Accessibility of antigen receptor loci to RAG is correlated with the presence of H3K4me3, which binds to a plant homeodomain (PHD) in the RAG-2 subunit and promotes V(D)J recombination. A point mutation in the PHD, W453A, eliminates binding of H3K4me3 and impairs recombination.
KEYWORDS: V(D)J recombination, epigenetic control, histone modification, lymphocyte development
ABSTRACT
Accessibility of antigen receptor loci to RAG is correlated with the presence of H3K4me3, which binds to a plant homeodomain (PHD) in the RAG-2 subunit and promotes V(D)J recombination. A point mutation in the PHD, W453A, eliminates binding of H3K4me3 and impairs recombination. The debilitating effect of the W453A mutation is ameliorated by second-site mutations that locate an inhibitory domain in the interval from residues 352 through 405 of RAG-2. Disruption of the inhibitory domain stimulates V(D)J recombination within extrachromosomal substrates and at endogenous antigen receptor loci. Association of RAG-1 and RAG-2 with chromatin at the IgH locus in B cell progenitors is dependent on recognition of H3K4me3 by the PHD. Strikingly, disruption of the inhibitory domain permits association of RAG with the IgH locus in the absence of H3K4me3 binding. Thus, the inhibitory domain acts as a gate that prohibits RAG from accessing the IgH locus unless RAG-2 is engaged by H3K4me3.
INTRODUCTION
The genes that encode antigen receptors are assembled from discrete variable (V), diversity (D), and joining (J) segments by V(D)J recombination. This process is initiated by the proteins recombination-activating gene 1 (RAG-1) and RAG-2, which together cleave participating gene segments at flanking recombination signal sequences (RSSs). There are two classes of RSSs, termed 12-RSS and 23-RSS, composed of conserved nonamer and heptamer elements separated by spacers of 12 bp or 23 bp, respectively. Rearrangement proceeds in the following sequence: (i) capture by RAG of a 12-RSS and a 23-RSS, resulting in synapsis of participating gene segments; (ii) nicking by RAG at the junction between each gene segment and its flanking RSS; (iii) transesterification to produce double-strand breaks; and (iv) joining of the participating gene segments by classical nonhomologous end joining (cNHEJ) (1, 2).
While a 12-RSS and a 23-RSS are sufficient to support DNA cleavage by RAG in vitro, V(D)J recombination in vivo is subject to higher-level constraints that restrict rearrangement to particular sets of gene segments in distinct lymphocyte lineages and developmental stages. In the B lineage, for example, V(D)J recombination occurs first at the immunoglobulin heavy-chain locus (IgH) and involves sequential DH-to-JH and VH-to-DJH joining (3). Upon productive rearrangement and expression of the immunoglobulin μ chain, further recombination at the IgH locus is suppressed, and rearrangement of light-chain loci is initiated (4). V(D)J recombination at an antigen receptor locus is invariably preceded by sterile germ line transcription from promoters whose activity is positively correlated with rearrangement (5, 6). At the IgH locus, μ0 transcripts initiate at the DQ52 promoter (7), and Iμ transcripts originate within the Eμ enhancer (8, 9). Activation of germ line transcription at antigen receptor loci is accompanied by alterations in chromatin organization and the establishment of chemical modifications characteristic of active chromatin, including hypermethylation of histone H3 at lysine 4 (H3K4me3) (10–15).
RAG-2 is 527 amino acid residues long. Only the N-terminal 387 amino acids, termed the core region, are required for RSS cleavage in vitro (16, 17), but removal of the noncore region, comprising residues 388 through 527, is associated with decreased recombination frequency (18) and increased aberrant recombination (19, 20) in vivo. The noncore region serves several regulatory functions, including cell cycle-dependent degradation (21–24), nuclear import (25), recognition of H3K4me3 (12, 13, 26), and autoinhibition (27). The binding of H3K4me3 to RAG-2 is mediated by a plant homeodomain (PHD) finger that spans residues 415 through 487 (26, 28). Engagement of H3K4me3 by the PHD finger stimulates recombination in vivo (12, 13) and cleavage of RSS substrates in vitro (27, 29, 30), conferring increases in substrate affinity and the catalytic rate (27, 30). Because the DNA-binding domains and catalytic core are largely contained within RAG-1 (31), these observations are consistent with the interpretation that H3K4me3 is an allosteric activator of the V(D)J recombinase. This interpretation was reinforced by the finding that binding of H3K4me3 to the RAG-2 PHD finger induces conformational changes in RAG-1 within the DNA-binding domains and a domain that acts as a scaffold for the catalytic core (32).
The ability of H3K4me3 to induce allosteric conformational changes and to stimulate RSS cleavage is abolished by a point mutation, W453A, in the RAG-2 PHD finger that eliminates binding of H3K4me3 and impairs V(D)J recombination in vivo (12, 26). In previous work, we identified mutations within the noncore region of RAG-2 but outside the PHD finger that could ameliorate the debilitating effect of the RAG-2 W453A mutation (27), thereby indicating the presence of an inhibitory domain. It remained unclear whether inhibition is accomplished by direct competition between the inhibitory domain and H3K4me3 for binding to the PHD finger or, alternatively, by interactions independent of the PHD finger. Disruption of the inhibitory domain conferred an increase in substrate affinity similar to that of wild-type (wt) RAG at maximal induction by H3K4me3; moreover, the affinity of the mutant for a substrate was not increased further by addition of H3K4me3 (27). In contrast, mutation of the inhibitory region spared the ability of H3K4me3 to stimulate RSS cleavage by increasing the kcat (27). These results suggested that the inhibitory domain does not act by simple competition with H3K4me3 for binding to the PHD and, moreover, predicted that relief of inhibition in vivo would preferentially stimulate substrate recognition over catalysis.
In the present study, we tested these predictions by examining the role of the inhibitory domain in regulating the accessibility of RAG-2 to endogenous antigen receptor genes. We mapped the inhibitory domain to the interval spanning amino acid residues 352 through 405 of RAG-2 and showed that inhibition is largely conferred by acidic residues within this interval. We found that inactivation of the inhibitory domain confers a gain-of-function recombination phenotype independent of the PHD finger, indicating that inhibition does not involve direct competition with H3K4me3 for binding to RAG-2. The stimulation of recombination observed upon inactivation of the inhibitory domain is accompanied by a parallel effect on the association of RAG with chromatin. In B progenitor cells, localization of wild-type RAG-1 and RAG-2 to the IgH locus is abolished by the W453A mutation, indicating that this pattern of chromatin localization is dependent on recognition of H3K4me3. Strikingly, disruption of the inhibitory domain permits association of RAG-2 and RAG-1 with the IgH locus even if RAG-2 is unable to bind H3K4me3. Thus, the RAG-2 inhibitory domain serves as a binary gate that permits the association of RAG-1 and RAG-2 with chromatin only if H3K4me3 is engaged by the RAG-2 PHD finger.
RESULTS
Definition of an inhibitory domain within the noncore region of RAG-2.
