Summary
Triggering of autoimmunity that leads to rheumatic disease has been suggested to depend upon gene–environment interactions occurring in epithelial barriers and associated immune cells. Genetic studies have identified associations of the FAM167A‐BLK locus with rheumatoid arthritis, systemic lupus erythematosus (SLE) and Sjögren's syndrome. While BLK (B lymphocyte kinase) has a well‐established role in B cells, family with sequence similarity to 167 member A (FAM167A) and its gene family remain uncharacterized. To begin to understand the role of FAM167A in rheumatic disease pathogenesis, we explored this gene family and cloned and investigated the gene products. Expression of quantitative trait locus analysis was performed in immune cells. FAM167A and FAM167B were cloned from human peripheral blood mononuclear cells (PBMC). Gene conservation and protein properties were analysed by online tools, mRNA expression measured in mouse organs by quantitative polymerase chain reaction (qPCR) and protein expression investigated in human tissues by immunohistochemistry. We found that autoimmune risk genotypes within the FAM167A‐BLK locus lead to increased expression of FAM167A. The FAM167 gene family includes two members, FAM167A and FAM167B, which are not homologous to any other annotated gene but are evolutionarily conserved. The encoded proteins, which we denote ‘disordered autoimmunity’ (DIORA)‐1 and DIORA‐2, respectively, are characterized by a high content of intrinsic disorder. Notably, DIORA‐1 has its highest expression in the lung, detectable in both bronchial epithelium and alveolar macrophages with an endosomal localization pattern. In summary, the FAM167A gene is associated with several rheumatic diseases and encodes a novel disordered protein, DIORA‐1, which is expressed highly in the lung, consistent with a potential role in disease pathogenesis.
Keywords: DIORA‐1, DIORA‐2, FAM167, rheumatoid arthritis, Sjögren's syndrome
Introduction
Autoimmune diseases develop in individuals with uncontrolled immune responses to self‐antigens, and 2–5% of the population in the developed world suffer from this group of disorders 1, 2, 3. Many of these are rheumatic diseases, in which the chronic inflammatory process leads to tissue destruction resulting in increased morbidity and, for most of these conditions, a shorter life expectancy. As no curative treatment yet exists, there is a critical need for a deeper understanding of the mechanisms underlying chronic autoimmune inflammation.
Although the pathogenesis of autoimmunity remains elusive, both environmental triggers and genetic factors have been linked to the onset of disease 4. Several studies indicate that environmental triggering may occur through the lung by, e.g. smoking in genetically predisposed individuals 2, 5, although the molecular mechanisms remain to be defined. The heritability in autoimmunity is well established, as family members of affected individuals have a significantly higher risk of developing an autoimmune disease 6, 7. Furthermore, many genome‐wide association studies (GWAS) have demonstrated associations of single nucleotide polymorphisms (SNPs) with autoimmune diseases 8. Among the non‐human leucocyte antigen (HLA) loci that have been associated consistently with rheumatic diseases such as rheumatoid arthritis, SLE and Sjögen's syndrome is the FAM167A‐BLK locus at chromosome 8p23.1 9, 10, 11. B lymphocyte kinase (BLK) encodes a tyrosine kinase of the Src family, is expressed primarily in B cells, acting downstream of the B cell receptor (BCR), and is important in B cell development 12. The autoimmunity‐associated BLK haplotype has been shown to result in altered mRNA and protein expression of BLK in naive B cell subsets 13. Alterations in BCR and nuclear factor kappa B (NF‐κB) signalling associated with SNPs in the BLK region have been reported, and support the hypothesis that the BLK gene is a causal factor behind the genetic disease association 14. However, somewhat surprisingly, BLK knock‐out mice show no overt immune phenotype 15.
Conversely, the function of BLK's neighbouring gene family with sequence similarity to 167, member A (FAM167A), is not known. The protein encoded by the open reading frame has not been characterized, and with the exception of studies on genetic association to inflammatory diseases, the FAM167A gene has not been described further. Interestingly, cis‐expression quantitative trait locus (eQTL) analysis has revealed an increased expression of FAM167A in cells carrying the SLE and Sjögren‐associated genotypes in the FAM167A‐BLK locus 10, 11, while the effect on BLK expression is moderate or even low 10, 13. Considering the higher expression of FAM167A from alleles associated with rheumatic disease, we cloned and characterized this unknown gene and its gene family, including analysis of conservation, domains/modules and specific features of the proteins encoded by the FAM167 genes, which we denote disordered autoimmunity (DIORA) proteins based on their properties and disease association. Further, we assessed their expression pattern and intracellular localization to begin to understand the function of the DIORA proteins and their potential role in autoimmune rheumatic disease.
Materials and methods
SNP and eQTL analysis
SNPs associated with rheumatoid arthritis, SLE and Sjögren's syndrome were chosen from the current literature, with the criteria of reaching genome‐wide significance (P < 5·0 × 10−8) and being the first reported and/or highest‐associated SNP within the region. To investigate linkage disequilibrium (LD), r 2 values for the SNPs were retrieved using LDlink (FIN population) 16.
