Abstract
Objectives
To characterise renal tissue metabolic pathway gene expression in different forms of glomerulonephritis.
Methods
Patients with nephrotic syndrome (NS), antineutrophil cytoplasmic antibody-associated vasculitis (AAV), systemic lupus erythematosus (SLE) and healthy living donors (LD) were studied. Clinically indicated renal biopsies were obtained at time of diagnosis and microdissected into glomerular and tubulointerstitial compartments. Microarray-derived differential gene expression of 88 genes representing critical enzymes of metabolic pathways and 25 genes related to immune cell markers was compared between disease groups. Correlation analyses measured relationships between metabolic pathways, kidney function and cytokine production.
Results
Reduced steady state levels of mRNA species were enriched in pathways of oxidative phosphorylation and increased in the pentose phosphate pathway (PP) with maximal perturbation in AAV and SLE followed by NS, and least in LD. Transcript regulation was isozymes specific with robust regulation in hexokinases, enolases and glucose transporters. Intercorrelation networks were observed between enzymes of the PPP (eg, transketolase) and macrophage markers (eg, CD68) (r=0.49, p<0.01). Increased PPP transcript levels were associated with reduced glomerular filtration rate in the glomerular (r=−0.49, p<0.01) and tubulointerstitial (r=−0.41, p<0.01) compartments. PPP expression and tumour necrosis factor activation were tightly co-expressed (r=0.70, p<0.01).
Conclusion
This study demonstrated concordant alterations of the renal transcriptome consistent with metabolic reprogramming across different forms of glomerulonephritis. Activation of the PPP was tightly linked with intrarenal macrophage marker expression, reduced kidney function and increased production of cytokines. Modulation of glucose metabolism may offer novel immune-modulatory therapeutic approaches in rare kidney diseases.
INTRODUCTION
Activated immune cells require alterations in metabolic activity to survive, proliferate and sustain effector responses. How intracellular metabolites regulate immune cells is an emerging field of study known as immuometabolism.1 In oncology, alteration of cancer cell metabolism to preferentially use glycolysis rather than the tricarboxylic acid (TCA) cycle for energy production is referred to as ‘aerobic glycolysis’ or the Warburg effect. Metabolic reprogramming of tumour cells towards enhanced glycolytic capacity is a defining characteristic of various malignancies and explains how tumours can be visualised by positron emission tomography studies coupled with radiolabelled fluorodeoxyglucose. In the context of immunity, similar alterations in metabolic pathways can promote effector functions in immune cell subsets to induce production of specific pro-inflammatory and anti-inflammatory cytokines.
Evidence of metabolic reprogramming in immune-mediated diseases is mostly limited to in vitro studies. Activation of hypoxia-inducible factor 1 alpha (HIF-1α) or stimulation of innate immune response receptors can upregulate pathways of glycolysis, promote differentiation of M1 macrophages and inform inflammatory responses via production of specific cytokines, including tumour necrosis factor (TNF).2–7 Some studies have provided in vivo evidence of immunometabolism in rheumatologic diseases. Metabolomic profiling of serum and synovial fluid has identified specific metabolites associated with rheumatoid arthritis.8–10 The pentose phosphate pathway (PPP) is a parallel pathway of glycolysis that may a play key role in specific inflammatory diseases. Defects in glycolytic flux due to upregulation of glucose-6-phosphate dehydrogenase (G6PD), an enzyme in the PPP, promote hyperproliferation and cytokine production in T cells from patients with rheumatoid arthritis.11 Activated metabolism with hyperactivation of the PPP has been demonstrated in circulating lymphocytes from patients with systemic lupus erythematosus (SLE), and metabolic inhibitors can ameliorate pathology in animal models of lupus.12–15
Nephrotic and nephritic syndromes represent a spectrum of glomerulonephropathies characterised in part by shared end-organ kidney damage with a significant degree of activation of ischaemic injury.16 To what extent immunometabolic changes contribute to different types of kidney disease is unknown. The objectives of this study were to compare metabolic pathways of gene transcription in renal tissue from patients with different forms of glomerulonephritis and to determine the cellular source of specific metabolic transcription signatures in these diseases.
