Abstract
Vestigial Like Family Member 4 (VGLL4) is a transcriptional cofactor of VGLL family, which includes VGLL1-4. Unlike other members of VGLL family, VGLL4 was described as a novel tumor suppressor containing two TDU motifs. VGLL4 executes its biological function through two TDU domains via interacting with TEA domain (TEAD) transcription factors. Lower expression of VGLL4 usually indicates poor survival in many cancers, such as lung cancer, gastric cancer, breast cancer, colorectal cancer, bladder cancer, pancreatic adenocarcinoma and esophageal squamous cancer. In cancer cells, the expression of VGLL4 is lower than that of normal tissues, moreover, expression level of VGLL4 is positively related to survival rate. VGLL4 is found to play an important role in several signal pathways, mainly acts as a tumor suppressor interacting with TEADs. In Hippo signaling pathway, VGLL4 competes with YAP in binding to TEADs and inhibits the downstream of YAP. In Wnt/β-catenin signaling pathway, VGLL4 negatively regulates Wnt/β-catenin signaling pathway via inhibiting β-catenin and TCF (T-cell factor). VGLL4 can also suppress epithelial-mesenchymal transition (EMT) and contribute to apoptosis signaling pathway.
Keywords: VGLL4, TDU, TEAD, YAP, Hippo, Wnt/β-catenin, EMT
Introduction
Transcription factors (TFs) are proteins that control the rate of transcription of genetic information from DNA to mRNA by binding to either enhancer or promoter regions of DNA adjacent to the genes which they regulate, their function is to regulate genes turning on and off in order to make sure genes are expressed in the right place at the right time and in the right amount throughout the life of the cells and the organisms, the transcription of the adjacent gene is either up-regulated or down-regulated [1-3]. Many transcription factors, especially proto-oncogenes or tumor suppressors, could regulate the cell cycle and determine the size of cells or organs and regulate the mitosis [4,5]. Transcription factors work alone or interact with other proteins forming a complex, promoting or blocking the recruitment of RNA polymerase (enzyme performs the transcription of genetic information from DNA to RNA) to specific genes [3,6,7]. Transcription factors contain at least one DNA-binding domain attaching to a specific sequence of DNA adjacent to the genes that they regulate [8,9]. Other proteins such as coactivators, histone acetyltransferases, histone deacetylases, chromatin remodelers, kinases, and methylases are also essential to gene regulation, but lack DNA-binding domains [10].
Transcriptional cofactors are proteins do not contain DNA-binding domain and exert their transcriptional regulatory functions through pairing with transcription factors to regulate transcription of a gene or set of genes [11]. Transcriptional cofactors are usually localized in the nucleus [11,12]. Most cofactors regulate gene expression by binding to an activator and inducing a conformational change that then allows the activator to bind to the DNA enhancer or promoter sequence [13,14]. After activator-coactivator complex binds to the enhancer, RNA polymerase II and other general transcription machinery are recruited to the DNA and transcription begins [15]. The mutation of transcriptional cofactor genes are linked to many diseases and disorders such as birth defects, cancers, neurodevelopmental disorders and intellectual disability [16,17].
VGLL family
Vestigial (VG) is described as a transcriptional coactivator related to the development of wings in Drosophila [18]. The first human homolog of VG was found by Vaudin et al. and named TONDU in 1999 [19]. Previous studies showed that VGLL proteins are transcriptional cofactors just like VG. VGLL proteins are regarded as a new group of TEAD-interacting partners participating in tumor genesis and metastasis. Four VGLL proteins have been found in mammals, named VGLL1-4, all of them do not contain DNA-binding domain, and they exert transcriptional regulatory functions through pairing with TEADs via their Tondu (TDU) domain(s) [19-22]. TEAD proteins are transcription factors associating with coactivators to induce the transcription of target genes [23]. TEAD family contains TEAD1~4 in mammals, all members of the TEAD family have a highly conserved DNA binding domain, the TEA/ATTS DNA-binding domain (TEAD), which has a consensus DNA sequence 5’-CATTCCA/T-3’ that is called the MCAT element [24]. TEAD1 is expressed in various tissues including skeletal muscle, pancreas, placenta, lung, and heart [25]. TEAD2 is selectively expressed in a subset of embryonic tissues including the cerebellum, testis, and distal portions of the forelimb and hindlimb buds, as well as the tail bud, but it is essentially absent in adult tissues [26]. TEAD3 is predominantly expressed in the placenta [27]. TEAD4 is preferentially expressed in the skeletal muscle [28]. VGLL1-3 had been identified as TEADs-related transcriptional coactivators required for cancer cells’ growth. VGLL1 promotes cell proliferation and exhibits high expression in basal-like breast cancer [29]. VGLL1-TEAD4 complex facilitates anchorage-independent cell proliferation in prostate cancer cell lines [30]. VGLL2 is fused in rhabdomyosarcoma [31], the interaction of VGLL2 and TEAD1 alters expression of myogenic genes [20,32]. VGLL3 is rich in soft tissue sarcoma [33]. The combination of VGLL3 and TEAD3 plays a role in determining age at maturation in Atlantic salmon [34]. However, in epithelial ovarian cancer, it is strange that the expression of VGLL3 is associated with a tumor suppressor phenotype [35]. Unlike VGLL1~3, VGLL4 is described as a tumor suppressor.
Structure of VGLL4
VGLL4 was initially identified as a transcript expressed in an adult human myeloid leukemia bone marrow-derived stem cell line [36]. VGLL4 is the only member of VGLL family expresses in heart and the biological function of VGLL4 was initially characterized in heart [37]. VGLL4 gene is on 3p25.3~3p25.2, includes 14 exons. Widely expression of VGLL4 genes was detected in human tissues [38]. VGLL4 protein is made up of 290 amino acids (METPLDVLSR AASLVHADDE KREAALRGEP RMQTLPVASA LSSHRTGPPP ISPSKRKFSM EPGDEDLDCD NDHVSKMSRI FNPHLNKTAN GDCRRDPRER SRSPIERAVA PTMSLHGSHL YTSLPSLGLE QPLALTKNSL DASRPAGLSP TLTPGERQQN RPSVITCASA GARNCNLSHC PIAHSGCAAP GPASYRRPPS AATTCDPVVE EHFRRSLGKN YKEPEPAPNS VSITGSVDDH FAKALGDTWL QIKAAKDGAS SSPESASRRG QPASPSAHMV SHSHSPSVVS), and Position 1 is an acetylation site, Position 52, 149, 153, 274 are phosphorylation sites [39-41]. In 2014, scientists from China described the crystal structure of VGLL4 for the first time [42]. The fundamental structure of VGLL4 contains one nuclear export signal, one conserved sequence and two TDU domains. Two TDU domains in the carboxyl terminal could combine with TEADs and execute the function of suppressing cancer cells’ growth and progression [22]. Comparing with other members of VGLL family, VGLL4 has an extra TDU motif in its carboxyl terminal domain and the extra TDU domain is considered to have special functions, deletion of the second TDU domain (aa242-252) completely abolished the inhibition function of VGLL4 while deletion of the first one (aa214-224) did not influence VGLL4’s function [22]. The amino terminal of VGLL4 protein could combine with Ubiquitin-specific protease 11 (USP11) , which increases VGLL4 protein stability by promoting its deubiquitination [43]. VGLL4 protein is mainly located in the nuclear, few in the cytoplasm, and the nuclear export signal motif combining with nuclear exportin chromosomal maintenance 1 (CRM-1,aslo known as Exportin 1) is essential for the exportation of VGLL4 protein from nucleus to cytoplasm [37,44].
