Abstract
α-Conotoxins preferentially antagonize muscle and neuronal nicotinic acetylcholine receptors (nAChRs). Native α-conotoxins have two disulfide links, CI–CIII and CII–CIV, and owing to the inherent properties of disulfide bonds, α-conotoxins have been systematically engineered to improve their chemical and biological properties. In this study, we explored the possibility of simplifying the disulfide framework of α-conotoxins Vc1.1, BuIA, ImI, and AuIB, by introducing [C2H,C8F] modification to the CI–CIII bond. We therefore explored the possibility of using hydrophobic packing of standard amino acid side chains to replace disulfide bonds as an alternative strategy to nonnatural amino acid cross-links. The impact of CI–CIII disulfide bond replacement on the conformation of the α-conotoxins was investigated using molecular dynamics (MD) simulations and nuclear magnetic resonance chemical shift index study. Two-electrode voltage clamp techniques and MD simulations were used to study the impact of disulfide bond deletion on the activities of the peptides at human neuronal nAChRs. All disulfide-deleted variants except ImI[C2H,C8F] had reduced potency for inhibiting nAChRs. Our results suggest that the CI–CIII disulfide bond is important to stabilize the secondary structure of α-conotoxins as well as their interaction with neuronal nAChR targets. Results from this study enrich our understanding of the function of the CI–CIII disulfide bond and are useful in guiding future structural engineering of the α-conotoxins.
Introduction
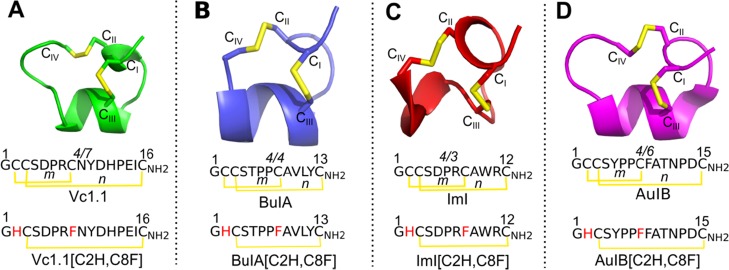
α-Conotoxins from the marine snail genus Conus are inhibitors of muscle and neuronal nicotinic acetylcholine receptors (nAChRs).1−3 The majority of native α-conotoxins have two internal disulfide cross-links between cysteines CI and CIII and CII and CIV. The number of residues between CII and CIII (m) and CIII and CIV (n) define different types of α-conotoxins, which are noted as m/n (Figure 1).1−3
Figure 1.
NMR solution structures and sequences of α-conotoxins Vc1.1 (green), BuIA (blue), ImI (red), and AuIB (magenta) and the sequences of their disulfide-deleted analogues. (A) Vc1.1, (B) BuIA, (C) ImI, and (D) AuIB are small disulfide-rich (yellow) peptides belonging to the 4/7, 4/4, 4/3, and 4/6 α-conotoxin subfamilies, respectively. The CI–CIII disulfide bond of the α-conotoxins is deleted by replacing the Cys residues with His at position 2 and Phe at position 8. *m and n indicate the number of residues between CII–CIII and CIII–CIV disulfide bonds, respectively.
Three α-conotoxins, Vc1.1, RgIA, and AuIB, have been shown to have a potent, long-lasting analgesic effect in rat models of chronic neuropathic and visceral pain.4−8 Although these α-conotoxins preferentially antagonize neuronal nAChRs, with RgIA and Vc1.1 targeting the α9α10 nAChR subtype4 and AuIB inhibiting the α3β4 nAChR subtype,9 the involvement of these nAChRs in pain transmission pathways remains unclear.10 These three conotoxins also potently inhibit high-voltage-activated (HVA; N- and R-type) calcium channels via the G-protein-coupled γ-aminobutyric acid B receptor (GABABR) in rat dorsal root ganglion neurons, providing another explanation to their analgesic properties.5,6 To date, Vc1.1 and AuIB have been reported to target the GABABR, whereas the activity of BuIA and ImI at GABABR is yet to be investigated. All four α-conotoxins Vc1.1, BuIA, ImI, and AuIB antagonize different nAChR subtypes, and therefore in the present study, the activity of the four conopeptides was evaluated on the neuronal nAChRs.
Despite their pharmacological potential, the application of conotoxins has been hindered by their short biological half-life, intrinsic disulfide bond shuffling, susceptibility to enzymatic degradation, poor absorption, and limited oral availability.11 Consequently, these conotoxins have been subjected to chemical modifications to improve the efficacy of synthesis and biological properties.12
Although the disulfide bridges are pivotal in stabilizing the structure of conotoxins, they are inherently unstable under reducing environments. Disulfide bond shuffling in conotoxins can result in heterogeneous peptide conformations, therefore increasing the production cost and concurrently lowering the peptide synthesis efficiency. Additionally, compounds with alternative disulfide connectivity typically have altered biochemical properties.13,14 One strategy to prevent disulfide bond shuffling of α-conotoxins is to replace one or both CI–CIII and CII–CIV disulfide bonds with nonnatural amino acid cross-links, such as lactam bridges,15 selenocysteines,16 dicarba bridges,17,18 cystathionine bridges,19 or dithiol amino acids.20 Replacement of disulfide bridges of α-conotoxins by lactam or dicarba bridges resulted in altered conformations and between 20- and 60-fold decreased activity or binding at nAChRs.15,17,18 Other strategies relying on selenocysteines, cystathionines, or dithiol amino acids had less impact on the structure, and the resulting compounds had similar or improved activity.16,19,20 Despite these successes, nonnatural amino acids are not optimal for the development of α-conotoxins as drugs because they cannot be produced recombinantly and they increase the cost of synthetic production. We have recently shown that an alternative strategy is to replace disulfide bonds with tight-binding standard amino acids, forming a hydrophobic mini-core.21 We showed that the [C2H,C8F] modification to the CI–CIII bond of the cyclic conotoxin Vc1.1 (cVc1.1) resulted in a nearly identical conformation and a decreased activity of more than twofold at α9α10 nAChR,21 contrasting with the complete loss of activity displayed by the dicarba replacement of the same disulfide bond.18 Therefore, [C2H,C8F] modification to the CI–CIII bond could be an efficient strategy to simplify the structure of the cyclic conotoxins. Then, a question arises regarding the feasibility of the application of [C2H,C8F] modification to the CI–CIII bond of the native α-conotoxins.
In this study, we investigated (i) the structure of 4/7(m/n)-Vc1.1, 4/4-BuIA, 4/3-ImI, and 4/6-AuIB disulfide-deleted [C2H,C8F] analogues, in which cysteine residues at positions 2 and 8 of the peptides are substituted with histidine and phenylalanine residues, respectively (Figure 1), using molecular dynamics (MD) simulation; (ii) the potency of Vc1.1[C2H,C8F] analogue at human (h)α9α10 and hα3β2 nAChRs, BuIA[C2H,C8F] at hα3β2 and hα3β4 nAChRs, ImI[C2H,C8F] at hα7 and hα9α10 nAChRs, and AuIB[C2H,C8F] at hα3β4 nAChRs expressed in Xenopus laevis oocytes; and (iii) the molecular docking models of the above [C2H,C8F] conotoxin analogues at their respective human nAChRs.