For analysis of eQTL effects, we used RNA expression data from peripheral blood mononuclear cells (PBMC) of genotyped patients with Sjögren's syndrome (n = 14) and healthy controls (n = 18) 17, CD19+ B cell mRNA expression and genotype data from 278 healthy volunteers 18 and CD14+ monocyte mRNA expression and genotype data from 414 healthy volunteers 19, applying the standard method in the MatrixEQTL R package 20. Microarray data for the PBMCs are deposited at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/), Accession number GSE48378.
Gene conservation analysis
PhylomeDB version 4 was used for generation of phylogenetic trees with human FAM167A as a seed sequence in human phylome 154, and the UCSC genome browser, GRCh37/hg19 (http://www.genome.ucsc.edu) was employed as a basis for generating positional graphs.
Protein sequence alignment and predictions
FAM167A/DIORA‐1 (Q96KS9) and FAM167B/DIORA‐2 (Q9BTA0) sequences were extracted from the Uniprot database (http://www.uniprot.org) and orthologue sequences found through a blast search. Multiple sequence alignments were generated with Clustal 21 and visualized in Jalview 22, utilizing the Clustal X colour scheme. The compiled prediction of disordered sequences within human DIORA‐1 and DIORA‐2 was extracted from D2P2 (http://d2p2.pro) 23, which includes the predictions by the Esprits, IUPred, PV2, PrDOS and the PONDR (VSL2b, VLXT) tools. Secondary structure prediction was performed for the human protein using YASPIN (http://www.ibi.vu.nl/programs/yaspinwww/help.php). Protein motifs were investigated using SCRATCH/the ABTMpro tool (http://scratch.proteomics.ics.uci.edu/) and NucPred (http://nucpred.bioinfo.se/nucpred/).
Animals and organ dissection
C57BL/6J mice (Jackson Laboratories, Bar Harbor, ME, USA) were killed at 8–10 weeks of age, and representative pieces of dissected organs were rinsed in phosphate‐buffered saline (PBS) and snap‐frozen on dry ice for later RNA extraction.
Quantitative polymerase chain reaction (qPCR)
Total RNA was extracted from representative organs pieces using the TRIzol reagent (Life Technologies, Carlsbad, CA, USA), following the protocol provided by the manufacturer. RNA from heart and skeletal muscles was extracted using an RNeasy Fibrous Tissue mini kit (Qiagen, Valencia, CA, USA) with on‐column RNase‐free DNase digestion (Qiagen). Reverse transcription was performed using the iScript cDNA Synthesis Kit (Bio‐Rad, Hercules, CA, USA), following the protocol provided.
Primers for amplification of mouse β‐Actin, Fam167a and Fam167b were as follows: β‐Actin forward: 5′‐GACGGCCAGGTCATCACTATTG‐3′, β‐Actin reverse: 5′‐AGGAAGGCTGGAAAAGAGCC‐3′; Fam167a forward: 5′‐GGCAAGCTGGAAGGCTTC‐3′, Fam167a reverse: 5′‐GCAGGTCTGCTCGATCTTCAAC‐3′; and Fam167b forward: 5′‐ GTGGCTCAGACGGGAGC‐3′, Fam167b reverse: 5′‐GGTCCACTTTCAGTCTGTGC‐3′. PCR was performed with a CFX384 real‐time system (Bio‐Rad) using the SYBR Green PCR Kit (Qiagen) and a two‐step protocol (95°C for 3 min, followed by 95°C for 10 s and 60°C for 45 s for 40 cycles). A standard curve was constructed using six twofold serial dilutions of pooled cDNA from RAW and A20 cell lines and mouse lung, spleen and adrenal glands in equal proportions. The expression levels of Fam167a and Fam167b were related to that of the housekeeping gene β‐Actin, calculated by the Bio‐Rad CFX ManagerTM software.
Immunohistochemistry of human lung tissues
Human lung biopsies (n = 5) and bronchoalveolar lavage (BAL) samples (n = 5) were taken during bronchoscopy of healthy volunteers, as described previously 24. The biopsies were fixed in 4% paraformaldehyde before paraffin embedding and cutting; 4–6‐µm sections were placed subsequently on SuperFrost® Plus slides and incubated overnight at 56°C, followed by deparaffinization in xylene, and rehydration through a graded ethanol series (100, 95, 70%) and PBS (Sigma‐Aldrich, Munich, Germany). Cytospin was used for collecting BAL cells on SuperFrost® Plus microscope slides followed by fixation in 4% paraformaldehyde.
Before staining, the slides were subjected to epitope retrieval with citrate buffer (pH 6.0) (Dako, Carpinteria, CA, USA) at 98°C for 30 min. The EnVision system (Dako) was used to stain tissue sections and BAL cells for DIORA‐1, following the manufacturer's protocol. Affinity‐purified rabbit anti‐DIORA‐1 antibody HPA030426 (1 µg/ml; Sigma) or rabbit immunoglobulin (Ig)G control (1 µg/ml; Dako) in antibody diluent (Dako) was used as primary antibody. The sections were counterstained with Mayer's haematoxylin and mounted under coverslips using Mountex (HistoLab, Gothenburg, Sweden).
Cloning of FAM167A and FAM167B
FAM167A and FAM167B were amplified from mRNA isolated from human PBMCs obtained from a healthy volunteer. Cloning primer sequences are available upon request. The PCR product was incorporated into the Gateway pDonor221, confirmed by sequencing, and thereafter subcloned into pcDNA 6.2/N‐YFP‐DEST.
