Abstract
Key contributions to protein structure and stability are provided by weakly polar interactions, which arise from asymmetric electronic distributions within amino acids and peptide bonds. Of particular interest are aromatic side chains whose directional π-systems commonly stabilize protein interiors and interfaces. Here, we consider aromatic–aromatic interactions within a model protein assembly: the dimer interface of insulin. Semi-classical simulations of aromatic–aromatic interactions at this interface suggested that substitution of residue TyrB26 by Trp would preserve native structure while enhancing dimerization (and hence hexamer stability). The crystal structure of a [TrpB26]insulin analog (determined as a T3Rf3 zinc hexamer at a resolution of 2.25 Å) was observed to be essentially identical to that of WT insulin. Remarkably and yet in general accordance with theoretical expectations, spectroscopic studies demonstrated a 150-fold increase in the in vitro lifetime of the variant hexamer, a critical pharmacokinetic parameter influencing design of long-acting formulations. Functional studies in diabetic rats indeed revealed prolonged action following subcutaneous injection. The potency of the TrpB26-modified analog was equal to or greater than an unmodified control. Thus, exploiting a general quantum-chemical feature of protein structure and stability, our results exemplify a mechanism-based approach to the optimization of a therapeutic protein assembly.
Keywords: molecular pharmacology, protein design, insulin, protein self-assembly, molecular dynamics
Introduction
Weakly polar interactions are ubiquitous among protein structures (1). Among such interactions, the relative packing of aromatic rings is of particular interest in relation to the organization of protein cores and subunit interfaces (2). Aromatic–aromatic interactions are governed by quantum-chemical properties, which underlie dispersion forces and give rise to asymmetric distribution of partial charges. Whereas aromatic stacking is prominent in nucleic acid structures, pairs of aromatic side chains in proteins more often exhibit edge-to-face (ETF)4 contacts (3). Can such contacts be exploited in therapeutic protein engineering? Here, we have analyzed aromatic–aromatic interactions in the insulin hexamer (4) as a basis for designing improved long-acting (basal) analogs. This class of analogs is central to the treatment of Type 1 and Type 2 diabetes mellitus (5).
Classical crystal structures of insulin hexamers (6, 7) immediately suggested a pathway of assembly (4). Pertinent to the mechanism of storage in the secretory granules of pancreatic β-cells (8), such assembly is also of pharmacologic importance (9). Insulin assembly both protects the hormone from degradation in pharmaceutical formulations and modulates its pharmacokinetic properties (10). Indeed, the first use of insulin analogs in diabetes therapy reflected efforts to destabilize the insulin hexamer and thereby accelerate absorption of monomers and dimers from the subcutaneous (SQ) depot (summarized in Fig. S1 (11, 12)). Because it is more straightforward to introduce unfavorable substitutions than favorable ones, such engineering (corresponding to rapid-acting analogs) was more successful than complementary efforts to enhance the thermodynamic (and kinetic) stability of the insulin hexamer (13). There are presently three rapid-acting insulin analogs in clinical use (14), but no basal products designed on the basis of enhanced hexamer assembly despite extensive efforts (15). These difficulties were circumvented by alternative mechanisms of protracted action (acylation and pH-dependent SQ precipitation (14); Fig. S2).
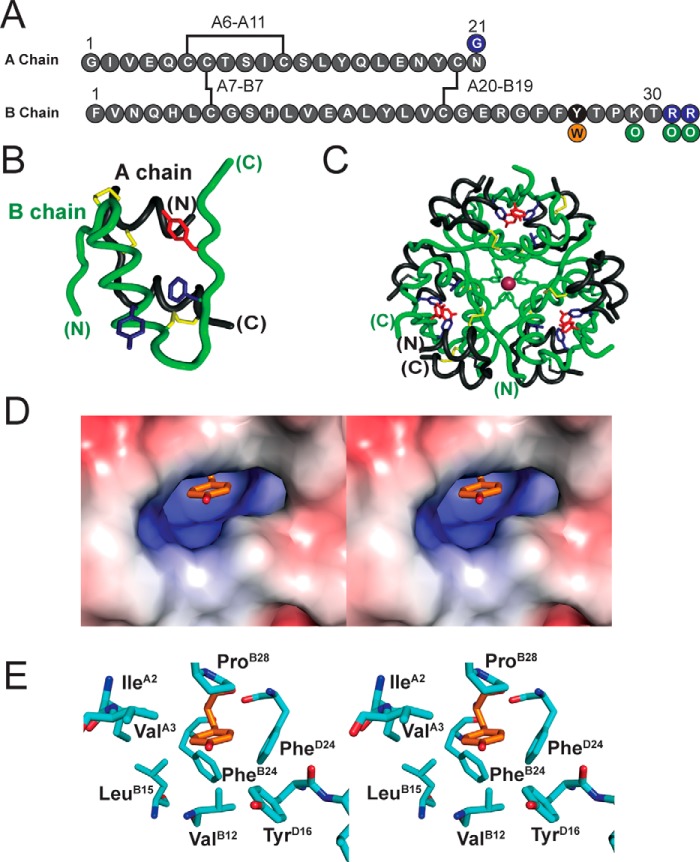
The dimer interface of insulin (repeated three times in the hexamer) contains a cluster of eight conserved aromatic rings (TyrB16, PheB24, PheB25, TyrB26, and their dimer-related mates in subunit D; Fig. 1, A–C). Of these, successive ETF contacts are formed by B16–D26, B24–B26, B24–D24, and B26–D16; the B25 side chain is peripheral to this network. Whereas variation at these sites is in general constrained by the structure of the hormone–receptor interface (16, 17), our attention focused on the B26 side chain because of its functional tolerance to diverse substitutions (18) and because of its partial exposure in the monomer, dimer, and hexamer (4) (Fig. 1, C and D). We sought to investigate whether variant B16–D26 and B26–D16 ETF contacts across the dimer interface might in principle modulate, in either direction, the strength of these interactions. We hypothesized that enhanced ETF contacts at this interface might provide the long-sought approach to stabilize the insulin hexamer and so improve the pharmacokinetc properties of basal formulations.
Figure 1.
Structural overview of insulin. A, sequence of insulin with disulfide bridges in black. TyrB26 is highlighted in black; the present TyrB26 → Trp substitution is orange; and modifications in pI-shifted clinical analog glargine in purple. Our semisynthetic pI-shifted analog contained Orn (green) instead of Lys or Arg. B, structure of an insulin monomer; the A chain is shown in black and B chain in green. TyrB26 is red, whereas PheB24 and TyrB16 are blue (PDB code 4INS). C, structure of zinc-coordinated insulin hexamer (T6 state), a trimer of dimers; TyrB26, PheB24, and TyrB16 are color-coded as in B. D, stereo view showing TyrB26 (sticks) in a cavity within insulin dimer (extracted from T3Rf3 hexamer, PDB code 1TRZ). E, corresponding stick model with residues labeled.
The present study had three parts. The first employed local modeling, using the standard CHARMM empirical energy function, to probe possible effects of a TyrB26 → Trp substitution on aromatic–aromatic interactions within the wildtype (WT) aromatic cluster. These molecular mechanics (MM) calculations suggested that substitution of TyrB26 by Trp could enhance dimer-related ETF contacts and yet otherwise preserve a native-like interface. We next prepared this analog to examine whether this substitution might indeed stabilize the insulin hexamer and retard its disassembly while preserving the biological activity of the monomeric hormone. The crystal structure of a [TrpB26]insulin analog (as a zinc–insulin hexamer) was essentially identical to that of WT insulin. Finally, we undertook studies in diabetic rats to obtain proof of principle that this approach could extend the duration of insulin action on SQ injection.
To our knowledge, our results represent the first exploitation of aromatic–aromatic interactions to enhance the physical and biological properties of a therapeutic protein. Because standard MM calculations employ a simplified model of aromatic systems (i.e. approximating their quantum-mechanical (QM) properties via partial atomic charges (19, 20)), ab initio QM simulations of the aromatic cluster and their incorporation in QM/MM simulations (21) promise to establish a rigorous foundation for therapeutic protein design, including further optimization through incorporation of modified or nonstandard amino acids (22–24). The present results suggest that insulin's conserved aromatic cluster can provide a natural laboratory for such foundational analysis and its therapeutic translation.
Results
Molecular mechanics calculations suggested that augmented aromatic–aromatic interactions are possible at the TrpB26 dimer interface
MM simulations were employed to estimate the strength of aromatic–aromatic interactions of TrpB26 at the insulin dimer interface in relationship to those of the native Tyr. These calculations employed the CHARMM empirical energy function in which aromatic rings contain partial atomic charges, parametrized to mimic the electrostatic properties of the π-system (19, 20). Working models of the variant dimer were obtained by local energy minimization (see “Experimental procedures”).
Energies of interaction between the eight aromatic residues at the insulin–dimer interface (TyrB16, PheB24, PheB25, TyrB26, and their symmetry-related mates) were calculated using local models in which TrpB26 was substituted within WT T2, R2, and TRf reference dimers (extracted from PDB structures 4INS, 1ZNJ, and 1TRZ) (25). The total interaction energy between B26 and the other aromatic residues at the TrpB26 interface, which was calculated using the full CHARMM potential energy function, was augmented by 2.0 and 0.8 kcal/mol relative to the minimized WT interface in the context of R2 and TRf structures, respectively, and diminished by 1.5 kcal/mol in the context of the T2 structure. Results are summarized in Table S1.
A minimal model was utilized to further evaluate the potential impact of a TrpB26 substitution on aromatic–aromatic interactions at the dimer interface. To this end, a structural model of the dimer interface (extracted from a T6 insulin hexamer; PDB code 4INS) was first built containing residue B26 (TyrB26 or TrpB26) and its nearest aromatic neighbors (PheB24, PheD24, or TyrD16). Simulations predicted the orientation of the B26 ring corresponding to free-energy minima of electrostatic interactions between the aromatic residues (the partial-charge parametrization of aromatic residues in CHARMM is shown in Fig. S3) (23). When substituted at position B26, Trp displayed improved electrostatic interactions with its three aromatic neighbors (relative to the WT Tyr) over a broad range of conformations (Fig. 2). This trend extended to conformations that are sterically permitted in the context of the WT insulin hexamer.
Figure 2.
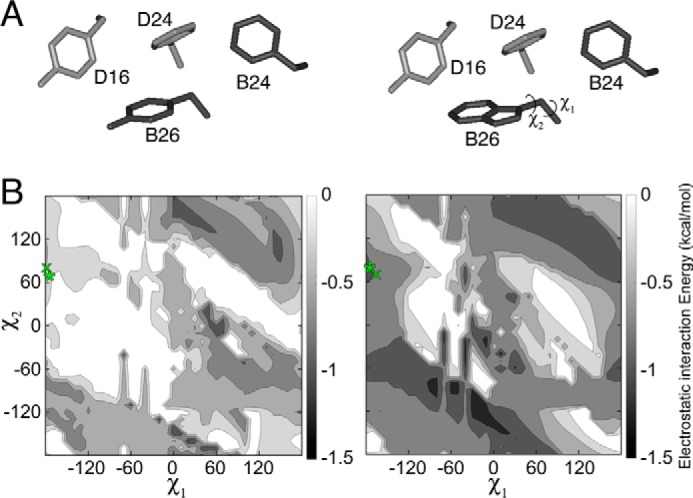
Molecular simulations of aromatic interactions in the insulin dimer. A, aromatic–aromatic interactions across insulin's dimer interface involve PheB24, TyrB16, PheD24 (sticks) and either TyrD26 (left) or TrpD26 (right). Residues were extracted from T6 structure 4INS. B, contour maps depicting empirical interaction energies between B26 (Tyr on left and Trp on right) at varying χ1 and χ2 angles and the other three residues shown in A. The orientation of TyrB26 or TrpB26 in the WT or variant crystal structure is indicated by a green “x”; orientation of TyrB26 or TrpB26 in the local model is indicated by a green asterisk.
TrpB26 analog exhibited markedly decreased hexamer dissociation rate
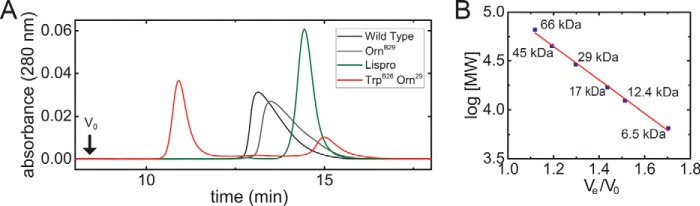
The effect of the TrpB26 substitution on the lifetime of insulin hexamers under formulation conditions was assessed in the context of [OrnB29]insulin, a structural equivalent of WT insulin that is amenable to production by trypsin-catalyzed semisynthesis (26). The lifetime of Co2+-substituted, phenol-stabilized (R6) hexamers of [TrpB26,OrnB29]insulin was assessed at equilibrium in relationship to native [TyrB26,OrnB29]insulin. Optical absorbance spectra of these analogs (characteristic of Co2+ with tetrahedral coordination) were similar to WT (Fig. 3, A and B). Assessment of R6 dissociation rates (summarized in Table 1) revealed a 150-fold increase in hexamer half-life of the TrpB26 analog relative to its parent [OrnB29]insulin (Fig. 3, C and D). This increase is remarkable given that the difference between the half-lives of a rapidly dissociating analog in clinical use (lispro)5 (27–29) and WT is <2-fold (Table 1).
