Abstract
A multidisciplinary
strategy, including both biochemical and biophysical
studies, was proposed here to evaluate the potential of lipid nanoaggregates
consisting of a mixture of a gemini–bolaamphiphilic lipid (C6C22C6) and the well-known helper lipid
1,2-dioleoyl-sn-glycero-3-phosphatidylethanolamine
(DOPE) to transfect plasmid DNA into living cells in an efficient
and safe way. For that purpose, several experimental techniques were
employed, such as zeta potential (phase analysis light scattering
methodology), agarose gel electrophoresis (pDNA compaction and pDNA
protection assays), small-angle X-ray scattering, cryo-transmission
electron microscopy, atomic force microscopy, fluorescence-assisted
cell sorting, luminometry, and cytotoxicity assays. The results revealed
that the cationic lipid and plasmid offer only 70 and 30% of their
nominal positive (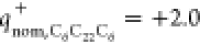 ) and negative charges (
) and negative charges (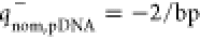 ),
respectively. Upon mixing with DOPE,
they form lipoplexes that self-aggregate in typical multilamellar
Lα lyotropic liquid-crystal nanostructures with sizes
in the range of 100–200 nm and low polydispersities, very suitably
fitted to remain in the bloodstream and cross the cell membrane. Interestingly,
these nanoaggregates were able to compact, protect (from the degrading
effect of DNase I), and transfect two DNA plasmids (pEGFP-C3, encoding
the green fluorescent protein, and pCMV-Luc, encoding luciferase)
into COS-7 cells, with an efficiency equal or even superior to that
of the universal control Lipo2000*, as long as the effective +/–
charge ratio was maintained higher than 1 but reasonably close to
electroneutrality. Moreover, this transfection process was not cytotoxic
because the viability of COS-7 cells remained at high levels, greater
than 80%. All of these features make the C6C22C6/DOPE nanosystem an optimal nonviral gene nanocarrier
in vitro and a potentially interesting candidate for future in vivo
experiments.
),
respectively. Upon mixing with DOPE,
they form lipoplexes that self-aggregate in typical multilamellar
Lα lyotropic liquid-crystal nanostructures with sizes
in the range of 100–200 nm and low polydispersities, very suitably
fitted to remain in the bloodstream and cross the cell membrane. Interestingly,
these nanoaggregates were able to compact, protect (from the degrading
effect of DNase I), and transfect two DNA plasmids (pEGFP-C3, encoding
the green fluorescent protein, and pCMV-Luc, encoding luciferase)
into COS-7 cells, with an efficiency equal or even superior to that
of the universal control Lipo2000*, as long as the effective +/–
charge ratio was maintained higher than 1 but reasonably close to
electroneutrality. Moreover, this transfection process was not cytotoxic
because the viability of COS-7 cells remained at high levels, greater
than 80%. All of these features make the C6C22C6/DOPE nanosystem an optimal nonviral gene nanocarrier
in vitro and a potentially interesting candidate for future in vivo
experiments.
Introduction
Nucleic acids are nowadays an attractive source of therapeutic agents because of their intimate structure–activity relationships and their highly specific mode of action, allowing exploitation of the cellular machinery in a predictable fashion to either stimulate or silence the expression of virtually any protein, with reduced toxicity and lessened side effects.1−4 These aspects are the basis of gene therapy, an exciting branch of medicine that aims to cure diseases at a molecular level by either repairing damaged cellular DNA5−7 or silencing the expression of abnormal genes.8−10 In the case of biomolecular drugs such as nucleic acids, the carrier becomes a decisive protagonist, allowing the drug to overcome the physiological barriers and reach its target in a fully functional form to carry out the designed therapeutic function, given that nucleic acids are easily degraded by nucleases in biological fluids and their membrane-crossing abilities and cellular uptake are seriously limited by their inherently large size, rigidity, and negative charge.11,12 Gene carriers or gene vectors are usually classified into two groups: viral and nonviral. Although viral gene vectors are normally more effective, they may cause serious adverse effects in the organism.13−15 Accordingly, researchers have mostly focused their efforts on the development of nonviral carriers capable of compacting, protecting, and transporting genetic materials in an efficient and safe way by combining high transfection efficiencies (TEs) with low cytotoxicity outputs.6,16−21
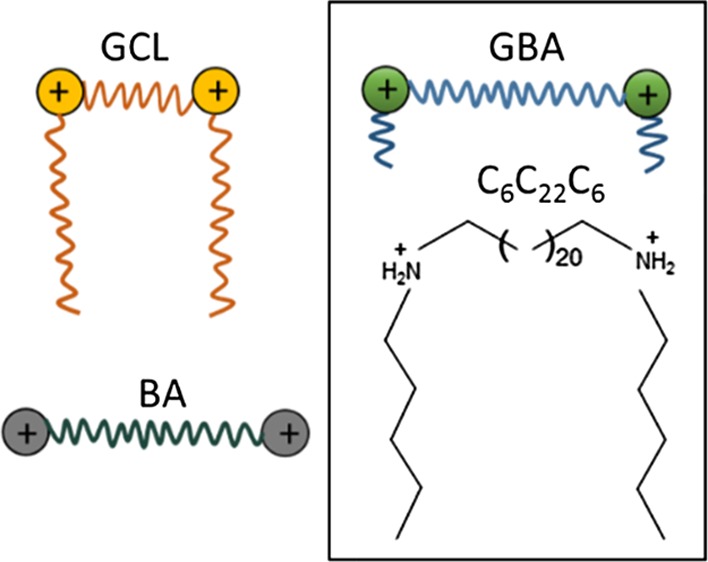
Among the nonviral gene vectors available, a wide series of gemini cationic lipids (GCLs), constituted by two hydrophobic chains and two hydrophilic cationic heads linked by a spacer, have proven in the last decade their capacity to compact, protect, and release DNA in the cytoplasm of living cells.6,22−24 Together with these GCLs, another class of lipids, known as bolaform or bolaamphiphilic (BA) lipids characterized by bearing two hydrophilic (ionic or nonionic) heads separated by one, two, or three long hydrophobic spacers (usually alkyl chains of 22–32 carbon atoms), have also exhibited potential as gene vectors.25−29 Their peculiar structure means that, in general terms, they tend to self-aggregate, forming monolayer-type membranes with less permeability, greater rigidity, and better durability than the lipid bilayer-type membranes typically formed from gemini lipids. In fact, BA lipids have already been successfully used as nonviral gene vectors in cases where at least one or both heads are positively charged.30−41
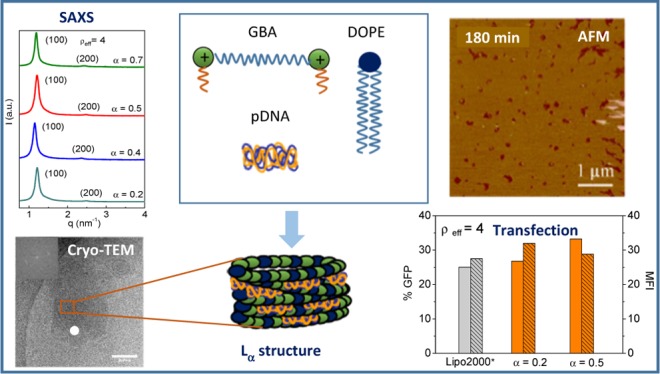
In this work, we have evaluated the potential of a so-called gemini–bolaamphiphilic (GBA) lipid as a novel gene vector. The GBA lipid was synthesized with a hybrid structure between that of a gemini lipid (GCL), insofar it contains two secondary amine-type cation heads each of them attached to short hydrophobic chains of 6 carbon atoms, and that of a BA lipid in the sense that the cationic heads are connected by a long spacer of 22 carbon atoms (referred to as C6C22C6, Scheme 1). The disproportion between the short length of the hydrophobic chains (6C) and that of the long spacer (22C) results in the latter, potentially playing a leading role first in the self-aggregation process and then in the formation of the lipoplex in the presence of DNA. As usual, with this type of nanocarrier, the cationic lipid was mixed with 1,2-dioleoyl-sn-glycero-3-phosphatidylethanolamine (DOPE), a well-known fusogenic and helper lipid that promotes cellular uptake.42−44 The C6C22C6/DOPE mixed lipid vector was used in this work to compact and transfect two DNA plasmids into COS-7 cells: one encoding the green fluorescent protein (GFP) (pEGFP-C3) and the other one encoding luciferase (pCMV-Luc). Accompanying this biochemical evaluation of the TE (fluorescence-assisted cell sorting (FACS) and luminometry) and cytotoxicity (alamarBlue assay), a biophysical study is included, comprising the size [dynamic light scattering (DLS)], structural [cryo-transmission electronic microscopy (cryo-TEM), small-angle X-ray scattering (SAXS), and atomic force microscopy (AFM)], and electrochemical (zeta potential and gel electrophoresis) characterizations of the optimal compositions of C6C22C6/DOPE–pDNA lipoplexes, as previously determined from their biochemical performance.