Regulation of VGLL4 expression
The expression of VGLL4 is influenced by miR-3662, miR-222, miR-130a and miR-130b (Figure 1). In HepG2 cells (a perpetual cell line which is derived from the liver tissue of a 15-year-old Caucasian American male with a well-differentiated hepatocellular carcinoma), miR-3662 expression was found to be increased by p53 [45]. AASLVHADDE KREAALRGEP RMQTLPVASA cg25619837 methylation probe and hsa-miR-3662 miRNA was significantly associated with gene expression level of VGLL4 [46], but the exact mechanisms need further investigation. Simultaneously, miR-222 also has great influence on the expression of VGLL4. In gastric cancer cells, VGLL4 is a direct target of miR-222, reduction expression of VGLL4 is accompanied with rising expression of miR-222 [47]. In addition, miR-130a, which is induced by YAP, could effectively repress VGLL4 expression and amplify the YAP signals, and inhibition of miR-130a could reverse organ size enlargement induced by Hippo pathway inactivation and block YAP-induced tumorigenesis. Mechanically, miR-130a specifically bind to 3’UTR of VGLL4, while miR-130a did not significantly reduce VGLL4 mRNA level, which suggests a mechanism of translation repression [48]. Recently, VGLL4 gene was found to be a direct target of miR-130b and VGLL4 suppression was crucial for miR-130b-induced bladder cancer cell proliferation, migration and invasion [49].
Figure 1.
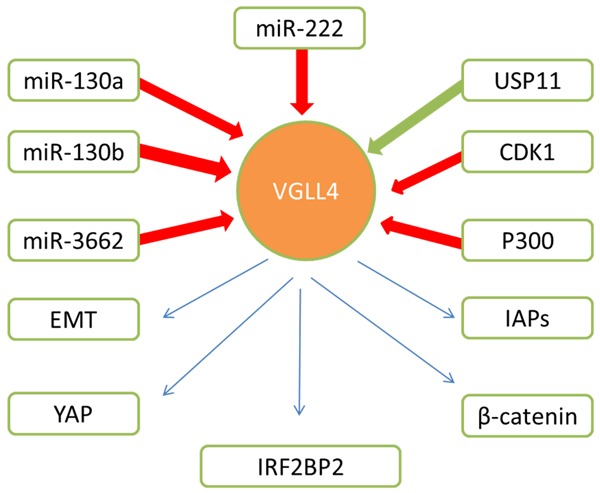
VGLL4 related factors. CDK1 phosphorylates VGLL4 affecting both YAP and β-catenin activity inhibiting tumor suppressing activity of VGLL4; USP11 interacts with VGLL4 and increases VGLL4 protein stability by promoting its deubiquitination; Histone actylase P300 acetylates VGLL4; miR-130a, miR130b, miR-222 and miR-3662 suppress expression of VGLL4; USP11 interacts with VGLL4 and increases VGLL4 protein stability; VGLL4 has influence on EMT, IAPs, IRF2BP2, Wnt/β-catenin pathway and YAP-induced Hippo pathway.
Post translational modification
Post translational modification of VGLL4 includes phosphorylation, acetylization and ubiquitylation (Figure 1). Cyclin-dependent kinase 1 (CDK1) is a key player in cell cycle regulation functioning as a serine/threonine kinase [50]. CDK1 interacts with a variety of target substrates forming complexes which influence cell cycle progression [51]. CDK1 also controls proliferation by limiting transcription factor activity [52]. VGLL4 is phosphorylated both in vivo and in vitro by CDK1 during G2/M arrest and normal mitosis. Ectopic expression of VGLL4 suppresses migration and proliferation of tumor cells, while the mitotic phosphorylation deficient mutant VGLL4-4A shows much stronger tumor suppressive activity compared with VGLL4 in pancreatic cancer tumorigenesis in vitro and in vivo. Taken together, CDK1-mediated mitotic phosphorylation of VGLL4 inhibits VGLL4 induced tumor-suppressing activity [41]. Histone actylase P300 could acetylate VGLL4, thus negatively regulates its binding to TEAD1. Furthermore, overexpression of an acetylation refractory VGLL4 mutant enhances TEAD1 degradation, revealing that VGLL4 inhibits TEAD1 not only via combing with it, but also promoting degradation of TEAD1 [53]. Ubiquitin-specific protease 11 (USP11), a deubiquitinating enzyme, is a member of the USP family and contains two internal ubiquitin-like domains and one N-terminal domain present in ubiquitin-specific proteases. Previous studies showed knockdown of USP11 promotes cell growth, migration, and invasion in a YAP-dependent manner, which indicates USP11 is a tumor suppressor [54]. Dramatically, USP11 interacts with VGLL4 and increases VGLL4 protein stability by promoting its deubiquitination [43].
Suppressing YAP-induced tumorigenesis
The Hippo signaling pathway, also known as the Salvador/Warts/Hippo (SWH) pathway, plays an important role in the regulation of cell proliferation and apoptosis. Hippo pathway also has critical role in stem cell and tissue specific progenitor cell self-renewal and expansion. Mutation of the Hippo gene results in uncontrollable organ size in animals, or a “hippopotamus”-like phenotype [55,56]. The Hippo pathway consists of a core kinase cascade, in which Hippo phosphorylates protein kinase Wts, once phosphorylated, Wts (LATS1/2 in mammals) becomes active [57,58]. Activated Wts phosphorylates and inactivates Yki (YAP/TAZ in mammals), which inhibits organ growth. Yki is a transcriptional coactivator, after activated, Yki binds to the transcription factor Scalloped (Sd) forming a complex. Yki-Sd complex localizes to the nucleus, increasing the expression of several genes that promote organ growth or inhibit apoptosis, such as cyclin E and diap1 [59]. Yki also activates expression of the bantam microRNA, a positive growth regulator, which specifically affects cell number [60,61]. In mammals, the two Yki orthologs are Yes-associated protein (YAP/YAP1/YAP65) and transcriptional coactivator with PDZ-binding motif (TAZ) [62]. YAP, a growth promoter both in vitro cell culture system and in vivo mouse models [63,64], is downstream essential effector of the Hippo pathway. Ectopic YAP expression is sufficient to drive cell proliferation, transformation, invasion and epithelial-mesenchymal transition (EMT) [65]. YAP has been found to be elevated in several human cancers, including breast cancer, colorectal cancer and liver cancer [66-68]. YAP/TAZ can bind to several transcription regulators, such as p73, Runx2 and TEADs [69], regulating the expression of downstream genes [70]. The interaction between YAP and all TEAD proteins was demonstrated both in vitro and in vivo, in both cases interaction of the two proteins increases TEAD transcriptional activity and activates downstream of YAP. The combination of YAP and TEAD forming a complex regulates the transcriptional output of Hippo pathway, eventually turns on the expression of downstream genes accelerating cell proliferation and inhibiting apoptosis [71,72] (Figure 2). Recently researches revealed that VGLL4 could bind to TEADs through its TDU domains and restrict the interaction between YAP and TEADs, thus inhibiting downstream oncogenes expression [22,73].
