Abstract
A current challenge in microbial pathogenesis is to identify biological control agents that may prevent and/or limit host invasion by microbial pathogens. In natura, hosts are often infected by multiple pathogens. However, most of the current studies have been performed under laboratory controlled conditions and by taking into account the interaction between a single commensal species and a single pathogenic species. The next step is therefore to explore the relationships between host–microbial communities (microbiota) and microbial members with potential pathogenic behavior (pathobiota) in a realistic ecological context. In the present study, we investigated such relationships within root-associated and leaf-associated bacterial communities of 163 ecologically contrasted Arabidopsis thaliana populations sampled across two seasons in southwest of France. In agreement with the theory of the invasion paradox, we observed a significant humped-back relationship between microbiota and pathobiota α-diversity that was robust between both seasons and plant organs. In most populations, we also observed a strong dynamics of microbiota composition between seasons. Accordingly, the potential pathobiota composition was explained by combinations of season-specific microbiota operational taxonomic units. This result suggests that the potential biomarkers controlling pathogen’s invasion are highly dynamic.
Introduction
In the last decade, a conspicuous effort has been made to characterize and understand the role of host-associated microorganisms. Animals and plants are currently considered as holobionts associated with the microorganisms that inhabit and evolve with them [1–3]. However, how commensal microbes interact with pathogens remains largely unknown. It has been recently reported that the human gut microbiota can protect against the potential overgrowth of indigenous opportunistic pathobionts (i.e., pathogenic species that inhabit the host at a low bacterial population size) [4] or pathogenic invaders [5] by niche competition and/or induction of the host immune system [6]. In plants, both rhizosphere-associated and phyllosphere-associated bacteria can directly or indirectly increase host resistance to phytopathogenic microorganisms via production of antimicrobial compounds or elicitation of plant defences [7–12]. For example, plant-associated strains belonging to the Sphingomonas genus can protect the model annual plant Arabidopsis thaliana from infections caused by the causal agent of bacterial spot Pseudomonas syringae [13]. Although these studies support the importance of certain microbes in protecting plants against infections, they mainly focused on the interaction between a single commensal species and a single pathogenic strain. Nevertheless, as previously observed in animals [14, 15], plants are often infected in natura by multiple pathogens [16–18]. Therefore, the current challenge in plant pathology is to investigate the relationships between resident microbial communities and pathogenic microbes at the community level, that is, between microbiota (defined here as the microbial communities inhabiting plants) and pathobiota (defined here as the complex of microorganisms with the potential to cause disease on a given plant host [19]).
Understanding how the native cortege of beneficial/commensal microbes interacts to protect plants against the invasion of a cortege of pathogenic species requires testing for theories in community ecology [20]. In plant community ecology, substantial efforts have been made in the last decades to understand the relationships between α-diversity of resident species and α-diversity of invasive species [21] by both applying spatial pattern studies and constructed community studies [22, 23]. In a large number of studies conducted on natural plant communities at a large spatial scale, species richness of both resident and invasive species were found to be positively correlated with the quality of the abiotic environment, thereby leading to a positive relationship between resident species diversity and invader success [22] (Supplementary Figure S1). On the other hand, as predicted by Elton’s theory (1958), constructed plant community studies performed in homogenous abiotic environments revealed a negative relationship between resident species diversity and invader success [22]. This negative relationship is explained by the negative impact of the increased resident species diversity on resource availability, which is in turn detrimental for the establishment of the invasive species (Supplementary Figure S1). These two contrasted phenomena lead to the invasion paradox (humped-back relationship between the diversity of invaders and the diversity of resident species, Supplementary Figure S1), which is therefore predicted to mainly result from the interplay between species diversity, resource availability, and niche dimensionality in ecologically relevant conditions [24]. In contrast to these studies performed on plant communities, most studies on microbial communities reported a negative relationship between the diversity of the resident microbial species and the invader success [24, 25]. The absence of studies conducted at a large spatial scale could explain the lack of evidences for a positive relationship between the α-diversity of microbial residents and the α-diversity of invaders.
Testing for the invasion paradox between microbiota and pathobiota therefore requires a characterization of the microbial communities across the range of native habitats encountered by a given plant species [17]. Accordingly, recent studies highlighted that the ecology of the habitats where both plants and microorganisms co-exist and evolve is a crucial variable to take into account when investigating the relationships between a host and its microbes participating in the holobiont system [26, 27]. In addition, in native habitats, plants are naturally exposed to pathogens whose attacks are influenced by local abiotic/biotic conditions [18].
In this study, we aimed to investigate the in situ relationships between bacterial microbiota and the potential bacterial pathobiota in 163 natural A. thaliana populations collected in southwest of France and inhabiting ecologically contrasted habitats. In particular, we aimed (i) to test for the invasion paradox and (ii) to identify the combinations of microbial species that can prevent and/or limit pathogen invasion. Because bacterial communities associated with plants can rapidly change within the host life cycle [26, 28–30] and can largely differ between plant compartments [27, 31, 32], we described both microbiota and potential pathobiota in the leaf and root compartments across two seasons within a single life cycle of A. thaliana.
Materials and methods
Identification of A. thaliana populations
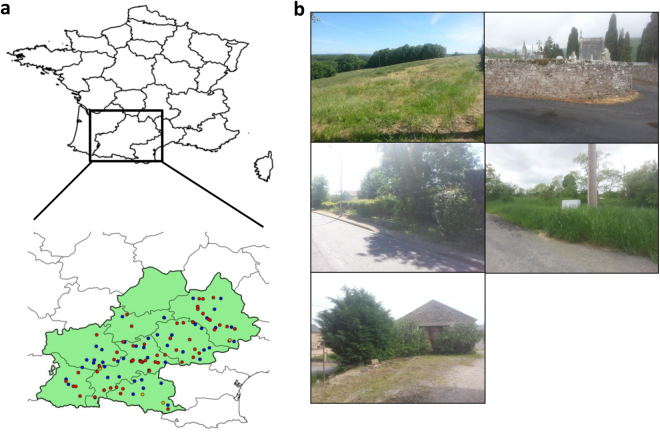
In this study, we focused on 163 natural A. thaliana populations identified in May 2014 in the Midi-Pyrénées region (Fig. 1a and Supplementary Table 1). These populations were chosen to maximize the diversity of habitats (such as climate, soil type, vegetation type, and degree of anthropogenic perturbation) encountered by A. thaliana (Fig. 1b). Because the 163 populations strongly differed in their main germination cohort in autumn 2014 (early November vs. early December; Supplementary Text), we defined three seasonal groups, hereafter named (i) “autumn” corresponding to 84 populations collected in November/December 2014, (ii) “spring with autumn” corresponding to 80 populations already sampled in autumn and additionally sampled in early-spring (February/March 2015), and (iii) “spring without autumn,” corresponding to 79 populations only sampled in early-spring (February/March 2015) (Supplementary Text).
Fig. 1.
Plant material. a Location of the 163 A. thaliana populations across the Midi-Pyrénées region (southwest of France). The average distance among populations was 99.9 km (median = 92.6 km, SD = 55.4 km). Blue dots represent the 80 populations collected in both autumn and spring, red dots represent the 79 populations collected in spring only and orange dots represent the 4 populations collected in autumn only. The map clearly shows that the range of sampling across the Midi-Pyrénées region was homogeneous during the two sampling seasons. b Diversity of habitats encountered by A. thaliana in the Midi-Pyrénées region
To avoid a confounding effect between the sampling date and geographical origin, populations were randomly collected during the sampling periods in autumn 2014 and early-spring 2015.
Sampling, generation of the gyrB amplicons, and sequences
The bacterial communities were characterized through amplification of a fraction of gyrB gene encoding for the bacterial gyrase β-subunit. This molecular marker has a deeper taxonomic resolution than other molecular markers designed on the hypervariable regions of the 16S rRNA gene [33, 34], thereby allowing to distinguish bacterial operational taxonomic units (OTUs) at the species level. Furthermore, this single-copy gene limits the overestimation of taxa carrying multiple copies of rrn operons. In this study, based on synthetic communities, we confirmed a better taxonomic resolution of the gyrB gene compared with the 16S rRNA gene (Supplementary Text and Supplementary Data 1).
