Abstract
Non-alcoholic fatty liver disease (NAFLD) is a chronic progressive liver disorder that begins with simple hepatic steatosis and progresses to non-alcoholic steatohepatitis, fibrosis, cirrhosis, and even liver cancer. As the global prevalence of NAFLD rises, it is increasingly important that we understand its pathogenesis and develop effective therapies for this chronic disease. Forkhead box O (FOXO) transcription factors are key downstream regulators in the insulin/insulin-like growth factor 1 (IGF1) signaling pathway, and have been implicated in a range of cellular functions including the regulation of glucose, triglyceride, and cholesterol homeostasis. The role of FOXOs in the modulation of immune response and inflammation is complex, with reports of both pro- and anti-inflammatory effects. FOXOs are reported to protect against hepatic fibrosis by inhibiting proliferation and transdifferentiation of hepatic stellate cells. Mice that are deficient in hepatic FOXOs are more susceptible to non-alcoholic steatohepatitis than wild-type controls. In summary, FOXOs play a critical role in maintaining metabolic and cellular homeostasis in the liver, and dysregulation of FOXOs may be involved in NAFLD development.
Keywords: Forkhead box O (FOXO), Non-alcoholic fatty liver disease (NAFLD), Insulin-like growth factor 1 (IGF1), Steatosis, Inflammation, Fibrosis
1. Introduction
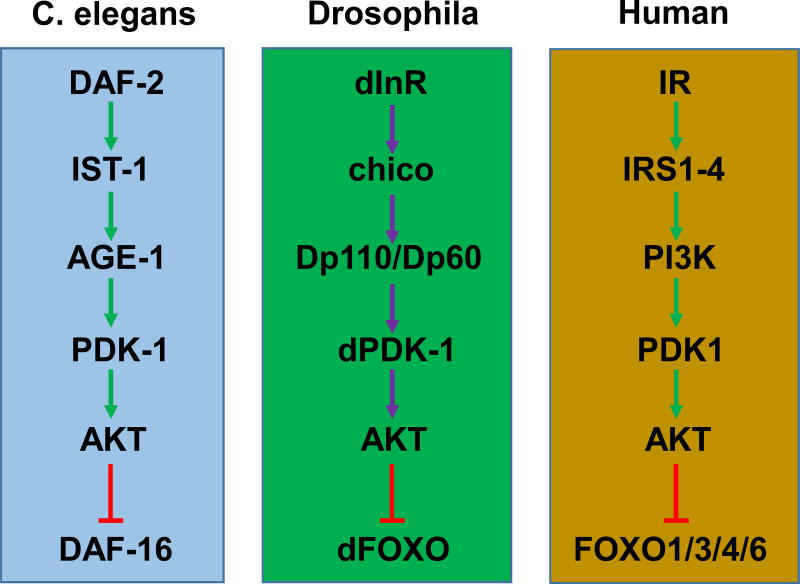
Forkhead box O (FOXO) transcription factors belong to the O subfamily of the forkhead box protein family.1 There is a single FOXO gene in Caenorhabditis elegans (DAF-16) and Drosophila (dFOXO), and four FOXO genes (FOXO1/3/4/6) in mammals. FOXO proteins are highly conserved, especially the forkhead box and transactivation domains, and RAC-alpha serine/threonine-protein kinase (AKT) conserves three major phosphorylation sites (Fig. 1). Mammals and other animals, such as Caenorhabditis elegans and Drosophila, share similar insulin/insulin-like growth factor (IGF) 1 signaling cascades (Fig. 2). Insulin/IGF1 activate insulin receptor/IGF1 receptor, which subsequently activate insulin receptor substrates through tyrosine phosphorylation. The activated insulin receptor substrates stimulate phosphoinositide 3-kinase, which converts Phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2] to phosphatidylinositol-3,4,5-trisphosphate [PI(3,4,5)P3]. This stimulates 3-phosphoinositide-dependent protein kinase 1 and mechanistic target of rapamycin complex 2, which activate AKT at Thr308 and Ser473, respectively.2–4 FOXOs are the immediate downstream effectors of AKT (Fig. 3).
Figure 1. The insulin/insulin-like signaling pathways are evolutionally conserved.
The FOXO transcription factors are regulated by the insulin/insulin-like signaling pathways that are well conserved in C. elegans, Drosophila, and mammals. Upon stimulation by insulin or insulin-like growth factors (IGFs), the insulin/IGFs receptors are activated, and subsequently the signaling cascade of IRS→PI3K→PDK1→AKT is activated as well. As a result, FOXOs are phosphorylated and inhibited by AKT.