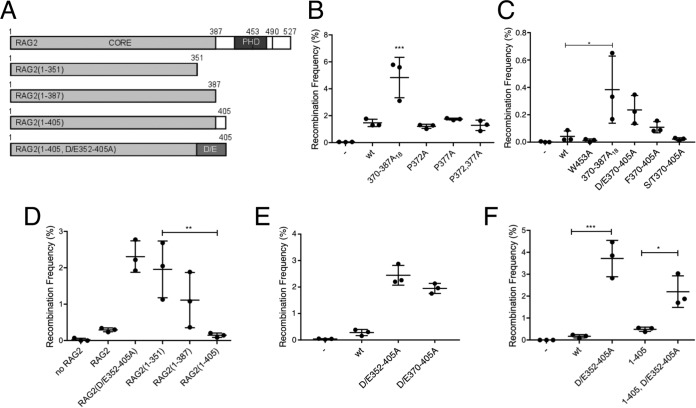
The RAG-2 W453A mutation disrupts the PHD finger, abolishes specific binding of the V(D)J recombinase to H3K4me3, and impairs recombination in vivo. We previously constructed second-site scanning alanine mutations between residues 370 and 405 of RAG-2 that stimulated activity of the RAG-2 W453A mutant. When assayed in the context of wild-type RAG-2, the resulting mutants, including RAG-2(388–405A18), exhibited a gain-of-function phenotype in extrachromosomal assays for recombination (27), indicating that the mutations disrupted an inhibitory domain. We employed a collection of substitution and truncation mutants (Fig. 1A) to more precisely define this domain. The interval between residues 370 and 405 is predominantly acidic, with interspersed proline, serine, threonine, and phenylalanine residues. We wished to determine the relative contributions of these residues to inhibition of RAG activity. This was of particular interest because structural analysis had suggested that the RAG-2 PHD finger may alternatively engage proline and H3K4me3 (26). Mutation of the proline (Fig. 1B; see Fig. S1A in the supplemental material), serine, and threonine or phenylalanine (Fig. 1C; see Fig. S1A in the supplemental material) residues conferred only modest increases in recombination of cotransfected extrachromosomal substrates. In contrast, neutralization of acidic residues in the 370-to-405 interval (D/E370–405A) increased recombination activity about 5-fold, to the level observed for RAG-2(370–387A18) (Fig. 1C). Because the predominance of acidic residues extends to residue 352, we determined whether inhibition of RAG activity could be further relieved by truncation into this region (Fig. 1A). Carboxy-terminal truncation of RAG-2 from residues 387 to 351 was associated with a more than 10-fold increase in recombination activity (Fig. 1D). Moreover, recombination activity was increased when neutralization of acidic residues was extended from residue 370 to residue 352 (Fig. 1E). Prior work suggested that further neutralization would impair recombination activity, as RAG-2(334–351A18) was inactive on an extrachromosomal substrate (27). In subsequent experiments to test relief of inhibition, we used the optimally active construct RAG-2(D/E352–405A) or the RAG-2(388–405A18) gain-of-function mutant.
FIG 1.
The inhibitory domain of RAG-2 functions independently of the PHD. (A) RAG-2 truncation mutants used in this study. Amino acid residues at boundaries of functional domains or at the C terminus are numbered. The canonical core (CORE) and PHD are indicated. The dashed and solid vertical lines denote W453 and T490, respectively. D/E designates an interval within which all acidic residues are mutated to alanine. (B) Effects of proline mutations within the inhibitory domain on signal joining. Recombination frequencies are plotted for three independent cotransfections of NIH 3T3 cells with the extrachromosomal substrate pJH200 (41), RAG-1, and the indicated RAG-2 variant. Means and standard deviations (SD) are indicated. The Brown-Forsythe test was used to confirm homogeneity of variances (P > 0.05). Significance was assessed by one-way analysis of variance (ANOVA) (***, P < 0.001). (C) Effects of phenylalanine, serine, and threonine mutations within the inhibitory domain on signal joining were assayed as for panel B. Significant difference, as assessed for panel B, is indicated (*, P < 0.05). (D) Residues 352 through 405 inhibit recombination in the absence of the PHD. Tests of statistical significance were as for panel B. Significant difference is indicated (**, P < 0.01). (E) Neutralization of acidic residues in the interval from residues 352 through 405 relieves inhibition. The data were analyzed as for panel B. Recombination frequencies associated with both mutations were significantly higher than for wild-type RAG2 (P < 0.0001). (F) The inhibitory domain suppresses recombination in the absence of the RAG-2 PHD. The data were analyzed as for panel B. *, P < 0.05; ***, P < 0.001. Not all significant differences are indicated.
Importantly, the stimulatory effects of truncation on recombination suggested that the inhibitory domain does not exert its suppressive effects solely through interaction with the PHD finger (Fig. 1D). To confirm this, we mutated the inhibitory domain in the context of a RAG-2 construct lacking the PHD finger. The resulting mutant, RAG-2(1–405, D/E352–405A), exhibited an approximately 4.5-fold increase in function relative to the RAG-2(1–405) mutant (Fig. 1F). None of these differences in activity could be attributed to changes in protein expression (see Fig. S1B to in the supplemental material F). These observations indicate that suppression of recombination by the inhibitory domain does not involve an interaction with the PHD finger, thereby disfavoring models for allosteric stimulation that invoke mutually exclusive binding of H3K4me3 or the inhibitory domain to the PHD.
The RAG-2 inhibitory domain suppresses recombination in B progenitor cells.
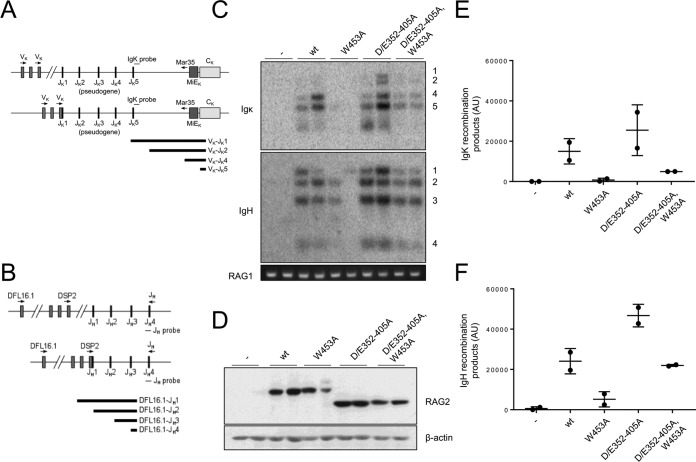
We asked whether disruption of the inhibitory domain would enhance rearrangement of endogenous immunoglobulin loci. Our initial experiments employed the RAG-2-deficient B progenitor cell line 63-12, which undergoes spontaneous V(D)J recombination upon introduction of RAG-2. Wild-type RAG-2 or RAG-2 mutants were introduced by retroviral transduction, and Vκ-to-Jκ (Fig. 2A) or DH-to-JH joining (Fig. 2B) was assayed. In cells transduced with wild-type RAG-2, we observed robust Vκ-to-Jκ joining (Fig. 2C, top, wt, and E) and DH-to-JH joining (Fig. 2C, bottom, wt, and F). The RAG-2 W453A mutation impaired recombination at both loci (Fig. 2C, W453A, E, and F), and this debilitating effect was reversed by secondary mutation of the inhibitory domain (Fig. 2C, D/E352–405A and W453A, E, and F). The observed effects of mutation on recombination were not explained by differences in expression of RAG-2 (Fig. 2D).
FIG 2.
