Abstract
Haematopoietic stem cells renew blood. Accumulation of DNA damage in these cells promotes their decline, while misrepair of this damage initiates malignancies. Here we describe the features and mutational landscape of DNA damage caused by acetaldehyde, an endogenous and alcohol-derived metabolite. This damage results in DNA double-stranded breaks that, despite stimulating recombination repair, also cause chromosome rearrangements. We combined transplantation of single haematopoietic stem cells with whole-genome sequencing to show that this DNA damage occurs in stem cells, leading to deletions and rearrangements that are indicative of microhomology-mediated end-joining repair. Moreover, deletion of p53 completely rescues the survival of aldehyde-stressed and mutated haematopoietic stem cells, but does not change the pattern or the intensity of genome instability within individual stem cells. These findings characterize the mutation of the stem-cell genome by an alcohol-derived and endogenous source of DNA damage. Furthermore, we identify how the choice of DNA-repair pathway and a stringent p53 response limit the transmission of aldehyde-induced mutations in stem cells.
The consumption of alcohol contributes to global mortality and cancer development1. Most of the toxic effects of alcohol are probably caused by its oxidation product acetaldehyde, which is highly reactive towards DNA2. The enzyme aldehyde dehydrogenase 2 (ALDH2) prevents acetaldehyde accumulation by oxidizing it efficiently to acetate, but around 540 million people carry a polymorphism in ALDH2 that encodes a dominant-negative variant of the enzyme3. Alcohol consumption in these individuals induces an aversive reaction and predisposes them to oesophageal cancer4. Nevertheless, ALDH2 deficiency is surprisingly well tolerated in humans. This could be because of the additional tier of protection provided by FANCD2, a DNA-crosslink-repair protein. In fact, genetic inactivation of Aldh2 and Fancd2 in mice leads to cancer and a profound haematopoietic phenotype5,6. In humans, deficiency in DNA-crosslink repair causes the inherited illness Fanconi anaemia, a devastating condition that leads to abnormal development, bone-marrow failure and cancer7. Acetaldehyde genotoxicity is likely to contribute to this phenotype, as Japanese children who are afflicted with Fanconi anaemia and carry the ALDH2 polymorphism display earlier-onset bone marrow failure8. Together, these data suggest that endogenous aldehydes are a ubiquitous source of DNA damage that impairs blood production.
It is likely that some of this damage occurs in haematopoietic stem cells (HSCs), which are responsible for lifelong blood production. HSC attrition is a feature of ageing, and mutagenesis in the remaining HSCs promotes dysfunctional haematopoiesis and leukaemia. Moreover, both humans and mice that lack DNA repair factors are prone to HSC loss, and in some cases, bone marrow failure9,10. HSCs employ DNA repair and respond to damage in a distinct manner compared to later progenitors11,12. While these observations point to a fundamental role for DNA repair in HSCs, recent work has highlighted that effective replication-stress responses maintain HSC function and integrity13. However, there is a key gap in our knowledge regarding the identity of the endogenous factors that damage DNA and lead to replication stress. Here we show that alcohol-derived and endogenous aldehydes damage the genomes of haematopoietic cells, and we characterize the surveillance and repair mechanisms that counteract this. We also establish a method that allows us to determine the mutational landscape of individual HSCs, and in doing so, provide new insight into the p53 response in mutagenized stem cells.
Ethanol stimulates homologous recombination repair
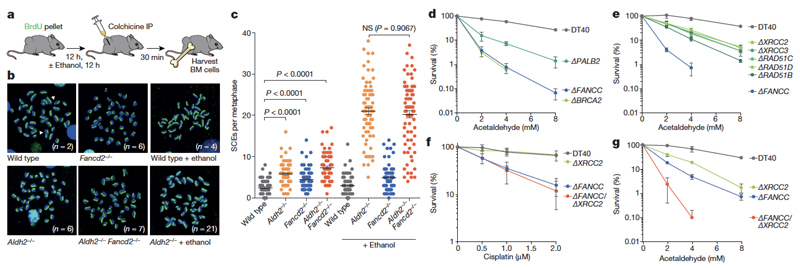
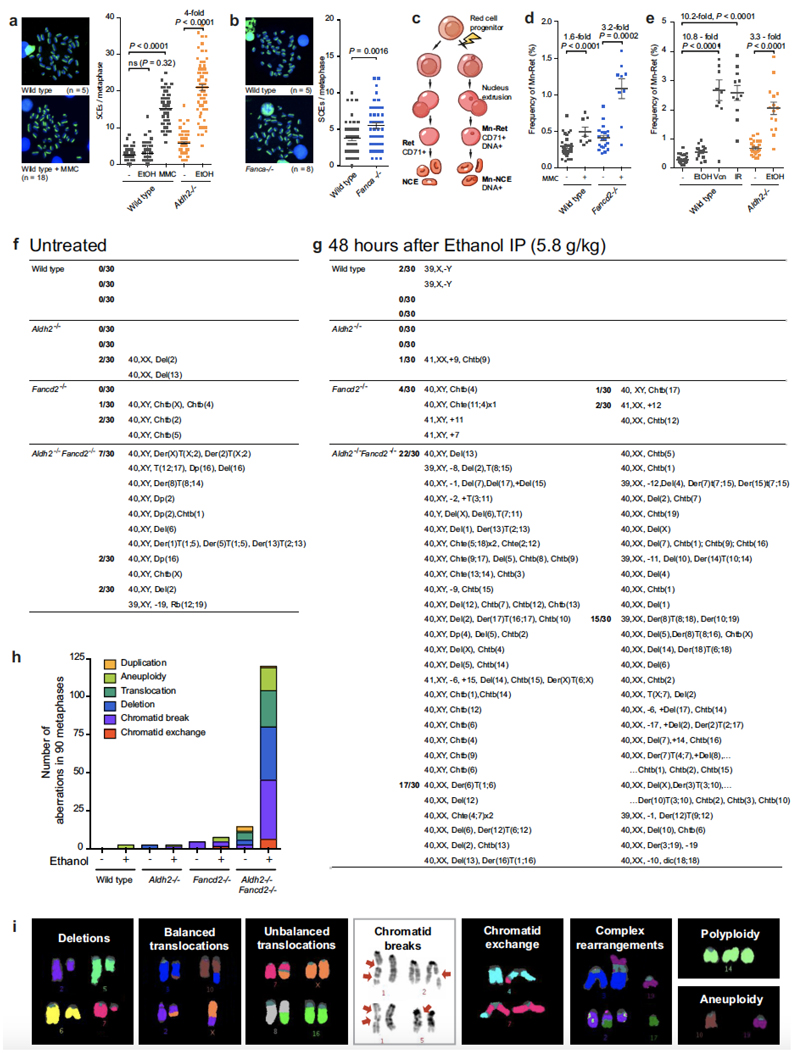
Aldh2−/−Fancd2−/− mice develop severe HSC attrition, causing spontaneous bone marrow failure, which can also be induced by exposing these mice to ethanol5,6. This genetic interaction suggests that in the absence of aldehyde catabolism (such as in Aldh2−/− mice), DNA repair is engaged to maintain blood homeostasis. To test this theory, we set out to monitor DNA repair activity in vivo. The Fanconi anaemia pathway repairs DNA crosslinks by using a replication-coupled excision mechanism that is completed by homologous recombination14,15. We therefore used a method to visualize sister-chromatid exchange (SCE) events in bone marrow cells of living mice; these represent recombination repair transactions coupled to replication (Fig. 1a). The number of SCE events is elevated 2.3-fold in Aldh2−/− mice, indicating that recombination repair is stimulated in response to endogenous aldehydes (Fig. 1b, c). Moreover, a single exposure to alcohol causes a fourfold increase in SCE events in Aldh2−/− mice (Fig. 1b, c, Extended Data Fig. 1a), suggesting that physiological acetaldehyde accumulation in blood cells is not sufficient to inactivate the homologous recombination repair factor BRCA216. Fancd2−/− mice do not show similar induction following exposure to ethanol; therefore, detoxification is the primary mechanism that prevents DNA damage by aldehydes and alcohol. Finally, the number of SCE events in Aldh2−/−Fancd2−/− mice is indistinguishable from that in Aldh2−/− mice, showing that homologous recombination repair occurs despite inactivation of FANCD2 (Fig. 1c, Extended Data Fig. 1b).
Figure 1. Ethanol induces potent homologous recombination in vivo.
a, Treatment of mice with BrdU for differential labelling of sister chromatids of bone marrow cells in vivo. Some mice were also treated with ethanol, a precursor of acetaldehyde. IP, intraperitoneal injection; BM, bone marrow. b, Representative images of bone-marrow metaphase spreads (n, number of SCEs per metaphase). c, Number of SCEs in the bone marrow of Aldh2−/−Fancd2−/− and control mice (triplicate experiments, 25 metaphases per mouse, n = 75; P calculated by two-sided Mann–Whitney test; data shown as mean and s.e.m.). NS, not significant. d–g, Clonogenic survival of DT40 DNA-repair mutants (triplicate experiments; data shown as mean and s.e.m.).
The repair of aldehyde-induced DNA damage is therefore not limited to the Fanconi anaemia crosslink-repair pathway. As the recombination machinery is essential for mouse development, we used the isogenic chicken B-cell line DT40, which has been used to define the involvement of homologous recombination in crosslink repair14. DT40 cells carrying disruptions of key homologous recombination genes show hypersensitivity to acetaldehyde (Fig. 1d, e), in a similar way to cells lacking the Fanconi anaemia gene FANCC. To test the relationship between the Fanconi anaemia and homologous recombination pathways, we analysed the sensitivity of cells deficient in both FANCC and XRCC2. These cells showed the same sensitivity to cisplatin as the single knockout cells (Fig. 1f), but were much more sensitive to acetaldehyde (Fig. 1g), indicating that homologous recombination repair confers additional acetaldehyde resistance beyond that provided by Fanconi anaemia crosslink repair. In summary, detoxification provides the dominant protection mechanism against endogenous aldehydes; however, when aldehydes damage DNA, cells use both DNA-crosslink and homologous recombination repair.
FANCD2 prevents alcohol-induced genomic instability
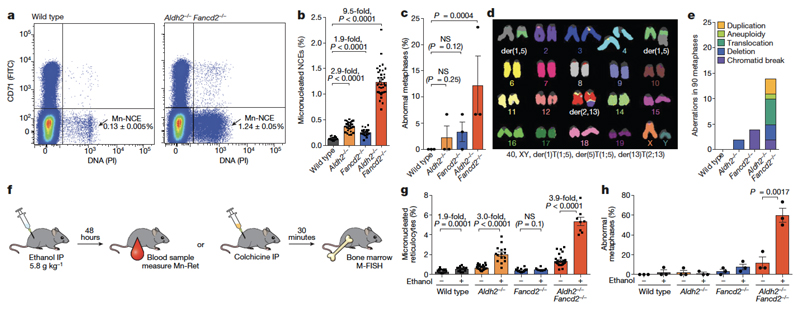
The active DNA-repair response in bone marrow cells indicates that even in the absence of FANCD2, there is an alternative repair response to both endogenous and ethanol-derived aldehydes. However, our previous work has shown that Aldh2−/−Fancd2−/− mice lose the ability to maintain blood production5,6. To determine whether this is due to the accumulation of damaged DNA, we examined haematopoietic cells for evidence of broken chromosomes. One marker of genetic instability is the formation of micronuclei, which are formed from lagging or broken chromosomes. Micronuclei are easily quantified in normochromic erythrocytes (NCEs) in vivo, because they persist following enucleation (Fig. 2a, Extended Data Fig. 1c). There is a significant increase in the proportion of NCEs with micronuclei in both Aldh2−/− (2.9-fold) and Fancd2−/− (1.9-fold) mice compared to wild-type controls, but the increase is much larger in Aldh2−/−Fancd2−/− mice (9.5-fold, Fig. 2b). These micronuclei could represent genomic instability during blood production. We therefore examined cells in metaphase obtained directly from the bone marrow of these mice with multiplex fluorescence in situ hybridization (M-FISH). More than 10% of Aldh2−/−Fancd2−/− bone-marrow cells carried chromosomal aberrations, encompassing all classes of cytogenetic change (Fig. 2c–e). These aberrations are not clonal events, because each karyotype was unique (Extended Data Fig. 1).
Figure 2. Spontaneous and ethanol-induced genomic instability in Aldh2−/−Fancd2−/− mice.
a, Quantification of micronucleated normochromic erythrocytes (Mn-NCE, CD71− PI+) by flow cytometry. b, Percentage of micronucleated normochromic erythrocytes (P calculated by two-sided Mann–Whitney test; data shown as mean and s.e.m.; n = 28, 28, 25 and 37 mice, left to right). c, Percentage of abnormal metaphases in bone marrow cells (P calculated by one-sided Fisher’s exact test; data shown as mean and s.e.m.; three mice per genotype, 30 metaphases per mouse). d, A Aldh2−/−Fancd2−/− metaphase, showing two translocations, see Extended Data Fig. 1f-i for the complete list of aberrations. e, Types of chromosomal aberrations (90 metaphases per genotype). f–h, Treatment of mice with ethanol to assess genomic instability (f) with the micronucleus assay (g) or M-FISH karyotyping (h). Percentage of micronucleated reticulocytes (Mn-Ret, CD71+ PI+) after ethanol treatment (g, P calculated by two-sided Mann–Whitney test; data shown as mean and s.e.m.; n = 29, 15, 25, 15, 20, 10, 28 and 9 mice, left to right). Abnormal metaphases in bone marrow cells after ethanol treatment (h, P calculated by one-sided Fisher’s exact test; data shown as mean and s.e.m.; 3 mice per genotype, 30 metaphases per mouse).
