Abstract
Posttranscriptional gene regulation by RNA-binding proteins, such as HuR (elavl1), fine-tune gene expression in T cells, leading to powerful effects on immune responses. HuR can stabilize target mRNAs and/or promote translation by interacting with their 3′ untranslated region adenylate and uridylate–rich elements. It was previously demonstrated that HuR facilitates Th2 cytokine expression by mRNA stabilization. However, its effects upon IL-2 homeostasis and CD4+ Th2 differentiation are not as well understood. We found that optimal translation of Il2ra (CD25) required interaction of its mRNA with HuR. Conditional HuR knockout in CD4+ T cells resulted in loss of IL-2 homeostasis and defects in JAK–STAT signaling, Th2 differentiation, and cytokine production. HuR-knockout CD4+ T cells from OVA-immunized mice also failed to proliferate in response to Ag. These results demonstrate that HuR plays a pivotal role in maintaining normal IL-2 homeostasis and initiating CD4+ Th2 differentiation.
INTRODUCTION
The control of T cell differentiation involves complex biological changes in multiple steps of gene regulation to orchestrate proper immune responses. Naive CD4+ T cells are capable of differentiating into several distinct lineages, including Th1, Th2, Th9, Th17, T follicular helper, and regulatory T cells. The fate of naive CD4+ T cell differentiation is determined by the strength of TCR signaling and the cytokines present during stimulation (1–3).
IL-2 is rapidly produced by T cells upon TCR stimulation, although its expression is only transient (4, 5). IL-2 itself acts in an autocrine and paracrine manner to regulate its own expression via signaling through IL-2R (4, 6). Intermediate (IL-2Rβ and common γ-chain) and high-affinity (IL-2Rα, IL-2Rβ and common γ-chain) IL-2Rs can transduce signals via the JAK–STAT pathway, primarily through phosphorylation of Stat5 (7, 8). This, in turn, activates Prdm1 (encoding Blimp1) transcription (9). Blimp1 acts as a transcriptional repressor, binding to the Il2 promoter and attenuating its transcription (9).
Over the past several decades, IL-2 has been shown to be required for optimal Th2 differentiation (10, 11). The effect of IL-2 on Th2 differentiation is not dependent on Gata3 (11, 12). IL-2 activates Th2 differentiation independently of its proliferation-inducing ability through p-Stat5, which augments Il4 locus accessibility (11, 13). IL-2/p-Stat5 signaling and Gata3 have a synergistic effect on IL-4 production (12), and the combination of IL-2/Stat5 and IL-4/Stat6/Gata3 signaling results in a strong positive-feedback mechanism to maintain Th2 lineage commitment (14–16).
Upon T cell activation, up to 50% of the dynamic changes that T cells experience occur at the posttranscriptional level (17, 18). Several trans-acting factors, including RNA-binding proteins (RBPs), microRNAs, and long noncoding RNAs (19), fine-tune T cell immune responses at this step of regulation (20, 21). A group of RBPs, which determine mRNA maturation, localization, stability, and translation, exhibit binding activity through recognizing adenylate and uridylate–rich elements (AREs) or U-rich elements that are often found in the 3′ untranslated region (UTR) of their target mRNAs (22, 23). Among these RBPs, HuR (elavl1) functions as a key regulating factor that modulates mRNA localization, stability, and translation (24). In resting cells, HuR localization is 90% nuclear (25); however, upon activation it is translocated to the cytoplasm where it mediates its functions (25). HuR is ubiquitously expressed and highly upregulated (~14-fold) in T cells post-activation (25). The mechanisms by which HuR facilitates mRNA stability and/or translation are not fully elucidated and may be due to the interplay of HuR with other destabilizing RBPs or microRNAs (26, 27). We demonstrated that HuR positively regulates the hallmark genes involved in Th2 and Th17 differentiation (28–31). IL-4, IL-13, and Gata3 expression are upregulated or downregulated when HuR is overexpressed or underexpressed in CD4+ T cells, respectively (31, 32). However, we found that HuR ablation in activated CD4+ T cells after Th2 lineage commitment resulted in paradoxical increases in Il4 and Il13 transcripts (30). This unexpected result led us to hypothesize that HuR may regulate Th2 transcripts differently before and after Th2 lineage commitment.
To further investigate HuR function in CD4+ T cells prior to activation, we generated distal lck-Cre ROSA HuRfl/fl mice to genetically ablate HuR in late-stage thymocytes. HuR-deleted CD4+ T cells have severely impaired production of IL-4, IL-5, and IL-13 upon activation. Nevertheless, they have strikingly increased IL-2 levels, with no change in IFN-γ expression. We established that the interaction of HuR with Il2ra mRNA is required for its optimal translation. Together, our data show that HuR is required for controlling normal IL-2 homeostasis, as well as for augmenting Th2 differentiation.
MATERIALS AND METHODS
Generation of distal lck-Cre ROSA HuRfl/fl
HuR floxed mice (HuRfl/fl) were established in the Atasoy laboratory, as previously described (30). Briefly, a vector was designed for homologous recombination in which the HuR gene was floxed with the insertion of loxP sites, resulting in the targeted deletion of exons 1 and 2 and a portion of the promoter region, additionally introducing a frame-shift mutation upon Cre-mediated recombination. The Neo gene was used as a selection marker, and flippase recognition target sites were used to delete the Neo gene after selection was completed by breeding to an flp recombinase mouse (see Ref. 30 for fuller details). To generate distal lck-Cre ROSA HuRfl/fl mice, HuRfl/fl mice were crossed to distal lck-Cre and ROSA-YFP mice. As outlined in Supplemental Fig. 1, a stop codon with loxP sites is inserted upstream of the YFP open reading frame (ORF), so that YFP protein is not expressed in cells that do not normally express Cre recombinase. However, Cre recombinase expression leads to targeted ablation of HuR gene, as well as removal of the stop codon upstream of YFP ORF, so YFP protein is expressed. Therefore, YFP protein fate maps cells in which HuR has been genetically ablated. All mice used were on a C57BL/6 background. All animal experiments and procedures were conducted in accordance with the guidelines set forth by the University of Missouri Animal Care and Use Committee.
CD4+ and naive CD4+ T cell isolation
Naive splenocytes and peripheral lymph nodes (LNs) were isolated from 8–12-wk-old distal lck-Cre ROSA HuRfl/fl or ROSA HuRfl/fl littermate control mice. CD4+ T cells were isolated using murine anti-CD4 (L3T4) MACS MicroBeads positive-column purification, following the manufacturer’s protocol (Miltenyi Biotec). Naive CD4+ T cells were isolated from peripheral LNs and spleens (SPs) of mice using a MACS Mouse Naive CD4+ T Cell Isolation Kit, following the manufacturer’s protocol.
Murine T cell activation in vitro
Isolated CD4+ T cells or naive CD4+ T cells from distal lck-Cre ROSA HuRfl/fl mice were separated according to their YFP expression (YFP+ HuR knockout [KO] versus YFP− endogenous control) using a MoFlo XDP sorter (Beckman Coulter). Naive CD4+ T cells or isolated CD4+ T cells from ROSA HuRfl/fl mice were used as wild-type (WT; exogenous) controls. Then cells were activated with plate bound anti-CD3 (5 μg/ml) and anti-CD28 (4 μg/ml) for 5 d in T cell media (RPMI 1640, 10% FCS, gentamicin, sodium pyruvate, L-glutamine, and 2-ME). Fresh media were added to the cells ondays2–4. Murine recombinant IL-4(100 U/ml) was added in some experiments, as indicated. For kinetic studies, cells were collected on days 0–5 and analyzed for RNA by RT-PCR and for protein by flow cytometry or Western blot.
Intracellular staining and flow cytometry
For cytokine staining, activated CD4+ T cells were restimulated with PMA (50ng/ml), ionomycin(1μg/ml), and brefeld in A(3μg/ml) for 5 h in T cell media at a concentration of 1 × 106 cells per milliliter. Cells (1–5 × 106) were then blocked with 2% normal mouse serum and Fc blocker (CD16/32) in 100 μl of FACS buffer for 15 min on ice. Cells were stained with surface marker Abs for 30 min on ice and washed with 1 ml of FACS buffer three times. Fixation was done with 100 μl of 2% paraformaldehyde in PBS for 15 min at room temperature (RT), and cells were washed once with FACS buffer. Cells were permeabilized with 100 μl of 0.2% saponin for 10 min on ice, and cytokine Abs (IL-2, IL-4, IL-5, IL-13, and IFN-γ) were added and allowed to incubate on ice for 30 min. Cells were washed with 1 ml of FACS buffer three times and analyzed by flow cytometry.