Scheme 1. Schematic Drawings of a GCL, a Cationic BA lipid, and the Resulting Hybrid Structure, that is, the Cationic GBA lipid Proposed and Synthesized in This Work and detailed structure of the C6C22C6 GBA lipid.
Results and Discussion
Multidisciplinary and complementary biophysical and biochemical studies, as those reported herein, are needed to assess, with a wider perspective, the ability of a given nanosystem to compact, protect, transport across cell membranes and deliver the nucleic acids into the cell interior, overcoming the physiological barriers that the nanocarrier–DNA complex may find along such a remarkable journey.
The electrochemical evaluation of the C6C22C6/DOPE–pDNA system, consisting of electrophoretic
mobility/zeta potential and agarose gel electrophoresis assays (compaction
and protection of pDNA against degradation by DNase I), is shown in Figures 1 and 2 at different molar compositions of the cationic lipid in
the mixed lipid (α). These three experiments allowed us to (i)
check whether the vector was capable of compacting pDNA and at what
lipoplex compositions this compaction would occur (inset in Figure 1); (ii) accurately
determine the electroneutrality of the system and thus the effective
charges of both cationic GBA lipid and plasmid DNA (Figure 1); and (iii) confirm that the
nanocarrier is able to protect adequately the plasmid against the
degrading effect of DNase I (Figure 2). The inset in Figure 1 reports an agarose gel electrophoresis experiment,
where pDNA was loaded as a control in the first lane of the gel and
C6C22C6/DOPE–pDNA lipoplexes
at different mass ratios (mL/mDNA = (mL+ + mL0)/mDNA, where mL+, mL0, and mDNA denote
the masses of the cationic gemini lipid, zwitterionic helper lipid,
and nucleic acid, respectively) were loaded in lanes 2–4. The
two fluorescence bands observed in lane 1 are characteristic of the
coiled and supercoiled forms of plasmid DNA, whereas the absence of
such bands in lanes 2–4, together with the fact that pDNA remains
immobile in the well of the gel, is a clear indication of the effective
compaction of pDNA by the C6C22C6/DOPE lipid vector. In particular, at α = 0.5, compaction is
already achieved at a mass ratio of mL/mDNA = 0.8. Additionally, this indicates
that electroneutrality in this lipoplex must be reached at compositions
below 0.8 when α = 0.5. Figure 1 also shows the plot of zeta potential versus mL/mDNA, displaying
a typical sigmoidal profile at different molar compositions. From
the electroneutrality ratios, (mL/mDNA)ϕ, that is, the mL/mDNA ratios at
which charge compensation occurs and the zeta potential is zero, the
effective charges of C6C22C6 (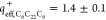 ) and pDNA (
) and pDNA (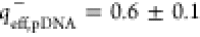 ) were calculated by a procedure
developed
by us and fully described elsewhere.45,46 The cationic
lipid and plasmid were found to exhibit only 70 and 30% of their nominal
positive (
) were calculated by a procedure
developed
by us and fully described elsewhere.45,46 The cationic
lipid and plasmid were found to exhibit only 70 and 30% of their nominal
positive (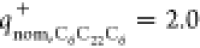 ) and
negative (
) and
negative (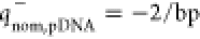 ) charges, respectively, which is a behavior
typically observed in this type of systems.46−48 It is well-known
that plasmids in supercoiled conformation are able to retain a significant
amount of Na+ cations under physiological conditions, ostensibly
reducing their anionic character. This fact may be advantageous for
our objectives because the lower the anionic character of the plasmid,
the smaller the amount of cationic vector necessary to compact it
and yield a net positive charge (necessary to cross the also negatively
charged cell membrane), thus reducing the overall cytotoxicity of
the nanocarrier. Such understanding of the effective charges allowed
us to work with effective charge ratios by using the following equation:
) charges, respectively, which is a behavior
typically observed in this type of systems.46−48 It is well-known
that plasmids in supercoiled conformation are able to retain a significant
amount of Na+ cations under physiological conditions, ostensibly
reducing their anionic character. This fact may be advantageous for
our objectives because the lower the anionic character of the plasmid,
the smaller the amount of cationic vector necessary to compact it
and yield a net positive charge (necessary to cross the also negatively
charged cell membrane), thus reducing the overall cytotoxicity of
the nanocarrier. Such understanding of the effective charges allowed
us to work with effective charge ratios by using the following equation:
| 1 |
Figure 1.
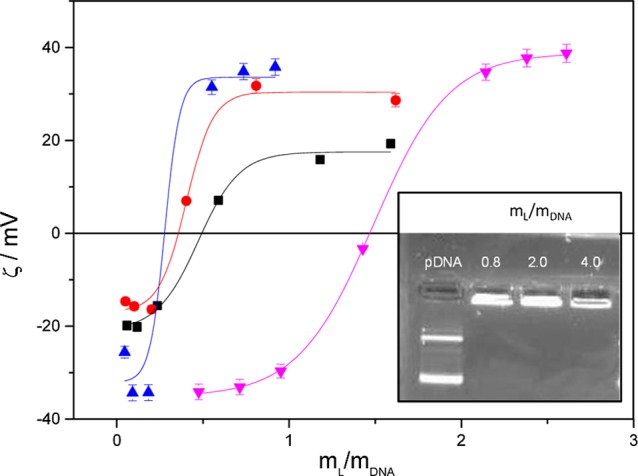
Plot of zeta potential as a function of mL/mDNA mass ratio of the C6C22C6/DOPE–DNA lipoplexes, constituted by ctDNA at several molar compositions of the cationic lipid in the mixed lipids (α = 0.5, pink line) and with pDNA at α = 0.2, 0.5, and 0.7 (blue, red, and black lines), respectively. Inset: agarose gel electrophoresis of C6C22C6/DOPE–pDNA at several mL/mDNA mass ratios and α = 0.5. Free pDNA (lane 1) was used as the control.
Figure 2.
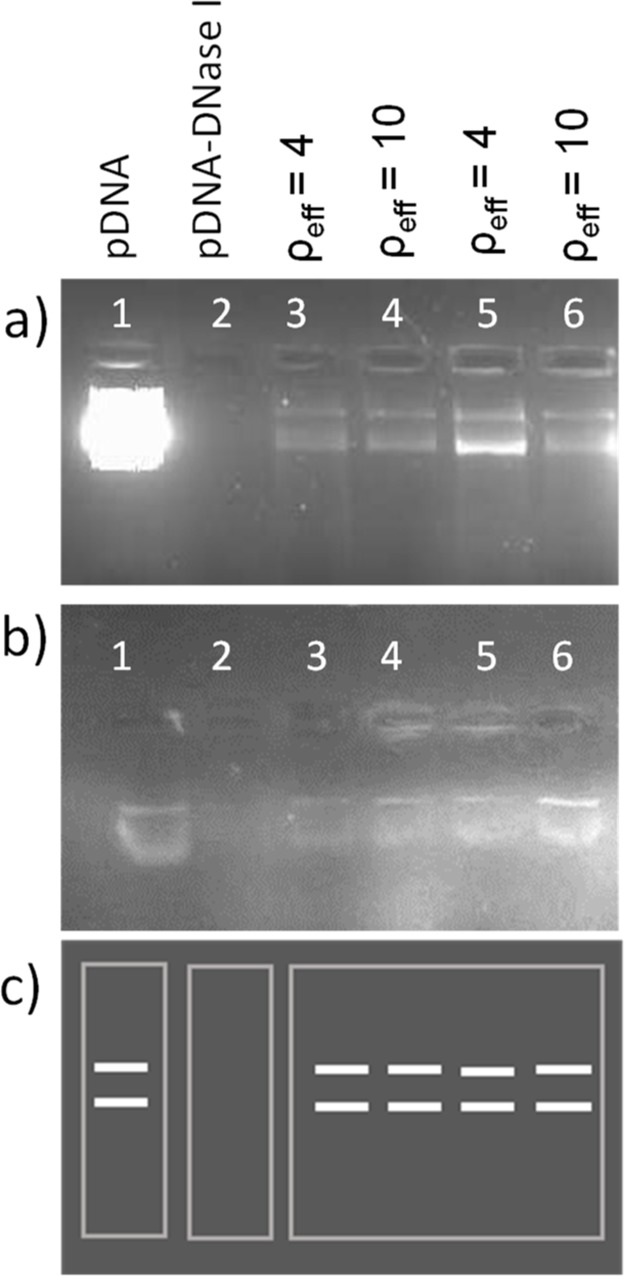
Protection assay of pDNA against degradation by DNase I (gel electrophoresis experiment): (a) pCMV-Luc plasmid, (b) pEGFP-C3 plasmid, and (c) a visual guide. In both experiments: lane 1, pDNA; lane 2, pDNA–DNase I; and lanes 3–6, C6C22C6/DOPE–pDNA lipoplexes at different molar compositions of the cationic lipid in the mixed lipids (α = 0.2 in lanes 3–4 and α = 0.5 in lanes 5–6) and different effective charge ratios of the lipoplex, ρeff = 4 (lanes 3 and 5) and ρeff = 10 (lanes 4 and 6).
