Abstract
B-cell epitope sequences from Zika virus (ZIKV) NS1 protein have been identified using epitope prediction tools. Mapping these sequences onto the NS1 surface reveals two major conformational epitopes and a single linear one. Despite an overall average sequence identity of ca. 55% between the NS1 from ZIKV and the four dengue virus (DENV) serotypes, epitope sequences were found to be highly conserved. Nevertheless, nonconserved epitope-flanking residues are responsible for a dramatically divergent electrostatic surface potential on the epitope regions of ZIKV and DENV2 serotypes. These findings suggest that strategies for differential diagnostics on the basis of short linear NS1 sequences are likely to fail due to immunological cross-reactions. Overall, results provide the molecular basis of differential discrimination between Zika and DENVs by NS1 monoclonal antibodies.
Introduction
Given the recent outbreak of Zika virus (ZIKV) in the Americas in 2015 and the association with microcephaly cases1,2 and neurological disorders, such as the Guillain–Barré syndrome, ZIKV was recently placed as a public health emergency of international concern by the World Health Organization.3 ZIKV is a flavivirus of the family Flaviviridae, transmitted by Aedes sp. mosquitoes,4 and closely related to dengue virus (DENV), West Nile virus, Japanese encephalitis virus, and yellow fever virus (YFV).5 To date, there is no prophylactic treatment nor vaccine available against ZIKV and disease control is limited to vector eradication strategies. The presumptive diagnosis is typically clinical, whereas confirmatory laboratory tests include classic virus isolation on cell culture and viral RNA detection by reverse transcriptase PCR (RT-PCR) in serum, saliva, and/or urine samples within the first 5 or 6 days of infection.6,7 ZIKV serology is usually performed by enzyme-linked immunosorbent assay (ELISA), using full-length viral proteins or linear peptides, however, due to high cross-reactivity of IgM and IgG antibodies between ZIKV and other related flavivirus, especially in endemic areas where cocirculation exists, confirmatory tests are necessary. Currently, confirmation testing is performed by plaque reduction neutralization test (PRNT), according to previously published protocols.8−11 At present, there are only a very few commercially available serology kits for ZIKV antibody detection; moreover, the specificity of these kits remains elusive, given the high cross-reactivity already reported among individuals who experienced consecutive flavivirus infections.12,13 On the basis of this, PRNT remains the “gold standard” for antiflavivirus differentiation.10 PRNT, however, is a high-cost technique that requires highly specialized laboratories and special regulations due to live virus manipulations. Although new protocols using recombinant viruses14 or reporter virus particles11 have been developed, these are still not available for ZIKV routine diagnosis.15 In this scenario, there is an urgent need for a low-cost unequivocal serological diagnostic method for ZIKV to be able to overcome the high cross-reactivity with other flaviviruses, such as DENV and yellow fever (especially on vaccinated individuals).16
The ZIKV positive-sense RNA genome comprises a single open reading frame encoding a unique polyprotein that is cleaved into three structural proteins (Capsid (C), premembrane (prM), and envelope (E), which form the virus particle, and seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5), which perform essential functions in genome replication, polyprotein cleavage, and modulation of cellular processes to viral advantage.17,18 Among the NS proteins, NS1 glycoprotein is a multifunctional virulence factor.19,20 Within infected cells, glycosylated NS1 is found as a membrane-associated dimer after translocation into the endoplasmic reticulum lumen, in which it colocalizes with dsRNA and the viral replication complex, being essential for viral genome replication.18 Infected cells also secrete NS1 as a hexameric lipoprotein,21 which interacts with complement-system proteins and has several immune-modulatory functions.18 This conformational dynamism of NS1 is also employed by the virus during the infection cycle to evade antiviral cellular immune responses from the host.22 The homodimer structure of ZIKV NS1 is presented in Figure 1 and encompasses a highly structured continuous extended β-sheet ladder on one surface. On the opposite face, there is a predominance of a nonstructured loop medley and a flexible loop, comprising the residues from 108 to 129.18 The loop surface of flaviviruses NS1 has been previously suggested to play a role in interactions of this protein with host factors and antibodies.23
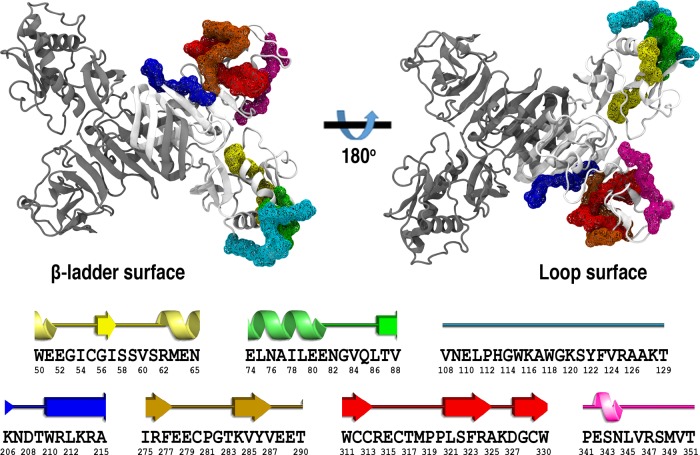
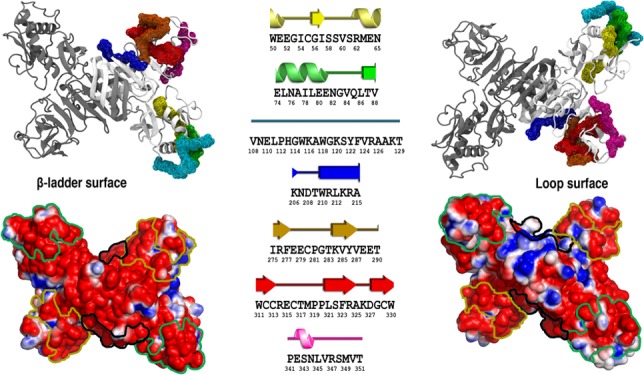
Figure 1.
Cartoon representation of the ZIKV NS1 homodimer structure. Protein chains are shown in white and gray. Putative epitopes are mapped as the wireframe surface and color coded as E1 in yellow, E2 in green, E3 in cyan, E4 in blue, E5 in orange, E6 in red, and E7 in magenta. The corresponding secondary structure assignment for each putative epitope is shown at the bottom, with their respective amino acid sequence (PDB ID 5K6K).