METHODS
Discovery cohort
Kidney biopsy samples from patients with glomerulonephritis and healthy donors were obtained from the European Renal cDNA Bank (ERCB) cohort. The ERCB is a multicentre study established to collect renal biopsy tissue for gene expression analysis at the time of a clinically indicated biopsy.17 Biopsies were obtained from patients after informed consent with approval of the local ethics committees. For this study, patients with nephrotic syndrome (NS, n=62) and with antineutrophil cytoplasmic antibody (ANCA)-associated vasculitis (AAV, n=23), a prototypical nephritic syndrome, were included in a discovery cohort. Three forms of NSs were studied: minimal change disease (MCD, n=14), membranous glomerulonephritis (MGN, n=21) and focal segmental glomerulosclerosis (FSGS, n=25). Two forms of AAV were included: granulomatosis with polyangiitis and microscopic polyangiitis (MPA). All patients with AAVhad a positive ANCA-antibody and diagnostic confirmation of disease by renal histology. Healthy tissue obtained from living transplant donors (LD, n=21) was used as a comparator group. Detailed histology from the ERCB cohort was not available, and clinical information recorded at the time of renal biopsy was limited but included use of glucocorticoids (yes/no, categorical variable) and glomerular filtration rate (GFR).18 To determine if gene expression signatures identified in the discovery cohort were unique to AAV or NS, relevant signatures were also queried in previously reported data from additional patients in the ERCB cohort, including patients with SLE (n=32) and patients who underwent tumour nephrectomy with donation of normal renal tissue adjacent to tumour (n=6).19
Validation cohort
An independent, validation cohort was studied consisting of microdissected renal biopsies from additional patients with AAV (n=57) from the ERCB cohort, additional LD (n=6), and patients with NS (n=107) recruited in the Nephrotic Syndrome Study Network (NEPTUNE). NEPTUNE is an ongoing multicentre prospective cohort study enrolling patients with glomerular diseases.20 Detailed clinical and histopathological data from patients with nephrotic syndrome enrolled in NEPTUNE, including quantification of tubulointerstitial fibrosis, has been previously published.21
Kidney tissue processing and transcriptional profiling
Kidney tissue was processed prior to transcriptional profiling as previously described.22 Briefly, collected renal tissue was stored in RNAlater (ThermoFisher) and manually microdissected into glomerular and tubulointerstitial compartments. Transcriptional data were used to assess reliability of microdissection, targeting 16-fold to 64-fold enrichment of glomerular-selective or tubulointerstitial-selective transcripts in each respective compartment. In the discovery cohort, RNA from each compartment was processed and analysed using Affymetrix GeneChip Human Genome U133A V.2.0 and U133 Plus V.2.0 platforms. In the validation cohort, samples were profiled on a Human Gene ST 2.1 array platform. Probe sets were annotated to Entrez Gene IDs using custom CDF V.19 generated from the University of Michigan Brain Array group, as previously described.23 Expression data were quantile normalised and batch corrected using COMBAT.24 Differential gene expression of selected gene transcripts was compared in the glomerular and tubulointerstitial compartments between patients with NS and AAV versus LD using the significance analysis of microarrays (SAM) method.25,26 Genes were defined as significantly differentially expressed with q-value <0.05. CEL files are accessible in GEO under reference numbers: GSE104948, GSE104954 and GSE108113.
Selected metabolic and inflammatory genes
A list of 88 genes related to metabolic pathways and 25 genes related to immune cell subset markers were selected a priori for analysis.27 Metabolic enzymes were selected to represent the following metabolic pathways: TCA cycle, glutaminolysis, fatty acid oxidation, glycolysis and the PPP. A composite gene expression score was created for each metabolic pathway by averaging z-scored transformed log2 expressed genes within the pathway. For the PPP, the composite score was created from the following genes: G6PD, PGLS, PGD, RPE, RPIA, TKT and TALDO1. A subset of enyzmes were categorised based on regulation by the transcription factor HIF-1α. A complete list of the selected genes is provided in online supplementary material.
Subset prediction from enrichment correlation analysis
Subset prediction from enrichment correlation (SPEC) was performed to determine the cell-specific source for PPP gene expression.28 Immune cell type-specific gene sets and renal cell type-specific gene sets were derived from previously published studies.29,30 Single-sample gene set enrichment analysis (ssGSEA) enrichment scores (ES) were calculated using each cell type-specific gene set and a PPP gene set curated from the KEGG metabolic pathways (V.5.2 from the Molecular Signatures Database). To determine how suitable a cell type gene set is for SPEC, cell type gene sets are randomly split in half (‘SET A and SET B’). ssGSEA is performed on all cell types and the number of times the ES from SET B is most strongly correlated with SET A rather than any other cell type is counted. The split experiment is replicated 100 times for each cell type gene set to generate a confidence metric in the findings. Correlation between each cell type ES and the PPP ES was calculated. Higher correlation indicates that a specific cell type contributes to PPP expression in the total population of samples.
Development of TNF activation score
Previously identified candidate genes causally downstream of TNF in kidney disease with at least three literature sources of evidence were selected as a gene set representative of TNF activation.31 A TNF activation score was generated in patient samples by transforming log2 expression profiles into z-scores and averaging z-scores of 138 TNF-dependent genes into a single metric for each patient sample.
Tissue immunofluorescence
The expression and localisation of immunometabolism-associated proteins was evaluated in paraffin-embedded kidney sections by indirect immunofluorescence in five patients with AAV. Kidney biopsies were classified based on histology as focal, crescentic, mixed or sclerotic, according to consensus guidelines.32 Briefly, after xylene/ethanol deparaffinisation, tissue sections were pretreated with Epitope Retrieval Solution (IHC World, Woodstock, Maryland, USA) for 40 min at 60°C. Subsequently, non-specific binding was blocked with 10% Normal Goat Serum (NGS) for 30 min at room temperature. Tissue slides were double stained with combinations of antibodies specific for CD68, TKT and HIF-1α (Abcam, Cambridge, Massachusetts, USA; BioLegend, San Diego, California, USA) overnight at 4°C in 1% NGS. Secondary antibodies were purchased from ThermoFisher Scientific (Grand Island, New York, USA), and tissue sections incubated accordingly for 1 hour at room temperature in 1% NGS. Hoechst-stained slides were then mounted with ProLong Gold Antifade Mountant (ThermoFisher), and visualised after curing overnight.