For CD25, IL-2Rβ, and CD132 detection, cells were stained as mentioned above but were not restimulated on day 5 postactivation. Cells were then permeabilized with 0.2% saponin and stained for intracytoplasmic proteins (cytokine Abs). For intranuclear protein staining (Blimp 1, cleaved caspase3, and Foxp3), a Foxp3 Fix/Perm Buffer Set (BD Biosciences) was used to fix and permeabilize the cells, following the manufacturer’s protocol.
Cells were analyzed using a CyAn ADP flow cytometer (Beckman Coulter). Summit 5.2 (Beckman Coulter) and FlowJo v10 (TreeStar) software were used for data analysis.
ELISA measurement for cytokine detection
On day 5 postactivation, cell culture supernatants were collected, and cytokine concentrations were measured using murine IL-2, IL-4, and IL-13 ELISA Ready-SET-Go! Kits (eBioscience). In some experiments, as indicated, 1 × 106 cells were restimulated with PMA (50 ng/ml) and ionomycin (1 μg/ml) in 1 ml of T cell media for 6 h. Culture supernatant was collected and used for cytokine detection with murineIL-2, IL-4 and IL-13 ELISA Ready-SET-Go! Kits (eBioscience).
Western blot analysis
Activated CD4+ T cells were collected and pelleted on day 4 postactivation, except where indicated. Cells were washed three times with ice-cold PBS and lysed in triple-detergent RIPA buffer (50 mM Tris-HCl [pH 8], 150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 1% SDS, 1 mM EDTA, 1× complete protease inhibitor, and 1× phosphatase inhibitor). Cell lysates (30–50 μg per lane) were loaded on 8–12% SDS-PAGE gels and transferred onto nitrocellulose membranes. The membranes were blocked with 5% BSA in TBST and probed with anti-STAT5 (1:250), anti–p-STAT5 pY694 (1:500), anti-STAT6 (1:500), anti–p-STAT6 pY641 (1:250) (BD Biosciences), anti–β-actin (1 μg/ml) (Sigma-Aldrich) and anti-HuR clone 3A2 (1 μg/ml). The secondary Abs used were sheep anti-mouse IgG HRP (1:5000) or goat anti-rabbit IgG HRP (1:5000). Anti-HuR 3A2 hybridomas were kindly provided by Joan Steitz (Yale School of Medicine).
RNA isolation and RT-PCR
RNA isolation was done using TRIzol extraction (Invitrogen), following the manufacturer’s protocol. Reverse transcription was performed using 0.5–1 μg of RNA with SuperScript III Reverse Transcriptase (Invitrogen). Quantitative PCR (qPCR) was done in triplicate using Platinum SYBR Green Universal (Invitrogen). Real-time PCR was analyzed using the comparative cycle threshold method, with GAPDH as an endogenous reference control and relative quantities calculated. Murine primers for qPCR were as follows (forward/reverse): Il2: 5′-CCCAAGCAG-GCCACAGAATTGAAA-3′ and 5′-AGTCAAATCCAGAACATG-CCGCAG-3′, Il4: 5′-AGATGGATGTGCCAAACGTCCTCA-3′ and 5′-AATATGCGAAGCACCTTGGAAGCC-3′, Il13: 5′-TGAGGA-GCTGAGCAACATCACACA-3′ and 5′-TGCGGTTACAGAGG-CCATGCAATA-3′, Ifng: 5′-GGCCATCAGCAACAACATAAGCGT-3′ and 5′-TGGGTTGTTGACCTCAAACTTGGC-3′, Gata3: 5′-TTTACCCTCCGGCTTCATCCTCCT-3′ and 5′-TGCACCTGA-TACTTGAGGCACTCT-3′, Il2ra (CD25): 5′-GCAATTTCGCC-GTTGAAGAG-3′ and 5′-TAGGGTGGAGAGAGTTCCATAC-3′, Elavl1 (HuR): 5′-ATGAAGACCACATGGCCGAAGACT-3′ and 5′-AGTTCACAAAGCCATAGCCCAAGC-3′, Prdm1 (Blimp1): 5′-CAGGTCTGCCACAAGAGATTTA-3′ and 5′-CACCTTGCA-TTGGTATGGTTTC-3′, and Gapdh: 5′-TCAACAGCAACTCCCA-CTCTTCCA-3′ and 5′-ACCCTGTTGCGTAGCCGTATTCA-3′.
mRNA stability measurement by actinomycin D
On day 4 postactivation, 3 μg/ml actinomycin D was added to the cell culture to stop nascent mRNA transcription. Cells were collected at 0, 1, 2, 3, 4, and 5 h after actinomycin D treatment, and the remaining RNA was isolated from the cells using TRIzol extraction. RT-PCR was performed as described previously. The amount of RNA at 0 h was set to 100%, and the percentage of remaining RNA at 0–5 h was plotted using a semilog scale.
Transcriptional analysis
Transcriptional activity was measured using the Click-iT Nascent RNA Capture Kit (Invitrogen). Briefly, on day 4 postactivation, activated CD4+ T cells were pulsed with EU for 5 h and harvested. RNA was isolated from the cells using TRIzol extraction. One microgram of RNA was used for the click reaction, following the manufacturer’s protocol. Twenty-five micrograms of Dynabeads was used for 500 ng of biotinylated labeled EU-RNA. Reverse transcription was performed using a SuperScript VILO cDNA Synthesis Kit (Invitrogen). qPCR was done using Platinum SYBR Green Universal (Invitrogen), as described.
Cell proliferation assay
Isolated CD4+ T cells were stained with Cell Proliferation Dye eFluor 670 (eBioscience), following the manufacturer’s protocol. In brief, 2 × 106 CD4+ T cells were washed three times with PBS and then resuspended in 500 μl of PBS at RT. Cell Proliferation Dye eFluor 670 was diluted with PBS to a final concentration of 10 μM. A total of 500 μl of the dye was added to 500 μl of the cell suspension while vortexing. Cells were then incubated at 37°C in the dark for 5 min. The reaction was stopped by adding 3 ml of cold FBS to the cell suspension and incubating on ice for 5 min. Cells were then spun down at 1200 rpm for 5 min at 4°C and washed three times with T cell media containing ≥10% FBS. Cells were then recounted, and 1 × 106 cells per milliliter were activated with plate-bound anti-CD3 (5 μg/ml) and anti-CD28 (4 μg/ml) for 3 d. Cells were then analyzed on day 3 by Cyan ADP flow cytometry (Beckman Coulter).
RNA immunoprecipitation
RNA immunoprecipitation was performedas previously described (33). Briefly, activated CD4+ T cells, under nonpolarizing conditions, were collected on day 4 postactivation. The cells were washed vigorously with ice-cold PBS and then lysed in polysomal lysis buffer (100 mM KCl, 5 mM MgCl2, 10 mM HEPES, 0.5% Nonidet P-40, 1 mM DTT, 100 U RnaseOUT, and 1× protease inhibitors). HuR (3A2) or IgG1 control Abs were precoated onto protein A Sepharose beads overnight at 4°C. Beads were then washed with NT-2 buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl2, 1 mM MgCl2, and 0.05% Nonidet P-40) before incubation with cell lysates. Equal amounts of lysates were added to the Ab-precoated beads and incubated for 4 h at 4°C. After 4 h, beads were washed seven times with NT-2 buffer and incubated with 20 U RNase-free DNase I (15 min, 37°C), followed by 100 μl of NT-2 buffer containing 0.1% SDS and 0.5 mg/ml proteinase K (30 min, 55°C). RNAs were then precipitated with phenol-chloroform. The isolated RNAs were reverse transcribed, and qPCR was performed to measure target transcript enrichment.
In vitro biotin pull-down assay
Primers containing the T7 sequence were designed to amplify different sections of Il2ra transcripts. Four biotinylated mRNA transcripts that are complementary to sequences in the Il2ra ORF and three sections of the Il2ra 3′UTR (Supplemental Fig. 2) were generated from cDNA of murine activated CD4+ T cells using a T7 in vitro transcription assay kit (Ambion). The biotinylated transcripts (1 μg) were incubated with protein lysates (40 μg) from activated murine CD4+ T cells to induce the association of HuR protein with biotinylated RNAs. Streptavidin beads (Dyna-beads M-280 streptavidin; Invitrogen) were added to the lysates, incubated for 30 min at RT, and washed three times with PBS. Protein loading buffer was added to the ribonucleoprotein (RNP) complex and heated at 95°C for 5 min. Then solutions were run on a protein gel. Western blot analysis was performed with HuR Ab. The following primers were used for biotin pull-down: Il2ra ORF (forward + T7): 5′-CCAAGCTTCTAATACGACTCACTATAGG-GTCTGTATGA-3′ and Il2ra ORF (reverse): 5′-TCTTCTGCTC-TTCCTCCATCTGT-3′, Il2ra first section (forward + T7): 5′-CCAAGCTTCTAATACGACTCACTATAGGGAATCACAAAG-3′ and Il2ra first section (reverse): 5′-CCTGCTGGGAAAGCATC-TAAGT-3′, Il2ra second section (forward + T7): 5′-CCAAGCTTC-TAATACGACTCACTATAGGGTCTGCGC-3′ and Il2ra second section (reverse): 5′-GTGTGGGCTAGAGATCAGCATAA-3′, and Il2ra third section (forward + T7): 5′-CCAAGCTTCTAATACGA-CTCACTATAGGGGATTCACAG-3′ and Il2ra third section (reverse): 5′-CCAAAGGCTTTCTTAATGTACACTAGTATCTAT-3′.
