ABSTRACT
The bacterium Vibrio cholerae is native to aquatic environments and can switch lifestyles to cause disease in humans. Lifestyle switching requires modulation of genetic systems for quorum sensing, intestinal colonization, and toxin production. Much of this regulation occurs at the level of gene expression and is controlled by transcription factors. In this work, we have mapped the binding of cAMP receptor protein (CRP) and RNA polymerase across the V. cholerae genome. We show that CRP is an integral component of the regulatory network that controls lifestyle switching. Focusing on a locus necessary for toxin transport, we demonstrate CRP-dependent regulation of gene expression in response to host colonization. Examination of further CRP-targeted genes reveals that this behavior is commonplace. Hence, CRP is a key regulator of many V. cholerae genes in response to lifestyle changes.
KEYWORDS: Vibrio, biochemistry, gene regulation, genome analysis
IMPORTANCE
Cholera is an infectious disease that is caused by the bacterium Vibrio cholerae. Best known for causing disease in humans, the bacterium is most commonly found in aquatic ecosystems. Hence, humans acquire cholera following ingestion of food or water contaminated with V. cholerae. Transition between an aquatic environment and a human host triggers a lifestyle switch that involves reprogramming of V. cholerae gene expression patterns. This process is controlled by a network of transcription factors. In this paper, we show that the cAMP receptor protein (CRP) is a key regulator of V. cholerae gene expression in response to lifestyle changes.
INTRODUCTION
Vibrio cholerae is a Gram-negative bacterium that causes the diarrheal disease cholera (1). Estimated to claim 3 to 5 million victims every year, cholera is endemic in regions of Asia and sub-Saharan Africa (1–4). Localized epidemics are also frequent; half a million cases have been attributed to the current outbreak in Yemen (2). Although notorious as a pathogen of humans, V. cholerae is native to aquatic environments (5). In this situation, the organism proliferates by colonizing crustaceans and other biota in their habitat (5–10). In particular, chitinous surfaces provide a substrate for biofilm formation and nutrients (5). Humans encounter V. cholerae following the ingestion of contaminated food or water (1). In response, the bacterium produces mucin-degrading enzymes and upregulates motility (5). This facilitates penetration of the intestinal mucosa (5). The subsequent attachment of V. cholerae cells to the intestinal epithelium requires toxin-coregulated pili (TCP) and accessory colonization factors (ACF) (11–13). Ultimately, disease results from the production of factors including cholera toxin (CTX), repeats in toxin (RTX), and hemolysin (HlyA) (42, 63–65).
Unsurprisingly, the expression of V. cholerae genes for quorum sensing, host colonization, and toxin production/export is precisely regulated (5). Most notably, an AraC/XylS family transcription factor called ToxT directly regulates the transcription of ctxAB, acfAD, and genes encoding the TCP (14). Production of ToxT is induced in the intestine and is codependent on two OmpR family regulators, ToxR and TcpP, which respond to extracellular signals that include osmolarity, pH, and bile (15–17). Genes encoding outer membrane porins OmpT and OmpU are also regulated by ToxR in a pathway that permits initial sensing of bile and subsequent resistance (18). Together, the aforementioned gene regulatory events comprise the ToxR regulon. Transcription factors with targets overlapping the ToxR regulon include VpsT, AphA, AphB, and the cyclic AMP (cAMP) receptor protein (CRP) (19–24). Best studied in Escherichia coli, CRP can activate transcription by binding targets centered either 41.5 or 61.5 bp upstream from a transcription start site (25). Since CRP binds DNA in response to the intracellular availability of cAMP, genes are controlled in response to nutrient availability (25–28). As such, CRP plays an integral role in the utilization of alternative carbon sources (29, 30). Hence, many E. coli genes that are differentially regulated in the intestine are controlled by CRP, including CTX-related toxins in pathogenic E. coli strains (26, 31). In V. cholerae, CRP is known to influence the ToxR regulon; CRP directly inhibits tcpP expression and activates the transcription of ompT (22, 32, 33). Remarkably, despite being a global regulator of transcription, direct control by CRP has only been demonstrated for seven V. cholerae genes (22, 31, 33–38). Furthermore, gene regulation by CRP during colonization of a host intestinal tract has never been studied. In this work, we have used chromatin immunoprecipitation (ChIP) coupled with DNA sequencing (ChIP-seq) to map the distribution of CRP across the V. cholerae genome. We show substantial overlap between the ToxR regulon and control of additional virulence factors not regulated by the ToxR system. Focusing on one such target, encoding RTX and its export system, we show that CRP is essential for specific induction of gene expression during intestinal colonization. Examination of additional CRP target genes reveals that similar effects are widespread.
RESULTS
Genome-wide distribution of CRP and RNA polymerase in Vibrio cholerae.
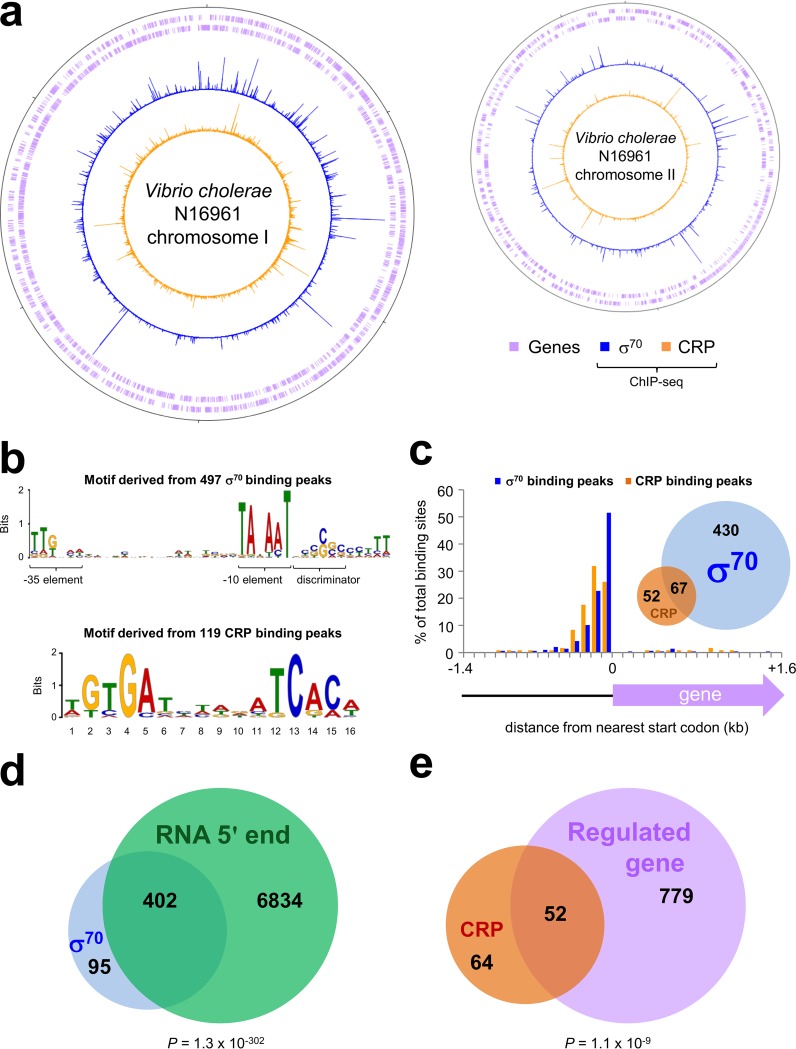
We used ChIP-seq to map global DNA binding by CRP and the RNA polymerase σ70 subunit in V. cholerae strain N16961 grown to mid-log phase in M9 minimal medium supplemented with 1% (wt/vol) fructose (39). The strain, isolated from a Bangladeshi patient in 1971, comprises 3,885 genes borne on two circular chromosomes of 2,961,146 bp (chromosome I) and 1,072,314 bp (chromosome II) (39). The binding profiles of CRP and σ70 are shown in Fig. 1a. In each plot, genes are illustrated by mauve lines (Fig. 1a, first and second tracks), σ70 binding in blue (Fig. 1a, third track), and CRP binding in orange (Fig. 1a, fourth track). We identified 497 binding peaks for σ70 and 119 binding peaks for CRP. The σ70 peaks were not distributed equitably; chromosome II accounts for 27% of the V. cholerae genome but aligned with 40% of the σ70 binding peaks. To assess the validity of our data, we examined the DNA sequence attributes of each peak. Hence, we used MEME (Multiple Em for Motif Elicitation) to identify sequence motifs associated with σ70 or CRP binding. The most statistically significant DNA motif associated with each group of peaks is shown in Fig. 1b. As expected, MEME recovered significant motifs matching the sequence of a housekeeping bacterial promoter (E = 4.5 × 10−29) (Fig. 1b, top) and the palindromic CRP binding sequence (E = 2.5 × 10−2) (Fig. 1b, bottom). For all peaks, we determined the distance to the nearest start codon and sorted these distances into 100-bp bins. The distribution of peaks among the bins is illustrated in Fig. 1c; σ70 most frequently binds the 100 bp preceding the 5′ end of a gene, while CRP binds further upstream. Of the 119 CRP binding peaks, 67 colocated with binding of σ70 (Fig. 1c, inset). We also compared our data with existing compendiums of the V. cholerae transcriptome (40, 41). Briefly, Papenfort and coworkers used differential RNA-seq to map transcription start sites (TSS) in V. cholerae (40). There was significant overlap with our data; 81% of the σ70 binding peaks matched a TSS (P = 1.3 × 10−302) (Fig. 1d). Thus, the combined data describe sigma factor preference, promoter sequence, and sites of transcription initiation for the majority of V. cholerae transcription units. In a separate study, Fong and Yildiz used DNA microarrays to detect changes in RNA levels resulting from crp deletion (41). Again, the overlap was significant, and 52 of the 119 CRP binding peaks identified a differentially expressed gene (P = 2.1 × 10−9) (Fig. 1e). Note that greater overlap of the CRP binding and gene regulatory data is not expected; many CRP-controlled promoters are active only under specific conditions, and most transcriptome changes will result from indirect effects of CRP (26–30).
FIG 1 .