Statistical analyses
Correlation analyses were performed using Pearson’s correlation. Kruskal-Wallis with post hoc Dunn’s test to account for multiple comparisons was used to compare metabolic pathway scores across multiple disease groups, and a p value <0.05 was considered significant for these analyses. Analyses were performed using GraphPad Prism V.7.0 (La Jolla, California, USA).
RESULTS
Alteration of metabolic pathways in the glomerular and tubulointerstitial compartments
Gene expression signatures for five major metabolic pathways were profiled in the glomerular and tubulointerstitial compartments between patients with NSs, AAV and healthy living donors. The clinical characteristics of the study population are presented in table 1.
Table 1.
Patient characteristics for discovery and validation cohorts
| Disease group | Discovery cohort | Validation cohort | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Living donor | Nephrotic syndrome | ANCA-associated vasculitis | Systemic lupus erythematosus | Tumour nephrectomy | Living donor | Nephrotic syndrome | ANCA-associated vasculitis | |
| Glomerular samples | 21 | 58 | 23 | 32 | 6 | 6 | 90 | 15 |
|
| ||||||||
| Tubulointerstitial samples | 21 | 47 | 21 | 32 | 0 | 5 | 107 | 57 |
|
| ||||||||
| Age, years (SD) | 47.3 (11.5) | 47.2 (17.7) | 58.0 (13.8) | 35.1 (13.3) | 66.4 (6.8) | 52.5 (6.9) | 47.1 (15.7) | 57.8 (9.8) |
|
| ||||||||
| Sex (% female) | 45 | 45 | 43 | 78 | 100 | 57 | 33 | 58 |
|
| ||||||||
| Glomerular filtration rate (SD) | 104 (31) | 83 (39) | 46 (31) | 64 (39) | 58 (10.5) | 108 (33) | 77 (32) | 31 (27) |
ANCA, antineutrophil cytoplasmic antibody.
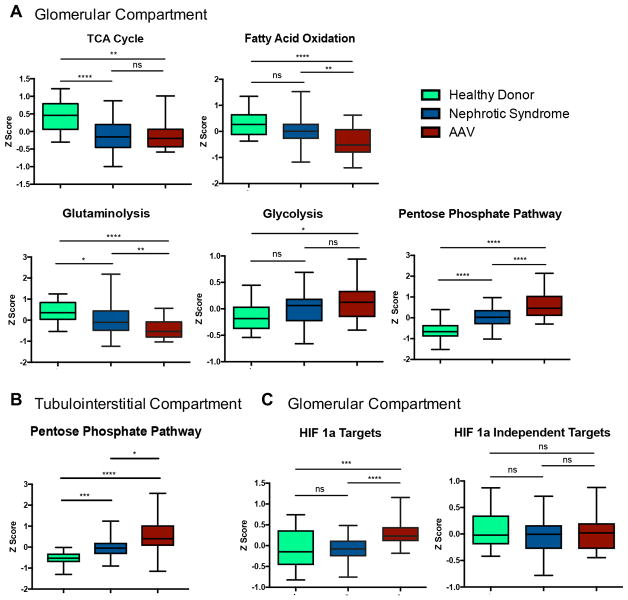
In the glomerular compartment of the discovery cohort, there was a coordinated pattern of altered gene expression related to specific metabolic pathways. Gene expression of the TCA cycle, fatty acid oxidation and glutaminolysis was repressed in NS relative to LD and further downregulated in patients with AAV (figure 1A). In contrast, there was significantly increased expression of the PPP in patients with NS and AAV relative to LD (p<0.001), with significantly higher expression in patients with AAV compared with NS (p<0.001). There was increased gene expression related to glycolysis in patients with NS and AAV compared with LD but this was not statistically significant. Differential expression of the PPP was also observed in the tubulointerstitial compartment of the discovery cohort (figure 1B). There was differential upregulation of glycolytic enzymes regulated by the transcription factor HIF-1α in patients with AAV compared with LD (p<0.001) and patients with NS (p<0.005), and there were no differences in expression of glycloytic enzymes not regulated by HIF-1α across the groups (figure 1C).
Figure 1.
Bar graphs of metabolic pathways showing disease-specific differential gene expression within the glomerular compartment in the discovery cohort (A). Bar graph of pentose phosphate pathway gene expression within the tubulointerstitial compartment in the discovery cohort (B). Differences in composite gene expression were compared in the discovery cohort between hypoxia-induced factor 1-alpha (HIF-1α) glycolytic targets and HIF-1α independent glycolytic enzymes in the glomerular compartment (C). *p<0.05; **p<0.01; ***p<0.005; ****p<0.001, NS, not significant. AAV, antineutrophil cytoplasmic antibody-associated vasculitis; TCA, tricarboxylic acid.
In the glomerular compartment of the discovery cohort, all seven enzymes of the PPP were significantly upregulated in patients with AAV compared with controls (fold change range 1.14–1.57, q<0.05). In the tubulointerstitial compartment, six enzymes of the PPP were significantly upregulated in patients with AAV compared with controls (fold change range 1.05–1.52; q<0.05) (table 2).