To characterize the bacterial communities, ~4 individuals per population and per season were sampled in situ at the rosette stage resulting in a total number of 1912 leaf and root samples (Supplementary Text). The epiphytic and endophytic bacterial components of either leaf or root samples were not separated. For each plant compartment, we therefore extracted the total DNA of both epiphytic and endophytic microbes (Supplementary Text). The amplicon gyrB was amplified with some modification of the protocol described in Barret et al. [33]. In particular, three internal tags were added at each 5′ and 3′ of the original primers to allow the multiplexing of three 96-well plates (Supplementary Text).
For each sample, PCR amplifications were repeated three times and technical replicates were pooled in a unique PCR plate. PCR products were purified by using Agencourt®AMPure® magnetic beads following the manufacturer’s instructions and purified amplicons were quantified with Nanodrop and appropriately diluted to obtain an equimolar concentration. Two microliters of equimolar PCR-purified products were used for a second PCR with the Illumina adaptors. The second PCR amplicons were then purified and quantified as described above to obtain a unique equimolar pool. The latter was quantified by real-time quantitative reverse transcriptase-polymerase chain reaction and then sequenced with Illumina MiSeq 2 × 250 v3 (Illumina Inc., San Diego, CA, USA) in the GeT-PlaGe Platform (Toulouse, France). MS-102-3003 MiSeq Reagent Kit v3 600 cycle was used for this purpose.
Bioinformatics analysis and data curation
Reads were demultiplexed by considering the three internal tags. After demultiplexing, the average number of sequences per sample was of 46,135 ± 19,820. Prior further analysis, the negative controls—consisting of the sterilized water used to clean up the samples during field sampling, the sterilized DNA free water used to elute the DNA after phenol/chloroform extraction, and the DNA free water used for PCR amplifications—were checked for the presence/absence of amplicons by blasting them against the gyrB database composed by 30,627 sequences ([33]; Supplementary Text). Because negative controls showed no trace of entire gyrB sequences, they were removed before clustering. Samples showing low sequence quality were also removed before clustering, resulting in a total of 1903 samples. Taxonomic affiliation of gyrB sequences was performed by using a Bayesian classifier [35] implemented in the classify.seqs command of mothur [36] against an in-house gyrB database containing 30,627 representative sequences with an 80% bootstrap confidence score. Clustering of sequences into OTUs was performed with Swarm [37] by using a clustering threshold (d) = 1. Only the OTUs that were composed by a minimum of five sequences across all samples were kept, resulting in a total of 278,333 OTUs. Then, we applied two steps of filtering. First, to control for sampling limitation within each sample, we estimated a Good’s coverage score for each sample [36]. Based on the distribution of the Good’s coverage score, only samples with a score more than 0.5 were considered. Second, only OTUs showing a minimum relative abundance of 1% in at least one sample were selected by using a home-made perl program. The final data set corresponded to a matrix of 1655 samples by 6627 OTUs. The 6627 OTUs obtained after the filtering of the data represented 2.4% of the totality of the OTUs but 55.6% of the totality of the reads (Supplementary Figure S2). In addition, the mean number of reads per OTU maintained after filtering was 51.3-fold higher than the one of the discarded OTUs (Supplementary Figure S2). This final data set was used to determine the matrix composed by the potential pathogenic species. More precisely, the potential pathobiota in both leaves and roots of the 163 A. thaliana populations was determined by using a list of the phytopathogenic bacteria established by the International Society of Plant Pathology Committee on the Taxonomy of Plant Pathogenic Bacteria (ISPP-CTPPB; Supplementary Data 2) [38, 39]. This potential pathobiota list (Supplementary Data Set 2) was composed by 199 bacterial species that were filtered on the microbiota matrix. The potential pathobiota matrix resulted in 11 bacterial species that were further curated. More precisely, we only considered: (i) the bacterial species already reported to colonize A. thaliana (Pseudomonas syringae, Pseudomonas viridiflava, Pantoea agglomerans, Sphingomonas melonis, and Xanthomonas campestris) [40, 41], (ii) the bacterial species pathogenic on Brassicaceae species (Pseudomonas marginalis, Streptomyces scabei, and Xanthomonas perforans) [42, 43], (iii) the causal agent of lamb’s lettuce spot Acidovorax valerianellae reported to be widely distributed in France [44]. The bacterial species Diaphorobacter oryzae and Janthinobacterium agaricidamnosum were removed from the potential pathobiota matrix because they have been reported previously only in non-plant habitats [45] and mushrooms [46], respectively. The resulted potential pathobiota matrix was composed by 1203 samples and 29 OTUs characterizing the nine bacterial species listed above (i.e., a given bacterial species can be represented by more than one OTU). The 29 potential pathogenic OTUs were removed from the microbiota matrix. Therefore, the final microbiota matrix used for further analysis was composed by 6598 OTUs.
Analysis of the α-diversity and β-diversity and characterization of the potential pathobiota
Shannon diversity and observed species richness were estimated on the final OTU matrix by using the summary.single function of mothur [36]. Indexes of α-diversity were also calculated by sample rarefaction of 300/600/900 iters. Results between non-rarefied and rarefied samples were similar and the complete non-rarefied data set was used for the calculation of microbiota diversity.
Due to sparsity of the OTU matrix, a Hellinger transformation [47] was performed by using the vegan R package [48] and the relative Hellinger distance was inferred with the decostand command in the vegan R package prior β-diversity analyses. Following Ramette [49], the resulting Hellinger distance matrix was reduced by running a principal coordinates analysis (PCoA) with the ape R package [50]. Because PCoA performed on Hellinger distance matrix based on rarefied data and on Jaccard similarity coefficient matrix led to similar patterns of ordination (Supplementary Figure S3), the PCoA coordinates from the non-rarefied Hellinger distance matrix were retrieved and used for statistical analysis described below.
Non-metric multidimensional scaling (NMDS) was also run on the Hellinger distance matrix. However, the values of stress were 0.431 and 0.293 for 2D and 3D NMDS ordination space, respectively. These stress values suggest a lack of fit between the ranks on the NMDS ordination configuration and the ranks in the original distance matrix [49].
Statistical analysis
Natural variation for the eight descriptors of microbiota (i.e., species richness, Shannon index, first and second PCoA axes) and potential pathobiota microbiota (i.e., species richness, Shannon index, first and second PCoA axes) was explored using different mixed models (Supplementary Text). A correction for the number of tests was performed to control the FDR at a nominal level of 5%.
In order to study the relationship between microbiota α-diversity and potential pathobiota α-diversity, linear and non-linear regressions were fitted using the “lm” and “nls” functions implemented in the R environment, respectively (Supplementary Text). Using a paired-sample t test, model selection was performed by comparing the goodness of fit between linear and non-linear models across the “diversity estimate × plant compartment × seasonal group” combinations. In order to confirm the significance of the humped-back curve observed between diversity estimates (species richness and Shannon index) of the microbiota and the potential pathobiota, the parameters of the non-linear model were compared to a null distribution of those parameters obtained by creating 100 random microbiota OTU matrices paired with 100 random pathobiota OTU matrices (Supplementary Text).
Although pathogen-focused network analysis has been used for investigating the relationships between whole microbiota and microbial pathogens, this method appears only suitable for studying monospecific interactions between a single pathogenic species and the rest of the microbial community members [51]. In order to include higher-order interactions in the study of the relationship between microbiota and potential pathobiota composition, a sparse partial least-square regression [52, 53] was therefore adopted to maximize the covariance between linear combinations of relative abundances of OTUs from the microbiota (matrix X) and linear combinations of relative abundances of species from the potential pathobiota (matrix Y) (Supplementary Text). Significance of the OTUs included in the linear combinations was estimated by a Jackknife resampling approach by leaving out 10% of the samples 1000 times (Supplementary Text).
Data availability
The raw FastQ reads were deposited in the Sequence Read Archive (SRA) of NCBI https://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP096011 under the study number SRP096011. The filtered matrix for both microbiota and potential pathobiota are available in Supplementary Data 4 and Supplementary Data 5, respectively. Raw values for both α-diversity and β-diversity used for statistical analysis are available in Supplementary Data 6.
Results
Characterization of the A. thaliana microbiota and potential pathobiota
We sampled 163 natural A. thaliana populations chosen to maximize the diversity of habitats encountered by A. thaliana in the Midi-Pyrénées region (Fig. 1). Due to differences in germination timing in autumn, about half of the populations were sampled both in autumn and in spring, whereas the other half of populations were sampled only in spring, thereby leading to three seasonal groups of populations, that is, “autumn,” “spring with autumn,” and “spring without autumn.”
