Abstract
Bacteroides thetaiotaomicron (Bt) is a prominent member of the human gut microbiota with an extensive capacity for glycan harvest. This bacterium expresses a five-protein complex in the outer membrane, called the starch utilization system (Sus), which binds, degrades, and imports starch into the cell. Sus is a model system for the many glycan-targeting polysaccharide utilization loci found in Bt and other members of the Bacteroidetes phylum. Our previous work has shown that SusG, a lipidated amylase in the outer membrane, explores the entire cell surface but diffuses more slowly as it interacts with starch. Here, we use a combination of single-molecule tracking, super-resolution imaging, reverse genetics, and proteomics to show that SusE and SusF, two proteins that bind starch, are immobile on the cell surface even when other members of the system are knocked out and under multiple different growth conditions. This observation suggests a new paradigm for protein complex formation: binding proteins form immobile complexes that transiently associate with a mobile enzyme partner.
Introduction
The human gut microbiome comprises thousands of different bacterial strains (1) that contribute to defense against pathogens, activation of the immune system, obesity, and malnutrition (2, 3, 4, 5). Many of these bacteria are required to break down complex plant-derived glycans that cannot be degraded by human digestive enzymes. Most glycan degradation depends on the collective action of the Gram-negative Bacteroidetes, a dominant bacterial phylum in the gut, which has evolved to degrade a vast array of carbohydrates (6). This is accomplished through the regulated expression of outer-membrane protein sets called starch-utilization (Sus)-like systems, which are composed of several proteins that bind, degrade, and import different glycans. Sus-like systems are encoded within discrete polysaccharide utilization loci (PULs), and each Sus-like system outer-membrane complex targets a distinct glycan substrate (7). The repertoire of PULs within these bacteria drives their metabolic lifestyle and fitness, dictating both the colonization and persistence of these microbes within the host gut (8, 9). A detailed understanding of the molecular mechanism of glycan transport is critical for the development of therapies that utilize diet to change the microbiota composition toward improved health.
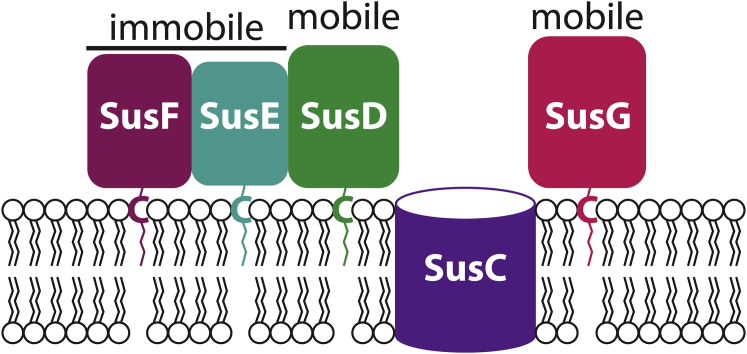
Sus-like systems contain proteins homologous in structure and function to several proteins originally described in the prototypical PUL, the Bacteroides thetaiotaomicron (Bt) Sus (10, 11). The Sus locus (susRABCDEFG) encodes five outer-membrane-localized proteins, SusCDEFG, which form the Sus complex at the cell surface (Fig. 1). Starch polysaccharides are initially bound by SusD, SusE, and SusF (12, 13) and are subsequently broken down into smaller oligosaccharides by the amylase SusG (14). These oligosaccharides are transported into the periplasm through the SusC TonB-dependent transporter, and then degraded to glucose by the SusA (15) and SusB (16) glycosidases.
Figure 1.
The outer membrane proteins of the Sus complex. SusC is a membrane-spanning β-barrel. SusDEFG are associated with the cell membrane through lipidation of a cysteine residue (C) near the N-terminus. The identity of the lipid groups is unknown. Single-molecule microscopy shows that SusD and SusG are mobile, whereas SusE and SusF are immobile. To see this figure in color, go online.
The Sus is a model system for understanding how complex polysaccharides are broken down by Bacteroidetes, which rely heavily on Sus-like systems for nutrient acquisition (17, 18). Nearly all sequenced gut Bacteroidetes genomes encode Sus-like systems that can metabolize a wide variety of substrates (7), yet little is known about how Sus proteins work together at the cell surface. The individual structures of the Bt Sus proteins SusCDEF have been elucidated, revealing that SusD has a single starch binding site, SusE and SusG each have two starch binding sites, and SusF has three starch binding sites (11). These binding sites do not have redundant functionality, but rather play discrete roles in glycan capture (19). Importantly, SusDEFG are not embedded in the membrane but are tethered to the cell surface via lipidation at an N-terminal cysteine, followed by a flexible linker of 15–20 amino acids such that each protein can be thought of as “floating” above the membrane like a balloon on a string (Fig. 1).
The SusDEFG proteins have discrete folds (Fig. S1). SusD (62 kDa) is a globular α-helical protein, whereas SusE (42 kDa) and SusF (52 kDa) are composed of three and four tandem Ig-like domains of ∼100 amino acids, respectively; besides the N-terminal domain of each protein, all domains have a single starch-binding site. There is a proline residue at nearly every domain junction, and we speculate that these prolines restrict the overall conformational flexibility of SusEF. SusG (78 kDa) comprises a globular catalytic domain and a carbohydrate-binding module (CBM) that protrudes from the catalytic site. The CBM is linked to the catalytic domain by two short linkers (residues 212–217 and 334–338) that are unlikely to impart structural flexibility, and thus, SusG, like SusE and SusF, is expected to maintain an extended conformation. Additionally, the proteins lacking the signal peptides could be expressed in Escherichia coli, were highly soluble, and bound starch. Therefore, an interaction with a protein partner, or the Bt membrane, is not essential for folding or for starch binding. Still, despite the detailed information in Fig. S1 regarding the individual structures (13, 14, 20), we could not predict how the proteins behave in the membrane environment or how they interact with each other, or with other proteins, on the cell surface.
We previously probed the movement of the glycoside hydrolase SusG in live anaerobic cells using single-molecule fluorescence imaging (21), and we discovered that this protein moves around the entire cell surface. SusG displays a noticeable decrease in its effective diffusion coefficient in the presence of the large polymer starch, as well as when it transiently associates with the SusCDEF proteins, indicating dynamic, starch-mediated Sus complex formation. Little is known about the organization and structure of the Bacteroides membrane, and our work with SusG was, to our knowledge, the first to examine cell-surface lipoprotein dynamics in live anaerobic cells. A key question, therefore, is whether other Sus surface components are similarly mobile, as answering this question would both inform a model of how the Sus proteins assemble in Bt and provide insight into the extent of protein mobility on the cell surface as bacteria interact with their environment.
