Summary
Transposable elements represent nearly half of mammalian genomes and are generally described as parasites or ‘junk DNA’. The LINE1 retrotransposon is the most abundant class and is thought to be deleterious for cells, yet is paradoxically highly expressed during early development. Here we report that LINE1 plays essential roles in mouse embryonic stem (ES) cells and pre-implantation embryos. In ES cells, LINE1 acts as a nuclear RNA scaffold that recruits Nucleolin and Kap1/Trim28 to repress Dux, the master activator of a transcriptional program specific to the 2-cell embryo. In parallel, LINE1 RNA mediates binding of Nucleolin and Kap1 to rDNA, promoting rRNA synthesis and ES cell self-renewal. In embryos, LINE1 RNA is required for Dux silencing, synthesis of rRNA and exit from the 2-cell stage. The results reveal an essential partnership between LINE1 RNA, Nucleolin, Kap1 and peri-nucleolar chromatin in the regulation of transcription, developmental potency and ES cell self-renewal.
Keywords: LINE1, LINE-1, retrotransposons, totipotency, pluripotency, Nucleolin, Kap1, rRNA, Dux, MERVL, hypertranscription, 2-cell stage, ES cells
In Brief
Highly expressed during early embryonic development, LINE1 element-derived RNA acts as a nuclear scaffold to facilitate essential gene expression programs
Introduction
Only about 1.5% of mammalian genomes is comprised of single-copy protein-coding sequences, whereas approximately half of their DNA derives from transposable elements (TEs). Despite their abundance, the roles and regulation of TEs have been understudied, in large part due to difficulty in mapping repetitive sequences to the genome. Nevertheless, TEs are now accepted as key drivers of genome evolution by rewiring gene regulatory networks, including in the human genome (Bourque, 2009).
The retrotransposon Long INterspersed Element 1 (LINE1) makes up the largest proportion of TE-derived sequences, and is the only class of autonomous TEs still active in human (Magiorkinis et al., 2015). LINE1-induced mutations have been linked to a growing number of diseases, including hematopoietic and neurological disorders, as well as several types of cancer [reviewed in (Burns, 2017)]. For this reason, LINE1 is generally thought to be silenced in differentiated cell types to avoid uncontrolled mutagenesis. However, the view of TEs such as LINE1 as strictly detrimental to cells may be too simplistic. LINE1 is expressed in normal neural progenitor cells, where it has been proposed to promote neuronal diversity (Muotri et al., 2005). LINE1 is also expressed in the pre-implantation embryo (Fadloun et al., 2013) and in the fetal germline (Ohno et al., 2013, Percharde et al., 2017b). A TE of a different class, Mouse Endogenous Retrovirus type L (MERVL), is also expressed in cleavage-stage embryos, where it drives the expression of many transcripts specific to zygotic genome activation (ZGA) and totipotency (Kigami et al., 2003, Macfarlan et al., 2012, Svoboda et al., 2004). Importantly, the rate of LINE1 retrotransposition in embryos and germ cells in vivo is low given the high levels of LINE1 RNA expression (Kano et al., 2009, Richardson et al., 2017, Newkirk et al., 2017). These observations raise the possibility that LINE1 RNA has as-yet undefined cellular roles, independent of retrotransposition.
We set out to test the hypothesis that LINE1 plays essential functions in mouse pluripotent cells. Our data point to a model whereby LINE1 and its chromatin partners are essential to orchestrate developmental progression during pre-implantation and for the self-renewal of ES cells.
Results
LINE1 RNA is nuclear localized and promotes ES cell self-renewal
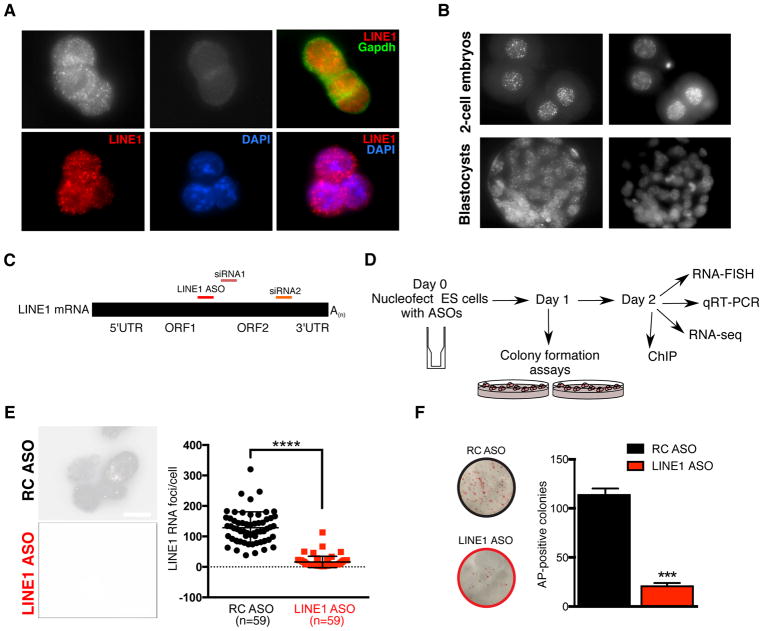
We first set out to investigate the expression and localization of LINE1 in mouse ES cells. LINE1 RNA is detected at high levels in a punctate pattern in the nuclei of ES cells, but not in the cytoplasm (Figure 1A), the opposite localization pattern of LINE1 protein, Orf1p (Figure S1A). LINE1 RNA is associated with euchromatin and generally excluded from heterochromatic foci (Figure 1A). These results are in agreement with the general localization of C0T-I repeat RNA, which includes LINE1 RNA (Hall et al., 2014). A similar nuclear localization of LINE1 RNA is detected in mouse 2-cell embryos and blastocysts [Figure 1B and (Fadloun et al., 2013)]. These results raised the possibility that LINE1 RNA plays a role in transcriptional regulation in ES cells.
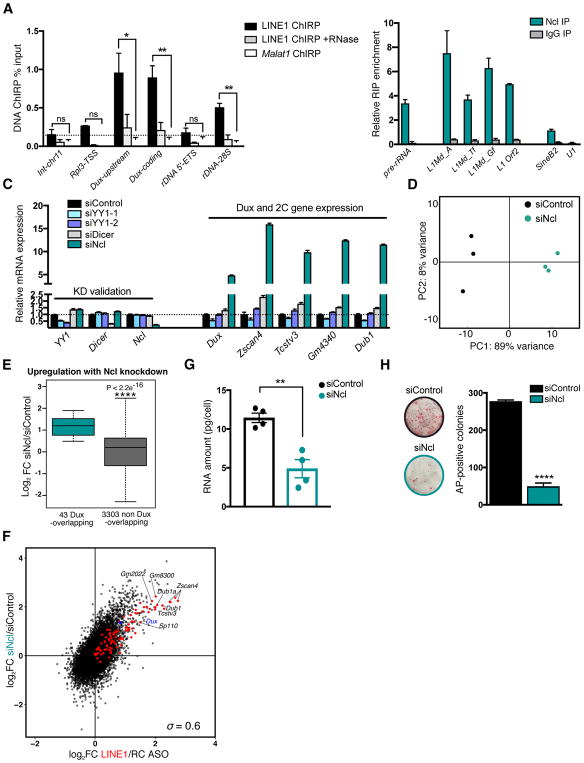
Figure 1. LINE1 RNA is nuclear localized in ES cells and is essential for self-renewal.
(A) LINE1 RNA FISH in ES cells, in the indicated conditions. Scale bar, 10μm.
(B) LINE1 RNA FISH in mouse 2-cell embryos and blastocysts. Scale bar, 20μm.
(C) Schematic of full-length LINE1 mRNA indicating the positions of the inter-ORF LINE1 ASO and the two independent siRNA sequences used in this study. The reverse complement (RC) of the LINE1 ASO is used throughout as a negative control.
(D) Workflow of LINE1 experiments.
(E) RNA FISH in ES cells showing nuclear LINE1 depletion 48h following nucleofection with LINE1 ASO. LINE1 RNA foci per cell were quantified in each condition from multiple fields, with mean +/− s.d indicated. Scale bar, 10μm.
(F) Representative images and quantification of the number of AP-positive colonies 5–6 days after initial plating. Data are mean +/− s.e.m., n=3 biological replicates. See also Figure S1.
We next developed a LINE1 RNA knockdown (KD) strategy using antisense oligos (ASOs) (Figure 1C). LINE1 ASOs lead to a significant reduction in nuclear LINE1 FISH signal, unlike control reverse complement (RC) ASO-treated ES cells (Figure 1D–E and S1B). We validated these results using two siRNAs as an independent KD method (Figure 1C and S1C). The lower level of knockdown using RNAi is to be expected given the greater ability of ASOs to knockdown nuclear RNAs (Lennox and Behlke, 2016). Surprisingly, knockdown of LINE1 results in a dramatic decrease in ES cell self-renewal (Figure 1F), a result validated by LINE1 RNAi (Figure S1D). In agreement, LINE1 KD ES cells exhibit a drastically reduced cell expansion rate and an altered cell cycle profile, with a significant decrease in the proportion of cells in S phase and an increase in cells in G2/M (Figure S1E–F). This is accompanied by only a modest increase in cell death (to ~1% of the total population, Figure S1G), and no changes to the overall levels of Oct4 or Nanog proteins (Fig S1H). Taken together, these data indicate that LINE RNA is required for the efficient propagation of ES cells.
LINE1 represses the 2C transcriptional program in ES cells
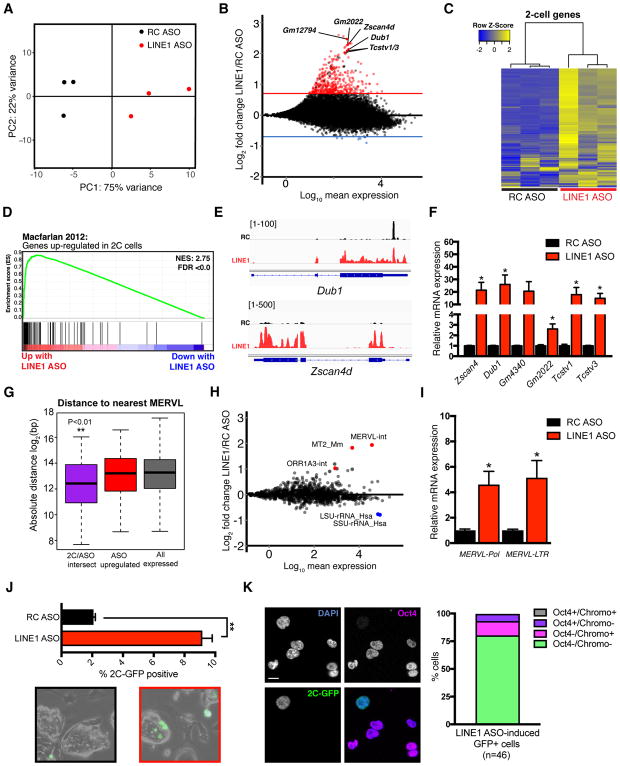
To analyze the transcriptional impact of LINE1 RNA KD, we performed RNA sequencing (RNA-seq), which revealed that LINE1 KD ES cells are reproducibly distinct from controls (Figure 2A and Figure S2A). Genes with a LINE1 element situated within or nearby show no evidence of downregulation, arguing for a direct effect of KD of LINE1 RNA itself (Figure S2B). Interestingly, LINE1 KD induces a significant upregulation of 414 transcripts (log2 fold-change >0.7, FDR <0.05), with only overall mild downregulation of single-copy genes (Figure 2B). Upregulated genes do not include markers of the three germ layers (Figure S2C–D), confirming that LINE1 KD does not induce precocious differentiation. Instead, there is a striking upregulation of genes transiently expressed at the 2-cell (2C) stage upon LINE1 KD (Figure 2C–D, S2E). The 2-cell stage encompasses the switch from maternal control to zygotic genome activation (ZGA), and is associated with a sharp, transient upregulation of many genes, termed 2-cell (2C) genes. Several 2C genes contain promoters originally derived from the TE MERVL, which is also sharply induced at this stage [reviewed in (Schoorlemmer et al., 2014)]. MERVL and 2C genes are rapidly repressed after the 2-cell stage, and in most ES cells (Macfarlan et al., 2011, Wu et al., 2016). 2C genes upregulated upon LINE1 KD include well-known markers such as Zscan4, Dub1, Gm4340, Tcstv1/3 and Zfp352 (Figure 2E–F and Table S1) along with the 2C-specific transposon MERVL (Figure 2G–I). 2C gene upregulation was additionally confirmed using LINE1 RNAi (Figure S2F). Inhibition of LINE1 retrotransposition using anti-retroviral drugs (Jones et al., 2008) does not phenocopy LINE1 RNA KD, indicating that its role in ES cells is independent of retrotransposition (Figure S2G–I). Moreover, LINE1 KD does not induce MERVL/2C expression in mouse embryonic fibroblasts (Figure S2J). We examined whether LINE1 KD induces conversion of ES cells to a 2C-like fate, using an ES cell line in which endogenous 2C-like cells are marked by GFP [(Ishiuchi et al., 2015, Macfarlan et al., 2012) and (Figure S2K)]. Depletion of LINE1 significantly increases the percentage of 2C-like cells that display the expected features: loss of chromocenters and lack of Oct4 protein (Figure 2J–K) or Nanog protein (data not shown). These results indicate that LINE1 acts to repress MERVL and the 2C transcriptional program in ES cells.
Figure 2. LINE1 knockdown causes upregulation of 2C genes and MERVL.
(A) PCA plot for all genes across all samples, showing that LINE1 KD ES cells have distinct gene expression profiles and are separated from controls along PC1.
(B) MA plot showing log2 fold-changes in the expression of each gene following LINE1 KD. Horizontal red or blue lines indicate FDR <0.05 and log2 fold-chance (FC) of > 0.7 or < −0.7, respectively. Select upregulated 2-cell (2C) genes are labeled in black.
(C) Heatmap showing expression changes of 142 2C genes as defined in (Macfarlan et al., 2012), upon LINE1 KD.
(D) GSEA for 2C genes as in (C) showing a preferential upregulation of nearly all genes upon LINE1 KD.
(E) Browser RNA-seq screenshots of 2C genes Zscan4d and Dub1 in RC or LINE1 ASO samples.
(F) qRT-PCR validation of 2C gene upregulation following LINE1 KD with ASOs. Data are mean +/− s.e.m., n=3 biological replicates.
(G). Distance analysis performed on the indicated sets of genes, calculating of log2 absolute distance in base-pairs (bp) to the nearest MERVL element. 2C/ASO intersect: 52 2C genes from (Macfarlan et al., 2012) also significantly upregulated with LINE KD; ASO upregulated: all significantly upregulated genes upon LINE1 KD. **P < 0.01, two-sided Wilcoxon rank sum test, calculated between 2C/ASO intersect and all expressed genes.
(H) MA plot showing log2 fold-changes in repeat expression following LINE1 KD. Upregulated MERVL repeats, and downregulated rRNA repeats, are indicated.
(I) qRT-PCR validation of MERVL expression following LINE1 KD. Data are mean +/− s.e.m., n=3 biological replicates.
(J) Percentage of 2C-like cells and representative micrographs in 2C-GFP reporter ES cells 48h after nucleofection with ASOs. Data are mean +/− s.e.m of 2 independent experiments.
(K) Immunofluorescence analysis of LINE1 KD-induced 2C-like cells. Graph depicts the percentage of GFP+ cells that have the expected features (loss of chromocenters and Oct4 protein). Scale bar, 10μm, n=number of cells. See also Figure S2 and Table S1.
