Abstract
Six strains of extremely halophilic and alkaliphilic euryarchaea were enriched and isolated in pure culture from surface brines and sediments of hypersaline alkaline lakes in various geographical locations with various forms of insoluble cellulose as growth substrate. The cells are mostly flat motile rods with a thin monolayer cell wall while growing on cellobiose. In contrast, the cells growing with cellulose are mostly nonmotile cocci covered with a thick external EPS layer. The isolates, designated AArcel, are obligate aerobic heterotrophs with a narrow substrate spectrum. All strains can use insoluble celluloses, cellobiose, a few soluble glucans and xylan as their carbon and energy source. They are extreme halophiles, growing within the range from 2.5 to 4.8 M total Na+ (optimum at 4 M) and obligate alkaliphiles, with the pH range for growth from 7.5 to 9.9 (optimum at 8.5–9). The core archaeal lipids of strain AArcel5T were dominated by C20–C20 dialkyl glycerol ether (DGE) (i.e. archaeol) and C20–C25 DGE in nearly equal proportion. The 16S rRNA gene analysis indicated that all six isolates belong to a single genomic species mostly related to the genera Saliphagus-Natribaculum-Halovarius. Taking together a substantial phenotypic difference of the new isolates from the closest relatives and the phylogenetic distance, it is concluded that the AArcel group represents a novel genus-level branch within the family Natrialbaceae for which the name Natronobiforma cellulositropha gen. nov., sp. nov. is proposed with AArcel5T as the type strain (JCM 31939T = UNIQEM U972T).
Keywords: Hypersaline, Soda lakes, Haloalkaliphilic, Natronoarchaea, Cellulotrophic
Introduction
Hypersaline habitats, such as inland salt and soda lakes and salterns with salt concentrations close to saturation are usually inhabited by a dense population of haloarchaea, which represent the extremely halophilic branch of the phylum Euryarchaeota. According to the current knowledge, haloarchaea are mostly aerobic heterotrophs, with a few exceptions of facultative anaerobes capable of utilizing simple soluble organic monomers [7], [8], [1], [16], [17], [6]. A few haloarchaeal species are capable of hydrolyzing polymeric substances, such as starch, proteins and olive oil [3], [5], [14], [19], [2]. However, the potential functioning of haloarchaea in the mineralization of insoluble organic polymers has not yet been considered and this function in hypersaline habitats is usually attributed to halophilic bacteria [1], [16]. In particular, next to nothing is known about the ability of haloarchaea to utilize native insoluble cellulose as a growth substrate. Glycosyl-hydrolase (GH) genes encoding putative cellulases have been noted in several haloarchaeal genomes and the presence of functional endoglucanases were demonstrated in two genera of neutrophilic haloarchaea, i.e. Halorhabdus and Haloarcula [10], [11], [26]. However, it remains to be investigated whether these archaea are actually capable of using native forms of cellulose as growth substrates.
So far, a single study focused on the functional aspect of cellulose degradation by haloarchaea has been published [22]. In that work, for the first time we were able to enrich and isolate in pure culture a number of haloarchaeal strains utilizing cellulose as the growth substrate. One of the most active groups included six natronarchaeal isolates from various alkaline hypersaline lakes which, to our knowledge, represent the first example of natronoarchaea with such a metabolic trait. This paper describes phenotypic and phylogenetic properties of the novel group and proposes to assign it into a novel genus and species Natronobiforma cellulotropha.
Material and methods
Samples
Surface sediments and near-bottom brines from various hypersaline alkaline inland lakes from Central Asia, Egypt and USA with salt concentration of 200–400 g l−1, pH from 9.3 to 11 and soluble carbonate alkalinity from 0.1 to 4 M were used to enrich for cellulotrophic natronoarchaea [22].
Enrichment, isolation and cultivation conditions
The alkaline (pH 9.5) base medium, containing 4 M total Na+ (2 M Na+ as sodium carbonates + 2 M NaCl) also included 1 g l−1 K2HPO4 and 5 g/l KCl and was supplemented after sterilization with 1 ml l−1 of trace metal solution and vitamin mix [18], 1 mM MgCl2, 2 mM (NH4)2SO4 and 20 mg l−1 of yeast extract. Addition of 200 mg l−1 of streptomycin served to inhibit growth of bacteria. Various forms of insoluble cellulose were used as the only carbon and energy source at a final concentration of 1 g l−1 (Table 1). Before inoculation, the sediments were resuspended 1:10 in the basic medium and after 5–10 min precipitation of the course fractions, a 1 ml portion from the top fraction containing mostly colloidal sediments and microbial cells was used to inoculate 20 ml cultures in 100 ml closed serum bottles placed on a rotary shaker at 37 °C and at 120 rpm. The development of cells was monitored by the visual extent of cellulose degradation, appearance of pink color and by microscopy. After visible cellulose degradation and biomass growth became evident (20–40 days), the cultures were serially diluted in the same medium but with amorphous cellulose as substrate and the maximal positive dilutions were plated onto a solid medium prepared by mixing 3 parts of the liquid medium (with additional solid NaCl addition to compensate for dilution with agar) and 2 parts of 5% extensively washed agar at 55 °C. After 2–6 weeks of incubation in closed plastic bags at 37 °C the colonies with clearance zones were transferred to the liquid media with amorphous cellulose and the positive cultures were further purified by several rounds of plating-liquid culture cultivation with amorphous cellulose. This, eventually, resulted in isolation of 6 pure cultures of cellulotrophic natronoarchaea with a common designation as AArcel (Table 1). The purity was checked microscopically (Zeiss Axioplan Imaging 2 microscope, Göttingen, Germany) and by the 16S rRNA gene sequencing.