Disruption of the RAG-2 inhibitory domain stimulates recombination at endogenous loci in pro-B cells. (A) Assay for Vκ-to-Jκ joining. The forward degenerate primer recognizes the seven most commonly used Vκ segments (44). The reverse primer hybridizes to a site 5′ of the κ intronic enhancer (MiEκ) (45). Variable (V), joining (J), and constant κ (Cκ) segments are indicated. The Southern hybridization probe used to detect recombination products (IgΚ probe) is indicated. Top line, unrearranged locus; bottom line, rearrangement to Jκ1. Below are shown the expected sizes of products of Vκ joining to Jκ1, Jκ2, Jκ4, and Jκ5. (B) Assay for DH-to-JH joining. Forward primers specific for the DSP2 or DFL16.1 family were used, as well as a reverse primer that hybridizes to a site 3′ of JH4. Top line, unrearranged locus; bottom line, rearrangement of DSP2 to JH1. Below are shown the expected sizes of products of DFL16.1 joining to JH1, JH2, JH3, and JH4. (C) Disruption of the RAG-2 inhibitory domain enhances recombination activity of RAG-2(W453A) in 63-12 pro-B cells. (Top) Recombination at the endogenous Igκ locus. Products of Vκ joining to Jκ1, -2, -4, or -5 are numbered on the right. −, empty virus. (Middle) DH-to-JH recombination at the endogenous IgH locus. Products of DH joining to JH1, -2, -3, or 4 are numbered on the right. (Bottom) A PCR amplicon from the RAG-1 locus as a control for input genomic DNA. (D) Immunodetection of RAG-2 variants (top) and actin (bottom) in the transduced 63-12 cells assayed for panel C. −, empty virus. (E) Quantitation of endogenous Igκ rearrangements assayed for panel C. AU, arbitrary units. Means and standard deviations are indicated. (F) Quantitation of endogenous DH-to-JH rearrangements assayed for panel C.
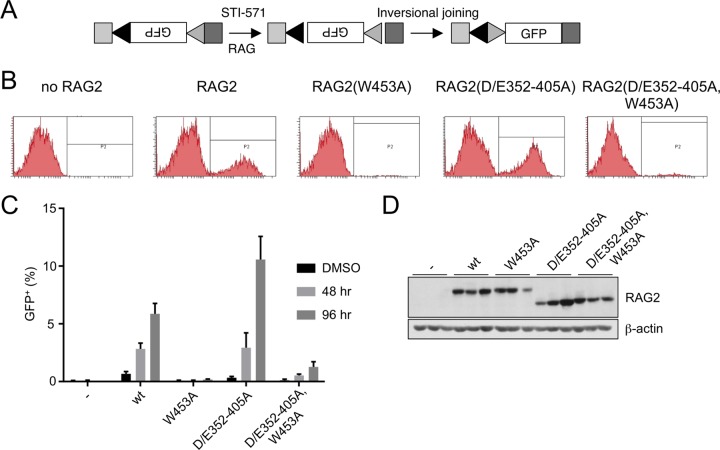
We introduced the same RAG-2 constructs into another RAG-2-deficient Abelson murine leukemia virus (AbMuLV)-transformed B progenitor cell line, R2K3, in which cell cycle arrest, RAG-1 expression, and V(D)J recombination are inducible by treatment with STI-571 (33). The R2K3 pre-B cell line carries an integrated recombination reporter that permits quantitation of recombination (Fig. 3A). After introduction of RAG-2 by retroviral transduction and cell cycle arrest by STI-571, inversional joining of recombination signal sequences within the reporter reorients the green fluorescent protein (GFP) cassette to permit its expression from an upstream promoter (Fig. 3B). As assessed by fluorescence, no recombination was observed in R2K3 cells lacking RAG-2, either before or after induction (Fig. 3B, no RAG-2, and C). Cells transduced with RAG-2 exhibited a low level of recombination, which increased robustly at 48 h and 96 h after induction by STI-571 (Fig. 3B, RAG-2, and C). Recombination was nearly abolished by the RAG-2 W453A mutation [Fig. 3B, RAG-2(W453A), and C], and this effect was modestly ameliorated by a second-site mutation in the inhibitory domain [Fig. 3B, RAG-2(D/E352-405A and W453A), and C]. Mutation of the inhibitory domain alone conferred a reproducible increase (about 2-fold at 96 h) in the frequency of recombination relative to the wild type (Fig. 3C; see Fig. S2A in the supplemental material). Thus, the stimulatory effect that we observed on recombination of endogenous gene segments upon mutation of the inhibitory domain was also exerted on an exogenous, integrated inversional substrate. Moreover, the effects of inhibitory domain and PHD finger mutations on recombination at endogenous Igκ and IgH loci in R2K3 cells were similar to those observed in the 63-12 cell line (see Fig. S2B in the supplemental material). The differential effects of these mutations on recombination could not be attributed to differences in expression of the RAG-2 protein (Fig. 3D; see Fig. S2C in the supplemental material) or to differences in μ0 or Iμ germ line transcripts at the IgH locus (see Fig. S2D and E in the supplemental material).
FIG 3.
The RAG-2 inhibitory domain suppresses recombination of an integrated substrate in the inducible R2K3 pro-B cell line. (A) The integrated substrate PMX-INV (33). An inverted GFP cassette is flanked by two RSSs (triangles) in the context of genomic sequence (light-gray and dark-gray boxes). After induction with STI-571 and DNA cleavage by RAG, joining inverts the reporter so that GFP is expressed. (B) Representative flow cytometric data for R2K3 cells transduced with the indicated RAG-2 variants 96 h after induction with STI-571; no RAG2, cells infected with empty retrovirus. GFP-positive cells lie within the gates labeled P2. (C) Percentages of cells expressing GFP 48 or 96 h after STI-571 induction plotted for R2K3 cells transduced with each of the indicated RAG-2 variants (means of three independent infections plus SD). DMSO (dimethyl sulfoxide), R2K3 cells 96 h after treatment with vehicle alone. −, cells infected with empty retrovirus. Pairwise comparisons (unpaired two-tailed t test) indicated significant differences between wild-type and RAG-2(D/E352–405A) at 96 h (P = 0.021), between RAG-2(453A) and the double mutant at 48 h and 96 h (P = 0.003 and 0.014, respectively), and between the wild type and RAG-2(W453A) at all time points (P = 0.009 in DMSO). (D) (Top) Immunodetection of myc-tagged RAG-2 variants in the transduced R2K3 cells assayed for panel C. Variants are indicated above the gel; −, infection with empty virus. The three lanes corresponding to each variant contained protein from three independent infections. (Bottom) Immunodetection of actin on the same membrane as above.
The RAG-2 inhibitory domain acts prior to repair.
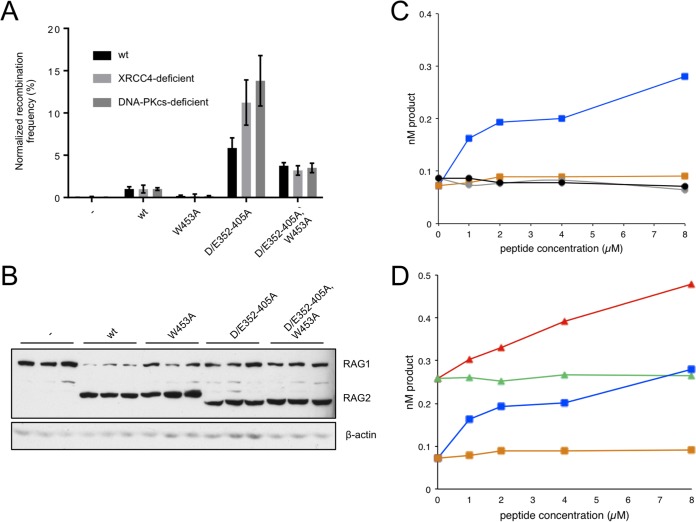
In the experiments described above, we defined an inhibitory domain in RAG-2 whose disruption stimulated V(D)J recombination within extrachromosomal and integrated substrates, as well as at endogenous loci. RAG participates at all stages of V(D)J recombination, including RSS binding, synapsis, DNA cleavage, and repair, the last step in collaboration with components of the NHEJ machinery. Our previous work indicated that inhibition is exerted at the stages of substrate recognition and DNA cleavage; we now wished to determine whether there was also an effect during repair. To do so, we employed cell lines deficient in the NHEJ factor XRCC4 or DNA-PKCS. If the inhibitory domain functions through an interaction with the NHEJ machinery, then in the absence of NHEJ, we would not expect inactivation of the inhibitory domain to stimulate recombination. In extrachromosomal assays for signal joint (Fig. 4A) formation the D/E352–405A mutation reversed the effect of the W453A mutation to similar extents in NHEJ-proficient and NHEJ-deficient cell lines. Moreover, RAG-2(D/E352–405A) exhibited a robust gain of function in all three cell lines (Fig. 4A). The effects of these mutations were not attributable to differences in protein expression (Fig. 4B). These results are consistent with the interpretation that the inhibitory domain participates in V(D)J recombination only prior to repair.
