Abstract
Translocated in liposarcoma (TLS) is an RNA-binding protein and a transcription-regulatory sensor of DNA damage. TLS binds promoter-associated noncoding RNA (pncRNA) and inhibits histone acetyltransferase (HAT) activity of CREB-binding protein (CBP)/E1A-binding protein P300 (p300) on the cyclin D1 (CCND1) gene. Although post-translational modifications of TLS, such as arginine methylation, are known to regulate TLS's nucleocytoplasmic shuttling and assembly in stress granules, its interactions with RNAs remain poorly characterized. Herein, using various biochemical assays, we confirmed the earlier observations that TLS is methylated by protein arginine methyltransferase 1 (PRMT1) in vitro. The arginine methylation of TLS disrupted binding to pncRNA and also prevented binding of TLS to and inhibition of CBP/p300. This result indicated that arginine methylation of TLS abrogates both binding to pncRNA and TLS-mediated inhibition of CBP/p300 HAT activities. We also report that an arginine residue within the Arg–Gly–Gly domain of TLS, Arg-476, serves as the major determinant for binding to pncRNA. Either methylation or mutation of Arg-476 of TLS significantly decreased pncRNA binding and thereby prevented a pncRNA-induced allosteric alteration in TLS that is required for its interaction with CBP/p300. Moreover, unlike WT TLS, an R476A TLS mutant did not inhibit CCND1 promoter activity in luciferase reporter assays. Taken together, we propose the hypothesis that arginine methylation of TLS regulates both TLS–nucleic acid and TLS–protein interactions and thereby participates in transcriptional regulation.
Keywords: fused in sarcoma (FUS); long noncoding RNA (long ncRNA, lncRNA); post-translational modification (PTM); RNA-protein interaction; cyclin D1; amyotrophic lateral sclerosis (ALS) (Lou Gehrig disease); histone acetylase; histone; RNA; gene expression; CBP; HAT activity; lncRNA; p300; TLS
Introduction
Translocated in liposarcoma (TLS;2 also known as fused in sarcoma (FUS)) is an RNA-binding protein that participates in a variety of cellular processes, including mRNA splicing, DNA repair, and transcriptional regulation (1, 2). TLS is also implicated in DNA damage responses and cell cycle arrest, and it prevents the propagation of DNA damage to facilitate DNA repair (3). Specifically, expression of the cell cycle gene CCND1 is inhibited by TLS after DNA damage by ionizing radiation (4). The inhibitory function of TLS is mediated by promoter-associated noncoding RNA-D (pncRNA-D) that is transcribed from the promoter region of CCND1 (4, 5). Binding with pncRNA-D induces allosteric alteration of TLS, which enhances interactions with CBP/p300 and leads to a decrease in histone acetyltransferase (HAT) activities of CBP/p300 and a reduction in the transcription of CCND1 (4).
Structurally, TLS comprises an N-terminal serine-tyrosine-glycine-glutamine (SYGQ)-rich domain, an RNA recognition motif, a C2-C2 zinc-finger domain, three arginine-glycine-glycine (RGG)-rich domains, and a C-terminal nuclear localization signal. The RGG domains of TLS are pivotal RNA-binding sites and display some selectivity in RNA binding (6, 7). Moreover, the domain recognition by nucleic acids can be regulated through the subtle amino acid alterations. For example, the RGG domain of TLS binds to G-quadruplex human telomere DNA and RNA, but the substitution of tyrosine with phenylalanine in the RGG domain abolishes the interaction with telomere DNA but not RNA (6). Furthermore, the majority of ALS-associated mutations are clustered in the C terminus of TLS, which impedes TLS nuclear localization and leads to the pathological aggregation in the cytoplasm (8). Our previous study revealed that the C terminus of TLS (RGG2–zinc finger–RGG3) is involved in the interaction with pncRNA (5). Hence, discriminating the pncRNA binding residues in TLS is crucial for understanding how lncRNA and TLS function together as a complex.
Post-translational modifications play roles in multiple biological processes, such as transcription, RNA processing, DNA repair, metabolism, and signal transduction (9). Arginine methylation is a common post-translational modification of RNA-binding proteins, although the associated regulatory mechanisms remain unclear. PRMT1 is a type I methyltransferase that mediates 90% of all asymmetric arginine dimethylations in mammalian cells (10). PRMT1 is characterized as a transcriptional coregulator, which methylates both histone and nonhistone proteins. For example, PRMT1 regulates histone H4 arginine 3 dimethylation, in which the histone marks can be recognized by TDRD3 for transcriptional activation (11). PRMT1 also methylates RIP40 and abolishes its corepressor activity for nuclear receptors (12). TLS is methylated by PRMT1 both in vitro and in vivo (13), and arginine residues within the three RGG domains of TLS are subjected to extensive asymmetric dimethylation by PRMT1 (14–17). Moreover, TLS methylation by PRMT1 also modulates the nuclear import of TLS via impeding transportin binding (8), suggesting that protein–protein interactions of TLS are probably regulated by arginine methylation. Ewing sarcoma (EWS) a structure similar to TLS, comprising three RGG domains, and exhibits the ability to bind to G-rich DNA and RNA, which fold into G-quadruplex structures (18). Moreover, the methylation of the RGG domain decreases the binding ability of EWS to bind to G-quadruplex DNA in vitro (18), suggesting that protein–nucleic acid interactions can be modulated through arginine methylation.
In this study, we have investigated interactions between pncRNA-D and methylated TLS and explored mechanisms by which TLS methylation regulates binding to long noncoding RNAs (lncRNAs) and also regulates CBP/p300 HAT activities. We performed in vitro methylation assays to monitor activities of PRMT1 in response to methylation and found that methylation repressed the binding of TLS to pncRNA-D. In addition, arginine residue Arg-476 in the RGG3 domain of TLS was identified as the pncRNA-D–binding residue, and methylation of the Arg-476 residue strongly inhibited pncRNA-D binding. In agreement with these findings, methylated TLS failed to bind pncRNA-D, resulting in a subsequent inhibition of interactions with CBP/p300, and restoration of CBP/p300 HAT activities from TLS-mediated suppression. Furthermore, a reporter assay revealed that substitution of Arg-476 with alanine in TLS disrupted its inhibition of CCND1-driven promoter activity. These findings are significant evidence that arginine methylation of TLS regulates the biological effects of noncoding RNA and affects the regulatory function of TLS on gene expression.
Results
Methylation of TLS by PRMT1 represses pncRNA-D binding in vitro
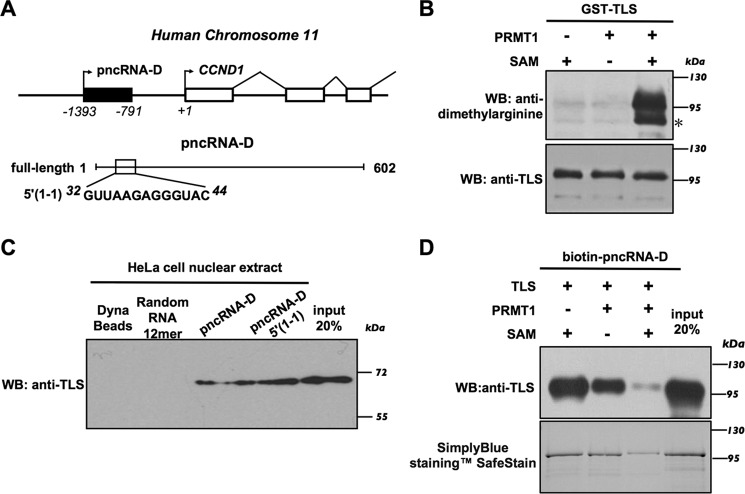
Among the lncRNAs transcribed from the promoter region of CCND1 (Fig. 1A), pncRNA-D is the transcript whose expression is most strongly induced after DNA damage (4, 5). In our previous study, full-length pncRNA-D was identified as a lncRNA of 602 nucleotides that is able to bind to TLS (5). To determine the effects of TLS methylation on RNA binding, we first performed in vitro methylation assays using bacterially expressed GST-TLS and Strep-PRMT1, followed by Western blot analysis with an anti-dimethylarginine antibody. These experiments showed that GST-TLS was methylated by PRMT1 in vitro in the presence of the methyl donor SAM (Fig. 1B). To examine the specificity of pncRNA-D binding, the HeLa cell nuclear extract pulldown assays were performed, and the RNA-bound endogenous TLS was detected using an anti-TLS antibody. The Dynabeads-bound, biotinylated full-length pncRNA-D showed binding to endogenous TLS, but TLS did not bind to a 12-mer synthetic oligonucleotide with randomized sequences (Fig. 1C). Consistent with a previous study (5), pncRNA-D fragment (5′(1-1); nucleotides 32–44) also exhibited a level of binding to endogenous TLS similar to that of the full-length pncRNA-D (Fig. 1C). The Dynabeads without RNA binding pulled down from the HeLa nuclear extract were used as a negative control.