We obtained 18,610,383 high-quality reads across 1655 samples, with on average ~10,136 reads per sample. After data filtering, we identified 6627 non-singleton bacterial OTUs. A large amount of these OTUs were specific to roots or leaves, as only ~8.1% OTUs (n = 540 OTUs) were shared between both plant compartments. However, the relative abundance of OTUs shared between leaf and root samples was 20.2 and 16.0 higher than the relative abundance of leaf-specific and root-specific OTUs, respectively. This suggests that generalist OTUs are dominant members of the Arabidopsis thaliana microbiota.
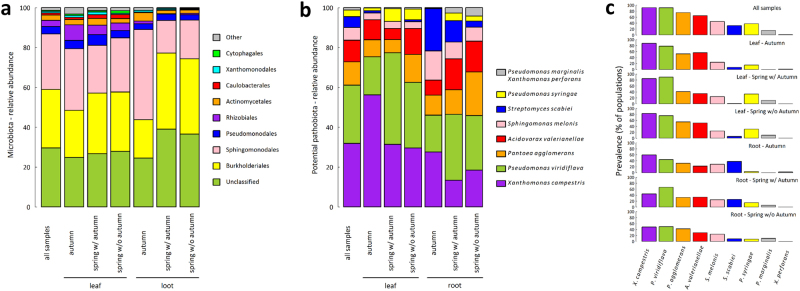
As commonly observed in A. thaliana and other plant species [27, 32, 54–56], bacterial communities were largely dominated by Proteobacteria (>80%). At the order level, Burkholderiales (29.3%) and Sphingomonodales (27.9%) were dominant (Fig. 2a). In comparison with autumn, samples collected in spring were enriched for Burkholderiales and depleted for Sphingomonodales in the root compartment (Fig. 2a). Germination timing in autumn did not impact the relative abundance of the bacterial orders in plants collected in spring (Fig. 2a).
Fig. 2.
Stacked barplots of the relative abundances of the major bacterial orders and bacterial species for microbiota and potential pathobiota of the A. thaliana populations collected in the Midi-Pyrénées region, respectively. a Stacked barplots representing the relative abundance for the ten most abundant bacterial orders of the microbiota. “All samples” n = 1655; leaf: “autumn” n = 314, “spring w/ autumn” n = 245, “spring wo/ autumn” n = 262; root: “autumn” n = 309, “spring w/ autumn” n = 267, “spring wo/ autumn” n = 258. In comparison with autumn, samples in spring were enriched for Burkholderiales (χ2 = 7.93, P < 0.01) and depleted for Sphingomonodales (χ2 = 18.18, P < 0.001) but only in the root compartment. b Stacked barplots representing the relative abundance of the nine bacterial species representing the potential pathobiota. “All samples” n = 1203; leaf: “autumn” n = 273, “spring w/ autumn” n = 209, “spring wo/ autumn” n = 213; root: “autumn” n = 180, “spring w/ autumn” n = 171, “spring wo/ autumn” n = 157. For the seasonal groups “autumn” and “spring w/autumn,” we observed a depletion of X. campestris (“autumn,” χ2 = 15.67, P < 0.01; “spring with autumn,” χ2 = 8.26, P < 0.05) and an enrichment of S. scabiei (“autumn,” χ2 = 18.61 P < 0.001; “spring with autumn,” χ2 = 9.31, P < 0.05) in the root compartment in comparison with the leaf compartment. In the leaf compartment, an enrichment of P. viridiflava between autumn and spring (χ2 = 15.20, P < 0.01) was associated with a depletion of X. campestris (χ2 = 11.50, P < 0.01). A correction for the number of tests was performed to control the false discovery rate at a nominal level of 5%. c Prevalence (number of populations in which the potential pathogen species was detected) of the nine bacterial species characterizing the potential pathobiota
Across all samples, we identified nine bacterial species described as phytopathogenic by the International Society of Plant Pathology Committee on the Taxonomy of Plant Pathogenic Bacteria [38, 39] (Fig. 2b). Among them, the three most abundant species representing more than 72% of the whole potential pathobiota were X. campestris (31.9%), P. viridiflava (29.2%), and P. agglomerans (11.7%) (Fig. 2b). For the seasonal groups “autumn” and “spring w/autumn,” we observed a depletion of X. campestris and an enrichment of S. scabiei in the root compartment in comparison with the leaf compartment (Fig. 2b). In the leaf compartment, an enrichment for P. viridiflava between autumn and spring was associated with a depletion for X. campestris (Fig. 2b). Similarly to the microbiota, no effect of germination timing in autumn was observed on the relative abundance of the potential pathogenic species for plants collected in spring (Fig. 2b).
It is noteworthy that the relationship between the relative abundance among samples and the prevalence among populations was weak for some potential pathogenic bacterial species (Fig. 2c). For example, although the relative abundance of P. syringae and S. scabiei among all samples ranged from 3.4% to 5.5%, respectively (Fig. 2b), these two species were present on average in more than 31% of the populations (Fig. 2c). Visual inspection of original data suggests that this pattern is mainly explained by the presence of few highly infected plants in many populations.
To confirm the pathogenic behavior of the potential pathobiota, representative bacterial strains of the three most abundant species were tested for their pathogenicity on their natural host A. thaliana and on a non-host plant (tobacco). For the P. syringae complex (including both P. syringae sensu stricto and P. viridiflava; Supplementary Text), we isolated 97 strains (74P. viridiflava strains and 23P. syringae sensu stricto strains; Supplementary Text, Supplementary Data 3, and Supplementary Figure S4). Their pathogenicity was assessed based on in planta bacterial growth, disease symptoms in A. thaliana and a hypersensitive response (HR) test on tobacco (Supplementary Text). We found that (i) 84 strains of the P. syringae complex induced a HR on tobacco (Supplementary Text and Supplementary Data 3), (ii) all the four strains tested for in planta growth were able to reach a population size of 106 colon-forming unit.cm−2 7 days post inoculation on A. thaliana (Supplementary Figure S5a and Supplementary Table S2), and (iii) seven out of eight strains tested were able to induce disease on at least one of the eight A. thaliana local accessions tested (Supplementary Text, Supplementary Figure S5b–S7, and Supplementary Table S3). For X. campestris, 59 strains were isolated (Supplementary Text and Supplementary Figure S8) and all of them induced disease symptoms on the A. thaliana Kas-1 accession (Supplementary Figure S9), which is susceptible to most X. campestris strains isolated from crops [57]. In addition, 52 of the 59 X. campestris induced a HR on tobacco (Supplementary Text and Supplementary Data 3). For P. agglomerans, we isolated a single strain (Supplementary Text and Supplementary Figure S10) that was able to induce disease symptoms on all the 23 A. thaliana local accessions tested (Supplementary Text, Supplementary Figure S11, S12, and Supplementary Table S4). Taken together, these results support that most of the strains identified here as part of the A. thaliana potential pathobiota have a pathogenic behavior on A. thaliana. In addition, the relative abundance of the potential pathobiota was significantly higher in A. thaliana individuals with visible disease symptoms (4.5%) than in asymptomatic A. thaliana individuals (1.6%) when sampled in situ (general linear model, F = 26.05, P < 0.001) (Supplementary Figure S13), strengthening the potential pathogenic behavior of the pathobiota identified in this study.