Besides playing a crucial role in the pathogenesis of the ZIKV infection, NS1 emerges as a potential candidate for differential diagnostics.16 NS1 is considered the major antigenic marker for flavivirus infections, and it has been used for the detection of DENV in patient sera.22,24−30 Despite structural conservation of NS1 among flaviviruses, the average sequence identity between ZIKV and DENV NS1 proteins is about 55%,16,31 and ZIKV-specific monoclonal antibodies (mAbs) have been isolated,16 showing that this protein contains regions capable of being specifically recognized. Additionally, some differences between the NS1 of ZIKV and other flaviviruses have been reported. A negatively charged glutamate residue located at position 315 of ZIKV NS1 has been pointed out as exclusive for ZIKV, being absent in other flaviviruses including DENV serotypes which have a serine residue at this position. As a consequence, differences are observed in the electrostatic surface potential of ZIKV NS1 compared to that of other flaviviruses.31 Another recent work reported the crystal structure of full-length ZIKV NS1 and revealed a region formed by the residues 108–129 on the wing-domain flexible loop, which had not been previously described in other flaviviruses.18 Nevertheless, complete mapping of B-cell ZIKV NS1 epitopes have not been reported. In the present work, we have assigned ZIKV NS1 putative B-cell epitopes and mapped the identified regions onto the corresponding three-dimensional structure. Comparisons of the identified B-cell ZIKV NS1 epitopes to their homologous regions in NS1 from DENV2 on the basis of sequence identity analysis and conformation assessment through molecular dynamics (MD) simulations provided a solid understanding of the molecular basis for differential antibody discrimination.
Results and Discussion
Epitope Prediction
Detection of antigen and pathogen specific antibodies is among the most used approaches for clinical diagnostics. Antigen specific antibody detection is used in the diagnosis of several diseases, and its application continues to expand. In addition, antibodies are the most effective mechanism of protection provided by the adaptive immune system. A critical and at many times limiting aspect for eliciting highly specific antibodies is the structural transiency of certain conformational B-cell epitopes. In addition, important epitopes are often cryptic, frequently buried within the protein, and only exposed when the virus binds to its receptor.32 Thus, when searching for B-cell epitopes in a given protein, it is important that they are located in accessible regions to allow proper recognition and interaction with their respective antibodies. Therefore, to perform in silico epitope prediction, the ZIKV NS1 protein structure (PDB ID: 5K6K)18 was used. Although the ZIKV NS1 structure is presented in a dimeric form, only the B chain was used in the epitope prediction analyses, as the three-dimensional coordinates for residues 113–120 have not been experimentally assigned for the chain A. Most methods of epitope prediction use properties related to surface exposure, such as flexibility or hydrophobicity.33 In this work, we have chosen three algorithms to assess the robustness of our epitope prediction rationale. We combined ElliPro that is based on the geometrical properties of the protein structure (antigenicity, solvent accessibility, and flexibility) and can predict linear and conformational epitopes,34 Epitopia that implements a machine-learning algorithm that was trained to discern antigenic features within a given protein and predicts the most probable conformational epitope regions,35 and DiscoTope that is based on amino acid statistics, spatial information, and surface accessibility and can predict residues that are part of conformational epitopes recognized by B-cells.36 A previous study has pointed out that consensus results from the three servers would improve the specificity and accuracy of epitope prediction, resulting in a lower number of false positive results and thus generating greater confidence.37 Considering consensus overlapping regions by at least two servers, 7 putative ZIKV NS1 regions have been selected, with epitope lengths ranging from 10 to 22 residues (Table 1). The identified regions include sequences that encompass the E315 residue and residues 108–129, which have been previously described due to the presence of exclusive features in ZIKV that were not found in other flaviviruses.18,31
Table 1. ZIKV NS1 Regions Identified in Silico as Putative Epitopes.
| ZIKV NS1 epitope | residue interval | sequence length | amino acid sequence |
|---|---|---|---|
| E1 | 50–65 | 16 | WEEGICGISSVSRMEN |
| E2 | 74–88 | 15 | ELNAILEENGVQLTV |
| E3 | 108–129 | 22 | VNELPHGWKAWGKSYFVRAAKT |
| E4 | 206–215 | 10 | KNDTWRLKRA |
| E5 | 275–290 | 16 | IRFEECPGTKVYVEET |
| E6 | 311–330 | 20 | WCCRECTMPPLSFRAKDGCW |
| E7 | 341–351 | 11 | PESNLVRSMVT |
Upon comparing the results obtained by each server, ElliPro predicted a higher number of B-cell epitopes, followed by Epitopia, whereas DiscoTope predicted only three putative epitopes (Table 2). The greater performance of Ellipro may be related to an algorithm that takes into account structural information as well as the identification of regions in the protein that protrude from the globular surface. Discotope presented a lower performance possibly because it uses a sequence-based prediction wherein individual residues are listed. Therefore, this suggests that at least for the NS1 protein of ZIKV, sequence information alone may lead to a poor prediction performance. The selected regions were mapped onto the ZIKV NS1 structure along with their corresponding secondary structure content, as shown in Figure 1. Among the seven identified putative epitopes, the structural mapping (Figure 1) suggests that those regions comprise two conformational epitopes (CE1 and CE2) and one linear epitope (LE1). CE1 consists of E1, E2, and E3 identified regions; CE2 consists of E5, E6, and E7 identified regions; whereas LE1 corresponds to the E4 region. The same epitope prediction scheme was carried out for the NS1 protein from DENV2 (PDB ID 4O6B(38)), as it is the only serotype structure available in the PDB. All three algorithms predicted the seven corresponding regions in the same manner as putative epitopes (data not shown for conciseness). This is not unexpected given that the two proteins share a nearly identical three-dimensional structure.
