were dried with N2 and treated with oxygen plasma (March Plasmod Plasma Etcher, March Instruments, CA) at 180 W for 30 s to remove any residual organic molecules bound to the surface while making the substrate more hydrophilic. QCM-D measurements were taken at the 3, 5, 7, and 11 overtones, and the presented data were measured and normalized at the third overtone (15 MHz). The samples were injected into the QCM-D chamber using a peristaltic pump (Ismatec Reglo Digital) with a flow rate of 50 μL/min at room temperature.
FRAP
Lateral lipid mobility was estimated using the FRAP technique. 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt) (Rh-PE) (Avanti Polar Lipid) (0.5 wt %) was included in the lipid precursor solution to fluorescently label the lipid bilayers. A commercially available flow chamber (stick-Slide I0.1 Luer, Ibidi, Munich, Germany) was used for bilayer formation using the SALB method. The SLBs were photobleached using a 532 nm, 100 mW laser beam that resulted in the formation of a ∼20 μm bleached spot. The bleaching period was ∼5 s, and the fluorescence recovery of the bilayer was measured using an inverted epifluorescence Eclipse TE 2000 microscope (Nikon) equipped with a 60× oil immersion objective (NA 1.49) combined with an Andor iXon + EMCCD camera (Andor Technology, Belfast, Northern Ireland). The images were recorded at 2 s intervals and analyzed using ImageJ software. The lipid diffusivity was determined by the Hankel transform method.64 For all fluorescence imaging experiments, glass coverslips (Menzel Glaser, Braunschweig, Germany) were used.
Cell Culture
The human hepatocarcinoma cell line (Huh 7.5) was obtained from Apath (NY). Cells were plated on a tissue culture dish (100 mm) containing 10 mL of Dulbecco’s Modified Eagles Medium (DMEM) (Hyclone), 10% fetal bovine serum (Hyclone), 1% penicillin, and streptomycin (Gibco) for several days to reach a confluency of 80%. Cells were detached from the plate by incubating the culture dish with 0.25% trypsin (Gibco) for ∼3–5 min. After the cell suspension was dispersed in the growth medium, the cell count of 2 × 104 was determined via a hemocytometer. A flow chamber (Sticky-Slide VI 0.4 Ibidi, Munich, Germany) was used for cell seeding. The cell growth area per channel was ∼0.6 cm2.
Evaluation of Huh 7.5 Cell Morphology
Cell morphology was visualized over a period of 24 h, wherein Huh 7.5 cells were fixed using 4% paraformaldehyde (Sigma) for 3 min. After washing with PBS three times, a 3% BSA blocking buffer was added and incubated for 1 h. Alexa Flour 555-labeled phalloidin (Life Technologies) was incubated with cells for 2 h at room temperature, to label filamentous actin (F-actin). The nucleus of each cell was labeled by addition of 10 μg/mL of 1,2,6-diamidino-2-phenylinodole, dihydrochloride (DAPI; Life Technologies) for 5 min. The fluorescence images were captured using a confocal microscope, LSM710 with a ZEN program (Carl Zeiss), and were analyzed using ImageJ software.
Quantification of Huh 7.5 Cell Surface Area
The optical images of Huh 7.5 cells were captured after 24 h using an inverted microscope (Nikon) with a 10× objective. The cell surface area was measured and analyzed using ImageJ software.
Determination of Cell Viability by LIVE/DEAD Kit
The qualitative investigation of Huh 7.5 cell viability was done using LIVE/DEAD Cell Viability assays (Life Technologies), following the manufacturer’s protocol. The proper volume of 2 μM calcein AM and 4 μM ethdium homodimer-1 (EthD-1) was prepared in DMEM media. The solution was added to each sample and incubated for 20 min at 37 °C. After incubation, the trapped calcein inside the living cells yielded a strong green fluorescence and was imaged using a 494–517 nm emission filter. For dead cells, EthD-1 was bonded with the nucleic acids inside the cell and produced a red fluorescence that was imaged using a 517–617 nm emission filter. An Eclipse TI-U microscope combined with an NIS-Elements Viewer program (Nikon, Japan) was used for fluorescence measurements.