Cell transfection and immunofluorescent microscopy
HeLa and SK‐MEL‐28 cells were grown on glass slides to a density of 10–30% before transfection with XtrmeGene9 (Sigma) using a 3 : 1 ratio of transfectant : DNA. After 20 h slides were rinsed in PBS and fixed for 12 min in 4% paraformaldehyde, stained with 4',6‐diamidino‐2‐phenylindole (DAPI) and mounted under coverslips using ProLong Gold (Thermo Fisher, Fremont, CA, USA). Slides were analysed in a fluorescence microscope (Leitz DM RBE Leica, Wetzlar, Germany).
Ethical approval
All parts of the study involving human subjects were approved by the Regional Ethics Committee Stockholm, and the subjects gave informed written consent. Studies involving mice were approved by the Stockholm Ethics Committee North.
Results
Disease associations with genetic polymorphisms of the FAM167A‐BLK locus
To compare the genetic signals from the FAM167A‐BLK locus in rheumatic diseases, we conducted a search of available genome‐wide association studies (GWAS) and publication databases for genetic studies investigating our locus of interest on chromosome 8. Studies showing disease associations reaching genome‐wide significance levels included rheumatoid arthritis, SLE and Sjögren's syndrome 9, 10, 11, with odds ratios (OR) ranging from 1·19 to 1·39 (Fig. 1a). Most of the disease‐associated SNPs in the FAM167A‐BLK locus fall into the intergenic region between the two genes that have opposite transcriptional directions, and most of them are positioned closer to the BLK gene, although the association signal stretches over both genes. An analysis of the linkage disequilibrium revealed a high r 2 (> 0·8) between the three main SNPs for each disease, indicating that it is the same genetic signal which is identified for all three diseases (Fig. 1b). To confirm whether the disease‐associated genotypes related to any difference in expression of FAM167A or BLK, we performed an eQTL analysis using microarray data from PBMCs of genotyped individuals. A substantial and significantly higher expression of FAM167A was noted for the disease‐associated polymorphisms (rs13277113: β = 0·35, P = 1·1 × 10−6), while the expression of BLK did not differ throughout genotypes (Fig. 1c and data not shown). Considering that eQTL effects are context‐dependent and may differ between cell types 19, the genotype‐related expression was also investigated in two data sets of sorted immune cells from PBMCs representing purified CD19+ B cells and CD14+ monocytes (Fig. 1d) 18, 19. The expression of FAM167A was related to the genotype of disease‐associated polymorphism in B cells, while an opposite eQTL of a lesser effect size, albeit highly significant, was observed for BLK. In monocytes, a significant eQTL effect was observed only for FAM167A. Altogether, these data demonstrate a significant genotype‐dependent differential regulation of the uncharacterized gene FAM167A, with higher expression in composite as well as discrete immune cell populations observed from alleles associated with autoimmune rheumatic diseases.
Figure 1.
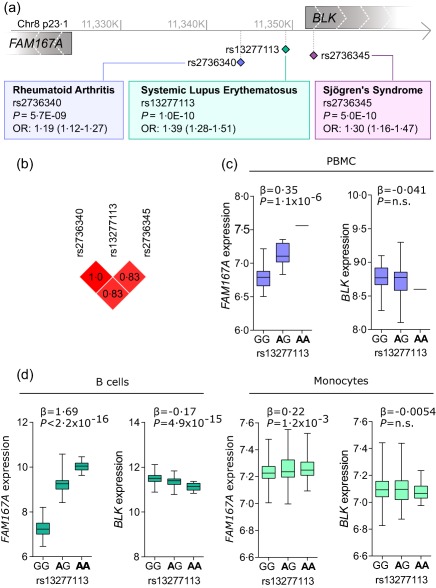
Rheumatic disease associations with polymorphisms of the FAM167A‐BLK locus. (a) Approximate location of the FAM167A and BLK genes on chromosome 8, and position of selected single nucleotide polymorphisns (SNP) described for rheumatoid arthritis 9, systemic lupus erythematosus 10 and Sjögren's syndrome 11. Transcriptional direction is indicated by hatched arrows. (b) Linkage disequilibrium (r 2) between the FAM167A‐BLK SNPs associated with rheumatoid arthritis, systemic lupus erythematosus and Sjögren's syndrome. Expression of FAM167A and BLK in (c) whole peripheral blood mononuclear cells, (d) sorted CD19+ B cells and CD14+ monocytes, stratified for the genotype of rs13277113. Bold text denotes the disease‐associated allele. For peripheral blood mononuclear cells (PBMC): GG n = 17, AG n = 14, AA n = 1; for B cells: GG n = 159, AG n = 102, AA n = 17; and for monocytes GG n = 238, AG n = 151, AA n = 25.
