Abstract
During cardiac development, DNA binding transcription factors and epigenetic modifiers regulate gene expression in cardiac progenitor cells (CPCs). We have previously shown that YY1 is essential for the commitment of mesodermal precursors into CPCs. However, the role of YY1 in the maintenance of CPC phenotype and their differentiation into cardiomyocytes is unknown. In this study, we found, by genome-wide transcriptional profiling and phenotypic assays, that YY1 overexpression prevents cardiomyogenic differentiation and maintains the proliferative capacity of CPCs. We show further that the ability of YY1 to regulate CPC phenotype is associate with its ability to modulate histone modifications specifically at a developmentally critical enhancer of Nkx2-5 and other key cardiac transcription factor such as Tbx5. Specifically, YY1 overexpression helps to maintain markers of gene activation such as the acetylation of histone H3 at lysine 9 (H3K9Ac) and lysine 27 (H3K27Ac) as well as tri-methylation at lysine 4 (H3K4Me3) at the Nkx2-5 cardiac enhancer. Furthermore, transcription factors associated proteins such as PoIII, p300, and Brg1 are also enriched at the Nkx2-5 enhancer with YY1 overexpression. The biological activities of YY1 in CPCs appear to be cell autonomous, based co-culture assays in differentiating embryonic stem cells. Altogether, these results demonstrate that YY1 overexpression is sufficient to maintain a CPC phenotype through its ability to sustain the presence of activating epigenetic/chromatin marks at key cardiac enhancers.
Keywords: cardiac progenitors, cardiogenesis, YY1, Nkx2-5, mesoderm, cardiomyocytes
Introduction
Understanding the mechanisms of cardiac lineage commitment is important not only for improving our knowledge of cardiogenesis per se, but also for identifying the pathophysiological basis of congenital heart disease, the most common form of human birth defects. In addition, a detailed understanding of cardiac lineage commitment from multipotent precursor cells may also be important for successful cell-based therapies in regenerative medicine 1–4. Recently, we and others have described the isolation and characterization of embryo- and embryonic stem cell (ESC)-derived cardiac progenitor cells (CPCs)5–9. These cells give rise to a variety of different cells types in the developing heart including cardiomyocytes, smooth muscle, endothelial cells, and cells of the conduction system. Through bioinformatic analysis of transcription factor binding sites within developmentally-important cardiac enhancers and confirmation by chromatin immunoprecipitation (ChIP) and genome-wide sequencing, we found that Ying Yang 1 (YY1) plays a critical role in the commitment of precardiac mesoderm into CPCs 10.
YY1, a DNA binding zinc finger transcription factor, is a member of the GLI-Kruppel family of nuclear proteins 11, 12. Binding sites for YY1 have been identified within the regulatory regions of several cardiac and skeletal muscles genes, including muscle creatine kinase, α-skeletal actin, α-myosin heavy chain, myosin light chain 2, and α-cardiac actin 13–18. Despite the well-described role of YY1 as a transcriptional repressor in differentiated cardiomyocytes, YY1 appears to activate the expression of CPC-associated transcription factors such as Nkx2-5, Isl-1, and Tbx5 10. YY1, in cooperation with GATA-4, has been shown to activate the B-type natriuretic peptide promoter in vitro 19 and induce periostin enhancer expression in the cardiac outflow tract 20. Furthermore, YY1 binding to a xenopus myosin light chain (xMLC) cardiac enhancer was necessary to induce full expression in transgenic embryos 21.
As an initiator protein that recruits other transcription factors and cofactors, YY1 can either activate or inhibit transcription in a context-dependent manner 22. In support of this, YY1 has been described to anchor polycomb group (PcG) proteins such as enhancer of zeste-2 (EZH2), embryonic ectoderm development (EED) and Suz12 to chromatin DNA and may substitute for polycomb protein pleiohomeotic during Drosophila development 23. In addition, YY1 anchoring to DNA is required for the Gata4-dependent transactivation of the Nkx2-5 gene 10. Thus far, no study has addressed the role of YY1 during CPC differentiation/maturation into cardiomyocytes. Recently, two global genomic analysis identified histone modifications across the genome during defined stages of cardiac differentiation leading to a better understanding of developmentally regulated chromatin transitions during lineage commitment 24, 25. While these studies elaborate the first epigenome of the differentiation of ESCs into cardiomyocytes, specific factors that promote the placement of these histone marks at cardiac enhancers was not specifically addressed.
In this study, we found that YY1 overexpression in ES cell-derived CPCs results in the maintenance of CPC phenotype as assessed by genome-wide transcriptional profiling and functional validation. We show that YY1 sustains the expression of CPC-associated genes by its ability to modulate chromatin activation marks at cardiac enhancers for Nkx2-5 and Tbx5. Specifically, YY1 coordinates the acetylation and methylation status of histone H3. In addition, YY1 recruits the transcription factors associated proteins p300 and Brg1 to cardiac genes. These results demonstrate a critical role of YY1 to regulate chromatin marks at a key developmental enhancers of Nkx2-5 and other cardiac genes.