Determination of Cell Number by Cell Counting Kit-8 (CCK-8)
Cell proliferation for days 1 and 3 was measured using a colorimetric assay CCK-8 (Dojindo Molecular Technologies, Japan). Based on the manufacturer’s protocol, a 10% CCK-8 reagent in complete media was added to the cells. After incubation for 1 h at 37 °C, the absorbance was measured at 450 nm using a microplate reader. The measured absorbance was converted to cell number according to a CCK-8 standard curve for Huh 7.5.
Determination of Human Albumin in Cell Culture Supernatants
Albumin secretions from the Huh 7.5 cells were measured using an enzyme-linked immunosorbent assay (ELISA) kit (Abcam). The cell culture media was collected after 24 h and was centrifuged at 10 000g for 10 min. The supernatant was transferred to a sterile Eppendorf tube and stored at −20 °C. Before the ELISA, the samples were kept at room temperature and the provided protocols were subsequently followed.
Immunofluorescent Stain for ECM Proteins
The primary antibodies, mouse monoclonal anti-Col I (Abcam) and mouse monoclonal IgG1 anti-FN (Santa Cruz), were incubated with conjugated Col I and FN, respectively, for 1 h at room temperature. The samples were then washed with PBS to remove unbounded antibodies. After washing, the secondary antibodies Alexa Fluor 488 Goat Anti-mouse IgG (H + L) (green-fluorescent dye, Life Technology) and Alexa Fluor 555 Rabbit Anti-Mouse IgG (H + L) (red-fluorescent dye, Life Technology) were added and incubated for 15 min to fluorescently label Col I and FN, respectively. Fluorescence images were captured with an inverted epifluorescence Eclipse TE 2000 microscope (Nikon) equipped with a 60× oil immersion objective (NA 1.49) combined with an Andor iXon + EMCCD camera (Andor Technology, Belfast, Northern Ireland). The data were analyzed using ImageJ software.
Acknowledgments
This work was supported by the National Research Foundation (NRF-NRFF2011-01).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.7b00158.
QCM-D analysis of BSA adsorption to Col I-functionalized SLB; bright field images of cells grown on different platforms (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Kasemo B. Biological surface science. Surf. Sci. 2002, 500, 656–677. 10.1016/S0039-6028(01)01809-X. [DOI] [Google Scholar]
- Krishnan V.; Kasuya Y.; Ji Q.; Sathish M.; Shrestha L. K.; Ishihara S.; Minami K.; Morita H.; Yamazaki T.; Hanagata N.; et al. Vortex-aligned fullerene nanowhiskers as a scaffold for orienting cell growth. ACS Appl. Mater. Interfaces 2015, 7, 15667–15673. 10.1021/acsami.5b04811. [DOI] [PubMed] [Google Scholar]
- Pandian G. N.; Sugiyama H. Nature-Inspired Design of Smart Biomaterials Using the Chemical Biology of Nucleic Acids. Bull. Chem. Soc. Jpn. 2016, 89, 843–868. 10.1246/bcsj.20160062. [DOI] [Google Scholar]
- Loessner D.; Meinert C.; Kaemmerer E.; Martine L. C.; Yue K.; Levett P. A.; Klein T. J.; Melchels F. P. W.; Khademhosseini A.; Hutmacher D. W. Functionalization, preparation and use of cell-laden gelatin methacryloyl-based hydrogels as modular tissue culture platforms. Nat. Protoc. 2016, 11, 727–746. 10.1038/nprot.2016.037. [DOI] [PubMed] [Google Scholar]
- Nawa E.; Yamamoto D.; Shioi A. Chemotactic Amoeboid-Like Shape Change of a Vesicle under a pH Gradient. Bull. Chem. Soc. Jpn. 2015, 88, 1536–1544. 10.1246/bcsj.20150190. [DOI] [Google Scholar]