LINE1 represses Dux, a master activator of the 2C program
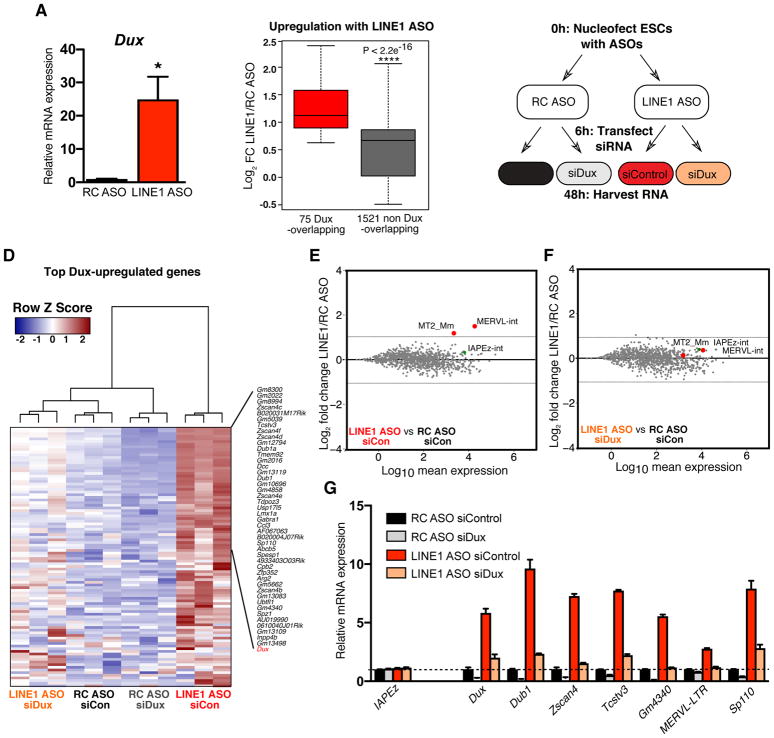
Next, we explored how LINE1 might repress the 2C state in ES cells. LINE1 KD induces a similar phenotype whether ES cells are grown in serum/LIF or 2i/LIF conditions [(Ying et al., 2008) and (Figure S3A–B)]. The expression of known repressors of the 2C state is not altered in LINE1 KD ES cells (Figure S3C). Interestingly, we found that the transcription factor Dux is significantly upregulated upon LINE1 KD (Figure 3A and Table S1). Recently, Dux was shown to bind directly to many 2C gene promoters and to be necessary and sufficient for 2C gene upregulation in ES cells and for pre-implantation development (De Iaco et al., 2017, Hendrickson et al., 2017, Whiddon et al., 2017). Moreover, CRISPR/Cas9-mediated deletion of Dux in zygotes impairs pre-implantation development, suggesting a potentially critical role in early development and ZGA (De Iaco et al., 2017). Analysis of RNA-seq data revealed that Dux target genes are amongst the most highly activated genes upon LINE1 KD (Figure 3B). Moreover, the repressive chromatin mark H3K9me2 is reduced at Dux and its downstream targets in LINE1 KD ES cells (Figure S3D–F). We next performed control or LINE1 KD using ASOs, with or without simultaneous KD of Dux using siRNAs (Figure 3C and S3G). RNA-seq revealed that Dux depletion significantly reduces the upregulation of Dux targets upon LINE1 KD (Figure S3H). Moreover, hierarchical clustering of Dux targets revealed that Dux KD rescues the effect of LINE1 KD (Figure 3D). Similarly, MERVL is no longer upregulated upon simultaneous KD of LINE1 and Dux (Figure 3E–G, Figure S3I). These data reveal that the upregulation of the 2C-program in LINE1 KD ES cells is Dux-dependent, raising the question of whether LINE1 acts directly or indirectly to restrict Dux expression in ES cells (see below).
Figure 3. The activation of the 2C program induced by LINE1 knockdown is Dux-dependent.
(A) qRT-PCR showing Dux upregulation with LINE1 KD. Data are mean +/− s.e.m., n=4 biological replicates.
(B) Boxplot analysis of significantly altered genes (FDR <0.05) upon LINE1 KD, showing that Dux targets are significantly more induced than non-targets. P-value is determined by two-sided Wilcoxon rank sum test.
(C) Workflow of LINE1/Dux knockdown experiments.
(D) Heatmap showing suppression of induction of Dux and top Dux target genes, defined as in (B), in LINE1 KD cells upon simultaneous Dux KD.
(E–F) MA plots of repeat expression changes in LINE1 vs RC ASO treatment, either with transfection of (E) siControl or (F) siDux.
(G) qRT-PCR validation of 2C gene and MERVL rescue following Dux depletion. Data are representative of 3 independent experiments, shown is mean +/− s.e.m., n=3 technical replicates. See also Figure S3 and Table S2.
LINE1 promotes ES self-renewal independently of its role in repressing Dux
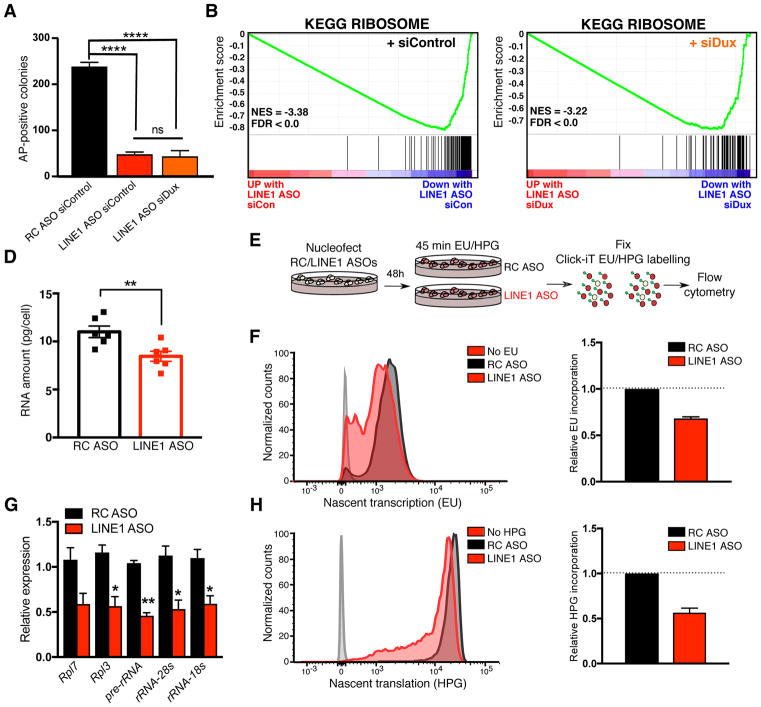
We subsequently investigated whether the induction of Dux is related to the decrease in self-renewal of LINE1 KD ES cells. We found that Dux KD does not rescue the self-renewal deficit observed upon LINE1 KD (Figure 4A), indicating that LINE1 plays additional, Dux-independent roles in ES cells. Transcripts upregulated upon LINE1 KD include p53 targets (Figure S4A). However, LINE1 KD in p53−/− ES cells (Sabapathy et al., 1997) both activates 2C gene expression (Figure S4B) and significantly decreases ES self-renewal (Figure S4C), similarly to wild-type ES cells. P53 activation is therefore not a primary cause of the self-renewal deficit of LINE1 KD ES cells.
Figure 4. LINE1 promotes translation and ES self-renewal independently of Dux.
(A) Colony-formation assay showing that Dux knockdown does not rescue self-renewal upon LINE1 KD. Data are mean +/− s.e.m., n=3 biological replicates.
(B–C) GSEA plot showing preferential downregulation of the KEGG ribosome pathway following LINE1 KD, with either co-transfection of (B) Control or (C) Dux siRNAs.
(D) RNA per cell in ES cells 48h after nucleofection with RC or LINE1 ASOs. Data are mean +/− s.e.m., n=6 independent batches of equal numbers of cells.
(E) Diagram of experiments labelling nascent RNA/proteins with EU/HPG following ASO nucleofection.
(F) Representative histogram (left) of nascent transcription in RC or LINE1 ASO-treated samples 24–48h after nucleofection, with cells incubated without EU shown as control, and quantification (right) showing the relative decrease in translation upon LINE1 KD. Data are = mean +/− s.e.m., n=2 independent experiments.
(G) qRT-PCR showing decrease in rRNA and ribosomal protein gene expression 48h after LINE1 KD, shown as mean +/− s.e.m, n=3 biological replicates.
(H) Representative histogram (left) and quantification (right) as in (G), but performed for HPG incubations 48h after LINE1 KD. Data are = mean +/− s.e.m., n=4 independent experiments. See also Figure S4.
While down-regulation of single-copy genes is mild in LINE1 KD ES cells (Figure 2B), functional annotations related to ribosomal biogenesis and translation are significantly enriched among downregulated genes (Figure 4B, S4D and Table S1). Moreover, ribosomal protein genes are reduced in expression upon LINE1 KD (Figure S4E–F) and remain downregulated upon simultaneous Dux depletion (Figure 4C and Table S2). Interestingly, ribosomal RNA (rRNA) is significantly downregulated upon LINE1 KD (Figure 2H). In agreement, total RNA levels per cell are reduced upon LINE1 KD (Figure 4D), along with nascent transcription, rRNA synthesis and nascent translation (Figure 4E–H). Direct chemical inhibition of rRNA synthesis (Haddach et al., 2012) leads to a similarly sharp reduction in the self-renewal of ES cells (Figure S4G–I). These results suggest that LINE1 promotes high levels of rRNA synthesis typical of and required by rapidly growing pluripotent cells (Guzman-Ayala et al., 2015). The self-renewal deficit of LINE1 KD ES cells is likely due to a combination of the reduction in nascent transcription, including of rRNA, and the cell cycle defects (Figure S1E–F).
LINE1 RNA binds Dux and rDNA loci in ES cells
We sought to reconcile the seemingly unrelated roles of LINE1 RNA in Dux repression and rRNA synthesis. The high abundance of nuclear LINE1 RNA (Figure 1A), as well as previous reports of its close association with chromatin (Hall et al., 2014) suggest that it may act similar to nuclear long non-coding RNAs (lncRNAs) to regulate gene expression. We therefore performed LINE1 Chromatin Immunoprecipitation by RNA Purification (ChIRP) (Chu et al., 2011). ChIRP probes designed against the length of L1spa RNA successfully capture LINE1 RNA, but not unrelated RNAs (Figure S5A). To assess the specificity of LINE1 RNA ChIRP, we performed parallel Malat1 ChIRP and verified its specific association with the Malat1 and Neat1 loci [(Figure S5B) and (Engreitz et al., 2014)]. LINE1 RNA is robustly detected at LINE1 DNA (Figure S5C), as might be expected from the previously reported close association of LINE1 RNA with chromosome domains from which it is transcribed (Hall et al., 2014). Interestingly, we found that LINE1 is significantly enriched at both Dux and rDNA loci compared to Malat1 RNA, but not at control regions (Figure 5A, Figure S5B). The LINE1 ChIRP signal is RNA-dependent, as RNase treatment largely prevents the recovery of LINE1 RNA-bound sites (Figure 5A). The repeated nature of LINE1 and the likelihood that unmapped LINE1 insertions exist in the genome may confound the source of the LINE1 ChIRP signal. Nevertheless, these data indicate that LINE1 RNA interacts with the Dux and rDNA loci, and raise the possibility that it cooperates with factors that regulate chromatin activity at these sites.
Figure 5. LINE1 RNA interacts with Nucleolin to coordinately repress Dux and activate rRNA synthesis.
(A) ChIRP enrichment in wild-type ES cells at the indicated DNA loci, using biotinylated probes against LINE1 RNA. Intergenic-chromosome 11 (int-chr11) and Rpl3-TSS are shown as negative regions. Data are mean +/− s.e.m., n=3 independent experiments.
(B) RIP in wild-type ES cells with Nucleolin (Ncl) or control IgG antibodies, showing Ncl association with the indicated RNAs. Pre-rRNA is shown as a positive control. Data are mean +/− s.e.m., n=2 independent experiments, and shown as % input normalized to Malat1 RNA.
(C) qRT-PCR showing knockdown of candidate 2C/Dux repressors alongside Dux and 2C gene expression. Data are representative of two independent experiments and are mean +/− s.e.m., n=3 technical replicates.
(D) PCA plot for all genes across all samples, showing that Ncl KD ES cells have distinct gene expression profiles and are separated from controls along PC1.
(E) Boxplot analysis of significantly altered genes (FDR <0.05) upon Ncl knockdown, revealing that Dux targets are significantly more induced than non-targets (see Fig. 3B). P-value is determined by two-sided Wilcoxon rank sum test.
(F) Scatter plot showing the log2 fold-change (FC) in the expression of all genes following LINE1 KD (x axis) or Ncl KD (y axis). Dux targets are indicated in red, with select 2C genes labelled in black. The Spearman’s correlation coefficient is indicated.
(G) RNA per cell in ES cells 48h after transfection with control or Ncl siRNAs. Data are mean +/− s.e.m., n=4 independent batches of equal numbers of cells.
(H) Colony-formation assay in ES cells following transfection with control or Ncl siRNAs. Data are mean +/− s.e.m., n=3 biological replicates. See also Figure S5 and Table S3.
Nucleolin depletion recapitulates LINE1 knockdown
Studies in the retrotransposition field have uncovered a number of proteins that interact with LINE1 RNA. One protein that caught our attention is Nucleolin, a well-known rDNA and rRNA binding protein required for rRNA synthesis and processing (Ginisty et al., 1998). Nucleolin was recovered in a search for proteins that interact with mouse LINE1 RNA, and this interaction appears to be conserved in human (Moldovan and Moran, 2015, Peddigari et al., 2013). Intriguingly, Nucleolin was also reported to bind to and repress the human DUX4 genomic repeat, D4Z4, in HeLa cells (Gabellini et al., 2002). Using RNA-immunoprecipitation (RIP)-qPCR, we verified that Nucleolin strongly associates, directly or indirectly, with LINE1 RNA in ES cells, at similar levels to pre-rRNA (Figure 5B). The Nucleolin RIP-qPCR signal is DNA-independent, indicating that the association of Nucleolin with LINE1 RNA detected by RIP is not secondary to binding to DNA (Figure S5D). In contrast, Nucleolin does not bind other RNAs such as the spliceosomal-RNA U1 (Figure 5B, S5D), despite its abundant nuclear expression (Figure S5E).
Next, we tested whether Nucleolin may be a repressor of Dux and the 2C program in ES cells. A targeted RNAi screen for putative 2C regulators revealed that KD of Nucleolin, but not of several other candidates tested, causes dramatic upregulation of Dux, MERVL and 2C genes (Figure 5C and S5F–H). RNA-seq analysis revealed that Dux targets and MERVL repeats are among the most highly upregulated transcripts upon Nucleolin KD (Figure 5D–E, S5I and Table S3). Moreover, there is a remarkably high similarity between the transcriptomes of ES cells depleted for LINE1 or Nucleolin (Figure 5F, Spearman ρ =0.6). These results indicate that Nucleolin KD largely recapitulates LINE1 KD at the transcriptional level. Nucleolin knockdown also mimics LINE1 KD ES cells with regards to inducing significant decreases in ribosomal protein gene transcription (Figure S5J), total RNA levels (Figure 5G) and ES cell self-renewal (Figure 5H). Similarly, Dux KD rescues 2C gene upregulation but not the self-renewal deficit induced by Nucleolin KD (Figure S5K–L). Together, our findings indicate that LINE1 RNA and Nucleolin protein interact and have overlapping functions in ES cells, including promotion of rRNA synthesis and repression of the Dux/2C program.