For a nanosystem to become a suitable gene vector, in addition to being able to efficiently compact the plasmid and generate a purely cationic entity, the system must be capable of protecting the nucleic acid against the degrading effect of the DNases present in human serum. This information is crucial when attempting to carry out in vivo studies. As can be seen in Figure 2, where the protection experiments against DNase I are presented, the mixed lipid system C6C22C6/DOPE efficiently protects the two plasmids used in this work, one encoding the GFP (pEGFP-C3) and the other one encoding luciferase (pCMV-Luc). Thus, lane 1 (naked pDNA) shows the characteristic fluorescence bands from the emission of ethidium bromide (EtBr) intercalated within the double helix of the plasmid DNA in its coiled and supercoiled forms, whereas lane 2 reflects the progress of the free plasmid along the gel in the absence of a vector after digestion treatment with DNase I. C6C22C6/DOPE–pDNA lipoplexes at different compositions and effective charge ratios were loaded in lanes 3–6 after digestion with DNase I and subsequent treatment with an anionic surfactant [Sodium dodecyl sulfate (SDS)], breaking the lipoplexes and freeing the plasmid. The absence of fluorescence bands in lane 2 and their presence in lanes 3–6 in both gels (Figure 2a,b) demonstrate that DNase I degraded the free plasmid in lane 2 but it did not have access to the plasmid compacted by the vector C6C22C6/DOPE at any of the studied cationic molar compositions of the mixed lipid (α = 0.2 and 0.5) and effective charge ratios (ρeff = 4 and 10).
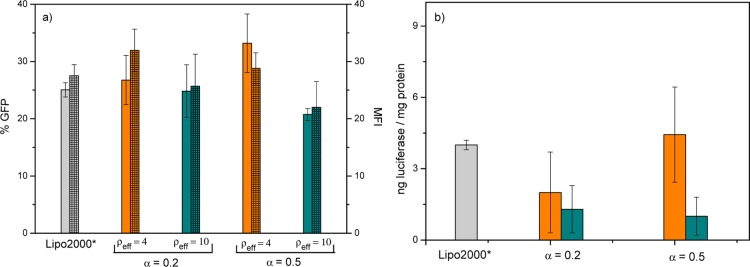
These observations allow us to conclude that the C6C22C6/DOPE lipidic system shows the required features to be, at least potentially, a suitable nonviral gene vector, given that it is capable of compacting, protecting from DNases, and transporting pDNA through the cell membrane into the cytoplasm. To verify whether this process could actually be carried out efficiently and safely, transfection experiments and cell viability assays were performed on COS-7 cells in the presence of 10% serum. Figure 3 shows the TE by means of the expression levels of (a) GFP (for plasmid pEGFP-C3), given in terms of % GFP (percentage of cells expressing GFP) and/or MFI (mean fluorescence intensity per cell) obtained from FACS experiments, and (b) luciferase (for plasmid pCMV-Luc), expressed in terms of nanograms of luciferase/milligrams of protein, as obtained from luminometry experiments. In both cases, the experiments were performed at two molar compositions of the mixed lipids, α = 0.2 and 0.5, and at two different effective charge ratios of the lipoplex, ρeff = 4 and 10, using Lipofectamine 2000 (Lipo2000*) as a positive control. Overall, the results presented in Figure 3a for plasmid pEGFP-C3 indicate that the TE levels are superior at ρeff = 4 to those at ρeff = 10, both at α = 0.2 and 0.5. In particular, the values of % GFP and MFI at ρeff = 4 and α = 0.2 or 0.5 are comparable to or even slightly higher than those with Lipo2000*, whereas at ρeff = 10, they are equal to or slightly lower than those of the control. On the other hand, Figure 3b shows that in the case of plasmid pCMV-Luc, there is a clear superiority in the nanograms of luciferase expressed per milligrams of protein obtained at ρeff = 4 than that at ρeff = 10 at the two α values studied, the levels of TE being comparable to those of the control only at α = 0.5.
Figure 3.
TE levels of C6C22C6/DOPE–pDNA lipoplexes in COS-7 cells at two molar compositions of the cationic lipid in the mixed lipids (α = 0.2 and 0.5): (a) expressed as % GFP (solid bars) and MFI (squared bars) for plasmid pEGFP-C3 and (b) expressed as nanograms of luciferase per milligrams of protein for plasmid pCMV-Luc. The experiments were performed in the presence of 10% serum (FBS). The orange and green bars correspond to the effective charge ratios of ρeff = 4 and 10 in the lipoplex, respectively. The gray bar corresponds to Lipo2000*, used here as the positive control.
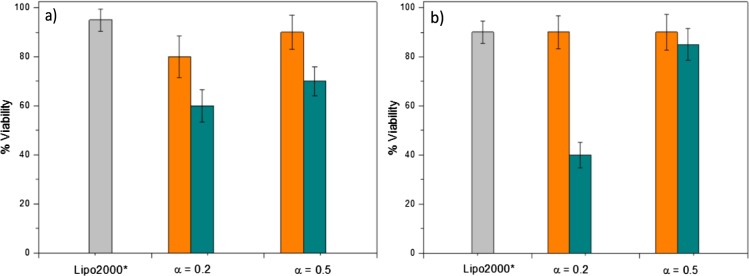
Gene therapy protocols require vectors not only to be efficient for the compaction, protection, transport, and delivery of plasmids inside the cell but also to perform these functions in a safe way. In this regard, the cytotoxicity of C6C22C6/DOPE–pDNA lipoplexes in COS-7 cells was evaluated by the alamarBlue assay at the same molar compositions and effective charge ratios used in the cell transfection experiments. Figure 4 reports the results obtained with the plasmids pEGFP-C3 and pCMV-Luc. Viability percentages above 60% were assumed to be reasonably acceptable, with values over 80% being the most desired. It is worth noting that the obtained cell viability levels are above 80% in both cases for ρeff = 4 at both molar ratios of α = 0.2 and 0.5. However, at ρeff = 10, the viability clearly falls below 80% and in one case even below 60%. Note that the cell viability of the Lipo2000* control is around 90%; therefore, it can be concluded that the nanovector used presents cellular viabilities comparable to those of the control only at the effective charge ratio of ρeff = 4. These results are consistent with other previously reported results48,49 and suggest that the viability of the cells is seriously compromised at high ρeff values, that is, at high cationic lipid contents, as one would expect.
Figure 4.
Cell viability of COS-7 cells in the presence of C6C22C6/DOPE–pDNA lipoplexes at two molar compositions of the cationic lipid in the mixed lipids (α = 0.2 and 0.5). The plasmids used were (a) pEGFP-C3 and (b) pCMV-Luc. The experiments were performed with 10% serum (FBS). The orange and green bars correspond to the effective charge ratios of ρeff = 4 and 10 in the lipoplex, respectively. The gray bar corresponds to Lipo2000*, used here as the positive control. The results were normalized to those obtained for untreated cells (100%).
So as to correlate the cellular activity and viability results with the structural characteristics of the studied lipoplexes, several key factors, such as the size, structure, morphology, and pDNA–mixed lipid interactions, were evaluated. For that purpose, DLS, SAXS, cryo-TEM, and AFM experiments were performed at the optimum effective charge ratio determined in the biochemical study (ρeff = 4) and at different compositions of the mixed lipid system. Table S1 in the Supporting Information summarizes the size (hydrodynamic diameter, Dh) and polydispersity (PDI) values for the C6C22C6/DOPE–pDNA lipoplexes at α = 0.2 and 0.5. The obtained sizes (in the range of 100–200 nm) and reasonably good homogeneity (low polydispersities of 0.2–0.3) are in the optimal range to cross the cell membrane and circulate in the bloodstream, making these lipoplexes potentially suitable DNA nanocarriers. No significant effect of the cationic lipid molar composition in the mixed lipid or of the effective charge ratio in the lipoplexes was observed on neither of these two parameters.
