Abstract
Background Steroid-sensitive nephrotic syndrome (SSNS) is a childhood disease with unclear pathophysiology and genetic architecture. We investigated the genomic basis of SSNS in children recruited in Europe and the biopsy-based North American NEPTUNE cohort.
Methods We performed three ancestry-matched, genome-wide association studies (GWAS) in 273 children with NS (Children Cohort Nephrosis and Virus [NEPHROVIR] cohort: 132 European, 56 African, and 85 Maghrebian) followed by independent replication in 112 European children, transethnic meta-analysis, and conditional analysis. GWAS alleles were used to perform glomerular cis-expression quantitative trait loci studies in 39 children in the NEPTUNE cohort and epidemiologic studies in GWAS and NEPTUNE (97 children) cohorts.
Results Transethnic meta-analysis identified one SSNS-associated single-nucleotide polymorphism (SNP) rs1063348 in the 3′ untranslated region of HLA-DQB1 (P=9.3×10−23). Conditional analysis identified two additional independent risk alleles upstream of HLA-DRB1 (rs28366266, P=3.7×10−11) and in the 3′ untranslated region of BTNL2 (rs9348883, P=9.4×10−7) within introns of HCG23 and LOC101929163. These three risk alleles were independent of the risk haplotype DRB1*07:01-DQA1*02:01-DQB1*02:02 identified in European patients. Increased burden of risk alleles across independent loci was associated with higher odds of SSNS. Increased burden of risk alleles across independent loci was associated with higher odds of SSNS, with younger age of onset across all cohorts, and with increased odds of complete remission across histologies in NEPTUNE children. rs1063348 associated with decreased glomerular expression of HLA-DRB1, HLA-DRB5, and HLA-DQB1.
Conclusions Transethnic GWAS empowered discovery of three independent risk SNPs for pediatric SSNS. Characterization of these SNPs provide an entry for understanding immune dysregulation in NS and introducing a genomically defined classification.
Keywords: nephrotic syndrome, genetic renal disease, gene expression, focal segmental glomerulosclerosis, pediatrics, genome-wide association study
Although a rare disease (incidence is two to seven patients per 100,000 children), idiopathic nephrotic syndrome (NS) is the most common glomerular disorder of childhood. The disease is acquired and mostly affects children younger than 10 years of age.1–3 There is substantial variability in pediatric idiopathic NS with regard to its ethnic variations, histologic appearance on light microscopy, and response to initial treatment with corticosteroids at standard doses.1–3 The majority of children have a form of steroid-sensitive nephrotic syndrome (SSNS) that generally has a good long-term renal prognosis compared with those with steroid-resistant NS. The most common glomerular histologies are minimal change disease and FSGS, with FSGS associated with decreased chances of complete remission, although these associations are not absolute.4 The reasons for this interindividual variability remain unknown.5
Immune dysregulation has been invoked as the major cause of SSNS, with the concept of podocyte damage as a secondary event in a primarily immune disease. This is supported by the full reversibility of proteinuria and foot process effacement within a few days of oral prednisone, the efficiency of immunosuppressive drugs targeting T and B cells to prevent relapses,6,7 and the association with other immune disorders, such as asthma and atopy.8 However, the morphologic changes in the glomerulus in SSNS (i.e., podocyte foot process effacement) may, in some patients, derive from a primarily intrinsic glomerular dysregulation.9–11
Genetic research in SSNS has historically focused on targeted analysis of candidate immune mediators and pathways, including permeability factor(s).12 These studies have had a number of limitations, including limited genotyping of candidate alleles and lack of replication (reviewed in ref. 13). Despite rare reports,14–16 the vast majority of patients with sporadic SSNS and patients with familial SSNS do not have a Mendelian mutation that explains their disease.17 A recent association study of 25,000 common exonic variants discovered two nonsynonymous single-nucleotide polymorphisms (SNPs) in HLA-DQA1 (in complete linkage disequilibrium [LD]) associated with SSNS in 214 Sri Lankan pediatric patients.18 Thus, SSNS seems to be a pathogenetically complex disease influenced by a complex interplay between host genetic risk factors and environmental stimuli.3,19 However, a genome-wide association study (GWAS) of SSNS in children has not yet been reported.
Here, we performed a transethnic GWAS of SSNS in children recruited in a French discovery cohort (NEPHROVIR) followed by replication in an Italian/Spanish cohort (ItSpa), meta-analysis, and conditional analysis. We then studied the significant risk alleles in children enrolled in the renal biopsy–based North American Nephrotic Syndrome Study Network (NEPTUNE) cohort to characterize their transcriptomic and clinical associations.
Methods
Patients
GWAS Cohorts
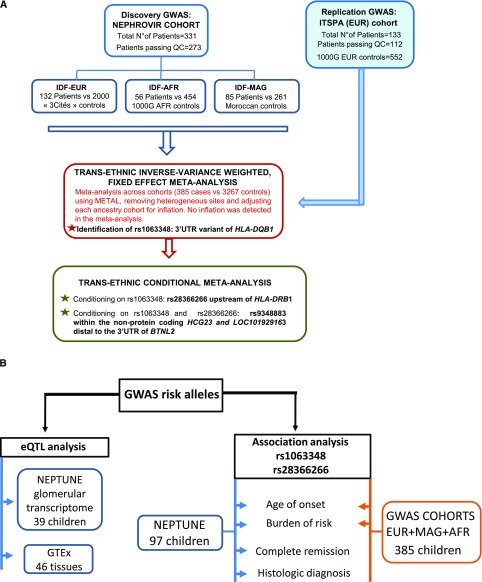
Patients with SSNS were enrolled in France, Italy, and Spain (Supplemental Material has descriptions and definitions). The discovery NEPHROVIR cohort (331 patients) was exclusively recruited in the Paris area (Ile-de-France),19 characterized by a high rate of multiethnic migrants, between December 2007 and January 2016. The replication Italian and Spanish cohort comprised 133 European whites (Figure 1) recruited between 2010 and 2016. The ethnically matched control cohorts are indicated in Figure 1A and Table 1.20,21
Figure 1.
Flowcharts. (A) Main outline of the sequential genome-wide association study (GWAS) strategy and transethnic analysis. (B) Flowchart of epidemiologic analyses. eQTL, expression quantitative trait loci; IDF-AFR, Ile-de-France African; IDF-EUR, Ile-de-France European; IDF-MAG, Ile-de-France Maghrebian; NEPHROVIR, Children Cohort Nephrosis and Virus; QC, quality control; NEPTUNE, Nephrotic Syndrome Study Network; 3′ UTR, 3′ untranslated region.
Table 1.
Demographic description of the NEPHROVIR and the ItSpa cohorts and the controls
| Characteristics | NEPHROVIR Patients | ItSpa Patients | 3 Cités Controls | 1000G EUR Controls | 1000G AFR Controls | Moroccan Controls |
|---|---|---|---|---|---|---|
| N of samples passing QC | 273 | 112 | 2000 | 552 | 454 | 261 |
| Sex | ||||||
| Men | 176 | 75 | 802 | 262 | 232 | 141 |
| Women | 97 | 37 | 1198 | 290 | 222 | 120 |
| Sex ratio, men/women | 1.81 | 2.02 | 0.67 | 0.9 | 1.05 | 1.18 |
| Age at diagnosis, yra | 4.30 [3.00–6.90] | 3.35 [2.47–5.0] | NA | NA | NA | NA |
| Ethnicity | ||||||
| Europe | 132 | 112 | 2000 | 552 | 0 | 0 |
| Maghreb | 85 | 0 | 0 | 0 | 0 | 261 |
| Africa | 56 | 0 | 0 | 0 | 454 | 0 |
The initial NEPHROVIR and ItSpa cohorts comprised 331 and 133 patients, respectively. Samples from 273 and 112 patients from these cohorts passed QC, respectively. NEPHROVIR, Children Cohort Nephrosis and Virus; ItSpa, Italian and Spanish; 1000G, 1000 Genomes project; EUR, European; AFR, African; QC, quality control; NA, not available.