Nucleolin and Kap1 bind to Dux and rDNA in a LINE1 RNA-dependent manner
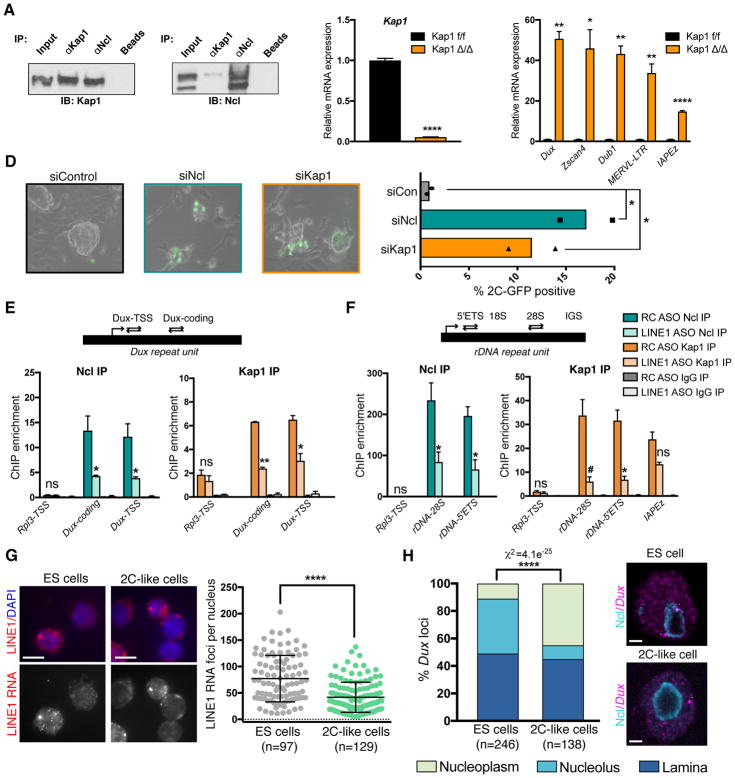
The only factor to date shown to directly bind and repress Dux in mouse ES cells is the co-repressor tripartite motif-containing protein 28 (TRIM28/Kap1) (De Iaco et al., 2017). We found that Nucleolin interacts with Kap1 in ES cells, suggesting that they may function together in a complex (Figure 6A). Using inducible mutant CreERT2;Kap1fl/fl ES cells (Rowe et al., 2010), we confirmed that Kap1 deletion induces a significant increase in Dux and 2C genes and MERVL (Figure 6B–C), and that Kap1 or Nucleolin KD both lead to over 10-fold increases in 2C-like cells in vitro (Figure 6D). Analysis of published RNA-seq data from Kap1-deleted ES cells (Ecco et al., 2016) also revealed a similarity in transcriptional changes to LINE1 KD, albeit less striking than the Nucleolin KD-LINE1 KD similarity (Figure S6A, Spearman ρ =0.34 and Figure 5F). We found that both Nucleolin and Kap1 bind Dux loci, and the levels of both proteins at Dux are significantly reduced upon LINE1 KD (Figure 6E). Moreover, efficient Nucleolin binding to rDNA is also dependent on LINE1 (Figure 6F). Surprisingly, Kap1 is also robustly recruited to rDNA in a LINE1-dependent manner (Figure 6F), and Kap1 deletion leads to similar reductions in nascent rRNA transcription as LINE1 or Nucleolin KD (Figure S6B). LINE1 KD does not affect the levels of Nucleolin or Kap1 RNA or protein (Figure S3C, S6C–E), nor their interaction (Figure S6F). Collectively, these data indicate that LINE1 is essential for the efficient chromatin binding of a Nucleolin-Kap1 complex that represses Dux and promotes rRNA expression.
Figure 6. LINE1 promotes binding of Nucleolin and Kap1 to Dux and rDNA.
(A) Co-IP showing association of endogenous Kap1 and Ncl proteins in ES cells.
(B–C) qRT-PCR confirming (B) Kap1 deletion and (C) upregulation of Dux, 2C genes, and ERVs. Data are mean +/− s.e.m, n=3 biological replicates.
(D) Representative micrographs (left) and percentage of 2C-like cells (right) in 2C-GFP reporter ES cells 3 days following siRNA KD of Ncl or Kap1, in n=2 independent experiments.
(E–F) ChIP assays for Ncl and Kap1 at (E) Dux and (F) rDNA, with or without LINE1 KD. Data are shown as % input normalized to enrichment at int-chr11 negative control region, and are mean +/− s.e.m., n=3 independent experiments for Ncl ChIP, and n=2 independent experiments, Kap1 ChIP.
(G) Example images and quantification of nuclear LINE1 RNA foci by RNA-FISH in ES cells (2C-GFP-) or 2C-like cells (2C-GFP+), representative of 2 experiments. Scale bar, 10μm, n=number of cells.
(H) Representative images and quantification of distinct localization patterns of Dux loci in ES vs 2C-like cells by IF for Ncl combined with Dux DNA-FISH, using 2C-GFP reporter ES cells. Nucleoli are labeled with Ncl antibodies. Example Dux loci are indicated (white arrows), showing two nucleolar loci in an ES cell vs one nucleoplasmic locus in a 2C-like cell. Statistics are calculated by Chi-squared test for the indicated number (n) of Dux loci. Scale bar, 2μm. See also Figure S6.
Subsequently we sought to interrogate what might be different in 2C-like cells, compared to standard ES cells, that allows for induction of Dux and the 2C program. While the levels or localization of Kap1 and Nucleolin are unchanged (Figure S6G), the nuclear abundance of LINE1 RNA is significantly reduced in 2C-like cells compared to ES cells (Figure 6G). In addition, 2C-like cells have significantly fewer nucleoli per cell compared to ES cells (Figure S6H). DNA FISH revealed that in ES cells the Dux loci are most often located in the peri-nucleolar or laminar regions (Figure 6H, S6I–K), both of which are thought to be transcriptionally repressive heterochromatin environments (Guetg and Santoro, 2012, Reddy et al., 2008). Intriguingly, the transition of ES cells to the 2C-like state is accompanied by a release of Dux loci from the Nucleolin-positive domain at peri-nuclelolar regions to the nucleoplasm (Figure 6H, S6K). Taken together, these data suggest that a combination of reduced abundance of LINE1 RNA with changes to the interaction of Dux with Nucleolin at peri-nucleolar heterochromatin may facilitate the entry of ES cells into a 2C-like state, a possibility that deserves future exploration.
LINE1 promotes silencing of the Dux/2-cell program, rRNA synthesis and developmental progression past the 2-cell stage
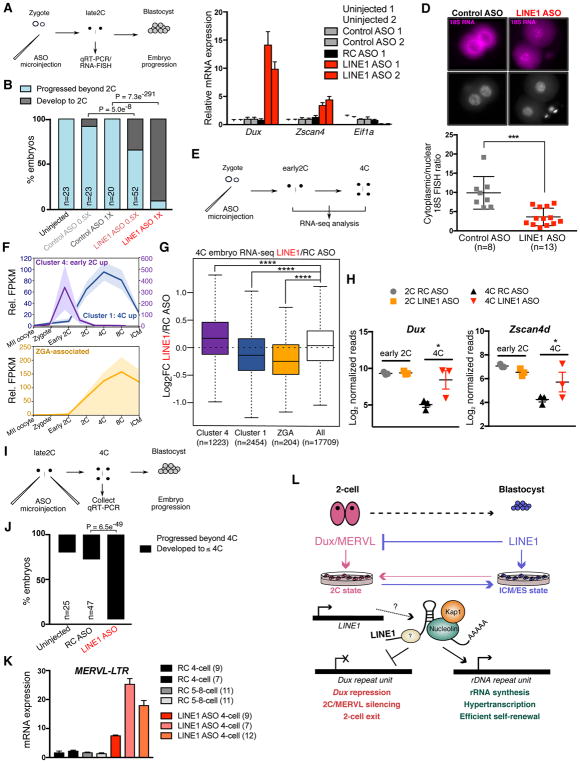
Lastly, we investigated the role of LINE1 in embryonic development (Figure 7A). Almost no LINE1 KD embryos reach the blastocyst stage and most arrest at the 2-cell stage (Figure 7B and S7A). Furthermore, 2-cell LINE1 KD embryos display high upregulation of Dux as well as its target 2-cell gene, Zscan4 (Figure 7C). In contrast, the expression of Eif1a, a marker of ZGA (Davis et al., 1996, Zeng and Schultz, 2005), is sharply reduced upon LINE1 KD. 2-cell embryos depleted of LINE1 exhibit significant reductions in chromatin accessibility (Figure S7B) in agreement with (Jachowicz et al., 2017), and display significantly increased levels of heterochromatin (Figure S7C). While control ASO-injected embryos show the expected strong cytoplasmic accumulation of 18S rRNA, in LINE1 ASO-injected embryos 18S rRNA is largely retained in the nucleus (Figure 7D). In agreement, chemical inhibition of rRNA synthesis from either the zygote (Lin et al., 2014) or 2-cell stage onwards (Figure S7D) is incompatible with pre-implantation development.
Figure 7. LINE1 regulates Dux silencing, rRNA synthesis and early development.
(A) Summary diagram of ASO microinjection experiments in panels (B–D).
(B) Developmental progression in the indicated number (n) of embryos following ASO microinjections, collected in 3 independent experiments. χ2 P-values were calculated for the developmental rate of embryos injected with LINE1 or Control ASOs.
(C) qRT-PCR for the indicated genes in late 2-cell (2C) embryos harvested 24h post injection. Data are mean +/− s.e.m., n=3 technical replicates, showing 2 independent experiments.
(D) 18S RNA FISH in late 2C embryos 24h following microinjection with Control or LINE1 ASOs, with nuclei stained by DAPI. Nuclear or cytoplasmic foci were counted in each embryo and plotted below as 18S cytoplasmic/nuclear ratio. Data show mean +/− s.d from the indicated number (n) of embryos from 2 independent experiments. Scale bar, 20μm.
(E) Summary diagram of 0.5X ASO microinjection experiments for RNA-seq in panels (F–H).
(F) RNA-seq data from (Wu et al., 2016), showing mean FPKM expression relative to MII oocyte for the indicated gene sets identified by K-means clustering, or for ZGA genes defined in (Zeng and Schultz, 2005). Shading denotes mean +/− s.e.m at each time point.
(G) Boxplot depicting the log2-fold change in 4-cell (4C) embryos upon LINE1 KD for the indicated number (n) of gene sets displayed in (G), compared to all expressed genes. P-value, two-sided Wilcoxon rank sum test.
(H) Examples of the expression of early 2C genes, Dux and Zscan4d, in the indicated embryo samples. P-value, toptable FDR, showing n=3 samples per condition.
(I) Summary diagram of late 2C ASO microinjection experiments in panels (J–K).
(J) Developmental progression in the indicated embryos following late 2C ASO microinjections. χ2 P-values were calculated for the developmental rate of the number (n) of embryos injected with 1X RC or LINE1 ASOs.
(K) qRT-PCR analysis of MERVL expression in 4C or 5–8C embryos following late 2C microinjections with ASOs. Data are mean +/− s.e.m n=3 technical replicates and are representative of 3 (LINE1 ASO) or 2 (RC ASO) independent experiments.
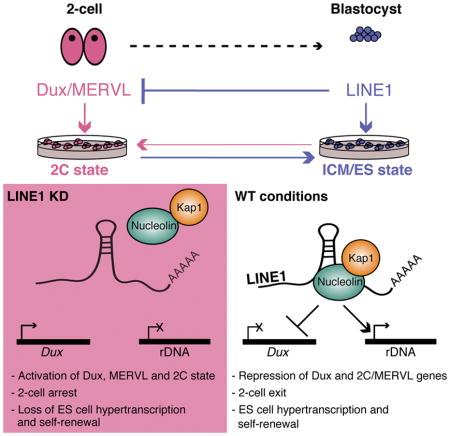
(L) Model for the role of LINE1 in early development and ES cells. LINE1 acts as an RNA-scaffold and binds to rDNA and Dux. LINE1 RNA-Ncl-Kap1 cooperate to turn off the 2C gene expression program and promote high levels of ribosome biosynthesis during early development. See also Figure S7 and Table S4.
We next performed RNA-seq on samples where zygotes were microinjected with lower concentrations (0.5X) of ASOs to allow some LINE1 KD embryos to progress to the 4-cell stage (Figure 7E). Very few transcriptional changes are detected in LINE1 KD embryos at the early 2-cell stage. In contrast, LINE1 KD 4-cell embryos are transcriptionally distinct from controls (Figure S7E, and Table S4). Using published data (Wu et al., 2016), we defined gene clusters associated with Dux/early 2-cell gene expression (cluster 4, Figure 7F and S7F), or with elevated expression maintained up to the 4-cell stage (cluster 1, Figure 7F and S7F). 4-cell LINE1 KD embryos display a significant upregulation of early 2-cell genes and reduced expression of 4-cell genes (Figures 7G–H and S7G). Genes associated with ZGA (Zeng and Schultz, 2005) are significantly decreased in 4-cell LINE1 KD embryos (Figure 7F–G), along with ribosomal genes (Figure S7H).
Finally, we performed LINE1 KD at the late 2-cell stage, when Dux has already been silenced and ZGA initiated (Figure 7I). Very few of these LINE1 KD embryos develop past the 4-cell stage (Figure 7J and Figure S7I). Moreover, 4-cell embryos depleted of LINE1 from the late 2-cell stage display upregulation of MERVL and 2-cell genes (Figure 7K and S7J). Overall, these data indicate that there is an ongoing requirement for LINE1 RNA for repression of the 2-cell program and developmental progression during early embryogenesis. Taken together, the data suggest that a failure to repress the Dux/2-cell program and defective ZGA and ribogenesis contribute to embryonic arrest upon LINE1 depletion.
Discussion
The expression of TEs such as LINE1 is generally thought to be detrimental to cells because it can cause mutations or apoptosis [e.g. (Malki et al., 2014, Burns, 2017)]. In contrast, we report here that the expression of LINE1 regulates exit from the 2-cell state by performing two main functions: repressing the 2C program induced by Dux, and activating rRNA synthesis to support rapid proliferation. These two functions are unified by the interaction of LINE1 RNA with Nucleolin, which we identify as a novel repressor of the Dux/2C program. Thus, rather than being a simple genomic parasite, LINE1 may best be viewed as a symbiont that is an integral part of the transcriptional networks that regulate cellular potency during early mammalian development (Figure 7L).
Nucleolin is most commonly associated with the positive regulation of rRNA synthesis (Ginisty et al., 1998). In recent years, Nucleolin has been shown to also have roles in chromatin remodeling, DNA replication, and DNA repair [reviewed in (Jia et al., 2017)]. Our work uncovers a novel function for Nucleolin, and it will be of interest to identify the functional elements in the LINE1 RNA by defining the regions that interact with Nucleolin and potentially other factors, like Kap1. Intriguingly, the region of mouse LINE1 RNA previously used to identify Nucleolin as an interactor was the interORF (Peddigari et al., 2013), the same region against which we designed the LINE1 ASO (Fig. 1C). It is therefore possible that the ASO disrupts the interaction between LINE1 RNA and Nucleolin, which in turn may contribute to destabilizing LINE1 RNA.
It remains unclear how LINE1 RNA-Nucleolin-Kap1 are targeted to the Dux cluster. LINE1-Nucleolin-Kap1 may have other interacting partners with DNA binding specificity. For example, YY1 has been implicated in targeting a repressive complex containing Nucleolin to the human DUX4 cluster (Gabellini et al., 2002), although we did not find a role for YY1 in Dux repression in ES cells (Figure 5C). Alternatively, KRAB-ZFP transcription factors are known to recruit Kap1 to repress TEs (Lupo et al., 2013, Wolf et al., 2015), a function that might have been coopted for Dux silencing. Finally, our data leave open the possibility that transcribed LINE1 DNA loci contribute in cis to Dux repression or rRNA activation via higher-order chromosome interactions (Figure 7L). Furthermore, while LINE1 is specifically detected at Dux and rRNA, it is likely to have other genomic targets that remain to be discovered.
Our data suggest that the LINE1-Nucleolin-Kap1 complex has both an activating function (at rDNA) and a repressive function (at Dux). While this might seem paradoxical, Nucleolin and Kap1 have been shown to have both activating and repressive effects. In addition to its role as activator of rDNA, Nucleolin has been reported to repress the expression of genes such as cMyc (Gonzalez et al., 2009), MMP13 (Samuel et al., 2008) or the D4Z4 repeat (Gabellini et al., 2002). Conversely, the co-repressor Kap1 also has activating functions both at the single gene level (Singh et al., 2015), as well as globally (Bunch et al., 2014). It is possible that the reduction in nascent rRNA synthesis in Kap1 mutant ES cells contributes their self-renewal defect, in addition to the previously reported de-repression of endogenous retroviruses (Rowe et al., 2010). Moreover, the localization of Dux loci to Nucleolin-positive peri-nucleolar regions in ES cells but not 2C-like cells provides a potential rationale at the level of nuclear 3D organization for the coordinate roles of LINE1/Nucleolin/Kap1 in activation of rDNA vs repression of Dux.
