Abstract
Bifidobacteria are beneficial anaerobes, and their O2 sensitivity levels differ among species as a function of unknown molecular mechanisms. Bifidobacterium longum subspecies infantis (B. infantis), a predominant colonizer of the gastrointestinal tract of infants, showed a hyper O2-sensitive growth profile with accompanying a production of H2O2. In this study, we characterized an NADPH oxidase as a key enzyme responsible for this microbe’s hyper O2 sensitivity. A dominant active elution peak of H2O2-forming NADPH oxidase activity was detected in the first step of column chromatography, and the purified NADPH oxidase (NPOX) was identified as a homolog of nitroreductase family proteins. The introduction of the gene encoding B. infantis NPOX (npoxA) into O2-tolerant Bifidobacterium minimum made the strain O2 sensitive and allowed it to produce H2O2. Knockout of the npoxA gene in B. infantis decreased the production of H2O2 and mitigated its B. infantis hyper O2 sensitivity. A transcript of B. infantis npoxA is induced by O2, suggesting that the aerobic production of toxic H2O2 is functionally conserved in B. infantis.
Introduction
Oxygen (O2) has a negative effect on the growth of anaerobes1,2; however, the habitats of anaerobes, such as the gastrointestinal tract, are frequently contaminated with O2 that has been dissolved in food and beverages. Therefore, anaerobes must possess systems to adapt to aerated environments for survival in nature3,4. Recent studies have identified an O2-inducible O2- and ROS-reducing enzyme complex in obligate anaerobes such as sulfate-reducing bacteria5,6, Clostridium7,8, and Bacteroides9. These findings indicate that these obligate anaerobes possess excellent systems to avoid oxidative damage for maintaining their obligate anaerobiosis in aerated environments.
Several gut anaerobes such as O2-sensitive bifidobacteria and lactic acid bacteria are known to produce H2O2, a toxic reactive oxygen species (ROS), after exposure to O2, which inhibits the cell growth10–12. These anaerobes do not harbor the above-mentioned genes encoding the ROS-reducing enzyme complex in their genomes. NAD(P)H peroxidase or alkylhydroperoxide (Ahp) reductase systems (AhpF-AhpC or thioredoxin reductase–AhpC system), which detoxify H2O2 to H2O using NAD(P)H in aerobes and facultatively anaerobes13–16, have been identified in these anaerobes17–19; however, exogenous H2O2 production has been detected in these anaerobes20,21. These results suggest that H2O2 production and decomposition are controlled in vivo and play a role in the biological functions of these bacteria in aerated environments. H2O2 is highly toxic for a bacterium’s own cells; thus, the reason for this H2O2 production remains unclear22,23.
Members of the genus Bifidobacterium are gram-positive, catalase-negative anaerobes that are known to be beneficial to human health24–26. The degree of O2 sensitivity differs among species and strains in the genus11,27–35. de Vries and Stouthamer (1969) proposed that O2-sensitive Bifidobacterium species produce H2O2 under aerobic growth conditions through a reaction catalyzed by NADH oxidase activity, which can be detected in cell extracts11. According to our studies using liquid shaking cultures at different O2 concentrations, bifidobacterial strains can be classified into four groups: O2 hypersensitive (5% O2-sensitive), O2-sensitive (10% O2-sensitive), O2-tolerant (21% O2-tolerant), and O2-hypertolerant (microaerophilic)33,36,37. H2O2 production has been detected in strains belonging to the O2-hypersensitive and O2-sensitive groups when cell growth was inhibited by O233. The majority of O2-tolerant and hyper O2-tolerant strains have been isolated from non-human sources such as a honeybee hindgut38 and bovine rumen39. Bifidobacterium asteroides (B. asteroides) was isolated from a honeybee hindgut and possesses a heme-catalase, which is a rare characteristic among bifidobacteria24,38,40,41. Several reports have revealed that the addition of catalase to the culture medium11,33 or the introduction of ROS-detoxifying enzymes into O2-sensitive Bifidobacterium strains42,43 improved their aerobic growth, indicating that H2O2 production strongly correlates with the inhibition of aerobic growth. With respect to the enzymes that produce H2O2 in bifidobacteria, an H2O2-forming NADH oxidase was identified in an O2-sensitive strain of Bifidobacterium bifidum (B. bifidum), a type species in the genus Bifidobacterium, and the purified enzyme was identified as a b-type dihydroorotate dehydrogenase (DHOD), which is the enzyme that catalyzes oxidation of dihydroorotate to orotate in pyrimidine biosynthesis44. This enzyme is suggested to be involved in H2O2 production in B. bifidum, but its function has not been confirmed in vivo because of the lack of a gene disruption technique for this bacterium.
In the present study, Bifidobacterium longum subspecies infantis (B. infantis) showed a hyper O2-sensitive growth profiles. B. infantis has been reported as a champion colonizer in the gut microbiota of human infants45 and is reported to confer benefits to premature infants in terms of promoting anti-inflammatory activity and decreasing the risk of necrotizing enterocolitis46,47. Use of B. infantis as a bioactive probiotic for infants is highly anticipated; therefore, we set out to identify the mechanisms underlying its hyper-O2 sensitivity. Here, we identified an enzyme that produces H2O2 in B. infantis. The role of this protein and the mechanisms and evolutionary significance of H2O2 production in this bacterium are discussed.
Results
B. infantis showed a hyper O2-sensitive growth profile
The degree of O2 sensitivity of B. infantis JCM1222T (=ATCC15697 = DSM20088) was tested at several O2 concentrations in liquid shaking cultures. B. infantis grew well under anoxic conditions, and the optical density at 660 nm (OD660) reached 8 to 10 in the final growth stage. This level of growth was one of the most productive observed in MRS medium among the tested Bifidobacterium species36,37. Although several O2-sensitive bifidobacterial strains, including B. bifidum and Bifidobacterium longum (B. longum), grew well in liquid shaking cultures at 5% O233, the growth of B. infantis was found to be significantly inhibited at 5% O2 (Fig. 1). Therefore, we classified B. infantis as O2-hypersensitive. H2O2 production in the culture medium was detected at 10% and 20% O2 (Table 1).
Figure 1.
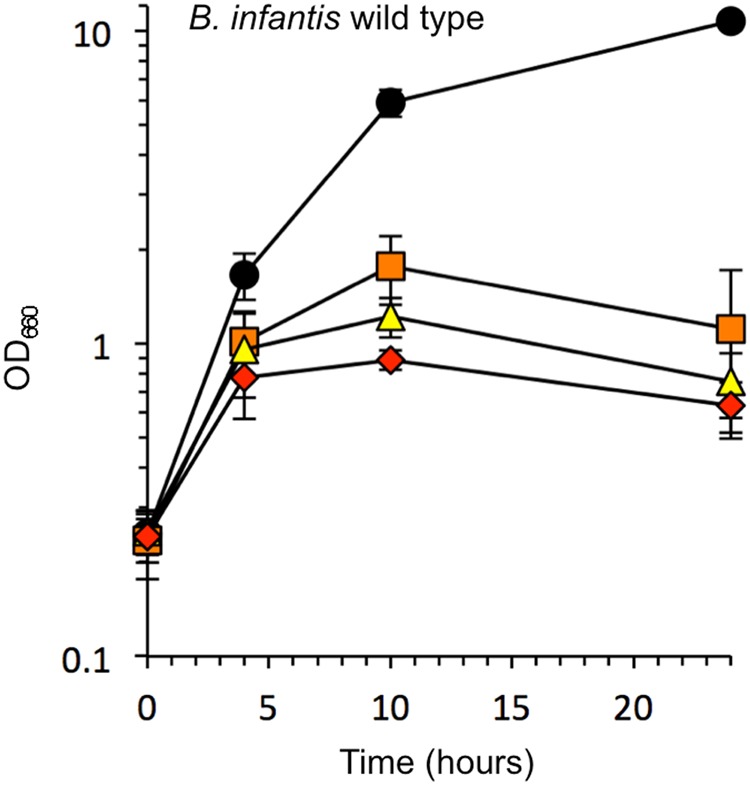
Growth of Bifidobacterium infantis JCM 1222T in liquid shake culture at various O2 concentrations. Circles, 100% N2 cultures; squares, 5% O2/95% N2 cultures; triangles, 10% O2/90% N2 cultures; diamonds, 20% O2/80% N2 cultures. Data represent the average ± SD of three independent cultures.