Table 2. Overview of the in Silico B-Cell Epitope Predictions for the Seven Identified Regions and Their Classification into Putative Conformational or Linear Epitopesa,b.
| ZIKV NS1 epitope | ElliPro | Epitopia | DiscoTope |
|---|---|---|---|
| CE1 | |||
| E1 | + | + | – |
| E2 | + | + | – |
| E3 | + | + | + |
| LE1 | |||
| E4 | + | – | + |
| CE2 | |||
| E5 | + | + | – |
| E6 | + | + | – |
| E7 | + | + | + |
ZIKV Peptide Microarray
To assess the reactivity of the identified ZIKV NS1 epitopes with specific IgG antibodies, a 15-mer peptide library spanning the entire ZIKV NS1 protein was tested against seven confirmed convalescent ZIKV positive patients and 6 ZIKV and DENV naïve human serum samples (control) in a peptide microarray. The ZIKV NS1 peptides were tested against each individual sample. Figure 2A shows the schematic representation of the assay, whereas Figure 2B shows the fluorescence readings for the putative epitopes. Values are presented as an average of individual measurements for each serum along with the corresponding standard error of the mean (SEM). SEM is a statistical method that takes into account the sample size and standard deviation to provide accuracy of the mean. (Given the small sample size (7 ZIKV positives and 6 controls), results from most statistical analyses, such as t-test, cannot be applicable with confidence). Accession numbers for the sequences used in the peptide microarray are listed in Table S1.
Figure 2.
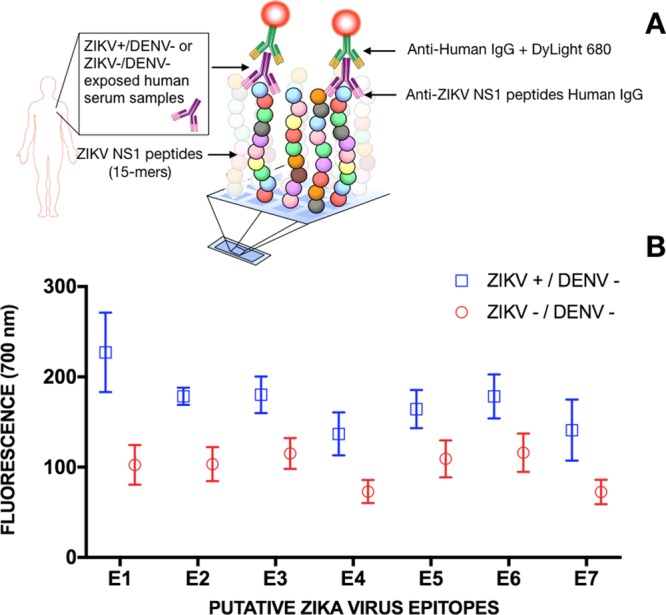
IgG response from sera of 7 confirmed ZIKV positive convalescent patients and 6 confirmed ZIKV and DENV negative patients to seven 15-mer peptides, whose sequences correspond to the predicted ZIKV NS1 epitopes. (A) The experimental detection scheme of the ZIKV NS1 15-mer peptide array. (B) AntiZIKV NS1 IgG detection for each ZIKV putative epitope expressed as the fluorescence average over the patients’ serum samples along with their corresponding standard mean error and statistical significance.
The results of the ZIKV peptide microarray technique show that all of the seven in silico predicted regions were recognized by IgG antibodies present in human serum samples from ZIKV-infected individuals. Although larger differences between positive and negative serum samples are observed for E1–E4, discrimination was possible for all cases. Inference about the amplitude of the differences cannot be made due to a small sample size and the fact that (intrinsically flexible) peptides are used as markers for antibody recognition and therefore may only be present in a recognizable conformation, part of the time. Nevertheless, these results confirm the immunogenicity of the identified regions and thus validate the computational prediction approach. (It is worth mentioning that for the limitations stated above, it is out of the scope of this work to draw conclusions regarding ZIKV immune response to these epitopes; this assay was used with the sole purpose of validating the computational predictions).
Sequence Identity Analysis
The identification of B-cell epitopes containing low sequence identity between ZIKV and DENV1–4 is of particular interest for the development of differential diagnostic tools between the two infections. Conversely, epitopes exhibiting high sequence identities between the two viruses and capable of eliciting cross-neutralization can potentially be used in multivalent vaccine formulation strategies. Alignment of the identified ZIKV NS1 putative epitopes with DENV NS1 revealed a considerably high identity with at least one strain (Figure 3). Therefore, all seven peptide sequences are unlikely to be successful for differential diagnosis between ZIKV and DENV patients. Interestingly, those seven regions comprise 31% of the entire NS1 protein and by excluding them from the alignment, the overall identity falls to ca. 16% (from ca. 55 to 39%, Figure 3). We consider sequence-bias prediction unlikely because the sequences of the seven epitopes are very different from each other. The evolutionary implications of having conserved B-cell epitopes between the two viruses are unknown. It suggests that these epitopes might be involved in important infection mechanisms because in principle, a lower identity would aid ZIKV to evade immune response from patients previously infected by DENV.
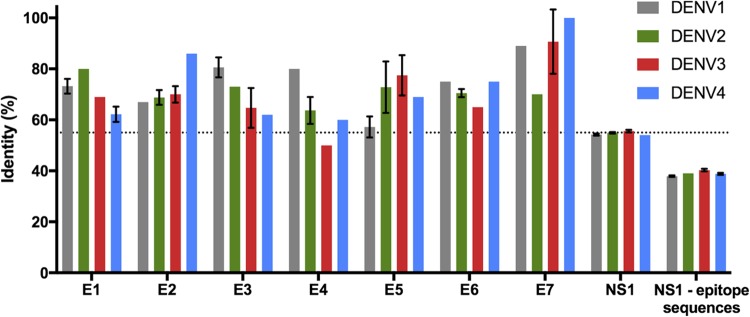
Figure 3.
Sequence identity comparison between ZIKV and DENV1–4 NS1 proteins. (Standard deviation bars are presented for the 10 most prevalent DENV strains. NS1-epitope sequences correspond to the entire NS1 sequence excluding the seven in silico predicted putative epitope regions). A dotted line represents the average identity between the full-length NS1 of the Brazilian strain with the 10 most prevalent strains of DENV1–4.
Nevertheless, it has been reported that antiZIKV NS1 mAbs were able to be virus type-specific, not showing cross-reactivity with DENV NS1.16 We have observed that individual linear sequences are conserved and clustered together in two regions, possibly forming two conformational epitopes. The high identities of the linear sequences of the epitopes of ZIKV are likely to cross-react with antibodies antiDENV and suggest that such differential antibody recognition takes place via conformational epitopes. Although one cannot overrule an eventual nonpredicted epitope on NS1 from ZIKV and DENV, we consider it unlikely given that the three algorithms used predicted epitopes on the same regions of the two proteins. Therefore, we postulate that such discrimination at the molecular-level is likely explained by either a contrasting tertiary arrangement of those regions on the protein surface and/or a distinct charge signature profile.