Results
YY1 regulates cardiac Nkx2-5 enhancer activity in vitro
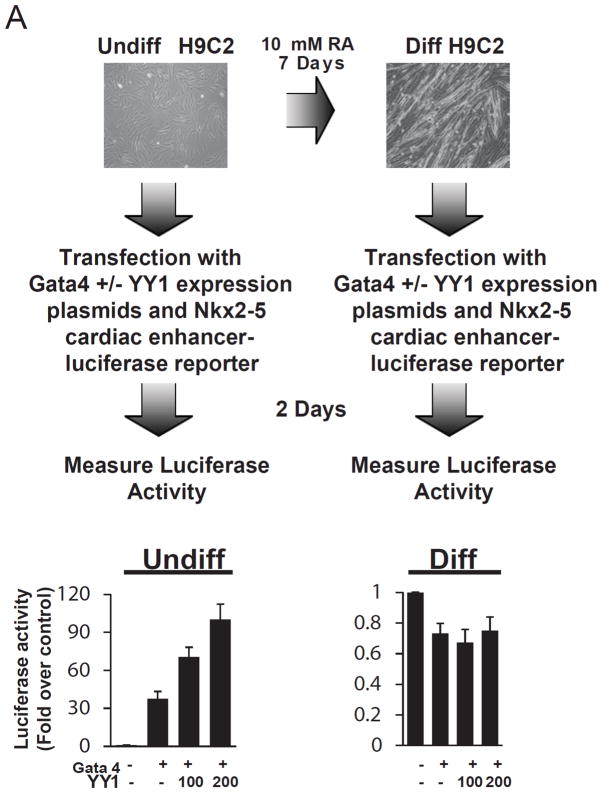
While YY1 is known to regulate the transcription of developmentally critical genes in a context dependent fashion, its ability to regulate the Nkx2-5 cardiac enhancer expression in different contexts has not been explored. To examine this, we utilized our previously described Nkx2-5-luciferase reporter and H9C2 cardiomyoblast cell line that was in vitro differentiated with 10 mM of retinoic acid for 7 days (Figure 1). As shown in Figure 1, YY1 and Gata4 collectively transactivate the Nkx2-5 cardiac enhancer in undifferentiated but not differentiated H9C2 cells (Figure 1). This suggests that YY1’s ability to promote of cardiac gene expression may be restricted to the a specific stage of development. To further investigate this in an in vivo context, we generated conditional loss of YY1 alleles in the developing heart by interbreeding floxed YY1 mice with the Nkx2-5 knock-in Cre mice (Nkx2-5-Cre) (Figures S1A) 26. At embryonic day 12.5, we observed no lethality due to the presence of homozygous YY1 floxed and the Nkx2-5-Cre alleles (Table S1). Moreover, histological analysis revealed no gross developmental defects in these embryos (Figure S1B). This phenotype is distinct from embryos with homozygous loss of YY1 in Mesp1-Cre descendants where a complete failure of CPC formation was observed 10. To determine the mechanism for the lack of YY1 requirement in cardiomyocyte differentiation/maturation, we investigated the expression level of YY1 in cardiac lineage cells and found that YY1 expression declines dramatically during normal cardiomyocyte differentiation (Figures S2). These data suggest that YY1 is required for cardiac development during the commitment and possibly, maintenance of CPCs, but is dispensable for their maturation into cardiomyocytes.
Figure 1.
Regulation of the Nkx2-5 cardiac enhancer by YY1 in vitro. Expression plasmids for YY1 and Gata4 and an Nkx2-5 cardiac enhancer-luciferase reporter were transfected into H9C2 cardiomyoblasts that were undifferentiated or differentiated in the presence of 10 μM of retinoic acid (RA) for 7 days. Following 2 days of incubation, the luciferase activity in each cell population was quantitated and normalized against an internal control.
YY1 gain-of-function maintains cardiac precursors in a progenitor-like state
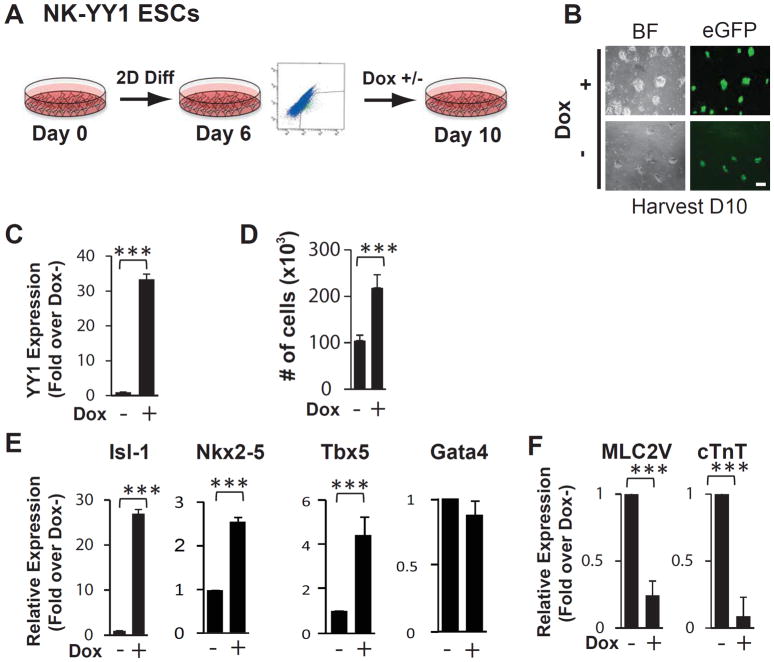
The lack of effect of YY1 deletion in differentiating cardiomyocytes in vivo prompted us to examine whether YY1 gain-of-function is associated with the maintenance of CPC phenotype. To address this, we employed our previously described doxycycline inducible YY1 overexpressing Nkx2-5 cardiac enhancer-eGFP ESCs line (NK-YY1) 10 and treated FACS-purified eGFP+ cells on day 6 of in vitro differentiation with or without doxycycline (Figure 2A, 2B). Four days after sorting and reseeding the eGFP+ cells, mRNA from each cell population was isolated and the expression of CPC and sarcomeric genes was quantitated by real-time PCR. We confirmed that doxycycline treatment led to the overexpression of YY1 (Figure 2C). We also found, interestingly, that YY1 overexpression increased the number of eGFP+ cells (Figures 2D) as well as the expression of a number of key CPC-associated transcription factors such as Isl-1, Nkx2-5, and Tbx5 (Figure 2E). Conversely, the expression of the cardiomyocyte markers MLC2v and cTnT was significantly decreased (Figure 2F). These data suggest that YY1 promotes proliferation of eGFP+ cells and their expression of cardiac transcription factors in association with a decrease in sarcomeric gene expression.
Figure 2.
YY1 overexpression inhibits cardiomyogenic differentiation of ES cell-derived CPCs. (A) Schematic diagram of in vitro cardiac differentiation of the NK-YY1 ESC line. NK-YY1 ESC line underwent monolayer differentiation in vitro for 6 days and eGFP+ cells were purified by FACS, replated, and cultured in the presence or absence of doxycycline to induce YY1 overexpression for an additional 4 days until day 10. (B) Bright field (BF) and fluorescence (GFP) microscopy analysis of FACS-purified eGFP+ cells after 4 days of YY1 overexpression. (C) Quantitative PCR analysis of YY1 expression in the presence or absence of doxycycline treatment for 4 days. (D) Quantification of cell number in each well after YY1 overexpression. (E) Quantitative PCR analysis of cardiac transcription factor expression after YY1 overexpression. (F) Quantitative PCR analysis of sarcomeric gene expression in YY1 overexpressed vs control cells. Three independent experiments were performed in triplicate. (***) P<0.001. Scale bar: 50μm.
