Abstract
Once relegated to the supply of energy and biosynthetic precursors, it has now become clear that metabolism is a specific mediator of nearly all physiologic processes. In the context of microbial pathogenesis, metabolism has expanded outside its canonical role in bacterial replication. Among human pathogens, this expansion has emerged perhaps nowhere more visibly than for Mycobacterium tuberculosis, the causative agent of tuberculosis. Unlike most pathogens, M. tuberculosis has evolved within humans, which are both host and reservoir. This makes unrestrained replication and perpetual quiescence equally incompatible strategies for survival as a species. In this Review, we summarize recent work that illustrates the diversity of metabolic functions that not only enable M. tuberculosis to establish and maintain a state of chronic infection within the host, but also facilitate survival in the face of drug pressure and, ultimately, completion of its life cycle.
INTRODUCTION
In the Lewis Carroll novel, Through the Looking Glass, the White Queen reminds Alice that “it’s a poor sort of memory that only works backward.” Nature similarly reminds scientists that it’s a poor sort of science that remains rooted in outdated concepts, no matter how well established1. For microbiologists, this lesson has proven especially invaluable in our understanding of pathogenicity. Early work viewed pathogenicity as little more than unrestricted growth of a microorganism in the host. However, growing evidence made it difficult to overlook the host as an essential and specific contributor to this process, resulting in a more refined view of pathogenicity as a complex, if not conditional, trait. Complicating this view even further was the discovery that pathogen virulence and replication can be dissociated from one another.
It should thus come as no surprise that our understanding of microbial pathogenicity is in the midst of yet another revision. Perhaps more surprising however is the nature of this revision. Long recognized for its broadly conserved composition and function, metabolism has recently emerged as an equally specific mediator of diverse biological phenotypes. Growing evidence has demonstrated that, despite consisting of a broadly conserved set of enzymes and reactions, metabolic networks have evolved to serve the specific biochemical needs of their hosts through a diverse array of configurations. For microorganisms, this diversity has begun to reveal specific and essential roles for metabolism apart from replication. Among pathogens, such roles have emerged perhaps most clearly for Mycobacterium tuberculosis.
M. tuberculosis is the causative agent of tuberculosis (TB) and leading cause of deaths due to an infectious disease, leading cause of deaths due to a transmissible disease, and leading cause of deaths due to drug resistance. DNA evidence indicates that M. tuberculosis has co-evolved with Homo sapiens as a species2. Within humans, M. tuberculosis resides mainly inside and amidst cells of the immune system that can contain, or eradicate, most other bacteria. Yet, humans serve as the only known natural host and reservoir of M. tuberculosis. M. tuberculosis has thus adapted its physiology to serve interdependent cell physiologic and pathogenic functions.
M. tuberculosis operates an erratic, though highly repetitive, life cycle (Box 1) that spans a diverse and heterogeneous range of niches and physiologic states, many of which are distinct from other pathogens. Given this unusual, if not unique, ecology, it is not entirely surprising to discover that M. tuberculosis has evolved an equally distinct set of metabolic capabilities to help facilitate its adaptation to and transit between hosts.
BOX 1. TB PATHOGENESIS: THE MYCOBACTERIAL CIRCLE OF LIFE.
Transmitted by aerosol, current WHO estimates indicate that Mycobacterium tuberculosis infects approximately a third of the global population108. Following infection, M. tuberculosis often enters a prolonged state of asymptomatic, latent infection that can last decades, if not the remaining lifetime of the host. In a minority of hosts, this latent infection reactivates to cause clinically symptomatic and contagious (active) disease109. Among immunocompetent individuals, this lifetime risk is estimated to be approximately 10%, but in immunocompromised hosts can be as high as 10% per year. In both immunocompetent and immunocompromised hosts however, latent infection is believed to harbor non- or slowly replicating populations of M. tuberculosis that serve as a reservoir for future disease, transmission and treatment relapse.
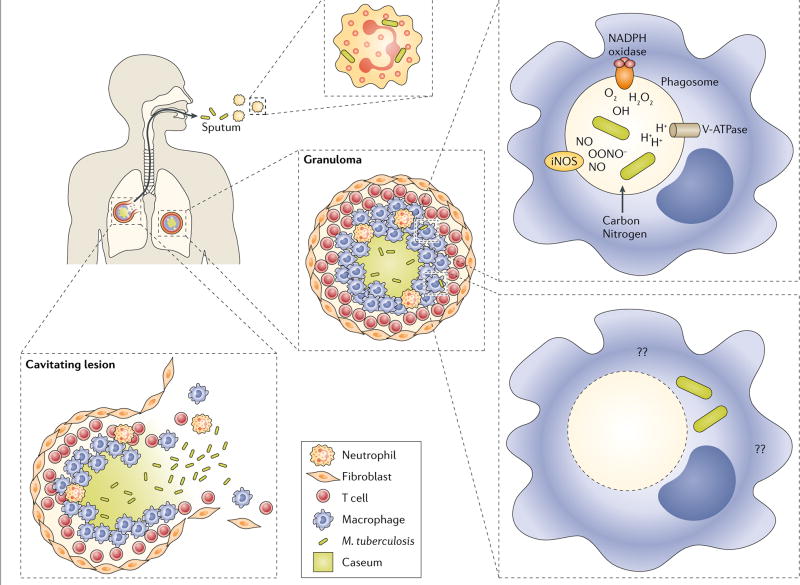
Following transmission to a new host via aerosol, M. tuberculosis is believed to first undergo phagocytosis in the lung by tissue resident alveolar macrophages and dendritic cells110M. tuberculosis is then thought to undergo a brief period of unrestricted intracellular replication, during which infected cells migrate to local draining lymph nodes. After reaching the regional lymph node, M. tuberculosis is believed to circulate through the bloodstream, where it encounters and infects additional host cells, ultimately re-seeding additional regions of the lung. With the onset of cellular immunity, local pro-inflammatory responses lead to the recruitment of additional monocytes and lymphocytes110, which, in turn, assemble around infected macrophages to contain M. tuberculosis within an organized cellular structure, known as a granuloma (figure). The granuloma is the pathological hallmark of tuberculosis (TB) and site in which M. tuberculosis is believed to persist in a prolonged state of slowed or arrested replication111–113.
Within the host, M. tuberculosis can infect numerous immune and non-immune cells. However, its primary host cell is the macrophage. Within macrophages, M. tuberculosis resides primarily inside phagosomes, apart from other microorganisms, where it can encounter a complex and dynamic range of host defenses, including acidification, reactive oxygen and nitrogen intermediates and antimicrobial peptides114,115. Interestingly, recent work has shown that M. tuberculosis can also escape to the cytosol where it likely encounters additional, and yet-to-be identified, environmental stringencies116,117. Irrespective of locale however, it is clear that M. tuberculosis has evolved in the face of diverse biochemical stringencies, many of which are capable of suppressing, but not eradicating, it.
M. tuberculosis is often contained within granulomas for extended intervals of time, often lasting decades. However, granuloma integrity can wane, resulting in cellular necrosis and
 that exposes the bacteria to a lipid rich, hypoxic extracellular environment118–120. Further loss of granuloma integrity, mediated in part via upregulation of matrix metalloproteinases, provides access to vascular nutrients and oxygen, followed by the eventual rupture of granulomas into the airways, releasing thousands of viable bacteria119,121.
that exposes the bacteria to a lipid rich, hypoxic extracellular environment118–120. Further loss of granuloma integrity, mediated in part via upregulation of matrix metalloproteinases, provides access to vascular nutrients and oxygen, followed by the eventual rupture of granulomas into the airways, releasing thousands of viable bacteria119,121.
Once released into the airways, M. tuberculosis ultimately completes its life cycle by forming infectious aerosols that enable transmission to a new host (figure). In sputum, M. tuberculosis has been shown to reside within neutrophils and bacteriologic and transcriptional profiling studies have identified distinct subpopulations of non-replicating bacteria as well subpopulations that fail to form colonies on agar plates and can only be recovered in liquid media122–125.
Here, we review the basic physiology of M. tuberculosis in the context of both its natural life cycle and pathogenicity in the host. We consider the specific roles played by metabolism in each of these processes. By doing so, we aim to not only reframe our understanding of pathogenicity along a biochemical axis but conversely also expand our understanding of metabolism along a physiologic axis.
METABOLIC REGULATION
Like most bacteria, M. tuberculosis can utilize a wide range of carbon sources to support in vitro growth. In vivo however, M. tuberculosis resides within intra- and extracellular niches whose nutrient composition is believed to be sparse and restrictive for replication3–5. At the same time, M. tuberculosis can re-initiate growth after prolonged periods of replicative quiescence, enabling transmission to new hosts and survival as a species. M. tuberculosis has thus adapted to a distinct, and potentially competing, set of metabolic pressures.
Co-catabolism of carbon substrates
Given these needs, it is not surprising that, despite encoding many of the same enzymes and pathways as other bacteria, M. tuberculosis has evolved distinct metabolic regulatory mechanisms of growth and replication. One widely conserved regulatory mechanism used by many bacteria to achieve maximal growth is carbon catabolite repression (CCR). CCR enables the selective and sequential use of carbon sources according to growth efficiency, such that growth is driven by metabolism of the carbon source capable of promoting fastest growth6. Moreover, this mechanism may confer a growth advantage in competitive settings6. It is perhaps for these reasons that classical CCR appears absent in M. tuberculosis, which often resides in biochemically stringent niches apart from other microorganisms. M. tuberculosis has instead evolved the capacity to co-catabolize multiple carbon substrates, such that it is able to grow faster and more extensively on carbon source mixtures than the total equivalent amount of any single component. Moreover, metabolic labelling experiments revealed that M. tuberculosis not only co-catabolized glucose and acetate, but this occurred in a compartmentalized and segregated manner allowing carbon flux through glycolysis and gluconeogenesis simultaneously7.