Here, we employ super-resolution imaging and single-molecule tracking to examine the starch-binding proteins SusE and SusF. In stark contrast to the highly mobile SusG, we find that SusE and SusF remain immobile at the cell surface despite changing environmental conditions and within strains with different genetic backgrounds. This is, to our knowledge, the first observation of lipoproteins that are significantly immobile (down to our 25 nm resolution). Similar high degrees of confinement have been observed in E. coli for some integral outer-membrane proteins, with confinement diameters from published studies ranging from 0.02 to 0.60 μm (22, 23), although our observations here show that the SusE and SusF confinement diameter is at most 0.025 μm, i.e., less than or equal to the localization precision of our super-resolution microscope. This immobility does not appear to be driven by the N-terminal sequence of the signal peptide or by lipidation, as swapping in the N-terminal sequence of SusE does not decrease SusG diffusion. Therefore, we hypothesize that the differences in mobility are due to interactions of the SusEF proteins with an unknown partner, possibly another protein or a component of the cell membrane. Overall, we present here, to our knowledge, a new model for outer-membrane protein collaboration on the surface of Gram-negative bacteria: assembly of a system of mobile proteins around select stationary protein centers.
Materials and Methods
Bacterial strains, mutagenesis, and cell growth
Bacterial strains in this study are listed in Table S1 and primers are listed in Table S2. Bacteroides thetaiotaomicron (Bt) was grown as previously described (21). Briefly, cells were cultured in medium containing 0.5% tryptone-yeast-extract-glucose and incubated at 37°C under anaerobic conditions in a Coy chamber. Approximately 24 h before imaging, cells were diluted into minimal medium containing 0.5% (wt/vol) carbohydrate (glucose or maltose). Mutations were performed as previously described (13) using a counter-selectable allelic exchange method. All mutants were created in a thymidine kinase deletion (Δtdk) mutant.
Imaging
Bt cells expressing fluorescent protein fusions were imaged on an Olympus IX71 inverted fluorescence microscope using a 1.40 numerical aperture 100× widefield oil-immersion objective. HaloTag labeling with TMR was performed as described previously (21), and TMR-labeled fusions were excited with a 561-nm laser (Coherent-Sapphire 561-50, Coherent, Santa Clara, CA). Fusions to PAmCherry were activated with a 406-nm laser (Coherent Cube 405-100) and excited with the 561-nm laser. Fluorescence emission was detected using a 512 × 512 pixel Photometrics Evolve electron-multiplying charge-coupled device camera. Images were collected continuously at a rate of 25 frames per second (fps) or at 1 fps for time-lapse imaging.
Data analysis
Single-molecule fitting and track construction were performed as previously described (24). The mean-square displacement (MSD) was calculated for each track, then trimmed so that only the first half of the MSD was retained to reduce the noise produced by averaging fewer steps at longer time lags (τ). MSD-versus-τ curves of length 10 or longer were used for further analysis (i.e., the original track had to be at least 20 steps long to be considered). The slope of each MSD curve was calculated by fitting a line to the first three points.
Western blotting, co-IP, and membrane fractionation
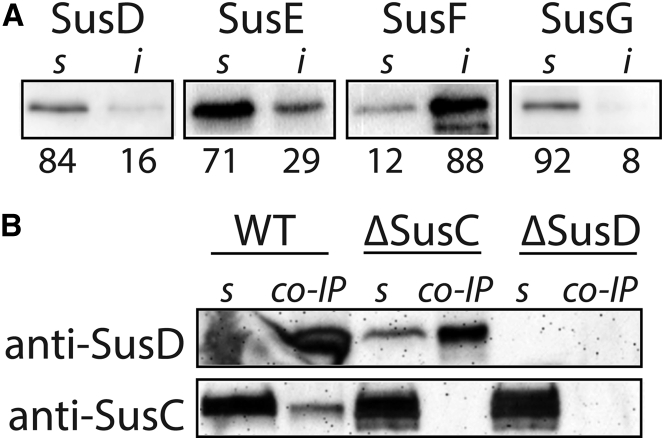
Bt membranes were collected by spinning from mid-log cells grown anaerobically in minimal medium containing 0.5% (w/v) maltose. Cross-linking was carried out anaerobically for 1 h on whole cells before fractionation. Cells were lysed by sonication and spun briefly to remove large debris. Membranes were collected by spinning for 30 min at 50,000 × g, washed with phosphate-buffered saline (PBS), then resuspended and incubated in PBS containing 1.5% (w/v) dodecyl maltoside. Solubilization was performed for 1 h, and the remaining insoluble protein was spun for 45 min at 100,000 × g. Co-immunoprecipitation (co-IP) was performed on solubilized membranes using the Crosslink Magnetic IP/Co-IP Kit (Pierce Protein Biology, Waltham, MA) according to manufacturer instructions. Sus outer-membrane lipoproteins were detected by Western blot using rabbit polyclonal primary antibodies and horseradish-peroxidase-conjugated goat anti-Rabbit IgG secondary antibody (Sigma-Aldrich, St. Louis, MO). Antibodies to SusD, SusE, and SusF were raised against the recombinantly expressed proteins (19, 25). The SusC antibodies were raised against the N-terminal plug domain of the protein (residues 118–242), which is conserved among many other SusC-like proteins in Bt. Therefore, the SusC antibodies cross-react with other SusC-like proteins present in our membrane preparations, as seen in Fig. 5 b.
Figure 5.
The Sus outer-membrane proteins vary in solubility and abundance. (A) The soluble (s) and insoluble (i) fractions of Sus outer-membrane lipoproteins after detergent extraction from fractionated Bt membranes. The relative density, as a percent of the total signal, is reported below each lane. (B) Co-IP of SusD from solubilized membranes of the three strains. Western blot on the co-IP samples was performed with custom SusD or SusC antibodies. Of note, anti-SusC cross-reacts to label other SusC-like proteins in Bt (Materials and Methods). The soluble (s) lane is the solubilized membranes used for the co-IP, and the co-IP lane indicates the sample that was eluted after immunoprecipitation.