Median [interquartile range].
Systems Genetics Cohort
NEPTUNE is a North American longitudinal cohort of patients with NS who underwent a clinically indicated kidney biopsy.22 Patients were not enrolled on the basis of age, response to immunosuppression, or histologic diagnosis. Blood was collected for whole-genome sequencing, an extra research biopsy core was collected for histologic examination and transcriptomics, and baseline and longitudinal phenotypic data were collected, including achievement of complete remission of proteinuria (UPCR≤300 mg/g at any visit). There were 97 pediatric patients eligible for epidemiologic studies; 39 of those were eligible for glomerular expression quantitative trait loci (eQTL) analysis (Supplemental Table 1).
Genotyping, Ancestry Matching, and Imputation
For the GWAS cohorts, isolation of DNA, genotyping, and quality control (QC) were performed with the use of standard procedures, and ancestry-matched controls were defined using standard approaches (Supplemental Figure 1, Supplemental Material). DNA from French patients (the NEPHROVIR cohort) and Maghrebian controls was analyzed with Illumina Human OmniExpress arrays. DNA from ItSpa patients was analyzed with Illumina Omni 2.5 arrays. Hybridization was performed by IntegraGen (Evry, France) using standard protocols, and genotype calls were obtained using Illumina Genome Studio software. Stringent QCs were applied to both SNPs and samples to remove any source of spurious association signal.
We imputed patients and controls together using only QC-passed SNPs on the Sanger Imputation Server.23 We used the Haplotype Reference Consortium panel as reference for European and Maghrebian cohorts23 and the 1000 Genomes project phase 3 data for the African cohort.21 Eligible children from the NEPTUNE cohort had previously undergone low-depth whole-genome sequencing, and this dataset was used for targeted lookups of GWAS alleles. One SNP did not pass quality metrics in the NEPTUNE data; it was imputed with high confidence (Supplemental Material).
GWAS
The GWAS were carried out using PLINK v1.07 under an additive genotypic model and using standard approaches (Supplemental Figure 2, Supplemental Material).24 Because the NEPHROVIR patients were of diverse ethnic origins, we performed ancestry-specific GWAS of 132 European (Ile-de-France European [IDF-EUR]) patients, 56 African (Ile-de-France African [IDF-AFR]) patients, and 85 Maghrebian (Ile-de-France Maghrebian [IDF-MAG]) patients. The threshold for significance was P<5×10−8. The replication phase consisted of a single SNP lookup of the lead NEPHROVIR-EUR SNP in the ItSpa cohort, with a threshold for significance of P<0.05 (Figure 1).
We then performed a transethnic, inverse variance–weighted, fixed effects meta-analysis across all four cohorts (385 patients versus 3267 controls) using METAL.25 After running METAL, we assessed heterogeneity using Cochran Q, and we considered a site to be heterogeneous if the Cochran Q test had a P value <0.10, a standard threshold for this test.26,27 We then removed these sites from consideration in the GWAS. The transethnic meta-analysis had a genome-wide significance threshold of 5×10−8.
In each ancestry cohort, stepwise conditional analysis at the lead GWAS SNP was performed using GCTA28 followed by transethnic meta-analysis with METAL as above. To account for genomic inflation in each ancestral cohort separately, we calculated λ by the median chi-squared statistic across all markers divided by 0.455. We then adjusted the summary statistics by this λ in each cohort when it was >1.0. P values were then calculated with these adjusted test statistics.29 For the first conditional analysis, we used a genome-wide significance threshold of 5×10−8 given the possibility that there could perhaps be some long-range translinkage with the lead GWAS SNP. Subsequent conditional analyses used an empirical significance threshold solely on the basis of the regions considered for association (additional details are in Results).
HLA Allelotype Analysis
SNP2HLA software was used to impute HLA allelotypes from SNP data in our two European patient-control cohorts (IDF-EUR versus 3 Cités and ItSpa versus the 1000 Genomes project European) using the T1DGC reference panel.30 The R package haplostats was then used to reconstruct HLA haplotypes. A logistic regression model was used to identify allelotypes or haplotypes enriched in patients, incorporating the first three principal components of genetic ancestry to correct for fine ethnic differences. Independence between the risk HLA haplotype and the three significant GWAS SNPs was tested by logistic regression.
eQTL Analysis (the NEPTUNE and the GTEx)
Glomerular-Specific eQTL
Affymetrix 2.1 ST array–quantitated transcriptomic data were generated from microdissected glomeruli.31 Genotypes were generated as previously described. A cis-eQTL study (genes within 1 Mb of the GWAS SNPs) was performed in 39 children from the NEPTUNE with these paired datasets (Supplemental Material).
Multitissue eQTL
Publicly available eQTL data from the GTEx portal (http://gtexportal.org; obtained 07/17) were analyzed to determine if any of the lead SSNS SNPs were eQTLs in any of the 46 available tissues included in this database, including immune cells.
Epidemiologic Studies
GWAS Cohorts
To determine the effect of combinations of risk alleles across independent GWAS loci, we used Fisher exact tests to compare the odds of specific risk-SNP combinations between patients and controls. Independent risk alleles were also tested for association with age of onset both individually and in combination (Supplemental Material).
NEPTUNE
Association of risk alleles with clinical and demographic traits was studied. In univariate and multivariate testing, independent risk alleles were tested for association with age of onset and achievement of complete remission both individually and in combination (Supplemental Material).
Results
GWAS Study Participants
NEPHROVIR included 331 patients, and the ItSpa cohort included 133 patients. Among those, 273 and 112 patients passed QC, respectively (Figure 1). After stringent QCs, we retained between 505,979 and 1,307,537 high-quality SNPs in each series. Demographic and clinical phenotypes were similar among these cohorts (Table 1). All samples of the ItSpa cohort were of European ancestry (Supplemental Figure 1, Supplemental Table 2).
NEPHROVIR Discovery Cohort
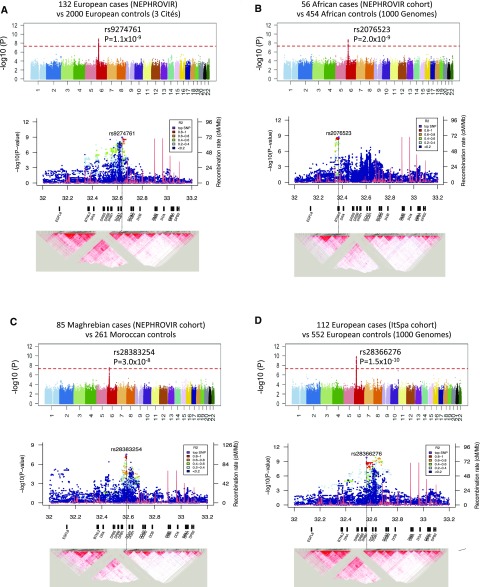
In the IDF-EUR cohort, the lead SNP (rs9274761; P=1.1×10−9; odds ratio [OR], 2.7; 95% confidence interval [95% CI] 2.0–3.8) was located in the HLA class 2 region between HLA-DQB1 and HLA-DQA2 (Figure 2A, Supplemental Table 3). In the IDF-AFR cohort, the lead SNP (rs2076523; P=2.0×10−9 OR, 4.8; 95% CI, 2.9–7.9) localized to the telomeric end of the HLA class 2 region was a missense variant K196E in exon 3 of butyrophilin like-2 (BTNL2) (Figure 2B, Supplemental Table 4). In the IDF-MAG cohort, a single SNP was discovered (rs28383254; P=3.0×10−8; OR, 5.7; 95% CI, 3.1–10.6) between HLA-DRB1 and HLA-DQA1 (Figure 2C, Supplemental Table 5).
Figure 2.