Figure 3.
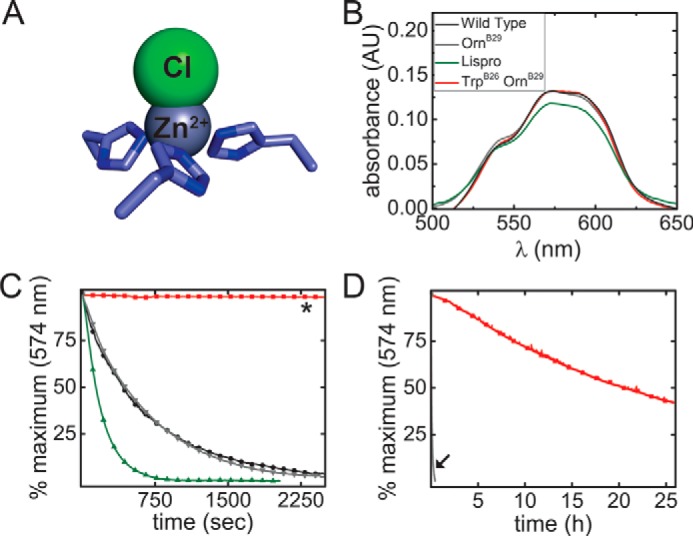
Hexamer dissociation of TrpB26 analog. A, tetrahedral Zn2+-coordination site in R6 insulin hexamer (ball-and-stick model): three HisB10 side chains and one chloride ion. B, absorbance spectra of d–d bands in corresponding Co2+ complex; the color code is indicated. C, hexamer dissociation curves as monitored at 574 nm after addition of excess EDTA; the color code is as in B. The lifetime of the [TrpB26,OrnB29]insulin hexamer was markedly prolonged (asterisk). D, dissociation of TrpB26,OrnB29 hexamer from 0 to 8000 s in relationship to that of parent [OrnB29]insulin (black arrow). Half-lives are given in Table 1.
Table 1.
Self-association properties of insulin analogs
| Analog | t½ hexamer dissociation | Calculated molecular mass by SECa |
|---|---|---|
| min ± S.D. | kDa | |
| Wildtype | 7.7 (± 1.3) | 9.7 |
| Lisprob | 4.6 (± 0.3) | 5.1 |
| OrnB29c | 8.2 (± 0.8) | 8.2 |
| TrpB26, OrnB29 | 1.2 (± 0.3) × 103 | 28.0, 4.0 |
a Proteins were made 0.6 mm in a buffer containing ZnCl2 at a ratio of 2 zinc ions per insulin hexamer and applied to SEC column as described under “Experimental procedures.” Masses were calculated from the plot in Fig. 4B.
b “Lispro” describes insulin analogs containing ProB28 →Lys and LysB29 →Pro substitutions. These substitutions impair dimerization (28, 29).
c Use of Orn simplified trypsin-catalyzed semisynthesis (33).
TrpB26 imposed kinetic barriers to the dissociation of hexamers into monomers
R6 dissociation kinetics were further examined by size exclusion chromatography (SEC). The TrpB26 analog (formulated in the presence of phenol and zinc ions) was injected onto an SEC column using a zinc- and phenol-free mobile phase. Subsequent dissociation of the R6 hexamers was monitored in the chromatograms (Fig. 4A, Table 1). Absence of a void volume signal (V0) indicated that none of the proteins formed large nonspecific aggregates. Whereas WT and [OrnB29]insulin eluted as a broad peak representing an association state intermediate between monomer and dimer (9.7 and 8.2 kDa, respectively) and insulin lispro eluted essentially as a monomer (5.1 kDa), [TrpB26,OrnB29]insulin eluted in two distinct peaks. The larger peak corresponded to a trimeric or tetrameric association state (molecular mass 28 kDa), and the smaller corresponded to a monomeric state (4 kDa; Fig. 4, A and B; see Fig. S4 for reference chromatograms of the monomer and hexamer). These findings suggest that oligomers intermediate between hexamer and dimer may delay dissociation of the TrpB26 analog; in particular, the absence of broad elution tails implies that TrpB26 imposes significant barriers to rapid dissociation.
Figure 4.
Size exclusion chromatography of TrpB26 hexamer. A, SEC chromatogram of insulin analogs in the presence of zinc and phenol. The void volume (V0, black arrow) was defined by thyroglobulin (molecular mass 669 kDa). B, plot of log(molecular weight) versus elution ratio (Ve/V0) of molecular weight standards. Linear relationship between log[MW] to elution ratio (Ve/V0) is indicated by the red line with coefficient of determination (R2) 0.996 and parameters log[molecular weight] = −1.71 × (Ve/V0) + 6.7012. Elution times of molecular weight standards are indicated by blue squares (labeled by molecular weight). Identity of molecular mass standards is as follows: 66 kDa, BSA; 45 kDa ovalbumin; 20 kDa, carbonic anhydrase, 17 kDa, myosin light chain; 12.4; cytochrome c, 6.5 IGF-I. Calculated Mr are given in Table 1.
TrpB26 analog retained native biological activity
The in vitro affinity of the TrpB26 analog for the lectin-purified insulin receptor (IR; isoform B holoreceptor) was determined to be 0.14(±0.02) nm, ∼50% that of [OrnB29]insulin (0.07(±0.02) nm) and WT insulin (0.08(±0.03) nm) (Fig. 5A). The potencies of these analogs, evaluated by intravenous (i.v.) injection in diabetic rats, were nonetheless, indistinguishable from WT insulin (Fig. 5B).
Figure 5.
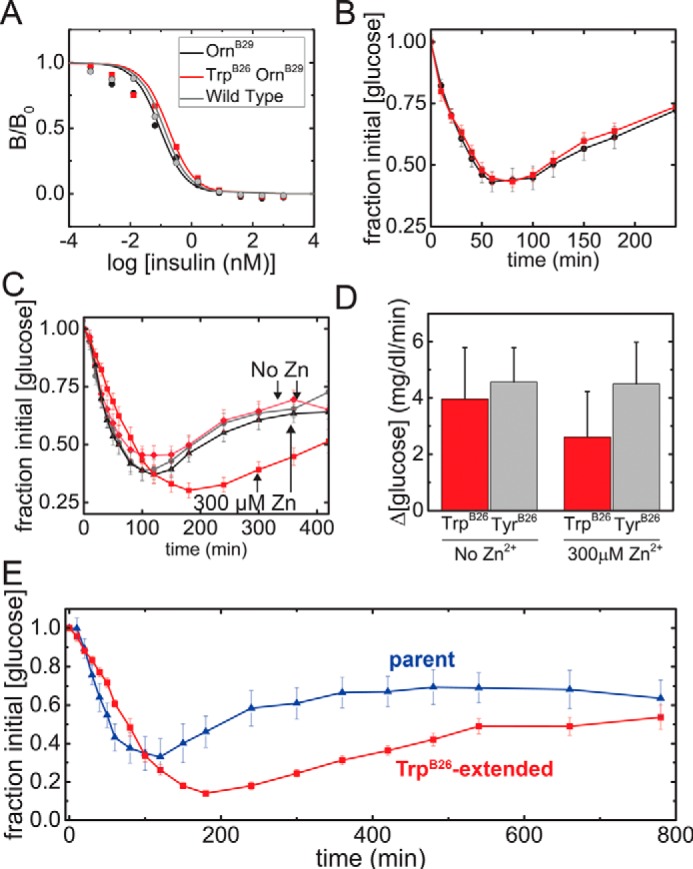
Pharmacology of TrpB26 analog. A, receptor-binding affinities (isoform B). The affinity of [TrpB26,OrnB29]insulin was reduced by 2-fold relative to [OrnB29]insulin (respective Kd estimates 0.14 (±0.03) and 0.07 (±0.02) nm). Color code is indicated in the inset. B, time course of [blood glucose] following i.v. injection in rats (n = 15); color code as in A. C, time course of [blood glucose] following SQ injection in the absence or presence of 0.3 mm ZnCl2 (n = 18). D, histogram summarizing the rate of fall of [blood glucose] over the first 30 min in C, black bars indicate S.D. E, time course of [blood glucose] following SQ injection of pI-shifted analogs: [GlyA21, OrnB29,OrnB31,Orn32]insulin, and its TrpB26 derivative (n = 6; color code in panel).
TrpB26 analog displayed a zinc-dependent delay in onset of biological activity
Hexamer assembly delays absorption of insulin from its SQ-injection site (11, 12). To assess the onset and duration of [TrpB26,OrnB29]insulin relative to [OrnB29]insulin, the pharmacodynamics (PD) profile of these proteins (made 0.15 mg/ml, corresponding to a monomer concentration of 27 μm and a putative hexamer concentration of 4.5 μm) were evaluated as zinc-free solutions or as pre-assembled phenol-stabilized R6 hexamers in the presence of excess zinc ions (0.30 mm ZnCl2; 70 zinc ions per hexamer). A zinc-dependent delay in onset of activity was observed on SQ injection of [TrpB26,OrnB29]insulin but not on injection of [OrnB29]insulin or WT insulin (Fig. 5C; see Fig. S5 for WT results). Whereas the latter PD profiles exhibited a nadir at about 120 min irrespective of the zinc ion concentration, the PD profile of [TrpB26,OrnB29]insulin occurred at (i) 120 min in the absence of zinc ions and (ii) 150 min in the presence of 0.30 mm zinc ions with corresponding delays in the rate of fall over the first 30 min (Fig. 5D). Together with the above in vitro results, these findings suggest that the prolonged lifetime of the TrpB26 R6 hexamer (as inferred from the above kinetic studies of the Co2+-substituted hexamer) are responsible for the inferred zinc-dependent delay in SQ absorption.
TrpB26 protracted the PD profile of a model pI-shifted analog
Insulin analogs with isoelectric points (pI) shifted to neutral pH generally exhibit prolonged activity due to precipitation in the SQ depot (30). To determine whether TrpB26 might further prolong the activity of such analogs, this substitution was introduced into a [GlyA21,OrnB29,OrnB31,OrnB32]insulin. This “glargine-like” framework was designed to recapitulate the pI shift of glargine with greater ease of semisynthesis.6 The proteins (formulated at 0.6 mm with 0.3 mm ZnCl2, corresponding to 3 zinc ions per hexamer) were each injected SQ in diabetic rats.
The pI-shifted parent analog displayed peak activity at about 120 min with blood glucose levels returning to baseline after about 360 min. By contrast, its TrpB26 derivative displayed a prolonged PD profile: peak activity occurred 180 min with slow return to baseline at >800 min (Fig. 5E; see Fig. S6 for i.v. potency). Such a marked delay in peak activity was not observed in the parent glargine-like analog or the TrpB26 derivative when administered in the absence of zinc (Fig. S7). These results suggest that TrpB26 may favorably be incorporated into current basal analogs as a complementary mechanism of prolonged SQ absorption.
Crystal structure of TrpB26 analog demonstrated native-like dimer interface
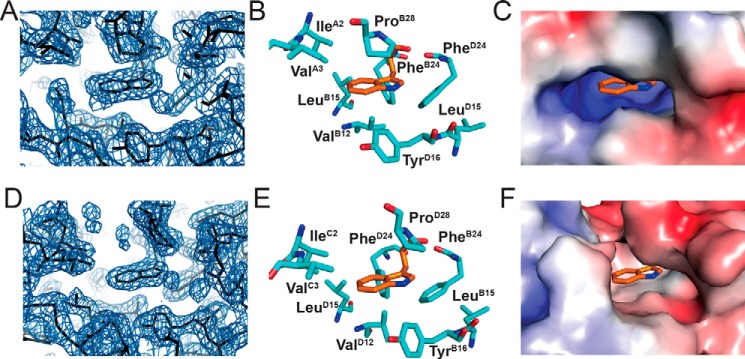
The crystal structure of [TrpB26,OrnB29]insulin was determined as a zinc-coordinated hexamer in the presence of phenol to a resolution of 2.25 Å. Diffraction and refinement statistics are provided in Table S2. The asymmetric unit constituted a “TRf” dimer7 (31, 32). The overall structures of the T and Rf protomers were essentially identical to those of WT insulin (Fig. S8) with respective r.m.s. deviations of 1.11 ± 0.30 and 1.36 ± 0.30. Additional r.m.s. deviations are given in Tables S3 and S4. Side chain packing near the B26 position was largely unperturbed. In both protomers the TrpB26 indole group was oriented with its six-member ring packing against conserved core residues IleA2, ValA3, and ValB12 (Fig. 6); the indole NH group is exposed to solvent in the TR dimer and T3Rf3 hexamer. The TrpB26 side chain in both R and T protomers also displayed dihedral angles within the range of the native Tyr in WT crystal structures (Tables S5 and S6, respectively) with a slight deviation in the positioning of the peptide backbone to accommodate the larger indole side chain.