Total membrane proteomic sample preparation
Using 5 mg/mL maltose as the carbon source, 1 L of Bt was grown to mid-log phase (OD600 0.65–0.75) at 37°C in minimal Bacteroides medium. Cells were centrifuged (10,000 × g for 15 min) then frozen in liquid N2. Cells were thawed in 20 mL of 20 mM KH2PO4 (pH 7.3), sonicated on ice, and intact cells were removed by centrifugation at 15,000 × g for 17 min. The supernatant was ultracentrifuged at 200,000 × g for 2 h at 4°C to pellet total membranes. The membrane pellet was resuspended in 20 mL of 20 mM KH2PO4 (pH 7.3), followed by a second round of ultracentrifugation. All steps from growth to membrane purification were repeated, and both preparations were submitted for mass spectrometry (runs 1 and 2; Table S3). For the first run, the final membrane pellet was resuspended in 7.5 mL of 20 mM KH2PO4 (pH 7.3) plus 0.1% Tween-20 and gently sonicated to completely resuspend the membranes. This sample was concentrated via a 5 kDa MWCO spin filter before proteomic analysis. For the second preparation only, half a tablet of cOmplete EDTA-free protease inhibitor (Roche, Basel, Switzerland) was included during cell lysis and the final membrane pellet was resuspended in 3 mL of 20 mM KH2PO4 (pH 7.3) plus 0.1% Tween-20; this sample did not require concentration.
Quantitative mass spectroscopy
The total membrane samples and co-IP samples were submitted to MS Bioworks LLC (Ann Arbor, MI) for Quant-works label-free unfractionated proteomic analysis, as described in (26). The protein abundances within each sample were determined by calculating for each protein the normalized spectral abundance factor, , where SpC is the spectral counts, MW is the protein molecular weight, and N is the total number of proteins.
Bt growth experiments
All Bt culturing was performed in a Coy anaerobic chamber (gas mix: 85% N2, 10% H2, and 5% CO2) at 37°C. Each strain was grown for 16 h from a freezer stock in tryptone-yeast extract-glucose medium and then back-diluted 1:100 into Bacteroides minimal medium supplemented with 5 mg/mL glucose. After 24 h, cultures were back-diluted 1:100 into Bacteroides minimal medium supplemented with 5 mg/mL of carbohydrate. Growth experiments were performed in triplicate. Plates were loaded into a Biostack automated plate-handling device coupled to a Powerwave HT absorbance reader (both from Biotek Instruments, Winooski, VT). Absorbance at 600 nm (OD600) was measured for each well at 20 min intervals. Data were processed using Gen5 software (BioTek), Microsoft Excel, and Prism.
Immunofluorescence
Bt strains were grown in Bacteroides medium with 5 mg/mL maltose to an OD600 of 0.6 and washed twice with PBS. Cells were resuspended in 0.25 mL PBS, to which 0.75 mL of 6% (v/v) formalin in PBS was added, and then incubated with rocking at 20°C for 1.5 h. Cells were washed twice with PBS, then resuspended in 0.5–1 mL blocking solution (2% (v/v) goat serum and 0.02% (w/v) NaN3 in PBS) and incubated for 16 h at 4°C. Cells were centrifuged and resuspended in 0.5 mL of a 1/100 dilution of custom rabbit antibody sera in blocking solution and incubated by rocking for 2 h at 20°C. Cells were washed with PBS and then resuspended in 0.4 mL of a 1/500 dilution of Alexa Fluor 488 goat anti-rabbit IgG (Life Technologies, Carlsbad, CA) in blocking solution and incubated with rocking for 1 h at 20°C. Cells were washed three times with an excess of PBS and then resuspended in 20 μL of PBS plus 1 μL of ProLong Gold antifade (Life Technologies). Cells were spotted on 0.8% (w/v) agarose pads and imaged using an Olympus IX70 inverted confocal microscope. Images were processed with Metamorph Software.
Results and Discussion
SusG dynamics are unchanged under aerobic conditions. Although we have previously shown that fusions of SusG to the enzymatic HaloTag can be labeled by the dye TMR for single-molecule imaging in living, anaerobic Bt (21), the fluorescent protein PAmCherry is advantageous, because its photoactivation allows control of the number of molecules that are fluorescent at one time. Bt is aerotolerant, and colonies are able to survive oxygen exposure for several days (27). We explored the possibility of imaging dormant cells under aerobic conditions to allow the use of PAmCherry, whose chromophore requires oxygen to mature. We found that SusG-HaloTag is mobile under aerobic conditions (Fig. S2, a and b). The single-molecule trajectories of the proteins can be quantitatively analyzed in plots of the MSD versus time lag (Fig. S2, c and d). Here, each trajectory is a curve with slope equal to four times the effective molecular diffusion coefficient, D. SusG-HaloTag protein dynamics are unchanged in the presence or absence of oxygen (Table 1). As before, we constructed SusG-PAmCherry by replacing the starch-binding CBM58 domain with PAmCherry (21). The SusG-PAmCherry fusion supports growth on starch (Fig. S3), and effective diffusion coefficients and distributions measured for SusG-PAmCherry in aerobic conditions were similar to those measured for the SusG-HaloTag fusion under anaerobic conditions (Table 1; (21)). The dynamics of SusG are therefore independent of both the identity of the fluorescent tag and the presence or absence of oxygen in the environment. The similar patterns of diffusion in aerobic and anaerobic conditions imply that the SusG dynamics are passive and likely a result of membrane fluidity, rather than dependent upon the cell’s redox status or on the proton motive force, as oxygen prevents maltooligosaccharide uptake by Bt. Because SusE and SusF are lipoproteins like SusG, and their glycan-binding activity is not oxygen-sensitive, we examined the dynamics of these proteins using PAmCherry fusions and in aerobic conditions.
Table 1.
Dynamics of Protein Constructs
| Protein Construct | Aerobic/Anaerobic | Tracks with D ≤ 0.001 (%) | Tracks with 0.001 < D ≤ 0.02 (%) | Tracks with 0.02 < D ≤ 0.05 (%) | Tracks with 0.05 < D ≤ 0.1 (%) | Tracks with D > 0.1 (%) | Total No. of Tracks | Tracks Shown in Fig. |
|---|---|---|---|---|---|---|---|---|
| SusG-HT | anaerobic | 25.7 | 52.1 | 17.4 | 3.3 | 1.5 | 1687 | Fig. S2d |
| SusG-HT | aerobic | 29.3 | 52.7 | 12.1 | 4.0 | 2.0 | 5788 | Fig. S2c |
| SusG-PAmCherry | aerobic | 29.3 | 44.1 | 14.4 | 8.6 | 3.5 | 256 | Fig. 2f |
| SusE-PAmCherry | aerobic | 43.2 | 50.2 | 5.5 | 1.1 | none | 273 | Fig. 2d |
| SusE-HT | anaerobic | 53.4 | 43.2 | 2.7 | none | 2.1 | 146 | Fig. S6f |
| SusE-HT | aerobic | 43.3 | 48.7 | 6.3 | 1.4 | 0.3 | 348 | Fig. S6e |
| SusF-PAmCherry | aerobic | 46.5 | 48.8 | 3.9 | 0.6 | 0.3 | 338 | Fig. 2e |
| SusD-PAmCherry (3-Ala linker) | aerobic | 21.8 | 55.5 | 16.4 | 4.6 | 1.8 | 110 | Fig. S7c |
| SusD-PAmCherry (20-Ala linker) | aerobic | 23.6 | 54.7 | 15.1 | 3.8 | 2.8 | 106 | Fig. S7d |
| SusE-Nterm-SusG-PAmCherry | aerobic | 23.8 | 42.9 | 15.5 | 11.9 | 5.6 | 84 | Fig. 4d |
Distributions of effective diffusion coefficients, D, are shown for each construct measured with single-molecule tracking. Values for D are in μm2/s.