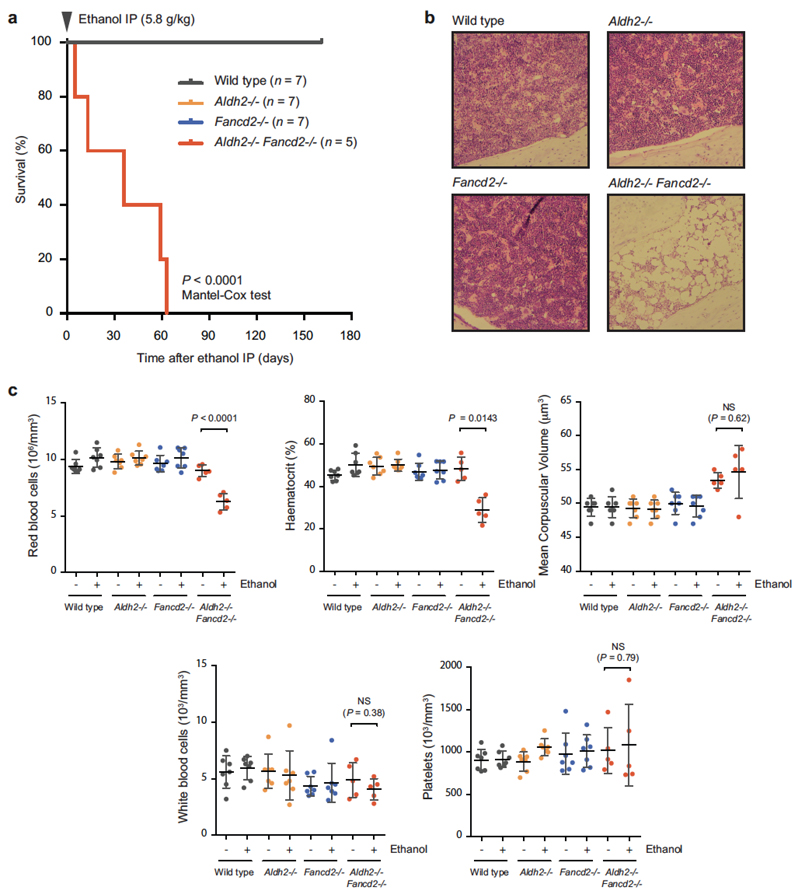
Next, we investigated whether this chromosome damage was exacerbated by exposure to ethanol. As a control, we exposed wild-type or Fancd2−/− mice to mitomycin C (Extended Data Fig. 1d). The experimental scheme (outlined in Fig. 2f) shows how we determined the prevalence of micronuclei in reticulocytes and aberrant metaphases following exposure to ethanol. A single dose of ethanol caused a marked increase in the proportion of reticulocytes containing micronuclei in Aldh2−/− mice. Notably, this induction was comparable to that observed in wild-type mice following exposure to agents known to induce genome instability, such as ionizing irradiation or vincristine (Extended Data Fig. 1e). However, there was a stronger induction of micronucleus formation in Aldh2−/−Fancd2−/− mice than in controls (Fig. 2g), which was accompanied by a striking increase in the number of abnormal metaphases, with almost 60% of metaphases having damaged chromosomes following ethanol exposure (Fig. 2h, Extended Data Fig. 1g–i). These mice rapidly lost the ability to produce blood and died from bone-marrow failure (Extended Data Fig. 2). These results show that, despite activation of homologous recombination, the Fanconi anaemia crosslink-repair pathway is essential for preventing chromosome breakage and loss of blood homeostasis in response to aldehydes.
Ku70 contributes to repair of aldehyde-induced breaks
The presence of chromosome breaks and translocations indicates that aldehydes cause double-stranded breaks, which could be processed by non-homologous end-joining (NHEJ) repair17. Previous studies in cell lines and nematodes have indicated that, in the absence of the Fanconi anaemia pathway, engagement of double-stranded breaks by NHEJ leads to further genomic instability18,19. Therefore, we investigated whether NHEJ and Fanconi anaemia repair are redundant in resolving endogenous DNA damage in HSCs, and whether there is a role for NHEJ in maintaining resistance to acetaldehyde.
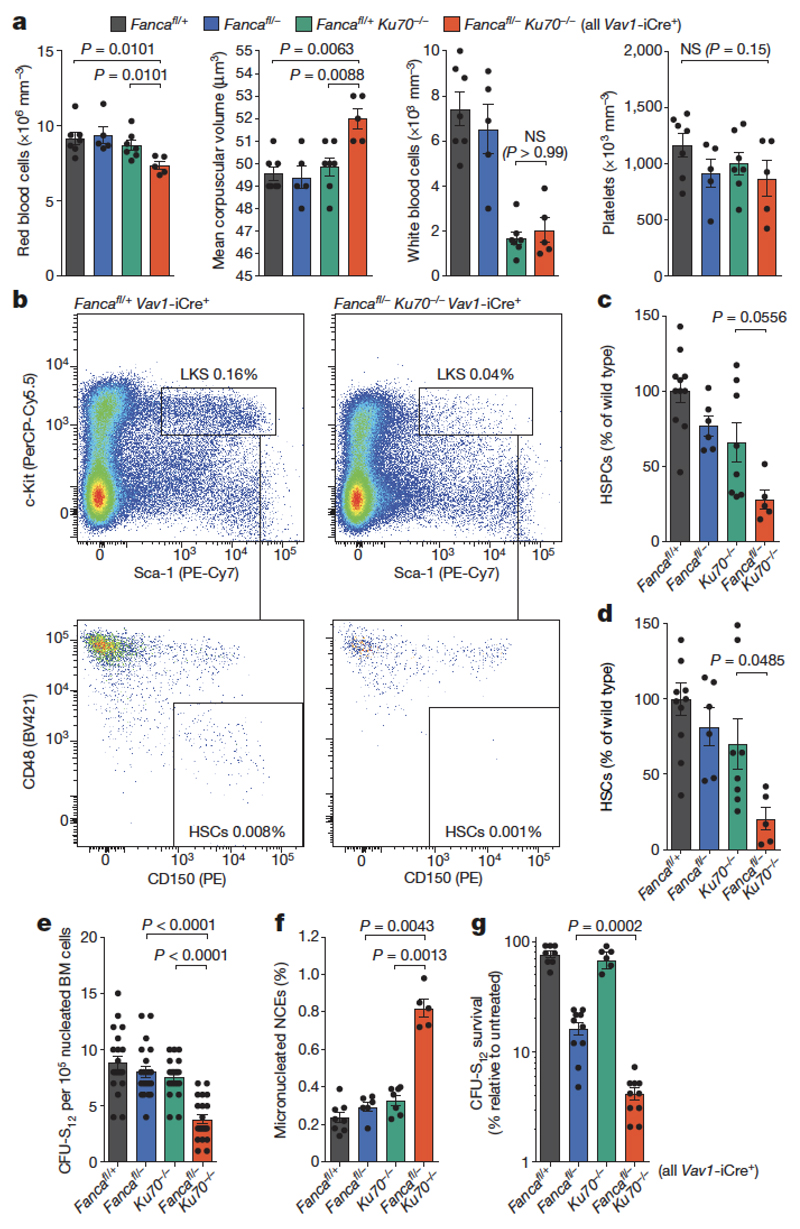
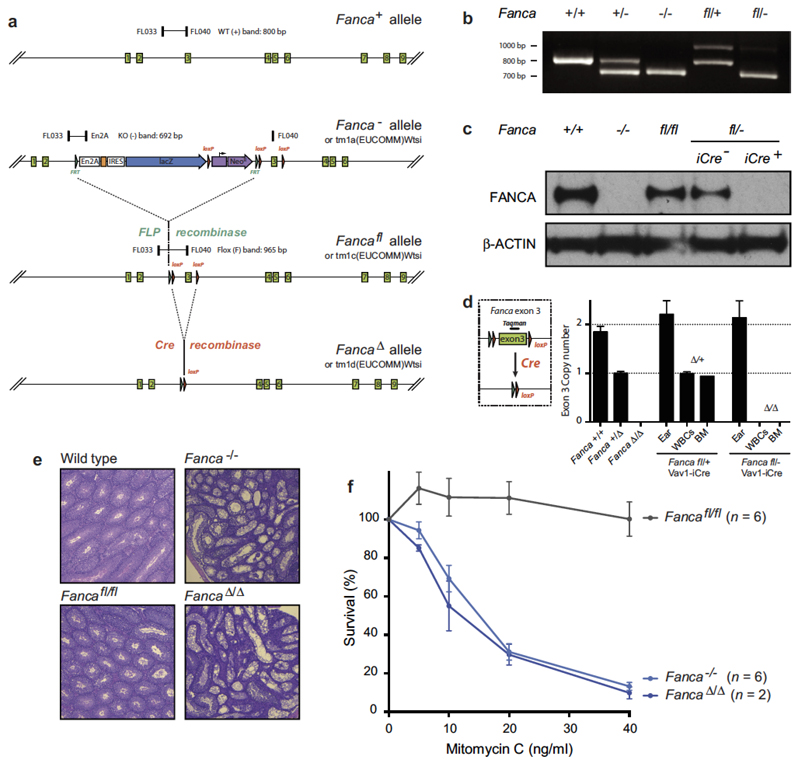
To do this, we crossed mice deficient in the known Fanconi anaemia repair gene Fanca with mice lacking the key NHEJ factor Ku70. We failed to obtain Fanca−/−Ku70−/− mice, indicating that there was a synthetic lethal interaction between Ku70-dependent NHEJ and Fanconi anaemia-crosslink repair (Supplementary Information Table 1). To bypass embryonic lethality, we generated blood-specific Fanca knockout mice (Extended Data Fig. 3) and crossed them with Ku70+/− mice to produce mice that had the double mutation in the blood compartment (Fancafl/-Ku70−/− Vav1-iCre). These mice were viable, indicating that the embryonic lethality of Fanca−/−Ku70−/− is not due to failed blood production (Supplementary Information Table 1). However, blood counts show that Fancafl/-Ku70−/− Vav1-iCre mice are anaemic (Fig. 3a) and have fewer HSCs compared to congenic controls (Fig. 3b–e). Fancafl/-Ku70−/− Vav1-iCre mice also display genomic instability, with increased frequency of micronuclei-containing NCEs (Fig. 3f). Finally, we tested whether Ku70 was required to maintain resistance of short-term (ST) HSCs to aldehydes by exposing bone marrow cells to acetaldehyde in vitro before injecting them into lethally irradiated recipients. Fancafl/-Ku70−/− Vav1-iCre ST-HSCs were much more sensitive to acetaldehyde than either of the single mutant ST-HSCs (Fig. 3g). These results indicate that in the mouse haematopoietic system, in the absence of Fanconi anaemia repair, NHEJ is required to provide resistance to endogenous and acetaldehyde-induced DNA damage. This result contrasts with the reported negative impact of active NHEJ on the viability of Fanconi anaemia-deficient chicken DT40 cells and worms.
Figure 3. NHEJ cooperates with the Fanconi anaemia pathway to maintain HSC integrity, genomic stability and cellular resistance to aldehydes.
a, Blood parameters of 8- to 12-week old mice (P calculated by two-sided Mann–Whitney test; data shown as mean and s.e.m.; n = 8, 6, 7 and 5 mice, left to right). RBC, red blood cells; MCV, mean corpuscular volume; WBC, white blood cells. b, Representative flow cytometry plot of HSPCs of Fancafl/-Ku70−/− Vav1-iCre mice and control. LKS, Lin−Kit+Sca-1+. c, d, Quantification of HSPCs (Lin−Kit+Sca-1+) and HSCs (Lin−Kit+Sca-1+CD48−CD150+) by flow cytometry (P calculated by two-tailed Student’s t-test; data shown as mean and s.e.m.; n as in a). e, Counts from colony-forming unit-spleen (CFU-S) assays in the bone marrow of Fancafl/-Ku70−/− Vav1-iCre and control mice. Each point represents the number of CFU-S12 in a single recipient (P calculated by two-sided Mann–Whitney test; data shown as mean and s.e.m.; n = 20 mice). f, Frequency of Mn-NCE (P calculated by two-sided Mann–Whitney test; data shown as mean and s.e.m.; n as in a). g, Survival of CFU-S12 after treatment with 4 mM acetaldehyde for 4 h, relative to untreated samples (P calculated by two-sided Mann–Whitney test; data shown as mean and s.e.m.; n = 10 mice).
Aldehyde-damaged HSCs are functionally compromised
Our results so far indicate that endogenous aldehydes give rise to double-stranded breaks in the absence of Fanconi anaemia repair, which are engaged by homologous recombination and NHEJ, but ultimately rearrange chromosomes in bone marrow cells. A key question is whether endogenous DNA damage and subsequent mutations accumulate in the HSC compartment. This is a critical question because there is evidence that HSCs differ in their DNA repair capacity and response compared to later progenitors11. Two obstacles had to be overcome in order to establish whether endogenous aldehydes mutate the genomes of these vital cells. First, the stochastic nature of DNA damage makes it unlikely that the same mutation will occur in multiple cells. Second, the scarcity of HSCs, especially in the case of Aldh2−/−Fancd2−/− mice, precludes the use of most conventional techniques to assess DNA damage. We also wanted to ascertain whether mutations arise in functional stem cells, and therefore avoided whole-genome amplification or short-term in vitro expansion of cells isolated by flow cytometry. Instead, we decided to define HSCs functionally and exploit the ability of a single HSC to reconstitute long-term blood production following transplantation into a lethally irradiated mouse20.
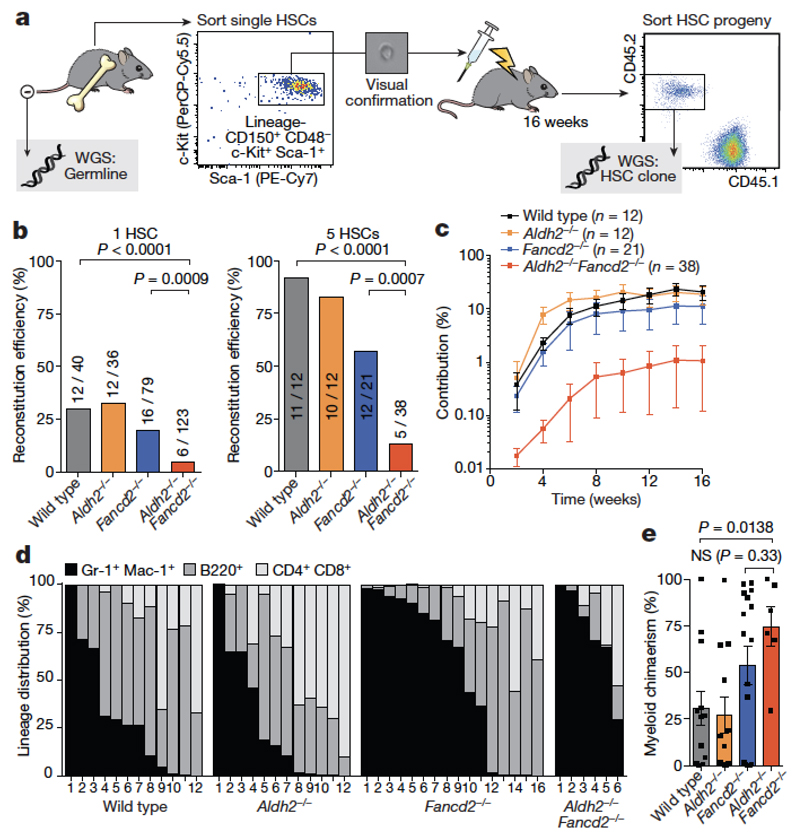
Our approach combines transplantation of a single HSC with whole-genome sequencing to obtain the mutational landscape of stem cells, while also allowing us to assess the functional capacity of mutant HSCs (Fig. 4a). We carried out transplants with one or five Aldh2−/−Fancd2−/− HSCs (Fig. 4b). These stem cells rarely engrafted (with a frequency of 4.8%), contributed less to haematopoiesis and were myeloid-biased compared to controls (Fig. 4c–e). These results indicate that Aldh2−/−Fancd2−/− HSCs are severely functionally compromised and share features with aged HSCs21,22.
