Alkene MOs are of interest for their potential roles in industrial biocatalysis, most notably for the stereoselective synthesis of epoxides. Wild-type bacteria that grow on alkenes have high activities for alkene oxidation but are problematic for biocatalysis, since they tend to consume the epoxide products. Using recombinant biocatalysts is the obvious alternative, but a major bottleneck is the low activities of recombinant alkene MOs. Here, we provide new high-activity recombinant biocatalysts for alkene oxidation, and we provide insights into how to further improve these systems.
KEYWORDS: monooxygenase, alkene, ethene, propene, Mycobacterium, biocatalysis, heterologous gene expression
ABSTRACT
Alkene monooxygenases (MOs) are soluble di-iron-containing enzymes found in bacteria that grow on alkenes. Here, we report improved heterologous expression systems for the propene MO (PmoABCD) and ethene MO (EtnABCD) from Mycobacterium chubuense strain NBB4. Strong functional expression of PmoABCD and EtnABCD was achieved in Mycobacterium smegmatis mc2155, yielding epoxidation activities (62 and 27 nmol/min/mg protein, respectively) higher than any reported to date for heterologous expression of a di-iron MO system. Both PmoABCD and EtnABCD were specialized for the oxidation of gaseous alkenes (C2 to C4), and their activity was much lower on liquid alkenes (C5 to C8). Despite intensive efforts to express the complete EtnABCD enzyme in Escherichia coli, this was not achieved, although recombinant EtnB and EtnD proteins could be purified individually in soluble form. The biochemical function of EtnD as an oxidoreductase was confirmed (1.36 μmol cytochrome c reduced/min/mg protein). Cloning the EtnABCD gene cluster into Pseudomonas putida KT2440 yielded detectable epoxidation of ethene (0.5 nmol/min/mg protein), and this could be stimulated (up to 1.1 nmol/min/mg protein) by the coexpression of cpn60 chaperonins from either Mycobacterium spp. or E. coli. Successful expression of the ethene MO in a Gram-negative host was validated by both whole-cell activity assays and peptide mass spectrometry of induced proteins seen on SDS-PAGE gels.
IMPORTANCE Alkene MOs are of interest for their potential roles in industrial biocatalysis, most notably for the stereoselective synthesis of epoxides. Wild-type bacteria that grow on alkenes have high activities for alkene oxidation but are problematic for biocatalysis, since they tend to consume the epoxide products. Using recombinant biocatalysts is the obvious alternative, but a major bottleneck is the low activities of recombinant alkene MOs. Here, we provide new high-activity recombinant biocatalysts for alkene oxidation, and we provide insights into how to further improve these systems.
INTRODUCTION
Gaseous hydrocarbons can be used as a sole source of carbon and energy by bacteria. The best studied of these are the methanotrophs (1–3), which are alphaproteobacteria and gammaproteobacteria that specialize in growth on methane. Growth on larger volatile hydrocarbons (C2 to C5) involves diverse proteobacteria (4–8) and actinobacteria (9–16); these bacteria differ fundamentally from methanotrophs in their physiology and biochemistry, especially in their more generalist approach to carbon sources. Both methanotrophs and other “gasotrophs” are important in biogeochemistry, since they influence the types and quantities of hydrocarbons emitted into the atmosphere from terrestrial and aquatic habitats (17–19).
The metabolic pathways operating in alkane- and alkene-oxidizing bacteria are diverse, but a common theme (at least in aerobes) is that the initial oxidation is always mediated by a monooxygenase (MO). The MOs are a fascinating enzyme family, not only due to their significance for biogeochemistry, but also due to their applications in bioremediation (20–22) and biocatalysis (23–26). Bacterial monooxygenases are diverse but can be broadly classified into six types according to their cofactor requirements and/or subcellular location, as follows: heme-dependent cytochrome p450s (27), soluble di-iron MOs (SDIMOs) (28), copper-containing membrane-located MOs (CuMMOs) (29), flavin-containing MOs (30), pterin-dependent MOs (31), and cofactor-independent MOs (32).
The SDIMOs are of special interest due to their high sequence diversity, broad substrate range, and useful applications (33–38); these enzymes can be divided into six distinct groups based on sequences and substrate range (37, 38). The group 1 and group 2 SDIMOs have six gene components and are phenol MOs (39) and toluene MOs (40), respectively. The group 3 SDIMOs have five or six gene components and include soluble methane monooxygenases (sMMOs) (41), plus a few MOs which oxidize larger gaseous alkanes (42, 43). The group 4 SDIMOs are four-component alkene monooxygenases from bacteria that grow on ethene (44, 45) and/or propene (46, 47). The group 5 and 6 SDIMOs have four components and act on gaseous alkanes (13, 48) and other organic compounds, like phenol (49) and acetone (50).
Despite their sequence and substrate diversity, all the SDIMOs have similar biochemistries (28, 41). Electrons are transferred from NADH to an oxidoreductase protein that contains flavin and iron-sulfur clusters, and thence to a catalytic hydroxylase (made of 2 to 3 proteins) that contains the binuclear iron active site (8, 51–53). In the active site, one oxygen atom in O2 is reduced to water, while the other is activated to a high-energy state and attacks the substrate. The catalytic activity is facilitated by a small cofactor-independent “coupling protein,” and depending on the SDIMO family, other proteins, such as ferredoxins, may also be part of the MO-enzyme complex.
Our understanding of SDIMO enzymology has been greatly facilitated by the ability to express these enzymes in hosts such as Escherichia coli (39, 40, 54, 55). Another major motivation for heterologous expression of SDIMOs is to facilitate their use as biocatalysts, since wild-type strains have disadvantages, such as slow or difficult growth, awkward regulation of gene expression, the presence of interfering enzyme activities, and further metabolism of the desired products (typically alcohols or epoxides). Unfortunately, some SDIMOs are resistant to this approach and make insoluble and/or inactive proteins in E. coli, requiring the use of more exotic heterologous hosts for functional expression (56–60). A notable recent breakthrough in this field was the finding that codon optimization and coexpression of chaperonins enabled functional expression of a group 5 SDIMO from Mycobacterium smegmatis in E. coli (61); this approach suggests a way forward for the expression of related 4-component SDIMOs, like the ethene and propene MOs.
Mycobacterium chubuense NBB4 is unique among hydrocarbon-oxidizing bacteria because it contains four different SDIMOs, in addition to a copper-containing MO, a P450, and an alkB homolog (38, 62, 63). Strain NBB4 has two group 4 SDIMOs (etnABCD and pmoABCD), an atypical group 3 SDIMO (smoXYB1C1Z), and a group 6 SDIMO (smoABCD). To date, there is experimental evidence that smoXYB1C1Z is a gaseous alkane/alkene MO (42) and that etnABCD is an ethene MO (64), but the other NBB4 SDIMOs are not experimentally characterized. Our previous studies of smoXYB1C1Z and etnABCD depended on the use of nonstandard host/vector systems, which were not straightforward to use, and gave recombinants with low MO activities. Ideally, a better host/vector system would be found for expressing these MOs, and especially the group 4 SDIMOs, which are of intense interest for biocatalysis due to their high enantioselectivity (65–67).
The alkene monooxygenases EtnABCD and PmoABCD of strain NBB4 have very similar predicted subunit sizes and similar overall predicted protein structure and function, based on their similarity to biochemically well-characterized SDIMOs, such as AmoABCD (58). The alkene MOs consist of a small hydroxylase subunit (EtnA or PmoA, 36 or 39 kDa), a coupling protein subunit (EtnB or PmoB, 12 kDa), a large hydroxylase subunit (EtnC or PmoC, 59 or 57 kDa), and a reductase subunit (EtnD or PmoD, 37 kDa). Some biochemical evidence for the alkene-inducible expression of PmoABCD and EtnABCD has been published (47, 62, 68), but to date, none of the individual subunits of these enzymes have been purified or characterized.
The aim of this study was to develop improved heterologous expression methods for alkene MOs, to enable better understanding of the biochemistry of these enzymes, and to create more robust systems for biocatalysis.
RESULTS
Expression of ethene MO and propene MO in M. smegmatis mc2155.
We previously reported that the ethene MO (EtnABCD) from Mycobacterium strain NBB4 could be functionally expressed in M. smegmatis mc2155 using a novel shuttle vector, pMycoFos (64). This vector was difficult to handle due to its large size (12.5 kb), so in order to obtain more detailed quantitative data on ethene MO expression, we developed a different cloning system (pUS116, 6.6 kb; see Fig. S1 in the supplemental material) for acetamide-inducible expression of the ethene MO in M. smegmatis. The pUS116 plasmid is similar to pJAM2 (69) and pLAM12 (70); its sequence and DNA are available from Addgene (https://www.addgene.org/84578/).
The etnABCD and pmoABCD gene clusters from strain NBB4 were amplified by PCR and cloned into pUS116 to yield plasmids pUS116-ETN and pUS116-PMO. Acetamide induction of mc2155 cultures carrying these two plasmids resulted in detectable MO activity (see next section). SDS-PAGE analysis revealed a very abundant protein at approximately 50 kDa in acetamide-induced cells of mc2155(pUS116-ETN) which was absent from induced cells containing the vector only (Fig. S3). This band is likely to be EtnC, which migrates faster than expected in SDS-PAGE gels (predicted mass, 59 kDa; see also Fig. S5). Another putative ethene MO protein was seen at approximately 37 kDa in the induced mc2155(pUS116-ETN) cells; this could be EtnA (predicted 36 kDa) and/or EtnD (predicted 37 kDa) The coupling protein EtnB (12 kDa) was not visible on SDS-PAGE gels.
Substrate range and specific activities of ethene MO and propene MO.
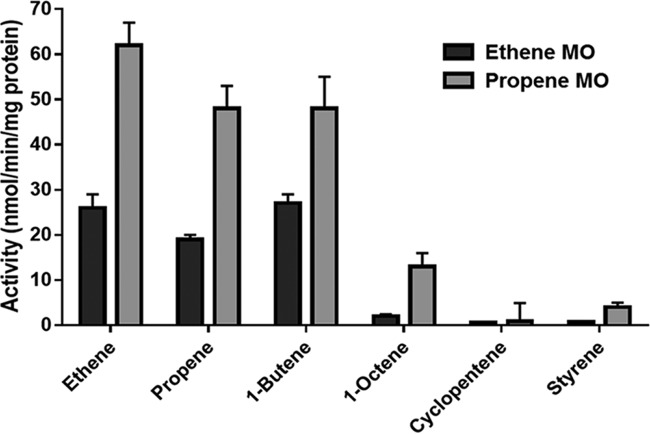
The activities of acetamide-induced mc2155(pUS116-ETN) and mc2155(pUS116-PMO) cells were tested against representative alkene substrates (Fig. 1). Both MOs were capable of oxidizing all the alkenes tested (C2 to C8), with the highest activity for ethene and lowest activity for cyclopentene. For all substrates tested, the propene MO demonstrated greater activity than the ethene MO, ranging from approximately 2-fold higher (ethene) to 7-fold higher (1-octene). The ethene-oxidizing activity of mc2155(pUS116-ETN) cells was approximately three-fold lower than the activity of wild-type ethene-grown NBB4 cells (62). The alkene-oxidizing activity of mc2155(pUS116-PMO) cells seen in this study was 3-fold higher than that of mc2155 cells expressing the PmoABCD enzyme of Mycobacterium strain M156 (57) and in fact was the highest activity reported to date for any heterologous system for SDIMO expression. Cells of mc2155 containing the vector only (pUS116) had no detectable activity on alkenes (data not shown).