Epitopes’ Conformational Arrangement and Electrostatic Surface Profiles
To gain insights into the dynamics and conformational profiles of ZIKV and DENV NS1 proteins, MD simulations were performed using the dimeric form of the protein from both viruses. The DENV serotype 2 was chosen as the representative of DENV strains, as it is the only dimeric structure currently available in the Protein Data Bank (PDB). On the basis of the MD results, no significant differences could be observed by comparing the global structural protein dynamics (Figure S1). The root-mean-square deviation, radius of gyration (Rg), and root-mean-square fluctuation for both systems showed equivalent behaviors for the NS1 dimers and each monomeric unit within the dimer. These findings are in agreement with the work by Brown et al.18 and Song et al.,31 showing that in its protein fold and domain arrangement, ZIKV NS1 is virtually identical to DENV2 NS1. Still, a small per-residue solvent accessibility (Figure S2) and fluctuational (Figure S1) differences may be observed in some of the seven predicted epitope regions, when comparing ZIKV and DENV2 NS1. However, differences in solvation on the epitope regions are subtle (Figure S3) and do not warrant a differential tertiary arrangement of such epitopes onto ZIKV and DENV2 NS1 surfaces.
Because the putative epitopes were shown to be conserved but not the remaining residues, the electrostatic surface potentials were calculated and plotted onto the molecular surface of representative frames of ZIKV and DENV2 NS1 extracted from the MD simulations (Figure 4). A significant divergence between ZIKV and DENV2 NS1 can be observed. Despite approximately 55% of sequence identity between the proteins, ZIKV NS1 shows a remarkably negatively charged profile, whereas DENV2 presents extensively positively charged regions, especially in the loop region. Accordingly, the same charge-distribution pattern was described by Brown et al. when evaluating the electrostatic surface potential of entire NS1 proteins from ZIKV and DENV.18 Because the outer face of NS1 is more likely to elicit an immune response, regardless of NS1 being on the cell surface or being secreted, this difference in electrostatic distribution between ZIKV and DENV has implications for immune system recognition and vaccine development and points toward different modes of flavivirus pathogenesis.
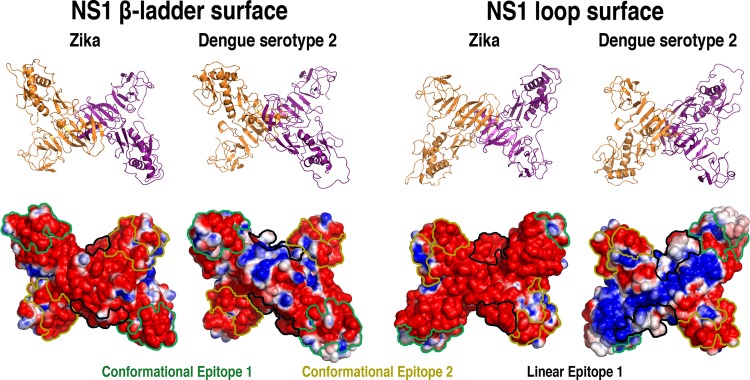
Figure 4.
Cartoon representations and electrostatic surface potential on both faces of glycosylated ZIKV and DENV2 NS1 proteins. The locations of the predicted epitopes are color-contoured (green: CE1, yellow: CE2, and black: LE1). Negatively charged regions are shown in red, positively charged regions in blue, and neutral regions in white. (Plotted potential ranged from −2 to +2 kJ mol–1 e–1.)
Most of these regions, however, occur away from the predicted epitopes (Figure 4) and shall be involved in other processes beyond antibody recognition. In addition, significant differences in the electrostatic potentials are also observed for the predicted epitopes (Figure 4). CE1 contains a higher amount of positively charged surface in the β-ladder view in DENV2 and significantly different conformational arrangements in the two proteins in the loop surface due to the Asn130-linked glycan orientation. For CE2, although the spatial distribution is quite similar in both surface views, the charge distribution differs between ZIKV and DENV2. LE1 also shows significant differences both in terms of conformation and electrostatic potential, being more exposed in the loop surfaces of both proteins but containing a considerably more positively charged surface in DENV2. Altogether, CE1 appears to have most conformational and charge-distribution differences between ZIKV and DENV2 (Figure 4) while being highly solvent-exposed in both (Figure S2), especially in the region between residues 108 and129 (Figure S3).
Conclusions
Putative ZIKV NS1 B-cell epitopes were mapped onto the NS1 protein surface. Our results revealed that: (i) ZIKV NS1 immunogenic regions are comprised by two conformational epitopes and a linear one; (ii) epitope sequences were found to be conserved between ZIKV and DENV; (iii) despite the high epitope identity and structural similarity, ZIKV and DENV2 NS1 proteins display a loop–surface interface with significantly different electrostatic potentials; (iv) these differences are suggested to explain the molecular basis for unique binding properties to host factors and specific protective monoclonal antibody recognition therefore providing a lead for rational epitope-focused design of diagnostic tools and structure-based vaccine antigens for ZIKV infection.
Materials and Methods
Epitope Prediction
Atomic coordinates for the Brazilian ZIKV NS1 strain obtained experimentally (PDB ID: 5K6K(18)) were used in three different structure-based tools for epitope prediction: ElliPro,34 Epitopia,35 and DiscoTope.36 Additionally, the solvent-accessible surface area was calculated using the postprocessing tool g_sas within GROMACS 5.1.2 package,39 to evaluate the exposure of each putative epitope to the solvent.
NS1 ZIKV Peptide Microarray
The publicly-available whole proteome sequences of 15 ZIKV strains (see accession numbers on Table S1) were retrieved from the NCBI database and translated into 15-mer peptides with a peptide-peptide overlap of 12 amino acids. The whole proteome array data will be published elsewhere. Peptide arrays were produced by the company PEPperPRINT GmbH (Heidelberg, Germany) in a laser printing process on glass slides, coated with a poly(ethylene glycol) methacrylate/poly(methyl methacrylate) graft copolymer, which were functionalized with a ßAla-ßAla-linker. In brief, a layer of amino acid particles containing Fmoc-amino acid pentafluorophenyl esters was printed layer after layer onto the functionalized glass slides, with intermittent melting (i.e., coupling) steps at 90 °C and chemical washing and capping steps.40 Peptides were generated in duplicates on the arrays, which were screened for IgG responses in human sera.