Figure 4. Single HSC transplantation reveals that Aldh2−/−Fancd2−/− HSCs are functionally compromised.
a, Transplantation of a single HSC for the generation of HSC clones in vivo. The HSC progeny (CD45.2+) were recovered after four months and analysed by whole-genome sequencing, alongside a germline reference. b, Percentage and number of irradiated recipients that were positive for reconstitution by one or five transplanted HSCs (P calculated by two-sided Fisher’s exact test). c, Contribution to blood production over time by five transplanted HSCs (data shown as mean and s.e.m.). d, Lineage composition of single-HSC clones four months after transplant; columns represent individual recipients or clones. e, Proportion of myeloid (Gr-1+Mac-1+) HSC-derived white blood cells (P calculated by two-sided Mann–Whitney test; data shown as mean and s.e.m.; n = 12, 12, 16 and 6 HSC clones, left to right).
Mutational landscape of aldehyde-damaged stem cells
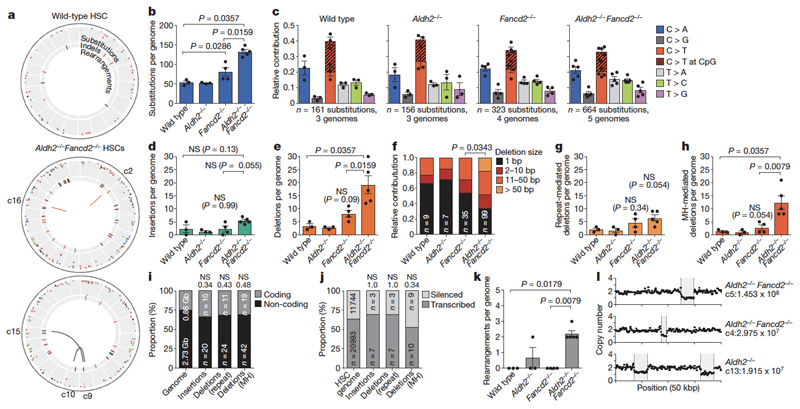
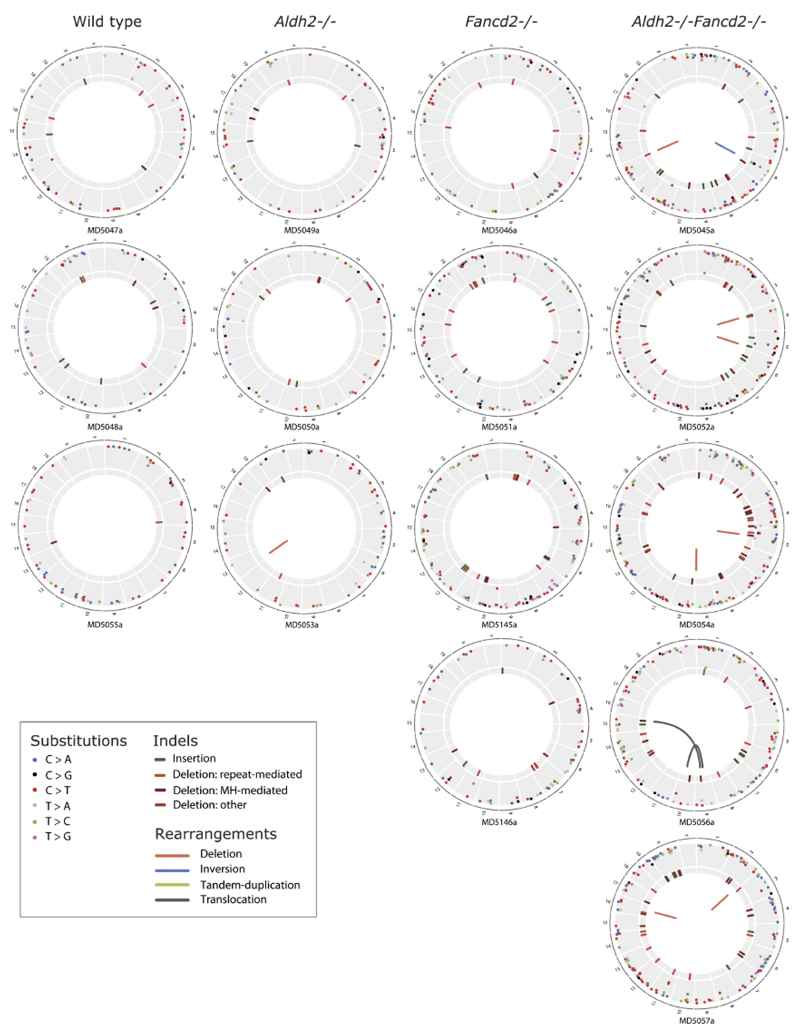
Our ultimate goal was to obtain clonal blood, which provided us with a physiological method to amplify stem-cell genomes. As outlined (Fig. 4a), four months after transplantation, we isolated the CD45.2+ HSC progeny and performed whole-genome sequencing at 20× coverage; tail DNA from the donor mouse served as the germline reference. This allowed us to detect heterozygous somatic changes, which are absent in the matched germline reference and represent mutations in the HSC. Genomes of Aldh2−/−Fancd2−/− HSCs were mutated with increased prevalence of indels, rearrangements and translocations (Fig. 5a, Extended Data Fig. 4).
Figure 5. Endogenous aldehydes mutate the HSC genome.
a, Circos plots showing mutations in three HSCs. All HSCs are shown in Extended Data Fig. 4. b, d, e, g, h, k, Mutations of different classes per genome (b, number of substitutions; P calculated by two-sided Mann–Whitney test; data shown as mean and s.e.m.; n = 3, 3, 4 and 5 HSC genomes, left to right). c, Point mutation classes in HSC genotypes. d, Number of insertions per genome. e, Number of deletions per genome. f, Distribution of the size of deletions (χ2 test, n shows number of deletions). g, Number of repeat-mediated deletions per genome. h, Number of microhomology (MH)-mediated deletions per genome. i, j, Indels in Aldh2−/−Fancd2−/− HSCs are randomly distributed: within or outside genes (i) (P calculated by hypergeometric distribution, n is number of indels), or between expressed or silenced genes (j) (P calculated by binomial distribution, n is number of indels). Numbers above columns, P values. k, Number of rearrangements per genome. l, Large copy-number losses in Aldh2−/−Fancd2−/− and Aldh2−/− HSCs at the indicated locations.
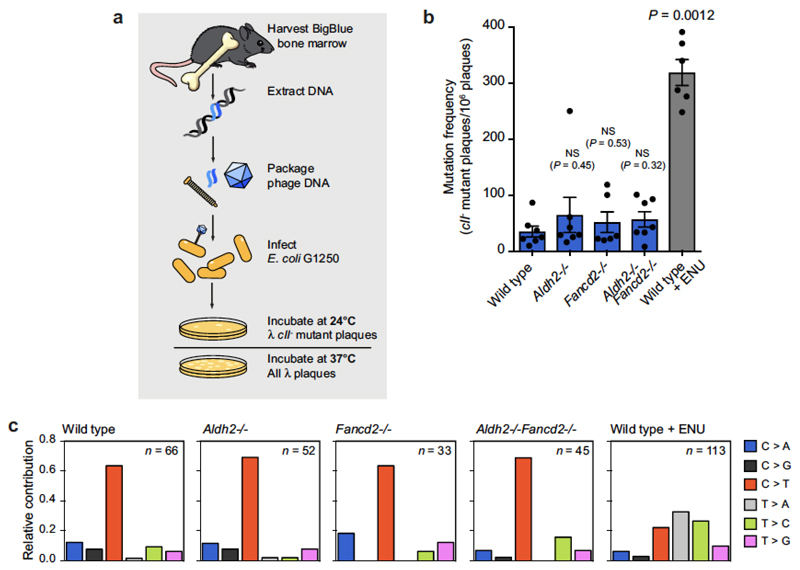
Although the number of single-base substitutions was significantly higher in Aldh2−/−Fancd2−/− genomes (Fig. 5b), the total numbers were low and no changes were detected in the type of substitutions (Fig. 5c). We also found no difference in the frequency or pattern of point mutations in bone marrow cells of Aldh2−/−Fancd2−/− mice using the Select-cII Big Blue in vivo reporter assay (Extended Data Fig. 5).
A limitation of our approach is that cells with the capacity to engraft may represent the least mutated HSCs. Nevertheless, we observed significant increases in the frequency of deletions, which were more prevalent (Fig. 5d, e) and larger (Fig. 5f) in Aldh2−/−Fancd2−/− genomes. The mean variant allele frequency (VAF) for all filtered indels was 0.47, establishing that these changes are of clonal origin. By examining the flanking regions, we found that microhomology-mediated deletions are the main contributors to the mutations observed in Aldh2−/−Fancd2−/− HSCs, indicative of NHEJ repair of double-stranded breaks23 (Fig. 5g, h). Additionally, the increase in the size of the deletions (Fig. 5f) suggests a role for alternative end-joining in the repair of some of these breaks, as alternative end-joining is characterized by increased resection in comparison to classical NHEJ24. Next, we analysed the location of indels across the genome, as recent work has suggested a role for the Fanconi anaemia pathway in preventing genomic instability at transcription–replication collisions25,26. However, we found no evidence of microhomology-mediated deletions being enriched at coding regions or transcribed genes (Fig. 5i, j), suggesting stochastic double-stranded break formation in Aldh2−/−Fancd2−/− HSCs.
The most striking change in Aldh2−/−Fancd2−/− HSCs was the presence of rearrangements that were not detected in most controls. Aldh2−/−Fancd2−/− stem cells contained on average two rearrangements per genome; in contrast, we observed only two large deletions among all ten control HSC genomes (Fig. 5k–l). In summary, these data provide the first whole-genome sequences obtained from single stem cells propagated in vivo. These stem cell genomes show that endogenous aldehydes induce a tapestry of inter-chromosomal changes that are mediated by mutagenic end-joining of DNA double-strand breaks.
A p53 response removes aldehyde-damaged HSCs
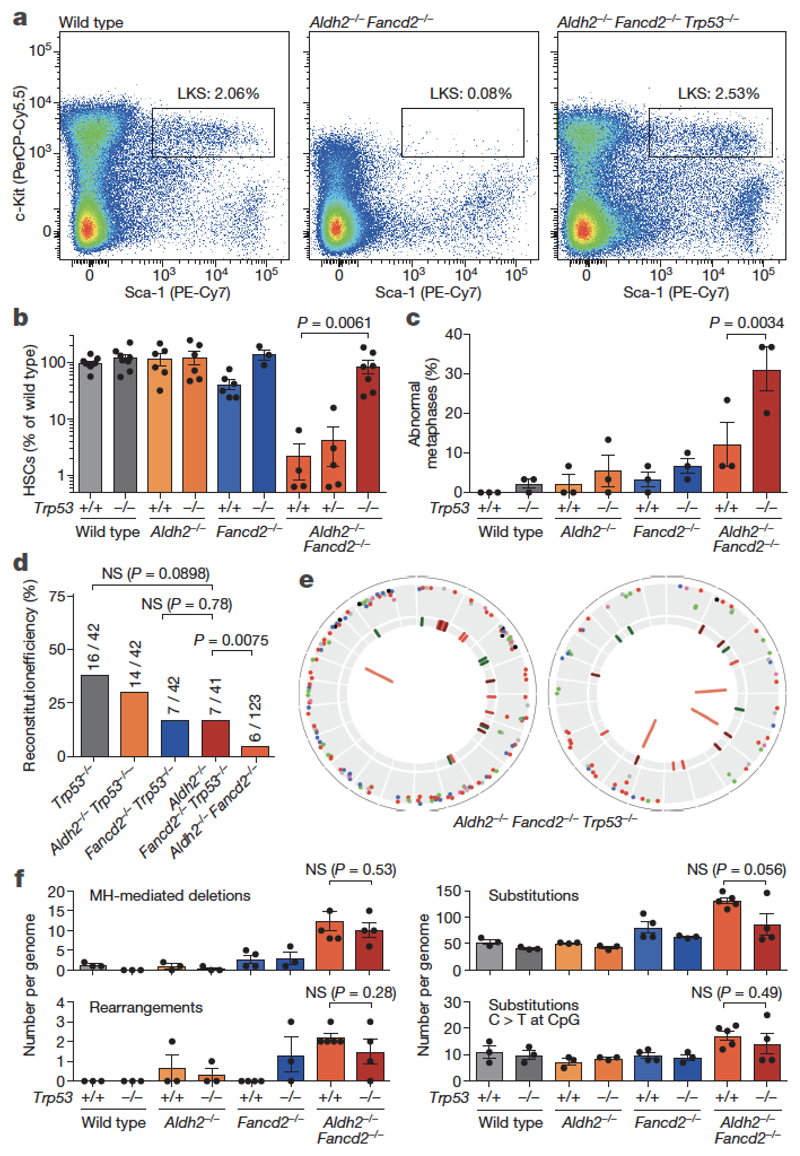
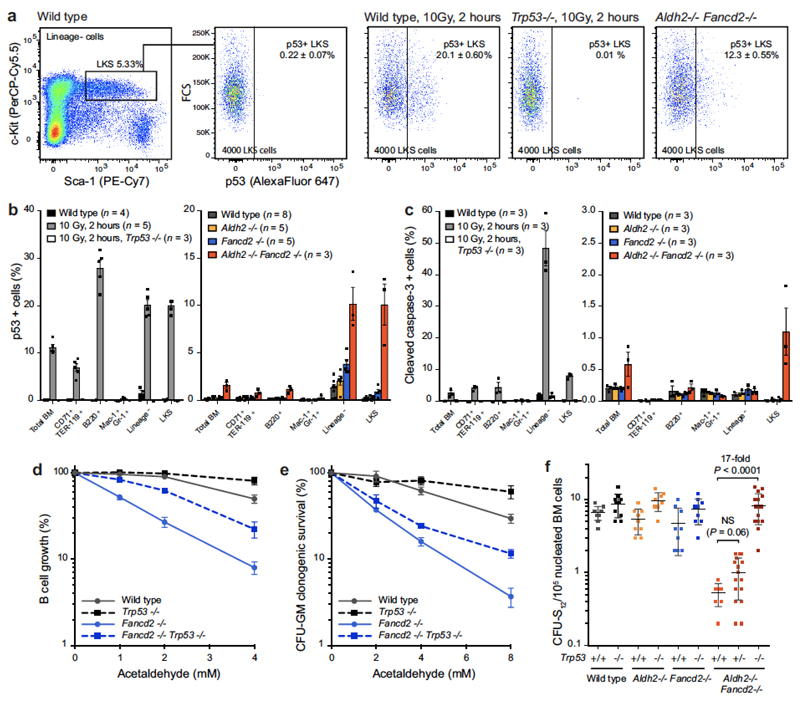
Strikingly, most Aldh2−/−Fancd2−/− HSCs failed to engraft (Fig. 4b). It is possible that HSCs that carry heavy DNA-damage burdens are eliminated, and selection pressure favours the survival of less damaged stem cells. It was therefore important to determine the mechanism of HSC loss in Aldh2−/−Fancd2−/− mice, and if attenuated, it would be important to determine the mutagenic consequences. p53 protein regulates the cellular response to DNA damage and, when activated, induces restorative processes or apoptosis. We found that Aldh2−/−Fancd2−/− haematopoietic stem and progenitor cells (HSPCs) accumulated p53 and cleaved caspase-3, indicating that endogenous-aldehyde stress activates the p53 response (Extended Data Fig. 6a–c). Furthermore, we found that genetic ablation of p53 partially suppressed the acetaldehyde hypersensitivity of Fancd2-deficient splenic B cells and granulocyte/macrophage colony forming units (Extended Data Fig. 6d, e).