Figure 6.
Crystal structure of [TrpB26,OrnB29]insulin. A, electron density of TrpB26 in T-state protomer showing the surrounding density in TRf asymmetric unit (contour level 2.0 Å). B, stick model corresponding to map in A; TrpB26 is orange. C, surfaces of residues surrounding TrpB26 (sticks) as indicated above. D–F, corresponding map and models of TrpB26 in the R state protomer.
Spectroscopic probes revealed native-like structure and thermodynamic stability of TrpB26 analogs in solution
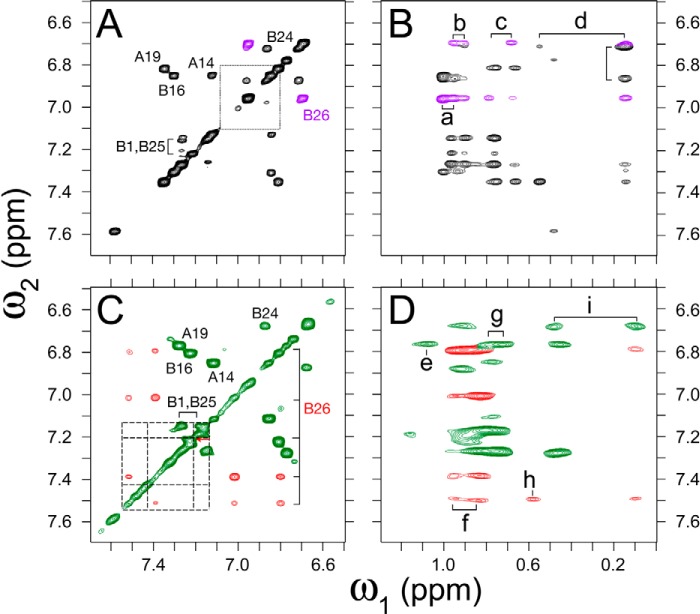
The native-like crystal structure of [TrpB26,OrnB29]insulin is in accordance with its unperturbed circular dichroism (CD) spectrum and thermodynamic stability under monomeric conditions (Fig. 7A). Free energies of unfolding (ΔGu 3.3 ± 0.1) kcal/mol at 25 °C as inferred from two-state modeling of chemical denaturation (33)) were indistinguishable due to small and compensating changes in transition midpoint and slope (m value) (33, 34) (Fig. 7B, Table 2). Further evidence that the crystal structure extends to the monomer in solution was provided by 2D 1H NMR studies of TrpB26 substituted within an engineered insulin monomer (lispro (35)). Whereas the spectrum of lispro (at pD 7.6 and 37 °C) exhibits sharp resonances for each aromatic spin system (Fig. 8A), as expected for a monomeric analog (35), the spectrum of its TrpB26 derivative exhibits broadening of resonances at the dimer interface (B16, B24–B26). The latter spin systems can be observed on TOCSY spectrum (Fig. 8C) but not in the corresponding DQF-COSY spectrum due to antiphase cancellation. Like the aromatic ring TyrB26 in spectra of insulin lispro (Fig. 8, A and B), the indole ring exhibited regiospecific nonlocal nuclear Overhauser enhancements (NOEs) from its six-member moiety to the methyl resonances of ValB12 and IleA2 (Fig. 8, B–D).
Figure 7.
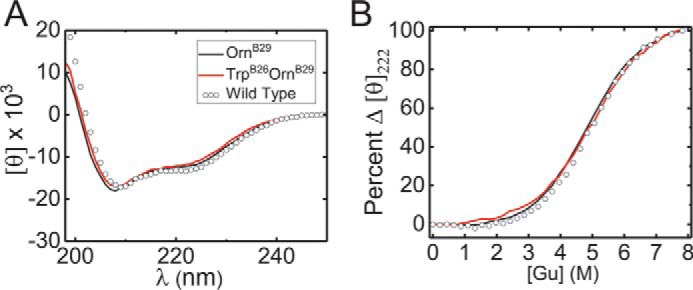
Thermodynamic stability of [TrpB26,OrnB29]insulin. A, CD spectra of [TrpB26,OrnB29]insulin, [OrnB29]insulin, and WT insulin. B, guanidine denaturation assays of insulin analogs monitored by ellipticity at 222 nm; color code as in A. Stabilities are given in Table 2.
Table 2.
Thermodynamic stabilities of insulin analogs
| Analog | ΔGua | Cmid | mb |
|---|---|---|---|
| kcal mol−1 | m | kcal mol−1 m−1 | |
| Wildtype | 3.4 ± 0.1c | 5.0 ± 0.1 | 0.68 ± 0.02 |
| OrnB29 | 3.3 ± 0.1 | 4.9 ± 0.1 | 0.67 ± 0.01 |
| TrpB26,OrnB29 | 3.3 ± 0.1 | 5.1 ± 0.2 | 0.64 ± 0.03 |
a Parameters were inferred from CD-detected guanidine denaturation data by application of a two-state model; uncertainties represent fitting errors for a given data set.
b The m-value (slope Δ(G)/Δ(M)) correlates with surface area exposed on denaturation.
c Analysis of replicates of [TrpB26,OrnB29]insulin, parent [OrnB29]insulin, and WT samples indicated that experimental standard errors were equal to or less than the above fitting errors: ±0.1 kcal mol−1 (ΔGu), ±0.1 m (Cmid), and ±0.01 kcal mol−1 m−1 (m).
Figure 8.
Homonuclear 2D NMR of a TrpB26 analog. 2D NMR studies of insulin analogs indicate similar TyrB26- and TrpB26 environments. A and B, spectra of parent monomer insulin lispro ([LysB28,ProB29]insulin): A, aromatic region of TOCSY spectrum with TyrB26 cross-peaks (magenta) shown relative to the Tyr spin system in free octapeptide GFFYTKPT (dotted lines); and B, region of NOESY spectrum showing contacts between aromatic protons (vertical axis, ω2) and methyl groups (horizontal axis, ω1). C and D, spectra of TrpB26 analog of insulin lispro: C, aromatic TOCSY spectrum highlighting TrpB26 cross-peaks (red) relative to the Trp spin system in free octapeptide GFFWTKPT (dashed lines) and D, region of NOESY spectrum corresponding to B. B26-related NOEs are shown in red. Cross-peak assignments: (a) γ-CH3 ValA3, (b) γ-CH3 ValB12, (c) γ-CH2, γ-CH3 IleA2, (d) δ-CH3 LeuB15, (e) γ-CH3 ValA3, (f) γ-CH3 ValB12, (g) γ-CH2, γ-CH3 IleA2, (h) δ-CH3 IleA2, and (i) δ-CH3 LeuB15. TOCSY mixing times in spectra A and C were 55 ms; NOESY mixing times in B and D were 150 ms.
The pattern of secondary shifts in the variant is similar to that in the parent monomer. In particular, the aromatic 1H NMR resonances of TrpB26 (red cross-peaks in Fig. 8C) exhibit upfield features (relative to Trp in the isolated B23–B30 octapeptide; dashed lines) similar to those of TyrB26 in the parent spectrum (purple cross-peaks in Fig. 8A versus dotted lines) (36). Dilution of the TrpB26 sample partially mitigated resonance broadening but preserved these trends in dispersion. Indole-specific NOEs indicated that the side chain assumes one predominant and asymmetric conformation within a native-like crevice between A- and B-chain α-helices (Table S7). Because TyrB26 undergoes rapid ring rotation about the Cβ–Cγ bond axis (“ring flips”), analogous side chain-specific NOEs (inferred in prior studies from molecular modeling) cannot be observed directly (Table S8).
MM calculations suggested improved aromatic–aromatic interactions within the variant crystal structure
The contribution of aromatic–aromatic interactions involving TrpB26 to the stability of the variant dimer interface of the T3Rf hexamer was evaluated through calculation of nonbonded interaction energies among aromatic residues B16, B24, B25, and B26 in the TRf dimer. These calculations, which employed the variant crystal structure, were in overall accordance with expectations based on our initial local MM-based modeling (above). In particular, based on aromatic–aromatic interactions alone, the TrpB26,OrnB29 dimer displayed an increase in interaction energy of 1.4 kcal/mol relative to WT TRf reference structure PDB 1TRZ; the results of these calculations are given in Table S9, a and b. Although the standard CHARMM empirical energy function, when applied to analyze either crystallographic and MM-minimized models of [TrpB26]insulin, suggested that the electrostatic properties of the Trp side chain were the primary contributors to the increased stability of the dimer, this physical interpretation may reflect the limitations of the partial-charge representation (23, 37). Indeed, preliminary ab initio QM simulations of a minimal model (consisting of two aromatic rings in vacuo) predicted that enhanced Van der Waals interactions may also make a significant contribution (Fig. S9) (see “Discussion”).
Discussion
The physical origins of protein stability and recognition define a foundational problem in biochemistry (38) with central application to molecular pharmacology (39). The zinc–insulin hexamer provides a favorable system for structure-based design due to its long history of crystallographic investigation (40). Indeed, the hexamer's rigidity, as interrogated by NMR spectroscopy (41), renders the overall structure robust to diverse amino acid substitutions (42, 43), even those that destabilize the dimer interface8 (32, 44). This rigid framework has often enabled analysis of discrete interactions without complications due to the long-range transmission of conformational change (31, 32).
Our studies, stimulated by the seminal recognition of aromatic–aromatic interactions by Burley and Petsko (2) more than 30 years ago, focused on the classical dimer interface, a basic building block of the hexamer (32). Long appreciated as “a thing of beauty” (4, 45), this interface contains eight aromatic residues, six of which engaged in a successive set of aromatic–aromatic interactions. Quantum-chemical simulations of model systems have suggested that nearest-neighbor interactions predominate even in the presence of multiple rings (46). Pairwise dissection of insulin's dimer interface has highlighted the potential opportunity to enhance its stability through substitution of TyrB26 by Trp. Whereas our crystallographic analysis verified that this substitution preserves native architecture, a [TrpB26]insulin analog exhibited a dramatic increase in hexamer lifetime in vitro. Results of animal testing demonstrated native intrinsic potency (i.e. on i.v. bolus injection) but with prolonged activity on SQ injection, presumably due to delayed dissociation of the variant zinc hexamers in the SQ depot.
Protein engineering of insulin analogs is constrained by the complexity of insulin's “conformational lifecycle”: from oxidative folding intermediates and self-assembly in the pancreatic β-cell (8) to adoption of an active, “open” conformation on receptor binding (Fig. 9, A and B) (16). Specific residues may play distinct roles at each stage. In particular, because interfaces within the insulin hexamer overlap the hormone's receptor-binding surface, essentially invariant among vertebrates (47), modifications often impair activity (13). A given WT residue may represent a compromise among competing structural tasks. A recent survey of 18 substitutions at position B26 demonstrated that Tyr is suboptimal with respect to IR-binding affinity but enhances self-assembly relative to more active alternatives (Ala, Ser, or Glu) (18). The latter side chains destabilize the “closed” dimer interface but are favorable at the solvated B26-related edge of the open hormone–receptor interface (Fig. 9C).
Figure 9.
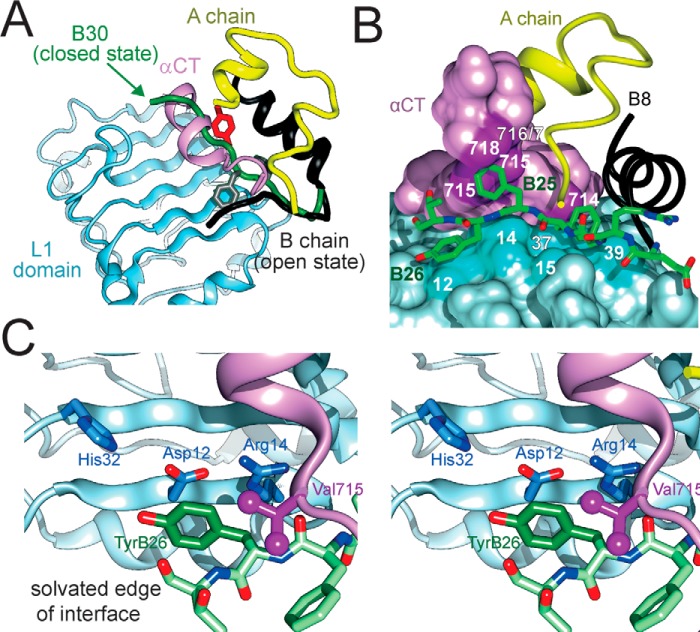
Binding surface of TyrB26 on an IR fragment. A, model of WT insulin (in classical T-state) overlaid on the structure of insulin bound to an IR fragment (PDB entry 4OGA). The L1 domain and part of the CR domain are shown in powder blue, whereas αCT is shown in purple. PheB24 and TyrB26 are, respectively, shown as gray and red sticks. The B-chain of IR-bound insulin is shown in dark gray (B6–B19) or black (B20–B27); the green tube indicates a classical location within overlay of residues B20–B30 (green arrow), thereby highlighting the steric clash of B26–B30 with αCT. Insertion of the insulin B20–B27 segment between L1 and αCT was associated with a small rotation of the B20–B23 β-turn and changes in main chain dihedral angles flanking B24. B, stick representation of B-chain residues B20–B27 packed between αCT and the L1 β2 strand. Color code in insulin segment: carbon atoms (green), nitrogen (blue), and oxygen (red). Residues B8–B19 are shown as a black ribbon, and the A-chain is shown as a yellow ribbon. Key contact surfaces of αCT with B24–B26 are highlighted in magenta and L1 with B24–B26 are highlighted in cyan. C, stereo view of the environment of TyrB26 within its binding site. Neighboring side chains in L1 and αCT are as labeled. This figure was adapted from Ref. 18 with permission of the authors.