Manhattan plots and haploblock reconstruction of the HLA class 2 region for genome-wide association studies of idiopathic steroid-sensitive nephrotic syndrome in four groups of patients and racially matched controls. The upper panels in A–D show the Manhattan plots, and the corresponding lower panels in A–D represent the haploblock reconstruction of the HLA class 2 region with the log-transformed P value on the y axis and the genomic position (in megabases) on the x axis. The top single-nucleotide polymorphism (SNP) is indicated in purple, and the SNPs in linkage disequilibrium with the top SNP are represented with the color code indicated in the box. The recombination rates across the region are also represented as a red line, and the different HLA genes are indicated below each plot. A, upper panel shows the Manhattan plot for 132 European patients (the NEPHROVIR cohort) and 2000 controls from the French 3 Cités cohort. B, upper panel shows the plot for 56 African patients (the NEPHROVIR cohort) and 454 racially matched controls from the 1000 Genomes project. C, upper panel shows the plot for 85 Maghrebian patients (the NEPHROVIR cohort) and 261 controls from Maghreb. D, upper panel shows the plot for 112 patients from southern Europe (Italy and Spain [ItSpa] cohort) and 552 racially matched controls from the 1000 Genomes project. The red horizontal dotted lines indicate the genome-wide significance level (5.0×10−8). The most significant SNP identifications and corresponding P values are indicated in each panel. Lower panels in A–D show that the haploblocks in which the top signals in both European and Maghrebian cohorts reside differ from that from the African cohort. Those in the European and Maghrebian cohorts fall within the haploblock delimited by recombination hotspots at 32.385 and 32.682 Mb, containing the most HLA-DR/DQ. By contrast, the top signals in Africans were located in a haploblock between 32.340 and 32.380 Mb, comprising butyrophilin like-2, HLA Complex Group 23, and the 5′ end of the Chromosome 6 Open Reading Frame 10.
ItSpa Replication Cohort
The lead SNP from the IDF-EUR cohort, rs9274761, replicated in the ItSpa replication cohort (P=2.0×10−7; OR, 3.5; 95% CI, 2.2–5.6). The most significant SNP in the latter cohort (rs28366276; P=1.5×10−10; OR, 5.5; 95% CI, 3.2–9.2) was between HLA-DRB1 and HLA-DQA1 (Figure 2D, Supplemental Table 6).
Per Ancestry Analysis of GWAS Loci at the HLA Class 2 Haploblock
Haploblock reconstruction of the HLA class 2 region revealed that the haploblocks in which the top signals in both European and Maghrebian cohorts reside differ from that of the African cohort (Figure 2).
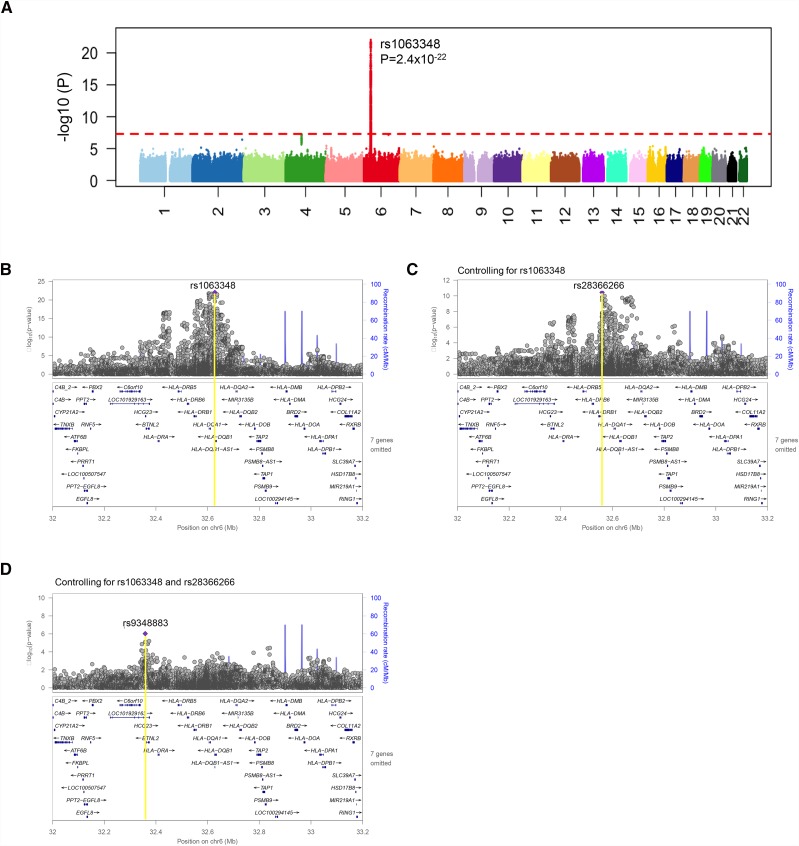
Transethnic Meta-Analysis and Conditional Analysis
Transethnic meta-analysis across the four cohorts (n=3652) (Tables 2 and 3) identified one SNP reaching genome-wide significance in the HLA class 2 region (rs1063348; P=9.3×10−23; OR, 3.33; 95% CI, 2.6–4.2) (Figure 3A, Supplemental Table 7). This SNP is in a 3′ untranslated region of HLA-DQB1 (Figures 3B and 4A). There were two SNPs that reached suggestive levels of significance: rs59882675 (P=5.9×10−8) in an intron of betacellulin on chromosome 4q13 and rs2858829 (P=6.8×10−8) in an intergenic region between Family With Sequence Similarity 26 Member F and Dermatan Sulfate Epimerase on chromosome 6q22.
Table 2.
Significant single-nucleotide polymorphisms, test statistics, and values from the trans-ethnic meta-analysis and conditional analyses
| SNP | Risk Allele | Other Allele | Risk AF | Meta-Analysis across Cohorts, n=3652 (385 Crossethnic Patients; 3267 Crossethnic Controls) | Meta-Analysis, Conditional on rs1063348, n=3652 (385 Crossethnic Patients; 3267 Crossethnic Controls) | Meta-Analysis, Conditional on rs1063348 and rs28366266, n=3652 (385 Crossethnic Patients; 3267 Crossethnic Controls) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR [95% CI] | P Value | Q Test | OR [95% CI] | P Value | Q Test | OR [95% CI] | P Value | Q Test | ||||
| rs1063348 (6:32627923) | Ga | A | 0.60 | 3.33 [2.62 to 4.23] | 9.30E−23 | 0.79 | — | — | — | — | — | — |
| rs28366266 (6:32559753) | Ca | T | 0.17 | 3.03 [2.40 to 3.85] | 4.10E−20 | 0.09 | 2.17 [1.73 to 2.74] | 3.66E−11 | 0.36 | — | — | — |
| rs9348883 (6:32358549) | Ta | A | 0.94 | 2.51 [1.49 to 4.22] | 4.70E−04 | 0.68 | 2.57 [1.53 to 4.31] | 3.40E−04 | 0.81 | 3.5 [2.13 to 5.83] | 9.40E−07 | 0.70 |
Genome-wide association study (GWAS) results for top SNPs of interest: rs1063348, top SNP in meta-analysis across cohorts; rs28366266, top SNP in meta-analysis conditional on rs1063348; and rs9348883, top SNP in meta-analysis conditioned on rs1063348 and rs28366266. For each SNP, we present the GWAS results in the crosspopulation meta-analysis of all individual cohorts taken together (n=3652, including 385 patients and 3267 controls) and the conditional crosspopulation meta-analysis conditioned on rs1063348 alone and then rs1063348 and rs28366266 together. ORs with 95% CIs and P values are reported for each SNP in each analysis. SNP, single-nucleotide polymorphism; AF, allele frequency; OR, odds ratio; 95% CI, 95% confidence interval; —, not tested.
Risk allele.
Table 3.