Table 2.
Glomerular compartment pentose phosphate pathway gene expression in ANCA-associated vasculitis compared with healthy donors in the discovery cohort
| Enzyme | Gene symbol | Glomerular compartment | Tubulointerstitial compartment | ||
|---|---|---|---|---|---|
|
|
|
||||
| Fold change | Q values | Fold change | Q values | ||
| Glucose-6-phosphate dehydrogenase | G6PD | 1.30 | <0.0001 | 1.10 | 0.0323 |
|
| |||||
| 6-Phosphogluconolactonase | PGLS | 1.59 | <0.0001 | 1.06 | 0.0855 |
|
| |||||
| Phosphogluconate dehydrogenase | PGD | 1.15 | 0.0210 | 1.16 | 0.0076 |
|
| |||||
| Ribulose-5-phosphate-3-epimerase | RPE | 1.19 | 0.0002 | 1.18 | <0.0001 |
|
| |||||
| Ribose-5-phosphate isomerase A | RPIA | 1.24 | <0.0001 | 1.19 | <0.0001 |
|
| |||||
| Transketolase | TKT | 1.71 | <0.0001 | 1.61 | <0.0001 |
|
| |||||
| Transaldolase 1 | TALDO1 | 1.28 | <0.0001 | 1.37 | <0.0001 |
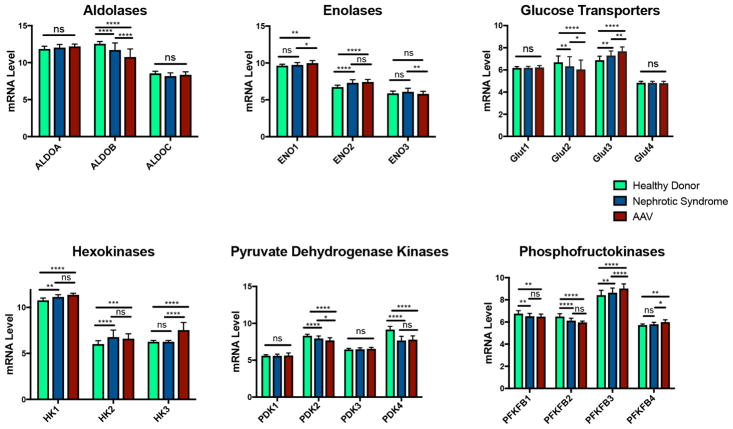
Differential expression of specific metabolic isozymes
Differential expression of selected, key regulatory metabolic isozymes was compared across LD, NS and AAV groups in the discovery cohort (figure 2). Specific isozymes were significantly upregulated (ENO2, HK1, HK2) or downregulated (PDK4, PFKB1, PFKFB2) in NS and AAV compared with LD without differences between NS and AAV. Other isozymes were significantly upregulated (GLUT3, PFKFB3) or downregulated (ALDOB, GLUT2, PDK2) in AAV>NS>LD. ENO1, HK3 and PFKB4 were only significantly upregulated in AAV compared with NS and LD. There were no significant differences across the groups for ALDOA, ALDOC, GLUT1, GLUT4, PDK1 and PDK3.
Figure 2.
Comparison of differences in glomerular gene expression (log2 mRNA levels) among selected metabolic isozymes in patients with nephrotic syndrome, antineutrophil cytoplasmic antibody-associated vasculitis and healthy donors in the discovery cohort. *p<0.05; **p<0.01; ***p<0.005; ****p<0.001, NS, not significant.
Pentose phosphate pathway gene expression signature in the validation cohort and in association with kidney function
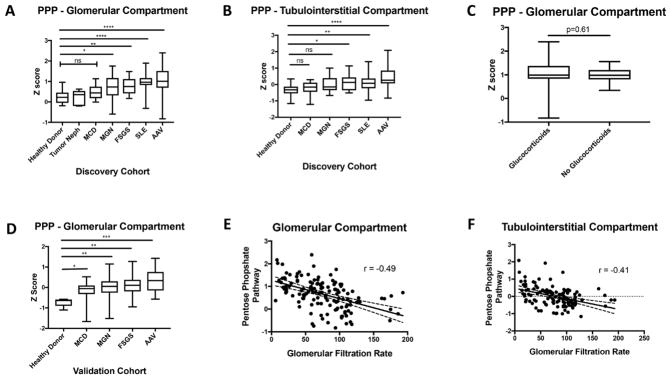
To confirm if increased PPP expression was unique to AAV, gene expression of the PPP was compared in the discovery cohort across a broader spectrum of diseases including SLE (figure 3A and B). PPP expression did not differ between LD, normal kidney biopsy sections from patients who underwent tumour nephrectomy and patients with MCD. PPP expression in SLE was significantly greater than LD (p<0.001) but was not different from AAV. Among the two types of NSs with potential inflammatory components (MGN, FSGS), there was increased PPP expression compared with LDs (p<0.01), with lower median PPP expression values than SLE or AAV. In an analysis restricted to patients with AAV or SLE, there was no significant difference in PPP expression score between patients categorised by concomitant glucocorticoid use at the time of biopsy versus those not treated; however, there was increased variability of PPP expression in those patients taking glucocorticoids (figure 3C). Differential expression of the PPP was confirmed in the validation cohort using samples from an independent group of patients with glomerulonephritis (figure 3D). There was a significant negative association between PPP expression and GFR in the glomerular compartment (r=−0.49, p<0.01) (figure 3E) and the tubulointerstitial compartment (r=−0.41, p<0.01) (figure 3F). In analyses restricted to patients with NS in the NEPTUNE cohort where detailed histology was available for review, GFR was negatively associated with PPP expression in the glomerular (r=−0.37, p<0.01) and the tubulointerstitial compartments (r=−0.29, p<0.01), and PPP expression in the tubulointerstitial compartment was significantly associated with an increased degree of tubulointerstitial fibrosis on pathology (r=0.22, p=0.03).