Table 1.
H2O2 accumulation of B. infantis and B. minimum under 0% to 20% O2 conditions.
| O2 (%) | H2O2 (µM)a | |||
|---|---|---|---|---|
| B. infantis wild type | B. infantis ∆npoxA | B. minimum pkkt427b | B. minimum + npoxA in pKKT427c | |
| 0 | NDd | ND | ND | ND |
| 5 | ND | ND | ND | ND |
| 10 | 29 ± 10 | (3 ± 1)e | ND | 172 ± 13 |
| 20 | 31 ± 6 | 14 ± 10 | ND | 169 ± 18 |
aH2O2 accumulation in the medium (24 h).
bVector pKKT427 was transformed into B. minimum.
cThe B. infantis npoxA gene was cloned in pKKT427 and transformed into B. minimum.
dND, not detected (less than 3 µM).
eData in parentheses are close to the reliable detection limit.
Data represent the average ± SD of three independent cultures.
Elution profiles of NAD(P)H oxidase activity in the first step of column chromatography
We previously identified the enzyme fractions that are involved in H2O2 production in an O2-sensitive B. bifidum strain using a Butyl-TOYOPEARL column44. In this study, we performed the same experiments for B. infantis. Proteins in the cell free extracts from B. infantis were fractionated according to their hydrophobic properties using a Butyl-TOYOPEARL column (Fig. 2A). One dominant elution peak of NADPH-dependent oxidase activity was detected, whereas NADH-dependent oxidase activity was detected as a minor elution peak. These major and minor peak fractions showed an H2O2-forming oxidase reaction in which stoichiometric production of H2O2 was detected by means of the reduction of O2. No significant NAD(P)H oxidase activity was detected in either the unbound fraction or the washed fractions in the first step of column chromatography.
Figure 2.
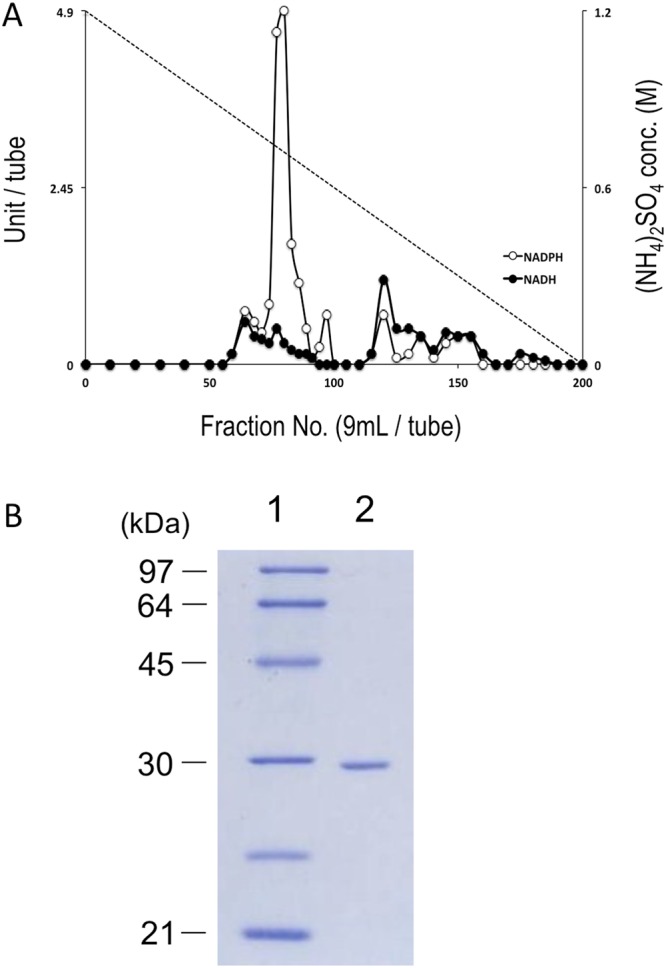
(A) Chromatographic elution profiles of B. infantis NADH and NADPH oxidases. Cell extracts, after treatment with streptomycin sulfate and ammonium sulfate, were applied to a Butyl-TOYOPEARL column equilibrated with 1.2 M ammonium sulfate in 50 mM potassium phosphate buffer (pH 7.0). After sample loading, the bound proteins were eluted with a linear gradient of ammonium sulfate, from 1.2 to 0 M, dissolved in the same buffer. Black circles, NADH oxidase activity; white circles, NADPH oxidase activity; dashed line, ammonium sulfate concentration (conc.). (B) SDS-PAGE of the purified B. infantis NPOX protein. After electrophoresis, the gel was stained with Coomassie brilliant blue. The protein standards (lane 1) and purified protein (lane 2) are indicated, along with the corresponding molecular masses (indicated on the left in kDa).
Purification and characterization of the NADPH oxidase in B. infantis
The fractions that showed NADPH oxidase activity were collected and purified by additional column chromatography steps. At each purification step, only one predominant active elution peak was detected. After the final step of affinity chromatography, the purified fractions appeared as a single band (29 kDa) by SDS-PAGE (Fig. 2B). The N-terminal sequence of the purified NADPH oxidase (NPOX) was sequenced, and the amino acid sequence (MVTNATIEALLGRRSIRKFK) showed 100% identity with the N-terminal sequence of the Blon_2447 gene product of B. infantis ATCC15697 in the genome database.
The gene encoding NPOX (designated as npoxA) was cloned from the B. infantis genome, and the amino acid sequence identity (% amino acid identity calculated from ClustalW alignment) showed 100% to that derived from BLON_RS12650 (Blon_2447). The translated product of B. infantis npoxA showed homology to proteins of the bacterial nitroreductase family and encodes a protein of 254 amino acids that shows ~35% identity to Escherichia coli oxygen-insensitive NADPH nitroreductase NfsA (accession number P17117)48, Bacillus subtilis NADPH-dependent nitroreductase NfrA1 (32% identity, accession number P39605)49, and Vibrio harveyi NADPH-flavin oxidoreductase FRP1 (35% identity, accession number Q56691)50. B. subtilis NfrA1 is reported to catalyze H2O2-forming NADH oxidase activity51, and V. harveyi FRP1 is reported to catalyze an FMN reductase reaction that is involved in a luciferase reaction that generates light in luminescent bacteria52. These NPOX homologs utilize NADH or NADPH or both as an electron donor to reduce substrates. The protein homologs are widely distributed in the genus Bifidobacterium. Homologous proteins were found in several B. longum strains (99% to 100%), Bifidobacterium breve (85% identity), B. bifidum (70% identity), Bifidobacterium minimum (B. minimum) (58% identity), and B. asteroides (52% identity). None of these proteins have been characterized in terms of function.