Summary statistics for the top SNPs from the trans-ethnic and conditional analyses within ancestry specific cohorts
| SNP | African Cohort, n=510 (56 IDF-AFR Patients; 454 1000G-AFR Controls) | European (French) Cohort, n=2132 (132 IDF-EUR Patients; 2000 3 Cités Controls) | Maghrebian Cohort, n=346 (85 IDF-MAG Patients; 261 Moroccan Controls) | European (Replication) Cohort, n=664 (112 ItSpa Patients; 552 1000G-EUR Controls) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR [95% CI]a | P Valueb | Risk AF | OR [95% CI]a | P Valueb | Risk AF | OR [95% CI]a | P Valueb | Risk AF | OR [95% CI]a | P Valueb | Risk AF | |
| rs1063348 (6:32627923) | 3.24 [1.93 to 5.45] | 3.06E−07 | 0.48 | 2.96 [2.04 to 4.29] | 1.17E−08 | 0.61 | 3.62 [1.97 to 6.68] | 3.70E−05 | 0.68 | 4.05 [2.42 to 6.79] | 1.06E−07 | 0.64 |
| rs28366266 (6:32559753) | 3.2 [1.78 to 5.76] | 1.10E−04 | 0.11 | 2.25 [1.59 to 3.2] | 5.64E−06 | 0.17 | 3.53 [1.99 to 6.28] | 1.74E−05 | 0.20 | 4.93 [2.94 to 8.27] | 1.34E−09 | 0.19 |
| rs9348883 (6:32358549) | 5.66 [0.96 to 33.4] | 5.60E−02 | 0.93 | 1.92 [0.82 to 4.59] | 1.30E−01 | 0.95 | 3.95 [0.88 to 17.67] | 6.10E−02 | 0.95 | 2.35 [1.08 to 5.14] | 3.21E−02 | 0.93 |
Genome-wide association study (GWAS) results within ancestry specific cohorts for top SNPs of interest: rs1063348, top SNP in meta-analysis across cohorts; rs28366266, top SNP in meta-analysis conditional on rs1063348; and rs9348883, top SNP in meta-analysis conditioned on rs1063348 and rs28366266. ORs with 95% CIs and P values are reported for each SNP in each analysis. SNP, single-nucleotide polymorphism; IDF-AFR, Ile-de-France African; 1000G-AFR, 1000 Genomes project African; IDF-EUR, Ile-de-France European; IDF-MAG, Ile-de-France Maghrebian; ItSpa, Italian and Spanish; 1000G-EUR, 1000 Genomes project European; OR, odds ratio; 95% CI, 95% confidence interval; AF, allele frequency.
Adjusted SEM by sqrt(λ) from each cohort.
P value adjusted by cohort-specific λ.
Figure 3.
Transethnic meta-analysis and conditional analyses. (A) Manhattan plot for the transethnic meta-analysis. One genome-wide significant single-nucleotide polymorphism (SNP) is identified at 6p21 in the HLA class 2 region (top SNP, rs1063348). There are two SNPs that reach suggestive levels of significance: rs59882675 (P=5.9×10−8) in an intron of betacellulin on chromosome 4q13 and rs2858829 (P=6.8×10−8) in an intergenic region between Family With Sequence Similarity 26 Member F and Dermatan Sulfate Epimerase on chromosome 6q22. (B) Locus zoom on the 6p21 region of the transethnic meta-analysis. The recombination rate across the region is represented as blue lines. The top SNP from this analysis is rs1063348 (6:32627923), with a P value of 9.3×10−23. The lower panel contains genes in this region, with the inclusion of rs1063348 in the 3′ untranslated region of HLA-DQB1. (C) 6p21 conditioned on rs1063348. Transethnic meta-analysis genome-wide association study (GWAS) P values in the 6p21 region conditional on rs1063348 are shown. The top SNP from this analysis is rs28366266 (6:32559753), with a P value of 3.7×10−11. The lower panel shows the location of rs28366266 upstream of HLA-DRB1. (D) 6p21 conditioned on top two significant SNPs. Transethnic meta-analysis GWAS P values in the 6p21 region conditional on both rs1063348 and rs28366266 are shown. The top SNP in this region from this analysis is rs9348883 (6:32358549), with a P value of 9.4×10−7. The lower panel shows the location of rs9348883 within HLA Complex Group 23 and LOC101929163, 3′ of butyrophilin like-2.
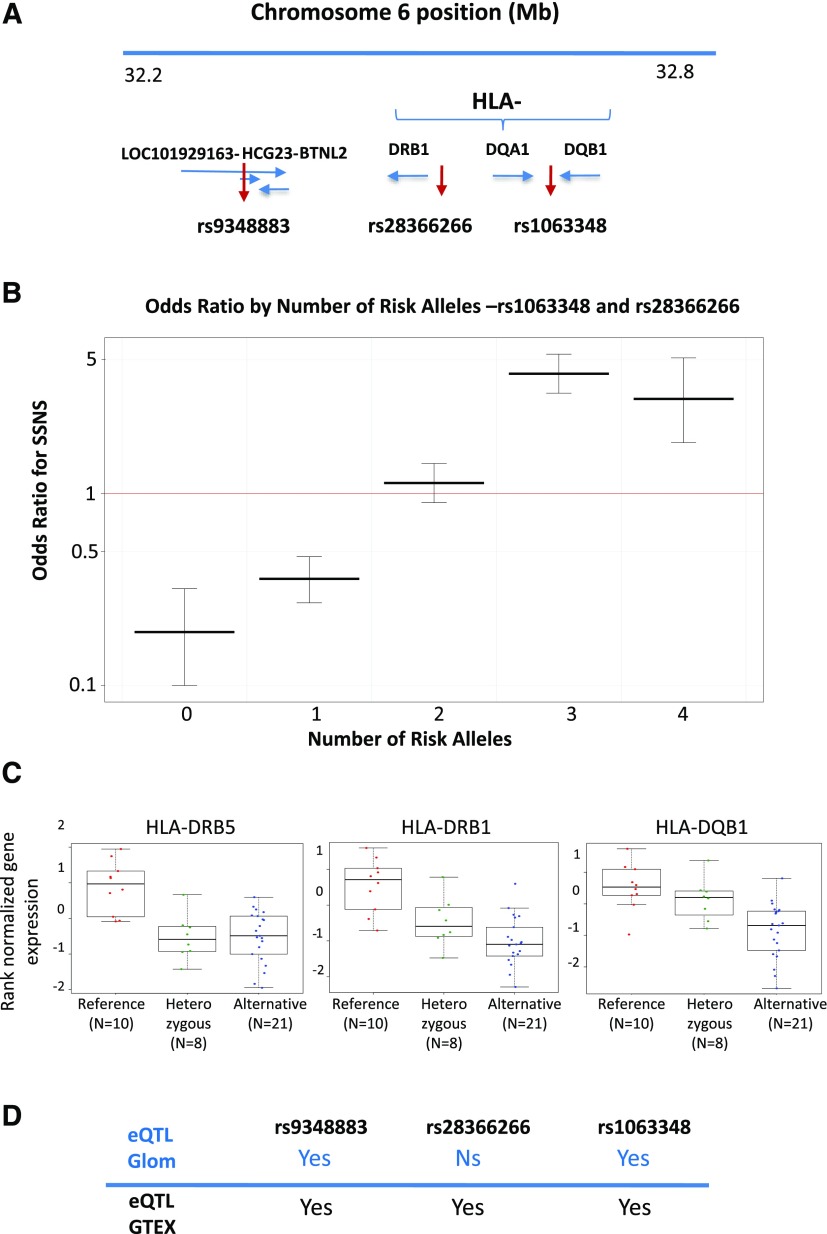
Figure 4.
Effect of the combination of risk alleles and expression quantitative trait loci (eQTL) results. (A) Position of the three lead single-nucleotide polymorphisms (SNPs) on chromosome 6, independently associated with SSNS. (B) Odds ratios of steroid-sensitive nephrotic syndrome (SSNS) as a function of increased burden of rs1063348 and rs28366266 risk alleles. Odds ratios for disease in cases with SSNS versus controls for patients with a particular number of risk alleles (computed using the Fisher exact test). (C) Box plot analysis of glomerular eQTL associated with rs1063348. In eligible Nephrotic Syndrome Study Network (NEPTUNE) pediatric patients (n=39) and under an additive model, rs1063348 was associated with significantly decreased glomerular expression of HLA-DRB1 (β=−0.7; FDR=9.9×10−4), HLA-DRB5 (β=−0.72; FDR=0.002), and HLA-DQB1 (β=−0.55; FDR=0.012). (D) Comparison of the NEPTUNE glomerular eQTL analysis with the GTEx database for the three top alleles. The lead SNP rs1063348 was associated with both glomerular eQTL and multitissue eQTL (rs9273462 in the GTEx). rs28366266 was eQTL in nonglomerular tissues, but association with glomerular eQTL was not significant in this study. rs934883 was eQTL for Ski2 like RNA helicase in glomerular tissue and numerous genes in a number of tissues in the GTEx. Ns, nonsignificant association with expression quantitative trait loci.