Figure 2.

VGLL4 in Hippo pathway. VGLL4 competes with YAP in binding to TEADs thus inhibiting downstream genes expression.
Suppressing Wnt/β-catenin signaling pathway
The Wnt signaling pathways are a group of signal transduction pathways made of proteins that pass signals into a cell through cell surface receptors. Wnt pathway was first identified for its role in carcinogenesis, then for its function in embryonic development, including body axis patterning, cell fate specification, cell proliferation and cell migration. Three Wnt signaling pathways have been characterized: the canonical Wnt pathway, the noncanonical planar cell polarity pathway and the noncanonical Wnt/calcium pathway [74]. The canonical Wnt pathway (Wnt/β-catenin pathway) is the Wnt pathway that causes an accumulation of β-catenin in the cytoplasm and its eventual translocation into the nucleus to act as a transcriptional coactivator of transcription factors that belong to the TCF/LEF (T-cell factor/lymphoid enhancing factor) family. Without Wnt, β-catenin would not accumulate in the cytoplasm since a destruction complex would normally degrade it by targeting it for ubiquitination, which subsequently sends it to the proteasome to be digested [75]. With Wnt taking part in, the destruction complex function becomes disrupted, which allows β-catenin to accumulate and localize to the nucleus and subsequently induce a cellular response via gene transduction alongside the TCF/LEF transcription factors and recruits other transcriptional coactivators. TCF/LEF interacts with other transcription factors and coactivators such as TEADs and β-catenin, facilitating target genes expression [75-77]. Whereas VGLL4 can combine with TEADs, thus inhibiting the interaction between TCF and TEADs, moreover, VGLL4 overexpression can suppress the expression level of β-catenin [44,78]. To sum up, VGLL4 negatively regulates Wnt/β-catenin signaling pathway via inhibiting β-catenin and TCF (Figure 3).
Figure 3.
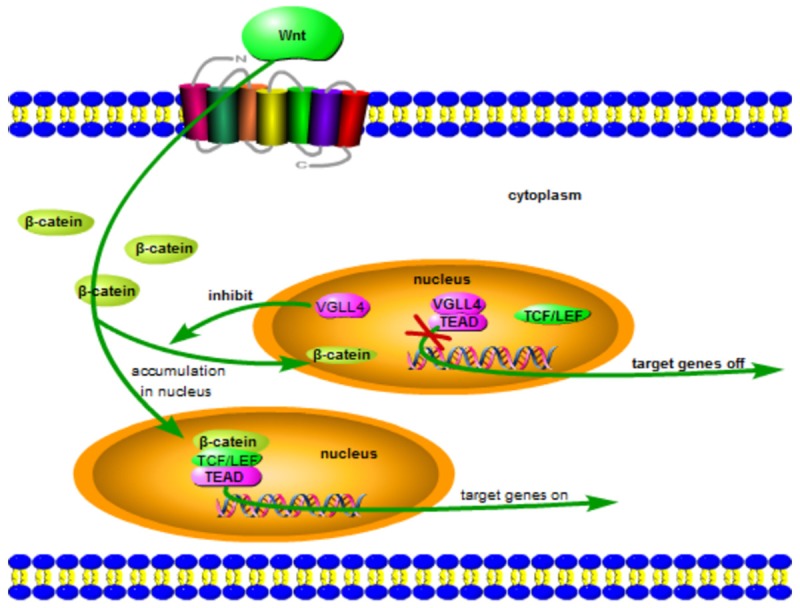
VGLL4 in Wnt/β-catenin pathway. VGLL4 down-regulatesβ-catenin suppressing its accumulation in nucleus, simultaneously, VGLL4 combines with TEADs thus inhibiting target genes of TCF/LEF.
Suppressing epithelial-mesenchymal transition
Epithelial-mesenchymal transition (EMT) is a process that epithelial cells lose their cell polarity and cell-cell adhesion, and gain invasive properties to become mesenchymal stem cells, which plays an important role in cancer progression and metastasis [79-81]. Loss of Ecadherin is a fundamental event in EMT, many transcription factors can repress E-cadherin directly or indirectly, which are known as EMT-TFs, such as SNAIL, TCF3, and ZEB. TCF3 can bind to E-cadherin promoter and repress its transcription [82]. TCF4/β-catenin complex could induce the EMT activator ZEB1 to regulate tumor invasiveness [83]. LEF1/β-catenin complex can repress E-cadherin mRNA, thus inducing the EMT [84]. Whereas VGLL4 overexpression increases the expression level of E-cadherin and decreases the expression level of β-catenin. Collectively, VGLL4 inhibits EMT in part through suppressing Wnt/β-catenin signaling pathway [44].
Contributing to apoptosis signaling pathway
Inhibitor of apoptosis proteins (IAPs) are a protein family function as inhibitors of programmed cell death. IAPs bind with caspases thereby inhibiting their activation and preventing apoptosis [85]. XIAP, cIAP1 and cIAP2 are members of IAPs family, VGLL4 was shown to interact with XIAP, cIAP1 and cIAP2, contributing to apoptosis signaling pathway via forcing the nuclear relocation of IAPs. VGLL4 executes its function of inhibiting IAPs by nuclear sequestration, the forced relocation of IAPs to nucleus by VGLL4 significantly reduced their ability to prevent Bax- and TNFα-induced apoptosis in vitro, which suggests that VGLL4 may play a role in the apoptotic pathways by regulating translation of IAPs between different cell compartments, however, VGLL4 does not play a role in signal transduction pathways involving TRAF2-cIAP1/2 interaction and VGLL4 overexpression has no influence on TNFa-induced NF-κB activity, considering that forced relocation of IAPs by VGLL4 significantly reduce their ability to prevent apoptosis, and TRAF2 (IAP-binding protein) coexpression abolished that effect of VGLL4 on IAP-mediated protection against apoptosis, collectively, VGLL4 requires an activation signal for IAP interaction, while the activation signal remains unknown [86].
Described as a tumor suppressor
VGLL4 was described as a tumor suppressor in many kinds of cancers, such as lung cancer [22], breast cancer [73], gastric cancer [42,44,47], colorectal cancer [78], bladder cancer [46,49], pancreatic adenocarcinoma [87] and esophageal squamous cancer [88]. Lower expression of VGLL4 usually indicates poor survival.