Global analysis of CRP and σ70 binding in Vibrio cholerae. (a) Genome-wide distribution of CRP and the RNA polymerase σ70 subunit in Vibrio cholerae strain N16961. Plots are shown for the two N16961 chromosomes. In each plot, the tick mark at the 12 o’clock position represents the first base pair (bp) of the chromosome and subsequent tick marks are spaced by 0.5 Mbp. In each plot, the first and second tracks (mauve lines) show the positions of genes, the third track (blue) is the σ70 binding profile, and the fourth track (orange) is the CRP binding profile. (b) DNA sequence motifs recovered from CRP and σ70 binding peaks. Top, DNA sequence motif identified by MEME present in DNA sequences associated with σ70 binding; bottom, DNA sequence motif generated from CRP binding peaks. (c) Locations of CRP and RNA polymerase binding peaks with respect to genes. Histogram depicts the distances between ChIP-seq binding peaks and the nearest 5′ end of a gene; data for CRP binding are in orange, and data for σ70 binding are in blue. Each binding peak was allocated to a series of 100-bp bins. Inset, Venn diagram that illustrates the number of overlapping CRP and σ70 binding peaks. (d) Overlap between σ70 DNA binding and transcription start sites. The Venn diagram illustrates numbers of overlapping σ70 binding sites (blue) and transcription start sites (green) (40). A σ70 binding peak centered within 50 bp of a transcription start was considered to overlap. To generate the P value, we used the chi-square test. To generate values for the expected overlap between the data sets, assuming no correlation, we randomized the positions of the σ70 peaks. (e) Overlap between CRP binding and CRP-regulated genes. The Venn diagram illustrates overlap between genes adjacent to CRP binding peaks (orange) and genes that were differently expressed in the absence of CRP (mauve) (41). To generate the P value, we used the chi-square test. To generate values for the expected overlap between the data sets, assuming no correlation, we randomly selected 831 genes from the V. cholerae genome and determined the number that were adjacent to CRP binding peaks.
Expression of the rtxBDE operon is repressed by CRP.
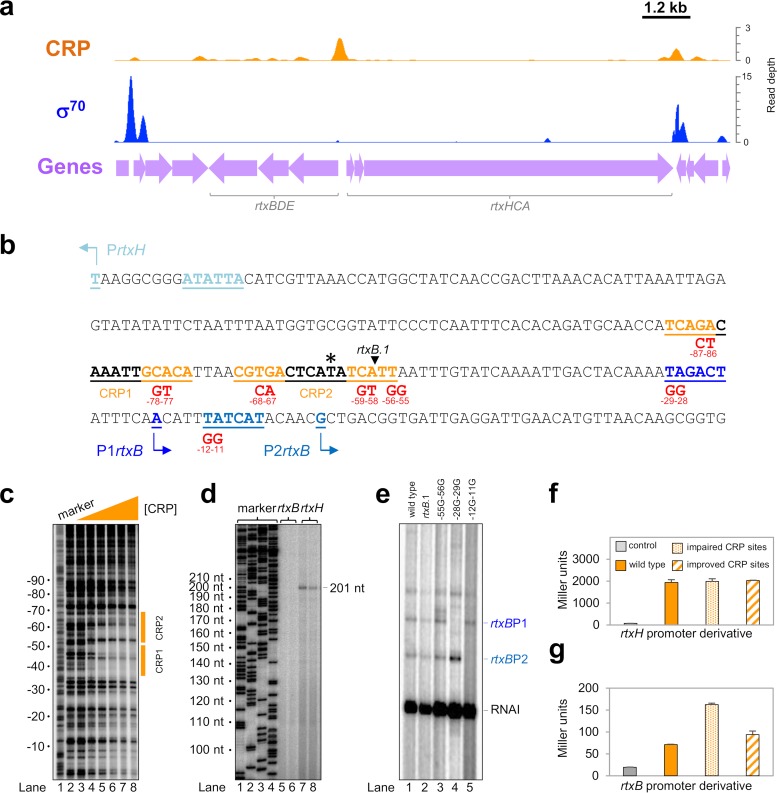
Many V. cholerae genes involved in pathogenicity were targeted by CRP (Table 1). We focused our attention on the gene cluster responsible for the cytotoxic activity of V. cholerae (42). The region comprises two operons called rtxHCA and rtxBDE. The rtxA gene encodes RTX toxin, while rtxH and rtxC encode a hypothetical protein and an acyltransferase, respectively. The divergent rtxBDE operon encodes components of the toxin secretion system. Figure 2a shows binding of CRP and σ70 to DNA between the two operons. The sequence of the intergenic region is shown in Fig. 2b. Two putative CRP sites (orange and underlined) overlap the center of the CRP ChIP-seq binding peak (Fig. 2b, asterisk). To confirm binding, we purified V. cholerae CRP for use in DNase I footprinting assays. The footprinting data are consistent with CRP binding to both of the putative sites (Fig. 2c). To identify promoters of rtxHCA and rtxBDE transcription, we examined our ChIP-seq data for σ70 and the TSS mapping data of Papenfort et al. (40). However, these were poorly informative; the σ70 binding levels were low and Papenfort et al. identified only a single intergenic promoter for rtxHCA. Indeed, the TSS data suggest that the region is prone to spurious intragenic transcription; the rtxHCA and rtxBDE genes contain a total of 21 internal promoters (40). To map canonical promoters in the rtxHCA-rtxBDE intergenic region, we used two approaches; mRNA primer extension and in vitro transcription. The results of the primer extension analysis are shown in Fig. 2d. We were unable to derive any extension products from rtxBDE transcripts, but a single 201-nucleotide extension product was generated from the rtxHCA transcript. The corresponding TSS aligns perfectly with the rtxHCA TSS identified by Papenfort et al. (labeled PrtxH in Fig. 2b) (40). A promoter −10 element is appropriately positioned upstream, and deletion of this sequence abolishes mRNA production (see Fig. S1a in the supplemental material). Since both RNA-seq and primer extension failed to identify promoters for rtxBDE, we reasoned that the operon must be repressed in vivo. Hence, the rtxBDE intergenic region was cloned upstream from the λoop terminator in plasmid pSR to create a template for in vitro transcription. The result of the experiment is shown in lane 1 of Fig. 2e. The control RNAI transcript is derived from the plasmid replication origin, and further transcripts could originate within the cloned intergenic DNA. Truncation of the rtxBDE intergenic region did not prevent synthesis of the additional RNAs (Fig. 2e, lane 2). Hence, the transcripts originate downstream from the truncation site marked by the inverted triangle in Fig. 2b. We made derivatives of the truncated DNA template with point mutations in all potential −10 hexamers. The mutations are illustrated in Fig. 2b. Two pairs of mutations, −29G −28G and −12G −11G, each prevent the production of a different transcript (Fig. 2e, lanes 3 to 5). The mutations have similar effects in vivo (Fig. S1b). We conclude that transcription originates from the promoters labeled P1rtxB and P2rtxB in Fig. 2b. Neither CRP site is appropriately positioned to activate PrtxH, P1rtxB, or P2rtxB. However, it has previously been shown that pairs of CRP binding sites upstream from promoters can repress transcription (43). Thus, we created derivatives of the intergenic region where the CRP sites were inactivated by the point mutations shown in Fig. 2b. We also altered the sites to match the consensus for CRP binding. The various DNA fragments were cloned in the appropriate orientation upstream from lacZ in plasmid pRW50T. The resulting DNA constructs were moved into V. cholerae strain N16961 by conjugation. Promoter activity was inferred by measuring β-galactosidase activity in lysates of the exconjugants. The data show that PrtxH activity is unaltered by any of the mutations (Fig. 2f). Conversely, the poorly active rtxBDE promoters have higher levels of activity when the CRP sites are mutated (Fig. 2g, stippled bar). Consistent with our observations, Fong and Yildiz reported repression of rtxBDE by CRP in their transcriptome analysis (41).
TABLE 1 .