Figure 1.
Arginine methylation of TLS inhibits the interaction with pncRNA-D. A, the pncRNA-D locus (top) and the high-affinity binding site of pncRNA-D (5′(1-1); from nucleotide 32 to 44) to TLS (bottom). B, methylation of GST-TLS by PRMT1 in vitro. The methylated TLS was detected by an anti-dimethylarginine antibody. An unrelated band is indicated with a star. C, endogenous TLS from HeLa cell nuclear extract bound to both full-length and 5′(1-1) of pncRNA-D. D, RNA pulldown assay using biotinylated pncRNA-D was performed with incubation with the unmethylated and methylated GST-TLS; 20% of the protein used for the RNA pulldown assays was loaded as input.
In subsequent total RNA-binding assays, we determined changes in global RNA binding before and after methylation of TLS. The recombinant GST-TLS protein was methylated in vitro before incubation with HeLa cell total RNA. Arginine methylation reduced the binding of TLS to the HeLa cell total RNA to 40% (Fig. S1). To determine whether arginine methylation of TLS also reduces its binding to pncRNA-D, biotinylated pncRNA-D pulldown assays were performed, and the result demonstrated that arginine methylation of TLS clearly decreased the binding to pncRNA-D (Fig. 1D). Consistent with this, SimplyBlue staining of the proteins that were pulled down confirmed the result that arginine methylation of TLS reduced its interactions with pncRNA-D. Our findings revealed that methylation of TLS by PRMT1 inhibits pncRNA-D binding.
Identification of Arg-476 residue as a pncRNA-D–binding site
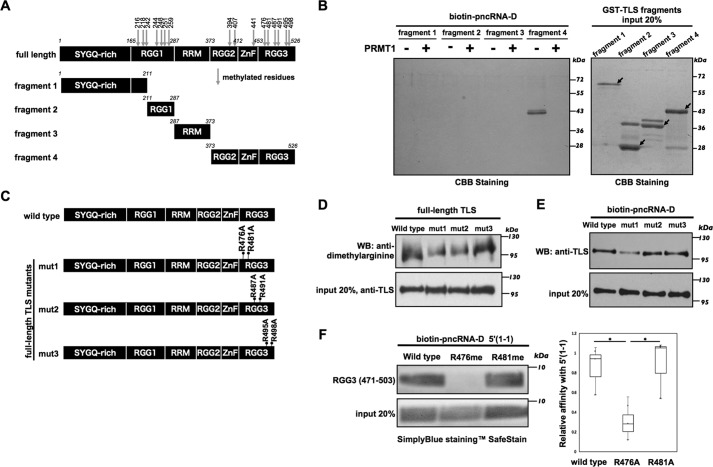
Our previous MS analyses identified 16 methylated arginine residues of TLS; 15 of the 16 arginine residues are found in RGG domains, and one is located in the zinc-finger domain of TLS (Fig. 2A) (19). The RGG domain is a common human RNA-binding domain, and arginine residues of TLS located at RGG domains are the preferred substrates for PRMTs (15). Thus, to determine which arginine residues contribute to pncRNA-D binding, we performed binding assays of TLS fragments. Initially, we divided TLS into four GST-tagged fragments bearing functional domains (Fig. 2A) and performed subsequent biotinylated pncRNA-D pulldown assays. The results revealed that among these domains, the RGG2–zinc finger–RGG3 (fragment 4) domain exhibited the strongest binding to pncRNA-D (Fig. 2B), as reported in our previous study (5). In agreement with the full-length GST-TLS–binding assay, methylated GST-TLS fragment 4 failed to bind pncRNA-D. More detailed analyses indicated that the arginine residue Arg-394 in the RGG2 domain was consistently dimethylated (19), and the mutation of the other arginine residue (Arg-407) did not affect binding with pncRNA-D (Fig. S2). Because the zinc-finger domain binds RNA with low affinity (20), we excluded it and the RGG2 domain from further analyses and focused on investigating arginine residues in the RGG3 domain.
Figure 2.
The Arg-476 residue contributes to the pncRNA-D–binding site. A, TLS methylation sites and the fragments. TLS contains a total of 16 arginine residues (arrows) that can be methylated. B, biotinylated pncRNA-D was pulled down with unmethylated or methylated TLS fragments 1–4. The pncRNA-D–bound TLS fragments were detected by Coomassie Brilliant Blue staining; 20% of the protein was loaded as input. C, GST-TLS constructs with the substitution of arginine by alanine are indicated as mut1 (R476A and R481A), mut2 (R487A and R491A), and mut3 (R495A and R498A). D, arginine methylation levels of GST-TLS mutants (mut1, mut2, and mut3) were detected by an anti-dimethylarginine antibody. E, biotinylated pncRNA-D was incubated with TLS mutants (mut1, mut2, and mut3). The pncRNA-D–bound TLS was detected by an anti-TLS antibody. F, biotinylated pncRNA-D fragment 5′(1-1) was incubated with synthesized peptides (RGG3, TLS amino acids 471–503) with one of their arginine residues methylated (R476me and R481me). Samples were stained with the SimplyBlueTM SafeStain for analysis. The intensity of the bands was measured by ImageJ and applied in the box plot analysis. n = 3. *, p < 0.05. 20% of the protein used for RNA pulldown assays was loaded as input.
To examine whether arginine residues in the RGG3 domain participates in pncRNA-D binding, we designed GST-TLS constructs in which pairs of arginine residues were substituted with alanine (Fig. 2C). We then compared methylation levels among those constructs using an anti-dimethylarginine antibody (Fig. 2D). In these experiments, all samples were detected by Western blotting analyses, but both mut1 (R476A/R481A) and mut2 (R487A/R491A) mutants showed decreased methylation levels compared with the other constructs. In agreement with these methylation data, pncRNA-D pulldown assays showed that the mut1 mutant had a low binding level to pncRNA-D (Fig. 2E). Although the mut2 mutant showed a decreased methylation level, its binding intensity to pncRNA-D fluctuated with protein concentration (data not shown), suggesting that this pair of arginine residues is recognized by PRMT1, but does not mainly regulate the pncRNA-D interaction. Accordingly, a mutant with all potential methylated arginines outside the RGG3 domain and two arginines in the RGG3 domain (R476A and R481A) as mutations showed almost no binding to pncRNA-D (Fig. S2B, mutRGG3-2). Therefore, we investigated the Arg-476 and Arg-481 residues in further analysis using a synthetic TLS RGG3 peptide (amino acids 472–503; Table 1) with a methylated Arg-476 or Arg-481 residue. In these experiments, the biotinylated 5′(1-1) was used for RNA pulldown assays. The results revealed that the TLS peptide bearing the methylated Arg-476 (R476me) showed much less binding to the biotin-5′(1-1) than the WT and the Arg-481–methylated (R481me) peptides (Fig. 2F).
Table 1.
Synthetic sequence information
Boldface Rme indicates methylated arginine residue.
| Peptide TLS RGG3(472–503) | Sequence |
|---|---|
| WT | RRGGRGGYDRGGYRGRGGDRGGFRGGRGGGDR |
| R476me | RRGGRmeGGYDRGGYRGRGGDRGGFRGGRGGGDR |
| R481me | RRGGRGGYDRmeGGYRGRGGDRGGFRGGRGGGDR |
Together, these data indicate that the C terminus of TLS interacts with pncRNA-D and that Arg-476 within the RGG3 domain is a major determinant for binding to pncRNA-D. Methylation of the Arg-476 residue of TLS dramatically decreases its interaction with pncRNA-D.
