Abstract
This study was designed to compare the performances of four different non-destructive methods of assessing onion quality, one of which was based on near-infrared spectroscopy, and three of which were based on spectral imaging. These methods involve a combination of wavelengths from visible to near-infrared with different acquisition systems that were applied to discriminate between pre-sorted onions by in situ measurements of the onion surface. Compared with the partial least squares discriminant analysis classification models associated with different methods, hyperspectral imaging (HSI) with both static horizontal and rotating orientation obtained a higher level of sensitivity and specificity with a lower classification error than did other methods. Moreover, models built with the reduced variables did not lower the model performances. Overall, these results demonstrate that HSI with selected wavelengths would be useful for further developing an improved real-time system for sorting onion bulbs.
Electronic supplementary material
The online version of this article (10.1007/s13197-018-3268-x) contains supplementary material, which is available to authorized users.
Keywords: Multispectral imaging, Hyperspectral imaging, Near-infrared spectroscopy, Principal component analysis, Partial least squares discriminant analysis, Allium cepa L.
Introduction
The onion is one of the most widely consumed vegetables in the world (Preedy and Watson 2014). After harvesting, field curing and drying, onion bulbs are kept in cold storage to maintain quality throughout the year and to satisfy consumers’ demand for extended availability (Rabinowitch and Currah 2002). During storage, onions are susceptible to various types of fungal and bacterial diseases, and the diseases show specific characteristic behaviors (Islam et al. 2017; Snowden 2010). The development of diseases is still unacceptably high and accounts for a large proportion of spoilage (pers. comm.).
Increased consumer awareness is making the marketing of onions more competitive. Consumers demand high-quality onions with clean and unblemished skins that are not split or loose (Brewster 2008) and are free of defects and diseases that can be achieved by non-destructive optical quality sorting techniques (de Oliveira et al. 2016; Zude 2008).
Near-infrared (NIR) spectroscopy is a non-destructive technique that is very simple and safe to use and has been intensively applied in the quality inspection of food over the past two decades (Patel et al. 2012). NIR spectroscopy measures one spatial point at a time, and it is suitable for the analysis of products which are high in water and carbohydrate content, as absorbance is higher for samples containing C–H, C–O and O–H molecules (Jha et al. 2010). Moreover, water has a strong absorption rate in the NIR region; thus, spectra from products with high water content (> 80%) are dominated by the spectral signature of water (Büning-Pfaue 2003).
Conversely, spectral imaging allows the simultaneous collection of spatial and spectral information about the product (Lu and Chen 1999; Vetrekar et al. 2015), which can be interpreted as a set of spectra on a two-dimensional area or a succession of images recorded at a number of specific wavelengths. If an application requires a real-time application, multispectral imaging (MSI) with several bands can be a practically viable solution. The processes involved in MSI are image acquisition, the algorithm for image processing and decision-making. MSI acquires images with a relatively higher spatial resolution at several wavelengths. In contrast, HSI often collects images with higher spectral resolution; thus, it provides additional information that MSI may have missed (Qin et al. 2013). The full wavelength spectrum of HSI data can be reduced to several key wavelengths for specific substances. Next, these key wavelengths can be implemented to develop an MSI system with enormous advantages.
Apart from different measurement techniques, the orientation of products during measurement also influences the results. Research has not been conducted to compare advanced techniques when considering different acquisition systems. Recently, Kuroki et al. (2017) investigated six different orientations of onions to predict rot levels using the transmittance of visible near-infrared spectroscopy. The best model with the highest R2 and the lowest error of prediction was obtained with the measurement at the equatorial region.
Sorting machines based on conventional NIR spectroscopy are available for the size and quality sorting of onions. These machines are working either on reflectance or transmittance mode. At the end of the storage period, during sorting, numerous misclassifications happen (pers. comm.), and misclassification of healthy bulbs leads to food loss and waste. Novel non-destructive techniques may be applied to improve accuracy in the sorting of onions. Hence, the objective of this study was to compare the classification efficiency of the NIR spectroscopic system and the NIR spectral imaging systems (MSI and HSI) with different acquisition techniques for classifying healthy and diseased onion bulbs. The comparison of these emerging analytical methods is worth investigating due to their interesting and direct applications for industrial process lines, which may lead to quality improvement and economic advantages.
Materials and methods
Plant material
An organically grown cultivar ‘Barito’ was selected for the experiment. At the end of the cold storage (~ 1 °C, ~ 80–90% Relative Humidity) period (6 months), a total of six sacks (~ 120 kg) were blindly sampled from the box and were sorted in an automated online sorting system. Bulbs (40–80 mm diameter) were sorted into healthy and diseased categories. From the healthy category, a total of 15 onions were blindly selected by hand based on a firm feeling and from the diseased category, a total of 30 onions were blindly selected. The selected bulbs were numbered immediately and were brought to the laboratory of Post-harvest Technology, Årslev, Denmark for further analysis.
Near-infrared spectroscopy
NIR spectroscopy was carried out using an AgriQuant (Q-Interline, Tølløse, Denmark) Fourier transform near-infrared (FT-NIR) spectrometer as described by Travers et al. (2014) (Online Resource 1a). The AgriQuant was operating in the 300–2500 nm range. The instrument was fitted with a fiber-optic cable and a direct contact probe working in reflectance mode with a diameter of approximately 10 mm. A white Teflon tile (polytetrafluoroethylene, PTFE) was used to provide a reference spectrum before measurement. The instrument was configured with PLCATS (Version 4.4) and InfraQuant 2.5 software. The spectral resolution was 30 cm−1. Two points were marked in each onion; point 1 is a random point at the maximum diameter (i.e., equatorial region) of the bulb, and point 2 is the opposite side of point 1. Measurements were performed at both points of each onion. As two measurements had been taken per onion, the data in each of these sets were averaged to provide one mean spectrum for each onion (Travers et al. 2014).