Polysomal fractionation analysis
CD4+ T cells activated under nonpolarizing conditions with plate-bound anti-CD3 and anti-CD28 were harvested on day 4 post-activation. Cycloheximide (0.1 mg/ml) was added to the cell cultures 15 min before the cells were collected. Cells were pelleted and washed three times with ice-cold PBS containing 0.1 mg/ml cycloheximide. Cytoplasmic extracts were carefully layered over 10–50% linear sucrose gradients in polysome buffer (10 mM HEPES [pH 7.5], 100 mM KCl, 2.5 mM MgCl2, 1 mM DTT, 50 U recombinant RNasin [Promega], and 0.1% IGEPAL CA-630 [Sigma-Aldrich]) and centrifuged at 17,000 rpm in a Beckman SW 32.1 Ti Rotor for 4 h at 4°C. Gradients were fractioned using an Isco gradient fractionation system equipped with a UA-6 detector. Light RNP fractions 40S, 60S, and 80S and heavy polysome fractions were monitored by the continuous UV-absorption profile at A254. Nine fractions were collected, and RNAs associated with each fraction were isolated using TRIzol extraction. RNAs from each fraction were reverse transcribed followed by qPCR. The percentage distribution of RNA in the 40S, 60S, 80S, and heavy polysome fractions was analyzed.
Statistical analysis
The p values were calculated using a two-tailed Student t test or one-way ANOVA with a Tukey multiple-comparisons test using Prism v7 (GraphPad).
RESULTS
HuR deletion in late-stage thymocytes does not interfere with thymic egression and peripheral T cell distribution
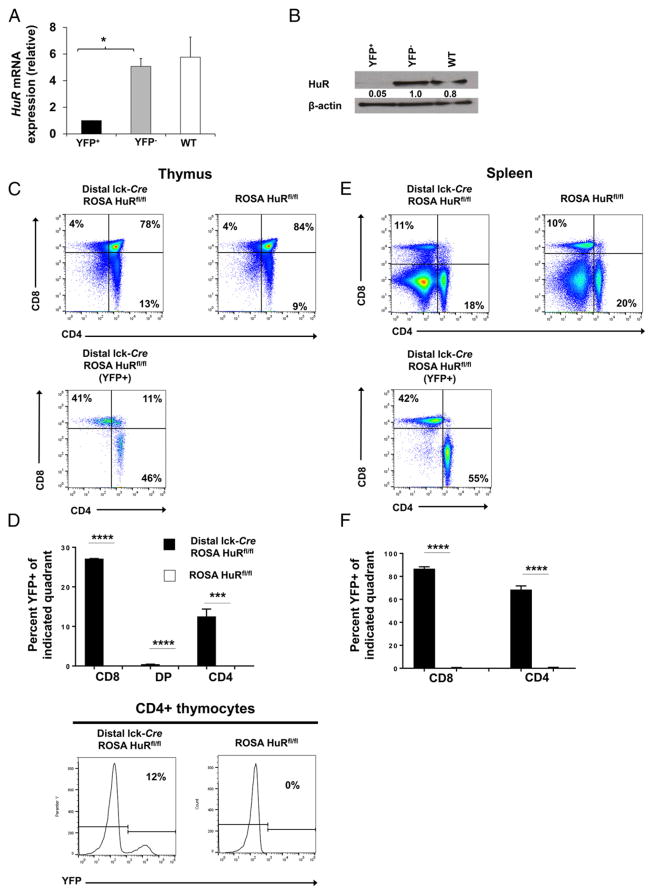
To study the role of HuR in CD4+ T cell cytokine regulation, we generated a novel conditional T cell–specific deletion of loxP-flanked Elavl1 alleles (HuRfl/fl). In these mice, Cre recombinase is expressed in late-stage thymocytes prior to T activation, under the control of the distal lck promoter (distal lck-Cre) (called distal lck-Cre HuRfl/fl in this article). The mice were crossed to ROSA YFP mice to obtain distal lck-Cre ROSA HuRfl/fl mice in which YFP expression can be used to identify target gene deletion. YFP+ CD4+ T cells from peripheral lymphoid organs of HuR-KO mice display nearly undetectable levels of HuR mRNA (~5-fold decrease) and protein (~95% reduction) compared with YFP− and WT CD4+ T cells (Fig. 1A, 1B). Conditional HuR-KO mice developed normally and had a normal peripheral T cell distribution (Fig. 1C, 1E). Of the YFP+ population in the thymus, ~0.5% were CD4+CD8+ (double-positive [DP]), 13% were CD4+, and 27% were CD8+ T cells (Fig. 1D). In the SP, these frequencies were even more striking, with ~68.5% of CD4+ T cells and 87% of CD8+ T cells lacking HuR (Fig. 1F). No histological abnormalities were observed in organs of HuR-deficient mice up to 1 y of age (data not shown). Therefore, HuR ablation in late-stage thymocytes does not alter thymic egression or peripheral T cell distribution.
FIGURE 1. Efficient HuR ablation in late-stage thymocytes does not alter thymic egress or peripheral T cell distribution.
(A and B) Isolated CD4+ T cells from the SP and LNs of HuR-KO mice (distal lck-Cre ROSA HuRfl/fl) were sorted according to their YFP expression (YFP+ versus YFP−); CD4+ T cells from WT HuRfl/fl mice were used as control cells. HuR levels were assessed by RT-PCR (A) and Western blot (B). (C) Frequency of DP, CD4 single-positive (CD4+ single-positive), and CD8 single-positive (CD8+ single-positive) cells among total thymocytes in HuR-KO mice (upper panels) and in YFP+ thymocytes (lower panel). (D) Frequency of YFP+ (HuR-deficient) cells among DP, CD4+ single-positive, and CD8+ single-positive thymocytes (upper panel) and representative line graphs of YFP expression (lower panel). (E) Frequency of DP, CD4+ single-positive, and CD8+ single-positive cells among total splenocytes (upper panel) and YFP+ splenocytes (lower panel). (F) Frequency of YFP+ (HuR-deficient) cells among DP, CD4+ single-positive, and CD8+ single-positive splenocytes. Data in (A) and (B) are representative of two independent experiments using at least two mice per group in each experiment. Data in (C)–(F) are representative of four independent experiments consisting of at least two mice per group in each experiment, along with representative flow cytometry plots. Bars represent mean + SEM of two (A and B) or four (D and F) independent experiments. *p < 0.05, ***p < 0.001, ****p < 0.0001, one-way ANOVA with the Tukey multiple-comparisons test or Student t test.