Figure 3.
Pentose phosphate pathway (PPP) gene expression is differentially regulated in a variety of glomerulonephritis and is associated with impaired kidney function. PPP gene expression differences among groups collected within the discovery cohort in the glomerular (A) and tubulointerstitial (B) compartments. Comparison of PPP gene expression within the glomerular compartment in the discovery cohort between patients with antineutrophil cytoplasmic antibody-associated vasculitis (AAV) or systemic lupus erythematosus (SLE) categorised by glucocorticoid use at the time of biopsy (C). Validation of PPP gene expression differences within the glomerular compartment in the validation cohort (D). Correlation between PPP expression and glomerular filtration rate in the glomerular (E) and tubulointerstitial (F) compartments in the discovery cohort. *p<0.05; **p<0.01; ***p<0.005; ****p<0.001, NS, not significant. FSGS, focal segmental glomerulosclerosis; MCD, minimal change disease; MGN, membranous glomerulonephritis.
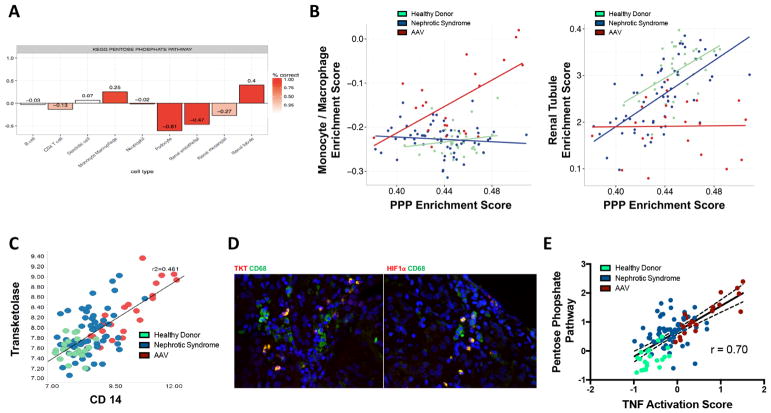
Cellular source of the pentose phosphate pathway gene expression signature
SPEC analysis of the discovery cohort glomerular compartment dataset indicated that PPP expression was specifically increased in monocyte/macrophages and renal tubular cells (figure 4A). Correlation analyses stratified by group (NS, LD, AAV) demonstrated positive correlation between monocyte/macrophage ES and PPP ES only in patients with AAV (r=0.62, p<0.01) and positive correlation between renal tubule ES and PPP ES only in LD (r=0.49, p<0.01) and patients with NS (r=0.63, p<0.01) (figure 4B). In the glomerular compartment in patients with LD, NS and AAV, there was positive correlation between transketolase (TKT), a representative enzyme of the PPP, and CD68, a representative macrophage marker (r=0.48; p<0.01) (figure 4C). In patients with AAV with crescentic findings on kidney biopsy (n=2), protein expression of TKT and HIF-1α colocalised with CD68+ macrophages, with no expression observed in renal parenchymal cells (figure 4D). In patients with AAV without crescentic histology (n=3), no protein expression of TKT or HIF-1α was observed (data not shown). To determine if PPP expression correlated with macrophage-related cytokine production, the PPP score from the glomerular compartment was correlated with a TNF activation score. There was strong, positive correlation (r=0.70, p<0.01) between the PPP and TNF activation scores, with the highest values observed in patients with AAV compared with NS and LD (figure 4E).
Figure 4.
Myeloid cells are likely a major source of activated pentose phosphate pathway (PPP) gene expression. Subset prediction from enrichment correlation (SPEC) predicts that renal tubule and monocyte/macrophages are the likely source of PPP in the discovery cohort with the intensity of the red bar indicates degree of confidence in the bar plot correlation of cell types and PPP expression (A). The monocyte/macrophage enrichment score correlates with the PPP enrichment score in antineutrophil cytoplasmic antibody-associated vasculitis (AAV), while the tubule enrichment score correlates with the PPP enrichment score in healthy living donor and nephrotic syndrome samples (B). Transketolase (TKT), a key regulatory enzyme within the PPP, correlates with CD14, a marker for monocyte/macrophages in the glomerular compartment in the discovery cohort (C). Tissue immunofluorescence demonstrates localisation of TKT and HIF-1α within monocytes/macrophages (CD14) in the glomerular compartment (D). The PPP gene expression score is strongly associated with increased expression of a tumour necrosis factor (TNF) activation score within the glomerular compartment (E).