Methylation of TLS inhibits pncRNA-D–mediated allosteric alterations
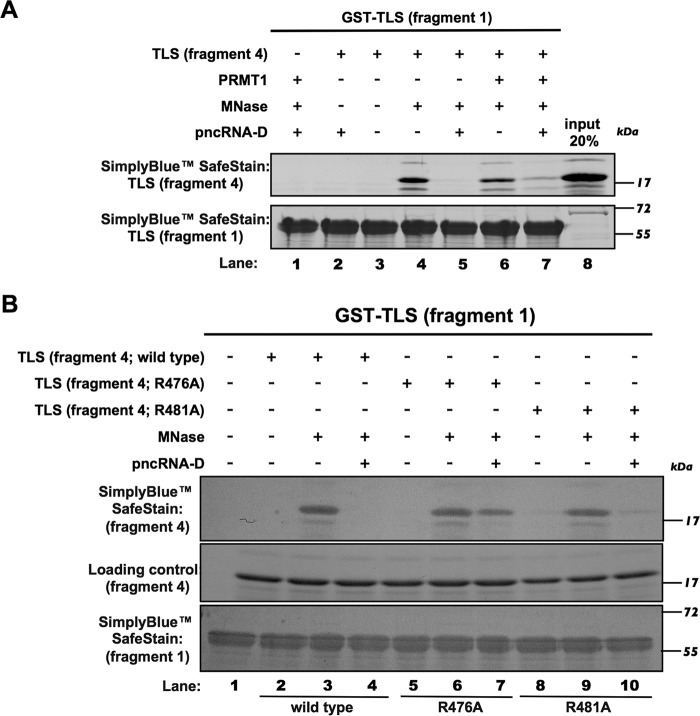
Our previous data showed interactions between the N and C termini of TLS in the absence of RNA (4). The allosteric alterations (dissociation of N and C termini) by RNA activate TLS, leading to the transcriptional repression of CCND1. Because methylation inhibits the RNA binding of TLS, the pncRNA-D–mediated allosteric alteration of TLS could be affected by arginine methylation. Therefore, we characterized the functional alteration of TLS in binding analyses of SYGQ-rich (fragment 1; N terminus) and RGG2–zinc finger–RGG3 (fragment 4; C terminus) domains. The GST tag was removed from GST-TLS fragment 4 using a cleavage enzyme, and the fragment 4 peptide was confirmed using an antibody against the RGG3 domain (data not shown). Bacterially expressed GST-TLS fragment 1 was purified from GSH-agarose beads and was used to pull down the tag-removed TLS fragment 4 to determine TLS-binding ability. It has been reported that bacterial RNAs from GST-TLS samples could activate TLS by disrupting the interactions between the N and C termini (4). Consistent with this finding, the subsequent experiment showed that the fragment 4 did not bind the bacterially expressed fragment 1 (Fig. 3A, lanes 2 and 3). However, after treatment with the calcium-dependent micrococcal nuclease (MNase), fragment 4 was able to bind fragment 1 (Fig. 3A, lane 4). Moreover, after inhibition of MNase activity by EGTA, these fragments dissociated in the presence of pncRNA-D (Fig. 3A, lane 5), although methylated fragment 4 remained bound to fragment 1 (Fig. 3A, lane 6), even in the presence of pncRNA-D (Fig. 3A, lane 7). Together, these data further indicate that the C terminus of TLS can act as a binding domain for pncRNA-D and that methylation inhibits interactions with pncRNA-D and disrupts RNA-mediated allosteric alteration of TLS.
Figure 3.
Methylated C-terminal end of TLS still binds to its N-terminal end in the presence of pncRNA-D. A, an N- and C-terminal TLS-binding assay was performed under the following conditions. Lane 1, GST-tagged fragment 1 (N-terminal end) following PRMT1 and MNase treatment without adding fragment 4 (C-terminal end) was used as a negative control. Lane 2, fragment 4 was incubated with pncRNA-D and then precipitated with fragment 1. Lane 3, GST-tagged fragment 1 was precipitated with fragment 4. Lane 4, fragment 4 was treated with MNase and then incubated with fragment 1. Lane 5, MNase-treated fragment 4 was incubated with fragment 1 in the presence of pncRNA-D after the inhibition of the MNase activity by EGTA. Lane 6, MNase-treated, methylated fragment 4 was incubated with fragment 1. Lane 7, MNase-treated, methylated fragment 4 was incubated with fragment 1 in the presence of pncRNA-D after the inhibition of the MNase activity. Lane 8, 20% of the protein of fragment 4 was loaded as input. B, GST-TLS fragment 1 was precipitated with TLS fragment 4 (WT, R476A, and R481A) (lanes 2, 5, and 8), fragment 4 with MNase treatment (lanes 3, 6, and 9), and fragment 4 in the presence of pncRNA-D after inhibition of MNase activity (lanes 4, 7, and 10). Samples were stained with the SimplyBlueTM SafeStain for analysis.
To examine whether the mutations in arginine residues affect the association of N- and C-terminal TLS fragments, we performed fragment-binding assays in which the fragment 4 contained either an R476A or R481A mutation. Both mutants of TLS fragment 4, as well as the WT, showed no binding with TLS fragment 1 due to bound bacterial RNAs (Fig. 3B, lanes 2, 5, and 8). TLS fragment 1 and 4 interactions were observed by treating with MNase (Fig. 3B, lanes 3, 6, and 9). After the inhibition of MNase activity, the WT and R481A mutant of fragment 4 exhibited dissociation from fragment 1 in the presence of pncRNA-D (Fig. 3B, lanes 4 and 10). However, the R476A mutant of fragment 4 still bound to fragment 1 in the presence of pncRNA-D (Fig. 3B, lane 7). These findings indicate that the substitution of Arg-476 with alanine disrupts the pncRNA-D–mediated allosteric alteration of TLS.
Methylation of TLS represses its binding to p300
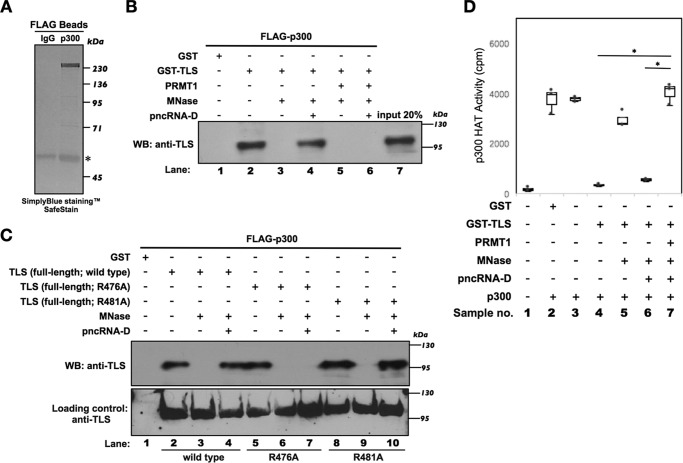
Because pncRNA-D did not block the associations of N- and C-terminal fragments of TLS when the C terminus was methylated, we examined the effects of this methylation on subsequent protein–protein interactions. We have already demonstrated the RNA-dependent binding of TLS to CBP/p300 (4). Because the RNA-mediated allosteric alteration of TLS is interfered with by arginine methylation, we hypothesize that the subsequent interactions between TLS and CBP/p300 could be blocked by arginine methylation. Thus, to examine the binding of TLS to CBP/p300 after methylation by PRMT1, we performed pulldown assays using the baculovirus-expressed human p300 protein (Fig. 4A). These experiments revealed that the binding of TLS to p300 (Fig. 4B, lane 2) was disrupted by MNase treatment (Fig. 4B, lane 3) and that the inhibition of MNase activity restores interactions between TLS and p300 in the presence of pncRNA-D (Fig. 4B, lane 4). However, methylated TLS failed to bind p300 (Fig. 4B, lane 5), even in the presence of pncRNA-D (Fig. 4B, lane 6). Collectively, these data demonstrated that methylation inhibits allosteric alteration of TLS by pncRNA-D and abolishes the interactions between TLS and CBP/p300.