Increased IL-2 and decreased Th2 cytokine expression occur in HuR-ablated CD4+ T cells when activated under nonpolarizing conditions
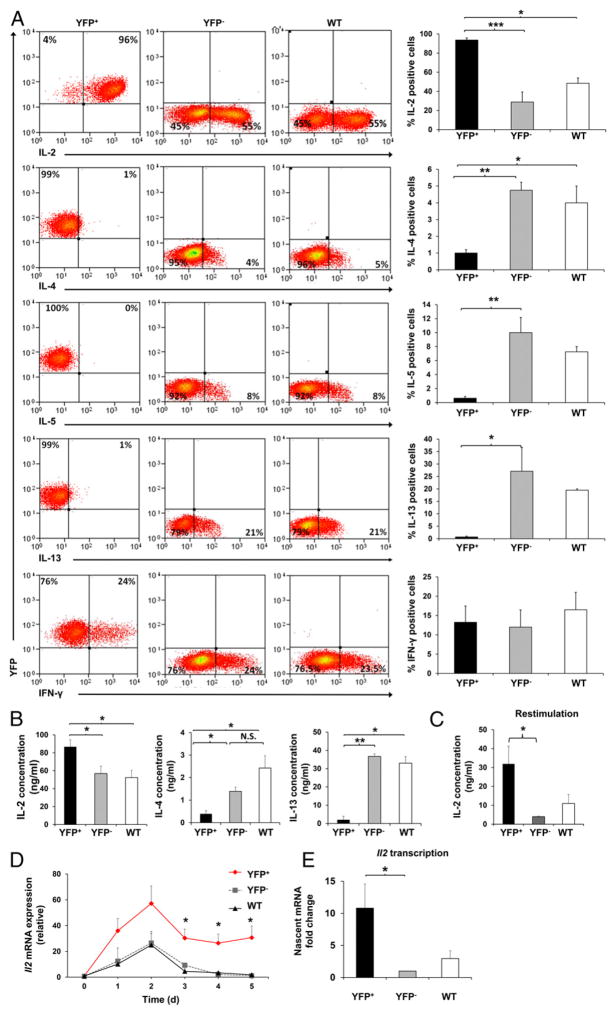
HuR is highly upregulated upon T cell activation. However, its kinetics during CD4+ T cell activation are not fully understood (25). Therefore, we examined HuR protein expression in vitro in WT CD4+ T cells activated under nonpolarizing conditions. HuR was rapidly upregulated, reaching peak expression at 48 h and returning to baseline on day 5 postactivation (data not shown). This suggested that HuR may facilitate T cell regulatory functions. We then assessed the role of HuR in T cell activation and cytokine regulation. Peripheral CD4+ T cells from distal lck-Cre ROSA HuRfl/fl (HuR-KO) and ROSA HuRfl/fl (littermate control) mice were enriched by column purification. The cells from HuR-KO mice were further sorted based on their YFP expression to obtain pure populations of YFP+ or YFP− cells (>95% purity). YFP+, YFP−, and WT CD4+ T cells represent HuR-KO, normal HuR (endogenous control), and HuR WT (ROSA HuRfl/fl WT control) cells, respectively. The cells were then activated under non-polarizing conditions and assessed for IL-2, IL-4, IL-5, IL-13, and IFN- γ expression on day 5 postactivation. Strikingly, the majority of YFP+ cells (up to 97%) were still positive for IL-2 compared with YFP− cells (~50%) and WT control cells (~50%) (Fig. 2A). In YFP+ cell culture supernatants, significant increases in IL-2 production were observed (Fig. 2B, left panel). The increases in IL-2 secretion in activated YFP+ cells were even more pronounced (up to 7-fold) when the cells were restimulated with PMA and ionomycin (Fig. 2C). Conversely, YFP+ cells produced scant amounts of the Th2 cytokines IL-4, IL-5, and IL-13 (Fig. 2A). Corresponding with a marked reduction in Th2 cytokine expression in YFP+ cells, the levels of IL-4 and IL-13 in the YFP+ culture supernatant were much lower than in YFP− cells and WT controls (Fig. 2B, middle and right panels). Therefore, we conclude that HuR negatively controls IL-2 expression and positively regulates Th2 cytokine production in activated CD4+ T cells.
FIGURE 2. Increases in IL-2 and decreases in Th2 cytokine expression in HuR-ablated CD4+ T cells.
(A) YFP+ (HuR-KO), YFP− (endogenous control), or WT (HuRfl/fl control with no Cre) CD4+ T cells were stimulated under nonpolarizing conditions with plate-bound anti-CD3 and anti-CD28. On day 5 of activation, cells were harvested, stimulated with PMA and ionomycin for 5 h, and stained for intracellular cytokines. (B) IL-2, IL-4, and IL-13 levels in the culture supernatant under nonpolarizing conditions on day 5 postactivation of YFP+, YFP−, and WT CD4+ T cells, as measured by ELISA. (C) IL-2 levels in activated YFP+, YFP−, and WT CD4+ T cells after restimulation with PMA and ionomycin for 5 h, as detected by ELISA. (D) RT-PCR analysis of Il2 mRNA in YFP+, YFP−, and WT CD4+ T cells activated under
HuR-deficient CD4+ T cells are incapable of sustaining high CD25 expression
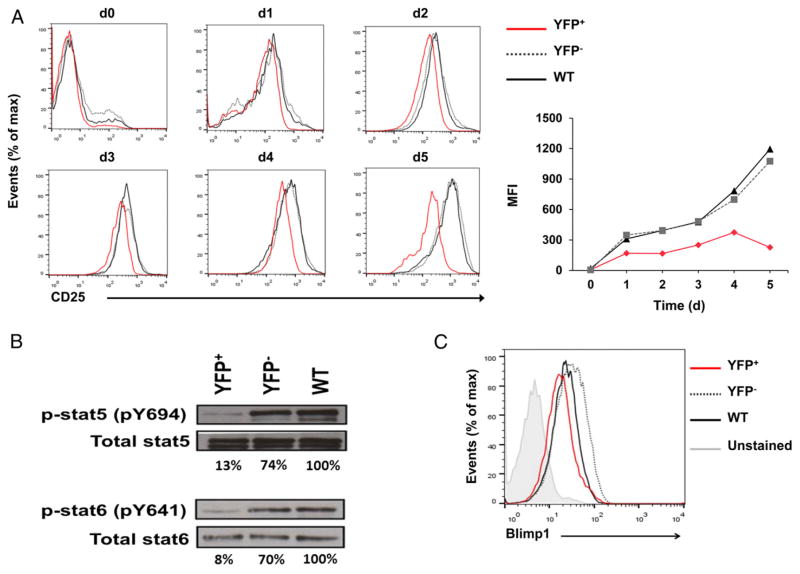
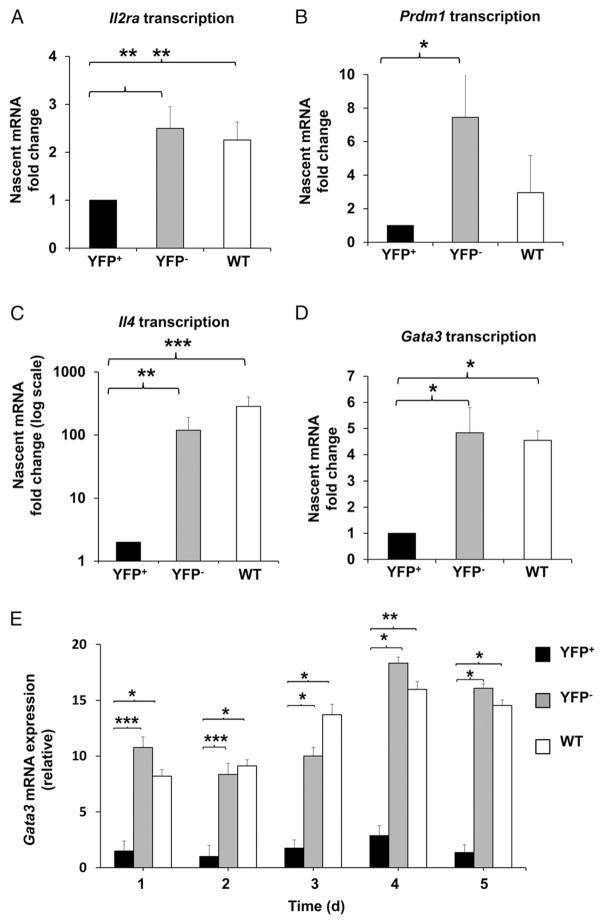
To determine the kinetics of IL-2 alteration in the absence of HuR, we measured Il2 mRNA levels in activated YFP+, YFP−, and WT control CD4+ T cells starting on day 0 (nonactivated) postactivation. In normal physiologic settings, IL-2 is rapidly and transiently expressed upon CD4+ T cell activation (5). The high levels of IL-2 expression are normally reduced to basal levels by 72 h postactivation, as seen in YFP− and WT control cells (Fig. 2D). However, Il2 mRNA levels in activated YFP+ cells were highly elevated (~30 fold) and remained significantly higher than the YFP− and WT controls, even after 72 h of activation (Fig. 2D). We hypothesized that HuR may act directly to inhibit IL-2 or indirectly by promoting the inhibitory pathway ofIL-2production. Because we observed from the Il2 mRNA kinetics data that YFP+ cells cannot completely turn off IL-2 expression to basal levels, it seemed more likely that the pathway regulating IL-2 production was defective. Therefore, we reasoned that HuR promotes the expression of key players involved in the modulation of IL-2 expression. To explore this possibility, we first determined the kinetics of CD25 (IL-2Rα) expression. Although the levels of CD25 expression in YFP+ CD4+ T cells were comparable to those in YFP− and WT cells up to 2 d postactivation, YFP+ cells were markedly incapable of sustaining high CD25 expression after day 3 (Fig. 3A). There was no change in IL-2Rβ (CD122) and a slight increase in the common γ-chain (CD132) expression in YFP+ cells compared with YFP− and WT control cells (data not shown). Additionally, we observed significant reductions in the signals downstream of IL-2R, p-Stat5, and Blimp1, with no change in total Stat5 in YFP+ cells (Fig. 3B, upper panel, Fig. 3C). Consistent with the augmentation of IL-2 expression and downregulation of IL-2–repressive signals found in YFP+ cells, Il2 transcription was strikingly increased, whereas Il2ra (CD25) and Prdm1 (Blimp1) transcription was markedly downregulated in YFP+ cells (Figs. 2E, 4A, 4B).