CRP binding peaks identified by ChIP-seq in the V. cholerae strain N16961 genome
| Genome component |
Peak centera | Site centerb | Site P valuec | Sequenced | Gene(s) close to CRP binding sitese |
CRP regulatedf |
|---|---|---|---|---|---|---|
| Chromosome I | 55771 | 55796.5 | 3.5E−03 | AGTGACTAAGCGTACA | 23Sa1 | ND |
| 99871 | 99790.5 | 8.3E−03 | TGTTACGAATATTACA | glpE<(VC0103) | Yes | |
| 134815 | 134755.5 | 5.6E−03 | TTTGTTTTGGATCGAT | VC0142a<>VC0143 | Yes | |
| 150732 | 150753.5 | 3.2E−07 | TGAGATTCAAATCACA | VC0159<>16Sb | Yes | |
| 177320 | 177327.5 | 3.2E−03 | CATAATCTGTATCAAA | VC0175 | Yes | |
| 242521 | 242505.5 | 7.1E−03 | TAAGGTTTAAGCCATT | (VC0237) | No | |
| 249227 | 249187.5 | 9.8E−03 | TTTGAAGGATGGCGTT | (VC0242) | No | |
| 264229 | 264210.5 | 5.1E−03 | TTAAATATGTATCATA | VC0258><VC0259 | No | |
| 274582 | NAg | NA | NA | (VC0269) | No | |
| 294153 | 294127.5 | 7.6E−03 | TGTAGGTGATATCTCA | VC0284 | No | |
| 454607 | 454517.5 | 3.4E−03 | TGTGTTGTTGCTCAAT | VC0423<>VC0424 | Yes | |
| 516492 | 516436.5 | 1.7E−03 | TGACAGTAATATCACT | VC0485<>VC0486 | No | |
| 522621 | 522571.5 | 5.3E−03 | AGTCCATTTGCTCACA | VC0489 | No | |
| 527963 | 527988.5 | 1.1E−03 | AGTTATTTTTTTCACT | VC0493<>VC0494 | No | |
| 569841 | 569895.5 | 4.9E−04 | AACGATTTTCCTCATA | VC0537<>VC0538 | Yes | |
| 657711 | 657766.5 | 5.2E−05 | TGTGACTCCCTTCGCA | VC0621 | No | |
| 676093 | 676049.5 | 2.4E−03 | AATGATATAAATCCAA | ompU<>greA | Yes | |
| 707829 | 707908.5 | 7.3E−03 | GCCGCTTGGCATCACA | VC0661<>VC0662 | No | |
| 712256 | 712206.5 | 1.6E−03 | TGCAATCTAAGTCATT | VC0665 | No | |
| 713923 | 714007.5 | 1.1E−03 | TTAGAATTTAATCGTA | VC0666 | No | |
| 748263 | 748338.5 | 4.1E−03 | GGCGAGATTACGCGTA | VC0699<>VC0700 | No | |
| 756779 | 756854.5 | 8.4E−07 | TGTGATAAAAGTCACT | VC0706 | No | |
| 762509 | 762522.5 | 1.8E−03 | TCTGACAATTATCTCG | VC0713 | Yes | |
| 788515 | 788501.5 | 6.9E−03 | TGTGAAATTTCACAAG | VC0734 | No | |
| 815345 | 815346.5 | 3.6E−07 | TGTGATATGATTCACA | engA | Yes | |
| 818079 | 818158.5 | 2.6E−03 | GGTTAATTAAGTCGCA | VC0765 | Yes | |
| 819917 | 819936.5 | 1.0E−02 | CGTCCGCAATATCAAA | VC0766<>VC0767 | No | |
| 880295 | 880357.5 | 5.3E−03 | TATGAGAAAGATAAAA | (VC0821) | No | |
| 888697 | 888746.5 | 2.1E−03 | TGCAATTAAGTTCTCA | tcpI<>tcpP | Yes | |
| 894874 | 894817.5 | 7.9E−03 | TATTATTGGATTCATT | (VC0833) | No | |
| 904634 | NA | NA | NA | (VC0842) | Yes | |
| 906219 | NA | NA | NA | acfA<>acfD | No | |
| 911066 | 911128.5 | 3.1E−03 | TATGATGAAAAACATT | VC0845><VC0846 | No | |
| 936058 | 936026.5 | 1.5E−03 | AAAGAGCTAAATCGTT | (VC0870) | No | |
| 999018 | 999093.5 | 9.6E−03 | CTTGGTTGTTTTCAAT | VC0932<>VC0934 | Yes | |
| 1011835 | 1011745.5 | 2.4E−03 | AGTGAGCTTGCCCAAG | (VC0947) | No | |
| 1037560 | 1037566.5 | 1.2E−03 | TTCGACGCATTTCAAA | VC0972 | Yes | |
| 1054013 | 1054031.5 | 1.4E−05 | CGTGATTTTTGTCGCG | tppB<>rfaH | No | |
| 1061159 | 1061198.5 | 1.5E−06 | GGTGATTAGGATCACA | nagA<>VC0995 | No | |
| 1090145 | 1090112.5 | 5.6E−04 | TGTGATGTTTGGCATC | VC1021 | No | |
| 1100204 | 1100264.5 | 4.0E−05 | TGTGATGCAAATCGAT | VC1034 | Yes | |
| 1139471 | 1139535.5 | 4.0E−03 | TCTGATTATTTTCAAG | VC1073 | No | |
| 1174954 | 1174946.5 | 6.7E−06 | TGTGGTTTATGTCACA | VC1104 | No | |
| 1198758 | 1198847.5 | 6.0E−05 | TGTGAGCTGTGGCACT | VC1130<>VC1131 | No | |
| 1212539 | 1212568.5 | 4.7E−03 | AGAGGCGAAATTCATT | VC1142<>clpS | Yes | |
| 1224786 | 1224787.5 | 4.2E−06 | TGTGATACTGGTCTCA | VC1152<>tfoX | No | |
| 1382925 | 1382894.5 | 5.9E−03 | TGTGAGAATTGTTAAT | VC1301 | Yes | |
| 1396772 | 1396717.5 | 1.2E−03 | ATTGATGTCACTCAAA | VC1313<>VC1314 | Yes | |
| 1408936 | 1408937.5 | 3.8E−03 | TTTTAACTGGTTCACA | VC1323<>VC1325 | Yes | |
| 1549042 | 1549038.5 | 6.9E−03 | TGTGCAATTTGTCTGA | rtxB<>rtxH | Yes | |
| 1568164 | 1568072.5 | 5.5E−03 | TATGAAAATGATGATA | ctxA | No | |
| 1683652 | 1683636.5 | 2.0E−03 | AGTGATGGGGTTAACA | VC1571<>VC1572 | No | |
| 1703584 | 1703620.5 | 6.4E−03 | TAATAAAAATGTCACA | VC1592 | No | |
| 1741600 | 1741668.5 | 4.7E−05 | TGTGATACGCTTCTCG | VC1620<>VC1621 | Yes | |
| 1776678 | 1776642.5 | 4.8E−03 | AGTGATTTATCACTAA | VC1649<>VC1650 | No | |
| 1789510 | 1789532.5 | 1.9E−05 | TATGACCAGTATCGCA | VC1656<>VC1658 | No | |
| 1903470 | 1903498.5 | 8.0E−04 | TTTGAGTTAATTCAAT | (VC1736) | Yes | |
| 1919651 | 1919579.5 | 6.2E−03 | TGTGCTAAATACAACG | (VC1771) | Yes | |
| 1922932 | NA | NA | NA | (VC1773) | No | |
| 1967295 | 1967278.5 | 2.1E−06 | CGAGATCTAAATCACA | VC1825<>VC1826 | No | |
| 1984776 | 1984846.5 | 3.3E−04 | TGAGAACTTTGTCAAA | VC1844 | Yes | |
| 1990074 | 1990055.5 | 4.4E−03 | GTCGAGACCACTCATA | VC1851 | No | |
| 1994054 | 1994088.5 | 5.4E−03 | ATTAATAAAAATCAAA | ompT<>dinG | Yes | |
| 2004839 | 2004771.5 | 2.9E−03 | TTTTAACAAAGTCACA | VC1864<>VC1865 | Yes | |
| 2055395 | 2055442.5 | 6.2E−03 | CATCAAATTTTTCACA | VC1904<>VC1905 | Yes | |
| 2059077 | 2059085.5 | 9.7E−03 | TGCCACGCAACGCTCA | cysB<>VC1909 | Yes | |
| 2168387 | 2168407.5 | 4.2E−03 | TTTGAGGAATTCCGCT | VC2013 | Yes | |
| 2190666 | 2190734.5 | 5.0E−03 | TGTGCGAATGTTAACA | VC2035 | No | |
| 2193110 | 2193078.5 | 1.1E−05 | AACGATATAAATCACA | VC2036<>VC2037 | Yes | |
| 2374476 | 2374498.5 | 3.0E−05 | TGTGAGCTTTATCATG | VC2219<>VC2220 | No | |
| 2433743 | 2433742.5 | 1.1E−06 | GGTGATTAAAATCACA | VC2278<>VC2279 | No | |
| 2457608 | 2457631.5 | 1.3E−06 | AGCGATTAAGATCACA | VC2303<>VC2305 | No | |
| 2537389 | 2537396.5 | 4.6E−03 | TGTGAATTCGGTGAAA | gltB | No | |
| 2550352 | 2550386.5 | 8.7E−03 | TGTTACTGGTATAACA | (VC2385) | No | |
| 2551356 | 2551299.5 | 3.5E−03 | AGTGATAAAAGTGAAG | (VC2386) | No | |
| 2558608 | 2558615.5 | 3.7E−03 | GATGAATTTATTCATC | VC2390 | Yes | |
| 2610382 | 2610307.5 | 9.8E−03 | GCTGATTCGCGTCTTG | VC2435<>tolC | No | |
| 2653838 | 2653780.5 | 7.5E−04 | CGCGAGTCTCTTCAAA | VC2473 | Yes | |
| 2667326 | 2667406.5 | 8.8E−03 | TAATATTCACGTCAAA | VC2486 | No | |
| 2699390 | 2699329.5 | 1.5E−03 | GGTGATGGTCGCCACT | pyrB | No | |
| 2743349 | 2743361.5 | 8.1E−04 | ATCGCGTCACATCACA | VC2561<>cpdB | No | |
| 2787939 | 2787903.5 | 3.0E−04 | TGAGATAAACCCCACA | VC2618 | Yes | |
| 2845246 | 2845280.5 | 5.7E−07 | TGTGATTTTCATCACG | VC2677 | No | |
| 2864757 | 2864763.5 | 9.2E−04 | ATAGATAAAACTCTCA | VC2698<>aspA | Yes | |
| 2933468 | 2933432.5 | 7.5E−04 | TTTGATTATCATCAAC | 16sg | ND | |
| 2936869 | 2936904.5 | 3.1E−04 | TTCGATACCAAGCACA | 23Sh | ND | |
| Chromosome II | 12067 | 12085.5 | 5.4E−08 | TGTGATCCGAATCACT | VCA0012<>VCA0013 | Yes |
| 86364 | 86274.5 | 5.8E−03 | GTCGAAATTCGCCACA | VCA0076 | No | |
| 99016 | 98927.5 | 2.0E−07 | TGTGATCTTTATCACT | VCA0089 | No | |
| 114856 | 114864.5 | 8.6E−03 | TTTAATAGATTTCTCA | VCA0104<>VCA0105 | No | |
| 152867 | 152849.5 | 7.4E−04 | TGTGATTGATGTGGCA | VCA0138 | No | |
| 181749 | 181688.5 | 2.5E−03 | TGAGAAAGCATTCAAA | VCA0164<>VCA0165 | No | |
| 217815 | 217798.5 | 6.0E−03 | TGTTATAAAAACCAAT | (VCA0200) | No | |
| 237015 | 237049.5 | 6.6E−03 | TAAGAATTATTTTACA | hlyB<>hlyA | No | |
| 247246 | 247185.5 | 7.5E−03 | TTGGCATAGCATCACA | VCA0224<>VCA0225 | Yes | |
| 267292 | 267253.5 | 3.7E−03 | TGATAGGTAGATCACC | VCA0246<>VCA0247 | No | |
| 300413 | 300391.5 | 8.4E−03 | TGCCCTATCTATCAAA | VCA0281 | Yes | |
| 334916 | 334914.5 | 4.3E−03 | ATTGACAGCTATCTAA | (VCA0334) | No | |
| 458259 | 458254.5 | 3.8E−03 | CGTGATTAAAAACGTC | VCA0523 | Yes | |
| 481906 | 481918.5 | 2.0E−03 | TTTCATAAAAGTCACG | VCA0544<>VCA0545 | Yes | |
| 492167 | 492243.5 | 6.9E−07 | TGTGATTGGAATCACT | VCA0554<>VCA0556 | No | |
| 564616 | NA | NA | NA | (VCA0628) | Yes | |
| 598381 | 598370.5 | 4.9E−04 | GTTGACAACAGTCACA | (VCA0662)<>VCA0663 | No | |
| 630430 | 630499.5 | 1.5E−03 | AATGATAGATAACACA | VCA0691 | Yes | |
| 687485 | 687472.5 | 3.3E−03 | CGTGATCGACATTAAA | (VCA0742)>VCA0743 | No | |
| 741821 | 741822.5 | 7.8E−06 | TGTGCTTTACATCACT | VCA0801 | Yes | |
| 784413 | 784364.5 | 2.1E−05 | TGTGATGCCGCTCGCA | VCA0840 | Yes | |
| 785337 | 785352.5 | 5.1E−04 | TTTGAACTTAGTCATT | VCA0843 | Yes | |
| 801056 | 801043.5 | 1.7E−05 | TGTGAAATGGCTCGCA | VCA0849 | Yes | |
| 832501 | 832514.5 | 6.2E−03 | TGCGACCTTGATTAAC | VCA0880 | Yes | |
| 849892 | 849906.5 | 3.0E−03 | GTTGACGCCTTTCTCA | VCA0896 | Yes | |
| 870862 | 870876.5 | 1.1E−03 | AATGATCAGGGGCAAA | VCA0917<>VCA0919 | Yes | |
| 874452 | 874411.5 | 4.1E−03 | TATAAATCAAATCATT | VCA0923 | Yes | |
| 897315 | 897352.5 | 4.8E−06 | AGCGAGCCAAATCACA | VCA0945<>VCA0946 | Yes | |
| 902411 | 902377.5 | 6.6E−03 | TGAAACACTTACCACT | VCA0952 | Yes | |
| 930851 | NA | NA | NA | (VCA0982)<>VCA0983 | No | |
| 963517 | 963536.5 | 8.2E−04 | TGTTAAGCAAATCGCA | VCA1013<>VCA1015 | No | |
| 994614 | 994588.5 | 2.5E−05 | CATGACACAGGTCACA | VCA1043<>(VCA1044) | Yes | |
| 1015957 | 1015902.5 | 2.0E−05 | TTTGACCATTATCACA | VCA1063 | No | |
Center of peak for CRP binding in ChIP-seq assays.