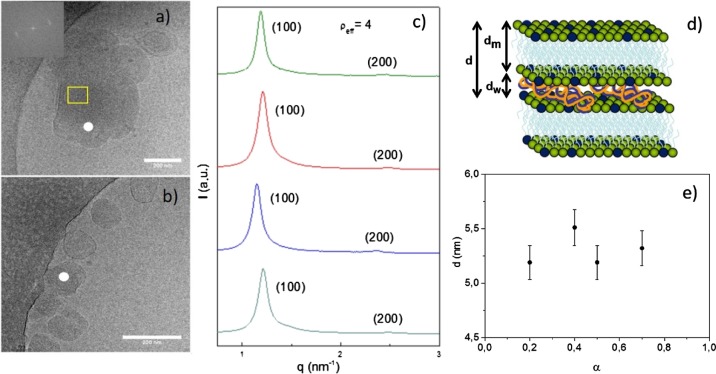
Figure 5 summarizes the results of cryo-TEM (panels a and b) and SAXS (panel c) experiments at ρeff = 4 and different molar compositions (α). Both experimental techniques revealed that these lipoplexes self-organize in multilamellar Lα phases of lyotropic liquid crystals, as previously reported for other mixed lipid-type nonviral gene vectors.49−51 Notice the presence of clear fingerprint patterns (indicated with a white point in the micrographs of panels a and b) regardless of the composition α in the mixed lipid system. The periodic repetition of lipid layers was studied by fast Fourier transform (FFT) analysis on certain zones of the micrographs (the yellow square in Figure 5a), where patterns of multilamellar stacking were found. The inset in Figure 5a shows, as an example, a FFT profile whose diffraction spots are typical of a lamellar structure. Additionally, the Bragg maxima reported in the SAXS diffractograms (Figure 5c) match adequately the hkl Miller indexes characteristic of multilamellar Lα structures, consisting of alternating layers of mixed lipids and supercoiled plasmid DNA in a sandwich-type fashion (see the scheme in Figure 5d), in total agreement with the cryo-TEM images. The interlayer distance (d) related to the q factor of the first maximum (d = 2π/q100) has an average value of 5.3 ± 0.3 nm, being not appreciably influenced by the composition of the mixed lipid system (α) within experimental error (Figure 5e). This distance, clearly shorter than the typical one (∼7 nm) found in other systems composed of gemini cationic/DOPE lipid mixtures,45,51 can be expressed as the sum of two thicknesses: that of the region where the hydrophobic chains of the lipids self-aggregate, traditionally denoted as dm, plus that of the aqueous region where the plasmid DNA is located, known as dw. Given the particular structure of the cationic lipid used (C6C22C6), two alternative arrangements can be proposed for the mixed lipid layer, both compatible with the SAXS results. The first one is a lipidic layer consisting of C6C22C6 cationic lipid molecules organized in a monolayer fashion mixed with DOPE helper lipid molecules in a bilayer disposition. In this case, the length of the C22 spacer of C6C22C6 determines the thickness of the monolayer, with the two hydrophobic C6 alkyl chains oriented inwardly with the two cationic quaternary ammonium heads exposed to both sides of the monolayer. This arrangement necessarily imposes an important interpenetration (of around seven methylene groups) of the hydrophobic chains of DOPE (18 carbon atoms) so that the system can self-organize in a bilayer fashion, matching well the d value of 5.3 nm imposed by the spacer length of the cationic GBA lipid. The second arrangement is the typical lipid bilayer consisting of a mixture of C6C22C6 and the DOPE helper lipid. In this case, the 22-carbon-atom spacer linking the cationic heads of C6C22C6 is forced to bend inwardly in a V-shaped manner so as to accommodate itself in the bilayer and the DOPE chains have to necessarily overlap extensively to fit the interlayer distance (5.3 nm) obtained from the diffractograms. Figure 6 shows a simplified 2D view of the Lα structure exhibited by these lipoplexes, with the two options discussed above. In both cases, either as a monolayer (Figure 6a) or as a lipid bilayer (Figure 6b), the thickness according to the Tanford’s model52 was estimated to be dm ≈ 3.0 nm, which would lead to a dw value of ca. 2.3 nm, which is enough to host the plasmid DNA. Several features can be highlighted: (i) both self-organization structures are compatible with the experimental thickness of d = 5.3 ± 0.3 nm and with the observation that d is not influenced by the DOPE content (Figure 5e); (ii) both arrangements have been previously reported for bolaform lipid systems27−29,33 and derived gemini cationic/DOPE lipid systems;45,47,51 and (iii) both arrangements provide an explanation of why the thickness of the lipid layer (d) is notably lower than that found in other GCL/DOPE mixed lipid systems forming a bilayer with no interpenetration of the lipid hydrophobic alkyl chains. Although the apparent lack of influence of the DOPE content on d confers the same weight to both structural options, the monolayer arrangement seems more feasible from a molecular interaction point of view, although further experiments would be necessary to confirm this.
Figure 5.
(a,b) Cryo-TEM micrographs of C6C22C6/DOPE–pDNA lipoplexes at ρeff = 4 and molar compositions of the cationic lipid in the mixed lipids of (a) α = 0.2 and (b) α = 0.5. The inset in (a) shows the diffraction spot from the FFT calculations over the selected area on the original micrograph. The white points indicate the lamellar structure with a multilamellar pattern. The scale bar is 200 nm. (c) SAXS diffractograms of C6C22C6/DOPE–pDNA lipoplexes at an effective charge ratio of ρeff = 4 and different molar compositions (α). (d) Three-dimensional scheme of the Lα multilamellar lyotropic liquid-crystal structure. (e) Plot of interlamellar distance (d) of this Lα multilamellar structure as a function of molar composition of the mixed lipids (α) at ρeff = 4.
Figure 6.
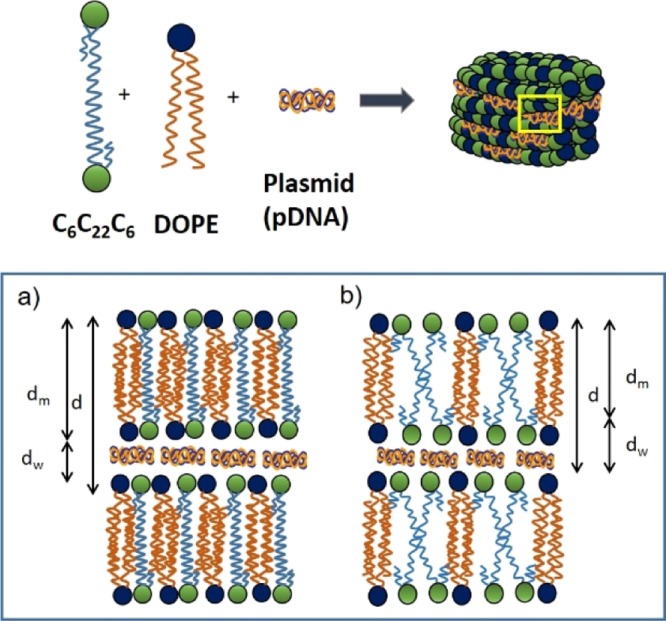
Schematic drawings of the cationic GBA lipid (C6C22C6), zwitterionic helper lipid (DOPE), and plasmid pDNAs (pEGFP-C3 or pCMV-Luc) self-organized in a Lα multilamellar lyotropic liquid-crystal structure. The inset at the bottom of the figure shows a magnified view of the yellow squared zone of the lipoplex, with two possible arrangements compatible with the SAXS and cryo-TEM results: (a) the 22C spacer of C6C22C6 forms a lipid monolayer and (b) C6C22C6 is organized in a typical lipid bilayer, with the spacer oriented inward and the bilayer with a V shape. In both cases, DOPE is organized in a lipid bilayer fashion, with the hydrophobic chains displaying extensive overlapping to fit the dimensions determined for the interlamellar distance (d).
