Abstract
tRNA molecules get heavily modified post-transcriptionally. The N-1 methylation of purines at position 9 of eukaryal and archaeal tRNA is catalyzed by the SPOUT methyltranferase Trm10. Remarkably, while certain Trm10 orthologs are specific for either guanosine or adenosine, others show a dual specificity. Structural and functional studies have been performed on guanosine- and adenosine-specific enzymes. Here we report the structure and biochemical analysis of the dual-specificity enzyme from Thermococcus kodakaraensis (TkTrm10). We report the first crystal structure of a construct of this enzyme, consisting of the N-terminal domain and the catalytic SPOUT domain. Moreover, crystal structures of the SPOUT domain, either in the apo form or bound to S-adenosyl-l-methionine or S-adenosyl-l-homocysteine reveal the conformational plasticity of two active site loops upon substrate binding. Kinetic analysis shows that TkTrm10 has a high affinity for its tRNA substrates, while the enzyme on its own has a very low methyltransferase activity. Mutation of either of two active site aspartate residues (Asp206 and Asp245) to Asn or Ala results in only modest effects on the N-1 methylation reaction, with a small shift toward a preference for m1G formation over m1A formation. Only a double D206A/D245A mutation severely impairs activity. These results are in line with the recent finding that the single active-site aspartate was dispensable for activity in the guanosine-specific Trm10 from yeast, and suggest that also dual-specificity Trm10 orthologs use a noncanonical tRNA methyltransferase mechanism without residues acting as general base catalysts.
Keywords: SPOUT, dual specificity, methyl transferase, tRNA modification
INTRODUCTION
RNA modification occurs in organisms belonging to the three domains of life (Bacteria, Eukarya, and Archaea). More than 100 types of RNA modifications have been identified up to now (Phizicky and Hopper 2010; Cantara et al. 2011; Machnicka et al. 2013), with the addition of a methyl group on the base or on the ribose of a nucleoside being the most frequently encountered modification. The RNA methyltransferases (MTases) that catalyze these methylations are classified in different groups (Czerwoniec et al. 2009). The Rossmann fold-like MTases (RFM) form the largest group, followed by the SpoU-TrmD (SPOUT) family (Anantharaman et al. 2002). These SPOUT-MTases bear a deep trefoil (topological) knot in the catalytic domain to which the methyl donor S-adenosyl-l-methionine (SAM) binds (Hori 2017). The large majority of the SPOUT-MTases exist as dimers with either perpendicular or antiparallel modes of dimerization (Tkaczuk et al. 2007). Many SPOUT enzymes are RNA 2′-O-methyltransferases, some acting on tRNA and others on rRNAs (Persson et al. 1997; Lövgren and Wikström 2001; Purta et al. 2006; Kuratani et al. 2008; Dunstan et al. 2009; Benítez-Páez et al. 2010). Other MTases of this family modify the base of the ribonucleoside (Byström and Björk 1982; Ero et al. 2008; Purta et al. 2008). Recently, also more uncommon SPOUT-enzyme activities have been discovered: a protein (arginine) MTase (Young et al. 2012) and an RNA aminocarboxypropyl-transferase (Meyer et al. 2016). Other families of SAM dependent RNA MTases exist, as revealed by the radical SAM MTases and the β barrel type MTases, although only a few members of these families are described in the literature (Yan and Fujimori 2011; Broderick et al. 2014; Kimura et al. 2014).
The Trm10 MTases belong to the SPOUT family, and contrary to most other known family members behave as monomers in solution. Initially Trm10 was identified as the enzyme catalyzing the formation of m1G9 in yeast tRNA (Jackman et al. 2003; Jackman and Alfonzo 2013; Swinehart et al. 2013). Interestingly, different Trm10 homologs display varying substrate specificities. Whereas human TrmT10A and TrmT10B catalyze the formation of m1G9 in cytoplasmic tRNA, the crenarchaeal Sulfolobus acidocaldarius Trm10 (SaTrm10) forms m1A9 (Kempenaers et al. 2010; Oerum et al. 2017a). Remarkably, the euryarchaeal Thermococcus kodakaraensis Trm10 (TkTrm10) displays a broadened specificity, forming m1A9 and m1G9 in tRNA (Kempenaers et al. 2010). The same dual specificity was observed for the third human Trm10 homolog TrmT10C, involved in methylation of mitochondrial tRNAs (Vilardo et al. 2012). This last Trm10 homolog is a component of the mitochondrial RNaseP complex (Holzmann et al. 2008), in which TrmT10C forms a subcomplex with the dehydrogenase HSD10. Association of TrmT10C with HSD10 is required for tRNA MTase activity (Vilardo and Rossmanith 2015; Oerum et al. 2017b).
Broadened specificity is not uncommon for tRNA-MTases. For example, the dual-specific enzyme RlmN of E. coli forms m2A in both 23S rRNA and tRNA (Benítez-Páez et al. 2012). Adjacent nucleotides in the tRNA molecule can also be a target for the same MTases. TrmI of Pyrococcus abyssi is such a region-specific enzyme forming m1A at positions 57 and 58 of the tRNA molecule (Roovers et al. 2004). The yeast enzyme Trm4 catalyzes the formation of m5C at positions 34, 40, 48, or 49 depending on the tRNAs (Motorin and Grosjean 1999). More intriguingly, there are tRNA MTases known to modify different types of bases. The first example is yeast Trm5 that can methylate G37 into m1G37, and I37 (where I is inosine) into m1I37 (Björk et al. 2001). Using an artificial system it was shown that NpmA of Streptoalloteichus tenebrarius, a 16S rRNA MTase forming m1A1408, possesses dual nucleobase specificity capable of catalyzing both m1A1408 and m1G1408 modifications (Zelinskaya et al. 2015). This last example of broadened specificity is very similar to the one found for TkTrm10 and human mitochondrial TrmT10C forming m1G9 and m1A9 in tRNA.
The protonation state of the N1 position of guanine and adenine are different at physiological pH, with the guanine N1 being protonated and the adenine N1 being unprotonated in their most common tautomeric form. Deprotonation of the N1-position of guanine, required for its methylation, has initially been proposed to involve an aspartate residue present in all Trm10 members that would act as a catalytic base (Shao et al. 2014). Remarkably, this residue is also present in the m1A-specific SaTrm10, while also a second aspartate residue is present in the active site of SaTrm10 and of the dual-specific TkTrm10. Mutation of both aspartate residues in SaTrm10 also leads to a reduced MTase activity (Van Laer et al. 2016). However, using detailed kinetic analysis it was recently demonstrated that mutation of the generally conserved aspartate residue (D210 in Saccharomyces cerevisiae) only leads to a small decrease in the catalytic rate of m1G formation and that it hence does not function as a catalytic base (Krishnamohan and Jackman 2017).
To get more insights into the catalytic mechanism and substrate specificity of the Trm10 enzymes in general, and the dual-specificity enzymes in particular, we set out to investigate the Trm10 ortholog from T. kodakaraensis. In this article we report the structure of TkTrm10, and the importance of the two active site aspartate residues of TkTrm10 is evaluated.