Evolutionary history and human genomic position of the FAM167 genes
To begin to characterize the FAM167A gene, we first searched for homologues and orthologues in other species. Using FAM167A as a seed sequence in the PhylomeDB database, we found that FAM167A‐like genes containing the Domain of Unknown Function (DUF) 3259 were present in mammalians, amphibians, reptiles, fish and tunicates, indicating that it is a highly conserved gene, with homologues present even in invertebrate species (Fig. 2a). We further identified two members of the FAM167 gene family, FAM167A and FAM167B, neither of which show homology to any other annotated human gene or known sequence motifs (Table 1). In humans, FAM167A is located on chromosome 8p23.1 next to BLK (Fig. 2d) and comprises four exons encoding a protein of 214 amino acids (Table 1). The FAM167B gene is located on chromosome 1p35.1 (Fig. 2e) and contains two exons coding for a smaller protein of 163 amino acids (Table 1). Of note, FAM167B neighbours the lymphocyte‐specific tyrosine kinase (LCK) gene, a homologue of BLK, which is expressed in T cells. Immediate upstream genes of FAM167A and B are also homologous [myotubularin‐related protein 9 (MTMR9) on chromosome 8 and myotubularin‐related protein 9‐like (MTMR9LP) on chromosome 1], suggesting that the whole region was duplicated during evolution. In all, the high conservation and presence of the FAM167 genes throughout species indicate that the gene family has an important, yet unknown, function.
Figure 2.
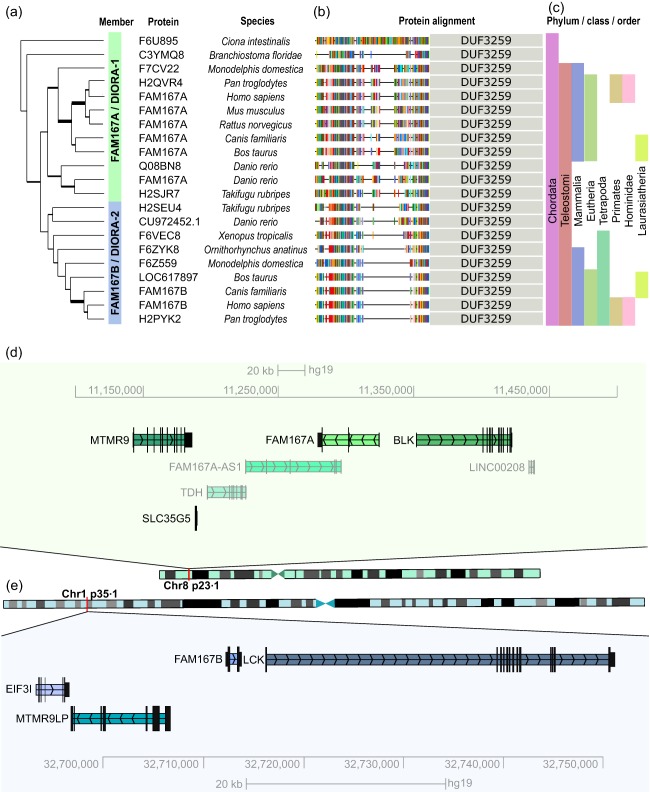
Evolutionary history and human genomic position of the FAM167 gene and disordered autoimmunity (DIORA) protein family. (a) Phylogenetic tree generated using FAM167A as seed sequence and (b) DIORA protein sections present in the different species (PhylomeDB). Homologues with predicted Domain of Unknown Function (DUF)3259 domains were included, the position of which is indicated for each species. Amino acid colouring according to the standard scheme of PhylomeDB. (c) Phylum/class/order of respective species. (d,e) Location of FAM167A and FAM167B and neighbouring genes on chromosomes 8p23.1 and 1p35.2–35.1, respectively, in the human genome [University of California Santa Cruz (UCSC) genome browser].
Table 1.
Predicted characteristics of the FAM167 genes and their encoded DIORA proteins
| FAM167A/DIORA‐1 | FAM167B/DIORA‐2 | |
|---|---|---|
| Synonyms* | C8orf13, D8S265 | C1orf90 |
| Paralogues † | 1 (FAM167B) | 1 (FAM167A) |
| Gene position † | 8p23.1 | 1p35.1 |
| Exons † | 4 | 2 |
| Transcripts/splice variants † | 7 (701–4094) bp | 1 (936 bp) |
| Protein coding transcripts † | 4 | 1 |
| Protein length ‡ | 214 aa | 163 aa |
| Differently spliced protein † | 91 aa | – |
| Molecular weight (full‐length) ‡ | 24·2 kD | 18·4 kD |
| Isoelectric point § | 5·93 | 5·28 |
| Atomic composition ¶ | C1052H1697N315O323S8 | C798H1295N239O245S8 |
| Negatively charged aa (Asp + Glu) ¶ | 33 | 26 |
| Positively charged aa (Arg + Lys) ¶ | 29 | 20 |
| Instability index (II) ¶ | 62·96 (unstable) | 66·36 (unstable) |
| Aliphatic index ¶ | 82·20 | 92·33 |
| Grand average of hydropathicity ¶ | −0·639 | −0·491 |
| Protein family** (aa) | PF11652 (130–214) | PF11652 (90–163) |
| DUF* (aa) | DUF3259 (130–214) | DUF3259 (90–163) |
| Coiled coil** (aa) | 134–154 | 73–93 |
| Disorder †† | 1–28; 57–109 | 10–25; 57–61 |
| Phosphorylation sites § | 6 | 1 |
| Transmembrane region ‡‡ | 0 | 0 |
| Nuclear localization signal §§ | 0 | 0 |
Information source: *GPSDB Gene/Protein Synonyms finder. †Ensembl. ‡UniProt. §PhosphositePlus. ¶ProtPram. **Pfam. ††D2P2. ‡‡SCRATCH/ABTMpro. §§NucPred.
aa = amino acid; bp = base pairs; DUF = Domain of Unknown Function; kD = kilodalton.