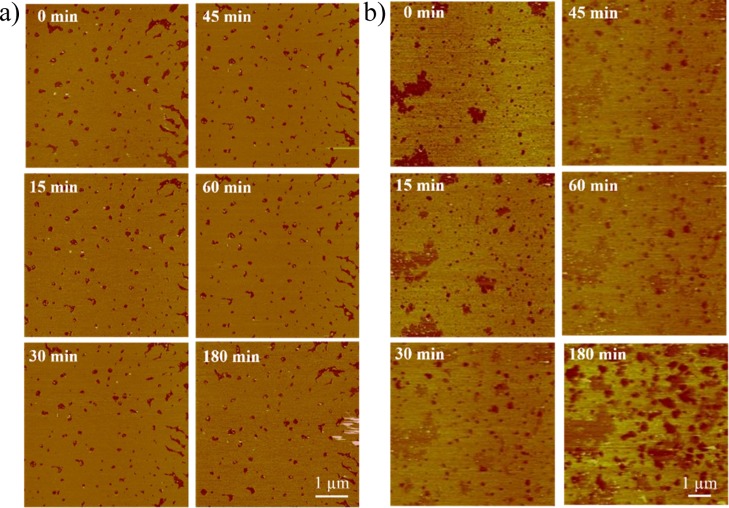
Undoubtedly, the way pDNA interacts with lipidic membranes is another important factor affecting the efficiency of the transfection process. AFM is known to be one of the techniques of choice to visualize these interactions. Accordingly, AFM experiments were performed for C6C22C6/DOPE samples (at α = 0.2 and 0.5) in the absence and presence of pDNA at ρeff = 4 and several incubation times (Figure 7). In Figure 7a, the topographic image of a supported lipid bilayer (SLB) of C6C22C6/DOPE at α = 0.2 is shown. The SLB does not fully cover the mica surface and exhibits a number of holes of ≥200 nm size (shown in red in the image). Such holes allowed us to deduce the thickness of the layer, showing a step height of 4.4 ± 0.4 nm from the mica surface to the top of the lipid layer. It is possible to observe nonadsorbed lipid structures as small yellow-white spikes. Upon injection of pDNA, no significant modification of the shape of the structures was observed at 60 min. After 180 min, although the margins of the holes remained almost unmodified, some aggregates were deposited on the SLB, as observed in the middle right region as white scratching marks. A different behavior was observed for the C6C22C6/DOPE SLB at α = 0.5. Figure 7b depicts the topographic image of this SLB, showing a structure similar to that in Figure 7a. In this case, the holes present a similar step height than before (4.6 ± 0.4 nm), but wider holes of 1 or 2 μm are observed. The mica surface in these wider holes is not flat but contains some small lipid aggregates on it. Upon injection of pDNA, some aggregates can be observed inside the wider holes but not on the lipid layer. This fact suggests that these aggregates are weakly attached to the lipid surface, thus possibly being swept away during scanning. This effect is visible up to 60 min of incubation with no significant changes on the edges of the small holes. After 180 min of incubation, the small holes become slightly wider and, in some regions, they merge to form wider structures. Such a different behavior of pDNA with lipid membranes suggests specific interactions of pDNA depending on the α value. The AFM studies revealed that pDNA modifies the SLB morphology in a more effective manner at α = 0.5 than at α = 0.2. Apparently, when pDNA molecules interact with the SLB at α = 0.5, a reduction of the lateral pressure between the lipids is observed, promoting the arrangement of the lipids into an SLB, even when the lipids are adsorbed on a solid surface such as the mica surface. This fact is evident for the rearrangement of the lipids in the SLB, that is, the fusion of holes, without solubilization of part of the SLB. When pDNA interacts with the SLB at α = 0.2, this effect is less evident. All of these findings allow us to conclude the following. First, pDNA molecules seem to be more shielded by mixed lipids when the amount of the GBA lipid is moderate. This feature correlates well with the enhanced protection of the plasmid from the degrading action of the DNase I enzyme, as pointed by the more intense fluorescence detected in the gels at α = 0.5 compared to that observed at α = 0.2 (Figure 2). Second, the pDNA molecules reduce the interface tension of the lipids more effectively at moderate lipid compositions, thus favoring more deformable lipid membranes. This feature, together with a larger accumulation of positive charges at α = 0.5, may promote the formation of multilayer structures, in agreement with the SAXS and cryo-TEM results (Figure 5) and also with the higher hydrodynamic sizes found for the lipoplexes at α = 0.5 than at α = 0.2 (see Table S1 in the Supporting Information). Finally, the pDNA–mixed lipid nanostructures seem to experience enhanced mobility at moderate lipid compositions, which may, in turn, favor the efficiency of the transfection process. This feature correlates well the slightly better TEs observed for lipoplexes at α = 0.5 compared to those at α = 0.2 (Figure 3).
Figure 7.
Topographic AFM images of the C6C22C6/DOPE SLB in the absence (t = 0 min) and presence of pDNA at ρeff = 4 and molar compositions of the cationic lipid in the mixed lipids of (a) α = 0.2 and (b) α = 0.5. The Z scale bar is 20 nm.
Conclusions
It can therefore be concluded that the cationic lipid C6C22C6 (with a hybrid structure between those of a gemini lipid and a BA lipid) mixed with DOPE and a plasmid DNA forms lipoplexes displaying a multilamellar nanostructure (Lα) with sizes in the range of 100–200 nm and low polydispersities, very suitable for its durability in the bloodstream (a key property for in vivo studies) and to cross the cell membrane (very important in transfection processes). The ability of the C6C22C6/DOPE nanosystem to compact, protect (from the degrading effect of DNase I), and transfect two DNA plasmids with different characteristics into COS-7 cells has been demonstrated, with an efficiency equal or even superior to that exhibited by the universal control Lipo2000*, as long as the effective charge ratio ρeff is maintained low. In addition, under these conditions, the viability of COS-7 cells was maintained at levels greater than 80%. Slightly better transfection performances were obtained at moderate GBA lipid contents in the lipid mixture, correlated with a better protection of the plasmid from the degrading action of the DNase I enzyme and the enhanced mobility of the pDNA–mixed lipid nanostructures at α = 0.5. In conclusion, C6C22C6/DOPE–pDNA lipoplexes are optimal candidates for nonviral gene nanocarriers because of their low toxicity and high biological activity in vitro, being therefore potentially interesting candidates for in vivo experiments.
Experimental Section
Materials
1,22-Bis(hexyl dimethyl ammonium)docosane (C6C22C6) was synthesized from the corresponding α,α′-dibromide. 1,22-Dibromodocosane was prepared by Kolbe electrolysis of 11-bromoundecanoic acid in methanol; 1,22-dibromodocosane (1.0 g) and anhydrous N,N-dimethyl hexyl amine (0.83 mL) in 50 mL of acetone were boiled under reflux for 96 h. The material obtained after removal of the solvent with a rotary evaporator was crystallized from ethanol–ether. The obtained crystals were then dried in a vacuum desiccator at ambient temperature to give the desired product (25% yield). 1H NMR (300 MHz, D2O, 25 °C): δ = 3.28 (m, 8H), δ = 3.11 (s, 12H), δ = 1.69 (brs, 8H), δ = 1.43–1.23 (m, 48H) and δ = 0.92 (m, 6H). MS (ESI): m/z calculated for [6-22-62+]2+, 283.83; found, 283.829; calculated for [6-22-62+ Br–]+, 645.57; found, 645.579. The zwitterionic lipid DOPE was purchased with highest purity from Avanti Polar Lipids, Inc., Alabaster, USA. The sodium salt of calf thymus DNA (ctDNA), as provided by Sigma-Aldrich (St. Louis, USA), was used as linear DNA to determine the effective charge of the GBA cationic vector (C6C22C6). The pEGFP-C3 plasmid DNA (4700 bp) encoding the GFP was extracted from competent Escherichia coli bacteria previously transformed with pEGFP-C3. The extraction was carried out using a GenElute HP Select Plasmid Gigaprep kit (Sigma-Aldrich). The pCMV-Luc VR1216 plasmid DNA (6934 bp) encoding luciferase (Clontech, Palo Alto, USA) was amplified in E. coli and isolated and purified using a QIAGEN Plasmid Giga kit (QIAGEN GMBH, Hilden, Germany). All reagents and solvents were of the highest grade commercially available and used without further purification.
Preparation of Lipoplexes
Appropriate amounts of C6C22C6 and DOPE were dissolved in chloroform to obtain the desired molar compositions of the cationic lipid (α) in the lipid mixtures. After briefly vortexing the corresponding solutions, the solvent was removed to yield dry lipid films. The resulting dry lipid films were then hydrated with 40 mM HEPES (pH 7.4) and homogenized by means of a combination of vortexing and sonication. By a sequential extrusion procedure explained in detail elsewhere,45,51 the multilamellar liposomes were transformed into the desired unilamellar ones. Appropriate amounts of a stock solution of pDNA prepared the day before were added to lipid suspensions to obtain the lipoplexes. pDNA concentrations were chosen to fit the optimum conditions for each experimental technique as follows: 1 mg/mL for zeta potential, 1 mg/mL for cryo-TEM, 200 μg/capillary (∼5 mg/mL) for SAXS, and 1 μg/well (2 μg/mL) for biological studies.
Zeta Potential and Particle Size
The phase analysis light scattering technique (Zeta PALS, Brookhaven Instruments Corp., Holtsville, USA) was employed to measure the electrophoretic mobility, which was then used to obtain the zeta potential of the nanoaggregates. The particle size was determined by a DLS method using a particle analyzer (Zeta Nano Series; Malvern Instruments, Barcelona, Spain). In both studies, samples were prepared with 40 mM HEPES buffer (pH 7.4) under experimental conditions of 25 °C, dispersant refractive index of 1.33 (water), viscosity of 0.9 cP, and dispersant dielectric constant of 78.5. Each zeta potential and particle size data point was taken as the average of 50 and 30 independent measurements, respectively. Measurements were carried out as a function of total lipid/DNA mass ratio ((mL+ + mL0)/mDNA) and at different molar fractions (α) of the cationic lipid in the GBA/DOPE mixed lipids.