RESULTS AND DISCUSSION
TkTrm10 is a slow tRNA methyltransferase displaying genuine dual specificity
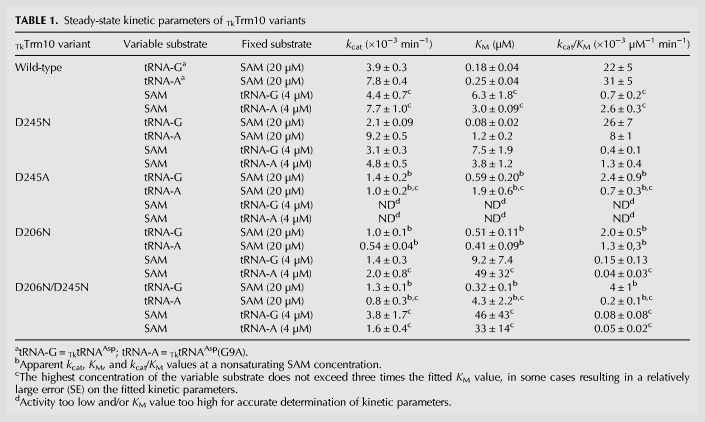
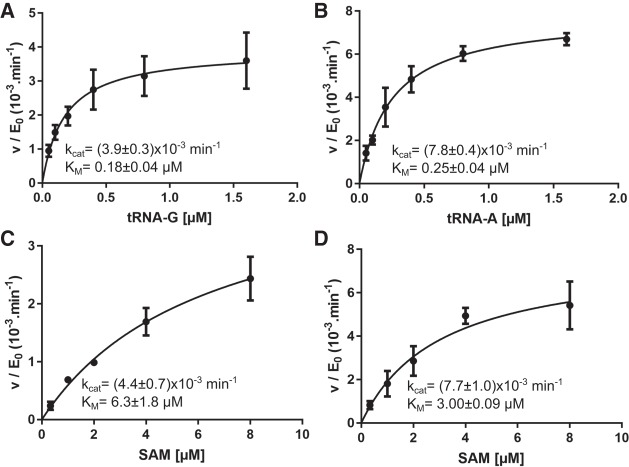
We have previously shown that TkTrm10 is a dual-specificity enzyme that catalyzes formation of both m1A and m1G at position 9 of certain tRNAs (Kempenaers et al. 2010). In order to gain more insight into the kinetic basis underlying this dual specificity we performed steady-state kinetic experiments. Hereto, we used in vitro transcribed tRNAAsp of T. kodakaraensis, which bears a guanosine at position 9 (later referred to as tRNA-G). For direct comparison we also made a mutant of TktRNAAsp where G9 has been changed into an adenosine (later referred to as tRNA-A). To determine the steady-state kinetic parameters, kcat and KM, we either varied the tRNA concentration at a fixed saturating concentration of SAM (20 µM), or we varied the SAM concentration at a saturating concentration of tRNA (4 µM). This steady-state analysis shows that TkTrm10 has KM values for its tRNA substrates in the submicromolar range [KM(tRNA-G) = 0.18 ± 0.04 µM, KM(tRNA-A) = 0.25 ± 0.04 µM] and a low micromolar KM for SAM (3–6 µM, depending on the tRNA substrate) (Table 1; Fig. 1). The high affinity for tRNA and micromolar affinity for SAM (KD = 9.4 ± 1.2 µM) is also confirmed by electrophoretic mobility shift assays (EMSA) and ITC measurements, respectively (Supplemental Fig. S1). The turnover number (kcat) of TkTrm10 is very low using either tRNA-G or tRNA-A as a substrate, with a slight preference for tRNA-A [kcat(tRNA-G) = (3.9 ± 0.3)10−3 min−1, kcat(tRNA-A) = (7.8 ± 0.4)10−3 min−1]. Together this proves that TkTrm10 is a genuine dual-specificity enzyme with very similar specificity constants for tRNA with either guanosine and adenosine at position 9 [kcat/KM(tRNA-G) = (22 ± 5)10−3 µM−1 min−1, kcat/KM(tRNA-A) = (31 ± 5)10−3 µM−1 min−1].
TABLE 1.
Steady-state kinetic parameters of TkTrm10 variants
FIGURE 1.
Steady-state (Michaelis–Menten) kinetics of the methyltransferase reaction catalyzed by wild-type TkTrm10. Curves are shown using either SAM at a fixed concentration of 20 µM and varying concentrations of tRNA-G (A) or tRNA-A (B), or using SAM as a variable substrate at a fixed concentration (4 µM) of tRNA-G (C) or tRNA-A (D). Every data point is the average (±SEM) of three independent measurements. The kcat and KM values (±SE) resulting from fitting on the Michaelis–Menten equation are given in the insets.
The low kcat value of TkTrm10 is remarkable, as this value is about 1000-fold lower than (single) turnover rates reported for yeast Trm10 or kcat values reported for the other N1-methyltranferases TrmD and Trm5 (Christian and Hou 2007; Krishnamohan and Jackman 2017). A first potential explanation could be that the low activity is a general feature of dual-specificity Trm10 orthologs, where catalytic efficiency has been sacrificed in favor of substrate promiscuity. Alternatively, it should be taken into consideration that TkTrm10 might require a so-far unidentified partner protein to attain full catalytic activity. Indeed, TrmT10C the only other characterized dual-specificity Trm10 ortholog so far, has also been shown to have a nearly negligible methyltransferase activity on its own, and binding to its partner protein HSD10 (or SDR5C1) is required for efficient tRNA modification (Vilardo et al. 2012). In analogy with TkTrm10, TrmT10C displays full tRNA binding capacity in absence of any partner protein. This opens up the intriguing possibility that the requirement for a partner protein to attain full catalytic activity could be a more general property of dual-specificity Trm10 enzymes. Further studies to identify such a partner protein in T. kodakaraensis are required to address this question.
Neither the N-terminal nor C-terminal domain of TkTrm10 contribute to its specificity
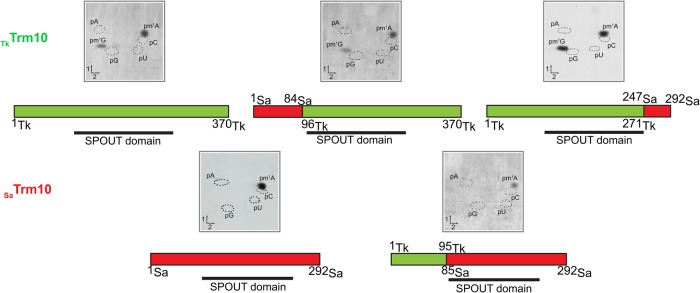
Sequence alignment of TkTrm10 with SaTrm10 (sequence identity = 29.5%; sequence similarity = 45%), of which the structure of the full-length protein has been solved, reveals that TkTrm10 also consists of three well defined domains, analogous to other enzymes of the Trm10 family (Supplemental Fig. S2). To determine whether the substrate specificity is entirely defined locally by the catalytic SPOUT domain, or also the N- and C-terminal domains contribute to specificity, domain swapping experiments were performed using the TkTrm10 (forming m1A and m1G) and SaTrm10 (forming m1A) enzymes. The rationale of choosing SaTrm10 in this experiment rather than yeast Trm10 (forming m1G) is the thermophilic character of both enzymes. Except for the variant of SaTrm10 containing the C-terminal domain of TkTrm10, all hybrids were successfully produced in E. coli and the purified proteins displayed tRNA methylation activity. As assessed via a 2D-TLC and shown in Figure 2, among the different chimeric proteins tested only the ones containing the SPOUT domain of TkTrm10 presented a dual specificity. This shows that the dual specificity is essentially conferred by elements present in the SPOUT domain.
FIGURE 2.
Domain swapping experiment of TkTrm10 and SaTrm10. MTase activity measurements of TkTrm10 and SaTrm10 chimeric proteins using total E. coli tRNA as substrate. E. coli tRNA was incubated in the presence of 30 µg of purified enzyme and [methyl-14C]-SAM (see Materials and Methods). tRNA was recovered and digested by nuclease P1. The resulting 5′-phosphate mononucleosides were analyzed by 2D-TLC followed by autoradiography. Circles in dotted lines show the migration of the four canonical nucleotides used as UV markers. The domain organization of the chimeric proteins is shown schematically with the Thermococcus part of the protein in green and the Sulfolobus part in red. The SPOUT domains are indicated. The numbers represent the N and C terminus or junction regions of TkTrm10 and SaTrm10.
The crystal structure of TkTrm10Δ26 shows the N-terminal and SPOUT domains
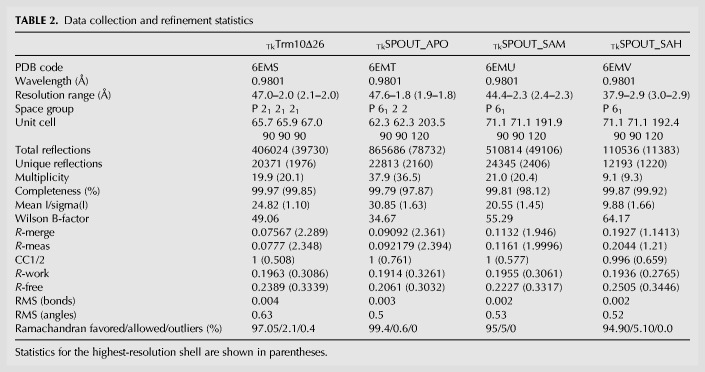
To gain insight into the domain organization, the tRNA binding mode and the mechanism of the dual specificity of TkTrm10, we set out to solve its crystal structure. Since the crystallization of the full length protein, consisting of 370 amino acid residues, was unsuccessful, we crystallized a C-terminal truncated construct lacking the last 26 residues (TkTrm10Δ26, res 1–344). The crystal structure of TkTrm10Δ26 was determined in the apo form using molecular replacement with the SaTrm10 structure (PDB code 5a7y) as a search model. The data collection, processing and refinement statistics are reported in Table 2. TkTrm10Δ26 is crystallized in space group P212121 with one copy in the asymmetric unit, and the structure was solved at a resolution of 2.0 Å. Clear electron density is present for residues 1–203, 218–235, and 245–266. The last 77 residues of TkTrm10Δ26 were not visible, either due to conformational flexibility or due to protein degradation during the course of crystallization. Apart from the missing residues at the C-terminal region, our model is also missing the first 19 residues of the N-terminal purification tag and the two internal regions ranging from residues 204–217 and residues 236–244, which are part of catalytic loops (see below).