FIG 1.
Substrate range and specific activities of recombinant mc2155 cells expressing propene MO and ethene MO. Activity was calculated based on endpoint assays of epoxide production in whole-cell suspensions. Bars represent averages of the results from three independent experiments, and the error bars show the standard deviation.
Discovery of etnH and its effect on ethene MO activity.
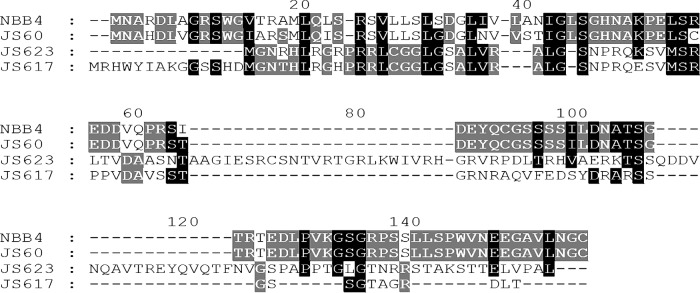
Inspection of the sequence of the etnABCD gene cluster of NBB4 revealed a small putative gene that had been overlooked in earlier annotations. This gene was designated etnH (for hypothetical). In NBB4, the etnH coding sequence overlapped slightly with both the upstream gene (etnE, coenzyme M transferase, 4-bp overlap) and the downstream gene (etnA, ethene MO beta-subunit, 11-bp overlap). A reasonable ribosome binding site (AAGGG) was found 5 bp upstream of the etnH start codon. BLAST searches yielded a single database homolog to EtnH (88% amino acid identity), which was encoded in the corresponding location between etnE and etnA in the alkene-oxidizing Mycobacterium sp. strain JS60. Manual inspection of other alkene MO gene clusters revealed small putative protein-coding regions between etnE and etnA in Mycobacterium strains JS617 and JS623, although the sequence identity between these inferred proteins and EtnH from NBB4 was low (10 to 14%) (Fig. 2). The sizes of the inferred EtnH proteins ranged from 89 to 133 amino acids. The putative EtnH proteins were of similar sizes to the MmoD subunit of sMMO, but the sequence identities to MmoD were very low (4 to 8%). Structure modeling using SWISS-MODEL predicted an EtnH structure matching a section of serine protease (structure database 1EA7), with 26% sequence identity over this region (Fig. S4).
FIG 2.
Alignment of predicted EtnH homologs from ethene MO gene clusters of various Mycobacterium species. The ClustalX program was used to make the alignment using default parameters, which was then visualized using GeneDoc.
A new plasmid construct (pUS116-HETN) was made in which the etnH gene was included in its natural location upstream of etnABCD, and the activities of mc2155(pUS116-HETN) versus mc2155(pUS116-ETN) cells were compared. With ethene as the substrate, the apparent specific activity of mc2155 cells expressing the etnHABCD gene cluster was 30 ± 3 nmol/min/mg protein, compared to 26 ± 5 nmol/min/mg protein for cells expressing etnABCD only. It therefore appears that etnH is not essential for MO activity, and the significance of this gene for ethene oxidation remains unknown. However, for further attempts to express the ethene MO in heterologous hosts (below), the etnH gene was included in all constructs.
Attempts to express the ethene MO in E. coli.
The ethene MO gene cluster (etnHABCD) was cloned into the pET15b plasmid to give the construct pET-ETN, which was transformed into E. coli BL21(DE3)(pLysS). After isopropyl-β-d-thiogalactopyranoside (IPTG) induction, these recombinants had no detectable MO activity, and only one putative MO protein band was visible in SDS-PAGE gels prepared from IPTG-induced cultures (40 kDa, likely to be EtnA, data not shown). The construct was modified by adding a more efficient ribosome binding site (RBS) to the EtnC gene (AGGAGG instead of GGAAG), to make plasmid pET-ETN+. Induced cells of BL21(DE3)(pLysS) containing pET-ETN+ still yielded no MO activity, but SDS-PAGE analysis showed that these cells did contain a new protein band (approximately 50 kDa) close to the expected size of EtnC (59 kDa), in addition to the putative EtnA band. Unfortunately, the putative EtnC band was not seen in the soluble cellular fraction, only in the SDS-solubilized whole-cell extract (Fig. S5), indicating that this protein was likely to have been misfolded.
Analysis of the etnHABCD operon of strain NBB4 using the Rare Codon Calculator (http://people.mbi.ucla.edu/sumchan/caltor.html) suggested that the failure to express ethene MO in E. coli could be due to the presence of rare E. coli codons, such as CCC (proline), AGG (arginine), CGG (arginine), and CGA (arginine). Since commercially available codon-optimizing E. coli strains, such as Rosetta2 (Novagen), could not supply all of these rare codons, we instead designed a synthetic codon-optimized etnHABCD gene cluster, as follows: overlapping ORFs (etnH-etnA) were separated, ribosome binding sites were changed to a consensus sequence, start codons were changed to ATG, stop codons were changed to TAA, and the genes were manually codon-optimized for E. coli at the putative problematic positions (above) with the assistance of an online optimization tool (Integrated DNA Technologies). The complete DNA sequences of the synthetic ethene MO gene sequences compared to the native sequences are shown in Fig. S1.
The synthetic etnHABCD genes were cloned into pET15b to create plasmid pET-sETN, which was transformed into BL21(DE3)(pLysS) cells. No evidence for MO activity (nitrobenzylpyridine [NBP] assay) or production of any of the MO proteins (SDS-PAGE) was seen in these cultures (data not shown), despite attempts to optimize variables, such as incubation temperature (20°C versus 37°C), inducer concentration (0.1 mM IPTG versus 1 mM IPTG), or the coexpression of molecular chaperones (plasmid pKJE7 or pGro7 [71]).
Expression of single ethene MO components in E. coli.
In order to further troubleshoot ethene MO expression, experiments were undertaken to express single-protein components of the enzyme in E. coli. We were not able to obtain the α-hydroxylase EtnC in E. coli in a soluble form, and since it is not possible to show activity for the whole hydroxylase component (predicted to be α2β2) without this subunit, we did not attempt to further purify the β-hydroxylase EtnA, which did appear to be soluble in E. coli (data not shown). The coupling protein (EtnB) was successfully cloned, expressed, and purified as a C-terminal His-tagged recombinant protein from E. coli BL21(DE3)(pLysS) cells, with no apparent solubility problems (Fig. S6). However, in the absence of functional purified ethene MO hydroxylase subunits, we could not directly assess the catalytic properties of EtnB.
The ethene MO reductase (EtnD) was insoluble and inactive when expressed in E. coli as a 6×His-tagged protein. Since previous workers (54) were successful in expressing soluble reductase (ThmD) from tetrahydrofuran monooxygenase, a group 5 SDIMO, using a maltose binding protein (MBP) fusion, we followed this strategy for EtnD using the vector pHisMAL, a derivative of pMAL-c5x which contains a 6×His tag for protein purification. We also followed the experimental procedures used for ThmD, which included supplementation with iron and sulfur, low-temperature incubation during protein expression, and a heat shock step prior to induction (believed to induce chaperonins to enhance protein folding).
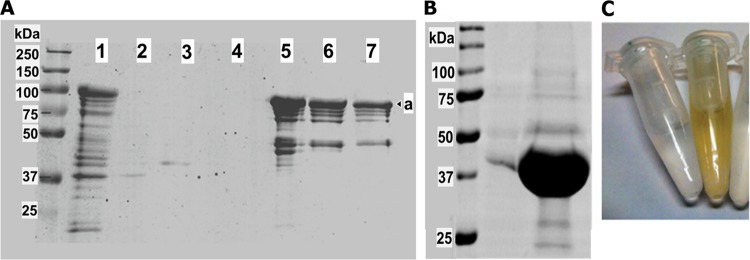
Purification of His-tagged proteins from BL21(DE3)(pHisMAL-etnD) cells yielded yellow elution fractions, suggesting the presence of a flavin adenine dinucleotide (FAD)-containing protein, as expected for EtnD. Upon SDS-PAGE analysis, a strong protein band at approximately 90 kDa was visible in induced cells and in the eluted column fractions (Fig. 3A), which was somewhat larger than expected for MBP-6×His-EtnD (81 kDa). The putative EtnD fusion protein was estimated to be approximately 50% pure after nickel-agarose column purification. After cleavage of the MBP tag and another round of column purification, a strong band of the expected size for EtnD (37 kDa) was produced, with an estimated purity of >95% (Fig. 3B). This purified protein was strongly yellow in color (Fig. 3C), as expected for a flavoprotein.
FIG 3.
SDS-PAGE analysis of EtnD purification from E. coli. (A) Sequential purification of the MBP-6×His-EtnD protein from total cell lysate (1) through column wash steps (2 to 4) to column elution steps (5 to 7), with “a” indicating the band predicted to be MBP-6×His-EtnD. (B) The EtnD protein after cleavage of the MBP tag and repurification. (C) Visual inspection of the final EtnD preparation (yellow) compared to other column fractions (clear).
The purified EtnD protein was tested for reductase activity using a cytochrome c reduction assay. No significant activity was seen when NADH or EtnD was omitted, but in the complete assay mixture, the rate of cytochrome c reduction was 2.46 nmol per minute (Fig. 4). The reaction mixtures contained 1.81 μg of protein, and thus the specific activity of the EtnD preparations was 1.36 μmol/min/mg protein. This level of activity is much lower than the TomA5 reductase from Burkholderia sp. strain G4 (512 μmol · min−1 · mg−1) (52) but is similar to that seen for the ThmD reductase from Pseudonocardia strain K1 (1.6 μmol · min−1 · mg−1) (54). The results confirm the predicted function of EtnD as the reductase component of the ethene MO enzyme.
FIG 4.