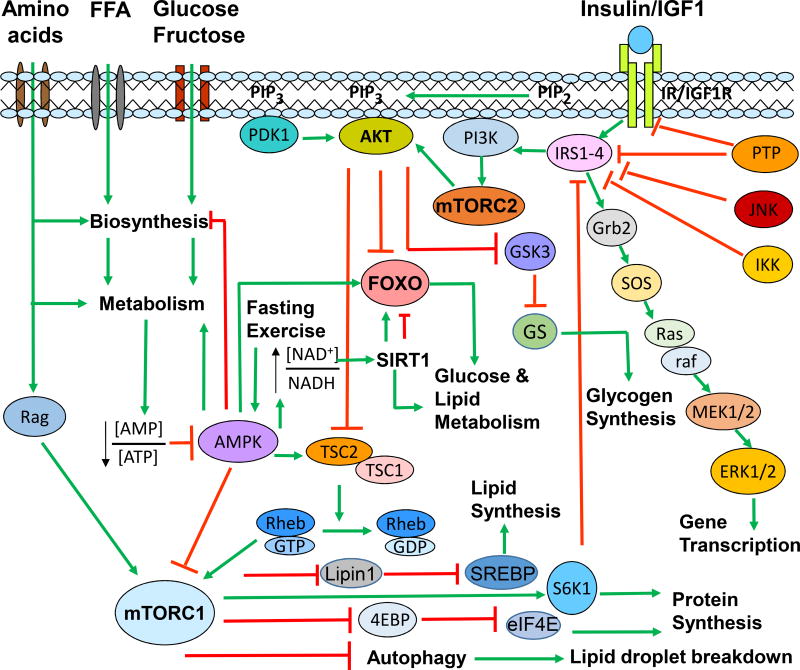
Figure 2. Insulin signaling and nutrient sensing pathways in hepatocytes.
Major signaling cascades in the insulin and amino acid signaling pathways are outlined in this simplified diagram. Insulin and nutrient signaling is normally integrated to maintain metabolic homeostasis. Insulin plays a critical role in glucose, lipid, and protein metabolism. Upon insulin stimulation, the insulin signaling cascade (IR→IRS→PI3K→PDK1/mTORC2→AKT) is activated. As a major kinase in the downstream of the insulin signaling, AKT controls hepatic glucose and lipid homeostasis. AKT activates glycogen synthesis by inhibiting GSK3 through phosphorylation. Meanwhile, AKT also inhibits the FOXO transcriptional activity for hepatic gluconeogenesis through phosphorylation and nuclear exclusion of FOXO. AKT also promotes lipid and protein synthesis through activation of mTORC1. In addition to insulin, amino acids also activate mTORC1 to promote protein synthesis and inhibit autophagy. mTORC1 stimulates lipogenesis through activation of SREBPs. FOXO is also modulated via deacetylation by SIRT1, an NAD+-dependent deacetylase. The energy sensor AMPK regulates metabolic homeostasis through activation of FOXO and inhibition of mTORC1.
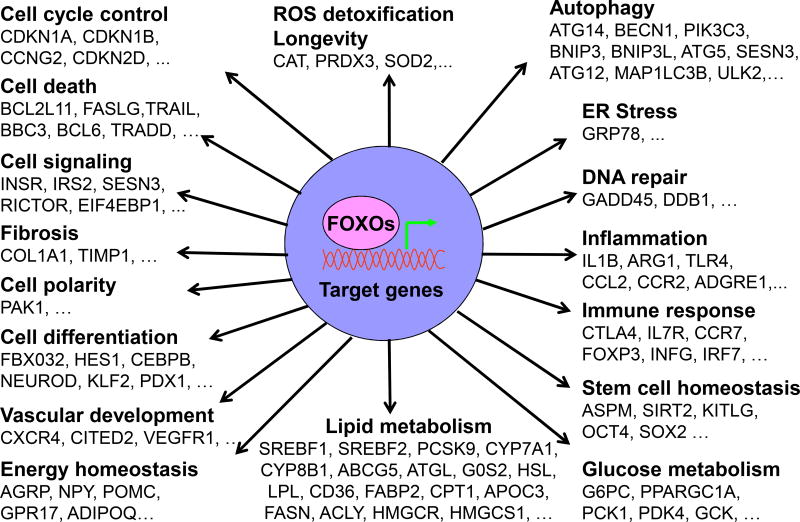
Figure 3. FOXOs have pleiotropic functions.
Major FOXO functions are highlighted here to indicate the involvement of FOXOs in multiple cellular processes including cell cycle control, cell differentiation, glucose and lipid metabolism, energy homeostasis, autophagy, ROS detoxification, ER stress, DNA repair, and immune response. Numerous genes have been identified as FOXO targets. Owing to the limited space, only a small number of the FOXO-regulated genes for each biological process are listed here.
FOXO transcriptional activity can be regulated by various post-translational modifications, though is predominantly regulated by phosphorylation and acetylation.5 AKT kinases play a critical role in FOXO inactivation by phosphorylating a few conserved serine/threonine sites of each FOXO (FOXO1-Thr24/Ser256/Ser319, FOXO3-Thr32/Ser253/Ser315, FOXO4-Thr32/Ser197/Ser262, FOXO6-Thr26/Ser184).6 In addition to AKT, there are a number of other kinases that can phosphorylate FOXOs, including adenosine monophosphate (AMP)-activated protein kinase (AMPK), c-Jun N-terminal kinase (JNK), extracellular signal-regulated kinase (ERK), p38 mitogen-activated protein kinase, mammalian sterile 20-like kinase 1, and protein kinase R-like endoplasmic reticulum kinase.7 In addition to phosphorylation, FOXOs can be acetylated by p300/ cyclic AMP response element-binding protein (CBP) acetyltransferases and deacetylated by sirtuin (SIRT) 1 and histone deacetylase 3.8–17
FOXOs have pleiotropic functions in animal systems, with effects on cell survival, anti-oxidative stress, autophagy, and metabolism (Fig. 4). In this short review, I will summarize our current understanding of liver FOXOs and their role in NAFLD development.