DIORA protein properties and sequence analysis
To explore the conservation, modules and specific features of the proteins encoded by the FAM167 genes, which we denote disordered autoimmunity (DIORA) proteins, representative species were chosen from the evolutionary tree and corresponding sequences extracted from UniProt. By aligning the DIORA‐1 and −2 protein sequences of distantly related species, we identified well‐conserved stretches near both the N‐ and C‐terminus of the proteins, potentially forming two modules (Figs 2 and 3). The C‐terminal parts of the proteins contain the DUF3259 element (Fig. 2b), based on which the genes were grouped into a family. A similarly well‐conserved sequence, TRRPSYLEW, is present in the N‐terminal portion of both DIORA‐1 and −2 (Fig. 3). In DIORA‐1, this is preceded by a highly conserved amino acid series of DDHLRSLKALTEKLRLE, while DIORA‐2 has a slightly shorter specific conserved stretch of LDSVKALTAKLQLQ. Both proteins contain a total of three cysteine residues, with the specific feature of a highly conserved C‐terminal cysteine. Both DIORA‐1 and −2 are predicted to contain a high degree of intrinsic disorder and elements of coiled‐coil (Fig. 4), but they lack distinguishing features such as nuclear localization signals and transmembrane domains (Fig. 3, Table 1). In summary, the DIORA proteins appear to belong to the group of intrinsically disordered proteins, and contain no described protein motifs.
Figure 3.
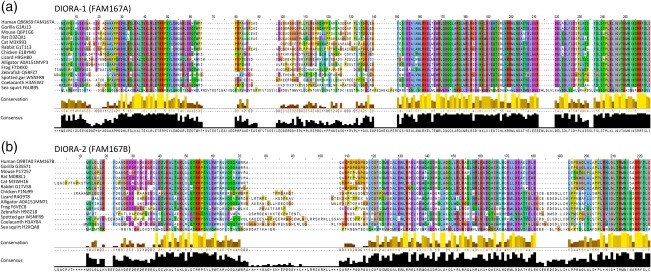
Disordered autoimmunity (DIORA) protein sequence alignments. (a) Alignment of the DIORA‐1 (FAM167A) and (b) DIORA‐2 (FAM167B) sequences for species ranging from human to sea squirt. Alignments were generated with Clustal and visualized in Jalview with the Clustal X colour scheme.
Figure 4.
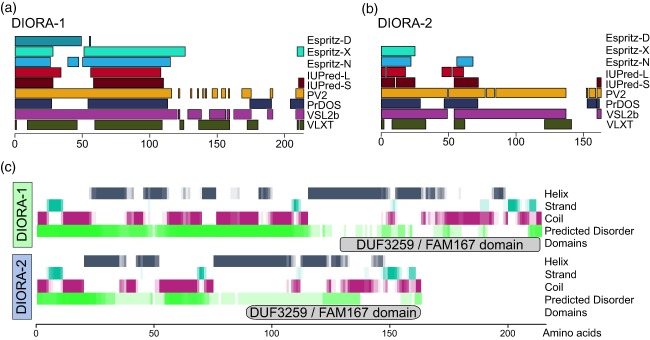
Secondary structure predictions in disordered autoimmunity (DIORA) proteins. The DIORA‐1 and the compiled prediction of disordered sequences within the (a) DIORA‐1 and (b) DIORA‐2 proteins by indicated tools. (c) The secondary structure prediction was generated for the human proteins using YASPIN and was aligned with the summary of the predicted disorder agreement. The localization of the Domain of Unknown Function (DUF)3259 domain is denoted.
Expression and intracellular localization of DIORA proteins
An important step in understanding protein function is characterization of its cellular expression and localization. Expression profiling by qPCR of an array of mouse organs revealed the highest expression of Fam167a in lung, but gene expression was also observed in spleen, skeletal muscle, brain, liver, thymus, lymph nodes and heart (Fig. 5a). Fam167b had its peak expression in adrenal glands, kidney and liver, with little or no expression observed in other analysed organs (Fig. 5b).
Figure 5.

Expression and cellular localization of FAM167 genes and encoded disordered autoimmunity (DIORA) proteins. (a,b) Quantification of Fam167a (a) and Fam167b (b) by SYBR quantitative polymerase chain reaction (qPCR). Organs were dissected from 10 mice and Fam167a expression was normalized to β‐actin expression. (c) Immunohistochemistry staining for DIORA‐1 in a human lung biopsy with isotype control. (d) Immunocytochemistry staining for DIORA‐1 of human bronchoalveolar lavage cells with isotype control. (e) Higher magnification of (d). Bars represents 100 μm. (f) HeLa cells transfected with yellow fluorescent protein (YFP)‐DIORA‐1 (green). Cell nuclei were stained by 4′,6‐diamidino‐2‐phenylindole (DAPI) (blue). (g) SK‐MEL‐28 cells transfected with YFP‐DIORA‐2 (green). Cell nuclei were stained by DAPI (blue). Bars represents 10 μm.