SusE and SusF are immobile in the membrane despite perturbations to the cellular environment
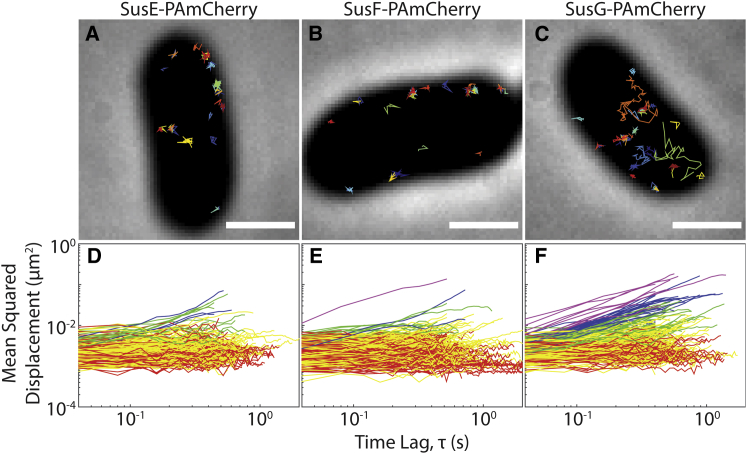
PAmCherry was fused to the C-terminus of SusE or SusF via a three-alanine linker. Cells expressing these fusions displayed wild-type growth kinetics on starch (Fig. S3, c and d), indicating that the fusions are functional. Immunofluorescence on fixed, non-permeabilized Bt cells using antibodies against SusE and SusF demonstrated that PAmCherry-labeled SusE and SusF localize to the outer surface of the cell, as expected (Fig. S4). To our surprise, and in contrast to our SusG fusions, the single-molecule trajectories of the SusE and SusF fusions are compact and confined to puncta, indicating that these constructs are immobile in the cell membrane over time spans ranging from a few seconds to tens of seconds (Figs. 2, a and b and S5; Movies S1 and S2). Of note, we observe mobile SusG and immobile SusE and SusF in identically treated cells, confirming that these dynamic observations are not due to unintended differences in the experimental conditions. The MSD curves show clear differences for the various Sus lipoproteins (Fig. 2, d–f; Table 1). Although 12.1% of SusG-PAmCherry molecules diffuse at a rate >0.05 μm2/s (Fig. 2 f, blue and purple curves), only 1.1% of SusE-PAmCherry and 0.9% of SusF-PAmCherry move this quickly (Fig. 2, d and e, blue and purple curves). Similarly, although 93.4% of SusE-PAmCherry and 95.3% of SusF-PAmCherry molecules have effective diffusion coefficients <0.02 μm2/s (Fig. 2, d and e, red and yellow curves), only 73.4% of SusG-PAmCherry molecules move this slowly (Fig. 2 f, red and yellow curves). To further confirm that Sus protein dynamics are unaffected by the presence of oxygen, we fused SusE to the HaloTag (HT) protein and imaged TMR-labeled SusE-HT under aerobic and anaerobic conditions. Like SusE-PAmCherry imaged aerobically, SusE-HT remained confined to puncta under both conditions (Fig. S6) and had a similar mobility profile (Table 1). The similarities within these different labeling and oxygen conditions further demonstrate that anaerobic conditions are not required to obtain physiologically relevant protein dynamics.
Figure 2.
SusE-PAmCherry and SusF-PAmCherry are highly confined, whereas SusG-PAmCherry explores the cell. (A–C) Each image shows a cell with 35 single-molecule tracks plotted in random colors. Scale bars, 1 μm. (D–F) MSD of all tracks lasting >20 frames is plotted for each protein fusion (red, effective diffusion coefficient (D) ≤ 0.001 μm2/s; yellow, 0.001 μm2/s < D ≤ 0.02 μm2/s; green, 0.02 μm2/s < D ≤ 0.05 μm2/s; blue, 0.05 μm2/s < D ≤ 0.1 μm2/s; purple, D > 0.1 μm2/s). To see this figure in color, go online.
We hypothesize that the enzymatic SusG diffuses in the outer membrane, whereas SusE and SusF remain stationary for optimal carbohydrate binding by the Sus system. To complete the picture, we therefore examined the mobility of a fusion of PAmCherry to SusD, the fourth carbohydrate-binding Sus outer-membrane protein. SusD-PAmCherry exhibited high mobility like SusG-PAmCherry (Fig. S7; Table 1). However, it should be noted that the SusD-PAmCherry strain exhibited delayed growth on starch, which suggests that this tag reduced the ability of SusD to effectively contribute to starch uptake. Therefore, it is difficult to discern whether the movement of SusD is due to the tag interfering with the interaction of SusD with its partner SusC, or if SusD is normally mobile. Changing the length of the linker between the C-terminus of SusD and PAmCherry from a 3-Ala linker to a 20-Ala linker did not resolve the growth defect (Fig. S3, e and f).