FIGURE 3. HuR deficiency alters the expression of components of the IL-2 signaling pathway.
(A) Flow cytometry of kinetic changes in CD25 protein expression of activated YFP+, YFP−, and WT CD4+ T cells on days 0–5. (B) p-Stat5, p-Stat6, total Stat5, and total Stat6 protein levels in activated YFP+, YFP−, and WT CD4+ T cells on day 4 postactivation. (C) Blimp1 protein expression in activated YFP+, YFP−, and WT CD4+ T cells on day 4 postactivation. Data are representative of two (B) or three (A and C) independent experiments. nonpolarizing conditions on days 0 to 5. (E) Transcriptional measurement using nascent RNA capture assay and RT-PCR analysis of Il2. Data are combined from three (B, C, and E) or four [(A), right panel and (D)] independent experiments, along with representative flow cytometry plots (A, left panel). Error bars represent mean + SEM of three (B, C, and E) or four [(A), right panel] independent experiments. The p values in (D) were calculated based on YFP+ versus YFP− and WT. *p < 0.05, **p < 0.01, ***p < 0.001, one-way ANOVA with the Tukey multiple-comparisons test.
FIGURE 4. Gene expression downstream of IL-2/p-Stat5 signaling is altered in the absence of HuR.
Transcriptional measurement using nascent RNA capture assay and RT-PCR analysis of Il2ra (A), Prdm1 (B), Il4 (C), and Gata3 (D) mRNAs in activated YFP+, YFP−, and WT CD4+ T cells. Data show relative amounts of nascent mRNAs on day 4 postactivation. (E) Steady-state Gata3 mRNA kinetics in activated YFP+, YFP−, and WT CD4+ T cells, as measured by RT-PCR. All data are from three or more independent experiments and represent mean + SEM. *p < 0.05, **p < 0.01, ***p < 0.001, one-way ANOVA with the Tukey multiple-comparisons test.
IL-2/p-Stat5 signals are required for IL-2 homeostasis and the initiation of Th2 differentiation (10, 11). To assess whether the reduction in Th2 cytokine production in HuR-KO cells was due to reduced IL-2/p-Stat5 signaling, we examined Il4 and Gata3 transcriptional activity. We showed that there were significant decreases in Il4 (~100-fold reduction) and Gata3 (~5-fold decrease) gene transcription (Fig. 4C, 4D). We also observed a significant reduction in Gata3 mRNA steady-state levels (Fig. 4E). Moreover, YFP+ cells have a prominent reduction in p-Stat6, which is known to promote Gata3 transcription downstream of IL-4R signaling (Fig. 3B). This may due, in part, to the reduction in IL-4 in the environment, which results in decreasing IL-4/p-Stat6 signaling (Fig. 2B, middle panel, Fig. 3B, lower panel). The defect in Th2 cytokine expression observed in YFP+ cells is not the result of the failure of the cells to express IL-4R, because YFP+ cells have normal or even slightly increased IL-4Rα- and common γ-chain expression (data not shown).
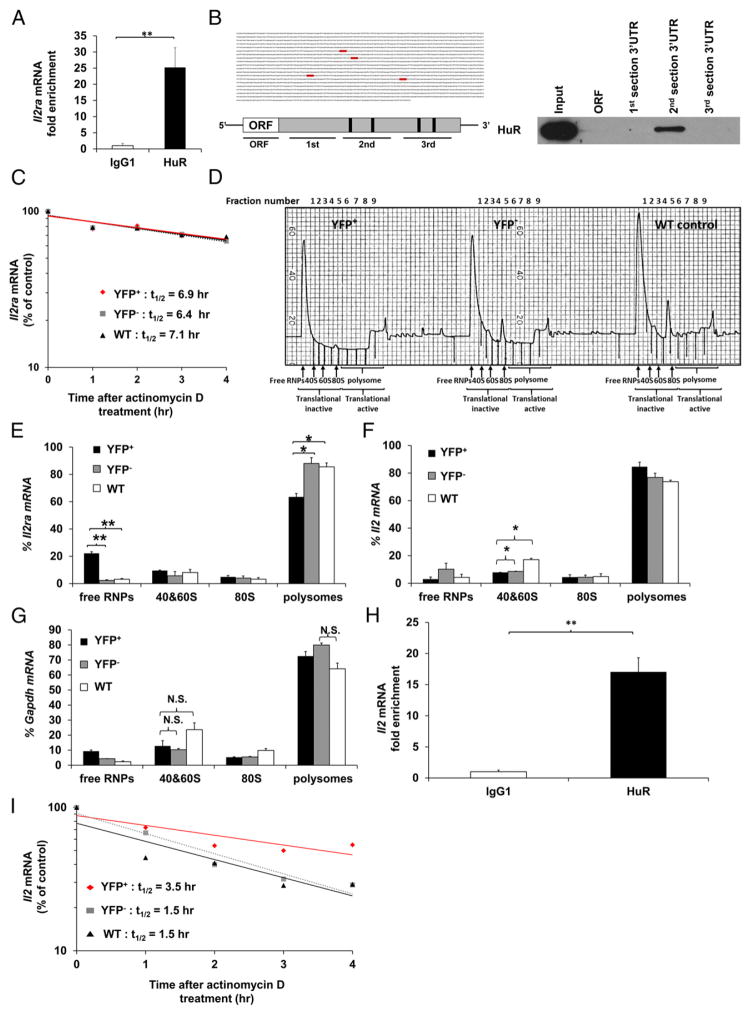
HuR binds directly to Il2ra 3′UTR mRNA and is required to promote optimal CD25 expression in activated CD4+ T cells
YFP+ cells failed to maximally upregulate CD25 expression postactivation compared with controls and were unable to downregulate IL-2 mRNA expression to basal levels (Figs. 2D, 3A). This implied that the negative-feedback loop, which inhibits IL-2 production through IL-2R, may be impaired in the absence of HuR. We hypothesized that HuR controls CD25 expression and IL-2 homeostasis by posttranscriptionally regulating Il2ra (encoding CD25) mRNA stability and/or translation. HuR can effect mRNA and protein expression by binding to the AREs present in the 3′UTR of its target mRNA transcripts and promoting their stability and/or translation. We first assessed the direct physical association of HuR and Il2ra mRNA by performing HuR RNA immunoprecipitation (RIP) assays. HuR RIP data demonstrated a significant Il2ra transcript enrichment in the anti–HuR RIP samples compared with isotype-matched IgG1 controls (Fig. 5A). To validate HuR binding sites on Il2ra transcripts, we used a computational program to search for putative HuR binding sites by determining ARE sequences. Four potential HuR binding sites on the Il2ra mRNA 3′UTR were identified (Fig. 5B, left panel; see Supplemental Fig. 2 for full size). We used a second independent method, biotin pull-down, to verify putative HuR binding sites on Il2ra mRNA. These data reveal direct binding of HuR to sequences in the Il2ra mRNA 3′UTR section 2, which contains two putative HuR binding sites. However, HuR did not associate with the ORF or 3′UTR sections 1 and 3 of Il2ra mRNA (Fig. 5B, right panel). Taken together, these results indicate that HuR physically interacts with ARE sequences in Il2ra 3′UTR mRNA.
FIGURE 5. HuR physically interacts with the Il2ra 3′UTR mRNA and enhances its translational efficiency in activated CD4+ T cells.
(A) RIP using HuR or IgG1 Ab, followed by RT-PCR to determine physical HuR mRNA targets. Data are fold enrichment of Il2ra mRNA in anti-HuR samples compared with IgG1 controls. (B) Putative HuR targets, ARE elements (gray), present in the 3′UTR of Il2ra mRNA (left panels) and biotin pull-down shows association of HuR with different portions of Il2ra mRNA (right panel). Data show HuR interaction with the second section of Il2ra 3′UTR mRNA containing the first two putative HuR binding sites. (C) Il2ra mRNA stability assay in activated YFP+, YFP−, and WT CD4+ T cells on day 4 postactivation. Data represent the percentage of mRNA remaining over time after actinomycin D treatment. (D) Absorbance profile for RNA separated by velocity sedimentation through a sucrose gradient. RNA was extracted from each fraction. Polysomal gradient analysis of Il2ra (E), Il2 (F), and Gapdh (G) mRNAs in activated YFP+, YFP−, and WT CD4+ T cells. Data are the percentage of Il2ra, Il2, and Gapdh mRNA distribution in 40S, 60S, 80S, and polysome fractions by RT-PCR. (H) RIP using HuR or IgG1 Ab, followed by RT-PCR to determine HuR mRNA targets. Data are fold enrichment of Il2 mRNA in anti-HuR samples compared with IgG1 controls. (I) Il2 mRNA stability assay in activated YFP+, YFP−, and WT CD4+ T cells on day 4 postactivation. Data are from three (A and H) or two (D, F, and G) independent experiments or are a representative of three (C and I) or two (B and D) independent experiments. Data are mean + SEM of three (A and H) or two (D, F, and G) independent experiments. *p < 0.05, **p < 0.01, two-tailed unpaired t test (A and H), one-way ANOVA with the Tukey multiple-comparisons test (E–G). N.S., not significant.