Figure 4.
Methylation of TLS could restore the inhibition of pncRNA-D–mediated histone acetyltransferase activity. A, the FLAG bead–purified, baculovirus-expressed p300 was detected by SimplyBlueTM SafeStain for analysis. Mouse IgG served as a negative control. Star, heavy chains. B, the interaction between p300 and TLS was examined by a precipitation assay as in Fig. 3A, followed by Western blot analysis using an anti-TLS antibody. C, the interaction between p300 and TLS mutants (R476A and R481A) was examined by a precipitation assay as in Fig. 3B, followed by Western blot analysis using an anti-TLS antibody. D, p300 HAT activity was measured by incubation with WT or methylated TLS, histones, and [3H]acetyl-CoA. WT and methylated TLS were sequentially treated with MNase and then incubated with pncRNA-D for comparison of HAT activity. HAT activity was calculated as counts/min (cpm). Background reaction controls were included containing histones and [3H]acetyl-CoA excluding the p300 to measure background labeling radioactive counts. Data are presented in a box plot. n = 3. *, p < 0.01.
Next, we examined the interactions between TLS mutants and p300 in the presence or absence of RNAs. As shown in Fig. 4C, the WT and TLS mutants (R476A and R481A) bound to p300 due to the presence of bacterial RNA (Fig. 4C, lanes 2, 5, and 8). The interactions were disrupted after MNase treatment (Fig. 4C, lanes 3, 6, and 9) and restored by the inhibition of MNase activity, followed by adding pncRNA-D (Fig. 4C, lanes 4 and 10). However, compared with the WT and the R481A mutant of TLS, the R476A mutant showed no restoration of the binding with p300 in the presence of pncRNA-D (Fig. 4C, lane 7). These data provided further evidence that R476A mutation impairs the pncRNA-D–mediated allosteric alteration of TLS, subsequently causing defects in p300 interactions.
Methylation represses the TLS HAT inhibitory activity
RNA-induced allosteric alteration of TLS binds to CBP/p300 and inhibits CBP/p300 HAT activities (4). Because methylation of TLS repressed its binding to p300, we postulated that methylated TLS could repress the TLS-induced inhibition of p300 HAT.
By HAT assays using a highly active human His-tagged p300 recombinant protein, we measured HAT activity according to counts of radioactivity of filter-bound histones. The HAT assays with p300 and TLS indicated a significant inhibition of p300 HAT activity by TLS (Fig. 4D, no. 4) compared with that of p300 alone (Fig. 4D, no. 3). Moreover, the HAT-inhibitory activity of GST-TLS was abolished by treatment with MNase (Fig. 4D, no. 5) and was restored by inhibiting MNase in the presence of pncRNA-D (Fig. 4D, no. 6). However, methylated TLS did not reduce p300 HAT activity either in the presence or absence of pncRNA-D (Fig. 4D, no. 7). These data suggested that the HAT-inhibitory activity of TLS is repressed by methylation. The GST protein alone had no effects on p300 HAT activity (Fig. 4D, no. 2).
R476A exhibits no inhibitory function on CCND1 promoter activity
In our previous study, TLS showed inhibitory function on the CCND1 promoter and repressed CCND1 transcription (4). Because the R476A mutant of TLS subsequently affects pncRNA-D binding, which leads to defects in the inhibition of CBP/p300 HAT activities, we postulated that R476A of TLS may be associated with a functional change in the regulation of CCND1 promoter activity compared with WT TLS.
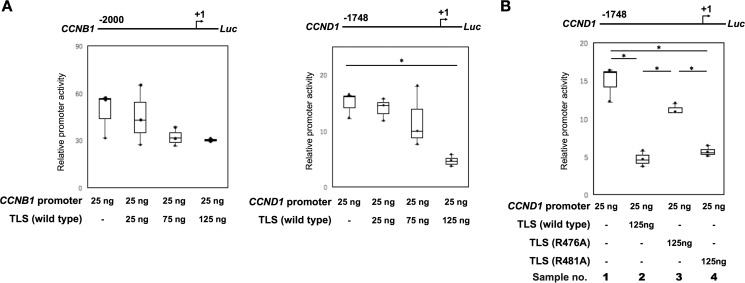
To examine the effects of TLS mutants on promoter activity, we performed a luciferase reporter assay. The CCND1-driven and the CCNB1-driven promoter plasmids were transfected into HeLa cells to measure the luciferase activities (Fig. 5A). The CCND1-driven promoter construct contains the pncRNA-D transcription region (from −1748 to +1). Co-transfection with TLS demonstrated that TLS inhibits CCND1 promoter activity in a concentration-dependent manner (Fig. 5A). TLS exerted much less inhibitory effect on CCNB1 promoter activity. Based on these data, we employed the most effective conditions of inhibition by TLS on CCND1 promoter for the TLS mutant analysis. As shown in Fig. 5B, TLS mutant R476A but not R481A exhibited a much smaller inhibitory effect on CCND1 promoter activity (Fig. 5B, no. 3 and 4), compared with WT TLS (Fig. 5B, no. 2). The sample containing CCND1 promoter plasmid without added transfection reagents was used as a negative control (data not shown).
Figure 5.
R476A mutation of TLS exhibits limited inhibitory function on CCND1-driven promoter. A, luciferase reporter assays for CCNB1 and CCND1 promoter activity in HeLa cells. B, HeLa cells were cotransfected with CCND1 promoter and Renilla control vectors, along with expression vector for TLS or TLS mutants (R476A and R481A). The transcriptional activity was determined by the ratio of the relative luciferase activity. Data are presented in a box plot. n = 3. *, p < 0.01.
Discussion
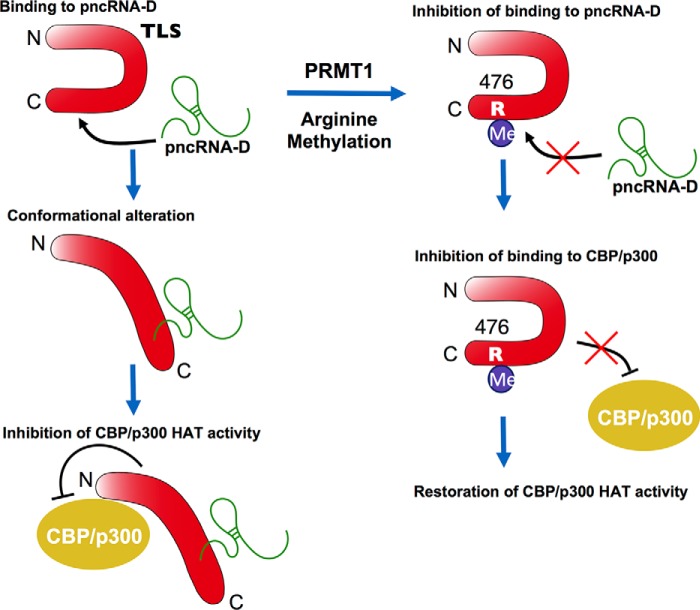
In this study, we demonstrated that PRMT1 methylates TLS in vitro and dramatically decreases pncRNA-D binding (Fig. 1). Upon lncRNA binding, TLS inhibits CBP/p300 HAT activities on the CCND1 promoter (4). The disruption of the binding of pncRNA-D to TLS causes defects of conformational alteration of TLS, reducing the binding to CBP/p300. Although the hypothesis remains still elusive, our luciferase assay clearly showed that the R476A TLS could not inhibit the CBP/p300 HAT activity. Therefore, we propose the model that arginine methylation of TLS abrogates TLS-mediated repression of CBP/p300 HAT activities (Fig. 6).
Figure 6.
Molecular mechanism of regulation of long noncoding RNA binding to TLS by arginine methylation with abrogation of TLS-induced inhibition of the CBP/p300 HAT activity. Binding of TLS to pncRNA-D induces conformational alteration of TLS. TLS inhibits the HAT activity of CBP/p300. PRMT1 catalyzes dimethylation on the Arg-476 of TLS. The methylation inhibits binding of TLS to pncRNA-D. The inhibition of the binding releases the CBP/p300 HAT activity from the inhibition by TLS.