LINE1 is expressed throughout pre-implantation development (Figure 1B and (Fadloun et al., 2013)), which implies that its presence alone may not suffice to repress Dux and the 2C program. However, analysis of RNA-seq data from early embryos reveals that Nucleolin and Kap1 are sharply induced at the late 2-cell stage, correlating with Dux silencing (Figure S7K). Thus, it is possible that, by promoting ZGA at the 2-cell stage, Dux induces several of its own repressors, which can then bind LINE1 RNA and silence the Dux loci. Moreover, there are large-scale changes to the organization of the nucleolus and its associated heterochromatin during these stages (Borsos and Torres-Padilla, 2016) that may pertain to the silencing of the Dux/2C program.
It has been proposed that the retrotransposition activity of LINE1 is essential for early mouse development, possibly by making cDNA copies of sperm-derived RNAs (Beraldi et al., 2006, Sciamanna et al., 2009). Using inhibitors that block LINE1 mobility [Figure S2G and (Jones et al., 2008)], we found that retrotransposition is not involved in the regulation of 2C gene expression nor of ES cell self-renewal (Figure S2H–I). Moreover, LINE1 ORF1 protein, which is required for retrotransposition, is predominantly cytoplasmic (Figure S1A), in contrast to the nuclear localization of LINE1 RNA. While we cannot at present exclude a non-canonical function for LINE1 ORF1 or ORF2 proteins in the developmental roles reported here, our data indicate that it acts as a chromatin-associated RNA that binds Nucleolin and Kap1 to regulate gene expression.
The partnership between LINE1 and Nucleolin in the regulation of rRNA synthesis indicates that LINE1 contributes to ES cell hypertranscription (Percharde et al., 2017a). This notion is supported by the fact that LINE1 KD leads to decrease in total RNA levels, nascent transcription, nascent translation and self-renewal. Although we do not detect LINE1 RNA binding at ribosomal protein genes, the synthesis of rRNA and ribosomal proteins genes is highly coordinated (Laferte et al., 2006). In addition, we have recently shown that reductions in translational output in ES cells rapidly induce a decrease in nascent transcription of highly-expressed genes, including rRNA and ribosomal protein genes (Bulut-Karslioglu et al., 2018). Our data support a model whereby LINE1-mediated induction of rRNA synthesis leads to global increases in ribosomal biogenesis, enabling rapid growth of the early embryo. We speculate that the LINE1-Nucleolin partnership may play roles in other stem/progenitor cells, where it might not necessarily act to repress Dux but may still promote ribogenesis and proliferation.
Our work raises the question of how a mechanism for control of developmental potency based on TEs might have evolved. Active TEs are under acute surveillance by cellular pathways that minimize transposition, including by Kap1 (Rowe et al., 2010). In part because of this, and in part because of a loss in ability to transpose due to an accumulation of mutations, the sequence of TEs is generally thought to be subject to a rapid rate of divergence. In fact, some mammalian species may have completely lost all retrotransposition-competent LINE1 elements in their genome, even though they can still express mutated LINE1 RNAs (Cantrell et al., 2008). Our results indicate that chromatin-associated LINE1 RNA regulates gene expression and developmental potency without requiring retrotransposition activity. This role of LINE1 as a chromatin-associated RNA therefore avoids the potential detrimental effects of LINE1 retrotransposition that have been reported in several disease states, including cancer (Burns, 2017). The interaction of LINE1 RNA with binding partners such as Nucleolin is expected to be mediated by RNA secondary structure, which is less constrained by primary sequence than protein-coding regions. Thus, rather than being a vulnerability, the regulation of early development by TEs may allow both robustness, due to the repeated nature of TEs, and adaptability, due to their rapid evolution and their potential to support transposition in conditions of stress. In this regard, it is interesting that the percentage of the genome occupied by LINE1 elements seems to have sharply increased with development of therian mammals [e.g., (Ivancevic et al., 2017)]. The exploration of the function of LINE1 in other species should shed light on the role of TEs in shaping the evolution of development.
STAR METHODS
CONTACT FOR REAGENT AND SOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Miguel Ramalho-Santos (mrsantos@ucsf.edu, mrsantos@lunenfeld.ca).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Animal experiments were performed with 5–8-week-old female and 2–6-month-old male C57BL/6 mice. Animals were maintained on 12 h light/dark cycle and provided with food and water ad libitum in individually ventilated units. Animal experiments in USA were in accordance with the guidelines of the UCSF Institutional Animal Care and Use Committee, protocol AN091331-03. Animal experiments in UK were approved by the University of Edinburgh’s Animal Welfare and Ethical Review Board (AWERB) and carried out under the authority of a UK Home Office Project License.
Mouse ES cell culture
Mouse E14Tg2A (E14) ES cells (male) were used for all experiments (Hooper et al., 1987), except for p53−/− ES cells (Sabapathy et al., 1997), which are V6.5-derived (male, a gift from Scott Oakes). ES cells were cultured on 0.1% gelatin-coated plates in ES-FBS culture medium (high glucose DMEM GlutaMAX with sodium pyruvate (Thermo Fisher Scientific), 15% FBS (Atlanta Biologicals), 0.1mM non-essential amino acids, 50 U/mL penicillin-streptomycin (UCSF Cell Culture Facility), 0.1mM 2-Mercaptoethanol (Millipore) and 1,000U/ml LIF supplement (ESGRO, Millipore). Where indicated, ES cells were grown in N2B27/2i/LIF conditions (DMEM/F-12, Neurobasal medium, 1x N2/B27 supplements, 1μM PD0325901, 3μM CHIR99021, LIF as above) according to (Ying et al., 2008) for at least 4 passages before being used for experiments. Deletion of Kap1 in CreERT2;Kap1fl/fl ES cells (undetermined sex, Rowe et al., 2010) was performed in ES/FBS conditions with 1μM 4-OHT overnight and analyzed 4 days later. Routine testing of E14 ES cells revealed absence of mycoplasma contamination. ES cells were not genotyped.
2C-GFP ES cell line
2C-GFP reporter ESCs were generated as in (Ishiuchi et al., 2015), using Addgene plasmid 69071, from parental E14 ES cells. 4×106 cells were nucleofected with 4μg linearized 2C-GFP plasmid, and plated at low density in 10cm2 plates. Selection was performed with 250μg/mL G418 (Mirus) commencing 36h after nucleofection and maintained for 8 days before individual colonies were picked and expanded. A clone with high, specific upregulation of 2C genes in sorted GFP+ cells (Figure S2K) was used for all experiments.
MEFs
E13.5 primary MEFs derived from pooled CD1 embryos (mixed sex) were cultured in MEF medium (as ES-FBS but without 2-Mercaptoethanol and LIF), and used within 4 passages of initial derivation.
METHOD DETAILS
ASO- and siRNA-Mediated Knockdown
A morpholino ASO targeting the inter-ORF region of LINE1 was designed with software available at Gene Tools LLC. The morpholino chemical backbone was chosen because of its stability, specificity and extensive use in pre-implantation mouse embryos [(e.g., (Lin et al., 2013)]. ASOs have been shown to be potent inhibitors of nuclear RNAs, such as lncRNAs (Lennox and Behlke, 2016). The LINE1 ASO was validated in silico to be perfectly homologous to at least 500 full-length LINE1 elements, using L1Base (Penzkofer et al., 2005). This sequence was verified by Blast as not having homologies to any known Refseq mRNAs. The reverse complement of the LINE1 ASO sequence does not target LINE1 RNA and was used as a control (RC ASO). In the indicated embryo experiments, a standard non-targeting control ASO (Gene Tools) was also used. Both lissaminated (cell line experiments) and non-lissaminated (embryo experiments) ASOs were utilized. ASOs were introduced into cells by nucleofection, utilizing an Amaxa Nucleofector 2b device and ES nucleofection kit (Lonza), according to the manufacturer’s instructions. 4–5 million cells were used per nucleofection together with 5nmol of the indicated ASO. Cells were plated in ES medium immediately following nucleofection and left to recover for 24–48h. For colony formation assays and expression analyses excluding co-transfections with siRNAs, Lissamine-positive ES cells were purified by FACS utilizing a BD FACSAria II (BD Biosciences) to enrich for successfully-nucleofected cells. Nucleofection efficiency was routinely 70% or above. All other experiments were performed on the bulk population of nucleofected cells without prior enrichment for ASO-positive populations. siRNA transfections were performed in ES cells with Lipofectamine 2000 (Thermo Fisher Scientific) as described previously (Percharde et al., 2012). ES cells were plated 5–7h before transfection at a density of 5×105 ES cells per 6-well culture area and transfected with 100 pmol siRNA, according to the manufacturer’s standard recommendations. A non-targeting siRNA (siGenome siControl #2, Dharmacon) was used as a control. For LINE1 siRNA knockdown, 2 independent siRNA sequences were designed using online tools available at Dharmacon and verified to have no perfect match to other mRNAs. For combined ASO/siRNA experiments, ES cells were first nucleofected with ASOs, then plated and left to recover for 7h. Next, the medium was exchanged for fresh ES-FBS without antibiotics and siRNA transfections were performed as above. The medium was exchanged the next morning and cells harvested for RNA extraction approximately 48h post initial nucleofection. All ASO and siRNA sequences are available in Supplemental data, Table S5.
Embryo Microinjection and Culture
Females were superovulated by administration of 7.5 I.U. of pregnant mare serum gonadotropin (PMSG; National Hormone Pituitary Program (NHPP), Harbor-UCLA Medical Center, CA, USA or Intervet UK) and 48h later with 7.5 I.U. of human chorionic gonadotropin (hCG; Sigma-Aldrich or Intervet UK). Embryo experiments were performed as described previously (Lin et al., 2013, Lin et al., 2014), and embryos were microinjected using a microinjector (FemtoJet 4i, Eppendorf) and an inverted microscope (Leica, DMi8) equipped with micromanipulators (Narishige). Zygotes were microinjected with Standard Control, RC, or LINE1 ASOs. ASO solutions were injected into the cytoplasm from a stock concentration of 1.5mM (1X) or 0.75mM (0.5X), utilizing 2–5 pl of solution per injection. Embryos were cultured in KSOM+AA medium (Millipore) at 37°C in 5% CO 2. For qRT-PCR analysis in late 2C embryos, 10–11 embryos were collected approximately 24h post injection for RNA isolation using the PicoPure RNA Extraction Kit (Arcturus), or were cultured for a further 3 days to monitor developmental progression. Embryo qRT-PCR data are normalized to Hprt expression. For embryo RNA-seq experiments, zygotes were microinjected with 0.5X ASO and cultured in KSOM+AA medium. Early 2C embryos were collected on approximately 33 h post hCG (~9 h post-microinjection). 4C embryos were collected after 2 days of in vitro culture. ASO injections at the late 2-cell stage were performed after collection approximately 43h post hCG administration, and 1X ASOs were microinjected into the cytoplasm of both blastomeres. Developmental progression to blastocyst was monitored, or 4C embryos were collected on the day following injection for RNA isolation and qRT-PCR analysis, as above. For inhibition of rRNA synthesis after ZGA, late 2C embryos were cultured in KSOM+AA with 1μM CX-5461 or 0.1% DMSO.
DNase-TUNEL Experiment
DNase I-TUNEL assays were performed in late 2C embryos. Embryos were permeabilized by 0.5% Triton X-100 in pre-extraction buffer (300mM sucrose, 25mM HEPES, 1M CaCl2, 50mM NaCl, 3mM MgCl2) for 5 min before digesting with 0.2U/μl of DNase I (NEB). Embryos were then fixed in 4% PFA. TUNEL Assays (Click-iT TUNEL Imaging assay, Thermo Fisher Scientific) were followed according to manufacturer’s instructions. The nuclear area was defined according to Hochest DNA staining and the intensity of nuclear TUNEL signal was quantified using Fiji software.
RNA FISH
Embryo RNA FISH was performed as previously described (Lin et al., 2014). Embryos were fixed in methanol, permeabilized in 70% ethanol and hybridized at 37°C using 48 single-molecule probes designed to span the length of LINE1 ORF2 RNA, designed from the sequence of L1spa (L1Md_Tf family) and expected to target the majority of transcribed LINE1 RNAs (Naas et al., 1998), or against 18S rRNA at 1:250 dilution. RNA-FISH in ES cells was performed on glass coverslips 48h after ASO or siRNA treatment, according to the manufacturer’s standard protocol (Biosearch Technologies). Fixation was performed in 1% paraformaldehyde (PFA) for 15 min, followed by permeabilization overnight in 70% ethanol at 4°C pr ior to hybridization. Hybridization was performed as above, using the probes against LINE1 RNA. Co-incubations were performed with Gapdh probes where indicated (purchased from Biosearch Tehnologies). For +RNase negative controls, coverslips were incubated in 2X saline-sodium citrate (SSC) buffer plus 10μg/mL RNAse A (Thermo Fisher Scientific) for 30 min at 37°C, then washed twice in 2X SSC, prior to LINE1 FISH hybridizations. For LINE1 RNA FISH in ES vs 2C-like cells, GFP+/− populations of 2C-GFP reporter cells were separated by FACS then plated onto separate wells on matrigel-coated Lab-Tek II Chambered Coverglass for 1h before fixation and hybridization as above. All images were collected on a Leica DMI 4000B inverted scope using a 100× oil-immersion objective, every 0.25μm using the Z-stack function of the MetaMorph software (Molecular Devices). All nuclei were counterstained with DAPI. RNA FISH foci were quantified with StarSearch online software [(Raj et al., 2008), Raj Lab, University of Pennsylvania], using identical threshold settings between different samples and images.
Cell-cycle and Self-renewal Assays
24h following nucleofection or siRNA transfection, ES cells were either trypsinized and counted manually (siRNAs) or isolated by FACS according to Lissamine fluorescence (ASOs); 1000 cells were plated per well of a 12-well plate in ES-FBS medium. Colonies were left to form over a period of 5–6 days and then fixed and stained for Alkaline Phosphatase (AP) according to the manufacturer’s recommendations (Sigma-Aldrich). The numbers of AP-positive colonies per well were manually counted. For proliferation assays, 1×106 ES cells were plated immediately following ASO nucleofection and then each day for the next four days the cells were trypsinized and counted, replating 1×106 cells each time back into culture. Cell-cycle analysis was performed according to (Bulut-Karslioglu et al., 2018) and the standard manufacturer’s protocol, incubating ES cells 48h after nucleofection with 10μM EdU for 1 h, followed by analysis of EdU incorporation and DNA content with the Click-iT EdU imaging kit (Life Technologies) and FxCycle-Violet, respectively.
RNA Extraction and Expression Analysis
For ASO-nucleofected samples, RNA was isolated from 300,000 FACS-purified ES cells using the RNeasy mini kit (Qiagen), according to manufacturer’s instructions. RNA was treated once on-column with DNAse I and once more off-column (RNAse-free DNase I from Thermo Fisher Scientific) to remove any residual DNA contamination. cDNA synthesis was performed from 1μg RNA with the High Capacity cDNA Reverse Transcription kit (Applied Biosystems) and qRT-PCR performed with SYBR green (KAPA) on an ABI-Prism PCR machine. qRT-PCR analysis following siRNA experiments or ASO/siRNA experiments were performed as above, but without prior FACS purifications. All gene expression data were normalized to two independent housekeeping genes (Rpl7, Ubb or H2A).
Retrotransposition Assay
ES cells were nucleofected with 5μg codon-optimized LINE1 CAG-ORFeus GF-P reporter plasmid (An et al., 2006) or LINE1 5′UTR-ORFeus GF-P reporter plasmid (Newkirk et al., 2017). The following morning, Stavudine (d4T, 20μM) or Tenofovir disoproxil fumarate (TDF, 4μM) (Jones et al., 2008) was added and cells cultured for a further 48h, when retrotransposition rates were determined by flow cytometry for GFP-positive cells, or cells harvested for RNA extraction as above. Colony formation assays in the presence of d4T and TDF were as above, with the addition of inhibitors performed 24h after plating and maintained throughout.