We therefore generated Aldh2−/−Fancd2−/−Trp53−/− triple-knockout mice. The severe HSC depletion of Aldh2−/−Fancd2−/− mice was completely rescued in the triple knockouts (Fig. 6a, b). In addition, the triple-knockout stem cells were functional, as the mice showed a complete rescue in the frequency of ST-HSCs upon bone-marrow transplantation (Extended Data Fig. 6f). Moreover, p53 deficiency fully restored the blood cytopenias of untreated Aldh2−/−Fancd2−/− mice and made these mice more resistant to alcohol exposure (Extended Data Fig. 7). Notably, Trp53 deletion did not rescue the embryonic lethality of Aldh2−/−Fancd2−/− embryos (in Aldh2−/− mothers), suggesting that a different checkpoint might mediate developmental failure (Supplementary Information Table 2).
Figure 6. A p53 response depletes aldehyde-damaged HSCs.
a, Representative flow cytometry plot of HSPCs (LKS). b, Quantification of HSCs as determined by flow cytometry (P calculated by two-sided Mann–Whitney test; data shown as mean and s.e.m.; n = 9, 8, 6, 6, 6, 3, 4, 5 and 7 mice, left to right). c, Frequency of abnormal metaphases in bone marrow cells (P calculated by two-sided Fisher’s exact test; data shown as mean and s.e.m.; 3 mice per genotype, 30 metaphases per mouse). See Extended Data Fig. 8b-d for a complete list of rearrangements. d, Proportion and number of irradiated recipients that were positive for reconstitution by transplantation of single HSCs (P calculated by two-sided Fisher’s exact test). e, Mutations in two Aldh2−/−Fancd2−/−Trp53−/− HSCs. f, Number of microhomology-mediated deletions, rearrangements, substitutions and clock substitutions per genome (P calculated by two-sided Mann–Whitney test; data shown as mean and s.e.m.; n = 3, 3, 3, 3, 4, 3, 5 and 4 HSC genomes, left to right).
We reasoned that the rescue of haematopoiesis in Aldh2−/−Fancd2−/−Trp53−/− mice must be occurring at the cost of genome integrity. Although the level of micronucleated NCEs in the blood of Aldh2−/−Fancd2−/−Trp53−/− mice appeared similar to that of Aldh2−/−Fancd2−/− mice (Extended Data Fig. 8a), we noticed a significant (P 0.0034) increase in chromosome rearrangements in Aldh2−/−Fancd2−/−Trp53−/− mice, as seen by M-FISH analysis of total bone marrow cells (Fig. 6c, Extended Data Fig. 8). However, neither of these analyses tell us whether genome stability is similarly compromised in Aldh2−/−Fancd2−/−Trp53−/− HSCs. We therefore performed transplantation of single HSCs combined with whole-genome sequencing, as described earlier, and observed that p53 deficiency partially rescued the engraftment defect of Aldh2−/−Fancd2−/− HSCs (Fig. 6d). Surprisingly, the genomes of Aldh2−/−Fancd2−/−Trp53−/− stem cells did not carry a greater mutation burden than those of Aldh2−/−Fancd2−/− HSCs (Fig. 6e, f). Indel and rearrangement calls were validated by targeted deep sequencing and PCR, respectively (Extended Data Figs 9, 10 and Methods). One plausible explanation for the lack of increased mutagenesis in Aldh2−/−Fancd2−/−Trp53−/− HSCs is the possibility that the very small number of HSCs in Aldh2−/−Fancd2−/− mice might have undergone more replicative cycles, thereby accruing a larger number of mutations. To address this, we quantified the frequency of ‘clock’ mutations (C to T at CpG sites), but this analysis showed no significant difference between Aldh2−/−Fancd2−/− and Aldh2−/−Fancd2−/−Trp53−/− HSCs (Fig. 6f). These results indicate that aldehyde-induced DNA damage induces p53 leading to HSC attrition, which is inconsistent with p53 being a negative regulator of Fanconi anaemia repair, as recently reported27. However, while Trp53 deletion completely rescues HSC depletion, this does not occur at the expense of genome stability in stem cells.
Discussion
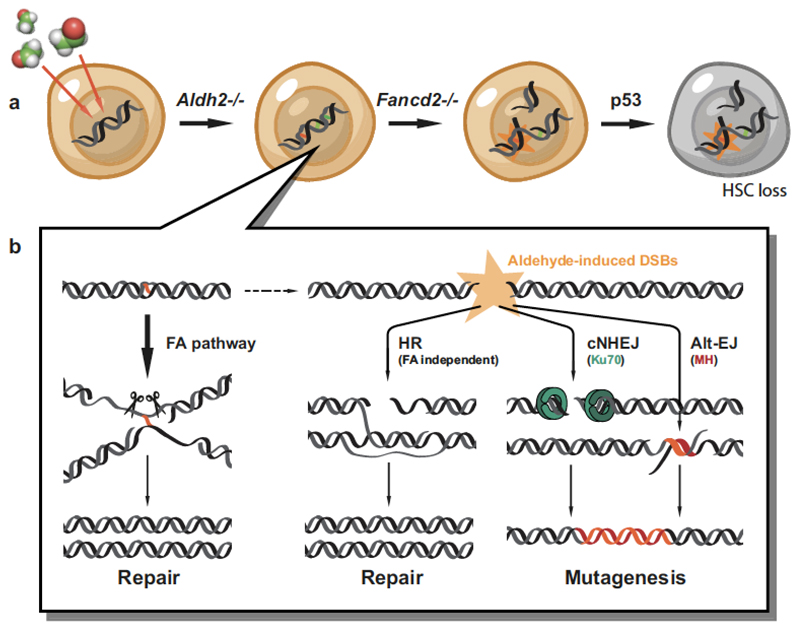
These results outline the mechanisms by which the mouse haematopoietic system and, more specifically, blood stem cells respond to an endogenous and alcohol-derived source of DNA damage. Primary protection against acetaldehyde is provided by Aldh2-mediated detoxification and, when this is lost or saturated, acetaldehyde damages DNA. The Fanconi anaemia pathway is the principal mechanism to counteract this damage, but NHEJ and homologous recombination can also deal with these lesions. These results therefore illustrate that coordinated pathway choice is critical for maintaining genome stability upon aldehyde exposure. The Fanconi anaemia pathway prevents aldehyde lesions from degenerating into double-stranded breaks, the illegitimate repair of which leads to a characteristic pattern of mutagenesis in HSCs (Extended Data Fig. 11). Aldehydes are capable of forming a diverse range of DNA lesions—from base adducts to DNA–DNA or DNA–protein crosslinks. The known molecular function of the Fanconi anaemia pathway suggests that the most physiologically toxic lesion caused by aldehydes may be a DNA interstrand crosslink. However, if it is indeed an interstrand crosslink, then the factors involved in translesion synthesis or homologous recombination processes are distinct from the previously described mechanism of interstrand-crosslink repair6,14,15,28. It will be important to resolve the nature of the lesion and the precise mechanics of its repair.
HSCs mutated by aldehydes are functionally compromised and display myeloid bias. The p53 response is critical in driving the loss in number and function of HSCs. Although Trp53 deletion rescues HSC defects, this, paradoxically, does not result in further genomic instability at the single HSC level. It is important to emphasize, however, that the pool of HSCs is larger, and therefore there is still an overall increase in mutation. Nevertheless, our work implies that the relationship between p53, DNA repair and genome stability is more complex in stem cells than previously appreciated. The central role for ALDH2 in removing genotoxic aldehydes has implications for the more than 540 million people who are deficient in ALDH2 activity. Alcohol exposure in such individuals may cause DNA double-strand breaks and chromosome rearrangements3. This large population may also be susceptible to alcohol-induced age-related blood disorders. More generally, this research provides a simple plausible explanation for the established epidemiological link between alcohol consumption and enhanced cancer risk4,29.
Methods
Mice
Aldh2−/−Fancd2−/− mice were generated on a C57BL/6 × 129S4S6/Sv F1 background. To this end, the previously reported Fancd2 allele (Fancd2tm1Hou; MGI ID: 2673422, a gift from M. Grompe) was back-crossed onto the C57BL/6Jo1a background for 10 generations and crossed with Aldh2+/− (C57BL/6N) mice to generate Aldh2+/−Fanca+/− mice on a pure C57BL/6 background. Likewise, the previously reported Aldh2 allele (Aldh2tm1a(EUCOMM)Wtsi; MGI ID: 4431566, EUCOMM6) was backcrossed from C57BL/6N onto 129S6/Sv for five generations and crossed with Fancd2+/− mice to generate Aldh2+/−Fancd2+/− mice on a 129S4S6/Sv background. Finally, Aldh2−/−Fancd2−/− and control mice were generated as F1 hybrids from crosses between Aldh2+/−Fancd2+/− females (129S4S6/Sv) and Aldh2+/−Fancd2+/− males (C57BL/6).
To generate Fanca−/−Ku70−/− mice on a pure C57BL/6 background, Fanca+/− mice (Fancatm1a(EUCOMM)Wtsi; MGI ID: 4434431, C57BL/6N, EUCOMM5) were crossed with Ku70+/− or Xrcc6+/− mice (Xrcc6tm1Fwa, MGI ID: 217995430), and the Fanca+/−Ku70+/− progeny were then intercrossed to generate all possible genotypes. Pups from these crosses were genotyped at between two and three weeks old. For the generation of Fancafl/-Ku70−/− Vav1-iCre+ tissue-specific double mutants (also on a pure C57BL/6 background), Fanca+/− mice were first crossed with FLP deletor mice31 to produce the Fanca floxed allele (Fancafl or Fancatm1c(EUCOMM)Wtsi). Recombination of the Frt sites was verified by PCR (using the primers FL033, GCCTTTGCTGCTCTAATTCCATGT; FL040, TCAGCTCACTGAGACGCAACCTTTT ACACT; and En2A, GCTTCACTGAGTCTCTGGCATCTC), and reconstitution of FANCA expression was verified by western blotting spleen extracts of Fancafl/fl mice (Extended Data Fig. 3). Fanca+/fl mice were then crossed with Ku70+/− mice to eventually produce Fancafl/flKu70+/− mice. Finally, these mice were crossed with Fanca+/−Ku70+/− Vav1-iCre to generate Fancafl/−Ku70−/−Vav1-iCre and control mice. The Vav1-iCre allele directs the expression of the iCre recombinase to HSCs and haematopoietic tissues32, and in this case yields the Fanca-null allele (FancaΔ or Fancatm1d(EUCOMM)Wtsi). FancaΔ/Δ mice phenocopy the Fanca−/− mice reported previously, as judged by FANCA expression, sterility and sensitivity to mitomycin C (Fig. 3, Extended Data Fig. 3).
Similarly to Aldh2−/−Fancd2−/− F1 mice, Aldh2−/−Fancd2−/−Trp53−/− mice were also generated in a C57BL/6 × 129S4S6/Sv F1 background. In brief, the Trp53 allele reported previously33 was backcrossed onto 129S6/Sv or C57BL/6J for six generations. Trp53+/− mice were then intercrossed with Aldh2−/−Fancd2+/− mice to establish parental (F0) strains on both genetic backgrounds, which were finally crossed to obtain Aldh2−/−Fancd2−/−Trp53−/− and control F1 mice.
For single HSC transplantation experiments, we used CD45.1 homozygous mice on a C57BL/6J × 129S6/Sv F1 background as recipients. CD45.1 (or Ptprca) had been serially backcrossed from B6.SJL onto 129S6/Sv for six generations, with selection at each generation by serotyping with anti-CD45.1 (A20, FITC, BioLegend) and anti-CD45.2 antibodies (104, PE-Cy7, BioLegend).
For the in vivo point-mutation assay, mice carrying BigBlue λLIZ shuttle vector repeats (Stratagene) were crossed with Aldh2−/−Fancd2+/− mice on a C57BL/6 × 129S4S6/Sv hybrid background. The resulting mice were intercrossed to obtain Aldh2−/−Fancd2−/− BigBlue λLIZ and control mice.
For the analysis of Mendelian segregation of alleles, sample size was determined by power analysis using http://biomath.info/power/chsq1gp.htm. Sufficient mice to detect a 50% reduction in expected frequency were used, using power of 0.8 and alpha 0.05. No statistical methods were used to predetermine sample size in the other animal experiments. No randomization was employed. The investigators were blinded to the genotypes of mice throughout the study and data were acquired by relying purely on identification numbers.
All animals were maintained in specific pathogen-free conditions. In individual experiments all mice were matched for gender and age (8–12 weeks). All animal experiments undertaken in this study were done so with the approval of the UK Home Office.
Ethanol treatment
For acute ethanol exposure, Aldh2−/−Fancd2−/− mice and appropriate controls were injected intraperitoneally with ethanol. The total dose of 5.8 g kg−1 was split into two injections separated by 4 h. Ethanol (96%, Sigma) was diluted to 28% v/v in saline, and administered twice at 13 ml kg−1. Mice were exsanguinated 48 h after the second injection and peripheral blood was analysed with the micronucleus assay. Alternatively, mice were injected with colchicine for the preparation of metaphase spreads for M-FISH. For SCE analysis, mice were injected with colchicine 12 h after the second ethanol injection.
For chronic ethanol treatment of Aldh2−/−Fancd2−/−Trp53−/− and control mice, ethanol was administered in drinking water for ten days as reported previously6. For the first five days, the drinking water supply was replaced by a solution of 10:15:75 blackcurrant Ribena:ethanol:water, followed by a 10:20:80 solution for the last five days. A 50 μl blood sample was taken from tail veins before alcohol exposure, and by cardiac puncture at the end of the experiment, to measure full blood counts. Femurs were dissected for histological analysis and to determine bone marrow cellularity.
Preparation of mouse bone marrow for metaphase spreads
Treated or untreated young mice (8–12 weeks of age) were injected intraperitoneally with 100 μl of colchicine (0.5% w/v in water, Sigma). After 30 min the mice were culled by cervical dislocation, femurs were harvested and placed in ice-cold PBS. Bone marrow cells were flushed with 10 ml of pre-warmed hypotonic solution (75 mM KCl, 37 °C) through a 70-μm cell strainer and incubated for 15 min in a water bath at 37 °C. After the incubation, 1 ml of fixative (3:1 methanol:acetic acid) was added dropwise to the hypotonic buffer, and mixed by gentle inversion of the tube. The tubes were spun down for 10 min at 250g and the supernatant was aspirated, leaving 50 μl and the cell pellet in the tube. The cells were resuspended by flicking the base of the tube very gently, 3 ml of fixative were added dropwise and the volume was made up to 10 ml by pipetting fixative down the side of the tube. The cells were incubated at room temperature for 30 min and stored at −20 °C until further use.