Protective self-assembly is of key pharmacologic importance
Insulin self-assembly protects the hormone from degradation and toxic misfolding in pancreatic β-cells9 (8, 48) and in pharmaceutical formulations (49). Because the zinc-insulin hexamer exhibits delayed SQ absorption relative to monomers and dimers, mutational destabilization of the hexamer (10, 12) led to development of rapid-acting insulin analogs (14). Efforts to stabilize the insulin hexamer, as a converse strategy to obtain protracted action, were less successful (13). Current long-acting insulin analogs rely instead on higher-order self-association of hexamers within the SQ depot (30) and binding acylated monomer to albumin as a circulating depot (highlighted in Fig. S2) (50, 51).
The problem of how to improve a protein interface is in general more subtle than its opposite, for destabilizing substitutions abound at conserved interfaces, whereas stabilizing substitutions can be rare (13). Structure-based candidate substitutions may encounter entropy–enthalpy compensation (EEC) (52, 53) or cause unintended biological perturbations (54). The challenge posed by insulin is magnified by the structural elegance of its self-assembly (as emphasized by Hodgkin and colleagues (55) in a classic review). Diverse structure-based strategies were previously undertaken with only limited success. Alternate strategies previously used to create basal insulin analogs are depicted in Fig. S2 and summarized in Table S10. One approach focused on the general nonpolar character of the dimer interface: additional hydrophobic substitutions were introduced in an effort to enhance this feature (56). Such designs were not successful and also impaired biological activity, although analogs were identified whose sparing solubility slowed SQ absorption (56). A second approach exploited the classical TR transition among insulin hexamers (57). At the pivot point of this allosteric transition (Fig. S10), an invariant glycine (GlyB8) was substituted by Ser in an effort to stabilize the more stable R state hexamer (58). This analog was unstable as a monomer (54) and exhibited reduced activity (58). Yet another approach sought to stabilize the hexamer by relieving electrostatic repulsion created by the internal clustering of six acidic side chains (GluB13): their isosteric substitution by Gln indeed promoted assembly of zinc-free hexamers but impaired biological activity (59). Overlap between the self-assembly surfaces of insulin and its receptor-binding surfaces thus compounded the optimization problem.
The present study revisited the architecture of the insulin hexamer in light of recent insights into the hormone's receptor-binding surface (18). Prominent roles are played by a quartet of aromatic residues in the C-terminal B-chain β-strand: TyrB16, PheB24, PheB25, and TyrB26 (33). These four residues, and the clustering of eight dimer-related side chains, have long been the focus of structure-activity relationships (60). Photoactivatable aromatic probes (para-azido-Phe) at any of these sites exhibited efficient cross-linking to the IR (61, 62). The co-crystal structure of an insulin monomer bound to a fragment of the IR ectodomain (the μIR model) revealed distinct binding sites at the surface of the L1 domain of the IR β-subunit (B16, B24, and B26) or its αCT element (B24 and B25). PheB24 packs within a nonpolar pocket near aromatic residues in L1 (residues Leu37 and Phe39) and αCT (residue Phe714); one wall of this pocket is defined by the aliphatic side chains of LeuB15, CysA20, and CysB19 as in free insulin (16). This environment differs in detail from those in the insulin dimer but exhibits analogous general features. Although aromaticity is not required to fill the B24-binding pocket (33), its specific size and shape constrain potential substitutions. TyrB16 lies at the periphery of L1 (near Phe39 and Lys40). Its substitution by Ala or other aromatic residues preserves activity (63). PheB25 occupies a cleft between αCT residues (Val715, Pro716, Arg717, and Pro718) that can only accommodate trigonal γ-carbons (22). B25-related aromatic–aromatic interactions are limited due to the peripheral location of this residue (Fig. S11).
We chose to focus on position B26 because of its broad functional tolerance of diverse substitutions (18). As illustrated above (Fig. 8C), TyrB26 binds at the solvated periphery of the μIR interface. Indeed, substitution of small, polar, or charged amino acids (such as Ala, Ser, or Glu) enhances receptor affinity, but at the price of impaired self-assembly and decreased thermodynamic stability with heightened susceptibility to physical degradation (16). These findings highlighted the evolutionary importance of the native dimer interface and dual role of TyrB26. We thus hypothesized that an aromatic substitution at the B26 position might enhance self-assembly without loss of biological activity.
The classical structure of the insulin dimer motivated study of successive aromatic–aromatic interactions as a physical mechanism of stability (6, 64). The increased stability of ETF aromatic–aromatic interactions involving Trp over those involving Tyr contributes to the increased stability of the TrpB26 hexamer. The larger size of the delocalized π-orbital of Trp in relationship to that of Tyr causes a stronger negative charge to accumulate on the “face” of the indole ring. For this reason, hydrogen atoms surrounding the aromatic rings of local residues form stronger electrostatic interactions with the face of the indole ring of Trp than with the phenol ring of Tyr (65, 66). Aromatic pairs involving Trp residues are less common than those involving Tyr or Phe (1). However, the strength of aromatic–aromatic interactions involving Trp is evidenced by functional importance of Trp-based interactions; an example is provided by a Trp–Tyr aromatic “lock” that stabilizes the active conformation of the ghrelin receptor (67). Indeed, comparison of electrostatic interactions of TrpB26 and TyrB26 within the minimized model of the B16/B24/B26 aromatic network of insulin revealed that interactions involving Trp were favored over those involving Tyr across a broad variety of orientations of the respective aromatic rings. Extension of the aromatic lock metaphor (introduced by Holst et al. (67) to describe conformational “trapping” in the GPCR structure) to the insulin hexamer highlights the kinetic effect of the B26 substitution on the rate of hexamer disassembly (as probed by the Co2+-EDTA sequestration assay), which was more dramatic than effects on equilibrium association (as probed by SEC). It would be of future interest to measure activation energies for disassembly. Insight into the structural origins of the prominent TrpB26-associated kinetic lock may be provided by activated molecular dynamics simulations of hexamer disassembly.
TrpB26 side chain exhibited an orientation similar to native Tyr
The TrpB26 side chain in the crystal structure of [TrpB26,OrnB29]insulin displayed an orientation similar to that of the native Tyr. A slight main chain shift in the B24–B28 β-strand (0.2 (±0.03) Å) was sufficient to enable native-like packing of the larger indole ring against the core of a protomer (Fig. S12). In both T and Rf subunits, the six-membered component of the indole side chain was oriented toward IleA2, ValA3, and ValB12. Based on the classical 3.5–6.5 inter-centroid distance, residue TrpB26 (of the T protomer) showed potential interactions with residues TyrD16 and PheD24. An interplanar angle of 78 degrees between TrpB26 and TyrD16 was indicative of classical aromatic–aromatic packing within proteins (2), which generally range from 50 to 90 degrees. TrpB26 and PheD24 displayed an interplanar angle of 36 degrees, however, this orientation is less common. Similarly, in the Rf protomer, TrpD26 packed near TyrB16 and PheB24. The interplanar angle between TrpD26 and TyrB16 was 62 degrees, whereas between TrpD26 and PheB24 was 48 degrees. Previous studies have suggested that Trp residues may form aromatic–aromatic interactions over longer distances than those formed by Tyr–Phe pairs (68). Thus, the two intra-chain aromatic pairs, TrpB26–PheB24 and TrpD26–PheD24 (respectively, separated by 7.8 and 7.1 Å) may interact more efficiently than the corresponding Tyr–Phe pairs in WT insulin (ring geometries are summarized in Table S11, a and b).
CHARMM calculations of aromatic–aromatic interactions across the dimer interface of [TrpB26,OrnB29]insulin revealed 1.4 kcal/mol increased interaction energy. Residue by residue analysis of each component of the aromatic network indicated that the increased strength of interaction across the dimer interface was the result of the interactions involving TrpD26 (see Table S9b). This result suggests that TyrB26 → Trp may only display stabilizing properties when in an R state protomer. If so, the T6 hexamer formed by [TrpB26]insulin would be expected to have dissociation kinetics similar to WT insulin, whereas the corresponding R6 hexamer may be expected to have markedly increased stability. The R → T transition, which is rapid in WT insulin on release of phenol, may represent the kinetic barrier responsible for the meta-stable association state observed in SEC experiments.
The packing of TrpB26/D26 within the respective cores of T- and Rf crystallographic protomers is similar in each case to the WT TyrB26/D26 and oriented such that the indole's nonpolar six-membered portion projects more deeply into a crevice between A- and B-chains than does its proximal heterocycle. Our 1H NMR studies of TrpB26 within an engineered monomer (35) provided evidence that this overall conformation does not require self-assembly. Analogous partial burial of TyrB26 and TrpB26 in the respective protein structures would in itself be expected to augment the variant's stability due to enhanced solvation free energy10 (69) (i.e. as predicted by water–octanol transfer studies of free Tyr and Trp (69); Table S12). Guanidine denaturation nonetheless, indicated that their stabilities are indistinguishable.11
We speculate that the predicted residue-specific differences in solvation free energy are attenuated by differences in protein dynamics leading to EEC (70). Although the two B26 side chains each exhibit upfield secondary 1H NMR shifts and analogous inter-residue NOEs, it is possible that the substitution is associated with local or nonlocal differences in protein dynamics. It would be of future interest to investigate dynamic features by amide proton 1H–2H exchange and heteronuclear NMR relaxation methods (70). Because TrpB26 promotes partial dimerization of insulin lispro under NMR conditions (as indicated by concentration-dependent 1H NMR resonance broadening), such studies may require use of an alternative monomeric template.
Given the ubiquity of EEC as a confounding general aspect of protein design (71), the profound effects of TrpB26 on the properties of the insulin hexamer seem all the more remarkable. We envision that EEC is circumvented in this case by the rigidity of the insulin hexamer (including the interlocked aromatic residues at its dimer interfaces) with efficient burial of the WT and variant B26 side chains (72). With similar internal structures and external solvation properties, the variant hexamer would gain (relative to WT) two advantages from the Tyr → Trp substitution: (i) greater B26-related solvation transfer free energy (73) (Table S12) augmented by (ii) an uncompensated enthalpic advantage arising from more favorable ETF aromatic interactions as next discussed.
Molecular mechanics calculations rationalized physical and pharmacologic properties
The structural rigidity of the R6 insulin hexamer (42, 74) motivated local MM-based modeling to assess the interactions contributing to the stability of the [TrpB26]insulin hexamers. Analysis of insulin oligomers by Raman spectroscopy revealed dampened conformational fluctuations of the R6 hexamer in relationship to lower-order oligomers and T6 hexamers (72). Moreover, the thermodynamic stability of the assembly was evidenced by the lack of conformational changes visualized by NMR spectroscopy over a temperature range of 10–80 °C (42). The resistance of the R6 hexamer to structural perturbation has also been shown in the context of mutant insulin analogs: native-like X-ray crystal structures have been reported of R6 containing a broad range of substitutions (Table S13) (17, 75, 76). Even substitutions that were shown to destabilize the dimer interface of insulin, such as the substitution of PheB24 by the nonaromatic cyclohexylalanine, were shown to have little impact on the global structure of the R6 hexamer. For this reason, the TyrB26 → Trp substitution was expected to affect only the local structure of the insulin hexamer. Thus, the effects of the mutation were amenable to initial analysis in simplified (eight-ring) models of the dimer interface of insulin.
Energy minimization of a local model of [TrpB26]insulin (i.e. as substituted into a WT insulin dimer extracted from a representative crystal structure of a T3Rf3 zinc hexamer) yielded a native-like framework with enhanced nearest-neighbor B26-related aromatic–aromatic interactions. The partial-charge (monopole) model of the aromatic rings in the CHARMM empirical energy function, parametrized in accordance with ab initio simulations (77), predicted an increase of 0.8 kcal/mol. Although this calculation could in principle have been confounded by transmitted conformational perturbations and did not consider potential changes in conformational entropy or solvation, its conservative features were verified by X-ray crystallography. That the structure of [TrpB26,OrnB29]insulin is essentially identical to WT suggests that local properties of TrpB26 directly underlie the observed increase in hexamer stability and lifetime.