Center of binding site identified by FIMO (Find Individual Motif Occurrences) using DNA motif recovered from the ChIP-seq data by MEME (Multiple Em for Motif Elicitation).
P value assigned to each site by FIMO describing the significance of the match to the motif generated by MEME.
DNA sequence of site identified by FIMO.
Parentheses indicate that the CRP site is located within that gene. Pairs of arrows represent divergent (<>) or convergent (><) genes. Single arrows indicate that gene pairs are in the same orientation on either the forward (>) or reverse (<) strand. Gene identification numbers are shown unless an alternative name for the gene is provided in the genome annotation or the wider literature. Genes regulated by ToxR or ToxT are underlined.
CRP-regulated genes described by Fong and Yildiz (41). ND, not detected: genes encoding stable rRNA species were not included in the transcriptome analysis and so no change in transcription could be detected.
NA, not applicable.
FIG 2 .
Repression of the rtxBDE operon by CRP. (a) The intergenic region between rtxBDE and rtxHCA is associated with CRP but not σ70. The graphs illustrate ChIP-seq data for CRP (orange) and σ70 (blue) binding to the rtx locus. Data have been smoothed in a 100-bp window. Genes are depicted by mauve arrows and labeled. (b) Sequence of the rtxBDE-rtxHCA gene regulatory region. The DNA sequence between rtxBDE and rtxHCA is shown. The center of the CRP binding peak identified in our ChIP-seq analysis is indicated by an asterisk. Putative CRP sites (orange) are underlined and labeled. The rtxH transcription start site (+1) is underlined and further highlighted by a bent arrow. The associated promoter −10 element is similarly colored and underlined. Two transcription start sites for rtxB are also labeled in the same way. The 5′ end of the rtxB.1 DNA fragment (see the legend to panel e) is indicated by an inverted black triangle. Point mutations used to inactivate CRP binding sites or promoter −10 elements are shown in red. (c) DNase I footprint of CRP binding to the rtxBDE-rtxHCA gene regulatory region. Results of a DNase I footprinting experiment using the rtxBDE intergenic region and purified V. cholerae CRP. The experiment is calibrated with a Maxam-Gilbert GA sequencing ladder, and positions relative to the P1rtxB transcription start site (+1) are labeled. The triangle indicates the addition of CRP at concentrations of 175, 350, 700, 1,400, 2,100, or 2,800 nM. The positions of the predicted CRP binding sites are shown by orange boxes. (d) Primer extension analysis of rtxH and rtxB promoter-derived transcripts. The gel shows arbitrary Sanger sequencing reactions for calibration (lanes 1 to 4) and primer extension products for rtxB (lanes 5 and 6) or rtxH (lanes 7 and 8) promoter-derived transcripts. (e) Transcripts derived from the rtxBDE intergenic region in vitro. The gel shows transcripts generated by V. cholerae RNA polymerase σ70 holoenzyme using the rtxBDE intergenic region, cloned in plasmid pSR, as a DNA template. The RNAI transcript is derived from the plasmid replication origin and serves as an internal control. The rtxB.1 derivative contains a truncated version of the rtxBDE intergenic region. The site of the truncation is marked in panel b. Mutations introduced to disrupt potential −10 hexamers are noted above the gel and are also shown in panel b. (f) Activity of PrtxH is not affected by CRP. Results of a β-galactosidase assay done using lysates of N16961 cells transformed with derivatives of the lacZ reporter plasmid, pRW50T, where lacZ expression is controlled by PrtxH. (g) Expression of rtxB is repressed by CRP. Results of a β-galactosidase assay done using lysates of N16961 cells transformed with derivatives of the lacZ reporter plasmid, pRW50T, where lacZ expression is controlled by P1rtxB and P2rtxB.
(a) Transcription from PrtxH requires identified promoter elements. (i) The results of primer extension assays that detect the rtxH transcript derived from plasmid pRW50T carrying the rtxH (panel ii, top) or truncated rtxH.1 (panel ii, bottom) DNA fragment. (ii) PrtxH is highlighted blue and CRP binding sites are shown in orange. (b) P1rtxB and P2rtxB make similar contributions to rtxB transcription. The figure shows β-galactosidase activity measurements for lysates of V. cholerae cells transformed with pRW50T derivatives carrying the full-length rtxB regulatory region (rtxB), a truncated derivative lacking CRP binding sites (rtxB.1), or versions of the truncated fragment with indicated promoter mutations. (c) Effects of crp and tcpA on zebrafish larva colonization. (i) Images from multiple zebrafish larvae colonized with the indicated V. cholerae strains. All strains were transformed with plasmid pMW-GFP to facilitate visualization of bacteria. (ii) Quantified fluorescence from multiple microscopy images. (d) Raw gel images. Download FIG S1, PDF file, 0.4 MB (404.3KB, pdf) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
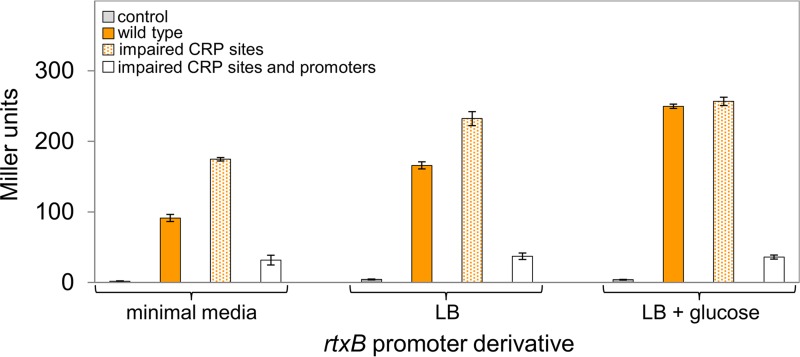
Expression of the rtxBDE operon responds to nutrient availability in a CRP-dependent manner.
The ability of CRP to bind DNA in vivo is regulated by nutrient availability. Hence, CRP binds to target sites when cells are grown in M9 minimal medium but binding is reduced in lysogeny broth (LB) and abolished upon the addition of glucose (25–28). As such, repression of rtxBDE should be relieved in rich medium. To test this, we used strain N16961 carrying the rtxB::lacZ fusions on pRW50T. The various strains were grown in M9 minimal medium, LB broth, or LB broth supplemented with 0.4% glucose. As expected, β-galactosidase activity due to the rtxB::lacZ fusion increased in LB broth and rose further upon the addition of glucose (Fig. 3, compare solid orange bars). Furthermore, inactivation of the CRP sites had a reduced effect in LB broth and no effect when glucose was present (Fig. 3, compare solid and stippled orange bars for each growth condition). Importantly, the sizes of changes in gene expression observed were similar to data for other CRP-regulated promoters (31). We confirmed that the observed gene expression was due to P1rtxB and P2rtxB. Hence, mutation of the promoter −10 elements greatly reduced lacZ expression (Fig. 3, open bars).
FIG 3 .
Nutrient availability controls rtxBDE expression in a CRP-dependent manner. Results of β-galactosidase assays done using lysates of N16961 cells transformed with derivatives of the lacZ reporter plasmid, pRW50T, carrying different rtxB::lacZ fusions. Cells were grown in M9 minimal medium, LB, or LB supplemented with 0.4% (vol/vol) glucose.
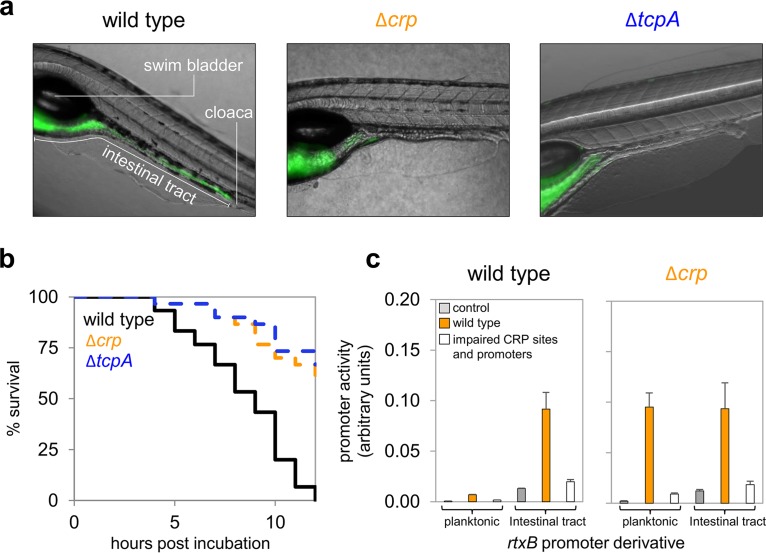
CRP plays an important role during colonization of an aquatic host.