Enzymatic properties
The purified enzyme showed high affinity for NADPH, rather than NADH, as the electron donor. The Km values for NADPH and NADH when O2 was used as an electron acceptor were 1.38 ± 0.12 and 113.3 ± 7.3 µM, respectively. The purified B. infantis NPOX showed a typical flavoprotein spectrum with absorbance maxima at 280, 373, and 444 nm (Fig. S2). The bound flavin was identified as FMN by HPLC analysis. In the NADPH oxidase reaction, stoichiometric production of H2O2 was detected following the reaction with O2; e.g., 1 µg enzyme consumed 28.3 nmol O2/5 min/mL, and 13.8 nmol O2/mL was produced after the addition of catalase. This result indicated that the purified enzyme reduces O2 by two reducing equivalents to H2O2. The purified enzyme used flavins and nitro compounds as electron acceptors under anoxic conditions (Table 2). The pH optimum for the NADPH oxidase reaction was found to be 5.5–6.0. The temperature optimum was approximately 30–40 °C, and activity significantly decreased at temperatures above 40 °C. The specific activity of this protein under air-saturated conditions at 37 °C was 13.8 ± 4.3 U/mg protein.
Table 2.
Identification of electron acceptors of B. infantis NPOX.
| Electron acceptora | Relative activity (%)b |
|---|---|
| O2 | 100 |
| DCIP | 358 |
| FAD | 229 |
| FMN | 74 |
| Nitrobenzene | 2090 |
| Nitrofurazone | 725 |
aRelative activity (%) was calculated in comparison with the activity of NADPH oxidase.
Data are the average of two independent measurements that varied by less than 5%.
Transformation of the B. infantis npoxA into an O2-tolerant B. minimum strain
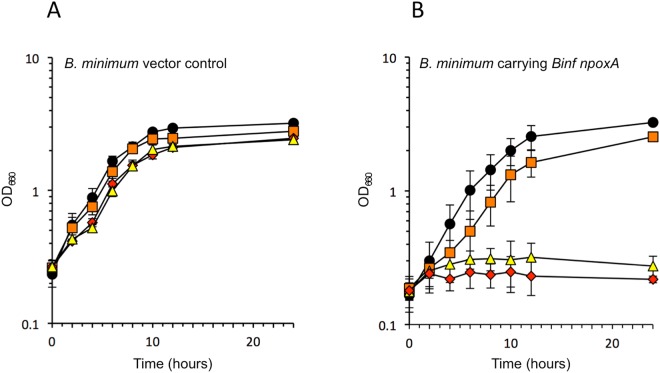
B. minimum DSM 20102T, which is an isolate from sewage53, showed an O2-tolerant profile32,37, and liquid shaking growth was not significantly inhibited in the presence of 20% O2 (Fig. 3A). Therefore, we classified B. minimum as a member of the O2-tolerant group and used this microbe for further study. The E. coli-Bifidobacterium shuttle vector pKKT427, originally developed for the transformation of B. longum strain 105-A54,55, was successfully transformed into B. minimum in this study; therefore, we used this vector to express B. infantis NPOX in B. minimum. B. infantis npoxA was cloned into pKKT427, and then the growth of transformants was tested at different O2 concentrations. The production of H2O2 was not detected in a B. minimum wild-type strain carrying pKKT427 in the presence of O2 (Table 1). However, B. minimum carrying npoxA in pKKT427 showed an O2-sensitive growth profile in the presence of 10% and 20% O2 with accompanying the production of H2O2 (Fig. 3B, Table 1). These results indicated that B. infantis NPOX is responsible for the O2 sensitivity of and H2O2 production by B. minimum.
Figure 3.
Growth of Bifidobacterium minimum DSM 20102T in liquid shake culture at various O2 concentrations with (B) or without (A) the B. infantis npoxA gene in the vector pKKT427. Circles, 100% N2 cultures; squares, 5% O2/95% N2 cultures; triangles, 10% O2/90% N2 cultures; diamonds, 20% O2/80% N2 cultures. Data represent the average ± SD of three independent cultures.
The growth of a B. infantis npoxA gene knockout strain
Single-gene transformation of the O2-tolerant B. minimum strain significantly increased H2O2 production and O2 sensitivity. To determine the function of the npoxA product in B. infantis, we constructed an npoxA deletion mutant (∆npoxA) (Fig. S1), and then the growth of this mutant was compared with that of a wild-type strain of B. infantis. B. infantis ∆npoxA grew well at 5% O2, and H2O2 production was significantly decreased at 10% and 20% O2 (Fig. 4, Table 1). These results indicated that the npoxA product contributes to both aerobic growth inhibition and H2O2 production in B. infantis. Candidates for the production of residual H2O2 were the minor active fractions of the NADH-dependent H2O2-forming oxidase (Fig. 2A).
Figure 4.
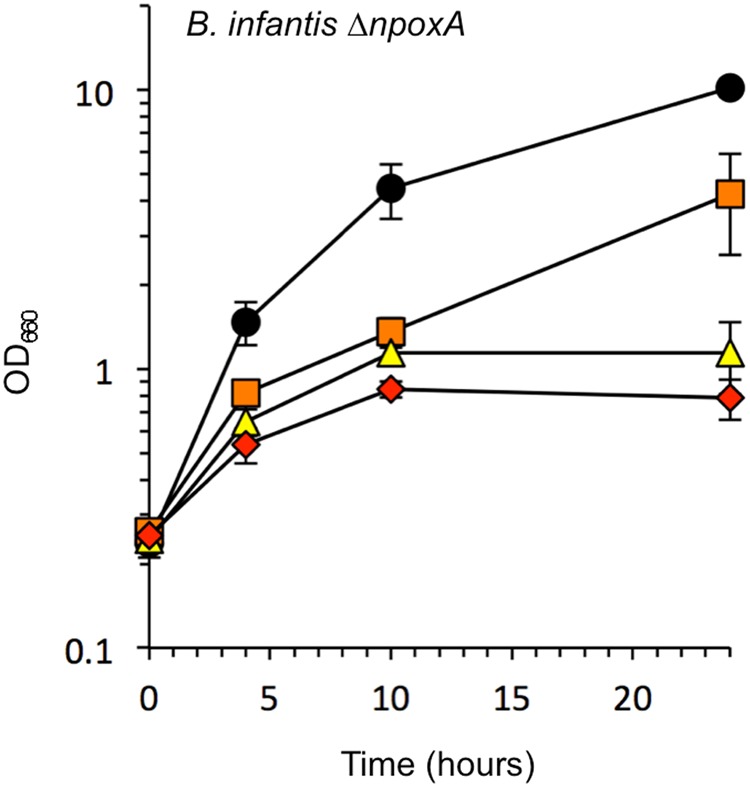
Growth of the ∆npoxA mutant of B. infantis in liquid shake culture at various O2 concentrations. Circles, 100% N2 cultures; squares, 5% O2/95% N2 cultures; triangles, 10% O2/90% N2 cultures; diamonds, 20% O2/80% N2 cultures. Data represent the average ± SD of four independent experiments.