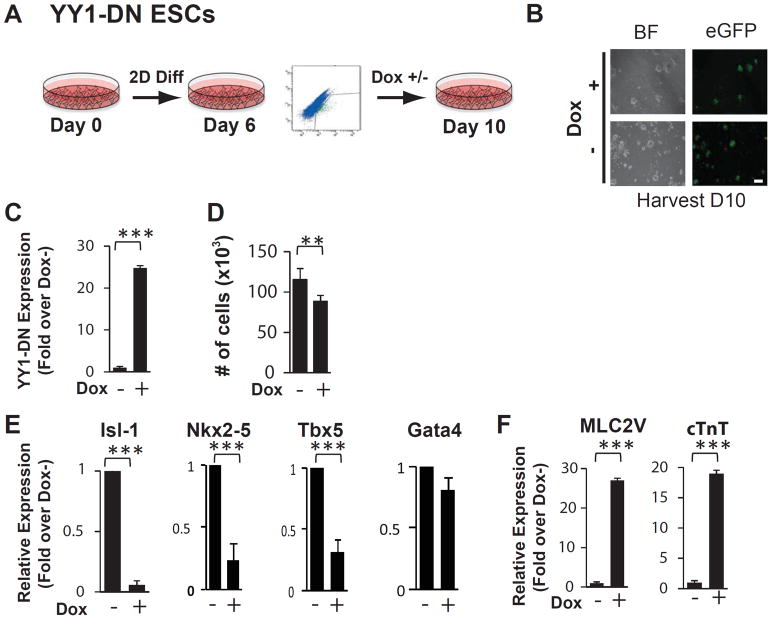
To determine whether these results are due specifically to YY1 overexpression, we employed a previously generated Nkx2-5-eGFP ES cell line that inducibly expresses the DNA binding domain of YY1 which acts in a dominant negative fashion (NK-YY1-DN) (Figure 3A, 3B) 10. We found that FACS-purified day 6 eGFP+ cells that have been treated with doxycycline for 4 days showed an increase in YY-DN expression (Figure 3C). We also found that the total number of cells decreased in the presence of YY1-DN overexpression (Figures 3D). Remarkably, the expression of CPC-associated transcription factors was significantly reduced by 3 to 10-fold (Figure 3E) while the expression of mature sarcomeric genes such as MLC2v and cTnT was significantly increased by 18 to 27-fold (Figure 3F). Together, these data strongly support the role of YY1 to maintain cardiac precursors in a cardiac progenitor cell state and prevent their maturation/differentiation into cardiomyocytes.
Figure 3.
Dominant negative YY1 expression promotes maturation of ESC-derived CPCs into differentiated cardiomyocytes. (A) Schematic diagram of in vitro cardiac differentiation of the NK-YY1-DN ESC line. NK-YY1-DN ESC line underwent monolayer differentiation in vitro for 6 days and eGFP+ cells were purified by FACS, replated, and cultured in the presence or absence of doxycycline to induce YY1-DN overexpression for 4 additional days until day 10. (B) Bright field (BF) and fluorescence (GFP) microscopy analysis of FACS-purified eGFP+ cells after 4 days of YY1-DN overexpression. (C) Quantitative PCR analysis of YY1-DN expression in the presence or absence of doxycycline. (D) Quantification of cell number in each well after YY1-DN overexpression. (E) Quantitative PCR analysis of cardiac transcription factor expression after YY1 overexpression. (F) Quantitative PCR analysis of sarcomeric gene expression in YY1-DN overexpressed vs control cells. Three independent experiments were performed in triplicate. (***) P<0.001, (**) P<0.05. Scale bar: 50μm.
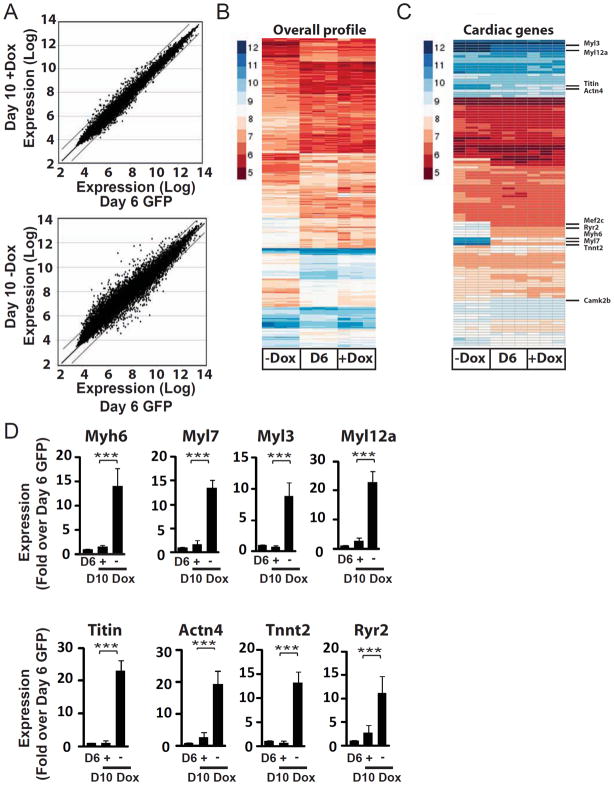
To further explore this hypothesis, we performed genome-wide transcriptional profiling to compare FACS-purified day 6 eGFP+ CPCs from in vitro differentiation of NK-YY1 ES cells with FACS-purified day 6 eGFP+ cells that have treated (or untreated) with doxycycline for 4 additional days. Based upon our prior experience, eGFP+ cells without YY1 overexpression differentiate predominantly into cardiomyocytes in four days. Interestingly, with YY1 overexpression, the transcriptional profile of YY1-treated cells at day 10 is highly similar to those from freshly isolated day 6 CPCs (Figure 4A-top panel). In contrast, the absence of YY1 overexpression resulted in a transcriptional profile that is distinct from day 6 CPCs (Figure 4A-bottom panel and Figure 4B) and exhibit significant increase in sarcomeric gene expression (e.g. Myl3, Myh6, Myl7, Myl12a, Titin, Actn4, and Tnnt2) (Figure 4C and Table S2). This increase in sarcomeric gene expression in cells without YY1 overexpression compared with cells overexpressing YY1 was confirmed by quantitative real time PCR analysis (Figure 4D). These data support the ability of YY1 to maintain a CPC-like transcriptional profile and prevent cardiomyogenic differentiation.
Figure 4.
YY1 overexpression maintains CPC gene expression. Microarray expression profiling of FACS-purified eGFP+ CPCs at day 6 of in vitro differentiation (Day 6 GFP) and purified eGFP+ cells that have been cultured for 4 additional days in the presence (Day 10 +Dox) or absence (Day 10 −Dox) of doxycycline. (A) Comparison of gene expression between Day 6 CPCs and Day 10 cells that have been treated with or without doxycycline. Genes showing greater than a 2-fold increase or decrease in expression are located outside the indicated lines. (B) Comparison of gene expression between Day 6 CPCs and Day 10 cells not treated with doxycycline. (C) Heat map showing the overall gene expression profile of Day 6 CPCs and CPCs that have been cultured for 4 days with or without doxycycline. (D) Quantitative PCR analysis of sarcomeric gene expression in differentiating CPCs with or without doxycycline treatment. P<0.001, (**) P<0.05.