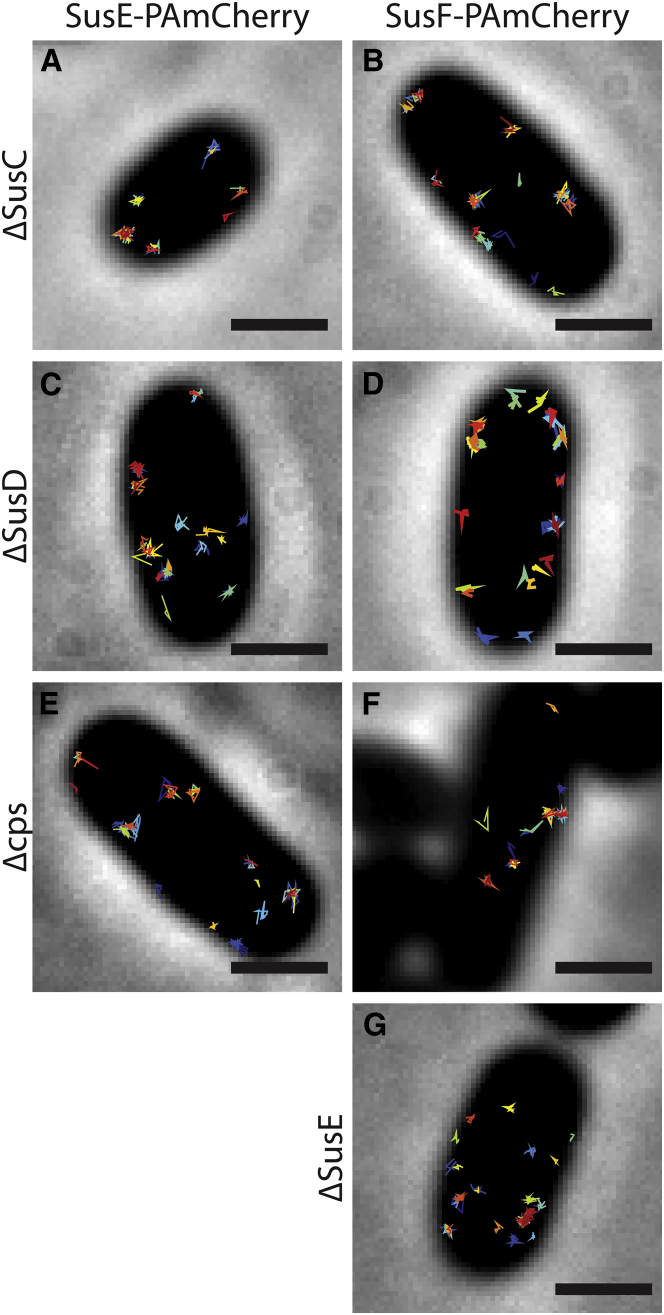
Because SusE and SusF remain stationary in the outer-cell membrane despite changes in oxygen and labeling conditions, we further measured the effect of the presence of other Sus proteins and capsular polysaccharide on the positioning of SusEF. Previous studies with formaldehyde cross-linking have demonstrated that SusC, SusD, and SusE interact (28), so we hypothesized that SusCD might be responsible for confining SusE and SusF in the membrane. However, both SusE and SusF remained immobile when SusC or SusD were deleted from the chromosome (Fig. 3, a–d). Furthermore, SusF remained immobile in a strain in which SusE was deleted (Fig. 3 g). Thus, the stationary character of SusE does not depend on SusCD, and the stationary character of SusF does not depend on SusCDE. Bt has a thick surface capsular polysaccharide layer (7). Although the precise monosaccharide and linkage composition of these capsules is unknown, we hypothesized that the glycan-binding proteins SusE and SusF may interact nonspecifically with the capsule or are otherwise influenced by the capsule organization on the cell surface. We examined SusEF dynamics in an acapsular strain of Bt (19) and found that both SusE-PAmCherry and SusF-PAmCherry remain immobile in the absence of the capsule (Δcps; Fig. 3, e and f). This is consistent with our previous demonstration that SusG protein dynamics are unchanged by the presence or absence of capsule (21). Thus, SusEF interactions with the capsule layer are not responsible for immobility.
Figure 3.
SusE-PAmCherry and SusF-PAmCherry remain highly confined even when other members of the Sus complex or the capsule (cps) machinery are knocked out. Each image (A–G) shows a single cell with 35 single-molecule tracks plotted in random colors. Scale bars, 1 μm. To see this figure in color, go online.
The SusE N-terminus does not confer immobility
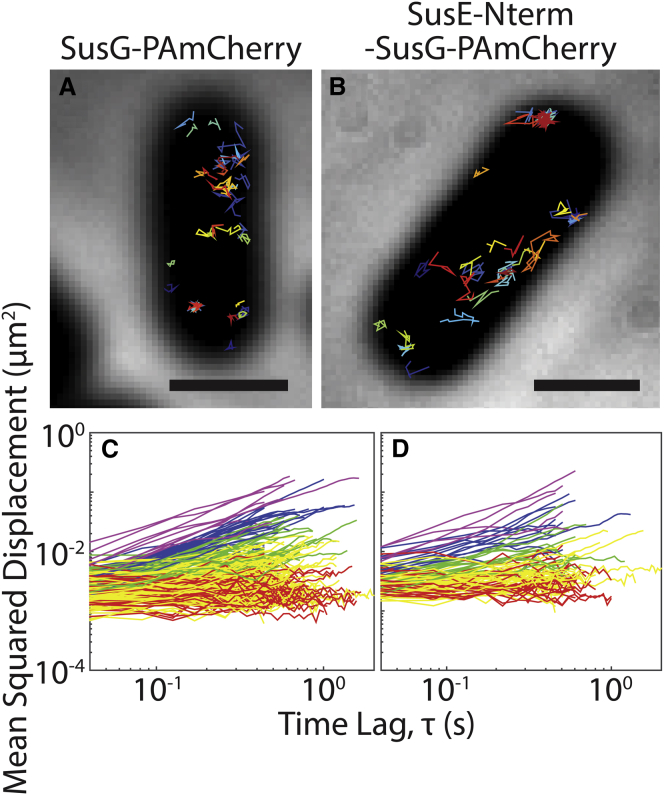
After cleavage of the signal peptide, mature SusDEFG are lipidated at an N-terminal cysteine and tethered to the outer leaflet of the outer membrane (28, 29). Previous structural studies of these lipoproteins demonstrate that they also contain a flexible disordered linker that further separates the N-terminal domain from the surface of the cell (11). To determine whether the N-terminal sequence of SusE confers immobility, we replaced the N-terminal region (M1–W44) of SusG—which contains the signal peptide through the flexible linker—with the analogous N-terminal region (M1–N28) of SusE. This “SusE-Nterm-SusG-PAmCherry” hybrid protein remained mobile, with dynamics indistinguishable from those of SusG-PAmCherry (Fig. 4; Table 1): 73.4% of SusG-PAmCherry molecules and 66.7% of SusE-Nterm-SusG-PAmCherry molecules diffuse at <0.02 μm2/s; 14.4% of SusG-PAmCherry molecules and 15.5% of SusE-Nterm-SusG-PAmCherry molecules diffuse at a rate between 0.02 and 0.05 μm2/s; and 12.1% of SusG-PAmCherry molecules and 17.5% of SusE-Nterm-SusG-PAmCherry molecules diffuse at >0.05 μm2/s. These results suggest that the immobility of SusE is not conferred by its N-terminus but is likely due to an unknown interaction with some membrane component or another protein. However, treatment of Bt cells with EDTA or lysozyme to disrupt lipopolysaccharide or peptidoglycan, respectively, failed to increase SusEF mobility (data not shown).