- Huang N.-C.; Ji Q.; Ariga K.; Hsu S.-h. Nanosheet transfection: effective transfer of naked DNA on silica glass. NPG Asia Mater. 2015, 7, e184 10.1038/am.2015.43. [DOI] [Google Scholar]
- Jackler G.; Steitz R.; Czeslik C. Effect of Temperature on the Adsorption of Lysozyme at the Silica/Water Interface Studied by Optical and Neutron Reflectometry. Langmuir 2002, 18, 6565–6570. 10.1021/la025605i. [DOI] [Google Scholar]
- Höök F.; Rodahl M.; Kasemo B.; Brzezinski P. Structural changes in hemoglobin during adsorption to solid surfaces: Effects of pH, ionic strength, and ligand binding. Proc. Natl. Acad. Sci. U.S.A. 1998, 95, 12271–12276. 10.1073/pnas.95.21.12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade J.; Hlady V.; Wei A. Adsorption of complex proteins at interfaces. Pure Appl. Chem. 1992, 64, 1777–1781. 10.1351/pac199264111777. [DOI] [Google Scholar]
- Rabe M.; Verdes D.; Seeger S. Understanding protein adsorption phenomena at solid surfaces. Adv. Colloid Interface Sci. 2011, 162, 87–106. 10.1016/j.cis.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Roach P.; Farrar D.; Perry C. C. Interpretation of protein adsorption: surface-induced conformational changes. J. Am. Chem. Soc. 2005, 127, 8168–8173. 10.1021/ja042898o. [DOI] [PubMed] [Google Scholar]
- Haynes C. A.; Norde W. Structures and stabilities of adsorbed proteins. J. Colloid Interface Sci. 1995, 169, 313–328. 10.1006/jcis.1995.1039. [DOI] [Google Scholar]
- Sackmann E. Supported membranes: Scientific and practical applications. Science 1996, 271, 43–48. 10.1126/science.271.5245.43. [DOI] [PubMed] [Google Scholar]
- Joubert J. R.; Smith K. A.; Johnson E.; Keogh J. P.; Wysocki V. H.; Gale B. K.; Conboy J. C.; Saavedra S. S. Stable, ligand-doped, poly (bis-SorbPC) lipid bilayer arrays for protein binding and detection. ACS Appl. Mater. Interfaces 2009, 1, 1310–1315. 10.1021/am900177p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicklein B.; Darder M.; Aranda P.; Ruiz-Hitzky E. Phospholipid–sepiolite biomimetic interfaces for the immobilization of enzymes. ACS Appl. Mater. Interfaces 2011, 3, 4339–4348. 10.1021/am201000k. [DOI] [PubMed] [Google Scholar]
- Glasmästar K.; Larsson C.; Höök F.; Kasemo B. Protein adsorption on supported phospholipid bilayers. J. Colloid Interface Sci. 2002, 246, 40–47. 10.1006/jcis.2001.8060. [DOI] [PubMed] [Google Scholar]
- Andersson A. S.; Glasmästar K.; Sutherland D.; Lidberg U.; Kasemo B. Cell adhesion on supported lipid bilayers. J. Biomed. Mater. Res., Part A 2003, 64, 622–629. 10.1002/jbm.a.10442. [DOI] [PubMed] [Google Scholar]
- Palekar R. U.; Myerson J. W.; Schlesinger P. H.; Sadler J. E.; Pan H.; Wickline S. A. Thrombin-targeted liposomes establish a sustained localized anticlotting barrier against acute thrombosis. Mol. Pharm. 2013, 10, 4168–4175. 10.1021/mp400210q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaizuka Y.; Douglass A. D.; Varma R.; Dustin M. L.; Vale R. D. Mechanisms for segregating T cell receptor and adhesion molecules during immunological synapse formation in Jurkat T cells. Proc. Natl. Acad. Sci. U.S.A. 2007, 104, 20296–20301. 10.1073/pnas.0710258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meulen S. A.; Dubacheva G. V.; Dogterom M.; Richter R. P.; Leunissen M. E. Quartz crystal microbalance with dissipation monitoring and spectroscopic ellipsometry measurements of the phospholipid bilayer anchoring stability and kinetics of hydrophobically modified DNA oligonucleotides. Langmuir 2014, 30, 6525–6533. 10.1021/la500940a. [DOI] [PubMed] [Google Scholar]