α-diversity of the A. thaliana microbiota and potential pathobiota
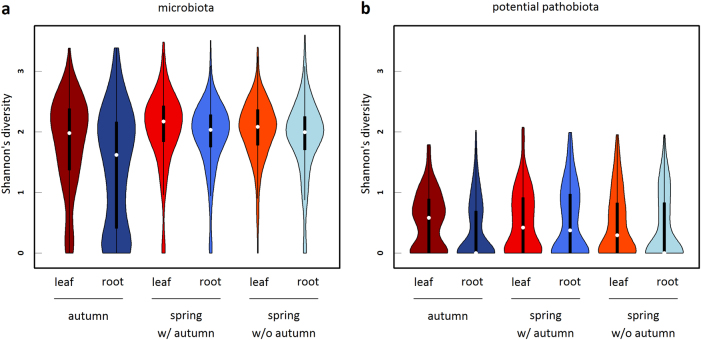
We investigated whether the level of α-diversity (species richness and Shannon index) estimated for each sample was driven by the effects of season, plant compartment, and population. Statistical results globally led to similar biological conclusions between species richness and Shannon index (Supplementary Tables S5–S12). Bacterial communities of the microbiota were on average less diverse in roots than in leaves, in particular in autumn (Fig. 3a and Supplementary Tables S5–S7). For the potential pathobiota, a similar pattern was observed in autumn but not in spring where the potential pathogenic communities were as diverse in roots as in leaves (Fig. 3b and Supplementary Tables S5–S7). More importantly, strong differences among populations in the dynamics of α-diversity between autumn and spring were observed for the microbiota and to a lesser extent for the potential pathobiota (Fig. 4 and Supplementary Table S5, S6).
Fig. 3.
Violin plots (i.e., box-and-whisker plot overlaid with a kernel density plot) representing the seasonal variation of the α-diversity inferred as Shannon index for both (a) microbiota and (b) potential pathobiota. Leaf and root samples are represented with a red and blue color scale, respectively. Microbiota: “autumn—leaf” n = 314, “autumn—root” n = 309, “spring w/ autumn—leaf” n = 245, “spring w/ autumn—root” n = 267, “spring w/o autumn—leaf” n = 262, “spring w/o autumn—root” n = 258. Pathobiota: “autumn—leaf” n = 273, “autumn—root” n = 180, “spring w/ autumn—leaf” n = 209, “spring w/ autumn—root” n = 171, “spring w/o autumn—leaf” n = 213, “spring w/o autumn—root” n = 157
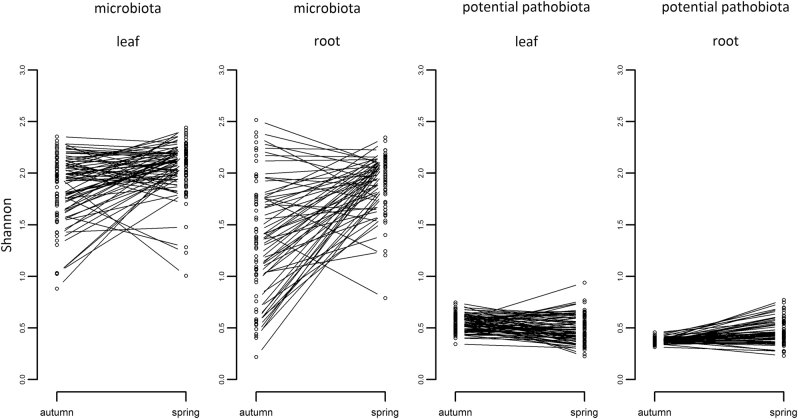
Fig. 4.
Variation among populations in the dynamics of α-diversity between autumn and spring. Each dot corresponds to the mean Shannon index (estimated as BLUPs, Supplementary Text) of a population. “Leaf” n = 74 populations and “root” n = 62 populations
Whatever the seasonal group considered, variation in α-diversity of the microbiota was first explained by differences among populations (~16.8% for species richness and ~26.2% for Shannon index) (Fig. 4 and Supplementary Tables S8–S11). In contrast, the main source of variation in the α-diversity of the potential pathobiota largely differs between the two seasons (Supplementary Tables S8–S11). Variation in α-diversity of the pathobiota in autumn and spring was first explained by the factor “plant compartment” (~12.5%) and the factor “population” (~13.8%), respectively (Supplementary Table S11). At the “plant compartment × seasonal group” level, the level of differentiation among populations for Shannon index was on average almost twice higher for the microbiota than for the potential pathobiota (~38.3% vs. ~14.5% of variance explained by the factor “population”) (Supplementary Table S12).
No effect of germination timing in autumn was observed on the microbiota and pathobiota α-diversity of plants collected in spring (Supplementary Table S7 and S11).
Relationships between microbiota and potential pathobiota α-diversity: testing for the invasion paradox
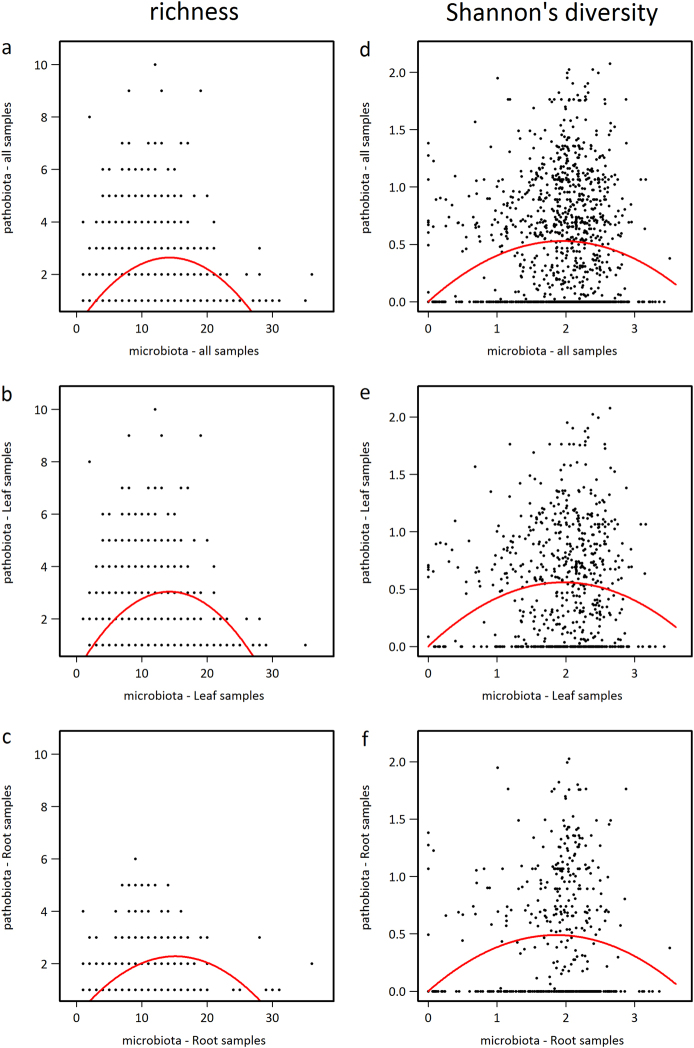
By considering all samples, we observed a highly significant humped-back relationship between the species richness of the potential pathobiota and the species richness of the microbiota (Fig. 5a, Supplementary Table S13, and Supplementary Figure S14). This humped-back relationship was robust whatever the “seasonal group × plant compartment” considered (Fig. 5b, c and Supplementary Table S13). A similar humped-back relationship was observed when considering Shannon index instead of species richness, with the exception of the root compartment in the seasonal group “spring w/ autumn” (Fig. 5d–f, Supplementary Table S13, and Supplementary Figure S14). Amongst all “seasonal group × plant compartment” combinations, up to ~20% of variation of potential pathobiota α-diversity was explained by variation of microbiota α-diversity (Supplementary Table S13). Importantly, the significance of the humped-back relationship was not affected by the threshold of the OTU filtering based on the minimum relative abundance of an OTU in at least one sample (Supplementary Figure S15).
Fig. 5.
Humped-back relationships between potential pathobiota α-diversity and microbiota α-diversity. a Species richness when considering all samples. b Species richness when considering samples from the leaf compartment. c Species richness when considering samples from the root compartment. d Shannon index when considering all samples. e Shannon index when considering samples from the leaf compartment. f Shannon index when considering samples from the root compartment. The red lines indicate a significant quadratic relationship, according to the following non-linear model: pathobiota’s diversity ~ k*microbiota’s diversity – q*microbiota’s diversity*microbiota’s diversity
The humped-back relationship observed in this study indicates that a poorly diversified potential pathobiota was associated with either a highly or a poorly diversified microbiota, whereas a highly diversified potential pathobiota was found in the presence of microbiota with an intermediate level of diversity. Altogether, these results are in line with theoretical expectations of the invasion paradox (Supplementary Figure S1).