Expression profiles of genes encoding B. infantis NADPH oxidase and alkylhydroperoxide reductase
To determine the effect of O2 on the expression of the gene encoding B. infantis NADPH oxidase (npoxA), we performed a Northern blot analysis. The result showed that B. infantis npoxA was significantly upregulated within 30 min after the start of 5% O2 aeration (Fig. 5B). We also determined the expression of genes encoding alkylhydroperoxide reductase (Ahp), which is an enzyme complex composed of AhpF and AhpC that detoxifies ROS. TrxR is an AhpF homolog in B. infantis, and ahpC-trxR genes are tandemly located in the genome (Fig. 5A). Northern analyses indicated that B. infantis ahpC was strongly upregulated within 10 minutes after the start of aeration (Fig. 5B). These results indicate that B. infantis express the genes for ROS production and for ROS detoxification in response to O2 stress.
Figure 5.
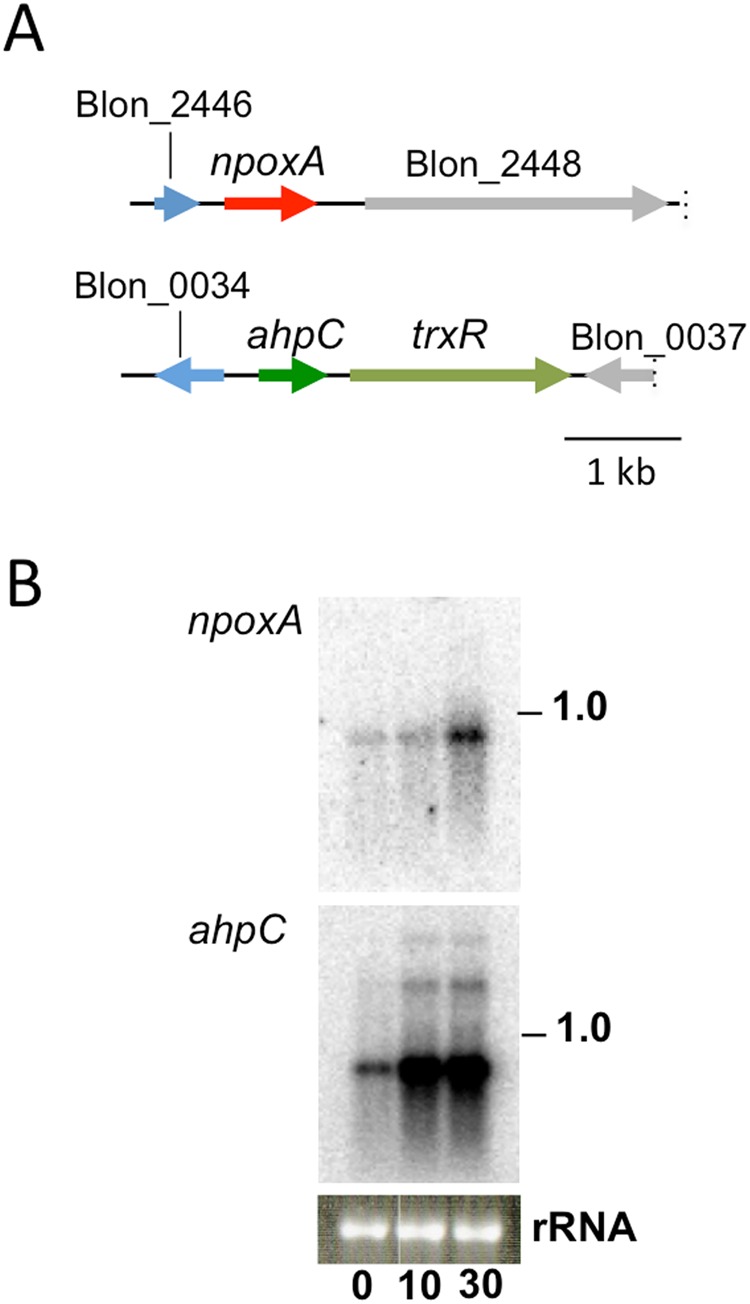
Genome structures and gene expression profiles of B. infantis npoxA and ahpC. (A) Genome structure of the npoxA (upper) and the ahpC (lower). (B) Northern blot analyses of 5 µg of B. infantis total RNA probed with npoxA or ahpC. 0, immediately before the start of aeration with 5% O2 in the mid-exponential phase; 10, after 10 min; 30, after 30 min. The estimated sizes of the observed transcripts are indicated on the right. Ethidium bromide staining of the ribosomal RNA (rRNA) to confirm equal RNA loading is shown below the autoradiogram. The membranes were reused for reprobing different probes.
Discussion
The growth of bifidobacterial strains is inhibited by O2, and this inhibition has been shown to occur in conjunction with H2O2 production. To date, b-type DHOD from an O2-sensitive strain of B. bifidum, has been identified as the only purified H2O2-forming NADH oxidase in the genus Bifidobacterium. In this study, we detected a dominant H2O2-forming NADPH oxidase activity peak in an O2-hypersensitive strain of B. infantis. This NADPH-specific peak was not detected in B. bifidum using the same hydrophobic column chromatography44, thus suggesting that the mechanisms of O2 sensitivity in these O2-sensitive strains differ from each other.
Purified B. infantis H2O2-forming NPOX showed high similarity (~30% identity) with proteins of the bacterial nitroreductase family. B. infantis NPOX exhibited nitroreductase- and flavin reductase-activity under anoxic conditions. The role of B. infantis H2O2-forming NPOX under anaerobic conditions is unclear; however, based on the O2-inducible gene expression profile, NPOX is expected to be functionally involved in the aerobic life of B. infantis. Similar expression of a transcript, from a gene that encodes an NPOX ortholog (Gene ID: BL0139), was observed by Oberg et al. in microarray data from the H2O2-sensitive strain B. longum D2975 and the H2O2-tolerant strain B. longum NCC270556. The NPOX homolog transcript was induced only in the H2O2-sensitive D2975 strain under oxidative stress. To clarify the expression of NPOX and its correlation with bifidobacterial O2 sensitivity, it would be desirable to compare the expression profiles and kinetics of NPOX homologs in other O2-tolerant and O2-sensitive strains37.
H2O2 is highly toxic to cells, and therefore, Bifidobacterium strains need to conserve systems to reduce the amount of H2O2 to avoid cell death. Alkylhydroperoxide reductase, a component of the AhpF-AhpC system that is known to decompose H2O2 efficiently, is widely conserved in bifidobacterial genomes. Although the function of this enzyme complex has not been characterized in the genus Bifidobacterium, the upregulation of ahpC (or the protein AhpC) in response to O2 or H2O2 stress has been reported in strains such as Bifidobacterium animalis subsp. lactis35 and B. longum56. These data suggested that ahpC is involved in the oxidative stress protection of bifidobacterial strains. In the present study, B. infantis induced ahpC within 10 minutes after exposure to O2. Interestingly, the induction of ahpC was more rapid than that of B. infantis npoxA (Fig. 5). This gene expression program is considered to be important for the strain to rapidly prepare the defense system against the generation of H2O2.