The general asymmetry of the variant TRf dimer in the crystallographic hexamer was associated with differences in the details of corresponding aromatic–aromatic interactions across the dimer interface. Although CHARMM calculations predicted an increase of a 1.4 kcal/mol in interaction energies (0.7 kcal/mol per protomer) in accordance with our initial modeling, residue by residue decomposition ascribed this increase primarily to TrpD26 (in the Rf protomer) and not to TrpB26 (in the T protomer). It is formally possible that TrpB26 is only stabilizing in an R state, but additional studies would be required to resolve this issue. Our cobalt-EDTA sequestration studies focused on the R6 state as the preferred storage vehicle in a pharmaceutical formulation. On SQ injection, rapid diffusion of phenolic ligands from the depot leads to a TR transition. That TrpB26 was found to delay subsequent absorption into the bloodstream suggests that this substitution also enhances the kinetic stability and lifetime of the T6 hexamer.
A seeming paradox is posed by the evolutionary exclusion of Trp at position B28 of vertebrate insulins despite the evident compatibility of this aromatic side chain with native structure and function. We speculate that this exclusion reflects the biological importance of the rapid disassembly of zinc insulin hexamers on their secretion by pancreatic β-cells. Whereas the enhanced thermodynamic and kinetic stabilities of [TrpB26]insulin hexamers would seem favorable for storage within secretory granules (as within pharmaceutical formulations) (8), delayed disassembly of the variant hexamers in the portal circulation would be predicted to reduce the hormone's bioavailability on first pass through the liver, as insulin dimers and hexamers cannot bind to the IR (78). Such delayed disassembly might also decrease the delivery of free zinc ions to the liver, recently predicted to constitute a regulatory signal in its own right (for review, see Ref. 79).
The resulting impairment in hormonal regulation of hepatic metabolism (and accompanying systemic hyperinsulinemia (80)) could in principle have imposed a selective disadvantage in the course of vertebrate evolution. Although to our knowledge, such a kinetics-based mechanism has not been observed in vivo, the converse, abnormally rapid clearance of insulin, has been found on release from zinc-deficient secretory granules; presumably zinc-free insulin oligomers more rapidly dissociate on dilution in the portal circulation and so are more efficiently cleared than WT zinc insulin hexamers (81). Similarly, the exclusion of Trp at position B26 of vertebrate IGFs may reflect a disadvantageous competition between self-assembly and binding to IGF-binding proteins, which are critical to the integrated physiology of IGF function (for review, see Ref. 82). Such functional complexity may impose hidden constraints on the evolution (and so divergence) of protein sequences. The conserved “aromatic triplet” of vertebrate insulins and IGFs provides a natural laboratory to uncover such evolutionary constraints (45). These considerations further suggest that structural features of a protein pertinent to its endogenous function may in general be distinct from those biophysical properties that may be re-engineered to optimize molecular pharmacology.
Comparison of [TrpB26]insulin and a corresponding iodo-Tyr analog
Design of [TrpB26]insulin was in part motivated by prior studies of an analog containing 3-iodo-Tyr at the B26 (17, 23, 83). The latter analog (“3I-Y-insulin”) exhibited a variety of favorable properties: increased affinity for the IR (83), increased thermodynamic stability, and augmented resistance to fibrillation (17). Moreover, when formulated as an R6 hexamer, this analog exhibited a decreased dissociation rate (17). Because modified amino acids raise the cost and complexity of protein manufacture, we wondered whether a natural amino acid might mimic, at least in part, the structure of 3I-Y-insulin and so confer favorable pharmacologic properties with conventional manufacturing.
This translational goal motivated our initial MM-based local modeling of [TrpB26]insulin. Analysis of the crystal structure of 3I-Y-insulin, determined as an R6 zinc hexamer (23), revealed that the large, nonpolar iodo-substituent packed within the core of the insulin protomer with preservation of a native-like dimer interface. Molecular dynamics simulations, undertaken with a multipolar electrostatic model of the modified aromatic ring (23), rationalized this conformation: the iodine atom efficiently filled a cryptic packing defect in WT insulin, lined by the conserved side chains of IleA2, ValA3, and TyrA19. The enhanced packing efficiency of the modified insulin and novel network of halogen-specific electrostatic interactions (“weakly polar” interactions (1)) appear to underlie the analog's increased thermodynamic stability and resistance to fibrillation. Whereas the standard partial-charge model of aromatic systems failed to account for the conformation of iodo-Tyr observed in the crystal structure, a multipolar electrostatic model rationalized thermodynamic stabilization of the dimer interface by this halogen “anchor” (23). Subtle changes in the geometry of aromatic–aromatic interactions were observed both in the simulations and in the crystal structure. Although activated MD simulations were not undertaken to probe the process of dimer dissociation, we presume that the above mechanisms of ground-state stabilization, enhanced core packing efficiency, and a halogen-specific weakly polar network, also underlie the increased barrier to dissociation as indicated by the prolonged lifetime of the variant R6 hexamer (23).
Although the profound QM effects of halo-aromatic substitutions, including weakly polar interactions and “halogen bonding” (84), cannot be recapitulated by natural amino acids, it seemed possible that enhanced core packing efficiency might be achieved by analogy to 3I-Y-insulin. Indeed, the steric profile of the asymmetric indole side chain of Trp is similar in size to 3-iodo-Tyr. Accordingly, we imagined that the offset six-membered portion of the bicyclic indole ring might pack within the core of insulin in a manner similar to the iodine atom. In this intuitive picture the extended π-system of TrpB26 was envisioned to interact with neighboring residues to recapitulate, at least in part, the favorable electrostatic properties of iodo-TyrB26 (23, 37).
The above line of reasoning led to the present set of studies. In accordance with our original intuition and local MM-based modeling, the crystal structures of 3I-Y-insulin and [TrpB26,OrnB29]insulin exhibited similar features. Nevertheless, salient differences between the quantum-chemical properties of iodo-Tyr and Trp might make the similar structures of these B26 analogs a fortuitous outcome of distinct mechanisms. Whereas effects of 3-iodo-Tyr on aromatic–aromatic interactions at the insulin dimer interface are subtle, presumably reflecting indirect inductive effects of iodo-substitutent (23), substitution of Tyr by Trp introduces a larger aromatic surface at this interface. It is possible that this feature underlies the more marked impact of TrpB26 on insulin oligomerization relative to 3-iodo-Tyr.
Aromatic–aromatic interactions exemplify limitations of classical models
The quantum origins of aromatic–aromatic interactions are complex, and so classical electrostatic models (such as partial-charge model of Tyr in the standard CHARMM empirical energy function) can be incomplete (85). Although QM calculations more fully capture this complexity, a tradeoff is encountered between rigor and computational feasibility, especially in complex systems such as proteins (86), but even in ab initio simulations of benzene–benzene interactions (46). Standard MM and MD methods thus employ parametrized force fields to approximate quantum-chemical interactions (21). Although parameters (e.g. partial charges assigned to an aromatic ring) have been chosen to provide reasonable protein models, such use of “monopolar” electrostatics neglects the polarizability of aromatic systems and so omits dispersion forces (87). Parametrized classical models may thus mischaracterize the strength or directionality of intermolecular interactions, particularly those involving more than one aromatic group or an aromatic group and a charged moiety. Although the pioneering studies of Burley and Petsko (88) suggested that the partial-charge model of aromatic rings in proteins was sufficient for characterizing aromatic–aromatic interactions, more recent work has highlighted its limitations with respect to delineating underlying physical mechanisms (85, 89). The limits of parametrized classical force fields are of particular importance when applications are sought in nonstandard systems (such as unnatural protein mutagenesis (90)) for which the parameters were not intended. Examples are provided by halogen-modified aromatic systems (widely employed in medicinal chemistry) (91), which are associated with marked changes in quantum-chemical properties (92). Formal QM/MM simulations of proteins may nonetheless be circumvented through force fields incorporating multipolar electrostatic models of aromatic–aromatic networks (85).
The correspondence between our initial local MM-based modeling and our experimental findings, however striking, may be coincidental (93): rigorous elucidation of the physics of the variant aromatic cluster at insulin's dimer interface may require application of free-energy MD-based simulations with explicit inclusion of water molecules (“molecular alchemy” (94)). This approach may also provide insights into whether or how EEC may be circumvented. Nevertheless, standard MM calculations can guide initial biochemical analysis of protein structure as a guide for protein engineering. In the present study such initial modeling reinforced our structural intuition by highlighting the plausibility that substitution of TyrB26 by Trp might preserve a native-like interface and in fact enhance its weakly polar properties. Because the rigid hexameric framework provided a favorable context for local modeling of amino acid substitutions at subunit interfaces and yet it is the monomer that functions as a hormone in the bloodstream, it would be of future interest to apply more sophisticated computational techniques to simulate the structure and dynamics of [TrpB26]insulin as a monomer in solution. Such predictions may in principle be tested through biophysical studies of an engineered insulin monomer containing TrpB26. Although this substitution is favorable in the context of the hexamer (and so of potential pharmacologic benefit), design of a monomeric NMR model will need to overcome the confounding effects of TrpB26, as defined in this experimental context, to promote self-association.
Concluding remarks
To our knowledge, this study represents the first exploitation of aromatic–aromatic interactions to enhance the biophysical properties of a therapeutic protein (95). Our approach may be broadly applicable in protein engineering (as ETF interactions are ubiquitous) and generalizable to nonstandard aromatic moieties. The latter would be likely to require QM/MM methods rather than classical force fields parametrized with partial charges (90). Overall effects of such substitutions on protein stability and self-assembly will require an integrated analysis of solvation free energies (86), changes in protein dynamics (96), and potential EEC (97). Three- and four-dimensional heteronuclear NMR experiments would be expected to provide higher-resolution information regarding the local and nonlocal interactions of the optimized aromatic system. In the present application, analyses of 15N relaxation and 1H–2H amide–proton exchange are expected to improve understanding of the molecular dynamics of the TrpB26-modified aromatic cluster and so provide a more rigorous biophysical context for its enhanced self-association properties (70, 96).
The present application to insulin demonstrates a direct relationship between stabilization of the insulin hexamer and prolonged activity of a basal analog. Continuous and flat 24-h insulin activity (“peakless” basal formulations) is of clinical interest in the treatment of Type 1 and Type 2 diabetes mellitus to reduce the risk of hypoglycemia (especially at night) at a given level of glycemic control (98). Because this mechanism is unrelated to present strategies to achieve protracted action, we envision that TrpB26-related enhancement of dimer-specific aromatic–aromatic interactions could favorably be introduced into current basal insulin formulations. In the future a combination of orthogonal molecular strategies might enable development of a once-a-week basal insulin therapy analogous to that of GLP-1 agonists (99). Optimization of weakly polar interactions may thus assume a central place in the toolkit of molecular pharmacology.
Experimental procedures
Materials
Insulin was purchased from BioDel® (Danbury, CT). Insulin glargine was obtained from Lantus® (Sanofi-Aventis, Paris, FR). Reagents for peptide synthesis were as described (100).
Preparation of insulin analogs
Variant insulins were prepared by semisynthesis (101). In brief, synthetic peptides were coupled to a tryptic fragment of insulin (des-octapeptide [B23-B30]insulin) in aqueous/organic solvent using trypsin as a catalyst. Following RP-HPLC purification, predicted molecular masses were confirmed by MS (33).
Hexamer disassembly assays
Disassembly of phenol-stabilized (R6) Co2+-substituted insulin hexamers was monitored as described (102). In brief, WT insulin or variants were made 0.6 mm in buffer containing 50 mm Tris-HCl (pH 7.4), 50 mm phenol, and 0.2 mm CoCl2 (18) and incubated overnight at room temperature to attain conformational equilibrium. Spectra (400–750 nm) were obtained to monitor tetrahedral Co2+ coordination (27) through its signature absorption band at 574 nm (27). Co2+ sequestration was initiated by addition of EDTA to a concentration of 2 mm. Dissociation was probed via attenuation of a 574-nm band (27); data were fit to a monoexponential decay equation (29).
Protein crystallography
Crystals were obtained by hanging drop vapor diffusion at room temperature in the presence of a 1:1.7 ratio of Zn2+ to protein monomer and a 3.5:1 ratio of phenol to protein monomer in Tris-HCl. Diffraction was observed using synchrotron radiation at a wavelength of 0.9795 Å at the Stanford Synchrotron Radiation Light Source (beamline BL7-1); crystals were flash frozen to 100 K. Structure determination was carried out using molecular replacement using CCP4 (103) and Phenix structure-determination suites (104). The resulting structure was validated using a PDB Redo server (105). The lattice contained one TRf dimer per asymmetric unit. The main chain conformations of the 97 residues in the refined model of the TRf dimer in the asymmetric unit (excluding 2 Thr, 1 Orn, and 1 Phe residues) each resided in a most favored Ramachandran region.
Receptor-binding assays
Analog affinities for detergent-solubilized IR-B holoreceptor were measured by a competitive-displacement assay (18). Successive dilutions of WT insulin or analogs were incubated overnight with wheat germ agglutinin-SPA beads (PerkinElmer Life Sciences), receptor, and radiolabeled tracer before counting (18). To obtain dissociation constants, competitive binding data were analyzed by nonlinear regression by the method of Wang (106).