TABLE 2.
Data collection and refinement statistics
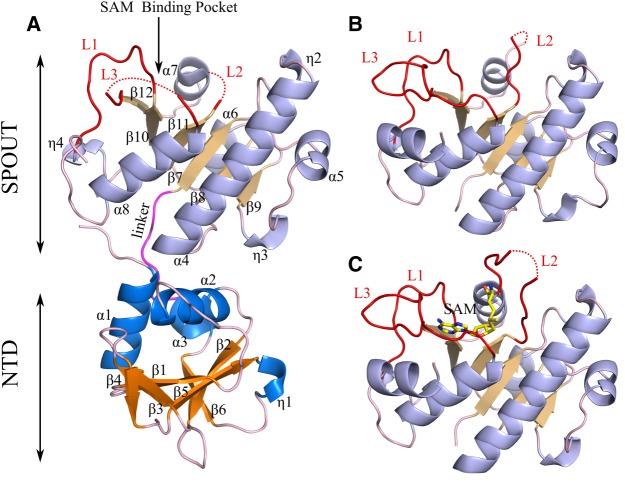
Crystal packing analysis using the PISA server (Read et al. 2011) indicates that TkTrm10Δ26 is a monomer, consistent with the elution volume in size exclusion chromatography (not shown). A monomeric arrangement is also in agreement with all the other currently characterized Trm10 orthologs, but differs from most other known SPOUT enzymes that occur as dimers (Tkaczuk et al. 2007). The overall fold of the protein is shown in Figure 3A. The TkTrm10Δ26 structure shows two distinct domains: an N-terminal domain (NTD) from residues 1–97 and a catalytic SPOUT domain from residues 97–266. The region spanning residues 267–344 of TkTrm10 is not observable in the crystal structure. In the structure of SaTrm10 (PDB ID 5a7y) the C-terminal residues formed an all helical domain, distinct from the NTD and SPOUT domains, but the sequence identity between SaTrm10 and TkTrm10 is low in this region.
FIGURE 3.
Overall fold of TkTrm10 constructs. (A) Crystal structure of TkTrm10Δ26 in cartoon representation. The α helices of the NTD and SPOUT domains are colored dark blue and light blue, respectively, while the β strands of the NTD and SPOUT domains are colored orange and light orange, respectively. The linker between the N-terminal and SPOUT domain is shown in pink. The three loop regions involved in forming the trefoil knot are shown in red and are labeled L1, L2, and L3. The missing regions of L2 and L3 are shown with red dotted curved lines. The SAM-binding pocket is indicated by an arrow. (B) Crystal structure of TkSPOUT_APO in cartoon representation. The missing L2 region is shown by a dotted red curved line. (C) Crystal structure of TkSPOUT_SAM in cartoon representation. The SAM molecule bound to the active site is shown as a stick model with carbon atoms colored yellow. The color code in panels B and C is the same as in A.
The NTD of TkTrm10Δ26 is formed by a mixed six-stranded β-sheet (β1 to β6), three α-helices (α1–α3) and one 310 (η1) helix. A search for structures similar to the NTD with the DALI server only finds the NTD of SaTrm10 suggesting this domain is unique for Trm10 (sequence identity NTD of TkTrm10 and SaTrm10 = 43%). In contrast with the NTD of SaTrm10, the NTD of TkTrm10 does not display a clear horseshoe shape, predominantly due to the replacement of two long and curved β-strands (β1 and β2) in the NTD of SaTrm10 by two times two shorter strands (β2/β3 and β4/β5, respectively) in TkTrm10 (Supplemental Fig. S3A). However, similar to the NTD of SaTrm10, the two helices α1 and α2, connected by a central β-strand (β1), are also present in the NTD of TkTrm10 and superpose well with the corresponding regions of SaTrm10 (Supplemental Fig. S3A).
The NTD is connected to the SPOUT domain by a flexible linker connecting α3 to β7. The SPOUT domain of TkTrm10 superposes onto that of SaTrm10 (PDB ID 5a7y) with an rmsd of 1.3 Å suggesting a highly similar structure (Supplemental Fig. S3B [sequence identity SPOUT of TkTrm10 and SaTrm10 = 30%]). The SPOUT domain of TkTrm10 consists of a six-stranded parallel β-sheet (β7–β12) surrounded by six α helices (α4–α9) and three 310 helices (η2–η4). A deep trefoil knot is formed at the C-terminal half of the SPOUT domain, constituted by β10–β12, α7–α8, η4 and the loops connecting β10 to η4 (loop L1, residues 181–191), β11 to α7 (loop L2, residues 202–215) and α8 to β12 (loop L3, residues 232–247). This knot structure creates the SAM binding pocket and the catalytic center of the protein. The two catalytic loops L2 and L3 are predominantly disordered in the apo structure.
The orientation of the SPOUT domain with respect to the NTD is very similar to SaTrm10, and superposition of TkTrm10Δ26 onto SaTrm10 (PDB ID 5a7y) yields an overall RMSD of 1.9 Å (Supplemental Fig. S3). The NTD interacts with the SPOUT domain via a number of hydrogen bonds and via hydrophobic interactions between residues of α1 and α2 on the one hand and α4 on the other hand, ultimately orienting the two domains in such a way that a highly positively charged surface spans both domains on one face of the protein (see further).
The crystal structures of the SPOUT domain of TkTrm10 with bound substrate and product show conformational flexibility of the active site loops
Subsequently we solved the crystal structures of the SPOUT domain (residues 97 to 272) of TkTrm10, both in the apo form (TkSPOUT_APO) or bound to the substrate SAM (TkSPOUT_SAM) or the product SAH (TkSPOUT_SAH). The data collection, processing and refinement statistics are reported in Table 2. The TkSPOUT_APO form crystallized in space group P6122 with one copy in the asymmetric unit and the structure was solved at 1.8Å resolution. TkSPOUT_SAM and TkSPOUT_SAH crystallized in space group P61 with three copies in the asymmetric unit and the structures were solved at a resolution of 2.3 and 2.9 Å, respectively.
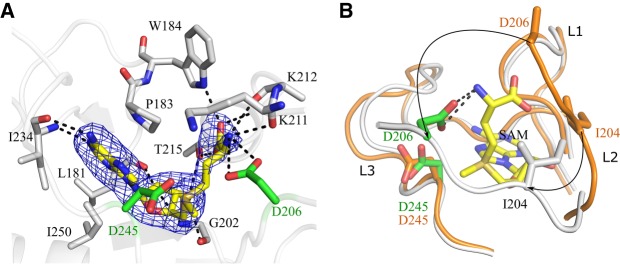
The electron density showed the presence of a SAM and SAH molecule in the active site of TkSPOUT_SAM and TkSPOUT_SAH, respectively. The SAM/SAH molecules bound to each protein molecule in the asymmetric unit of either TkSPOUT_SAM and TkSPOUT_SAH superpose well with a slight flexibility in the carboxylic end of the SAM/SAH molecule (Supplemental Figs. S4, S5). In addition, all the protomers in the asymmetric unit of TkSPOUT_SAM and TkSPOUT_SAH structures superpose very well onto each other (Supplemental Figs. S4, S5). Therefore, chain A of TkSPOUT_SAM and TkSPOUT_SAH were used for further interpretation. As expected, the overall folds of TkSPOUT_APO, TkSPOUT_SAM, and TkSPOUT_SAH superpose very well with the SPOUT domain of TkTrm10Δ26, with an RMSD value of 0.5, 0.5, and 0.6 Å, respectively. However, differences in the conformation of the catalytic loops L2 and L3 are evident among all these structures. Unlike in TkTrm10Δ26, the L3 loop and a part of the L2 loop are visible in TkSPOUT_APO (Fig. 3B). Ligand binding in the TkSPOUT_SAM and TkSPOUT_SAH structures leads to further ordering and major conformational changes of the active site loop L2 (Fig. 4; Supplemental Fig. S6). Also, the L3 loop shows a slight inward movement in the SAM/SAH-bound form when compared to the APO form. These loop movements place a number of additional residues into the active site (Fig. 4).