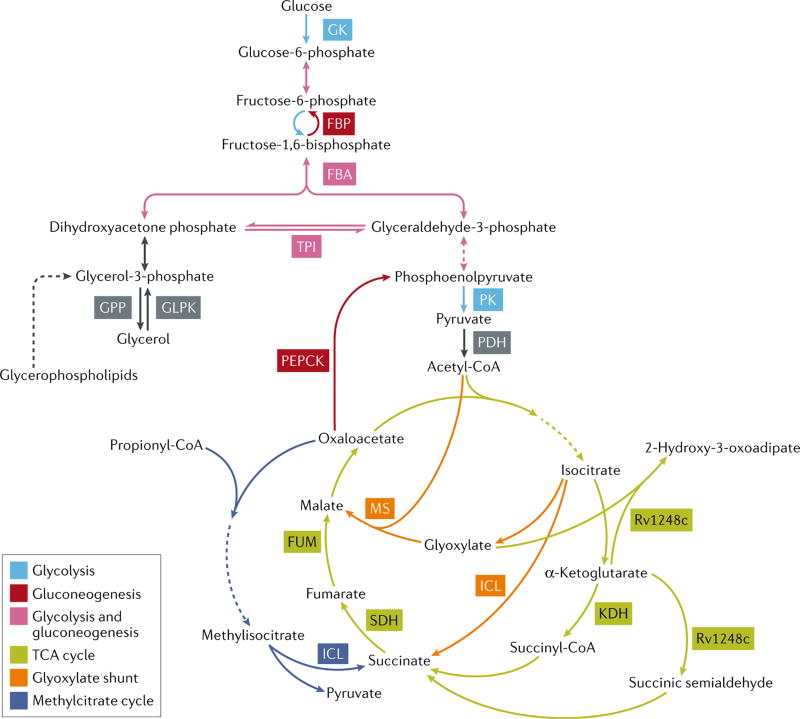
Genetic studies of M. tuberculosis mutants lacking enzymes that catalyze critical reactions in both glycolysis and gluconeogenesis, such as fructose-1,6-bisphophate aldolase (FBA) and triosephosphate isomerase (TPI)8,9 (Figure 1), have provided similar evidence for the existence of carbon co-catabolism. Genes encoding these enzymes could only be deleted from the M. tuberculosis genome when two carbon sources, a glycolytic and a gluconeogenic substrate, that enter metabolism at both sides of the interrupted metabolic step were provided in the culture medium.
Figure 1. Enzymes required for growth and/or persistence of M. tuberculosis.
Schematic representation of central carbon metabolism pathways and key enzymes in M. tuberculosis. Enzymatic steps dedicated to glycolysis are depicted in light blue, reversible steps of glycolysis/gluconeogenesis are depicted in pink, dedicated steps of gluconeogenesis are depicted in red, the TCA cycle is depicted in green, the glyoxylate shunt is depicted in orange, and the methylcitrate cycle is depicted in dark blue. Pathways of glycerol and glyerolphospholipid metabolism are depicted in grey. GK, glucokinase; GLPK, glycerol kinase; GPP, glycerol phosphate phosphatase; FBP, fructose bisphosphatase; FBA, fructose bisphosphate aldolase; TPI, triosephosphate isomerase; PK, pyruvate kinase; PDH, pyruvate dehydrogenase; KDH, ketoglutarate dehydrogenase; ICL, isocitrate lyase; SDH, succinate dehydrogenase; FUM, fumarase; MS, malate synthase; PEPCK, phosphoenolpyruvate caboxykinase.
Pyruvate kinase, which catalyzes the final and rate-limiting reaction in glycolysis, converting phosphoenolpyruvate to pyruvate10,11 (Figure 1) was conversely shown to be essential for co-catabolism of glucose and fatty acid carbon sources. Moreover, genetic deletion of pyruvate kinase in M. tuberculosis resulted in allosteric inhibition of the

 enzyme isocitrate dehydrogenase arising from accumulation of the pyruvate kinase substrate phosphoenolpyruvate, when cultured on a mixture of glucose and fatty acid12. This inhibition, in turn, reduced flux through the TCA cycle, and impaired the ability of M. tuberculosis to co-catabolize glucose and short chain fatty acids.
enzyme isocitrate dehydrogenase arising from accumulation of the pyruvate kinase substrate phosphoenolpyruvate, when cultured on a mixture of glucose and fatty acid12. This inhibition, in turn, reduced flux through the TCA cycle, and impaired the ability of M. tuberculosis to co-catabolize glucose and short chain fatty acids.
How and when M. tuberculosis segregates metabolism of individual carbon sources from one another and achieves their functional compartmentalization remain important but unanswered questions. Interestingly, both FBA and TPI mutants were found to be severely attenuated in a mouse model of TB8,9, suggesting that M. tuberculosis may not have access to a balanced diet of glucose and fatty acids in vivo. In vitro studies of the FBA mutant however demonstrated that growth on single carbon source could only be rescued over a specific range of carbon source ratios, suggesting the existence of important but unrecognized metabolic differences in model systems relevant to disease pathogenesis8. Accordingly, although the pyruvate kinase mutant was not attenuated in mice, in vitro studies had shown that the growth defect of pyruvate kinase-deficient M. tuberculosis on mixtures of glucose and fatty acids could be rescued by addition of glutamate12, a nutrient thought to be available to M. tuberculosis in a macrophage-like human cell line, mouse lungs and guinea pig granulomas13,14.
In addition to facilitating growth in nutrient limited environments, recent work using in silico modeling of metabolomics and transcriptomic data suggested that co-catabolism may also help buffer the metabolism of M. tuberculosis against perturbations by the immune system or chemotherapy15.
cAMP: classical regulator, alternative roles
While lacking classical CCR, recent work suggests that M. tuberculosis appears to have retained the capacity to regulate nutrient utilization in a cAMP-dependent manner. For example, a screen for small molecules capable of inhibiting intracellular growth identified several compounds that required the presence of cholesterol to inhibit growth of M. tuberculosis in liquid culture16. These compounds were subsequently found to increase intrabacterial levels of cAMP, a canonical effector of catabolite repression in E. coli 17. Moreover, it was found that the activity of these compounds could be overcome by the addition of acetate, suggesting that cAMP levels could regulate cholesterol uptake or metabolism. Evidence for the former has since emerged with the discovery of a gene, lipid uptake coordinator A (lucA), linking cholesterol and fatty acid import via stabilization of accessory subunits of both the Mce4 cholesterol and Mce1 fatty acid importer complexes18.
Metabolic recycling
In addition to optimizing utilization of multiple extracellular nutrients, M. tuberculosis can, when needed, coordinate this metabolism with utilization of its internal nutrient stores. For example, work by Larrouy-Maumus showed that M. tuberculosis can recycle the lipid polar heads from glycerophospholipds via a D,L-glycerol 3-phosphate phosphatase, generating inorganic phosphate and glycerol which serves as substrate of the glycerol kinase GlpK19 (Figure 1). While the physiological role of this recycling pathway remains to be identified, the importance of recycling has been demonstrated for other metabolites. M. tuberculosis has been shown to be capable of reclaiming trehalose released from turnover of its cell wall glycolipid trehalose monomycolate (TMM) and this process is required to establish infection in mice20. Moreover, exposure of M. tuberculosis to hypoxia, which is common in many the of intra- and extracellular niches of M. tuberculosis21, triggered a reduction in nutrient uptake and linked increase in trehalose mycolate catabolism to prepare M. tuberculosis for subsequent re-initiation of peptidoglycan biosynthesis and re-entry into cell cycle as discussed below22.
PRINCIPLES OF PERSISTENCE
Despite their traditional role as replicative ‘fuel’, metabolic enzymes have independently begun to emerge as distinct and essential mediators that enable M. tuberculosis to persist in the face of immune and drug induced pressure23–26. M. tuberculosis has evolved a life cycle in which its replication is actively slowed or arrested by host immunity, yet remains competent to resume and facilitate transmission to a new host after decades of containment27. This state of slowed or arrested replication further underlies persistence, which is a form of non-heritable antibiotic resistance or tolerance widely believed to mitigate the activity of most conventional antibiotics28 and a subject of extensive interest.
Nutritional homeostasis
Evidence for a specific role of metabolic enzymes in persistence first emerged from seminal studies by McKinney and colleagues29. These studies identified isocitrate lyase 1, one of two isocitrate lyases (ICLs) in M. tuberculosis, as dispensable for standard in vitro growth but selectively essential for survival in immune activated, rather than resting, macrophages, and maintenance, rather than establishment, of an infection in mice. Based on the canonical role of ICL in the
 (Figure 1), these phenotypes were interpreted to indicate that survival of persistent M. tuberculosis required the specific assimilation of even-chain fatty acid substrates into 2-carbon acetyl coenzyme A (CoA) units for
(Figure 1), these phenotypes were interpreted to indicate that survival of persistent M. tuberculosis required the specific assimilation of even-chain fatty acid substrates into 2-carbon acetyl coenzyme A (CoA) units for
 of TCA cycle and/or gluconeogenic intermediates.
of TCA cycle and/or gluconeogenic intermediates.
Studies of the
 functions of ICLs in M. tuberculosis later identified a similarly essential role for pckA, which encodes the gluconeogenic enzyme phosphoenolpyruvate carboxykinase (PEPCK)30. PEPCK was found to be dispensable for growth and survival in vitro, but essential for growth during acute infection. Moreover, transcriptional silencing of pckA during the chronic phase of infection resulted in clearance of M. tuberculosis from mouse lungs and spleens. These findings thus established that metabolic enzymes mediate specific and essential roles in persistence that are biologically dissociated from replication.
functions of ICLs in M. tuberculosis later identified a similarly essential role for pckA, which encodes the gluconeogenic enzyme phosphoenolpyruvate carboxykinase (PEPCK)30. PEPCK was found to be dispensable for growth and survival in vitro, but essential for growth during acute infection. Moreover, transcriptional silencing of pckA during the chronic phase of infection resulted in clearance of M. tuberculosis from mouse lungs and spleens. These findings thus established that metabolic enzymes mediate specific and essential roles in persistence that are biologically dissociated from replication.
In conjunction with the advent of genome-scale transposon mutagenesis and conditionally regulated genetic silencing technologies, an inventory of metabolic enzymes required for persistence in mice has since begun to emerge31–33. To date, this inventory has chiefly consisted of enzymes of central pathways also required for in vitro growth and/or establishment of a chronic infection in mice8,23,25,30,34–40. Nevertheless, the existence of enzymes that are selectively required for survival during the chronic, rather than acute, phase of infection in mice, including the metabolic stress response regulator, RelA, suggests the existence of a persistence-specific metabolic program41.