Multispectral imaging
Spectral imaging at the visible to NIR (VIS–NIR) region with static horizontal orientation was carried out with a multispectral imaging (MSI) system. VideometerLab equipment (Videometer A/S, Hørsholm, Denmark) at 18 different wavelengths (405, 435, 450, 470, 505, 525, 570, 590, 630, 645, 660, 700, 780, 850, 870, 890, 940 and 970 nm) working on reflectance mode was used to capture images (Online Resource 1b). In this setup, onions were placed horizontally inside the acquisition system facing point 1 (i.e., equatorial region) towards the camera, and the reflection was recorded. The Videometer consisted of a high-resolution 1280 × 960, 45 μm monochrome grayscale CCD camera mounted at the top of the sphere, specially assembled 18 high power LED light sources, an integrating sphere with a matte white coating to ensure that the light is scattered evenly with a uniform, diffuse light at illumination. A computer equipped with a data acquisition system and the VideometerLab software version 3.0.30 (Videometer A/S, Hørsholm, Denmark) was used to acquire images, image correction, and analysis. The equipment was calibrated using bright, dark and geometric references prior to measurement.
Hyperspectral imaging (VIS–NIR)
Spectral imaging at the visible to NIR (VIS–NIR) region with a rotating orientation was performed with a hyperspectral imaging (HSI-VIS–NIR) system. Onions were placed in a customized industrial sorting machine equipped with a CMOS camera and LEDs covering the spectral region from 425 to 968 nm and captured with 216 hyperspectral bands. The onions were rotated continuously during data acquisition, making it possible to capture its entire surface apart from the distal ends of the onion, thus recording a data cube of almost the whole surface. This setup resulted in high-quality images as diffused illumination reduced surface glare and high-speed recordings with low exposure time avoided motion blur. The spatial resolution in this set up was 0.625 mm/pixel.
Hyperspectral imaging (SWIR)
Spectral imaging at the short wave infrared (SWIR) region with a horizontal static orientation was performed with a hyperspectral imaging (HSI-SWIR) system. A conveyor belt setup equipped with an InGaAs camera was used to capture hyperspectral images of the onions from 1016 to 1742 nm with 216 hyperspectral bands. Two halogen lamps were used as light sources, and they caused specular reflections on certain parts of the onion surface, causing saturation of the sensor and making the capture of certain areas of the surface unusable. Onions were placed on the conveyor belt in a similar way to capture the surface around point 1 at the equatorial region, as performed in an MSI system. Reflection from onions in this setup could only be captured one side at a time. The spatial resolution in this setup was 0.3125 mm/pixel.
Optics in both setups consisted of VIS–NIR or SWIR TechSpec fore-lenses (Edmund Optics, USA) along with the VIS–NIR V10E or SWIR V17E ImSpector spectrographs (Specim, Finland), and a white Teflon tile was used as a white reference. A dark reference was obtained by blocking the sensor input. The photograph and schematic diagram of the HSI-(VIS–NIR), and HSI-(SWIR) instruments are shown in Online Resource 1c, and Online Resource 1d respectively.
Visual quality assessment
The internal quality of the onions was evaluated visually. Onions were cut from the neck to the base, and diseases were registered by visual assessment of the symptoms as shown by Snowden (2010) and Schwartz and Mohan (2007). Before cutting, the weight and diameter of each onion was recorded.
Total soluble solids (TSS)
One to two drops of onion juice was taken from the two outer fleshy scales of the onion, and soluble solids (% TSS) were measured on a refractometer.
Hypercube processing
In HSI, only a single line was scanned in each frame, and the frame also contained a spectrum for each point on the line. Merging several line scans or frames of a subject yielded a hyperspectral data cube with two spatial axes and one spectral axis. Thus, HSI data obtained from the ith measurement can be represented by a hypercube , where the j index corresponds to the line scan spatial direction, the k index corresponds to the spatial dimension obtained by the movement of the sample, and the n index represents the spectral dimension. Each of the measured hyperspectral cubes was first converted to reflection coefficients with the white reference and the black reference as given by Eq. (1).
| 1 |
A region of interest (ROI) mask corresponding to the location of the onion was obtained by thresholding the spatial dimension as described by Eq. (2):
| 2 |
where corresponds to a wavelength of 864 and 1604 nm for VIS–NIR and SWIR, respectively, and an empirically determined threshold of 0.2 was used.
To correct for uneven illumination, the spectral dimension of the reflection coefficients was normalized with its sum, as described by Eq. (3):
| 3 |
Finally, the average normalized spectrum was calculated for each sample from the region of interest and the normalized reflection coefficients as described by Eq. (4):
| 4 |
Hypercubes were processed using routines built in-house in R (R Core Development Team 2016, version 3.3.1).
Spectral pre-processing
Savitzky–Golay (SAV-GOL) is a signal smoothing and derivative calculation method. It has three arguments: a width of the filter, a polynomial order, and the derivative order. The Standard Normal Variate (SNV) normalization method is a weighted type of normalization, which calculates the standard deviation of all the pooled variables for the given sample. Multiplicative signal correction (MSC) corrects the additive and multiplicative effects caused by non-uniform scattering across the spectra.
Before pre-processing, FT-NIR spectra at the range < 748 and > 1165 nm were removed due to having a weak signal and high noise at both ends of the detector’s range (Travers et al. 2014). Figure 1 presents raw spectra and pre-processed spectra from four different methods. Savitzky–Golay spectral smoothing (3-point, 1st order polynomial, no derivative) and SNV was applied to the data set in a similar way as that described by Travers et al. (2014). MSI, HSI-(VIS–NIR) and HSI-(SWIR) were pre-processed using MSC as suggested by Wang et al. (2013), that is, by regressing a measured spectrum against the mean spectrum and then correcting the measured spectrum using the slope of this fit. All pre-processed spectra were mean-centered by subtracting the mean from the data values separately for every variable.