FIG 4.
The inhibitory domain acts prior to repair. (A) Assay of RAG-2 mutants in NHEJ-deficient cells. Signal joining was assayed in wild-type Chinese hamster ovary (CHO) cells and in derivative cell lines deficient in XRCC4 or DNA-PKCS. Wild-type RAG-2 and RAG-2 mutants are designated below the graph. For each cell line, recombination frequencies were normalized to those observed for wild-type RAG-2 and plotted as normalized means ± SD (n = 3). (B) Immunodetection of RAG-1 and RAG-2 variants in transfected CHO cells. (Top) Detection of myc-tagged RAG-1 and RAG-2. (Bottom) Immunodetection of actin on the same membrane as above. (C) Comparison of coupled cleavage in reaction mixtures containing wild-type RAG-2 or RAG-2(W453A). Accumulation of hairpin product at 1 h (nM product) is plotted as a function of the concentration of H3K4me0 or H3K4me3. Blue squares, wild-type RAG-2 with H3K4me3; orange squares, wild-type RAG-2 with H3K4me0; gray circles, RAG-2(W453A) with H3K4me3; black circles; RAG-2(W453A) with H3K4me0. (Republished from reference 32.) (D) Comparison of coupled cleavage in reaction mixtures containing wild-type RAG-2 or RAG-2(D/E352-405A). Red triangles, RAG-2(D/E352–405A) with H3K4me3; green triangles, RAG-2(D/E352–405A) with H3K4me0; blue squares, wild-type RAG-2 with H3K4me3; orange squares, wild-type RAG-2 with H3K4me0.
Prior work suggested that mutation of acidic residues within this region increases the use of alternative NHEJ (alt-NHEJ) (34), which is characterized by excessive deletions and microhomology (1). The use of microhomology is not, however, restricted to alt-NHEJ (35). In wild-type cells, all but one of the recovered signal junctions produced by RAG-2 variants were precise (see Fig. S3A in the supplemental material), suggesting that the robust increase in recombination frequency associated with the D/E352–405 mutation does not result from the use of alt-NHEJ. In wild-type cells, similar use of microhomology and deletion at coding joints was observed for all the RAG-2 variants assayed (see Fig. S4A, D, and E in the supplemental material). While we were able to obtain few junctions in assays of wild-type RAG-2 in repair-deficient cell lines, the junctions obtained from cells expressing the RAG-2 inhibitory domain mutants were characterized by an increase in excessive deletion that was not accompanied by an increased use of microhomology (see Fig. S3 and S4 in the supplemental material). We conclude that the use of modes of DNA repair other than NHEJ does not account for the increase in recombination frequency observed upon disruption of the inhibitory domain.
To confirm that the inhibitory domain acts prior to repair, equivalent amounts of active wt RAG-2 and RAG-2(D/E352–405A), as determined by burst kinetic analysis (27), were assayed in vitro for coupled cleavage of a radiolabeled 12-RSS in the presence of unlabeled 23-RSS substrate and increasing amounts of a peptide spanning H3K4me3 or an unmethylated control (H3K4me0) (Fig. 4C and D). We previously reported (27, 32) that wild-type RAG is stimulated in a dose-dependent fashion by H3K4me3 but not by H3K4me0, while RAG-2(W453A) is unresponsive to H3K4me3 (Fig. 4C). In the absence of H3K4me3, the basal activity of the RAG-2(D/E352–405A) inhibitory domain mutant was elevated to a level similar to that of maximally stimulated wild-type RAG-2, indicating that the inhibitory domain exerts a suppressive effect at the level of DNA cleavage (Fig. 4D). Consistent with previous observations (27), the activity of the inhibitory domain mutant could be further stimulated by addition of H3K4me3 (Fig. 4D), suggesting that RAG retains at least partial responsiveness to H3K4me3 in the presence of the D/E352–405A mutation.
The RAG-2 inhibitory domain is a gate that restricts access of RAG to chromatin.
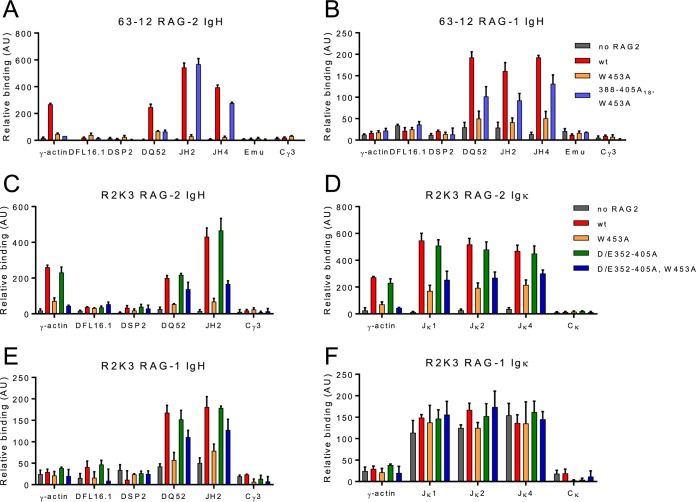
The distribution of RAG-2 over chromatin is positively correlated with the density of H3K4me3, both within and outside antigen receptor loci (36). Based on the observation that H3K4me3 recognition by RAG-2 is essential for efficient V(D)J recombination and the close correlation between genome-wide H3K4me3 levels and RAG-2 binding, it is widely believed that the interaction of RAG-2 with H3K4me3 is a key element in bringing RAG-1 and RAG-2 to endogenous antigen receptor loci. The fact that this requirement was bypassed by inactivation of the inhibitory domain suggested that the RAG-2 inhibitory domain might function in modulating access of RAG to chromatin. To test this, we used chromatin immunoprecipitation (ChIP) to probe the distribution of RAG-2 or RAG-1 over immunoglobulin loci in 63-12 or R2K3 B progenitor cells expressing wild-type RAG-2 or RAG-2 mutants.