Table 1.
Cellulotrophic natronoarchae isolated from hypersaline alkaline lakes.
| Strain | Isolated from: |
Chemical parameters of brines |
Cellulose form used in the enrichment | |||
|---|---|---|---|---|---|---|
| Lake | Area | Salt (g l−1) | pH | Alkalinity (M) | ||
| AArcel 2 | Bitter-1 | Kulunda Steppe Altai, Russia | 330 | 10.3 | 4.0 | Amorphous |
| AArcel 4 | Soda crystallizer | 380 | 9.6 | 3.1 | Avicel | |
| AArcel 5T | Tanatar-1 | 400 | 11.0 | 4.9 | Sigma 20 μm | |
| AArcel 9 | Mixed from 3 lakes | 330–400 | 9.6–11.0 | 3.1–4.9 | Filter paper | |
| AArcel 6 | Shar-Burdiin, Hotontyn | n-e Mongolia | 220–360 | 9.6–9.9 | 0.9–1.2 | Amorphous |
| AArcel 8-1 | Owens lake | California | 180 | 9.7 | 1.0 | Amorphous |
Phenotypic characterization
For the total cell electron microscopy, the cells were centrifuged and resuspended in 3 M NaCl, fixed with paraformaldehyde (final concentration 3%, v/v) for 2 h at room temperature, then washed again with the same NaCl solutions. The fixed cells were positively contrasted with 1% (w/v) uranyl acetate. For thin sectioning, the cell pellets were fixed in 1% (w/v) OsO4 containing 3.0 M NaCl for 1 week at 4 °C, washed and resuspended in 3 M NaCl, stained overnight with 1% (w/v) uranyl acetate, dehydrated in ethanol series, and embedded in Epon resin. Thin sections were post-stained with 1% (w/v) lead citrate.
The core membrane lipids were obtained by acid hydrolysis (5% HCl in methanol by reflux for 3 h) of the freeze-dried cells and subsequent analysis by HPLC-MS for GDGTs and archaeol derivatives according to [24]. Intact polar lipids were obtained by Bligh Dyer extraction of freeze-dried cells and subsequent HPLC-MS analysis as described in Ref. [20].
Genome sequencing
Fragment genomic libraries of strains Aarcel5T and Aarcel2 were prepared by NEBNext® Ultra™ kit (New England Biolabs, USA) according to the manufacturer’s instructions and sequenced on Illumina Miseq™ System using 2*150 bp paired-end read cartridge. Preliminary assembly and gene prediction was performed by CLC Genomics Workbench 10.5 (Qiagen, Germany) with recommended parameters. Draft genome assemblies were used only for obtaining 16S rRNA and rpoB′ gene sequences and will be published upon the improvement of assemblies by sequencing of long insert (jumping) genomic libraries.
Phylogenetic analysis
16S rRNA and rpoB′ gene sequences were extracted from the draft genome assemblies and deposited in the Genbank under the accession numbers: (MG938052-MG938053 for 16S rRNA and MG940906 and MG940907 for rpoB′ genes of strains AArcel5 and AArcel2, respectively).
To perform 16S rRNA gene sequence-based phylogenetic analysis, the sequences of all type species of the Natrialbaceae genera were obtained from the Genbank and aligned together with complete sequences of strains AArcel5T and AArcel2 and nearly complete sequences of strains AArcel9 and AArcel8-1 in Muscle, implemented in Mega 6 package [23]. The phylogenetic analysis was performed in Mega 6 using Maximum Likelihood algorithm and the General Time Reversible (GTR) model (G + I, 4 categories) [15]. Partial sequences of AArcel4 and AArcel6 were identical to AArcel5 sequence and were not analyzed but placed on the tree together with the type strain.
To perform the rpoB'-based phylogenetic analysis, full-length nucleotide sequences of the rpoB' gene of type species from the Natrialbaceae were obtained from GenBank or IMG and aligned using the G-INS-i method in MAFFT server v7 [9]. Phylogenetic tree was constructed in Mega 6 using Maximum Likelihood algorithm with GTR model (G + I, 4 categories. For phylogeny reconstruction based on RpoB′ proteins, the nucleotide sequences were translated, aligned in MAFFT server v7 using the G-INS-i algorithm, and the tree was constructed using Maximum Likelihood algorithm with the LG model. To estimate the rpoB′ gene distances of all validly published Natrialbaceae representatives, the pairwise distances matrix based on percentage of sequence identities was constructed using Mega 6.