DISCUSSION
Under conditions of cellular homeostasis, the TCA cycle serves as the most efficient source of energy production in humans. However, under conditions of cellular stress, including inflammatory microenvironments, glucose can become a preferred metabolic substrate. This study demonstrated concordant alterations of the renal transcriptome consistent with metabolic reprogramming across different forms of glomerulonpehritis. Gene expression profiling of renal tissue from the glomerular compartment revealed downregulation of pathways of cellular homeostasis, including the TCA cycle, glutaminolysis and fatty acid oxidation, and upregulation of pathways of glucose metabolism, including the PPP. Significant upregulation of HIF-1α-related gene transcripts and colocalisation of HIF-1α and CD68 by tissue immunofluorescence in the glomeruli of patients with AAV suggests that this transcription factor plays a critical role in the regulation of glycolytic pathways in glomerulonephritis.16,33
Activation of the PPP in both the glomerular and tubulointerstitial compartments in a discovery and validation cohort was the most striking finding in this study. Increased expression of enzymes of the PPP was demonstrated in NSs compared with healthy living donors with the highest levels of PPP expression seen in inflammatory kidney diseases. In patients with NS, increased expression of the PPP in the tubulointerstitial compartment was significantly associated with reduced kidney function and increased intensity of tubulointerstitial fibrosis. Although renal disease in AAV is typically defined by glomerular involvement, similar alterations of PPP enzyme transcription were observed in renal biopsies from patients with AAV in both the glomerular and tubulointerstitial compartments. Global alterations of the renal transcriptome across different anatomic compartments are therefore associated with renal disease in AAV. Similar levels of PPP expression in kidney biopsies from patients with AAV or SLE indicate that alterations of metabolic pathways might be shared across different forms of glomerulonephritis.
Several lines of evidence suggest that monocyte/macrophages are likely a major contributor to PPP expression in these diseases. Increased PPP expression was observed in the NS subtypes where inflammatory features on histology are most pronounced, including MGN and FSGS compared with MCD. Computational analyses showed that PPP expression strongly correlated with monocyte/macrophage surface markers, especially in patients with AAV, and protein expression of PPP enzymes colocalised to macrophages within the glomerular compartment by tissue immunofluorescence. One function of the PPP is to generate NADPH and maintain redox balance, which may be particularly important to cellular survival in activated macrophages undergoing oxidative burst. Another function of the PPP is to generate nucleic acid precursors. Production of biomass through the PPP could facilitate generation of the necessary messenger RNA and protein to enable effector functions. Activation of the PPP is known to induce pro-inflammatory cytokine production in macrophages,5 and in this study, strong correlation was observed between PPP expression and TNF activation within the glomerular compartment, particularly in AAV.
In addition to regulation of important metabolic pathways, differential expression of key, regulatory metabolic isozymes was observed across the conditions studied. These findings may inform future functional studies of metabolic pathways in renal disease. PFKFB3, which was upregulated in both NSs and AAV, has been specifically associated with the Warburg effect in tumour cells because its activity increases the rate of glycolysis. 34 Among glucose transporters, which facilitate glucose passage across plasma membranes, there was upregulation of GLUT3 and downregulation of GLUT2 in both NS and AAV. GLUT3 is the highest affinity glucose receptor and therefore may play a key role in facilitating glucose metabolism in these conditions.35 Hexokinases regulate the first step in glycolysis, and significant increased expression of HK3 was observed in patients with AAV. HK3 is the predominant hexokinase in myeloid cells and is upregulated in peripheral blood samples from patients with AAV in a prior transcriptomic study.36,37 The functions of HK3 are poorly characterised, making it an attractive candidate for future functional studies.
This study has some important potential limitations to consider. Concomitant use of glucocorticoids can affect gene expression and information about glucocorticoid dose at the time of biopsy was not available; however, no significant difference between PPP scores were observed when adjusting for glucocorticoid use as a categorical variable. Detailed pathological descriptions from renal biopsies in the ECRB cohort was not available across the cohort, precluding comparison of the renal transcriptome with histological characteristics of disease. Urinary metabolites were not studied; however, alterations of glycolysis-related transcripts in animal models of diabetes have predicted changes in glycolytic metabolites in renal cortex and urine.38 Finally, subgroup comparisons were limited by small sample sizes.
Distinct alterations in cellular metabolism were observed in the renal transcriptome from patients with different forms of glomerulonephritis, including NSs and systemic inflammatory diseases such as AAV and SLE. Global patterns of gene expression are indicative of increased utilisation of glucose and decreased oxidative phosphorylation, especially in patients with inflammatory kidney diseases. Metabolic reprogramming of cells within affected renal tissue may constitute a form of shared molecular pathology across different types of glomerulonephritis. The strong correlations between markers of glycolysis, macrophage-related markers and inflammatory cytokines observed in this study further suggest that altered immunometabolism may also play a role in the pathophysiology across a spectrum of kidney diseases. Validation of these findings in prospective, observational cohorts with assessment of potential associations between metabolic gene expression signature, detained renal histology and long-term clinical outcomes is warranted. Modulation of glucose metabolism could offer novel approaches to the treatment of these rare syndromes.