Gel Electrophoresis
DNA Compaction Assay
Lipoplexes along with uncomplexed pDNA were loaded on 1% agarose gels (with 0.7 μL of GelRed) and run for 30 min at 80 mV in 1× TAE buffer. Fully complexed lipoplexes appeared as a fluorescence band in the wells of the gel, whereas uncomplexed pDNA appeared outside the wells. The fluorescence spectroscopy conditions were excitation at 302–312 nm and emission at 600 nm. The fluorescence intensity of each band was measured using the commercial Quantity One software and a Gel Doc XR instrument (Bio-Rad).
DNA Protection Assay
DNase I (1 U/μg pDNA) was added to each mixed lipid sample and stirred for 30 min at 37 °C. Then, 20 μL of 0.25 M EDTA was added to inactive DNase I, and the samples were incubated for 15 min. Next, 15 μL of 25% SDS was added and incubated for 5 min. Samples were electrophoresed for 40 min under 80 mV in 1% agarose gel with 1 μL of EtBr. The fluorescence spectroscopy conditions were excitation at 482 nm and emission at 616 nm. The integrity of the plasmid at each composition was compared with that of the untreated DNA as a control.
Small-Angle X-Ray Scattering
SAXS experiments were carried out on a beamline NCD11 at the ALBA Synchrotron (Barcelona, Spain). The energy of the incident beam was 12.6 keV (λ = 0.995 Å). Samples were placed in sealed glass capillaries. The scattered X-rays were detected on a Quantum 210r CCD detector, converted to one-dimensional scattering by radial averaging, and represented as a function of momentum transfer vector. SAXS experiments were performed for C6C22C6/DOPE–pDNA lipoplexes at an effective charge ratio (GBA lipid/DNA) of ρeff = 4 and at several cationic lipid compositions of the mixed lipids, α = 0.2, 0.4, 0.5, and 0.7. Measurements for each composition were run in duplicate in two independent capillaries.
Cryo-TEM
Cryo-TEM experiments were carried out following the standard procedure.53−55 In these experiments, perforated Quantifoil R1.2/1.3 (a hole diameter of 1.2 μm) on a 400-mesh copper grid was used. The images were obtained using a JEOL JEM 2011 cryo-electron microscope operated at 200 kV under low-dose conditions and using different degrees of defocus (500–700 nm) to obtain the adequate phase contrast. Images were recorded with a Gatan 794 Multiscan digital camera. Finally, the CCD images were processed and analyzed with DigitalMicrograph. Cryo-TEM experiments were performed with C6C22C6/DOPE–pDNA lipoplexes at ρeff = 4 and at two cationic lipid compositions in the mixed lipid, α = 0.2 and 0.5.
Atomic Force Microscopy
Supported planar bilayers for AFM experiments were prepared as follows: 60 μL of a mixed lipid sample with the desired composition in 40 mM HEPES (pH 7.4) was incubated at 37 °C for 45 min on a freshly cleaved mica surface to ensure liposome extension and subsequent formation of an SLB. After this period, the sample was gently rinsed with buffer to eliminate all nonadsorbed mixed lipids and mounted immediately on the top of the AFM scanner. A minimum of 30 min was required to stabilize the sample in the liquid to minimize any drift effects during scanning. AFM intermittent contact mode images in liquid at 24.0 ± 0.2 °C were obtained using a commercial NanoScope IV MultiMode microscope (Bruker, AXS Corporation, Madison, USA) with MSNL-10 silicon tips with a nominal spring constant of 30 pN/nm. The individual spring constants of the cantilevers were measured using the thermal tune method, yielding values similar to the nominal spring constant value. During scanning, the vertical force was maintained at the minimum value, maximizing the amplitude set point value while keeping the vibration amplitude as low as possible. Images were scanned at a scan angle of 0° and a scan rate of 1.5 Hz. After injection of a pDNA solution (0.1 μg/μL) and scanning of the first image, the tip was retracted from the surface of the sample until the next image was obtained to exclude side effects due to the proximity of the tip to the SLB. All images were processed by the NanoScope Analysis software (Bruker AXS Corporation, Santa Barbara, USA).
Cell Culture
COS-7 (African green monkey kidney) cells (American Type Collection, Rockville, USA) were maintained at 37 °C under 5% CO2 in a complete medium constituted by Dulbecco’s modified Eagle medium—high glucose + GlutaMAX (Gibco BRL Life Technologies) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (FBS), penicillin (100 U/mL), and streptomycin (100 μg/mL).
In Vitro TE
Two methods were used to evaluate the TE: luminometry for the pDNA encoding luciferase and FACS for the pDNA encoding the GFP. In both methods, each measurement was carried out in triplicate in three wells from three independent cultures. Lipofectamine (Lipo2000*) was used as the positive control (1.5 μL of Lipo2000*/μg of DNA).
Luminometry
Cells were seeded in a complete medium in 48-well plates and incubated for 24 h at 37 °C in 5% CO2. The medium was then removed, and 0.3 mL of the same medium and 0.2 mL of the complexes were added to each well. After 4 h of incubation, the medium was replaced by a complete medium and the cells were further incubated for 48 h in a medium containing 10% FBS. The cells were washed with phosphate-buffered saline (PBS) and lysate with 100 μL of reporter lysis buffer (Promega, Madison, USA) at room temperature for 10 min, followed by two freeze–thaw cycles. The lysate cells were centrifuged for 2 min at 12 000g to pellet the debris. Then, 20 μL of the supernatant was assayed for total luciferase activity using the luciferase assay reagent (Promega). A luminometer (Sirius-2, Berthold Detection Systems, Innogenetics, Diagnóstica y Terapéutica, Barcelona, Spain) was used to measure the luciferase activity. The protein content of the lysates was measured by the DC protein assay reagent (Bio-Rad, Hercules, CA, USA) using bovine serum albumin as the standard. Data are expressed as nanograms of luciferase per milligrams of protein.
Fluorescence-Assisted Cell Sorting
Cells were seeded in a medium in 48-well plates and incubated for 24 h at 37 °C in 5% CO2. The medium was removed, and 200 μL of trypsin (1×) was added to each well. After 2 min of incubation at 37 °C, 400 μL of complete medium was added to neutralize the trypsin. The samples were transferred to cytometer tubes and centrifuged for 5 min at 1450 rpm and 4 °C. Finally, the pellets were resuspended in 500 μL of a buffer containing PBS with 0.5% BSA and 2.5 mM EDTA. Then, cells were sorted using a flow cytometer. FACS analysis was performed using a Calibur 345 cytometer equipped with a 488 nm laser and the BD CellQuest Pro software. The cells were first gated using a forward scatter versus side scatter strategy to exclude any debris (low events) and then specifically analyzed by their 530 nm emission (FL1-H channel; the axis FL1-H shows the relative intensity of the GFP fluorescence). The data were analyzed using the FlowJo LLC data software. TEs were quantified by means of the percentage of GFP cells, that is, the percentage of cells in which GFP expression was observed, and the average intensity of fluorescence per cell (MFI).
Cell Viability
The cell viability was quantified by a modified alamarBlue assay. Briefly, 1 mL of 10% (v/v) alamarBlue dye in Dulbecco’s modified Eagle medium, supplemented with 10% (v/v) FBS medium, was added to each well 48 h after transfection. After 2 h of incubation at 37 °C, 200 μL of the supernatant was assayed by measuring the absorbance at 570 and 600 nm. Wells containing medium and alamarBlue dye without cells were used as blanks. The cell viability was calculated according to the following formula: (A570 – A600)treated cells × 100/(A570 – A600)control cells. Each sample was measured in three independent wells, and Lipo2000* was used as the positive control (1.5 μL of Lipo2000*/μg of DNA).
Acknowledgments
Financial support from the Ministerio de Economia y Competitividad of Spain (projects CTQ2012-30821, CTQ2015-65972-R, CTQ2015-64425-C2-2-R, and CTQ2014-55208-P), Madrid Regional Government (S2013/MIT-2807), Xunta de Galicia (GR 2007/085; IN607C 2016/03 and Centro Singular de Investigación de Galicia accreditation 2016–2019, ED431G/09), the European Regional Development Fund (ERDF), and Universidad Complutense de Madrid, Spain (project UCMA05-33-010), is gratefully acknowledged. SAXS experiments were performed on the NCD11 beamline at the ALBA Synchrotron Light Facility with the collaboration of the ALBA staff. Cryo-TEM experiments were performed at the Servei de Microscopia of Univ. Autónoma of Barcelona (Spain). A.G.-M. acknowledges receipt of a Ramón y Cajal Fellowship from the Spanish MINECO. The authors also thank C. Aicart-Ramos for performing the plasmid DNA amplification at the Departamento de Bioquímica y Biología Molecular I (Universidad Complutense de Madrid, Spain).
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsomega.7b01657.