YY1 regulates chromatin modifications at the Nkx2-5 cardiac enhancer
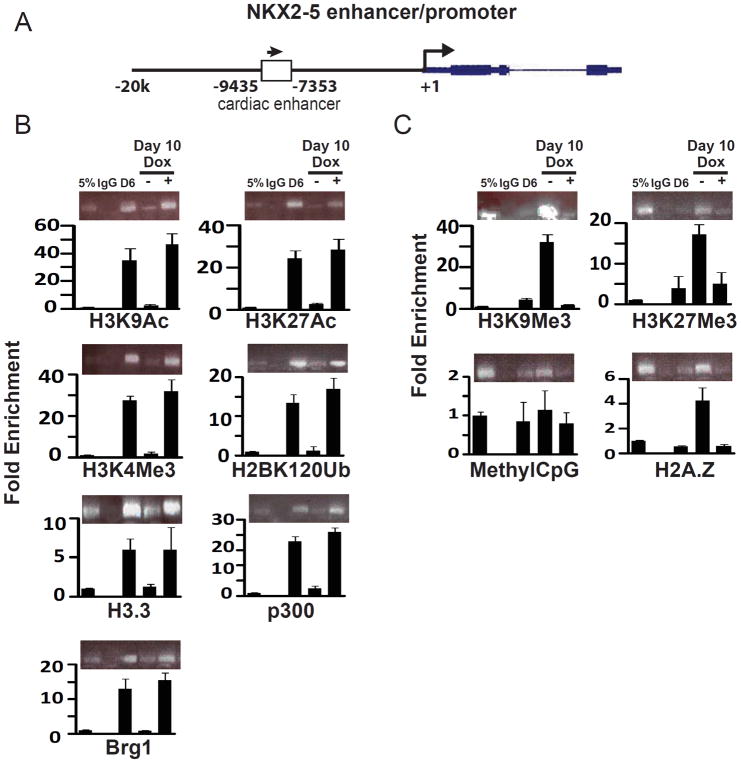
The ability of YY1 overexpression to maintain CPC phenotype in day 6 eGFP+ CPCs suggests its ability to alter epigenetic modifications at the Nkx2-5 enhancer. To investigate this, we isolated FACS-purified day 6 eGFP+ cells from NK-YY1 ESCs and performed chromatin immunoprecipitation (ChIP) using multiple chromatin modification-related antibodies on these cells, as well as on cells that were further cultured with or without YY1 overexpression for four additional days. We found, reassuringly, that chromatin modifications associated with an active or poised transcriptional state, including H3K9Ac, H3K27Ac, H3K4Me3, and H2BK120Ub, are all up-regulated in the presence of YY1 overexpression at or near the Nkx2-5 cardiac enhancer (Figure 5B) but not in regions away from the enhancer (Figure S3). Consistent with this finding, proteins such as H3.3, p300, and Brg1 that are typically found at transcriptionally active sites are also increased in the presence of YY1 overexpression. On the other hand, when YY1 is overexpressed, chromatin modifications that are associated with transcriptional repression are decreased at the Nkx2-5 cardiac enhancer (Figure 5C), suggesting that YY1 maintains Nkx2-5 enhancer expression by sustaining the presence of active chromatin marks.
Figure 5.
Determination of the effect of YY1 overexpression on chromatin modifications at the Nkx2-5 cardiac enhancer. (A) Genomic diagram of the NKX2-5 locus and the location of the primer pairs used for chromatin immunoprecipitation (ChIP). (B) ChIP assays for chromatin marks and transcription factors associated with transcriptional activation. (e.g. H3K9Ac, K3K27Ac, K3K4Me3, H3BK120Ub, H3.3, p300, and Brg1). Day 6 FACS-purified eGFP+ cells and purified eGFP+ cells that have been treated for 4 additional days with or without YY1 overexpression were subjected to ChIP using antibodies binding to the indicated chromatin mark. The pulled down DNA was analyzed for the presence of the Nkx2-5 cardiac enhancer (arrow). (C) ChIP experiments were performed as described in (B) using antibodies against the indicated chromatin marks (e.g. H3K9Me3, K3K27Me3, H2A.Z, and methylCpG). The presence of these chromatin marks at the Nkx2-5 cardiac enhancer was assessed by PCR analysis.
YY1 regulates chromatin modifications at enhancer regions of other cardiac genes
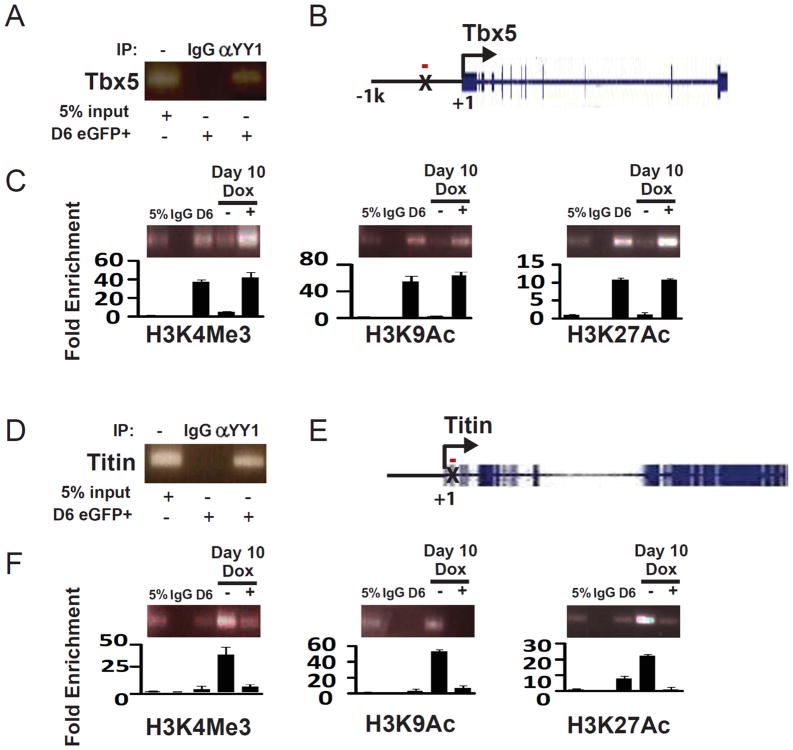
We have previously identified multiple sites of YY1 binding in enhancer regions of cardiac transcription factors and sarcomeric proteins using genome-wide ChIP-sequencing 10. Among those candidates, we found YY1 binding sites at the TBX5 and TITIN loci but the expression of these two genes appears to be differentially regulated when YY1 is overexpressed (Figure 2E and 4C). To determine whether YY1 overexpression can regulate chromatin modification at these YY1 binding sites, we assessed for the presence of active chromatin marks (e.g. H3K4Me3, H3K9Ac, and H3K27Ac) at these gene loci during CPC differentiation (Figure 6 and S4). We found, interestingly, that YY1 overexpression led to the maintenance of active chromatin marks at the YY1 binding site of the TBX5 locus while suppressing the presence of these marks at the TITIN locus (Figure 6C and 6F), consistent with the effect of YY1 on Tbx5 and Titin gene expression. Furthermore, the effect of YY1 overexpression on chromatin marks was specific to YY1 binding sites at these gene loci as the level of chromatin modification was unaffected in regions away from YY1 binding sites (Figure S4C, S4F).