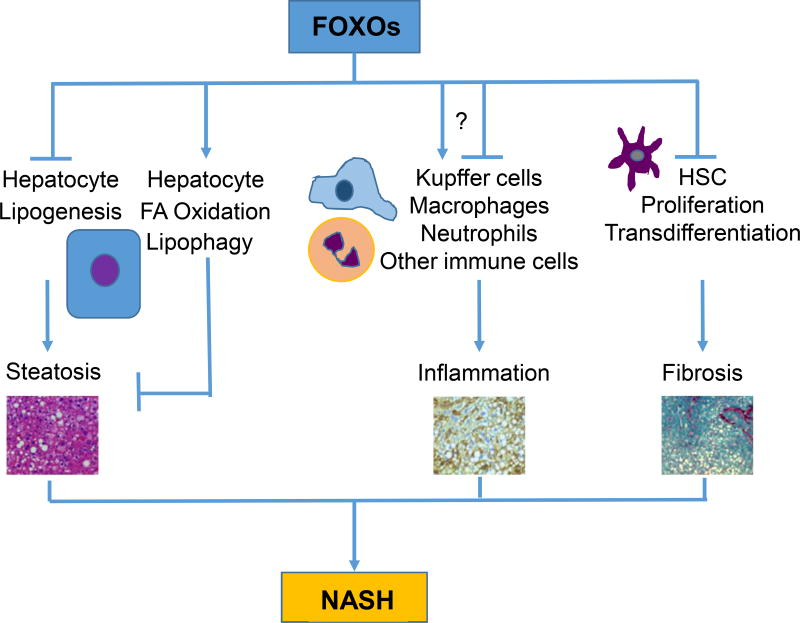
Figure 4. A working model depicting the involvement of FOXOs in the pathogenesis of NASH.
This is a very simplistic view of FOXOs in the development of NASH from the perspective of three major cell types in the liver – hepatocytes, Kupffer cells, and hepatic stellate cells (HSCs). Crosstalks between these and other cell types are not illustrated here. In hepatocytes, FOXOs suppress the development of steatosis by promoting lipophagy and fatty acid oxidation and inhibiting triglyceride and cholesterol biosynthesis. In immune cells including Kupffer cells and circulated macrophages, the role of FOXOs is not very clear as both pro- and anti-inflammation activities of FOXOs have been reported in the literature. Additional studies are needed to clarify the role of FOXOs in hepatic immune cells. In hepatic stellate cells (HSCs), FOXO1 has been shown to suppress HSC proliferation and transdifferentiation, thus inhibiting hepatic fibrosis.
2. FOXOs in glucose and lipid metabolism
The interplay between FOXO transcription factors and insulin and nutrient signaling pathways means that FOXOs play an important role in both glucose and lipid metabolism (Fig. 3).18–40 The role of FOXOs in the regulation of genes that are critically involved in glucose, triglyceride, and cholesterol metabolism is summarized below.
2.1. FOXOs in hepatic glucose metabolism
FOXOs have been shown to play a critical role in hepatic glucose homeostasis. Knockout of either FoxO1 alone or FoxO1/3/4 altogether specifically in mouse liver leads to lower blood glucose levels under both fasting and non-fasting conditions.21,25,26,35,36,40 FoxO6 whole body knockout mice also exhibit lower levels of fasting and non-fasting blood glucose.18 In response to starvation, FOXOs transcriptionally activate the hepatic gluconeogenic program by inducing a number of genes including phosphoenolpyruvate carboxykinase 1, glucose-6-phosphatase catalytic subunit, and pyruvate dehydrogenase kinase 4.24,26,35,36,38,40,41 Meanwhile, FOXOs also inhibit glycolysis, likely through suppression of glucokinase and pyruvate kinase gene expression (Fig. 4).24,26,35,36,38,41 By doing so, FOXOs help maintain normal blood glucose levels during starvation. However, under insulin resistant or diabetic conditions, with the tight control of insulin signaling lacking, FOXOs continuously activate hepatic gluconeogenesis and thereby promote hyperglycemia.26,41
2.2. FOXOs in hepatic triglyceride metabolism
FOXOs play a critical role in triglyceride homeostasis by regulating de novo lipogenesis, fatty acid oxidation, import of free fatty acids from the blood circulation, and export of triglyceride-rich very low density lipoproteins to the blood circulation (Fig. 4). In the regulation of de novo lipogenesis, FOXOs suppress the lipogenic master regulator sterol regulatory element binding protein (SREBP) 1 at the transcriptional level. As a result, a number of genes involved in fatty acid biosynthesis are also modulated by FOXOs, including acetyl-CoA carboxylase alpha, fatty acid synthase, adenosine triphosphate citrate lyase, malic enzyme 1, mitochondrial glycerol-3-phosphate acyltransferase, and stearoyl-CoA desaturase 1.20,21,23,29,31,36–38 Moreover, FOXOs activate lipolysis and fatty acid oxidation genes including adipose triacylglycerol lipase, hormone-sensitive lipase, lipoprotein lipase, and carnitine palmitoyltransferase 1.21,31,37,38,42 Interestingly, FOXO1 also suppresses expression of the G0/G1 switch-2 gene that encodes an inhibitor of adipose triacylglycerol lipase.37 FOXO1 has been shown to upregulate fatty acid transporters such as Leukocyte differentiation antigen CD36.43 In addition, FOXOs promote lipid droplet breakdown through activation of lipophagy, an autophagy process that degrades lipid droplets for energy production. A number of autophagy-related genes including autophagy related 5 (ATG5), ATG12, ATG14, beclin 1, phosphatidylinositol 3-kinase catalytic subunit type 3 (PIK3C3), and sestrin 3 are regulated by FOXOs. A role of autophagy in the promotion of lipid metabolism in the liver has been suggested by numerous studies;34,44–49 however, the underlying mechanism remains largely unclear.