- Persson F.; Fritzsche J.; Mir K. U.; Modesti M.; Westerlund F.; Tegenfeldt J. O. Lipid-based passivation in nanofluidics. Nano Lett. 2012, 12, 2260–2265. 10.1021/nl204535h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler K. S.; Durfee P. N.; Theron C.; Ashley C. E.; Carnes E. C.; Brinker C. J. Protocells: Modular Mesoporous Silica Nanoparticle-Supported Lipid Bilayers for Drug Delivery. Small 2016, 12, 2173–2185. 10.1002/smll.201502119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri K.; Nakamura M.; Koumoto K. Magnetoresponsive smart capsules formed with polyelectrolytes, lipid bilayers and magnetic nanoparticles. ACS Appl. Mater. Interfaces 2010, 2, 768–773. 10.1021/am900784a. [DOI] [PubMed] [Google Scholar]
- Wang T.; Zhen Y.; Ma X.; Wei B.; Wang N. Phospholipid bilayer-coated aluminum nanoparticles as an effective vaccine adjuvant-delivery system. ACS Appl. Mater. Interfaces 2015, 7, 6391–6396. 10.1021/acsami.5b00348. [DOI] [PubMed] [Google Scholar]
- Svedhem S.; Dahlborg D.; Ekeroth J.; Kelly J.; Höök F.; Gold J. In situ peptide-modified supported lipid bilayers for controlled cell attachment. Langmuir 2003, 19, 6730–6736. 10.1021/la034172w. [DOI] [Google Scholar]
- Thid D.; Holm K.; Eriksson P. S.; Ekeroth J.; Kasemo B.; Gold J. Supported phospholipid bilayers as a platform for neural progenitor cell culture. J. Biomed. Mater. Res., Part A 2008, 84, 940–953. 10.1002/jbm.a.31358. [DOI] [PubMed] [Google Scholar]
- Lee I.-C.; Liu Y.-C.; Tsai H.-A.; Shen C.-N.; Chang Y.-C. Promoting the selection and maintenance of fetal liver stem/progenitor cell colonies by layer-by-layer polypeptide tethered supported lipid bilayer. ACS Appl. Mater. Interfaces 2014, 6, 20654–20663. 10.1021/am503928u. [DOI] [PubMed] [Google Scholar]
- Lee I.-C.; Wu Y.-C. Assembly of Polyelectrolyte Multilayer Films on Supported Lipid Bilayers To Induce Neural Stem/Progenitor Cell Differentiation into Functional Neurons. ACS Appl. Mater. Interfaces 2014, 6, 14439–14450. 10.1021/am503750w. [DOI] [PubMed] [Google Scholar]
- Eyre D. R. Collagen: molecular diversity in the body’s protein scaffold. Science 1980, 207, 1315–1322. 10.1126/science.7355290. [DOI] [PubMed] [Google Scholar]
- Pankov R.; Yamada K. M. Fibronectin at a glance. J. Cell Sci. 2002, 115, 3861–3863. 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- Hynes R. O. The extracellular matrix: not just pretty fibrils. Science 2009, 326, 1216–1219. 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai N.; Wong C. C.; Gong Y. X.; Tan S. C.; Chan V.; Liao K. Modulating cell adhesion dynamics on carbon nanotube monolayer engineered with extracellular matrix proteins. ACS Appl. Mater. Interfaces 2010, 2, 1038–1047. 10.1021/am9008117. [DOI] [PubMed] [Google Scholar]
- Parenteau-Bareil R.; Gauvin R.; Berthod F. Collagen-based biomaterials for tissue engineering applications. Materials 2010, 3, 1863–1887. 10.3390/ma3031863. [DOI] [Google Scholar]
- Place E. S.; Evans N. D.; Stevens M. M. Complexity in biomaterials for tissue engineering. Nat. Mater. 2009, 8, 457–470. 10.1038/nmat2441. [DOI] [PubMed] [Google Scholar]
- Choi B.; Kim S.; Lin B.; Wu B. M.; Lee M. Cartilaginous extracellular matrix-modified chitosan hydrogels for cartilage tissue engineering. ACS Appl. Mater. Interfaces 2014, 6, 20110–20121. 10.1021/am505723k. [DOI] [PubMed] [Google Scholar]