SCE assay for mouse bone marrow
The staining of metaphase spreads for the quantification of SCEs was adapted from published protocols34,35. A 50 mg BrdU slow-release pellet (Innovative Research of America) was surgically implanted subcutaneously into 8-to-12-week-old mice. Unchallenged mice were injected with colchicine 24 h later and metaphases were prepared after 30 min. Mice challenged with ethanol were injected intraperitoneally with ethanol 8 h and 12 h after implantation of the BrdU pellet. A total ethanol dose of 5.8 g kg−1 was split between these two doses as described previously. Metaphases were prepared as outlined above. Cells were then dropped from a height of 30 cm onto chilled, humidified slides. The slides were then dried for 1 h at 62 °C in a hybridization oven. Cells were washed in 2× SSC for 5 min at room temperature. Cells were stained for 15 min at room temperature with 1 μg ml−1 Hoechst 33258 pentahydrate (H3569, Molecular Probes) in 2× SSC. The slides were then transferred to a Petri dish with 2× SSC and exposed to UV irradiation for 30 min in a Stratalinker Crosslinker (Stratagene). The slides were then dehydrated by passing them through an ethanol series (70%, 96% and 100%) and placed in PBS for 5 min at room temperature. The DNA was denatured by exposure to 0.07 N NaOH for 2 min at room temperature. The slides were then washed three times in PBS for 5 min. The slides were then blocked in PBS, 1% BSA, 0.5% Tween-20 for 1 h at room temperature and stained overnight with a FITC-conjugated mouse anti-BrdU antibody (Clone B44, BD Biosciences) diluted 1:1 in PBS, 3% BSA, 0.5% Tween-20 at room temperature. The slides were then washed three times with PBS, 1% BSA, 0.5% Tween-20 for 5 min at room temperature and stained with goat anti-mouse Alexa Fluor-488 secondary antibody (A-11001, Life Technologies) diluted 1:500 in PBS, 1% BSA, 0.5% Tween-20 for 6 h at room temperature. The slides were then washed three times in PBS, 1% BSA, 0.5% Tween-20 for 15 min and stained with Hoechst 33342 trihydrochloride (H3570, Molecular Probes) diluted 1:2000 in PBS for 15 min at room temperature. The slides were then washed three times in PBS for 10 min on each occasion, washed once in water for 5 min, mounted with ProLong Gold Antifade Mountant (P36930, Molecular Probes) and coverslips were lowered onto the slides. Thirty metaphases were captured per sample using a Zeiss LSM 780 confocal microscope (Zeiss). The number of sister-chromatid exchanges per metaphase was then counted blind.
M-FISH karyotyping
For M-FISH, chromosome-specific DNA libraries were generated from flow-sorted chromosomes provided by the Flow Cytometry Core Facility of the Wellcome Trust Sanger Institute, using the GenomePlex Complete whole-genome amplification kit (Sigma–Aldrich). A mouse 21-colour painting probe was prepared following the pooling strategy36. Five mouse-chromosome pools were each labelled with ATTO 425-, ATTO 488-, Cy3-, Cy5- and Texas Red-dUTPs (Jena Bioscience), respectively, using the GenomePlex WGA reamplification kit (Sigma–Aldrich) and a dNTP mixture as described previously37. The labelled products were pooled and sonicated to achieve a size range of 200–1,000 bp, optimal for use in chromosome painting. The sonicated DNA was ethanol-precipitated together with mouse Cot-1 DNA (Thermo Fisher Scientific), and resuspended in a hybridization buffer (50% formamide, 2× SSC, 10% dextran sulfate, 0.5 M phosphate buffer, 1× Denhardt’s solution, pH 7.4). Bone marrow cells suspended in fixative as described above (3:1 methanol:acetic acid) were dropped onto precleaned microscope slides, followed by fixation in acetone (Sigma–Aldrich) for 10 min and dehydration through an ethanol series (70%, 90% and 100%). Metaphase spreads on slides were denatured by immersion in an alkaline denaturation solution (0.5 M NaOH, 1.0 M NaCl) for 2 min, followed by rinsing in 1 M Tris-HCl (pH 7.4) solution for 3 min, 1× PBS for 3 min and dehydration through a 70%, 90% and 100% ethanol series. The M-FISH probes were denatured at 65 °C for 10 min before being applied onto the denatured slides. The hybridization area was sealed with a 22 mm × 22 mm coverslip and rubber cement. Hybridization was carried out in a 37 °C incubator for approximately 44–48 h. The post-hybridization washes included a 5-min stringent wash in 0.5× SSC at 75 °C, followed by a 5 min rinse in 2× SSC containing 0.05% Tween-20 (VWR) and a 2 min rinse in 1× PBS, both at room temperature. Finally, slides were mounted with SlowFade Gold mounting solution containing DAPI (Thermo Fisher Scientific). M-FISH images were visualized on a Zeiss AxioImager D1 fluorescent microscope equipped with narrow band-pass filters for DAPI, DEAC, FITC, Cy3, Texas Red and Cy5 fluorescence and an ORCA-EA CCD camera (Hamamatsu). M-FISH digital images were captured using the SmartCapture software (Digital Scientific UK), and processed using the SmartType Karyotyper software (Digital Scientific UK). Thirty metaphases for each sample were karyotyped by M-FISH.
DT40 clonogenic survival
DT40 cells were grown in RPMI Medium 1640 (Life Technologies, 61870), supplemented with 7% fetal bovine serum (FBS, Life Techonologies, 10270), 3% chicken serum (Life Technologies, 16110), 50 µM β-mercaptoethanol and penicillin/streptomycin, at 37 °C in a 5% CO2 incubator. Sensitivity assays were performed as previously described6. In brief, 105 cells were incubated with drug-containing medium in a sealed FACS tube at 37 °C for 2 h (acetaldehyde) or 1 h (mitomycin C or cisplatin). Dilutions were plated in 6-well plates containing semi-solid medium (4000 cP methyl cellulose (M0512 Sigma), DMEM/F-12 powder (Life Technologies, 32500-043), 7% FBS, 3% chicken serum, 50 µM β-mercaptoethanol and penicillin/streptomycin). Plates were incubated for 7–10 days, after which timed colonies were counted manually. Survival is plotted as a percentage relative to untreated cells. Each data point represents the mean of three independent experiments each carried out in quadruplicate.
Sensitivity assays of primary mouse B cells
These assays were performed as described previously5. Lymphocytes purified from the spleen using Lympholyte M (Cederlane) were stimulated with LPS (L4391, Sigma) at a final concentration of 40 μg ml−1. Cells (4 × 105) were then plated with acetaldehyde in one well of a 24-well plate. After seven days, viable cells were counted using trypan blue exclusion from 100 images on a Vi-Cell XR cell viability counter (Beckman Coulter). Each data point represents the mean of three independent experiments each carried out in quadruplicate.
Survival assays of colony-forming units (CFU)
Bone marrow cells were isolated using IMDM medium, and single cell suspensions were obtained by passing the bone marrow through a 70-μm cell strainer (Falcon). Nucleated cells were counted by diluting cells tenfold in a 3% solution of acetic acid with methylene blue (Stem Cell Technologies) using a Vi-Cell XR cell viability counter (Beckman Coulter). Cells were resuspended to make up 1.5 ml of IMDM containing 30 × 106 cells and 250 μl of each suspension was mixed with 250 μl of IMDM containing 2× acetaldehyde to give final concentrations of 0, 1, 2, 4 and 8 mM acetaldehyde. The cells were incubated at 37 °C for 4 h in sealed tubes, after which two tenfold serial dilutions were made. 400 μl of cells were then added to 4 ml of MethoCult M3534 (StemCell Technologies), and the total volume of each dilution was plated in two wells of a six-well plate two wells each containing 106, 105 and 104 cells, respectively. After seven days of culture at 37 °C with 5% CO2, the colonies were counted and the relative survival was plotted. Each data point represents the average of experimental duplicates carried out on three mice of each genotype.
Flow cytometry
The micronucleus assay was performed essentially as described previously38. Treated or untreated mice (8–12 weeks of age) were bled and 62 μl blood was mixed with 338 μl PBS supplemented with 1,000 U ml−1 of heparin (Calbiochem). 360 μl of blood suspension was then added to 3.6 ml of methanol at −80°C and stored at −80°C for at least 12 h. 1 ml of fixed blood cells was then washed with 6 ml of bicarbonate buffer (0.9% NaCl, 5.3 mM NaHCO3). The cells were resuspended in 150 μl of bicarbonate buffer and 20 μl of this suspension was used for subsequent staining. 72 μl of bicarbonate buffer, 1 μl of FITC-conjugated CD71 antibody (GenTex, clone R17217.1.4) and 7 μl RNase A (Sigma) were premixed and added to 20 μl of each cell suspension. The cells were stained at 4 °C for 45 min, followed by addition of 1 ml bicarbonate buffer and centrifugation. Finally, cell pellets were resuspended in 500 μl bicarbonate buffer supplemented with 5 μg ml−1 propidium iodide (Sigma). The samples were analysed immediately on an LSRII FACS analyser (BD) and the data analysed with FlowJo v10.0.7.
For HSC quantification, bone marrow cells were isolated from tibiae and femurs with staining buffer (PBS supplemented with 2.5% FCS) and strained through 70-μm meshes. Red cells were lysed by resuspending the cells in 10 ml red cell lysis buffer (MACS Miltenyi Biotec) for 10 min at room temperature. After centrifugation, the cell pellet was resuspended in staining buffer and nucleated cells were counted with 3% acetic acid (StemCell Technologies) on a Vi-Cell XR cell viability counter (Beckman Coulter). Bone marrow cells (10 × 106 cells) were resuspended in 200 μl of staining buffer containing the following antibody solution: FITC-conjugated lineage cocktail with antibodies against CD4 (clone H129.19, BD Pharmingen), CD3e (clone 145-2C11, eBioscience), Ly-6G/Gr-1 (clone RB6-8C5, eBioscience), CD11b/Mac-1 (clone M1/70, BD Pharmingen), CD45R/B220 (clone RA3-6B2, BD Pharmingen), Fcε R1α (clone MAR-1, eBioscience), CD8a (clone 53-6.7, BD Pharmingen), CD11c (clone N418, eBioscience), TER-119 (clone Ter119, BD Pharmingen) and CD41 (FITC, clone MWReg30, BD Pharmigen); c-Kit (PerCP-Cy5.5, clone 2B8, eBioscience), Sca-1 (PE-Cy7, clone D7, eBioscience), CD150 (PE, clone TC15-12F12.2, BioLegend) and CD48 (biotin, clone HM48-1, BioLegend). The samples were incubated for 15 min at 4 °C and washed with 2 ml buffer. The cell pellets were resuspended in 200 μl staining buffer containing streptavidin-BV421 and incubated for another 15 min at 4 °C. Finally, cells were washed, resuspended in 500 µl staining buffer, data were acquired on a Fortessa FACS analyser (Becton Dickinson) and analysed with FlowJo v10.0.7. LKS cells were defined as lineage− CD41−Sca-1+ Kit+ and HSCs were defined as LKS CD48− CD150+.
To assess engraftment of single HSCs into irradiated recipients, 50 μl of blood was obtained from the tail vein of recipient mice every two weeks. RBCs were lysed by the addition of 1 ml of ammonium chloride lysis buffer (155 mM NH4Cl, 10 mM KHCO3, 0.1 mM Na2EDTA, pH 7.2) and incubated for 10 min at room temperature. After centrifugation, the cell pellets were resuspended in 100 µl of staining buffer containing antibodies against: CD4 (FITC, clone H129.19, BD Pharmingen), CD8a (FITC, clone 53-6.7, BD Pharmingen), CD45R/B220 (PerCP-Cy5.5, clone RA3-6B2, BioLegend), CD11b/Mac-1 (PE, clone M1/70, BD Pharmingen), Ly-6G/Gr-1 (PE, clone 1A8, BD Pharmingen), TER-119 (PE-Cy7, clone TER-119, BioLegend), CD45.1 (BV421, clone A20, BioLegend) and CD45.2 (APC, clone 104, BioLegend). After incubation, cells were washed with 3 ml of staining buffer before being resuspended in 250 μl of the same buffer. Samples were run on the Fortessa analyser (BD) with the HTS module and the multilineage chimaerism was calculated using FlowJo v10.0.7. TER-119 was used to exclude RBC debris and chimaerism was calculated for each of the WBC lineages (CD45+ total WBCs, B220+ B cells, CD4+CD8+ T cells and Gr-1+ Mac-1+ myeloid cells) as the proportion of cells derived from the single HSC (CD45.2+) over the total number of CD45+ cells, which includes cells derived from the recipient or carrier cells (CD45.1+).
For intracellular staining of p53 and cleaved caspase-3, total bone marrow cells were stained with the lineage-depletion kit (130-090-858, MACS Miltenyi Biotec) following the manufacturer’s instructions and passed through LS magnetic columns. Lineage-depleted cells were spun down for 5 min at 1,200 r.p.m. and the pellets were resuspended in 200 μl MACS buffer with the antibodies described above for HSC quantification. In parallel, 3 × 106 total bone marrow cells were stained with antibodies against committed lineages: CD45R/B220 (PE, clone RA3-6B2, BD Pharmingen) and IgM (FITC, clone II/41, BD Pharmingen) for B cell progenitors; TER-119 (FITC, clone TER-119, BD Pharmingen) and CD71 (PE, clone C2, BD Pharmingen) for erythroid maturation; and CD11b/Mac-1 (PE, clone M1/70, BD Pharmingen) and Ly-6G/Gr-1 (FITC, clone 1A8, eBioscience) for monocyte/granulocyte progenitors. After antibody staining, cells were washed, then fixed and permeabilized with BD Cytofix/Cytoperm solution (554722, BD Pharmingen) following the manufacturer’s instructions. Finally, cells were stained with either anti-p53 (AlexaFluor647, clone 1C12, Cell Signalling) or anti-cleaved-caspase-3 (AlexaFluor647, clone D3E9, Cell Signalling) antibodies.
Gating strategies are described in Supplementary Fig. 2.
Blood counts
Total blood was collected in K3EDTA MiniCollect tubes (Greiner bio-one) and analysed on a VetABC analyser (Horiba).
Western blot
The FANCA antibody (Cell Signalling, D1L2Z) was used at 1:1000 in 5% w/v BSA, 1× TBS, 0.1% Tween-20 at 4 °C with gentle shaking, overnight. The β-actin antibody (Abcam, 8227) was used at 1:3,000 in the same conditions. Swine anti-rabbit immunoglobulins HRP (Dako) was used as secondary antibody at 1:2,000 for 1 h at room temperature.