Arginine methylation is one of the most abundant protein modifications and is frequently observed in heterogeneous nuclear ribonucleoprotein (21). Arginine methylation of TLS is involved in biological processes such as DNA damage response and transcriptional regulation. The present study revealed that the TLS-mediated inhibition of the p300 HAT activity can be regulated by arginine methylation (Fig. 4D). To investigate the molecular mechanism behind the restoration of CBP/p300 HAT activities by arginine methylation, RNA pulldown assays were performed, and the results revealed that methylated TLS exhibits almost no binding to pncRNA-D (Fig. 1D). Methylation of TLS seems to abolish binding not only to pncRNA-D but also to other RNA species, because we observed that methylated TLS displayed about 60% less binding to HeLa cell RNA fractions (Fig. S1). This highly reduced RNA binding may be due to the hypermethylation of TLS by PRMT1 in vitro. TLS binds to RNA with limited sequence specificity, but the preference of G-rich RNA binding partly results from arginine hydrogen bonding to guanine (20). Our previous study indicated that pncRNA-D containing the GGUG and GGU motifs could confer the binding preference to TLS (5). Therefore, disruption of the protein–RNA interactions by arginine methylation is considered to involve the feature that the addition of methyl groups to the guanidino nitrogen atoms of arginine may remove a potential hydrogen bond donor and have a negative effect on protein–RNA interactions.
Recently, it has been reported that the RGG domains of TLS play a pivotal role in RNA binding (20). It has also been shown that arginine methylation of TLS is mainly detected in the RGG3 domain (22). TLS-CHOP, a fusion protein produced by chromosomal translocation, exhibits no binding to PRMT1 and is hypomethylated, suggesting that the C terminus of TLS is necessary for mediating interactions with PRMT1 (23). Consistent with these findings, our data revealed that the RGG3 domain contributes to the pncRNA-D binding, and substitution of arginine residues with alanine in this domain produces a protein that displays a remarkable reduction of methylation levels (Fig. 2). However, our previous study indicated that the RGG2–zinc finger–RGG3 domain exhibits affinity with pncRNA-D that is higher than that with the RGG3 domain alone (5), suggesting that the RGG2 and zinc-finger domains enhance the binding cooperatively. Indeed, the zinc-finger domain has been demonstrated to enhance the protein–RNA interactions, but this domain alone shows no responsibility for the RNA binding activity of TLS (20, 24).
Our mass spectrometric analyses identified 16 methylated arginine residues in TLS (19). The arginine methylation pattern of TLS is variable, although four arginine residues (Arg-216, Arg-218, Arg-242, and Arg-394) exhibit consistent dimethylation. The biological function of these four dimethylated arginine residues of TLS remains unresolved, but two arginine residues (Arg-216 and Arg-218) are located in a consensus motif for the Akt recognition motif, indicating a possible cross-talk between arginine methylation and serine phosphorylation. The same Akt substrate motif is found in forkhead box O (FOXO) protein, and arginine methylation by PRMT1 within the motif blocks Akt-mediated phosphorylation and inhibits FOXO cytoplasmic localization and protein degradation (25). We found that substitution of the four arginine residues (Arg-216, Arg-218, Arg-242, and Arg-394) with alanine exhibited no binding alterations with pncRNA-D compared with WT TLS (data not shown), suggesting that these residues are involved in other biological activities rather than RNA binding. Herein, unlike the arginine residues examined in this study, we speculate that these four arginine residues might determine the protein structure and stability.
Allosteric alteration is one way that proteins control their functions. This process is triggered through interaction of proteins with ligands, including other proteins and nucleic acids. RNA-mediated conformational alteration of TLS is crucial for the activation of TLS. It is conceivable that the loss of RNA binding to methylated TLS results in defects in the TLS conformational alteration. For example, it has been reported that TLS binds to chromatin in an RNA-dependent manner, and PRMT1 knockdown reduces TLS arginine methylation level and increases the amount of chromatin-bound TLS (22). Taken together with our findings, it could be concluded that RNA-mediated TLS conformational alteration is required for subsequent protein–protein and protein–nucleic acid interactions and that methylation of TLS interferes with these processes.
TLS, EWS, and TAF15 (TET/FET) are members of the ribonucleoprotein family of RNA-binding proteins that share structural similarity in containing three RGG domains that are flanked by other functional domains. TET proteins may regulate gene expression in either a coactivation or a corepression manner through interactions with their binding partners. Moreover, the transcriptional regulation of the TET proteins may be controlled by arginine methylation. For example, EWS binds CBP/p300 for HNF-4–mediated transcriptional activation, but the coactivator activity of EWS is repressed by PRMT1 (26). In addition, deletion of the RGG domains of EWS abolishes interaction with PRMT1 (26), suggesting that the C terminus of EWS is required for PRMT1 recognition. Methylation of TAF15 by PRMT1 has been shown to positively regulate the expression of TAF15 target genes (27). All of the TET proteins have been shown to bind CBP/p300, and the interactions are RNA-mediated (4), suggesting similarities in the regulation of gene expression. Along with our findings, the C terminus of TET proteins is required for PRMT1 recognition, and gene expression could be regulated by methylation of the TET proteins.
Transcription is supposed to be regulated by the interactions between divergent transcriptional factors and cofactors. Our present and previous studies showed that TLS specifically binds and inhibits CBP/p300 HAT activities, inducing a subsequent transcriptional repression of CCND1, indicating that TLS works as a corepressor (4) (Fig. 5B). We also reported that TLS complex with PRMT1 promotes transcription from the survivin promoter (19). Moreover, survivin promoter activity was slightly increased with TLS alone, suggesting that TLS might also act as a coactivator. These findings show that TLS should work as either a corepressor or a coactivator, depending on the promoter.
It has been demonstrated that TLS promotes cell cycle arrest in the G1 phase by reducing the expression of CCND1 (4, 28). Knockdown of PRMT1 significantly decreases the cancer cells in the S phase of the cell cycle (29). The repression of arginine methylation has also been reported as a possible therapeutic target against cancers (29–31). The dysregulation of PRMTs probably leads to hypermethylation of TLS and disruption of the inhibitory activity of TLS against CBP/p300 HAT activities, causing the overexpression of CCND1, which accelerates the proliferation of cancer cells. Therefore, our findings could serve as a model system that allows further experiments to examine the molecular mechanisms relating to the methylation of TLS with cell cycle regulation and therapeutics against cancers.
In conclusion, we have demonstrated that arginine methylation on TLS inhibits its binding to lncRNA. Binding of TLS to pncRNA-D leads to conformational alterations that facilitate inhibitory interactions with CBP/p300. The CBP/p300 HAT activities are significantly inhibited by the binding of TLS but are restored by TLS methylation, which disrupts RNA–protein interactions. These data present a novel link of lncRNA activity to epigenetic regulations utilizing protein methylations in the human genome.