Composition and structure of A. thaliana microbiota and potential pathobiota
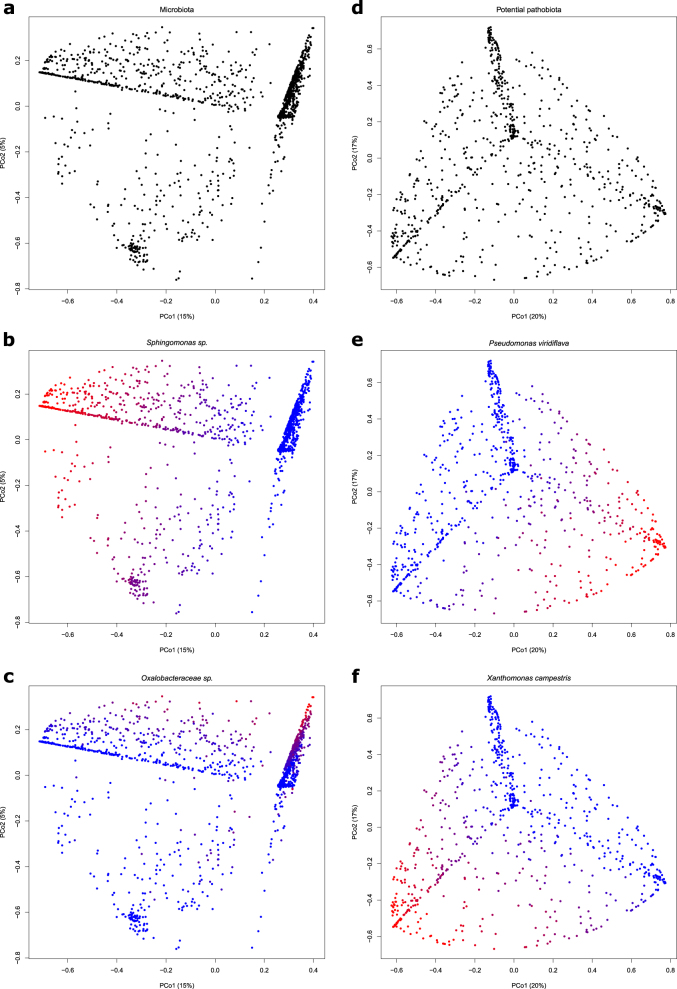
For the microbiota, the first two PCoA axes explained 20% of the β-diversity (Fig. 6). The microbiota was structured according to a pattern of two perpendicular branches (Fig. 6a). The relative abundance variation of the most abundant OTU, that is, an OTU belonging to the genus Sphingomonas, explained up to 20.7% of the variation along the first branch (Fig. 6b). The relative abundance variation of the second most abundant OTU (an OTU belonging to the family Oxalobacteraceae) was related to the variation along the second branch (Fig. 6c). For the potential pathobiota structured according to a pattern of three branches (Fig. 6d), the first two PCoA axes explained 37% of the β-diversity (Fig. 6d). The variation along two of these branches was related strongly but independently to the relative abundance variations of P. viridiflava and X. campestris (Fig. 6e, f).
Fig. 6.
Bacterial composition and structure of A. thaliana illustrated by principal coordinates (PCoA) plots based on Hellinger dissimilarity matrices for microbiota (left panels) and potential pathobiota (right panels). n = 1655 samples for the microbiota matrix and n = 1203 samples for the potential pathobiota matrix. a PCoA plot of microbiota for all samples indicates that bacterial composition is mainly structured along two axes. The (b) and (c) plots illustrate the relationships between the microbiota composition and the abundance of the two most abundant OTUs, that is, Sphingomona ssp., and Oxalobacteraceae unclassified sp., respectively. d PCoA plot of potential pathobiota for all samples indicates that bacterial composition is mainly structured along three axes. The (e) and (f) plots illustrate the relationships between the potential pathobiota composition and the abundance of the two most abundant potential pathogenic OTUs, that is, Pseudomonas viridiflava and Xanthomonas campestris, respectively. Red-to-blue color gradient indicates high-to-low OTU relative abundance per sample
We observed strong differences among populations for the dynamics of β-diversity of the microbiota between autumn and spring, with the factor “season × population” explaining 53.8% and 43.2% of the variation along the first and second PCoA axes, respectively (Supplementary Figure S16 and Supplementary Table S5, S6). The dynamics of β-diversity of the potential pathobiota between autumn and spring was also dependent on the considered population but to a much lesser extent than what was observed for the microbiota β-diversity (i.e., 9.8% and 7.6% of the variation along the first and second PCoA axes, respectively) (Supplementary Table S5, S6).
Within each seasonal group, the variation of microbiota along the PCoA axes was largely explained by differences among populations (up to ~81%) (Supplementary Figure S17 and Supplementary Table S8–S11), while the variation of the potential pathobiota β-diversity was first explained by the factors “population” (~16.2%) and “plant compartment” (~12.6%) when considering the first and second PCoA axis, respectively (Supplementary Table S11). At the “plant compartment × seasonal group” level, the level of differentiation among populations for β-diversity was on average higher for the microbiota than for the potential pathobiota (~76.5% vs. ~10.8% of the first PCoA axis variation explained by the factor “population”) (Supplementary Table S12 and Supplementary Figure S17).
An effect of germination timing in autumn on the β-diversity of plants collected in spring was not observed on microbiota but on potential pathobiota. In the leaf compartment, variation along the first PCoA axis was driven by differences among populations with early autumn germinants (i.e., “spring with autumn” seasonal group), while variation along the second PCoA axis was driven by differences among populations with late autumn germinants (i.e., “spring without autumn” seasonal group) (Supplementary Table S12 and Supplementary Figure S17).
Relationship between microbiota and potential pathobiota β-diversity
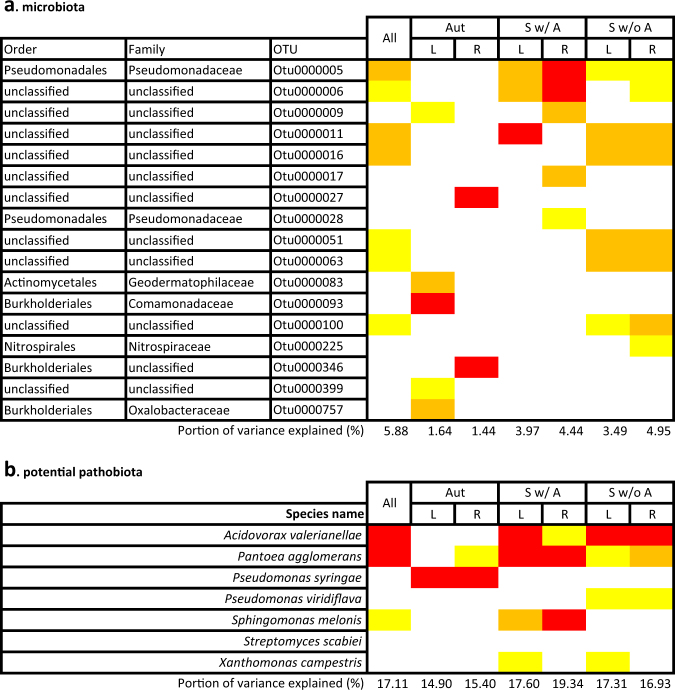
For each “seasonal group × plant compartment” combination, a non-negligible percentage of variation of the potential pathobiota composition (16.9% on average) was explained by a minor fraction of the variation of the microbiota composition (3.3% on average) (Fig. 7). The identity of the microbiota OTUs associated with variation of the potential pathobiota composition largely differs between seasons, plant compartment (in particular in autumn) and germination cohorts in autumn (Figs. 7 and 8). Among the 17 candidate OTUs of the microbiota, the fifth most abundant microbiota OTU (Pseudomonas moraviensis) was the most prevalent microbiota OTU across the six “seasonal group × plant compartment” combinations. However, a large fraction of the candidate microbiota OTUs (58.8%) corresponds to unclassified OTUs at the order level.
Fig. 7.