Rat studies
Male Lewis rats (mean mass ∼300 g) were rendered diabetic by streptozotocin. Effects of insulin analogs formulated in Lilly® buffer (18) on blood glucose concentration following SQ injection were assessed in relationship to WT or [OrnB29]insulin (18). [OrnB29]Insulin and [TrpB26,OrnB29]insulin were made in the above buffer. GlyA21, OrnB29, OrnB31, OrnB32, TrpB26 and GlyA21, OrnB29, OrnB31, and OrnB32 were dissolved in diluted HCl (pH 4) containing meta-cresol, glycerol, and a 1:2 ratio of ZnCl2:insulin monomer. Rats were injected SQ with 3.44 nmol of insulin or insulin analogs (∼12–13.7 nmol).
Animals used in this study were housed in the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC)-accredited facilities of Case Western Reserve University (CWRU) School of Medicine. All procedures were approved by the Institutional Animal Care and Use Committee (IACUC) Office at CWRU, which provided Standard Operating Procedures and reference materials for animal use (in accordance with the NIH Guide for the Care and Use of Laboratory Animals). The animal health program for all laboratory animals was directed by the CWRU Animal Resource Center. Animal care and use was further monitored for Training and Compliance issues by Veterinary Services.
Size exclusion chromatography
Analogs were made 0.6 mm in 10 mm Tris-HCl (pH 7.4), 1.6% glycerol (v/v), 0.3 mm ZnCl2, and 7 mm phenol (33). Insulin samples (20 μl) were loaded on an Enrich® SEC70 column (10 × 300 mm with fractionation range 3–70 kDa); the mobile phase consisted of 10 mm Tris-HCl (pH 7.4), 140 mm NaCl, and 0.02% sodium azide. Elution times were monitored by absorbance at 280 nm. Molecular masses and void volume (V0) were inferred in reference to standard proteins (18).
Spectroscopy
CD spectra were acquired in 10 mm potassium phosphate (pH 7.4) and 50 mm KCl (33). Free energies of unfolding (ΔGu) were inferred at 25 °C from two-state modeling of protein denaturation by guanidine HCl (33, 34). 1H NMR spectra were acquired at 700 MHz at pH 8.0 or pD 7.6 (direct meter reading) at 37 °C (36). Chemical shifts of aromatic protons in residues B24–B24 (Phe-Phe-Tyr or Phe-Phe-Trp) were evaluated in relationship to corresponding chemical shifts in respective octapeptides B23–B30, presumed to represent random-coil shifts.
Molecular mechanics calculations
Calculations were performed using CHARMM (kindly provided by Prof. M. Karplus). Its standard empirical energy function was employed (in whose development aromatic rings were parametrized by partial atomic charges in accordance with ab initio QM simulations (21)). Representative WT insulin dimers were obtained from PDB entries 4INS, 1ZNJ, and 1TRZ. These structures and corresponding TrpB26 homology models were subjected to local energy minimization (100 steps of steepest descent followed by Adopted Basis Newton-Raphson with gradient tolerance tolg 0.0008/10,000 steps). Minimizations were halted either at 1,000 steps or when the above tolerance was reached. Changes in conformation were allowed only to eight side chains (TyrB16, PheB24, PheB25, TyrB26 (or TrpB26), and their dimer-related mates; the remaining atoms in the respective dimers were fixed. Total interaction energies and respective electrostatic components were obtained between the side chain of residue B26 and the neighboring three aromatic side chains (PheB24 and dimer-related TyrD16 and PheD24). Following this survey of crystallographic dimers, such energies were further evaluated in a simplified molecular model that contained only the side chains of residues TyrB26 (or TrpB26) and the same neighboring three aromatic residues as extracted from PDB entry 4INS; this yielded the electrostatic interaction energy map shown in Fig. 2, in which B26 χ1 and χ2 dihedral angles were systematically varied without energy minimization.
Quantum mechanics calculations
Electron density and molecular electrostatic potential of Tyr and Trp side chains were calculated with B3LYP and 6-31G(d) basis sets using Gaussian utility Cubegen in Gaussian09 (77). The isosurface map was generated using Jmol (107). Ab initio energies of interaction between pairs of isolated aromatic rings were determined by calculating interaction energies using the MP2 method with the aug-cc-pVDZ basis set in Gaussian 09 (77).
Author contributions
N. K. R., N. F. P., and M. A. W. conceptualization; N. K. R., N. P. W., A. N. T., and V. C. Y. data curation; N. K. R. validation; N. K. R., N. P. W., A. N. T., V. C. Y., and F. I.-B. investigation; N. K. R. and M. A. W. writing-original draft; N. P. W. and V. C. Y. formal analysis; N. F. P., V. C. Y., and M. A. W. supervision; N. F. P. and M. A. W. funding acquisition; N. F. P. methodology; N. F. P., F. I.-B., and M. A. W. writing-review and editing; F. I.-B. and M. A. W. resources.
Supplementary Material
Acknowledgments
We thank K. El-Hage, M. Lawrence, M. Meuwly, and V. Pandyarajan for helpful discussion; K. Carr, R. Grabowski, P. Macklis, and M. Swain for assistance with rat studies; V. Kumar and F. van den Akker for advice regarding X-ray crystallography; J. Whittaker and L. Whittaker for advice regarding IR-binding assays; P. De Meyts (Novo Nordisk) for the gift of radiolabeled insulin; and M. C. Lawrence for assistance preparing Fig. 9. Use of the Stanford Synchrotron Radiation Lightsource at the SLAC National Accelerator Laboratory is supported by United States Department of Energy (DOE), Office of Science and Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and National Institutes of Health NIGMS Grant P41GM103393.
This work was supported in part by National Institutes of Health Grant R01 DK040949 (to M. A. W.). M. A. W. has equity in Thermalin, Inc. (Cleveland, OH) where he serves as Chief Innovation Officer; he has also been a consultant to Merck Research Laboratories and DEKA Research & Development Corp. N. F. B. P. is a consultant to Thermalin, Inc. F. I.-B. serves has equity in Thermalin, Inc. and is a consultant to Sanofi and Novo Nordisk. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We dedicate this article to the memory of the late Prof. Colin Ward (Eliza and Walter Hall Institute, Melbourne, Australia).
This article contains Figs. S1–S12 and Tables S1–S13.
The atomic coordinates and structure factors (code 2CK2) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Insulin lispro, containing substitutions ProB28 → Lys and LysB29 → Pro, exhibited an attenuated 574-nm band (69); this is associated with a greater proportion of spectroscopically attenuated T3Rf3 hexamer and invisible T6 hexamer (Fig. 3B).
This analog simulated pI-shifted insulin glargine, which contains two additional arginine residues at the C-terminal B chain (B31 and B32) and a substitution of AsnA21 by Gly (see Fig. 1A) (29, 69). Use of Orn permitted semisynthesis.
One protomer in the (dimeric) asymmetric unit adopted the classical “T” conformation (Fig. 1B); the other adopted in the frayed R state (31, 32) in which residues B3–B8 extend the B-chain α-helix.
An exception is provided by a substitution at the primary zinc-binding site (HisB10 → Asp), which converts the hexamer into a novel dodecamer with zinc-coordination mediated by HisB5 (26). The structure of each protomer is native-like.
An experiment of nature provided in vivo evidence for the relationship between zinc coordination and protection from toxic insulin misfolding: in the rodent species Octogon degus, substitution of HisB10 by Asn is associated with senile amyloidosis of the islets due to insulin fibrillation (70), resulting in DM (48).
The identification of a destabilizing Trp → Tyr mutation in Escherichia coli thioesterase-I demonstrated, in breach, the improved cavity-filling properties of Trp relative to Tyr (72).
That [TrpB26,OrnB29]insulin exhibited a greater exposure of nonpolar surfaces on denaturation in guanidine HCl than did [OrnB29]insulin was suggested by a change in m values in two-state modeling (Table 2).
- ETF
- edge-to-face
- αCT
- carboxyl-terminal domain of IR
- μIR
- domain-minimized insulin micro-receptor
- EEC
- entropy-enthalpy compensation
- IR
- insulin receptor
- L1
- leucine-rich domain of IR
- MD
- molecular dynamics
- MM
- molecular mechanics
- PD
- pharmacodynamics
- PDB
- Protein Data Bank
- r.m.s. deviation
- root mean square deviation
- SQ
- subcutaneous
- QM
- quantum-mechanical
- SEC
- size exclusion chromatography
- IGF
- insulin-like growth factor
- TOCSY
- total correlated spectroscopy
- i.v.
- intravenous
- Orn
- ornithine
- WT
- insulin pertains to the human sequence unless otherwise stated.
References
- 1. Burley S. K., and Petsko G. A. (1988) Weakly polar interaction in proteins. Adv. Protein Chem. 39, 125–189 [DOI] [PubMed] [Google Scholar]
- 2. Burley S. K., and Petsko G. A. (1985) Aromatic-aromatic interaction: a mechanism of protein structure stabilization. Science 229, 23–28 10.1126/science.3892686 [DOI] [PubMed] [Google Scholar]
- 3. Waters M. L. (2002) Aromatic interactions in model systems. Curr. Opin. Chem. Biol. 6, 736–741 10.1016/S1367-5931(02)00359-9 [DOI] [PubMed] [Google Scholar]
- 4. Blundell T. L., Dodson G. G., Hodgkin D. C., and Mercola D. A. (1972) Insulin: the structure in the crystal and its reflection in chemistry and biology. Adv. Protein Chem. 26, 279–402 10.1016/S0065-3233(08)60143-6 [DOI] [Google Scholar]
- 5. Salsali A., and Nathan M. (2006) A review of types 1 and 2 diabetes mellitus and their treatment with insulin. Am. J. Ther. 13, 349–361 10.1097/00045391-200607000-00012 [DOI] [PubMed] [Google Scholar]
- 6. Adams M. J., Blundell T. L., Dodson E. J., Dodson G. G., Vijayan M., Baker E. N., Hardine M. M., Hodgkin D. C., Rimer B., and Sheet S. (1969) Structure of rhombohedral 2 zinc insulin crystals. Nature 224, 491–495 10.1038/224491a0 [DOI] [Google Scholar]
- 7. Bentley G., Dodson E., Dodson G., Hodgkin D., and Mercola D. (1976) Structure of insulin in 4-zinc insulin. Nature 261, 166–168 10.1038/261166a0 [DOI] [PubMed] [Google Scholar]
- 8. Dodson G., and Steiner D. (1998) The role of assembly in insulin's biosynthesis. Curr. Opin. Struct. Biol. 8, 189–194 10.1016/S0959-440X(98)80037-7 [DOI] [PubMed] [Google Scholar]
- 9. Brange J., and Langkjaer L. (1997) Insulin formulation and delivery. Pharm. Biotechnol. 10, 343–409 [DOI] [PubMed] [Google Scholar]
- 10. Brange J., Owens D. R., Kang S., and Volund A. (1990) Monomeric insulins and their experimental and clinical implications. Diab. Care 13, 923–954 10.2337/diacare.13.9.923 [DOI] [PubMed] [Google Scholar]
- 11. DeFelippis M. R., Chance R. E., and Frank B. H. (2001) Insulin self-association and the relationship to pharmacokinetics and pharmacodynamics. Crit. Rev. Ther. Drug Carrier Syst. 18, 201–264 [PubMed] [Google Scholar]
- 12. Berger M., Halban P. A., Girardier L., Seydoux J., Offord R., and Renold A. E. (1979) Absorption kinetics of subcutaneously injected insulin. Diabetologia 17, 97–99 10.1007/BF01222209 [DOI] [PubMed] [Google Scholar]
- 13. Brange J., and Vølund A. (1999) Insulin analogs with improved pharmacokinetic profiles. Adv. Drug Deliv. Rev. 35, 307–335 10.1016/S0169-409X(98)00079-9 [DOI] [PubMed] [Google Scholar]
- 14. Mayer J. P., Zhang F., and DiMarchi R. D. (2007) Insulin structure and function. Biopolymers 88, 687–713 10.1002/bip.20734 [DOI] [PubMed] [Google Scholar]
- 15. Markussen J., Jonassen I., Havelund S., Brandt J., Kurtzhals P., Hansen P. H., and Kaarsholm N. C. (September 16, 2003) U. S. patent US 10620651, Novo Nordisk AS [Google Scholar]