Histological analysis
Tissue samples were fixed in 10% neutral-buffered formalin for at least 24 h. The femur samples were then decalcified and embedded in paraffin, and 4 μm sections were cut before staining with haematoxylin and eosin using standard methods.
In vivo point mutation assay
The λ select-cII (BigBlue) mutagenesis assay was performed following the manufacturer’s instructions. This assay allows the detection of point mutations in vivo and is based on the ability of coliphage λ to multiply either through the lytic or lysogeneic cycles in Escherichia coli.
In brief, the RecoverEase DNA-isolation kit (Stratagene) was used to extract genomic DNA from the bone marrow of 8-to-12-week old Aldh2−/−Fancd2−/− and control mice that carry Big Blue λLIZ repeats. The shuttle vector that was recovered from mouse genomic DNA was packaged into phage with the Transpack packaging extract (Stratagene), which was then used to infect E. coli G1250 (Stratagene).
To assess the frequency of mutations within the cII gene, infected E. coli were diluted in TB1 top agar, spread on ten 100-mm TB1-agar plates and incubated at 24 °C for 48 h. The plaques were enumerated, picked and replated to confirm that they were lytic. This provided the number of mutated phage within the sample. To determine the total number of phage undergoing the lysogenic cycle and the mutation frequency, 10-and-50-fold dilutions of the stock of infected E. coli in TB1 top agar were spread on two 100-mm TB1-agar plates and incubated at 37 °C for 24 h. Incubation at 37 °C switches all phage to the lytic cycle, and therefore allows the total number of replication-competent phage to be assessed. The mutation frequency was then calculated as the number of cII-mutated plaques (24 °C)/total number of plaques (37 °C).
As a positive control for the in vivo λ select-cII mutagenesis assay, mice were exposed to 150 mg kg−1 N-ethyl-N-nitrosourea (ENU, Sigma) in a single intraperitoneal injection on one occasion. Mice were allowed to recover and were euthanized 3 weeks later.
CFU-S12 assays
CFU-S12 assays were performed as described previously5, except that CFU-S colonies were counted after 12 days. In brief, to assess the frequency of CFU-S in mutant mice, total bone marrow was flushed from the femora and tibiae of mutant mice and appropriate controls. Nucleated cells were enumerated using a solution of 3% acetic acid and methylene blue and injected intravenously into 20 recipient mice that had been lethally irradiated. After 12 days the spleens were fixed in Bouin’s solution (Sigma), and the number of colonies were counted and expressed relative to the number of total bone marrow cells injected.
To assess the survival of CFU-S12 after exposure to acetaldehyde, we treated total bone marrow cells with 4 mM acetaldehyde for 4 h in vitro before injecting them into lethally irradiated recipient mice. After 12 days, the number of CFU-S were counted. Survival was expressed relative to the untreated control for each genotype. Each data point represents the mean CFU-S survival in ten recipient mice, expressed relative to untreated samples of the corresponding genotype.
Single HSC transplants
The single-stem-cell transplants were performed as described previously39,40. CD45.1 recipient mice (C57BL/6J × 129S6/Sv F1, 8-to-12-weeks old) were fed with water supplemented with the antibiotic enrofloxacin (Baytril, Bayer Corporation) for seven days before irradiation and for the duration of the experiment. The lethal radiation was delivered as a split dose of 1,000 rad (500 rad each, 3 h apart) using a 137Cs GSR C1m source (GSR Gmbh).
The lethally irradiated recipients were injected with single HSCs sorted from Aldh2−/−Fancd2−/− and control mice on a C57BL/6Jo1a × 129S4S6/Sv F1 background (CD45.2, 8-to-12 weeks old). Bone marrow cells from these mice were extracted with IMDM medium (GIBCO), filtered through a 70-μm strainer, spun down for 5 min at 1,200 r.p.m. and resuspended in 6 ml IMDM medium at room temperature. These cells were then overlaid onto 6 ml Lympholyte M (Cederlane) in a 15-ml Falcon tube and spun down for 20 min at 1,400g at room temperature with the brake off. The interface containing the mononucleated cells was transferred into another 15-ml tube and topped up with ice-cold MACS buffer (PBS pH 7.2, 0.5% BSA, 2 mM EDTA). After 5 min of centrifugation at 1,200 r.p.m., the cell pellets were resuspended in 320 μl of MACS buffer, stained with the lineage-depletion kit (130-090-858, MACS Miltenyi Biotec) following the manufacturer’s instructions and passed through LS magnetic columns. Lineage-depleted cells were spun down for 5 min at 1,200 r.p.m. and the pellets were resuspended in 200 μl MACS buffer with the antibodies described previously (see Flow cytometry), except that anti-CD48 was directly conjugated to BV421 (clone HM48-1, BioLegend). The cells were resuspended in 500 μl of MACS buffer and run on a Synergy sorter (Sony Biotechnology Inc.).
Single HSCs, defined as lineage−c-Kit+Sca-1+CD41−CD48−CD150+, were sorted into 100 μl StemSpan SFEM medium (StemCell Technologies) in each well of a round bottom, 96-well plate (Costar) using a Synergy cell sorter (Sony Biotechnology). The plates were spun down for 5 min at 180g and the presence of a single cell per well was confirmed visually. The full content of selected wells was loaded into insulin syringes (29G, 0.5-inch needle) containing 2 × 105 carrier cells in 300 μl Hank’s balanced salt solution (StemCell technlogies). The contents of the syringe were used to dislodge the single HSC from the bottom of the well, avoiding the creation of bubbles. The entire volume (400 μl) was injected into the tail veins of irradiated recipients and chimaerism was measured every 2 weeks using flow cytometry. Recipients were considered reconstituted by the single stem cell if chimaerism for CD45.2+ WBCs was ≥0.1%.
Whole-genome sequencing of mouse HSC clones
Single HSCs were allowed to expand in vivo for four months to guarantee that the transplanted cell was a stem cell. Recipient mice that were positive for reconstitution were euthanized four months after transplantation, and the blood, bone marrow, spleen and thymus were collected. All tissues were prepared for flow cytometry as described above and stained with CD45.1 (FITC, clone A20, BioLegend) and CD45.2 (APC, clone 104, BioLegend) antibodies. CD45.2+ cells were sorted from each tissue on a Synergy cell sorter (Sony Biotechnology), spun down and frozen at −80 °C. Genomic DNA was then extracted from the CD45.2+ bone-marrow cells and from a tail biopsy that had been collected at 2 weeks of age from the same mouse that provided the single HSC. Genomic DNA was extracted with the Puregene Cell and Tissue kit (Qiagen) following the manufacturer’s instructions.
Whole-genome sequencing was performed as described previously41. In brief, short-insert 500-bp genomic libraries were constructed according to Illumina library protocols and 100-base paired-end sequencing was performed on HiSeq 2000 or HiSeq X genome analysers to an average of 20× coverage. Short-insert paired-end reads were aligned to the reference mouse genome (NCBIM38) using BWA-MEM (http://bio-bwa.sourceforge.net/). For each HSC clone, the matched tail sample was used as the reference. We sequenced two of the Aldh2−/−Fancd2−/− HSC clones to 40× coverage, together with their matched germ-line references. We noted a big overlap between the variants found at 20× and 40× coverage (data not shown), showing that doubling the coverage did not actually uncover many more mutations. Any additional calls found when the coverage was increased to 40× were predominantly subclonal, with a mean VAF of 0.22. Therefore, we concluded that 20× whole-genome sequencing provided sufficient coverage to allow us to uncover mutations present in the transplanted HSC.
Substitutions, indels and structural changes were called with the CaVEMan, Pindel and BRASS algorithms, which are described in detail elsewhere42. In addition to previously reported filtering41, we asked that all variants were unique to each HSC clone and not found in unrelated HSC or tail samples, or in unrelated mouse strains. We did not apply a VAF filter for clonality because when we examined the VAF distribution of the final datasets, these were centered around 0.5 (Extended Data Fig. 9). For the transcriptional analysis, previously reported HSC RNA-seq data was aligned with Bowtie2 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml against NCBIM3843, and the overlap between indels and transcribed genes was calculated in R. To calculate the fraction of the genome covered by genes, positions of genes were retrieved from Ensembl. Overlapping regions between two genes were taken into account and counted only once.
Validation of indel and rearrangement calls
Indel calls of less than 50 bp were validated using multiplex PCR and targeted re-sequencing. We used 13 multiplex-primer combinations to capture and simultaneously amplify 172 (50%) of the previously identified indels. Primers were designed using MPprimer to capture regions of between 190 and 250 bp in size44 (sequences available upon request). The first of round multiplex-PCR amplifications was performed with tailed gene primers and was individually barcoded by a second round of PCR with pre-validated MiSeq-ready primers45, using a high fidelity polymerase (Q5 Hot Start HF, New England Biolabs). The PCR reaction conditions were as follows: Group 1, 100 ng DNA input in 25 μl PCR reaction, 95 °C for 2 min, six cycles of 98 °C for 20 s, 65 °C for 60 s, 60 °C for 60 s, 55 °C for 60 s, 50 °C for 60 s and 70 °C for 60 s, the reaction was then held at 4 °C until addition of barcoded second-round primers, followed by 19 cycles of 98 °C for 20 s, 62 °C for 15 s and 72 °C for 30 s, then 72 °C 60 s; Group 2, 10 ng DNA input in 5 μl PCR reaction, 95 °C for 2 min, seven cycles of 95 °C for 20 s, 58 °C for 17 min and 70 °C for 60 s, then held at 4 °C until addition of barcoded second-round primers, followed by 23 cycles of 98 °C for 20 s, 62 °C for 15 s and 72 °C for 30 s, then 72 °C 60 s. Each sample was pooled, size selected by SPRI (×0.8) and quantified before being stored at −20 °C until sequencing. Two MiSeq runs (300-bp paired-end) were used for variant confirmation, and reads were mapped with BWA. We analysed positions where the coverage was higher than 100× (159/172). With this approach, we were able to validate 91.2% of the original calls: 14/159 (8.8%) calls had VAF values <0.1 and were deemed false positives. The normal distribution of VAFs around 0.5 is consistent with most indels being clonal in origin.
For validation of rearrangements, we designed nested PCRs surrounding the breakpoints determined by the BRASS algorithm (primer sequences available upon request). PCR reactions were carried out in 20 μl, using 10 ng (HSC clones or tails) or 400 ng (donor bone marrow) of genomic DNA and GoTaq G2 Hot Start Polymerase (M7401, Promega). The first round of PCR amplifications was performed at 95 °C for 2 min, 35 cycles of 95 °C for 30 s, 55 °C for 20 s and 72 °C for 30 s, then 72 °C for 5 min. The reactions were diluted to 1 in 50 and 1 μl of the diluted reaction was used as template for a second round of PCR with nested primers, to increase specificity and sensitivity, performed at 95 °C for 2 min, 35 cycles of 95 °C for 30 s, 60 °C for 20 s and 72 °C for 30 s, then 72 °C for 5 min. The reactions were analysed on 2% agarose gels, bands of the expected sizes were excised and the identities of all products were confirmed by Sanger sequencing.
Using this approach, we found that 16/27 (59%) of rearrangements could be detected in the bone marrow of donor mice at the time the HSCs were transplanted (Extended Data Fig. 10). Any rearrangements present before transplantation must be clonal (that is, would not have arisen after the transplant). The failure to amplify the remaining 11 rearrangements by PCR does not mean that these are sub-clonal (that is, post-transplant) events. PCR amplification will depend on how much the transplanted HSC was contributing to blood production in the donor animal at the time of the transplant, as well as the sensitivity of each PCR. Therefore, we inferred clonality for the remaining calls by looking at loss of copy number (in the case of deletions, see Fig. 5l) and the number of reads involved in the rearrangement at the breakpoint for copy number-neutral changes.
Statistical analysis
Sample number (n) indicates the number of independent biological samples in each experiment. Sample numbers and experimental repeats are indicated in figure legends or Methods. Normality of data distribution was tested using the D’Agostino–Pearson omnibus normality test and variance was estimated before deciding on a statistical test. Unless otherwise stated in the figure legend, data are shown as the mean ± s.e.m. and the two-sided nonparametric Mann–Whitney test was used to assess statistical significance. Analysis was performed using GraphPad Prism.
Extended Data
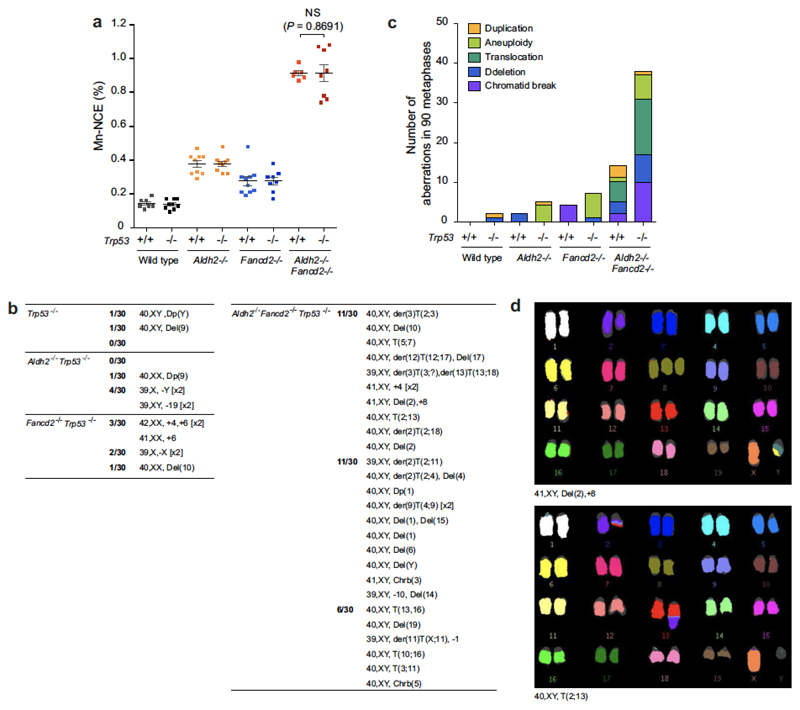
Extended Data Figure 1. Ethanol-induced genomic instability.
a, Left, representative images of bone marrow metaphase spreads from wild-type mice treated with mitomycin C (MMC); n shows the number of SCE events per metaphase. Right, comparison between number of SCEs in the bone marrow of wild-type and Aldh2−/− mice treated with ethanol (5.8 g kg−1) or MMC (1 mg kg−1). Triplicate experiments, 25 metaphases per mouse, n = 75; P calculated by two-sided Mann–Whitney test; data shown as mean and s.e.m. Ethanol causes a strong homologous recombination response in Aldh2−/− mice, comparable to that observed in wild-type mice exposed to MMC. b, Left, representative images of bone marrow metaphase spreads from wild-type and Fanca−/− mice; n shows the number of SCE events per metaphase. Right, quantification of SCEs (duplicate experiments, 25 metaphases per mouse, n = 50; P calculated by two-sided Mann–Whitney test; data shown as mean and s.e.m.). Mice deficient in cross-link repair (Fanca−/−, or Fancd2−/− in Fig. 1a) show a small but significant increase in the number of spontaneous SCE events, indicating that a homologous recombination repair response occurs in the absence of the Fanconi anaemia pathway. c, Scheme depicting the formation of micronucleated erythrocytes. Micronuclei (Mn) generated by fragmentation or mis-segregation of chromosomes during erythrocyte maturation remain in the erythrocyte after extrusion of the main nucleus. These fragments can be detected by a DNA stain (PI+). During maturation, red-cell progenitors lose CD71 expression. Therefore, peripheral CD71+ red cells represent immature, short-lived reticulocytes (Ret) and CD71− cells represent mature, long-lived normochromic erythrocytes (NCEs). d, Proof-of-principle experiment showing the induction of micronucleated reticulocytes 48 h after MMC treatment (1 mg kg−1). P calculated by two-sided Mann–Whitney test; data shown as mean and s.e.m.; n = 29, 8, 20 and 9 mice, left to right. e, Treatment of Aldh2−/− mice with ethanol (5.8 g kg−1) leads to potent micronucleus formation. This induction is comparable to that observed in wild-type mice that were treated with the aneugen vincristine (Vcn, 0.2 mg kg−1, 48 h) or clastogenic γ-irradiation (IR, 400 rad, 48 h)46. P calculated by two-sided Mann–Whitney test; data shown as mean and s.e.m.; n = 29, 15, 10, 11, 25 and 15 mice. f, List of chromosomal aberrations observed in the bone marrow of 8-to-12-week-old untreated Aldh2−/−Fancd2−/− and control mice. g, List of chromosomal aberrations observed in the bone marrow of 8-to-12-week-old Aldh2−/−Fancd2−/− and control mice 48 h after ethanol treatment (5.8 g kg−1, injected intraperitoneally, IP). In f and g, three mice and 30 metaphases per mouse were analysed per condition, and the numbers represent the fraction of abnormal metaphases per mouse. h, Bar chart classifying the type of aberrations for each genotype (90 metaphases per condition). i, Examples of different types of chromosomal aberrations.