After conditioning on rs1063348, subsequent meta-analysis identified an independent significant SNP upstream of HLA-DRB1 (rs28366266; P=3.7×10−11; OR 2.2, 95% CI, 1.7–2.7) (Figures 3C and 4A, Supplemental Table 8A). There were no other loci outside the HLA region that exceeded or approached significance in this conditional analysis. Thus, when subsequently conditioning on rs1063348 and rs28366266, we based our empirical significance threshold solely on an approximately 12-Mb region of chromosome 6 (6:32000000–33200000) around rs28366266. By performing a conservative Bonferroni correction for the 19,024 variants in this window (i.e., not taking LD between variants into account), our significance threshold was thus P=2.6×10−6. In this way, we discovered a significant SNP (rs9348883; P=9.4×10−7; OR, 3.5; 95% CI, 2.1–5.8) within the introns of two nonprotein coding genes HLA Complex Gene 23 (HCG23) and LOC101929163 in close proximity to the 3′ untranslated region of BTNL2 (Figures 3D and 4A, Supplemental Table 8B).
In the GWAS cohorts, we studied whether a higher burden of risk alleles rs1063348 and rs28366266 was associated with increased odds of SSNS in cases versus controls (Figure 4B, Supplemental Table 9).We found that those with three or four risk alleles had significantly higher odds of being patients (three risk alleles: OR, 4.2; 95% confidence interval [95% CI], 3.4 to 5.4; four risk alleles: OR, 3.1; 95% CI, 1.9 to 5.1). The addition of rs9348883 to this burden analysis did not significantly alter the odds of SSNS.
HLA Allelotype Analysis
We imputed HLA allelotypes and reconstructed haplotypes from SNP data in our two European patients-control cohorts. The allelotypes HLA-DQA1*02:01, DQB1*02:02, and DRB1*07:01, defining a common haplotype, were significantly enriched in both IDF-EUR (P=7.2×10−4; OR, 2.1; 95% CI, 1.4–3.3) and ItSpa (P=1.0×10−8; OR, 5.7; 95% CI, 3.1–10.3) patients. However, in a logistic regression model including this haplotype with the three independent SNPs identified in the meta-analysis (rs1063348, rs28366266, and rs9348883), only the top SNPs remained significant and not the haplotype (Supplemental Table 10). Thus, noncoding variants may play an important role in predisposing to SSNS, presumably by modifying gene expression.
Post-GWAS Analyses
The independent risk SNPs discovered in the GWAS cohort were further characterized in the NEPTUNE cohort via association studies with glomerular transcriptomics, demographic characteristics, and clinical phenotypes.
Glomerular eQTL
Via a cis-eQTL analysis (within 1 Mb of the SNP) of the 39 NEPTUNE patients with glomerular transcriptomic data (Supplemental Material) and under an additive model, the G allele of rs1063348 was associated with significantly decreased glomerular expression of HLA-DRB1 (β=−0.7; FDR=9.9×10−4), HLA-DRB5 (β=−0.72; FDR=0.002), and HLA-DQB1 (β=−0.55; FDR=0.012) (Figure 4C). rs28366266 did not have glomerular cis-eQTLs that reached statistical significance (Supplemental Table 11). The risk allele of rs934883 (“T”) was associated with significantly increased expression of Ski2 like RNA helicase (β=1.1; FDR=0.049).
We used GTEx to determine whether these risk alleles regulated gene expression in other tissues that could contribute to SSNS. We hypothesized that immune cells could have transcripts significantly regulated by the GWAS alleles given the established role of the immune system in the pathogenesis of SSNS. The lead SNPs at the two independent loci in the HLA region, rs1063348 and rs28366266, were significant multitissue eQTLs for numerous HLA genes, with a direction of effect consistent to that in the glomerulus. rs1063348 was a significant multitissue eQTL for decreased expression of HLA-DQB1 and HLA-DRB (including whole-blood and EBV-transformed lymphocytes). rs28366266 had a similar relationship with HLA-DQB1 andHLA-DRB1, but it was not an eQTL for HLA-DRB5 in the GTEx data (Figure 4D). rs9348883 also is an eQTL for numerous genes in a number of tissues in the GTEx. These included Zinc Finger and BTB Domain Containing 12 and MHC Class I Polypeptide-Related Sequence B in whole blood and HLA-DRB6 and NOTCH4 in adipose tissue.
Comparison with Previously Identified SNPs
We evaluated whether the previously reported SSNS risk alleles in HLA-DQA118 explained the loci that we discovered from the GWAS and whether they were eQTLs (Supplemental Material). The two SNPs (rs1129740/rs107160) were only in strong LD with rs1063348 (Supplemental Table 12), and when conditioning on them, the other two loci remained significant (Supplemental Figure 3). rs1129740/rs1071630 also shared a similar glomerular eQTL profile as rs1063348, with significant associations with HLA-DRB1, HLA-DRB5, and HLA-DQB1 (Supplemental Table 13).
Epidemiologic Analyses
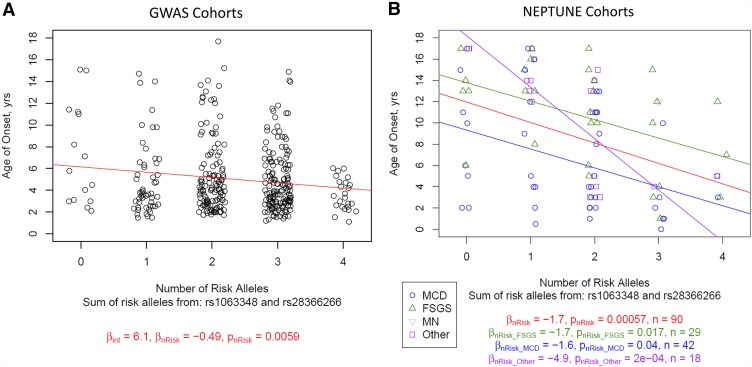
In the transethnic GWAS cohorts, two of the three lead SNPs were significantly associated with decreased age of onset (rs1063348 and rs28366266). When considering rs1063348 and rs28366266 in combination, presence of each additional risk allele was associated with a 6-month decrease in age of onset of disease (P=0.006) (Figure 5A). A similar suggestive association was observed in the population stratified by ethnicity (Supplemental Figure 4).
Figure 5.
Age of disease onset in genome-wide association study (GWAS) and Nephrotic Syndrome Study Network (NEPTUNE) cohorts as a function of increased burden of rs1063348 and rs28366266 risk alleles. (A) GWAS cohorts. Linear regression of age of onset by summed risk alleles from rs1063348 and rs28366266. Lines and values correspond to linear regression predictions. Note a significant relationship with more risk alleles and lower age of onset. (B) The NEPTUNE cohort. Linear regression of age of onset by summed risk alleles from rs1063348 and rs28366266, stratified by histologic diagnosis. Lines and values correspond to linear regression predictions, with colors corresponding to overall regression (red), regression in patients diagnosed with FSGS (green), regression in patients diagnosed with minimal change disease (MCD; blue), and regression in patients with other proteinuric glomerulopathies (purple). There were not enough patients diagnosed with membranous nephropathy to perform linear regression alone. Note a significant relationship with more risk alleles and lower age of onset.