Although largely overlooked as a vector for V. cholerae, a recent study found that up to 87% of fish species were colonized by the bacterium in certain localities (7). Indeed, it has been suggested that colonization of fish has sustained the epidemicity of cholera in India (8). Since fish and humans have similar gut mucosa, the former have emerged as a model to study intestinal colonization (9, 10). The zebrafish larva model is particularly useful; bacteria are added to saltwater solutions in which larvae are free swimming and colonization follows without intervention (10). Given that changes in nutrient availability are associated with host colonization, we examined the role of CRP in this process. Figure 4a shows representative images of zebrafish larvae infected with V. cholerae strain E7946 or derivatives. Further images for each strain are shown in Fig. S1c. All strains express green fluorescent protein to facilitate their visualization. Infections due to the wild-type strain are disseminated throughout the intestinal tract (Fig. 4a, left). Conversely, infections caused by cells lacking CRP or TCP are limited to the upper intestinal tract and fail to colonize the midintestine and posterior intestine (Fig. 4a, middle and right). Quantification of fluorescence in microscopy images revealed 3-fold reductions for the Δcrp and ΔtcpA strains relative to the amount in the wild type (Fig. S1c). We also monitored survival of the larvae during incubation with the bacteria (Fig. 4b). All larvae infected with wild-type V. cholerae were dead by the end of the time course (Fig. 4b, black line). Conversely, infections caused by strains lacking CRP or TCP were not usually fatal (Fig. 4b, orange and blue lines). Hence, both CRP and TCP are important for colonization of fish (22).
FIG 4 .
CRP is required for efficient host colonization and dependent induction of rtxBDE. (a) Colonization of the zebrafish larva intestinal tract by V. cholerae strain E7946 and derivatives. The three panels show representative fluorescence microscopy images overlaid on light microscopy images of zebrafish larvae colonized with the indicated V. cholerae strains. All bacterial strains were transformed with plasmid pMW-GFP and express green fluorescent protein (GFP) to facilitate detection. Further images are shown in Fig. S1c in the supplemental material. (b) Survival of zebrafish larvae following infection with V. cholerae strain E7946 and derivatives. (c) Expression of rtxBDE is induced by zebrafish larva colonization. Results of a β-galactosidase assay done using lysates of bacterial cells growing planktonically in E3 medium or obtained from the zebrafish intestinal tract. Strains are indicated and were transformed with derivatives of pRW50T encoding different rtxB::lacZ fusions.
Induction of the rtxBDE operon during host colonization is mediated by CRP.
We next considered the possibility that transcription from the rtxBDE promoters might be triggered during colonization of the intestinal tract. To test this, zebrafish larvae were colonized with either wild-type or Δcrp derivatives of V. cholerae carrying rtxB::lacZ fusions in plasmid pRW50T. After colonization, planktonic bacteria were recovered from the water and larvae were sacrificed to release the intestinal bacteria. The levels of β-galactosidase expression were then determined from lysates of the two populations. The data obtained for wild-type V. cholerae are shown in the left panel of Fig. 4c. Low rtxBDE expression was measured for planktonic V. cholerae. However, rtxBDE expression increased substantially during colonization of the larval intestinal tract. As expected, this increase in expression required P1rtxB and P2rtxB (Fig. 4c, compare orange and open bars). In cells lacking CRP, the expression of rtxBDE was uncoupled from host colonization. Hence, high rtxBDE expression was measured in planktonic as well as intestinal populations (Fig. 4c, right). Note that the differences in gene expression observed are not due to the different colonization properties of the Δcrp strain; deregulation of rtxBDE occurs in planktonic populations rather than within the larvae. Furthermore, any differences in bacterial cell numbers were accounted for by normalization.
CRP modulates the expression of many V. cholerae genes during host colonization.
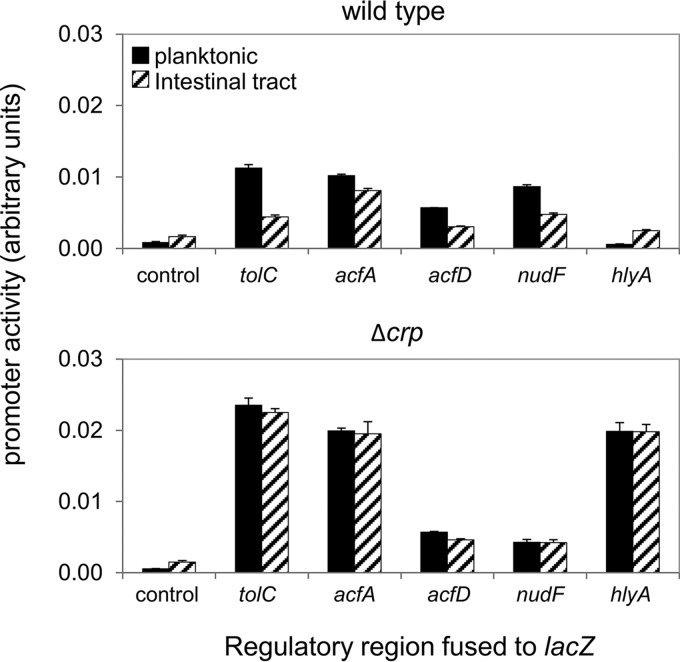
We reasoned that other CRP-targeted promoters would lose the ability to differentiate between aquatic environments and the host intestinal tract when CRP was absent. To test this, the promoters of the following five genes were selected, using our ChIP-seq data as a guide: tolC (encoding an outer membrane channel important for bile tolerance), acfA (encoding accessory colonization factor A), acfD (encoding accessory colonization factor D), nudF (encoding a pyrophosphatase), and hlyA (encoding hemolysin) (44). The promoter region of each gene was cloned upstream from lacZ in plasmid pRW50T, and the β-galactosidase activity was determined (Fig. 5a). The data show that all of the genes were expressed at different levels in planktonic (solid bars) and intestinal (striped bars) populations. The experiment was repeated in cells lacking Δcrp (Fig. 5b). The expression of all genes was rendered insensitive to host colonization (Fig. 5b, compare solid and striped bars).
FIG 5 .
CRP couples the expression of many V. cholerae genes to host colonization. Results of β-galactosidase assays done using lysates of bacterial cells grown planktonically in E3 medium or obtained from the zebrafish intestinal tract. Strains are indicated and were transformed with derivatives of pRW50T encoding different LacZ fusions. Significant differences between levels of β-galactosidase activity in planktonic and intestinal populations were observed in all cases for wild-type cells (P values determined using a two-tailed Student’s t test were 0.0014, 0.0011, 0.0003, 0.0005, and 0.0037 for the tolC, acfA, acfD, nudF, and hlyA promoters, respectively). For cells lacking CRP, a significant, albeit much smaller difference was only apparent for the acfD promoter (P = 0.0036).
DISCUSSION
The ability of V. cholerae to persist in environmental reservoirs, colonize the intestinal tract, and cause disease requires careful coordination of gene expression (5). This process is best characterized for key virulence factors that collectively reside in the ToxR regulon (5, 12, 14). In this paper, we have investigated the role of CRP. We show that CRP targets five of the nine ToxR regulon transcription units. Hence, we identified binding sites for CRP adjacent to ompU, acfA, and acfD, in addition to the known targets ompT and tcpPH. Other genes involved in V. cholerae pathogenicity were also targets for CRP. These included rtxBDE, tolC, and hlyA. Previous transcriptome analysis led to speculation that CRP may modulate the expression of V. cholerae virulence factors in response to host colonization (33). Here, we have tested this prediction using the zebrafish larva colonization model (9). For all genes examined, differential expression between planktonic and intestinal populations required CRP. We have paid particularly close attention to genes encoding the RTX toxin export machinery. Our data are consistent with repression of rtxBDE by CRP that is relieved within the intestinal tract and other nutrient-rich environments. Previous host colonization studies support our model. In particular, Mandlik and colleagues previously monitored global transcription in V. cholerae using RNA-seq (45). Their data demonstrate induction of rtxBDE within the intestinal tracts of both mice and rabbits. Furthermore, the same study detected repression of rtxBDE in M9 minimal medium compared to its expression in LB broth. Similarly, Boardman et al. noted repression of rtxBDE in nutrient-poor environments (46). We argue that CRP mediates these effects directly by binding sites upstream from the rtxBDE promoters (Fig. 2 to 4). Curiously, while the RTX toxin export machinery is expressed only upon nutrient upshift, the divergent rtxHCA genes that encode the RTX toxin appear constitutively transcribed (Fig. 2). Indeed, posttranscriptional control by the VqmR small RNA (sRNA) has previously been shown to regulate RTX toxin expression (45). We speculate that this allows the system to exist in a poised state so that toxin export is only triggered within a host organism.
Our data show that CRP is required for control of V. cholerae genes in addition to the rtxBDE operon in response to the intestinal environment (Fig. 5). Hence, CRP is integral to the regulatory network that controls V. cholerae lifestyle switching. This is intriguing, given that oral rehydration therapies (ORT) used to treat the effects of cholera contain glucose (26, 47). Hence, it is possible that ORT modification could be used to modulate the ability of V. cholerae to colonize a host and cause disease (26, 47). Complete dissection of the role CRP plays during V. cholerae lifestyle switching should provide an evidence base for any such ORT modification. In this regard, our findings provide an important starting point for further studies.
MATERIALS AND METHODS
Strains, plasmids, and oligonucleotides.
V. cholerae strains N16961 and E7946 are described by Heidelberg et al. and Miller et al., respectively (39, 48). The Δcrp derivative of E7946 was constructed by MuGENT using PCR oligonucleotides listed in Table S1 in the supplemental material (49). The E. coli K-12 strains JCB387 and DH5α are described by Page et al. (50) and Taylor et al. (51) and were used for general cloning and conjugation, respectively. Plasmid pRW50T was constructed by excision of the DNA fragment comprising cynX from pRW50 (52) using NheI and BstEII. The oriT region was amplified from plasmid RK2 (53) in such a way that oriT was flanked by NheI and BstEII restriction sites to facilitate ligation at the locus from which cynX was excised. Plasmid pDCRP-Vc is a derivative of pDCRP (54) that encodes the CRP protein of V. cholerae rather than that of E. coli. More-detailed descriptions of strains and plasmids, along with sequences of oligonucleotides, are provided in Table S1.
Strains, plasmids, and oligonucleotides. Download TABLE S1, DOCX file, 0.03 MB (30.2KB, docx) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ChIP and DNA sequencing.