Extended Data Figure 2. A single dose of ethanol precipitates bone-marrow failure in Aldh2−/−Fancd2−/− mice.
a, A single dose of ethanol (5.8 g kg−1, injected intraperitoneally) leads to anaemia in Aldh2−/−Fancd2−/− mice one to two months after treatment (P calculated by Mantel-Cox test; n = number of mice). b, Haematoxylin and eosin staining of bone marrow sections 30 days after ethanol treatment (original magnification, ×100). c, Full blood-count analysis for Aldh2−/−Fancd2−/− and control mice, before injection and terminal bleeds after ethanol treatment (P calculated by paired t-test; data shown as mean and s.e.m.; n = number of mice, as in a).
Extended Data Figure 3. Generation of a conditional Fanca allele.
a, Mice carrying the previously reported Fanca− allele (Fancatm1a(EUCOMM)Wtsi) were crossed with mice carrying the FLP recombinase, yielding the Fancafl allele (Fancatm1c(EUCOMM)Wtsi). This allele restores FANCA expression as shown by western blot (Fig. 3). Cre-mediated recombination of Fancafl yields the FancaΔ allele (Fancatm1d(EUCOMM)Wtsi), which lacks exon 3 and leads to loss of FANCA protein (Fig. 3). b, Genotyping PCRs for the wild-type, Fanca− and Fancafl alleles with primers FL033, FL040 and En2A; showing bands of the expected sizes. c, Western blot (single experiment) showing complete absence of FANCA protein in the spleens of Fanca−/− and Fancafl/− Vav1-iCre mice. For gel source data, see Supplementary Fig. 1. d, Determination of the number of exon 3 copies by quantitative (q)PCR. Wild-type, Fanca+/Δ and FancaΔ/Δ mice carry 2, 1 and 0 copies, respectively. Fancafl Vav1-iCre mice show tissue-specific deletion of exon 3 in white blood cells (WBCs) and bone marrow (n = 4 technical replicates; bars: mean, s.d.). e, Microscopic analysis of haematoxylin and eosin-stained sections of testes (original magnification, ×50) from wild-type, Fanca−/−, Fancafl/fl and FancaΔ/Δ males at 12 weeks, showing impaired spermatogenesis in testes of Fanca−/− and FancaΔ/Δ mice (one experiment). f, Sensitivity assay of transformed mouse-embryonic fibroblasts (MEFs) derived from Fanca−/−, Fancafl/fl and FancaΔ/Δ embryos, showing hypersensitivity of both Fanca−/− and FancaΔ/Δ cells to the cross-linking agent mitomycin C (n = number of experiments, each carried out in quadruplicate; bars: mean, s.e.m.).
Extended Data Figure 4. Endogenous aldehydes mutate the HSC genome.
Circos plots showing the mutations observed in all sequenced HSC clones (wild type, n = 3; Aldh2−/−, n = 3; Fancd2−/−, n = 4; and Aldh2−/−Fancd2−/−, n = 5 HSC genomes). Substitutions, indels and rearrangements are plotted.
Extended Data Figure 5. Detection of point mutations in mice with the BigBlue reporter system.
a, Chromosome 4 of the BigBlue reporter mouse harbours a λ-phage transgene that contains the mutational target. The phage DNA can be recovered from mouse tissues, packaged into phage and used to infect bacteria. Phage cII mutants can be detected by the ability of these phage to form plaques at 24 °C. b, Quantification of the frequency of cII− -mutant phage recovered from the bone marrow of young Aldh2−/−Fancd2−/− and control mice carrying the BigBlue transgene. ENU-treated mice serve as positive controls for the assay (P calculated by two-sided Mann–Whitney test; data shown as mean and s.e.m.; n = 7, 7, 6, 7 and 6 mice, left to right). c, Relative contribution of the indicated mutation classes to the point-mutation spectra of cII−-mutant phage isolated from the bone marrow. The ENU-mutation spectrum is characterized by T to A transversions and T to C transitions. n is the number of sequenced cII− mutant phage.
Extended Data Figure 6. Aldehyde-induced stress elicits a p53 response.
a, Representative flow cytometry plots for the quantification of p53+ LKS cells from 8-to-12-week-old Aldh2−/−Fancd2−/− and control mice. Cells were collected from wild-type and Trp53−/− mice 2 h after 10 Gy irradiation as positive and negative controls, respectively, for the assay. b, Quantification of the frequency of p53+ cells in different bone-marrow populations. c, Quantification of the frequency of cleaved-caspase-3+ cells in different bone marrow populations by flow cytometry. In b and c, irradiated wild-type and Trp53−/− mice were used as controls. Owing to the low numbers of LKS CD48− CD150+ cells in Aldh2−/−Fancd2−/− mice, the number of p53+ or cleaved-caspase-3+ HSCs could not be determined (data shown as mean and s.e.m.; n = number of mice). d, e, Survival of B cells and myeloid progenitors (CFU-GM) following exposure to acetaldehyde in vitro. Cells were obtained from Fancd2−/−Trp53−/− and control mice. Each point represents the mean of three independent experiments, each carried out in quadruplicate; data shown as mean and s.e.m. f, Frequency of CFU-S12 in the bone marrow of Aldh2−/−Fancd2−/−Trp53−/− and control mice. Each point represents the number of CFU-S12 in the spleen of a single recipient (P calculated by two-sided Mann–Whitney test; data shown as mean and s.e.m.; n = 10–15 mice).
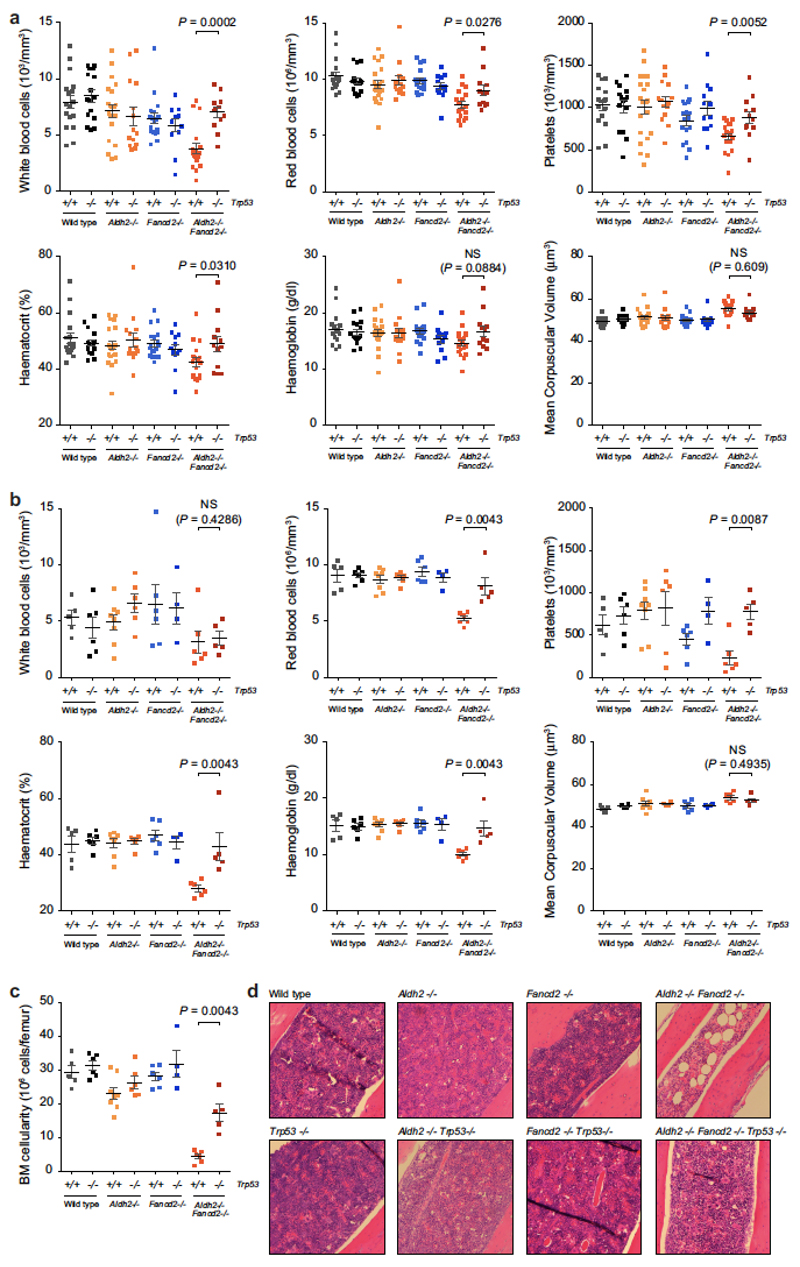
Extended Data Figure 7. p53 deficiency suppresses peripheral-blood cytopenias and ethanol-induced bone-marrow failure in Aldh2−/−Fancd2−/− mice.
a, Full blood count analysis of Aldh2−/−Fancd2−/−Trp53−/− and control mice (8-to-12 weeks old, on a C57BL/6 × 129S4S6/Sv F1 background). A significant increase in the number of white blood cells, red blood cells, platelets and haematocrit was observed in Aldh2−/−Fancd2−/−Trp53−/− mice compared to Aldh2−/−Fancd2−/− mice (P calculated by two-sided Mann–Whitney test; data shown as mean and s.e.m.; n = 17, 16, 21, 14, 18, 12, 18 and 12 mice, left to right). b, Aldh2−/−Fancd2−/−, Aldh2−/−Fancd2−/−Trp53−/− and control mice were treated with ethanol in their drinking water for 10 days as described previously6. Full blood-count analyses were carried out after 10 days of ethanol treatment. c, Bone marrow cellularity after 10 days of ethanol treatment. In b, c, P calculated by two-sided Mann–Whitney test; data shown as mean and s.e.m.; n = 5, 6, 8, 6, 6, 4, 6 and 5 mice, left to right). d, Haematoxylin and eosin staining of bone-marrow sections 10 days after ethanol treatment (original magnification, ×100).
Extended Data Figure 8. Genomic instability in Aldh2−/−Fancd2−/−Trp53−/− mice.
a, Quantification of micronucleated NCEs in the blood of Aldh2−/−Fancd2−/−Trp53−/− and control mice (P calculated by two-sided Mann–Whitney test; data shown as mean and s.e.m.; n = 8 mice). b, List of chromosomal aberrations observed in the bone marrow of 8-to-12 week-old untreated Aldh2−/−Fancd2−/−Trp53−/− and control mice. Three mice and 30 metaphases per mouse were analysed per genotype; the numbers represent the fraction of abnormal metaphases per mouse. c, Bar chart classifying the types of aberrations for each genotype (90 metaphases per condition). d, Examples of two metaphases from an Aldh2−/−Fancd2−/−Trp53−/− mouse.
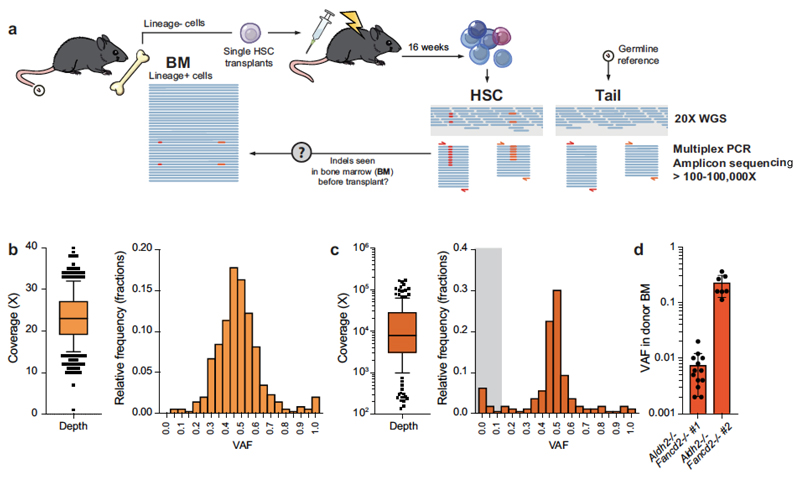
Extended Data Figure 9. Validation of indels by targeted deep sequencing.
a, Scheme depicting the generation of HSC clones by transplantation of single stem cells, subsequent whole-genome sequencing and validation of indel calls by amplicon deep sequencing. On the basis of the indel location from 20× whole-genome sequencing, we designed multiplex PCRs and deep sequenced the PCR products to higher coverage (100–100,000×) to confirm that the calls were not sequencing artefacts. In addition, we attempted to detect indels in DNA samples of bone-marrow cells from the mice that provided the transplanted HSCs. b, Coverage depth and VAF of the filtered set of indel calls from whole-genome sequencing (n = 342 indels; box plot shows the mean, box edges represent the first and third quartiles, whiskers extend over 10–90% of data). c, Coverage depth and VAF of the indel calls from deep sequencing validation (n = 159 locations; box plot shows the mean, box edges represent the first and third quartiles, whiskers extend over 10–90% of data). One hundred and fifty-nine locations had coverage greater than 100× and were used for the analysis. We could validate the presence of 91.2% of the initial calls; 14/159 (8.8%) calls had VAF <0.1 and were deemed false positives (indicated by grey shading). Note that the VAF distribution is centred tightly around 0.5, confirming the clonal nature of most indels. d, We used targeted deep sequencing to look for indel calls in bone-marrow samples from the mice that provided the transplanted HSCs. In most cases, the calls were below the detection limit of the assay (VAF <0.0001). However, we could detect indels from two Aldh2−/−Fancd2−/− HSCs, indicative of ‘clonal haematopoiesis’ in these mice (accounting for 0.7 and 21.4% of blood production, respectively). Data shown as mean and s.e.m.; n = 13 and 7 indels.