RNA-sequencing
Sample preparation for RNA-seq in ES cells was performed as previously described (Percharde et al., 2017b). RNA was extracted utilizing the RNeasy mini kit as for qRT-PCR, then 800ng DNAse-treated total RNA was used per library preparation according to the NEBNext Ultra Directional Library Prep Kit for Illumina (NEB). Embryo RNA-seq was performed in pools of 14–18 early 2C or 4C embryos per sample, with total RNA isolated using the PicoPure RNA Extraction Kit (Arcturus). Libraries were generated using the SMARTer Stranded Total RNA-seq Kit v2 (Takara) and 500pg total RNA input per sample. In all cases, three replicates were sequenced per condition at the UCSF Center for Advanced Technology on an Illumina HiSeq 2500 or HiSeq 4000, with 50bp single end reads.
Immunofluorescence
ES cells were plated onto matrigel-coated 8-well chambers for 1h, fixed for 10 min in 4% PFA, and blocked and permeabilized in one step in IF buffer (PBS, 10% donkey serum, 2.5% BSA) plus 0.4% Triton X-100. Incubations were performed overnight at 4°C, using the following antibodies and dilutions in IF buffer: GFP (1:100), Oct4 (1:100), Nanog (1:200), Kap1 (1:500), Nucleolin (1:1000 x), Orf1p (Di Giacomo et al., 2014) (1:500). The next day, slides were washed 3 times in PBS, incubated for 60 min in the relevant fluorescently-conjugated secondary antibodies (1:1000 in IF buffer, Life Technologies), and washed again as before. Slides were mounted with Vectashield containing DAPI and imaged on a Leica SP5 upright confocal microscope at 63X magnification under oil immersion. Embryo immunofluorescence (IF) experiments were performed as above, except blocking was performed in 5% BSA and incubation with primary antibodies was at 37°C for 1.5 hr. with antibodies against H3K9me2 or H3K9me3 (1:50), and nuclei counterstained with DAPI. Images of stained embryos were acquired by a spinning disk confocal (CSU-W1, Yokogawa) on an upright microscope frame (BX-63, Olympus) using a 60x silicon oil immersion objective (UPLSAPO 60XS2, Olympus) with additional 2x intermediate magnification. IF signal intensity was quantified using Fiji software.
RNA Immunoprecipitation (RIP)
RIP experiments were performed on nuclear extracts according to a standard Abcam protocol, with the following modifications. 1μg anti-Nucleolin or control IgG antibodies were pre-bound to 30μL Protein A Dynabeads (Thermo Fisher Scientific) and incubated rotating for at least 3h at 4°C. Beads were next collected on a DynaM ag (Thermo Fisher Scientific) and resuspended in RIP buffer (150mM KCl, 25mM Tris pH 7.4, 5mM EDTA, 0.5mM DTT, 0.5% NP40, protease and RNase inhibitors) containing 500ng/μL tRNA (Thermo Fisher Scientific) and 1 mg/mL RNase-free BSA (bioWORLD) to block for 30 min, then collected and used immediately in RNA immunoprecipitation. Where indicated, 40U of Turbo DNase (Thermo Fisher Scientific) was added to nuclear extracts and incubated for 30 min at 37 °C, then quenched with 10mM EDTA, befo re continuing with immunoprecipitations. Prior to immunoprecipitation, ES nuclear extracts were also blocked for 30 min with 20μl Protein A Dynabeads at 4°C, 30 min. Cleared nuclear lysates were incubated together with antibody-bound blocked beads overnight at 4°C. The next day, lysates were washed four times in RIP buffer, once in PBS, and RNA was extracted from beads using Trizol and standard phenol-chloroform extraction. The aqueous phase containing the RNA was loaded onto RNeasy mini columns (Qiagen) with 2x volume of 100% ethanol and RNA was purified according to the standard protocol. RNA was DNAse I treated twice as before and used to generate cDNA for qRT-PCR.
Nascent Transcription and Translation Assays
For nascent transcription assays, ES cells were cultured for 45 min in normal ES medium supplemented with 1mM EU. For nascent translation assays, cells were incubated in HPG medium (as ES-FBS medium with the following substitutions: Methionine- and Cysteine-free DMEM, 1mg/mL BSA instead of FBS) for 30 min, followed by HPG medium plus 50μM HPG for 45 min. Following incubations, cells were collected by trypsinization, fixed, and permeabilized, and nascent RNA or proteins were labelled using the Click-iT Alexa Fluor 488 RNA or Protein Synthesis Assay Kit (Thermo Fisher Scientific). All samples were analyzed on a LSR II Flow Cytometer (BD Biosciences). Relative EU or HPG incorporation was quantified by comparing the EU/HPG median fluorescence intensity between LINE1 and RC nucleofected samples, and carried out 24–48h after nucleofection.
Nascent RNA capture followed by qRT-PCR
EU incorporation was performed as above, with the exception that incubations were with 0.4mM EU for 30 min. Where indicated, CX-5461 was added in the morning prior to EU addition, for a total of 8h treatment. ES cells were washed, collected by trypsinization, counted, and 2×105 were used to extract RNA. Nascent RNAs was captured according to standard protocols within the Click-iT Nascent RNA Capture Kit (Invitrogen), and used in qRT-PCR assays with the indicated primer sets (see Table S5 for sequences).
Chromatin Isolation by RNA Precipitation
ChIRP was performed according to (Yin et al., 2015) with some modifications. 59nt DNA probes were biotinylated through terminal transferase (NEB) with Bio-N6-ddATP (ENZO) as substrate. E14 ES cells were harvested by trypsin digestion and crosslinked with 2mM dithiobis(succinimidyl propionate), (DSP, Thermo Scientific) in PBS at room temperature for 30 min with gentle end to end rotation. Formaldehyde was added to a final concentration of 3.7% to crosslink for 10 min further, then quenched with 250mM Glycine at room temperature for 5 min. ES cells were centrifuged and the pellet was washed with ice-cold PBS for 3 times, then snap frozen in liquid nitrogen and stored at −80°C. Crosslinked cells (~1×107) were resuspended with 500μl DNase I digestion solution (20mM Tris-HCl, pH7.5, 5mM MgCl2, 0.5mM CaCl2, 0.5% Triton X-100) with 1/20 volume of vanadyl ribonucleoside complex (VRC, NEB), 2.5μl protease inhibitors and 2.5μl 200mM PMSF. Dnase I was added to a final concentration of 12 U/ml; the reaction was rotated for 10 min at 37°C and stopped with 20mM EDTA. Chromatin was pelleted, washed once with nuclear lysis buffer (NLB, 50mM Tris-HCl, pH7.5, 10mM EDTA, 1% SDS, inhibitors) and sonicated in NLB (5 cycles of: 25% amplitude, 6 sec on, 15 sec off, Vibra-Cell™ Ultrasonic Liquid Processors). Insoluble material was removed by centrifugation and the supernatant used for ChIRP experiments. For the Rnase treatment control, samples were treated with 10μg/ml Rnase A/T1 at 37°C for 20 min. For hybridization, samples were incubated with 20pmol probes per 200μl lysate, supplemented with ¼ volume of 5x hybridization buffer (50mM Tris-HCl, pH7.5, 10mM EDTA, 1.5M NaCl, 50% formamide). The hybridization was conducted at 39°C rotating for 3h. 50μl pre-balanced streptavidin M280 beads were then added and the incubation continued for an additional 3h. The beads were washed 5 times total with 0.2× SSC wash buffer (0.1×SSC, 1% SDS) at 42°C. For the Rnase treatment control, after 3 times of washing, the beads were treated once with 10μg/ml Rnase A/T1 at 37°C in Rnase digestion buffer (50mM Tris-HCl, pH7.5, 75mM NaCl, 1mM DTT), before washing two more times. To elute, the beads were washed once with SDS elution buffer (50mM Tris-HCl, 5mM MgCl2, 75mM NaCl, 1% SDS) at 39°C for 20 min, and once with elution buffer (50mM Tris-HCl, 5mM MgCl2, 75mM NaCl, 0.1% Triton X-100) at 39°C for 5 min. DNA was eluted from the beads by RNAse H treatment in two sequential incubations with Rnase H (NEB) at 37°C for 20 min, and with SDS elution buffer at room temperature, 2 min, combining all eluents. Crosslinking was reversed by treatment with 0.1μg/μl protease K, 150mM NaCl, and 10mM EDTA at 65°C overnight and the DNA was purified using the MinElute PCR Purification Kit (QIAGEN). ChIRP enrichments were analyzed by qPCR of the purified DNA. For RNA-ChIRP analysis, beads were boiled in NLB after the 0.1x SSC washes, then further reverse-crosslinked by boiling at 95°C for 30 min in the presence of 1mM DTT. Reverse crosslinked RNA was purified using Trizol and processed for qRT-PCR analysis as for other RIP RNA samples.
DNA FISH
Co-staining of Dux DNA-FISH and immunofluorescence followed a protocol modified from what was previously described (Guan et al., 2017). The sequences of 22 ssDNA oligos to tile the 5kb genomic region of one Dux repeat unit were designed via OligoArray2.1 (Rouillard et al., 2003), synthesized by IDT with 5′ amino modifications, pooled together in equimolar ratio, and covalently labeled with Cy3. For immunofluorescence combined with Dux DNA-FISH experiments, 2C-GFP ES cells were seeded onto Lab-Tek II Chambered Coverglass coated with Matrigel for 1h, then fixed 5 min with 4% paraformaldehyde followed by an ice-cold MeOH wash for 5 min on ice. Samples were then incubated with 80% (v/v) formamide at 80°C for 10 min followed by a PBS wash for 1 min. Cells were next incubated in IF blocking buffer (2.5% BSA, 0.4% Triton in PBS) at room temperature for 30 min. The hybridization of oligo probes to the Dux repeat region was achieved in 5 min at room temperature in a solution containing 50% (v/v) formamide, 2x SSC, and 1μM oligo probes followed by a wash of 40% (v/v) formamide at room temperature for 3 min. Samples were next incubated with primary antibodies against Nucleolin and GFP in IF blocking buffer for 30 min at room temperature followed by incubation with secondary antibodies (Alexa Fluor, 1:3000) for 30 min at room temperature, and nuclei counterstained with DAPI. Fluorescence images were taken on an inverted microscope with 100x (N.A.=1.4) oil objective using X-Cite LED illumination in DAPI, GFP, RFP, and Cy5 channels, utilizing an Andor Zyla sCMOS camera for detection (Nikon Imaging Center at UCSF). The image stacks were acquired with a spinning disk confocal module with 0.25μm in z-step. For scoring of Dux localization, all images were analyzed in Fiji, identifying individual z-stacks containing one locus, then comparing across Nucleolin or DAPI channels to identify overlap or co-localization with nucleoli, edges of nuclei (lamina), or nucleoplasm. Positive GFP signal was used to identify 2C-like cells and score localization for each Dux locus in each ES vs 2C-like cell. Data were collected and quantified from two separate FISH experiments across multiple individual chambers.
Chromatin Immunoprecipitation (ChIP)
Low-cell histone ChIP was performed starting from 5×105 Lissamine-positive ES cells purified by FACS 48h after ASO nucleofection. Cells were fixed in suspension for 8 min in 1% formaldehyde, washed and the pellets snap frozen. Chromatin was prepared utilizing reagents and the protocol from Low Cell Number ChIP Kit (Diagenode). Cell pellets were thawed, lysed and sonicated using a Covaris S220 focused-ultrasonicator for 8 cycles with the following conditions: 60 sec per cycle, duty cycle 2%, 200 cycles per burst, intensity 3. IPs and washes were performed according to kit protocols, utilizing 1×105 cells per IP. For transcription factor ChIP, chromatin fixation and preparation of nuclear extracts were performed as described previously (Stock et al., 2007). ES cells were fixed and washed as above, then incubated in swelling buffer (25mM HEPES pH 7.9, 1.5mM MgCl2, 10mM KCl, 0.1% NP-40) for 30 min on ice with frequent vortexing and passed 5 times through a 25G needle to shear cell membranes. Nuclei were pelleted by centrifugation then resuspended in sonication buffer (50mM HEPES pH 7.9, 140mM NaCl, 1mM EDTA, 1% Triton X-100, 0.1% Sodium deoxycholate, 0.1% SDS) and incubated on ice a further 30 min before proceeding to sonication. Swelling and sonication buffers were supplemented with the following inhibitors fresh each time: 1x Halt Protease Inhibitor Cocktail (Thermo Fisher Scientific), 5mM sodium fluoride, 5mM sodium butyrate, 1mM PMSF, along with 10mM N-ethylmaleimide (NEM) (Thompson et al., 2015). Chromatin samples were sonicated using a Covaris S220 focused-ultrasonicator for 14 cycles with the following conditions: 60 sec per cycle, duty cycle 5%, 200 cycles per burst, intensity 4. Resultant DNA fragments were of an average length of 200–500bp. Immunoprecipitations were carried out overnight at 4°C with 20 μg chromatin, 3μg Kap1/control antibody or 0.4μg Nucleolin antibody, and 30μL Protein A/G Dynabeads per IP. ChIP washes were performed the following day as follows: twice in low-salt buffer (20mM Tris-HCl pH 8, 0.1% SDS, 1% Trixon X-100, 2mM EDTA, 150mM NaCl), once in high-salt buffer (as above but with 500mM NaCl), once in LiCl wash buffer (10mM Tris-HCl pH 8, 250mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1mM EDTA), once in TE buffer, all for 7 min at 4°C for each wash. Finally, Dynabeads containing immune complexes along with input samples were resuspended in elution buffer (50mM sodium bicarbonate, 50mM Tris-Hcl pH 8, 1% SDS, 1mM EDTA) containing 100ug/mL RNAse A (Thermo Fisher Scientific) and incubated for 30 min at 37°C. Crosslinks were next reversed by additi on of 0.5mg/mL Proteinase K (Ambion) and incubation at 55°C shaking for at least 2h. DNA was purified with Q IAquick PCR purification columns (Qiagen) and ChIP enrichments analyzed by qPCR. ChIP enrichments were calculated as % input and normalized to enrichment at the negative control region, intergenic chr11, int-chr11 for each experiment. Primer sequences are listed in Table S5.
Western blotting
Whole cell extracts were prepared from ES cells in ice-cold RIPA buffer containing protease-inhibitors (as for ChIP but minus NEM). Proteins were separated on 4–15% Mini-Protean TGX SDS Page gels (BioRad) and transferred to PVDF membranes. Blocking was performed for 45 min in 5% milk/PBS-T buffer followed by incubation overnight with primary antibodies at 4°C. The following day, membranes were incubated with the appropriate anti-mouse/rabbit secondary antibodies conjugated to HRP (Jackson) for 1h, and proteins were detected by ECL or ECL Plus reagent and autoradiography.
Co-immunoprecipitation (Co-IP)
Co-IP assays were performed on nuclear extracts. Cell pellets were resuspended in hypotonic buffer (10mM HEPES pH 7.9, 5mM MgCl2, 0.25M Sucrose, 0.1% NP40, protease inhibitors). Cell suspensions were passed through an 18G needle, incubated for 10 min on ice and centrifuged. The resulting pellet was resuspended in nuclear extraction buffer (10mM HEPES pH7.9, 1mM MgCl2, 0.1mM EDTA, 0.5M NaCl, 0.5% Triton X-100, 25% glycerol, protease inhibitors). The nuclear suspension was passed through an 18 G needle, incubated on ice for 30 min, sonicated in a Bioruptor (Diagenode) for 5 min at 30 sec on, 30 sec off and centrifuged at 4000 rpm for 10 min in a benchtop centrifuge at 4°C. Supernatants conta ining nuclear extracts were quantified using a Pierce BCA protein assay kit (Thermo Fisher Scientific), and 100μg nuclear extracts were immunoprecipitated using the indicated primary antibodies, rotating overnight at 4°C. The following day, immune complexes were bound to Protein A/G Dynabeads (Thermo Fisher Scientific), washed 3 times in nuclear extraction buffer at 4°C 1 0 min, boiled in LDS Sample Buffer (Thermo Fisher Scientific) and used for western blotting analysis.