Acknowledgments
The authors acknowledge Mariana Kaplan, Chief of the Systemic Autoimmunity Branch within Intramural NIAMS, for assistance in the conduct of this study. The authors also acknowledge all participating centers of the European Renal cDNA Bank, the Vasculitis Clinical Research Consortium and the Nephrotic Syndrome Study Network Consortium.
Funding The Vasculitis Clinical Research Consortium (VCRC), U54 AR057319, and the Nephrotic Syndrome Study Network Consortium (NEPTUNE), U54 DK083912, are part of the National Institutes of Health (NIH) Rare Disease Clinical Research Network (RDCRN), supported through collaboration between the Office of Rare Diseases Research (ORDR), NCATS and the National Institute of Diabetes, Digestive, and Kidney Diseases (NIDDK) and the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS). This research was also supported through the Intramural Research Program at the NIAMS. Additional funding and/or programmatic support for this project has also been provided by the Else Kröner-Fresenius Foundation (ERCB), VCRC, University of Michigan, the NephCure Kidney International and the Halpin Foundation and the Applied Systems Biology Core at the University of Michigan George M O’Brien Kidney Translational Core Center. Dr Taroni is supported by the University of Pennsylvania Training Program in Rheumatic Diseases (NIAMS T32AR007442).
Footnotes
Part of this manuscript was presented at the 2016 ACR/ARHP Annual Meeting.
Contributors PCG, SE, JK, MK and PAM designed the study. YLL, MTL, W-JJ, BG, CDC collected the data and performed the experiments. PCG, SE, JNT, HP, CSG, JK, MK and PAM performed the statistical analysis. PCG, SE, JNT, YLL, LM, HP, CSG, JK, MK and PAM analysed the data. PCG, SE, JNT, YLL, JK, MK and PAM wrote the manuscript. All authors approved the final version of the manuscript.
Competing interests None declared.
Patient consent Not required.
Ethics approval The study was approved by the ethics committees for the European Renal cDNA Bank (ERCB) and the Nephrotic Syndrome Study Network Consortium (NEPTUNE). Ethics approval for the gene expression studies was provided by the University of Michigan (HUM0002468).
Provenance and peer review Not commissioned; externally peer reviewed.
References
- 1.O’Neill LA, Kishton RJ, Rathmell J. A guide to immunometabolism for immunologists. Nat Rev Immunol. 2016;16:553–65. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tannahill GM, Curtis AM, Adamik J, et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496:238–42. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galván-Peña S, O’Neill LA. Metabolic reprograming in macrophage polarization. Front Immunol. 2014;5:420. doi: 10.3389/fimmu.2014.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moon JS, Hisata S, Park MA, et al. mTORC1-Induced HK1-Dependent Glycolysis Regulates NLRP3 Inflammasome Activation. Cell Rep. 2015;12:102–15. doi: 10.1016/j.celrep.2015.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Haschemi A, Kosma P, Gille L, et al. The sedoheptulose kinase CARKL directs macrophage polarization through control of glucose metabolism. Cell Metab. 2012;15:813–26. doi: 10.1016/j.cmet.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jha AK, Huang SC, Sergushichev A, et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity. 2015;42:419–30. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 7.O’Neill LA, Pearce EJ. Immunometabolism governs dendritic cell and macrophage function. J Exp Med. 2016;213:15–23. doi: 10.1084/jem.20151570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim S, Hwang J, Xuan J, et al. Global metabolite profiling of synovial fluid for the specific diagnosis of rheumatoid arthritis from other inflammatory arthritis. PLoS One. 2014;9:e97501. doi: 10.1371/journal.pone.0097501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang M, Chen T, Feng H, et al. Serum metabolic signatures of four types of human arthritis. J Proteome Res. 2013;12:3769–79. doi: 10.1021/pr400415a. [DOI] [PubMed] [Google Scholar]
- 10.Weyand CM, Goronzy JJ. Immunometabolism in early and late stages of rheumatoid arthritis. Nat Rev Rheumatol. 2017;13:291–301. doi: 10.1038/nrrheum.2017.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Z, Fujii H, Mohan SV, et al. Phosphofructokinase deficiency impairs ATP generation, autophagy, and redox balance in rheumatoid arthritis T cells. J Exp Med. 2013;210:2119–34. doi: 10.1084/jem.20130252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morel L. Immunometabolism in systemic lupus erythematosus. Nat Rev Rheumatol. 2017;13:280–90. doi: 10.1038/nrrheum.2017.43. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez DR, Telarico T, Bonilla E, et al. Activation of mammalian target of rapamycin controls the loss of TCRzeta in lupus T cells through HRES-1/Rab4-regulated lysosomal degradation. J Immunol. 2009;182:2063–73. doi: 10.4049/jimmunol.0803600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perl A, Hanczko R, Lai ZW, et al. Comprehensive metabolome analyses reveal N-acetylcysteine-responsive accumulation of kynurenine in systemic lupus erythematosus: implications for activation of the mechanistic target of rapamycin. Metabolomics. 