Results and discussion
Phenotypic properties
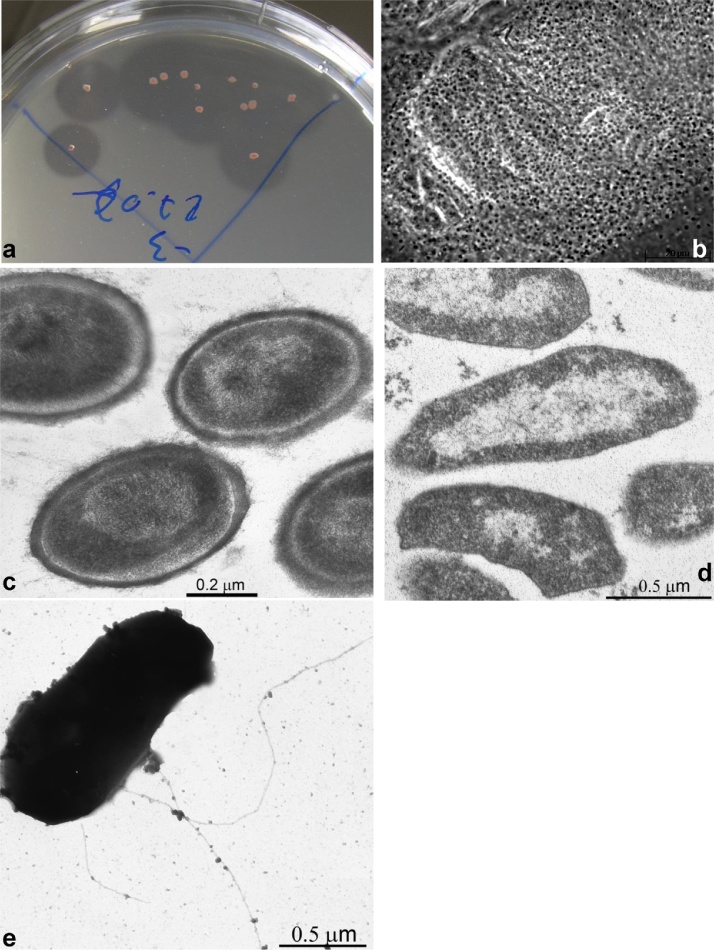
On plates with amorphous cellulose, all strains formed pin-point pink colonies after 4–6 weeks incubation with a large clearance around them, indicative of cellulose hydrolysis (Fig. 1a). Growth in liquid culture with all forms of celluloses started with a massive attachment of cells to the solid phase cellulose surface, followed by gradual dissolution of cellulose and appearance of cells in the liquid phase. A dramatic change in cell morphology was observed in those two phases. The cells aggregated with the cellulose particles were non-motile cocci (Fig. 1b) covered with a thick electron dense external layer (Fig. 1c), while the free suspended cells in the second growth phase were dominated by motile thin flat rods with a thin cell wall (Fig. 1d,e). The cells in the colonies resembled the coccoid cells from the first phase in liquid cultures, while the cells grown on cellobiose in liquid culture were similar to those from the second growth phase on cellulose.
Fig. 1.
Cell morphology of strain AArcel5 growing on amorphous cellulose at pH 9.5, 4 M total Na+ and 37 °C. (a) colonies; (b) phase contrast and (c) electron microscopy of thin sections of cells during absorption phase on cellulose; (d) electron microscopy of thin sections and (e) and whole free suspended cells from the second growth phase on cellulose.
The polar membrane lipids were analyzed in the type strain AArcel5T grown with cellobiose at 37 °C. 4 M total Na+ and pH 9.3 harvested in the mid-exponential growth phase. The core membrane lipids were represented by two dominant components: archaeol (C20–C20 dialkyl glycerol ether (DGE), 58% of the total) and extended archaeol (C20–C25 DGE, 40% of the total). Traces of the monoglycerol ether (MGE) lipids (1-C20 MGE, 2-C20 MGE, and 2-C25 MGE) were also detected. The intact polar lipids were dominated (in order of abundance) by phosphatidylglycerophosphate methylester (PGP-Me), phosphatidylglycerol (PG), a phosphatidylglycose (GL-PG), a diglycosyl (2GL), and phosphatidylglycerophosphate (PGP) (Supplementary Fig. S1).
The AArcel strains are obligately aerobic saccharolytic archaea with a limited range of substrates supporting growth. They are the first natronoarchaea reported as being specialized in the utilization of native insoluble celluloses (but they cannot use an artificial soluble analogue carboxymethyl cellulose CMC) [22]. All strains can grow with xylan (from birch and beech) and barley beta-glucan. AArcel strains 2, 4, and 5 also utilized lichenan, glucomannan and β-1,4-mannan, but growth was much slower. Amorphous chitin also seems to be a substrate for these 3 strains, but the growth was unstable: the transition from cellulose to chitin was only randomly successful, failing on many occasions. Apparently growth on this polymer is not optimal for these archaea. Alpha-glucans, such as starch and starch-like polymers, were not utilized. Among the sugars, only two dimers were used — cellobiose and maltose (less actively). Sugars not utilized included: glucose, fructose, galactose, mannose, rhamnose, arabinose, raffinose, sucrose, trehalose, maltose, glucosamin, N-acetylglucosamine, glucouronic acid, halacturonic acid, lactose, ribose, xylose, melezitose and melibiose. Sugar alcohols which tested negative included glycerol, sorbitol and mannitol. Negative organic acids were C2–C8 saturated fatty acids, lactate, pyruvate, succinate, malate and fumarate. The organic nitrogen compounds not utilized were glutamate, aspartate meat and casein peptons and yeast extract. Ammonium was utilized as the N-source with cellobiose as carbon and energy substrate, while nitrate and urea were negative. Anaerobic fermentative growth with arginine, cellobiose or maltose was not observed, nor was anaerobic respiration (with cellobiose as the electron donor) with nitrate, sulfur, fumarate or DMSO as electron acceptors.