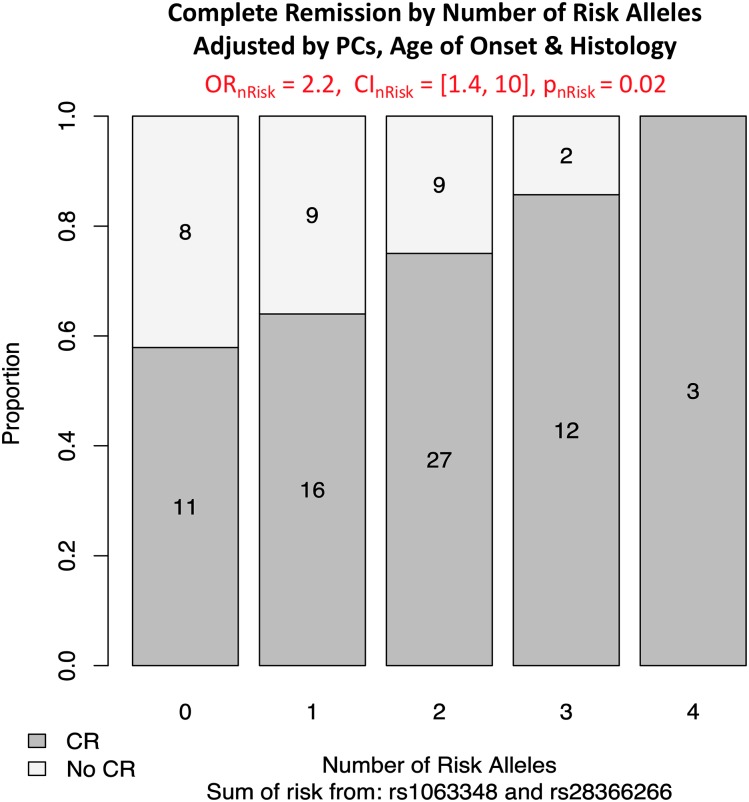
Studying the NEPTUNE cohort provided an opportunity to replicate initial discoveries, while also extending the phenotypic spectrum of these risk alleles in a non-SSNS cohort. In particular, we hypothesized that these risk alleles were associated with a distinct subtype of NS defined by younger age of onset and achievement of remission independent of histology. Because we found that these three risk alleles were not associated with a specific histology (Supplemental Table 14), we performed subsequent association analyses in children across all histologic diagnoses. As in the GWAS cohorts, an analysis combining rs1063348 and rs28366266 identified significant association with younger age of onset (Figure 5B). Each additional risk allele (from zero to a maximum of four) was associated with a 2-year decreased age of disease onset (P=4.8×10−5). The same relationship was observed in an analysis stratified by histology (Figure 5B) and for children who achieved complete remission (Supplemental Figure 5). Finally, in a multivariate analysis, each additional rs1063348 and rs28366266 risk allele was associated with a 2.2 greater odds of achieving complete remission (95% CI, 1.4 to 10; P=0.02) (Figure 6). This was adjusted for histologic diagnosis, four principal components of genetic ancestry, and age of onset.
Figure 6.
Complete remission (CR) in the Nephrotic Syndrome Study Network (NEPTUNE) cohort as a function of increased burden of rs1063348 and rs28366266 risk alleles. Logistic regression of CR by summed risk alleles from rs1063348 and rs28366266 in the NEPTUNE cohort adjusted for status, age of disease onset, and histologic diagnosis. Odds ratio (OR), 95% confidence interval (95% CI), and P value are from logistic regression for the number of risk alleles. The bar plot shows the proportion of CR for each risk allele category. There is a significant association between number of risk alleles and the proportion of CR, with more risk alleles corresponding to increased odds of achieving CR.
Discussion
Here, we used an integrative genomics approach in three independent pediatric NS cohorts to not only discover risk alleles for SSNS but also, immediately characterize their transcriptional and epidemiologic effects across diverse tissues and cohorts, respectively. We began with a comprehensive analytic pipeline for GWAS consisting of (1) a genome-wide SNP array followed by imputation in both discovery and replication cohorts, (2) transethnic meta-analysis, and (3) conditional analysis. In this way, we discovered three significant independent SSNS-associated alleles with risk that was shared across ethnicities. Although we cannot pinpoint the causal allele or gene, we are using a “closest gene” approach to refer to the loci tagged by these GWAS alleles as “HLA-DQB1,” “HLA-DRB1,”and “BTNL2-HCG23-LOC101929163.” Each of these genes has been previously implicated in other immune-mediated nonglomerular human diseases.32–35 Furthermore, we found significantly increased odds of SSNS with an increased burden of the rs1063348 and rs28366266 risk alleles per individual. This behavior is reminiscent of findings in other immune-related diseases, such as membranous nephropathy, type 1 diabetes, and celiac disease, in which independent SNPs affect disease risk via additive and nonadditive effects.36,37 Altogether, this supports our understanding of the central role of immune dysregulation in SSNS, provides specific targets for further inquiry, and brings even more evidence of the pleiotropic effects of genetic variation in loci relating to immune regulation.38
The two most significant loci were within the HLA class 2 region at the 3′ untranslated region of HLA-DQB1 and upstream of HLA-DRB1. The heterodimeric HLA molecules that present peptide antigens on the surface of antigen-presenting cells (APCs) for recognition by T cell receptors can also be induced on podocytes, sometimes considered as nonhematopoietic professional APCs.39 Of note, HLA-DQB1 and HLA-DRB1 loci are strongly associated with EBV infection, which has been previously hypothesized to be involved in NS pathogenesis.40,41 Engagement of HLA class 2 on the podocyte surface might also regulate podocyte function and survival as it does for APCs.42,43 Given that the two main SSNS targets are putatively the podocyte and the lymphocyte,7,9 we used eQTL analysis to gain insight into transcriptional effects of these SNPs on gene expression across tissues.
The HLA-DQB1 lead SNP (rs1063348) was an eQTL for multiple HLA transcripts in both the glomerulus in children with NS and across multiple tissues from the deceased donors in the GTEx project, including blood and EBV-transformed lymphocytes. This risk allele was associated with significantly decreased expression across tissues. The same eQTLs were observed for the previously implicated exonic HLA-DQA1 variants18 (in very strong LD with our lead SNP). The HLA-DRB1 SNP (rs28366266) shared similar multitissue eQTL behavior for multiple HLA transcripts. Altogether, these observations raise a number of questions about the identity of the functional variant(s) at this locus as well as the tissues in which it is mediating its effect on NS. Although beyond the scope of this study, future studies to elucidate the mechanism, the target tissue, and the relative role of different variants will be aided by expansion of glomerular and blood eQTL datasets in patients and controls combined with in silico nanodissection44 and single-cell RNA-Seq–derived deconvolution methods to pinpoint the specific cells affected by these eQTLs.45 Finally, as the methods mature, future single-cell RNA-Seq studies using human kidney samples and peripheral blood from patients with NS, similar to those done in other tissues,46 will also advance our understanding of the cell type specificity of these eQTLs.
The lead SNP at the third independent locus was within introns of HCG23 and LOC101920163 (a minimally characterized long noncoding RNA) and in close proximity to the 3′ untranslated region of BTNL2, a gene not previously implicated in SSNS. BTNL2 modulates T cell activity by suppressing T cell proliferation and cytokine production as well as by inducing FoxP3 expression and Treg differentiation.47,48 The LOC1019291163-BTNL2 gene region has been associated with susceptibility to many other autoimmune diseases: for example, membranous nephropathy, Crohn disease, and vitiligo.35,49,50
We identified the risk haplotype DRB1*07:01-DQA1*02:01-DQB1*02:02 in European patients. Similar findings were recently reported in the study by Adeyemo et al.,51 which showed that DRB1*07:01, DQA1*02:01, and DQB1*02:01 were the classic HLA alleles the most significantly associated with early-onset NS in black children. The HLA-DRB1*07:01-HLA-DQA1*02:01-HLA-DQB1*02:02 haplotype is associated with a high risk of Escherichia coli–derived asparaginase hypersensitivity in childhood acute lymphoblastic leukemia,52 which is reminiscent of allergic trigger in some patients with SSNS. Importantly, the three independent lead SNPs identified in the post-GWAS meta-analysis remained significant after conditioning on the risk haplotype. This suggests that the genetic control of childhood SSNS may involve both coding classic allelotypes for presentation to the immune system of as yet unknown antigens and regulatory SNPs that may act distantly on HLA (and non-HLA) genes as shown by our eQTL studies and in other cancer and autoimmune diseases.53,54 The fact that the lead SNPs and the risk haplotype are conserved across ethnicities reinforces their contribution to the disease pathogenesis.