We next sought to determine how HuR might be regulating Il2ra mRNA transcripts. mRNA stability assays were performed in YFP+, YFP−, and WT activated CD4+ T cells. As seen in Fig. 5C, Il2ra mRNA stability in YFP+ cells is comparable to YFP− and WT control cells. It has been previously reported that HuR can solely regulate translation of its target transcripts without interfering with their stability (34). To that end, we examined whether HuR functions by augmenting Il2ra translation by performing polysomal gradient analysis. The absorbance profile for RNA separated by velocity sedimentation through sucrose gradient fractionation contains the low and high m.w. profiles (Fig. 5D). Interestingly, the polysomal distributions were altered in YFP+ cells compared with YFP− cells and WT controls (Fig. 5D). We discovered a defect in the recruitment of Il2ra mRNA to the high m.w. polysomes in the absence of HuR, as seen by increased Il2ra mRNA in the free RNP complex (free RNPs: inactive ribosomes) and reduced Il2ra mRNA associated with heavy polysomes (active ribosomes) (Fig. 5E). The Il2 and Gapdh mRNA distributions were unaffected by HuR KO (Fig. 5F, 5G). These results suggested that HuR directly binds to the ARE sequences in the 3′UTR of Il2ra mRNA and is required for maximizing Il2ra mRNA translation but not stability.
We also observed that HuR interacts with Il2 mRNA, as shown by HuR RIP (Fig. 5H). A significant increase in Il2 mRNA stability was seen in YFP+ cells compared with YFP− cells and WT controls (Fig. 5I). However, the mechanisms by which Il2 mRNA stability is increased in YFP+ cells remain to be determined.
Exogenous IL-4 does not rescue Th2 cytokine expression in HuR-deficient cells
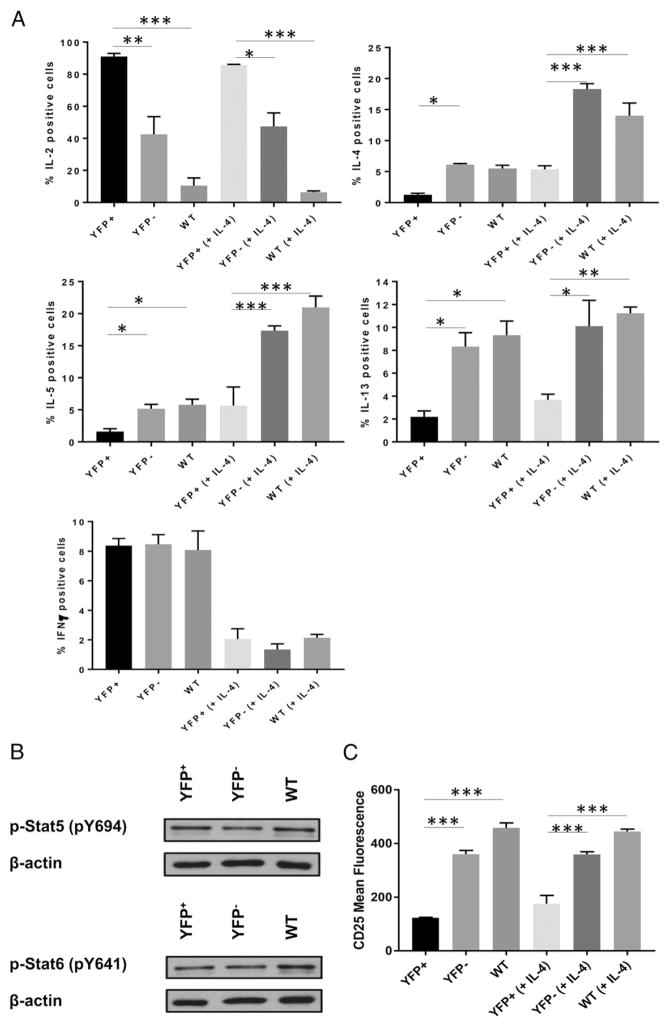
We then determined whether impairment of Th2 cytokine expression in HuR-deficient cells is due to cell-intrinsic defects. YFP+, YFP−, and WT CD4+ T cells were activated in the presence of rIL-4. YFP+ cells had significantly decreased IL-4, IL-5, and IL-13 compared with YFP− cells and WT controls (Fig. 6A), whereas increases in IL-2 persisted. Additionally, the defects in p-Stat5 and p-Stat6 observed in HuR-KO cells were restored with rIL-4 (Fig. 6B). The CD25defect still persisted (Fig. 6C). High IL-2expression was still observed in HuR-KO cells (data not shown). Therefore, we conclude that defects in Th2 cytokine production in YFP+ cells are cell intrinsic, because the failure of YFP+ cells to differentiate into the Th2 lineage persisted after addition of rIL-4.
FIGURE 6. Exogenous IL-4 cannot rescue Th2 cytokine expression in HuR-deficient cells.
(A) YFP+, YFP−, or WT CD4+ T cells were stimulated or not with 100 U/ml rIL-4. Five days postactivation, cells were harvested and restimulated with PMA and ionomycin for 5 h, and cytokine production was assessed by intracellular cytokine staining. (B) p-Stat5 and p-Stat6 levels in YFP+, YFP−, and WT CD4+ T cells activated in the presence of IL-4. (C) CD25 expression in YFP+, YFP−, and WT CD4+ T cells activated in the presence or absence of 1000 U/ml rIL-4 for 5 d. Data (mean + SEM) are a representative of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, one-way ANOVA with the Tukey multiple-comparisons test.
HuR deficiency results in impaired CD4+ T cell proliferation in response to Ag
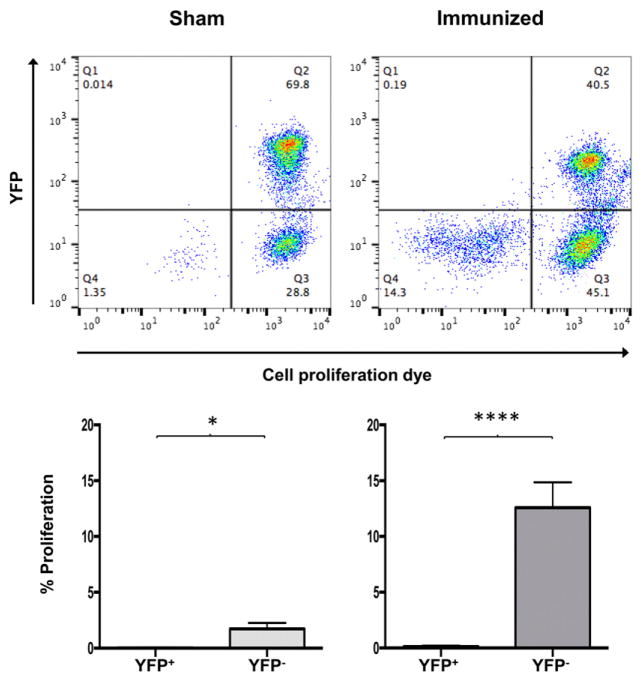
To determine whether deficiency in proliferation also occurs when T cells are stimulated with Ag, mice were immunized with whole OVA protein and boosted on day 10. On day 17, CD4+ T cells were harvested from SP and LNs, loaded with proliferation dye, and restimulated in the presence of OVA peptide–loaded APCs on day 17. After 3 d, YFP+ T cells showed a significant reduction in proliferation in OVA-immunized (98%) mice and sham immunization controls (99%) (Fig. 7). We conclude that HuR deficiency hinders Ag-specific proliferation.
FIGURE 7. HuR deficiency results in impaired CD4+ T cell Ag-induced proliferation.
Mice were immunized with 50 μg of whole OVA protein and boosted with 50 μg of OVA protein on day 10. SP and LNs were harvested on day 17, and CD4+ T cells were stained with Cell Proliferation Dye eFluor 660 and cultured in the presence of OVA peptide–loaded APCs for 3 d. Data show the percentage proliferation of YFP+ or YFP− CD4+ T cells in OVA protein–immunized or sham controls. Data are representative of three independent experiments (upper panel) or are combined from three experiments (lower panels). *p < 0.05, ****p < 0.0001, two-tailed unpaired t test.
We also examined the effect of HuR upon CD4+ T cell apoptosis by detecting cleaved caspase3 in activated YFP+, YFP−, and WT CD4+ T cells under nonpolarizing conditions. There were no differences in apoptosis among the various groups (data not shown). These data suggest that HuR promotes Ag-specific T cell proliferation, perhaps through influencing TCR signaling or memory induction, and its ablation does not alter cell apoptosis.