In contrast to the expanding inventory of enzymes described above, the specific functions and mechanisms that define their essentiality remain understudied. For example, as stated above, the canonical role of ICL in the glyoxylate shunt initially implicated anaplerosis of TCA cycle intermediates and gluconeogenesis as essential features of persistence. However, this view was later complicated by the discovery that ICL functions as both an isocitrate and methylisocitrate lyase in M. tuberculosis42, and that loss of the latter activity, which functions to both detoxify and assimilate odd chain fatty acids, was sufficient to produce an in vitro bactericidal phenotype similar to that observed in mice43. To complicate matters further, the canonical enzymatic activity of ICL was also found to mediate key functions required for adaptation to hypoxia and defense against oxidative stress24,44. Notwithstanding, additional evidence for the critical role of gluconeogenesis in establishing infection in the mouse model is provided by the severe attenuation of M. tuberculosis that lacks its two fructose bisphosphatases45.
In vitro biochemical and metabolic labeling studies demonstrated that PEPCK in M. tuberculosis, a canonical enzyme of gluconeogenesis, could catalyze the reverse, or anaplerotic, reaction under reducing conditions and in macrophage-like cell lines13,46. In this regard, it is interesting to note that deletion of the only two glucokinases, PPGK and GLKA, in M. tuberculosis selectively impaired growth on glucose, but not acetate, and survival during the chronic, but not acute, phase of infection in mice. Moreover, this survival defect was not restored in a phagocyte oxidase-deficient mouse, arguing against a primary role in generation of the anti-oxidant NADPH37.
Looking across a broader range of genetically defined mutants, an emerging property among some metabolic enzymes required for M. tuberculosis persistence is their ability to regulate levels of chemically reactive or biologically toxic intermediates. Metabolic intermediates are often viewed as innocuous transients en route to more functional biological end-products. However, the accumulation of some intermediates has been found to mediate the toxicity of other metabolites, such as the allosteric inhibitory activity of the propionyl CoA-derived intermediate, 2-methylcitrate, on the gluconeogenic enzyme, fructose-1,6-bisphosphatase, which may contribute to the apparent metabolic toxicity of cholesterol and odd-chain fatty acids43,47,48. The chemical structures of other intermediates are often highly labile, if not reactive, and, in some cases, associated with biologically toxic activities, such as the ability of propionyl-CoA itself to react with and titrate pools of the TCA cycle intermediate, oxaloacetate43.
Building on this theme, mounting evidence has demonstrated that the bactericidal phenotype of some metabolic enzyme deletion or depletion strains may, in part, be due to the chemical reactivity of their substrates. Genetic deletion of malate synthase, for example, resulted in sterilization of M. tuberculosis in vitro when cultured on fatty acids, and during both the acute and chronic phases of infection in mice38. Metabolomic profiling revealed that malate synthase-deficient M. tuberculosis cultured on fatty acids accumulated high levels of its aldehyde substrate, glyoxylate, acetyl phosphate, acetoacetyl-CoA, butyryl-CoA, acetoacetate, and β-hydroxybutyrate. These changes were indicative of a glyoxylate-induced titration of oxaloacetate, resulting in a metabolic state of acetate overload, and
 . Accordingly, reduction of intrabacterial glyoxylate levels using a chemical inhibitor of ICL, which generates glyoxylate as a product, restored growth of malate synthase-deficient M. tuberculosis, despite inhibiting entry of carbon into the glyoxylate shunt. Genetic inactivation of fumarase (FUM) was found to be similarly bactericidal to M. tuberculosis during the acute and chronic phases of infection in a mouse model of TB39. Moreover, in vitro depletion of FUM resulted in marked accumulation of its substrate fumarate, a chemically reactive
. Accordingly, reduction of intrabacterial glyoxylate levels using a chemical inhibitor of ICL, which generates glyoxylate as a product, restored growth of malate synthase-deficient M. tuberculosis, despite inhibiting entry of carbon into the glyoxylate shunt. Genetic inactivation of fumarase (FUM) was found to be similarly bactericidal to M. tuberculosis during the acute and chronic phases of infection in a mouse model of TB39. Moreover, in vitro depletion of FUM resulted in marked accumulation of its substrate fumarate, a chemically reactive
 , which was shown to react broadly with protein and metabolite thiols and impair anti-oxidant defenses and viability of M. tuberculosis. In a third example, studies of Rv1248c, a multifunctional enzyme, which is involved in the metabolism of α-ketoglutarate and which was previously annotated as the E1 component of the α-ketoglutarate dehydrogenase complex, revealed a selectively essential role during the chronic phase of infection. This defect could be attributed, in part, to an inability to metabolize an increased flux of the chemically reactive intermediates glyoxylate and succinic semialdehyde arising from the oxidative arm of the TCA cycle23.
, which was shown to react broadly with protein and metabolite thiols and impair anti-oxidant defenses and viability of M. tuberculosis. In a third example, studies of Rv1248c, a multifunctional enzyme, which is involved in the metabolism of α-ketoglutarate and which was previously annotated as the E1 component of the α-ketoglutarate dehydrogenase complex, revealed a selectively essential role during the chronic phase of infection. This defect could be attributed, in part, to an inability to metabolize an increased flux of the chemically reactive intermediates glyoxylate and succinic semialdehyde arising from the oxidative arm of the TCA cycle23.
While untested, it is interesting to consider analogous roles for enzymes such as triose phosphate isomerase (TPI) and fructose bisphosphate aldolase (FBA) in the regulation of the reactive aldehyde, methylglyoxal, a spontaneous and enzymatic product that can arise from accumulation of the shared glycolytic and gluconeogenic intermediate, dihydroxyacetone phosphate8,9. From the standpoint of drug development, enzymes whose inhibition has the potential to result in the accumulation of reactive substrates or intermediates may merit particular attention due to their dominant negative toxic gain of function.
In addition to enzymes of central carbon metabolism, a number of more clearly defined physiologic functions required for growth and/or persistence in the host have emerged from studies of genes associated with more discrete or isolated metabolic activities. These include genes involved in the biosynthesis of enzymatic co-factors such as biotin (bioA), NAD (nadE) and pyridoxine (pdx1), all of which are essential for replication and persistence in vitro and/or in vivo49–52. Among metabolic enzymes, the persistence defect of metA-deficient M. tuberculosis is perhaps the most clearly defined. MetA catalyzes the first committed step in methionine biosynthesis53. Deletion of metA results in a rapid bactericidal death in vitro, which can be chemically rescued with methionine supplementation alone, and is associated with survival defects in both the acute and chronic phases of infection. These results thus define a specific and essential role for methionine or methionine-derived metabolites for in vivo growth and survival, which potentially involve regulation of protein biosynthesis, nucleic acid modification, and lipid biosynthesis. Moreover, these examples collectively help to elucidate the homeostatic nutritional requirements for M. tuberculosis to persist in the host.
Anti-oxidant defense
In addition to fueling the nutritional requirements of persistent or non- or slowly replicating M. tuberculosis, metabolic enzymes have also emerged as key mediators of anti-oxidant defense (Figure 2). Studies of alkyl hydroperoxide reductase C (AhpC), a member of the peroxiredoxin family of nonheme peroxidases, first revealed a four-protein NADH-dependent peroxidase and peroxynitrite reductase (PNR-P) system, which included the E2 (Dlat) and E3 (Lpd) components of the pyruvate dehydrogenase and α-ketoglutarate dehydrogenase complexes; with Lpd running in reverse as the E1, and DlaT as E2 components in this system26,54. Subsequent work revealed that this system could also be fueled by the oxidative decarboxylation of alpha-ketoacids via the activities of Rv1248c (for α-ketoglutarate as described above) or the annotated E1 component of its pyruvate dehydrogenase complex, AceE (for pyruvate), perhaps indicating an evolutionary coupling between metabolite detoxification and anti-oxidant defense23.
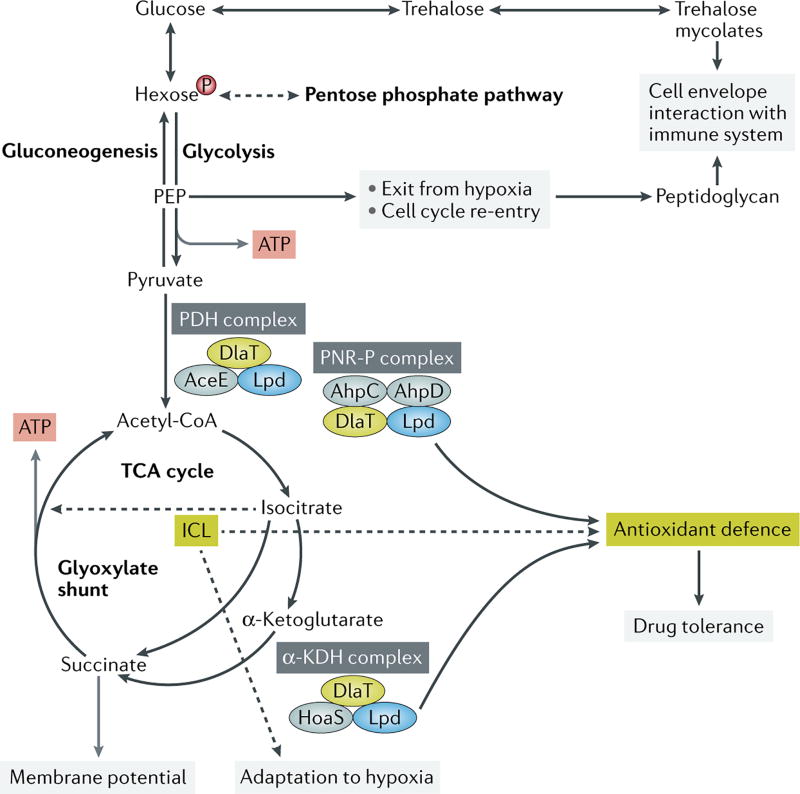
Figure 2. Effects of metabolism beyond fulfilling nutritional demands on the physiology of M. tuberculosis.
The pentose phosphate pathway supports peptidoglycan biosynthesis. Trehalose recycling from trehalose mycolates in the cell envelope facilitates exit from hypoxia and re-entry into active replication through the generation of glycolytic intermediates including phosphoenol pyruvate (PEP). The pyruvate dehydrogenase (PDH) complex and the α-ketoglutarate (KDH) complex share enzymes with the peroxynitrite reductase and peroxidase (PNR-P) complex and provide antioxidant defense. Isocitrate lyase (ICL) serves antioxidant functions and facilitates drug tolerance. The TCA metabolite succinate controls membrane potential.