Immunoprecipitations with monoclonal anti-CRP and anti-σ70 antibodies (Neoclone, Madison, WI) were done as described by Haycocks et al. (26) using lysates of strain N16961 (39). Lysates were prepared from mid-log-phase cells cultured in M9 minimal medium supplemented with 1% (wt/vol) fructose. Libraries were prepared using immunoprecipitated protein-DNA complexes immobilized with protein A-Sepharose. DNA fragments were then given blunt ends, poly(A) tails, and bar codes. This was done using an NEB quick blunting and ligation kit, the Klenow fragment (5′-3′ exo-; NEB), and NEXTflex chromatin immunoprecipitation-DNA sequencing (ChIP-seq) barcodes (Bioo Scientific). Following elution of complexes from the protein-A Sepharose, cross-links were reversed, and bar-coded libraries were amplified by PCR. The number of PCR cycles was determined empirically for each library. After amplification, the library concentration was quantified using Qubit analysis and real-time PCR. Equimolar library concentrations were pooled and sequenced using an Illumina MiSeq instrument.
Bioinformatics.
The Fastq files obtained after DNA sequencing were converted into Fastq Sanger format, using FastqGroomer, and aligned to GenBank reference sequences (accession numbers AE003852.1 and AE003853.1) using BWA (Burroughs-Wheeler Aligner) for Illumina. The reference sequences correspond to chromosome I and chromosome II, respectively, of V. cholerae strain N16961. The resulting SAM (Sequence Alignment Map) files were converted to BAM (Binary Alignment Map) format using SAM-to-BAM. For each experiment, coverage per base was determined using multiBamSummary. Subsequent processing was done using R. Data were normalized to the same average read depth, and mean coverage per base was determined for each pair of biological replicates. Signals due to nonspecifically immunoprecipitated DNA present in a mock experiment were subtracted from the final binding profiles. To select peaks for CRP or σ70 binding, we used Artemis to generate a coverage plot and selected peaks. The peak centers were set as the center of the region passing the cutoff rounded to the nearest integer. Peaks for CRP and σ70 were defined as overlapping if the peak centers were within 250 bp of each other.
Proteins.
The V. cholerae CRP protein was expressed in E. coli strain M182Δcrp and purified using cAMP-agarose as described previously (54). The V. cholerae RNA polymerase was purified using a method derived from that of Burgess and Jendrisak (55). Briefly, V. cholerae strain N16961 was grown to mid-log phase in 8 liters of LB medium. Cells were harvested by centrifugation and resuspended in 100 ml of lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 2 mM MgCl2, 0.1 mM dithiothreitol [DTT], 2 mM EDTA, 1 mM 2-mercaptoethanol, 5% glycerol, 0.2% Triton X-100, and 0.25 mg/ml lysozyme). One protease inhibitor cocktail tablet (Roche) was added per 20 ml of buffer. Cell lysis and DNA shearing were done using four 30-s pulses, at 20% output, with a Misonix, Inc., XL2020 tip sonicator. Lysates were cleared by centrifugation at 39,000 × g for 45 min at 4°C. Following filtration (0.45-µm filter), polymin P and ammonium sulfate precipitations were done as described in Burgess and Jendrisak (55). Precipitated protein was resuspended in TGED buffer (10 mM Tris-HCl, pH 7.9, 5% glycerol, 0.1 mM EDTA, and 0.1 mM DTT) containing 100 mM NaCl and passed through a HiPrep heparin FF column (GE Healthcare). The column was washed with 0.1 M NaCl TGED, and RNA polymerase was eluted in TGED using a gradient to 1 M NaCl. RNA polymerase-containing fractions were pooled and protein precipitated using ammonium sulfate. After resuspension in TGED, RNA polymerase was further purified using a Mono Q HR column (GE Healthcare). Column washing and protein elution were as described in the previous step. RNA polymerase-containing fractions were pooled and dialyzed against −80°C storage buffer (TGED, 0.1 M NaCl, 50% glycerol).
DNase I footprinting and in vitro transcription.
For electrophoretic mobility shift assay (EMSA) experiments, DNA fragments were prepared using PCR as described by Shimada et al. (56), with oligonucleotides listed in Table S1. Protein binding and subsequent electrophoresis were done as described by Chintakayala et al. (57). For footprinting experiments, DNA fragments were prepared as described by Grainger et al. (58). Protein binding, DNA digestion, and electrophoresis were done as described by Singh and Grainger (59). Briefly, DNA fragments were labeled at one end using [γ-32P]ATP and T4 polynucleotide kinase and used at a final concentration of ~10 nM in footprinting reactions. All reaction mixtures contained an excess of herring sperm DNA (12.5 µg ml−1) as a nonspecific competitor. Our in vitro transcription assays were done as described by Haycocks et al. (26). DNase I-digested DNA and in vitro-generated RNA transcripts were analyzed on 6% DNA sequencing gels (Molecular Dynamics). The results were visualized using a Fuji phosphor screen and Bio-Rad Molecular Imager FX. Raw gel images are in Fig. S1d.
Primer extension assays.
Transcript start sites were mapped by primer extension, as described previously (59), using RNA purified from a V. cholerae strain carrying the appropriately oriented rtxBDE-rtxHCA intergenic region cloned in pRW50T. The 5′-end-labeled primer D49724, which anneals downstream from the HindIII site in pRW50, was used in all experiments. Primer extension products were analyzed on denaturing 6% polyacrylamide gels calibrated with size standards derived from M13mp18 phage DNA sequencing reactions. Gels were visualized using a Fuji phosphor screen and Bio-Rad Molecular Imager FX.
β-Galactosidase assays.
β-Galactosidase assays using lysates of liquid V. cholerae cultures were done as described previously (26) following the protocol of Miller (60). For experiments with zebrafish larva, colonization by V. cholerae was first instigated as described below. E3 medium was prepared as a 1 liter 50× stock containing 14.6 g NaCl, 0.65 g KCl, 2.20 g CaCl2, 4.05 g MgSO4, and 23.85 g HEPES adjusted to pH 7. A 1× dilution was prepared using ddH2O. Following infection, the larvae were euthanized with 2.5 mg/ml tricaine and the E3 medium was agitated to resuspended bacteria that had sunk to the bottom of the well. The V. cholerae-containing E3 medium was then transferred to a sterile bijou and the larvae to a sterile 1.5-ml dolphin microcentrifuge tube. Fish were washed with E3 medium by gentle pipetting to remove residual bacteria. Larvae were then homogenized to release bacterial cells using a hand-held motorized homogenizer, and E3 was added so that the homogenate had a volume similar to that of the isolated medium. Two drops each of toluene and 1% (wt/vol) sodium deoxycholate were added to each sample, and the resulting cell lysates were assayed for β-galactosidase activity. To normalize for cell numbers, 0.5 µl of each cell suspension was diluted in 1.5 ml of E3 medium prior to cell lysis. One hundred microliters of this suspension was spread on LB agar plates containing 5 µg/ml tetracycline, 100 µg/ml streptomycin, 50 µg/ml spectinomycin, and 40 µg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). This allowed for confident selection of V. cholerae cells containing pRW50T derivatives that were enumerated by counting the number of colonies formed after overnight incubation at 37°C. All assay values are the means of the results of three independent experiments with a standard deviation equivalent to <10% of the mean β-galactosidase activity.
Zebrafish larva colonization and survival assays.
Adult zebrafish were kept at pH 7.5 and 26°C in a recirculating tank system, with light/dark cycles of 14/10 h, at the University of Birmingham aquatic facility. Zebrafish care, breeding, and experimentation were done according to the Animal (Scientific Procedures) Act 1986 (61) under home office project license 40/3681. Zebrafish embryos derived from the wild-type AB strain (62) were harvested in petri dishes containing water from the fish system. After harvesting, between 50 and 60 embryos were transferred to 90-mm petri dishes containing 25 ml of E3 medium supplemented with 0.03% (vol/vol) methylene blue and 0.02 mg/ml 1-phenyl 2-thiourea. The embryos were incubated at 32°C for 4 days with light/dark cycles of 14/10 h. The incubation medium was regularly replaced to minimize microbial contamination. On day 3 of the incubation, required strains of V. cholerae were streaked to generate single colonies that were used to inoculate 5 ml of M9 minimal medium. The resulting cultures were incubated overnight at 37°C with shaking. One milliliter of the overnight culture was transferred to 5 ml of fresh M9 minimal medium the following day. The resulting culture was incubated at 37°C with shaking until mid-log phase. Cells were then harvested by centrifugation and washed three times with 5 ml of E3 buffer by sequential resuspension and centrifugation. After washing, the cells were resuspended in 5 ml of E3 medium, the optical density was determined, and 106 cells were transferred into each well of a 24-well cell culture plate. The larvae were sedated by adding 166 µl of 40-mg/ml tricaine to the petri dish. Five larvae were then transferred to each well of the culture plate, which was incubated at 30°C overnight. Death was determined by loss of movement and heartbeat in opaque larvae that had settled at the bottom of the well.
Microscopy.
Zebrafish embryos colonized with V. cholerae were imaged using a Zeiss Axio Observer Z1 microscope with 10× objective for fluorescence and differential interference contrast. Prior to visualization, embryos were immobilized in 0.4% low-melting-point agarose in E3 buffer and 160 µg/ml tricaine. Imaging was done at 32°C and humidity maintained at 80% using an OkoLab stage. The ImageJ image processing package (NIH) software was used to visualize the images and merge the fields.
Data availability.
DNA sequencing reads are stored in ArrayExpress under accession number E-MTAB-6472. Genome Browser files (Data Set S1 to S6) and instructions (Text S1) are provided in the supplemental material.
N16961 chromosome I GenBank file. Download DATA SET S1, TXT file, 6 MB (6.2MB, txt) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
N16961 chromosome II GenBank file. Download DATA SET S2, TXT file, 2.2 MB (2.2MB, txt) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
CRP ChIP-seq Artemis graph file for N16961 chromosome I. Download DATA SET S3, TXT file, 17.5 MB (18MB, txt) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
CRP ChIP-seq Artemis graph file for N16961 chromosome II. Download DATA SET S4, TXT file, 6.4 MB (6.5MB, txt) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RNA polymerase σ70 ChIP-seq Artemis graph file for N16961 chromosome I. Download DATA SET S5, TXT file, 10.7 MB (10.9MB, txt) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RNA polymerase σ70 ChIP-seq Artemis graph file for N16961 chromosome II. Download DATA SET S6, TXT file, 4.2 MB (4.3MB, txt) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Instructions for viewing ChIP-seq data in the Artemis genome browser. Download TEXT S1, PDF file, 0.6 MB (667.8KB, pdf) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
We thank Jenny Ritchie, Brendan Wren, Joe Wade, Chris Thomas, Rachel Kettles, and Doug Browning for advice and support.