In lung cancer cells, the expression of VGLL4 is significantly lower than that of normal tissues. In vitro, ectopic expression of VGLL4 inhibits the growth of lung cancer cells; in de novo mouse models, VGLL4 significantly suppresses lung cancer progression. Moreover, ectopic VGLL4 expression significantly reduces the TEAD4-dependent luciferase activity, while VGLL4 mutant without two TDU domains obviously loss inhibitory function, consistently, VGLL4 knockdown significantly increases TEADs’ transcriptional activity in HEK-293T, mechanistically, TEAD4 is highly expressed in lung cancer cells, VGLL4 competes with YAP in binding to TEAD4 and inhibits the growth of cancer cells through two TDU domains [22]. YAP is the downstream essential effector of Hippo pathway, nuclear YAP binds to and activates transcription factors TEADs eventually turns on expression of downstream genes [89]. The interaction between VGLL4 and TEAD4 represses YAP-induced target genes expression eventually suppressing the growth of lung cancer cells.
In breast cancer, VGLL4 is also a tumor suppressor gene which combines with YAP and inhibits the downstream genes of YAP. The median overall survival and relapse-free survival within five years of diagnosis is positively correlated with expression of VGLL4 [73]. VGLL4 selectively represses YAP dependent genes induction and tumorigenic phenotypes through competing with YAP in binding to TEAD1. Co-IP assay shows that TEAD1 and VGLL4 co-precipitated but VGLL4 with the deletion of TDU2 and VGLL4 with the deletion of both TDU2 and TDU1 could not bind to TEAD1. VGLL4 interacts with TEAD1 through its second TDU domain at its C-terminus, overexpression of VGLL4 or VGLL4 with a TDU1 deletion completely abolished luciferase activity associated with the TEAD response element. In contrast, VGLL4 with TDU2 deleted or TDU1 and TDU2 deleted failed to block this activity, which demonstrates that TDU2 domain of VGLL4 is sufficient and necessary to inhibit YAP activity [73].
In gastric cancer, the expression of VGLL4 in tumor tissues is significantly lower than that of normal tissues and peritumoral tissues, and VGLL4 expression is associated with tumor size, TNM stage, serosal invasion, vascular invasion, and lymph node metastasis. In addition, down-regulation of VGLL4 results in a worse 5-year survival for gastric cancer patients, overexpression of VGLL4 can suppress oncogenic properties and inhibit migration and invasion of gastric cancer cell lines both in vitro and vivo [42,44,47]. VGLL4 overexpression increases the expression of E-cadherin meanwhile decreases the expression of β-catenin, while knockdown of VGLL4 suppresses the expression of E-cadherin accompanied by increasing expression of β-catenin [44]. It has been demonstrated that Wnt signaling pathway activation can induce EMT [84], considering that LEF1/β-catenin complex can downregulate E-cadherin mRNA thus inducing the EMT, and VGLL4 overexpression decreases the expression of β-catenin, collectively, VGLL4 inhibits EMT in part through suppressing Wnt/β-catenin signaling pathway in gastric cancer [44,84]. Beyond that, a miR-222/VGLL4/YAP-TEAD1 regulatory loop promoting proliferation and invasion of gastric cancer cells was found. VGLL4 is a direct target of miR-222 in gastric cancer cells, the reduction of VGLL4 is accompanied with miR-222 expression increase, miR-222 directly targets VGLL4 and exerts its function via suppressing VGLL4 expression, thus resulting in YAP-TEAD1 activation. TEAD1 transcriptionally enhances miR-222 expression to maintain the miR-222/VGLL4/YAP-TEAD1 regulatory loop contributing to proliferation and invasion of gastric cancer cells through physically binding to miR-222 promoter then accelerating miR-222 expression [47]. Additionally, a peptide mimicking VGLL4 function called “Super-TDU” acting as a YAP antagonist therapy against gastric cancer in mice was found, revealing that VGLL4 suppressed gastric cancer growth in vitro by competing with YAP for TEAD binding [42].
In colorectal cancer, the expression of VGLL4 is significantly down-regulated and VGLL4 expression is positively associated with patient survival, moreover, knockdown of VGLL4 accelerates proliferation and tumor formation in colorectal cancer cells. VGLL4 inhibits colorectal cancer growth in vitro and in vivo. A designed peptide mimicking the function of VGLL4 effectively inhibited colorectal cancer progression in a de novo mouse model. Functionally, VGLL4 targets a TCF4-TEAD4 complex to regulate both Wnt/β-catenin signaling pathway and Hippo signaling pathway, the Hippo pathway transcription factor TEAD4 directly associates with the Wnt pathway transcription factor TCF4 via their DNA-binding domains, forming a complex on target genes. VGLL4 combines with this TEAD4-TCF4 complex to interfere the functional interplay between TEAD4 and TCF4, suppressing the expression of downstream genes, thus influencing both Wnt/β-catenin signaling pathway and YAP-induced signaling pathway [78].
In esophageal squamous cell carcinoma, the expression of VGLL4 is down-regulated, whereas forced expression of VGLL4 inhibits cell growth and migration, and knockdown of VGLL4 promotes the tumorigenecity of esophageal squamous cell carcinoma cells. Simultaneously, overexpression of VGLL4 suppresses the expression of connective tissue growth factor, while knockdown of VGLL4 promotes the expression of connective tissue growth factor. VGLL4 regulated the growth and motility of esophageal squamous cell carcinoma cells part through repressing the expression of connective tissue growth factor, but the exact mechanism needs further exploration [88,90]. It has been reported that connective tissue growth factor is overexpressed in esophageal squamous cell carcinoma and promotes tumorigenicity through β-catenin-TCF/LEF signaling [90]. Collectively, VGLL4 may restrict connective tissue growth factor through inhibiting Wnt/β -pathway.
In pancreatic cancer, VGLL4 is considered as a candidate tumor repressor, and VGLL4 gene mutation is significantly associated with poor patient survival [87]. In bladder cancer, lower expression of VGLL4 is also associated with a worse survival outcome [46,48]. While in prostate cancer, it is strange that VGLL4 up-regulation is associated with poor prognosis [91].
In physiological processes and non-oncologic diseases
VGLL4 overexpression in hESCs (human embryonic stem cells) significantly decreases cell death in response to dissociation stress. Moreover, the increasing expression of VGLL4 enhances hESC colony formation from single cells. VGLL4 overexpression promotes survival of hESCs in the context of dissociation stress by decreasing Caspase activation. Conversely, reduction in VGLL4 by shRNA knockdown results in an increase in Caspase activation, and impairs the ability of hESCs to respond to the pro-survival effects of Rock inhibition [92]. In cardiac myocytes, VGLL4 is a negative modulator of transcriptional enhancer factor 1 (TEF1) and myocyte enhancer factor 2 (MEF2). A mammalian two-hybrid assay showed VGLL4 interacts with both TEF-1 and MEF2 forming a bridge between TEF-1 and MEF2 through its two TDU domains in cardiac myocytes. Mechanistically, VGLL4 interacts with TEF-1 suppressing the activation of α-skeletal actin promoter and interferes with the activity of a MEF2-dependent myosin light chain promoter. Two TDU motifs of VGLL4 allow TEF-1 and MEF2 to bind to VGLL4 at the same time, additionally, neither TDU motif alone is sufficient to restore full interaction with TEF-1 and MEF2 [37]. In muscle, VGLL4 is described as a novel partner of interferon response factor 2 binding protein 2 (IRF2BP2). IRF2BP2 combines with TEAD1 activating vascular endothelial growth factor A (VEGFA) expression, while IRF2BP2 with TEAD4 is not sufficient to activate VEGFA promoter, however, IRF2BP2 promotes VEGFA expression through interacting with TEAD4-VGLL4 complex and TDU1 of VGLL4 is required for IRF2BP2 interaction [93]. In non-oncologic diseases, several genome-wide association studies (GWAS) indicates that expression of VGLL4 is significantly related anorexia nervosa [94], neuroticism [95], comorbid depressive syndrome and alcohol dependence [96], but exact mechanisms remain to be explored.