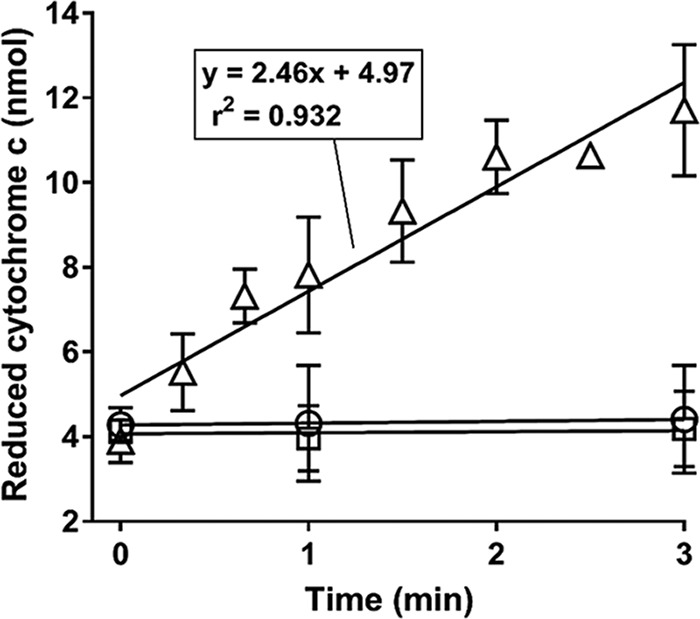
Cytochrome c reduction activity of EtnD purified from E. coli. △, complete assay mixture; ○, assay mixture lacking EtnD; □, assay mixture lacking NADH. Symbols indicate the mean of three (○, □) or five (△) independent experiments, and error bars show the standard deviation.
Expression of ethene MO in P. putida KT2440.
P. putida KT2440 was chosen as an alternative expression host for ethene MO due to its rapid growth, ease of genetic manipulation, and high GC content; the high GC content may help in translating genes from Mycobacterium species. In addition, KT2440 is derived from a hydrocarbon-degrading ancestor (72), and thus, it may contain accessory factors which facilitate oxygenase expression. Broad-host-range plasmids containing the natural (nETN) or synthetic (sETN) ethene MO gene clusters were constructed (pBBR1-nETN and pBBR1-sETN, respectively); the expression of cloned genes in these plasmids is constitutive, from the depressed lac promoter. When pBBR1-nETN and pBBR1-sETN were electroporated into P. putida KT2440, transformant colonies grew much more slowly on LB-kanamycin (LB-Km) agar plates than colonies containing the vector only (48 h versus 16 h to give 2-mm colonies), which suggests a burden imposed by successful transcription and/or translation of at least some MO proteins (in contrast, E. coli TOP10 colonies containing these constructs grew at the same rate as those transformed with vector only). Qualitative tests indicated no MO activity in KT2440(pBBR1-sETN) cells, so further work focused solely on cells containing pBBR1-nETN.
The monooxygenase activities of KT2440(pBBR1-nETN) recombinants were very inconsistent when grown in broth, ranging from undetectable up to 0.9 nmol min−1 · mg−1 protein in different experiments. This result agrees with earlier observations (57) that plasmids containing alkene MO genes can be unstable, and it prompted us to instead perform epoxidation assays directly on pooled cells scraped from the initial transformation plates. This yielded more reproducible data and gave apparent specific activities for ethene epoxidation of 0.5 ± 0.2 nmol/min/mg protein (Fig. 5). The epoxide was not detected in cells carrying the vector only or when KT2440(pBBR1-nETN) cells were incubated without ethene (data not shown). Cotransformation of compatible plasmids expressing chaperonin proteins stimulated the epoxidation activity. The GroEL chaperonin of E. coli stimulated epoxidation activity by 120%, while the MimG chaperonin of M. smegmatis stimulated activity by 50%. These experiments are significant, since they show that recombinant expression of complete functional alkene monooxygenases is possible in Gram-negative hosts.
FIG 5.
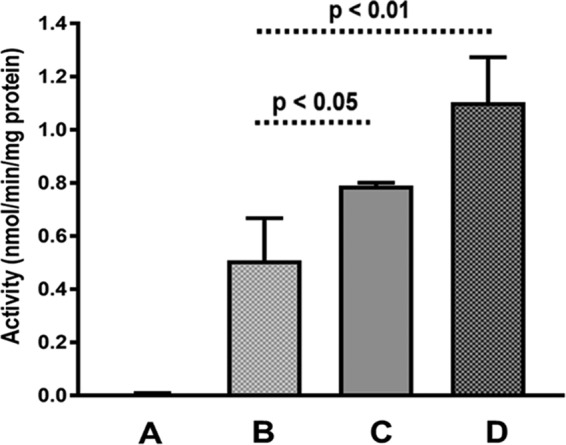
Alkene monooxygenase activity in P. putida KT2440 cells containing different plasmids. (A) pBBR1MCS-2. (B) pBBR1-nETN. (C) pBBR1-nETN plus pUCP-MimG. (D) pBBR1-nETN plus pUCP-GroEL. Activity was measured via an NBP assay of epoxide production from ethene in resting cell suspensions. Bars represent averages of the results from three independent experiments, and the error bars show the standard deviation. P values show the significance of differences between test conditions, measured as single-tailed t tests.
The expression of EtnABCD in KT2440 cells was analyzed further by SDS-PAGE (Fig. 6), and the identities of particular upregulated and downregulated proteins (relative to cells carrying the vector only) were determined by mass spectrometry (Table 1). This analysis confirmed the soluble expression of each of the subunits of the ethene monooxygenase, which were found in upregulated bands (e, f, g, and k) at approximately the expected sizes. In the case of upregulated band e at approximately 60 kDa, this was predominantly KT2440 chaperonin protein, with only traces of EtnC present; it is likely that the majority of the EtnC protein migrated faster than predicted (see also Fig. S3 and S5) and was present in a band that was less-obviously upregulated and not subjected to mass spectrometry (MS) analysis.
FIG 6.
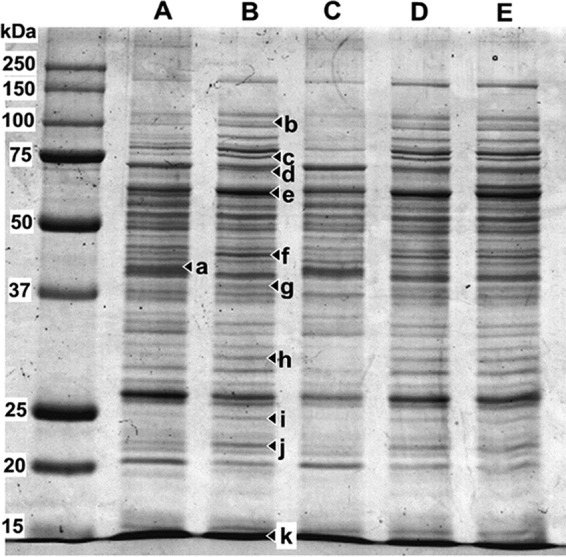
SDS-PAGE analysis of P. putida KT2440 cells carrying different plasmids. (A) pBBR1MCS-2. (B) pBBR1-nETN. (C) pUCP24. (D) pBBR1-nETN plus pUCP-MimG. (E) pBBR1-nETN plus pUCP-GroEL. Bands a to k were excised for further analysis by LC-MS peptide fingerprinting.
TABLE 1.
Upregulated and downregulated proteins in KT2440(pBBR1-nETN) cells identified by LC-MS of gel bands
| Gel band | Identified proteina | Gene (strain) | Accession no. | MASCOT scoreb | No. of sequencesc | emPAId |
|---|---|---|---|---|---|---|
| a | Phosphoglycerate kinase | pgk (KT2440) | PGK_PSEPK | 797 | 11 | 3.53 |
| b | Elongation factor G1 | fusA (KT2440) | EFG1_PSEPK | 2,504 | 22 | 1.89 |
| c | Chaperone | htpG (KT2440) | HTPG_PSEPK | 2,590 | 44 | 15.85 |
| d | Chaperone | cpn60 (KT2440) | CH60_PSEPK | 948 | 21 | 3.33 |
| e | Chaperone | cpn60 (KT2440) | CH60_PSEPK | 6,933 | 33 | 18.87 |
| e | Ethene MO alpha-subunit | etnC (NBB4) | ACZ56346 | 61 | 1 | 0.06 |
| f | Elongation factor TU-B | tufB (KT2440) | EFTU2_PSEPK | 1,394 | 23 | 10.25 |
| f | Ethene MO reductase | etnD (NBB4) | ACZ56347 | 298 | 6 | 0.82 |
| g | Glutaminase-asparaginase | ansB (KT2440) | ASPQ_PSEPK | 497 | 9 | 1.61 |
| g | Ethene MO beta-subunit | etnA (NBB4) | ACZ56344 | 304 | 7 | 0.75 |
| h | 50S ribosomal protein | rplB (P. putida F1) | RL3_PSEP1 | 567 | 10 | 5.9 |
| i | Cytidylate kinase | cmk (KT2440) | KCY_PSEPK | 400 | 7 | 2.14 |
| j | Heat shock cofactor | grpE (KT2440) | GRPE_PSEPK | 154 | 7 | 0.83 |
| k | Nucleoside diphosphate kinase | ndk (KT2440) | NDK_PSEPK | 712 | 11 | 31.33 |
| k | Ethene MO coupling protein | etnB (NBB4) | ACZ56345 | 581 | 5 | 7.96 |
All protein bands except band a were upregulated with respect to cells containing vector only, while band a was downregulated. For bands containing only KT2440 proteins, just the best matching (i.e., most abundant) protein is shown. For bands containing NBB4 proteins, the best matches from both the host strain (KT2440) and from NBB4 are shown; this better reflects the fact that the SDS-PAGE gel does not fully resolve the proteins, and the NBB4 proteins are mixed with host proteins in these gel slices.
MASCOT score calculated as −10 × log10(p), where p is the probability of the result. A probability of 10−20 thus becomes a score of 200. Scores greater than 13 correspond to a P value of <0.05.
Number of individual peptides from the protein that were identified by LC-MS.
emPAI, exponentially modified protein abundance index, calculated as 10(N-observed/N-observable) − 1, where N-observed is the number of experimentally observed peptides and N-observable is the calculated number of observable peptides for each protein.
Native KT2440 proteins involved in translation (elongation factors and 50S ribosomal protein, bands b, f, and h) and protein folding (chaperonins and heat shock proteins, bands c, d, e, and j) appeared to be upregulated in cells expressing EtnABCD, as were glutaminase-asparaginase (band g) and cytidylate kinase (band I). Phosphoglycerate kinase (band a) was notably downregulated in cells expressing EtnABCD. In cells carrying the accessory plasmid pUCP-GroEL, a new band corresponding to the size of GroEL (60 kDa) was seen (compare lanes D and E), although this was not verified by liquid chromatography-mass spectrometry (LC-MS).
DISCUSSION
Development of a robust heterologous expression system in M. smegmatis mc2155 has enabled us to perform a quantitative analysis of the activities and substrate ranges of two recombinant alkene monooxygenases of interest for biocatalysis. The epoxidation activities detected are the highest reported to date for recombinant cells expressing any kind of SDIMO enzyme. Although the complete alkene MOs were nonfunctional in E. coli, we succeeded in purifying the EtnB and EtnD components of the ethene MO and we functionally characterized EtnD. We have provided rigorous proof that alkene MOs from actinobacteria can be functionally expressed in a Gram-negative host (P. putida), which enabled new insights into the impact of alkene MO expression on the host cells; this will support further work to enhance MO expression systems.