Results and Discussion
Structural Impact of the Replacement of Disulfide Bond CI–CIII
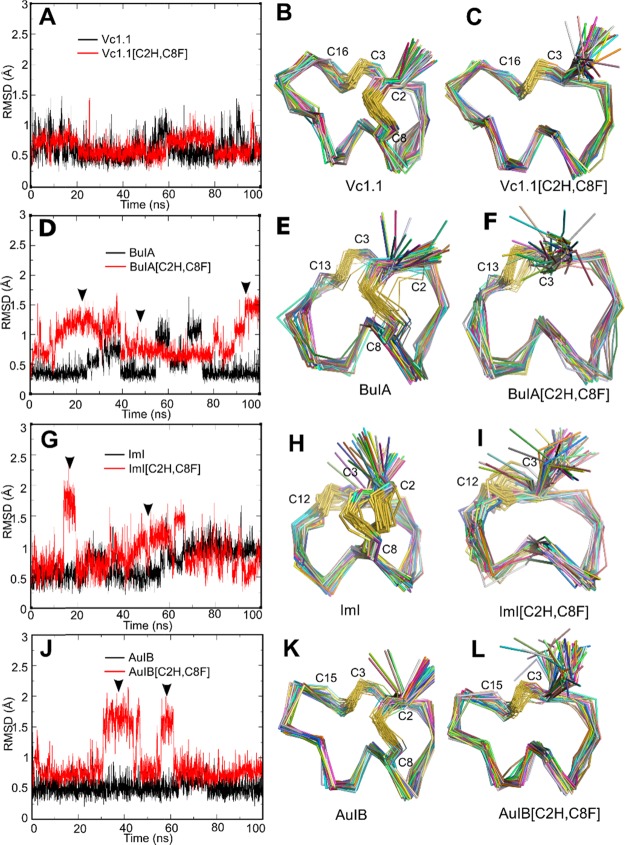
The backbone conformation of the [C2H,C8F] conotoxin variants remained similar to that of their parent peptide during MD simulation, but some transient instabilities were observed. The backbone conformation of both Vc1.1[C2H,C8F] and Vc1.1 was very stable during MD simulations, with the backbone root-mean-square deviation (rmsd) values ranging from 0.5 to 1 Å (Figure 2A), suggesting that the mutant and wild-type peptides might have similar backbone conformation (Figure 2B,C). The backbone conformation of AuIB appeared to be also stable, with rmsd values ranging between 0.25 and 0.75 Å (Figure 2J). The substitution of both AuIB C2 and C8 resulted in a deviation of less than 1 Å for ∼70% of the simulation, with short-lived metastable conformations characterized by 1.7 Å rmsd from the parent peptide appearing twice during the 100 ns simulation. This suggests a probably more critical role of the CI–CIII disulfide bond in stabilizing the structure of AuIB than that of Vc1.1.
Figure 2.
Structural comparison of the wild-type α-conotoxins Vc1.1, BuIA, ImI, and AuIB and their disulfide-deleted analogues from 100 ns MD simulations. (A,D,G,J) rmsd for backbone Cα of the wild-type (black line) and mutant (red line) conotoxins. (B,C,E,F,H,I,K,L) Backbone conformation of the peptides extracted from the last 50 ns MD trajectories with equal time intervals. ▼ indicates the time phase when rmsd significantly fluctuated. Values of the Hα chemical shift deviation from the wild-type peptide are used to evaluate the mutational effect on the peptide secondary structure.
Short-lived metastable conformations were also observed in the simulations of BuIA, with 80% of explored conformations being less than 1 Å rmsd from the nuclear magnetic resonance (NMR) solution structure, and two short-lived periods where the conformation was ∼1.2 Å rmsd (Figure 2D). The simulation of BuIA suggests that the fold of some α-conotoxins might be unstable in water, which is not completely surprising because most peptides of similar size are disordered in solution. The [C2H,C8F] substitution destabilized the fold of BuIA in the first 40 ns, as evidenced by the unstable rmsd values. These values were nevertheless below 1.5 Å and then stabilized at around 0.75 Å for nearly 50 ns, suggesting that the conformation at that time could be as stable as the conformation of BuIA. The range of conformations explored during the simulation remained similar to those of the parent peptide (Figure 2E,F).
The MD simulation of ImI was not stable, with two apparent metastable conformations characterized by rmsd values of 0.5 and 1.0 Å (Figure 2G). Similar to the other three peptides, the replacement of the first cysteine resulted in some destabilization, which can be seen in the greater fluctuation of the rmsd of the variant compared to the parent peptide. The panel of conformations adopted by ImI[C2H,C8F] was <1 Å rmsd for 70% of the simulation, and the range of conformations was globally similar to those observed during ImI MD simulation (Figure 2H,I).
MD simulations of the wild-type and variant conotoxins indicated that the removal of the disulfide bond formed between Cys2 and Cys8 might result in no conformational change for Vc1.1 and ImI, whereas modest impact could be observed for AuIB and BuIA, with 0.2 and 0.3 Å rmsd change, respectively.
For all peptides apart from Vc1.1, the removal of the disulfide conformation was inferred to result in destabilization of the native fold, with more frequent exploration of alternative conformations. The smallest peptide without a disulfide bond displaying a defined fold in solution is Trp cage, which has 20 amino acid residues.22 The α-conotoxins studied here are noticeably smaller (12 and 15 residues), and their conformational stability despite their small size is attributed to their two disulfide bond cross-links. It is therefore remarkable that the present MD simulations suggest that some of these variants with only one disulfide bond could still display a defined fold.
Structural differences between conotoxins are contributed by their individual sequence of amino acids and more importantly their innate disulfide frameworks. In comparison to Vc1.1, AuIB, and BuIA, ImI has the shortest n loop and consequently the shortest helix. This decreased secondary structure content resulted in a lower stability of ImI compared to the other three peptides, as evidenced by the fluctuations of rmsd. The lower stability of ImI can also be attributed to the presence of only one proline residue, whereas Vc1.1, AuIB, and BuIA have 2, 3, and 2 proline residues, respectively. Proline residues have a fixed φ backbone dihedral angle, and they therefore contribute to decreased peptide flexibility. Taken together, ImI has the least rigid peptide backbone compared to Vc1.1, AuIB, and BuIA.