Figure 4.
SusG-PAmCherry (A and C) remains mobile when the 44 N-terminal amino acids are replaced with the 28 N-terminal amino acids from SusE (B and D), changing the lipidation signal. (A and B) Each image shows a cell with 35 single-molecule tracks plotted in random colors. Scale bars, 1 μm. (C and D) The MSD of all tracks lasting >20 frames is plotted for each protein fusion (red, effective diffusion coefficient (D) ≤ 0.001 μm2/s; yellow, 0.001 μm2/s < D ≤ 0.02 μm2/s; green, 0.02 μm2/s < D ≤ 0.05 μm2/s; blue, 0.05 μm2/s < D ≤ 0.1 μm2/s; purple: D > 0.1 μm2/s). To see this figure in color, go online.
Overall, though our experiments do not identify the mechanism that immobilizes SusEF, they demonstrate that the immobility of SusEF in the Bt outer membrane is an important property that remains robust to perturbations: SusEF remain stationary in the membrane independent of fluorescent tag identity or oxygen concentration and despite numerous perturbations, including knockouts of other Sus system proteins. This strong propensity of several proteins to remain stationary while their putative binding partners are mobile in the bacterial cell membrane presents a new model for the cooperative action of an outer-membrane protein system.
SusCDE can be captured in an outer-membrane complex
Previous work has captured a SusCDE interaction via formaldehyde cross-linking of cells followed by native polyacrylamide gel electrophoresis (28). To further examine this interaction and any others that may take place within this complex, we performed co-IP of SusD using custom anti-SusD with solubilized Bt membranes. This solubilization efficiently releases SusD, SusE, and SusG; however, SusF remains in the insoluble fraction (Fig. 5 a), even upon prolonged incubation with detergent (Fig. S8). SusF is an otherwise soluble protein when expressed recombinantly without its lipidation site, so its enrichment in the insoluble membrane fraction suggests that it interacts strongly with the membrane either directly or via binding to an unknown insoluble molecule. Although it is tempting to speculate that the immobility of both SusE and SusF is due to their interactions in the membrane, SusE is mostly solubilized, whereas SusF is not. Thus, it is unclear whether the enrichment of SusF in the insoluble fraction is related to its stationary nature, or whether these two observations are independent characteristics of this protein.
Based on previous reports, we hypothesized that some of the Sus proteins would interact through transient or weak protein-protein interactions (28). To detect Sus complex formation, we performed quantitative proteomics on the total membrane fraction from Bt before solubilization, as well as on SusD and SusE co-IP elutions from cells with or without formaldehyde cross-linking (Tables 2 and S4). For these experiments, we report the normalized spectral abundance factor (NSAF), a measure of how much of the total spectrum for a sample can be assigned to a particular protein, normalized by the molecular weight of that protein (30). Under native (non-cross-linking) conditions, co-IP of SusD brings along SusC, as demonstrated via Western blot (Fig. 5 b), and the relative abundance of SusD/SusC is ∼1:0.6 (Table 2). However, very little SusE is captured as a co-eluent in the native SusD pulldowns; the relative abundance of SusD/SusE is at most 1:0.1. This low SusD/SusE ratio, and the fact that it was only detected in one native experiment, suggests that SusE interacts only weakly with SusD or SusC in this experiment. When Bt cells were treated with formaldehyde to covalently link weakly or transiently interacting proteins, more SusE was immunoprecipitated along with SusD. For reasons that remain unclear, a lower amount of SusC was captured with SusD when formaldehyde was added. It is possible that SusE and SusC compete for interactions with SusD such that capturing more SusE limits the ability of SusC to interact with SusD. Reciprocally, co-IP of SusE under native conditions did not capture additional Sus proteins, but formaldehyde treatment resulted in co-elution of SusD and SusC (Tables 2 and S4). This need for formaldehyde suggests that the interaction of SusE with the SusCD complex is weak and/or transient.
Table 2.
Relative Sus Outer-Membrane Protein Abundances Identified by Proteomics Were Calculated from NSAF values
| Total Membrane Proteome No. 1 | Total Membrane Proteome No. 2 | SusD Co-IP Native No. 1 | SusD Co-IP Native No. 2 | SusD Co-IP Formaldehyde | SusE Co-IP Native | SusE Co-IP Formaldehyde | |
|---|---|---|---|---|---|---|---|
| SusC | 0.80 | 0.85 | 0.58 | 0.60 | 0.32 | NA | 0.07 |
| SusD | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | NA | 0.21 |
| SusE | 0.41 | 0.52 | NA | 0.11 | 0.22 | 1.00 | 1.00 |
| SusF | 0.41 | 0.58 | NA | NA | NA | NA | NA |
| SusG | 0.23 | 0.26 | NA | NA | NA | NA | NA |
Total membrane samples 1 and 2 are from Bt cells grown on maltose to induce Sus expression. Because of differences in sample concentration, these replicates are not averaged. Co-IP reactions were performed on solubilized fractionated membranes from maltose-grown cells. Co-IP results represent pooled triplicates.
In these experiments, the lack of SusG or SusF captured via co-IP was not necessarily unexpected. We have previously suggested that SusG interacts with the Sus complex transiently and therefore is unlikely to be pulled down with the complex natively (21). However, here SusG is still not captured with SusD or SusE in formaldehyde-treated cells. One possibility is that formaldehyde does not cross-link SusG because of the absence or spacing of appropriate chemical reactive groups at the interaction interface (31). Another important distinction is that for co-IP and proteomics, sus expression was induced in Bt cells with the small sugar maltose and not on starch, as this polysaccharide is difficult to remove from cells after culturing for the downstream experiments. Thus, the entire Sus complex may not associate unless the large starch polysaccharide is present. In addition, SusF may not be captured via co-IP because it is not efficiently solubilized from the membrane, yet it is possible that SusF in its native environment on the cell surface interacts with the other Sus proteins. How the Sus proteins interact with each other in the membrane is still unknown, though our work demonstrates that SusCDE interact, and our previous work suggests that SusG interacts transiently with these proteins during starch catabolism (21).