2.3. FOXOs in hepatic cholesterol metabolism
FOXOs also regulate a number of genes involved in cholesterol biosynthesis and metabolism (Fig. 4). SREBP-2, the master regulator of cholesterol biosynthesis, is a direct target of FOXOs, especially FOXO3.32 Hepatic FoxO1/3/4 triple knockouts show increased expression of the SREBP-2 gene.32 As expected, a number of SREBP-2 target genes including 3-hydroxy-3-methylglutaryl-CoA reductase and 3-hydroxy-3-methylglutaryl-CoA synthase 1 are also suppressed by FOXOs.21,23,32,36,38 In addition to cholesterol biosynthesis, FOXO1 regulates cholesterol conversion to bile acids by modulating bile acid biosynthetic genes including cytochrome P450 family 7 subfamily A polypeptide 1 (CYP7A1), CYP7B1, and CYP8B1, although there are inconsistent findings with regard to the role of FOXO1 in the CYP7A1 gene regulation. FOXO1 also upregulates the genes encoding biliary cholesterol transporters—ATP binding cassette subfamily G member 5 and member 8.50–56
In addition, FOXOs regulate low-density lipoprotein (LDL)-cholesterol homeostasis. Normally, LDL-cholesterol is degraded through a LDL receptor (LDLR)-mediated clearance process; however, when the level of proprotein convertase subtilisin/kexin type 9 (PCSK9) is elevated, the interaction between PCSK9 and LDLR leads to the degradation of LDLR and causes an increase in LDL-cholesterol.57 Interestingly, the PCSK9 gene is suppressed by FOXO3 and SIRT 6. When FOXO3 or SIRT 6 is deficient in the liver, circulating LDL-cholesterol levels are elevated.58
3. FOXOs in non-alcoholic steatohepatitis
As FOXOs play a critical role in glucose and lipid homeostasis, it is not surprising that dysregulation of hepatic FOXOs may lead to metabolic disorders. Studies of FoxO gene knockouts and overexpression in mice have provided strong evidence regarding the role of FOXOs in hepatic steatosis. On a regular diet, deletion of FoxO1/3 or FoxO1/3/4 genes in mouse liver leads to mild or moderate hepatic steatosis, respectively.29,31,36 Overexpression of a constitutively active FOXO1 transgene reduces hepatic triglyceride content.37,38 When challenged by high-fat diets, FoxO1/3/4 liver-specific knockout mice develop very severe hepatic steatosis, especially on a high-fat plus cholesterol diet.29
FOXOs have been shown to modulate inflammation through regulation of a number of genes including interleukin 1 beta, toll-like receptor 4, C-C motif chemokine ligand 2, C-C motif chemokine receptor 2, and adhesion G protein-coupled receptor E1 (also named EMR1 or F4/80) (Fig. 4). Overexpression of constitutively active FOXO1 mutant in macrophages mediated by a LysM-Cre induces the expression of the C-C motif chemokine receptor 2 gene and increases the number of proinflammatory M1-type macrophages in mouse adipose tissue59 (though whether similar changes occur in hepatic macrophages or Kupffer cells is unclear). Mice that are deficient in FoxO1/3/4 specifically in hepatocytes are susceptible to high-fat plus cholesterol diet-induced inflammation and liver injury.29 It has been reported that FOXO1 expression and activity is elevated in patients with steatohepatitis.60 More studies are needed to clarify the role of FOXOs in human non-alcoholic steatohepatitis.