Figure 6.
Regulation of chromatin marks at the YY1 binding sites in the TBX5 and TITIN loci. (A) Confirmation of YY1 binding to the TBX5 locus in FACS-purified day 6 eGFP+ cells by anti-YY1 ChIP-PCR. (B) Diagram of the TBX5 locus. The YY1 binding site is indicated by X. The red bar represents the position of the amplicon for ChIP experiments. (C) Cells were in vitro differentiated and FACS-purified as described in Figure 2A. ChIP experiments were performed using antibodies against H3K9Ac, K3K27Ac, and H3K4Me3 and IgG as a negative control. The difference in chromatin marks at the YY1 binding site with or without YY1 overexpression was quantitated by real-time PCR. (D) Confirmation of YY1 binding to the TITIN locus in FACS-purified day 6 eGFP+ cells by anti-YY1 ChIP. (E) Diagram of the TITIN locus. The YY1 binding site is indicated by X. The red bar represents the position of the amplicon for ChIP experiments. (F) Cells were in vitro differentiated and FACS-purified as described in Figure 2A. ChIP experiments were performed using antibodies against H3K9Ac, K3K27Ac, and K3K4Me3 and IgG as a negative control. The difference in chromatin marks at the YY1 binding site with or without YY1 overexpression was quantitated by real-time PCR.
YY1 functions in a cell autonomous fashion
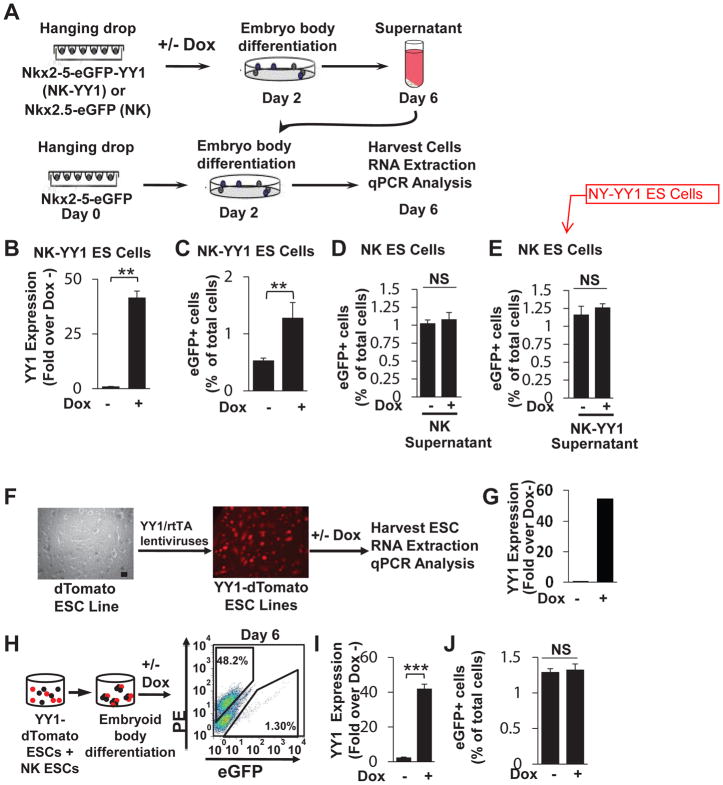
To further examine the mechanism involved in YY1 expansion of the CPC pool, we explored whether a non-cell autonomous process may be involved whereby YY1 either induces the secretion of soluble factors in an autocrine or paracrine fashion or mediates direct cell-to-cell interaction to promote cardiogenic commitment. We employed the ESC line that expresses eGFP under the regulatory sequence of the 2.1 kb Nkx2-5 cardiac-specific enhancer (NK). To determine whether a soluble factor is involved we treated differentiating NK ES cells with the supernatant from in vitro differentiated, doxycycline treated NK-YY1 ES cells or control NK ES cells (Figure 7A). We first confirmed the doxycycline-dependent increase in YY1 expression in differentiating NK-YY1 ES cells (Figure 7B) and the corresponding increase in cardiac differentiation (as assessed by the percentage of eGFP+ cells) (Figure 7C). We then added this and control NK ES cell supernatant to differentiating NK ES cells and found that the supernatants with YY1 overexpression had no effect on cardiac differentiation when compared with control supernatant (Figure 7D–E). These results suggest that YY1 promotion of CPC phenotype does not involve the secretion of a soluble factor.
Figure 7.
YY1 functions in a cell-autonomous manner. (A) A schematic diagram of the experimental design. Both NK and NK-YY1 ESCs were in vitro differentiated in the presence or absence of doxycycline treatment. At day 6, the cultured supernatant from each cell line/treatment condition was collected and added to a parallel differentiated NK ESC line at day 2 of in vitro differentiation. Supernatant-treated NK ESCs were then harvested at day 6 and the percentage of eGFP+ cells was quantified by flow cytometric analysis. (B) Confirmation of YY1 overexpression in differentiated NK-YY1 ESCs following doxycycline treatment. (C) Quantification of the YY1-induced increase in eGFP+ cells by flow cytometric analysis. (D) Quantification of the percentage of eGFP+ cells in NK ESC supernatant with or without doxycycline (+/− Dox)-treated NK ESCs at day 6 of differentiation. (E) Same as in D except that supernatants from NK-YY1 ESC (+/− Dox) have been added. (F) Generation of doxycycline-inducible YY1 overexpressing dTomato+ ESC lines (YY1-dTomato) by infection of dTomato+ ESCs with doxycycline-inducible YY1 overexpressing lentivirus. (G) Quantitative PCR analysis of YY1 overexpression in undifferentiated YY1-dTomato ESCs in the presence/absence of doxycycline. (H) Co-differentiation of YY1-dTomato and NK ESCs (1:1 ratio) in an embryoid body mixing experiment. Flow cytometric analysis was performed on day 6 of in vitro differentiation. Note that dTomato+ cells represent approximately half (48.2%) of the total cells, confirming the lack of a proliferation bias with YY1 overexpression. (I) Quantitative PCR analysis of YY1 overexpression with or without doxycycline treatment. (J) Flow cytometric analysis of the percentage of eGFP+ cells in the presence/absence of doxycycline. (***) P 0.001, (**) P 0.05. Scale bar: 50μm.