Considering that polymorphisms of FAM167A have been associated with several rheumatic diseases and that lung has been suggested as an organ where triggering autoimmune reactions may take place 4, 25, we also wanted to confirm expression of FAM167A in human lung. Staining for DIORA‐1 by immunohistochemistry in lung biopsies (Fig. 5c) and BAL cells (Fig. 5d) demonstrated intracellular protein expression in both the bronchial epithelium and alveolar macrophages. The alveolar macrophage staining appeared cytoplasmic, with a speckled pattern (Fig. 5e). To confirm the intracellular localization of the DIORA proteins, we cloned the FAM167A and FAM167B genes and inserted them into a YFP expression vector for transfection and expression in HeLa and SK‐MEL‐28 cells, respectively. The choice of cell lines was based on suitability for localization studies and endogenous expression of FAM167A and B. For both the DIORA‐1 and −2 proteins, a speckled cytoplasmic localization pattern was observed (Fig. 5f,g). Altogether, our findings demonstrate that DIORA proteins are cytoplasmic, and that DIORA‐1 is expressed highly by bronchial epithelium and lung alveolar macrophages, suggesting a potential role in rheumatic disease pathogenesis.
Discussion
SNPs of the FAM167A‐BLK locus have been associated with several autoimmune diseases, including Sjögren's syndrome, SLE and rheumatoid arthritis 9, 10, 11. While a role for BLK downstream of the B cell receptor is established, the function of FAM167A is not known. In view of our own and others’ observation of cis‐eQTL regulation of FAM167A expression by the disease‐associated genotypes 10, 11, we aimed to characterize this gene and its gene family.
Gathering and analysing information from public databases, we observed that the FAM167 genes have been conserved throughout evolution, and can be tracked back to invertebrate tunicates, suggesting that they have an important, still unexplored, function. Interestingly, the Src tyrosine kinase BLK is positioned downstream of FAM167A, while FAM167B is close to the Src tyrosine kinase LCK. The genes upstream of FAM167A and B also share homology, pointing to an ancestral duplication event that gave rise to the two variants in the gene family.
The cis‐eQTL effect observed for disease‐associated polymorphisms of the FAM167A‐BLK locus affects predominantly FAM167A expression, with a higher FAM167A expression associated with the disease risk genotypes. An eQTL effect, however, has also been reported for BLK, leading to a somewhat decreased expression of BLK from alleles associated with rheumatic disease 10. In our study, we were unable to confirm the BLK eQTL effect in PBMC, which could relate to us using PBMC when previous observations were made in Epstein–Barr virus (EBV)‐transformed B cells or sorted cells 26, or to the lower number of samples included in our study. Indeed, significant, and opposite, eQTL effects on FAM167A and BLK were observed in purified primary CD19+ B cells for the disease‐associated polymorphisms, while an eQTL effect was observed only for FAM167A in circulating CD14+ monocytes, albeit at much lower expression levels than in B cells. Notably, circulating CD14+ monocytes develop from a different embryonic origin than tissue‐resident macrophages 27. Interestingly, FAM167A and BLK are positioned within a common large inversion polymorphism region of chromosome 8 28, 29 and recent reports also implicate a role in autoimmune diseases for additional genes within and surrounding this region by identification of novel genetic signals 30 and cis‐eQTL 30, as well as sex‐influenced expression differences mediated by disease‐associated polymorphisms 31.
Comparison of the FAM167A‐ and FAM167B‐encoded DIORA‐1 and −2 proteins from distantly related animals revealed highly conserved residues spanning long stretches at both ends of the proteins, suggesting that these modules are of high functional importance for the proteins. Notably, the middle of the DIORA‐1 proteins contains a non‐conserved region that is absent in mammalian DIORA‐2 proteins, defining this region as the one being most different between the two proteins. We infer that this middle region contains a linker that accounts for the different length of the two proteins in humans, and that it may intersperse two conserved modules.
A prominent feature of both DIORA proteins is the high content of sequences with predicted disorder. Most proteins are made up of both structured and intrinsically disordered regions (IDR), with proteins containing entirely disordered sequences referred to as intrinsically disordered proteins (IDPs) 32, 33. These often interact with other proteins, regulating cellular processes such as signalling, transcription and translation 33. In addition to their regulatory functions, IDPs participate in the ordered assembly of macromolecular structures and in the binding and transport of small molecules 33. Further, IDPs have also been implicated in a number of diseases. Many key oncogenic proteins, including c‐Myc, p53 and BRCA1, have large IDRs 34 and in neurodegenerative disorders, aggregation of intrinsically unstructured proteins into amyloid is thought to be responsible for cell dysfunction 35. Myelin basic protein (MBP), which is involved in the pathogenesis of multiple sclerosis, is an example of an intrinsically disordered protein linked to autoimmune disease 36, now joined by DIORA‐1.