In this study, knockout of npoxA in B. infantis enabled the strain to grow under microaerobic conditions, with a decrease in H2O2 production under aerobic conditions. Improvement of bacterial O2 sensitivity by single-gene knockout has been demonstrated in the obligate anaerobe Bacteroides fragilis, with deletion of oxe enabling the strain to grow under micro-oxic conditions (as high as 2% O2)57. Although the function of the oxe gene product has not yet been identified, a mutation in oxe enhanced the H2O2-scavenging activity of the mutant relative to that of the wild type. The reason for the production of a toxic H2O2 in anaerobes has been unknown; however, it is obvious that both B. fragilis and B. infantis control the production and degradation of H2O2 according to their own genetic programs.
We conclude here that B. infantis induces the expression of NPOX in response to an increase in O2, leading to the production of H2O2 to maintain anaerobiosis. B. infantis ∆npoxA still produced H2O2 at elevated O2 concentrations, indicating the presence of another H2O2-forming oxidase in vivo. A b-type DHOD, that is conserved in the genome of B. infantis, is a likely candidate. Further characterization of the residual H2O2 production in B. infantis, as well as analysis of the enzyme kinetics of the O2-tolerant B. minimum NPOX homolog, will be needed to describe the mechanisms of anaerobiosis in these strains.
Methods
Bacterial strain and growth conditions
B. infantis (Bifidobacterium longum subsp. infantis) JCM 1222T (=ATCC 15697T, DSM 20088T) was used in this study. B. infantis was grown anaerobically at 37 °C in modified MRS medium (pH 6.7). The culturing conditions under several O2 concentrations were as described previously33.
Chemicals
All chemicals were of analytical grade. ß-NADPH, ß-NADH, FAD, FMN, riboflavin, nitrobenzene, nitrofurazone, and 2,6-dichlorobenzenoneindophenol (DCIP) were from Sigma. Water was prepared with an Ellix-10 Milli-Q ultra-pure water system (Millipore, Tokyo, Japan).
Enzyme purification
Microaerobically grown B. infantis cells (cultured statically but stirred with a magnetic stirrer in 3 liters of medium in a 5-liter flask with a cotton plug) were harvested for enzyme purification. NAD(P)H oxidase activity was assayed spectrophotometrically in 1 ml of air saturated 50 mM potassium phosphate buffer (pH 6.5) at 37 °C. The reaction was started by the addition of enzyme solution, and the decrease in absorbance at 340 nm (ε340 = 6,220 M−1·cm−1) was monitored with a spectrophotometer (HITACHI U-3300, Hitachi, Japan). One unit of activity was defined as the amount of enzyme that catalyzes the oxidation of 1 µmol NAD(P)H per minute.
Microaerobically grown B. infantis cells (80 g) were suspended in 240 ml of 50 mM potassium phosphate buffer, pH 6.5, containing 0.1 mM DTT, 0.2 mM PMSF, and 5 mM EDTA, and then the cells were disrupted by treatment with a French pressure cell at 140 MPa. All purification procedures were carried out at 4 °C or on ice. Cell free extracts (CFE) were obtained by removing cell debris by centrifugation at 39,000 g for 15 min. The cytoplasmic solution obtained by ultracentrifugation at 100,000 g for 2 h was treated with 10% streptomycin sulfate (dissolved in 10 mM potassium phosphate buffer, pH 7.0) to remove nucleic acids. After centrifugation at 39,000 g for 15 min, the supernatant was fractionated by the stepwise addition of solid ammonium sulfate (20%). The supernatant obtained after centrifugation at 30,000 g was dissolved in 50 mM potassium phosphate buffer, pH 7.0, containing 1 M ammonium sulfate, 5 mM EDTA, 0.2 mM phenylmethanesulfonyl fluoride (PMSF), and 0.1 mM dithiothreitol (DTT). After centrifugation at 39.000 g for 15 min, the supernatant was applied on a hydrophobic interaction chromatography using Butyl-TOYOPEARL 650 s (TOSOH, Tokyo, Japan) column chromatography. The enzyme was eluted with a linear gradient from 1.2 M to 0 M ammonium sulfate dissolved in the same buffer. After the Butyl-TOYOPEARL chromatography, active fractions of NADPH oxidase activity were obtained. The solution after Butyl-TOYOPEARL chromatography was dialyzed against 50 mM potassium phosphate buffer, pH 6.5, containing 0.5 mM EDTA, 0.2 mM PMSF, and 0.1 mM DTT for 6 h, and this procedure was repeated 3 times using freshly prepared buffer each time. The enzyme solution was then applied to DEAE Sephacel (GE Healthcare, USA) column chromatography. The column was sequentially washed with the same buffer containing 0 mM and 150 mM NaCl, and eluted with buffer containing 250 mM NaCl. The active fractions were pooled and dialyzed for 12 h against 10 mM potassium phosphate buffer, pH 6.5, containing 0.5 mM EDTA and 0.1 mM DTT and subjected to hydroxyapatite (WAKO, Japan) column chromatography. After applying the enzyme solution to the hydroxyapatite column, the protein was sequentially washed with 10 mM and 50 mM potassium phosphate buffer, pH 6.5, and eluted with 100 mM potassium phosphate buffer, pH 6.5. The active fractions were collected and concentrated using Amicon Ultra centrifugal filter units (30,000 Da cutoff; MILLIPORE, Cork, Ireland). The concentrated enzyme was applied to POLOS HQ/H (PerSeptive Biosystems, Framingham, USA) column chromatography. The column was sequentially washed with 50 mM potassium phosphate buffer (pH 6.5), then eluted with a linear gradient of 0 mM–250 mM NaCl. The active fraction was collected and concentrated using Amicon Ultra. After this chromatography, the enzyme purity in the active fractions was checked by SDS-PAGE with Coomassie Brilliant Blue G-250 staining or silver staining. By using this enzyme concentrator, the basal buffer of the enzyme solution was changed to 50 mM potassium phosphate buffer, pH 6.5, by diluting the concentrated enzyme solution 100-fold with the new buffer, concentrating to the original volume, and then repeating the dilution and concentration steps; the second concentrated enzyme solution was subjected to enzyme assay. Protein concentration was determined by the dye-binding assay58.
The purified enzyme was subjected to SDS-PAGE and electroblotted onto polyvinylidene difluoride membranes (NIPPON GENETICS, Tokyo, Japan). The N-terminal amino acid sequence was determined by the Edman degradation method using the peptide sequencer described previously7. The UV-Vis absorption spectrum was recorded on a Hitachi U3300 spectrophotometer (Hitachi, Tokyo, Japan) using a 1 cm path length quartz cuvette.
The bound flavins were extracted and determined by HPLC analysis with fluorescence detector according to a previously described method using a CAPCELL PACK C18 column (4.6 mm I.D. × 150 mm L; SHISEIDO CO., LTD. Tokyo, Japan) with 5 mM ammonium acetate in methanol as the mobile phase7. Riboflavin, FAD and FMN were used as standards.
Enzyme properties
The optimal pH of the purified enzyme was determined in 50 mM potassium phosphate buffer in the pH range of pH 5.0 – pH 8.0 at 37 °C, which is the optimum growth temperature for B. infantis. The temperature optimum of the purified enzyme was determined in 50 mM potassium phosphate buffer, pH 6.5, over a temperature range from 25 to 60 °C.
H2O2 production by the B. infantis NAD(P)H oxidase reaction was detected while monitoring O2 production using an oxygen electrode by adding catalase into the reaction vessel8,33,44. Catalase catalyzes the stoichiometric conversion of 1 mol H2O2 to 1 mol H2O and 1/2 mol O2. The H2O2-forming type NAD(P)H oxidase produces 50% O2 after the addition of catalase to the total amount of O2 consumed by the NAD(P)H oxidase reaction.