The specific activity for epoxidation of recombinant M. smegmatis cells that we report here is substantially higher than any other heterologous expression system for alkene MOs (or indeed any kind of SDIMO) reported to date (Table 2); this is significant, since the low activity of recombinants has been one of the major bottlenecks for applying these enzymes in industrial biocatalysis. While the activities reported here are only 30 to 50% of those seen in wild-type alkene-oxidizing bacteria, wild-type bacteria are not easily applicable to industrial use, since they readily consume epoxides, are slow-growing, and require specific alkenes to induce the MO. The higher activities reported here for mc2155 cells expressing the PmoABCD enzyme of Mycobacterium strain NBB4 compared to the same host expressing PmoABCD of Mycobacterium strain M156 (57) are most likely due to the improved expression properties of the cloning vector pUS116, since the propene MO enzymes of NBB4 and M156 share very high sequence identity (91% in the PmoC subunit).
TABLE 2.
Comparison of SDIMO activities in whole cells of wild-type and recombinant bacteria
| Strain | Enzyme | Substrate | Activity (nmol/min/mg protein)a | Reference |
|---|---|---|---|---|
| Wild-type bacteria | ||||
| Burkholderia sp. strain G4 | TomA0A1A2A3A4A5 | Phenol | 466 | 85 |
| Pseudomonas sp. strain OX1 | TouABCDEF | Naphthalene | 408 | 84 |
| Gordonia sp. strain TY-5 | PrmABCD | Propane | 157 | 13 |
| Mycobacterium sp. strain JS60 | EtnABCD | Ethene | 118 | 45 |
| Xanthobacter sp. strain Py2 | XamoABCDEF | Propene | 110 | 87 |
| Mycobacterium chubuense NBB4 | EtnABCD | Ethene | 93 | 88 |
| Rhodococcus sp. strain AD45 | IsoABCDEF | Isoprene | 76 | 89 |
| Rhodococcus sp. strain B276 | AmoABCD | Propene | 59 | 90 |
| Methylosinus sp. strain OB3b | MmoXYBZDC | Propene | 52 | 91 |
| Nocardioides sp. strain JS614 | EtnABCD | Ethene | 39 | 92 |
| Recombinant bacteria | ||||
| M. smegmatis mc2155 | PmoABCD | Ethene | 62 | This study |
| M. smegmatis mc2155 | EtnABCD | Ethene | 27 | This study |
| M. smegmatis mc2155 | PmoABCD | Propene | 16 | 57 |
| E. coli JM109 | TouABCDEF | Toluene | 7 | 92 |
| Methylomicrobium sp. strain BG8 | MmoXYBZDC | Methane | 3 | 93 |
| Streptomyces sp. strain TK24 | AmoABCD | Ethene | 2b | 58 |
| M. smegmatis mc2155 | MimABCD | Phenol | 1c | 60 |
| E. coli Rosetta 2 | MimABCD | Phenol | 1 | 61 |
| P. putida KT2440 | EtnABCD | Ethene | 1 | This study |
| E. coli TG1 | TomA0A1A2A3A4A5 | Toluene | 1 | 94 |
Specific activity in whole cells to the nearest integer.
Activity measured in cell extracts rather than whole cells.
Activity calculated from endpoint assay, assuming 50% protein in cell dry weight.
The accuracy of the specific activity calculations reported here could be compromised if nonepoxide products (e.g., alcohols) were made from the alkenes. We do not believe this to be the case, based on previous literature on preferred substrates of alkene MOs, the fact that alkene functional groups were available at high concentrations in the assay, and the lack of other metabolites seen in previous GC analyses of alkene-oxidizing NBB4 cells. The use of endpoint assays is another possible source of error in specific activity calculations, but we believe these can be justified since excess alkene substrates were provided, the cell densities were not dramatically high, and glucose was provided as a cosubstrate to ensure sustained alkene oxidation (but not growth; no nitrogen source was present). If substrates were in fact depleted and/or oxidation rates lowered prior to the endpoint assay, this would result in underestimation of the activities, and thus, the specific activities calculated here must be considered accurate or conservative estimates and not exaggerated claims. It is also worth noting that previous estimates of alkene MO activity in other studies may have been hampered by the toxicity of accumulated epoxides; an advantage of the NBP assay used in this study is that it traps reactive epoxides in a nontoxic form.
The experiments described here allowed confirmation of the substrate range of the two alkene monooxygenases of Mycobacterium strain NBB4 in a way which was not possible using wild-type cells. This is especially significant in the case of this isolate, which is unusual in its possession of many diverse MO systems, which complicates the task of assigning specific enzymes to specific substrates (62). It is now possible to say conclusively that the EtnABCD and PmoABCD enzymes have very closely overlapping substrate ranges; all the gaseous alkenes (C2 to C4) are good substrates for both enzymes, and both enzymes show a dramatic decrease in activity between C4 and C8 alkenes. For any single substrate, mc2155 cells expressing the PmoABCD enzyme consistently gave higher activities than cells expressing EtnABCD; however, despite this, we directed most of our subsequent efforts at EtnABCD expression, since this alkene MO has higher enantioselectivity than PmoABCD or the related AmoABCD (57, 65, 66, 73). The pattern of epoxidation activity observed here differed from that seen earlier in wild-type NBB4 cells grown on ethene (65); this could be because NBB4 cells consume particular epoxides, or because other MOs in NBB4 are coincidentally induced by ethene and contribute to alkene oxidation. In either case, this is evidence for the value of our heterologous expression approach to pinpoint the substrate range of the alkene MO enzymes.
We have successfully expressed and purified the reductase (EtnD) and coupling protein (EtnB) from the ethene MO of NBB4 in E. coli; these are important steps toward developing a cell-free system for stereoselective alkene epoxidation, and also toward gaining a better understanding of the biochemistry of this enzyme. The EtnC subunit proved recalcitrant to soluble expression in E. coli, despite efforts to coinduce chaperonins (GroE, DnaJK, and GrpE), and to test different expression conditions (temperature, inducer concentration, and accessory plasmids, like pLysS). EtnC contains the catalytic di-iron center, and it is possible that thus-far-unknown accessory proteins are required for its correct assembly; there is precedent for this in related proteins, such as ribonucleotide reductase (74). It is notable that the SDIMOs that can be functionally expressed in E. coli (e.g., toluene/o-xylene MO [ToMO] and toluene ortho-monooxygenase [TOM]) have two more subunits than the alkene MOs, and one of these extra subunits is a ferredoxin, similar to one of the accessory proteins of ribonucleotide reductase. On the other hand, sMMO has 6 subunits but is resistant to expression in E. coli. The genome sequences do not provide any obvious answers; for example, in strain NBB4, the ethene MO and propene MO gene clusters are not associated with any ferredoxins or flavodoxins or other obvious helper proteins. These gene clusters also lack chaperonin genes, which are frequently found immediately adjacent to SDIMO gene clusters (13, 61, 75, 76). Strain NBB4 does possess chaperonin genes adjacent to its group 3 and 6 SDIMO gene clusters; could this be a prerequisite to later acquisition of the alkene monooxygenases? This hypothesis is attractive, but imperfect, since Nocardioides sp. strain JS614 has a functional ethene MO but no other SDIMOs or SDIMO-associated chaperonins in its genome.
The SDIMO literature includes substantial controversy surrounding heterologous expression experiments. Successful expression of the Rhodococcus sp. strain B-276 alkene MO AmoABCD in E. coli was seen using trifluoropropene as the substrate (46), but later workers could not replicate this using the natural substrate propene (58). The success of sMMO expression in Pseudomonas spp. and other Gram-negative hosts has also been debated (77, 78), and while there are reports in the nonacademic literature of successful sMMO expression in E. coli (e.g., patent US20170183638 and see http://2014.igem.org/Team:Braunschweig), it remains to be seen whether these claims will hold up to rigorous peer review and replication in independent laboratories. In light of these issues, our work here with alkene MO expression in P. putida KT2440 is especially significant, since we have rigorous evidence from both activity assays and from peptide mass spectrometry that confirm that all of the components of the ethene MO are expressed in a soluble and functional form in this host. It appears that KT2440 and mc2155 share some factor or property that E. coli is lacking which enables heterologous alkene MO expression.
We have shown that the expression of either Mycobacterium or E. coli chaperonins stimulates the epoxidation activity of ethene MO in P. putida; this confirms that GroEL (cpn60) homologs are helpful for heterologous expression of an alkene MO (group 4 SDIMO) and brings these enzymes into line with the behavior of gaseous alkane-oxidizing SDIMOs (groups 3, 5, and 6). The importance of protein folding is emphasized by our finding that the expression of ethene MO in strain KT2440 appears to result in the upregulation of three host chaperonins (cpn60, htpG, and grpE), but the issue of which chaperonins perform this function (if any) in the wild-type hosts remains unresolved. The expression of etnABCD in KT2440 resulted in the downregulation of some core enzymes, such as Pgk; this may reflect the diversion of cellular energy from NADH toward NADPH, which is needed for oxidative stress responses (79), or it could be due to the sensitivity of Pgk to oxidative damage (80). Further work using more high-resolution quantitative methods (e.g., iTRAQ LC-tandem MS [LC-MS/MS]) is needed to better understand the impact of alkene MOs on host cells; this approach may hold the key to successful high-level expression of these enzymes in preferred heterologous hosts, such as E. coli.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Mycobacterium chubuense strain NBB4 was the source of the ethene and propene monooxygenase gene clusters; this isolate has been described previously (38, 62), and the sequences of its chromosome and two large plasmids are available in GenBank (accession numbers NC_018022, NC_018023, and NC_018027). E. coli TOP 10 cells (Invitrogen) were used for cloning procedures and for the expression of proteins from pMAL vectors. E. coli BL21(DE3) cells (NEB) were used for protein expression from pET vectors. Other expression hosts used included Mycobacterium smegmatis mc2155 (81) and Pseudomonas putida KT2440 (72). The vectors pET15b+ (Novagen) and pMAL-c2x (NEB) were used for the expression in E. coli. An in-house modified version of pMAL-c2x was used (pHisMAL), which contained an N-terminal 6×His tag. pHisMAL was constructed by annealing oligonucleotides EFL17 and EFL18 (Table 3), digesting with NdeI, and ligating into NdeI-cut pMAL. The broad-host-range vector pBBR1MCS-2 (82) was used for cloning and expression in Pseudomonas spp., while the acetamidase-inducible shuttle/expression plasmid pUS116 was used for cloning and expression in Mycobacterium species. Plasmid pUS116 (Fig. S1) was made by excision of the gp60 and gp61 genes from pJV53 (70) by digestion with BstBI and NheI and religation of the plasmid backbone. A summary of all the plasmids used and made during this study is shown in Table 4.