4. FOXOs in fibrosis
Human NAFLD is a progressive liver disease that begins with simple steatosis, transitions to hepatic inflammation, and later develops fibrosis as extracellular matrix proteins such as collagen gradually accumulate in the liver. Hepatic stellate cells (HSCs) are believed to play a crucial role in the development of liver fibrosis.61 FOXO1 has been shown to inhibit proliferation and transdifferentiation of HSCs, partly through the regulation of cyclin-dependent kinase inhibitor 1B and superoxide dismutase 2.62 After a bile duct ligation, FoxO1+/− mice are more predisposed to hepatic fibrosis than wild-type mice.62 Using the immortalized human HSC cell line LX-2, it has been shown that FOXO1 and FOXO3 are also involved in the tumor necrosis factor-related apoptosis-inducing ligand-mediated apoptosis of HSCs.63 In addition to their effect on HSCs, FOXO1/3/4 in hepatocytes play a protective role in diet-induced liver fibrosis. When hepatic FoxO1/3/4 genes are deleted in mice, expression of fibrogenic genes including type I collagen alpha 1 and tissue inhibitor of metalloproteinase 1 is greatly elevated after the knockout mice are challenged with either a high-fat or high-fat plus cholesterol diet.29
5. Conclusions
As FOXOs have been implicated in longevity in different organisms,5,64–67 their salutary functions in the liver, including maintaining glucose, triglyceride, and cholesterol homeostasis, and modulating inflammation and fibrosis, may contribute to the prolonged lifespan and protection against NAFLD (Fig. 5). Importantly, FOXO activity needs to be controlled according to dynamic environmental cues, as over- or under-activation may lead to undesirable consequences. For example, under insulin resistant conditions, FOXOs are constitutively active, resulting in elevated hepatic glucose output and M1-type macrophage activation.21,30,40,59,60,68–71 Additional studies are needed to fully understand the role of FOXOs in normal hepatic function and NAFLD development.
Supplementary Material
Supplementary Figure. 1. FOXO family members have high similarity in protein sequences. Human FOXO1/3/4/6 protein sequences from the NCBI database were used for the multiple sequence alignment analysis using an online tool – Clustal Omega – on the EMBO-EBI website. The known protein domains in FOXOs are underlined and labeled. The conserved AKT phosphorylation sites are highlighted in red.
Acknowledgments
This work was supported in part by the USA National Institutes of Health (NIH) grants including DK091592 and DK107682 from the National Institute of Diabetes and Digestive and Kidney Diseases and AA024550 from the National Institute on Alcohol Abuse and Alcoholism, by the Showalter Scholar award from Indiana University School of Medicine and Showalter Trust, by Indiana Clinical and Translational Sciences Institute grant ULITR001108 from the NIH National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award.
Footnotes
Edited by Peiling Zhu and Genshu Wang.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The author declares that he has no conflict of interest.
References
- 1.Carlsson P, Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev Biol. 2002;250:1–23. doi: 10.1006/dbio.2002.0780. [DOI] [PubMed] [Google Scholar]
- 2.Pajvani UB, Accili D. The new biology of diabetes. Diabetologia. 2015;58:2459–2468. doi: 10.1007/s00125-015-3722-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Webb AE, Brunet A. FOXO transcription factors: key regulators of cellular quality control. Trends Biochem Sci. 2014;39:159–169. doi: 10.1016/j.tibs.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arden KC. FOXO animal models reveal a variety of diverse roles for FOXO transcription factors. Oncogene. 2008;27:2345–2350. doi: 10.1038/onc.2008.27. [DOI] [PubMed] [Google Scholar]
- 5.Calnan DR, Brunet A. The FoxO code. Oncogene. 2008;27:2276–2288. doi: 10.1038/onc.2008.21. [DOI] [PubMed] [Google Scholar]
- 6.Manning BD, Toker A. AKT/PKB Signaling: Navigating the Network. Cell. 2017;169:381–405. doi: 10.1016/j.cell.2017.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klotz LO, Sánchez-Ramos C, Prieto-Arroyo I, Urbánek P, Steinbrenner H, Monsalve M. Redox regulation of FoxO transcription factors. Redox Biol. 2015;6:51–72. doi: 10.1016/j.redox.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daitoku H, Sakamaki J, Fukamizu A. Regulation of FoxO transcription factors by acetylation and protein-protein interactions. Biochim Biophys Acta. 2011;1813:1954–1960. doi: 10.1016/j.bbamcr.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Matsuzaki H, Daitoku H, Hatta M, Aoyama H, Yoshimochi K, Fukamizu A. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc Natl Acad Sci U S A. 2005;102:11278–11283. doi: 10.1073/pnas.0502738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pramanik KC, Fofaria NM, Gupta P, Srivastava SK. CBP-mediated FOXO-1 acetylation inhibits pancreatic tumor growth by targeting SirT. Mol Cancer Ther. 2014;13:687–698. doi: 10.1158/1535-7163.MCT-13-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mihaylova MM, Vasquez DS, Ravnskjaer K, et al. Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell. 2011;145:607–621. doi: 10.1016/j.cell.2011.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perrot V, Rechler MM. The coactivator p300 directly acetylates the forkhead transcription factor Foxo1 and stimulates Foxo1-induced transcription. Mol Endocrinol. 2005;19:2283–2298. doi: 10.1210/me.2004-0292. [DOI] [PubMed] [Google Scholar]