The substrate specificity of the purified enzyme was assayed spectrophotometrically at 37 °C using 50 mM potassium phosphate buffer, pH 6.5. Electron acceptors for a purified enzyme were investigated by adding various substrates (final concentration, 50 µM) to the anaerobic cuvette together with the enzyme solution. For all acceptors except O2, the reactions were carried out under anoxic conditions by purging the anaerobic glass cuvette containing the reaction buffer with O2-free argon gas. NAD(P)H:O2 oxidoreductase activity was assayed under air saturated conditioin. NAD(P)H (final 150 µM) was used as an electron donor for the enzyme. For the reactions of NAD(P)H:acceptor oxidoreductase, one unit of activity was defined as the amount of enzyme that catalyzes the reduction of 1 µmol of NAD(P)H (ε340 = 6,220 M−1 cm−1) per minute. For the assay of DCIP, one unit of activity was defined as the amount of enzyme that catalyzes the reduction of 1 µmol of DCIP (ε600 = 22,000 M−1·cm−1) per minute. The values for relative activities (%) and the specific activities (U/mg protein) are the average of two independent measurements that varied by less than 5%. The apparent Km values for NADH and NADPH were determined by varying the concentrations of both NAD(P)H in 50 mM potassium phosphate buffer, pH 6.5 using the Enzyme Kinetics Module 1.3 (SigmaPlot 11, SYSTAT Software, Chicago, IL). Initial rates were determined from linear plots of NAD(P)H reduction.
Northern hybridization
Northern hybridization was performed as described previously8. Total RNA (5 µg) extracted from B. infantis cells harvested before or after the 5% O2 stress was loaded on agarose gels and blotted onto nylon membranes (Hybond N+, GE Healthcare, Cicago, IL, USA). The membrane was then probed with the 32P-labeled gene probe of B. infantis npoxA. The membrane was subsequently stripped of the probe and reprobed with a probe of B. infantis ahpC. Northern hybridization using the same membrane was repeated twice to confirm the results. The following oligonucleotide primer pairs were used to amplify the probes for B. infantis npoxA: 5′-ATGGTTACCAACGCAACAAT and 52-CTAATCCAGACGGAAGCCCT, B. infantis ahpC: 5′-ATGACTCTTCTGCAGCATGA and 52-TCACAGCTGGCCGACGAGGT. Before hybridization with a different probe, the membrane was washed at 90 °C for 2 minutes in a stripping buffer (5% SDS in 50 mM Tris-HCl, pH7.5) and reused for the next northern analysis by reprobing with a different gene probe.
Expression of B. infantisnpoxA in B. minimum
B. infantis npoxA including its promoter region was amplified by PCR using the forward primer (5′-TAAAAGCTTTGGATGATGGTTTCTGTTGG) included a HindIII restriction site, and the reverse primer (5′-TAAGCGGCCGCCTAATCCAGACGGAAGCCCTG) included a NotI restriction site. The purified PCR product was digested with HindIII and NotI and ligated into HindIII- and NotI-digested pKKT427 vector54, resulting in the plasmid pBInpoxA, which was transformed into B. minimum by electroporation. B. minimum cells carrying pBInpoxA or cells carrying a control vector pKKT427 were tested the growth in MRS medium containing spectinomycin (Sp) (80 µg/ml) under several O2 concentrations.
Gene knockout of npoxA
A plasmid pKO403Cm-∆npoxA for the knockout of the B. infantis npoxA gene was constructed as follows (Fig. S1). The PCR primers were designed according to the npoxA gene sequence (BLON_RS12650) and the flanking genomic regions of B. infantis JCM1222T (=ATCC15697) (GenBank Accession no. NC_011593). To obtain the npoxA deletion DNA fragments, upstream (1.35 kb) and downstream (1.36 kb) regions of the putative npoxA gene of B. infantis were amplified by PCR using the primers shown in Fig. S1B. The produced DNA fragments were connected to the upstream and downstream regions of the Sp resistance gene (Spr) by Golden Gate Assembly59 with SapI as the Type IIS restriction enzyme. The obtained fragments were cloned into pKO403Cm, a Bifidobacterium-Escherichia coli shuttle vector60, which carries a Golden Gate cloning site, chloramphenicol (Cm) resistance gene (Cmr), and temperature-sensitive replicon origin60. B. infantis cells were transformed with pKO403Cm-∆npoxA by electroporation55. After transformation, the cells were spread and cultured at 42 °C on MRS plates containing Sp (50 µg/ml). The transformants were transferred onto two MRS agar plates containing either Sp or Cm. The obtained Sp-resistant/Cm-sensitive transformants were selected as candidates for the double-crossover mutant. The deletion of npoxA was confirmed by PCR using Primer-1 and Primer-2 (Fig. S1C) and nucleotide sequencing to confirm the precise disruption.
Electronic supplementary material
Acknowledgements
We thank many colleagues for advice, discussion, and technical assistance, especially Mr. Ken Makino and Mr. Susumu Ijyuin. This work was supported by a grant-in-aid for scientific research from the Japan Society for the Promotion of Science (No. 20580086 to S.K.).