In 63-12 cells, wild-type RAG-2 is localized to DQ52 and the JH cluster (Fig. 5A), as well as over actively transcribed non-Ig loci (Fig. 5A; see Fig. S5A, wt, in the supplemental material), consistent with previous observations (36). The RAG-2 W453A mutation, which abolishes binding to H3K4me3 (12), eliminated association of RAG-2 with the IgH locus and other active loci (Fig. 5A; see Fig. S5A, W453A, in the supplemental material). Strikingly, disruption of the inhibitory domain in the context of the W453A mutation allowed RAG-2 access to the IgH locus in the absence of H3K4me3 binding (Fig. 5A, 388–405A18 and W453A). In contrast, association of the RAG-2 W453A mutant with active non-Ig loci was not restored by second-site mutation of the inhibitory domain, consistent with the interpretation that off-target binding of RAG-2 to chromatin is mediated principally, if not wholly, by H3K4me3. In 63-12 cells expressing wild-type RAG-2, RAG-1 was associated with the IgH locus at DQ52 and the JH cluster but, unlike RAG-2, was not found at the γ-actin locus (Fig. 5B, wt). Mutation of the RAG-2 PHD finger greatly reduced the binding of RAG-1 to the IgH locus, suggesting that this association is established or maintained by the interaction of H3K4me3 with RAG-2 (Fig. 5B, W453A). Introduction of a second-site mutation in the RAG-2 inhibitory domain reversed the effect of the W453A mutation, allowing RAG-1 to bind the IgH locus in the absence of an interaction between RAG-2 and H3K4me3.
FIG 5.
The RAG-2 inhibitory domain gates access to the IgH locus. (A and B) ChIP of RAG-2 (A) and RAG-1 (B) at the IgH locus in 63-12 cells expressing wt RAG-2 or the indicated mutant. Association of RAG-2 or RAG-1 with each of the genomic regions defined below the graph was assayed by quantitative PCR (qPCR). Means and SD are indicated; n = 2. (C and D) ChIP of RAG-2 (C) and RAG-1 (D) at the IgH locus in R2K3 cells expressing wt RAG2 or the indicated mutant, displayed as in panels A and B. (E and F) ChIP of RAG-2 (E) or RAG-1 (F) at the Igκ locus in R2K3 cells expressing wt RAG2 or the indicated mutant, displayed as for panels A and B.
Similar patterns of RAG-2 and RAG-1 association with the IgH locus and of RAG-2 with active non-Ig loci were observed in R2K3 cells expressing wild-type RAG-2 (Fig. 5C and E; see Fig. S5B, wt, in the supplemental material), and these patterns were unaffected by mutation of the inhibitory domain alone (Fig. 5C and E; see Fig. S5B, D/E352–405A, in the supplemental material). As was observed for 63-12 cells, the RAG-2 W453A mutation greatly reduced binding of both RAG-2 and RAG-1 to all regions of chromatin examined (Fig. 5C and E; see Fig. S5B, W453A, in the supplemental material), while a second mutation in the RAG-2 inhibitory domain partially restored association of RAG-2 and RAG-1 with the IgH locus (Fig. 5C and E; see Fig. S5B, D/E352–405A and W453A, in the supplemental material). We have not further explored the basis for incomplete rescue of RAG-2 binding to IgH by this double mutation. RAG-2 and RAG-1 were associated with the Jκ cluster of the Igκ locus in R2K3 cells expressing wild-type RAG-2 (Fig. 5D and F). While the W453A mutation reduced binding of RAG-2 to the Igκ locus (Fig. 5D, W453A), the reduction was not as great as that observed at the IgH locus; moreover, disruption of the inhibitory domain did not reverse the effect of the W453A mutation on RAG-2 binding at the Igκ locus (Fig. 5D, D/E352–405A and W453A). These results were not due to differences in H3K4 trimethylation (see Fig. S5C in the supplemental material). Association of RAG-1 with the Jκ cluster was unimpaired by the RAG-2 W453A mutation (Fig. 5F, W453A). Thus, binding of RAG-1 to the Igκ locus is independent of H3K4me3 engagement by RAG-2, consistent with the observation that RAG-2 is not required for recruitment of RAG-1 to Igκ (36). Taken together these observations indicate (i) that the binding of RAG-1 and RAG-2 to the IgH locus is dependent, directly or indirectly, on engagement of H3K4me3 by the RAG-2 PHD finger; (ii) that this requirement is imposed by the RAG-2 inhibitory domain; (iii) that disruption of the inhibitory domain removes this restriction, allowing RAG-2 and RAG-1 to access the IgH locus in a manner independent of H3K4me3 binding; and (iv) that the ability of the inhibitory domain to modulate access of RAG to chromatin is locus specific, in that disruption of the inhibitory domain permits association of RAG-2 with IgH, but not Igκ, in the absence of H3K4me3 binding.
DISCUSSION
The accessibility hypothesis and establishment of adaptive immune specificity.
Because the unit of responsiveness in the adaptive immune response is the individual B or T lymphocyte, the specificity of adaptive responses requires a close correspondence between receptor specificity and a responsive cell. This mapping, in turn, is enforced by mechanisms that ensure the productive rearrangement of antigen receptor loci in their appropriate lymphoid lineages and that prohibit the expression of multiple receptor specificities within the same cell. Some 30 years ago, these considerations prompted the accessibility hypothesis, which posits that locus- and allele-specific access of RAG to antigen receptor loci is governed by developmentally regulated alterations in the chromatin structure (3).
Much work directed toward confirmation of the accessibility hypothesis has addressed mechanisms that act at the level of antigen receptor genes and their surrounding chromatin to restrict access to RAG. Consistent with the accessibility hypothesis, a body of observations has established (i) that sterile transcription of antigen receptor loci accompanies rearrangement (5), (ii) that any mutation found to abolish germ line transcription also impairs rearrangement (37), (iii) that loci poised for rearrangement are also associated with epigenetic marks of active chromatin (36), and (iv) that recombination can be modulated by manipulation of histone modifications (38).
Allosteric activation and autoinhibition.
The observation that RAG-2 is allosterically activated by a specific marker of active chromatin, H3K4me3, has directed attention to the role the V(D)J recombinase itself may play in governing its own access to antigen receptor loci (12, 13, 26). In this communication, we show that residues spanning positions 352 through 405 of RAG-2, which encompass a previously identified inhibitory domain, gate access of RAG to chromatin. The existence of this domain was first suggested by the differential dependence of wild-type and core RAG-2 on recognition of H3K4me3 in extrachromosomal assays for recombination (12, 27). The ability of core RAG-2 to bypass the requirement for H3K4me3 binding in these assays was consistent with the interpretation that the core truncation of RAG-2 at residue 387 disrupted an inhibitory region. These observations, moreover, provided a rationale for models of allosteric activation in which autoinhibition is relieved by binding of H3K4me3 to the RAG-2 PHD finger (27, 39). Indeed, H3K4me3 acts allosterically on RAG to increase substrate affinity and the catalytic rate (27, 30, 39), and these effects can be mimicked by disruption of the inhibitory domain (27).
Nonetheless, RAG retains the capacity for further stimulation of catalysis by H3K4me3 even when the RAG-2 inhibitory domain is disrupted (27). This suggests that the stimulatory effect of H3K4me3 on the catalytic rate is at least in part exerted independently of the inhibitory domain. Conversely, the inhibitory domain suppresses RAG activity in vivo even in the absence of the PHD finger, eliminating models of autoinhibition that require direct interaction of the inhibitory domain and the PHD finger. Binding of H3K4me3 to the PHD finger is associated with increased accessibility of the inhibitory domain to proteolysis (32), suggesting that in the absence of H3K4me3, the inhibitory domain is masked. While the interacting surface that shelters the inhibitory domain in the absence of H3K4me3 has not been identified, one candidate is a highly positively charged surface within RAG-1 that interacts with substrate DNA (31, 40).