2015;11:1157–74. doi: 10.1007/s11306-015-0772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin Y, Choi SC, Xu Z, et al. Normalization of CD4+ T cell metabolism reverses lupus. Sci Transl Med. 2015;7:274ra18. doi: 10.1126/scitranslmed.aaa0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shved N, Warsow G, Eichinger F, et al. Transcriptome-based network analysis reveals renal cell type-specific dysregulation of hypoxia-associated transcripts. Sci Rep. 2017;7:8576. doi: 10.1038/s41598-017-08492-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lindenmeyer MT, Kretzler M, Boucherot A, et al. Interstitial vascular rarefaction and reduced VEGF-A expression in human diabetic nephropathy. J Am Soc Nephrol. 2007;18:1765–76. doi: 10.1681/ASN.2006121304. [DOI] [PubMed] [Google Scholar]
- 18.Levey AS, Coresh J, Greene T, et al. Using standardized serum creatinine values in the modification of diet in renal disease study equation for estimating glomerular filtration rate. Ann Intern Med. 2006;145:247–54. doi: 10.7326/0003-4819-145-4-200608150-00004. [DOI] [PubMed] [Google Scholar]
- 19.Ju W, Greene CS, Eichinger F, et al. Defining cell-type specificity at the transcriptional level in human disease. Genome Res. 2013;23:1862–73. doi: 10.1101/gr.155697.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gadegbeku CA, Gipson DS, Holzman LB, et al. Design of the Nephrotic Syndrome Study Network (NEPTUNE) to evaluate primary glomerular nephropathy by a multidisciplinary approach. Kidney Int. 2013;83:749–56. doi: 10.1038/ki.2012.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mariani LH, Martini S, Barisoni L, et al. Interstitial fibrosis scored on whole-slide digital imaging of kidney biopsies is a predictor of outcome in proteinuric glomerulopathies. Nephrol Dial Transplant. 2018;33:310–8. doi: 10.1093/ndt/gfw443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen CD, Frach K, Schlöndorff D, et al. Quantitative gene expression analysis in renal biopsies: a novel protocol for a high-throughput multicenter application. Kidney Int. 2002;61:133–40. doi: 10.1046/j.1523-1755.2002.00113.x. [DOI] [PubMed] [Google Scholar]
- 23.Dai M, Wang P, Boyd AD, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 25.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmid H, Boucherot A, Yasuda Y, et al. Modular activation of nuclear factor-kappaB transcriptional programs in human diabetic nephropathy. Diabetes. 2006;55:2993–3003. doi: 10.2337/db06-0477. [DOI] [PubMed] [Google Scholar]
- 27.Caspi R, Billington R, Ferrer L, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2016;44:D471–D480. doi: 10.1093/nar/gkv1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bolen CR, Uduman M, Kleinstein SH. Cell subset prediction for blood genomic studies. BMC Bioinformatics. 2011;12:258. doi: 10.1186/1471-2105-12-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Renal tubule cells. http://nano.princeton.edu/standard/view/renal-tubule/
- 31.Eddy S, Mariani L, Martini S, et al. An Integrative genomics approach to predict patients with TNF activation in progressive nephrotic syndrome. Kidney Precision meeting abstract. http://www.niddk.nih.gov/news/events-calendar/Documents/abstract.pdf.
- 32.Berden AE, Ferrario F, Hagen EC, et al. Histopathologic classification of ANCA-associated glomerulonephritis. J Am Soc Nephrol. 2010;21:1628–36. doi: 10.1681/ASN.2010050477. [DOI] [PubMed] [Google Scholar]
- 33.Marín-Hernández A, Gallardo-Pérez JC, Ralph SJ, et al. HIF-1 alpha modulates energy metabolism in cancer cells by inducing over-expression of specific glycolytic isoforms. Mini Rev Med Chem. 2009;9:1084–101. doi: 10.2174/138955709788922610. [DOI] [PubMed] [Google Scholar]
- 34.Atsumi T, Chesney J, Metz C, et al. High expression of inducible 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (iPFK-2; PFKFB3) in human cancers. Cancer Res. 2002;62:5881–7. [PubMed] [Google Scholar]
- 35.Simpson IA, Dwyer D, Malide D, et al. The facilitative glucose transporter GLUT3: 20 years of distinction. Am J Physiol Endocrinol Metab. 2008;295:E242–E253. doi: 10.1152/ajpendo.90388.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Federzoni EA, Humbert M, Torbett BE, et al. CEBPA-dependent HK3 and KLF5 expression in primary AML and during AML differentiation. Sci Rep. 2014;4:4261. doi: 10.1038/srep04261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheadle C, Berger AE, Andrade F, et al. Transcription of proteinase 3 and related myelopoiesis genes in peripheral blood mononuclear cells of patients with active Wegener’s granulomatosis. Arthritis Rheum. 2010;62:1744–54. doi: 10.1002/art.27398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sas KM, Kayampilly P, Byun J, et al. Tissue-specific metabolic reprogramming drives nutrient flux in diabetic complications. JCI Insight. 2016;1:e86976. doi: 10.1172/jci.insight.86976. [DOI] [PMC free article] [PubMed] [Google Scholar]