Two crystal structures of homologous Sus-like complexes from Bt have been determined, as reported in Glenwright (32). In the first Sus-like complex, which may target peptides, when the SusCD-like pair was isolated from solubilized membranes, two additional lipoproteins, encoded within the same locus and akin to SusEF, co-purified as part of this complex. The intimate association of these four proteins, BT2261-2264, is one example of Sus-like proteins that may associate stably. However, when this group isolated the second SusCD-like complex, BT1762/3, only the SusCD-like pair was captured, not the additional lipoprotein, BT1761, nor the enzyme, BT1760, also encoded within the PUL (32). Taken together with our data presented here, it is likely that the proteins within different Sus-like systems assemble with different affinities, stabilities, and dynamics depending on the nature of the individual proteins and the substrate that the system imports.
Quantitative proteomics on total Bt membranes measure the relative abundance of all proteins within the Sus and other homologous Sus-like systems. Although Sus is abundant in the membrane proteome, several other Sus-like systems and SusCD pairs are also highly expressed (Table S3). Some of these highly represented systems likely target other necessary nutrients such as vitamins or iron. The relative abundances of the five Sus lipoproteins in the membrane fraction of Bt are not equal: SusD and SusC are the most abundant, and SusE, SusF, and SusG have lower and more variable abundances. The SusD/SusC and SusD/SusE ratios differ between the native and cross-linked co-IPs compared to the total relative abundance of these proteins in the Bt outer membrane, likely because our co-IP experiments cannot capture every Sus protein assembly that exists on the cell surface. Our observation that less SusC and more SusE co-immunoprecipitate with SusD in the presence of formaldehyde may suggest that the Sus proteins cycle on and off of each other. This idea was first suggested by the Salyers lab, who reported that less SusC associates with SusD when SusE is present (28), as visualized via native polyacrylamide gel electrophoresis of solubilized Bt membranes. Therefore, we hypothesize that some Sus outer membrane proteins interact weakly and transiently, and not simultaneously in one distinct complex. In other words, there is some dynamic equilibrium between Sus proteins that may produce various combinations of SusCDEFG on the cell surface. The differences in the dynamics among the Sus proteins (Fig. 1), where SusG and SusD are mobile whereas SusE and SusF are not, likely contribute significantly to the formation of multiple “versions” of the complex.
Conclusions
We observed here for the first time, to our knowledge, that the protein components in Bt that collaborate to capture starch at the cell surface have distinctly different mobilities. In particular, we found that SusE and SusF remain highly confined to puncta, a behavior that differs from that of SusG and SusD, which diffuse across the Bt cell surface. These differences are surprising because the Sus outer membrane proteins have similar lipidation signals and are simultaneously expressed from the same locus. Although the mechanism that drives mobility or immobility is unclear, the immobility of SusE and SusF is robust to changes in the fluorescent label, to perturbations to Sus protein expression, and to the deletion of cell-surface capsular polysaccharides. The imperturbability of this phenotype suggests that it plays a role in the protein interactions involved in Sus complex formation.
In our previously published study of SusG dynamics (21), we noted that SusG (78 kDa) slows substantially in the presence of corn amylopectin, which has a molecular mass of 107–108 Da (33). Here, however, we observe that SusE (42 kDa) and SusF (52 kDa) are immobile even without exposure to starch; the experiments reported here are all performed in the presence of the small disaccharide maltose and/or the monosaccharide glucose. Of note, none of the Sus proteins binds maltose, but this small sugar effectively upregulates the sus operon, allowing us to observe Sus proteins on the cell surface. Therefore, neither the size of the SusEF proteins, alone or as fusions with PAmCherry (25 kDa) or Halo tag (33 kDa), nor the presence of maltose is likely to affect protein dynamics.
The weak and/or transient interactions involved in Sus complex formation are revealed here by several experimental approaches. The proteomic data suggests that the Sus proteins are present in different abundances on the cell surface: similar amounts of SusC and SusD are found in the membrane, whereas SusEFG are much less abundant. This difference suggests that these proteins do not engage in a static, high-affinity complex with equal components of all the proteins. Additionally, the co-IP experiments support a model of SusCDE interacting weakly—perhaps transiently—and in different stoichiometries. The single-molecule trajectories of SusG, which is highly mobile on the cell surface, corroborate this interpretation. Furthermore, although the SusD fusion has a growth defect, SusD-PAmCherry is also mobile. The movement of SusG and SusD may demonstrate that transient binding and unbinding of these proteins occurs during catabolism. Although the SusC/SusD ratio is ∼1:1 in the total-membrane proteomic samples, suggesting the proteins are present in the membrane at about equal amounts, we capture about two times more SusD than SusC during co-IP with SusD antibodies. The difference between the 1:1 SusC/SusD ratio seen by spectral abundance and the 1:2 ratio in co-IP supports the hypothesis of a transient interaction, and it is therefore likely that the affinity of SusC and SusD is weaker than in other SusCD-like pairs. For instance, during a recent structure determination of two other SusCD-like pairs, the SusC-like and SusD-like proteins were co-purified in a 1:1 complex at almost every domain junction (32).
Overall, this work supports a model whereby protein complex formation is a function of both protein affinity and mobility. Interaction among the Sus proteins may occur at immobile SusEF centers that nucleate the formation of transient SusCDEF or SusCDEFG complexes, motivating the need for more biophysical characterization to complement existing biochemical tools in assessing protein complexes on the bacterial cell surface. Thus, we present, to our knowledge, a new paradigm for the formation of enzyme complexes in the outer membrane of Gram-negative bacteria cells: within the Bt Sus protein system, SusE and SusF form stationary starch binding sites at which starch degradation occurs by transient binding of mobile SusG.
Author Contributions
All authors designed the research. H.H.T. performed single-molecule imaging experiments and data analysis. M.H.F. engineered the strains and performed molecular biology experiments and data analysis. All authors discussed all the results. The manuscript was written and edited by all authors. All authors read and approved the final manuscript.
Acknowledgments
Funding for this research was provided by the National Institutes of Health (NIH R01 GM118475-01 to N.M.K.). Further support was provided by National Science Foundation grant DB-1306434 to H.H.T. and Army Research Office grant W911NF-16-1-0450 to J.S.B. M.H.F. was partially supported by a predoctoral fellowship from the Cellular Biotechnology Training Program (T32GM008353). Yi Liao and David Rowland provided advice for data analysis.
Editor: Catherine Galbraith.
Footnotes
Hannah H. Tuson and Matthew H. Foley contributed equally to this work.
Eight figures, four tables, and two movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)35095-6.
Contributor Information
Nicole M. Koropatkin, Email: nkoropat@umich.edu.
Julie S. Biteen, Email: jsbiteen@umich.edu.
Supporting Material
Movies are acquired via time-lapse imaging, with one 40 ms frame acquired every 1 second. Scale bar = 1 μm.