To examine whether YY1 overexpression enhances cell-to-cell interactions that indirectly promote the maintenance of CPC phenotype, we employed a “mixing” experiment that involves co-differentiation of dTomato-labeled doxycycline-inducible YY1 overexpressing ES cells (dTomato-YY1) with NK ES cells (Figure 7F–J). We hypothesized that if YY1 increases cell-to-cell interactions that indirectly promotes the maintenance of the CPC phenotype, we should detect an increase in the fraction of eGFP+ cells in differentiating NK ES cells. To generate the dTomato-YY1 ES cell line, we infected a previously derived dTomato expressing ES cells line with a doxycycline-inducible YY1 overexpressing lentivirus to generate multiple clonally-derived dTomato-YY1 ES cell lines (Figure 7F). The doxycycline-dependent overexpression of YY1 was confirmed in undifferentiated dTomato-YY1 ES cells and one such line with ~55-fold increase in YY1 overexpression was further studied (Figure 7G). Undifferentiated dTomato-YY1 ES cells from this line were added to NK ES cells in a 1:1 ratio and differentiated in vitro as embryoid bodies (Figure 7H). With doxycycline induction, the differentiated embryoid bodies showed a 40-fold increase in YY1 expression by qPCR analysis (Figure 7I). Despite the increase in YY1 expression, the percentage of dTomato−/−eGFP+ cells derived from NK ES cells was not increased (Figure 7J), supporting the lack of involvement of YY1-induced cell-to-cell interactions to promote the maintenance of the CPC phenotype. Together, these data support a cell autonomous role for YY1 to regulate chromatin modification and CPC genes expression.
Discussion
Despite decades of investigation in cardiogenesis, our understanding of the molecular regulation of early CPC renewal and differentiation remains incomplete. To gain additional insights into this biological process, we postulated that YY1 may play a critical role in the maintenance of CPC phenotype during cardiogenesis. To explore this, we evaluated the consequence of YY1 overexpression on cardiac gene expression and found that YY1 promotes the proliferation and the expression of cardiac transcription factors at the expense of sarcomeric gene expression. This finding was corroborated by genome-wide transcriptional profiling showing that YY1 overexpression maintains the expression of CPC-specific genes. We further investigated the modulation of chromatin marks by YY1 and uncovered the ability of YY1 to increase transcriptional activation marks at the Nkx2-5 cardiac enhancer as well as enhancer of other cardiac genes. Finally, we show that YY1 regulates the induction and maintenance of the ES cell-derived CPC population in a cell-autonomous manner. Taken together, these results identify YY1 as the first gene that is sufficient to maintain CPC phenotype via modification of chromatin marks associated with the expression Nkx2-5 and other critical cardiac transcription factors.
YY1 plays a role in the maintenance of CPC phenotype
Previously, we have identified an essential requirement for YY1 to regulate CPC commitment in vivo, as evidenced by the absence of CPC formation in Mesp1-Cre;YY1 Flox/Flox embryos at embryonic day 7.5 10. In the current study, we investigated the role of YY1 during CPC differentiation and cardiomyocyte maturation and found an absence of an immediate role for YY1 during this process (Figure S1). We show that the lack of requirement for YY1 coincided with a dramatic down-regulation of YY1 expression during cardiomyocyte maturation, a finding that points to a potentially inhibitory role for YY1 in cardiac differentiation (Figure S2). Indeed, earlier studies in adult cardiomyocytes have supported a repressive role of YY1 in sarcomeric gene expression 27, 28. Consistent with this, we found that overexpression of YY1 in differentiating CPCs leads to downregulation of a number of sarcomeric genes such as α- and β-myosin chain, cardiac troponin T, and titin (Figure 4D). These and other studies (Figures 2 and 3) demonstrate that YY1 expression is sufficient for the maintenance of a CPC phenotype. Of note, our previous study10 showed that YY1 overexpression from day 0 to 10 of ESC differentiation also led to an increase in CPCs but the increase in cardiovascular differentiation into cardiomyocytes, smooth muscle cells, and endothelial cells requires the cessation of YY1 overexpression between day 6 to 10 of differentiation. Hence, the role of YY1 during cardiac development appears to be biphasic – expression of YY1 in precardiac mesoderm facilitates the induction of CPC phenotype while expression in differentiating CPCs results in inhibition of cardiovascular cell lineage differentiation. This observation is reminiscent of the role of the WNT/β-catenin pathway in different stages of cardiogenesis 29–32. Further studies will be needed to elucidate the direct and indirect targets involved in the maintenance of CPC phenotype by YY1.
YY1 modulates the presence of chromatin marks at key enhancers of cardiac genes
Our finding that YY1 overexpression can maintain a CPC transcriptional profile at a genome-wide level suggests the ability of YY1 to regulate chromatin modifications at key enhancers of cardiac genes. Specifically, we found that overexpression of YY1 increased histone marks such as H3K9Ac, H2K27Ac, H3K4Me3, H2BK120Ub that are associated with transcriptional activation (Figure 5B) 33. Conversely, YY1 overexpression reduced chromatin marks such as H3K9Me3 and H3K27Me3 associated with transcriptional repression (Figure 5C). These data support the role of YY1 to maintain the expression of CPC genes such as Nkx2-5 and Tbx5 by maintaining the presence of activating chromatin marks at cardiac enhancers in these gene loci. While YY1 served to activate the expression of CPC genes, we also found it can inhibit the expression of sarcomeric genes that are associated with mature cardiomyocytes. In particular, we found that YY1 decreased histone activation marks such as H3K4Me3, H3K9Ac, and H3K27Ac near the transcription start site of Titin. These findings are supportive of a context-dependent role for YY1 to regulate cardiac gene expression. It should noted that while activating chromatin marks at the Nkx2.5 enhancer appear to down-regulate with cardiomyocyte differentiation (Figure 5B) the expression of endogenous Nkx2.5 persists. This is most likely due to transcriptional activities from other enhancers of Nkx2.5 (e.g. at the −3 to −4 kb upstream of transcriptional start site).