Immunostaining of Peptide Microarrays
Peptide microarrays were placed in incubation trays (PEPperPRINT GmbH; Heidelberg, Germany) and blocked for 30 min at room temperature at 120 rpm orbital shaking, with western blot blocking buffer MB-070 (Rockland). Then, sera were diluted by 1:1000 in phosphate-buffered saline (PBS) buffer with 0.05% Tween 20, pH 7.4 (PBS with Tween 20 (PBST)), and 10% blocking buffer, incubating the sera for 16 h at 4 °C and 50 rpm orbital shaking. Peptide microarrays were washed three times with PBST, followed by incubation with a 1:2500 dilution of the secondary antibody (goat anti-human IgG Fc specific DyLight 680; Rockland) together with the control antibody diluted by 1:500 (anti-c-Myc antibody, PEPperPRINT, Germany) for 30 min at room temperature and 120 rpm orbital shaking. The peptide microarrays were washed for 3 × 10 s with PBST and rinsed with deionized water. After drying in a stream of air, fluorescent images were acquired using an Odyssey Imaging System (LI-COR) at 700 nm, with a resolution of 21 μm and a scanning sensitivity of 7. Image analysis and quantification was performed with PepSlide Analyzer software (Sicasys Software GmbH; Heidelberg, Germany).
Samples used for the peptide array experiments were collected from individuals with acute febrile illnesses enrolled in a prospective cohort study from May 2015 to May 2016. The cohort was established in an urgent health care clinic in the Recife metropolitan region as part of the International Research Consortium on Dengue Risk Assessment, Management, and Surveillance-IDAMS.41,42 The ages of patients from whom sera were used for the assays varied from 9 to 57 years, wherein 16 were females and 13 were males. Sample collection was performed on the first day of recruitment (Day 1 – acute sample), which following the IDAMS protocol corresponds to the period within the first 72 h of the febrile period, and in the convalescent phase (days 10–30 after recruitment – convalescent sample). For molecular viral diagnosis, quantitative real-time PCR (qRT-PCR) for DENV and ZIKV was performed following modified previously reported protocols.43,44 As for serology, samples were assayed for antiDENV IgM and IgG and antiZIKV IgM through ELISA. The Panbio Dengue Capture ELISA was used for the antiDENV IgM and IgG assays, following the manufacturer’s protocol. The antiZIKV IgM MAC-ELISA protocol was that of the Centers for Diseases Control and Prevention (CDC).43,45 According to assay results, each sample was classified as: (1) ZIKV Positive/DENV Negative, if they were positive for ZIKV qRT-PCR and negative for DENV qRT-PCR in the acute phase and/or positive for antiZIKV IgM with titers > 2 times those for antiDENV IgM in the convalescent phase in the MAC-ELISA, and negative for antiDENV IgG in the acute phase in the Panbio assay; (2) ZIKV Negative/DENV Negative, if they were negative for DENV and ZIKV qRT-PCRs in the acute phase and negative for antiDENV and antiZIKV IgM in the acute phase in MAC-ELISA and negative for antiDENV IgG in both acute and convalescent phases in the Panbio assay. The Panbio indirect antiDENV IgM and IgG test was not performed. Additionally, one serum sample used in this study was collected from a 21 year old male individual enrolled in the prospective cohort study PRONEX established in the city of Recife, Brazil. The sample was collected on day 2 after the onset of symptoms and its serological status determined as negative for DENV IgM and IgG ELISAs and positive in the ZIKV IgM ELISA.
Sequence Identity Analysis
ZIKV NS1 selected regions’ sequences were aligned with DENV serotypes (DENV1, DENV2, DENV3, and DENV4) using the basic local alignment search tool provided by National Center for Biotechnology Information – NCBI46 to obtain their sequence identity content. The same procedure was done with the complete sequence and for the sequence excluding the putative epitopes. Accession codes: DENV1 (AHK09916.1, AHK09915.1, AKA43760.1, AHK09902.1, AHK09902.1, AHK09909.1, AHK09914.1, AHK09919.1, AMO26073.1, AKK23335.1), DENV2 (AAD11533.1, AAA42947.1, AAL58460.1, AAD11532.1, AEZ68639.1, AJP06043.1, AAD11531.1, AAR17682.1, CAA35218.1, AHK09925.1), DENV3 (AHK09941.1, AAB52248.2, AHK09929.1, AAC04352.2, AAB52247.1, AAR17683.1, AHK09939.1, AAB52244.2, AAB2245.2, AHK09940.1), DENV4 (AHK09946.1, AHG23296.1, AKQ00032.1, AKQ00028.1, AAW51420.1, AHG23273.1, AHG23297.1, AHG23295.1, AEA50927.1, ACL99035.1).
NS1 Glycosylation, Modeling, and MD Simulations
NS1 structures for Dengue serotype 2 and ZIKV were retrieved from PDB ID 4O6B(38) and 5K6K,18 respectively. The missing loops in those structures were modeled using ModLoop tool,47,48 whereas the N-glycosylation motifs located at Asn130 (containing Asn130-Asn131-Ser132 in ZIKV and Asn130-Gln131-Thr132 in DENV2) and Asn207 (containing Asn207-Asp208-Thr209 in both proteins) were modified using glycosciences modeling tools,49 respectively, with a complex-type and with a high-mannose type oligosaccharide50 (Figure S3). The electrostatic potential surfaces were calculated using APBS.51 All MD calculations and remaining analyses were performed using GROMACS simulation suite, version 5.1.2.39 All atomistic simulations were performed using GROMOS 54A7 force field for the protein part52 and GROMOS 53A6GLYC parameters set for the glycan moieties.53,54 The glycosylated NS1 structures were explicitly solvated in 18 nm × 18 nm × 18 nm cubic boxes, using periodic boundary conditions in x, y, and z directions and the SPC water model,55 employing at least 14 Å distance from the outside of each structure and the box edge. To neutralize the charge of these systems, Na+ counterions were added, as necessary. The LINCS method56 was applied to constrain covalent bond lengths, allowing an integration step of 2 fs after an initial energy minimization using steepest descent algorithm. A short-range cutoff radius of 1.4 nm was used for all nonbonded interactions. Long-range electrostatic interactions were calculated through the reaction-field method,57 with ε = 66. The system was coupled to a thermostat with a reference temperature of 310 K using the velocity-rescale scheme,58 with a relaxation time of 0.4 ps. Pressure was maintained constant at 1 bar by isotropic coordinate scaling using Parrinello–Rahman barostat,59,60 with a coupling constant of 1.0 ps and a compressibility of 4.5 × 10–5 bar. A 1 ns MD simulation with positional restraints on all heavy atoms, with a force constant of 1000 kJ mol –1, was performed as an equilibration step and was not taken into account to calculate the average ensemble properties. Subsequently, all systems were further extended to 200 ns (without positional restraints). Representative structures were selected from the equilibrated part of trajectories (last 50 ns) by clustering all the frames generated within this time scale (employing g_cluster tool from GROMACS suite, with a 0.4 nm cutoff). Thus, each of ZIKV and DENV2 NS1 MD simulations provided a single conformational cluster and the representative frames used for electrostatic surface potential analyses.