FIGURE 4.
Active site arrangement and conformational changes in the SAM-bound SPOUT domain of TkTrm10 (TkSPOUT_SAM). (A) The SAM-binding pocket of TkTrm10. Several residues surrounding or interacting with SAM are shown as sticks with carbon atoms colored gray. Two active site residues conserved in archaeal m1A and m1A/m1G-specific Trm10 orthologs (Asp206 and Asp245 in TkTrm10) are colored green. The polar interactions with SAM are indicated with dotted black lines. The difference electron density map (Fo–Fc, contoured at 2σ) around SAM is shown as a blue colored mesh. (B) Superposition of TkSPOUT_APO (orange) and TkSPOUT_SAM (gray). The arrows denote the movements of Asp206 and Ile204 within the active site loop L2 upon SAM binding. SAM is represented as sticks with carbon atoms colored yellow. The interaction formed by Asp 206 and the –NH2 group of SAM is shown by the dotted lines. The color code of TkSPOUT_SAM is kept the same as in panel A.
Overall, the substrate SAM and the product SAH are bound in a binding pocket created by the trefoil knot formed by the three loops L1, L2, and L3, and both ligands adopt the typical bent conformation, which is shown to be an essential conformation for MTase activity in SPOUT enzymes (Fig. 4; Supplemental Figs. S4–S6; Elkins et al. 2003; Schubert et al. 2003; Nureki et al. 2004; Somme et al. 2014). Since most of the interactions are identical in TkSPOUT_SAM and TkSPOUT_SAH, we only describe the interactions with SAM. The adenine moiety of SAM is deeply buried in a hydrophobic environment, where the adenine moiety is stacked between Pro183 and Ile250. Additionally, the amino group at C6 and the N1 atom of the adenine base make interactions with the main chain carbonyl and amino group of Ile234. The ribose sugar moiety of SAM adopts a C3′ endo conformation, similar to the other available Trm10 structures. The 2′ OH group of the ribose moiety interacts with the main chain carbonyl group of Leu181 and main chain amino group of Gly202, while the 3′ OH interacts with the main chain carbonyl and amino groups of Gly202 and the side chain hydroxyl group of Thr215. The amino end of SAM interacts with the main chain carbonyl groups of Lys211 and Lys212 and the side chain of Asp206. The carboxyl moiety of SAM interacts with the side chains of Trp184 and Thr215, the main chain amino groups of Thr214 and Thr215, and the main chain carbonyl group of Lys212. The reactive methyl group is positioned at 3.7 and 3.6 Å from the side chains of Asp245 and Asp206, respectively. Both of these Asp residues are also present in the active site of the m1A-specific SaTrm10 (Van Laer et al. 2016). Furthermore, Asp206 is highly conserved in all Trm10 proteins (Supplemental Figs. S2, S7). However, while it was initially proposed that the latter aspartate residue could play the role of general base during m1G formation (Shao et al. 2014), this hypothesis was recently refuted since it was shown that mutation of this residue has only a minor effect on the activity of the m1G-specific enzyme from Saccharomyces cerevisiae (Krishnamohan and Jackman 2017).
Substrate docking reveals a potential binding mode of the target nucleosides
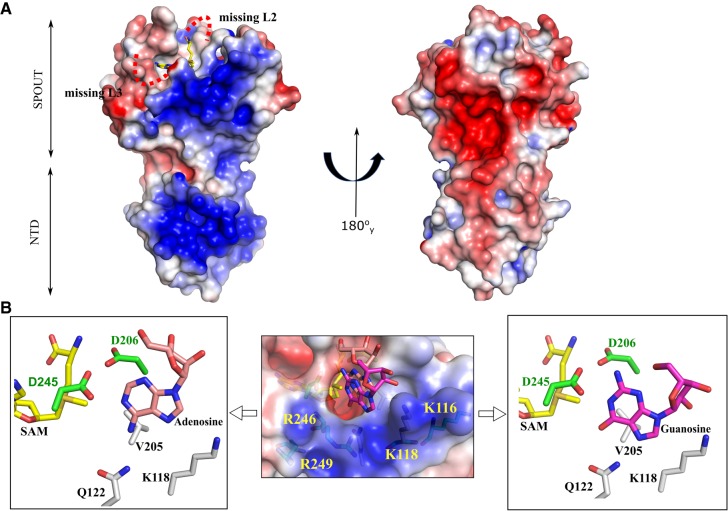
Mapping the electrostatic potential on the solvent accessible surface of the TkTrm10Δ26 and TkSPOUT_SAM structures shows a large positively charged surface spanning both the NTD and the SPOUT domain on one face of the protein (Fig. 5A). This charge distribution is similar to that observed on the SaTrm10 FL structure, where the positive charge further expands to the C-terminal domain (Supplemental Fig. S8). The accessible surface representation of TkSPOUT_SAM also reveals a cavity centered around the methionine moiety of SAM, which likely represents the site of entry for tRNA methylation (Fig. 5B). A model for the tRNAiMet bound to SaTrm10 has previously been proposed, where the binding site for the substrate tRNA spans the whole positively charged surface from NTD to CTD (Van Laer et al. 2016). Superposition of the tRNAiMet-SaTrm10 model on the TkTrm10Δ26 structure shows that a similar tRNA binding mode is possible for TkTrm10 (Supplemental Fig. S8). Within this positively charged surface four residues, Arg246, Arg249, Lys116, and Lys118, are located near the SAM binding pocket (Fig. 5B). In addition, three consecutive lysines (K211–K213) are also present in L2, which is disordered in the apo form but predominantly ordered in the SPOUT_SAM/SAH structures. These residues are also positioned within the positively charged patch of the protein and might change conformation upon interacting with the tRNA substrate.
FIGURE 5.
Electrostatic surface potential of TkTrm10 constructs and possible tRNA binding site. (A) Electrostatic potential mapped on the solvent accessible surface of TkTrm10Δ26 in two orientations. The missing loops in the structure are shown by red dotted curved lines in the left panel. For reference, SAM, shown in yellow sticks, is placed in the active site. (B) The middle panel shows the electrostatic potential mapped on the solvent accessible surface of the TkSPOUT_SAM structure with a docked guanosine and adenosine molecule. SAM, guanosine, and adenosine are shown as sticks with carbon atoms colored yellow, magenta, and salmon, respectively. The positively charged residues are labeled on the surface. The left and right panels show a zoomed-in view of, respectively, the docked adenosine and guanosine in the TkSPOUT_SAM active site. The surrounding residues discussed in this study are shown.
To gain further insight into the catalytic center of the methyl transfer reaction, we resorted to docking of a guanosine and adenosine moiety into the active site of the TkSPOUT_SAM structure using AutoDock 4.0 (Morris and Huey 2009). The grid map in the docking protocol is constrained to the surroundings of the active site cavity. The coordinates of guanosine and adenosine were obtained from the ternary complex of H. influenza TrmD in complex with sinefungin and tRNA (G37 of tRNA) (PDB ID 4yvi) and from the ternary complex of the human m1A58 methyltransferase with SAH and tRNA (A58 of tRNA) (PDB ID 5CCB), respectively. Calculations from 20 Autodock runs for guanosine gave binding conformations with an average free energy of −3.5 kcal/mol, while that of adenosine gave binding conformations with an average free energy of −3.2 kcal/mol. For guanosine docking, all 20 conformations were superimposable and are properly oriented to accept a methyl group at the N-1 position, while for adenosine only six out of 20 conformations are superimposable and at the same time also properly oriented to accept the methyl group. One of these properly oriented minimum energy conformations of guanosine and adenosine were used to generate the final models of the ternary complexes consisting of the TkSPOUT_SAM structure and the docked nucleosides (Fig. 5B). Although the docking models should be treated with some precaution, especially considering that further conformational changes could take place upon tRNA binding, they show that the N-1 atoms of guanosine and adenosine are properly oriented to accept the methyl group from the methionine moiety of the substrate SAM, by stacking of the nucleobase onto the highly conserved Val205. The side chains of Asp206 and Asp245 are located at a distance of 3.9/3.5 and 3.5/4.0 Å to the N1 atom of guanosine and adenosine, respectively. The Asp245 and Asp206 side chains are also located within interaction distance of the exocyclic amino group at position 2 of the guanine base. In addition, Gln122 is properly oriented to form a hydrogen bond with the C6-carbonyl group of the guanine base or, provided some small conformational changes, with the C6-amino group of adenosine. These residues might thus be important for substrate binding and/or catalysis.