Interestingly, we observed the highest expression of mouse Fam167a in lung tissue. In human lung biopsies, the corresponding protein DIORA‐1 was found in alveolar macrophages and bronchial epithelium. In rheumatoid arthritis, triggering of autoimmunity has been suggested to occur in the lungs after exposure to environmental agents, such as cigarette smoke, textile dust and silica 25. The environmental agents defined for rheumatoid arthritis have been shown to act by driving both post‐translational modifications of autoantigens and the local accumulation and activation of antigen‐presenting cells. One possibility is that DIORA‐1, with its high expression in the alveolar macrophages and bronchial epithelium, plays a role in this process. Interstitial lung disease may develop in a subset of patients with rheumatoid arthritis, SLE and Sjögren's syndrome 37, 38, 39, 40, suggesting the additional possibility that polymorphisms of FAM167A may contribute to pulmonary involvement in rheumatic diseases.
In contrast to Fam167a, Fam167b was expressed predominantly in the adrenal gland, and its role in organ function will be investigated in future studies. Here, using YFP‐tagged fusion proteins in cellular transfection experiments, we show that the DIORA‐1 and −2 proteins localize to the cytoplasm, presenting in a dot‐like pattern compatible with, e.g. endosomal‐related vesicles.
In summary, our study presents a comprehensive description and analysis of the FAM167 gene family and its corresponding proteins, which we propose to denote ‘disordered autoimmunity’ (DIORA)‐1 and −2 based on their characteristics and disease association. Importantly, our study reveals that FAM167A, the expression of which is regulated by disease‐associated polymorphisms present in the FAM167A‐BLK locus, is expressed predominantly in lung – an organ central to autoimmune rheumatic disease. Altogether, our data uncover FAM167A/DIORA‐1 as a protein of interest for further investigations in the context of rheumatic disease pathogenesis.
Disclosure
The authors declare no competing interests.
Author contributions
L. M., G. E. T. and M. W. H. conceived the study. J. G. recruited patients. L. M., G. E., L. M., L. A. and J. I. R. S. performed experiments, analysed the data and generated figures and table with input from A. E. and M. W. H. L. M. wrote the manuscript together with M. W. H. All authors edited and approved the final manuscript.
Ethics approval
Studies of rodent tissues were approved by the Ethical committee Stockholm North. Studies of human tissue were approved by the Regional Ethics Committees in Stockholm, and participants gave informed written consent.
Acknowledgements
We thank Professor Julian C. Knight and his research group at the University of Oxford for sharing genotype and expression data from primary B cells and monocytes. We thank Vijole Ottosson and Amina Ossoinak for excellent technical assistance, and gratefully acknowledge Dr Aurélie Ambrosi, Karolinska Institutet, for contributing to the writing of the paper. The study was supported by grants from the Swedish Research Council, the Swedish Rheumatism association, the King Gustaf the V:th 80‐year foundation, the Heart–Lung Foundation, the Stockholm County Council, The Swedish Cancer Foundation and Karolinska Institutet.
References
- 1. Cooper GS, Stroehla BC. The epidemiology of autoimmune diseases. Autoimmun Rev 2003; 2:119–25. [DOI] [PubMed] [Google Scholar]
- 2. Klareskog L, Malmstrom V, Lundberg K, Padyukov L, Alfredsson L. Smoking, citrullination and genetic variability in the immunopathogenesis of rheumatoid arthritis. Semin Immunol 2011; 23:92–8. [DOI] [PubMed] [Google Scholar]
- 3. Ambrosi A, Wahren‐Herlenius M. Update on the immunobiology of Sjogren's syndrome. Curr Opin Rheumatol 2015; 27:468–75. [DOI] [PubMed] [Google Scholar]
- 4. Catrina AI, Joshua V, Klareskog L, Malmstrom V. Mechanisms involved in triggering rheumatoid arthritis. Immunol Rev 2016; 269:162–74. [DOI] [PubMed] [Google Scholar]
- 5. Parks CG, de Souza Espindola Santos A, Barbhaiya M, Costenbader KH. Understanding the role of environmental factors in the development of systemic lupus erythematosus. Best Pract Res Clin Rheumatol 2017; 31:306–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bogdanos DP, Smyk DS, Rigopoulou EI et al Twin studies in autoimmune disease: genetics, gender and environment. J Autoimmun 2012; 38:J156–69. [DOI] [PubMed] [Google Scholar]
- 7. Selmi C, Lu Q, Humble MC. Heritability versus the role of the environment in autoimmunity. J Autoimmun 2012; 39:249–52. [DOI] [PubMed] [Google Scholar]
- 8. Wahren‐Herlenius M, Dorner T. Immunopathogenic mechanisms of systemic autoimmune disease. Lancet 2013; 382:819–31. [DOI] [PubMed] [Google Scholar]
- 9. Gregersen PK, Amos CI, Lee AT et al REL, encoding a member of the NF‐kappaB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nat Genet 2009; 41:820–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hom G, Graham RR, Modrek B et al Association of systemic lupus erythematosus with C8orf13‐BLK and ITGAM‐ITGAX. N Engl J Med 2008; 358:900–9. [DOI] [PubMed] [Google Scholar]