Acknowledgments
This work was supported by grants from FACEPE, CAPES, CNPq and CuraZika Foundation. Computational resources were provided by the Brazilian National Supercomputing Center (LNCC).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.7b00608.
MD structural data analyses; glycosylation sequence used in NS1 proteins; and accession numbers for the ZIKV sequences used in the peptide array (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Lover A. A. Zika virus and microcephaly. Lancet Infect. Dis. 2016, 16, 1331–1332. 10.1016/S1473-3099(16)30462-5. [DOI] [PubMed] [Google Scholar]
- de Araújo T. V.; Rodrigues L. C.; de Alencar Ximenes R. A.; de Barros Miranda-Filho D.; Montarroyos U. R.; de Melo A. P.; Valongueiro S.; de Albuquerque M. F.; Souza W. V.; Braga C.; Filho S. P.; Cordeiro M. T.; Vazquez E.; Di Cavalcanti Souza Cruz D.; Henriques C. M.; Bezerra L. C.; da Silva Castanha P. M.; Dhalia R.; Marques-Júnior E. T.; Martelli C. M. Association between Zika virus infection and microcephaly in Brazil, January to May, 2016: preliminary report of a case-control study. Lancet Infect. Dis. 2016, 16, 1356–1363. 10.1016/S1473-3099(16)30318-8. [DOI] [PubMed] [Google Scholar]
- Lessler J.; Chaisson L. H.; Kucirka L. M.; Bi Q.; Grantz K.; Salje H.; Carcelen A. C.; Ott C. T.; Sheffield J. S.; Ferguson N. M.; Cummings D. A.; Metcalf C. J.; Rodriguez-Barraquer I. Assessing the global threat from Zika virus. Science 2016, 353, aaf8160 10.1126/science.aaf8160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chouin-Carneiro T.; Vega-Rua A.; Vazeille M.; Yebakima A.; Girod R.; Goindin D.; Dupont-Rouzeyrol M.; Lourenco-de-Oliveira R.; Failloux A. B. Differential Susceptibilities of Aedes aegypti and Aedes albopictus from the Americas to Zika Virus. PLoS Neglected Trop. Dis. 2016, 10, e0004543 10.1371/journal.pntd.0004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear H. M.; Diamond M. S. Zika Virus: New Clinical Syndromes and Its Emergence in the Western Hemisphere. J. Virol. 2016, 90, 4864–4875. 10.1128/JVI.00252-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes B. E.; Smalley C. A.; Tiner B. L.; Beasley D. W.; Milligan G. N.; Reece L. M.; Hombach J.; Barrett A. D. Research and development of Zika virus vaccines. npj Vaccines 2016, 1, 16007 10.1038/npjvaccines.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waggoner J. J.; Pinsky B. A. Zika Virus: Diagnostics for an Emerging Pandemic Threat. J. Clin. Microbiol. 2016, 54, 860–867. 10.1128/JCM.00279-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. J.; Martin D. A.; Karabatsos N.; Roehrig J. T. Detection of anti-arboviral immunoglobulin G by using a monoclonal antibody-based capture enzyme-linked immunosorbent assay. J. Clin. Microbiol. 2000, 38, 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin D. A.; Muth D. A.; Brown T.; Johnson A. J.; Karabatsos N.; Roehrig J. T. Standardization of immunoglobulin M capture enzyme-linked immunosorbent assays for routine diagnosis of arboviral infections. J. Clin. Microbiol. 2000, 38, 1823–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuno G. Serodiagnosis of flaviviral infections and vaccinations in humans. Adv. Virus Res. 2003, 61, 3–65. [DOI] [PubMed] [Google Scholar]
- Maeda A.; Maeda J. Review of diagnostic plaque reduction neutralization tests for flavivirus infection. Vet. J. 2013, 195, 33–40. 10.1016/j.tvjl.2012.08.019. [DOI] [PubMed] [Google Scholar]
- Wong S. J.; Furuya A.; Zou J.; Xie X.; Dupuis Ii A. P.; Kramer L. D.; Shi P.-Y. A Multiplex Microsphere Immunoassay for Zika Virus Diagnosis. EBioMedicine 2017, 16, 136–140. 10.1016/j.ebiom.2017.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumita L. M.; Rodrigues J. P.; Ferreira N. E.; Felix A. C.; Souza N. C. S.; Machado C. M.; Júnior H. F. d. A. Detection of human anti-zika virus IgG by elisa using an antigen from in vitro infected vero cells: preliminary results. Rev. Inst. Med. Trop. Sao Paulo 2016, 58, 89. 10.1590/s1678-9946201658089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B. W.; Kosoy O.; Hunsperger E.; Beltran M.; Delorey M.; Guirakhoo F.; Monath T. Evaluation of chimeric Japanese encephalitis and dengue viruses for use in diagnostic plaque reduction neutralization tests. Clin. Vaccine Immunol. 2009, 16, 1052–1059. 10.1128/CVI.00095-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musso D.; Gubler D. J. Zika Virus. Clin. Microbiol. Rev. 2016, 29, 487–524. 10.1128/CMR.00072-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stettler K.; Beltramello M.; Espinosa D. A.; Graham V.; Cassotta A.; Bianchi S.; Vanzetta F.; Minola A.; Jaconi S.; Mele F.; Foglierini M.; Pedotti M.; Simonelli L.; Dowall S.; Atkinson B.; Percivalle E.; Simmons C. P.; Varani L.; Blum J.; Baldanti F.; Cameroni E.; Hewson R.; Harris E.; Lanzavecchia A.; Sallusto F.; Corti D. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science 2016, 353, 823–826. 10.1126/science.aaf8505. [DOI] [PubMed] [Google Scholar]
- Kuno G.; Chang G. J. Full-length sequencing and genomic characterization of Bagaza, Kedougou, and Zika viruses. Arch. Virol. 2007, 152, 687–696. 10.1007/s00705-006-0903-z. [DOI] [PubMed] [Google Scholar]