Membrane bioenergetics
From a biochemical perspective, no essential function has been more strongly implicated for metabolic enzymes in persistence of M. tuberculosis by in vitro, animal and clinical evidence than maintenance of membrane function. This evidence stems, in large part, from studies of hypoxic M. tuberculosis. Though classified as an obligate aerobe, growing evidence indicates that M. tuberculosis resides mainly in intra- and extracellular niches that are often either directly (ambient O2<1%) or functionally hypoxic (such as imposed by nitric oxide)55. Microbiologic studies have shown that, although M. tuberculosis is unable to survive abrupt changes in O2 concentration, it can persist in a non- or slowly replicating, antibiotic tolerant state for up to decades at O2 tensions as low as 0.03% if allowed to adapt gradually56. From a metabolic perspective, 13C tracing experiments conducted in two independent studies revealed that hypoxia induced a metabolic remodeling of the TCA cycle in M. tuberculosis, resulting in the production of succinate as a secreted endproduct44,57. This increased production of succinate was subsequently shown to be essential for viability because secretion of succinate as an electrogenic substrate is used to maintain membrane potential, ATP synthesis and anaplerosis at a rate proportional to the respiratory capacity44. Similar evidence for the essentiality of membrane function later emerged from studies on the essentiality of the membrane anchor subunit of succinate dehydrogenase (Sdh1; Rv0249c) in vitro and in infected mice58. The arguably most compelling line of evidence supporting the essentiality of membrane function homeostasis however is the clinical efficacy of bedaquiline, a potent and species-selective inhibitor of the ATP synthase in M. tuberculosis, and pretomanid (PA-824) (and its related nitroimidazo-oxazole, delamanid (OPC-67683)), which are believed to kill hypoxic M. tuberculosis by poisoning its respiratory chain through the intrabacterial generation of nitric oxide59–61. Moreover, both bedaquiline and the nitro-imidazoles have been shown to be capable of killing non-replicating M. tuberculosis populations in vitro, in animal models and in people62,63.
METABOLIC REGULATION OF THE CELL CYCLE
Persistence in a latently infected host only contributes to survival of a bacterial species if it leads to active disease and transmission to a new host at some point and with sufficient frequency. The transition from latency to active disease requires re-initiation of bacterial growth. Yet, knowledge of the specific requirements for cell cycle re-entry has remained largely lacking. As alluded to above, a recent study by Eoh and colleagues reported that the adaptation of M. tuberculosis to hypoxia was accompanied by an accumulation of glycolytic and pentose phosphate pathway intermediates, which was accompanied by linked decreases in the downstream glycolytic intermediate, phosphoenolpyruvate, and an upstream glucose disaccharide, trehalose22. These changes were found to derive from catabolism of cell surface trehalose mycolates by the treS-encoded trehalose synthase, and more importantly, facilitate the re-initiation of de novo synthesis of peptidoglycan, inhibition of which was shown to be essential for cell cycle re-entry and, by extension, viability. This study thus demonstrated the existence of an anticipatory metabolic regulatory response that was initiated at the time of cell cycle exit but required for re-entry.
METABOLIC REGULATION OF IMMUNOREACTIVITY
Somewhat paradoxically, M. tuberculosis can elicit an inflammatory response that is sufficient to cause liquefactive tissue necrosis and facilitate transmission in some hosts, yet can also remain clinically asymptomatic for decades, if not lifetime of the host, in a state of persistent infection. This duality is due, in part, to the dynamic and immunoreactive nature of its cell surface lipids64,65.
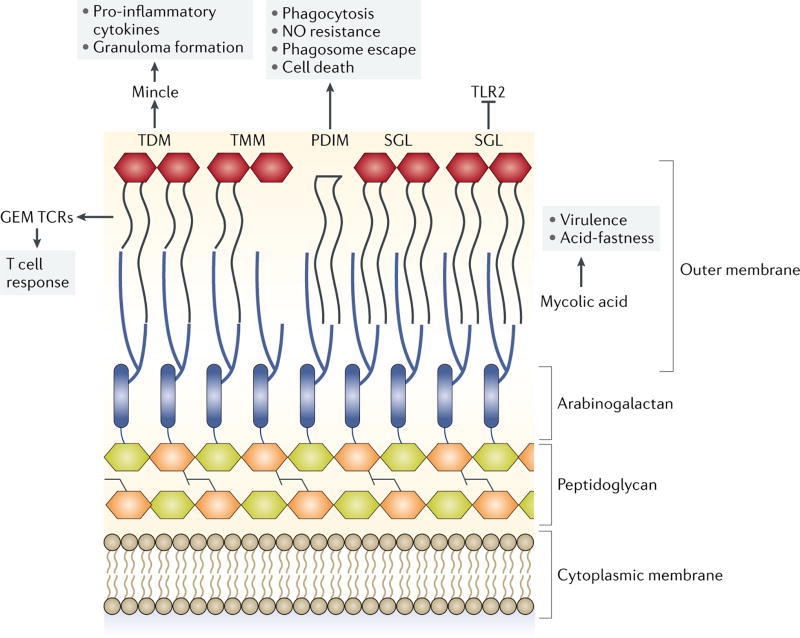
Lipids account for approximately 60% of the M. tuberculosis cell envelope by dry weight, and are diverse in structure and key determinants of its pathogenicity (reviewed in66). Moreover, growing evidence has suggested direct links between metabolic adaptations of M. tuberculosis to its host niche and the composition and immunoreactivity of its cell surface lipids (Figure 3). The most abundant cell wall glycolipid in M. tuberculosis, cord factor (trehalose 6,6’-dimycolate, TDM) activates the C-type lectin Mincle on macrophages to stimulate proinflammatory cytokine secretion and granuloma formation67. TDM-coated beads induce granulomas in mice with phenotypes similar to human TB granulomas, including foamy macrophage formation, fibrosis, and necrosis68,69. In response to host-imposed environmental stimuli M. tuberculosis controls the composition and quantity of immune stimulatory molecules. For example, during hypoxia, M. tuberculosis repurposes cell envelope trehalose mycolates, both trehalose monomycolate (TMM) and TDM, rendering the pathogen less immunostimulatory22,70.
Figure 3. Immunoreactive cell envelope lipids of M. tuberculosis.
Trehalose dimycolate (TDM) activates Mincle on macrophages to stimulate proinflammatory cytokine secretion and granuloma formation. Mycolic acids confer acid fastness and are crucial for virulence. The mycolic acid chains are antigenic determinants for the conserved human germline encoded mycolyl lipid-reactive (GEM) T cell receptors (TCRs) and may modulate T cell responses. Phthiocerol dimycocerosate (PDIM) has been shown to contribute to virulence by multiple mechanisms, including by stimulating phagocytosis, protecting against nitric oxide (NO), promoting escape from the phagosome and inducing host cell death. Sulfolipids and sulfoglycolipids (SGLs) are immunomodulatory and required for virulence. SGLs can antagonize TLR2 and thereby interfere with recognition by the immune system.
Mycolic acids
Changes in the composition of mycolic acids in response to various environmental stimuli have similarly been shown to be central to the adaptation of M. tuberculosis to the in vivo environment. M. tuberculosis lacking Pca, the enzyme that catalyzes the formation of the proximal cyclopropane ring of immunoreactive alpha-mycolic acids, for example, exhibited alterations in
 morphology (a biomarker of virulence), an altered lipid profile and was unable to persist during chronic mouse infection71. Eukaryotic-like Ser/Thr protein kinases (STPK) have similarly been shown to control the chain length of mycolic acids via phosphorylation of the β-ketoacyl-acyl carrier protein (ACP)-synthase KasB72. Phosphorylation-mediated inhibition of KasB activity results in the production of shorter mycolic acids, loss of
morphology (a biomarker of virulence), an altered lipid profile and was unable to persist during chronic mouse infection71. Eukaryotic-like Ser/Thr protein kinases (STPK) have similarly been shown to control the chain length of mycolic acids via phosphorylation of the β-ketoacyl-acyl carrier protein (ACP)-synthase KasB72. Phosphorylation-mediated inhibition of KasB activity results in the production of shorter mycolic acids, loss of
 and severe attenuation in mice. Although the environmental signals that control STPK activity and specific kinases responsible for KasB phosphorylation remain to be determined, this regulation likely contributes to the ability of M. tuberculosis to persist during latent infection as evidenced by the reduced, but persistent, bacterial burden and loss of acid fastness observed in tissues of chronically infected mice 73. Recent work revealed that the mycolate chains serve as antigenic determinants for the conserved human germline-encoded mycolyl lipid-reactive (GEM) T cell receptors (TCRs) and that structural alterations of the mycolate chains may modulate GEM-TCR activity74.
and severe attenuation in mice. Although the environmental signals that control STPK activity and specific kinases responsible for KasB phosphorylation remain to be determined, this regulation likely contributes to the ability of M. tuberculosis to persist during latent infection as evidenced by the reduced, but persistent, bacterial burden and loss of acid fastness observed in tissues of chronically infected mice 73. Recent work revealed that the mycolate chains serve as antigenic determinants for the conserved human germline-encoded mycolyl lipid-reactive (GEM) T cell receptors (TCRs) and that structural alterations of the mycolate chains may modulate GEM-TCR activity74.
Phthiocerol dimycocerosates and sulfoglycolipids
In addition to mycolic acid composition, M. tuberculosis also controls the abundance and size of two other bioactive lipids, phthiocerol dimycocerosate (PDIM) and sulfolipid-1 (SL-1) in response to the availability of their common precursor, methyl malonyl-CoA (MMCoA)75. Moreover, growth on odd-chain fatty acids, potentially derived from cholesterol in vivo, promoted increased flux of MMCoA through lipid biosynthetic pathways, resulting in increased levels of SL-1 and PDIM; in vivo perturbation of SL-1 and PDIM biosynthesis attenuated M. tuberculosis virulence in mice76,77.
PDIM is a virulence lipid believed to enhance host cell phagocytosis by targeting the host lipid membrane, confer protection against nitric oxide, interfere with recognition and recruitment of immune cells and promote escape from the phagosome and host cell death78–81. Moreover, PDIM appears specifically important in the context of innate pulmonary immunity as PDIM deficiency attenuated growth of M. tuberculosis in mouse lungs, but not in other organs and is not required for persistence in the lungs of mice82. By contrast, sulfolipids are long-studied virulence factors whose immunoregulatory activities have only recently begun to emerge. Moreover, recent studies of M. tuberculosis mutants with increased capacity to activate NF-κB identified sulfoglycolipids as competitive inhibitors of TLR2 and immune recognition of M. tuberculosis83.