Experimental procedures
Antibodies and reagents
Mouse anti-TLS/FUS antibody (15/TLS; 611384) was purchased from BD Biosciences. Rabbit anti-GST antibody (X5; sc-459) was purchased from Santa Cruz Biotechnology, Inc. Rabbit anti-asymmetric dimethyl-arginine antibody (catalog no. 39232) was purchased from Active Motif. Anti-mono- and dimethyl-arginine antibody (7E6; ab412) was purchased from Abcam. Insect cells expressing His-tagged human histone acetyltransferase p300 (AG-40T-0023) were purchased from AdipoGen Life Sciences. SAM was purchased from Sigma-Aldrich. EGTA was purchased from Sigma-Aldrich. The RNase inhibitor (SIN-201) was purchased from Toyobo Life Science, and PreScission Protease (catalog no. 27084301) was purchased from GE Healthcare. Histone from calf thymus (type II-A) was purchased from Sigma-Aldrich.
cDNA constructs
The vector pASK-IBA-5plus (IBA) was used to construct plasmids expressing human Strep-tagged PRMT1. Specifically, the full-length TLS DNA sequence was inserted into the pGEX6p-1 vector via its multiple cloning sites (BamHI/XhoI), and the GST-TLS construct was generated. Subsequently, full-length human p300 was cloned into pFastBAC vector (Thermo Fisher) and was modified with an in-frame C-terminal FLAG tag. The resulting GST-TLS mutants were constructed by replacing Arg with Ala in pGEX-TLS using KOD Plus mutagenesis kits (Toyobo). The full-length pncRNA-D sequence was inserted into pcDNA3.1(+) vector (Thermo Fisher) via its multiple cloning sites (BamHI/XhoI) and linearized by enzyme cleavage (XhoI) for downstream in vitro transcription. The CCND1-driven promoter sequence (from −1748 to +1) was inserted into pGL4.10[luc2] vector (Promega) via its multiple cloning sites (KpnI/HindIII) for a reporter assay. The CCNB1-driven promoter sequence (from −2000 to +1) was inserted into pGL4.10[luc2] vector via the same multiple cloning sites as CCND1 for the reporter assay. The full-length TLS DNA sequence was inserted into the pcDNA3.1(+) vector via its multiple cloning sites (BamHI/XhoI), and the resulting TLS mutants were constructed by replacing Arg with Ala using KOD Plus-mutagenesis kits. Primer pairs are listed in Table 2.
Table 2.
Primer pairs for site-directed mutagenesis
| Primer pair | Sequence |
|---|---|
| GST-TLS mutant primer pairs | |
| mut1 | |
| Forward | 5′-cgtggtggcgcaggaggctatgatgcaggcggcta-3′ |
| Reverse | 5′-acgatcatccccgtagttaccccccatgtgagagcca-3′ |
| mut2 | |
| Forward | 5′-ggctaccggggcgccggcggggacgctggaggc-3′ |
| Reverse | 5′-gcctcgatcatagcctcctctgccaccacgacg-3′ |
| mut3 | |
| Forward | 5′-cggaggcttcgccgggggcgccggtggtggg-3′ |
| Reverse | 5′-cggtccccgccgcggccccggtagc-3′ |
| TLS mutant primer pairs for luciferase assay | |
| R476A | |
| Forward | 5′-cgtggtggcgcaggaggctatgatcgaggcggcta-3′ |
| Reverse | 5′-acgatcatccccgtagttaccccccatgtgagagcca-3′ |
| R481A | |
| Forward | 5′-cgtggtggcagaggaggctatgatgcaggcggcta-3′ |
| Reverse | 5′-acgatcatccccgtagttaccccccatgtgagagcca-3′ |
Recombinant protein overexpression and purification
Full-length GST-TLS and TLS fragments were overexpressed and purified as described previously (32). Briefly, constructs were transfected into Escherichia coli strain Y1090, and transformants were grown at 37 °C in Luria–Bertani medium containing ampicillin (0.1 mg/ml). Protein expression was induced with 1 mm isopropyl β-d-1-thiogalactopyranoside at an OD of 0.5 for 3 h at 20 °C. Cells were then harvested by centrifugation (3,500 × g for 15 min), and cell pellets were lysed by sonication in whole-cell extract (WCE) buffer containing 25 mm HEPES (pH 7.9), 50 mm NaCl, 2.5 mm MgCl2, 0.1 mm EDTA, 0.05% Triton X-100, and proteinase inhibitors. After ultracentrifugation at 40,000 × g for 40 min, supernatants containing the expressed proteins were purified using glutathione-agarose beads (Sigma-Aldrich). Strep-PRMT1 was then induced by incubating with 200 ng/ml anhydrotetracycline (IBA) for 4.5 h at 28 °C, and the protein was collected and purified using Strep-Tactin Sepharose (IBA). To express full-length p300, Sf9 insect cells (Thermo Fisher) were grown in Grace's insect medium (Thermo Fisher) containing 10% fetal bovine serum. Suspensions of 1–2 × 106 cells/ml were infected with passage 3 virus stock at a multiplicity of infection of 0.5. Cells were then harvested after 72 h and lysed by sonication. Protein concentrations were determined using BCA protein assay kits (Thermo Fisher).
In vitro methylation
In vitro methylation reactions were performed as described previously (33). Briefly, purified GST-TLS (3 μg) was incubated with purified Strep-tagged PRMT1 (3 μg) in the absence or presence of SAM (2 mm) for 2 h at 30 °C. Reactions were stopped by the addition of SDS sample buffer containing 0.25 m Tris-HCl (pH 6.8), 4% SDS, 10% 2-mercaptoethanol, and 20% glycerol, and the mixtures were then boiled at 100 °C for 2 min followed by SDS-PAGE and Western blot analysis.
RNA pulldown assays
RNA pulldown assays were performed as described previously (5). Briefly, in vitro transcription of full-length pncRNA-D was performed using MEGAscript kits (Ambion). The linearized pcDNA3.1(+) plasmid containing the full-length pncRNA-D sequence was used for a transcription template. To obtain the biotinylated pncRNA-D, the Bio-16-UTP (2 μl, 20 nmol; Ambion) was added to the reaction mixture. Dynabeads-M280 (Thermo Fisher) were then washed with PBS containing 0.02% Tween 20, and 10 pmol of biotinylated pncRNA-D was added to the beads and incubated for 15 min at room temperature with rotation. Subsequently, beads were incubated with WT or methylated purified protein (or protein lysate from mutant pulldown experiments) for 1 h at 4 °C with rotation. Beads were then washed three times in 1 ml of WCE buffer and resuspended in SDS sample buffer and boiled at 100 °C for 2 min. Finally, the Dynabeads were removed, and supernatants were applied for Western blotting and silver-staining analyses. For pncRNA-D fragment 5′(1-1) pulldown assays, Dynabeads-M280 were prepared in PBS with 0.02% Tween 20 and were resuspended in WCE buffer (10 μl) with 2 μl of RNase inhibitor mixture containing 10 μg of tRNA. Biotinylated RNA oligonucleotide of 5′(1-1) (100 μm) was added to the Dynabeads suspensions and incubated at room temperature for 15 min with rotation. The peptide (5 μg) was added to the Dynabeads mixtures and initially incubated at room temperature for 10 min, followed with additional incubation for 50 min at 4 °C. After washing three times with 1 ml of WCE buffer, the Dynabeads were resuspended in SDS sample buffer and boiled at 100 °C for 1 min. Samples were analyzed with 15% SDS-polyacrylamide gel and stained with the SimplyBlueTM SafeStain (Thermo Fisher). Images in TIF format were quantified using the ImageJ software.
Proteinase digestion
GST tags were eliminated using PreScission proteinase (GE Healthcare) according to the manufacturer's instructions. Briefly, 1-ml lysates containing GST-TLS fragment 4 were incubated with 400 μl of GSH-agarose beads (200-μl bed volume) and purified for 2 h at 4 °C with rotation, followed by three washes with 1 ml of WCE buffer and then two washes in 1 ml of digestion buffer containing 50 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, and 1 mm DTT (pH 7.0). Aliquots of agarose beads were resuspended in 200 μl of digestion buffer containing 20 units of enzyme and incubated at 4 °C for 18 h with rotation. Digested mixtures were then spin-filtered (20,000 × g, 1 min) to collect the protein. The presence of digested proteins was confirmed using SDS-PAGE, SimplyBlue stainingTM SafeStain (Thermo Fisher), and/or Western blotting.
N- and C-terminal TLS-binding assays
The GST tag of GST-TLS fragment 4 was removed before the following treatments. The TLS fragment 4 was treated with MNase (40 units) at 37 °C for 1 h, and the reaction was terminated by the addition of EGTA (10 mm). Subsequently, both methylated and unmethylated TLS fragment 4 (C terminus; 10 μg) were incubated with GST-TLS fragment 1 (fragment 1; N terminus), which was purified from 100-μl lysates in the presence or absence of 10 pmol of pncRNA-D for 2 h at 4 °C with rotation. For the mutation binding assays, the GST tag of WT and mutants of GST-TLS fragment 4 was removed before the MNase treatment. Incubation with pncRNA-D (10 pmol) was performed on ice for 30 min after inhibition of MNase activity by EGTA (10 mm). The fragment 4 (WT and mutants) was then incubated overnight at 4 °C with GST-TLS fragment 1, which was purified from 50 μl of lysate. All samples were washed three times in 1-ml aliquots of WCE buffer and then suspended in SDS sample buffer for SDS-PAGE and SimplyBlueTM SafeStain (Thermo Fisher) analyses.