Conclusion and prospects
VGLL4 is a member of transcriptional cofactors, just like other transcriptional cofactors, VGLL4 interacts with transcription factors such as TEADs, controlling the rate of transcription of genetic information from DNA to mRNA and regulating downstream of specific genes. VGLL4 acts as a tumor suppressor mainly competing with YAP in binding to TEADs. VGLL4 also reduces Wnt/β-catenin signaling pathway through restricting β-catenin and TCF. Simultaneously, VGLL4 suppresses Epithelial-mesenchymal transition (EMT) and forces the nuclear relocation of IAPs. Tumor suppressing function of VGLL4 has been demonstrated, however, regulation of VGLL4 remains elusively, which includes regulation at transcriptional level and post-translational modification. The mutation of transcriptional cofactor genes have been linked to diseases and disorders such as birth defects, cancers, neurodevelopmental disorders and intellectual disability, we can replace cofactors with a synthetic ligand that allows for control over an increase or decrease in gene expression. It is inspiring that a peptide mimicking VGLL4 function could act as a YAP antagonist therapy against gastric cancer and colorectal cancer. With the development of theory and technology, it will hopefully provide better targets for future drug therapies based on cofactors. Considering that expression of VGLL4 gene is influenced by several miRNAs, and function of VGLL4 protein is affected by post-translational modification, it may be effective to promote VGLL4 gene expression via inhibiting relative miRNA expression and enhance VGLL4 protein stabilization through dephosphorylation, deacetylation and deubiquitination. Finally, the regulation networks of tumor genesis, migration and invasion are so complicated, which part plays decisive role remains to be explored, for example, VGLL4 could compete with YAP interacting with TEADs, however, many factors can facilitate YAP-induced tumorigenesis and other signaling pathways can also participate in. Which factor is the most decisive role, which signaling pathway function as dominant role and how they get the balance are still unknown in VGLL4-induced regulation network.
Acknowledgements
This project was supported by Shanghai Science and Technology Commission (NO. 17411967200) and Shanghai Municipal Commission of Health and Family Planning (NO. 201640097). We give special thanks to other members in our research group for their valuable suggestion.
Disclosure of conflict of interest
None.
References
- 1.Karin M. Too many transcription factors: positive and negative interactions. New Biol. 1990;2:126–131. [PubMed] [Google Scholar]
- 2.Latchman DS. Transcription factors: an overview. Int J Biochem Cell Biol. 1997;29:1305. doi: 10.1016/s1357-2725(97)00085-x. [DOI] [PubMed] [Google Scholar]
- 3.Nikolov DB, Burley SK. RNA polymerase II transcription initiation: a structural view. Proc Natl Acad Sci U S A. 1997;94:15–22. doi: 10.1073/pnas.94.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wheaton K, Atadja P, Riabowol K. Regulation of transcription factor activity during cellular aging. Biochem Cell Biol. 1996;74:523–534. doi: 10.1139/o96-056. [DOI] [PubMed] [Google Scholar]
- 5.Meyyappan M, Atadja PW, Riabowol KT. Regulation of gene expression and transcription factor binding activity during cellular aging. Biol Signals. 1996;5:130–138. doi: 10.1159/000109183. [DOI] [PubMed] [Google Scholar]
- 6.Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 7.Lee TI, Young RA. Transcription of eukaryotic protein-coding genes. Annu Rev Genet. 2000;34:77. doi: 10.1146/annurev.genet.34.1.77. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell PJ, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 9.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 10.Brivanlou AH, Darnell JE Jr. Signal transduction and the control of gene expression. Science. 2002;295:813–818. doi: 10.1126/science.1066355. [DOI] [PubMed] [Google Scholar]
- 11.Courey AJ. Mechanisms in transcriptional regulation. Blackwell Publishing. 2008 [Google Scholar]
- 12.Vosnakis N, Koch M, Scheer E, Kessler P, Mély Y, Didier P, Tora L. Coactivators and general transcription factors have two distinct dynamic populations dependent ontranscription. EMBO J. 2017;36:2710–2725. doi: 10.15252/embj.201696035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spiegelman BM, Heinrich R. Biological control through regulated transcriptional coactivators. Cell. 2004;119:157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 14.Scholes NS, Weinzierl RO. Molecular dynamics of “Fuzzy” transcriptional activator-coactivator interactions. PLoS Comput Biol. 2016;12:e1004935. doi: 10.1371/journal.pcbi.1004935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit Rev Biochem Mol Biol. 2006;41:105. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 16.Uzman A. Molecular cell biology (4th edition) Biochemistry & Molecular Biology Education. 2001;29:126–128. [Google Scholar]
- 17.Kumar R, O’Malley BW. NR coregulators and human diseases. World Scientific. 2008 [Google Scholar]
- 18.Morgan TH, Sturtevant AH Carnegie Institution of Washington. Contributions to the genetics of Drosophila melanogaster. 1919;22 [Google Scholar]
- 19.Vaudin P, Delanoue R, Davidson I, Silber J, Zider A. TONDU (TDU), a novel human protein related to the product of vestigial (vg) gene of Drosophila melanogaster interacts with vertebrate TEF factors and substitutes for Vg function in wing formation. Development. 1999;126:4807–4816. doi: 10.1242/dev.126.21.4807. [DOI] [PubMed] [Google Scholar]
- 20.Maeda T, Chapman DL, Stewart AF. Mammalian vestigial-like 2, a cofactor of TEF-1 and MEF2 transcription factors that promotes skeletal muscle differentiation. J Biol Chem. 2002;277:48889–48898. doi: 10.1074/jbc.M206858200. [DOI] [PubMed] [Google Scholar]
- 21.Hélias-Rodzewicz Z, Pérot G, Chibon F, Ferreira C, Lagarde P, Terrier P, Coindre JM, Aurias A. YAP1 and VGLL3, encoding two cofactors of TEAD transcription factors, are amplified and overexpressed in a subset of soft tissue sarcomas. Genes Chromosomes Cancer. 2010;49:1161–1171. doi: 10.1002/gcc.20825. [DOI] [PubMed] [Google Scholar]