Fig. 1.
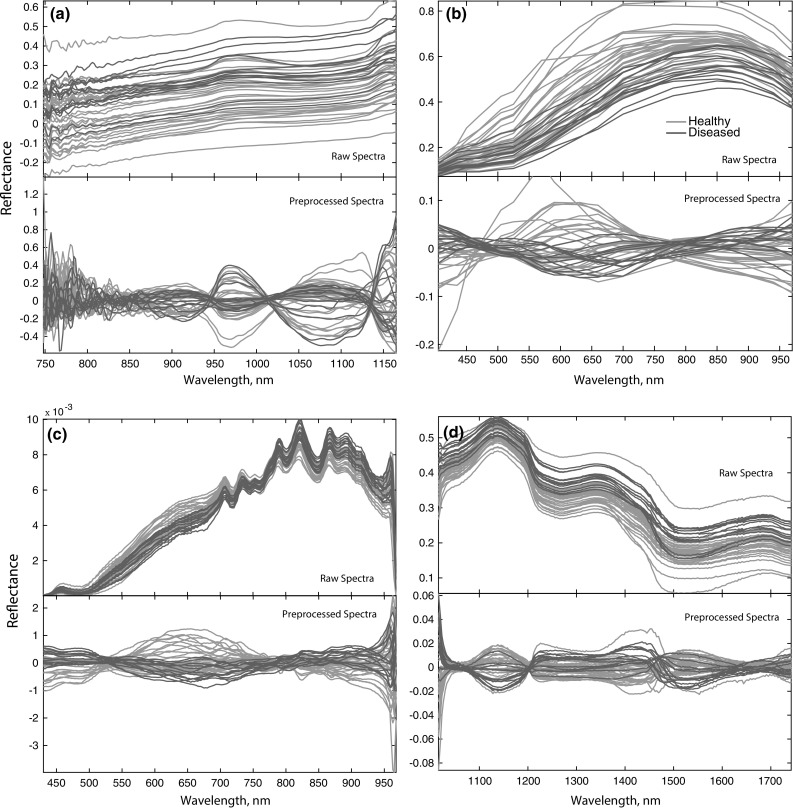
Raw spectra and Pre-processed spectra. a FT-NIR spectra, b MSI spectra, c HSI-(VIS–NIR) spectra, d HSI-(SWIR) spectra
Model building
Principal component analysis (PCA) was carried out to express the major information contained in the original variables with a lower number of variables, called principal components (PCs), which describe the main sources of variation in the data (Vetrekar et al. 2015). Partial least squares discriminant analysis (PLS-DA) models were built to differentiate between healthy and diseased onions (Roggo et al. 2007). Seventy percent of the samples were used to calibrate the model, and 30% were used for prediction. The models were cross-validated using leave-one-out cross-validation (Borras et al. 2014), and the optimal number of latent variables was chosen on the basis of the minimum value of the cross-validated classification error average.
A series of parameters can be used to evaluate the performance of classification models, such as sensitivity, specificity, and classification error. Sensitivity is the ability of the model to correctly classify healthy onions using the values of true positive (TP) and false negative (FN) and was calculated using Eq. (5). Specificity is the capacity of the model to correctly identify diseased onions relative to the values of true negative (TN) and false positive (FP) and was calculated using Eq. (6).
| 5 |
| 6 |
To identify important wavelengths for the models, Variable Important in Projection (VIP) scores (Zhang et al. 2017) and a Selectivity Ratio (SR) (Zhang et al. 2017) were calculated. Spectral pre-processing and multivariate data analysis and model building (PCA, PLS-DA) were carried out using PLS Toolbox (PLS_Toolbox V802, Eigenvector Research Inc., USA) in a MATLAB environment (version 9.3.0, MathWorks, Natick, MA, USA).
Results and discussion
Onion quality
The weight of healthy onions varied from 97 to 190 g, while the weight of diseased onion varied from 101 to 191 g (Online Resource 2). Diameters of onions were between 40 and 80 mm, as onions were selected only in that size range. The TSS of healthy onions varied from 7 to 7.5%, while it varied from 3.4 to 6% for diseased onions. The DM content of onion bulbs is an important parameter that partly defines storage life, pungency, and firmness (Sinclair et al. 1995). DM and TSS were found to be significantly correlated with the percentage of diseased bulbs (Ko et al. 2002). A wide variation in TSS in diseased onions in this study indicates a variation in stages of disease development.
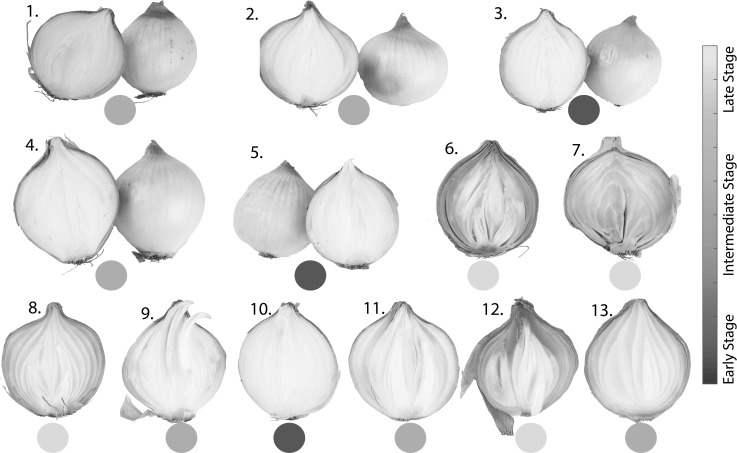
Figure 2 presents the images of diseased onions from the visual quality assessment. From the visual quality assessment, 13 out of 30 onions were identified as diseased; thus, 17 non-diseased onions were added to the group of healthy onions in further analysis. Diseased onions are numbered from 1 to 13 (Fig. 2). It can be observed from Fig. 2 that the level of infection varies from the early stage to the late stage. Different types of diseases were identified. Gray mold rot is a fungal disease caused by Botrytis spp., sour skin rot is a bacterial disease caused by Pseudomonas cepacia, neck rot is a fungal disease caused by Botrytis spp., and basal rot, which is also a fungal disease, is caused by Fusarium spp. Diseased onions 3, 5 and 10 were shown to be at an early stage of infection; among those, 3 and 5 were infected with gray mold. Onions 1, 2, 4, 9, 11 and 13 were shown to be at an intermediate stage of infection. Among those onions, onions 1, 2 and 4 were also infected with gray mold. Onions 6, 8 and 13 were infected with bacteria (sour skin), while onion 7 was infected with both bacteria and fungi (sour skin and basal rot). Onions 6, 7, 8 and 12 showed the late stage of infection. Both 11 and 12 were infected with neck rot disease.