Our observations, however, remain consistent with the interpretation that H3K4me3 stimulates RAG activity by relieving suppression of substrate binding and the catalytic rate by the inhibitory domain. If this is true, then H3K4me3 must also be able to exert an additional stimulatory effect on catalysis that is not abolished by disruption of the inhibitory domain. Alternatively, the stimulatory effects of H3K4me3 may be exerted independently of the stimulatory effects produced by disruption of the RAG-2 inhibitory domain. The former interpretation is favored by the fact that disruption of the inhibitory domain mimics the stimulatory effect of H3K4me3 on substrate affinity and the catalytic rate, as well as the observation that engagement of H3K4me3 by the RAG-2 PHD finger induces conformational changes in the inhibitory domain associated with alterations in the conformation of the RAG-1 RSS-binding and catalytic regions (32). Nonetheless, the available evidence does not formally distinguish between these possibilities.
Removal of the canonical noncore region of RAG-2, which overlaps the inhibitory domain, has been associated with decreased V(D)J recombination (18), while we have shown that selective mutation of the inhibitory domain yields a gain-of-function phenotype. The difference likely lies in the fact that en bloc removal of the noncore region disrupts a number of regulatory functions, in addition to the inhibitory domain, including cell cycle-dependent control (21), nuclear import (25), and recognition of H3K4me3 (12, 13).
Neutralization of acidic residues in the region of RAG-2 that we identified as an inhibitory domain has been associated with destabilization of RAG signal end complexes (SECs) and genomic instability (34). Our results provide a biochemical framework in which to rationalize these effects. For example, if formation of a stable SEC were to require disengagement of RAG from H3K4me3, then mutations that mimic the effect of H3K4me3 binding, such as those identified here, might destabilize the SEC or hinder its formation, resulting in reduced genomic stability.
A gating function in RAG that imposes specificity for active chromatin.
We now show that disruption of the RAG-2 inhibitory domain confers two unambiguous phenotypes: (i) increased frequency of V(D)J recombination and (ii) association of RAG-1 and RAG-2 with the IgH locus independently of RAG-2 binding to H3K4me3. Taken together, these observations indicate that the RAG-2 inhibitory domain enforces coupling between the accessibility of the IgH locus to RAG and the engagement of H3K4me3 by the RAG-2 PHD. Thus, the chromatin state of antigen receptor loci is not sufficient to determine access of the V(D)J recombinase. Rather, the V(D)J recombinase itself plays an active role in modulating its access.
Disruption of the RAG-2 inhibitory domain robustly restored association of RAG-1 and RAG-2 with the IgH locus in the absence of H3K4me3 binding but only modestly reversed the debilitating effect of a PHD mutation on recombination. The differential effects of inhibition on locus association and recombination are consistent with our prior observation that relief of inhibition uncouples substrate affinity, but not the catalytic rate, from stimulation by H3K4me3 and with the prediction that relief of inhibition in vivo would preferentially stimulate substrate recognition over catalysis.
The pattern of RAG recruitment to the Igκ locus, however, was quite different. Specifically, substantial RAG-1 recruitment was supported even by RAG-2(W453A), indicating that RAG-2 recognition of H3K4me3 was not required. These observations are wholly consistent with a previously established distinction between IgH and Igκ with respect to RAG-2 dependence of RAG-1 binding: association of RAG-1 with IgH was shown to require RAG-2, while RAG-1 was able to bind Igκ in the absence of RAG-2 (36). Importantly, despite the association of RAG-1 with Igκ in pro-B cells expressing RAG-2(W453A), Igκ rearrangements were not observed, consistent with an essential role for H3K4me3 in stimulating catalysis.
Our observations suggest a model for association of RAG with IgH in which (i) RAG, prior to engagement with H3K4me3, resides predominantly in a low-affinity state with respect to RSS binding; (ii) upon engagement of H3K4me3 by the RAG-2 PHD finger, the conformational distribution of RAG is shifted to a high-affinity state; (iii) RAG, now in a high-affinity conformation, binds stably to a neighboring RSS. Under this model, the W453A mutation impairs association of RAG-1 and RAG-2 with the IgH locus because, in the absence of H3K4me3 binding, the affinity of RAG for an RSS remains low. Against the background of the W453A mutation, a second mutation that disrupts the inhibitory domain restores specific association of RAG-1 and RAG-2 with the IgH locus; according to this stepwise model for chromatin association, the requirement for H3K4me3 binding would be bypassed because inactivation of the RAG-2 inhibitory domain confers constitutive high-affinity RSS binding. The inability of inactivating inhibitory domain mutations to restore the association of RAG-2(W453A) with active chromatin at non-Ig loci is consistent with the requirement of this model that specific association of RAG with the IgH locus involves high-affinity RSS binding.
This parsimonious model, however, incompletely explains the positioning of RAG at antigen receptor loci. For example, although inactivation of the inhibitory domain restores association of RAG-2(W453A) with the IgH locus, the distribution of RAG within the locus is not coextensive with RSSs, as is evident from the lack of RAG over DSP2 and DFL16. Moreover, mutation of the inhibitory domain failed to restore association of RAG-2(W453A) with Igκ despite the presence of RSS arrays at that locus. This suggests that in the absence of a functional PHD the RAG-2 inhibitory domain may prohibit access of RAG to the IgH locus by blocking interaction between RAG and a structural feature other than an RSS. We note that this possibility and the sequential model invoking RSS recognition are not mutually exclusive, and both are amenable to activation through an intermediate step involving engagement of H3K4me3 by RAG-2.
Omenn syndrome is an immunodeficiency disorder in which pathogenic oligoclonal T cell expansion is typically accompanied by profound B lymphopenia. Most cases of Omenn syndrome can be ascribed to hypomorphic mutations in RAG-1 or RAG-2, and in RAG-2, these mutations are overrepresented within the PHD finger (31). The selective dependence of IgH-RAG interactions on H3K4me3 recognition may shed light on the relative severity of the B lymphoid developmental defect in Omenn syndrome. Mutations in RAG-2 that abolish recognition of H3K4me3 would be expected to impair access of RAG to the IgH locus but not to the Igκ locus. If association of RAG with the T cell receptor (TCR) β and TCR α loci in humans were similarly insensitive to disruption of the PHD, as suggested by the ability of RAG-1 to bind to the corresponding mouse loci in the absence of RAG-2 (36), then impairment of H3K4me3 recognition would be expected to have a more debilitating effect on B cell development than on that of T cells.
MATERIALS AND METHODS
Cell culture.
NIH 3T3 and HEK 293T cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (FBS) and 1× penicillin-streptomycin-glutamine (PSG). R2K3 and 63-12 cell lines were propagated in RPMI 1640 medium supplemented with 10% FBS, 50 μΜ 2-mercaptoethanol, 1× PSG, 0.7× minimal essential medium (MEM) nonessential amino acid solution, 1 mM sodium pyruvate, 10 mM HEPES. R2K3 cells with integrated PMX-INV recombination substrate were kindly provided by Barry Sleckman (Weill Cornell Medicine) (33). All the cells were maintained at 37°C in 5% CO2.
Antibodies.
Commercial antibodies against the following proteins were used in this study: actin (clone AC-40; Sigma-Aldrich; catalog no. A3853); c-myc (clone 9E10; Santa Cruz Biotechnology; catalog no. sc-40); RAG-1 (Abcam; catalog no. ab172637); H3K4me3 (Active Motif; catalog no. 39915); mouse IgG (GE Healthcare; catalog no. NA931). Antibody against RAG-2 was a gift from David Schatz (Yale University). Immunoblotting was performed in 5% nonfat dry milk in PBST (1× phosphate-buffered saline [PBS], 0.1% Tween).