Taken together, these observations thus define the composition and immunoreactivity of cell surface lipids as specific and essential determinants of M. tuberculosis virulence and establish a multifactorial link between metabolism, lipid biosynthesis and immunoreactivity in the pathogenesis of TB.
METABOLIC REGULATION OF DRUG TOLERANCE
Studies of M. tuberculosis cells that were exposed to growth-limiting stresses resembling the host environment or recovered from the sputa of patients with active pulmonary TB have long established an association between bacterial dormancy, lipid inclusions and antibiotic tolerance. However, only recent work has begun to elucidate the metabolic basis of this relationship. Work by Baek and colleagues specifically showed that hypoxia induced a redirection of carbon flux away from central metabolic pathways into the storage molecule triacylglycerol (TAG)84. Intracellular TAG accumulation is associated with a reduction in growth rate and increased tolerance to antibiotics in vitro and during mouse infection. M. tuberculosis utilizes the TAG transporter LprG to regulate intracellular TAG levels by transporting it out of the cytoplasm into the cell wall85. Inside hypoxic, lipid-rich macrophages M. tuberculosis imports fatty acids that are derived from host TAG and incorporates them into its own TAG supply86. The synthesis, accumulation and subcellular distribution of TAG thus appear critical for the cellular homeostasis of M. tuberculosis and link lipid metabolism with growth rate regulation, virulence and antibiotic tolerance.
Several other recent observations have similarly implicated the metabolic state of M. tuberculosis as a specific determinant of drug tolerance. First, ICL has been identified as mediator of antibiotic tolerance, functioning through its role in antioxidant defense rather than through its canonical activity in fatty acid metabolism24. Second, the glutamine amidotransferase GatCAB maintains translational fidelity, yet it fails to do so perfectly. Clinical M. tuberculosis isolates with mutations in a subunit of GatCAB showed increased mistranslation, which was associated with increased rifampicin tolerance, likely mediated by mistranslation of RNA polymerase subunit B87,88. Third, host-derived stresses can induce drug tolerance and impair drug efficacy against intracellular M. tuberculosis89. M. tuberculosis residing in activated myeloid cells isolated from infected mice showed a greater degree of drug tolerance than M. tuberculosis in resting cells89. Moreover, transcriptional profiling of drug treated intracellular M. tuberculosis led to a model predicting that host stress-induced remodeling of bacterial physiology results in a metabolically quiescent state that cross-protects M. tuberculosis against environmental and antibiotic injury.
Additional evidence in support of metabolism as a specific and regulated mediator of tolerance comes from the transcriptional analysis of M. tuberculosis exposed to the ATP synthase inhibitor bedaquiline90. This identified a regulatory network, which is controlled by two transcription factors and which coordinates multiple mechanisms that drive M. tuberculosis into a bedaquiline-tolerant state90. Specific nutrient availability may control this adaptive regulatory network because killing of M. tuberculosis by bedaquiline was enhanced when grown on fatty acids in contrast to carbon sources that promote ATP synthesis via glycolysis91. Intracellular metabolic reprograming resulting in the utilization of fatty acids is likely the reason for the observed increased bactericidal activity of bedaquiline against intracellular M. tuberculosis92.
Collectively, these observations argue that antibiotic tolerance is not solely a product of stochastic variability among minor bacterial subpopulations, but also a deterministically encoded metabolic capability of the majority population, though the specific mechanisms which mediate this capability remain to be determined28.
METABOLIC ENZYMES AS DRUG TARGETS
In 1939, the Nobel Prize in Physiology or Medicine was awarded to Gerhard Domagk for discovering the antibacterial effects of prontosil, the first broadly used therapeutic with selective antibacterial activity. Prontosil interferes with the bacterial synthesis of tetrahydrofolic acid93. Trimethoprim, an inhibitor of dihydrofolate reductase that is still in clinical use, was developed as an antibiotic in the 1960s94. Despite this precedent for the clinical value of inhibiting bacterial metabolism, many metabolic enzymes have long been perceived to be difficult if not impossible targets for antibacterial drug discovery. In part, because the substrates of many metabolic enzymes, particularly those in central carbon metabolism, tend to be small and hydrophilic. The active sites of these enzymes are therefore often dominated by properties that score poorly in druggability predictions95. Recent studies have nevertheless begun to reveal their potential as clinically potent and selective targets96,97.
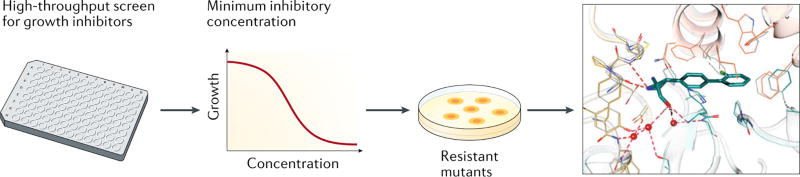
Screens for compounds that inhibit growth of M. tuberculosis in liquid culture identified two compounds, a sulfonale97 and an azetidine derivative, BRD459296. Selection and sequencing of resistant mutants suggested heterotetrameric tryptophan synthase (TrpAB), which catalyzes the last two steps in the biosynthesis of tryptophan, as a candidate target for both compounds (Figure 4). Additional genetic and biochemical studies confirmed that inhibition of TrpAB indeed is the mechanism which inhibits growth of M. tuberculosis. Structural analyses revealed that both the sulfanole and BRD4592 do not interact directly with the active site of TrpAB, but instead bind to the interface between its α- and β-subunits. Elegant and detailed mechanistic analyses strongly suggest that BRD4592 inhibits TrpAB activity not by perturbing the active site structure from a distance, but instead by reducing the enzymes flexibility and locking it in a closed conformation96. BRD4592 restricts growth of M. marinum in zebrafish embryos and the sulfanole showed activity in the mouse model of TB97.
Figure 4. Identification of a small-molecule allosteric inhibitor of tryptophan synthase (TrpAB) in M. tuberculosis.
High-throughput screening identified compounds that inhibit the growth of against M. tuberculosis. Compounds with a low minimum inhibitory concentration (MIC) were used to select for resistant mutants. This suggested tryptophane synthase as target. The co-crystal structure shows that the inhibitor binds to TrpAB outside of the active site of the enzyme. Structural image adapted with permission from ref96.
In contrast to the enzymes of central metabolism, those required for fatty acid biosynthesis have long been recognized as attractive targets for TB drug development. Among these are enzymes needed to synthesize mycolic acids, long-chain fatty acids that are the hallmark of the mycobacterial cell envelope. Several inhibitors of mycolic acid synthesis are of clinical value and none more so than isoniazid, which inhibits the essential enoyl–acyl-carrier protein (ACP) reductase, InhA, and has been a cornerstone of TB chemotherapy since the 1950s98. Isoniazid is a prodrug that requires activation by the catalase KatG. Resistance due to loss-of-function mutations in katG occurs frequently99, which limits the value of isoniazid. One strategy to overcome the high frequency of resistance is to develop InhA inhibitors that do not require KatG100; another is the inhibition of other enzymes required for mycolic acid biosynthesis. One such enzyme is polyketide synthase 13 (Pks13), an essential enzyme required for the condensation of mycolic acid precursors101. The therapeutic potential of inhibiting Pks13 was recently demonstrated in a study that used a structure-guided approach to advance a previously identified inhibitor to the extent that is activity could be tested in vivo. The most optimized inhibitor, TAM16, was highly potent in vitro against a panel of drug-sensitive and drug-resistant M. tuberculosis strains (minimum inhibitory concentrations ranged from 0.05 to 0.42 µM) and exhibits very attractive physicochemical, toxicological and pharmacological properties102. In mice, TAM16 showed similar efficacy as isoniazid, both alone and in combination with rifampicin, another first-line drug. In vitro, its frequency of resistance was approximately 100-fold lower than that of isoniazid.
What general conclusions can be drawn from the recent work on TrpAB and Pks13? For one, these studies confirm several principles that have guided recent antibacterial drug discovery efforts. These include that (i) screening for whole-cell activity can overcome one of the main reasons for failure (the inability of many small molecules to enter bacteria), (ii) pursuing new targets avoids cross-resistance with existing drugs, and (iii) a detailed understanding of the mechanism of action of an inhibitor can greatly facilitate its development. The work on TrpAB further emphasizes an intrinsic limitation of druggability predictions when they are based on active site evaluations alone and suggests that searching for allosteric inhibitors can be an attractive strategy for targeting metabolic enzymes. The latter conclusion is also supported by advances in cancer drug development, which resulted in allosteric modulators of several metabolic enzymes, including pyruvate kinase, glutaminase C, and isocitrate dehydrogenase103. Finally, it is worth recognizing that M. tuberculosis can grow without TrpAB when fed with sufficient amounts of tryptophan. The beneficial effect that chemical inhibition of TrpAB has on M. tuberculosis infection in mice strongly suggests that conditionally essential enzymes can serve as targets for drug development. However, it remains important to integrate efforts that develop inhibitors against this type of target with experiments that determine the concentrations of the complementing metabolites in human samples, ideally isolated from TB patients and uninfected controls.
OUTLOOK
Looking back, it has become apparent that metabolism serves the pathogenicity of M. tuberculosis through multiple roles beyond that of replicative fuel. These include cell intrinsic functions, such as nutritional homeostasis, membrane function, anti-oxidant defense, and cell cycle transit, which are required to maintain viability, as well as extrinsic functions, such as regulation of immunoreactivity and antibiotic tolerance, which are required for survival within the host niche. In retrospect, the discovery of such roles is not entirely surprising as metabolism is the biochemical foundation of all physiologic processes. What is more surprising however is the specificity through which metabolism has met these needs. Growing evidence, both within and outside of microbial physiology, has demonstrated that metabolism serves cellular physiology in ways that are qualitatively and quantitatively specific, and may provide vulnerability to species- and phenotype-specific modulation.
Looking ahead, it will be of particular importance to understand the underlying biochemical functions or principles that explain the diverse phenotypic roles of metabolism in the pathogenicity of M. tuberculosis. Toward this end, the advent of new technologies has made it increasingly possible to replace sequence homology-based inferences with direct experimental measurements. Such technologies promise to not only expand specific knowledge of the pathogenicity of M. tuberculosis but also enable computational modeling-based technologies to address more complex questions that go beyond homology-based reconstructions104–107. One such fundamental question is the biochemical objective of persistent M. tuberculosis. Awaiting such answers, it is important to be reminded by the White Queen of the importance of “(believing) as many as six impossible things before breakfast”.
ToC blurb.