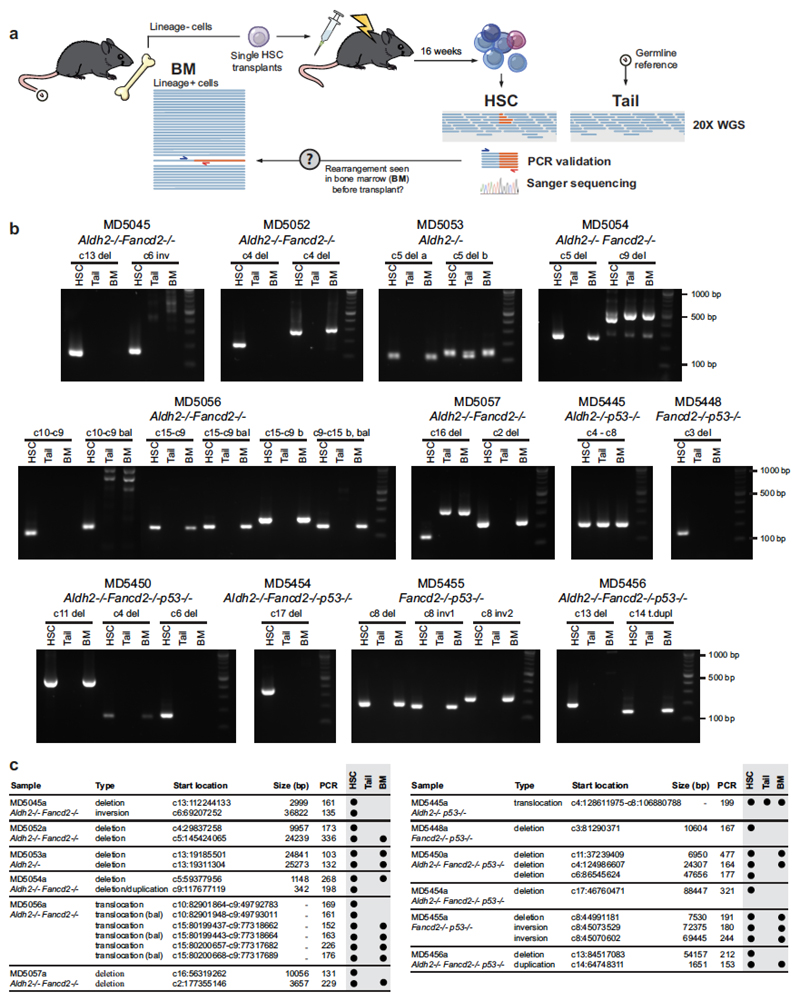
Extended Data Figure 10. Validation of rearrangements by PCR.
a, Scheme depicting the generation of HSC clones by transplantation of single stem cells, subsequent whole-genome sequencing and validation of rearrangement calls by PCR. We designed primers for nested PCRs flanking the breakpoints calculated by the BRASS algorithm, and the identity of the products was confirmed by Sanger sequencing. In addition, we attempted to detect the rearrangements in DNA samples of bone-marrow cells from the mice that provided the transplanted HSCs, demonstrating that these changes did not arise during clonal expansion and were present in the stem cell at the time of transplantation. b, Agarose gels (one experiment) showing presence of specific PCR amplification from DNA of HSC clones, absence in matched germline samples from the tail of the same mouse and, in some cases, detection in bone-marrow tissue that predates the transplants. PCR amplification in these samples is dependent on the contribution of the transplanted HSC to blood production, and the sensitivity of each PCR. Gel source data is shown in Supplementary Fig. 1. c, List summarizing the rearrangements found in 28 HSC clones and the results from b. All 27 rearrangements could be detected by PCR and confirmed by Sanger sequencing. 16/27 (59%) rearrangements could be detected before transplantation.
Extended Data Figure 11. Mechanisms to maintain genetic integrity and suppress mutagenesis by endogenous aldehydes in HSCs.
a, Aldehyde catabolism and FA-pathway-mediated DNA repair constitute two distinct tiers of protection against aldehyde damage. Loss of this protection leads to the accumulation of DNA damage and mutagenesis. Passage of mutated genetic information is prevented by the activation of p53, leading to HSC loss. b, In the absence of a functional Fanconi anaemia pathway, aldehyde lesions degenerate into DNA double-strand breaks that can be repaired through error-free recombination. However, this mechanism is not sufficient to fully compensate for Fanconi anaemia inactivation, leading to the engagement of both classical and alternative end-joining, and subsequent mutagenic repair.
Supplementary Material
Supplementary Information is available in the online version of the paper.
Acknowledgements
We thank D. Kent for technical advice with single-HSC transplants; R. Berks, A. Middleton, J. Wiles, C. Knox, X. Gong, J. Roe, J. Willems, the ARES staff and Biomed for their help with mouse work; M. Daly, F. Zhang, V. Romashova and M. Balmont for help with flow cytometry; and J. Sale, C. Rada, M. Taylor, Y. L. Wu and members of the Patel laboratory for critical reading of the manuscript. The Human Research Tissue Bank (supported by the NIHR Cambridge Biomedical Research Centre) processed histology. K.J.P. is supported by the MRC and the Jeffrey Cheah Foundation. G.P.C. and L.M. were supported by CRUK. J.I.G. is supported by the Wellcome Trust and King’s College, Cambridge.
Footnotes
Data availability
Whole-genome sequencing data have been deposited in the EMBL European Nucleotide Archive (ENA) under the accession code ERP009447. All other data are available upon reasonable request from the authors.
Author Contributions J.I.G., G.P.C. and K.J.P. conceived the study and wrote the manuscript. J.I.G. conducted the majority of the experiments and analysed the data. G.P.C. assisted in the characterization of genomic instability in Aldh2−/−Fancd2−/− mice and single HSC transplantation. F.L. analysed the survival of chicken DT40 cells, performed western blotting and assisted with the analysis of micronucleus samples. L.M. assisted with single cell transplantation and performed the BigBlue in vivo point-mutation analysis. S.L and F.Y. performed the M-FISH karyotyping of mouse metaphases. N.P. performed validations of indels by targeted deep sequencing. G.G., S.R., S.N.-Z. and M.S. provided assistance with the analysis and interpretation of sequencing data.
Author Information Reprints and permissions information is available at www.nature.com/reprints. Readers are welcome to comment on the online version of the paper. Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The authors declare no competing financial interests.
References
- 1.Roswall N, Weiderpass E. Alcohol as a risk factor for cancer: existing evidence in a global perspective. J Prev Med Public Health. 2015;48:1–9. doi: 10.3961/jpmph.14.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang M, et al. Identification of DNA adducts of acetaldehyde. Chem Res Toxicol. 2000;13:1149–1157. doi: 10.1021/tx000118t. [DOI] [PubMed] [Google Scholar]
- 3.Lai CL, et al. Dominance of the inactive Asian variant over activity and protein contents of mitochondrial aldehyde dehydrogenase 2 in human liver. Alcohol Clin Exp Res. 2014;38:44–50. doi: 10.1111/acer.12215. [DOI] [PubMed] [Google Scholar]
- 4.Yokoyama A, Omori T. Genetic polymorphisms of alcohol and aldehyde dehydrogenases and risk for esophageal and head and neck cancers. Jpn J Clin Oncol. 2003;33:111–121. doi: 10.1093/jjco/hyg026. [DOI] [PubMed] [Google Scholar]
- 5.Garaycoechea JI, et al. Genotoxic consequences of endogenous aldehydes on mouse haematopoietic stem cell function. Nature. 2012;489:571–575. doi: 10.1038/nature11368. [DOI] [PubMed] [Google Scholar]
- 6.Langevin F, Crossan GP, Rosado IV, Arends MJ, Patel KJ. Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature. 2011;475:53–58. doi: 10.1038/nature10192. [DOI] [PubMed] [Google Scholar]
- 7.Garaycoechea JI, Patel KJ. Why does the bone marrow fail in Fanconi anemia? Blood. 2014;123:26–34. doi: 10.1182/blood-2013-09-427740. [DOI] [PubMed] [Google Scholar]
- 8.Hira A, et al. Variant ALDH2 is associated with accelerated progression of bone marrow failure in Japanese Fanconi anemia patients. Blood. 2013;122:3206–3209. doi: 10.1182/blood-2013-06-507962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nijnik A, et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447:686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- 10.Rossi DJ, et al. Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature. 2007;447:725–729. doi: 10.1038/nature05862. [DOI] [PubMed] [Google Scholar]
- 11.Milyavsky M, et al. A distinctive DNA damage response in human hematopoietic stem cells reveals an apoptosis-independent role for p53 in self-renewal. Cell Stem Cell. 2010;7:186–197. doi: 10.1016/j.stem.2010.05.016. [DOI] [PubMed] [Google Scholar]
- 12.Mohrin M, et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 2010;7:174–185. doi: 10.1016/j.stem.2010.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flach J, et al. Replication stress is a potent driver of functional decline in ageing haematopoietic stem cells. Nature. 2014;512:198–202. doi: 10.1038/nature13619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niedzwiedz W, et al. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Long DT, Räschle M, Joukov V, Walter JC. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science. 2011;333:84–87. doi: 10.1126/science.1204258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tan SLW, et al. A class of environmental and endogenous toxins induces BRCA2 haploinsufficiency and genome instability. Cell. 2017;169:1105–1118.e15. doi: 10.1016/j.cell.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bunting SF, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adamo A, et al. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell. 2010;39:25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 19.Pace P, et al. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 2010;329:219–223. doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- 20.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 21.Dykstra B, Olthof S, Schreuder J, Ritsema M, de Haan G. Clonal analysis reveals multiple functional defects of aged murine hematopoietic stem cells. J Exp Med. 2011;208:2691–2703. doi: 10.1084/jem.20111490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pang WW, et al. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc Natl Acad Sci USA. 2011;108:20012–20017. doi: 10.1073/pnas.1116110108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghezraoui H, et al. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol Cell. 2014;55:829–842. doi: 10.1016/j.molcel.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wyatt DW, et al. Essential roles for Polymerase θ-mediated end joining in the repair of chromosome breaks. Mol Cell. 2016;63:662–673. doi: 10.1016/j.molcel.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.García-Rubio ML, et al. The Fanconi anemia pathway protects genome integrity from R-loops. PLoS Genet. 2015;11:e1005674. doi: 10.1371/journal.pgen.1005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwab RA, et al. The Fanconi anemia pathway maintains genome stability by coordinating replication and transcription. Mol Cell. 2015;60:351–361. doi: 10.1016/j.molcel.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaber S, Toufektchan E, Lejour V, Bardot B, Toledo F. p53 downregulates the Fanconi anaemia DNA repair pathway. Nat Commun. 2016;7:11091. doi: 10.1038/ncomms11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oberbeck N, et al. Maternal aldehyde elimination during pregnancy preserves the fetal genome. Mol Cell. 2014;55:807–817. doi: 10.1016/j.molcel.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawashima N, et al. Aldehyde dehydrogenase-2 polymorphism contributes to the progression of bone marrow failure in children with idiopathic aplastic anaemia. Br J Haematol. 2015;168:460–463. doi: 10.1111/bjh.13122. [DOI] [PubMed] [Google Scholar]
- 30.Gu Y, et al. Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity. 1997;7:653–665. doi: 10.1016/S1074-7613(00)80386-6. [DOI] [PubMed] [Google Scholar]
- 31.Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. doi: 10.1002/1526-968X(200011/12)28:3/4<106::AID-GENE30>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 32.de Boer J, et al. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur J Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- 33.Donehower LA, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 34.Giri SD, Chatterjee A. Modulation of mitomycin C-induced sister chromatid exchanges and cell cycle delay by buthionine sulfoximine and reduced glutathione in mouse bone marrow cells in vivo. Mutat Res. 1998;413:227–234. doi: 10.1016/S1383-5718(98)00023-0. [DOI] [PubMed] [Google Scholar]
- 35.Orsburn B, et al. Differential requirement for H2AX and 53BP1 in organismal development and genome maintenance in the absence of poly(ADP)ribosyl polymerase 1. Mol Cell Biol. 2010;30:2341–2352. doi: 10.1128/MCB.00091-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geigl JB, Uhrig S, Speicher MR. Multiplex-fluorescence in situ hybridization for chromosome karyotyping. Nat Protoc. 2006;1:1172–1184. doi: 10.1038/nprot.2006.160. [DOI] [PubMed] [Google Scholar]
- 37.Gribble SM, et al. Massively parallel sequencing reveals the complex structure of an irradiated human chromosome on a mouse background in the Tc1 model of Down syndrome. PLoS One. 2013;8:e60482. doi: 10.1371/journal.pone.0060482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reinholdt L, Ashley T, Schimenti J, Shima N. Forward genetic screens for meiotic and mitotic recombination-defective mutants in mice. Methods Mol Biol. 2004;262:87–107. doi: 10.1385/1-59259-761-0:087. [DOI] [PubMed] [Google Scholar]
- 39.Kent D, Dykstra B, Eaves C. Isolation and assessment of long-term reconstituting hematopoietic stem cells from adult mouse bone marrow. Curr Protoc Stem Cell Biol. 2007 doi: 10.1002/9780470151808.sc02a04s3. [DOI] [PubMed] [Google Scholar]
- 40.Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13:102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Behjati S, et al. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature. 2014;513:422–425. doi: 10.1038/nature13448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nik-Zainal S, et al. The genome as a record of environmental exposure. Mutagenesis. 2015;30:763–770. doi: 10.1093/mutage/gev073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cabezas-Wallscheid N, et al. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell. 2014;15:507–522. doi: 10.1016/j.stem.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 44.Shen Z, et al. MPprimer: a program for reliable multiplex PCR primer design. BMC Bioinformatics. 2010;11:143. doi: 10.1186/1471-2105-11-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quail MA, et al. A large genome center’s improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krishna G, Fiedler R, Theiss JC. Simultaneous evaluation of clastogenicity, aneugenicity and toxicity in the mouse micronucleus assay using immunofluorescence. Mutat Res. 1992;282:159–167. doi: 10.1016/0165-7992(92)90090-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.