Assays for extrachromosomal recombination.
Assays for recombination of extrachromosomal substrates pJH200 and pJH290 (41) were performed as described previously (27), with modifications. Briefly, 10 μg of maltose-binding protein (MBP)–RAG1–myc–His, 10 μg of MBP-RAG2-myc-His (wild type or variant, as indicated), and 4 μg either pJH200 or pJH290 were transfected into NIH 3T3 cells. After 48 h, plasmid DNA was recovered by a modified Hirt extraction (Qiagen; catalog no. 27104). DNA (3 μl; about 40 μg) was transformed into 50 μl DH5α Max Efficiency cells (Thermo Fisher; catalog no. 18258012). An aliquot (1.7%) of the transformation mixture was plated on LB agar containing 50 μg/ml ampicillin, and the remainder was plated on LB agar containing 50 μg/ml ampicillin and 20 μg/ml chloramphenicol. The plates containing ampicillin alone were scored after 16 h at 37°C, while plates containing ampicillin and chloramphenicol were scored at 20 h.
Immunoblotting.
Protein extraction from cell pellets was performed by addition of 150 μl of 60 mM Tris (pH 7.6), 1% SDS and incubation at 100°C for 30 min. For immunoblotting, 60 to 90 μg protein was fractionated by SDS-8% PAGE and transferred to a 0.45-μm nitrocellulose membrane. The membranes were blocked for at least 45 min with 5% nonfat dry milk in PBST (1× PBS, 0.1% Tween).
Assays for germ line transcription.
RNA was isolated from R2K3 cells at 48 h after arrest by STI-571, and cDNA was synthesized by random-hexamer priming from total RNA. Sequences corresponding to Iμ or μ0 transcripts were detected by PCR using primers listed in Table S2 in the supplemental material. Amplification of cDNA for actin beta (ACTB) was performed as a control (42).
Chromatin immunoprecipitation.
Transduced B progenitor cells were treated with 3 μM STI571 for 21 h. ChIP was performed as described for RAG-1 and RAG-2 (36) and for H3K4me3 (43). Input and immunoprecipitated DNA was quantified by PicoGreen staining (Thermo Fisher). Each ChIP was performed in duplicate, and each real-time PCR was performed in duplicate. For analysis of H3K4me3 enrichment, 200 pg of DNA was used. The relative abundances of amplicons in the immunoprecipitated DNA relative to input were analyzed by real-time PCR using the primers listed in Table S3 in the supplemental material. The enrichment (IP/Inputcorr) of RAG-1 or RAG-2 in specific regions was calculated as described previously (36).
Expression constructs, assays for recombination of endogenous immunoglobulin gene segments, assays for coupled cleavage, cloning methods, oligonucleotide reagents, primers, and probes used to assay rearrangement, and primers used in ChIP analyses are described in Tables S1 to S3 in the supplemental material.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant R01CA160256 to S.D. and by the Intramural Research Program of the NIA (National Institute on Aging).
We are grateful to John Bettridge for conducting the coupled cleavage assays. Anti-RAG-1 and -RAG-2 antibodies were the kind gift of David Schatz (Yale University). We thank our colleagues in the Johns Hopkins Department of Molecular Biology and Genetics for stimulating discussions.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00159-18.
REFERENCES
- 1.Deriano L, Roth DB. 2013. Modernizing the nonhomologous end-joining repertoire: alternative and classical NHEJ share the stage. Annu Rev Genet 47:433–455. doi: 10.1146/annurev-genet-110711-155540. [DOI] [PubMed] [Google Scholar]
- 2.Gellert M. 2002. V(D)J Recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem 71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 3.Alt FW, Yancopoulos GD, Blackwell TK, Wood C, Thomas E, Boss M, Coffman R, Rosenberg N, Tonegawa S, Baltimore D. 1984. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J 3:1209–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jung D, Giallourakis C, Mostoslavsky R, Alt FW. 2006. Mechanism and control of V(D)J recomination at the immunoglobulin heavy chain locus. Annu Rev Immunol 24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 5.Van Ness BG, Weigert M, Coleclough C, Mather EL, Kelley DE, Perry RP. 1981. Transcription of the unrearranged mouse Cκ locus: sequence of the initiation region and comparison of activity with a rearranged Vκ-Cκ gene. Cell 27:593–602. doi: 10.1016/0092-8674(81)90401-3. [DOI] [PubMed] [Google Scholar]
- 6.Yancopoulos G, Alt FW. 1985. Developmentally controlled and tissue-specific expression of unrearranged VH gene segments. Cell 40:271–281. doi: 10.1016/0092-8674(85)90141-2. [DOI] [PubMed] [Google Scholar]
- 7.Alessandrini A, Desiderio SV. 1991. Coordination of immunoglobulin DJH transcription and D-to-JH rearrangement by promoter-enhancer approximation. Mol Cell Biol 11:2096–2107. doi: 10.1128/MCB.11.4.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lennon GG, Perry RP. 1985. Cmu-containing transcripts initiate heterogeneously within the IgH enhancer region and contain a novel 5′-nontranslatable exon. Nature 318:475–478. doi: 10.1038/318475a0. [DOI] [PubMed] [Google Scholar]
- 9.Su L-K, Kadesch T. 1990. The immunoglobulin heavy-chain enhancer functions as the promoter for Imu sterile transcription. Mol Cell Biol 10:2619–2624. doi: 10.1128/MCB.10.6.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakraborty T, Chowdhury D, Keyes A, Jani A, Subrahmanyam R, Ivanova I, Sen R. 2007. Repeat organization and epigenetic regulation of the DH-Cμ domain of the immunoglobulin heavy-chain gene locus. Mol Cell 27:842–850. doi: 10.1016/j.molcel.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 11.Goldmit M, Ji Y, Skok J, Roldan E, Jung S, Cedar H, Bergman Y. 2005. Epigenetic ontogeny of the Igk locus during B cell development. Nat Immunol 6:198–203. doi: 10.1038/ni1154. [DOI] [PubMed] [Google Scholar]
- 12.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. 2007. A plant homeodomain in Rag-2 that binds hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity 27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matthews AGW, Kuo AJ, Ramón-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, Shi Y, Kutateladze TG, Yang W, Gozani O, Oettinger MA. 2007. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature 450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morshead KB, Ciccone DN, Taverna SD, Allis CD, Oettinger MA. 2003. Antigen receptor loci poised for V(D)J rearrangement are broadly associated with BRG1 and flanked by peaks of histone H3 dimethylated at lysine 4. Proc Natl Acad Sci U S A 100:11577–11582. doi: 10.1073/pnas.1932643100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subrahmanyam R, Du H, Ivanova I, Chakraborty T, Ji Y, Zhang Y, Alt FW, Schatz DG, Sen R. 2012. Localized epigenetic changes induced by DH recombination restricts recombinase to DJH junctions. Nat Immunol 13:1205–1212. doi: 10.1038/ni.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cuomo CA, Oettinger MA. 1994. Analysis of regions of RAG-2 important for V(D)J recombination. Nucleic Acids Res 22:1810–1814. doi: 10.1093/nar/22.10.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadofsky MJ, Hesse JE, Gellert M. 1994. Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res 22:1805–1809. doi: 10.1093/nar/22.10.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steen SB, Han JO, Mundy C, Oettinger MA, Roth DB. 1999. Roles of the “dispensable” portions of RAG-1 and RAG-2 in V(D)J recombination. Mol Cell Biol 19:3010–3017. doi: 10.1128/MCB.19.4.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sekiguchi J, Whitlow S, Alt FW. 2001. Increased accumulation of hybrid V(D)J joins in cells expressing truncated versus full-length RAGs. Mol Cell 8:1383–1390. doi: 10.1016/S1097-2765(01)00423-3. [DOI] [PubMed] [Google Scholar]