A strength of our study is that the GWAS cohorts only included children with SSNS, resulting in a younger and relatively narrow distribution of ages of onset, whereas the children in the North American NEPTUNE had minimal change disease, FSGS, and MN as well as differing responses to immunosuppression. Yet, despite these substantial demographic and clinical differences between the children within and across these cohorts, we discovered shared epidemiologic associations. An increased number of risk alleles at HLA-DRB1/-DQB1 were still associated with an earlier age of disease onset in all of these populations. In the NEPTUNE children, this was independent of histologic diagnosis and achievement of complete remission. Furthermore, in the NEPTUNE patients, there was a significant increase in odds of achieving complete remission for children with a greater number of these risk alleles across histologies, races, and ages of onset. Taken together, we suggest that pediatric NS associated with risk alleles at these HLA-DRB1 and HLA-DQB1 loci represents a clinically distinct subtype of this condition characterized by an early disease onset and a relatively benign disease course, which seems to be defined more accurately by genetic susceptibility than by histologic appearance.
In conclusion, although SSNS is a rare disease, we were empowered to make novel biologic and clinical discoveries in this disease through an integrative strategy of transethnic GWAS, glomerular eQTL, and clinical association. The transethnic significant loci identify common risk variants uniting SSNS biology across ancestries. Integrating our results with previous knowledge of these loci more firmly establishes SSNS as another primary glomerular disease with genetic susceptibility that arises, at least in part, from immune dysregulation. The next steps are to understand the role for the HLA class 2 molecules in SSNS pathophysiology as modifiers of immunity, investigate further the pathophysiologic relevance and clinical correlates of the non-HLA loci, and study epidemiologic associations of these risk alleles in larger and independent cohorts. Furthering our genomic understanding of pediatric SSNS and other forms of this condition in these ways may provide meaningful biologic and clinical insights.
Disclosures
None.
Supplementary Material
Acknowledgments
This study was funded by European Research Council grant ERC-2012-ADG_20120314 (grant agreement 322947) and Agence Nationale pour la Recherche “Genetransnephrose” grant ANR-16-CE17-004-01. M.G.S. is supported by the Charles Woodson Clinical Research Fund and National Institutes of Health grant R01-DK108805. The Nephrotic Syndrome Study Network Consortium (NEPTUNE; U54-DK-083912) is a part of the National Center for Advancing Translational Sciences (NCATS) Rare Disease Clinical Research Network (RDCRN) supported through a collaboration between the Office of Rare Diseases Research (ORDR), the NCATS, and the National Institute of Diabetes, Digestive, and Kidney Diseases. The RDCRN is an initiative of the ORDR of the NCATS. Additional funding and/or programmatic support for this project has also been provided by the University of Michigan, NephCure Kidney International, and the Halpin Foundation. The NEPHROVIR cohort has been supported by two grants from the Programme Hospitalier de Recherche Clinique: grants PHRC 2007-AOM07018 and PHRC 2011-AOM11002. The NEPHROVIR network is coordinated by the Pediatric Nephrology Unit of Robert Debré Hospital, the “Unité de Recherche Clinique de l’Est Parisien,” and the “Délégation de la Recherche Clinique de la Région Ile-de-France.” M.V. and M.C. are supported by the Associazone per la Cura del bambino Nefropatico Organizzazione Non Lucrativa di Utilità Sociale.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related editorial, “Leveraging Ancestral Heterogeneity to Map Shared Genetic Risk Loci in Pediatric Steroid-Sensitive Nephrotic Syndrome,” on pages 1793–1794.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2017111185/-/DCSupplemental.
References
- 1.Vivarelli M, Massella L, Ruggiero B, Emma F: Minimal change disease. Clin J Am Soc Nephrol 12: 332–345, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banh TH, Hussain-Shamsy N, Patel V, Vasilevska-Ristovska J, Borges K, Sibbald C, et al.: Ethnic differences in incidence and outcomes of childhood nephrotic syndrome. Clin J Am Soc Nephrol 11: 1760–1768, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dossier C, Lapidus N, Bayer F, Sellier-Leclerc AL, Boyer O, de Pontual L, et al.: Epidemiology of idiopathic nephrotic syndrome in children: Endemic or epidemic? Pediatr Nephrol 31: 2299–2308, 2016 [DOI] [PubMed] [Google Scholar]
- 4.Gipson DS, Troost JP, Lafayette RA, Hladunewich MA, Trachtman H, Gadegbeku CA, et al.: Complete remission in the nephrotic syndrome study network. Clin J Am Soc Nephrol 11: 81–89, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hjorten R, Anwar Z, Reidy KJ: Long-term outcomes of childhood onset nephroticsyndrome. Front Pediatr 4: 53, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elie V, Fakhoury M, Deschênes G, Jacqz-Aigrain E: Physiopathology of idiopathic nephrotic syndrome: Lessons from glucocorticoids and epigenetic perspectives. Pediatr Nephrol 27: 1249–1256, 2012 [DOI] [PubMed] [Google Scholar]
- 7.Colucci M, Corpetti G, Emma F, Vivarelli M: Immunology of idiopathic nephrotic syndrome. Pediatr Nephrol 33: 573–584, 2018 [DOI] [PubMed] [Google Scholar]
- 8.Wei C-C, Lin C-L, Shen T-C, Sung F-C: Occurrence of common allergic diseases in children with idiopathic nephrotic syndrome. J Epidemiol 25: 370–377, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mathieson PW: Proteinuria and immunity--an overstated relationship? N Engl J Med 359: 2492–2494, 2008 [DOI] [PubMed] [Google Scholar]
- 10.Faul C, Donnelly M, Merscher-Gomez S, Chang YH, Franz S, Delfgaauw J, et al.: The actin cytoskeleton of kidney podocytes is a direct target of the antiproteinuric effect of cyclosporine A. Nat Med 14: 931–938, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fornoni A, Sageshima J, Wei C, Merscher-Gomez S, Aguillon-Prada R, Jauregui AN, et al.: Rituximab targets podocytes in recurrent focal segmental glomerulosclerosis. Sci Transl Med 3: 85ra46, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davin JC: The glomerular permeability factors in idiopathic nephrotic syndrome. Pediatr Nephrol 31: 207–215, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karp AM, Gbadegesin RA: Genetics of childhood steroid-sensitive nephrotic syndrome. Pediatr Nephrol 32: 1481–1488, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghiggeri GM, Dagnino M, Parodi S, Zennaro C, Amoroso A, Pugliese F, et al.: Discordant evolution of nephrotic syndrome in mono- and dizygotic twins. Pediatr Nephrol 21: 419–422, 2006 [DOI] [PubMed] [Google Scholar]
- 15.Gee HY, Ashraf S, Wan X, Vega-Warner V, Esteve-Rudd J, Lovric S, et al.: Mutations in EMP2 cause childhood-onset nephrotic syndrome. Am J Hum Genet 94: 884–890, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gee HY, Zhang F, Ashraf S, Kohl S, Sadowski CE, Vega-Warner V, et al.: KANK deficiency leads to podocyte dysfunction and nephrotic syndrome. J Clin Invest 125: 2375–2384, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorval G, Gribouval O, Martinez-Barquero V, Machuca E, Tête MJ, Baudouin V, et al.: Clinical and genetic heterogeneity in familial steroid-sensitive nephrotic syndrome. Pediatr Nephrol 33: 473–483, 2018 [DOI] [PubMed] [Google Scholar]