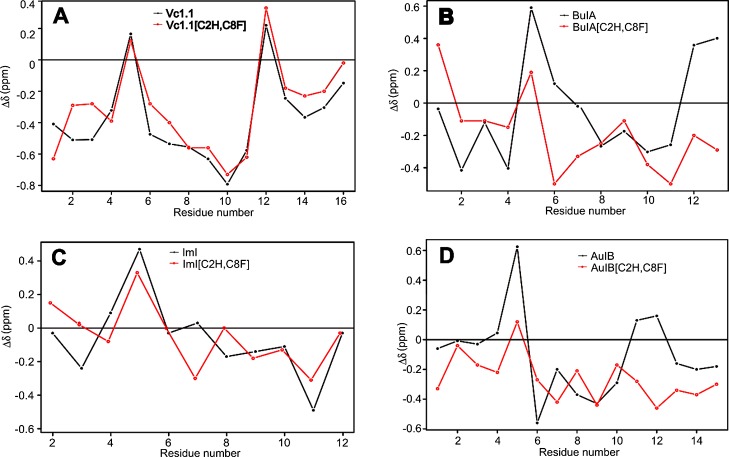
The Hα secondary chemical shifts of Vc1.1 and its disulfide-deleted analogue are nearly identical, with a correlation coefficient R2 of 0.91, demonstrating that the two peptides have similar structures (Figure 3A). NMR spectrometry data thus supported the results from MD simulations regarding the conformational stability and similarity of Vc1.1 and Vc1.1[C2H,C8F]. By contrast, the removal of the disulfide bridge greatly affected the structure of BuIA (R2 = 0.23), with significant Hα secondary chemical shift deviation from the wild-type peptide in the first half portion and also in the C-terminal tail of the peptide (Figure 3B). Interestingly, the secondary shifts indicate an increased α-helical content in the second half of the peptide of disulfide-deleted BuIA. For ImI, minimal Hα secondary chemical shift deviation (R2 = 0.73) was observed between the wild-type and disulfide-deleted peptides (Figure 3C), indicating that their structures are similar, as predicted by the MD simulations. For AuIB, the Hα secondary chemical shifts for positions 6, 7, 8, and 10 deviate between the wild-type and the disulfide-deleted variant by 0.5 ppm, suggesting a small change of conformation (Figure 3D). The Hα secondary chemical shifts from positions 11 to 15 are more negative for the variant, suggesting a helical structure. The deletion of the CI–CIII disulfide bond of BuIA and AuIB resulted in higher rmsd values during MD simulations and supported by NMR spectroscopy analysis, which indicated that their C-terminus has an increased helical content compared to the parent peptides.
Figure 3.
Superimposed Hα secondary chemical shifts of wild-type α-conotoxins Vc1.1, BuIA, ImI, and AuIB (black line) and their disulfide-deleted analogues (red line). The correlation coefficients (R2 score) for (A) Vc1.1 vs Vc1.1[C2H,C8F], (B) BuIA vs BuIA[C2H,C8F], (C) ImI vs ImI[C2H,C8F], and (D) AuIB vs AuIB[C2H,C8F] are 0.91, 0.23, 0.73, and 0.55, respectively.
Only Vc1.1[C2H,C8F] showed an overall Hα secondary chemical shift R2 value of more than 0.9, suggesting that the removal of the Cys residues at positions 2 and 8 essentially maintains the peptide secondary structure. The secondary structures of BuIA, ImI, and AuIB, however, are highly dependent on the cysteine bond between residues Cys2 and Cys8. Previously, we have demonstrated the involvement of introduced residues, His2 in electrostatic interaction and Phe8 in the formation of the peptide hydrophobic core, both important in stabilizing the secondary structure of cVc1.1[C2H,C8F].21 Thus, here we assumed that the varying stability between Vc1.1[C2H,C8F], ImI[C2H,C8F], and AuIB[C2H,C8F] is primarily due to their sequence differences.
The backbone root-mean-square fluctuation (RMSF) of the wild-type peptide and its disulfide-deleted analogues was calculated to evaluate their stability in solution (Figure S1). Removal of the disulfide bond resulted in significant backbone fluctuation for the first four residues of Vc1.1[C2H,C8F], whereas the RMSF for residues between 5 and 16 is similar between Vc1.1 and its disulfide-deleted analogue. Interestingly, the backbone RMSF of BuIA is comparable to that of BuIA[C2H,C8F], despite their large structural conformational difference. In contrast to BuIA, the RMSF for AuIB[C2H,C8F] is substantially larger than that of the wild-type. The backbone RMSF for both the N- and C-termini of ImI[C2H,C8F] is larger than that of the wild-type for positions 1–3 and 10–12.
Impaired Inhibitory Effect of Disulfide-Deleted α-Conotoxin Analogues at Human nAChRs
Removal of the CI–CIII disulfide bond of Vc1.1, BuIA, and AuIB (10 μM) substantially reduced the ability of the peptides to inhibit acetylcholine (ACh)-evoked currents of their respective human nAChR targets expressed in X. laevis oocytes (see Figure 4).
Figure 4.
Activity of Vc1.1, BuIA, ImI, and AuIB and their disulfide-deleted analogues (10 μM) at human nAChR subtypes. Superimposed representative ACh-evoked currents recorded from X. laevis oocytes expressing hα9α10 (A,E), hα3β2 (B,C), hα3β4 (D,G), and hα7 (F) nAChRs in the absence (black trace) and presence (red trace) of 10 μM Vc1.1 and Vc1.1[C2H,C8F] (A,B), BuIA and BuIA[C2H,C8F] (C,D), ImI and ImI[C2H,C8F] (E,F), and AuIB and AuIB[C2H,C8F] (G).
Despite the high degree of secondary structural similarity between Vc1.1 and Vc1.1[C2H,C8F], inhibition of hα9α10 nAChR by the [C2H,C8F] mutant was detected only at 30 μM, with negligible activity at 1 and 10 μM (Figure 5A). Similarly, there was minimal inhibition of hα3β2 nAChR by 1 and 10 μM Vc1.1[C2H,C8F], and substantial inhibition was only observed at 30 μM (Figure 5B). By contrast, wild-type Vc1.1 is a potent inhibitor of hα9α10 and hα3β2 nAChRs, with complete inhibition observed at 10 μM. Previously engineered [C2H,C8F] mutant of cVc1.1 was reported to retain the structural integrity of the wild-type counterpart but with only twofold loss of potency at inhibiting hα9α10 nAChR.21 In this study, we show dramatic loss of Vc1.1[C2H,C8F] activity at hα9α10 nAChR, although it is structurally similar to Vc1.1, suggesting that the CI–CIII disulfide bond may be directly involved in the interaction between the peptide and the nAChR. Arguably, the difference in potency between the cyclized and linear [C2H,C8F] mutants could be due to the presence of the short linker, joining the N- and C-termini of the cyclized peptide. The linker was originally introduced as a scaffold to stabilize the three-dimensional structure of cVc1.1, and it may have inadvertently compensated the loss of cVc1.1[C2H,C8F] potency.
Figure 5.