DLS data (hydrodynamic diameters, Dh, and polydispersities, PDI) of C6C22C6/DOPE–pDNA lipoplexes, 1H NMR spectrum (300 MHz, D2O, 25 °C), electrospray ionization mass spectrum, and data of the electrospray ionization mass spectrum of C6C22C6 (PDF)
Author Contributions
The manuscript was written through the contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Junquera E.; Aicart E. Recent progress in gene therapy to deliver nucleic acids with multivalent cationic vectors. Adv. Colloid Interface Sci. 2016, 233, 161–175. 10.1016/j.cis.2015.07.003. [DOI] [PubMed] [Google Scholar]
- Verma I. M.; Weitzman M. D. Gene therapy: twenty-first century medicine. Annu. Rev. Biochem. 2005, 74, 711–738. 10.1146/annurev.biochem.74.050304.091637. [DOI] [PubMed] [Google Scholar]
- Khakshoor O.; Kool E. T. Chemistry of nucleic acids: impacts in multiple fields. Chem. Commun. 2011, 47, 7018–7024. 10.1039/c1cc11021g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalinska J. B.; Gewirtz A. M. Nucleic-acid therapeutics: basic principles and recent applications. Nat. Rev. Drug Discovery 2002, 1, 503. 10.1038/nrd837. [DOI] [PubMed] [Google Scholar]
- Dias R. S.; Lindman B.. DNA Interaction with Polymers and Surfactants; Wiley & Sons: Hoboken, NJ, 2008. [Google Scholar]
- Junquera E.; Aicart E. Cationic lipids as transfecting agents of DNA in gene therapy. Curr. Top. Med. Chem. 2014, 14, 649–663. 10.2174/1568026614666140118203128. [DOI] [PubMed] [Google Scholar]
- Islam M. A.; Park T.-E.; Singh B.; Maharjan S.; Firdous J.; Cho M.-H.; Kang S.-K.; Yun C.-H.; Choi Y.-J.; Cho C.-S. Major degradable polycations as carriers for DNA and siRNA. J. Controlled Release 2014, 193, 74–89. 10.1016/j.jconrel.2014.05.055. [DOI] [PubMed] [Google Scholar]
- Devi G. R. siRNA-based approaches in cancer therapy. Cancer Gene Ther. 2006, 13, 819–829. 10.1038/sj.cgt.7700931. [DOI] [PubMed] [Google Scholar]
- Kesharwani P.; Gajbhiye V.; Jain N. K. A review of nanocarriers for the delivery of small interfering RNA. Biomaterials 2012, 33, 7138–7150. 10.1016/j.biomaterials.2012.06.068. [DOI] [PubMed] [Google Scholar]
- Khurana B.; Goyal A. K.; Budhiraja A.; Arora D.; Vyas S. P. siRNA delivery using nanocarriers—an efficient tool for gene silencing. Curr. Gene Ther. 2010, 10, 139–155. 10.2174/156652310791111010. [DOI] [PubMed] [Google Scholar]
- Dominska M.; Dykxhoorn D. M. Breaking down the barriers: siRNA delivery and endosome escape. J. Cell Sci. 2010, 123, 1183–1189. 10.1242/jcs.066399. [DOI] [PubMed] [Google Scholar]
- Zuhorn I. S.; Engberts J. B. F. N.; Hoekstra D. Gene delivery by cationic lipid vectors: overcoming cellular barriers. Eur. Biophys. J. 2007, 36, 349–362. 10.1007/s00249-006-0092-4. [DOI] [PubMed] [Google Scholar]
- Sakurai H.; Kawabata K.; Sakurai F.; Nakagawa S.; Mizuguchi H. Innate immune response induced by gene delivery vectors. Int. J. Pharm. 2008, 354, 9–15. 10.1016/j.ijpharm.2007.06.012. [DOI] [PubMed] [Google Scholar]
- Foldvari M.; Badea I.; Wettig S.; Verrall R.; Bagonluri M. Structural characterization of novel gemini non-viral DNA delivery systems for cutaneous gene therapy. J. Exp. Nanosci. 2006, 1, 165–176. 10.1080/17458080500411965. [DOI] [Google Scholar]
- Verma I. M.; Somia N. Gene therapy—promises, problems and prospects. Nature 1997, 389, 239–242. 10.1038/38410. [DOI] [PubMed] [Google Scholar]
- Yin H.; Kanasty R. L.; Eltoukhy A. A.; Vegas A. J.; Dorkin J. R.; Anderson D. G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- Mintzer M. A.; Simanek E. E. Nonviral vectors for gene delivery. Chem. Rev. 2008, 109, 259–302. 10.1021/cr800409e. [DOI] [PubMed] [Google Scholar]
- Martínez-Negro M.; Kumar K.; Barrán-Berdón A. L.; Datta S.; Kondaiah P.; Junquera E.; Bhattacharya S.; Aicart E. Efficient cellular knockdown mediated by siRNA nanovectors of gemini cationic lipids having delocalizable headgroups and oligo-oxyethylene spacers. ACS Appl. Mater. Interfaces 2016, 8, 22113–22126. 10.1021/acsami.6b08823. [DOI] [PubMed] [Google Scholar]
- Angelov B.; Angelova A.; Filippov S. K.; Karlsson G.; Terrill N.; Lesieur S.; Štěpánek P. Topology and internal structure of PEGylated lipid nanocarriers for neuronal transfection: synchrotron radiation SAXS and cryo-TEM studies. Soft Matter 2011, 7, 9714–9720. 10.1039/c1sm06447a. [DOI] [Google Scholar]
- Angelov B.; Angelova A.; Filippov S. K.; Narayanan T.; Drechsler M.; Štěpánek P.; Couvreur P.; Lesieur S. DNA/Fusogenic Lipid Nanocarrier Assembly: Millisecond Structural Dynamics. J. Phys. Chem. Lett. 2013, 4, 1959–1964. 10.1021/jz400857z. [DOI] [PubMed] [Google Scholar]
- Angelova A.; Garamus V. M.; Angelov B.; Tian Z.; Li Y.; Zou A. Advances in structural design of lipid-based nanoparticle carriers for delivery of macromolecular drugs, phytochemicals and anti-tumor agents. Adv. Colloid Interface Sci. 2017, 249, 331. 10.1016/j.cis.2017.04.006. [DOI] [PubMed] [Google Scholar]
- Niculescu-Duvaz D.; Heyes J.; Springer C. Structure-activity relationship in cationic lipid mediated gene transfection. Curr. Med. Chem. 2003, 10, 1233–1261. 10.2174/0929867033457476. [DOI] [PubMed] [Google Scholar]
- Ahmed T.; Kamel A. O.; Wettig S. D. Interactions between DNA and Gemini surfactant: impact on gene therapy: part I. Nanomedicine 2016, 11, 289–306. 10.2217/nnm.15.203. [DOI] [PubMed] [Google Scholar]
- Zhi D.; Zhang S.; Cui S.; Zhao Y.; Wang Y.; Zhao D. The headgroup evolution of cationic lipids for gene delivery. Bioconjugate Chem. 2013, 24, 487–519. 10.1021/bc300381s. [DOI] [PubMed] [Google Scholar]
- Meister A.; Blume A. Self-assembly of bipolar amphiphiles. Curr. Opin. Colloid Interface Sci. 2007, 12, 138–147. 10.1016/j.cocis.2007.05.003. [DOI] [Google Scholar]
- Mamusa M.; Resta C.; Barbero F.; Carta D.; Codoni D.; Hatzixanthis K.; McArthur M.; Berti D. Interaction between a cationic bolaamphiphile and DNA: The route towards nanovectors for oligonucleotide antimicrobials. Colloids Surf., B 2016, 143, 139–147. 10.1016/j.colsurfb.2016.03.031. [DOI] [PubMed] [Google Scholar]
- Fuhrhop J. H.; Fritsch D. Bolaamphiphiles form ultrathin, porous and unsymmetric monolayer lipid membranes. Acc. Chem. Res. 1986, 19, 130–137. 10.1021/ar00125a002. [DOI] [Google Scholar]
- Fuhrhop J. H.; Wang T. Bolaamphiphiles. Chem. Rev. 2004, 104, 2901–2938. 10.1021/cr030602b. [DOI] [PubMed] [Google Scholar]
- Mao G.; Tsao Y.-H.; Tirrell M.; Davis H. T.; Hessel V.; van Esch J.; Ringsdorf H. Monolayers of bolaform amphiphiles: Influence of alkyl chain length and counterions. Langmuir 1994, 10, 4174–4184. 10.1021/la00023a044. [DOI] [Google Scholar]
- Philosof-Mazor L.; Dakwar G. R.; Popov M.; Kolusheva S.; Shames A.; Linder C.; Greenberg S.; Heldman E.; Stepensky D.; Jelinek R. Bolaamphiphilic vesicles encapsulating iron oxide nanoparticles: New vehicles for magnetically targeted drug delivery. Int. J. Pharm. 2013, 450, 241–249. 10.1016/j.ijpharm.2013.04.017. [DOI] [PubMed] [Google Scholar]