- 22.Zhang W, Gao Y, Li P, Shi Z, Guo T, Li F, Han X, Feng Y, Zheng C, Wang Z, Li F, Chen H, Zhou Z, Zhang L, Ji H. VGLL4 functions as a new tumor suppressor in lung cancer by negatively regulating the YAP-TEAD transcriptional complex. Cell Res. 2014;24:331–343. doi: 10.1038/cr.2014.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao JH, Davidson I, Matthes H, Garnier JM, Chambon P. Cloning, expression, and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- 24.Bürglin TR. The TEA domain: a novel, highly conserved DNA-binding motif. Cell. 1991;66:11. doi: 10.1016/0092-8674(91)90132-i. [DOI] [PubMed] [Google Scholar]
- 25.Anbanandam A, Albarado DC, Nguyen CT, Halder G, Gao X, Veeraraghavan S. Insights into transcription enhancer factor 1 (TEF-1) activity from the solution structure of the TEA domain. Proc Natl Acad Sci U S A. 2006;103:17225–17230. doi: 10.1073/pnas.0607171103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yasunami M, Suzuki K, Houtani T, Sugimoto T, Ohkubo H. Molecular characterization of cDNA encoding a novel protein related to transcriptional enhancer factor-1 from neural precursor cells. J Biol Chem. 1995;270:18649–18654. doi: 10.1074/jbc.270.31.18649. [DOI] [PubMed] [Google Scholar]
- 27.Mann CJ, Osborn DP, Hughes SM. Vestigiallike-2b (VITO-1b) and Tead-3a (Tef-5a) expression in zebrafish skeletal muscle, brain and notochord. Gene Expr Patterns. 2007;7:827–836. doi: 10.1016/j.modgep.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yagi R, Kohn MJ, Karavanova I, Kaneko KJ, Vullhorst D, Depamphilis ML, Buonanno A. Transcription factor TEAD4 specifies the trophectoderm lineage at the beginning of mammalian development. Development. 2007;134:3827. doi: 10.1242/dev.010223. [DOI] [PubMed] [Google Scholar]
- 29.Castilla MA, Lopez-Garcia MA, Atienza MR, Rosa-Rosa JM, Diaz-Martin J, Pecero ML, Vieites B, Romero-Perez L, Benitez J, Calcabrini A, Palacios J. VGLL1 expression is associated with a triple-negative basal-like phenotype in breast cancer. Endocr Relat Cancer. 2014;21:587–599. doi: 10.1530/ERC-13-0485. [DOI] [PubMed] [Google Scholar]
- 30.Pobbati AV, Chan SW, Lee I, Song H, Hong W. Structural and functional similarity between the Vgll1-TEAD and the YAP-TEAD complexes. Structure. 2012;20:1135–1140. doi: 10.1016/j.str.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 31.Alaggio R, Zhang L, Sung YS, Huang SC, Chen CL, Bisogno G, Zin A, Agaram NP, LaQuaglia MP, Wexler LH, Antonescu CR. A molecular study of pediatric spindle and sclerosing rhabdomyosarcoma: Identification of novel and recurrent VGLL2-related fusions in infantile cases. Am J Surg Pathol. 2016;40:224–235. doi: 10.1097/PAS.0000000000000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Günther S, Mielcarek M, Krüger M, Braun T. VITO-1 is an essential cofactor of TEF1-dependent muscle-specific gene regulation. Nucleic Acids Res. 2004;32:791–802. doi: 10.1093/nar/gkh248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Antonescu CR, Zhang L, Nielsen GP, Rosenberg AE, Dal Cin P, Fletcher CD. Consistent t(1;10) with rearrangements of TGFBR3 and MGEA5 in both myxoinflammatory fibroblastic sarcoma and hemosiderotic fibrolipomatous tumor. Genes Chromosomes Cancer. 2011;50:757–764. doi: 10.1002/gcc.20897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christensen KA, Gutierrez AP, Lubieniecki KP, Davidson WS. TEAD3, implicated by association to grilsing in Atlantic salmon. Aquaculture. 2017;479:571–578. [Google Scholar]
- 35.Gambaro K, Quinn MC, Wojnarowicz PM, Arcand SL, de Ladurantaye M, Barres V, Ripeau JS, Killary AM, Davis EC, Lavoie J, Provencher DM, Mes-Masson AM, Chevrette M, Tonin PN. VGLL3 expression is associated with a tumor suppressor phenotype in epithelial ovarian cancer. Mol Oncol. 2013;7:513–530. doi: 10.1016/j.molonc.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagase T, Seki N, Tanaka A, Ishikawa K, Nomura N. Prediction of the coding sequences of unidentified human genes. IV. The coding sequences of 40 new genes (KIAA0121-KIAA0160) deduced by analysis of cDNA clones from human cell line KG-1 (supplement) DNA Res. 1995;2:167–74. 199–210. doi: 10.1093/dnares/2.4.167. [DOI] [PubMed] [Google Scholar]
- 37.Chen HH, Mullett SJ, Stewart AF. Vgl-4, a novel member of the vestigial-like family of transcription cofactors, regulates alpha1-adrenergic activation of gene expression in cardiac myocytes. J Biol Chem. 2004;279:30800–30806. doi: 10.1074/jbc.M400154200. [DOI] [PubMed] [Google Scholar]
- 38.Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J, Habuka M, Tahmasebpoor S, Danielsson A, Edlund K, Asplund A, Sjostedt E, Lundberg E, Szigyarto CA, Skogs M, Takanen JO, Berling H, Tegel H, Mulder J, Nilsson P, Schwenk JM, Lindskog C, Danielsson F, Mardinoglu A, Sivertsson A, von Feilitzen K, Forsberg M, Zwahlen M, Olsson I, Navani S, Huss M, Nielsen J, Ponten F, Uhlen M. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strausberg RL, Feingold EA, Grouse LH, Derge JG, Klausner RD, Collins FS, Wagner L, Shenmen CM, Schuler GD, Altschul SF. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc Natl Acad Sci U S A. 2002;99:16899. doi: 10.1073/pnas.242603899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerhard DS, Wagner L, Feingold EA, Shenmen CM, Grouse LH, Schuler G, Klein SL, Old S, Rasooly R, Good P. The status, quality, and expansion of the NIH full-length cDNA project: the Mammalian Gene Collection (MGC) Genome Res. 2004;14:2121. doi: 10.1101/gr.2596504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng Y, Stauffer S, Zhou J, Chen X, Chen Y, Dong J. Cyclin-dependent kinase 1 (CDK1)-mediated mitotic phosphorylation of the transcriptional co-repressor Vgll4 inhibits its tumor-suppressing activity. J Biol Chem. 2017;292:15028–15038. doi: 10.1074/jbc.M117.796284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiao S, Wang H, Shi Z, Dong A, Zhang W, Song X, He F, Wang Y, Zhang Z, Wang W, Wang X, Guo T, Li P, Zhao Y, Ji H, Zhang L, Zhou Z. A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell. 2014;25:406. doi: 10.1016/j.ccr.2014.01.010. [DOI] [PubMed] [Google Scholar]
- 43.Zhang E, Shen B, Mu X, Qin Y, Zhang F, Liu Y, Xiao J, Zhang P, Wang C, Tan M. Ubiquitinspecific protease 11 (USP11) functions as a tumor suppressor through deubiquitinating and stabilizing VGLL4 protein. Am J Cancer Res. 2016;6:2901. [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Wang Z, Zhang W, Qian K, Liao G, Xu W, Zhang S. VGLL4 inhibits EMT in part through suppressing Wnt/β-catenin signaling pathway in gastric cancer. Med Oncol. 2015;32:83. doi: 10.1007/s12032-015-0539-5. [DOI] [PubMed] [Google Scholar]