- Brown W. C.; Akey D. L.; Konwerski J. R.; Tarrasch J. T.; Skiniotis G.; Kuhn R. J.; Smith J. L. Extended surface for membrane association in Zika virus NS1 structure. Nat. Struct. Mol. Biol. 2016, 23, 865–867. 10.1038/nsmb.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller D. A.; Young P. R. The flavivirus NS1 protein: molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antiviral Res. 2013, 98, 192–208. 10.1016/j.antiviral.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Watterson D.; Modhiran N.; Young P. R. The many faces of the flavivirus NS1 protein offer a multitude of options for inhibitor design. Antiviral Res. 2016, 130, 7–18. 10.1016/j.antiviral.2016.02.014. [DOI] [PubMed] [Google Scholar]
- Gutsche I.; Coulibaly F.; Voss J. E.; Salmon J.; d’Alayer J.; Ermonval M.; Larquet E.; Charneau P.; Krey T.; Megret F.; Guittet E.; Rey F. A.; Flamand M. Secreted dengue virus nonstructural protein NS1 is an atypical barrel-shaped high-density lipoprotein. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 8003–8008. 10.1073/pnas.1017338108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young P. R.; Hilditch P. A.; Bletchly C.; Halloran W. An antigen capture enzyme-linked immunosorbent assay reveals high levels of the dengue virus protein NS1 in the sera of infected patients. J. Clin. Microbiol. 2000, 38, 1053–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edeling M. A.; Diamond M. S.; Fremont D. H. Structural basis of Flavivirus NS1 assembly and antibody recognition. Proc. Natl. Acad. Sci. U.S.A. 2014, 111, 4285–4290. 10.1073/pnas.1322036111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller D. A.; Young P. R. The flavivirus NS1 protein: molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antiviral Res. 2013, 98, 192–208. 10.1016/j.antiviral.2013.03.008. [DOI] [PubMed] [Google Scholar]
- Cecchetto J.; Fernandes F. C.; Lopes R.; Bueno P. R. The capacitive sensing of NS1 Flavivirus biomarker. Biosens. Bioelectron. 2017, 87, 949–956. 10.1016/j.bios.2016.08.097. [DOI] [PubMed] [Google Scholar]
- Cavalcanti I. T.; Guedes M. I.; Sotomayor M. D.; Yamanaka H.; Dutra R. F. A label-free immunosensor based on recordable compact disk chip for early diagnostic of the dengue virus infection. Biochem. Eng. J. 2012, 67, 225–230. 10.1016/j.bej.2012.06.016. [DOI] [Google Scholar]
- Dias A. C. M. S.; Gomes-Filho S. L.; Silva M. M.; Dutra R. F. A sensor tip based on carbon nanotube-ink printed electrode for the dengue virus NS1 protein. Biosens. Bioelectron. 2013, 44, 216–221. 10.1016/j.bios.2012.12.033. [DOI] [PubMed] [Google Scholar]
- Figueiredo A.; Vieira N. C.; Dos Santos J. F.; Janegitz B. C.; Aoki S. M.; Junior P. P.; Lovato R. L.; Nogueira M. L.; Zucolotto V.; Guimarães F. E. Electrical detection of dengue biomarker using egg yolk immunoglobulin as the biological recognition element. Sci. Rep. 2015, 5, 7865 10.1038/srep07865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M. M. S.; Dias A.; Cordeiro M.; Marques E.; Goulart M.; Dutra R. A thiophene-modified screen printed electrode for detection of dengue virus NS1 protein. Talanta 2014, 128, 505–510. 10.1016/j.talanta.2014.06.009. [DOI] [PubMed] [Google Scholar]
- Su C.-C.; Wu T.-Z.; Chen L.-K.; Yang H.-H.; Tai D.-F. Development of immunochips for the detection of dengue viral antigens. Anal. Chim. Acta 2003, 479, 117–123. 10.1016/S0003-2670(02)01529-5. [DOI] [Google Scholar]
- Song H.; Qi J.; Haywood J.; Shi Y.; Gao G. F. Zika virus NS1 structure reveals diversity of electrostatic surfaces among flaviviruses. Nat. Struct. Mol. Biol. 2016, 23, 456–458. 10.1038/nsmb.3213. [DOI] [PubMed] [Google Scholar]
- Viana I. F.; Soares T. A.; Lima L. F.; Marques E. T.; Krieger M. A.; Dhalia R.; Lins R. D. De novo design of immunoreactive conformation-specific HIV-1 epitopes based on Top7 scaffold. RSC Adv. 2013, 3, 11790–11800. 10.1039/c3ra41562g. [DOI] [Google Scholar]
- Flower D. R. Designing immunogenic peptides. Nat. Chem. Biol. 2013, 9, 749–753. 10.1038/nchembio.1383. [DOI] [PubMed] [Google Scholar]
- Ponomarenko J.; Bui H.-H.; Li W.; Fusseder N.; Bourne P. E.; Sette A.; Peters B. ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinf. 2008, 9, 514 10.1186/1471-2105-9-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein N. D.; Mayrose I.; Martz E.; Pupko T. Epitopia: a web-server for predicting B-cell epitopes. BMC Bioinf. 2009, 10, 287 10.1186/1471-2105-10-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haste Andersen P.; Nielsen M.; Lund O. Prediction of residues in discontinuous B-cell epitopes using protein 3D structures. Protein Sci. 2006, 15, 2558–2567. 10.1110/ps.062405906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borley D. W.; Mahapatra M.; Paton D. J.; Esnouf R. M.; Stuart D. I.; Fry E. E. Evaluation and use of in-silico structure-based epitope prediction with foot-and-mouth disease virus. PLoS One 2013, 8, e61122 10.1371/journal.pone.0061122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey D. L.; Brown W. C.; Dutta S.; Konwerski J.; Jose J.; Jurkiw T. J.; DelProposto J.; Ogata C. M.; Skiniotis G.; Kuhn R. J.; Smith J. L. Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system. Science 2014, 343, 881–885. 10.1126/science.1247749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abraham M. J.; Murtola T.; Schulz R.; Páll S.; Smith J. C.; Hess B.; Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- Stadler V.; Felgenhauer T.; Beyer M.; Fernandez S.; Leibe K.; Guttler S.; Groning M.; Konig K.; Torralba G.; Hausmann M.; Lindenstruth V.; Nesterov A.; Block I.; Pipkorn R.; Poustka A.; Bischoff F. R.; Breitling F. Combinatorial synthesis of peptide arrays with a laser printer. Angew. Chem., Int. Ed. Engl. 2008, 47, 7132–7135. 10.1002/anie.200801616. [DOI] [PubMed] [Google Scholar]