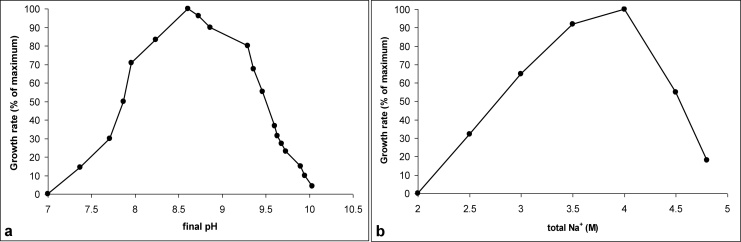
These archaea belong to the group of extreme halophiles, growing optimally at 4 M total Na+ (Fig. 2a) and are moderate alkaliphiles with a pH optimum around 9 (Fig. 2b). It is important to stress that, when working with alkaliphiles, the pH profiling should be performed with an obligatory check of the pH change during growth [21]. For this particular group of natronoarchaea mineralizing sugars and producing metabolic acids, we observed a drop of the pH from 11 to 9.5 even when using a highly buffered sodium carbonate system. In our experience, if a natronarchaeon is growing at pH 6 it will not grow at pH 10, which leads to the impression that in the case, for example, of Natribaculum breve, the final pH check at the highest pH range was not performed [12]. The type strain AArcel5T grew within a relatively wide temperature range from 20 to 53 °C with an optimum at 40 °C. However, it should be mentioned that the growth at high temperature (above 43 °C) was only possible at the lowest growth pH (8.5). The most probable explanation is that a combination of high temperature and high alkalinity results in instability of the haloarchaeal S-layer.
Fig. 2.
Influence of pH at 4 M total Na+ (a) and Na+ at pH 9 (b) on growth of strain AArcel5 with cellobiose at 37 °C. The results are mean values from two biological replicate experiments.
Phylogenetic analysis
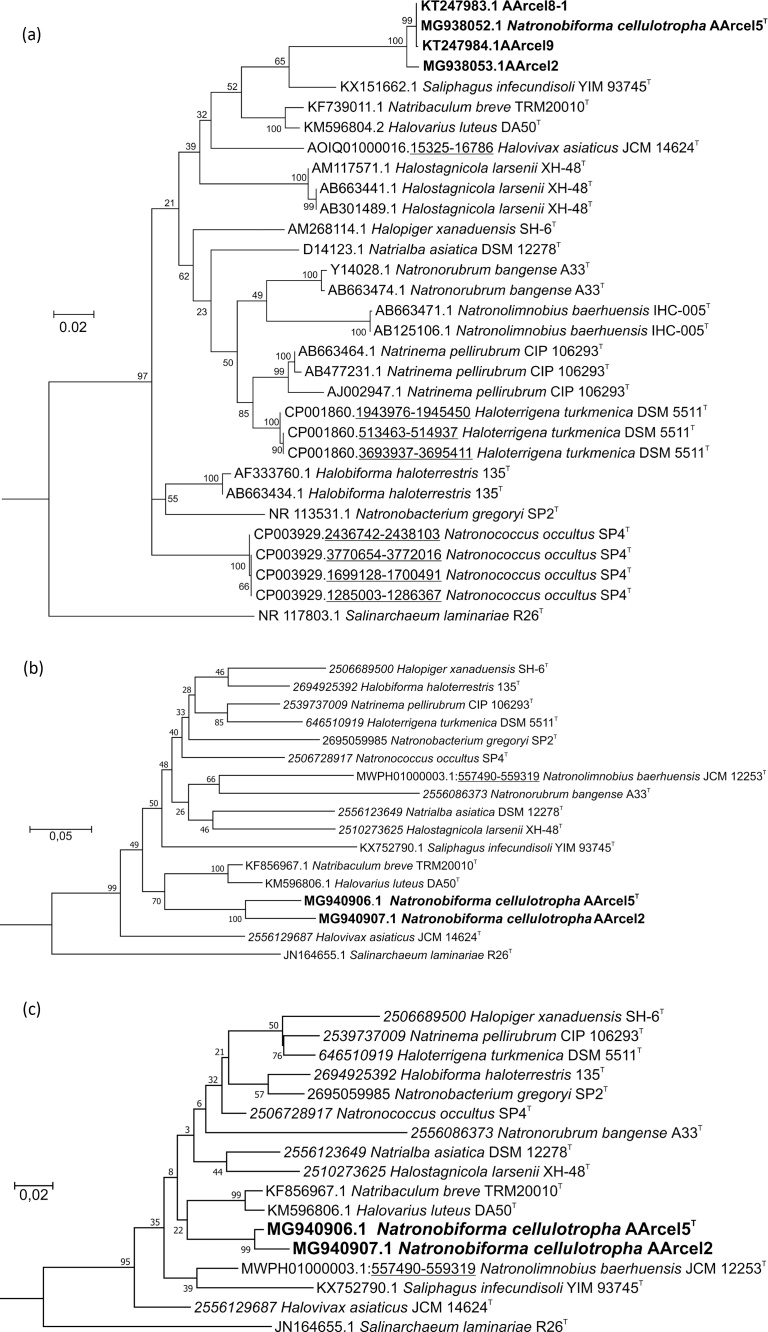
The results of 16S rRNA gene sequence analysis demonstrated that the AArcel isolates formed a separate, single species-level group within the family Natrialbaceae with a recently described neutrophilic Saliphagus infecundisoli as the closest relative (Fig. 3a). Strain AArcel2 was most remote from the type strain AArcel5T (99.1% of sequence identity), but still within the currently recognized species border. Calculated of the Average Nucleotide Identity (ANI) between these two genome-sequenced strains gave a value close to the statistically average species border (95%). Taking into account practically identical phenotypes, it can be concluded that all six AArcel cellulotrophic isolates belong to a single species.