Inhibition of human nAChR subtypes by Vc1.1, BuIA, ImI, and AuIB and their disulfide-deleted analogues. The bar graph of the relative ACh-evoked current amplitude mediated by hα9α10 (A,E), hα3β2 (B,C), hα3β4 (D,G), and hα7 (F) in the presence of 1, 10, and 30 μM Vc1.1 and Vc1.1[C2H,C8F] (A,B), BuIA and BuIA[C2H,C8F] (C,D), ImI and ImI[C2H,C8F] (E,F), and AuIB and AuIB[C2H,C8F] (G). Whole-cell currents mediated by hα9α10 and hα3β2 nAChRs were activated by 6 μM ACh, whereas those mediated by hα7 and hα3β4 were activated by 100 and 300 μM ACh, respectively. Data expressed as mean ± SEM, n = 5 to 16 (unpaired Student’s t-test; *p < 0.05, **p < 0.0001 vs the relative current amplitude of 1.0).
Disulfide deletion of BuIA similarly resulted in a significant loss of potency at hα3β2 and hα3β4 nAChRs (Figure 5C,D). For both human nAChR subtypes, 1 μM BuIA inhibited >90% of the ACh-evoked current amplitude compared to ∼70% at 30 μM BuIA[C2H,C8F]. AuIB[C2H,C8F] exhibited no activity at hα3β4 nAChR, with no inhibition of the ACh-evoked current amplitude in the presence of 30 μM AuIB[C2H,C8F] (Figure 5G).
The potency of ImI[C2H,C8F] at inhibiting ACh-evoked currents mediated by hα9α10 and hα7 nAChRs was also reduced, albeit to a lesser extent compared to the [C2H,C8F] mutants of Vc1.1, BuIA, and AuIB. Compared to ImI, ImI[C2H,C8F] remains largely active at inhibiting both human nAChR subtypes (Figure 5E,F), which is consistent with the observed subtle secondary structural change between the two peptides. Although the conformational shift of ImI[C2H,C8F] is larger (R2 = 0.73) than that of Vc1.1[C2H,C8F] (R2 = 0.91), the mutant ImI peptide retains comparable potency to ImI at 10 and 30 μM peptide concentrations tested, in inhibiting hα9α10 and hα7 nAChRs. By contrast, only 30 μM Vc1.1[C2H,C8F] had discernible inhibitory activity at both nAChRs. The concentration-dependent activity of ImI and ImI[C2H,C8F] at the hα7 nAChR subtype was determined, giving a half-maximal inhibitory concentration (IC50) of 497 ± 32 nM (as reported previously)23 and 9.59 ± 0.62 μM, respectively (Figure S2).
Despite α-conotoxin disulfide-deleted analogues of Vc1.1, ImI, and AuIB being structurally similar to their wild-type counterparts, functional screening of their potency at human nAChRs revealed contradicting results. AuIB[C2H,C8F] did not inhibit hα3β4 nAChR (up to 30 μM), whereas [C2H,C8F] analogues of Vc1.1 and BuIA have significantly reduced potency at human nAChR subtypes. By contrast, relatively minimal impact on the activity of ImI[C2H,C8F] at hα9α10 and hα7 nAChRs was observed.
In another study, oxidation of the first disulfide bond of ImI was identified as more important for binding than that of the second disulfide bond.24 A more recent study reported that downsized α-conotoxins having only the first loop cyclized by the CI–CIII disulfide bond were still active at nAChRs. By contrast, cyclization of the second loop using the CII–CIV disulfide bond resulted in inactive conotoxins.25 More generally, most attempts at replacing only the first disulfide bond using nonnatural cross-linking residues resulted in a significant decrease in activity.15,17,18 An alternative strategy for simplifying the structure of α-conotoxins with minimal impact on activity could therefore be the substitution of the second disulfide bond, which could be achieved using the methodology presented here.
Binding Mode of α-Conotoxins and Their [C2H,C8F] Mutants at Human nAChR Principal Subunit C-Loop
MD simulations on models of the α-conotoxin–nAChR complex were performed to gain a better understanding on the interaction between the peptides and their target human nAChRs. The binding pocket for α-conotoxins at nAChRs is formed at the extracellular interface between adjacent principal (+) α subunit and complementary (−) α/β subunit. Specifically, (+) subunit loops A–C and loops D–F of the (−) subunit contribute to α-conotoxin recognition by nAChRs.
Our generated docking models revealed that for all four wild-type α-conotoxins (Figure 6A,D,G,J), the CI–CIII disulfide bond is buried in the binding site and formed direct contact with the disulfide bond formed between residues Cys192 and Cys193 of the (+) subunit C-loop. By contrast, less interaction was observed between the [C2H,C8F] mutants and the nAChRs, resulting in larger movement of the C-loop (Figure 6C,F,I,L). The opening of the hα9α10 nAChR C-loop bound with Vc1.1[C2H,C8F] and the opening of the hα3β4 nAChR C-loop bound with AuIB[C2H,C8F] were significantly right shifted to a comparable extent of ∼3 Å. The C-loop opening of the AuIB[C2H,C8F]-bound hα3β4 nAChR was the most right shifted, resulting in increased solvent exposure of the peptide and consequently substantial loss of potency at inhibiting the nAChR. By contrast, the opening of the hα3β2 nAChR C-loop-bound BuIA[C2H,C8F] and the opening of the α7 nAChR C-loop-bound ImI[C2H,C8F] were only slightly right shifted.
Figure 6.
Molecular docking of wild-type α-conotoxins Vc1.1, BuIA, ImI, and AuIB and their disulfide-deleted analogues (orange)at homology models of human nAChRs (the principal (+) subunit in green; the complementary (−) subunit in cyan). (A,B) Vc1.1 and Vc1.1[C2H,C8F] bound to the α10(+)α9(−) binding site of hα9α10 nAChR. (D,E) BuIA and BuIA[C2H,C8F] bound to the α3(+)β2(−) binding site of hα3β2 nAChR. (G,H) ImI and ImI[C2H,C8F] bound to the α10(+)α9(−) binding site of hα7 nAChR. (J,K) AuIB and AuIB[C2H,C8F] bound to the α3(+)β4(−) binding site of hα3β4 nAChR. The dashed lines show the H bond formed between pairwise interacting residues of the conotoxins and nAChRs. (C,F,I,L) Opening probability distribution for the C-loop of the human nAChRs for the wild-type α-conotoxin (blue) and the mutants (orange). The arrow indicates right shift movement of the nAChR C-loop.
The weaker binding of Vc1.1[C2H,C8F] and AuIB[C2H,C8F] to their nAChR targets mainly resulted from the solvent-semiexposed side chain of the introduced His2 residue and the bulky benzyl side chain of the Phe8 residue. As a result, Vc1.1[C2H,C8F] and AuIB[C2H,C8F] have fewer contacts with the tip of the C-loop, which could be a contributing factor to their reduced potency at inhibiting their target nAChRs despite their minor secondary structure perturbation. The loss of Vc1.1[C2H,C8F] potency can also be attributed to the absence of hydrogen bond between peptide His12 and hα9α10 nAChR Glu195 residues because of the increased opening of the C-loop. In addition, the loss of AuIB[C2H,C8F] potency can also be attributed to the dramatic increase in backbone fluctuation, decrease in backbone stability as well as conformational change in the bound state.