TABLE 3.
Oligonucleotides used in this study
| Oligonucleotide name | Sequence | Description |
|---|---|---|
| VMC2 | CGTAGGAGGATTTAAATTTAGGAACCTCGGAG | etnA reverse primer for cloning into pET15b |
| VMC9 | GAGCATTTAAATGGCTGTTAGTCACATTC | etnA forward primer for cloning into pET15b |
| VMC4 | CCGATTTAAATTTAGTCGCGGAATCGTTC | etnB reverse primer for cloning into pET15b |
| VMC10 | CAAGGTATTTAAATGTTGTCATCTGCCACGC | etnB forward primer for cloning into pET15b |
| VMC6 | GGAATTTAAATTTAGCGGCGTCGGAGGATG | etnC reverse primer for cloning into pET15b |
| VMC11 | ACGATTTAAATGGCTAATCCCACTATCGAAG | etnC forward primer for cloning into pET15b |
| VMC7 | TCCATTTAAATATGGGTGACACAGTAACCGTAC | etnD forward primer for cloning into pHisMAL |
| VMC8 | CTCATTTAAATCTATCCAGACGGAACG | etnD reverse primer for cloning into pET15b and pHisMAL |
| VMC12 | TCCATTTAAATGGGTGACACAGTAACCGTAC | etnD forward primer for cloning into pET15b |
| VMC15 | CTCACTATAGGGGAATTGTGAGCGG | pET vector forward primer for clone screening |
| VMC16 | CCCTCAAGACCCGTTTAGAGGC | pET vector reverse primer for clone screening |
| VMC20 | CAATACCAAGAAGGAGGAGAAACATGGC | Forward oligonucleotide containing strong RBS for etnC |
| VMC21 | GCCATGTTTCTCCTCCTTCTTGGTATTG | Reverse oligonucleotide containing strong RBS for etnC |
| VMC25 | CAGCCATATTTAAATGGCTGCCGC | etnHABCD reverse primer for cloning into pET15b or pUS116 |
| VMC39 | CAAGGATTTAAATGAACGCACGCGATCTCG | etnHABCD forward primer for cloning into pET15b or pUS116 |
| NW128 | GCGATTTAAATCGACAAGAACGACGCTTTCGG | pmoABCD forward primer for cloning into pUS116 |
| NW129 | GTGATTTAAATTCTCCCACTCGCCCATGCTG | pmoABCD reverse primer for cloning into pUS116 |
| EFL17 | AATCATTGCATATGGGCAGCAGCCATCATCATC ATCATCACAGCAGCGGGCATATGAAGTC | Forward oligonucleotide encoding 6×His tag to add to pMAL |
| EFL18 | AAAGACTTCATATGCCCGCTGCTGTGATGATGA TGATGATGGCTGCTGCCCATATGCAATGA | Forward oligonucleotide encoding 6×His tag to add to pMAL |
| MAL69 | TGCGGAGCTCTAAAAGGAGGAGCAAAATG | Synthetic etnHABCD forward primer for cloning into pET15b |
| MAL70 | TCGTGCGGCCGCTTATTATCCAGACGG | Synthetic etnHABCD reverse primer for cloning into pET15b |
| MAL71 | GGTTCACTTTTATCCCTGCGC | Synthetic etnD forward primer for clone screening |
| MAL72 | GATATTCGTCGATACTCCGTGG | Synthetic etnH reverse primer for clone screening |
| MAL77 | GACTTCTAGACTGAGTCTCAAAACCAAGG | Synthetic etnHABCD forward primer for cloning into pBBR1MCS-2 |
| MAL78 | TGCTGAGCTCTCGTATGTTGTGTGG | Synthetic etnHABCD reverse primer for cloning into pBBR1MCS-2 |
| MAL79 | CGTAGGTACCTAACTCCAAGGGCAGCAATG | etnHABCD forward primer for cloning into pBBR1MCS-2 |
| MAL80 | CTTCAGGCCTCCGACCGTCAGTCCG | etnHABCD reverse primer for cloning into pBBR1MCS-2 |
| MAL85 | CGGCTCCTTCTTCATTGACC | etnH reverse primer for clone screening |
| MAL86 | CCTGCGGACGTCTCTTCA | etnD forward primer for clone screening |
TABLE 4.
Plasmids used in this study
| Plasmid name | Relevant characteristicsa | Reference or source |
|---|---|---|
| pET15b | Vector for cloning and expression in E. coli, Apr, IPTG inducible | Novagen |
| pET28c | Vector for cloning and expression in E. coli, Kmr, IPTG inducible, pBR322 replicon | Novagen |
| pUS116 | Shuttle vector for cloning in E. coli and expression in mycobacteria, Kmr, acetamide inducible, pUC19 and pAL5000 replicons | Derived from pJV53 (70) |
| pBBR1MCS-2 | Vector for cloning and expression in Gram-negative bacteria, Kmr, constitutive expression, pBBR replicon | 38 |
| pGro7 | Provides chaperonin GroE from E. coli, Cmr, inducible by arabinose, p15A replicon | TaKaRa Bio, Inc. |
| pKJE7 | Provides chaperonins DnaJ, DnaK, and GrpE from E. coli, Cmr, inducible by arabinose, p15A replicon | TaKaRa Bio, Inc. |
| pUS116-ETN | pUS116 containing etnABCD genes of Mycobacterium NBB4 | This study |
| pUS116-HETN | pUS116 containing etnHABCD genes of Mycobacterium NBB4 | This study |
| pUS116-PMO | pUS116 containing pmoABCD genes of Mycobacterium NBB4 | This study |
| pET-etnA | pET15b+ containing etnA gene | This study |
| pET-etnB | pET15b+ containing etnB gene | This study |
| pET-etnC | pET15b+ containing etnC gene | This study |
| pET-etnD | pET15b+ containing etnD gene | This study |
| pET-ETN | pET15b+ containing etnABCD gene cluster | This study |
| pET-ETN+ | pET-ETN with strong RBS added upstream of etnC | This study |
| pET-sETN | pET28c containing codon-optimized etnHABCD gene cluster | This study |
| pMAL-c2x | Vector for IPTG-inducible cytoplasmic expression of MBP fusions, Apr, pBR322 replicon | NEB |
| pHisMAL | pMAL-c5x with in-frame 6×His tag added at 5′ end of MBP gene | This study |
| pHisMAL-etnD | pHisMAL containing etnD fused in-frame to 6×His tag and MBP | This study |
| pBBR1-ETN | pBBR1MCS-2 containing etnHABCD gene cluster | This study |
| pBBR1-sETN | pBBR1MCS-2 containing codon-optimized etnHABCD gene cluster | This study |
Apr, ampicillin resistant; Kmr, kanamycin resistant; Cmr, chloramphenicol resistant; MBP, maltose-binding protein.
Molecular biology methods.
PCR was used to amplify the ethene MO and propene MO gene clusters or individual MO genes from Mycobacterium NBB4 genomic DNA (primers given in Table 3). PCR was conducted with high-fidelity Phusion polymerase (NEB). PCR products were purified before and after restriction digestion using QIAquick columns (Qiagen). Plasmids were extracted using the alkaline lysis method (83) and then purified after digestion using the SureClean kit (Bioline). Where necessary, digested PCR products or plasmids were blunted using T4 DNA polymerase (NEB). The yields and sizes of DNA from plasmid preparations, PCR, and digestions were checked by agarose gel electrophoresis on 1% agarose gels made in 0.5% Tris-borate-EDTA (TBE) buffer (83), poststained with GelRed (Biotium). Ligations were done with T4 DNA ligase (NEB). Ligation mixtures and purified plasmids were transformed into E. coli by heat shock of chemically competent cells. Plasmids were transformed into P. putida KT2440 and M. smegmatis mc2155 by electroporation (Bio-Rad XCell machine), with 2-mm-gap electrocuvettes and electroporator settings of 2.5 kV, 25 μF, and 200 Ω (for KT2440) or 1,000 Ω (for mc2155).
Construction of expression plasmids.
For cloning into pET15b(+) and pUS116, the PCR products of the etn and pmo genes (primer pairs VMC2-VMC9, VMC4-VMC10, VMC6-VMC11, VMC8-VMC12, VMC25-VMC39, and NW128-NW129) were digested with SwaI and ligated to blunted BamHI-cut vectors. The resulting plasmids were named pET-ETN, pUS116-ETN, pET-etnA, pET-etnB, pET-etnC, and pET-etnD (Table 4). A synthetic etnHABCD gene cluster was synthesized (by IDT) with E. coli codon usage, all nonoverlapping genes, and strong RBSs throughout (Fig. S2). The synthetic DNA was reamplified with primers MAL69-MAL70, digested with SacI-NotI, and ligated into similarly digested pET28c to create pET-sETN. For the expression of an MBP-EtnD fusion protein, the etnD gene was amplified from NBB4 DNA (primers VMC7-VMC8), digested with SwaI, and inserted in the blunted BamHI site of pHisMAL to create pHisMAL-etnD.
For the expression of ethene MO in strain KT2440, two different plasmids, pBBR1-nETN and pBBR1-sETN, were constructed. The PCR-amplified etnHABCD gene cluster (primers MAL79-MAL80) was digested with KpnI and StuI and ligated into KpnI-EcoRV-cut pBBR1MCS-2 to make pBBR1-nETN. The synthetic codon-optimized ethene MO sequence (see above) was reamplified with primers MAL77-MAL78, digested with SacI-XbaI, and ligated to similarly digested pBBR1MCS-2 to make pBBR1-sETN.
During all the above-described work, the correct plasmid constructs were detected by colony PCR of ampicillin-resistant (Apr) E. coli clones, using one primer in the insert DNA and one primer in the vector backbone (Table 3). The structure of all new plasmids was confirmed by restriction digestion of the whole plasmids using multiple enzymes, and by sequencing of PCR products amplified across both ligation junctions. All structures and sequences were as expected.
Expression of ethene monooxygenase in heterologous hosts.
E. coli cultures containing pET15b-based constructs were grown for 16 h in 5 ml LB broth containing 100 μg/ml ampicillin at 37°C and then diluted 1:100 in 500 ml fresh medium of the same type (500 ml), and incubation continued until the optical density at 600 nm (OD600) reached 0.5. Cultures were then induced with 1 mM IPTG and incubated for a further 4 h at either 20°C or 37°C. Cells were collected by centrifugation (5,000 × g, 4°C, 15 min) and either assayed immediately for MO activity or stored at −80°C for protein analysis. E. coli cultures containing pHisMAL-etnD were grown and induced in the same manner, except that the medium included 1 mM l-cysteine and 0.3 mM ferrous sulfate, a heat shock was added (42°C, 30 min) prior to induction, and IPTG induction was done for 7 h at 20°C. For protein expression in M. smegmatis mc2155, recombinants were grown at 30°C in 500 ml minimal salts medium (MSM) (84) containing 2% succinate, 2% acetamide, 0.5% Tween 80, and 20 μg/ml kanamycin. After the OD600 reached 0.7 (typically 72 h), cells were harvested by centrifugation and used immediately for activity assays, or they were frozen at −80°C for later protein analysis.