Fig. 2.
Visual quality assessment of onion diseases (numbers represent individual onion for measurement)
Diseased bulbs were externally sound, except the one with basal rot. In the case of gray mold rot, when the bulbs were cut from neck to base, one to three outer fleshy scales were found to be soft, water-soaked (mushy) and brownish in color. Onion bulbs infected with sour scale had a characteristic sour smell and water-soaked scales. These characteristics are in accordance with Snowden (2010). Onions 9 and 10 were infected with unknown diseases and they showed scales with mushy consistency but without any smell. In the group of healthy onions, all onions were externally and internally sound and had no visual symptoms of diseases.
Exploratory data analysis
As a starting point for the analysis of the spectra, principal component analysis (PCA) was performed with the data sets. Based on FT-NIR data, a PCA was conducted with two PCs. The first principal component (PC1) accounted for 62% of the total variance, while the second principal component (PC2) accounted for 17% of the total variance. No pattern could be observed from the score plot (Fig. 3a) using two PCs. Moreover, an additional PC3 could not separate the two clusters (Online Resource 3).
Fig. 3.
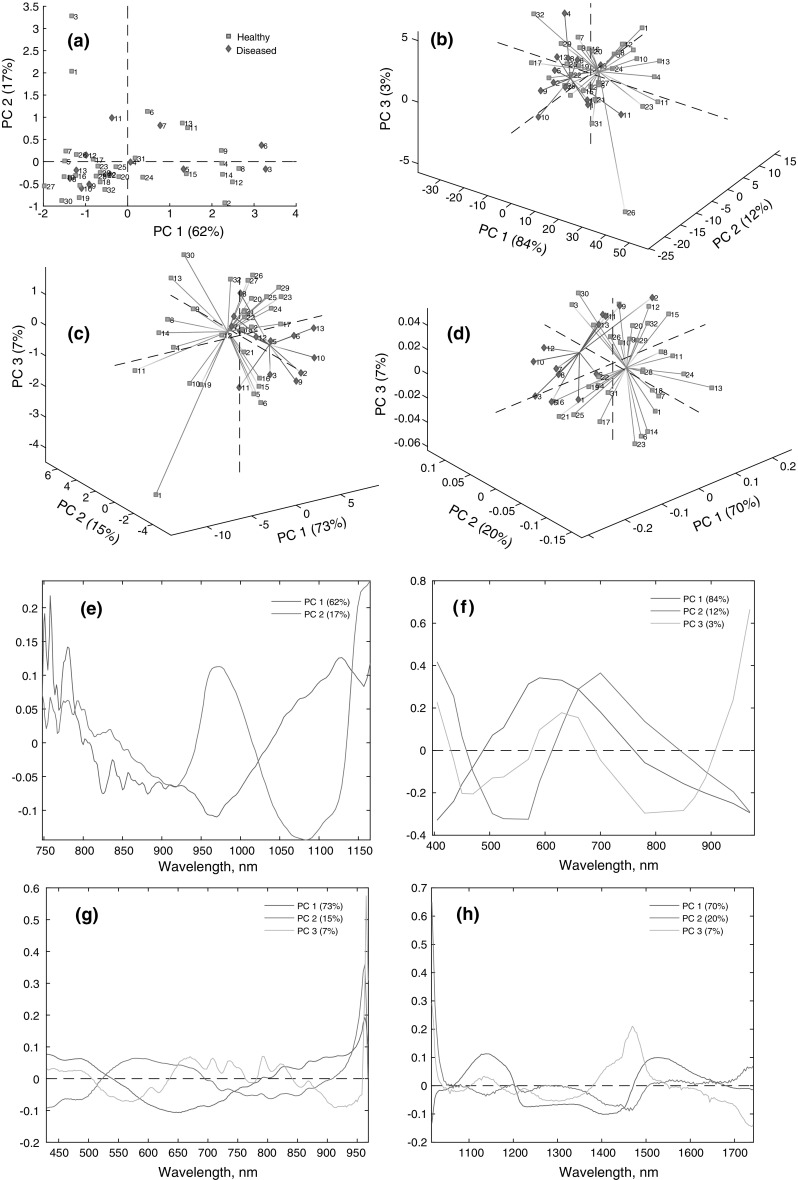
Principal component analysis-score plots. a FT-NIR, b MSI, c HSI-(VIS–NIR), d HSI-(SWIR); principal component analysis-loading plots. e FT-NIR, f MSI, g HSI-(VIS–NIR), h HSI-(SWIR)
A PCA was performed with three PCs for MSI data. The score plot shows the formation of clusters of diseased onions along PC1 and PC2 (Fig. 3b). PC1, PC2, and PC3 explained 84, 12 and 3% of the original variance, respectively. It can be seen from the score plot that diseased onions 6, 8 and 13 are similar; all of those were infected with sour skin. Additionally, diseased onions 2 and 9 lie very close to each other in the score plot; those were infected with gray mold. In addition, diseased onions 2 and 9 were shown to be at an intermediate stage of the disease. Similarly, diseased onions 11 and 12 lie close to each other, and those bulbs were infected with neck rot (Fig. 2). Wavelengths of 405, 435, 470 and 780–970 nm contributed to the samples that were positioned at the left side of PC1, where most of the diseased onions were situated. Conversely, wavelengths of 570–700 nm contributed to the samples that are positioned on the right side of PC1 (Fig. 3f).