Author Contributions
K.T., Ta.S., J.K., S.U. performed the experiments and analysis. I.N. performed the knockout mutation. Y.K. and To.S. supervised genetic experiments and provided useful comments. Y.N. conceived and provided useful comments. K.T. and S.K. wrote the manuscript. S.K. conceived and designed this study. All authors discussed the results and commented on the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-29030-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.McCord JM, Keele BB, Jr., Fridovich I. An enzyme-based theory of obligate anaerobiosis: the physiological function of superoxide dismutase. Proc. Natl. Acad. Sci. USA. 1971;68:1024–1027. doi: 10.1073/pnas.68.5.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Morris JG. Oxygen and the obligate anaerobe. J. Appl. Bacteriol. 1976;40:229–244. doi: 10.1111/j.1365-2672.1976.tb04171.x. [DOI] [PubMed] [Google Scholar]
- 3.Pan N, Imlay JA. How does oxygen inhibit central metabolism in the obligate anaerobe Bacteroides thetaiotaomicron. Mol. Microbiol. 2001;39:1562–1571. doi: 10.1046/j.1365-2958.2001.02343.x. [DOI] [PubMed] [Google Scholar]
- 4.Baughn AD, Malamy MH. The strict anaerobe Bacteroides fragilis grows in and benefits from nanomolar concentrations of oxygen. Nature. 2004;427:441–444. doi: 10.1038/nature02285. [DOI] [PubMed] [Google Scholar]
- 5.Gomes CM, et al. Studies on the redox centers of the terminal oxidase from Desulfovibrio gigas and evidence for its interaction with rubredoxin. J. Biol. Chem. 1997;272:22502–22508. doi: 10.1074/jbc.272.36.22502. [DOI] [PubMed] [Google Scholar]
- 6.Lumppio HL, Shenvi NV, Summers AO, Voordouw G, Kurtz DM., Jr. Rubrerythrin and rubredoxin oxidoreductase in Desulfovibrio vulgaris: a novel oxidative stress protection system. J. Bacteriol. 2001;183:101–108. doi: 10.1128/JB.183.1.101-108.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawasaki S, et al. Adaptive responses to oxygen stress in obligatory anaerobes Clostridium acetobutylicum and Clostridium aminovalericum. Appl. Environ. Microbiol. 2005;71:8442–8450. doi: 10.1128/AEM.71.12.8442-8450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawasaki S, Sakai Y, Takahashi T, Suzuki I, Niimura Y. O2 and reactive oxygen species detoxification complex composed of O2-responsive NADH:rubredoxin oxidoreductase – flavoprotein A2 – desulfoferrodoxin operon enzymes, rubperoxin, and rubredoxin in Clostridium acetobutylicum. Appl. Environ. Microbiol. 2009;75:1021–1029. doi: 10.1128/AEM.01425-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mishra S, Imlay JA. An anaerobic bacterium, Bacteroides thetaiotaomicron, uses a consortium of enzymes to scavenge hydrogen peroxide. Mol. Microbiol. 2013;90:1356–1371. doi: 10.1111/mmi.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whittenbury R. Hydrogen peroxide formation and catalase activity in the lactic acid bacteria. J. Gen. Microbiol. 1964;35:13–26. doi: 10.1099/00221287-35-1-13. [DOI] [PubMed] [Google Scholar]
- 11.de Vries W, Stouthamer AH. Factors determining the degree of anaerobiosis of Bifidobacterium strains. Arch. Mikrobiol. 1969;65:275–287. doi: 10.1007/BF00407109. [DOI] [PubMed] [Google Scholar]
- 12.Thomas EL, Pera KA. Oxygen metabolism of Streptococcus mutans: uptake of oxygen and release of superoxide and hydrogen peroxide. J. Bacteriol. 1983;154:1236–1244. doi: 10.1128/jb.154.3.1236-1244.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niimura Y, Poole LB, Massey V. Amphibacillus xylanus NADH oxidase and Salmonella typhimurium alkylhydroperoxide reductase flavoprotein components show extremely high scavenging activity for both alkyl hydroperoxide and hydrogen peroxide in the presence of S. typhimurium alkyl-hydroperoxide reductase 22-kDa protein component. J. Biol. Chem. 1995;270:25645–25650. doi: 10.1074/jbc.270.43.25645. [DOI] [PubMed] [Google Scholar]
- 14.Poole LB, Ellis HR. Flavin-dependent alkyl hydroperoxide reductase from Salmonella typhimurium. 1. Purification and enzymatic activities of overexpressed AhpF and AhpC proteins. Biochemistry. 1996;35:56–64. doi: 10.1021/bi951887s. [DOI] [PubMed] [Google Scholar]
- 15.Baker LM, Raudonikiene A, Hoffman PS, Poole LB. Essential thioredoxin-dependent peroxiredoxin system from Helicobacter pylori: genetic and kinetic characterization. J. Bacteriol. 2001;183:1961–1973. doi: 10.1128/JB.183.6.1961-1973.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seaver LC, Imlay JA. Alkyl hydroperoxide reductase in the primary scavenger of endogenous hydrogen peroxide in. Escherichia coli. J. Bacteriol. 2001;183:7173–7181. doi: 10.1128/JB.183.24.7173-7181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higuchi M, et al. Functions of two types of NADH oxidases in energy metabolism and oxidative stress of Streptococcus mutans. J. Bacteriol. 1999;181:5940–5947. doi: 10.1128/jb.181.19.5940-5947.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klijn A, Mercenier A, Arigoni F. Lessons from the genomes of bifidobacteria. FEMS Microbiol. Rev. 2005;29:491–509. doi: 10.1016/j.fmrre.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 19.Xiao M, et al. Oxidative stress-related responses of Bifidobacterium longum subsp. longum BBMN68 at the proteomic level after exposure to oxygen. Microbiology. 2011;157:1573–1588. doi: 10.1099/mic.0.044297-0. [DOI] [PubMed] [Google Scholar]
- 20.Pericone CD, Overweg K, Hermans PW, Weiser JN. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect. Immun. 2000;68:3990–3997. doi: 10.1128/IAI.68.7.3990-3997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pridmore RD, Pittet AC, Praplan F, Cavadini C. Hydrogen peroxide production by Lactobacillus johnsonii NCC 533 and its role in anti- Salmonella activity. FEMS Microbiol. Lett. 2008;283:210–215. doi: 10.1111/j.1574-6968.2008.01176.x. [DOI] [PubMed] [Google Scholar]
- 22.Imlay JA. Pathways of oxidative damage. Annu. Rev. Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 23.Mishra S, Imlay J. Why do bacteria use so many enzymes to scavenge hydrogen peroxide? Arch. Biochem. Biophys. 2012;525:145–160. doi: 10.1016/j.abb.2012.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones, D. & Collins, M. D. Irregular, nonsporing Gram-positive rods, p. 1261–1434. In Sneath, P. H. A, Mair, N. S., M. Sharpe, E. & Holt, J. G. (eds), Bergey’s manual of systematic bacteriology. Williams and Wilkins Co, Baltimore. (1986).
- 25.Leahy SC, Higgins DG, Fitzgerald GF, van Sinderen D. Getting better with bifidobacteria. J. Appl. Microbiol. 2005;98:1303–1315. doi: 10.1111/j.1365-2672.2005.02600.x. [DOI] [PubMed] [Google Scholar]
- 26.Riviere A, Selak M, Lantin D, Leroy F, Vuyst LD. Bifidobacteria and butyrate producing colon bacteria: importance and strategies for their stimulation in the human gut. Front. Microbiol. 2016;7:979. doi: 10.3389/fmicb.2016.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimamura S, et al. Relationship between oxygen sensitivity and oxygen metabolism of Bifidobacterium species. J. Dairy. Sci. 1992;75:3296–3306. doi: 10.3168/jds.S0022-0302(92)78105-3. [DOI] [PubMed] [Google Scholar]
- 28.Meile L, et al. Bifidobacterium lactis sp. nov., a moderately oxygen-tolerant species isolated from fermented milk. Syst. Appl. Microbiol. 1997;20:57–64. doi: 10.1016/S0723-2020(97)80048-3. [DOI] [Google Scholar]
- 29.Ahn JB, Hwang HJ, Park JH. Physiological responses of oxygen-tolerant anaerobic Bifidobacterium longum under oxygen. J. Microbiol. Biotechnol. 2001;11:443–451. [Google Scholar]
- 30.Talwalkar A, Kailasapathy K. Metabolic and biochemical responses of probiotic bacteria to oxygen. J. Dairy. Sci. 2003;86:2537–2546. doi: 10.3168/jds.S0022-0302(03)73848-X. [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez R, Blancas A, Santillana R, Azaola A, Wacher C. Growth and final product formation by Bifidobacterium infantis in aerated fermentations. Appl. Microbiol. Biotechnol. 2004;65:606–610. doi: 10.1007/s00253-004-1603-9. [DOI] [PubMed] [Google Scholar]
- 32.Simpson PJ, Stanton C, Fitzgerald GF, Ross RP. Intrinsic tolerance of Bifidobacterium species to heat and oxygen and survival following spray drying and storage. J. Appl. Microbiol. 2005;99:493–501. doi: 10.1111/j.1365-2672.2005.02648.x. [DOI] [PubMed] [Google Scholar]
- 33.Kawasaki S, Mimura T, Satoh T, Takeda K, Niimura Y. Response of microaerophilic Bifidobacterium species, B. boum and B. thermophilum, to oxygen. Appl. Environ. Microbiol. 2006;72:6854–6858. doi: 10.1128/AEM.01216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kawasaki S, et al. Effect of CO2 on colony development by Bifidobacterium species. Appl. Environ. Microbiol. 2007;73:7796–7798. doi: 10.1128/AEM.01163-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruiz L, et al. Molecular clues to understand the aerotolerance phenotype of Bifidobacterium animalis subsp. lactis. Appl. Environ. Microbiol. 2012;78:644–650. doi: 10.1128/AEM.05455-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawasaki, S. Response of Bifidobacterium species to oxygen, p 103–110. In Sonomoto, K. & Yokota, A. (eds), Lactic Acid Bacteria and Bifidobacteria, Caister Academic Press, Norfolk, UK. (2011).