On the other hand, the minor conformational change for the C-loop of hα3β2 nAChR/BuIA[C2H,C8F] and the C-loop of hα7 nAChR/ImI[C2H,C8F] might explain their relatively smaller decrease in activity than that of Vc1.1[C2H,C8F] and AuIB[C2H,C8F]. Loss of BuIA[C2H,C8F] activity at human nAChRs can be attributed to the large local conformational perturbation from BuIA, as evident from MD simulations and Hα chemical shift analysis. Although the secondary structure of BuIA[C2H,C8F] was most affected, the peptide still retained some activity at inhibiting hα3β2 and hα3β4 nAChRs. The relatively smaller loss of activity for BuIA[C2H,C8F] might originate from the smaller conformational perturbation to the C-loop and the comparable backbone RMSF to the wild-type. In contrast to BuIA[C2H,C8F], the secondary structure of ImI[C2H,C8F] is similar to that of ImI in both unbound and bound states; therefore, the CI–CIII disulfide bond can be postulated to have a minor influence on the structure and inhibitory activity of ImI at hα9α10 and hα7 nAChRs.
Additionally, the bound-state rmsd of Vc1.1, BuIA, ImI, and AuIB and their disulfide-deleted analogues was calculated to qualitatively evaluate the influence of the conformational change to their nAChR binding (Figure S3). In the bound state, the backbone rmsd for Vc1.1[C2H,C8F] and ImI[C2H,C8F] is similar to that of the wild-type, whereas the rmsd for BuIA[C2H,C8F] and AuIB[C2H,C8F] is substantially larger compared to that of the wild-type. Overall, these results are consistent with MD simulations of the unbound state.
In summary, our results demonstrate that the CI–CIII bond of the α-conotoxins regardless of their cysteine framework classification has a critical role of not only in stabilizing their secondary structure and maintaining their stability but also in their binding to multiple human nAChRs. Most importantly, we provide evidence to support the fact that this disulfide bond is not just a redundant feature but has been selectively conserved within the α-conotoxins as a structural scaffold for them to be biologically active.
Methods
Homology Modeling
Models of Vc1.1/Vc1.1[C2H,C8F]-bound human α9α10 nAChRs, ImI/ImI[C2H,C8F]-bound human α9α10 and α7 nAChRs, BuIA/BuIA[C2H,C8F]-bound human α3β4 nAChRs and α3β2 nAChRs, and AuIB/AuIB[C2H,C8F]-bound human α3β4 nAChRs were built using Modeller (version 9v12), as described previously.26 The sequences of human α1, α3, α4, α6, α7, α9, α10, β2, β3, and β4 nAChR subunits were retrieved from the UniProt database.27 The crystal structures of Aplysia californica AChBP (ACh binding protein) in complex with α-conotoxin PnIA[A10L,D14K] (PDB code 2BR8)28 and the extracellular domains of mouse α1 (PDB code 2QC1)29 and human α9 (PDB code 4D01)30 nAChR subunits were used as templates to build 200 models of each α-conotoxin/nAChR complexes. Models with the lowest discrete optimized protein energy score31 were selected for further structural refinement using MD simulations.
MD Simulations of α-Conotoxins
The designs of α-conotoxins Vc1.1, BuIA, ImI, and AuIB tested in this study are summarized in Figure 1. Their structures were modeled by substituting the corresponding residues in the structure of α-conotoxins (Vc1.1 PDB code 2H8S,32 BuIA PDB code 2I28,33 ImI PDB code 2C9T,34 and AuIB PDB code 1MXN(35)) using Modeller (version 9v12).36,37
The protonation states of α-conotoxin His, Asp, and Glu residues were predicted using the PropKa 3.1 method.38 The models and the first NMR structure were minimized and refined using MD simulations performed with the Amber 14 package and ff14SB force field.39,40 The peptides were solvated in a truncated octahedral TIP3P water box containing ∼3000 water molecules. Sodium ions were added to neutralize the systems. The systems were first minimized with 3000 steps of steepest descent and then 3000 steps of conjugate gradient with the solute restrained to their position by a harmonic force of 100 kcal/mol·Å2. A second minimization was then performed but with all position restraints withdrawn. The systems were then gradually heated up from 50 to 300 K in the NVT ensemble over 100 ps with the solute restrained to their position using a 5 kcal/mol·Å2 harmonic force potential. The MD simulations were then carried out in the NPT ensemble, and the position restraints were gradually removed over 100 ps. The production runs were conducted over 100 ns simulation time with pressure coupling set at 1 atm and a constant temperature of 300 K. The MD simulations used a time step of 2 fs, and all bonds involving hydrogen atoms were maintained to their standard length using the SHAKE algorithm.41 The particle mesh Ewald method was used to model long-range electrostatic interactions.42 MD trajectories were analyzed using visual molecular dynamics,43 and molecules were drawn using PyMol (Schrödinger, LLC). Procedures and parameters set up are the same for the MD simulation of the α-conotoxin-bound nAChRs.
α-Conotoxin Synthesis
All α-conotoxins used in this study were assembled on the Rink amide methylbenzhydrylamine resin using solid-phase peptide synthesis with a neutralization/2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate activation procedure for Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry as described previously.44 Cleavage was achieved by treatment with a mixture of trifluoroacetic acid, phenol, water, and triisopropylsilane as scavengers in the ratio of 88:5:5:2, at room temperature (20–25 °C) for 2 h. Trifluoroacetic acid was evaporated at low pressure in a rotary evaporator. Peptides were precipitated with ice-cold ether, filtered, dissolved in 50% buffer A/B (buffer A consists of 99.95% H2O/0.05% trifluoroacetic acid and buffer B consists of 90% CH3CN/10% H2O/0.045% trifluoroacetic acid), and lyophilized. Crude peptides were purified by reversed-phase high-performance liquid chromatography (RP-HPLC) on a Phenomenex C18 column using a gradient of 0–100% methanol for 80 min, with the eluent monitored at 214/280 nm. Electrospray mass spectrometry confirmed the molecular mass of the peptides before they were pooled and lyophilized for oxidation. The cysteines in the peptides were oxidized in 0.1 M NH4HCO3 (pH 8–8.5) at a concentration of 1 mg/mL, and the mixture was stirred at room temperature for 48 h. Owing to its poor solubility in water, the reduced ImI[C2H,C8F] peptide was dissolved in 20% CH3CN/80% H2O prior to adding NH4HCO3. The oxidized peptides were then purified by RP-HPLC using a gradient of 0–40% buffer B over 40 min. Analytical RP-HPLC and electrospray mass spectrometry were used to confirm the purity and molecular mass of the synthesized peptides.