- 13.Banks AS, Kim-Muller JY, Mastracci TL, et al. Dissociation of the glucose and lipid regulatory functions of FoxO1 by targeted knockin of acetylation-defective alleles in mice. Cell Metab. 2011;14:587–597. doi: 10.1016/j.cmet.2011.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frescas D, Valenti L, Accili D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J Biol Chem. 2005;280:20589–20595. doi: 10.1074/jbc.M412357200. [DOI] [PubMed] [Google Scholar]
- 15.Daitoku H, Hatta M, Matsuzaki H, et al. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci U S A. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jing E, Gesta S, Kahn CR. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab. 2007;6:105–114. doi: 10.1016/j.cmet.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang F, Tong Q. SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1's repressive interaction with PPARgamma. Mol Biol Cell. 2009;20:801–808. doi: 10.1091/mbc.E08-06-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calabuig-Navarro V, Yamauchi J, Lee S, et al. Forkhead box O6 (FoxO6) depletion attenuates hepatic gluconeogenesis and protects against fat-induced glucose disorder in mice. J Biol Chem. 2015;290:15581–15594. doi: 10.1074/jbc.M115.650994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng Z, Guo S, Copps K, et al. Foxo1 integrates insulin signaling with mitochondrial function in the liver. Nat Med. 2009;15:1307–1311. doi: 10.1038/nm.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng X, Zhang W, O-Sullivan I, et al. FoxO1 inhibits sterol regulatory element-binding protein-1c (SREBP-1c) gene expression via transcription factors Sp1 and SREBP-1c. J Biol Chem. 2012;287:20132–20143. doi: 10.1074/jbc.M112.347211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong XC, Copps KD, Guo S, et al. Inactivation of hepatic Foxo1 by insulin signaling is required for adaptive nutrient homeostasis and endocrine growth regulation. Cell Metab. 2008;8:65–76. doi: 10.1016/j.cmet.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gross DN, Wan M, Birnbaum MJ. The role of FOXO in the regulation of metabolism. Curr Diab Rep. 2009;9:208–214. doi: 10.1007/s11892-009-0034-5. [DOI] [PubMed] [Google Scholar]
- 23.Haeusler RA, Han S, Accili D. Hepatic FoxO1 ablation exacerbates lipid abnormalities during hyperglycemia. J Biol Chem. 2010;285:26861–26868. doi: 10.1074/jbc.M110.134023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haeusler RA, Hartil K, Vaitheesvaran B, et al. Integrated control of hepatic lipogenesis versus glucose production requires FoxO transcription factors. Nat Commun. 2014;5:5190. doi: 10.1038/ncomms6190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haeusler RA, Kaestner KH, Accili D. FoxOs function synergistically to promote glucose production. J Biol Chem. 2010;285:35245–35248. doi: 10.1074/jbc.C110.175851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O-Sullivan I, Zhang W, Wasserman DH, et al. FoxO1 integrates direct and indirect effects of insulin on hepatic glucose production and glucose utilization. Nat Commun. 2015;6:7079. doi: 10.1038/ncomms8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imae M, Fu Z, Yoshida A, Noguchi T, Kato H. Nutritional and hormonal factors control the gene expression of FoxOs, the mammalian homologues of DAF-16. J Mol Endocrinol. 2003;30:253–262. doi: 10.1677/jme.0.0300253. [DOI] [PubMed] [Google Scholar]
- 28.Lee S, Dong HH. FoxO integration of insulin signaling with glucose and lipid metabolism. J Endocrinol. 2017;233:R67–R79. doi: 10.1530/JOE-17-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan X, Zhang Y, Kim HG, Liangpunsakul S, Dong XC. FOXO transcription factors protect against the diet-induced fatty liver disease. Sci Rep. 2017;7:44597. doi: 10.1038/srep44597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qu S, Altomonte J, Perdomo G, et al. Aberrant Forkhead box O1 function is associated with impaired hepatic metabolism. Endocrinology. 2006;147:5641–5652. doi: 10.1210/en.2006-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tao R, Wei D, Gao H, Liu Y, DePinho RA, Dong XC. Hepatic FoxOs regulate lipid metabolism via modulation of expression of the nicotinamide phosphoribosyltransferase gene. J Biol Chem. 2011;286:14681–14690. doi: 10.1074/jbc.M110.201061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tao R, Xiong X, DePinho RA, Deng CX, Dong XC. Hepatic SREBP-2 and cholesterol biosynthesis are regulated by FoxO3 and Sirt6. J Lipid Res. 2013;54:2745–2753. doi: 10.1194/jlr.M039339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tikhanovich I, Cox J, Weinman SA. Forkhead box class O transcription factors in liver function and disease. J Gastroenterol Hepatol. 