As an obligate human pathogen, Mycobacterium tuberculosis has evolved to survive and thrive in biochemically challenging niches in its host. Ehrt, Schnappinger and Rhee review the unique metabolic features that enable M. tuberculosis pathogenesis and persistence but also represent drug targets.
Acknowledgments
This work was supported by grants R01AI063446 (NIAID) and U19AI111143 (Tri-Institutional TB Research Unit, part of the NIAID TBRU Network)……..
Glossary terms
- TCA cycle
The tricarboxylic acid cycle is a biochemical energy-generating pathway for the final steps of the oxidation of carbohydrates and fatty acids
- Glyoxylate shunt
An anaplerotic pathway that converts isocitrate into malate and succinate bypassing the two decarboxylation steps of the tricarboxylic acid cycle
- Anaplerosis
The process of replenishing metabolite pools
- Cataplerosis
The process of extracting metabolites for biosynthetic reactions
- Ketoacidosis
The accumulation of high concentrations of the ketones acetoacetate and beta-hydroxybutyrate
- Dienophile
A compound that readily reacts with an unsaturated hydrocarbon (diene)
- Cording
The aggregation of mycobacteria in a structure that resembles cords. This is due to the glycolipid trehalose dimycolate, also called cord factor
- Acid-fastness
The physical property of the mycolic acid-containing mycobacterial cell envelope to resist decolorization by acid/ethanol
- Caseation
The necrotic death of cells in the center of a granuloma resulting in an acellular mass that resembles a soft crumbly cheese;
Footnotes
Author contributions
All authors researched data for the article, contributed substantially to discussion of the content, wrote the article and reviewed and edited the manuscript before submission.
Competing interests
The authors declare no competing interests.
References
- 1.Yaffe MB. Looking ahead to the past. Sci Signal. 2010;3:eg7–eg7. doi: 10.1126/scisignal.3138eg7. [DOI] [PubMed] [Google Scholar]
- 2.Gutierrez MC, et al. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathog. 2005;1:e5. doi: 10.1371/journal.ppat.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pavelka MS, Chen B, Kelley CL, Collins FM, Jacobs WR., Jr Vaccine efficacy of a lysine auxotroph of Mycobacterium tuberculosis. Infect Immun. 2003;71:4190–4192. doi: 10.1128/IAI.71.7.4190-4192.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hondalus MK, et al. Attenuation of and protection induced by a leucine auxotroph of Mycobacterium tuberculosis. Infect Immun. 2000;68:2888–2898. doi: 10.1128/iai.68.5.2888-2898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sambandamurthy VK, et al. A pantothenate auxotroph of Mycobacterium tuberculosis is highly attenuated and protects mice against tuberculosis. Nat Med. 2002;8:1171–1174. doi: 10.1038/nm765. [DOI] [PubMed] [Google Scholar]
- 6.Görke B, Stülke J. Carbon catabolite repression in bacteria: many ways to make the most out of nutrients. Nat Rev Micro. 2008;6:613–624. doi: 10.1038/nrmicro1932. [DOI] [PubMed] [Google Scholar]
- 7.de Carvalho LPS, et al. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chemistry & Biology. 2010;17:1122–1131. doi: 10.1016/j.chembiol.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 8.Puckett S, et al. Inactivation of Fructose-1,6-Bisphosphate Aldolase Prevents Optimal Co-catabolism of Glycolytic and Gluconeogenic Carbon Substrates in Mycobacterium tuberculosis. PLoS Pathog. 2014;10:e1004144. doi: 10.1371/journal.ppat.1004144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trujillo C, et al. Triosephosphate Isomerase Is Dispensable In Vitro yet Essential for Mycobacterium tuberculosis To Establish Infection. mBio. 2014;5:e00085–14–e00085–14. doi: 10.1128/mBio.00085-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kayne FJ. 11 Pyruvate Kinase. The Enzymes. 1973;8:353–382. [Google Scholar]
- 11.Zhong W, et al. Allosteric pyruvate kinase-based “logic gate” synergistically senses energy and sugar levels in Mycobacterium tuberculosis. Nat Commun. 2017:1–14. doi: 10.1038/s41467-017-02086-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noy T, et al. Central Role of Pyruvate Kinase in Carbon Co-catabolism of Mycobacterium tuberculosis. J Biol Chem. 2016;291:7060–7069. doi: 10.1074/jbc.M115.707430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beste DJV, et al. C-Flux Spectral Analysis of Host-Pathogen Metabolism Reveals a Mixed Diet for Intracellular Mycobacterium tuberculosis. Chemistry & Biology. 2013:1–10. doi: 10.1016/j.chembiol.2013.06.012. This study showed that intracellular M. tuberculosis has access to glycolytic C3 substrates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Somashekar BS, et al. Metabolic Profiling of Lung Granuloma in Mycobacterium tuberculosisInfected Guinea Pigs: Ex vivo 1H Magic Angle Spinning NMR Studies. J. Proteome Res. 2011;10:4186–4195. doi: 10.1021/pr2003352. [DOI] [PubMed] [Google Scholar]
- 15.Zimmermann M, et al. Integration of Metabolomics and Transcriptomics Reveals a Complex Diet of Mycobacterium tuberculosis during Early Macrophage Infection. mSystems. 2017;2:e00057–17. doi: 10.1128/mSystems.00057-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanderven BC, et al. Novel Inhibitors of Cholesterol Degradation in Mycobacterium tuberculosis Reveal How the Bacterium’s Metabolism Is Constrained by the Intracellular Environment. PLoS Pathog. 2015;11:1–20. doi: 10.1371/journal.ppat.1004679. This work provided a framework to exploit carbon metabolism for TB drug development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson RM, et al. Chemical activation of adenylyl cyclase Rv1625c inhibits growth of Mycobacterium tuberculosis on cholesterol and modulates intramacrophage signaling. Mol Microbiol. 2017;105:294–308. doi: 10.1111/mmi.13701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nazarova EV, et al. Rv3723/LucA coordinates fatty acid and cholesterol uptake in Mycobacterium tuberculosis. Elife. 2017;6:e26969. doi: 10.7554/eLife.26969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larrouy-Maumus G, et al. Discovery of a glycerol 3-phosphate phosphatase reveals glycerophospholipid polar head recycling in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2013;110:11320–11325. doi: 10.1073/pnas.1221597110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalscheuer R, Weinrick B, Veeraraghavan U, Besra GS, Jacobs WR. Trehalose-recycling ABC transporter LpqY-SugA-SugB-SugC is essential for virulence of Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2010;107:21761–21766. doi: 10.1073/pnas.1014642108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rustad TR, Sherrid AM, Minch KJ, Sherman DR. Hypoxia: a window into Mycobacterium tuberculosislatency. Cell Microbiol. 2009;11:1151–1159. doi: 10.1111/j.1462-5822.2009.01325.x. [DOI] [PubMed] [Google Scholar]
- 22.Eoh H, et al. Metabolic anticipation in Mycobacterium tuberculosis. Nat Microbiol. 2017;2:17084. doi: 10.1038/nmicrobiol.2017.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maksymiuk C, Balakrishnan A, Bryk R, Rhee KY, Nathan CF. E1 of α-ketoglutarate dehydrogenase defends Mycobacterium tuberculosis against glutamate anaplerosis and nitroxidative stress. Proc Natl Acad Sci USA. 2015;112:E5834–43. doi: 10.1073/pnas.1510932112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nandakumar M, Nathan C, Rhee KY. Isocitrate lyase mediates broad antibiotic tolerance in Mycobacterium tuberculosis. Nat Commun. 2014;5:4306. doi: 10.1038/ncomms5306. [DOI] [PubMed] [Google Scholar]
- 25.Venugopal A, et al. Virulence of Mycobacterium tuberculosis depends on lipoamide dehydrogenase, a member of three multienzyme complexes. Cell Host Microbe. 2011;9:21–31. doi: 10.1016/j.chom.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bryk R, Lima CD, Erdjument-Bromage H, Tempst P, Nathan C. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science. 2002;295:1073–1077. doi: 10.1126/science.1067798. [DOI] [PubMed] [Google Scholar]
- 27.Nathan C. Taming Tuberculosis: A Challenge for Science and Society. Cell Host Microbe. 2009;5:220–224. doi: 10.1016/j.chom.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 28.Nathan C. Fresh approaches to anti-infective therapies. Sci Transl Med. 2012;4:140sr2. doi: 10.1126/scitranslmed.3003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKinney J, et al. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406:735–738. doi: 10.1038/35021074. A seminal study that identified for the first time a gene required for persistence of M. tuberculosis in mice. [DOI] [PubMed] [Google Scholar]
- 30.Marrero J, Rhee KY, Schnappinger D, Pethe K, Ehrt S. Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc Natl Acad Sci USA. 2010;107:9819–9824. doi: 10.1073/pnas.1000715107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ehrt S, Rhee K, Schnappinger D. Mycobacterial genes essential for the pathogen's survival in the host. Immunol. Rev. 2015;264:319–326. doi: 10.1111/imr.12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang YJ, et al. Tryptophan Biosynthesis Protects Mycobacteria from CD4 T-Cell-Mediated Killing. Cell. 2013;155:1296–1308. doi: 10.1016/j.cell.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci USA. 2003;100:12989–12994. doi: 10.1073/pnas.2134250100. The first study that screened a transposon mutant library to identify M. tuberculosis genes that are required for virulence in the mouse model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muñoz-Elías E, McKinney J. Mycobacterium tuberculosis isocitrate lyases 1 and 2 are jointly required for in vivo growth and virulence. Nat Med. 2005;11:638–644. doi: 10.1038/nm1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blumenthal A, Trujillo C, Ehrt S, Schnappinger D. Simultaneous analysis of multiple Mycobacterium tuberculosis knockdown mutants in vitro and in vivo. PLoS ONE. 2010;5:e15667. doi: 10.1371/journal.pone.0015667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pandey AK, Sassetti CM. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci USA. 2008;105:4376–4380. doi: 10.1073/pnas.0711159105. This work reveals that M. tuberculosis requires cholesterol for persistence in vivo. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marrero J, Trujillo C, Rhee KY, Ehrt S. Glucose Phosphorylation Is Required for Mycobacterium tuberculosis Persistence in Mice. PLoS Pathog. 2013;9:e1003116. doi: 10.1371/journal.ppat.1003116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puckett S, et al. Glyoxylate detoxification is an essential function of malate synthase required for carbon assimilation in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2017;114:E2225–E2232. doi: 10.1073/pnas.1617655114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruecker N, et al. Fumarase Deficiency Causes Protein and Metabolite Succination and Intoxicates Mycobacterium tuberculosis. Cell Chem Biol. 2017;24:306–315. doi: 10.1016/j.chembiol.2017.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhee KY, et al. Central carbon metabolism in Mycobacterium tuberculosis: an unexpected frontier. Trends in Microbiology. 2011;19:307–314. doi: 10.1016/j.tim.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dahl JL, et al. The role of Rel Mtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc Natl Acad Sci USA. 2003;100:10026–10031. doi: 10.1073/pnas.1631248100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gould TA, van de Langemheen H, Muñoz-Elías EJ, Mckinney JD, Sacchettini JC. Dual role of isocitrate lyase 1 in the glyoxylate and methylcitrate cycles in Mycobacterium tuberculosis. Mol Microbiol. 2006;61:940–947. doi: 10.1111/j.1365-2958.2006.05297.x. [DOI] [PubMed] [Google Scholar]