- 16. Menting J. G., Yang Y., Chan S. J., Phillips N. B., Smith B. J., Whittaker J., Wickramasinghe N. P., Whittaker L., Pandyarajan V., Wan Z., Yadav S. P., Carroll J. M., Srokes N., Roberts C. T., Ismail-Beigi F., et al. (2014) Protective hinge in insulin opens to enable its receptor engagement. Proc. Natl. Acad. Sci. U.S.A. 111, 1–48 10.1073/iti0114111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pandyarajan V., Smith B. J., Phillips N. B., Whittaker L., Cox G. P., Wickramasinghe N., Menting J. G., Wan Z.-l., Whittaker J., Ismail-Beigi F., Lawrence M. C., and Weiss M. A. (2014) Aromatic anchor at an invariant hormone-receptor interface function of insulin residue B24 with application to protein design. J. Biol. Chem. 289, 34709–34727 10.1074/jbc.M114.608562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pandyarajan V., Phillips N. B., Rege N., Lawrence M. C., Whittaker J., and Weiss M. A. (2016) Contribution of TyrB26 to the function and stability of insulin: structure-activity relationships at a conserved hormone-receptor interface. J. Biol. Chem. 291, 12978–12990 10.1074/jbc.M115.708347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brooks B. R., Bruccoleri R. E., Olafson B. D., States D. J., Swaminathan S. A., and Karplus M. (1983) CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4, 187–217 10.1002/jcc.540040211 [DOI] [Google Scholar]
- 20. Kramer C., Gedeck P., and Meuwly M. (2012) Atomic multipoles: electrostatic potential fit, local reference axis systems, and conformational dependence. J. Comput. Chem. 33, 1673–1688 10.1002/jcc.22996 [DOI] [PubMed] [Google Scholar]
- 21. Brooks B. R., Brooks C. L. 3rd, Mackerell A. D. Jr., Nilsson L., Petrella R. J., Roux B., Won Y., Archontis G., Bartels C., Boresch S., Caflisch A., Caves L., Cui Q., Dinner A. R., Feig M., et al. (2009) CHARMM: the biomolecular simulation program. J. Comput. Chem. 30, 1545–1614 10.1002/jcc.21287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nakagawa S. H., and Tager H. S. (1986) Role of the phenylalanine B25 side chain in directing insulin interaction with its receptor: steric and conformational effects. J. Biol. Chem. 261, 7332–7341 [PubMed] [Google Scholar]
- 23. El Hage K., Pandyarajan V., Phillips N. B., Smith B. J., Menting J. G., Whittaker J., Lawrence M. C., Meuwly M., and Weiss M. A. (2016) Extending halogen-based medicinal chemistry to proteins: iodo-insulin as a case study. J. Biol. Chem. 291, 27023–27041 10.1074/jbc.M116.761015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lieblich S. A., Fang K. Y., Cahn J. K., Rawson J., LeBon J., Ku H. T., and Tirrell D. A. (2017) 4S-Hydroxylation of insulin at ProB28 accelerates hexamer dissociation and delays fibrillation. J. Am. Chem. Soc. 139, 8384–8387 10.1021/jacs.7b00794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ciszak E., and Smith G. D. (1994) Crystallographic evidence for dual coordination around zinc in the T3R3 human insulin hexamer. Biochemistry 33, 1512–1517 10.1021/bi00172a030 [DOI] [PubMed] [Google Scholar]
- 26. Riemen M. W., Pon L. A., and Carpenter F. H. (1983) Preparation of semisynthetic insulin analogues from bis(tert-butyloxycarbonyl)-desoctapeptide-insulin phenylhydrazide: importance of the aromatic region B24–B26. Biochemistry 22, 1507–1515 10.1021/bi00275a027 [DOI] [PubMed] [Google Scholar]
- 27. Roy M., Brader M. L., Lee R. W., Kaarsholm N. C., Hansen J. F., and Dunn M. F. (1989) Spectroscopic signatures of the T to R conformational transition in the insulin hexamer. J. Biol. Chem. 264, 19081–19085 [PubMed] [Google Scholar]
- 28. Brems D. N., Alter L. A., Beckage M. J., Chance R. E., DiMarchi R. D., Green L. K., Long H. B., Pekar A. H., Shields J. E., and Frank B. H. (1992) Altering the association properties of insulin by amino acid replacement. Protein Eng. 5, 527–533 10.1093/protein/5.6.527 [DOI] [PubMed] [Google Scholar]
- 29. Birnbaum D. T., Kilcomons M. A., DeFelippis M. R., and Beals J. M. (1997) Assembly and dissociation of human insulin and LysB28ProB29-insulin hexamers: a comparison study. Pharm. Res. 14, 25–36 10.1023/A:1012095115151 [DOI] [PubMed] [Google Scholar]
- 30. Gillies P. S., Figgitt D. P., and Lamb H. M. (2000) Insulin glargine. Drugs 59, 253–260; discussin 261–262 10.2165/00003495-200059020-00009 [DOI] [PubMed] [Google Scholar]
- 31. Chothia C., Lesk A. M., Dodson G. G., and Hodgkin D. C. (1983) Transmission of conformational change in insulin. Nature 302, 500–505 10.1038/302500a0 [DOI] [PubMed] [Google Scholar]
- 32. Dodson G. G., Dodson E. J., Turkenburg J. P., and Bing X. (1993) Molecular recognition in insulin assembly. Biochem. Soc. Trans. 21, 609–614 10.1042/bst0210609 [DOI] [PubMed] [Google Scholar]
- 33. Pandyarajan V., Phillips N. B., Cox G. P., Yang Y., Whittaker J., Ismail-Beigi F., and Weiss M. A. (2014) Biophysical optimization of a therapeutic protein by non-standard mutagenesis: studies of an iodo-insulin derivative. J. Biol. Chem. 289, 23367–23381 10.1074/jbc.M114.588277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sosnick T. R., Fang X., and Shelton V. M. (2000) Application of circular dichroism to study RNA folding transitions. Methods Enzymol. 317, 393–409 10.1016/S0076-6879(00)17026-0 [DOI] [PubMed] [Google Scholar]
- 35. Holleman F., and Hoekstra J. B. (1997) Insulin lispro. N. Engl. J. Med. 337, 176–183 10.1056/NEJM199707173370307 [DOI] [PubMed] [Google Scholar]
- 36. Hua Q. X., Jia W., and Weiss M. A. (2011) Conformational dynamics of insulin. Front. Endocrinol. 2, 48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. El Hage K., Bereau T., Jakobsen S., and Meuwly M. (2016) Impact of quadrupolar electrostatics on atoms adjacent to the σ-hole in condensed-phase simulations. J. Chem. Theory Comp. 12, 3008–3019 10.1021/acs.jctc.6b00202 [DOI] [PubMed] [Google Scholar]
- 38. Sikosek T., and Chan H. S. (2014) Biophysics of protein evolution and evolutionary protein biophysics. J. R. Soc. Interface 11, 20140419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baron R., and McCammon J. A. (2013) Molecular recognition and ligand association. Annu. Rev. Phys. Chem. 64, 151–175 10.1146/annurev-physchem-040412-110047 [DOI] [PubMed] [Google Scholar]
- 40. Blundell T. L., Cutfield J. F., Cutfield S. M., Dodson E. J., Dodson G. G., Hodgkin D. C., Mercola D. A., and Vijayan M. (1971) Atomic positions in rhombohedral 2-zinc insulin crystals. Nature 231, 506–511 10.1038/231506a0 [DOI] [PubMed] [Google Scholar]
- 41. Chang X., Jorgensen A. M., Bardrum P., and Led J. J. (1997) Solution structures of the R6 human insulin hexamer. Biochemistry 36, 9409–9422 10.1021/bi9631069 [DOI] [PubMed] [Google Scholar]
- 42. Jacoby E., Hua Q. X., Stern A. S., Frank B. H., and Weiss M. A. (1996) Structure and dynamics of a protein assembly: 1H-NMR studies of the 36 kDa R6 insulin hexamer. J. Mol. Biol. 258, 136–157 10.1006/jmbi.1996.0239 [DOI] [PubMed] [Google Scholar]
- 43. Keller D., Clausen R., Josefsen K., and Led J. J. (2001) Flexibility and bioactivity of insulin: an NMR investigation of the solution structure and folding of an unusually flexible human insulin mutant with increased biological activity. Biochemistry 40, 10732–10740 10.1021/bi0108150 [DOI] [PubMed] [Google Scholar]
- 44. Brange J., Ribel U., Hansen J. F., Dodson G., Hansen M. T., Havelund S., Melberg S. G., Norris F., Norris K., and Snel L. (1988) Monomeric insulins obtained by protein engineering and their medical implications. Nature 333, 679–682 10.1038/333679a0 [DOI] [PubMed] [Google Scholar]
- 45. Weiss M. A., and Lawrence M. C. (2018) A thing of beauty: structure and function of insulin's “aromatic triplet.” Diabetes Obes. Metab. 20, 51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sinnokrot M. O., and Sherrill C. D. (2006) High-accuracy quantum mechanical studies of π-π interactions in benzene dimers. J. Phys. Chem. A 110, 10656–10668 10.1021/jp0610416 [DOI] [PubMed] [Google Scholar]
- 47. Conlon J. M. (2001) Evolution of the insulin molecule: insights into structure-activity and phylogenetic relationships. Peptides 22, 1183–1193 10.1016/S0196-9781(01)00423-5 [DOI] [PubMed] [Google Scholar]
- 48. Jekl V., Hauptman K., and Knotek Z. (2011) Diseases in pet degus: a retrospective study in 300 animals. J. Small Anim. Prac. 52, 107–112 10.1111/j.1748-5827.2010.01028.x [DOI] [PubMed] [Google Scholar]
- 49. Brange J., and Havelund S. (1984) Stabilized insulin preparations and method for their production. in U. S. Patent and Trademark Office patent full-text database, A61K 3726 Ed., Novo Industries A/S [Google Scholar]
- 50. Jonassen I., Havelund S., Hoeg-Jensen T., Steensgaard D. B., Wahlund P. O., and Ribel U. (2012) Design of the novel protraction mechanism of insulin degludec, an ultra-long-acting basal insulin. Pharm. Res. 29, 2104–2114 10.1007/s11095-012-0739-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kurtzhals P., Havelund S., Jonassen I., Kiehr B., Larsen U. D., Ribel U., and Markussen J. (1995) Albumin binding of insulins acylated with fatty acids: characterization of the ligand-protein interaction and correlation between binding affinity and timing of the insulin effect in vivo. Biochem. J. 312, 725–731 10.1042/bj3120725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chodera J. D., and Mobley D. L. (2013) Entropy-enthalpy compensation: role and ramifications in biomolecular ligand recognition and design. Annu. Rev. Biophys. 42, 121–142 10.1146/annurev-biophys-083012-130318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Birnbaum D. T., Dodd S. W., Saxberg B. E., Varshavsky A. D., and Beals J. M. (1996) Hierarchical modeling of phenolic ligand binding to 2Zn-insulin hexamers. Biochemistry 35, 5366–5378 10.1021/bi9600557 [DOI] [PubMed] [Google Scholar]
- 54. Hua Q. X., Nakagawa S., Hu S. Q., Jia W., Wang S., and Weiss M. A. (2006) Toward the active conformation of insulin: stereospecific modulation of a structural switch in the B chain. J. Biol. Chem. 281, 24900–24909 10.1074/jbc.M602691200 [DOI] [PubMed] [Google Scholar]
- 55. Blundell T. L., Cutfield J. F., Cutfield S. M., Dodson E. J., Dodson G. G., Hodgkin D. C., and Mercola D. A. (1972) Three-dimensional atomic structure of insulin and its relationship to activity. Diabetes 21, 492–505 10.2337/diab.21.2.S492 [DOI] [PubMed] [Google Scholar]
- 56. Markussen J., Hougaard P., Ribel U., Sørensen A. R., and Sørensen E. (1987) Soluble, prolonged-acting insulin derivatives: I. degree of protraction and crystallizability of insulins substituted in the termini of the B-chain. Protein Eng. 1, 205–213 10.1093/protein/1.3.205 [DOI] [PubMed] [Google Scholar]
- 57. Weiss M. A. (2009) The structure and function of insulin: decoding the TR transition. Vitam. Horm. 80, 33–49 10.1016/S0083-6729(08)00602-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Markussen J., Diers I., Engesgaard A., Hansen M. T., Hougaard P., Langkjaer L., Norris K., Ribel U., Søorensen A. R., and Sørensen E. (1987) Soluble, prolonged-acting insulin derivatives: II. degree of protraction and crystallizability of insulins substituted in positions A17, B8, B13, B27, and B30. Protein Eng. 1, 215–223 10.1093/protein/1.3.215 [DOI] [PubMed] [Google Scholar]
- 59. Bentley G. A., Brange J., Derewenda Z., Dodson E. J., Dodson G. G., Markussen J., Wilkinson A. J., Wollmer A., and Xiao B. (1992) Role of B13 Glu in insulin assembly: the hexamer structure of recombinant mutant (B13 Glu → Gln) insulin. J. Mol. Biol. 228, 1163–1176 10.1016/0022-2836(92)90323-C [DOI] [PubMed] [Google Scholar]
- 60. Mirmira R. G., Nakagawa S. H., and Tager H. S. (1991) Importance of the character and configuration of residues B24, B25, and B26 in insulin-receptor interactions. J. Biol. Chem. 266, 1428–1436 [PubMed] [Google Scholar]