Relationships between (a) microbiota β-diversity and (b) potential pathobiota β-diversity based on a sparse partial least-square regression (sPLSR). Only OTUs with a loading value above 0.2 in more than 75% of the 1000 Jackknife resampled matrices were considered as significant. The color gradient indicates the strength of the loading values for both microbiota OTUs and potential pathobiota OTUs (yellow: 0.2 < loadings < 0.35; orange: 0.35 < loadings < 0.5; red: loadings > 0.5). “All,” “Aut,” “Sw/ A,” and “Sw/o A” stand for all samples and samples from the three seasonal groups (i.e., “autumn” populations, “spring w/ autumn” populations, and “spring w/o autumn” populations), “L” and “R” stand for leaf and root, respectively. “All” n = 1655; “Aut—L” n = 314, “S w/ A—L” n = 245, “S w/o A—L” n = 262; “Aut—R” n = 309, “S w/ A—R” n = 267, and “S w/o A—R” n = 258
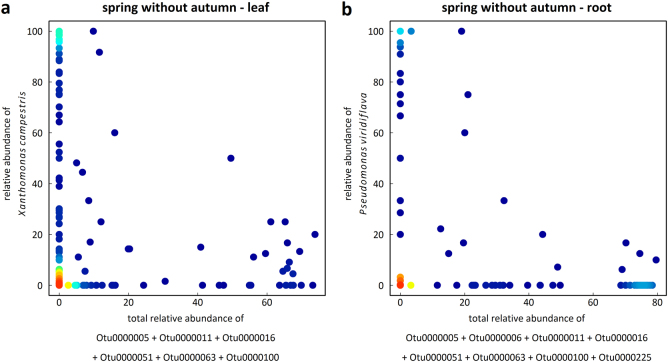
Fig. 8.
2D density plots illustrating the relationships between the relative abundance of the two most abundant bacterial species of the potential pathobiota and the total relative abundance of the microbiota OTUs identified by sparse partial least-square regression (sPLSR). a Example with X. campestris in the seasonal group “spring without autumn” in the leaf compartment (Pearson’s r = −0.15, P = 0.0187, n = 262 samples). b Example with P. viridiflava in the seasonal group “spring without autumn” in the root compartment (Pearson’s r = −0.17, P = 0.0006, n = 258 samples). Red-to-blue color gradient represents high-to-low density gradient
Discussion
The characterization of the A. thaliana potential pathobiota in an ecological context
Several evidences suggest that the potential pathobiota was well characterized in our set of 163 natural populations of A. thaliana, thereby allowing investigating the bacterial microbiota–potential pathobiota relationships. First, based on a deeper taxonomic resolution, the gyrB marker allows us to distinguish bacterial OTUs at the species level, thereby facilitating the identification of the OTUs that potentially belong to the pathobiota. For example, in this study, we discriminated in the genus Pseudomonas, bacterial species that are potentially pathogenic (e.g., Pseudomonas viridiflava) from bacterial species belonging to the microbiota (e.g., Pseudomonas moraviensis). In addition, according to the gyrB community profiling, X. campestris and P. viridiflava were the two most abundant species of the A. thaliana potential pathobiota. These observations are congruent with a previous bacterial community profiling obtained with a 16S rRNA region of 1.4 kb from a field experiment on wild-type and mutant lines of A. thaliana [41]. Second, we detected in situ a strong relationship between the presence of disease symptoms and the relative abundance of the potential pathobiota. Third, pathogenicity tests on both host and non-host plants confirmed that most of the strains belonging to the three most abundant species—composing almost three quarters of the whole potential pathobiota—have a pathogenic behavior. Finally, we found a significant and strong positive relationship between relative abundance and α-diversity of the pathobiota (Supplementary Table S14). The latter result suggests that plants are more often infected by consortia of pathogenic species than by a single pathogenic lineage, which is in line with similar observations obtained in humans and animals [58]. For example, multiple closely related Borrelia genospecies have been found to display a positive co-occurrence in ticks [59]. Studies on plant pathogens also suggest that co-infection is a frequent process mediated by niche functionality of the colonizing species [16]. More broadly, primary infecting pathogenic lineages can open the route for opportunistic lineages, as previously demonstrated for brassica black rot disease in which leaf infection by Xanthomonas is followed by the infection of a soft-rotting bacterium responsible for the rotting disease [60].
Putting the characterization of bacterial communities in an ecological genomics framework
In this study, we found that the in situ microbiota and potential pathobiota of A. thaliana were affected by the combined effects of season, plant compartment, and population. In particular, we observed a strong dynamics in community succession of the microbiota between seasons, reinforcing the need to study the dynamics of bacterial communities over the entire plant life cycle (from seed to seed) across a large range of native habitats [17]. In addition, our study revealed that seasonal community succession of the microbiota largely differed among the 163 populations. Because α-diversity and β-diversity varied at a very small geographic scale (Supplementary Figure S18), understanding among-population variation of seasonal community succession will require a thorough characterization of abiotic and biotic factors known to influence microbiota variation in plant species, such as soil conditions [54, 56], micro-local climatic conditions [61], and plant community composition [62, 63].
In contrast to the microbiota, we found that plant compartment mainly influenced the composition of the potential pathobiota as well as the relative abundance of the most abundant pathogenic species. Our results are in accordance with previous studies reporting that pathogenic species such as P. syringae sensu lato and X. campestris evolved specific strategies (e.g., entry by stomata, hydatodes and wounds) to infect the leaf compartment in a wide range of crops [64]. On the other hand, the most abundant OTUs of the microbiota were shared between leaves and roots. In agreement, the use of a gnotobiotic A. thaliana plant system allows to demonstrate potential reciprocal relocation between root and leaf microbiota members [65]. In the same study, whole-genome sequencing and functional analysis of bacteria associated with both leaves and roots of A. thaliana highlighted a clear taxonomy and functional overlap of the bacterial populations inhabiting this plant species [65]. Altogether, these results are in contrast with previous studies on human microbiota demonstrating a remarkable organization of microbes into body site niches [6, 58]. This discrepancy might originate from a lack of specialized plant tissues constituting strictly specialized niches for commensal bacteria as observed in human organs.
The invasion paradox is mediated by distinct microbiota composition between seasons and plant organs
In agreement with theoretical expectations on the invasion paradox [21, 23, 66], we observed a humped-back relationship between potential pathobiota α-diversity and microbiota α-diversity. Such a pattern may have been observed due to the large range of habitats where A. thaliana plants have been collected, thereby increasing the range of dimensionality of in planta ecological niches available for microbes. Besides the interplay between diversity and niche dimensionality, other hypotheses can contribute to the explanation of the humped-back relationship. An increase in microbiota diversity can be associated with an increase of antagonistic (e.g., production of antimicrobial compounds) and predator (e.g., grazing) species [67], leading to a negative relationship between potential pathobiota α-diversity and microbiota α-diversity. Concerning the positive relationship between poorly diversified potential pathobiota and microbiota, several non-exclusive hypotheses can be advanced. First, ecological disturbance can increase the random establishment of a single pathogenic species with negative consequences on resident species diversity [68]. Second, by producing virulence proteins, a pathogen can exclusively invade a given plant compartment [69]. Third, both microbiota and potential pathobiota can be poorly diversified because other microbial communities (such as fungal and oomycete communities) exploit most of the resources available in the plant, leading to niche inter-kingdom competition [26].
Interestingly, the pattern of invasion paradox was robust between compartments but also between seasons, despite a strong seasonal community succession of the microbiota in most populations. While this observation reinforces the importance of α-diversity in A. thaliana populations to maintain a microbiota balance to prevent pathogen occurrence, it also suggests that the potential pathobiota composition was associated with season-specific combinations of microbiota OTUs. Accordingly, the identity of the microbiota OTUs associated with variation of the potential pathobiota composition largely differs between seasons. This dynamics in the potential biomarkers (i.e., specific bacteria taxa preventing pathogen spread) controlling pathogen’s invasion may result from niche overlap and other mechanisms regulating microbe–microbe interactions that drastically influence the bacterial composition. A better understanding of the processes underlying the observed invasion paradox will require the isolation of a large number of strains representative of the microbiota found in the 163 A. thaliana populations. This step will especially be relevant for the numerous microbiota OTUs that are unclassified at the order level but associated with variation of the potential pathobiota composition.
However, we should acknowledge that a substantial fraction of the variation of the A. thaliana pathobiota was not explained by the microbiota variation. This fraction of unexplained pathobiota variation may result from (i) habitat-specific combinations of bacterial species controlling the potential pathobiota, (ii) the control of bacterial pathogen species by fungal or oomycete microbes [26], and (iii) the genetics of A. thaliana shaping natural variation in both pathogen’s abundance and prevalence [17]. Teasing apart the relative roles of these putative factors will require a thorough complementary ecological and genomic characterization of our natural populations of A. thaliana.