Fig. 3.
Phylogeny of the AArcel strains. (a) Maximum Likelihood 16S rRNA gene sequence-based phylogenetic tree showing position of the AArcel strains (in bold) within the family Natrialbaceae. Branch lengths (see scale) correspond to the number of substitutions per site with corrections, associated with the model (GTR, G + I, 4 categories). All positions with less than 95% site coverage were eliminated. Totally 1359 positions were used in the alignment of 32 sequences (except for the partial AArcel4 and AArcel6 sequences, 100% identical to AArcel5T). Numbers at nodes indicate bootstrap values of 1000 repetitions. Halomarina oriensis strain JCM 16495 (AB663390.1) was used as an outgroup. (b) Maximum Likelihood rpoB' gene sequence-based tree showing position of the AArcel2 and AArcel5 strains (in bold) within family Natrialbaceae. Totally 1827 positions were used in the alignment of 18 sequences. Halomarina oriensis JCM 16495 (KJ870934.1) was used as an outgroup. (c): Maximum Likelihood RpoB' protein sequence-based tree showing position AArcel2 and AArcel5 strains (in bold) within the family Natrialbaceae. Totally 608 positions were used in the alignment of 18 sequences. Halomarina oriensis JCM 16495 (KJ870934.1) was used as an outgroup. Sequences with accession numbers in italic were obtained from IMG, in roman – from the Genbank.
Another marker, widely used for phylogenetic reconstructions of Halobacteria, is the RNA-polymerase subunit B′ gene [13]. The phylogenetic trees based on comparative analysis of rpoB′ gene and RpoB′ protein sequences of the type species from Natrialbaceae revealed that AArcel5T and AArcel2 formed a branch positioned separately from the closely related genera (Fig. 3b,c). The rpoB′ gene sequences of AArcel5 and AArcel2 strains were 92.2% similar to each other and had 82.5–88.8% identity with the members of Natrialbaceae (Supplementary Table S1 and Fig. S2). Phylogenetic analysis of the rpoB'-gene of all validly published Natrialbaceae was done to reveal all inter- and intra-genus clustering (Supplementary Fig. S2). It showed that only Halovivax, Halostagnicola, Natrialba, Natronococcus, Saliphagus and Salinarchaeum genera formed monophyletic clusters. These genera are similar to AArcel5T-AArcel2 in the intrageneric/intergeneric distances, calculated basing on the percentage of sequence identities (Supplementary Table S1): Halovivax 89.9–97.8%/82.4–89.6%; Halostagnicola 94.4%/81.2–89%; Natrialba 88.6–98.6%/82–89%; Natronococcus 91.4–93%/82.4–90.8%; Saliphagus nd/81.5–87.1% and Salinarchaeum 92.4%/80.2–85.1%.
Furthermore, phenotypic comparison shows a clear physiological differentiation of the AArcel isolates from the three closest relatives in Natrialbaceae (Table 2).
Table 2.
Comparative property of cellulotrophic natronoarchaea with the nearest phylogenetic relatives in Natrialbaceae: Saliphagus infecundisoli[25], Natribaculum breve[13] and Halovivax asiaticus[4]. PGP-Me – phosphatidylglycerophosphate methylester; PG – phosphatidylglycerols; GL-PG – phosphatidylglycose; 2GL – diglycosyl; PGS – phosphatidylglycerol sulfate; PGP – phosphatidylglycerophosphate; GL – glycolipid; PL – phospholipid; glycolipids: TGD-1 (galactosyl mannosyl glucosyl diether), S2-DGD (disulfated mannosyl glucosyl diether). Antibiotics: s, streptomycin; k, kanamycin; a, ampicillin; t, tetracyclin; v, vancomycin; g, gentamycin; r, rifampicin;. e, erythromycin, c, chrolarmphenicol.