DISCUSSION
Little is known about how posttranscriptional events affect T cell fates and function or Ag-dependent proliferation. We found that the RBP HuR is critical for controlling IL-2 homeostasis and reinforces the previously reported role for HuR in Th2 differentiation. We discovered that HuR interacts with and promotes Il2ra (CD25) mRNA translation. Previously, we identified Il2 as a HuR target using RNA sequencing (35). Our data suggest that HuR may regulate IL-2 expression by promoting CD25 expression. The decrease in high-affinity IL-2R expression in HuR-KO cells results in a significant reduction in signaling events, such as Stat5 phosphorylation and Blimp1 expression, downstream of the receptor. Although HuR-KO CD4+ T cells expressed comparable levels of intermediate-affinity IL-2R (IL-2Rβ- and common γ-chain) compared with the controls, the inhibitory signal received from these receptors was insufficient to optimize p-Stat5 and Blimp1 expression and, thus, suppress Il2 transcription. These data suggested that optimal HuR-mediated CD25 expression in activated CD4+ T cells may be required for controlling IL-2 homeostasis.
During the course of our investigations, the Kontoyiannis laboratory conditionally ablated HuR in thymocytes using the proximal lck-Cre system (36), which is expressed early in thymocytes. These mice had issues with T cell development, apoptosis, activation, and thymic egress. Therefore, we selected the distal lck-Cre system to avoid repercussions on T cell development, because deletion of HuR would occur later during T cell development.
The role of HuR in Th2 differentiation seems to be time dependent. Our data suggest that HuR is required for the initiation phase, but not the maintenance phase, of Th2 differentiation. HuR ablation in CD4+ T cells after Th2 lineage commitment in OX40-Cre HuRfl/fl mice causes an unexpected upregulation of Th2 cytokine transcripts (30). In contrast, HuR-KO CD4+ T cells from distal lck-CreROSA HuRfl/fl mice show profoundly diminished Th2 cytokine expression. IL-2/Stat5 signaling has been shown previously to augment stable Il4 locus accessibility to promote Il4 transcription (10, 11, 37). However, the IL-2 signal alone cannot replace the function of IL-4 to initiate Th2 differentiation (10, 11, 37). In our model, inadequate CD25 expression in HuR-KO cells results in a significant reduction in p-Stat5, which leads to a marked decrease in Il4 transcription and Th2 cytokine production. We also observed significant reductions in p-Stat6 and Gata3, both of which are required for IL-4–dependent Th2 differentiation. The decrease in Stat6 phosphorylation is not due to defects in IL-4R expression in HuR-KO cells. Interestingly, c-Maf–deficient cells have an inability to sustain high CD25 expression, similar to that observed in HuR-KO cells, and also have significantly reduced levels of p-Stat5, which leads to diminished Th2-associated cytokine expression (38). We propose that high-affinity IL-2R may be important for HuR-dependent optimization of IL-2/Stat5 signaling and promotion of Th2 differentiation.
Interestingly, we observed a paradoxical increase in Il2 stability, a HuR target transcript, in the absence of HuR. Thus, there may very well be CD25-independent mechanisms that contribute to runaway IL-2 production. We are currently investigating this possibility. Previously, HuR has been shown to interact with Il2 mRNA, but HuR binding does not alter Il2 transcript localization, stability, or translation (39). Although HuR interacts with Il2 mRNA, it does not associate with the canonical ARE in the Il2 3′UTR (39). The mechanisms by which HuR regulates the Il2 mRNA transcript through its direct binding require further investigation.
The addition of rIL-4 does not rescue Th2 cytokine production, but it promotes phosphorylation of Stat5 and Stat6 in HuR-deficient cells. This is in line with previous reports that IL-4 can promote Stat5 phosphorylation (40–42). The activation of Stat5 may be the result of IL-4 signaling through Jak3, which is associated with the common γ-chain. However, despite the increased signaling through Stat5 and Stat6, HuR-deficient cells cannot differentiate into Th2 cells when they have received rIL-4. These results suggest that the inability of HuR-deficient cells to efficiently become Th2 cells is due to cell-intrinsic defects.
HuR-deficient cells also showed a marked inability to proliferate, whether in response to Ag or nonspecific activation. One of the most well-known functions of IL-2 is its ability to promote T cell proliferation. Our data strongly suggest that HuR plays a role in T cell Ag recognition or memory T cell formation. Further study is needed to understand the underlying mechanisms.
We propose that HuR–CD25 interactions may function as a linchpin in controlling IL-2 homeostasis, Th2 differentiation, and Ag-specific proliferation. Activation of CD4+ T cells in the absence of HuR leads to a series of cascading failures that result in abnormal IL-2 homeostasis and aberrant Th2 cytokine expression. This may be due, in part, to the inability of CD4+ T cells to optimize Il2ra translation, leading to decreases in CD25 expression. In addition, HuR may promote Th2 differentiation by at least two possible mechanisms. First, HuR promotes stable CD25 expression, which induces adequate IL-2 signaling required for Th2 differentiation. Second, HuR stabilizes Th2 hallmark transcripts, including Gata3 and Il13, which promotes Th2 initiation (30–32). Therefore, it is possible that the direct regulation of Th2 cytokine transcript stability by HuR may contribute to the intrinsic defects observed in Th2 differentiation in HuR-deleted cells. Given the importance of IL-2 and CD25 in CD4+ T cell differentiation, we are currently investigating the role of HuR in other CD4+ T cell lineages, such as regulatory T cells and T follicular helper cells. These findings may also have potential applications in tolerance and anergy, given the central role that IL-2 homeostasis plays in these fields of immunity.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants AI079341 and AI080870. J.L.M. and M.G. were supported by the National Institute on Aging–Intramural Research Program, National Institutes of Health.
We thank Dr. Steven F. Ziegler, Dr. Mark Ansel, and Dr. Chyi-Song Hsieh for the suggestions on this work.
Abbreviations used in this article
- ARE
adenylate and uridylate–rich element
- DP
double-positive
- KO
knockout
- LN
lymph node
- ORF
open reading frame
- qPCR
quantitative PCR
- RBP
RNA-binding protein
- RIP
RNA immunoprecipitation
- RNP
ribonucleoprotein
- RT
room temperature
- SP
spleen
- UTR
untranslated region
- WT
wild-type
Footnotes
The online version of this article contains supplemental material.
DISCLOSURES
The authors have no financial conflicts of interest.