- 61. Xu B., Hu S. Q., Chu Y. C., Huang K., Nakagawa S. H., Whittaker J., Katsoyannis P. G., and Weiss M. A. (2004) Diabetes-associated mutations in insulin: consecutive residues in the B chain contact distinct domains of the insulin receptor. Biochemistry 43, 8356–8372 10.1021/bi0497796 [DOI] [PubMed] [Google Scholar]
- 62. Huang K., Xu B., Hu S. Q., Chu Y. C., Hua Q. X., Qu Y., Li B., Wang S., Wang R. Y., Nakagawa S. H., Theede A. M., Whittaker J., De Meyts P., Katsoyannis P. G., and Weiss M. A. (2004) How insulin binds: the B-chain α-helix contacts the L1 β-helix of the insulin receptor. J. Mol. Biol. 341, 529–550 10.1016/j.jmb.2004.05.023 [DOI] [PubMed] [Google Scholar]
- 63. Hu S. Q., Burke G. T., and Katsoyannis P. G. (1993) Contribution of the B16 and B26 tyrosine residues to the biological activity of insulin. J. Protein Chem. 12, 741–747 10.1007/BF01024932 [DOI] [PubMed] [Google Scholar]
- 64. Baker E. N., Blundell T. L., Cutfield J. F., Cutfield S. M., Dodson E. J., Dodson G. G., Hodgkin D. M., Hubbard R. E., Isaacs N. W., and Reynolds C. D. (1988) The structure of 2Zn pig insulin crystals at 1.5-Å resolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 319, 369–456 10.1098/rstb.1988.0058 [DOI] [PubMed] [Google Scholar]
- 65. Guvench O., and Brooks C. L. 3rd. (2005) Tryptophan side chain electrostatic interactions determine edge-to-face vs parallel-displaced tryptophan side chain geometries in the designed β-hairpin “trpzip2.” J. Am. Chem. Soc. 127, 4668–4674 10.1021/ja043492e [DOI] [PubMed] [Google Scholar]
- 66. Samanta U., Pal D., and Chakrabarti P. (1999) Packing of aromatic rings against tryptophan residues in proteins. Acta Crystallogr. D 55, 1421–1427 10.1107/S090744499900726X [DOI] [PubMed] [Google Scholar]
- 67. Holst B., Nygaard R., Valentin-Hansen L., Bach A., Engelstoft M. S., Petersen P. S., Frimurer T. M., and Schwartz T. W. (2010) A conserved aromatic lock for the tryptophan rotameric switch in TM-VI of seven-transmembrane receptors. J. Biol. Chem. 285, 3973–3985 10.1074/jbc.M109.064725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bhattacharyya R., Samanta U., and Chakrabarti P. (2002) Aromatic-aromatic interactions in and around α-helices. Protein Eng. 15, 91–100 10.1093/protein/15.2.91 [DOI] [PubMed] [Google Scholar]
- 69. Lee L. C., Chou Y. L., Chen H. H., Lee Y. L., and Shaw J. F. (2009) Functional role of a non-active site residue Trp(23) on the enzyme activity of Escherichia coli thioesterase I/protease I/lysophospholipase L(1). Biochim. Biophys. Acta 1794, 1467–1473 10.1016/j.bbapap.2009.06.008 [DOI] [PubMed] [Google Scholar]
- 70. Glidden M. D., Yang Y., Phillips N. B., Carr K., Smith N. A., Wickramasinghe N. P., Smith B. J., Ismail-Beigi F., Lawrence M. C., and Weiss M. A. (2017) Solution structure and dynamics of a single-chain insulin analog. J. Biol. Chem. 293, 69–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Frederick K. K., Marlow M. S., Valentine K. G., and Wand A. J. (2007) Conformational entropy in molecular recognition by proteins. Nature 448, 325–329 10.1038/nature05959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Dong J., Wan Z., Popov M., Carey P. R., and Weiss M. A. (2003) Insulin assembly damps conformational fluctuations: Raman analysis of amide I line widths in native states and fibrils. J. Mol. Biol. 330, 431–442 10.1016/S0022-2836(03)00536-9 [DOI] [PubMed] [Google Scholar]
- 73. Wimley W. C., and White S. H. (1996) Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat. Struct. Mol. Biol. 3, 842 10.1038/nsb1096-842 [DOI] [PubMed] [Google Scholar]
- 74. Hassiepen U., Federwisch M., Mülders T., and Wollmer A. (1999) The lifetime of insulin hexamers. Biophys. J. 77, 1638–1654 10.1016/S0006-3495(99)77012-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Derewenda U., Derewenda Z., Dodson G., and Brange J. (1987) The crystal-structure of the B-12 Ile human insulin prepared by site-directed mutatgenesis. Protein Eng. 222, 425–433 [Google Scholar]
- 76. Liu M., Wan Z. L., Chu Y. C., Aladdin H., Klaproth B., Choquette M., Hua Q. X., Mackin R. B., Rao J. S., De Meyts P., Katsoyannis P. G., Arvan P., and Weiss M. A. (2009) Crystal structure of a “non-foldable” insulin: impaired folding efficiency and ER stress despite native activity. J. Biol. Chem. 284, 35259–35272 10.1074/jbc.M109.046888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Frisch M. J., Trucks G. W., Schlegel H. B., Scuseria G. E., Robb M. A., Cheeseman J. R., Scalmani G., Barone V., Mennucci B., Petersson G. A., Nakatsuji H., Caricato M., Li X., Hratchian H. P., Izmaylov A. F., et al. (2009) Gaussian 09, revision A.02., Gaussian, Inc., Wallingford, CT [Google Scholar]
- 78. Shoelson S. E., Lu Z. X., Parlautan L., Lynch C. S., and Weiss M. A. (1992) Mutations at the dimer, hexamer, and receptor-binding, surfaces of insulin independently affect insulin-insulin and insulin-receptor interactions. Biochemistry 31, 1757–1767 10.1021/bi00121a025 [DOI] [PubMed] [Google Scholar]
- 79. O'Halloran T. V., Kebede M., Philips S. J., and Attie A. D. (2013) Zinc, insulin, and the liver: a ménage à trois. J. Clin. Investig. 123, 4136–4139 10.1172/JCI72325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Michael M. D., Kulkarni R. N., Postic C., Previs S. F., Shulman G. I., Magnuson M. A., and Kahn C. R. (2000) Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol. Cell 6, 87–97 10.1016/S1097-2765(05)00015-8 [DOI] [PubMed] [Google Scholar]
- 81. Tamaki M., Fujitani Y., Hara A., Uchida T., Tamura Y., Takeno K., Kawaguchi M., Watanabe T., Ogihara T., Fukunaka, A. et al. (2013) The diabetes-susceptible gene SLC30A8/ZnT8 regulates hepatic insulin clearance. J. Clin. Investig. 123, 4513–4524 10.1172/JCI68807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Forbes B., McCarthy P., and Norton R. (2012) Insulin-like growth factor binding proteins: a structural perspective. Front. Endocrinol. 3, 38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Heidenreich K. A., Yip C. C., Frank B. H., and Olefsky J. M. (1985) The preparation and characterization of mono-iodinated photoreactive analogs of insulin. Biochem. Biophys. Res. Commun. 126, 1138–1145 10.1016/0006-291X(85)90304-3 [DOI] [PubMed] [Google Scholar]
- 84. Scholfield M. R., Zanden C. M., Carter M., and Ho P. S. (2013) Halogen bonding (X-bonding): a biological perspective. Protein Sci. 22, 139–152 10.1002/pro.2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Marshall G. R. (2013) Limiting assumptions in molecular modeling: electrostatics. J. Comput. Aided Mol. Des. 27, 107–114 10.1007/s10822-013-9634-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. MacKerell A. D., Bashford D., Bellott M., Dunbrack R. L., Evanseck J. D., Field M. J., Fischer S., Gao J., Guo H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F. T., Mattos C., et al. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 10.1021/jp973084f [DOI] [PubMed] [Google Scholar]
- 87. MacKerell A. D., Brooks B., Brooks C. L., Nilsson L., Roux B., Won Y., and Karplus M. (2002) CHARMM: the energy function and its parameterization. in Encyclopedia of Computational Chemistry (Schleyer P. v. R., Allinger N. L., Clark T., Gasteiger J., Kollman P. A., Schaefer H. F. III, Schreiner P. R., eds) pp. 271–277, John Wiley & Sons, Ltd., Chichester, United Kingdom [Google Scholar]
- 88. Burley S., and Petsko G. (1986) Dimerization energetics of benzene and aromatic amino acid side chains. J. Am. Chem. Soc. 108, 7995–8001 10.1021/ja00285a019 [DOI] [Google Scholar]
- 89. Kamerlin S. C., Haranczyk M., and Warshel A. (2009) Progress in ab initio QM/MM free-energy simulations of electrostatic energies in proteins: accelerated QM/MM studies of pKa, redox reactions and solvation free energies. J. Phys. Chem. B 113, 1253–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Reha D., Kabelác M., Ryjácek F., Sponer, Sponer J. E., Suhai M., and Hobza P. (2002) Intercalators: 1. nature of stacking interactions between intercalators (ethidium, daunomycin, ellipticine, and 4′,6-diaminide-2-phenylindole) and DNA base pairs: ab initio quantum chemical, density functional theory, and empirical potential study. J. Am. Chem. Soc. 124, 3366–3376 10.1021/ja011490d [DOI] [PubMed] [Google Scholar]
- 91. Steward L. E., and Chamberlin A. R. (1998) Protein engineering with non-standard amino acids. Methods Mol. Biol. 77, 325–354 [DOI] [PubMed] [Google Scholar]
- 92. Dougherty D. A. (2000) Unnatural amino acids as probes of protein structure and function. Curr. Opin. Chem. Biol. 4, 645–652 10.1016/S1367-5931(00)00148-4 [DOI] [PubMed] [Google Scholar]
- 93. van Gunsteren W. F., Daura X., Hansen N., Mark A. E., Oostenbrink C., Riniker S., and Smith L. J. (2018) Validation of molecular simulation: an overview of issues. Angew. Chem. Int. Ed. 57, 884–902 10.1002/anie.201702945 [DOI] [PubMed] [Google Scholar]
- 94. Aleksandrov A., Thompson D., and Simonson T. (2010) Alchemical free energy simulations for biological complexes: powerful but temperamental. J. Mol. Recognit. 23, 117–127 [DOI] [PubMed] [Google Scholar]
- 95. Kannan N., and Vishveshwara S. (2000) Aromatic clusters: a determinant of thermal stability of thermophilic proteins. Protein Eng. 13, 753–761 10.1093/protein/13.11.753 [DOI] [PubMed] [Google Scholar]
- 96. Szyperski T., Luginbühl P., Otting G., Güntert P., and Wüthrich K. (1993) Protein dynamics studied by rotating frame nitrogen-15 spin relaxation times. J. Biomol. NMR 3, 151–164 [DOI] [PubMed] [Google Scholar]
- 97. Freire E. (2009) A thermodynamic approach to the affinity optimization of drug candidates. Chem. Biol. Drug Des. 74, 468–472 10.1111/j.1747-0285.2009.00880.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Cryer P. E. (2002) Hypoglycaemia: the limiting factor in the glycaemic management of type I and type II diabetes. Diabetologia 45, 937–948 10.1007/s00125-002-0822-9 [DOI] [PubMed] [Google Scholar]
- 99. Christensen M., and Knop F. K. (2010) Once-weekly GLP-1 agonists: how do they differ from exenatide and liraglutide? Curr. Diab. Rep. 10, 124–132 10.1007/s11892-010-0102-x [DOI] [PubMed] [Google Scholar]
- 100. Wan Z. L., Huang K., Hu S. Q., Whittaker J., and Weiss M. A. (2008) The structure of a mutant insulin uncouples receptor binding from protein allostery: an electrostatic block to the TR transition. J. Biol. Chem. 283, 21198–21210 10.1074/jbc.M800235200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Inouye K., Watanabe K., Morihara K., Tochino Y., Kanaya T., Emura J., and Sakakibara S. (1979) Enzyme-assisted semisynthesis of human insulin. J. Am. Chem. Soc. 101, 751–752 10.1021/ja00497a051 [DOI] [Google Scholar]
- 102. Rahuel-Clermont S., French C. A., Kaarsholm N. C., Dunn M. F., and Chou C. I. (1997) Mechanisms of stabilization of the insulin hexamer through allosteric ligand interactions. Biochemistry 36, 5837–5845 10.1021/bi963038q [DOI] [PubMed] [Google Scholar]
- 103. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., Grosse-Kunstleve R. W., et al. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221 10.1107/S0108768110007202,10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Joosten R. P., Womack T., Vriend G., and Bricogne G. (2009) Re-refinement from deposited X-ray data can deliver improved models for most PDB entries. Acta Crystallogr. D 65, 176–185 10.1107/S0907444908037591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wang Z. X. (1995) An exact mathematical expression for describing competitive binding of two different ligands to a protein molecule. FEBS Lett. 360, 111–114 10.1016/0014-5793(95)00062-E [DOI] [PubMed] [Google Scholar]
- 107. Willighagen E., and Howard M. (2007) Fast and scriptable molecular graphics in web browsers without Java3D. Nat. Proceed. 10.1038/npre.2007.50.1 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.