- Popov M.; Hammad I. A.; Bachar T.; Grinberg S.; Linder C.; Stepensky D.; Heldman E. Delivery of analgesic peptides to the brain by nano-sized bolaamphiphilic vesicles made of monolayer membranes. Eur. J. Pharm. Biopharm. 2013, 85, 381–389. 10.1016/j.ejpb.2013.06.005. [DOI] [PubMed] [Google Scholar]
- Hwang S. R.; Kim K. Nano-enabled delivery systems across the blood–brain barrier. Arch. Pharmacal Res. 2014, 37, 24–30. 10.1007/s12272-013-0272-6. [DOI] [PubMed] [Google Scholar]
- Fariya M.; Jain A.; Dhawan V.; Shah S.; Nagarsenker M. S. Bolaamphiphiles: a pharmaceutical review. Adv. Pharm. Bull. 2014, 4, 483–491. 10.5681/apb.2014.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zana R.Bolaform and dimeric (gemini) surfactants. Specialist Surfactants; Springer, 1997; pp 81–103. [Google Scholar]
- Akhtar S.; Benter I. Toxicogenomics of non-viral drug delivery systems for RNAi: Potential impact on siRNA-mediated gene silencing activity and specificity. Adv. Drug Delivery Rev. 2007, 59, 164–182. 10.1016/j.addr.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Denoyelle S.; Polidori A.; Brunelle M.; Vuillaume P. Y.; Laurent S.; ElAzhary Y.; Pucci B. Synthesis and preliminary biological studies of hemifluorinated bifunctional bolaamphiphiles designed for gene delivery. New J. Chem. 2006, 30, 629–646. 10.1039/b513944a. [DOI] [Google Scholar]
- Jain N.; Arntz Y.; Goldschmidt V.; Duportail G.; Mély Y.; Klymchenko A. S. New unsymmetrical bolaamphiphiles: Synthesis, assembly with DNA, and application for gene delivery. Bioconjugate Chem. 2010, 21, 2110–2118. 10.1021/bc100334t. [DOI] [PubMed] [Google Scholar]
- Kim T.; Afonin K. A.; Viard M.; Koyfman A. Y.; Sparks S.; Heldman E.; Grinberg S.; Linder C.; Blumenthal R. P.; Shapiro B. A. In silico, in vitro, and in vivo studies indicate the potential use of bolaamphiphiles for therapeutic siRNAs delivery. Mol. Ther.--Nucleic Acidss 2013, 2, e80 10.1038/mtna.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao D. D.; Vorhies J. S.; Senzer N.; Nemunaitis J. siRNA vs. shRNA: Similarities and differences. Adv. Drug Delivery Rev. 2009, 61, 746–759. 10.1016/j.addr.2009.04.004. [DOI] [PubMed] [Google Scholar]
- Weissig V.; Torchilin V. P. Cationic bolasomes with delocalized charge centers as mitochondria-specific DNA delivery systems. Adv. Drug Delivery Rev. 2001, 49, 127–149. 10.1016/s0169-409x(01)00131-4. [DOI] [PubMed] [Google Scholar]
- Stern A.; Guidotti M.; Shaubi E.; Popov M.; Linder C.; Heldman E.; Grinberg S. Steric environment around acetylcholine head groups of bolaamphiphilic nanovesicles influences the release rate of encapsulated compounds. Int. J. Nanomed. 2014, 9, 561–574. 10.2147/IJN.S53563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson J.; Arvidson G.; Karlsson G.; Almgren M. Complexes between cationic liposomes and DNA visualized by cryo-TEM. Biochim. Biophys. Acta 1995, 1235, 305–312. 10.1016/0005-2736(95)80018-b. [DOI] [PubMed] [Google Scholar]
- Lasic D. D.; Strey H.; Stuart M. C. A.; Podgornik R.; Frederik P. M. The structure of DNA–liposome complexes. J. Am. Chem. Soc. 1997, 119, 832–833. 10.1021/ja962713g. [DOI] [Google Scholar]
- Feitosa E.; Alves F. R.; Niemiec A.; Oliveira M. E. C. D. R.; Castanheira E. M. S.; Baptista A. L. F. Cationic liposomes in mixed didodecyldimethylammonium bromide and dioctadecyldimethylammonium bromide aqueous dispersions studied by differential scanning calorimetry, nile red fluorescence, and turbidity. Langmuir 2006, 22, 3579–3585. 10.1021/la053238f. [DOI] [PubMed] [Google Scholar]
- Misra S. K.; Muñoz-Úbeda M.; Datta S.; Barrán-Berdón A. L.; Aicart-Ramos C.; Castro-Hartmann P.; Kondaiah P.; Junquera E.; Bhattacharya S.; Aicart E. Effects of a delocalizable cation on the headgroup of gemini lipids on the lipoplex-type nanoaggregates directly formed from plasmid DNA. Biomacromolecules 2013, 14, 3951–3963. 10.1021/bm401079h. [DOI] [PubMed] [Google Scholar]
- Muñoz-Úbeda M.; Misra S. K.; Barrán-Berdón A. L.; Aicart-Ramos C.; Sierra M. B.; Biswas J.; Kondaiah P.; Junquera E.; Bhattacharya S.; Aicart E. Why is less cationic lipid required to prepare lipoplexes from plasmid DNA than linear DNA in gene therapy?. J. Am. Chem. Soc. 2011, 133, 18014–18017. 10.1021/ja204693f. [DOI] [PubMed] [Google Scholar]
- Barrán-Berdón A. L.; Misra S. K.; Datta S.; Muñoz-Úbeda M.; Kondaiah P.; Junquera E.; Bhattacharya S.; Aicart E. Cationic gemini lipids containing polyoxyethylene spacers as improved transfecting agents of plasmid DNA in cancer cells. J. Mater. Chem. B 2014, 2, 4640–4652. 10.1039/c4tb00389f. [DOI] [PubMed] [Google Scholar]
- Kumar K.; Barrán-Berdón A. L.; Datta S.; Muñoz-Úbeda M.; Aicart-Ramos C.; Kondaiah P.; Junquera E.; Bhattacharya S.; Aicart E. A delocalizable cationic headgroup together with an oligo-oxyethylene spacer in gemini cationic lipids improves their biological activity as vectors of plasmid DNA. J. Mater. Chem. B 2015, 3, 1495–1506. 10.1039/c4tb01948b. [DOI] [PubMed] [Google Scholar]
- Barrán-Berdón A. L.; Martínez-Negro M.; García-Río L.; Domènech O.; de Ilarduya C. T.; Aicart E.; Junquera E. A biophysical study of gene nanocarriers formed by anionic/zwitterionic mixed lipids and pillar[5]arene polycationic macrocycles. J. Mater. Chem. B 2017, 5, 3122–3131. 10.1039/c6tb02939f. [DOI] [PubMed] [Google Scholar]
- Blanco J. L. J.; Ortega-Caballero F.; Blanco-Fernández L.; Carmona T.; Marcelo G.; Martínez-Negro M.; Aicart E.; Junquera E.; Mendicuti F.; de Ilarduya C. T.; Mellet C. O.; Fernández J. M. G. Trehalose-based Janus cyclooligosaccharides: the “click” synthesis and DNA-directed assembly into pH-sensitive transfectious nanoparticles. Chem. Commun. 2016, 52, 10117–10120. 10.1039/c6cc04791b. [DOI] [PubMed] [Google Scholar]
- Muñoz-Úbeda M.; Misra S. K.; Barrán-Berdón A. L.; Datta S.; Aicart-Ramos C.; Castro-Hartmann P.; Kondaiah P.; Junquera E.; Bhattacharya S.; Aicart E. How does the spacer length of cationic gemini lipids influence the lipoplex formation with plasmid DNA? Physicochemical and biochemical characterizations and their relevance in gene therapy. Biomacromolecules 2012, 13, 3926–3937. 10.1021/bm301066w. [DOI] [PubMed] [Google Scholar]
- Tanford C.The Hydrophobic Effect: Formation of Micelles and Biological Membranes; Wiley & Sons: New York, 1980. [Google Scholar]
- Bednar J.; Woodcock C. L.. Chromatin; Academic Press Inc: San Diego, CA, 1999; Vol. 304, pp 191–213. [Google Scholar]
- Dubochet J.; Adrian M.; Chang J.-J.; Homo J.-C.; Lepault J.; McDowall A. W.; Schultz P. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 1988, 21, 129–228. 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- Dubochet J.; Zuber B.; Eltsov M.; Bouchet-Marquis C.; Al-Amoudi A.; Livolant F. How to “read” a vitreous section. Methods Cell Biol. 2007, 79, 385–406. 10.1016/s0091-679x(06)79015-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.