- 11. Lessard CJ, Li H, Adrianto I et al Variants at multiple loci implicated in both innate and adaptive immune responses are associated with Sjogren's syndrome. Nat Genet 2013; 45:1284–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Samuelson EM, Laird RM, Maue AC, Rochford R, Hayes SM. Blk haploinsufficiency impairs the development, but enhances the functional responses, of MZ B cells. Immunol Cell Biol 2012; 90:620–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Simpfendorfer KR, Olsson LM, Manjarrez Orduno N et al The autoimmunity‐associated BLK haplotype exhibits cis‐regulatory effects on mRNA and protein expression that are prominently observed in B cells early in development. Hum Mol Genet 2012; 21:3918–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gourh P, Agarwal SK, Martin E et al Association of the C8orf13‐BLK region with systemic sclerosis in North‐American and European populations. J Autoimmun 2010; 34:155–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Texido G, Su IH, Mecklenbrauker I et al The B‐cell‐specific Src‐family kinase Blk is dispensable for B‐cell development and activation. Mol Cell Biol 2000; 20:1227–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Machiela MJ, Chanock SJ. LDassoc: an online tool for interactively exploring genome‐wide association study results and prioritizing variants for functional investigation. Bioinformatics 2018; 34:887–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brauner S, Folkersen L, Kvarnstrom M et al H1N1 vaccination in Sjogren's syndrome triggers polyclonal B cell activation and promotes autoantibody production. Ann Rheum Dis 2017; 76:1755–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fairfax BP, Makino S, Radhakrishnan J et al Genetics of gene expression in primary immune cells identifies cell type‐specific master regulators and roles of HLA alleles. Nat Genet 2012; 44:502–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fairfax BP, Humburg P, Makino S et al Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science 2014; 343:1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 2012; 28:1353–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li W, Cowley A, Uludag M et al The EMBL‐EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res 2015; 43:W580–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview version 2 – a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009; 25:1189–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oates ME, Romero P, Ishida T et al D(2)P(2): database of disordered protein predictions. Nucleic Acids Res 2013; 41:D508–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wiken M, Grunewald J, Eklund A, Wahlstrom J. Higher monocyte expression of TLR2 and TLR4, and enhanced pro‐inflammatory synergy of TLR2 with NOD2 stimulation in sarcoidosis. J Clin Immunol 2009; 29:78–89. [DOI] [PubMed] [Google Scholar]
- 25. Malmstrom V, Catrina AI, Klareskog L. The immunopathogenesis of seropositive rheumatoid arthritis: from triggering to targeting. Nat Rev Immunol 2017; 17:60–75. [DOI] [PubMed] [Google Scholar]
- 26. Thalayasingam N, Nair N, Skelton AJ et al CD4+ and B lymphocyte expression quantitative traits at rheumatoid arthritis risk loci in patients with untreated early arthritis: implications for causal gene identification. Arthritis Rheumatol 2018; 70:361–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gomez Perdiguero E, Klapproth K, Schulz C et al Tissue‐resident macrophages originate from yolk‐sac‐derived erythro‐myeloid progenitors. Nature 2015; 518:547–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Antonacci F, Kidd JM, Marques‐Bonet T et al Characterization of six human disease‐associated inversion polymorphisms. Hum Mol Genet 2009; 18:2555–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Salm MP, Horswell SD, Hutchison CE et al The origin, global distribution, and functional impact of the human 8p23 inversion polymorphism. Genome Res 2012; 22:1144–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Demirci FY, Wang X, Morris DL et al Multiple signals at the extended 8p23 locus are associated with susceptibility to systemic lupus erythematosus. J Med Genet 2017; 54:381–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Linden M, Ramirez Sepulveda JI, James T et al Sex influences eQTL effects of SLE and Sjogren's syndrome‐associated genetic polymorphisms. Biol Sex Differ 2017; 8:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van der Lee R, Buljan M, Lang B et al Classification of intrinsically disordered regions and proteins. Chem Rev 2014; 114:6589–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry 2002; 41:6573–82. [DOI] [PubMed] [Google Scholar]
- 34. Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys 2008; 37:215–46. [DOI] [PubMed] [Google Scholar]
- 35. Jarosz DF, Khurana V. Specification of physiologic and disease states by distinct proteins and protein conformations. Cell 2017; 171:1001–14. [DOI] [PubMed] [Google Scholar]
- 36. Vassall KA, Bamm VV, Harauz G. MyelStones: the executive roles of myelin basic protein in myelin assembly and destabilization in multiple sclerosis. Biochem J 2015; 472:17–32. [DOI] [PubMed] [Google Scholar]
- 37. Doyle TJ, Dellaripa PF. Lung manifestations in the rheumatic diseases. Chest 2017; 152:1283–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ramirez Sepulveda JI, Kvarnstrom M, Brauner S, Baldini C, Wahren‐Herlenius M. Difference in clinical presentation between women and men in incident primary Sjogren's syndrome. Biol Sex Differ 2017; 8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ramirez Sepulveda JI, Kvarnstrom M, Eriksson P et al Long‐term follow‐up in primary Sjogren's syndrome reveals differences in clinical presentation between female and male patients. Biol Sex Differ 2017; 8:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kvarnstrom M, Ottosson V, Nordmark B, Wahren‐Herlenius M. Incident cases of primary Sjogren's syndrome during a 5‐year period in Stockholm County: a descriptive study of the patients and their characteristics. Scand J Rheumatol 2015; 44:135–42. [DOI] [PubMed] [Google Scholar]