- 45.Yang Y, Liu W, Ding R, Xiong L, Dou R, Zhang Y, Guo Z. Comprehensive expression profiling and functional network analysis of p53-regulated MicroRNAs in HepG2 cells treated with doxorubicin. PLoS One. 2016;11:e0149227. doi: 10.1371/journal.pone.0149227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shivakumar M, Lee Y, Bang L, Garg T, Sohn KA, Kim D. Identification of epigenetic interactions between miRNA and DNA methylation associated with gene expression as potential prognostic markers in bladder cancer. BMC Med Genomics. 2017;10:30. doi: 10.1186/s12920-017-0269-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li N, Yu N, Wang J, Xi H, Lu W, Xu H, Deng M, Zheng G, Liu H. miR-222/VGLL4/YAPTEAD1 regulatory loop promotes proliferation and invasion of gastric cancer cells. Am J Cancer Res. 2015;5:1158. [PMC free article] [PubMed] [Google Scholar]
- 48.Shen S, Guo X, Yan H, Lu Y, Ji X, Li L, Liang T, Zhou D, Feng XH, Zhao JC, Yu J, Gong XG, Zhang L, Zhao B. A miR-130a-YAP positive feedback loop promotes organ size and tumorigenesis. Cell Res. 2015;25:997–1012. doi: 10.1038/cr.2015.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu X, Kong C, Zhang Z. miR-130b promotes bladder cancer cell proliferation, migration and invasion by targeting VGLL4. Oncol Rep. 2018;39:2324–2332. doi: 10.3892/or.2018.6300. [DOI] [PubMed] [Google Scholar]
- 50.Young PG. The Cell Cycle: Principles of Control by David O Morgan. Quarterly Review of Biology. 2008;83:113. [Google Scholar]
- 51.Enserink JM, Kolodner RD. An overview of Cdk1-controlled targets and processes. Cell Div. 2010;5:11. doi: 10.1186/1747-1028-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benanti JA. Create, activate, destroy, repeat: Cdk1 controls proliferation by limiting transcription factor activity. Curr Genet. 2016;62:271–276. doi: 10.1007/s00294-015-0535-5. [DOI] [PubMed] [Google Scholar]
- 53.Lin Z, Guo H, Cao Y, Zohrabian S, Zhou P, Ma Q, Vandusen N, Guo Y, Zhang J, Stevens SM. Acetylation of VGLL4 regulates Hippo-YAP signaling and postnatal cardiac growth. Dev Cell. 2016;39:466. doi: 10.1016/j.devcel.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 55.Saucedo LJ, Edgar BA. Filling out the Hippo pathway. Nat Rev Mol Cell Biol. 2007;8:613–621. doi: 10.1038/nrm2221. [DOI] [PubMed] [Google Scholar]
- 56.Zhao B, Tumaneng K, Guan KL. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat Cell Biol. 2011;13:877–883. doi: 10.1038/ncb2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Q, Li S, Mana-Capelli S, Roth Flach RJ, Danai LV, Amcheslavsky A, Nie Y, Kaneko S, Yao X, Chen X, Cotton JL, Mao J, McCollum D, Jiang J, Czech MP, Xu L, Ip YT. The conserved misshapen-warts-Yorkie pathway acts in enteroblasts to regulate intestinal stem cells in drosophila. Dev Cell. 2014;31:291–304. doi: 10.1016/j.devcel.2014.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meng Z, Moroishi T, Mottier-Pavie V, Plouffe SW, Hansen CG, Hong AW, Park HW, Mo JS, Lu W, Lu S, Flores F, Yu FX, Halder G, Guan KL. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the hippo pathway. Nat Commun. 2015;6:8357. doi: 10.1038/ncomms9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang J, Wu S, Barrera J, Matthews K, Pan D. The hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the drosophila homolog of YAP. Cell. 2005;122:421–434. doi: 10.1016/j.cell.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 60.Thompson BJ, Cohen SM. The hippo pathway regulates the bantam microRNA to control cell proliferation and apoptosis in drosophila. Cell. 2006;126:767–774. doi: 10.1016/j.cell.2006.07.013. [DOI] [PubMed] [Google Scholar]
- 61.Nolo R, Morrison CM, Tao C, Zhang X, Halder G. The bantam microRNA is a target of the hippo tumor-suppressor pathway. Curr Biol. 2006;16:1895. doi: 10.1016/j.cub.2006.08.057. [DOI] [PubMed] [Google Scholar]
- 62.Wang K, Degerny CM, Yang X. YAP, TAZ, and Yorkie: a conserved family of signal-responsive transcriptional coregulators in animal development and human disease. Biochem Cell Biol. 2009;87:77–91. doi: 10.1139/O08-114. [DOI] [PubMed] [Google Scholar]
- 63.Ota M, Sasaki H. Mammalian tead proteins regulate cell proliferation and contact inhibition as transcriptional mediators of hippo signaling. Development. 2008;135:4059. doi: 10.1242/dev.027151. [DOI] [PubMed] [Google Scholar]
- 64.Hong W, Guan KL. The YAP and TAZ transcription coactivators: key downstream effectors of the mammalian Hippo pathway. Seminars in Cell & Developmental Biology. 2012;23:785. doi: 10.1016/j.semcdb.2012.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, Lai ZC, Guan KL. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zender L, Spector MS, Xue W, Flemming P, Cordoncardo C, Silke J, Fan ST, Luk JM, Wigler M, Hannon GJ. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Steinhardt AA, Gayyed MF, Klein AP, Dong J, Maitra A, Pan D, Montgomery EA, Anders RA. Expression of yes-associated protein in common solid tumors. Hum Pathol. 2008;39:1582–1589. doi: 10.1016/j.humpath.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kango-Singh M, Singh A. Regulation of organ size: insights from the drosophila hippo signaling pathway. Dev Dyn. 2009;238:1627–1637. doi: 10.1002/dvdy.21996. [DOI] [PubMed] [Google Scholar]
- 69.Badouel C, Garg A, Mcneill H. Herding hippos: regulating growth in flies and man. Curr Opin Cell Biol. 2009;21:837. doi: 10.1016/j.ceb.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 70.Liu M, Zhao S, Lin Q, Wang XP. YAP regulates the expression of Hoxa1 and Hoxc13 in mouse and human oral and skin epithelial tissues. Mol Cell Biol. 2015;35:1449–1461. doi: 10.1128/MCB.00765-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vassilev A, Kaneko KJ, Shu H, Zhao Y, Depamphilis ML. TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/yes-associated protein localized in the cytoplasm. Genes Dev. 2001;15:1229–1241. doi: 10.1101/gad.888601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koontz Laura M, Yi LC, Feng Y, Zheng Y, Yu J, Bo H, Qian C, Wu S, Pan D. The hippo effector yorkie controls normal tissue growth by antagonizing scalloped-mediated default repression. Dev Cell. 2013;25:388. doi: 10.1016/j.devcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang Y, Shen H, Withers HG, Yang N, Denson KE, Mussell AL, Truskinovsky A, Fan Q, Gelman IH, Frangou C, Zhang J. VGLL4 selectively represses YAP-Dependent gene induction and tumorigenic phenotypes in breast cancer. Sci Rep. 2017;7:6190. doi: 10.1038/s41598-017-06227-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nusse R, Varmus HE. Wnt genes. Cell. 1992;69:1073–1087. doi: 10.1016/0092-8674(92)90630-u. [DOI] [PubMed] [Google Scholar]