- 43.Eoh H, Rhee KY. Methylcitrate cycle defines the bactericidal essentiality of isocitrate lyase for survival of Mycobacterium tuberculosison fatty acids. Proc Natl Acad Sci USA. 2014;111:4976–4981. doi: 10.1073/pnas.1400390111. This study dissected the specific biochemical mechanism underlying the essentiality of isocitrate lyase for growth and survival of M. tuberculosis with fatty acids as carbon source. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eoh H, Rhee KY. Multifunctional essentiality of succinate metabolism in adaptation to hypoxia in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2013;110:6554–6559. doi: 10.1073/pnas.1219375110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ganapathy U, et al. Two enzymes with redundant fructose bisphosphatase activity sustain gluconeogenesis and virulence in Mycobacterium tuberculosis. Nat Commun. 2015;6:7912. doi: 10.1038/ncomms8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Machová I, et al. Mycobacterium tuberculosis phosphoenolpyruvate carboxykinase is regulated by redox mechanisms and interaction with thioredoxin. J Biol Chem. 2014;289:13066–13078. doi: 10.1074/jbc.M113.536748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee W, VanderVen BC, Fahey RJ, Russell DG. Intracellular Mycobacterium tuberculosis Exploits Host-derived Fatty Acids to Limit Metabolic Stress. J Biol Chem. 2013;288:6788–6800. doi: 10.1074/jbc.M112.445056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas ST, VanderVen BC, Sherman DR, Russell DG, Sampson NS. Pathway Profiling in Mycobacterium tuberculosis: Elucidation of cholesterol-derived catabolite and enzymes that catalyze its metabolism. J Biol Chem. 2011;286:43668–43678. doi: 10.1074/jbc.M111.313643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim J-H, et al. A genetic strategy to identify targets for the development of drugs that prevent bacterial persistence. Proc Natl Acad Sci USA. 2013;110:19095–19100. doi: 10.1073/pnas.1315860110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Woong Park S, et al. Evaluating the Sensitivity of Mycobacterium tuberculosis to Biotin Deprivation Using Regulated Gene Expression. PLoS Pathog. 2011;7:e1002264. doi: 10.1371/journal.ppat.1002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dick T, Manjunatha U, Kappes B, Gengenbacher M. Vitamin B6 biosynthesis is essential for survival and virulence of Mycobacterium tuberculosis. Mol Microbiol. 2010;78:980–988. doi: 10.1111/j.1365-2958.2010.07381.x. [DOI] [PubMed] [Google Scholar]
- 52.Boshoff HIM, et al. Biosynthesis and recycling of nicotinamide cofactors in Mycobacterium tuberculosis. An essential role for NAD in nonreplicating bacilli. J Biol Chem. 2008;283:19329–19341. doi: 10.1074/jbc.M800694200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berney M, et al. Essential roles of methionine and S-adenosylmethionine in the autarkic lifestyle of Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2015;112:10008–10013. doi: 10.1073/pnas.1513033112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bryk R, Griffin P, Nathan C. Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature. 2000;407:211–215. doi: 10.1038/35025109. [DOI] [PubMed] [Google Scholar]
- 55.Hartman TE, Gardete S, Rhee KY, Jansen RS, Wang Z. Metabolic Perspectives on Persistence. Microbiology Spectrum. 2017;5:1–17. doi: 10.1128/microbiolspec.tbtb2-0026-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wayne L, Hayes L. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 1996;64:2062. doi: 10.1128/iai.64.6.2062-2069.1996. A seminal study that established a model for nonreplication persistence of M. tuberculosis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watanabe S, et al. Fumarate reductase activity maintains an energized membrane in anaerobic Mycobacterium tuberculosis. PLoS Pathog. 2011;7:e1002287. doi: 10.1371/journal.ppat.1002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hartman T, et al. Succinate Dehydrogenase is the Regulator of Respiration in Mycobacterium tuberculosis. PLoS Pathog. 2014;10:e1004510. doi: 10.1371/journal.ppat.1004510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dawson R, et al. Efficiency and safety of the combination of moxifloxacin, pretomanid (PA-824), and pyrazinamide during the first 8 weeks of antituberculosis treatment: a phase 2b, open-label, partly randomised trial in patients with drug-susceptible or drug-resistant pulmonary tuberculosis. Lancet. 2015;385:1738–1747. doi: 10.1016/S0140-6736(14)62002-X. [DOI] [PubMed] [Google Scholar]
- 60.Diacon AH, et al. The diarylquinoline TMC207 for multidrug-resistant tuberculosis. N Engl J Med. 2009;360:2397–2405. doi: 10.1056/NEJMoa0808427. [DOI] [PubMed] [Google Scholar]
- 61.Gler MT, et al. Delamanid for multidrug-resistant pulmonary tuberculosis. N Engl J Med. 2012;366:2151–2160. doi: 10.1056/NEJMoa1112433. [DOI] [PubMed] [Google Scholar]
- 62.Andries K. A Diarylquinoline Drug Active on the ATP Synthase of Mycobacterium tuberculosis. Science. 2005;307:223–227. doi: 10.1126/science.1106753. This paper reported bedaquiline, which in 2012 became the first new anti-TB drug with a novel mechanism of action since rifampicin was approved in 1974. [DOI] [PubMed] [Google Scholar]
- 63.Singh R, et al. PA-824 kills nonreplicating Mycobacterium tuberculosis by intracellular NO release. Science. 2008;322:1392–1395. doi: 10.1126/science.1164571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stamm CE, Collins AC, Shiloh MU. Sensing of Mycobacterium tuberculosis and consequences to both host and bacillus. Immunol. Rev. 2015;264:204–219. doi: 10.1111/imr.12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ishikawa E, et al. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J Exp Med. 2009;206:2879–2888. doi: 10.1084/jem.20091750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Neyrolles O, Guilhot C. Recent advances in deciphering the contribution of Mycobacterium tuberculosis lipids to pathogenesis. Tuberculosis. 2011;91:187–195. doi: 10.1016/j.tube.2011.01.002. A comprehensive review of the cell envelope lipids of M. tuberculosis and their contribution to virulence. [DOI] [PubMed] [Google Scholar]
- 67.Ishikawa E, et al. Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C-type lectin Mincle. J Exp Med. 2009;206:2879–2888. doi: 10.1084/jem.20091750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Geisel RE, Sakamoto K, Russell DG, Rhoades ER. In vivo activity of released cell wall lipids of Mycobacterium bovis bacillus Calmette-Guérin is due principally to trehalose mycolates. J Immunol. 2005;174:5007–5015. doi: 10.4049/jimmunol.174.8.5007. [DOI] [PubMed] [Google Scholar]
- 69.Rhoades E, et al. Identification and macrophage-activating activity of glycolipids released from intracellular Mycobacterium bovis BCG. Mol Microbiol. 2003;48:875–888. doi: 10.1046/j.1365-2958.2003.03473.x. [DOI] [PubMed] [Google Scholar]
- 70.Galagan JE, et al. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature. 2013;499:178–183. doi: 10.1038/nature12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Glickman MS, Cox JS, Jacobs WR. A novel mycolic acid cyclopropane synthetase is required for cording, persistence, and virulence of Mycobacterium tuberculosis. Mol Cell. 2000;5:717–727. doi: 10.1016/s1097-2765(00)80250-6. A foundational study that describes the importance fo cyclopropanation of mycolic acids for long-term persistence and virulence of M. tuberculosis. [DOI] [PubMed] [Google Scholar]
- 72.Vilchèze C, et al. Phosphorylation of KasB Regulates Virulence and Acid-Fastness in Mycobacterium tuberculosis. PLoS Pathog. 2014;10:e1004115–18. doi: 10.1371/journal.ppat.1004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seiler P, et al. Cell-wall alterations as an attribute of Mycobacterium tuberculosis in latent infection. J Infect Dis. 2003;188:1326–1331. doi: 10.1086/378563. [DOI] [PubMed] [Google Scholar]
- 74.Chancellor A, et al. CD1b–restricted GEM T cell responses are modulated by Mycobacterium tuberculosis mycolic acid meromycolate chains. Proc Natl Acad Sci USA. 2017;162:201708252–9. doi: 10.1073/pnas.1708252114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jain M, et al. Lipidomics reveals control of Mycobacterium tuberculosis virulence lipids via metabolic coupling. Proc Natl Acad Sci USA. 2007;104:5133–5138. doi: 10.1073/pnas.0610634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang X, Nesbitt NM, Dubnau E, Smith I, Sampson NS. Cholesterol Metabolism Increases the Metabolic Pool of Propionate in Mycobacterium tuberculosis. Biochemistry. 2009;48:3819–3821. doi: 10.1021/bi9005418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Griffin JE, et al. Cholesterol Catabolism by Mycobacterium tuberculosis Requires Transcriptional and Metabolic Adaptations. Chemistry & Biology. 2012;19:218–227. doi: 10.1016/j.chembiol.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rousseau C, et al. Production of phthiocerol dimycocerosates protects Mycobacterium tuberculosis from the cidal activity of reactive nitrogen intermediates produced by macrophages and modulates the early immune response to infection. Cell Microbiol. 2004;6:277–287. doi: 10.1046/j.1462-5822.2004.00368.x. [DOI] [PubMed] [Google Scholar]