The ability of YY1 to regulate the presence of activating chromatin marks at the Nkx2-5 cardiac enhancer suggests that YY1 may serve as a pioneer factor. The concept of a pioneer factor, initially introduced by Cirrulo et al. 34 and reviewed recently by Smale et al. 35 relates to the hypothesis that enhancers for tissue-specific genes are bound by transcription factors long before the gene is transcribed 36–38. These pioneer factors are located preferentially on chromosomes that possess unmethylated CpG dinucleotides, histone modifications, and/or remodeling enzymes 35. YY1 may play the role of a pioneer factor in the cardiac enhancer of Nkx2-5 leading to the recruitment of other transcription factors and chromatin modifiers to this site. In support of this hypothesis, YY1 has been shown to interact with histone modifier enzymes as a part of the PRC2 polycomb complex 17, 39–43. In addition, our ChIP assays demonstrated that YY1 overexpression recruits Brg1 and p300 to the NKX2-5 cardiac enhancer (Figure 5). Taken together, these data support a unique role for YY1 to regulate the chromatin environment for cardiac genes.
Conclusion
In summary, we identified a mechanism of CPC regulation by YY1, namely the ability of YY1 to modulate chromatin modifications in the vicinity of the Nkx2-5 enhancer and other cardiac markers. Specifically, YY1 coordinates the acetylation and methylation status of histone H3. In addition, YY1 recruits p300 and Brg1 to the Nkx2-5 cardiac enhancer. Genome-wide transcriptional profiling analysis demonstrates that YY1 overexpression maintains CPCs in an immature state. These results define a mechanistic role for YY1 to modulate cardiogenesis as a chromatin regulator of Nkx2-5 and other cardiac genes.
Materials and Methods
ESC and H9C2 cell culture
YY1 WT and YY1 DN inducible ES cell lines was generated previously 44. ES cell lines were cultured on a monolayer of mitomycin-inactivated mouse embryonic fibroblasts and maintained in CJ7 media as previously described 5. H9C2 cells were obtained from ATCC and maintained in Dulbeccos’s Modified Eagle’s Medium supplemented with 10% FBS and 5000 i.u./mL penicillin/streptomycin according to vendor supplied protocol.
ESC in vitro differentiation
All in vitro differentiation assays were performed according to Huang et al. 45.
Statistical Analysis
Statistical analyses for all comparative studies were performed using the 2-tailed Student’s t-test. For microarray data, statistical analyses are described within that section.
Detailed Materials and Methods can be found in Supplementary Information.
Supplementary Material
Acknowledgments
We thank Laura Prickett-Rice for her assistance with the flow cytometry and John Hutchinson and Oliver Hofmann for their assistance with microarrays analysis. Financial support was provided by a Postdoctoral Fellowship Award American Heart Association postdoctoral fellowship (S.G, A.S.) and by NIH Director’s New Innovator Award, the NHLBI, and the Harvard Stem Cell Institute (S.M.W.)
Non-standard Abbreviations and Acronyms
- ChIP-seq
chromatin immunoprecipitation sequencing
- CPCs
cardiac progenitor cells
- TF
transcription factor
- TFBS
transcription factor binding site
- TSS
transcription start site
- YY1
Yin Yang 1
Footnotes
Author Contributions
SG and SW designed the experiments; SG performed the experiments; SG and SW performed data analysis; SG, GL, AS, RS and SW prepared the manuscript.
Disclosures
None.
References
- 1.Braam SR, Passier R, Mummery CL. Cardiomyocytes from human pluripotent stem cells in regenerative medicine and drug discovery. Trends Pharmacol Sci. 2009;30:536–545. doi: 10.1016/j.tips.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Evans SM, Mummery C, Doevendans PA. Progenitor cells for cardiac repair. Semin Cell Dev Biol. 2007;18:153–160. doi: 10.1016/j.semcdb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 3.Harvey RP. Regenerative medicine: Heart redevelopment. Nature. 2010;467:39–40. doi: 10.1038/467039a. [DOI] [PubMed] [Google Scholar]
- 4.Parmacek MS, Epstein JA. Pursuing cardiac progenitors: regeneration redux. Cell. 2005;120:295–298. doi: 10.1016/j.cell.2005.01.025. [DOI] [PubMed] [Google Scholar]
- 5.Wu SM, Fujiwara Y, Cibulsky SM, et al. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell. 2006;127:1137–1150. doi: 10.1016/j.cell.2006.10.028. [DOI] [PubMed] [Google Scholar]
- 6.Moretti A, Caron L, Nakano A, et al. Multipotent embryonic isl1+ progenitor cells lead to cardiac, smooth muscle, and endothelial cell diversification. Cell. 2006;127:1151–1165. doi: 10.1016/j.cell.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 7.Cai CL, Liang X, Shi Y, et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang L, Soonpaa MH, Adler ED, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–528. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 9.Kattman SJ, Huber TL, Keller GM. Multipotent flk-1+ cardiovascular progenitor cells give rise to the cardiomyocyte, endothelial, and vascular smooth muscle lineages. Dev Cell. 2006;11:723–732. doi: 10.1016/j.devcel.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Gregoire S, Karra R, Passer D, et al. Essential and unexpected role of yin yang 1 to promote mesodermal cardiac differentiation. Circ Res. 2013;112:900–910. doi: 10.1161/CIRCRESAHA.113.259259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi Y, Seto E, Chang LS, et al. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 12.Seto E, Shi Y, Shenk T. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature. 1991;354:241–245. doi: 10.1038/354241a0. [DOI] [PubMed] [Google Scholar]
- 13.Lee TC, Zhang Y, Schwartz RJ. Bifunctional transcriptional properties of YY1 in regulating muscle actin and c-myc gene expression during myogenesis. Oncogene. 1994;9:1047–1052. [PubMed] [Google Scholar]
- 14.Vincent CK, Gualberto A, Patel CV, et al. Different regulatory sequences control creatine kinase-M gene expression in directly injected skeletal and cardiac muscle. Mol Cell Biol. 1993;13:1264–1272. doi: 10.1128/mcb.13.2.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen CY, Schwartz RJ. Competition between negative acting YY1 versus positive acting serum response factor and tinman homologue Nkx-2.5 regulates cardiac alpha-actin promoter activity. Mol Endocrinol. 1997;11:812–822. doi: 10.1210/mend.11.6.0015. [DOI] [PubMed] [Google Scholar]