| Property | “Natronobiforma cellulositropha” (6 strains) | Saliphagus infecundisoli | Natribaculum breve | Halovivax asiaticus |
|---|---|---|---|---|
| Cell morphology | Thin flat motile rods on cellobiose; cocci with thick cell wall on cellulose | Cocci | Motile pleomorphic rods | Pleomorphic nonmotile, from rods to discs |
| Pigmentation | Pink | Pink | Red | Pale-pink |
| Anaerobic growth | - (With cellobiose as substrate) | – | Contradictorya | – |
| Growth substrates: | ||||
| Polymers | Insoluble cellulose, xylane, chitin (3 strains), β-1,4 glucans and mannan | Starch, dextrin | Starchb, gelatin hydrolysis | Proteolyticc |
| Sugars | Cellobiose, maltose | Glucose, mannose, raffinose, sucrose, trehalose | Glucose | Lactose, raffinose, xylose, trehalose |
| Amino acids | – | Glutamate, aspartate, ornithine, lysine | ||
| Organic acids | – | Pyruvate, succinate | Pyruvate | Acetate |
| Esterase activity | – | Tween-20 | – | Tween-80 |
| Catalase/oxidase | +/+ | +/+ | +/Weak | |
| Antibiotic resistance | s, k, a, t, v, g, e, p (50–100 mg l−1) r, c (<50 mg l−1) | a, v, g, e, c, p (discs) | s, k, a, t, v, g, e, c, p, r (discs) | s, k, a, t, v, g, e, c, p (discs) |
| Salinity range (opt.) M Na+ | 2.5–4.8 (4.0) | 2–6 (2.5–3.0) | 0.9–5.1 (2.6) | 1.6–4.8 (2.5) |
| Mg requirement | Low | Low | Low | Low |
| pH range (opt.) | 7.5–9.9 (8.5–9.0) | 6.0–8.5 (7.0–7.5) | 6.0–10e (7.0–7.5) |
6.5–8.5 |
| Temperature (°C) | 18–53d (opt. 43) | 25–50 (opt. 37) | 30–62 (opt. 37) | 25–45 (opt. 37) |
| Core lipids | C20–C20, C20–C25 | nd | nd | C20–C20, C20–C25 |
| Polar lipids | PG, PGP-Me, GL-PG, 2GL, PGP | PG, PGP-Me, PGS, three GL | PG, PGP-Me, TGD-1, S2-DGD | PG, PGP-Me, two PL, four GL |
| G + C, mol% | 65.4–65.5 | 64.4 | 63.9 | 60.3 |
| Habitat | Hypersaline alkaline lakes (s–w Siberia, n–e Mongolia, California) | Saline soil (China) | Saline soil (China) | Hypersaline lake (Inner Mongolia) |
nd – no data.
It is stated that it grows anaerobically with nitrate, and next – that it can not reduce nitrate to nitrite or nitrite to N2.
Since this organism did not utilize maltose, its capability to grow with starch is questionable.
Growth on polymers was not investigated.
At pH 8.5.
The final pH is not measured, the high pH limit is not justified.
In conclusion, the six AArcel strains isolated from hypersaline alkaline lakes represent a first example of natronoarchaea specialized in utilization of native insoluble celluloses as growth substrate. Taking into account their unique phenotypic properties and the phylogenetic distances (based on two conservative phylogenetic markers) from the nearest genera in Natrialbaceae, we propose to classify the group as a novel genus and species Natronobiforma cellulotropha with strain AArcel5 as the type strain. The novel genus and species protologue (diagnosis) is provided in Table 3.
Table 3.
Natronobiforma cellulositropha: protologue.
| Parameter | Genus: Natronobiforma gen. nov. | Species: Natronobiforma cellulositropha sp. nov. |
| Date created | 2018-03-04 | 2018-03-04 |
| Taxon number (TXNR) | TA00433 | TA00433 |
| Author (AUTE) | Dimitry Y. Sorokin | |
| Species name (SPNA) | Natronobiforma cellulotropha | |
| Genus name (GENA) | Natronobiforma | |
| Specific epithet (SPEP) | “Cellulotropha” from Natronobiforma cellulotropha | |
| Species status (SPST) | sp. nov. | |
| Etymology (GETY/SPTY) | Natronobiforma (Na.tro.no.bi.for' ma Gr. neutral n. natron, arbitrarily derived from the Arabic n. natrun or natron, soda; L. adv. num. bis, twice; L. fem. n. forma, form, shape; N.L. fem. n. Natronobiforma, the dimorphic natronoarchaeon | Cellulositropha (cel.lu.lo.si.tro' pha N.L. n. cellulosum, cellulose; N.L. fem. n. from Gr. n. fem. trophê, nourishment, food; N.L. fem. adj. cellulositropha, utilizer of cellulose) |
| Authors (AUT) | Dimitry Y. Sorokin, Tatiana V. Khijniak, Nadezhda A. Kostrikina, Alexander G. Elcheninov, Stepan V. Toshchakov, Nicole J. Bale, Jaap S. Sinninghe Damstéd, Ilya V. Kublanov | |
| Title (TITL) | Natronobiforma cellulotropha gen. nov., sp. nov., a novel haloalkaliphilic member of the family Natrialbaceae (class Halobacteria) from hypersaline alkaline lakes | |
| Journal (JOUR) | Systematic and Applied Microbiology | |
| Corresponding author (COAU). | Dimitry Y. Sorokin | |
| E-mail of corresponding author (EMAU) | d.sorokin@tudelft; soroc@inmi.ru | |
| Designation of the type strain (TYPE) | AArcel5 | |
| Strain collection numbers (COLN) | JCM31939; UNIQEM U972 | |
| 16S rRNA gene accession number (16 SR) | KT247980 | |
| Alternative house-keeping genes: gene [accession numbers] (HKGN) | rpoB′ [MG940906] | |
| Genome status (GSTA) | Draft | |
| GC mol% (GGCM) | 65.4–65.5 (genomes of AArcel5T and AArcel2) | |
| Country of origin (COUN) | Russian Federation | |
| Region of origin (REGI) | Altai region | |
| Date of isolation (DATI) | 2013-08-15 | |
| Source of isolation (SOUR) | Surface sediments and brines of hypersaline alkaline lakes | Surface sediments from hypersaline soda lake Tanatar-1 |