2013;28(Suppl 1):125–131. doi: 10.1111/jgh.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiong X, Tao R, DePinho RA, Dong XC. The autophagy-related gene 14 (Atg14) is regulated by forkhead box O transcription factors and circadian rhythms and plays a critical role in hepatic autophagy and lipid metabolism. J Biol Chem. 2012;287:39107–39114. doi: 10.1074/jbc.M112.412569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiong X, Tao R, DePinho RA, Dong XC. Deletion of hepatic FoxO1/3/4 genes in mice significantly impacts on glucose metabolism through downregulation of gluconeogenesis and upregulation of glycolysis. PLoS One. 2013;8:e74340. doi: 10.1371/journal.pone.0074340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang K, Li L, Qi Y, et al. Hepatic suppression of Foxo1 and Foxo3 causes hypoglycemia and hyperlipidemia in mice. Endocrinology. 2012;153:631–646. doi: 10.1210/en.2011-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang W, Bu SY, Mashek MT, et al. Integrated regulation of hepatic lipid and glucose metabolism by adipose triacylglycerol lipase and FoxO proteins. Cell rep. 2016;15:349–359. doi: 10.1016/j.celrep.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang W, Patil S, Chauhan B, et al. FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J Biol Chem. 2006;281:10105–10117. doi: 10.1074/jbc.M600272200. [DOI] [PubMed] [Google Scholar]
- 39.Zhu J, Mounzih K, Chehab EF, Mitro N, Saez E, Chehab FF. Effects of FoxO4 overexpression on cholesterol biosynthesis, triacylglycerol accumulation, and glucose uptake. J Lipid Res. 2010;51:1312–1324. doi: 10.1194/jlr.M001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsumoto M, Pocai A, Rossetti L, Depinho RA, Accili D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab. 2007;6:208–216. doi: 10.1016/j.cmet.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 41.Dong X, Park S, Lin X, Copps K, Yi X, White MF. Irs1 and Irs2 signaling is essential for hepatic glucose homeostasis and systemic growth. J Clin Invest. 2006;116:101–114. doi: 10.1172/JCI25735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kamei Y, Mizukami J, Miura S, et al. A forkhead transcription factor FKHR up-regulates lipoprotein lipase expression in skeletal muscle. FEBS Lett. 2003;536:232–236. doi: 10.1016/s0014-5793(03)00062-0. [DOI] [PubMed] [Google Scholar]
- 43.Bastie CC, Nahlé Z, McLoughlin T, et al. FoxO1 stimulates fatty acid uptake and oxidation in muscle cells through CD36-dependent and -independent mechanisms. J Biol Chem. 2005;280:14222–14229. doi: 10.1074/jbc.M413625200. [DOI] [PubMed] [Google Scholar]
- 44.Jaber N, Dou Z, Chen JS, et al. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci U S A. 2012;109:2003–2008. doi: 10.1073/pnas.1112848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang X, Petyaykina K, Tao R, Xiong X, Dong XC, Liangpunsakul S. The inhibitory effect of ethanol on Sestrin3 in the pathogenesis of ethanol-induced liver injury. Am J Physiol Gastrointest Liver Physiol. 2014;307:G58–G65. doi: 10.1152/ajpgi.00373.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sinha RA, Singh BK, Zhou J, et al. Loss of ULK1 increases RPS6KB1-NCOR1 repression of NR1H/LXR-mediated Scd1 transcription and augments lipotoxicity in hepatic cells. Autophagy. 2017;13:169–186. doi: 10.1080/15548627.2016.1235123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin CW, Zhang H, Li M, et al. Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. J Hepatol. 2013;58:993–999. doi: 10.1016/j.jhep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider JL, Suh Y, Cuervo AM. Deficient chaperone-mediated autophagy in liver leads to metabolic dysregulation. Cell Metab. 2014;20:417–432. doi: 10.1016/j.cmet.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singh R, Kaushik S, Wang Y, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biddinger SB, Haas JT, Yu BB, et al. Hepatic insulin resistance directly promotes formation of cholesterol gallstones. Nat Med. 2008;14:778–782. doi: 10.1038/nm1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li T, Ma H, Park YJ, et al. Forkhead box transcription factor O1 inhibits cholesterol 7alpha-hydroxylase in human hepatocytes and in high fat diet-fed mice. Biochim Biophys Acta. 2009;1791:991–996. doi: 10.1016/j.bbalip.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Haeusler RA, Pratt-Hyatt M, Welch CL, Klaassen CD, Accili D. Impaired generation of 12-hydroxylated bile acids links hepatic insulin signaling with dyslipidemia. Cell Metab. 2012;15:65–74. doi: 10.1016/j.cmet.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li T, Kong X, Owsley E, Ellis E, Strom S, Chiang JY. Insulin regulation of cholesterol 7alpha-hydroxylase expression in human hepatocytes: roles of forkhead box O1 and sterol regulatory element-binding protein 1c. J Biol Chem. 2006;281:28745–28754. doi: 10.1074/jbc.M605815200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Park WH, Pak YK. Insulin-dependent suppression of cholesterol 7α-hydroxylase is a possible link between glucose and cholesterol metabolisms. Exp Mol Med. 2011;43:571–579. doi: 10.3858/emm.2011.43.10.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dansen TB, Kops GJ, Denis S, et al. Regulation of sterol carrier protein gene expression by the forkhead transcription factor FOXO3a. J Lipid Res. 2004;45:81–88. doi: 10.1194/jlr.M300111-JLR200. [DOI] [PubMed] [Google Scholar]