Purification of ethene MO subunits from E. coli.
Cell pellets from 500 ml of culture were resuspended in 5 ml immobilized-metal affinity chromatography (IMAC) binding buffer (20 mM Na2HPO4 [pH 8.0], 0.5 M sodium chloride, 20 mM imidazole) containing lysozyme (0.1 mg/ml), DNase (10 μg/ml), and protease inhibitor cocktail (catalog no. P2714; Sigma-Aldrich) and incubated for 1 h at 20°C. Cells were disrupted by sonication, and then a clarified cell extract was obtained by centrifugation (5,000 × g, 4°C, 15 min) and filtration (0.45-μm pore size). The cell extract was diluted 1:1 with IMAC binding buffer and applied to a 1-ml Ni-Sepharose HisTrap affinity column (GE Life Sciences). The column was then washed with 10 ml of IMAC wash buffer (20 mM Na2HPO4 [pH 8.0], 0.5 M sodium chloride, 80 mM imidazole), and then His-tagged proteins were eluted with 5 ml of IMAC elution buffer (20 mM Na2HPO4 [pH 8.0], 0.5 M sodium chloride, 500 mM imidazole). Fractions were analyzed by SDS-PAGE, and the fractions containing recombinant proteins of interest were pooled, transferred to a 3-ml dialysis cassette (3,500 molecular weight cutoff), and dialyzed at 4°C for 3 h in 1 liter of buffer (25 mM morpholinepropanesulfonic acid [MOPS] [pH 7.0] with 5% [vol/vol] glycerol). Dialyzed proteins were subsequently concentrated using a Vivaspin 6 desalting column and stored at −80°C for later analysis. In the case of the 6×His-MBP-EtnD fusion protein, the above-described procedure was modified as follows: a thrombin cleavage step (25°C, 16 h) was done after the dialysis, a second round of IMAC purification was performed, with EtnD eluted in IMAC wash buffer (the His-tagged MBP is retained on the column), and then the dialysis was repeated before concentrating with the Vivaspin column.
Protein analysis by SDS-PAGE and mass spectrometry.
Whole-cell lysates and purified protein fractions were analyzed via SDS-PAGE to determine protein expression. SDS-PAGE was done according to standard protocols (85) with the Mini-Protean kit (Bio-Rad) and 10% (wt/vol) acrylamide gels, stained with Coomassie brilliant blue. The molecular mass of the proteins was estimated by comparison with a ladder (Precision Plus Protein standards; Bio-Rad). SDS-PAGE bands of particular interest were processed for peptide mass fingerprinting as follows. Gel bands were excised using sterile blades, washed in destain solution (40% [vol/vol] acetonitrile, 24 mM ammonium bicarbonate [pH 7.8]), drained, vacuum-dried, rehydrated in trypsin solution (12 ng/μl trypsin in 40 mM ammonium bicarbonate) for 1 h at 4°C, and then incubated 16 h at 37°C. Peptides were concentrated and desalted with C18 PerfectPure tips (Eppendorf) and eluted in matrix (8 mg/ml α-cyano-4-hydroxycinnamic acid, 70% [vol/vol] acetonitrile, 1% [vol/vol] formic acid) directly onto target plates. Peptide mass maps were generated by matrix-assisted laser desorption ionization–time of flight MS (MALDI-TOF MS) using a Voyager DE-STR spectrometer (Applied Biosystems), with mass calibration using trypsin autolysis peaks m/z 2,211.11 and m/z 842.51. Fingerprint data were used to search NCBI via MASCOT (Matrix Science). Search parameters included ±0.2 Da peptide mass tolerance and one missed cleavage per peptide, with identifications based on MASCOT score, E value, number of peptide mass matches, and total percentage sequence coverage of peptides.
Alkene oxidation assays.
Induced cells were washed and resuspended in 20 mM K2HPO4 buffer (pH 7) at an OD600 of 10 (E. coli and M. smegmatis) or an OD600 of 40 (P. putida). Assays were run in 16-ml crimp-sealed glass vials, each of which contained an open 1.5-ml glass vial containing 1 ml of 100 mM nitrobenzylpyridine (NBP). A cell suspension (0.9 ml) was added to the 16-ml vial (but outside the 1.5-ml vial), and then 0.1 ml of 20% glucose was added to the cells to provide an energy source. Alkene substrates (10% of headspace for gases or 50 mM for liquids) were injected into the outer vial, and the vials were incubated with shaking at 30°C for 16 h. The NBP was removed, heated at 90°C for 1 h, and then added to a 500-μl trimethylamine-acetone solution (1:1), which developed a purple color if epoxide was present. The absorbance of the epoxide-NBP adducts at 600 nm was converted into concentrations of the epoxides via the extinction coefficients (65); then, specific activity was calculated based on the time elapsed and the amount of protein in the assay. Protein concentrations were calculated from the measured OD600 data by applying standard curves, in which protein was measured based on UV absorbance measurement of alkali-lysed cells (see reference 86 for details). For M. smegmatis cells, protein concentration (in micrograms per milliliter) was measured as 99 × the OD600, while for P. putida cells, protein concentration (in micrograms per milliliter) was measured as 69 × the OD600.
Cytochrome c activity assay.
The reductase activity of EtnD was determined using cytochrome c as an electron acceptor, similar to methods described previously (54). Purified protein was added to a solution of 100 μM cytochrome c and 400 μM NADH in buffer (25 mM MOPS [pH 7.0], 5% [vol/vol] glycerol) in a spectrophotometer cuvette and incubated at 25°C. Activity was measured by the change in absorbance at 550 nm over 10 min. Reduced cytochrome c concentrations were calculated using the extinction coefficient ε = 21.1 mM−1 · cm−1 (54). The Prism GraphPad software was used to graph the data (cytochrome c reduction versus time) and determine the reaction rate, based on a linear fit through the initial portion of the data. Specific activity was calculated by dividing the reaction rate by the amount of protein in the assay (1.5 mg).
Supplementary Material
ACKNOWLEDGMENTS
Funding to support the postdoctoral fellows and research assistants involved in this work (V.M., N.L.W., and E.F.L.) came from ARC Discovery grants DP0877315 and DP120101155 from the Australian Research Council. Consumable costs for students (M.V.S., R.H., and M.-A.L.) were provided by The University of Sydney. M.-A.L. was supported by an Australian Postgraduate Award scholarship.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00397-18.
REFERENCES
- 1.Chistoserdova L. 2015. Methylotrophs in natural habitats: current insights through metagenomics. Appl Microbiol Biotechnol 99:5763–5779. doi: 10.1007/s00253-015-6713-z. [DOI] [PubMed] [Google Scholar]
- 2.Hanson R, Hanson T. 1996. Methanotrophic bacteria. Microbiol Rev 60:439–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang H, Chen Y, Jiang PX, Zhang C, Smith TJ, Murrell JC, Xing XH. 2010. Methanotrophs: multifunctional bacteria with promising applications in environmental bioengineering. Biochem Eng J 49:277–288. doi: 10.1016/j.bej.2010.01.003. [DOI] [Google Scholar]
- 4.van Ginkel CG, de Bont JAM. 1986. Isolation and characterization of alkene-utilizing Xanthobacter spp. Arch Microbiol 145:403–407. doi: 10.1007/BF00470879. [DOI] [Google Scholar]
- 5.Arp DJ. 1999. Butane metabolism by butane-grown 'Pseudomonas butanovora'. Microbiology 145:1173–1180. doi: 10.1099/13500872-145-5-1173. [DOI] [PubMed] [Google Scholar]
- 6.Johnson EL, Hyman MR. 2006. Propane and n-butane oxidation by Pseudomonas putida GPo1. Appl Environ Microbiol 72:950–952. doi: 10.1128/AEM.72.1.950-952.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Danko AS, Luo M, Bagwell CE, Brigmon RL, Freedman DL. 2004. Involvement of linear plasmids in aerobic biodegradation of vinyl chloride. Appl Environ Microbiol 70:6092–6097. doi: 10.1128/AEM.70.10.6092-6097.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Small FJ, Ensign SA. 1997. Alkene monooxygenase from Xanthobacter strain Py2–purification and characterization of a four-component system central to the bacterial metabolism of aliphatic alkenes. J Biol Chem 272:24913–24920. doi: 10.1074/jbc.272.40.24913. [DOI] [PubMed] [Google Scholar]
- 9.van Hylckama Vlieg JE, Leemhuis H, Spelberg JHL, Janssen DB. 2000. Characterization of the gene cluster involved in isoprene metabolism in Rhodococcus sp. strain AD45. J Bacteriol 182:1956–1963. doi: 10.1128/JB.182.7.1956-1963.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnston A, Crombie AT, El Khawand M, Sims L, Whited GM, McGenity TJ, Murrell JC. 2017. Identification and characterisation of isoprene-degrading bacteria in an estuarine environment. Environ Microbiol 19:3526–3537. doi: 10.1111/1462-2920.13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Bont JAM. 1976. Oxidation of ethylene by soil bacteria. Antonie Van Leeuwenhoek 42:59–71. doi: 10.1007/BF00399449. [DOI] [PubMed] [Google Scholar]
- 12.Coleman NV, Spain JC. 2003. Distribution of the coenzyme M pathway of epoxide metabolism among ethene- and vinyl chloride-degrading Mycobacterium strains. Appl Environ Microbiol 69:6041–6046. doi: 10.1128/AEM.69.10.6041-6046.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotani T, Yamamoto T, Yurimoto H, Sakai Y, Kato N. 2003. Propane monooxygenase and NAD+-dependent secondary alcohol dehydrogenase in propane metabolism by Gordonia sp. strain TY-5. J Bacteriol 185:7120–7128. doi: 10.1128/JB.185.24.7120-7128.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yagi O, Hashimoto A, Iwasaki K, Nakajima M. 1999. Aerobic degradation of 1,1,1-trichloroethane by Mycobacterium spp. isolated from soil. Appl Environ Microbiol 65:4693–4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamamura N, Yeager CM, Arp DJ. 2001. Two distinct monooxygenases for alkane oxidation in Nocardioides sp. strain CF8. Appl Environ Microbiol 67:4992–4998. doi: 10.1128/AEM.67.11.4992-4998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Ginkel CG, De Jong E, Tilanus JWR, De Bont JAM. 1987. Microbial oxidation of isoprene, a biogenic foliage volatile and of 1,3-butadiene, an anthropogenic gas. FEMS Microbiol Ecol 45:275–279. doi: 10.1016/0378-1097(87)90004-8. [DOI] [Google Scholar]