Electronic supplementary material
Acknowledgements
This work was funded by the Région Midi-Pyrénées (CLIMARES project), the LABEX TULIP (ANR-10-LABX-41, ANR-11-IDEX-0002-02), and the Métaprogramme MEM (INRA, Metabar programme). Part of this work was carried out on the Toulouse Plant–Microbe Phenotyping facility (http://tpmp.inra.fr), which is part of the Laboratoire des Interactions Plantes Micro-organismes, LIPM—UMR INRA441/CNRS2594. We are also grateful to the staff of the LIPM greenhouse for their assistance during growth chamber experiments.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0152-7) contains supplementary material, which is available to authorized users.
References
- 1.Bordenstein SR, Theis KR. Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol. 2015;13:1–23. doi: 10.1371/journal.pbio.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vandenkoornhuyse P, Quaiser A, Duhamel M, Le Van A, Dufresne A. The importance of the microbiome of the plant holobiont. New Phytol. 2015;206:1196–206. doi: 10.1111/nph.13312. [DOI] [PubMed] [Google Scholar]
- 3.Youle M, Knowlton N, Rohwer F, Gordon J, Relman DA. Superorganisms and holobionts. Microbe Mag. 2013;8:152–3. doi: 10.1128/microbe.8.152.1. [DOI] [Google Scholar]
- 4.Vayssier-Taussat M, Albina E, Citti C, Cosson JF, Jacques MA, Lebrun MH, et al. Shifting the paradigm from pathogens to pathobiome: new concepts in the light of meta-omics. Front Cell Infect Microbiol. 2014;4:29. doi: 10.3389/fcimb.2014.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamada N, Chen GY, Inohara N, Nunez G. Control of pathogens and pathobionts by the gut microbiota. Nat Immunol. 2013;14:685–90. doi: 10.1038/ni.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–41. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haney CH, Samuel BS, Bush J, Ausubel FM. Associations with rhizosphere bacteria can confer an adaptive advantage to plants. Nat Plants. 2015;1:15051. doi: 10.1038/nplants.2015.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendes R, Garbeva P, Raaijmakers JM. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. 2013;37:634–63. doi: 10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
- 9.Ritpitakphong U, Falquet L, Vimoltust A, Berger A, Métraux JP, L’Haridon F. The microbiome of the leaf surface of Arabidopsis protects against a fungal pathogen. New Phytol. 2016;210:1033–43. doi: 10.1111/nph.13808. [DOI] [PubMed] [Google Scholar]
- 10.Santhanam R, Luu VT, Weinhold A, Goldberg J, Oh Y, Baldwin IT. Native root-associated bacteria rescue a plant from a sudden-wilt disease that emerged during continuous cropping. Proc Natl Acad Sci USA. 2015;112:5013–20. doi: 10.1073/pnas.1505765112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berg G, Grube M, Schloter M, Smalla K. Unraveling the plant microbiome: looking back and future perspectives. Front Microbiol. 2014;5:148. doi: 10.3389/fmicb.2014.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bodenhausen N, Bortfeld-Miller M, Ackermann M, Vorholt JA. A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PLoS Genet. 2014;10:e1004283. doi: 10.1371/journal.pgen.1004283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Innerebner G, Knief C, Vorholt JA. Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl Environ Microbiol. 2011;77:3202–10. doi: 10.1128/AEM.00133-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Short FL, Murdoch SL, Ryan RP. Polybacterial human disease: the ills of social networking. Trends Microbiol. 2014;22:508–16. doi: 10.1016/j.tim.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singer M. Pathogen–pathogen interaction. Virulence. 2010;1:10–18. doi: 10.4161/viru.1.1.9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamichhane JR, Venturi V. Synergisms between microbial pathogens in plant disease complexes: a growing trend. Front Plant Sci. 2015;6:385. doi: 10.3389/fpls.2015.00385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roux F, Bergelson J. The Genetics underlying natural variation in the biotic interactions of Arabidopsis thaliana: the challenges of linking evolutionary genetics and community ecology. Curr Top Dev Biol. 2016;119:111–56. doi: 10.1016/bs.ctdb.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Bartoli C, Roux F, Lamichhane JR. Molecular mechanisms underlying the emergence of bacterial pathogens: an ecological perspective. Mol Plant Pathol. 2016;17:303–10. doi: 10.1111/mpp.12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartoli C, Roux F. Genome-wide association studies in plant pathosystems: toward an ecological genomics approach. Front Plant Sci. 2017;8:763. doi: 10.3389/fpls.2017.00763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mallon CA, Le Roux X, van Doorn GS, Dini-Andreote F, Poly F, Salles JF. The impact of failure: unsuccessful bacterial invasions steer the soil microbial community away from the invader’s niche. ISME J. 2018. 10.1038/s41396-017-0003-y. [DOI] [PMC free article] [PubMed]
- 21.Fridley JD, Stachowicz JJ, Naeem S, Sax DF, Seabloom EW, Smith MD, et al. The invasion paradox: reconciling pattern and process. Ecology. 2007;88:3–17. doi: 10.1890/0012-9658(2007)88[3:TIPRPA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 22.Naeem S, Knops JMH, Tilman D, Howe KM, Kennedy T, Gale S. Plant diversity increases resistance to invasion in the absence of covarying extrinsic factors. Oikos. 2000;91:97–108. doi: 10.1034/j.1600-0706.2000.910108.x. [DOI] [Google Scholar]
- 23.Tomasetto F, Duncan RP, Hulme PE. Environmental gradients shift the direction of the relationship between native and alien plant species richness. Divers Distrib. 2013;19:49–59. doi: 10.1111/j.1472-4642.2012.00939.x. [DOI] [Google Scholar]
- 24.Mallon CA, van Elsas JD, Salles JF. Microbial invasions: the process, patterns, and mechanisms. Trends Microbiol. 2015;23:719–29. doi: 10.1016/j.tim.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 25.Eisenhauer N, Schulz W, Scheu S, Jousset A. Niche dimensionality links biodiversity and invisibility of microbial communities. Funct Ecol. 2013;27:282–8. doi: 10.1111/j.1365-2435.2012.02060.x. [DOI] [Google Scholar]
- 26.Agler MT, Ruhe J, Kroll S, Morhenn C, Kim ST, Weigel D, et al. Microbial hub taxa link host and abiotic factors to plant microbiome wariation. PLoS Biol. 2016;14:e1002352. doi: 10.1371/journal.pbio.1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wagner MR, Lundberg DS, del Rio TG, Tringe SG, Dangl JL, Mitchell-Olds T. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat Commun. 2016;7:12151. doi: 10.1038/ncomms12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaparro JM, Badri DV, Vivanco JM. Rhizosphere microbiome assemblage is affected by development. ISME J. 2014;8:790–803. doi: 10.1038/ismej.2013.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Copeland JK, Yuan L, Layeghifard M, Wang PW, Guttman DS. Seasonal community succession of the phyllosphere microbiome. Mol Plant Microbe Interact. 2015;28:274–85. doi: 10.1094/MPMI-10-14-0331-FI. [DOI] [PubMed] [Google Scholar]
- 30.Schlaeppi K, Dombrowski N, Oter RG, Ver Loren van Themaat E, Schulze-Lefert P. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc Natl Acad Sci USA. 2014;111:585–92. doi: 10.1073/pnas.1321597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bodenhausen N, Horton MW, Bergelson J. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS ONE. 2013;8:e56329. doi: 10.1371/journal.pone.0056329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coleman-Derr D, Desgarennes D, Fonseca-Garcia C, Gross S, Clingenpeel S, Woyke T, et al. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol. 2016;209:798–811. doi: 10.1111/nph.13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barret M, Briand M, Bonneau S, Préveaux A, Valière S, Bouchez O, et al. Emergence shapes the structure of the seed microbiota. Appl Environ Microbiol. 2015;81:1257–66. doi: 10.1128/AEM.03722-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rezki S, Campion C, Iacomi-Vasilescu B, Preveaux A, Toualbia Y, Bonneau S, et al. Differences in stability of seed-associated microbial assemblages in response to invasion by phytopathogenic microorganims. Peer J. 2016;4:e1923. doi: 10.7717/peerj.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–41. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahé F, Rognes T, Quince C, De Vargas C, Dunthorn M. Swarmv2: highly-scalable and high-resolution amplicon clustering. Peer J. 2015;3:e1420. doi: 10.7717/peerj.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bull CT, De Boer SH, Denny TP, Firrao G, Fischer-Le S, et al. Comprehensive list of names of plant pathogenic bacteria, 1980–2007. J Plant Pathol. 2010;92:551–92. [Google Scholar]