NMR Study of α-Conotoxin Mutants
NMR spectra of the mutants were recorded on a Bruker AVANCE 500 MHz spectrometer. Samples were dissolved in 90% H2O/10% D2O at pH 4.25 at a concentration of 2 mg/mL. Total correction spectroscopy (TOCSY) and nuclear Overhauser enhancement spectroscopy (NOESY) data were collected at 290 K. The Hα chemical shifts were assigned by analyzing the TOCSY and NOESY spectra using CcpNmr software.45
In Vitro cRNA Synthesis
Plasmid pMXT construct of the human α7 nAChR subunit was linearized with BamHI, and plasmid pT7TS constructs of human α3, α9, α10, β2, and β4 nAChR subunits were linearized with XbaI restriction enzymes (NEB, Ipswich, MA) for in vitro cRNA transcription using SP6 (hα7) and T7 (hα3, α9, α10, β2, and β4) mMessage mMachine transcription kits (AMBION, Foster City, CA).
Oocyte Preparation and Microinjection
All procedures were approved by the University of Sydney Animal Ethics Committee. Stage V–VI oocytes were obtained from X. laevis, defolliculated with 1.5 mg/mL collagenase type II (Worthington Biochemical Corp., Lakewood, NJ) at room temperature for 1–2 h in OR-2 solution containing (in mM) 82.5 NaCl, 2 KCl, 1 MgCl2, and 5 HEPES at pH 7.4. The oocytes were injected with 5 ng cRNA for hα3β2, hα3β4, and hα7 nAChRs or 35 ng cRNA for hα9α10 nAChR (concentration confirmed spectrophotometrically and by gel electrophoresis) using glass pipettes pulled from glass capillaries (3-000-203 GX, Drummond Scientific Co., Broomall, PA). The oocytes were incubated at 18 °C in sterile ND96 solution composed of (in mM) 96 NaCl, 2 KCl, 1 CaCl2, 1 MgCl2, and 5 HEPES at pH 7.4, supplemented with 5% FBS, 50 mg/L gentamicin (GIBCO, Grand Island, NY), and 10 000 U/mL penicillin–streptomycin (GIBCO, Grand Island, NY).
Oocyte Two-Electrode Voltage Clamp Recording and Data Analysis
Electrophysiological recordings were carried out 2–5 days post-cRNA microinjection. Two-electrode voltage clamp recordings of X. laevis oocytes expressing human nAChRs were performed at room temperature (21–24 °C) using a GeneClamp 500B amplifier and pCLAMP9 software interface (Molecular Devices, Sunnyvale, CA) at a holding potential of −80 mV. Voltage-recording and current-injecting electrodes were pulled from GC150T-7.5 borosilicate glass (Harvard Apparatus, Holliston, MA) and filled with 3 M KCl, giving resistances of 0.3–1 MΩ.
The oocytes were perfused with ND96 solution using a continuous Legato 270 push/pull syringe pump perfusion system (KD Scientific, Holliston, MA) at a rate of 2 mL/min. For oocytes expressing hα9α10 nAChRs, 50 nL of 50 mM BAPTA was injected using a glass pipette 1 h before recording and perfused with ND115 solution containing (in mM) 115 NaCl, 2.5 KCl, 1.8 CaCl2, and 10 HEPES at pH 7.4. Owing to the Ca2+ permeability of hα9α10 nAChRs, BAPTA injection was carried out to prevent the activation of endogenous calcium-activated chloride channels in X. laevis oocytes.
Initially, oocytes were briefly washed with bath solution (ND96 or ND115) followed by three applications of ACh at half-maximal excitatory concentration (EC50) of 6 μM for hα9α10 and hα3β2, 100 μM for hα7, and 300 μM for hα3β4 nAChRs. The oocytes were washed with bath solution for 3 min between ACh applications. The oocytes were incubated with peptides for 5 min with the perfusion system turned off, followed by coapplication of ACh and peptide with flowing bath solution. All peptide solutions were prepared in ND96/ND115 + 0.1% bovine serum albumin. Peak current amplitudes before (ACh alone) and after (ACh + peptide) peptide incubation were measured using Clampfit software (Molecular Devices, Sunnyvale, CA, USA) where the ratio of ACh + peptide-evoked current amplitude to ACh alone-evoked current amplitude was used to assess the activity of the peptides at human nAChRs. All electrophysiological data were pooled (n = 5 to 16) and represent mean ± standard error of the mean (SEM). Data sets were compared using an unpaired Student’s t-test. Differences were regarded statistically significant when p < 0.05. The IC50 was determined from the concentration–response curve fitted to a nonlinear regression function and reported with error of the fit. Data analysis was performed using GraphPad Prism 5 (GraphPad Software, La Jolla, CA).
Acknowledgments
This work was supported by the grant from the National Natural Science Foundation of China (NSFC) (no. 81502977 for R.Y.), the Special Foundation for Qingdao Basic Research Program (no. 15-9-1-85-jch for R.Y.), and the Australian Research Council (Discovery Project Grant DP150103990 for D.J.A. and Q.K.).
Glossary
Abbreviations
- AChBP
acetylcholine binding protein
- DRG
dorsal root ganglion
- GABABR
γ-aminobutyric acid B receptor
- HBTU
neutralization/2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate
- MBHA
rink amide methylbenzhydrylamine
- MD
molecular dynamics
- nAChR
nicotinic acetylcholine receptor
- RP-HPLC
reversed-phase HPLC
- AuIB
α-conotoxin AuIB
- BuIA
α-conotoxin BuIA
- ImI
α-conotoxin ImI
- Vc1.1
α-conotoxin Vc1.1
- IC50
half-maximal inhibitory concentration
- RMSF
root mean square fluctuation
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.7b00639.
Stability of the wild-type α-conotoxins Vc1.1, BuIA, ImI, and AuIB and their disulfide-deleted analogues; concentration–response relationships for the inhibition of human α7 nAChR subtype by ImI and ImI[C2H,C8F]; and comparison of the backbone rmsd of the wild-type α-conotoxins Vc1.1, BuIA, ImI, and AuIB (black line) and their disulfide-deleted analogues (red line) in the nAChR-bound state (PDF)
Author Contributions
N.T. and H.-S.T. contributed equally to this work. N.T. performed peptide synthesis and structure characterization and contributed to writing part of the paper on peptide synthesis; H.-S.T. conducted the electrophysiological experiments, analyzed the data, and contributed to writing the paper; X.J. performed NMR structural study of the peptide; Q.K. participated in analyzing the modeling data; R.Y. conducted computational modeling and analyzed the data; and D.J.A. and R.Y. conceived the idea of the project, provided the financial support and resources, and contributed to writing the paper.
The authors declare no competing financial interest.