- Huang C.-J.; Cho N.-J.; Hsu C.-J.; Tseng P.-Y.; Frank C. W.; Chang Y.-C. Type I collagen-functionalized supported lipid bilayer as a cell culture platform. Biomacromolecules 2010, 11, 1231–1240. 10.1021/bm901445r. [DOI] [PubMed] [Google Scholar]
- Huang C.-J.; Tseng P.-Y.; Chang Y.-C. Effects of extracellular matrix protein functionalized fluid membrane on cell adhesion and matrix remodeling. Biomaterials 2010, 31, 7183–7195. 10.1016/j.biomaterials.2010.05.076. [DOI] [PubMed] [Google Scholar]
- Marqués-Gallego P.; de Kroon A. I. Ligation strategies for targeting liposomal nanocarriers. BioMed Res. Int. 2014, 2014, 129458 10.1155/2014/129458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabaei S. R.; Choi J.-H.; Haw Zan G.; Zhdanov V. P.; Cho N.-J. Solvent-Assisted Lipid Bilayer Formation on Silicon Dioxide and Gold. Langmuir 2014, 30, 10363–10373. 10.1021/la501534f. [DOI] [PubMed] [Google Scholar]
- Tabaei S. R.; Jackman J. A.; Kim S.-O.; Zhdanov V. P.; Cho N.-J. Solvent-Assisted Lipid Self-Assembly at Hydrophilic Surfaces: Factors Influencing the Formation of Supported Membranes. Langmuir 2015, 31, 3125–3134. 10.1021/la5048497. [DOI] [PubMed] [Google Scholar]
- Jackman J. A.; Tabaei S. R.; Zhao Z.; Yorulmaz S.; Cho N.-J. Self-assembly formation of lipid bilayer coatings on bare aluminum oxide: overcoming the force of interfacial water. ACS Appl. Mater. Interfaces 2015, 7, 959–968. 10.1021/am507651h. [DOI] [PubMed] [Google Scholar]
- Tabaei S. R.; Ng W. B.; Cho S.-J.; Cho N.-J. Controlling the Formation of Phospholipid Monolayer, Bilayer, and Intact Vesicle Layer on Graphene. ACS Appl. Mater. Interfaces 2016, 8, 11875–11880. 10.1021/acsami.6b02837. [DOI] [PubMed] [Google Scholar]
- Tabaei S. R.; Jackman J. A.; Kim S.-O.; Liedberg B.; Knoll W.; Parikh A. N.; Cho N.-J. Formation of Cholesterol-Rich Supported Membranes Using Solvent-Assisted Lipid Self-Assembly. Langmuir 2014, 30, 13345–13352. 10.1021/la5034433. [DOI] [PubMed] [Google Scholar]
- Tabaei S. R.; Jackman J. A.; Liedberg B.; Parikh A. N.; Cho N.-J. Observation of Stripe Superstructure in the β-Two-Phase Coexistence Region of Cholesterol–Phospholipid Mixtures in Supported Membranes. J. Am. Chem. Soc. 2014, 136, 16962–16965. 10.1021/ja5082537. [DOI] [PubMed] [Google Scholar]
- Dixon M. C. Quartz crystal microbalance with dissipation monitoring: enabling real-time characterization of biological materials and their interactions. J. Biomol. Tech. 2008, 19, 151. [PMC free article] [PubMed] [Google Scholar]
- Nordin D.; Yarkoni O.; Savinykh N.; Donlon L.; Frankel D. Revealing the selective interactions of fibronectin with lipid bilayers. Soft Matter 2011, 7, 10666–10675. 10.1039/c1sm06291c. [DOI] [Google Scholar]
- Pelta J.; Berry H.; Fadda G.; Pauthe E.; Lairez D. Statistical conformation of human plasma fibronectin. Biochemistry 2000, 39, 5146–5154. 10.1021/bi992770x. [DOI] [PubMed] [Google Scholar]
- Kleinman H. K.; McGoodwin E. B.; Martin G. R.; Klebe R. J.; Fietzek P. P.; Woolley D. E. Localization of the binding site for cell attachment in the alpha1 (I) chain of collagen. J. Biol. Chem. 1978, 253, 5642–5646. [PubMed] [Google Scholar]
- Jokinen J.; Dadu E.; Nykvist P.; Käpylä J.; White D. J.; Ivaska J.; Vehviläinen P.; Reunanen H.; Larjava H.; Häkkinen L. Integrin-mediated cell adhesion to type I collagen fibrils. J. Biol. Chem. 2004, 279, 31956–31963. 10.1074/jbc.M401409200. [DOI] [PubMed] [Google Scholar]