- 79.Astarie-Dequeker C, et al. Phthiocerol Dimycocerosates of M. tuberculosis Participate in Macrophage Invasion by Inducing Changes in the Organization of Plasma Membrane Lipids. PLoS Pathog. 2009;5:e1000289–17. doi: 10.1371/journal.ppat.1000289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cambier CJ, Falkow S, Ramakrishnan L. Host Evasion and Exploitation Schemes of Mycobacterium tuberculosis. Cell. 2014;159:1497–1509. doi: 10.1016/j.cell.2014.11.024. [DOI] [PubMed] [Google Scholar]
- 81.Quigley J, et al. The Cell Wall Lipid PDIM Contributes to Phagosomal Escape and Host Cell Exit of Mycobacterium tuberculosis. mBio. 2017;8:e00148–17–12. doi: 10.1128/mBio.00148-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cox JS, Chen B, McNeil M, Jacobs WR. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 83.Blanc L, et al. Mycobacterium tuberculosis inhibits human innate immune responses via the production of TLR2 antagonist glycolipids. Proc Natl Acad Sci USA. 2017;4:201707840–6. doi: 10.1073/pnas.1707840114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Baek S-H, Li AH, Sassetti CM. Metabolic regulation of mycobacterial growth and antibiotic sensitivity. 9, e1001065 (2011) PLoS Biol. 2011;9:e1001065. doi: 10.1371/journal.pbio.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Martinot AJ, et al. Mycobacterial Metabolic Syndrome: LprG and Rv1410 Regulate Triacylglyceride Levels, Growth Rate and Virulence in Mycobacterium tuberculosis. PLoS Pathog. 2016;12:e1005351. doi: 10.1371/journal.ppat.1005351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Daniel J, Maamar H, Deb C, Sirakova TD, Kolattukudy PE. Mycobacterium tuberculosis Uses Host Triacylglycerol to Accumulate Lipid Droplets and Acquires a Dormancy-Like Phenotype in Lipid-Loaded Macrophages. PLoS Pathog. 2011;7:e1002093. doi: 10.1371/journal.ppat.1002093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Javid B, et al. Mycobacterial mistranslation is necessary and sufficient for rifampicin phenotypic resistance. Proc Natl Acad Sci USA. 2014;111:1132–1137. doi: 10.1073/pnas.1317580111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Su H-W, et al. The essential mycobacterial amidotransferase GatCAB is a modulator of specific translational fidelity. Nat Microbiol. 2016;1:16147. doi: 10.1038/nmicrobiol.2016.147. [DOI] [PubMed] [Google Scholar]
- 89.Liu Y, et al. Immune activation of the host cell induces drug tolerance in Mycobacterium tuberculosis both in vitro and in vivo. J Exp Med. 2016;213:809–825. doi: 10.1084/jem.20151248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Peterson EJR, Ma S, Sherman DR, Baliga NS. Network analysis identifies Rv0324 and Rv0880 as regulators of bedaquiline tolerance in Mycobacterium tuberculosis. Nat Microbiol. 2016;1:16078. doi: 10.1038/nmicrobiol.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Koul A, et al. Delayed bactericidal response of Mycobacterium tuberculosis to bedaquiline involves remodelling of bacterial metabolism. Nat Commun. 2014;5:3369. doi: 10.1038/ncomms4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dhillon J, Andries K, Phillips PPJ, Mitchison DA. Bactericidal activity of the diarylquinoline TMC207 against Mycobacterium tuberculosis outside and within cells. Tuberculosis. 2010;90:301–305. doi: 10.1016/j.tube.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 93.Greenwood D. Antibiot Chemother (Ninth Edition) 2010:2–9. [Google Scholar]
- 94.Bushby SR, Hitchings GH. Trimethoprim, a sulphonamide potentiator. Br J Pharmacol Chemother. 1968;33:72–90. doi: 10.1111/j.1476-5381.1968.tb00475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Volkamer A, Kuhn D, Grombacher T, Rippmann F, Rarey M. Combining Global and Local Measures for Structure-Based Druggability Predictions. J. Chem. Inf. Model. 2012;52:360–372. doi: 10.1021/ci200454v. [DOI] [PubMed] [Google Scholar]
- 96.Wellington S, et al. A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase. Nat Chem Biol. 2017;13:943–950. doi: 10.1038/nchembio.2420. This study provides a thorough and elegant mechanistic analysis of novel TrpAB inhibitors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Abrahams KA, et al. Inhibiting mycobacterial tryptophan synthase by targeting the inter-subunit interface. Scientific Reports. 2017;7:9430. doi: 10.1038/s41598-017-09642-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bernstein J, Lott WA, Steinberg BA, Yale HL. Chemotherapy of experimental tuberculosis. V. Isonicotinic acid hydrazide (nydrazid) and related compounds. Am Rev Tuberc. 1952;65:357–364. doi: 10.1164/art.1952.65.4.357. [DOI] [PubMed] [Google Scholar]
- 99.Vilchèze C, Jacobs WR. Resistance to Isoniazid and Ethionamide in Mycobacterium tuberculosis: Genes, Mutations, and Causalities. Microbiol Spec. 2014;2:MGM2–0014–2013. doi: 10.1128/microbiolspec.MGM2-0014-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Soutter HH, et al. Discovery of cofactor-specific, bactericidal Mycobacterium tuberculosis InhA inhibitors using DNA-encoded library technology. Proc Natl Acad Sci USA. 2016;113:E7880–E7889. doi: 10.1073/pnas.1610978113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Portevin D, et al. A polyketide synthase catalyzes the last condensation step of mycolic acid biosynthesis in mycobacteria and related organisms. Proc Natl Acad Sci USA. 2004;101:314–319. doi: 10.1073/pnas.0305439101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Aggarwal A, et al. Development of a Novel Lead that Targets M. tuberculosis Polyketide Synthase 13. Cell. 2017;170:249–259.e25. doi: 10.1016/j.cell.2017.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.DeLaBarre B, Hurov J, Cianchetta G, Murray S, Dang L. Action at a distance: allostery and the development of drugs to target cancer cell metabolism. Chemistry & Biology. 2014;21:1143–1161. doi: 10.1016/j.chembiol.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 104.Reaves ML, Rabinowitz JD. Metabolomics in systems microbiology. Current Opinion in Biotechnology. 2011;22:17–25. doi: 10.1016/j.copbio.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yus E, et al. Impact of genome reduction on bacterial metabolism and its regulation. Science. 2009;326:1263–1268. doi: 10.1126/science.1177263. [DOI] [PubMed] [Google Scholar]
- 106.Lofthouse EK, et al. Systems-Based Approaches to Probing Metabolic Variation within the Mycobacterium tuberculosis Complex. PLoS ONE. 2013;8:e75913–14. doi: 10.1371/journal.pone.0075913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Garay CD, Dreyfuss JM, Galagan JE. Metabolic modeling predicts metabolite changes in Mycobacterium tuberculosis. BMC Systems Biology. 2015:1–16. doi: 10.1186/s12918-015-0206-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.World Health Organization. Global tuberculosis report 2016. 2016. [Google Scholar]
- 109.Lin P, Flynn J. Understanding latent tuberculosis: a moving target. J Immunol. 2010;185:15. doi: 10.4049/jimmunol.0903856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ernst JD. The immunological life cycle of tuberculosis. Nat Rev Immunol. 2012:1–11. doi: 10.1038/nri3259. [DOI] [PubMed] [Google Scholar]
- 111.Russell DG. Mycobacterium tuberculosis: here today, and here tomorrow. Nat Rev Mol Cell Biol. 2001;2:569–577. doi: 10.1038/35085034. [DOI] [PubMed] [Google Scholar]
- 112.Barry CE, et al. The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat Rev Micro. 2009;7:845–855. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Philips JA, Ernst JD. Tuberculosis Pathogenesis and Immunity. Annu Rev Pathol Mech Dis. 2012;7:353–384. doi: 10.1146/annurev-pathol-011811-132458. [DOI] [PubMed] [Google Scholar]
- 114.Stallings CL, Glickman MS. Is Mycobacterium tuberculosis stressed out? A critical assessment of the genetic evidence. Microbes Infect. 2010;12:1091–1101. doi: 10.1016/j.micinf.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ehrt S, Schnappinger D. Mycobacterial survival strategies in the phagosome: defence against host stresses. Cell Microbiol. 2009;11:1170–1178. doi: 10.1111/j.1462-5822.2009.01335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.van der Wel N, et al. M. tuberculosis and M. leprae Translocate from the Phagolysosome to the Cytosol in Myeloid Cells. Cell. 2007;129:1287–1298. doi: 10.1016/j.cell.2007.05.059. [DOI] [PubMed] [Google Scholar]
- 117.Houben ENG, et al. Composition of the type VII secretion system membrane complex. Mol Microbiol. 2012;86:472–484. doi: 10.1111/j.1365-2958.2012.08206.x. [DOI] [PubMed] [Google Scholar]
- 118.Boshoff H, Barry C. Tuberculosis—metabolism and respiration in the absence of growth. Nat Rev Micro. 2005;3:70–80. doi: 10.1038/nrmicro1065. [DOI] [PubMed] [Google Scholar]
- 119.Russell DG. Who puts the tubercle in tuberculosis? Nat Rev Micro. 2006;5:39–47. doi: 10.1038/nrmicro1538. [DOI] [PubMed] [Google Scholar]
- 120.Via LE, et al. Tuberculous Granulomas Are Hypoxic in Guinea Pigs, Rabbits, and Nonhuman Primates. Infect Immun. 2008;76:2333–2340. doi: 10.1128/IAI.01515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Elkington PT, D'Armiento JM, Friedland JS. Tuberculosis immunopathology: the neglected role of extracellular matrix destruction. Sci Transl Med. 2011;3:71ps6. doi: 10.1126/scitranslmed.3001847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Garton NJ, et al. Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculous sputum. PLoS Med. 2008;5:e75. doi: 10.1371/journal.pmed.0050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Eum S-Y, et al. Neutrophils are the predominant infected phagocytic cells in the airways of patients with active pulmonary TB. Chest. 2010;137:122–128. doi: 10.1378/chest.09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mukamolova GV, Turapov O, Malkin J, Woltmann G, Barer MR. Resuscitation-promoting factors reveal an occult population of tubercle Bacilli in Sputum. Am J Respir Crit Care Med. 2010;181:174–180. doi: 10.1164/rccm.200905-0661OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chengalroyen MD, et al. Detection and Quantification of Differentially Culturable Tubercle Bacteria in Sputum from Patients with Tuberculosis. Am J Respir Crit Care Med. 2016;194:1532–1540. doi: 10.1164/rccm.201604-0769OC. [DOI] [PMC free article] [PubMed] [Google Scholar]