A PCA with HSI-(VIS–NIR) data was conducted with three PCs. PC1, PC2 and PC3 describe 73, 15 and 7% of the total variance, respectively (Fig. 3c). Diseased onions 2 and 9 are positioned together in the score plot, and 3, 5 and 10 are positioned in the same plane. This is in accordance with previous observations during the visual quality assessment (Fig. 2). Diseased onions 7 and 8 show similar behavior in the score plot (Fig. 3c), and those were at the late stage of infection. The score plot also reveals that diseased onions 6 and 13 are similar, and those were infected with sour skin. Wavelengths of 771–965 nm are responsible for the samples where most of the diseased onions are positioned, while wavelengths of 552–726 nm are responsible for the samples where most of the healthy onions are positioned (Fig. 3g).
A PCA with three PCs was conducted on the HSI-(SWIR) data. Clusters of healthy and diseased onions can be seen along PC1 and PC3 (Fig. 3d). Wavelengths from 1067 to 1202 nm and 1472 to 1678 nm contribute to PC1, which explains 70% of the total variance. Wavelengths 1016–1047 and 1502–1739 nm contribute in PC2, which explains 20% of the total variance. Wavelengths 1016–1040, 1384–1546 and 1104–1168 nm contribute to PC3, which explains 7% of the total variance (Fig. 3h). The score plot shows that diseased onions 2, 9, 11 and 13 are similar (Fig. 3d), which is in accordance with previous observations. All three of these onions were shown to be at an intermediate stage of infection during the visual quality assessment. Diseased onions 3 and 5 are positioned together, and those were identified with gray mold. In addition, diseased onions 7 and 8 are positioned together in the score plot (Fig. 3d). Both of those were infected with sour skin. Moreover, diseased onions 6, 7, 8 and 12 were shown to be at the late stage of infection (Fig. 2).
Though certain clusters of diseased onions were interpretable in terms of specific diseases and degrees of infections in score plots with spectral imaging, healthy and diseased onions were still mixed. However, the principal component analysis suggests that it could be possible to classify onions accurately using a supervised classification model with spectral imaging.
Classification models
Partial least squares discriminant analysis (PLS-DA) models were built to compare the efficiency of different methods in classifying healthy and diseased onions. The optimal model for FT-NIR data was built with two latent variables, explaining a total of 72% of the variation in the spectra () and 25% of the variation of the vector of classes (). The PLS-DA model with MSI data was built with two latent variables, explaining a total of 97% of the variation in the spectra () and 33% of the variation in the vector of classes (). For HSI-(VIS–NIR) and HSI-(SWIR) data, the optimal PLS-DA model was also calibrated with two latent variables. PLS-DA models for the HSI-(VIS–NIR) and the HSI-(SWIR) data explained a total of 83 and 84% of the variation in the spectra (), respectively and 58 and 38% of the variation of the vector of classes (), respectively. Table 1 summarizes the PLS-DA model statistics obtained with the FT-NIR, MSI, and HSI data. High sensitivity and specificity levels in prediction were obtained using both types of HSI data to compare to MSI and FT-NIR data.
Table 1.
Partial least squares discriminant analysis classification model statistics
| Whole spectra | Reduced spectra | |||||
|---|---|---|---|---|---|---|
| FT-NIR | MSI | HSI-(VIS–NIR) | HSI-(SWIR) | HSI-(VIS–NIR) | HSI-(SWIR) | |
| Calibration | ||||||
| Sensitivity (%) | 90 | 70 | 80 | 90 | 80 | 80 |
| Specificity (%) | 52 | 82 | 91 | 78 | 87 | 82 |
| Class error (%) | 28 | 23 | 14 | 15 | 16 | 18 |
| Cross validation | ||||||
| Sensitivity (%) | 80 | 70 | 70 | 80 | 80 | 80 |
| Specificity (%) | 39 | 73 | 87 | 78 | 87 | 78 |
| Class error (%) | 40 | 28 | 21 | 20 | 16 | 20 |
| Prediction | ||||||
| Sensitivity (%) | 33 | 33 | 66 | 100 | 66 | 100 |
| Specificity (%) | 11 | 22 | 33 | 66 | 44 | 77 |
| Class error (%) | 77 | 72 | 50 | 16 | 44 | 11 |
FT-NIR Fourier transform near-infrared, MSI multispectral imaging, HSI-(VIS–NIR) hyperspectral imaging (visible near-infrared), HSI-(SWIR), hyperspectral imaging (short wave infrared)
False positive results in classification will reduce the profit for the industry/farmer, while false negative results may lead to consumer dissatisfaction and rejection. The performance of PLS-DA models built with MSI, HSI-(VIS–NIR) and HSI-(SWIR) was better than FT-NIR. The model with FT-NIR achieved 80% correct classification for healthy onions, while 39% correct classification was achieved for diseased onions in cross-validation, with 28, 40 and 77% as classification error in calibration, cross-validation, and prediction, respectively. The PLS-DA calibration model with MSI data showed 70% correct classification for healthy onions, while it showed 82% correct classification for diseased onions. The PLS-DA cross-validation model obtained 70 and 73% correct classification for healthy and diseased onions, respectively. In prediction, 22% were correctly classified as diseased onions, while FT-NIR achieved 11% correct classification. The model with HSI-(VIS–NIR) obtained 80% correct classification of healthy onions in calibration, 70% in cross-validation and 66% in prediction; an improvement in the results was achieved with HSI-(SWIR) data in calibrated (90%), cross-validated (80%) and in the predicted model (100%). On the other hand, 91, 87 and 33% correct classification for diseased onions were achieved with VIS–NIR data in calibration, cross-validation, and prediction, respectively, while 78% correct classification was achieved with SWIR data in both calibration and cross-validation, respectively. Sixty-six percent correct classification was achieved with SWIR data in prediction. The results are slightly better than the findings reported by Wang et al. (2012). The authors introduced onions vertically (facing with the neck towards the camera), and HSI at 950–1650 nm with linear discriminant analysis (LDA) was able to correctly predict 63% of onion bulbs infected with sour skin. In the present study, VIS–NIR showed a classification error of 14, 21, and 50%, while 15, 20 and 16% with SWIR data respectively in calibration, cross-validation, and prediction.