- 16.MacLellan WR, Lee TC, Schwartz RJ, et al. Transforming growth factor-beta response elements of the skeletal alpha-actin gene. Combinatorial action of serum response factor, YY1, and the SV40 enhancer-binding protein, TEF-1. J Biol Chem. 1994;269:16754–16760. [PubMed] [Google Scholar]
- 17.Mariner PD, Luckey SW, Long CS, et al. Yin Yang 1 represses alpha-myosin heavy chain gene expression in pathologic cardiac hypertrophy. Biochem Biophys Res Commun. 2005;326:79–86. doi: 10.1016/j.bbrc.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Lee TC, Shi Y, Schwartz RJ. Displacement of BrdUrd-induced YY1 by serum response factor activates skeletal alpha-actin transcription in embryonic myoblasts. Proc Natl Acad Sci U S A. 1992;89:9814–9818. doi: 10.1073/pnas.89.20.9814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhalla SS, Robitaille L, Nemer M. Cooperative activation by GATA-4 and YY1 of the cardiac B-type natriuretic peptide promoter. J Biol Chem. 2001;276:11439–11445. doi: 10.1074/jbc.M100208200. [DOI] [PubMed] [Google Scholar]
- 20.Lindsley A, Snider P, Zhou H, et al. Identification and characterization of a novel Schwann and outflow tract endocardial cushion lineage-restricted periostin enhancer. Dev Biol. 2007;307:340–355. doi: 10.1016/j.ydbio.2007.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Latinkic BV, Kotecha S, Mohun TJ. Induction of cardiomyocytes by GATA4 in Xenopus ectodermal explants. Development. 2003;130:3865–3876. doi: 10.1242/dev.00599. [DOI] [PubMed] [Google Scholar]
- 22.Gordon S, Akopyan G, Garban H, et al. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25:1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 23.Atchison L, Ghias A, Wilkinson F, et al. Transcription factor YY1 functions as a PcG protein in vivo. Embo J. 2003;22:1347–1358. doi: 10.1093/emboj/cdg124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paige SL, Thomas S, Stoick-Cooper CL, et al. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell. 2012;151:221–232. doi: 10.1016/j.cell.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wamstad JA, Alexander JM, Truty RM, et al. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell. 2012;151:206–220. doi: 10.1016/j.cell.2012.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moses KA, DeMayo F, Braun RM, et al. Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis. 2001;31:176–180. doi: 10.1002/gene.10022. [DOI] [PubMed] [Google Scholar]
- 27.Sucharov CC, Helmke SM, Langer SJ, et al. The Ku protein complex interacts with YY1, is up-regulated in human heart failure, and represses alpha myosin heavy-chain gene expression. Mol Cell Biol. 2004;24:8705–8715. doi: 10.1128/MCB.24.19.8705-8715.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sucharov CC, Mariner P, Long C, et al. Yin Yang 1 is increased in human heart failure and represses the activity of the human alpha-myosin heavy chain promoter. J Biol Chem. 2003;278:31233–31239. doi: 10.1074/jbc.M301917200. [DOI] [PubMed] [Google Scholar]
- 29.Kwon C, Cordes KR, Srivastava D. Wnt/beta-catenin signaling acts at multiple developmental stages to promote mammalian cardiogenesis. Cell Cycle. 2008;7:3815–3818. doi: 10.4161/cc.7.24.7189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura T, Sano M, Songyang Z, et al. A Wnt- and beta -catenin-dependent pathway for mammalian cardiac myogenesis. Proc Natl Acad Sci U S A. 2003;100:5834–5839. doi: 10.1073/pnas.0935626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pal R, Khanna A. Role of smad- and wnt-dependent pathways in embryonic cardiac development. Stem Cells Dev. 2006;15:29–39. doi: 10.1089/scd.2006.15.29. [DOI] [PubMed] [Google Scholar]
- 32.Qyang Y, Martin-Puig S, Chiravuri M, et al. The renewal and differentiation of Isl1+ cardiovascular progenitors are controlled by a Wnt/beta-catenin pathway. Cell Stem Cell. 2007;1:165–179. doi: 10.1016/j.stem.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 33.Gardner KE, Allis CD, Strahl BD. Operating on chromatin, a colorful language where context matters. J Mol Biol. 2011;409:36–46. doi: 10.1016/j.jmb.2011.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cirillo LA, Lin FR, Cuesta I, et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 35.Smale ST. Pioneer factors in embryonic stem cells and differentiation. Curr Opin Genet Dev. 2010;20:519–526. doi: 10.1016/j.gde.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taube JH, Allton K, Duncan SA, et al. Foxa1 functions as a pioneer transcription factor at transposable elements to activate Afp during differentiation of embryonic stem cells. J Biol Chem. 2010;285:16135–16144. doi: 10.1074/jbc.M109.088096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zaret KS, Watts J, Xu J, et al. Pioneer factors, genetic competence, and inductive signaling: programming liver and pancreas progenitors from the endoderm. Cold Spring Harb Symp Quant Biol. 2008;73:119–126. doi: 10.1101/sqb.2008.73.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu J, Watts JA, Pope SD, et al. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 2009;23:2824–2838. doi: 10.1101/gad.1861209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas MJ, Seto E. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene. 1999;236:197–208. doi: 10.1016/s0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- 40.Caretti G, Di Padova M, Micales B, et al. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai Y, Jin J, Yao T, et al. YY1 functions with INO80 to activate transcription. Nat Struct Mol Biol. 2007;14:872–874. doi: 10.1038/nsmb1276. [DOI] [PubMed] [Google Scholar]
- 42.Sucharov CC, Langer S, Bristow M, et al. Shuttling of HDAC5 in H9C2 cells regulates YY1 function through CaMKIV/PKD and PP2A. Am J Physiol Cell Physiol. 2006;291:C1029–1037. doi: 10.1152/ajpcell.00059.2006. [DOI] [PubMed] [Google Scholar]
- 43.Sucharov CC, Dockstader K, McKinsey TA. YY1 protects cardiac myocytes from pathologic hypertrophy by interacting with HDAC5. Mol Biol Cell. 2008;19:4141–4153. doi: 10.1091/mbc.E07-12-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gregoire S, Karra R, Passer D, et al. Essential and Unexpected Role of YY1 to Promote Mesodermal Cardiac Differentiation. Circ Res. doi: 10.1161/CIRCRESAHA.113.259259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang X, Wu SM. Isolation and functional characterization of pluripotent stem cell-derived cardiac progenitor cells. Curr Protoc Stem Cell Biol. Chapter 1(Unit 1F):10. doi: 10.1002/9780470151808.sc01f10s14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.