- 37.Kawasaki, S., Watanabe, M., Fukiya, S. & Yokota, A. Stress responses of Bifidobacteria: oxygen and bile acid as the stressors, p 131–143. In Mattarelli, P., Biavati, B., Holzapfel, W. & Wood, B. J. B. (eds), The bifidobacteria and related organisms, Academic Press, Oxford, UK. (2017).
- 38.Scardovi V, Trovatelli LD. New species of bifido bacteria from Apis mellifica L. and Apis indica F. A contribution to the taxonomy and biochemistry of the genus Bifidobacterium. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg. 1969;123:64–88. [PubMed] [Google Scholar]
- 39.Scardovi V, Trovatelli LD, Biavati B, Zani G. Bifidobacterium cuniculi, Bifidobacterium choerinum, Bifidobacterium boum, and Bifidobacterium pseudocatenulatum: four new species and their deoxyribonucleic acid homology relationships. Int. J. Syst. Bacteriol. 1979;29:291–311. doi: 10.1099/00207713-29-4-291. [DOI] [Google Scholar]
- 40.Hayashi K, et al. Purification and characterization of oxygen-inducible haem catalase from oxygen-tolerant Bifidobacterium asteroides. Microbiology. 2013;159:89–95. doi: 10.1099/mic.0.059741-0. [DOI] [PubMed] [Google Scholar]
- 41.Bottacini F, et al. Bifidobacterium asteroides PRL2011 genome analysis reveals clues for colonization of the insect Gut. PLoS One. 2012;7:e44229. doi: 10.1371/journal.pone.0044229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He J, Sakaguchi K, Suzuki T. Acquired tolerance to oxidative stress in Bifidobacterium longum 105-A via expression of a catalase gene. Appl. Environ. Microbiol. 2012;78:2988–2990. doi: 10.1128/AEM.07093-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuo F, et al. Homologous overexpression of alkyl hydroperoxide reductase subunit C (ahpC) protects Bifidobacterium longum strain NCC2705 from oxidative stress. Res. Microbiol. 2014;165:581–589. doi: 10.1016/j.resmic.2014.05.040. [DOI] [PubMed] [Google Scholar]
- 44.Kawasaki S, Satoh T, Todoroki M, Niimura Y. b-type dihydroorotate dehydrogenase is purified as a H2O2-forming NADH oxidase from Bifidobacterium bifidum. Appl. Environ. Microbiol. 2009;75:629–636. doi: 10.1128/AEM.02111-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Underwood MA, German JB, Lebrilla CB, Mills DA. Bifidobacterium longum subspecies infantis: champion colonizer of the infant gut. Pediatr. Res. 2015;77:229–235. doi: 10.1038/pr.2014.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sela DA, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. USA. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Underwood MA, et al. Bifidobacterium longum subsp. infantis in experimental necrotizing enterocolitis: alterations in inflammation, innate immune response, and the microbiota. Pediatr. Res. 2014;76:326–333. doi: 10.1038/pr.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zenno S, et al. Biochemical characterization of NfsA, the Escherichia coli major nitroreductase exhibiting a high amino acid sequence homology to Frp, a Vibrio harveyi flavin oxidoreductase. J. Bacteriol. 1996;178:4508–4514. doi: 10.1128/jb.178.15.4508-4514.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zenno S, Kobori T, Tanokura M, Saigo K. Purification and characterization of NfrA1, a Bacillus subtilis nitro/flavin reductase capable of interacting with the bacterial luciferase. Biosci. Biotechnol. Biochem. 1998;62:1978–1987. doi: 10.1271/bbb.62.1978. [DOI] [PubMed] [Google Scholar]
- 50.Lei B, Liu M, Huang S, Tu SC. Vibrio harveyi NADPH-flavin oxidoreductase: cloning, sequencing and overexpression of the gene and purification and characterization of the cloned enzyme. J. Bacteriol. 1994;176:3552–3558. doi: 10.1128/jb.176.12.3552-3558.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cortial S, et al. NADH oxidase activity of Bacillus subtilis nitroreductase NfrA1: insight into its biological role. FEBS Lett. 2010;584:3916–3922. doi: 10.1016/j.febslet.2010.08.019. [DOI] [PubMed] [Google Scholar]
- 52.Li X, Chow DC, Tu SC. Thermodynamic analysis of the binding of oxidized and reduced FMN cofactor to Vibrio harveyi NADPH-FMN oxidoreductase FRP apoenzyme. Biochemistry. 2006;45:14781–14787. doi: 10.1021/bi0610956. [DOI] [PubMed] [Google Scholar]
- 53.Scardovi V, Trovatelli LD. Bifidobacterium animalis (Mitsuoka) comb. nov. and the “minimum” and “subtile” groups of new bifidobacteria found in sewage. Int. J. Syst. Bacteriol. 1974;24:21–28. doi: 10.1099/00207713-24-1-21. [DOI] [Google Scholar]
- 54.Tanaka K, Samura K, Kano Y. Structural and functional analysis of pTB6 from Bifidobacterium longum. Biosci. Biotechnol. Biochem. 2005;69:422–425. doi: 10.1271/bbb.69.422. [DOI] [PubMed] [Google Scholar]
- 55.Yasui K, et al. Improvement of bacterial transformation efficiency using plasmid artificial modification. Nucleic. Acids. Res. 2009;37:e3. doi: 10.1093/nar/gkn884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oberg TS, Ward RE, Steele JL, Broadbent JR. Transcriptome analysis of Bifidobacterium longum strains that show a differential response to hydrogen peroxide stress. J. Biotechnol. 2015;212:58–64. doi: 10.1016/j.jbiotec.2015.06.405. [DOI] [PubMed] [Google Scholar]
- 57.Meehan BM, Baughn AD, Gallegos R, Malamy MH. Inactivation of a single gene enables microaerobic growth of the obligate anaerobe Bacteroides fragilis. Proc. Natl. Acad. Sci. USA. 2012;109:12153–12158. doi: 10.1073/pnas.1203796109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 59.Engler C, Gruetzner R, Kandzia R, Marillonnet S. Golden Gate Shuffling: A One-Pot DNA shuffling method based on type IIs restriction enzymes. PLoS ONE. 2009;4(5):e5553. doi: 10.1371/journal.pone.0005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sakaguchi K, He J, Tani S, Kano Y, Suzuki T. A targeted gene knockout method using a newly constructed temperature-sensitive plasmid mediated homologous recombination in Bifidobacterium longum. Appl. Microbiol. Biotechnol. 2012;95:499–509. doi: 10.1007/s00253-012-4090-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.