Compared to other techniques, models with HSI-(SWIR) were shown to have a better performance in classifying healthy and diseased onions. The only drawback of applying HSI to the processing line is the large amount of data from the hyperspectral images, which increases the complexity of data analysis and slows the speed of processing (Ferrari et al. 2015). However, due to the large data size of each hyperspectral image, data dimensionality reduction is necessary in order to develop MSI for real-time monitoring of large sets of samples in the processing line (Calvini et al. 2016).
An efficient way to reduce data dimensionality is to identify major regions on the spectrum responsible for the classification of healthy and diseased onions using VIP and SR. In the PLS-DA model, the variables that show a VIP value > 1 are the most important variables (Gerretzen et al. 2016). Similarly, a high selectivity ratio indicates the major region that has the ability to discriminate between two groups of onions. For the PLS-DA models with HSI-(VIS–NIR) data and HSI-(SWIR) data, a threshold of VIP > 1.5 and VIP > 1.4 was used, respectively. A threshold of SR > 1.09 and SR > 0.8 was used for models with HSI-(VIS–NIR) and HSI-(SWIR) data, respectively. The PLS-DA VIP, SR, and selected variables are shown in Fig. 4.
Fig. 4.
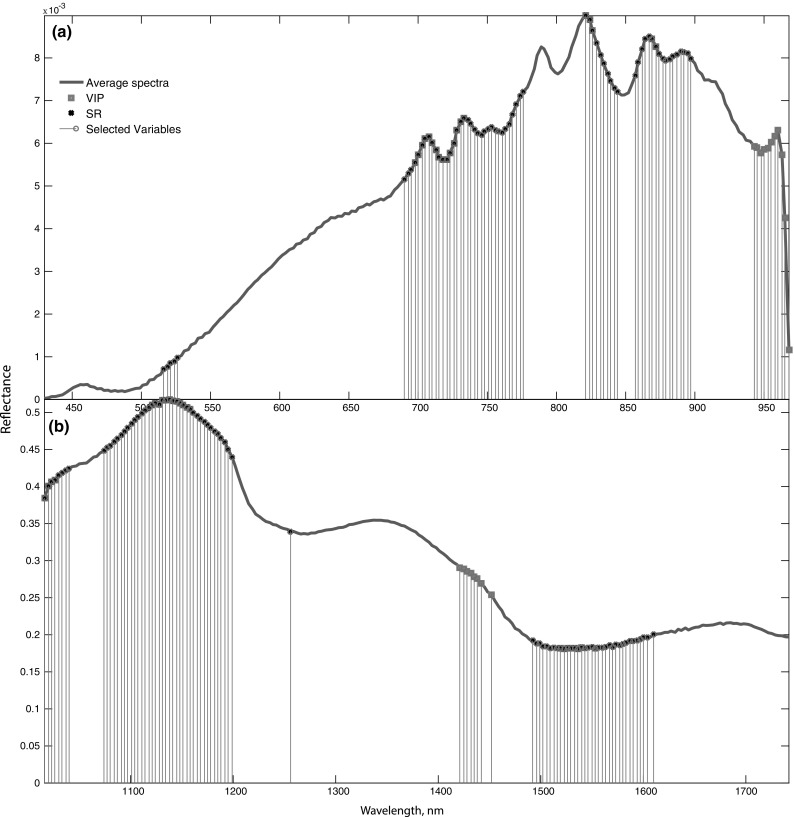
Variable Important in projection (VIP), selectivity ratio (SR) and selected variables. a HSI-(VIS–NIR), b HSI-(SWIR)
New PLS-DA models for HSI were built with the selected variables to compare the model performances with that of the one built with whole spectra (Table 1). The optimal model for HSI-(VIS–NIR) data was built with two latent variables, explaining a total of 87% of the variation in the spectra (X) and 49% of the variation of the vector of classes (y). A PLS-DA model with HSI-(SWIR) data was also built with two latent variables, explaining a total of 85% of the variation in the spectra (X) and 39% of the variation of the vector of classes (y). The performance of calibration models built with the selected variables was slightly lower than the model with whole spectra. However, in cross-validation and in prediction, the performance of the model with selected variables was either similar to or better than that of the models with whole spectra. For instance, sensitivity in prediction in the model with selected variables was almost the same as that of the previous models, while higher specificity was achieved in the model with selected variables. Several wavelengths 516–526, 690–776, 821–844, 857–897 and 943–968 nm were identified as important variables responsible for the classification with HSI-(VIS–NIR) data (Fig. 4a). In the literature, the difference in spectral features between healthy, neck rot, and sour skin was found at 670–740, 780–870, and 950–1170 nm (Chugunov and Li 2015).