- 75.Staal FJ, Clevers H. Tcf/Lef transcription factors during T-cell development: unique and overlapping functions. Hematol J. 2000;1:3–6. doi: 10.1038/sj.thj.6200001. [DOI] [PubMed] [Google Scholar]
- 76.Kramps T, Peter O, Brunner E, Nellen D, Froesch B, Chatterjee S, Murone M, Züllig S, Basler K. Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin-TCF complex. Cell. 2002;109:47–60. doi: 10.1016/s0092-8674(02)00679-7. [DOI] [PubMed] [Google Scholar]
- 77.MacDonald BT, Tamai K, He X. Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev Cell. 2009;17:9. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li C, Miao H, Zhang L, Li L, Hao Q, Shi J, Zhou Z. VGLL4 targets a TCF4-TEAD4 complex to coregulate Wnt and Hippo signalling in colorectal cancer. Nat Commun. 2017;8:14058. doi: 10.1038/ncomms14058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 80.Wu Y, Zhou BP. New insights of epithelialmesenchymal transition in cancer metastasis. Acta Biochim Biophys Sin (Shanghai) 2008;40:643–650. doi: 10.1111/j.1745-7270.2008.00443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen T, You Y, Jiang H, Wang ZZ. Epithelialmesenchymal transition (EMT): a biological process in the development, stem cell differentiation and tumorigenesis. J Cell Physiol. 2017;232:3261–3272. doi: 10.1002/jcp.25797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cuevas EP, Eraso P, Mazón MJ, Santos V, Morenobueno G, Cano A, Portillo F. LOXL2 drives epithelial-mesenchymal transition via activation of IRE1-XBP1 signalling pathway. Sci Rep. 2017;7:44988. doi: 10.1038/srep44988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sánchez-Tilló E, de Barrios O, Siles L, Cuatrecasas M, Castells A, Postigo A. β-catenin/TCF4 complex induces the epithelial-to-mesenchymal transition (EMT)-activator ZEB1 to regulate tumor invasiveness. Proc Natl Acad Sci U S A. 2011;108:19204–9. doi: 10.1073/pnas.1108977108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jamora C, Dasgupta R, Kocieniewski P, Fuchs E. Links between signal transduction, transcription and adhesion in epithelial bud development. Nature. 2003;422:317. doi: 10.1038/nature01458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gyrdhansen M, Meier P. IAPs: from caspase inhibitors to modulators of NF-κB, inflammation and cancer. Nat Rev Cancer. 2010;10:561. doi: 10.1038/nrc2889. [DOI] [PubMed] [Google Scholar]
- 86.Jin HS, Park HS, Shin JH, Kim DH, Jun SH, Lee CJ, Lee TH. A novel inhibitor of apoptosis protein (IAP)-interacting protein, vestigial-like (Vgl)-4, counteracts apoptosis-inhibitory function of IAPs by nuclear sequestration. Biochem Biophys Res Commun. 2011;412:454–459. doi: 10.1016/j.bbrc.2011.07.117. [DOI] [PubMed] [Google Scholar]
- 87.Merrett N. Sleeping beauty mutagenesis reveals cooperating mutations and pathways in pancreatic adenocarcinoma. Proc Natl Acad Sci U S A. 2012;109:5934–41. doi: 10.1073/pnas.1202490109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jiang W, Yao F, He J, Lv B, Fang W, Zhu W, He G, Chen J, He J. Downregulation of VGLL4 in the progression of esophageal squamous cell carcinoma. Tumour Biol. 2015;36:1289–1297. doi: 10.1007/s13277-014-2701-7. [DOI] [PubMed] [Google Scholar]
- 89.Zhao B, Lei QY, Guan KL. The Hippo-YAP pathway: new connections between regulation of organ size and cancer. Curr Opin Cell Biol. 2008;20:638–646. doi: 10.1016/j.ceb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Deng YZ, Chen PP, Wang Y, Yin D, Koeffler HP, Li B, Tong XJ, Xie D. Connective tissue growth factor is overexpressed in esophageal squamous cell carcinoma and promotes tumorigenicity through beta-catenin-T-cell factor/Lef signaling. J Biol Chem. 2007;282:36571–36581. doi: 10.1074/jbc.M704141200. [DOI] [PubMed] [Google Scholar]
- 91.Lyu P, Zhang SD, Yuen HF, McCrudden CM, Wen Q, Chan KW, Kwok HF. Identification of TWIST-interacting genes in prostate cancer. Sci China Life Sci. 2017;60:386–396. doi: 10.1007/s11427-016-0262-6. [DOI] [PubMed] [Google Scholar]
- 92.Tajonar A, Maehr R, Hu G, Sneddon JB, Rivera-Feliciano J, Cohen DE, Elledge SJ, Melton DA. Brief report: VGLL4 is a novel regulator of survival in human embryonic stem cells. Stem Cells. 2013;31:2833–2841. doi: 10.1002/stem.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Teng AC, Kuraitis D, Deeke SA, Ahmadi A, Dugan SG, Cheng BL, Crowson MG, Burgon PG, Suuronen EJ, Chen HH. IRF2BP2 is a skeletal and cardiac muscle-enriched ischemia-inducible activator of VEGFA expression. FASEB J. 2010;24:4825–4834. doi: 10.1096/fj.10-167049. [DOI] [PubMed] [Google Scholar]
- 94.Clarke TK, Crist RC, Doyle GA, Weiss AR, Brandt H, Crawford S, Crow S, Fichter MM, Halmi KA, Johnson C, Kaplan AS, La Via M, Mitchell JE, Strober M, Rotondo A, Treasure J, Woodside DB, Keel P, Klump KL, Lilenfeld L, Plotnicov K, Magistretti PJ, Bergen AW, Kaye WH, Schork NJ, Berrettini WH. Characterization of genetic variation in the VGLL4 gene in anorexia nervosa. Psychiatr Genet. 2014;24:183–184. doi: 10.1097/YPG.0000000000000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kim SE, Kim HN, Yun YJ, Heo SG, Cho J, Kwon MJ, Chang Y, Ryu S, Shin H, Shin C, Cho NH, Sung YA, Kim HL. Meta-analysis of genomewide SNP- and pathway-based associations for facets of neuroticism. J Hum Genet. 2017;62:903–909. doi: 10.1038/jhg.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Edwards AC, Aliev F, Bierut LJ, Bucholz KK, Edenberg H, Hesselbrock V, Kramer J, Kuperman S, Nurnberger JI Jr, Schuckit MA. Genome-wide association study of comorbid depressive syndrome and alcohol dependence. Psychiatr Genet. 2012;22:31. doi: 10.1097/YPG.0b013e32834acd07. [DOI] [PMC free article] [PubMed] [Google Scholar]