TLS and p300-binding assays
Reactions were performed by capturing baculovirus-expressed FLAG-tagged p300 on anti-FLAG-M2 affinity agarose beads (Sigma-Aldrich). Affinity agarose beads (25 μl) were incubated with 50-μl aliquots of p300 lysates at 4 °C for 1 h with rotation. After washing three times with 1 ml of WCE buffer, the beads were incubated with unmethylated or methylated TLS in the presence of pncRNA-D (10 pmol). For the mutation-binding assays, on-bead FLAG-tagged p300 (50 μl of lysate) was incubated with full-length TLS (WT and mutants), which was purified from 100 μl of lysate for 2 h at 4 °C with rotation. Incubation with pncRNA-D (10 pmol) was performed on ice for 30 min after inhibition of MNase activity by EGTA (10 mm). The beads were then washed three times with 1 ml of WCE buffer, and reactions were terminated by adding SDS sample buffer. Supernatants were then used for Western blotting analyses.
HAT activity assays
Pulldown HAT assays were performed as described previously (4), with modifications. Briefly, samples of unmethylated and methylated GST-TLS were eluted from GST beads and then incubated with or without pncRNA-D for 1 h. Samples were washed three times in HAT assay buffer containing 50 mm Tris-HCl (pH 8.0), 10% glycerol, 0.1 mm EDTA, and 1 mm DTT and then incubated with 100 ng of His-tagged p300 (AdipoGen Life Sciences) on ice for 1 h. Histones (10 μg; type II-A; Sigma-Aldrich) and [3H]acetyl-CoA (0.5 μCi; PerkinElmer) were then added, and reaction mixtures were incubated at 30 °C for 30 min. After brief centrifugation, supernatants were spotted onto p81 Whatman filters (GE Healthcare). Filters were then dried at room temperature for 20 min and were washed in 50 mm sodium carbonate/bicarbonate buffer (pH 9.3) three times for 20 min each. Subsequently, filters were rinsed twice for 20 s in acetone and dried for 20 min at room temperature. Dried filters were then placed into scintillation vials with 2 ml of Ultima Gold XR liquid (Packard BioScience). Finally, [3H]acetylated histones were quantified according to radioactive counts/min using a liquid scintillation counter (Beckman). Background reaction controls were included containing histones and [3H]acetyl-CoA excluding the p300 to measure background labeling radioactive counts.
MNase treatment
GST-TLS fragment 4 was purified using GSH-agarose beads, and the agarose beads were treated with 40 units of MNase in buffer containing 20 mm Tris-HCl (pH 8.0), 5 mm NaCl, and 2.5 mm CaCl2 at 37 °C for 1 h. Reactions were terminated by the addition of 10 mm EGTA.
Western blot analysis
Western blot analyses were performed as described previously (5). Briefly, after electroblotting of proteins from SDS-PAGE gels, nitrocellulose membranes (Bio-Rad) were incubated with antibodies at 1:1,000 dilution for 2 h at room temperature. Membranes were washed three times with PBS containing 0.02% Tween 20 (PBST) for 5 min each and then incubated with secondary anti-mouse (Dako, P0161) or anti-rabbit horseradish peroxidase–conjugated IgG (Cell Signaling, 70745) antibodies diluted 1:4,000 in 1% skim milk in PBST. Signals were detected using SuperSignal West Pico substrate (Thermo Fisher).
Luciferase reporter assay
HeLa cells were cultured as described previously (19). Transfection was performed using Lipofectamine 3000 (Invitrogen) according to the manufacturer's protocol. HeLa cells were seeded in a 96-well plate at 1.0 × 104 cells/well. Twenty-four hours later, cells were transfected with a plasmid mixture containing 25 ng of CCND1-driven firefly luciferase reporter construct, 25 ng of Renilla luciferase construct (pRL-TK; Promega), and 125 ng of TLS or 125 ng of mutated TLS expression plasmid. Total amounts of DNA (175 ng) were kept constant in all experiments by using corresponding empty vectors. The cells were harvested and lysed 48 h after transfection, and luciferase activity was determined using the Dual-Luciferase assay kit (Promega). Luciferase activities were normalized to Renilla luciferase activities. The promoter reporter DNA without adding transfection reagent was used as negative control. Data are presented in a box-and-whisker plot of three independent results.
Statistics
Data are expressed as mean ± S.D. Pairwise differences were identified using Student's t test and were considered significant when p < 0.05.
Author contributions
R. K. and W. C. conceived the concept and designed the experiments. W. C. conducted the RNA pulldown assay and Western blot analysis, plasmid construction, protein-binding assay, and HAT assay. W. C. and R. Y. conducted the luciferase reporter gene assay. N. U. purified GST-TLS. W. C., R. Y., and R. K. wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
This work was supported in part by a grant-in-aid for “Support Project of Strategic Research Center in Private Universities” from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) to Saitama Medical University Research Center for Genomic Medicine and by KAKENHI from JSPS to MEXT of Japan (KibanB Grants 22390057 and 25293073, KibanC Grant 18K06939). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1 and S2.
- TLS
- translocated in liposarcoma
- FUS
- fused in sarcoma
- GST
- glutathione S-transferase
- HAT
- histone acetyltransferase
- MNase
- micrococcal nuclease
- lncRNA
- long noncoding RNA
- pncRNA
- promoter-associated ncRNA
- PRMT
- protein arginine methyltransferase
- WB
- Western blotting
- WCE
- whole-cell extract
- CBP
- CREB-binding protein
- CREB
- cAMP-response element–binding protein
- SAM
- S-adenosylmethionine
- EWS
- Ewing sarcoma.