The major constituents of an onion are water, protein, and carbohydrates (Brewster 2008). It has been reported that pectic enzymes are involved in the degradation of pectic constituents of cell walls, which is caused by pathogenic microorganisms that are related to several onion diseases, including bacterial soft rot (Obi and Umezurike 1981). Additionally, Botrytis cinerea, one of the pathogenic microorganisms responsible for onion diseases, has been reported to produce pectic degrading enzymes (Aboaba 2009). A pectinase enzyme hydrolyzes the pectin and provides a mushy consistency/water-soaked appearance (Jay et al. 2008). Results from the analysis of the loading plots, PLS-DA-VIP plots, and SR plots indicate that the spectral difference between healthy and diseased onions could be associated with the different compositions and proportions of water, sugar, and cellulose in healthy and diseased onions and their degradation products in diseased onions (Chugunov and Li 2015; Williams and Norris 1987; Workman 2000).
Reducing data dimensionality by identifying the optimal regions in the spectra in this study will be helpful in the industrial adaptation of this method. However, although VIP and SR are not truly variable selection methods, they are used as tools for assessing the relevance of the spectral variables. The new PLS-DA model built with the reduced variables selected using VIP and SR resulted in small or no decreases in model performance.
Furthermore, it seems that the choices of the orientation of the onion during MSI, HSI-(VIS–NIR) and HSI-(SWIR) were satisfactory. There is a possibility of overlooking valuable information about the bulb if the bulb is placed with the neck facing to the camera. Thus, it is better to place the onion with the equatorial region facing towards the camera. In such a setup, the basal plates do not overlap with the system optical axis. Kuroki et al. (2017) suggested a similar orientation for the best model to predict the percentage of rot in onions using the transmittance mode of hyperspectral imaging. The acquisition system in MSI and HSI-(SWIR) were in accordance with that study. In addition, the use of rotating rollers in our study is promising as it provided scans of the whole onion surface.
Conclusion
This work compared spectroscopic techniques, both point-based and image-based, for the classification of healthy and diseased onions. The results from this particular study indicate that hyperspectral imaging at the SWIR range together with multivariate analysis and a chemometric model could become a promising non-destructive method for classifying healthy and diseased onions. The results also suggest that the direct measurement of onions using a hyperspectral imaging system with several selective wavelengths, either scanning the whole surface using rotating rollers or taking several scans of the onions, facing the equatorial region towards the sensor by placing it on a sample holder and shifting sides could be adapted as an automated method for quality sorting in the onion industry.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Photograph of a) NIR spectroscopic system (FT-NIR instrument), b) VideometerLab instrument (multispectral imaging system); photograph and schematic diagram of c) hyperspectral imaging system (HSI-VIS-NIR instrument), d) hyperspectral imaging system (HSI-SWIR instrument). (PDF 1662 kb)
Bulb weight, diameter and total soluble solids (TSS) of onions. (PDF 16 kb)
Principal component analysis of FT-NIR data. Score plot with three principal components. (PDF 66 kb)
Acknowledgements
This work was funded by the Innovation Fund Denmark in the project ‘Strategies and Technologies to Reduce Postharvest Losses of Potatoes and Vegetables’ J. No. 1382-00057B. Onions for the experiment were grown, stored and primarily sorted at Axel Månsson A/S.
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s13197-018-3268-x) contains supplementary material, which is available to authorized users.
References
- Aboaba S. The role of pectinase enzyme in the development of soft rot caused by Pseudomonas fluorescens in the purple variety of onions (Allium cepa) Afr J Microbiol Res. 2009;3(4):163–167. [Google Scholar]
- Borras E, Amigo JM, van den Berg F, Boque R, Busto O. Fast and robust discrimination of almonds (Prunus amygdalus) with respect to their bitterness by using near infrared and partial least squares-discriminant analysis. Food Chem. 2014;153:15–19. doi: 10.1016/j.foodchem.2013.12.032. [DOI] [PubMed] [Google Scholar]
- Brewster JL. Onions and Other Vegetable Alliums. Wallingford: CABI; 2008. [Google Scholar]
- Büning-Pfaue H. Analysis of water in food by near infrared spectroscopy. Food Chem. 2003;82(1):107–115. doi: 10.1016/S0308-8146(02)00583-6. [DOI] [Google Scholar]
- Calvini R, Foca G, Ulrici A. Data dimensionality reduction and data fusion for fast characterization of green coffee samples using hyperspectral sensors. Anal Bioanal Chem. 2016;408(26):7351–7366. doi: 10.1007/s00216-016-9713-7. [DOI] [PubMed] [Google Scholar]
- Chugunov S, Li C. Monte Carlo simulation of light propagation in healthy and diseased onion bulbs with multiple layers. Comput Electron Agric. 2015;117:91–101. doi: 10.1016/j.compag.2015.07.015. [DOI] [Google Scholar]
- de Oliveira EM, Leme DS, Barbosa BHG, Rodarte MP, Pereira RGFA. A computer vision system for coffee beans classification based on computational intelligence techniques. J Food Eng. 2016;171:22–27. doi: 10.1016/j.jfoodeng.2015.10.009. [DOI] [Google Scholar]
- Ferrari C, Foca G, Calvini R, Ulrici A. Fast exploration and classification of large hyperspectral image datasets for early bruise detection on apples. Chemom Intell Lab Syst. 2015;146:108–119. doi: 10.1016/j.chemolab.2015.05.016. [DOI] [Google Scholar]