This work was funded by BBSRC grant number BB/N005961/1 awarded to D.C.G. and A.-M.K. and NIH grant number AI055058 to A.C. The Islamic Development Bank supported J.M.-R. with the award of a Ph.D. scholarship.
Footnotes
Citation Manneh-Roussel J, Haycocks JRJ, Magán A, Perez-Soto N, Voelz K, Camilli A, Krachler A-M, Grainger DC. 2018. cAMP receptor protein controls Vibrio cholerae gene expression in response to host colonization. mBio 9:e00966-18. https://doi.org/10.1128/mBio.00966-18.
REFERENCES
- 1.Clemens JD, Nair GB, Ahmed T, Qadri F, Holmgren J. 2017. Cholera. Lancet 390:1539–1549. doi: 10.1016/S0140-6736(17)30559-7. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization 2017. Cholera vaccines: WHO position paper—August 2017. Wkly Epidemiol Rec 92:477–498. http://apps.who.int/iris/bitstream/handle/10665/258763/WER9234.pdf;jsessionid=C7D00D4E8D44A2BC072C8340C5032E29?sequence=1.28845659 [Google Scholar]
- 3.World Health Organization 2013. Cholera, 2012. Wkly Epidemiol Rec 88:321–334. http://www.who.int/wer/2013/wer8831.pdf. [PubMed] [Google Scholar]
- 4.UPMC Centre for Health Security 2014. The costs and burden of infectious diseases. http://www.idcostcalc.org/index.html. Accessed 25 June 2018.
- 5.Nelson EJ, Harris JB, Morris JG Jr, Calderwood SB, Camilli A. 2009. Cholera transmission: the host, pathogen and bacteriophage dynamic. Nat Rev Microbiol 7:693–702. doi: 10.1038/nrmicro2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halpern M, Izhaki I. 2017. Fish as hosts of Vibrio cholerae. Front Microbiol 8:282. doi: 10.3389/fmicb.2017.00282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Senderovich Y, Izhaki I, Halpern M. 2010. Fish as reservoirs and vectors of Vibrio cholerae. PLoS One 5:e8607. doi: 10.1371/journal.pone.0008607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pandit CG, Hora SL. 1951. The probable role of the hilsa fish, Hilsa ilisa (Ham) in maintaining cholera endemicity in India. Indian J Med Sci 15:343–356. [Google Scholar]
- 9.Gomez D, Sunyer JO, Salinas I. 2013. The mucosal immune system of fish: the evolution of tolerating commensals while fighting pathogens. Fish Shellfish Immunol 35:1729–1739. doi: 10.1016/j.fsi.2013.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Runft DL, Mitchell KC, Abuaita BH, Allen JP, Bajer S, Ginsburg K, Neely MN, Withey JH. 2014. Zebrafish as a natural host model for Vibrio cholerae colonization and transmission. Appl Environ Microbiol 80:1710–1717. doi: 10.1128/AEM.03580-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor RK, Miller VL, Furlong DB, Mekalanos JJ. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc Natl Acad Sci U S A 84:2833–2837. doi: 10.1073/pnas.84.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrington DA, Hall RH, Losonsky G, Mekalanos JJ, Taylor RK, Levine MM. 1988. Toxin, toxin-coregulated pili, and the toxR regulon are essential for Vibrio cholerae pathogenesis in humans. J Exp Med 168:1487–1492. doi: 10.1084/jem.168.4.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peterson KM, Mekalanos JJ. 1988. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect Immun 56:2822–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies BW, Bogard RW, Young TS, Mekalanos JJ. 2012. Coordinated regulation of accessory genetic elements produces cyclic di-nucleotides for V. cholerae virulence. Cell 149:358–370. doi: 10.1016/j.cell.2012.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skorupski K, Taylor RK. 1997. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol Microbiol 25:1003–1009. doi: 10.1046/j.1365-2958.1997.5481909.x. [DOI] [PubMed] [Google Scholar]
- 16.Midgett CR, Almagro-Moreno S, Pellegrini M, Taylor RK, Skorupski K, Kull FJ. 2017. Bile salts and alkaline pH reciprocally modulate the interaction between the periplasmic domains of Vibrio cholerae ToxR and ToxS. Mol Microbiol 105:258–272. doi: 10.1111/mmi.13699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang M, Liu Z, Hughes C, Stern AM, Wang H, Zhong Z, Kan B, Fenical W, Zhu J. 2013. Bile salt-induced intermolecular disulfide bond formation activates Vibrio cholerae virulence. Proc Natl Acad Sci U S A 110:2348–2353. doi: 10.1073/pnas.1218039110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Provenzano D, Klose KE. 2000. Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc Natl Acad Sci U S A 97:10220–10224. doi: 10.1073/pnas.170219997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krasteva PV, Fong JC, Shikuma NJ, Beyhan S, Navarro MV, Yildiz FH, Sondermann H. 2010. Vibrio cholerae VpsT regulates matrix production and motility by directly sensing cyclic di-GMP. Science 327:866–868. doi: 10.1126/science.1181185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skorupski K, Taylor RK. 1999. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol Microbiol 31:763–771. doi: 10.1046/j.1365-2958.1999.01215.x. [DOI] [PubMed] [Google Scholar]
- 21.Kovacikova G, Skorupski K. 1999. A Vibrio cholerae LysR homolog, AphB, cooperates with AphA at the tcpPH promoter to activate expression of the ToxR virulence cascade. J Bacteriol 181:4250–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skorupski K, Taylor RK. 1997. Cyclic AMP and its receptor protein negatively regulate the coordinate expression of cholera toxin and toxin-coregulated pilus in Vibrio cholerae. Proc Natl Acad Sci U S A 94:265–270. doi: 10.1073/pnas.94.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kovacikova G, Skorupski K. 2001. Overlapping binding sites for the virulence gene regulators AphA, AphB and cAMP-CRP at the Vibrio cholerae tcpPH promoter. Mol Microbiol 41:393–407. doi: 10.1046/j.1365-2958.2001.02518.x. [DOI] [PubMed] [Google Scholar]
- 24.Srivastava D, Harris RC, Waters CM. 2011. Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J Bacteriol 193:6331–6341. doi: 10.1128/JB.05167-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Busby S, Ebright RH. 1999. Transcription activation by catabolite activator protein (CAP). J Mol Biol 293:199–213. doi: 10.1006/jmbi.1999.3161. [DOI] [PubMed] [Google Scholar]
- 26.Haycocks JR, Sharma P, Stringer AM, Wade JT, Grainger DC. 2015. The molecular basis for control of ETEC enterotoxin expression in response to environment and host. PLoS Pathog 11:e1004605. doi: 10.1371/journal.ppat.1004605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wade JT, Belyaeva TA, Hyde EI, Busby SJ. 2001. A simple mechanism for co-dependence on two activators at an Escherichia coli promoter. EMBO J 20:7160–7167. doi: 10.1093/emboj/20.24.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landis L, Xu JJ, Johnson RC. 1999. The cAMP receptor protein CRP can function as an osmoregulator of transcription in Escherichia coli. Genes Dev 13:3081–3091. doi: 10.1101/gad.13.23.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gosset G, Zhang Z, Nayyar S, Cuevas WA, Saier MH Jr.. 2004. Transcriptome analysis of Crp-dependent catabolite control of gene expression in Escherichia coli. J Bacteriol 186:3516–3524. doi: 10.1128/JB.186.11.3516-3524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grainger DC, Hurd D, Harrison M, Holdstock J, Busby SJ. 2005. Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc Natl Acad Sci U S A 102:17693–17698. doi: 10.1073/pnas.0506687102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang DE, Smalley DJ, Tucker DL, Leatham MP, Norris WE, Stevenson SJ, Anderson AB, Grissom JE, Laux DC, Cohen PS, Conway T. 2004. Carbon nutrition of Escherichia coli in the mouse intestine. Proc Natl Acad Sci U S A 101:7427–7432. doi: 10.1073/pnas.0307888101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li CC, Crawford JA, DiRita VJ, Kaper JB. 2000. Molecular cloning and transcriptional regulation of ompT, a ToxR-repressed gene in Vibrio cholerae. Mol Microbiol 35:189–203. doi: 10.1046/j.1365-2958.2000.01699.x. [DOI] [PubMed] [Google Scholar]
- 33.Liang W, Pascual-Montano A, Silva AJ, Benitez JA. 2007. The cyclic AMP receptor protein modulates quorum sensing, motility and multiple genes that affect intestinal colonization in Vibrio cholerae. Microbiology 153:2964–2975. doi: 10.1099/mic.0.2007/006668-0. [DOI] [PubMed] [Google Scholar]
- 34.Li CC, Merrell DS, Camilli A, Kaper JB. 2002. ToxR interferes with CRP-dependent transcriptional activation of ompT in Vibrio cholerae. Mol Microbiol 43:1577–1589. doi: 10.1046/j.1365-2958.2002.02845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen B, Liang W, Wu R, Liang P, Kan B. 2013. Phenotype microarray screening of carbon sources used by Vibrio cholerae identifies genes regulated by the cAMP receptor protein. Can J Microbiol 59:472–478. doi: 10.1139/cjm-2013-0084. [DOI] [PubMed] [Google Scholar]
- 36.Wu R, Zhao M, Li J, Gao H, Kan B, Liang W. 2015. Direct regulation of the natural competence regulator gene tfoX by cyclic AMP (cAMP) and cAMP receptor protein (CRP) in vibrios. Sci Rep 5:14921. doi: 10.1038/srep14921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kariisa AT, Grube A, Tamayo R. 2015. Two nucleotide second messengers regulate the production of the Vibrio cholerae colonization factor GbpA. BMC Microbiol 15:166. doi: 10.1186/s12866-015-0506-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou YY, Zhang HZ, Liang WL, Zhang LJ, Zhu J, Kan B. 2013. Plasticity of regulation of mannitol phosphotransferase system operon by CRP-cAMP complex in Vibrio cholerae. Biomed Environ Sci 26:831–840. doi: 10.3967/bes2013.006. [DOI] [PubMed] [Google Scholar]