- 17.Whalen SC. 2005. Biogeochemistry of methane exchange between natural wetlands and the atmosphere. Environ Eng Sci 22:73–94. doi: 10.1089/ees.2005.22.73. [DOI] [Google Scholar]
- 18.Le Mer J, Roger P. 2001. Production, oxidation, emission and consumption of methane by soils: a review. Eur J Soil Biol 37:25–50. doi: 10.1016/S1164-5563(01)01067-6. [DOI] [Google Scholar]
- 19.Broadgate WJ, Malin G, Kupper FC, Thompson A, Liss PS. 2004. Isoprene and other non-methane hydrocarbons from seaweeds: a source of reactive hydrocarbons to the atmosphere. Mar Chem 88:61–73. doi: 10.1016/j.marchem.2004.03.002. [DOI] [Google Scholar]
- 20.Shennan J. 2006. Utilisation of C2-C4 gaseous hydrocarbons and isoprene by microorganisms. J Chem Technol Biotechnol 81:237–256. doi: 10.1002/jctb.1388. [DOI] [Google Scholar]
- 21.Mattes TE, Alexander AK, Coleman NV. 2010. Aerobic biodegradation of the chloroethenes: pathways, enzymes, ecology, and evolution. FEMS Microbiol Rev 34:445–475. doi: 10.1111/j.1574-6976.2010.00210.x. [DOI] [PubMed] [Google Scholar]
- 22.Sullivan JP, Dickinson DP, Chase HA. 1998. Methanotrophs, Methylosinus trichosporium OB3b, sMMO, and their application to bioremediation. Crit Rev Microbiol 24:335–373. doi: 10.1080/10408419891294217. [DOI] [PubMed] [Google Scholar]
- 23.Behrendorff J, Huang WL, Gillam EMJ. 2015. Directed evolution of cytochrome P450 enzymes for biocatalysis: exploiting the catalytic versatility of enzymes with relaxed substrate specificity. Biochem J 467:1–15. doi: 10.1042/BJ20141493. [DOI] [PubMed] [Google Scholar]
- 24.de Gonzalo G, Mihovilovic MD, Fraaije MW. 2010. Recent developments in the application of Baeyer-Villiger monooxygenases as biocatalysts. Chembiochem 11:2208–2231. doi: 10.1002/cbic.201000395. [DOI] [PubMed] [Google Scholar]
- 25.Torres Pazmiño DE, Winkler M, Glieder A, Fraaije MW. 2010. Monooxygenases as biocatalysts: classification, mechanistic aspects and biotechnological applications. J Biotechnol 146:9–24. doi: 10.1016/j.jbiotec.2010.01.021. [DOI] [PubMed] [Google Scholar]
- 26.Leak DJ, Sheldon RA, Woodley JM, Adlercreutz P. 2009. Biocatalysts for selective introduction of oxygen. Biocat Biotrans 27:1–26. doi: 10.1080/10242420802393519. [DOI] [Google Scholar]
- 27.Girvan HM, Munro AW. 2016. Applications of microbial cytochrome P450 enzymes in biotechnology and synthetic biology. Curr Opin Chem Biol 31:136–145. doi: 10.1016/j.cbpa.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 28.Leahy JG, Batchelor PJ, Morcomb SM. 2003. Evolution of the soluble diiron monooxygenases. FEMS Microbiol Rev 27:449–479. doi: 10.1016/S0168-6445(03)00023-8. [DOI] [PubMed] [Google Scholar]
- 29.Balasubramanian R, Smith SM, Rawat S, Yatsunyk LA, Stemmler TL, Rosenzweig AC. 2010. Oxidation of methane by a biological dicopper centre. Nature 465:115–119. doi: 10.1038/nature08992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Berkel WJH, Kamerbeek NM, Fraaije MW. 2006. Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J Biotechnol 124:670–689. doi: 10.1016/j.jbiotec.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 31.Zhao GS, Xia TH, Song J, Jensen RA. 1994. Pseudomonas aeruginosa possesses homologs of mammalian phenylalanine hydroxylase and 4-alpha-carbinolamine dehydratase/DCoH as part of a 3-component gene cluster. Proc Natl Acad Sci U S A 91:1366–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fetzner S. 2002. Oxygenases without requirement for cofactors or metal ions. Appl Microbiol Biotechnol 60:243–257. doi: 10.1007/s00253-002-1123-4. [DOI] [PubMed] [Google Scholar]
- 33.He Y, Mathieu J, da Silva MLB, Li MY, Alvarez PJJ. 2018. 1,4-Dioxane-degrading consortia can be enriched from uncontaminated soils: prevalence of Mycobacterium and soluble di-iron monooxygenase genes. Microb Biotechnol 11:189–198. doi: 10.1111/1751-7915.12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crombie AT, Murrell JC. 2014. Trace-gas metabolic versatility of the facultative methanotroph Methylocella silvestris. Nature 510:148–151. doi: 10.1038/nature13192. [DOI] [PubMed] [Google Scholar]
- 35.Li MY, Mathieu J, Yang Y, Fiorenza S, Deng Y, He ZL, Zhou JZ, Alvarez PJJ. 2013. Widespread distribution of soluble di-iron monooxygenase (SDIMO) genes in arctic groundwater impacted by 1,4-dioxane. Environ Sci Technol 47:9950–9958. doi: 10.1021/es402228x. [DOI] [PubMed] [Google Scholar]
- 36.Miqueletto PB, Andreote FD, Dias AC, Ferreira JC, Dos Santos Neto EV, de Oliveira VM. 2011. Cultivation-independent methods applied to the microbial prospection of oil and gas in soil from a sedimentary basin in Brazil. AMB Express 1:35–35. doi: 10.1186/2191-0855-1-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holmes AJ, Coleman NV. 2008. Evolutionary ecology and multidisciplinary approaches to prospecting for monooxygenases as biocatalysts. Antonie Van Leeuwenhoek 94:75–84. doi: 10.1007/s10482-008-9227-1. [DOI] [PubMed] [Google Scholar]
- 38.Coleman NV, Bui NB, Holmes AJ. 2006. Soluble di-iron monooxygenase gene diversity in soils, sediments and ethene enrichments. Environ Microbiol 8:1228–1239. doi: 10.1111/j.1462-2920.2006.01015.x. [DOI] [PubMed] [Google Scholar]
- 39.Nordlund I, Powlowski J, Shingler V. 1990. Complete nucleotide sequence and polypeptide analysis of multicomponent phenol hydroxylase from Pseudomonas sp. strain CF600. J Bacteriol 172:6826–6833. doi: 10.1128/jb.172.12.6826-6833.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bertoni G, Martino M, Galli E, Barbieri P. 1998. Analysis of the gene cluster encoding toluene/o-xylene monooxygenase from Pseudomonas stutzeri OX1. Appl Environ Microbiol 64:3626–3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sirajuddin S, Rosenzweig AC. 2015. Enzymatic oxidation of methane. Biochemistry 54:2283–2294. doi: 10.1021/acs.biochem.5b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin KE, Ozsvar J, Coleman NV. 2014. SmoXYB1C1Z of Mycobacterium sp. strain NBB4: a soluble methane monooxygenase (sMMO)-like enzyme, active on C2 to C4 alkanes and alkenes. Appl Environ Microbiol 80:5801–5806. doi: 10.1128/AEM.01338-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sluis MK, Sayavedra-Soto LA, Arp DJ. 2002. Molecular analysis of the soluble butane monooxygenase from 'Pseudomonas butanovora'. Microbiology 148:3617–3629. doi: 10.1099/00221287-148-11-3617. [DOI] [PubMed] [Google Scholar]
- 44.Mattes TE, Coleman NV, Spain JC, Gossett JM. 2005. Physiological and molecular genetic analyses of vinyl chloride and ethene biodegradation in Nocardioides sp. strain JS614. Arch Microbiol 183:95–106. doi: 10.1007/s00203-004-0749-2. [DOI] [PubMed] [Google Scholar]
- 45.Coleman NV, Spain JC. 2003. Epoxyalkane:coenzyme M transferase in the ethene and vinyl chloride biodegradation pathways of Mycobacterium strain JS60. J Bacteriol 185:5536–5545. doi: 10.1128/JB.185.18.5536-5545.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saeki H, Furuhashi K. 1994. Cloning and characterization of a Nocardia corallina B-276 gene cluster encoding alkene monooxygenase. J Ferment Bioeng 78:399–406. doi: 10.1016/0922-338X(94)90037-X. [DOI] [Google Scholar]
- 47.Woodland MP, Matthews CS, Leak DJ. 1995. Properties of a soluble propene monooxygenase from Mycobacterium sp. (strain M156). Arch Microbiol 163:231–234. doi: 10.1007/BF00305358. [DOI] [Google Scholar]
- 48.Kotani T, Kawashima Y, Yurimoto H, Kato N, Sakai Y. 2006. Gene structure and regulation of alkane monooxygenases in propane-utilizing Mycobacterium sp. TY-6 and Pseudonocardia sp. TY-7. J Biosci Bioeng 102:184–192. doi: 10.1263/jbb.102.184. [DOI] [PubMed] [Google Scholar]
- 49.Furuya T, Hirose S, Osanai H, Semba H, Kino K. 2011. Identification of the monooxygenase gene clusters responsible for the regioselective oxidation of phenol to hydroquinone in mycobacteria. Appl Environ Microbiol 77:1214–1220. doi: 10.1128/AEM.02316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Furuya T, Nakao T, Kino K. 2015. Catalytic function of the mycobacterial binuclear iron monooxygenase in acetone metabolism. FEMS Microbiol Lett 362:fnv136. doi: 10.1093/femsle/fnv136. [DOI] [PubMed] [Google Scholar]
- 51.Lund J, Woodland MP, Dalton H. 1985. Electron transfer reactions in the soluble methane monooxygenase of Methylococcus capsulatus Bath. Eur J Biochem 147:297–305. doi: 10.1111/j.1432-1033.1985.tb08750.x. [DOI] [PubMed] [Google Scholar]
- 52.Newman LM, Wackett LP. 1995. Purification and characterization of toluene 2-monooxygenase from Burkholderia cepacia G4. Biochemistry 34:14066–14076. doi: 10.1021/bi00043a012. [DOI] [PubMed] [Google Scholar]
- 53.Pikus JD, Studts JM, Achim C, Kauffmann KE, Munck E, Steffan RJ, McClay K, Fox BG. 1996. Recombinant toluene-4-monooxygenase: catalytic and Mossbauer studies of the purified diiron and Rieske components of a four-protein complex. Biochemistry 35:9106–9119. doi: 10.1021/bi960456m. [DOI] [PubMed] [Google Scholar]
- 54.Oppenheimer M, Pierce BS, Crawford JA, Ray K, Helm RF, Sobrado P. 2010. Recombinant expression, purification, and characterization of ThmD, the oxidoreductase component of tetrahydrofuran monooxygenase. Arch Biochem Biophys 496:123–131. doi: 10.1016/j.abb.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 55.West CA, Salmond GPC, Dalton H, Murrell JC. 1992. Functional expression in Escherichia coli of protein B and protein C from soluble methane monooxygenase of Methylococcus capsulatus (Bath). J Gen Microbiol 138:1301–1307. doi: 10.1099/00221287-138-7-1301. [DOI] [PubMed] [Google Scholar]