QUANTIFICATION AND STATISTICAL ANALYSIS
RNA-seq analysis
RNA-seq reads were quality-checked, trimmed, and aligned to reference genome mm9 using Tophat2 (Kim et al., 2013) and default settings, apart from for TE analysis where reads were aligned using Tophat2 setting g -1 to map each TE to one random location. Reads were counted for each gene or TE family using the Subread package, FeatureCounts (Liao et al., 2013), and data normalized utilizing DEseq2 in R/Bioconductor (Love et al., 2014). All other RNA-seq analyses and statistics were performed in R/Bioconductor utilizing custom R scripts. For transcriptional analysis of Dux targets, these were identified as the set of genes detected in our dataset that are most highly induced by Dux over-expression in ES cells (Hendrickson et al., 2017), which we verified to be directly bound by Dux in over 70% of cases (Hendrickson et al., 2017).
Statistical analysis
All statistical analyses were performed with Graphpad Prism 7.0 software, except genome-wide data analyses which were performed in R/Bioconductor. Details of individual tests are outlined within each figure legend, including number and type of replication performed (n) and the reported error either as standard deviation (s.d) or standard error of the mean (s.e.m). All statistics are * P <0.05, ** P <0.01, *** P < 0.001, **** P <0.0001 and calculated by two-tailed Student’s t-test unless specifically noted otherwise in the legend. Welch’s correction was applied to t-tests whenever the variance was unequal between conditions.
DATA AND SOFTWARE AVAILABILITY
The accession number for the sequencing data reported in this paper is GEO: GSE100939.
Supplementary Material
Toptable analysis of differentially expressed genes and TEs upon LINE1 KD with ASOs
(A) Representative immunofluorescence staining showing cytoplasmic LINE1 Orf1p in ES cells. Scale bar, 10μm.
(B) Representative FACS plots showing percentage Lissamine-positive ES cells 48h after nucleofection with Lissaminated RC or LINE1 ASOs
(C) Representative images and quantification of LINE1 RNA FISH in ES cells 48h after transfection with either Control or two independent siRNAs against LINE1. Data are mean +/− s.d for the indicated number (n) of cells. Scale bar, 10μm.
(D) Colony formation assays in ES cells following transfection with control (RC) or LINE1 siRNAs. Data are mean +/− s.e.m, n =3 biological replicates.
(E) Growth curves of ES cells following nucleofection with RC or LINE1 ASOs, shown as mean +/− s.e.m, n=3 biological replicates.
(F) Cell-cycle analysis 48h after RC/LINE1 ASO nucleofection, with representative FACS plots and quantification of n=3 biological replicates. **P < 0.01, Chi-squared test.
(G) Cell viability analysis in Lissamine-positive ES cells, measured by Sytox-blue staining. Dead cells (permeabilized with 4% paraformaldehyde for 30 min) were used as a positive control for Sytox-blue staining. Shown are representative FACS plots plus quantification from n=3 independent experiments, +/− s.e.m.
(H) Western blot (left) and immunofluorescence (right) analysis showing no overall change to Oct4/Nanog protein levels in ES cells 48h after LINE1 KD. Scale, 10μm.
(A) LINE1 KD ES cells show a distinct gene expression profile, as evidenced by unsupervised hierarchical clustering of samples using Pearson Correlation coefficients.
(B) Boxplot showing log2FC in LINE1- vs RC ASO-nucleofected samples of the indicated number (n) of genes. Note that categories are non-inclusive, i.e., L1 <100kb refers to genes <100kb but >10kb away from a full-length LINE1 element. P values are calculated by two-sided Wilcoxon rank sum test.
(C) Expression analysis of pluripotency genes and markers from all 3 germ layers, taken from RNA-seq data following LINE1 KD. Data are mean +/− s.e.m, n=3 biological replicates in each condition. Endo, endoderm, Meso, mesoderm, Ecto, ectoderm, plurip, pluripotency.
(D) qRT-PCR validation of germ layer marker expression following ASO treatment. Data are mean +/− s.e.m, n=4 biological replicates
(E) Venn diagrams indicating the overlap between top upregulated genes with LINE1 KD (log2 fold-change > 1.0) and the indicated datasets. Datasets are as follows, ‘Up in 2C-like cells’: significantly upregulated genes in 2C-tdTomato+ cells (log2 fold-change > 1.0), ‘Up with CAF-1 depletion’: overlap of top 200 genes upregulated upon knockdown of both subunits of CAF-1 [(p60, p150), (Ishiuchi et al., 2015)], ‘Up in Zscan4+/MERVL+ cells’: genes upregulated in Zscan4/MERVL-double-positive cells [(log2 fold-change > 1.0), (Eckersley-Maslin et al., 2016)]. P-values were calculated by Fisher’s exact test, assuming a universe of 18,000 expressed genes.
(F) 2C gene expression 48h after transfection of ES cells with Control or LINE1 siRNAs. Data are mean +/− s.e.m, n=3 biological replicates.
(G) LINE1 retrotransposition assay, where retrotransposition mediated by a codon-optimized LINE1 transgene (ORFeus) (An et al., 2006, Newkirk et al., 2017) results in GFP expression. Treatment with LINE1 reverse transcriptase inhibitors, Stavudine (d4T, 20μM) or Tenofovir disoproxil fumarate (TDF, 4μM) inhibits LINE1 retrotransposition. Data are number of GFP-positive cells per 300,000 live cells, representative of 2 independent experiments.
(H) Colony formation assay of ES cells treated with reverse transcriptase inhibitors, indicating that inhibition of LINE1 retrotransposition does not affect self-renewal. Data are mean +/− s.e.m, n=4 biological replicates. (I) 2C gene expression after 48h treatment with reverse transcriptase inhibitors, data are mean +/− s.d, n=2 biological replicates
(J) Expression of 2C genes or TEs 48h after LINE1 KD in MEF cells. Data are mean +/− s.e.m, n=3 biological replicates.
(K) Validation of clonal 2C-GFP reporter ES cells, by qRT-PCR analysis performed for Dux and 2C genes in sorted 2C-GFP positive vs negative cells. Data are mean +/− s.d n=2 biological replicates. n.s., not significant, n.d., not detected.
(A) qRT-PCR analysis showing Dux and 2C upregulation in ES cells cultured in N2B27/2i/LIF conditions upon LINE1 KD. Data are mean +/− s.e.m, n=3 technical replicates, representative of 3 experiments.
(B) Colony formation assay performed in nucleofected ES cells cultured in N2B27/2i/LIF conditions. Data are mean +/− s.e.m, n=3 biological replicates.
(C) Expression analysis of previously reported 2C/MERVL repressors following LINE1 KD, taken from RNA-seq data. Data are mean +/− s.e.m, n=3 biological replicates in each condition.
(D–F) ChIP assays for the histone marks (D) H3K9me2, (E) H3K9me3, or (F) H3K27me3 performed 48h after nucleofection with ASOs and sorting for Lissamine+ cells. Data are mean +/− s.e.m n=2 technical replicates, representative of at least 2 independent experiments.
(G) Principal Component Analysis (PCA) plot for all genes across all samples, confirming ASO/siRNA RNA-seq samples have distinct gene expression profiles.
(H) Boxplot analysis of Dux-target genes following LINE1 KD and co-transfection of either siControl or siDux (n=100 genes expressed in our data, from the original list of 200 most-upregulated genes). P-value is determined by two-sided Wilcoxon rank sum test.
(I) Graph of MERVL repeat expression in RNA-seq samples, showing that simultaneous Dux knockdown rescues MERVL upregulation upon LINE1 KD.
Toptable analysis of differentially expressed genes and TEs upon LINE1 KD with ASOs, with or without KD of Dux.
Toptable analysis of differentially expressed genes and TEs upon Nucleolin KD.
Toptable analysis of differentially expressed genes in early-2C or 4C embryos upon LINE1 KD with ASOs.
Oligo and probe sequences used in this study.
(A) GSEA analysis of RNA-seq data in LINE1 vs RC nucleofected ES cells, showing a preferential upregulation of p53 targets upon LINE1 KD.
(B) qRT-PCR analysis of 2C/MERVL gene expression in p53−/− ES cells analyzed 24–48h after nucleofection with RC/LINE1 A18SOs. Data are mean +/− s.e.m, n=3 biological replicates, normalized to RC ASO 24h. #P <0.01, LINE1 ASO 24h vs RC ASO 48h.
(C) Colony formation assay in p53−/− ES cells, showing that LINE1 KD still inhibits self-renewal. Data are mean +/− s.e.m, n=3 biological replicates.
(D) GSEA analysis as in (A), demonstrating a preferential downregulation of ribosomal genes upon LINE1 KD.
(E) Heatmap analysis of n=86 ribosomal genes in RC or LINE1 ASO RNA-seq samples, with samples grouped by unsupervised hierarchical clustering.
(F) Boxplot depicting downregulation of Rpl/Rps genes compared to all expressed genes upon LINE1 KD. P-value calculated by two-sided Wilcoxon rank sum test.
(G) Nascent RNA expression of the indicated ribosomal RNA and protein-coding genes following 8h treatment with CX-5461. Data are mean +/− s.e.m, n=2 biological replicates.
(H) Colony formation assay performed in ES cells plated after 8 h treatment with CX-5461. Data are mean +/− s.e.m, n=3 biological replicates.
(I) Growth curves of ES cells with or without 8h CX-5461 treatment. Data are mean +/− s.e.m, n=3 biological replicates.
(A) Analysis of the amounts of the indicated RNAs pulled down by LINE1 or Malat1 ChIRP probes, expressed as RNA-ChIRP % input. Data are mean +/− s.e.m, n=3 independent experiments.
(B) ChIRP-qPCR analysis for ChIRP positive control, Malat1, confirming that Malat1 RNA, but not LINE1 RNA, is highly associated with Malat1 and Neat1 genomic loci. Data are mean +/− s.e.m, n=3 independent experiments.
(C) ChIRP-qPCR analysis with control or LINE1 primers, indicating that LINE1 RNA is found associated with LINE1 DNA. Data are mean +/− s.e.m, n=3 independent experiments.
(D) RIP assay performed with or without DNase treatment before immunoprecipitations, indicating that the Nucleolin (Ncl)-LINE1 RNA association is DNase-independent. Data are representative of 2 independent experiments and shown as mean +/− s.e.m, n=3 technical replicates.
(E) Nuclear enrichment of the indicated RNAs taken form RIP input expression data. RNA expression is shown relative to cytoplasmic RNA, Gapdh. Data are mean +/− s.e.m, n=3 independent experiments.
(F) qRT-PCR showing knockdown of candidate 2C/Dux repressors alongside Dux and 2C gene expression. Data are representative of two independent experiments and are mean +/− s.e.m., n=3 technical replicates. (G–H) Confirmation of Ncl knockdown, (G) at the RNA level, by qRT-PCR, and (H) at the protein level, by Western blotting. Images in (H) were spliced together to remove unrelated lanes.
(I) Expression of MERVL repeats in RNA-seq data upon knockdown of Ncl, showing mean +/− s.e.m, n=3 biological replicates.
(J) GSEA analysis of RNA-seq data upon Ncl knockdown, showing a preferential downregulation of ribosomal genes.
(K) Colony formation assays performed in ES cells plated 24h after transfection with the indicated siRNAs. Data are mean +/− s.e.m, n=3 biological replicates.
(L) qRT-PCR analysis performed in ES cells 48h after transfection with the indicated siRNAs, showing that the induction of 2C genes upon Ncl KD depends on Dux expression. Data are mean +/− s.e.m, n=3 technical replicates, representative of three experiments.
(A) Scatter graph comparing gene expression changes upon LINE1 KD (this study) with changes upon Kap1 deletion (Ecco et al., 2016), with the indicated Spearman’s correlation coefficient. Dux targets are shown in red.
(B) Nascent RNA expression for ribosomal protein coding genes or rRNA upon KD of Ncl, LINE1 or Kap1. Data are mean +/− s.e.m, n=3 biological replicates.
(C–D) qRT-PCR confirmation that KD of LINE1 by (C) ASOs or (D) siRNAs does not alter Ncl RNA expression. Data are mean +/− s.e.m, n=3 biological replicates, n.s., not significant.
(E) Western blot analysis of protein extracts from ES cells treated with RC or LINE1 ASOs, confirming that Kap1 and Ncl protein levels are unaffected.
(F) Co-IP assays performed in ES nuclear extracts 48h after nucleofection with RC/LINE1 ASOs, demonstrating that Ncl and Kap1 still interact in the absence of LINE1 RNA.
(G) Immunofluorescence analysis of Ncl and Kap1 proteins in 2C-GFP cells, showing no change between ES and 2C-like states. Green arrows in (G–H) denote 2C-GFP positive cells. Scale, 10μm.
(H) Representative immunofluorescence images and quantification of the number of nucleoli per cell in ES vs 2C-like cells. 2C-like cells were identified by positive 2C-GFP signal and absence of Oct4. ****P<0.0001, Chi-squared test. Scale, 10μm, n=number of cells.
(I) Representative co-immunofluorescence/DNA-FISH images and example scoring for each Dux localization pattern. Each locus was scored as associated with the lamina (L), nucleolus (N), or nucleoplasm (P). Nucleoli were stained with anti-Ncl, and 2C-GFP cells with anti-GFP antibodies. Scale, 2μm.
(J) Example images of Dux localization in unsorted 2C-GFP cells, showing identification of 2C-like cells by GFP-staining and nucleoli by anti-Ncl. Dux loci are indicated by white arrows, and a 2C-like cell by a green arrow. Scale, 10μm.
(K) Quantification of the proportion of ES or 2C-like cells containing at least one nucleoplasmic Dux locus vs no nucleoplasmic loci. Statistics are calculated by Chi-squared test for the indicated number (n) of cells.
(A) Developmental progression to blastocyst of the indicated Uninjected, Control ASO or LINE1 ASO injected embryos. Data shown are the percent of all embryos from 3 independent experiments at the indicated developmental stage, n=number of embryos.
(B) Analysis of chromatin accessibility in late 2C embryos following zygote ASO microinjections by DNase-TUNEL experiments. Intensities are calculated per nucleus and data shown as mean +/− s.d for the number (n) of nuclei, in data combined from 2 independent experiments for each assay. Nuclei are counterstained by Hoechst. Scale, 20μm.
(C) Immunofluorescence for global repressive chromatin marks H3K9me2/3, quantified as in (C). Scale, 20μm.
(D) Rates of developmental progression to blastocyst in embryos cultured from the 2C stage with or without CX-5461. P-values in (A–B and I) are calculated by Chi-squared test, n=number of embryos.
(E) PCA plot of all genes across all samples, showing that samples are separated firstly by developmental stage, and secondly by ASO microinjection, according to global gene expression profiles.
(F) FPKM-normalized RNA-seq data from (Wu et al., 2016) demonstrating the 6 separate gene clusters identified by K-means clustering. Cluster 4 contains early-2C genes such as Dux itself.
(G) RNA-seq normalized, log2-transformed expression for late 2C genes (which peak in expression in late-2C/4C stage), Ddit4l and Dub1a, in the indicated embryo samples. N=3 independent batches of embryos were sequenced per condition. *FDR <0.05, ****FDR<0.0001, Toptable analysis.
(H) GSEA analysis showing that ribosomal genes are preferentially downregulated in 4C embryos after LINE1 KD.
(I) Developmental progression rates to blastocyst in embryos following late-2C microinjection with ASOs, in embryos collected from 2 independent experiments, n=number of embryos.
(J) qRT-PCR expression analysis of the indicated 2C genes in 4C embryos after late-2C ASO microinjections. Data are mean+/− s.e.m, n=3 technical replicates, for the indicated independent batches of embryos.
(K) Expression data from (Wu et al., 2016) illustrating the expression levels of Kap1/Ncl during pre-implantation development. Data are FPKM, normalized to expression level in the zygote.
Highlights.