Role of active site residues in dual specificity of TkTrm10
Site-directed mutagenesis was used to study the role of the TkTrm10 active site residues Asp206, Asp245, and Gln122 in catalysis and substrate specificity.
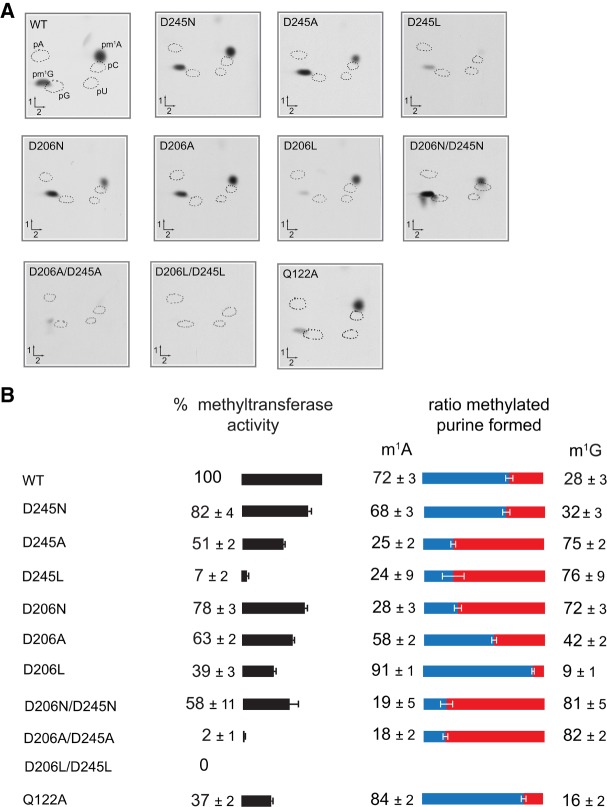
The residues Asp206 and Asp245 were replaced by either alanine, to evaluate their total contribution to catalysis, or by asparagine, to evaluate any role in proton exchange. Additionally, Asp245 was replaced by a leucine residue since the m1G-specific Trm10s possess a Leu at the position equivalent to Asp245 (Supplemental Fig. S2). Similarly, the effect of substitution of Asp206 by Leu was investigated. First, we used a semiquantitative approach to assess the methyltransferase activity, by measuring the incorporation of the 14C-methyl group from labeled SAM in bulk E. coli tRNA after 30 min and TCA precipitation. Subsequently, the ratio of m1A/m1G formation was measured for each mutant and compared to the WT using a TLC assay (Fig. 6).
FIGURE 6.
Methyl transferase (MTase) activities of mutant TkTrm10 compared to the WT. E. coli tRNA was incubated in the presence of 30 µg of purified enzyme and [methyl-14C]-SAM (see Materials and Methods) after which either total radioactivity incorporated in tRNA was counted after TCA precipitation or the 5′-phosphate mononucleosides, obtained after digestion of the tRNA by nuclease P1, were separated by 2D-TLC. The radioactive nucleotides were then identified by autoradiography. (A) Autoradiograms of the 5′-phosphate mononucleosides separated by 2D-TLC. Circles in dotted lines show the migration of the four canonical nucleotides used as UV markers. The TkTrm10 variant is indicated at the top left corner of each autoradiogram. The radioactivity of the spots corresponding to pm1A and pm1G was measured by scintillation counting. (B) Total radioactivity incorporated in tRNA by the different TkTrm10 variants related to WT (left panel) and the pm1A/pm1G ratio (right panel). Values given are the average ± SD of three independent experiments.
When Asp245 was replaced by Asn or Ala, only a relatively small reduction of total activity was observed (respectively 82 ± 4 % and 51 ± 2% of activity left). The m1A/m1G formation ratio was 72/28 for the WT enzyme, while it was 68/32 for the D245N mutation and 25/75 for the D245A mutant (Fig. 6). To further quantify the effect of the mutations we determined the steady-state kinetic parameters using in vitro transcribed T. kodakaraensis tRNAs (tRNA-G and tRNA-A, see above). The D245N mutation shows no effect on kcat for either tRNA-A and tRNA-G, thus excluding a role of D245 in proton transfer (Table 1; Supplemental Fig. S9). While the D245N mutation has no effect on the KM of SAM and results in a slightly lowered KM for tRNA-G, it results in an almost fivefold higher KM for tRNA-A. Assuming that KM is a measure of affinity, this result is somewhat unexpected since in our docking models D245 could interact with the 2-amino group of guanosine, while this is not the case for adenosine (Fig. 5). The D245A mutant shows an increased KM value for SAM, such that no saturation is reached at 30 µM and no kinetic parameters could be obtained with SAM as a variable substrate. However, apparent kcat and KM values were obtained using tRNA-A and tRNA-G as variable substrates (using SAM at a subsaturating concentration of 20 µM). Although overall the effects of the D245A mutation on tRNA modification are rather small, these are more pronounced on the kcat and KM value for tRNA-A (Table 1; Supplemental Fig. S9). This results in an inversion of the specificity toward a more m1G-specific enzyme, as also observed in our TLC-assay. Finally, the D245L variant resulted in an almost inactive enzyme (7 ± 2% activity left) (Fig. 6). The remaining activity is too low for a quantitative assessment of the kinetic parameters. Considering that the D245N and D245A mutations have only minor effects on total activity, the loss of activity for the D245L mutant is probably due to severe steric hindrance upon SAM binding. An ITC experiment indeed shows that SAM binding is completely impaired in the D245L mutant (Supplemental Fig. S1). Alternatively, it can also not be excluded that the D245L mutation causes a local perturbation of the TkTrm10 active site structure.
A small effect on the methyltransferase activity was also observed when Asp206 was replaced by Asn or Ala (respectively 78 ± 3% and 63 ± 2% of activity left). Comparable to the D245 mutations, the D206N and D206A mutations shift the m1A/m1G formation ratio toward m1G, which is most pronounced for the D206N mutation (28/72 compared to 72/28 for the wild-type enzyme) (Fig. 6). In contrast the D206L mutation has a larger impact on total activity (39 ± 3% activity remaining), probably at least partially due to impaired SAM binding as observed in ITC (KD = 68 ± 19 µM, Supplemental Fig. S1). Determination of the steady-state kinetic parameters for D206N confirm these findings. Using tRNA as a variable substrate we find a small yet more pronounced effect on the apparent kcat value for tRNA-A methylation compared to tRNA-G methylation (Table 1; Supplemental Fig. S9). While these data show that Asp206 only plays a minor role in the methyltransferase reaction, as also found for the m1G-specific Trm10 from yeast (Krishnamohan and Jackman 2017), they confirm the change in m1A/m1G specificity observed via TLC. Again, a more pronounced effect of D206 mutations on m1A formation compared to m1G formation is unexpected from our docking models where D206 is within interaction distance of the 2-amino group of guanosine. Due to the low activity of the enzymes and the insensitivity of our kinetic assay (where the concentration of enzyme needs to be maintained below the substrate concentration), the kinetic traces for D206A and D206L were too noisy for accurate fitting.
Since the consequences of the replacement of D206 and D245 by either Ala or Asn are not very pronounced, this prompted us to analyze double mutants where both D206 and D245 were replaced by Asn, Ala or Leu (Fig. 6). The D206N/D245N double mutant showed a rather slightly decreased activity (58 ± 11% activity remaining) with an m1A/m1G ratio (19/81) comparable to the D206N single mutant. On the other hand, the D206A/D245A and D206L/D245L double mutants were dramatically affected (2 ± 1% and 0% activity remaining, respectively). While the activity drop of the D206L/D245L can be fully attributed to a severely impaired SAM binding, the D206A/D245A mutant still binds tRNA and SAM (albeit with lower affinity, KD = 20.4 ± 2.9 µM), as assessed by EMSA and ITC (Supplemental Fig. S1). While the activity of the D206A/D245A and D206L/D245L mutants is too low to allow a full kinetic analysis, we did determine the kinetic parameters of the D206N/D245N mutant. The KM values show that the mutations affect SAM binding (Table 1), again consistent with ITC data (KD = 19.3 ± 3.4 µM, Supplemental Fig. S1). The kinetic parameters using tRNA-G and tRNA-A as variable substrates confirm that m1A formation is most affected by the mutation (Table 1; Supplemental Fig. S9), both owing to a larger effect on kcat and KM, consistent with the TLC assay and agreeing with a combined effect of the D206N and D245N mutations.