- 20.Talukder SR, Dudley DD, Alt FW, Takahama Y, Akamatsu Y. 2004. Increased frequency of aberrant V(D)J recombination products in core RAG-expressing mice. Nucleic Acids Res 32:4539–4549. doi: 10.1093/nar/gkh778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang H, Chang F-C, Ross AE, Lee J, Nakayama K, Nakayama K, Desiderio S. 2005. Ubiquitylation of RAG-2 by Skp2-SCF links destruction of the V(D)J recombinase to the cell cycle. Mol Cell 18:699–709. doi: 10.1016/j.molcel.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 22.Lee J, Desiderio S. 1999. Cyclin A/CDK2 regulates V(D)J recombination by coordinating RAG-2 accumulation and DNA repair. Immunity 11:771–781. doi: 10.1016/S1074-7613(00)80151-X. [DOI] [PubMed] [Google Scholar]
- 23.Li Z, Dordai DI, Lee J, Desiderio S. 1996. A conserved degradation signal regulates RAG-2 accumulation during cell division and links V(D)J recombination to the cell cycle. Immunity 5:575–589. doi: 10.1016/S1074-7613(00)80272-1. [DOI] [PubMed] [Google Scholar]
- 24.Zhang L, Reynolds TL, Shan X, Desiderio S. 2011. Coupling of V(D)J recombination to the cell cycle suppresses genomic instability and lymphoid tumorigenesis. Immunity 34:163–174. doi: 10.1016/j.immuni.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross AE, Vuica M, Desiderio S. 2003. Overlapping signals for protein degradation and nuclear localization define a role for intrinsic RAG-2 nuclear uptake in dividing cells. Mol Cell Biol 23:5308–5319. doi: 10.1128/MCB.23.15.5308-5319.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramón-Maiques S, Kuo AJ, Carney D, Matthews AGW, Oettinger MA, Gozani O, Yang W. 2007. The plant homeodomain finger of RAG2 recognizes histone H3 methylated at both lysine-4 and arginine-2. Proc Natl Acad Sci U S A 104:18993–18998. doi: 10.1073/pnas.0709170104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu C, Ward A, Bettridge J, Liu Y, Desiderio S. 2015. An autoregulatory mechanism imposes allosteric control on the V(D)J recombinase by histone H3 methylation. Cell Rep 10:29–38. doi: 10.1016/j.celrep.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Callebaut I, Mornon J-P. 1998. The V(D)J recombination activating protein RAG2 consists of a six-bladed propeller and a PHD fingerlike domain, as revealed by sequence analysis. Cell Mol Life Sci 54:880–891. doi: 10.1007/s000180050216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grundy GJ, Ramón-Maiques S, Dimitriadis EK, Kotova S, Biertümpfel C, Heymann JB, Steven AC, Gellert M, Yang W. 2009. Initial stages of V(D)J recombination: the organization of RAG1/2 and RSS DNA in the postcleavage complex. Mol Cell 35:217–227. doi: 10.1016/j.molcel.2009.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimazaki N, Tsai AG, Lieber MR. 2009. H3K4me3 stimulates the V(D)J RAG complex for both nicking and hairpinning in trans in addition to tethering in cis: implications for translocations. Mol Cell 34:535–544. doi: 10.1016/j.molcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim M-S, Lapkouski M, Yang W, Gellert M. 2015. Crystal structure of the V(D)J recombinase RAG1-RAG2. Nature 518:507–511. doi: 10.1038/nature14174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bettridge J, Na CH, Pandey A, Desiderio S. 2017. H3K4me3 induces allosteric conformational changes in the DNA-binding and catalytic regions of the V(D)J recombinase. Proc Natl Acad Sci U S A 114:1904–1909. doi: 10.1073/pnas.1615727114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gapud EJ, Lee B-S, Mahowald GK, Bassing CH, Sleckman BP. 2011. Repair of chromosomal RAG-mediated DNA breaks by mutant RAG proteins lacking phosphatidylinositol 3-like kinase consensus phosphorylation sites. J Immunol 187:1826–1834. doi: 10.4049/jimmunol.1101388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coussens MA, Wendland RL, Deriano L, Lindsay CR, Arnal SM, Roth DB. 2013. RAG2's acidic hinge restricts repair-pathway choice and promotes genomic stability. Cell Rep 4:870–878. doi: 10.1016/j.celrep.2013.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pannunzio NR, Li S, Watanabe G, Lieber MR. 2014. Non-homologous end joining often uses microhomology: implications for alternative end joining. DNA Repair 17:74–80. doi: 10.1016/j.dnarep.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. 2010. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell 141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bolland DJ, Wood AL, Afshar R, Featherstone K, Oltz EM, Corcoran AE. 2007. Antisense intergenic transcription precedes Igh D-to-J recombination and is controlled by the intronic enhancer Emu. Mol Cell Biol 27:5523–5533. doi: 10.1128/MCB.02407-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osipovich O, Milley R, Meade A, Tachibana M, Shinkai Y, Krangel MS, Oltz EM. 2004. Targeted inhibition of V(D)J recombination by a histone methyltransferase. Nat Immunol 5:309–316. doi: 10.1038/ni1042. [DOI] [PubMed] [Google Scholar]
- 39.Grundy GJ, Yang W, Gellert M. 2010. Autoinhibition of DNA cleavage mediated by RAG1 and RAG2 is overcome by an epigenetic signal in V(D)J recombination. Proc Natl Acad Sci U S A 107:22487–22492. doi: 10.1073/pnas.1014958107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ru H, Chambers MG, Fu T-M, Tong AB, Liao M, Wu H. 2015. Molecular mechanism of V(D)J recombination from synaptic RAG1-RAG2 complex structures. Cell 163:1138–1152. doi: 10.1016/j.cell.2015.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hesse JE, Lieber MR, Gellert M, Mizuuchi K. 1987. Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V-(D) J AIgnals. Cell 49:775–783. doi: 10.1016/0092-8674(87)90615-5. [DOI] [PubMed] [Google Scholar]
- 42.Liu L-L, Zhao H, Ma T-F, Ge F, Chen C-S, Zhang Y-P. 2015. Identification of AIzation of RT-qPCR expression studies in human breast cancer cell lines treated with and without transient transfection. PLoS One 10:e0117058. doi: 10.1371/journal.pone.0117058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chakraborty T, Perlot T, Subrahmanyam R, Jani A, Goff PH, Zhang Y, Ivanova I, Alt FW, Sen R. 2009. A 220-nucleotide deletion of the intronic enhancer reveals an epigenetic hierarchy in immunoglobulin heavy chain locus activation. J Exp Med 206:1019–1027. doi: 10.1084/jem.20081621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aoki-Ota M, Torkamani A, Ota T, Schork N, Nemazee D. 2012. Skewed primary Igk repertoire and V-J joining in C57BL/6 mice: implications for recombination accessibility and receptor editing. J Immunol 188:2305–2315. doi: 10.4049/jimmunol.1103484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inlay M, Alt FW, Baltimore D, Xu Y. 2002. Essential roles of the κ light chain intronic enhancer and 3′ enhancer in κ rearrangement and demethylation. Nat Immunol 3:463–468. doi: 10.1038/ni790. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.