Supplementary Material
References
- Dutton J. L.; Craik D. J. alpha-Conotoxins nicotinic acetylcholine receptor antagonists as pharmacological tools and potential drug leads. Curr. Med. Chem. 2001, 8, 327–344. 10.2174/0929867013373453. [DOI] [PubMed] [Google Scholar]
- Craik D. J.; Adams D. J. Chemical modification of conotoxins to improve stability and activity. ACS Chem. Biol. 2007, 2, 457–468. 10.1021/cb700091j. [DOI] [PubMed] [Google Scholar]
- Azam L.; McIntosh J. M. Alpha-conotoxins as pharmacological probes of nicotinic acetylcholine receptors. Acta Pharmacol. Sin. 2009, 30, 771–783. 10.1038/aps.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincler M.; Wittenauer S.; Parker R.; Ellison M.; Olivera B. M.; McIntosh J. M. Molecular mechanism for analgesia involving specific antagonism of α9α10 nicotinic acetylcholine receptors. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 17880–17884. 10.1073/pnas.0608715103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaghan B.; Haythornthwaite A.; Berecki G.; Clark R. J.; Craik D. J.; Adams D. J. Analgesic α-conotoxins Vc1.1 and Rg1A inhibit N-type calcium channels in rat sensory neurons via GABAB receptor activation. J. Neurosci. 2008, 28, 10943–10951. 10.1523/jneurosci.3594-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napier I. A.; Klimis H.; Rycroft B. K.; Jin A. H.; Alewood P. F.; Motin L.; Adams D. J.; Christie M. J. Intrathecal α-conotoxins Vc1.1, AuIB and MII acting on distinct nicotinic receptor subtypes reverse signs of neuropathic pain. Neuropharmacology 2012, 62, 2202–2207. 10.1016/j.neuropharm.2012.01.016. [DOI] [PubMed] [Google Scholar]
- Di Cesare Mannelli L.; Cinci L.; Micheli L.; Zanardelli M.; Pacini A.; McIntosh M. J.; Ghelardini C. α-Conotoxin RgIA protects against the development of nerve injury-induced chronic pain and prevents both neuronal and glial derangement. Pain 2014, 155, 1986–1995. 10.1016/j.pain.2014.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadi S.; Christie M. J. α9-Nicotinic acetylcholine receptors contribute to the maintenance of chronic mechanical hyperalgesia, but not thermal or mechanical allodynia. Mol. Pain 2014, 10, 64. 10.1186/1744-8069-10-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S.; Kulak J. M.; Cartier G. E.; Jacobsen R. B.; Yoshikami D.; Olivera B. M.; McIntosh J. M. α-Conotoxin AuIB selectively blocks α3β4 nicotinic acetylcholine receptors and nicotine-evoked norepinephrine release. J. Neurosci. 1998, 18, 8571–8579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevin S. T.; Clark R. J.; Klimis H.; Christie M. J.; Craik D. J.; Adams D. J. Are α9α10 nicotinic acetylcholine receptors a pain target for α-conotoxins?. Mol. Pharmacol. 2007, 72, 1406–1410. 10.1124/mol.107.040568. [DOI] [PubMed] [Google Scholar]
- Carstens B. B.; Clark R. J.; Daly N. L.; Harvey P. J.; Kaas Q.; Craik D. J. Engineering of conotoxins for the treatment of pain. Curr. Pharm. Des. 2011, 17, 4242–4253. 10.2174/138161211798999401. [DOI] [PubMed] [Google Scholar]
- Schroeder C. I.; Craik D. J. Therapeutic potential of conopeptides. Future Med. Chem. 2012, 4, 1243–1255. 10.4155/fmc.12.70. [DOI] [PubMed] [Google Scholar]
- Grishin A. A.; Wang C.-I. A.; Muttenthaler M.; Alewood P. F.; Lewis R. J.; Adams D. J. α-Conotoxin AuIB isomers exhibit distinct inhibitory mechanisms and differential sensitivity to stoichiometry of α3β4 nicotinic acetylcholine receptors. J. Biol. Chem. 2010, 285, 22254–22263. 10.1074/jbc.m110.111880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tietze A. A.; Tietze D.; Ohlenschläger O.; Leipold E.; Ullrich F.; Kühl T.; Mischo A.; Buntkowsky G.; Görlach M.; Heinemann S. H.; Imhof D. Structurally diverse μ-conotoxin PIIIA isomers block sodium channel NaV1.4. Angew. Chem., Int. Ed. 2012, 51, 4058–4061. 10.1002/anie.201107011. [DOI] [PubMed] [Google Scholar]
- Hargittai B.; Solé N. A.; Groebe D. R.; Abramson S. N.; Barany G. Chemical syntheses and biological activities of lactam analogues of α-conotoxin SI. J. Med. Chem. 2000, 43, 4787–4792. 10.1021/jm990635c. [DOI] [PubMed] [Google Scholar]
- Armishaw C. J.; Daly N. L.; Nevin S. T.; Adams D. J.; Craik D. J.; Alewood P. F. α-Selenoconotoxins, a new class of potent α7 neuronal nicotinic receptor antagonists. J. Biol. Chem. 2006, 281, 14136–14143. 10.1074/jbc.m512419200. [DOI] [PubMed] [Google Scholar]
- MacRaild C. A.; Illesinghe J.; van Lierop B. J.; Townsend A. L.; Chebib M.; Livett B. G.; Robinson A. J.; Norton R. S. Structure and activity of (2,8)-dicarba-(3,12)-cystino α-ImI, an α-conotoxin containing a nonreducible cystine analogue. J. Med. Chem. 2009, 52, 755–762. 10.1021/jm8011504. [DOI] [PubMed] [Google Scholar]
- Van Lierop B. J.; Robinson S. D.; Kompella S. N.; Belgi A.; McArthur J. R.; Hung A.; MacRaild C. A.; Adams D. J.; Norton R. S.; Robinson A. J. Dicarba α-conotoxin Vc1.1 analogues with differential selectivity for nicotinic acetylcholine and GABAB receptors. ACS Chem. Biol. 2013, 8, 1815–1821. 10.1021/cb4002393. [DOI] [PubMed] [Google Scholar]
- Dekan Z.; Vetter I.; Daly N. L.; Craik D. J.; Lewis R. J.; Alewood P. F. α-Conotoxin ImI incorporating stable cystathionine bridges maintains full potency and identical three-dimensional structure. J. Am. Chem. Soc. 2011, 133, 15866–15869. 10.1021/ja206408q. [DOI] [PubMed] [Google Scholar]
- Chen S.; Gopalakrishnan R.; Schaer T.; Marger F.; Hovius R.; Bertrand D.; Pojer F.; Heinis C. Dithiol amino acids can structurally shape and enhance the ligand-binding properties of polypeptides. Nat. Chem. 2014, 6, 1009–1016. 10.1038/nchem.2043. [DOI] [PubMed] [Google Scholar]
- Yu R.; Seymour V. A. L.; Berecki G.; Jia X.; Akcan M.; Adams D. J.; Kaas Q.; Craik D. J. Less is More: Design of a highly stable disulfide-deleted mutant of analgesic cyclic α-conotoxin Vc1.1. Sci. Rep. 2015, 5, 13264. 10.1038/srep13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidigh J. W.; Fesinmeyer R. M.; Andersen N. H. Designing a 20-residue protein. Nat. Struct. Biol. 2002, 9, 425–430. 10.1038/nsb798. [DOI] [PubMed] [Google Scholar]