References
- 1. Lagier-Tourenne C., Polymenidou M., and Cleveland D. W. (2010) TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum. Mol. Genet. 19, R46–R64 10.1093/hmg/ddq137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhou Y., Liu S., Oztürk A., and Hicks G. G. (2014) FUS-regulated RNA metabolism and DNA damage repair: implications for amyotrophic lateral sclerosis and frontotemporal dementia pathogenesis. Rare Dis. 2, e29515 10.4161/rdis.29515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhou B. B., and Elledge S. J. (2000) The DNA damage response: putting checkpoints in perspective. Nature 408, 433–439 10.1038/35044005 [DOI] [PubMed] [Google Scholar]
- 4. Wang X., Arai S., Song X., Reichart D., Du K., Pascual G., Tempst P., Rosenfeld M. G., Glass C. K., and Kurokawa R. (2008) Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454, 126–130 10.1038/nature06992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yoneda R., Suzuki S., Mashima T., Kondo K., Nagata T., Katahira M., and Kurokawa R. (2016) The binding specificity of Translocated in LipoSarcoma/FUsed in Sarcoma with lncRNA transcribed from the promoter region of cyclin D1. Cell Biosci. 6, 4 10.1186/s13578-016-0068-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ramos A., Hollingworth D., and Pastore A. (2003) G-quartet-dependent recognition between the FMRP RGG box and RNA. RNA 9, 1198–1207 10.1261/rna.5960503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Phan A. T., Kuryavyi V., Darnell J. C., Serganov A., Majumdar A., Ilin S., Raslin T., Polonskaia A., Chen C., Clain D., Darnell R. B., and Patel D. J. (2011) Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat. Struct. Mol. Biol. 18, 796–804 10.1038/nsmb.2064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dormann D., Madl T., Valori C. F., Bentmann E., Tahirovic S., Abou-Ajram C., Kremmer E., Ansorge O., Mackenzie I. R., Neumann M., and Haass C. (2012) Arginine methylation next to the PY-NLS modulates transportin binding and nuclear import of FUS. EMBO J. 31, 4258–4275 10.1038/emboj.2012.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bedford M. T., and Clarke S. G. (2009) Protein arginine methylation in mammals: who, what, and why. Mol. Cell 33, 1–13 10.1016/j.molcel.2008.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tang J., Frankel A., Cook R. J., Kim S., Paik W. K., Williams K. R., Clarke S., and Herschman H. R. (2000) PRMT1 is the predominant type I protein arginine methyltransferase in mammalian cells. J. Biol. Chem. 275, 7723–7730 10.1074/jbc.275.11.7723 [DOI] [PubMed] [Google Scholar]
- 11. Yang Y., Lu Y., Espejo A., Wu J., Xu W., Liang S., and Bedford M. T. (2010) TDRD3 is an effector molecule for arginine-methylated histone marks. Mol. Cell 40, 1016–1023 10.1016/j.molcel.2010.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mostaqul Huq M. D., Gupta P., and Wei L. N. (2008) Post-translational modifications of nuclear co-repressor RIP140: a therapeutic target for metabolic diseases. Curr. Med. Chem. 15, 386–392 10.2174/092986708783497382 [DOI] [PubMed] [Google Scholar]
- 13. Yamaguchi A., and Kitajo K. (2012) The effect of PRMT1-mediated arginine methylation on the subcellular localization, stress granules, and detergent-insoluble aggregates of FUS/TLS. PLoS One 7, e49267 10.1371/journal.pone.0049267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Belyanskaya L. L., Gehrig P. M., and Gehring H. (2001) Exposure on cell surface and extensive arginine methylation of ewing sarcoma (EWS) protein. J. Biol. Chem. 276, 18681–18687 10.1074/jbc.M011446200 [DOI] [PubMed] [Google Scholar]
- 15. Rappsilber J., Friesen W. J., Paushkin S., Dreyfuss G., and Mann M. (2003) Detection of arginine dimethylated peptides by parallel precursor ion scanning mass spectrometry in positive ion mode. Anal. Chem. 75, 3107–3114 10.1021/ac026283q [DOI] [PubMed] [Google Scholar]
- 16. Pahlich S., Bschir K., Chiavi C., Belyanskaya L., and Gehring H. (2005) Different methylation characteristics of protein arginine methyltransferase 1 and 3 toward the Ewing sarcoma protein and a peptide. Proteins 61, 164–175 10.1002/prot.20579 [DOI] [PubMed] [Google Scholar]
- 17. Hung C. J., Lee Y. J., Chen D. H., and Li C. (2009) Proteomic analysis of methylarginine-containing proteins in HeLa cells by two-dimensional gel electrophoresis and immunoblotting with a methylarginine-specific antibody. Protein J. 28, 139–147 10.1007/s10930-009-9174-3 [DOI] [PubMed] [Google Scholar]
- 18. Takahama K., Kino K., Arai S., Kurokawa R., and Oyoshi T. (2011) Identification of Ewing's sarcoma protein as a G-quadruplex DNA- and RNA-binding protein. FEBS J. 278, 988–998 10.1111/j.1742-4658.2011.08020.x [DOI] [PubMed] [Google Scholar]
- 19. Du K., Arai S., Kawamura T., Matsushita A., and Kurokawa R. (2011) TLS and PRMT1 synergistically coactivate transcription at the survivin promoter through TLS arginine methylation. Biochem. Biophys. Res. Commun. 404, 991–996 10.1016/j.bbrc.2010.12.097 [DOI] [PubMed] [Google Scholar]
- 20. Ozdilek B. A., Thompson V. F., Ahmed N. S., White C. I., Batey R. T., and Schwartz J. C. (2017) Intrinsically disordered RGG/RG domains mediate degenerate specificity in RNA binding. Nucleic Acids Res. 45, 7984–7996 10.1093/nar/gkx460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Boffa L. C., Karn J., Vidali G., and Allfrey V. G. (1977) Distribution of NG, NG,-dimethylarginine in nuclear protein fractions. Biochem. Biophys. Res. Commun. 74, 969–976 10.1016/0006-291X(77)91613-8 [DOI] [PubMed] [Google Scholar]
- 22. Yang L. (2015) Physiological function of FUS: an RNA binding protein in motor neuron disease, Ph.D. thesis, University of Kentucky, Lexington, KY [Google Scholar]
- 23. Fujimoto K., Arai S., Matsubara M., Du K., Araki Y., Matsushita A., and Kurokawa R. (2013) Implicated role of liposarcoma related fusion oncoprotein TLS-CHOP in the dysregulation of arginine-specific methylation through PRMT1. Cell Biol. 1, 18–23 10.11648/j.cb.20130102.11 [DOI] [Google Scholar]
- 24. Burdach J., O'Connell M. R., Mackay J. P., and Crossley M. (2012) Two-timing zinc finger transcription factors liaising with RNA. Trends Biochem. Sci. 37, 199–205 10.1016/j.tibs.2012.02.001 [DOI] [PubMed] [Google Scholar]
- 25. Yamagata K., Daitoku H., Takahashi Y., Namiki K., Hisatake K., Kako K., Mukai H., Kasuya Y., and Fukamizu A. (2008) Arginine methylation of FOXO transcription factors inhibits their phosphorylation by Akt. Mol. Cell 32, 221–231 10.1016/j.molcel.2008.09.013 [DOI] [PubMed] [Google Scholar]
- 26. Araya N., Hiraga H., Kako K., Arao Y., Kato S., and Fukamizu A. (2005) Transcriptional down-regulation through nuclear exclusion of EWS methylated by PRMT1. Biochem. Biophys. Res. Commun. 329, 653–660 10.1016/j.bbrc.2005.02.018 [DOI] [PubMed] [Google Scholar]
- 27. Jobert L., Argentini M., and Tora L. (2009) PRMT1 mediated methylation of TAF15 is required for its positive gene regulatory function. Exp. Cell Res. 315, 1273–1286 10.1016/j.yexcr.2008.12.008 [DOI] [PubMed] [Google Scholar]
- 28. Brooke G. N., Culley R. L., Dart D. A., Mann D. J., Gaughan L., McCracken S. R., Robson C. N., Spencer-Dene B., Gamble S. C., Powell S. M., Wait R., Waxman J., Walker M. M., and Bevan C. L. (2011) FUS/TLS is a novel mediator of androgen-dependent cell-cycle progression and prostate cancer growth. Cancer Res. 71, 914–924 10.1158/0008-5472.CAN-10-0874 [DOI] [PubMed] [Google Scholar]
- 29. Yoshimatsu M., Toyokawa G., Hayami S., Unoki M., Tsunoda T., Field H. I., Kelly J. D., Neal D. E., Maehara Y., Ponder B. A., Nakamura Y., and Hamamoto R. (2011) Dysregulation of PRMT1 and PRMT6, Type I arginine methyltransferases, is involved in various types of human cancers. Int. J. Cancer 128, 562–573 10.1002/ijc.25366 [DOI] [PubMed] [Google Scholar]
- 30. Scaramuzzino C., Monaghan J., Milioto C., Lanson N. A. Jr., Maltare A., Aggarwal T., Casci I., Fackelmayer F. O., Pennuto M., and Pandey U. B. (2013) Protein arginine methyltransferase 1 and 8 interact with FUS to modify its sub-cellular distribution and toxicity in vitro and in vivo. PLoS One 8, e61576 10.1371/journal.pone.0061576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Morettin A., Baldwin R. M., and Côté J. (2015) Arginine methyltransferases as novel therapeutic targets for breast cancer. Mutagenesis 30, 177–189 10.1093/mutage/geu039 [DOI] [PubMed] [Google Scholar]
- 32. Takahama K., Takada A., Tada S., Shimizu M., Sayama K., Kurokawa R., and Oyoshi T. (2013) Regulation of telomere length by G-quadruplex telomere DNA- and TERRA-binding protein TLS/FUS. Chem. Biol. 20, 341–350 10.1016/j.chembiol.2013.02.013 [DOI] [PubMed] [Google Scholar]
- 33. Fujimoto K., and Kurokawa R. (2014) Development of a mouse monoclonal antibody for the detection of asymmetric dimethylarginine of Translocated in LipoSarcoma/FUsed in Sarcoma and its application in analyzing methylated TLS. Cell Biosci. 4, 77 10.1186/2045-3701-4-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.