Together these results rule out a role of D206 and D245 in a proton transfer step of the methyltransferase reaction, and they show that both residues individually only play a marginal role in catalysis. Both residues have a quite subtle influence on substrate specificity, and mutation of either residue shift the specificity of the TkTrm10 enzyme toward a preference of m1G formation over m1A formation. The more severe effect of the D206A/D245A double mutation on activity could point toward a role of both Asp residues in proper positioning of the purine base for acceptance of a methyl group from SAM. These results thus conflict with the initial findings and proposals that the residue corresponding to Asp206 would act as a general base in m1G formation by yeast Trm10 (Shao et al. 2014), while a role of both the residues corresponding to Asp206 and Asp245 was proposed in m1A formation by S. acidocaldarius Trm10 (Van Laer et al. 2016). This discrepancy could be caused by the use of less quantitative methods to assess enzyme activity in the latter two studies. Indeed, using a detailed kinetic analysis, Krishnamohan and Jackman (2017) recently proved that D210 (D206 in TkTrm10) does not play the role of a general base in the m1G formation by S. cerevisiae Trm10, suggesting a noncanonical tRNA methyltransferase mechanism for the m1G-specific Trm10 enzymes. Our results corroborate and extend these conclusions for the m1A/m1G dual-specificity Trm10 enzymes, where neither of the two conserved active site Asp residues play a role in proton transfer, and thus prompts reconsideration of the catalytic mechanism of all Trm10 orthologs.
Finally, the generally conserved Gln122 of TkTrm10 was replaced by an alanine residue. This mutation resulted in a rather mild reduction of the activity (37 ± 2% activity left; see Fig. 6). Moreover, TLC analysis revealed only a rather small effect on the m1A/m1G specificity (84/16 ratio for Q122A compared to 72/28 ratio for the wild-type enzyme). Our finding that Gln122 has only a rather small contribution to catalysis again contrasts with the consequences of a similar mutation in yeast SpTrm10 (Q118A) that resulted in total loss of m1G-forming activity (Shao et al. 2014).
Conclusions
Despite the structural similarity of the catalytic domain of eukaryotic, crenarchaeal, and euryarchaeal Trm10, these enzymes display different specificities. In analogy to other m1G-forming enzymes, such as TrmD and Trm5, a universally conserved Asp residue (equivalent to Asp206 in TkTrm10) has initially been proposed as a catalytic base for m1G formation. A second Asp residue is present in archaeal Trm10 (equivalent to Asp245 in TkTrm10), and these two Asp residues were found to be important for m1A formation in SaTrm10. However, recently a study appeared that rules out the role of Asp210 (equivalent to Asp206 in TkTrm10) as a general base in m1G formation in S. cerevisiae (Krishnamohan and Jackman 2017). So far, very little is known about the mechanism of dual-specificity m1A/m1G-forming Trm10 orthologs. Here we report the first structures of such a dual-specificity enzyme, which also possesses two Asp residues in its active site. Although, both residues are ideally placed in the active site to interact with the target nucleobases, mutation of neither of these residues to asparagine has a very pronounced effect on activity. Also, the double D206N/D245N mutation has a negligible effect on catalysis. These findings rule out an important catalytic role for both residues and suggest that TkTrm10-catalyzed formation of m1A and m1G occurs without the implication of a general base. Only a double D206A/D245A mutation has a severe effect on the catalytic rate, which cannot solely be attributed to impaired substrate (SAM) binding, suggesting a more subtle role of both Asp residues in catalysis, e.g. by inducing proper alignment of the substrates in the active site. Such a role in orientation of the substrates in the active site is also reflected in a shift in substrate specificity in the D206 and D245 mutants, both favoring m1G formation in comparison to the wild-type enzyme. These results are thus in line with the recent findings for the m1G-specific enzyme of S. cerevisiae, and suggest that all Trm10 orthologs use a noncanonical mechanism to catalyze their methyltransfer reaction, without the use of an enzymatic general base catalyst.
MATERIALS AND METHODS
Cloning, expression, and protein purification
The open reading frame coding for the His-tagged Trm10 recombinant protein and shortened versions, corresponding to residues 1–344 (TkTrm10Δ26) and 97–272 (SPOUT domain), were cloned in pET28b and expressed in E. coli strain Rosetta (DE3) (Novagen). Freshly transformed cells were grown to an OD600 of 0.5–0.6 at 37°C in 1 L of Luria broth. One millimolar IPTG (isopropyl-β-d-thiogalactopyranoside) was then added to induce recombinant protein expression (37°C, 3 h). Cells were harvested, and the pellet was resuspended in 40 mL of buffer A (Tris-HCl 50 mM pH 8.0, 1 M NaCl) prior to cell disruption by sonication. The lysate was cleared by centrifugation (12,000 rpm, 20 min), and heat-treated for 30 min at 70°C. After centrifugation (12,000 rpm, 20 min), the cleared supernatant was applied to a Chelating-Sepharose fast flow column charged with Ni2+ previously equilibrated with buffer A. The column was washed with the same buffer, and the protein was eluted with a linear gradient (150 mL; from 0 to 0.5 M) of imidazole in buffer A. The fractions containing TkTrm10 were pooled. Further, the protein was passed through an S75 (10/300) Superdex column preequilibrated in 50 mM Tris, 1 M NaCl pH 8.0, and the homogeneous protein fractions at the peak of interest were pooled. After addition of 2 mM DTT and 10% glycerol the protein was flash frozen.
The genes resulting in the chimeric proteins were synthesized by GeneArt. All the described variants of the TKTrm10 protein were generated by site-directed mutagenesis using the QuikChange Kit (Agilent) and oligonucleotides obtained from Sigma-Aldrich.
T7 in vitro transcription of tRNA genes
Plasmid pUC18-tRNAAsp (G9A) was generated by site directed mutagenesis (QuikChange Kit of Agilent) and oligonucleotides (Sigma-Aldrich) using pUC18-tRNAAsp as a template (Kempenaers et al. 2010). Large scale tRNA production was performed according to the instructions of the RiboMAX Kit of Promega. The tRNA transcripts were purified using G-25 columns (GE Healthcare).
tRNA MTase assays and kinetic analysis
A semi-quantitative method to follow tRNA methylation consisted in measuring the amount of 14C transferred to total E. coli tRNA using adenosyl-l-methionine,S-[methyl-14C] as the methyl donor. The reaction mixture (300 µL) consisted of 50 mM Tris-HCl pH 8.0, 400 mM NaCl, 5 mM MgCl2, 10 mM DTT, 60 µg total tRNA, 25 nCi adenosyl-l-methionine, S-[methyl-14C] (50 mCi/mmol; PerkinElmer) and enzyme. The solution was incubated for 30 min at 60°C. The reaction was stopped by phenol extraction and the nucleic acids were TCA-precipitated. Radioactive methylated tRNA was captured on a Whatman GF/C filter and washed three times with ethanol prior to the measurement of radioactivity in a scintillation counter. For 2D-TLC, the tRNA was ethanol precipitated after phenol extraction and thereafter hydrolyzed by P1 nuclease, and autoradiography was performed as described in Kempenaers et al. (2010). All measurements were performed in triplicate. Values are given as the mean (±SD).
Steady-state kinetic analyses were based on the procedure described by Hou and Masuda (2015). In 50 µL of reaction buffer, containing 50 mM Tris·HCl, pH 8, 5 mM MgCl2 and 10 mM DTT, either various concentrations of the tRNA transcript were mixed with a saturating concentration of adenosyl-l-methionine, S-[methyl-3H] (20 µM; 2600 dpm/pmol), or various concentrations of adenosyl-l-methionine,S-[methyl-3H] (2600 dpm/pmol) were mixed with a saturating concentration of the tRNA transcript (4 µM). The reaction was started by addition of the T. kodakaraensis Trm10 enzyme at a concentration of 220 µM that was optimized to ensure linear increase in product formation during the entire time of the assay. After 15 min of incubation at 60°C the reaction was stopped by phenol extraction, the nucleic acids were precipitated by 5% (wt/vol) trichloroacetic acid, captured on a Whatman GF/C filter and washed three times with ethanol. The amount of methyl transfer was quantified using a scintillation counter. Each data point is the average (±SEM) of three independent measurements, except otherwise stated. The Michaelis–Menten equation was fitted using GraphPad prism 7 software to determine KM (±SE) and kcat (±SE)
Electrophoretic mobility shift assay (EMSA)
A total of 2 × 104 cpm of radioactive [α32P]ATP transcript from tRNAAsp of T. kodakaraensis was incubated in the presence of various quantities of enzyme in 50 mM Tris pH 8.0, 5 mM MgCl2, 5 mM DTT and 1 µg of polyC, at 60°C for 30 min (Kempenaers et al. 2010). The binding reaction (with a total volume of 20 µL) was stopped by the addition of 4 µL of stop solution (0.05% bromophenol blue in 30% glycerol), and the mixture was separated by polyacrylamide gel electrophoresis (on a 6% PAGE gel with dimensions 190 mm × 160 mm × 1.5 mm) in TB buffer at room temperature. The electrophoresis was performed at a voltage of 180 V until the samples entered the gel and further at 150 V until the end of the run (1½ h) and autoradiography was performed overnight.