LINE1 RNA is abundant and nuclear in mouse ES cells and preimplantation embryos
LINE1 knockdown inhibits ES cell self-renewal and induces transition to a 2C state
LINE1 RNA recruits Nucleolin/Kap1 to repress Dux and activate rRNA synthesis
In embryos, LINE1 inhibition causes persistence of the 2C program and impaired ZGA
Acknowledgments
We thank J. Boeke for the LINE1 CAG-ORFeus GF-P plasmid, W. An for the CMV-5′UTR-ORFeus GF-P plasmid and D. He for technical assistance with RIP-qPCR. We are grateful to R. Blelloch, M. Conti, D. Lim and members of the Santos lab for input or critical reading of the manuscript. We thank D. O’Carroll for the LINE1 ORF1p antibody, and D. Trono for the CreERT2;Kap1fl/fl ES cells. C-JL thanks R. Smith for managing the mouse colony and technical support, B. Nashun for the Dnase I-TUNEL assay protocol, and D. Soong for setting scanning conditions for confocal microscopy. C-JL is a Royal Society of Edinburgh Personal Research Fellow funded by the Scottish Government. Animal work partially undertaken in the MRC Centre for Reproductive Health, University of Edinburgh is funded by the MRC Centre grant MR/N022556/1 to C-JL. XS and YY were supported by grants 31471219 and 31630095 from the National Natural Science Foundation of China. BH is a Chan Zuckerberg Biohub Investigator. This work was supported by CIRM postdoctoral fellowship TG2-01153 to MP, a CIRM Bridges Fellowship to GP, a W.M. Keck Foundation Medical Research Grant to BH, and NIH grants R01GM113014 and R01GM123556 and a pilot grant from the UCSF Resource Allocation Program to MR-S.
Footnotes
Author Contributions
MP and MR-S conceived of the project. MP designed and performed all the experiments in ES cells with the below exceptions, and performed all bioinformatic analyses. C-JL performed all embryo experiments. YY performed ChIRP experiments under the supervision of XS. JG performed Dux DNA-FISH localization experiments, supervised by BH and analyzed by MP and JG. AB-K performed Co-IP assays and western blots in LINE1-depleted extracts. GP performed some of the ASO and siRNA knockdown experiments, supervised by MP. SB provided technical assistance with ES cell manipulations. MP and MR-S wrote the manuscript with input from all authors.
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- An W, Han JS, Wheelan SJ, Davis ES, Coombes CE, Ye P, Triplett C, Boeke JD. Active retrotransposition by a synthetic L1 element in mice. Proc Natl Acad Sci U S A. 2006;103:18662–7. doi: 10.1073/pnas.0605300103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beraldi R, Pittoggi C, Sciamanna I, Mattei E, Spadafora C. Expression of LINE-1 retroposons is essential for murine preimplantation development. Mol Reprod Dev. 2006;73:279–87. doi: 10.1002/mrd.20423. [DOI] [PubMed] [Google Scholar]
- Borsos M, Torres-Padilla ME. Building up the nucleus: nuclear organization in the establishment of totipotency and pluripotency during mammalian development. Genes Dev. 2016;30:611–21. doi: 10.1101/gad.273805.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourque G. Transposable elements in gene regulation and in the evolution of vertebrate genomes. Curr Opin Genet Dev. 2009;19:607–12. doi: 10.1016/j.gde.2009.10.013. [DOI] [PubMed] [Google Scholar]
- Bulut-Karslioglu A, Macrae TA, Oses-Prieto JA, Covarrubias S, Percharde M, Ku G, Diaz A, Mcmanus MT, Burlingame AL, Ramalho-Santos M. The Transcriptionally Permissive Chromatin State of Embryonic Stem Cells Is Acutely Tuned to Translational Output. Cell Stem Cell. 2018;22:369–383. e8. doi: 10.1016/j.stem.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunch H, Zheng X, Burkholder A, Dillon ST, Motola S, Birrane G, Ebmeier CC, Levine S, Fargo D, Hu G, et al. TRIM28 regulates RNA polymerase II promoter-proximal pausing and pause release. Nat Struct Mol Biol. 2014;21:876–83. doi: 10.1038/nsmb.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KH. Transposable elements in cancer. Nat Rev Cancer. 2017;17:415–424. doi: 10.1038/nrc.2017.35. [DOI] [PubMed] [Google Scholar]
- Cantrell MA, Scott L, Brown CJ, Martinez AR, Wichman HA. Loss of LINE-1 activity in the megabats. Genetics. 2008;178:393–404. doi: 10.1534/genetics.107.080275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Qu K, Zhong FL, Artandi SE, Chang HY. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–78. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis W, Jr, De Sousa PA, Schultz RM. Transient expression of translation initiation factor eIF-4C during the 2-cell stage of the preimplantation mouse embryo: identification by mRNA differential display and the role of DNA replication in zygotic gene activation. Dev Biol. 1996;174:190–201. doi: 10.1006/dbio.1996.0065. [DOI] [PubMed] [Google Scholar]
- De Iaco A, Planet E, Coluccio A, Verp S, Duc J, Trono D. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet. 2017;49:941–945. doi: 10.1038/ng.3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giacomo M, Comazzetto S, Sampath SC, Sampath SC, O’carroll D. G9a co-suppresses LINE1 elements in spermatogonia. Epigenetics Chromatin. 2014;7:24. doi: 10.1186/1756-8935-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecco G, Cassano M, Kauzlaric A, Duc J, Coluccio A, Offner S, Imbeault M, Rowe HM, Turelli P, Trono D. Transposable Elements and Their KRAB-ZFP Controllers Regulate Gene Expression in Adult Tissues. Dev Cell. 2016;36:611–23. doi: 10.1016/j.devcel.2016.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckersley-Maslin MA, Svensson V, Krueger C, Stubbs TM, Giehr P, Krueger F, Miragaia RJ, Kyriakopoulos C, Berrens RV, Milagre I, et al. MERVL/Zscan4 Network Activation Results in Transient Genome-wide DNA Demethylation of mESCs. Cell Rep. 2016;17:179–92. doi: 10.1016/j.celrep.2016.08.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Sirokman K, Mcdonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, Lander ES. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014;159:188–99. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadloun A, Le Gras S, Jost B, Ziegler-Birling C, Takahashi H, Gorab E, Carninci P, Torres-Padilla ME. Chromatin signatures and retrotransposon profiling in mouse embryos reveal regulation of LINE-1 by RNA. Nat Struct Mol Biol. 2013;20:332–8. doi: 10.1038/nsmb.2495. [DOI] [PubMed] [Google Scholar]
- Gabellini D, Green MR, Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell. 2002;110:339–48. doi: 10.1016/s0092-8674(02)00826-7. [DOI] [PubMed] [Google Scholar]
- Ginisty H, Amalric F, Bouvet P. Nucleolin functions in the first step of ribosomal RNA processing. EMBO J. 1998;17:1476–86. doi: 10.1093/emboj/17.5.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez V, Guo K, Hurley L, Sun D. Identification and characterization of nucleolin as a c-myc G-quadruplex-binding protein. J Biol Chem. 2009;284:23622–35. doi: 10.1074/jbc.M109.018028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J, Liu H, Shi X, Feng S, Huang B. Tracking Multiple Genomic Elements Using Correlative CRISPR Imaging and Sequential DNA FISH. Biophys J. 2017;112:1077–1084. doi: 10.1016/j.bpj.2017.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guetg C, Santoro R. Formation of nuclear heterochromatin: the nucleolar point of view. Epigenetics. 2012;7:811–4. doi: 10.4161/epi.21072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman-Ayala M, Sachs M, Koh FM, Onodera C, Bulut-Karslioglu A, Lin CJ, Wong P, Nitta R, Song JS, Ramalho-Santos M. Chd1 is essential for the high transcriptional output and rapid growth of the mouse epiblast. Development. 2015;142:118–27. doi: 10.1242/dev.114843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddach M, Schwaebe MK, Michaux J, Nagasawa J, O’brien SE, Whitten JP, Pierre F, Kerdoncuff P, Darjania L, Stansfield R, et al. Discovery of CX-5461, the First Direct and Selective Inhibitor of RNA Polymerase I, for Cancer Therapeutics. ACS Med Chem Lett. 2012;3:602–6. doi: 10.1021/ml300110s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Carone DM, Gomez AV, Kolpa HJ, Byron M, Mehta N, Fackelmayer FO, Lawrence JB. Stable C0T-1 repeat RNA is abundant and is associated with euchromatic interphase chromosomes. Cell. 2014;156:907–19. doi: 10.1016/j.cell.2014.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson PG, Dorais JA, Grow EJ, Whiddon JL, Lim JW, Wike CL, Weaver BD, Pflueger C, Emery BR, Wilcox AL, et al. Conserved roles of mouse DUX and human DUX4 in activating cleavage-stage genes and MERVL/HERVL retrotransposons. Nat Genet. 2017;49:925–934. doi: 10.1038/ng.3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper M, Hardy K, Handyside A, Hunter S, Monk M. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. 1987;326:292–5. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- Ishiuchi T, Enriquez-Gasca R, Mizutani E, Boskovic A, Ziegler-Birling C, Rodriguez-Terrones D, Wakayama T, Vaquerizas JM, Torres-Padilla ME. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat Struct Mol Biol. 2015;22:662–71. doi: 10.1038/nsmb.3066. [DOI] [PubMed] [Google Scholar]
- Ivancevic A, Kortschak D, Bertozzi T, Adelson D. Re-evaluating inheritance in genome evolution: widespread transfer of LINEs between species. 2017 bioRxiv. [Google Scholar]
- Jachowicz JW, Bing X, Pontabry J, Boskovic A, Rando OJ, Torres-Padilla ME. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo. Nat Genet. 2017;49:1502–1510. doi: 10.1038/ng.3945. [DOI] [PubMed] [Google Scholar]
- Jia W, Yao Z, Zhao J, Guan Q, Gao L. New perspectives of physiological and pathological functions of nucleolin (NCL) Life Sci. 2017 doi: 10.1016/j.lfs.2017.07.025. [DOI] [PubMed] [Google Scholar]
- Jones RB, Garrison KE, Wong JC, Duan EH, Nixon DF, Ostrowski MA. Nucleoside analogue reverse transcriptase inhibitors differentially inhibit human LINE-1 retrotransposition. PLoS One. 2008;3:e1547. doi: 10.1371/journal.pone.0001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, Ostertag EM, Kazazian HH., Jr L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–12. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kigami D, Minami N, Takayama H, Imai H. MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biol Reprod. 2003;68:651–4. doi: 10.1095/biolreprod.102.007906. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laferte A, Favry E, Sentenac A, Riva M, Carles C, Chedin S. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 2006;20:2030–40. doi: 10.1101/gad.386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennox KA, Behlke MA. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016;44:863–77. doi: 10.1093/nar/gkv1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CJ, Conti M, Ramalho-Santos M. Histone variant H3.3 maintains a decondensed chromatin state essential for mouse preimplantation development. Development. 2013;140:3624–34. doi: 10.1242/dev.095513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CJ, Koh FM, Wong P, Conti M, Ramalho-Santos M. Hira-mediated H3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Dev Cell. 2014;30:268–79. doi: 10.1016/j.devcel.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo A, Cesaro E, Montano G, Zurlo D, Izzo P, Costanzo P. KRAB-Zinc Finger Proteins: A Repressor Family Displaying Multiple Biological Functions. Curr Genomics. 2013;14:268–78. doi: 10.2174/13892029113149990002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Agarwal S, Driscoll S, Lettieri K, Wang J, Andrews SE, Franco L, Rosenfeld MG, Ren B, et al. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev. 2011;25:594–607. doi: 10.1101/gad.2008511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. doi: 10.1038/nature11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magiorkinis G, Blanco-Melo D, Belshaw R. The decline of human endogenous retroviruses: extinction and survival. Retrovirology. 2015;12:8. doi: 10.1186/s12977-015-0136-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malki S, Van Der Heijden GW, O’donnell KA, Martin SL, Bortvin A. A role for retrotransposon LINE-1 in fetal oocyte attrition in mice. Dev Cell. 2014;29:521–33. doi: 10.1016/j.devcel.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan JB, Moran JV. The Zinc-Finger Antiviral Protein ZAP Inhibits LINE and Alu Retrotransposition. PLoS Genet. 2015;11:e1005121. doi: 10.1371/journal.pgen.1005121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–10. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- Naas TP, Deberardinis RJ, Moran JV, Ostertag EM, Kingsmore SF, Seldin MF, Hayashizaki Y, Martin SL, Kazazian HH. An actively retrotransposing, novel subfamily of mouse L1 elements. EMBO J. 1998;17:590–7. doi: 10.1093/emboj/17.2.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newkirk SJ, Lee S, Grandi FC, Gaysinskaya V, Rosser JM, Vanden Berg N, Hogarth CA, Marchetto MCN, Muotri AR, Griswold MD, et al. Intact piRNA pathway prevents L1 mobilization in male meiosis. Proc Natl Acad Sci U S A. 2017 doi: 10.1073/pnas.1701069114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno R, Nakayama M, Naruse C, Okashita N, Takano O, Tachibana M, Asano M, Saitou M, Seki Y. A replication-dependent passive mechanism modulates DNA demethylation in mouse primordial germ cells. Development. 2013;140:2892–903. doi: 10.1242/dev.093229. [DOI] [PubMed] [Google Scholar]
- Peddigari S, Li PW, Rabe JL, Martin SL. hnRNPL and nucleolin bind LINE-1 RNA and function as host factors to modulate retrotransposition. Nucleic Acids Res. 2013;41:575–85. doi: 10.1093/nar/gks1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzkofer T, Dandekar T, Zemojtel T. L1Base: from functional annotation to prediction of active LINE-1 elements. Nucleic Acids Res. 2005;33:D498–500. doi: 10.1093/nar/gki044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percharde M, Bulut-Karslioglu A, Ramalho-Santos M. Hypertranscription in Development, Stem Cells, and Regeneration. Dev Cell. 2017a;40:9–21. doi: 10.1016/j.devcel.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percharde M, Lavial F, Ng JH, Kumar V, Tomaz RA, Martin N, Yeo JC, Gil J, Prabhakar S, Ng HH, et al. Ncoa3 functions as an essential Esrrb coactivator to sustain embryonic stem cell self-renewal and reprogramming. Genes Dev. 2012;26:2286–98. doi: 10.1101/gad.195545.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percharde M, Wong P, Ramalho-Santos M. Global Hypertranscription in the Mouse Embryonic Germline. Cell Rep. 2017b;19:1987–1996. doi: 10.1016/j.celrep.2017.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Van Den Bogaard P, Rifkin SA, Van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–9. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature. 2008;452:243–7. doi: 10.1038/nature06727. [DOI] [PubMed] [Google Scholar]
- Richardson SR, Gerdes P, Gerhardt DJ, Sanchez-Luque FJ, Bodea GO, Munoz-Lopez M, Jesuadian JS, Kempen MHC, Carreira PE, Jeddeloh JA, et al. Heritable L1 retrotransposition in the mouse primordial germline and early embryo. Genome Res. 2017 doi: 10.1101/gr.219022.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillard JM, Zuker M, Gulari E. OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res. 2003;31:3057–62. doi: 10.1093/nar/gkg426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature. 2010;463:237–40. doi: 10.1038/nature08674. [DOI] [PubMed] [Google Scholar]
- Sabapathy K, Klemm M, Jaenisch R, Wagner EF. Regulation of ES cell differentiation by functional and conformational modulation of p53. EMBO J. 1997;16:6217–29. doi: 10.1093/emboj/16.20.6217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel S, Twizere JC, Beifuss KK, Bernstein LR. Nucleolin binds specifically to an AP-1 DNA sequence and represses AP1-dependent transactivation of the matrix metalloproteinase-13 gene. Mol Carcinog. 2008;47:34–46. doi: 10.1002/mc.20358. [DOI] [PubMed] [Google Scholar]
- Schoorlemmer J, Perez-Palacios R, Climent M, Guallar D, Muniesa P. Regulation of Mouse Retroelement MuERV-L/MERVL Expression by REX1 and Epigenetic Control of Stem Cell Potency. Front Oncol. 2014;4:14. doi: 10.3389/fonc.2014.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciamanna I, Vitullo P, Curatolo A, Spadafora C. Retrotransposons, reverse transcriptase and the genesis of new genetic information. Gene. 2009;448:180–6. doi: 10.1016/j.gene.2009.07.011. [DOI] [PubMed] [Google Scholar]
- Singh K, Cassano M, Planet E, Sebastian S, Jang SM, Sohi G, Faralli H, Choi J, Youn HD, Dilworth FJ, et al. A KAP1 phosphorylation switch controls MyoD function during skeletal muscle differentiation. Genes Dev. 2015;29:513–25. doi: 10.1101/gad.254532.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–35. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Svoboda P, Stein P, Anger M, Bernstein E, Hannon GJ, Schultz RM. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev Biol. 2004;269:276–85. doi: 10.1016/j.ydbio.2004.01.028. [DOI] [PubMed] [Google Scholar]
- Thompson PJ, Dulberg V, Moon KM, Foster LJ, Chen C, Karimi MM, Lorincz MC. hnRNP K coordinates transcriptional silencing by SETDB1 in embryonic stem cells. PLoS Genet. 2015;11:e1004933. doi: 10.1371/journal.pgen.1004933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiddon JL, Langford AT, Wong CJ, Zhong JW, Tapscott SJ. Conservation and innovation in the DUX4-family gene network. Nat Genet. 2017;49:935–940. doi: 10.1038/ng.3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf G, Greenberg D, Macfarlan TS. Spotting the enemy within: Targeted silencing of foreign DNA in mammalian genomes by the Kruppel-associated box zinc finger protein family. Mob DNA. 2015;6:17. doi: 10.1186/s13100-015-0050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Huang B, Chen H, Yin Q, Liu Y, Xiang Y, Zhang B, Liu B, Wang Q, Xia W, et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 2016;534:652–7. doi: 10.1038/nature18606. [DOI] [PubMed] [Google Scholar]
- Yin Y, Yan P, Lu J, Song G, Zhu Y, Li Z, Zhao Y, Shen B, Huang X, Zhu H, et al. Opposing Roles for the lncRNA Haunt and Its Genomic Locus in Regulating HOXA Gene Activation during Embryonic Stem Cell Differentiation. Cell Stem Cell. 2015;16:504–16. doi: 10.1016/j.stem.2015.03.007. [DOI] [PubMed] [Google Scholar]
- Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–23. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F, Schultz RM. RNA transcript profiling during zygotic gene activation in the preimplantation mouse embryo. Dev Biol. 2005;283:40–57. doi: 10.1016/j.ydbio.2005.03.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Toptable analysis of differentially expressed genes and TEs upon LINE1 KD with ASOs
(A) Representative immunofluorescence staining showing cytoplasmic LINE1 Orf1p in ES cells. Scale bar, 10μm.