Movies are acquired via time-lapse imaging, with one 40 ms frame acquired every 1 s. Scale bar, 1 μm.
References
- 1.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaiko G.E., Stappenbeck T.S. Host-microbe interactions shaping the gastrointestinal environment. Trends Immunol. 2014;35:538–548. doi: 10.1016/j.it.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sekirov I., Russell S.L., Finlay B.B. Gut microbiota in health and disease. Physiol. Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 4.Bäckhed F., Ley R.E., Gordon J.I. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 5.Bäckhed F., Ding H., Gordon J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El Kaoutari A., Armougom F., Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 2013;11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 7.Martens E.C., Koropatkin N.M., Gordon J.I. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J. Biol. Chem. 2009;284:24673–24677. doi: 10.1074/jbc.R109.022848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McNulty N.P., Wu M., Gordon J.I. Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biol. 2013;11:e1001637. doi: 10.1371/journal.pbio.1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martens E.C., Chiang H.C., Gordon J.I. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe. 2008;4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tancula E., Feldhaus M.J., Salyers A.A. Location and characterization of genes involved in binding of starch to the surface of Bacteroides thetaiotaomicron. J. Bacteriol. 1992;174:5609–5616. doi: 10.1128/jb.174.17.5609-5616.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foley M.H., Cockburn D.W., Koropatkin N.M. The Sus operon: a model system for starch uptake by the human gut Bacteroidetes. Cell. Mol. Life Sci. 2016;73:2603–2617. doi: 10.1007/s00018-016-2242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shipman J.A., Berleman J.E., Salyers A.A. Characterization of four outer membrane proteins involved in binding starch to the cell surface of Bacteroides thetaiotaomicron. J. Bacteriol. 2000;182:5365–5372. doi: 10.1128/jb.182.19.5365-5372.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koropatkin N.M., Martens E.C., Smith T.J. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure. 2008;16:1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koropatkin N.M., Smith T.J. SusG: a unique cell-membrane-associated α-amylase from a prominent human gut symbiont targets complex starch molecules. Structure. 2010;18:200–215. doi: 10.1016/j.str.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 15.D’Elia J.N., Salyers A.A. Contribution of a neopullulanase, a pullulanase, and an α-glucosidase to growth of Bacteroides thetaiotaomicron on starch. J. Bacteriol. 1996;178:7173–7179. doi: 10.1128/jb.178.24.7173-7179.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitamura M., Okuyama M., Yao M. Structural and functional analysis of a glycoside hydrolase family 97 enzyme from Bacteroides thetaiotaomicron. J. Biol. Chem. 2008;283:36328–36337. doi: 10.1074/jbc.M806115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sonnenburg J.L., Xu J., Gordon J.I. Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science. 2005;307:1955–1959. doi: 10.1126/science.1109051. [DOI] [PubMed] [Google Scholar]
- 18.Martens E.C., Lowe E.C., Gordon J.I. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 2011;9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cameron E.A., Kwiatkowski K.J., Martens E.C. Multifunctional nutrient-binding proteins adapt human symbiotic bacteria for glycan competition in the gut by separately promoting enhanced sensing and catalysis. MBio. 2014;5 doi: 10.1128/mBio.01441-14. e01441–e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cameron E.A., Maynard M.A., Martens E.C. Multidomain carbohydrate-binding proteins involved in Bacteroides thetaiotaomicron starch metabolism. J. Biol. Chem. 2012;287:34614–34625. doi: 10.1074/jbc.M112.397380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karunatilaka K.S., Cameron E.A., Biteen J.S. Superresolution imaging captures carbohydrate utilization dynamics in human gut symbionts. MBio. 2014;5 doi: 10.1128/mBio.02172-14. e02172–e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kleanthous C., Rassam P., Baumann C.G. Protein-protein interactions and the spatiotemporal dynamics of bacterial outer membrane proteins. Curr. Opin. Struct. Biol. 2015;35:109–115. doi: 10.1016/j.sbi.2015.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lill Y., Jordan L.D., Ritchie K. Confined mobility of TonB and FepA in Escherichia coli membranes. PLoS One. 2016;11:e0160862. doi: 10.1371/journal.pone.0160862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao Y., Schroeder J.W., Biteen J.S. Single-molecule motions and interactions in live cells reveal target search dynamics in mismatch repair. Proc. Natl. Acad. Sci. USA. 2015;112:E6898–E6906. doi: 10.1073/pnas.1507386112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogers T.E., Pudlo N.A., Martens E.C. Dynamic responses of Bacteroides thetaiotaomicron during growth on glycan mixtures. Mol. Microbiol. 2013;88:876–890. doi: 10.1111/mmi.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cockburn D.W., Orlovsky N.I., Koropatkin N.M. Molecular details of a starch utilization pathway in the human gut symbiont Eubacterium rectale. Mol. Microbiol. 2015;95:209–230. doi: 10.1111/mmi.12859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooper A.J., Kalinowski A.P., Salyers A.A. Construction and characterization of a Bacteroides thetaiotaomicron recA mutant: transfer of Bacteroides integrated conjugative elements is RecA independent. J. Bacteriol. 1997;179:6221–6227. doi: 10.1128/jb.179.20.6221-6227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho K.H., Salyers A.A. Biochemical analysis of interactions between outer membrane proteins that contribute to starch utilization by Bacteroides thetaiotaomicron. J. Bacteriol. 2001;183:7224–7230. doi: 10.1128/JB.183.24.7224-7230.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shipman J.A., Cho K.H., Salyers A.A. Physiological characterization of SusG, an outer membrane protein essential for starch utilization by Bacteroides thetaiotaomicron. J. Bacteriol. 1999;181:7206–7211. doi: 10.1128/jb.181.23.7206-7211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Old W.M., Meyer-Arendt K., Ahn N.G. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Hoffman E.A., Frey B.L., Auble D.T. Formaldehyde crosslinking: a tool for the study of chromatin complexes. J. Biol. Chem. 2015;290:26404–26411. doi: 10.1074/jbc.R115.651679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glenwright A.J., Pothula K.R., van den Berg B. Structural basis for nutrient acquisition by dominant members of the human gut microbiota. Nature. 2017;541:407–411. doi: 10.1038/nature20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manelius R., Buléon A., Bertoft E. The substitution pattern in cationised and oxidised potato starch granules. Carbohydr. Res. 2000;329:621–633. doi: 10.1016/s0008-6215(00)00206-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movies are acquired via time-lapse imaging, with one 40 ms frame acquired every 1 second. Scale bar = 1 μm.
Movies are acquired via time-lapse imaging, with one 40 ms frame acquired every 1 s. Scale bar, 1 μm.