- Gerretzen J, Szymańska E, Bart J, Davies AN, van Manen H-J, van den Heuvel ER, Jansen JJ, Buydens LMC. Boosting model performance and interpretation by entangling preprocessing selection and variable selection. Anal Chim Acta. 2016;938:44–52. doi: 10.1016/j.aca.2016.08.022. [DOI] [PubMed] [Google Scholar]
- Islam MN, Wang A, Skov Pedersen J, Edelenbos M. Microclimate tools to monitor quality changes in stored onions. Acta Hortic. 2017;1154:229–234. doi: 10.17660/ActaHortic.2017.1154.30. [DOI] [Google Scholar]
- Jay JM, Loessner MJ, Golden DA. Modern Food Microbiology. New York: Springer; 2008. [Google Scholar]
- Jha SN, Narsaiah K, Sharma AD, Singh M, Bansal S, Kumar R. Quality parameters of mango and potential of non-destructive techniques for their measurement—a review. J Food Sci Technol. 2010;47(1):1–14. doi: 10.1007/s13197-010-0004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko S-S, Chang W-N, Wang J-F, Cherng S-J, Shanmugasundaram S. Storage variability among short-day onion cultivars under high temperature and high relative humidity, and its relationship with disease incidence and bulb characteristics. J Am Soc Hortic Sci. 2002;127(5):848–854. [Google Scholar]
- Kuroki S, Nishino M, Nakano S, Deguchi Y, Itoh H. Positioning in spectral measurement dominates estimation performance of internal rot in onion bulbs. Postharvest Biol Technol. 2017;128:18–23. doi: 10.1016/j.postharvbio.2017.02.001. [DOI] [Google Scholar]
- Lu R, Chen Y-R (1999) Published hyperspectral imaging for safety inspection of food and agricultural products. In: Photonics east (ISAM, VVDC, IEMB). International society for optics and photonics, pp 121–133
- Obi SK, Umezurike GM. Pectic enzyme activities of bacteria associated with rotted onions (Allium cepa) Appl Environ Microbiol. 1981;42(4):585–589. doi: 10.1128/aem.42.4.585-589.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel KK, Kar A, Jha SN, Khan MA. Machine vision system: a tool for quality inspection of food and agricultural products. J Food Sci Technol. 2012;49(2):123–141. doi: 10.1007/s13197-011-0321-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preedy VR, Watson RR. The mediterranean diet: an evidence-based approach. New York: Elsevier; 2014. [Google Scholar]
- Qin J, Chao K, Kim MS, Lu R, Burks TF. Hyperspectral and multispectral imaging for evaluating food safety and quality. J Food Eng. 2013;118(2):157–171. doi: 10.1016/j.jfoodeng.2013.04.001. [DOI] [Google Scholar]
- Rabinowitch HD, Currah L. Allium crop science: recent advances. Wallingford: CABI; 2002. [Google Scholar]
- Roggo Y, Chalus P, Maurer L, Lema-Martinez C, Edmond A, Jent N. A review of near infrared spectroscopy and chemometrics in pharmaceutical technologies. J Pharm Biomed Anal. 2007;44(3):683–700. doi: 10.1016/j.jpba.2007.03.023. [DOI] [PubMed] [Google Scholar]
- Schwartz HF, Mohan SK. Compendium of onion and garlic diseases and pests. St. Paul: APS Press; 2007. [Google Scholar]
- Sinclair PJ, Blakeney AB, Barlow E. Relationships between bulb dry matter content, soluble solids concentration and non-structural carbohydrate composition in the onion (Allium cepa) J Sci Food Agric. 1995;69(2):203–209. doi: 10.1002/jsfa.2740690210. [DOI] [Google Scholar]
- Snowden AL. Post-harvest diseases and disorders of fruits and vegetables. Boca Raton: CRC Press; 2010. [Google Scholar]
- Travers S, Bertelsen MG, Petersen KK, Kucheryavskiy SV. Predicting pear (cv. Clara Frijs) dry matter and soluble solids content with near infrared spectroscopy. LWT Food. Sci Technol. 2014;59(2, Part 1):1107–1113. doi: 10.1002/jsfa.6343. [DOI] [PubMed] [Google Scholar]
- Vetrekar NT, Gad RS, Fernandes I, Parab JS, Desai AR, Pawar JD, Naik GM, Umapathy S. Non-invasive hyperspectral imaging approach for fruit quality control application and classification: case study of apple, chikoo, guava fruits. J Food Sci Technol. 2015;52(11):6978–6989. doi: 10.1007/s13197-015-1838-8. [DOI] [Google Scholar]
- Wang W, Li C, Tollner EW, Gitaitis RD, Rains GC. Shortwave infrared hyperspectral imaging for detecting sour skin (Burkholderia cepacia)-infected onions. J Food Eng. 2012;109(1):38–48. doi: 10.1016/j.jfoodeng.2011.10.001. [DOI] [Google Scholar]
- Wang H, Li C, Wang M. Quantitative determination of onion internal quality using reflectance, interactance, and transmittance modes of hyperspectral imaging. Trans ASABE. 2013;56(4):1623–1635. [Google Scholar]
- Williams P, Norris K. Near-infrared technology in the agricultural and food industries. St. Paul: American Association of Cereal Chemists, Inc.; 1987. [Google Scholar]
- Workman J. The handbook of organic compounds, three-volume set: NIR, IR, R, and UV–Vis spectra featuring polymers and surfactants. New York: Elsevier; 2000. [Google Scholar]
- Zhang C, Jiang H, Liu F, He Y. Application of near-infrared hyperspectral imaging with variable selection methods to determine and visualize caffeine content of coffee beans. Food Bioprocess Technol. 2017;10(1):213–221. doi: 10.1007/s11947-016-1809-8. [DOI] [Google Scholar]
- Zude M. Optical monitoring of fresh and processed agricultural crops. Milton Park: Taylor & Francis; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Photograph of a) NIR spectroscopic system (FT-NIR instrument), b) VideometerLab instrument (multispectral imaging system); photograph and schematic diagram of c) hyperspectral imaging system (HSI-VIS-NIR instrument), d) hyperspectral imaging system (HSI-SWIR instrument). (PDF 1662 kb)
Bulb weight, diameter and total soluble solids (TSS) of onions. (PDF 16 kb)
Principal component analysis of FT-NIR data. Score plot with three principal components. (PDF 66 kb)