Isothermal titration calorimetry
Binding affinities of wild-type and mutant TkTrm10 for SAM were determined by isothermal titration calorimetry (ITC), using the MicroCal iTC200 system (GE Healthcare) with a reference power of 10 microcal/sec. All measurements were performed in 50 mM Tris pH 8.0, 1 M NaCl at 25°C. The ligand SAM was titrated from the injection syringe to the protein (TkTrm10) solution in the sample cell. Each experiment consisted of one test injection of 0.5 µL (not included in analysis) and 20 injections of 2 µL of SAM with a duration of 4 sec. The stirring speed was 750 rpm and the interval between injections was 120 sec. The protein concentrations in the cell were 122 µM (wild-type TkTrm10), 130 µM (D206A/D245A), 130 µM (D206N/D245N), 300 µM (D206L), and 150 µM (D245L). The corresponding SAM concentrations in the syringe were 1.2 mM (wild-type TkTrm10), 1.5 mM (D206A/D245A), 2 mM (D206N/D245N), 4mM (D206L), and 1.5 mM (D245L). Origin 7.0 was used for data integration and fitting to a single binding site model using the standard Marquardt nonlinear regression method as provided in the Microcal Origin routines.
Crystallization of TkTrm10 constructs
Initial crystallization screens of the different TkTrm10 constructs were performed using a vapor diffusion sitting drop setup using various Hampton Crystallization screen kits with a PHENIX crystallization robot.
For crystallization of the TkTrm10Δ26 construct, the protein solution was concentrated to 14 mg/mL. 1.5 µL of protein solution was mixed with an equal volume of the crystallization solution in a sitting drop vapor diffusion crystallization plate at 293 K. Single crystals were obtained in a crystallization condition consisting of 18% PEG3350 and 0.1 M Tris pH 7.5 after several months. Crystals were harvested and flash cooled with liquid nitrogen in a cryoprotectant reagent containing the mother liquor supplemented with 20% PEG 200.
Crystals of the SPOUT domain of TkTrm10 (in which Cys120 was replaced by Ala to facilitate crystallization) in apo form were obtained at a protein concentration of 15 mg/mL using a crystallization solution containing 20% PEG 3350 and 8%v/v Tascimate pH 5.0. Crystals of the SAM- or SAH-bound SPOUT domain were obtained by preincubating the protein solution with 2 mM of SAH/SAM and using a crystallization buffer containing 10% PEG6000, 0.2 M LiCl, and 0.1 M NaAc pH 5.5. All the crystals were cryo-protected by the addition of 22%–25% glycerol to the mother liquor. For the ligand-bound protein the cryoprotectant was also supplemented with either 1 mM of SAM or SAH
Data collection and refinement
For the TkTrm10Δ26 crystals, initial data were collected at 2.3 Å resolution on a Rigaku MicroMax-007 HF home source. The data set was processed with XDS and XscALE (Kabsch 2010). The structure was solved via molecular replacement using PHASER within the PHENIX software package (Adams et al. 2010), and using residues 1–247 of the S. acidocaldarius Trm10 structure (PDB code 5a7y) as search model. Additional model building was performed using the Phenix Autobuild tool of the PHENIX software package (Adams et al. 2010). The resulting model from the phenix.autobuild contained 207 amino acid residues in 8 fragments. The model was further manually built with COOT (Emsley and Cowtan 2004), alternated with refinement in PHENIX.REFINE. The model was subsequently refined against a higher resolution data set collected at 100 K at beamline Proxima 2A of the Soleil synchrotron (France) on the same crystal, making sure to use the same set of reflections for cross-validation. TLS refinement was implemented in the refinement steps, using three individual TLS groups (Painter and Merritt 2006). This model was further refined at a resolution of 2.0 Å with a final Rwork/Rfree value of 0.19/0.24. This final model was used for further analysis.
Data collection on the crystals of the TkSPOUT domain, either in the apo form or bound to SAM or SAH, was performed at beamline Proxima 2A of the Soleil synchrotron (France). The phase problem for TkSPOUT_SAM and TkSPOUT_APO was solved via molecular replacement using PHASER (McCoy 2006; McCoy et al. 2007) within the PHENIX software package and using the SPOUT domain of the TkTrm10Δ26 structure as a search model, while the TkSPOUT_SAM structure (without ligands) was used as a search model to solve the structure of TkSPOUT_SAH. TLS refinement was implemented for each structure with 8, 13, and 10 TLS group for TkSPOUT_APO, TkSPOUT_SAM, and TkSPOUT_SAH, respectively. TkSPOUT_APO, TkSPOUT_SAM, and TkSPOUT_SAH were refined at a resolution of 1.8 Å, 2.3 Å, and 2.9 Å resolution and with a final Rwork/Rfree value of 0.19/0.21, 0.19/0.22, and 0.22/0.28, respectively.
Substrate docking
The crystal structure of TkSPOUT_SAM was preprocessed with AutoDock tools to remove all the solvent molecules (Morris and Huey 2009). SAM was kept at its original position during the docking procedure. Polar hydrogen and Kollman charges were added and the files were converted to PDBQT format. The coordinates of guanosine were obtained from the ternary complex of H. influenza TrmD in complex with sinefungin and substrate tRNA (G37 of tRNA) (PDB ID 4yvi); the coordinates of adenosine were obtained from the ternary complex of the human m1A58 methyltransferase with SAH and tRNA (A58 of tRNA) (PDB ID 5CCB). These coordinates were also preprocessed with Autodock tools, and polar hydrogen was added before the autodock experiment. During the docking process, the torsional angle around C1′-N9 was allowed to rotate. The docking space was constrained by defining the grid space covering the active site cavity. The conformation of active site residues was kept rigid during the procedure. The calculations were performed using a Lamarckian genetic algorithm and a maximum of 10 conformations. The docking result with minimum energy is used to show the interaction with the active site.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank the staff of the Beamline Proxima II of the Soleil Synchrotron (France). This work was supported by grants of Fonds Wetenschappelijk Onderzoek-Vlaanderen (R.K.S. and W.V.), Hercules foundation, a Strategic Research Program Financing of the VUB (W.V.), BioStruct-X by the European Community's Seventh Framework Programme (W.V.), Fonds D. et A. Van Buuren and the Fonds J. Brachet (L.D.). We also acknowledge M. Kempenaers and J. Somme for their contribution to the early stages of this work.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.064345.117.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66: 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Koonin EV, Aravind L. 2002. SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. J Mol Microbiol Biotechnol 4: 71–75. [PubMed] [Google Scholar]
- Benítez-Páez A, Villarroya M, Douthwaite S, Gabaldón T, Armengod ME. 2010. YibK is the 2′-O-methyltransferase TrmL that modifies the wobble nucleotide in Escherichia coli tRNALeu isoacceptors. RNA 16: 2131–2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benítez-Páez A, Villarroya M, Armengod M-E. 2012. The Escherichia coli RlmN methyltransferase is a dual-specificity enzyme that modifies both rRNA and tRNA and controls translational accuracy. RNA 18: 1783–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björk GR, Kerstin J, Kristina N, Johansson JO, Marcus Byström AS, Persson OP. 2001. A primordial tRNA modification required for the evolution of life? EMBO J 20: 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick JB, Duffus BR, Duschene KS, Shepard EM. 2014. Radical S-adenosylmethionine enzymes. Chem Rev 114: 4229–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byström AS, Björk GR. 1982. Chromosomal location and cloning of the gene (trmD) responsible for the synthesis of tRNA (m1G) methyltransferase in Escherichia coli K-12. Mol Gen Genet 188: 440–446. [DOI] [PubMed] [Google Scholar]
- Cantara WA, Crain PF, Rozenski J, McCloskey JA, Harris KA, Zhang X, Vendeix FAP, Fabris D, Agris PF. 2011. The RNA modification database, RNAMDB: 2011 update. Nucleic Acids Res 39: D195–D201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian T, Hou Y-M. 2007. Distinct determinants of tRNA recognition by the TrmD and Trm5 methyl transferases. J Mol Biol 373: 623–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerwoniec A, Kasprzak JM, Kaminska KH, Rother K, Purta E, Bujnicki JM. 2009. Folds and functions of domains in RNA modification enzymes. In DNA and RNA modification enzymes: structure, mechanism, function and evolution (ed. Grosjean H), pp. 289–302. Landes Bioscience, Austin, Texas. [Google Scholar]