- 18.Gbadegesin RA, Adeyemo A, Webb NJ, Greenbaum LA, Abeyagunawardena A, Thalgahagoda S, et al.; Mid-West Pediatric Nephrology Consortium : HLA-DQA1 and PLCG2 are candidate risk loci for childhood-onset steroid-sensitive nephrotic syndrome. J Am Soc Nephrol 26: 1701–1710, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dossier C, Sellier-Leclerc AL, Rousseau A, Michel Y, Gautheret-Dejean A, Englender M, et al.: Prevalence of herpesviruses at onset of idiopathic nephrotic syndrome. Pediatr Nephrol 29: 2325–2331, 2014 [DOI] [PubMed] [Google Scholar]
- 20.3C Study Group : Vascular factors and risk of dementia: Design of the Three-City Study and baseline characteristics of the study population. Neuroepidemiology 22: 316–325, 2003 [DOI] [PubMed] [Google Scholar]
- 21.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al.; 1000 Genomes Project Consortium : An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gadegbeku CA, Gipson DS, Holzman LB, Ojo AO, Song PX, Barisoni L, et al.: Design of the Nephrotic Syndrome Study Network (NEPTUNE) to evaluate primary glomerular nephropathy by a multidisciplinary approach. Kidney Int 83: 749–756, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, et al.; Haplotype Reference Consortium : A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 48: 1279–1283, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al.; PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willer CJ, Li Y, Abecasis GR: METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26: 2190–2191, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evangelou E, Ioannidis JP: Meta-analysis methods for genome-wide association studies and beyond. Nat Rev Genet 14: 379–389, 2013 [DOI] [PubMed] [Google Scholar]
- 27.Cochran WG: The combination of estimates from different experiments. Biometrics 10: 101–129, 1954 [Google Scholar]
- 28.Yang J, Lee SH, Goddard ME, Visscher PM: GCTA: A tool for genome-wide complex trait analysis. Am J Hum Genet 88: 76–82, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devlin B, Roeder K, Wasserman L: Genomic control, a new approach to genetic-based association studies. Theor Popul Biol 60: 155–166, 2001 [DOI] [PubMed] [Google Scholar]
- 30.Jia X, Han B, Onengut-Gumuscu S, Chen WM, Concannon PJ, Rich SS, et al.: Imputing amino acid polymorphisms in human leukocyte antigens. PLoS One 8: e64683, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sampson MG, Robertson CC, Martini S, Mariani LH, Lemley KV, Gillies CE, et al.; Nephrotic Syndrome Study Network : Integrative genomics identifies novel associations with APOL1 risk genotypes in black NEPTUNE subjects. J Am Soc Nephrol 27: 814–823, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolin A, Lahtela EL, Anttila V, Petrek M, Grunewald J, van Moorsel CHM, et al.; SNP variants in Major Histocompatibility Complex are associated with sarcoidosis susceptibility-A joint analysis in four European populations. Front Immunol 8: 422, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prescott NJ, Lehne B, Stone K, Lee JC, Taylor K, Knight J, et al. ; UK IBD Genetics Consortium : Pooled sequencing of 531 genes in inflammatory bowel disease identifies an associated rare variant in BTNL2 and implicates other immune related genes. PLoS Genet 11: e1004955, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Julià A, Domènech E, Chaparro M, García-Sánchez V, Gomollón F, Panés J, et al.: A genome-wide association study identifies a novel locus at 6q22.1 associated with ulcerative colitis. Hum Mol Genet 23: 6927–6934, 2014 [DOI] [PubMed] [Google Scholar]
- 35.Stanescu HC, Arcos-Burgos M, Medlar A, Bockenhauer D, Kottgen A, Dragomirescu L, et al.: Risk HLA-DQA1 and PLA(2)R1 alleles in idiopathic membranous nephropathy. N Engl J Med 364: 616–626, 2011 [DOI] [PubMed] [Google Scholar]
- 36.Lenz TL, Deutsch AJ, Han B, Hu X, Okada Y, Eyre S, et al.: Widespread non-additive and interaction effects within HLA loci modulate the risk of autoimmune diseases. Nat Genet 47: 1085–1090, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu X, Deutsch AJ, Lenz TL, Onengut-Gumuscu S, Han B, Chen WM, et al.: Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk. Nat Genet 47: 898–905, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiryluk K, Li Y, Scolari F, Sanna-Cherchi S, Choi M, Verbitsky M, et al.: Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat Genet 46: 1187–1196, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goldwich A, Burkard M, Olke M, Daniel C, Amann K, Hugo C, et al.: Podocytes are nonhematopoietic professional antigen-presenting cells. J Am Soc Nephrol 24: 906–916, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rubicz R, Yolken R, Drigalenko E, Carless MA, Dyer TD, Bauman L, et al.: A genome-wide integrative genomic study localizes genetic factors influencing antibodies against Epstein-Barr virus nuclear antigen 1 (EBNA-1). PLoS Genet 9: e1003147, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dossier C, Jamin A, Deschênes G: Idiopathic nephrotic syndrome: The EBV hypothesis. Pediatr Res 81: 233–239, 2017 [DOI] [PubMed] [Google Scholar]
- 42.Haylett RS, Koch N, Rink L: MHC class II molecules activate NFAT and the ERK group of MAPK through distinct signaling pathways in B cells. Eur J Immunol 39: 1947–1955, 2009 [DOI] [PubMed] [Google Scholar]
- 43.de Almeida DE, Holoshitz J: MHC molecules in health and disease: At the cusp of a paradigm shift. Self Nonself 2: 43–48, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ju W, Greene CS, Eichinger F, Nair V, Hodgin JB, Bitzer M, et al.: Defining cell-type specificity at the transcriptional level in human disease. Genome Res 23: 1862–1873, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gillies CE, Putler R, Menon R, Otto E, Yasutake K, Nair V, et al. : An eQTL landscape of kidney tissue in human nephrotic syndrome. bioRxiv doi: 10.1101/281162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, et al.: Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science 356: eaah4573, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen T, Liu XK, Zhang Y, Dong C: BTNL2, a butyrophilin-like molecule that functions to inhibit T cell activation. J Immunol 176: 7354–7360, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swanson RM, Gavin MA, Escobar SS, Rottman JB, Lipsky BP, Dube S, et al.: Butyrophilin-like 2 modulates B7 costimulation to induce Foxp3 expression and regulatory T cell development in mature T cells. J Immunol 190: 2027–2035, 2013 [DOI] [PubMed] [Google Scholar]
- 49.Yang SK, Hong M, Zhao W, Jung Y, Baek J, Tayebi N, et al.: Genome-wide association study of Crohn’s disease in Koreans revealed three new susceptibility loci and common attributes of genetic susceptibility across ethnic populations. Gut 63: 80–87, 2014 [DOI] [PubMed] [Google Scholar]
- 50.Jin Y, Birlea SA, Fain PR, Gowan K, Riccardi SL, Holland PJ, et al.: Variant of TYR and autoimmunity susceptibility loci in generalized vitiligo. N Engl J Med 362: 1686–1697, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adeyemo A, Esezobor C, Solarin A, Abeyagunawardena A, Kari JA, El Desoky S, et al.: HLA-DQA1 and APOL1 as risk loci for childhood-onset steroid-sensitive and steroid-resistant nephrotic syndrome. Am J Kidney Dis 71: 399–406, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kutszegi N, Yang X, Gézsi A, Schermann G, Erdélyi DJ, Semsei ÁF, et al.: HLA-DRB1*07:01-HLA-DQA1*02:01-HLA-DQB1*02:02 haplotype is associated with a high risk of asparaginase hypersensitivity in acute lymphoblastic leukemia. Haematologica 102: 1578–1586, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He H, Li W, Liyanarachchi S, Srinivas M, Wang Y, Akagi K, et al.: Multiple functional variants in long-range enhancer elements contribute to the risk of SNP rs965513 in thyroid cancer. Proc Natl Acad Sci U S A 112: 6128–6133, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, et al.: Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518: 337–343, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.