- Jaenisch T.; Sakuntabhai A.; Wilder-Smith A. DengueTools, Dengue research funded by the European Commission-scientific strategies of three European dengue research consortia. PLoS Neglected Trop. Dis. 2013, 7, e2320 10.1371/journal.pntd.0002320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch T.; Tam D. T.; Kieu N. T.; Van Ngoc T.; Nam N. T.; Van Kinh N.; Yacoub S.; Chanpheaktra N.; Kumar V.; See L. L.; Sathar J.; Sandoval E. P.; Alfaro G. M.; Laksono I. S.; Mahendradhata Y.; Sarker M.; Ahmed F.; Caprara A.; Benevides B. S.; Marques E. T.; Magalhaes T.; Brasil P.; Netto M.; Tami A.; Bethencourt S. E.; Guzman M.; Simmons C.; Quyen N. T.; Merson L.; Dung N. T.; Beck D.; Wirths M.; Wolbers M.; Lam P. K.; Rosenberger K.; Wills B. Clinical evaluation of dengue and identification of risk factors for severe disease: protocol for a multicentre study in 8 countries. BMC Infect. Dis. 2016, 16, 120. 10.1186/s12879-016-1440-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti R. S.; Kosoy O. L.; Laven J. J.; Panella A. J.; Velez J. O.; Lambert A. J.; Campbell G. L. Chikungunya virus in US travelers returning from India, 2006. Emerging Infect. Dis. 2007, 13, 764–767. 10.3201/eid1305.070015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanciotti R. S.; Kosoy O. L.; Laven J. J.; Velez J. O.; Lambert A. J.; Johnson A. J.; Stanfield S. M.; Duffy M. R. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerging Infect. Dis. 2008, 14, 1232–1239. 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordeiro M. T.; Brito C. A.; Pena L. J.; Castanha P. M.; Gil L. H.; Lopes K. G.; Dhalia R.; Meneses J. A.; Ishigami A. C.; Mello L. M.; Alencar L. X.; Guarines K. M.; Rodrigues L. C.; Marques E. T. Results of a Zika Virus (ZIKV) Immunoglobulin M-Specific Diagnostic Assay Are Highly Correlated With Detection of Neutralizing Anti-ZIKV Antibodies in Neonates With Congenital Disease. J. Infect. Dis. 2016, 214, 1897–1904. 10.1093/infdis/jiw477. [DOI] [PubMed] [Google Scholar]
- Altschul S. F.; Madden T. L.; Schäffer A. A.; Zhang J.; Zhang Z.; Miller W.; Lipman D. J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiser A.; Do R. K.; Sali A. Modeling of loops in protein structures. Protein Sci. 2000, 9, 1753–1773. 10.1110/ps.9.9.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiser A.; Sali A. ModLoop: automated modeling of loops in protein structures. Bioinformatics 2003, 19, 2500–2501. 10.1093/bioinformatics/btg362. [DOI] [PubMed] [Google Scholar]
- Lütteke T.; Bohne-Lang A.; Loss A.; Goetz T.; Frank M.; von der Lieth C.-W. GLYCOSCIENCES. de: an Internet portal to support glycomics and glycobiology research. Glycobiology 2006, 16, 71R–81R. 10.1093/glycob/cwj049. [DOI] [PubMed] [Google Scholar]
- Pryor M. J.; Wright P. J. Glycosylation mutants of dengue virus NS1 protein. J. Gen. Virol. 1994, 75, 1183–1187. 10.1099/0022-1317-75-5-1183. [DOI] [PubMed] [Google Scholar]
- Baker N. A.; Sept D.; Joseph S.; Holst M. J.; McCammon J. A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 2001, 98, 10037–10041. 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid N.; Eichenberger A. P.; Choutko A.; Riniker S.; Winger M.; Mark A. E.; van Gunsteren W. F. Definition and testing of the GROMOS force-field versions 54A7 and 54B7. Eur. Biophys. J. 2011, 40, 843–856. 10.1007/s00249-011-0700-9. [DOI] [PubMed] [Google Scholar]
- Pol-Fachin L.; Rusu V. H.; Verli H.; Lins R. D. GROMOS 53A6GLYC, an Improved GROMOS Force Field for Hexopyranose-Based Carbohydrates. J. Chem. Theory Comput. 2012, 8, 4681–4690. 10.1021/ct300479h. [DOI] [PubMed] [Google Scholar]
- Pol-Fachin L.; Verli H.; Lins R. D. Extension and validation of the GROMOS 53A6(GLYC) parameter set for glycoproteins. J. Comput. Chem. 2014, 35, 2087–2095. 10.1002/jcc.23721. [DOI] [PubMed] [Google Scholar]
- Berendsen H.; Grigera J.; Straatsma T. The missing term in effective pair potentials. J. Phys. Chem. 1987, 91, 6269–6271. 10.1021/j100308a038. [DOI] [Google Scholar]
- Hess B.; Bekker H.; Berendsen H. J.; Fraaije J. G. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997, 18, 1463–1472. . [DOI] [Google Scholar]
- Tironi I. G.; Sperb R.; Smith P. E.; van Gunsteren W. F. A generalized reaction field method for molecular dynamics simulations. J. Chem. Phys. 1995, 102, 5451–5459. 10.1063/1.469273. [DOI] [Google Scholar]
- Bussi G.; Donadio D.; Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Parrinello M.; Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. 10.1063/1.328693. [DOI] [Google Scholar]
- Nosé S.; Klein M. Constant pressure molecular dynamics for molecular systems. Mol. Phys. 1983, 50, 1055–1076. 10.1080/00268978300102851. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.