(B) Representative FACS plots showing percentage Lissamine-positive ES cells 48h after nucleofection with Lissaminated RC or LINE1 ASOs
(C) Representative images and quantification of LINE1 RNA FISH in ES cells 48h after transfection with either Control or two independent siRNAs against LINE1. Data are mean +/− s.d for the indicated number (n) of cells. Scale bar, 10μm.
(D) Colony formation assays in ES cells following transfection with control (RC) or LINE1 siRNAs. Data are mean +/− s.e.m, n =3 biological replicates.
(E) Growth curves of ES cells following nucleofection with RC or LINE1 ASOs, shown as mean +/− s.e.m, n=3 biological replicates.
(F) Cell-cycle analysis 48h after RC/LINE1 ASO nucleofection, with representative FACS plots and quantification of n=3 biological replicates. **P < 0.01, Chi-squared test.
(G) Cell viability analysis in Lissamine-positive ES cells, measured by Sytox-blue staining. Dead cells (permeabilized with 4% paraformaldehyde for 30 min) were used as a positive control for Sytox-blue staining. Shown are representative FACS plots plus quantification from n=3 independent experiments, +/− s.e.m.
(H) Western blot (left) and immunofluorescence (right) analysis showing no overall change to Oct4/Nanog protein levels in ES cells 48h after LINE1 KD. Scale, 10μm.
(A) LINE1 KD ES cells show a distinct gene expression profile, as evidenced by unsupervised hierarchical clustering of samples using Pearson Correlation coefficients.
(B) Boxplot showing log2FC in LINE1- vs RC ASO-nucleofected samples of the indicated number (n) of genes. Note that categories are non-inclusive, i.e., L1 <100kb refers to genes <100kb but >10kb away from a full-length LINE1 element. P values are calculated by two-sided Wilcoxon rank sum test.
(C) Expression analysis of pluripotency genes and markers from all 3 germ layers, taken from RNA-seq data following LINE1 KD. Data are mean +/− s.e.m, n=3 biological replicates in each condition. Endo, endoderm, Meso, mesoderm, Ecto, ectoderm, plurip, pluripotency.
(D) qRT-PCR validation of germ layer marker expression following ASO treatment. Data are mean +/− s.e.m, n=4 biological replicates
(E) Venn diagrams indicating the overlap between top upregulated genes with LINE1 KD (log2 fold-change > 1.0) and the indicated datasets. Datasets are as follows, ‘Up in 2C-like cells’: significantly upregulated genes in 2C-tdTomato+ cells (log2 fold-change > 1.0), ‘Up with CAF-1 depletion’: overlap of top 200 genes upregulated upon knockdown of both subunits of CAF-1 [(p60, p150), (Ishiuchi et al., 2015)], ‘Up in Zscan4+/MERVL+ cells’: genes upregulated in Zscan4/MERVL-double-positive cells [(log2 fold-change > 1.0), (Eckersley-Maslin et al., 2016)]. P-values were calculated by Fisher’s exact test, assuming a universe of 18,000 expressed genes.
(F) 2C gene expression 48h after transfection of ES cells with Control or LINE1 siRNAs. Data are mean +/− s.e.m, n=3 biological replicates.
(G) LINE1 retrotransposition assay, where retrotransposition mediated by a codon-optimized LINE1 transgene (ORFeus) (An et al., 2006, Newkirk et al., 2017) results in GFP expression. Treatment with LINE1 reverse transcriptase inhibitors, Stavudine (d4T, 20μM) or Tenofovir disoproxil fumarate (TDF, 4μM) inhibits LINE1 retrotransposition. Data are number of GFP-positive cells per 300,000 live cells, representative of 2 independent experiments.
(H) Colony formation assay of ES cells treated with reverse transcriptase inhibitors, indicating that inhibition of LINE1 retrotransposition does not affect self-renewal. Data are mean +/− s.e.m, n=4 biological replicates. (I) 2C gene expression after 48h treatment with reverse transcriptase inhibitors, data are mean +/− s.d, n=2 biological replicates
(J) Expression of 2C genes or TEs 48h after LINE1 KD in MEF cells. Data are mean +/− s.e.m, n=3 biological replicates.
(K) Validation of clonal 2C-GFP reporter ES cells, by qRT-PCR analysis performed for Dux and 2C genes in sorted 2C-GFP positive vs negative cells. Data are mean +/− s.d n=2 biological replicates. n.s., not significant, n.d., not detected.
(A) qRT-PCR analysis showing Dux and 2C upregulation in ES cells cultured in N2B27/2i/LIF conditions upon LINE1 KD. Data are mean +/− s.e.m, n=3 technical replicates, representative of 3 experiments.
(B) Colony formation assay performed in nucleofected ES cells cultured in N2B27/2i/LIF conditions. Data are mean +/− s.e.m, n=3 biological replicates.
(C) Expression analysis of previously reported 2C/MERVL repressors following LINE1 KD, taken from RNA-seq data. Data are mean +/− s.e.m, n=3 biological replicates in each condition.
(D–F) ChIP assays for the histone marks (D) H3K9me2, (E) H3K9me3, or (F) H3K27me3 performed 48h after nucleofection with ASOs and sorting for Lissamine+ cells. Data are mean +/− s.e.m n=2 technical replicates, representative of at least 2 independent experiments.
(G) Principal Component Analysis (PCA) plot for all genes across all samples, confirming ASO/siRNA RNA-seq samples have distinct gene expression profiles.
(H) Boxplot analysis of Dux-target genes following LINE1 KD and co-transfection of either siControl or siDux (n=100 genes expressed in our data, from the original list of 200 most-upregulated genes). P-value is determined by two-sided Wilcoxon rank sum test.
(I) Graph of MERVL repeat expression in RNA-seq samples, showing that simultaneous Dux knockdown rescues MERVL upregulation upon LINE1 KD.
Toptable analysis of differentially expressed genes and TEs upon LINE1 KD with ASOs, with or without KD of Dux.
Toptable analysis of differentially expressed genes and TEs upon Nucleolin KD.
Toptable analysis of differentially expressed genes in early-2C or 4C embryos upon LINE1 KD with ASOs.
Oligo and probe sequences used in this study.
(A) GSEA analysis of RNA-seq data in LINE1 vs RC nucleofected ES cells, showing a preferential upregulation of p53 targets upon LINE1 KD.
(B) qRT-PCR analysis of 2C/MERVL gene expression in p53−/− ES cells analyzed 24–48h after nucleofection with RC/LINE1 A18SOs. Data are mean +/− s.e.m, n=3 biological replicates, normalized to RC ASO 24h. #P <0.01, LINE1 ASO 24h vs RC ASO 48h.
(C) Colony formation assay in p53−/− ES cells, showing that LINE1 KD still inhibits self-renewal. Data are mean +/− s.e.m, n=3 biological replicates.
(D) GSEA analysis as in (A), demonstrating a preferential downregulation of ribosomal genes upon LINE1 KD.
(E) Heatmap analysis of n=86 ribosomal genes in RC or LINE1 ASO RNA-seq samples, with samples grouped by unsupervised hierarchical clustering.
(F) Boxplot depicting downregulation of Rpl/Rps genes compared to all expressed genes upon LINE1 KD. P-value calculated by two-sided Wilcoxon rank sum test.
(G) Nascent RNA expression of the indicated ribosomal RNA and protein-coding genes following 8h treatment with CX-5461. Data are mean +/− s.e.m, n=2 biological replicates.
(H) Colony formation assay performed in ES cells plated after 8 h treatment with CX-5461. Data are mean +/− s.e.m, n=3 biological replicates.
(I) Growth curves of ES cells with or without 8h CX-5461 treatment. Data are mean +/− s.e.m, n=3 biological replicates.
(A) Analysis of the amounts of the indicated RNAs pulled down by LINE1 or Malat1 ChIRP probes, expressed as RNA-ChIRP % input. Data are mean +/− s.e.m, n=3 independent experiments.
(B) ChIRP-qPCR analysis for ChIRP positive control, Malat1, confirming that Malat1 RNA, but not LINE1 RNA, is highly associated with Malat1 and Neat1 genomic loci. Data are mean +/− s.e.m, n=3 independent experiments.
(C) ChIRP-qPCR analysis with control or LINE1 primers, indicating that LINE1 RNA is found associated with LINE1 DNA. Data are mean +/− s.e.m, n=3 independent experiments.
(D) RIP assay performed with or without DNase treatment before immunoprecipitations, indicating that the Nucleolin (Ncl)-LINE1 RNA association is DNase-independent. Data are representative of 2 independent experiments and shown as mean +/− s.e.m, n=3 technical replicates.
(E) Nuclear enrichment of the indicated RNAs taken form RIP input expression data. RNA expression is shown relative to cytoplasmic RNA, Gapdh. Data are mean +/− s.e.m, n=3 independent experiments.
(F) qRT-PCR showing knockdown of candidate 2C/Dux repressors alongside Dux and 2C gene expression. Data are representative of two independent experiments and are mean +/− s.e.m., n=3 technical replicates. (G–H) Confirmation of Ncl knockdown, (G) at the RNA level, by qRT-PCR, and (H) at the protein level, by Western blotting. Images in (H) were spliced together to remove unrelated lanes.
(I) Expression of MERVL repeats in RNA-seq data upon knockdown of Ncl, showing mean +/− s.e.m, n=3 biological replicates.
(J) GSEA analysis of RNA-seq data upon Ncl knockdown, showing a preferential downregulation of ribosomal genes.
(K) Colony formation assays performed in ES cells plated 24h after transfection with the indicated siRNAs. Data are mean +/− s.e.m, n=3 biological replicates.
(L) qRT-PCR analysis performed in ES cells 48h after transfection with the indicated siRNAs, showing that the induction of 2C genes upon Ncl KD depends on Dux expression. Data are mean +/− s.e.m, n=3 technical replicates, representative of three experiments.
(A) Scatter graph comparing gene expression changes upon LINE1 KD (this study) with changes upon Kap1 deletion (Ecco et al., 2016), with the indicated Spearman’s correlation coefficient. Dux targets are shown in red.
(B) Nascent RNA expression for ribosomal protein coding genes or rRNA upon KD of Ncl, LINE1 or Kap1. Data are mean +/− s.e.m, n=3 biological replicates.
(C–D) qRT-PCR confirmation that KD of LINE1 by (C) ASOs or (D) siRNAs does not alter Ncl RNA expression. Data are mean +/− s.e.m, n=3 biological replicates, n.s., not significant.
(E) Western blot analysis of protein extracts from ES cells treated with RC or LINE1 ASOs, confirming that Kap1 and Ncl protein levels are unaffected.
(F) Co-IP assays performed in ES nuclear extracts 48h after nucleofection with RC/LINE1 ASOs, demonstrating that Ncl and Kap1 still interact in the absence of LINE1 RNA.
(G) Immunofluorescence analysis of Ncl and Kap1 proteins in 2C-GFP cells, showing no change between ES and 2C-like states. Green arrows in (G–H) denote 2C-GFP positive cells. Scale, 10μm.
(H) Representative immunofluorescence images and quantification of the number of nucleoli per cell in ES vs 2C-like cells. 2C-like cells were identified by positive 2C-GFP signal and absence of Oct4. ****P<0.0001, Chi-squared test. Scale, 10μm, n=number of cells.
(I) Representative co-immunofluorescence/DNA-FISH images and example scoring for each Dux localization pattern. Each locus was scored as associated with the lamina (L), nucleolus (N), or nucleoplasm (P). Nucleoli were stained with anti-Ncl, and 2C-GFP cells with anti-GFP antibodies. Scale, 2μm.
(J) Example images of Dux localization in unsorted 2C-GFP cells, showing identification of 2C-like cells by GFP-staining and nucleoli by anti-Ncl. Dux loci are indicated by white arrows, and a 2C-like cell by a green arrow. Scale, 10μm.
(K) Quantification of the proportion of ES or 2C-like cells containing at least one nucleoplasmic Dux locus vs no nucleoplasmic loci. Statistics are calculated by Chi-squared test for the indicated number (n) of cells.
(A) Developmental progression to blastocyst of the indicated Uninjected, Control ASO or LINE1 ASO injected embryos. Data shown are the percent of all embryos from 3 independent experiments at the indicated developmental stage, n=number of embryos.
(B) Analysis of chromatin accessibility in late 2C embryos following zygote ASO microinjections by DNase-TUNEL experiments. Intensities are calculated per nucleus and data shown as mean +/− s.d for the number (n) of nuclei, in data combined from 2 independent experiments for each assay. Nuclei are counterstained by Hoechst. Scale, 20μm.
(C) Immunofluorescence for global repressive chromatin marks H3K9me2/3, quantified as in (C). Scale, 20μm.
(D) Rates of developmental progression to blastocyst in embryos cultured from the 2C stage with or without CX-5461. P-values in (A–B and I) are calculated by Chi-squared test, n=number of embryos.
(E) PCA plot of all genes across all samples, showing that samples are separated firstly by developmental stage, and secondly by ASO microinjection, according to global gene expression profiles.
(F) FPKM-normalized RNA-seq data from (Wu et al., 2016) demonstrating the 6 separate gene clusters identified by K-means clustering. Cluster 4 contains early-2C genes such as Dux itself.
(G) RNA-seq normalized, log2-transformed expression for late 2C genes (which peak in expression in late-2C/4C stage), Ddit4l and Dub1a, in the indicated embryo samples. N=3 independent batches of embryos were sequenced per condition. *FDR <0.05, ****FDR<0.0001, Toptable analysis.
(H) GSEA analysis showing that ribosomal genes are preferentially downregulated in 4C embryos after LINE1 KD.
(I) Developmental progression rates to blastocyst in embryos following late-2C microinjection with ASOs, in embryos collected from 2 independent experiments, n=number of embryos.
(J) qRT-PCR expression analysis of the indicated 2C genes in 4C embryos after late-2C ASO microinjections. Data are mean+/− s.e.m, n=3 technical replicates, for the indicated independent batches of embryos.
(K) Expression data from (Wu et al., 2016) illustrating the expression levels of Kap1/Ncl during pre-implantation development. Data are FPKM, normalized to expression level in the zygote.