- 39.Heidelberg JF, Eisen JA, Nelson WC, Clayton RA, Gwinn ML, Dodson RJ, Haft DH, Hickey EK, Peterson JD, Umayam L, Gill SR, Nelson KE, Read TD, Tettelin H, Richardson D, Ermolaeva MD, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann RD, Nierman WC, White O, Salzberg SL, Smith HO, Colwell RR, Mekalanos JJ, Venter JC, Fraser CM. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papenfort K, Förstner KU, Cong JP, Sharma CM, Bassler BL. 2015. Differential RNA-seq of Vibrio cholerae identifies the VqmR small RNA as a regulator of biofilm formation. Proc Natl Acad Sci U S A 112:E766–E775. doi: 10.1073/pnas.1500203112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fong JC, Yildiz FH. 2008. Interplay between cyclic AMP-cyclic AMP receptor protein and cyclic di-GMP signaling in Vibrio cholerae biofilm formation. J Bacteriol 190:6646–6659. doi: 10.1128/JB.00466-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin W, Fullner KJ, Clayton R, Sexton JA, Rogers MB, Calia KE, Calderwood SB, Fraser C, Mekalanos JJ. 1999. Identification of a Vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. Proc Natl Acad Sci U S A 96:1071–1076. doi: 10.1073/pnas.96.3.1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee DJ, Busby SJ. 2012. Repression by cyclic AMP receptor protein at a distance. mBio 3:e00289-12. doi: 10.1128/mBio.00289-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bina JE, Mekalanos JJ. 2001. Vibrio cholerae tolC is required for bile resistance and colonization. Infect Immun 69:4681–4685. doi: 10.1128/IAI.69.7.4681-4685.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mandlik A, Livny J, Robins WP, Ritchie JM, Mekalanos JJ, Waldor MK. 2011. RNA-Seq-based monitoring of infection-linked changes in Vibrio cholerae gene expression. Cell Host Microbe 10:165–174. doi: 10.1016/j.chom.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boardman BK, Meehan BM, Fullner Satchell KJ. 2007. Growth phase regulation of Vibrio cholerae RTX toxin export. J Bacteriol 189:1827–1835. doi: 10.1128/JB.01766-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kühn J, Finger F, Bertuzzo E, Borgeaud S, Gatto M, Rinaldo A, Blokesch M. 2014. Glucose- but not rice-based oral rehydration therapy enhances the production of virulence determinants in the human pathogen Vibrio cholerae. PLoS Negl Trop Dis 8:e3347. doi: 10.1371/journal.pntd.0003347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller VL, DiRita VJ, Mekalanos JJ. 1989. Identification of toxS, a regulatory gene whose product enhances toxR-mediated activation of the cholera toxin promoter. J Bacteriol 171:1288–1293. doi: 10.1128/jb.171.3.1288-1293.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dalia AB, McDonough E, Camilli A. 2014. Multiplex genome editing by natural transformation. Proc Natl Acad Sci U S A 111:8937–8942. doi: 10.1073/pnas.1406478111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Page L, Griffiths L, Cole JA. 1990. Different physiological roles of two independent pathways for nitrite reduction to ammonia by enteric bacteria. Arch Microbiol 154:349–354. doi: 10.1007/BF00276530. [DOI] [PubMed] [Google Scholar]
- 51.Taylor RG, Walker DC, McInnes RR. 1993. E. coli host strains significantly affect the quality of small scale plasmid DNA preparations used for sequencing. Nucleic Acids Res 21:1677–1678. doi: 10.1093/nar/21.7.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lodge J, Fear J, Busby S, Gunasekaran P, Kamini NR. 1992. Broad host range plasmids carrying the Escherichia coli lactose and galactose operons. FEMS Microbiol Lett 74:271–276. [DOI] [PubMed] [Google Scholar]
- 53.Sen D, Van der Auwera GA, Rogers LM, Thomas CM, Brown CJ, Top EM. 2011. Broad-host-range plasmids from agricultural soils have IncP-1 backbones with diverse accessory genes. Appl Environ Microbiol 77:7975–7983. doi: 10.1128/AEM.05439-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rhodius VA, Busby SJ. 2000. Interactions between activating region 3 of the Escherichia coli cyclic AMP receptor protein and region 4 of the RNA polymerase sigma(70) subunit: application of suppression genetics. J Mol Biol 299:311–324. doi: 10.1006/jmbi.2000.3737. [DOI] [PubMed] [Google Scholar]
- 55.Burgess RR, Jendrisak JJ. 1975. A procedure for the rapid, large-scale purification of Escherichia coli DNA-dependent RNA polymerase involving polymin P precipitation and DNA-cellulose chromatography. Biochemistry 14:4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- 56.Shimada T, Ishihama A, Busby SJ, Grainger DC. 2008. The Escherichia coli RutR transcription factor binds at targets within genes as well as intergenic regions. Nucleic Acids Res 36:3950–3955. doi: 10.1093/nar/gkn339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chintakayala K, Singh SS, Rossiter AE, Shahapure R, Dame RT, Grainger DC. 2013. E. coli Fis protein insulates the cbpA gene from uncontrolled transcription. PLoS Genet 9:e1003152. doi: 10.1371/journal.pgen.1003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grainger DC, Belyaeva TA, Lee DJ, Hyde EI, Busby SJ. 2004. Transcription activation at the Escherichia coli melAB promoter: interactions of MelR with the C-terminal domain of the RNA polymerase alpha subunit. Mol Microbiol 51:1311–1320. doi: 10.1111/j.1365-2958.2003.03930.x. [DOI] [PubMed] [Google Scholar]
- 59.Singh SS, Grainger DC. 2013. H-NS can facilitate specific DNA-binding by RNA polymerase in AT-rich gene regulatory regions. PLoS Genet 9:e1003589. doi: 10.1371/journal.pgen.1003589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 61.UK Parliament 1986. Animal (Scientific Procedures) Act 1986. Chapter 14. https://www.legislation.gov.uk/ukpga/1986/14/resources. [Google Scholar]
- 62.Brown KH, Dobrinski KP, Lee AS, Gokcumen O, Mills RE, Shi X, Chong WW, Chen JY, Yoo P, David S, Peterson SM, Raj T, Choy KW, Stranger BE, Williamson RE, Zon LI, Freeman JL, Lee C. 2012. Extensive genetic diversity and substructuring among zebrafish strains revealed through copy number variant analysis. Proc Natl Acad Sci U S A 109:529–534. doi: 10.1073/pnas.1112163109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mekalanos JJ, Swartz DJ, Pearson GD, Harford N, Groyne F, de Wilde M. 1983. Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature 306:551–557. doi: 10.1038/306551a0. [DOI] [PubMed] [Google Scholar]
- 64.Rader AE, Murphy JR. 1988. Nucleotide sequences and comparison of the hemolysin determinants of Vibrio cholerae El Tor RV79(Hly+) and RV79(Hly−) and classical 569B(Hly−). Infect Immun 56:1414–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Manning PA, Brown MH, Heuzenroeder MW. 1984. Cloning of the structural gene (hly) for the haemolysin of Vibrio cholerae El Tor strain 017. Gene 31:225–231. doi: 10.1016/0378-1119(84)90213-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a) Transcription from PrtxH requires identified promoter elements. (i) The results of primer extension assays that detect the rtxH transcript derived from plasmid pRW50T carrying the rtxH (panel ii, top) or truncated rtxH.1 (panel ii, bottom) DNA fragment. (ii) PrtxH is highlighted blue and CRP binding sites are shown in orange. (b) P1rtxB and P2rtxB make similar contributions to rtxB transcription. The figure shows β-galactosidase activity measurements for lysates of V. cholerae cells transformed with pRW50T derivatives carrying the full-length rtxB regulatory region (rtxB), a truncated derivative lacking CRP binding sites (rtxB.1), or versions of the truncated fragment with indicated promoter mutations. (c) Effects of crp and tcpA on zebrafish larva colonization. (i) Images from multiple zebrafish larvae colonized with the indicated V. cholerae strains. All strains were transformed with plasmid pMW-GFP to facilitate visualization of bacteria. (ii) Quantified fluorescence from multiple microscopy images. (d) Raw gel images. Download FIG S1, PDF file, 0.4 MB (404.3KB, pdf) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Strains, plasmids, and oligonucleotides. Download TABLE S1, DOCX file, 0.03 MB (30.2KB, docx) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
N16961 chromosome I GenBank file. Download DATA SET S1, TXT file, 6 MB (6.2MB, txt) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
N16961 chromosome II GenBank file. Download DATA SET S2, TXT file, 2.2 MB (2.2MB, txt) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
CRP ChIP-seq Artemis graph file for N16961 chromosome I. Download DATA SET S3, TXT file, 17.5 MB (18MB, txt) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
CRP ChIP-seq Artemis graph file for N16961 chromosome II. Download DATA SET S4, TXT file, 6.4 MB (6.5MB, txt) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RNA polymerase σ70 ChIP-seq Artemis graph file for N16961 chromosome I. Download DATA SET S5, TXT file, 10.7 MB (10.9MB, txt) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RNA polymerase σ70 ChIP-seq Artemis graph file for N16961 chromosome II. Download DATA SET S6, TXT file, 4.2 MB (4.3MB, txt) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Instructions for viewing ChIP-seq data in the Artemis genome browser. Download TEXT S1, PDF file, 0.6 MB (667.8KB, pdf) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
DNA sequencing reads are stored in ArrayExpress under accession number E-MTAB-6472. Genome Browser files (Data Set S1 to S6) and instructions (Text S1) are provided in the supplemental material.
N16961 chromosome I GenBank file. Download DATA SET S1, TXT file, 6 MB (6.2MB, txt) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
N16961 chromosome II GenBank file. Download DATA SET S2, TXT file, 2.2 MB (2.2MB, txt) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
CRP ChIP-seq Artemis graph file for N16961 chromosome I. Download DATA SET S3, TXT file, 17.5 MB (18MB, txt) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
CRP ChIP-seq Artemis graph file for N16961 chromosome II. Download DATA SET S4, TXT file, 6.4 MB (6.5MB, txt) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RNA polymerase σ70 ChIP-seq Artemis graph file for N16961 chromosome I. Download DATA SET S5, TXT file, 10.7 MB (10.9MB, txt) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
RNA polymerase σ70 ChIP-seq Artemis graph file for N16961 chromosome II. Download DATA SET S6, TXT file, 4.2 MB (4.3MB, txt) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Instructions for viewing ChIP-seq data in the Artemis genome browser. Download TEXT S1, PDF file, 0.6 MB (667.8KB, pdf) .
Copyright © 2018 Manneh-Roussel et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.