- Knight C. G.; Morton L. F.; Peachey A. R.; Tuckwell D. S.; Farndale R. W.; Barnes M. J. The Collagen-binding A-domains of Integrins α1β1 and α2β1recognize the same specific amino acid sequence, GFOGER, in native (Triple-helical) collagens. J. Biol. Chem. 2000, 275, 35–40. 10.1074/jbc.275.1.35. [DOI] [PubMed] [Google Scholar]
- Kim S.-H.; Turnbull J.; Guimond S. Extracellular matrix and cell signalling: the dynamic cooperation of integrin, proteoglycan and growth factor receptor. J. Endocrinol. 2011, 209, 139–151. 10.1530/JOE-10-0377. [DOI] [PubMed] [Google Scholar]
- Stupack D. G.; Cheresh D. A. Get a ligand, get a life: integrins, signaling and cell survival. J. Cell Sci. 2002, 115, 3729–3738. 10.1242/jcs.00071. [DOI] [PubMed] [Google Scholar]
- Schwartz M. A.; Assoian R. K. Integrins and cell proliferation. J. Cell Sci. 2001, 114, 2553–2560. [DOI] [PubMed] [Google Scholar]
- Frith J. E.; Mills R. J.; Hudson J. E.; Cooper-White J. J. Tailored integrin–extracellular matrix interactions to direct human mesenchymal stem cell differentiation. Stem Cells Dev. 2012, 21, 2442–2456. 10.1089/scd.2011.0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood J. D.; Cheresh D. A. Role of integrins in cell invasion and migration. Nat. Rev. Cancer 2002, 2, 91–100. 10.1038/nrc727. [DOI] [PubMed] [Google Scholar]
- Martinez-Hernandez A.; Amenta P. S. The hepatic extracellular matrix. I. Components and distribution in normal liver. Virchows Arch. A: Pathol. Anat. Histopathol. 1993, 423, 1–11. 10.1007/BF01606425. [DOI] [PubMed] [Google Scholar]
- Sauerbrey G. Verwendung von Schwingquarzen zur Wägung dünner Schichten und zur Mikrowägung. Z. Phys. 1959, 155, 206–222. 10.1007/BF01337937. [DOI] [Google Scholar]
- Ruoslahti E.; Pierschbacher M. D. New perspectives in cell adhesion: RGD and integrins. Science 1987, 238, 491–497. 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- Nakanishi K.; Sakiyama T.; Kumada Y.; Imamura K.; Imanaka H. Recent advances in controlled immobilization of proteins onto the surface of the solid substrate and its possible application to proteomics. Curr. Proteomics 2008, 5, 161–175. 10.2174/157016408785909622. [DOI] [Google Scholar]
- Rusmini F.; Zhong Z.; Feijen J. Protein immobilization strategies for protein biochips. Biomacromolecules 2007, 8, 1775–1789. 10.1021/bm061197b. [DOI] [PubMed] [Google Scholar]
- Ishiyama M.; Miyazono Y.; Sasamoto K.; Ohkura Y.; Ueno K. A highly water-soluble disulfonated tetrazolium salt as a chromogenic indicator for NADH as well as cell viability. Talanta 1997, 44, 1299–1305. 10.1016/S0039-9140(97)00017-9. [DOI] [PubMed] [Google Scholar]
- Tominaga H.; Ishiyama M.; Ohseto F.; Sasamoto K.; Hamamoto T.; Suzuki K.; Watanabe M. A water-soluble tetrazolium salt useful for colorimetric cell viability assay. Anal. Commun. 1999, 36, 47–50. 10.1039/a809656b. [DOI] [Google Scholar]
- Juliano R. L.; Haskill S. Signal transduction from the extracellular matrix. J. Cell Biol. 1993, 120, 577. 10.1083/jcb.120.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson P.; Jonsson M. P.; Tegenfeldt J. O.; Höök F. A method improving the accuracy of fluorescence recovery after photobleaching analysis. Biophys. J. 2008, 95, 5334–5348. 10.1529/biophysj.108.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.