- Wan J.; Huang J. X.; Vetter I.; Mobli M.; Lawson J.; Tae H.-S.; Abraham N.; Paul B.; Cooper M. A.; Adams D. J.; Lewis R. J.; Alewood P. F. α-Conotoxin dendrimers have enhanced potency and selectivity for homomeric nicotinic acetylcholine receptors. J. Am. Chem. Soc. 2015, 137, 3209–3212. 10.1021/jacs.5b00244. [DOI] [PubMed] [Google Scholar]
- Lamthanh H.; Jegou-Matheron C.; Servent D.; Ménez A.; Lancelin J.-M. Minimal conformation of the α-conotoxin ImI for the α7 neuronal nicotinic acetylcholine receptor recognition: correlated CD, NMR and binding studies. FEBS Lett. 1999, 454, 293–298. 10.1016/s0014-5793(99)00831-5. [DOI] [PubMed] [Google Scholar]
- Carstens B. B.; Berecki G.; Daniel J. T.; Lee H. S.; Jackson K. A. V.; Tae H.-S.; Sadeghi M.; Castro J.; O’Donnell T.; Deiteren A.; Brierley S. M.; Craik D. J.; Adams D. J.; Clark R. J. Structure–Activity studies of cysteine-rich α-conotoxins that inhibit high-voltage-activated calcium channels via GABAB receptor activation reveal a minimal functional motif. Angew. Chem., Int. Ed. 2016, 55, 4692–4696. 10.1002/anie.201600297. [DOI] [PubMed] [Google Scholar]
- Yu R.; Craik D. J.; Kaas Q. Blockade of neuronal α7-nAChR by α-conotoxin ImI explained by computational scanning and energy calculations. PLoS Comput. Biol. 2011, 7, e1002011 10.1371/journal.pcbi.1002011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrane M.; Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database 2011, 2011, bar009. 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celie P. H. N.; Kasheverov I. E.; Mordvintsev D. Y.; Hogg R. C.; van Nierop P.; van Elk R.; van Rossum-Fikkert S. E.; Zhmak M. N.; Bertrand D.; Tsetlin V.; Sixma T. K.; Smit A. B. Crystal structure of nicotinic acetylcholine receptor homolog AChBP in complex with an α-conotoxin PnIA variant. Nat. Struct. Mol. Biol. 2005, 12, 582–588. 10.1038/nsmb951. [DOI] [PubMed] [Google Scholar]
- Dellisanti C. D.; Yao Y.; Stroud J. C.; Wang Z.-Z.; Chen L. Crystal structure of the extracellular domain of nAChR α1 bound to α-bungarotoxin at 1.94 A resolution. Nat. Neurosci. 2007, 10, 953–962. 10.1038/nn1942. [DOI] [PubMed] [Google Scholar]
- Zouridakis M.; Giastas P.; Zarkadas E.; Chroni-Tzartou D.; Bregestovski P.; Tzartos S. J. Crystal structures of free and antagonist-bound states of human α9 nicotinic receptor extracellular domain. Nat. Struct. Mol. Biol. 2014, 21, 976–980. 10.1038/nsmb.2900. [DOI] [PubMed] [Google Scholar]
- Shen M.-Y.; Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006, 15, 2507–2524. 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark R. J.; Fischer H.; Nevin S. T.; Adams D. J.; Craik D. J. The synthesis, structural characterization, and receptor specificity of the α-conotoxin Vc1.1. J. Biol. Chem. 2006, 281, 23254–23263. 10.1074/jbc.m604550200. [DOI] [PubMed] [Google Scholar]
- Chi S.-W.; Kim D.-H.; Olivera B. M.; McIntosh J. M.; Han K.-H. NMR structure determination of α-conotoxin BuIA, a novel neuronal nicotinic acetylcholine receptor antagonist with an unusual 4/4 disulfide scaffold. Biochem. Biophys. Res. Commun. 2006, 349, 1228–1234. 10.1016/j.bbrc.2006.08.164. [DOI] [PubMed] [Google Scholar]
- Ulens C.; Hogg R. C.; Celie P. H.; Bertrand D.; Tsetlin V.; Smit A. B.; Sixma T. K. Structural determinants of selective α-conotoxin binding to a nicotinic acetylcholine receptor homolog AChBP. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 3615–3620. 10.1073/pnas.0507889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton J. L.; Bansal P. S.; Hogg R. C.; Adams D. J.; Alewood P. F.; Craik D. J. A new level of conotoxin diversity, a non-native disulfide bond connectivity in α-conotoxin AuIB reduces structural definition but increases biological activity. J. Biol. Chem. 2002, 277, 48849–48857. 10.1074/jbc.m208842200. [DOI] [PubMed] [Google Scholar]
- Šali A.; Blundell T. L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Webb B.; Sali A.. Comparative protein structure modeling using MODELLER. Current Protocols in Bioinformatics; John Wiley & Sons, Inc., 2016; Vol. 86, pp 2.9.1–2.9.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M. H. M.; Søndergaard C. R.; Rostkowski M.; Jensen J. H. PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theory Comput. 2011, 7, 525–537. 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- Hornak V.; Abel R.; Okur A.; Strockbine B.; Roitberg A.; Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins: Struct., Funct., Bioinf. 2006, 65, 712–725. 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case D. A.; Betz R. M.; Cerutti D. S.; Cheatham T. E. III; Darden T. A.; Duke R. E.; Giese T. J.; Gohlke H.; Goetz A. W.; Homeyer N.; Izadi S.; Janowski P.; Kaus J.; Kovalenko A.; Lee T. S.; LeGrand S.; Li P.; Lin C.; Luchko T.; Luo R.; Madej B.; Mermelstein D.; Merz K. M.; Monard G.; Nguyen H.; Nguyen H. T.; Omelyan I.; Onufriev A.; Roe D. R.; Roitberg A.; Sagui C.; Simmerling C. L.; Botello-Smith W. M.; Swails J.; Walker R. C.; Wang J.; Wolf R. M.; Wu X.; Xiao L.; Kollman P. A.. AMBER; University of California: San Francisco, 2016.
- Miyamoto S.; Kollman P. A. Settle: An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 1992, 13, 952–962. 10.1002/jcc.540130805. [DOI] [Google Scholar]
- Darden T.; York D.; Pedersen L. Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. 10.1063/1.464397. [DOI] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. VMD: visual molecular dynamics. J. Mol. Graphics 1996, 14, 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Yu R.; Kompella S. N.; Adams D. J.; Craik D. J.; Kaas Q. Determination of the α-conotoxin Vc1.1 binding site on the α9α10 nicotinic acetylcholine receptor. J. Med. Chem. 2013, 56, 3557–3567. 10.1021/jm400041h. [DOI] [PubMed] [Google Scholar]
- Chignola F.; Mari S.; Stevens T. J.; Fogh R. H.; Mannella V.; Boucher W.; Musco G. The CCPN Metabolomics Project: a fast protocol for metabolite identification by 2D-NMR. Bioinformatics 2011, 27, 885–886. 10.1093/bioinformatics/btr013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.