- 56.Li T, Ma H, Chiang JY. TGFbeta1, TNFalpha, and insulin signaling crosstalk in regulation of the rat cholesterol 7alpha-hydroxylase gene expression. J Lipid Res. 2008;49:1981–1989. doi: 10.1194/jlr.M800140-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cohen JC, Hobbs HH. Genetics. Simple genetics for a complex disease. Science. 2013;340:689–690. doi: 10.1126/science.1239101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tao R, Xiong X, DePinho RA, Deng CX, Dong XC. FoxO3 transcription factor and Sirt6 deacetylase regulate low density lipoprotein (LDL)-cholesterol homeostasis via control of the proprotein convertase subtilisin/kexin type 9 (Pcsk9) gene expression. J Biol Chem. 2013;288:29252–29259. doi: 10.1074/jbc.M113.481473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kawano Y, Nakae J, Watanabe N, et al. Loss of Pdk1-Foxo1 signaling in myeloid cells predisposes to adipose tissue inflammation and insulin resistance. Diabetes. 2012;61:1935–1948. doi: 10.2337/db11-0770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Valenti L, Rametta R, Dongiovanni P, et al. Increased expression and activity of the transcription factor FOXO1 in nonalcoholic steatohepatitis. Diabetes. 2008;57:1355–1362. doi: 10.2337/db07-0714. [DOI] [PubMed] [Google Scholar]
- 61.Tsuchida T, Friedman SL. Mechanisms of hepatic stellate cell activation. Nat Rev Gastroenterol Hepatol. 2017;14:397–411. doi: 10.1038/nrgastro.2017.38. [DOI] [PubMed] [Google Scholar]
- 62.Adachi M, Osawa Y, Uchinami H, Kitamura T, Accili D, Brenner DA. The forkhead transcription factor FoxO1 regulates proliferation and transdifferentiation of hepatic stellate cells. Gastroenterology. 2007;132:1434–1446. doi: 10.1053/j.gastro.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 63.Park SJ, Sohn HY, Yoon J, Park SI. Down-regulation of FoxO-dependent c-FLIP expression mediates TRAIL-induced apoptosis in activated hepatic stellate cells. Cell Signal. 2009;21:1495–1503. doi: 10.1016/j.cellsig.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 64.Morris BJ, Willcox DC, Donlon TA, Willcox BJ. FOXO3: a major gene for human longevity--a mini-review. Gerontology. 2015;61:515–525. doi: 10.1159/000375235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schaible R, Sussman M. FOXO in aging: did evolutionary diversification of FOXO function distract it from prolonging life? Bioessays. 2013;35:1101–1110. doi: 10.1002/bies.201300078. [DOI] [PubMed] [Google Scholar]
- 66.Mathew R, Pal Bhadra M, Bhadra U. Insulin/insulin-like growth factor-1 signalling (IIS) based regulation of lifespan across species. Biogerontology. 2017;18:35–53. doi: 10.1007/s10522-016-9670-8. [DOI] [PubMed] [Google Scholar]
- 67.Martins R, Lithgow GJ, Link W. Long live FOXO: unraveling the role of FOXO proteins in aging and longevity. Aging Cell. 2016;15:196–207. doi: 10.1111/acel.12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cook JR, Matsumoto M, Banks AS, Kitamura T, Tsuchiya K, Accili D. A mutant allele encoding DNA binding-deficient FoxO1 differentially regulates hepatic glucose and lipid metabolism. Diabetes. 2015;64:1951–1965. doi: 10.2337/db14-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Su D, Coudriet GM, Hyun Kim D, et al. FoxO1 links insulin resistance to proinflammatory cytokine IL-1beta production in macrophages. Diabetes. 2009;58:2624–2633. doi: 10.2337/db09-0232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fan W, Morinaga H, Kim JJ, et al. FoxO1 regulates Tlr4 inflammatory pathway signalling in macrophages. EMBO J. 2010;29:4223–4236. doi: 10.1038/emboj.2010.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miao H, Zhang Y, Lu Z, Liu Q, Gan L. FOXO1 involvement in insulin resistance-related pro-inflammatory cytokine production in hepatocytes. Inflamm Res. 2012;61:349–358. doi: 10.1007/s00011-011-0417-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure. 1. FOXO family members have high similarity in protein sequences. Human FOXO1/3/4/6 protein sequences from the NCBI database were used for the multiple sequence alignment analysis using an online tool – Clustal Omega – on the EMBO-EBI website. The known protein domains in FOXOs are underlined and labeled. The conserved AKT phosphorylation sites are highlighted in red.