References
- 1.Constant S, Pfeiffer C, Woodard A, Pasqualini T, Bottomly K. Extent of T cell receptor ligation can determine the functional differentiation of naive CD4+ T cells. J Exp Med. 1995;182:1591–1596. doi: 10.1084/jem.182.5.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hosken NA, Shibuya K, Heath AW, Murphy KM, O’Garra A. The effect of antigen dose on CD4+ T helper cell phenotype development in a T cell receptor-alpha beta-transgenic model. J Exp Med. 1995;182:1579–1584. doi: 10.1084/jem.182.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Panhuys N, Klauschen F, Germain RN. T-cell-receptor-dependent signal intensity dominantly controls CD4(+) T cell polarization in vivo. Immunity. 2014;41:63–74. doi: 10.1016/j.immuni.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Villarino AV, Tato CM, Stumhofer JS, Yao Z, Cui YK, Hennighausen L, O’Shea JJ, Hunter CA. Helper T cell IL-2 production is limited by negative feedback and STAT-dependent cytokine signals. J Exp Med. 2007;204:65–71. doi: 10.1084/jem.20061198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sojka DK, Bruniquel D, Schwartz RH, Singh NJ. IL-2 secretion by CD4+ T cells in vivo is rapid, transient, and influenced by TCR-specific competition. J Immunol. 2004;172:6136–6143. doi: 10.4049/jimmunol.172.10.6136. [DOI] [PubMed] [Google Scholar]
- 6.Boyman O, Sprent J. The role of interleukin-2 during homeostasis and activation of the immune system. Nat Rev Immunol. 2012;12:180–190. doi: 10.1038/nri3156. [DOI] [PubMed] [Google Scholar]
- 7.Lin JX, Leonard WJ. Signaling from the IL-2 receptor to the nucleus. Cytokine Growth Factor Rev. 1997;8:313–332. doi: 10.1016/s1359-6101(97)00021-x. [DOI] [PubMed] [Google Scholar]
- 8.Lin JX, Leonard WJ. The role of Stat5a and Stat5b in signaling by IL-2 family cytokines. Oncogene. 2000;19:2566–2576. doi: 10.1038/sj.onc.1203523. [DOI] [PubMed] [Google Scholar]
- 9.Gong D, Malek TR. Cytokine-dependent Blimp-1 expression in activated T cells inhibits IL-2 production. J Immunol. 2007;178:242–252. doi: 10.4049/jimmunol.178.1.242. [DOI] [PubMed] [Google Scholar]
- 10.Zhu J, Cote-Sierra J, Guo L, Paul WE. Stat5 activation plays a critical role in Th2 differentiation. Immunity. 2003;19:739–748. doi: 10.1016/s1074-7613(03)00292-9. [DOI] [PubMed] [Google Scholar]
- 11.Cote-Sierra J, Foucras G, Guo L, Chiodetti L, Young HA, Hu-Li J, Zhu J, Paul WE. Interleukin 2 plays a central role in Th2 differentiation. Proc Natl Acad Sci USA. 2004;101:3880–3885. doi: 10.1073/pnas.0400339101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maier E, Duschl A, Horejs-Hoeck J. STAT6-dependent and -independent mechanisms in Th2 polarization. Eur J Immunol. 2012;42:2827–2833. doi: 10.1002/eji.201242433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kagami S, Nakajima H, Suto A, Hirose K, Suzuki K, Morita S, Kato I, Saito Y, Kitamura T, Iwamoto I. Stat5a regulates T helper cell differentiation by several distinct mechanisms. Blood. 2001;97:2358–2365. doi: 10.1182/blood.v97.8.2358. [DOI] [PubMed] [Google Scholar]
- 14.Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu Rev Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 15.Paul WE. What determines Th2 differentiation, in vitro and in vivo? Immunol Cell Biol. 2010;88:236–239. doi: 10.1038/icb.2010.2. [DOI] [PubMed] [Google Scholar]
- 16.Paul WE, Zhu J. How are T(H)2-type immune responses initiated and amplified? Nat Rev Immunol. 2010;10:225–235. doi: 10.1038/nri2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheadle C, Fan J, Cho-Chung YS, Werner T, Ray J, Do L, Gorospe M, Becker KG. Control of gene expression during T cell activation: alternate regulation of mRNA transcription and mRNA stability. BMC Genomics. 2005;6:75. doi: 10.1186/1471-2164-6-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheadle C, Fan J, Cho-Chung YS, Werner T, Ray J, Do L, Gorospe M, Becker KG. Stability regulation of mRNA and the control of gene expression. Ann N Y Acad Sci. 2005;1058:196–204. doi: 10.1196/annals.1359.026. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska EA, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raghavan A, Dhalla M, Bakheet T, Ogilvie RL, Vlasova IA, Khabar KS, Williams BR, Bohjanen PR. Patterns of coordinate down-regulation of ARE-containing transcripts following immune cell activation. Genomics. 2004;84:1002–1013. doi: 10.1016/j.ygeno.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 21.Raghavan A, Ogilvie RL, Reilly C, Abelson ML, Raghavan S, Vasdewani J, Krathwohl M, Bohjanen PR. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res. 2002;30:5529–5538. doi: 10.1093/nar/gkf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakheet T, Williams BR, Khabar KS. ARED 3.0: the large and diverse AU-rich transcriptome. Nucleic Acids Res. 2006;34:D111–D114. doi: 10.1093/nar/gkj052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen CY, Xu N, Shyu AB. Highly selective actions of HuR in antagonizing AU-rich element-mediated mRNA destabilization. Mol Cell Biol. 2002;22:7268–7278. doi: 10.1128/MCB.22.20.7268-7278.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Atasoy U, Watson J, Patel D, Keene JD. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J Cell Sci. 1998;111:3145–3156. doi: 10.1242/jcs.111.21.3145. [DOI] [PubMed] [Google Scholar]
- 26.Meisner NC, Filipowicz W. Properties of the regulatory RNA-binding protein HuR and its role in controlling miRNA repression. Adv Exp Med Biol. 2011;700:106–123. doi: 10.1007/978-1-4419-7823-3_10. [DOI] [PubMed] [Google Scholar]
- 27.Tiedje C, Ronkina N, Tehrani M, Dhamija S, Laass K, Holtmann H, Kotlyarov A, Gaestel M. The p38/MK2-driven exchange between tristetraprolin and HuR regulates AU-rich element-dependent translation. PLoS Genet. 2012;8:e1002977. doi: 10.1371/journal.pgen.1002977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan J, Ishmael FT, Fang X, Myers A, Cheadle C, Huang SK, Atasoy U, Gorospe M, Stellato C. Chemokine transcripts as targets of the RNA-binding protein HuR in human airway epithelium. J Immunol. 2011;186:2482–2494. doi: 10.4049/jimmunol.0903634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J, Cascio J, Magee JD, Techasintana P, Gubin MM, Dahm GM, Calaluce R, Yu S, Atasoy U. Posttranscriptional gene regulation of IL-17 by the RNA-binding protein HuR is required for initiation of experimental autoimmune encephalomyelitis. J Immunol. 2013;191:5441–5450. doi: 10.4049/jimmunol.1301188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gubin MM, Techasintana P, Magee JD, Dahm GM, Calaluce R, Martindale JL, Whitney MS, Franklin CL, Besch-Williford C, Hollingsworth JW, et al. Conditional knockout of the RNA-binding protein HuR in CD4+ T cells reveals a gene dosage effect on cytokine production. Mol Med. 2014;20:93–108. doi: 10.2119/molmed.2013.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stellato C, Gubin MM, Magee JD, Fang X, Fan J, Tartar DM, Chen J, Dahm GM, Calaluce R, Mori F, et al. Coordinate regulation of GATA-3 and Th2 cytokine gene expression by the RNA-binding protein HuR. J Immunol. 2011;187:441–449. doi: 10.4049/jimmunol.1001881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yarovinsky TO, Butler NS, Monick MM, Hunninghake GW. Early exposure to IL-4 stabilizes IL-4 mRNA in CD4+ T cells via RNA-binding protein HuR. J Immunol. 2006;177:4426–4435. doi: 10.4049/jimmunol.177.7.4426. [DOI] [PubMed] [Google Scholar]
- 33.Dahm GM, Gubin MM, Magee JD, Techasintana P, Calaluce R, Atasoy U. Method for the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts using RIP-Chip. J Vis Exp. 2012;67:3851. doi: 10.3791/3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson P. Post-transcriptional control of cytokine production. Nat Immunol. 2008;9:353–359. doi: 10.1038/ni1584. [DOI] [PubMed] [Google Scholar]
- 35.Techasintana P, Davis JW, Gubin MM, Magee JD, Atasoy U. Transcriptomic-wide discovery of direct and indirect HuR RNA targets in activated CD4+ T cells. PLoS One. 2015;10:e0129321. doi: 10.1371/journal.pone.0129321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Papadaki O, Milatos S, Grammenoudi S, Mukherjee N, Keene JD, Kontoyiannis DL. Control of thymic T cell maturation, deletion and egress by the RNA-binding protein HuR. J Immunol. 2009;182:6779–6788. doi: 10.4049/jimmunol.0900377. [DOI] [PubMed] [Google Scholar]
- 37.Yamane H, Zhu J, Paul WE. Independent roles for IL-2 and GATA-3 in stimulating naive CD4+ T cells to generate a Th2-inducing cytokine environment. J Exp Med. 2005;202:793–804. doi: 10.1084/jem.20051304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hwang ES, I, White A, Ho IC. An IL-4-independent and CD25-mediated function of c-maf in promoting the production of Th2 cytokines. Proc Natl Acad Sci USA. 2002;99:13026–13030. doi: 10.1073/pnas.202474499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seko Y, Azmi H, Fariss R, Ragheb JA. Selective cytoplasmic translocation of HuR and site-specific binding to the interleukin-2 mRNA are not sufficient for CD28-mediated stabilization of the mRNA. J Biol Chem. 2004;279:33359–33367. doi: 10.1074/jbc.M312306200. [DOI] [PubMed] [Google Scholar]
- 40.Castro A, Sengupta TK, Ruiz DC, Yang E, Ivashkiv LB. IL-4 selectively inhibits IL-2-triggered Stat5 activation, but not proliferation, in human T cells. J Immunol. 1999;162:1261–1269. [PubMed] [Google Scholar]
- 41.Lischke A, Moriggl R, Brändlein S, Berchtold S, Kammer W, Sebald W, Groner B, Liu X, Hennighausen L, Friedrich K. The interleukin-4 receptor activates STAT5 by a mechanism that relies upon common gamma-chain. J Biol Chem. 1998;273:31222–31229. doi: 10.1074/jbc.273.47.31222. [DOI] [PubMed] [Google Scholar]
- 42.Friedrich K, Kammer W, Erhardt I, Brändlein S, Sebald W, Moriggl R. Activation of STAT5 by IL-4 relies on Janus kinase function but not on receptor tyrosine phosphorylation, and can contribute to both cell proliferation and gene regulation. Int Immunol. 1999;11:1283–1294. doi: 10.1093/intimm/11.8.1283. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.