- 39.Bull CT, Coutinho TA, Denny TP, Firrao G, Fischer-Le Sau M, Li X, et al. List of new names of plant pathogenic bacteria (2011–2) J Plant Pathol. 2014;96:223–6. [Google Scholar]
- 40.Jakob K, Goss EM, Araki H, Van T, Kreitman M, Bergelson J. Pseudomonas viridiflava and P. syringae—natural pathogens of Arabidopsis thaliana. Mol Plant Microb Interact. 2002;15:1195–203. doi: 10.1094/MPMI.2002.15.12.1195. [DOI] [PubMed] [Google Scholar]
- 41.Kniskern JM, Traw MB, Bergelson J. Salicylic acid and jasmonic acid signalling defense pathways reduce natural bacterial diversity on Arabidopsis thaliana. Mol Plant Microb Inter. 2007;12:1512–22. doi: 10.1094/MPMI-20-12-1512. [DOI] [PubMed] [Google Scholar]
- 42.Charron CS, Sams CE. Impact of glucosinolate content in broccoli (Brassica oleracea (italic group)) on growth of Pseudomonas marginalis, a causal agent of bacterial soft rot. Plant Dis. 2002;86:629–32. doi: 10.1094/PDIS.2002.86.6.629. [DOI] [PubMed] [Google Scholar]
- 43.Lerat S, Simao-Beaunoir AM, Beaulieu C. Genetic and physiological determinants of Stremptomyces scabei pathogenicity. Mol Plant Pathol. 2009;10:579–55. doi: 10.1111/j.1364-3703.2009.00561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gardan L, Stead DE, Dauga C, Gillis M. Acidovorax valerianellae sp. nov., a novel pathogen of lamb’s lettuce [Valerianella locusta (L.) Latter] Int J Syst Evol Microbiol. 2003;53:795–800. doi: 10.1099/ijs.0.02404-0. [DOI] [PubMed] [Google Scholar]
- 45.Pham VH, Park SJ, Roh Y, Roh DH, Rhee SK. Diaphorobacter oryzae sp. nov., isolated from a thiosulfate-oxidizing enrichment culture. Int J Syst Evol Microbiol. 2009;59:218–21. doi: 10.1099/ijs.0.001669-0. [DOI] [PubMed] [Google Scholar]
- 46.Lincoln SP, Fermor TR, Tindall BJ. Janthinobacterium agaracidamnosum sp. nov., a soft rot pathogen of Agaricus bisporus. Int J Syst Bacteriol. 1999;4:1577–89. doi: 10.1099/00207713-49-4-1577. [DOI] [PubMed] [Google Scholar]
- 47.Legendre P, Gallagher ED. Ecologically meaningful transformations for ordination of species data. Oecologia. 2001;129:271–80. doi: 10.1007/s004420100716. [DOI] [PubMed] [Google Scholar]
- 48.Jari Oksanen F, Blanchet G, Kindt R, Legendre P, O’Hara RG, Simpson G, et al. Vegan: community ecology package. R package version 1.16-0; 2009. citeulike-article-id:6657659.
- 49.Ramette A. Multivariate analyses in microbial ecology. FEMS Microbiol Ecol. 2007;62:142–60. doi: 10.1111/j.1574-6941.2007.00375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–90. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 51.Poudel R, Jumpponen A, Schlatter DC, Paulitz TC, Gardener BB, Kinkel LL, et al. Microbiome networks: a systems framework for identifying candidate microbial assemblages for disease management. Phytopathology. 2016;106:1083–96. doi: 10.1094/PHYTO-02-16-0058-FI. [DOI] [PubMed] [Google Scholar]
- 52.Carrascal LM, Galván I, Gordo O. Partial least squares regression as an alternative to current regression methods used in ecology. Oikos. 2009;118:681–90. doi: 10.1111/j.1600-0706.2008.16881.x. [DOI] [Google Scholar]
- 53.Lê Cao KA, Rossouw D, Robert-Granié C, Besse P. Sparse PLS: variable selection when integrating omics data. Stat Appl Gen Mol Biol. 2008;7:35. doi: 10.2202/1544-6115.1390. [DOI] [PubMed] [Google Scholar]
- 54.Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 2013;64:807–38. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 55.Horton MW, Bodenhausen N, Beilsmith K, Meng D, Muegge BD, Subramanian S, et al. Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nat Commun. 2014;5:5320. doi: 10.1038/ncomms6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huard-Chauveau C, Perchepied L, Debieu M, Rivas S, Kroj T, Kars I, Bergelson J, Roux F, Roby D. An atypical kinase under balancing selection confers broad-spectrum disease resistance in Arabidopsis. PLoS Genet. 2013;9:e1003766. doi: 10.1371/journal.pgen.1003766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, et al. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol. 2012;8:e1002606. doi: 10.1371/journal.pcbi.1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herrman C, Gern L, Voordouw MJ. Species co-occurrence patterns among lyme borreliosis pathogens in the tick vector Ixodes ricinus. Appl Environ Microbiol. 2013;79:7273–80. doi: 10.1128/AEM.02158-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Williams PH. Black rot: a continuing. Plant Dis. 1980;64:736–42. doi: 10.1094/PD-64-736. [DOI] [Google Scholar]
- 61.Classen AT, Sundqvist MK, Henning JA, Newman GS, Moore JAM, Cregger MA, et al. Direct and indirect effects of climate change on soil microbial and soil microbial-plant interactions: what lies ahead? Ecosphere. 2015;6:130. doi: 10.1890/ES15-00217.1. [DOI] [Google Scholar]
- 62.Aleklett K, Leff JW, Fierer N, Hart M. Wild plant species growing closely connected in a subalpine meadow host distinct root-associated bacterial communities. Peer J. 2015;3:e804. doi: 10.7717/peerj.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Geremia RA, Pușcaș M, Zinger L, Bonneville JM, Choler P. Contrasting microbial biogeographical patterns between anthropogenic subalpine grasslands and natural alpine grasslands. New Phytol. 2016;209:1196–207. doi: 10.1111/nph.13690. [DOI] [PubMed] [Google Scholar]
- 64.Mansfield J, Genin S, Magori S, Citovsky V, Sriariyanum M, Ronald, et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol Plant Pathol. 2012;13:614–29. doi: 10.1111/j.1364-3703.2012.00804.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bai Y, Müller DB, Srinivas G, Garrido-Oter R, Potthoff E, Rott M, et al. Functional overlap of the Arabidopsis leaf and root microbiota. Nature. 2015;528:364–9. doi: 10.1038/nature16192. [DOI] [PubMed] [Google Scholar]
- 66.Levine JM, D’Antonio CM. Elton revisited: a review of evidence linking diversity and invasibility. Oikos. 1999;87:15–26. doi: 10.2307/3546992. [DOI] [Google Scholar]
- 67.Mallon CA, Poly F, Le Roux X, Marring I, van Elsas JD, Salles JF. Resource pulses can alleviate the biodiversity–invasion relationship in soil microbial communities. Ecology. 2015;96:915–26. doi: 10.1890/14-1001.1. [DOI] [PubMed] [Google Scholar]
- 68.Kinnunen M, Dechesne A, Proctor C, Hammes F, Johnson D, Quintela-Baluja M, et al. A conceptual framework for invasion in microbial communities. ISME J. 2016;10:2773–5. doi: 10.1038/ismej.2016.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hacquard S, Spaepen S, Garrido-Oter R, Schulze-Lefert P. Interplay between innate immunity and the plant microbiota. Annu Rev Phytopathol. 2017;55:565–89. doi: 10.1146/annurev-phyto-080516-035623. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw FastQ reads were deposited in the Sequence Read Archive (SRA) of NCBI https://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP096011 under the study number SRP096011. The filtered matrix for both microbiota and potential pathobiota are available in Supplementary Data 4 and Supplementary Data 5, respectively. Raw values for both α-diversity and β-diversity used for statistical analysis are available in Supplementary Data 6.