- 56.Champreda V, Zhou N-Y, Leak DJ. 2004. Heterologous expression of alkene monooxygenase components from Xanthobacter autotrophicus Py2 and reconstitution of the active complex. FEMS Microbiol Lett 239:309–318. doi: 10.1016/j.femsle.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 57.Chan Kwo Chion CK, Askew SE, Leak DJ. 2005. Cloning, expression, and site-directed mutagenesis of the propene monooxygenase genes from Mycobacterium sp. strain M156. Appl Environ Microbiol 71:1909–1914. doi: 10.1128/AEM.71.4.1909-1914.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Smith TJ, Lloyd JS, Gallagher SC, Fosdike WLJ, Murrell JC, Dalton H. 1999. Heterologous expression of alkene monooxygenase from Rhodococcus rhodochrous B-276. Eur J Biochem 260:446–452. doi: 10.1046/j.1432-1327.1999.00179.x. [DOI] [PubMed] [Google Scholar]
- 59.Lock M, Nichol T, Murrell JC, Smith TJ. 2017. Mutagenesis and expression of methane monooxygenase to alter regioselectivity with aromatic substrates. FEMS Microbiol Lett 364:6. doi: 10.1093/femsle/fnx137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Furuya T, Hayashi M, Semba H, Kino K. 2013. The mycobacterial binuclear iron monooxygenases require a specific chaperonin-like protein for functional expression in a heterologous host. FEBS J 280:817–826. doi: 10.1111/febs.12070. [DOI] [PubMed] [Google Scholar]
- 61.Furuya T, Hayashi M, Kino K. 2013. Reconstitution of active mycobacterial binuclear iron monooxygenase complex in Escherichia coli. Appl Environ Microbiol 79:6033–6039. doi: 10.1128/AEM.01856-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coleman NV, Yau S, Wilson NL, Nolan LM, Migocki MD, Ly M-a, Crossett B, Holmes AJ. 2011. Untangling the multiple monooxygenases of Mycobacterium chubuense strain NBB4, a versatile hydrocarbon degrader. Environ Microbiol Rep 3:297–307. doi: 10.1111/j.1758-2229.2010.00225.x. [DOI] [PubMed] [Google Scholar]
- 63.Coleman NV, Le NB, Ly MA, Ogawa HE, McCarl V, Wilson NL, Holmes AJ. 2012. Hydrocarbon monooxygenase in Mycobacterium: recombinant expression of a member of the ammonia monooxygenase superfamily. ISME J 6:171–182. doi: 10.1038/ismej.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ly MA, Liew EF, Le NB, Coleman NV. 2011. Construction and evaluation of pMycoFos, a fosmid shuttle vector for Mycobacterium spp. with inducible gene expression and copy number control. J Microbiol Methods 86:320–326. doi: 10.1016/j.mimet.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 65.Cheung S, McCarl V, Holmes AJ, Coleman NV, Rutledge PJ. 2012. Substrate range and enantioselectivity of epoxidation reactions mediated by the ethene-oxidising Mycobacterium strain NBB4. Appl Microbiol Biotechnol 97:1131–1140. doi: 10.1007/s00253-012-3975-6. [DOI] [PubMed] [Google Scholar]
- 66.Owens CR, Karceski JK, Mattes TE. 2009. Gaseous alkene biotransformation and enantioselective epoxyalkane formation by Nocardioides sp. strain JS614. Appl Microbiol Biotechnol 84:685–692. doi: 10.1007/s00253-009-2019-3. [DOI] [PubMed] [Google Scholar]
- 67.Weijers CAGM, van Ginkel CG, de Bont JAM. 1988. Enantiomeric composition of lower epoxyalkanes produced by methane-, alkane-, and alkene-utilizing bacteria. Enzyme Microb Technol 10:214–218. doi: 10.1016/0141-0229(88)90069-5. [DOI] [Google Scholar]
- 68.Chuang AS, Mattes TE. 2007. Identification of polypeptides expressed in response to vinyl chloride, ethene, and epoxyethane in Nocardioides sp. strain JS614 by using peptide mass fingerprinting. Appl Environ Microbiol 73:4368–4372. doi: 10.1128/AEM.00086-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Triccas JA, Parish T, Britton WJ, Gicquel B. 1998. An inducible expression system permitting the efficient purification of a recombinant antigen from Mycobacterium smegmatis. FEMS Microbiol Lett 167:151–156. doi: 10.1111/j.1574-6968.1998.tb13221.x. [DOI] [PubMed] [Google Scholar]
- 70.van Kessel JC, Hatfull GF. 2007. Recombineering in Mycobacterium tuberculosis. Nat Methods 4:147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 71.Nishihara K, Kanemori M, Kitagawa M, Yanagi H, Yura T. 1998. Chaperone coexpression plasmids: differential and synergistic roles of DnaK-DnaJ-GrpE and GroEL-GroES in assisting folding of an allergen of Japanese cedar pollen, Cryj2, in Escherichia coli. Appl Environ Microbiol 64:1694–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bagdasarian M, Lurz R, Ruckert B, Franklin FCH, Bagdasarian MM, Frey J, Timmis KN. 1981. Specific-purpose plasmid cloning vectors. 2 Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 16:237–247. [DOI] [PubMed] [Google Scholar]
- 73.Takagi M, Uemura N, Furuhashi K. 1990. Microbial transformation processes of aliphatic hydrocarbons. Ann NY Acad Sci 613:697–701. doi: 10.1111/j.1749-6632.1990.tb18248.x. [DOI] [Google Scholar]
- 74.Huang M, Parker MJ, Stubbe J. 2014. Choosing the right metal: case studies of class I ribonucleotide reductases. J Biol Chem 289:28104–28111. doi: 10.1074/jbc.R114.596684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stafford GP, Scanlan J, McDonald IR, Murrell JC. 2003. rpoN, mmoR and mmoG, genes involved in regulating the expression of soluble methane monooxygenase in Methylosinus trichosporium OB3b. Microbiology 149:1771–1784. doi: 10.1099/mic.0.26060-0. [DOI] [PubMed] [Google Scholar]
- 76.Kurth EG, Doughty DM, Bottomley PJ, Arpi DJ, Sayavedra-Sotol LA. 2008. Involvement of BmoR and BmoG in n-alkane metabolism in ‘Pseudomonas butanovora’. Microbiology 154:139–147. doi: 10.1099/mic.0.2007/012724-0. [DOI] [PubMed] [Google Scholar]
- 77.Wood TK. 2002. Active expression of soluble methane monooxygenase from Methylosinus trichosporium OB3b in heterologous hosts. Microbiology 148:3328–3329. doi: 10.1099/00221287-148-11-3328. [DOI] [PubMed] [Google Scholar]
- 78.Murrell JC. 2002. Expression of soluble methane monooxygenase genes. Microbiology 148:3329–3330. doi: 10.1099/00221287-148-11-3329. [DOI] [PubMed] [Google Scholar]
- 79.Lemire J, Alhasawi A, Appanna VP, Tharmalingam S, Appanna VD. 2017. Metabolic defence against oxidative stress: the road less travelled so far. J Appl Microbiol 123:798–809. doi: 10.1111/jam.13509. [DOI] [PubMed] [Google Scholar]
- 80.Tsukamoto Y, Fukushima Y, Hara S, Hisabori T. 2013. Redox control of the activity of phosphoglycerate kinase in Synechocystis sp. PCC6803. Plant Cell Physiol 54:484–491. doi: 10.1093/pcp/pct002. [DOI] [PubMed] [Google Scholar]
- 81.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol 4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 82.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM Jr, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 83.Sambrook J, Russell DW. 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 84.Coleman NV, Mattes TE, Gossett JM, Spain JC. 2002. Biodegradation of cis-dichloroethene as the sole carbon source by a beta-proteobacterium. Appl Environ Microbiol 68:2726–2730. doi: 10.1128/AEM.68.6.2726-2730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Simpson RJ. 2006. SDS-PAGE of proteins. Cold Spring Harb Protoc 2006:pdb.prot4313. doi: 10.1101/pdb.prot4313. [DOI] [Google Scholar]
- 86.Coleman NV, Mattes TE, Gossett JM, Spain JC. 2002. Phylogenetic and kinetic diversity of aerobic vinyl chloride-assimilating bacteria from contaminated sites. Appl Environ Microbiol 68:6162–6171. doi: 10.1128/AEM.68.12.6162-6171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bertoni G, Bolognese F, Galli E, Barbieri P. 1996. Cloning of the genes for and characterization of the early stages of toluene and o-xylene catabolism in Pseudomonas stutzeri OX1. Appl Environ Microbiol 62:3704–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Le NB, Coleman NV. 2011. Biodegradation of vinyl chloride, cis-dichloroethene and 1,2-dichloroethane in the alkene/alkane-oxidising Mycobacterium strain NBB4. Biodegradation 22:1095–1108. doi: 10.1007/s10532-011-9466-0. [DOI] [PubMed] [Google Scholar]
- 89.van Hylckama Vlieg JET, Kingma J, van den Wijngaard AJ, Janssen DB. 1998. A glutathione S-transferase with activity towards cis-dichloroepoxyethane is involved in isoprene utilization by Rhodococcus sp. strain AD45. Appl Environ Microbiol 64:2800–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Saeki H, Akira M, Furuhashi K, Averhoff B, Gottschalk G.. Degradation of trichloroethene by a linear-plasmid-encoded alkene monooxygenase in Rhodococcus corallinus (Nocardia corallina) B-276. Microbiology 145:1721–1730. [DOI] [PubMed] [Google Scholar]
- 91.Oldenhuis R, Oedzes JY, van der Waarde JJ, Janssen DB. 1991. Kinetics of chlorinated hydrocarbon degradation by Methylosinus trichosporium OB3b and toxicity of trichloroethylene. Appl Environ Microbiol 57:7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bertoni G, Bolognese F, Galli E, Barbieri P. 1996. Cloning of the genes for and characterization of the early stages of toluene and o-xylene catabolism in Pseudomonas stutzeri OX1. Appl Environ Microbiol 62:3704–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lloyd JS, De Marco P, Dalton H, Murrell JC. 1999. Heterologous expression of soluble methane monooxygenase genes in methanotrophs containing only particulate methane monooxygenase. Arch Microbiol 171:364–370. doi: 10.1007/s002030050723. [DOI] [PubMed] [Google Scholar]
- 94.Rui LY, Kwon YM, Fishman A, Reardon KF, Wood TK. 2004. Saturation mutagenesis of toluene ortho-monooxygenase of Burkholderia cepacia G4 for enhanced 1-naphthol synthesis and chloroform degradation. Appl Environ Microbiol 70:3246–3252. doi: 10.1128/AEM.70.6.3246-3252.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.