| Sampling dates (DATS) | 2013-07-07 | |
| Geographic location (GEOL) | South Siberia, N–E Mongolia, California | S–W Siberia, Kulunda Steppe |
| Latitude (LATI) | 51°39′N | |
| Longtitude (LONG) | 79°48′E | |
| Depth (DEPT) | 0.1 m | |
| Temperature of the sample (TEMS) | 25 °C | |
| pH of the sample (PHSA) | 11.0 | |
| Salinity of the sample (SALS) | 40% | |
| Number of strains in study (NSTR) | 6 | |
| Source of isolation of non-type strains (SAMP) | Hypersaline alkaline lakes in Russia, Mongolia and California | |
| Growth medium, incubation conditions (CULT) | Alkaline medim containing 4 M Na+ with pH 9–9.5 and cellulose as substrate | 4 M total Na+, equal mix of sodium carbonate and NaCl on the basis of Na molarity, pH 9.5; incubation – 37 °C; amorphous cellulose or cellobiose as C and energy source |
| Conditions of preservation (PRES) | Deep freezing in 15% glycerol (v/v) | |
| Gram stain (GRAM) | Negative | |
| Cell shape (CSHA) | Pleomorphic, from flat motile rods to nonmotile coccoid cells | |
| Cell size (CSZI) | 0.5–0.8 μm in diameter, length is variable | |
| Motility (MOTY) | Motile | |
| Motility type (MOTK) | Flagellar | |
| Type of flagellation (TFLA) | Variable, from single subpolar to several peretrichous flagella | |
| Sporulation (SPOR) | None | |
| Colony morphology (COLM) | Pink, up to 2 mm | |
| Temperature range for growth (TEMR) | 20–53 °C | |
| Lowest temperature for growth (TEML) | 20 | |
| Highest temperature for growth(TEMH) | 53 | |
| Optimal temperature for growth (TEMO) | 43 | |
| Lowest pH for growth (PHLO) | 7.5 | |
| Highest pH for growth (PHHI) | 9.9 | |
| Optimum pH for growth (PHOP) | 9.0 | |
| pH category (PHCA) | Alkaliphile (optimum > 8.5) | |
| Lowest NaCl concentration for growth (SALL) | 2.5 | |
| Highest NaCl concentration for growth (SALH) | 4.8 | |
| Optimum salt concentration for growth (SALO) | 4.0 | |
| Other salts important for growth | Sodium carbonates | |
| Salinity category (SALC) | Extreme halophilic (optimum & gt; 15% NaCl) | |
| Relation to oxygene (OREL) | Aerobe | |
| O2 conditions for strain testing (OCON) | Aerobic | |
| Carbon source used (class) (CSUC) | Carbohydrates | |
| Specific compounds (CSUC) | Cellulose, xylan, mannan, cellobiose, maltose | |
| Nitrogen source (NSOU) | Ammonium | |
| Terminal electron acceptor (ELAC) | O2 | |
| Energy metabolism (EMET) | Chemoorganotrophic | |
| Phospholipids (PHOS) | Core membrane lipids are archaeol (C20–C20 DGE) and C20–C25 DGE in equal proportion | Phosphatidylglycerophosphate methylester (PGP-Me), phosphatidylglycerol (PG), phosphatidylglycerol sulfate (PGS) and phosphatidylglycerophosphate (PGP) |
| Glycolipids (GLYC) | Phosphatidylglycose (GL-PG), diglycosyl (2GL) | |
| Habitat (HABT) | Hypersaline alkaline lakes | |
| Extraordinary feautres (EXTR) | Growth with native insoluble cellulose | Fast growth with insoluble native celluloses; more than 30 GH glucosyl-hydrolases genes in the genome |
Funding
This work was supported by the Russian Science Foundation (grant 16-14-00121) and by the Russian Academy of Sciences and Federal Agency of Scientific Organizations (project 0104-2018-0033). This project also received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement No. 694569 – MICROLIPIDS).
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.syapm.2018.04.002.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Andrei A.S., Banciu H.L., Oren A. Living with salt: metabolic and phylogenetic diversity of archaea inhabiting saline ecosystems. FEMS Microbiol. Lett. 2012;330:1–9. doi: 10.1111/j.1574-6968.2012.02526.x. [DOI] [PubMed] [Google Scholar]
- 2.Amoozegar M.A., Siroosi M., Atashgahi S., Smidt H., Ventosa A. Systematics of haloarchaea and biotechnological potential of their hydrolytic enzymes. Microbiology. 2017;163:623–645. doi: 10.1099/mic.0.000463. [DOI] [PubMed] [Google Scholar]
- 3.Bhatnagar T., Boutaiba S., Hacene H., Cayol J.-L., Fardeau M.-L., Ollivier B., Baratti J.C. Lipolytic activity from Halobacteria: screening and hydrolase production. FEMS Microbiol. Lett. 2005;248:133–140. doi: 10.1016/j.femsle.2005.05.044. [DOI] [PubMed] [Google Scholar]
- 4.Castillo A.M., Gutiérrez M.C., Kamekura M., Ma Y., Cowan D.A., Jones B.E., Grant W.D., Ventosa A. Halovivax asiaticus gen. nov., sp. nov., a novel extremely halophilic archaeon isolated from Inner Mongolia, China. Int. J. Syst. Evol. Microbiol. 2006;56:765–770. doi: 10.1099/ijs.0.63954-0. [DOI] [PubMed] [Google Scholar]
- 5.Enache M., Kamekura M. Hydrolytic enzymes of halophilic microorganisms and their economic values. Rom. J. Biochem. 2010;47:47–59. [Google Scholar]
- 6.Grant B.D., Jones B.E. Bacteria, archaea and viruses of soda lakes. In: Schagerl M., editor. Soda lakes of East Africa. Springer; Switzerland: 2016. pp. 97–147. [Google Scholar]
- 7.Halophiles and hypersaline environments. Current research and future trends. Ventosa, A., Oren, A., Ma, Y. (Eds.) (2011) Springer, Heidelberg, Dordrecht, London, New York, 387 pp.