- Dunstan MS, Hang PC, Zelinskaya NV, Honek JF, Conn GL. 2009. Structure of the thiostrepton resistance methyltransferase•S-adenosyl-l-methionine complex and its interaction with ribosomal RNA. J Biol Chem 284: 17013–17020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkins PA, Watts JM, Zalacain M, Van Thiel A, Vitazka PR, Redlak M, Andraos-Selim C, Rastinejad F, Holmes WM. 2003. Insights into catalysis by a knotted TrmD tRNA methyltransferase. J Mol Biol 333: 931–949. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- Ero R, Peil L, Liiv A, Remme J. 2008. Identification of pseudouridine methyltransferase in Escherichia coli. RNA 14: 2223–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzmann J, Frank P, Lö E, Bennett KL, Gerner C, Rossmanith W. 2008. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell 135: 462–474. [DOI] [PubMed] [Google Scholar]
- Hori H. 2017. Transfer RNA methyltransferases with a SpoU-TrmD (SPOUT) fold and their modified nucleosides in tRNA. Biomolecules 7: 1–24. [Google Scholar]
- Hou YM, Masuda I. 2015. Kinetic analysis of tRNA methyltransferases. Methods Enzymol 560: 91–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman JE, Alfonzo JD. 2013. Transfer RNA modifications: nature's combinatorial chemistry playground. Wiley Interdiscip Rev RNA 4: 35–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman JE, Montange RK, Malik HS, Phizicky EM. 2003. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA 9: 574–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. 2010. Xds. Acta Crystallogr D Biol Crystallogr 66: 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempenaers M, Roovers M, Oudjama Y, Tkaczuk KL, Bujnicki JM, Droogmans L. 2010. New archaeal methyltransferases forming 1-methyladenosine or 1-methyladenosine and 1-methylguanosine at position 9 of tRNA. Nucleic Acids Res 38: 6533–6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Miyauchi K, Ikeuchi Y, Thiaville PC, De Crécy-Lagard V, Suzuki T. 2014. Discovery of the β-barrel–type RNA methyltransferase responsible for N6-methylation of N6-threonylcarbamoyladenosine in tRNAs. Nucleic Acids Res 42: 9350–9365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamohan A, Jackman JE. 2017. Mechanistic features of the atypical tRNA m1G9 SPOUT methyltransferase, Trm10. Nucleic Acids Res 41: 9019–9029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuratani M, Bessho Y, Nishimoto M, Grosjean H, Yokoyama S. 2008. Crystal structure and mutational study of a unique SpoU family archaeal methylase that forms 2′-O-methylcytidine at position 56 of tRNA. J Mol Biol 375: 1064–1075. [DOI] [PubMed] [Google Scholar]
- Lövgren JM, Wikström PM. 2001. The rlmB gene is essential for formation of Gm2251 in 23S rRNA but not for ribosome maturation in Escherichia coli. J Bacteriol 183: 6957–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka MA, Milanowska K, Oglou OO, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. 2013. MODOMICS: A database of RNA modification pathways—2013 update. Nucleic Acids Res 41: 262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ. 2006. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr 63: 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer B, Wurm JP, Sharma S, Immer C, Pogoryelov D, Kötter P, Lafontaine DLJ, Wöhnert J, Entian KD. 2016. Ribosome biogenesis factor Tsr3 is the aminocarboxypropyl transferase responsible for 18S rRNA hypermodification in yeast and humans. Nucleic Acids Res 44: 4304–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris G, Huey R. 2009. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30: 2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y, Grosjean H. 1999. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA 5: 1105–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nureki O, Watanabe K, Fukai S, Ishii R, Endo Y, Hori H, Yokoyama S. 2004. Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme. Structure 12: 593–602. [DOI] [PubMed] [Google Scholar]
- Oerum S, Dégut C, Barraud P, Tisné C. 2017a. m1A post-transcriptional modification in tRNAs. Biomolecules 7: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oerum S, Roovers M, Leichsenring M, Acquaviva-Bourdain C, Beermann F, Gemperle-Britschgi C, Fouilhoux A, Korwitz-Reichelt A, Bailey HJ, Droogmans L, et al. 2017b. Novel patient missense mutations in the HSD17B10 gene affect dehydrogenase and mitochondrial tRNA modification functions of the encoded protein. Biochim Biophys Acta 1863: 3294–3302. [DOI] [PubMed] [Google Scholar]
- Painter J, Merritt EA. 2006. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr 62: 439–450. [DOI] [PubMed] [Google Scholar]
- Persson BC, Jäger G, Gustafsson C. 1997. The spoU gene of Escherichia coli, the fourth gene of the spoT operon, is essential for tRNA (Gm18) 2′-O-methyltransferase activity. Nucleic Acids Res 25: 4093–4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK. 2010. tRNA biology charges to the front. Genes Dev 24: 1832–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purta E, van Vliet F, Tkaczuk KL, Dunin-Horkawicz S, Mori H, Droogmans L, Bujnicki JM. 2006. The yfhQ gene of Escherichia coli encodes a tRNA:Cm32/Um32 methyltransferase. BMC Mol Biol 7: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Douthwaite S. 2008. YbeA is the m3Ψ methyltransferase RlmH that targets nucleotide 1915 in 23S rRNA. RNA 14: 2234–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read RJ, Adams PD, Arendall WB III, Brunger AT, Emsley P, Joosten RP, Kleywegt GJ, Krissinel EB, Lütteke T, Otwinowski Z, et al. 2011. A new generation of crystallographic validation tools for the Protein Data Bank. Structure 19: 1395–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roovers M, Wouters J, Bujnicki JM, Tricot C, Stalon V, Grosjean H, Droogmans L. 2004. A primordial RNA modification enzyme: the case of tRNA (m1A) methyltransferase. Nucleic Acids Res 32: 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert HL, Blumenthal RM, Cheng X. 2003. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci 28: 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Yan W, Peng J, Zuo X, Zou Y, Li F, Gong D, Ma R, Wu J, Shi Y, et al. 2014. Crystal structure of tRNA m1G9 methyltransferase Trm10: insight into the catalytic mechanism and recognition of tRNA substrate. Nucleic Acids Res 42: 509–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somme J, Van Laer B, Roovers M, Steyaert J, Versées W, Droogmans L. 2014. Characterization of two homologous 2′-O-methyltransferases showing different specificities for their tRNA substrates. RNA 20: 1257–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinehart WE, Henderson JC, Jackman JE. 2013. Unexpected expansion of tRNA substrate recognition by the yeast m1G9 methyltransferase Trm10. RNA 19: 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkaczuk K, Dunin-Horkawicz S, Purta E, Bujnicki JM. 2007. Structural and evolutionary bioinformatics of the SPOUT superfamily of methyltransferases. BMC Bioinformatics 8: 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Laer B, Roovers M, Wauters L, Kasprzak JM, Dyzma M, Deyaert E, Kumar Singh R, Feller A, Bujnicki JM, Droogmans L, et al. 2016. Structural and functional insights into tRNA binding and adenosine N1-methylation by an archaeal Trm10 homologue. Nucleic Acids Res 44: 940–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardo E, Rossmanith W. 2015. Molecular insights into HSD10 disease: impact of SDR5C1 mutations on the human mitochondrial RNase P complex. Nucleic Acids Res 43: 5112–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardo E, Nachbagauer C, Buzet A, Taschner A, Holzmann J, Rossmanith W. 2012. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase-extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res 40: 11583–11593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F, Fujimori DG. 2011. RNA methylation by radical SAM enzymes RlmN and Cfr proceeds via methylene transfer and hydride shift. Proc Natl Acad Sci 108: 3930–3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young BD, Weiss DI, Zurita-Lopez CI, Webb KJ, Clarke SG, McBride AE. 2012. Identification of methylated proteins in the yeast small ribosomal subunit: a role for SPOUT methyltransferases in protein arginine methylation. Biochemistry 51: 5091–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelinskaya N, Witek MA, Conn GL. 2015. The pathogen-derived aminoglycoside resistance 16S rRNA methyltransferase NpmA possesses dual m1A1408/m1G1408 specificity. Antimicrob Agents Chemother 59: 7862–7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.