- 8.Halophiles, Part III. In: Extremophiles Handbook, Horikoshi, K. (Ed.) (2011) vol. 1, Springer, Tokyo, pp. 255–402.
- 9.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2017 doi: 10.1093/bib/bbx108. bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li T.X., Yu H.-Y. Halostable cellulase with organic solvent tolerance from Haloarcula sp. LLSG7 and its application in bioethanol fermentation using agricultural wastes. J. Ind. Microbiol. Biotechnol. 2013;13:1357–1365. doi: 10.1007/s10295-013-1340-0. [DOI] [PubMed] [Google Scholar]
- 11.Li X., Yu H.-Y. Characterization of a halostable endoglucanase with organic solvent-tolerant property from Haloarcula sp. G10. Int. J. Biol. Macromol. 2013;62:101–106. doi: 10.1016/j.ijbiomac.2013.08.047. [DOI] [PubMed] [Google Scholar]
- 12.Liu Q., Ren M., Zhang L.-L. Natribaculum breve gen. nov., sp. nov. and Natribaculum longum sp. nov., halophilic archaea isolated from saline soil. Int. J. Syst. Evol. Microbiol. 2015;65:604–608. doi: 10.1099/ijs.0.060541-0. [DOI] [PubMed] [Google Scholar]
- 13.Minegishi H., Kamekura M., Itoh T., Echigo A., Usami R., Hashimoto T. Further refinement of the phylogeny of the Halobacteriaceae based on the full-length RNA polymerase subunit B′ (rpoB′) gene. Int. J. Syst. Evol. Microbiol. 2011;60:2398–2408. doi: 10.1099/ijs.0.017160-0. [DOI] [PubMed] [Google Scholar]
- 14.Moshfegh A.R.M., Shahverdi G., Zarrini M.A. Faramarzi Biochemical characterization of an extracellular polyextremophilic α-amylase from the halophilic archaeon Halorubrum xinjiangense. Extremophiles. 2013;17:677–687. doi: 10.1007/s00792-013-0551-7. [DOI] [PubMed] [Google Scholar]
- 15.Nei M., Kumar S. Oxford University Press; New York: 2000. Molecular Evolution and Phylogenetics. [Google Scholar]
- 16.Oren A. Halophilic microbial communities and their environments. Curr. Opin. Biotechnol. 2015;33:119–124. doi: 10.1016/j.copbio.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Oren A. Life at high salt concentrations. In: Rosenberg E., editor. The Prokaryotes. Ecophysiology and Biochemistry. 4th ed. Springer; NewYork: 2013. pp. 429–440. [Google Scholar]
- 18.Pfennig N., Lippert K.D. Über das Vitamin B12-Bedürfnis phototropher Schwefelbakterien. Arch. Mikrobiol. 1966;55:245–256. [Google Scholar]
- 19.Selim S., Hagagy N., Azis M.A., Meleigy T.S., Pessione E. Thermostable alkaline halophilic-protease production by Natronolimnobius innermongolicus WN18. Nat. Prod. Res. 2014;28:1476–1479. doi: 10.1080/14786419.2014.907288. [DOI] [PubMed] [Google Scholar]
- 20.Sinninghe Damsté J.S., Rijpstra W.I.C., Hopmans E.C., Jung M.Y., Kim J.G. Intact polar and core glycerol dibiphytanyl glycerol tetraether lipids of group I. 1a and I. 1b Thaumarchaeota in soil. Appl. Environ. Microbiol. 2012;78:6866–6874. doi: 10.1128/AEM.01681-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sorokin D.Y. Is there a limit for high-pH growth? Int. J. Syst. Evol. Microbiol. 2005;55:1405–1406. doi: 10.1099/ijs.0.63737-0. [DOI] [PubMed] [Google Scholar]
- 22.Sorokin D.Y., Toschakov S.V., Kolganova T.V., Kublanov I.V. Halo(natrono)archae isolated from hypersaline lakes utilize cellulose and chitin as growth substrates. Front. Microbiol. 2015;6 doi: 10.3389/fmicb.2015.00942. article 942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weijers J.W.H., Panoto E., van Bleijswijk J., Schouten S., Balk M., Stams A.J.M., Rijpstra W.I.C., Sinninghe Damsté J.S. Constraints on the biological source(s) of the orphan branched tetraether membrane lipids. Geomicrobiol. J. 2009;26:402–414. [Google Scholar]
- 25.Yin X.-Q., Liu B.-B., Chu X., Salam N., Li X., Yang Z.-W., Zhang Y., Xiao M., Li W.-J. Saliphagus infecundisoli gen. nov., sp. nov., an extremely halophilic archaeon isolated from a saline soil. Int. J. Syst. Evol. Microbiol. 2018;67:4154–4160. doi: 10.1099/ijsem.0.002270. [DOI] [PubMed] [Google Scholar]
- 26.Zhang S., Datta J., Eichler N., Ivanova S.D., Axen C.A., Kerfeld E. Rubin Identification of a haloalkaliphilic and thermostable cellulose with improved ionic liquid tolerance. Green Chem. 2011;13:2083–2090. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.