GINT1 is shown to be an indispensable glycosyltransferase that facilitates the glycosylation of GlcNAccontaining glycosylinositol phosphorylceramides in rice, with a minor role in Arabidopsis.
Abstract
Glycosylinositol phosphorylceramides (GIPCs), which have a ceramide core linked to a glycan headgroup of varying structures, are the major sphingolipids in the plant plasma membrane. Recently, we identified the major biosynthetic genes for GIPC glycosylation in Arabidopsis (Arabidopsis thaliana) and demonstrated that the glycan headgroup is essential for plant viability. However, the function of GIPCs and the significance of their structural variation are poorly understood. Here, we characterized the Arabidopsis glycosyltransferase GLUCOSAMINE INOSITOLPHOSPHORYLCERAMIDE TRANSFERASE1 (GINT1) and showed that it is responsible for the glycosylation of a subgroup of GIPCs found in seeds and pollen that contain GlcNAc and GlcN [collectively GlcN(Ac)]. In Arabidopsis gint1 plants, loss of the GlcN(Ac) GIPCs did not affect vegetative growth, although seed germination was less sensitive to abiotic stress than in wild-type plants. However, in rice, where GlcN(Ac) containing GIPCs are the major GIPC subgroup in vegetative tissue, loss of GINT1 was seedling lethal. Furthermore, we could produce, de novo, “rice-like” GlcN(Ac) GIPCs in Arabidopsis leaves, which allowed us to test the function of different sugars in the GIPC headgroup. This study describes a monocot GIPC biosynthetic enzyme and shows that its Arabidopsis homolog has the same biochemical function. We also identify a possible role for GIPCs in maintaining cell-cell adhesion.
Plant sphingolipids are highly complex, diverse structural lipids. Glycosylinositol phosphorylceramides (GIPCs) represent the major sphingolipids in the plant plasma membrane. Approximately 64% of sphingolipids and ∼25% of total plasma membrane lipids are GIPCs, which interact with sterols to form membrane microdomains (Markham and Jaworski, 2007; Cacas et al., 2016). Besides serving as a membrane structural component, GIPCs are bioactive molecules involved in cell signaling and regulation (Dunn et al., 2004).
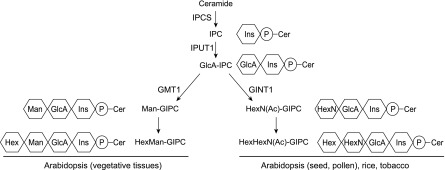
The core structure of plant GIPCs consists of inositol phosphorylceramide (IPC) linked to GlcA. Additional sugar units can be added, such as glucosamine (GlcN), N-acetyl-glucosamine (GlcNAc), Man, Gal, and arabinose (Markham et al., 2006, 2013; Buré et al., 2011). The glycan patterns vary among plant species and tissues, indicating that different GIPC glycosylation pathways and enzymes are involved. Generally, there are two types of glycan structure: hexose(s) linked to GlcA-IPC in Arabidopsis (Arabidopsis thaliana) vegetative tissue and GlcN(Ac) linked to GlcA-IPC in rice (Oryza sativa), tobacco (Nicotiana tabacum), and reproductive tissues (seeds and pollen) of Arabidopsis, as shown in Figure 1.
Figure 1.
Representative structures and synthetic pathways of GIPC sugar headgroups in plants. The first (Ins) and second (GlcA) carbohydrate residues are conserved in plants, but the third residue (Hex, HexN, or HexNAc) is variable in plant tissues/species. This study identifies the (N-acetyl)hexosamine-specific transferase GINT1. IPCS, inositolphosphorylceramide synthase (ceramide phosphoinosityl transferase); IPUT1, inositolphosphorylceramide glucuronosyl transferase; GMT1, GIPC mannosyl transferase; P, phosphate group; Ins, inositol; Cer, ceramide.
The headgroup structure and biosynthesis of plant GIPCs has not been fully elucidated. However, recently, characterization of GIPC biosynthetic enzymes was reported (Fig. 1). GIPCs are initially formed by the addition of an inositol phosphate to the ceramide through the action of IPC synthase (Wang et al., 2008). IPCs then go through several glycosylation steps in the Golgi. INOSITOL PHOSPHORYLCERAMIDE GLUCURONOSYLTRANSFERASE1 (IPUT1), the first glycosyltransferase (GT) identified, transfers GlcA from UDP-GlcA to IPC (Rennie et al., 2014), forming GlcA-IPC. GOLGI-LOCALIZED NUCLEOTIDE SUGAR TRANSPORTER1 (GONST1) is responsible for importing GDP-Man into the Golgi specifically for GIPC biosynthesis, and GIPC MANNOSYL-TRANSFERASE1 (GMT1) uses this substrate to transfer Man onto GlcA-IPC (Mortimer et al., 2013; Fang et al., 2016). gmt1, gonst1, and pollen-rescued iput1 show severe dwarfism and constitutive accumulation of salicylic acid, suggesting that the headgroup plays an important role in plant growth and development, as well as in defense responses (Mortimer et al., 2013; Fang et al., 2016; Tartaglio et al., 2017). More recently, we showed that the headgroup of GIPCs can serve as the binding site for pathogenic toxins and as a result determine the host selectivity of some classes of plant pathogens (Lenarčič et al., 2017).
In this work, we characterized the GT64 protein GLUCOSAMINE INOSITOLPHOSPHORYLCERAMIDE TRANSFERASE1 (GINT1) and showed that it is responsible for the addition of GlcNAc to GIPCs. In Arabidopsis, in which GlcN(Ac)-containing GIPCs are dominant in seeds and pollen (Luttgeharm et al., 2015), gint1 plants grow normally under standard conditions. However, GlcN(Ac)-containing GIPCs are the dominant form in almost all tissue types in rice, and loss of OsGINT1 results in seedling lethality.
RESULTS
Identification of a Putative GIPC GT in GT64
We recently characterized AtGMT1 (At3g55830), a member of the Arabidopsis CAZy GT64 family, as a GIPC mannosyltransferase (Fang et al., 2016). Here, we chose to investigate At5g04500, the homolog of AtGMT1, which we now propose to rename GLUCOSAMINE INOSITOLPHOSPHORYLCERAMIDE TRANSFERASE1 (AtGINT1). The GT64 family has three members in Arabidopsis, which show low percentage sequence similarity to each other (29% shared sequence identity between AtGMT1 and AtGINT1; Fang et al., 2016). AtGINT1 is 767 amino acids long (compared to AtGMT1, which is 334 amino acids) and has orthologs across the plant kingdom, from charophyte to angiosperms (Supplemental Fig. S1). AtGINT1 also contains a predicted exostosin (EXT) domain (Inter-Pro; Finn et al., 2017). In humans, EXTs have both a GT64 and a GT47 domain and are required for the biosynthesis of heparan-sulfate, a glycosaminoglycan, in which the major repeating disaccharide unit consists of GlcA linked to GlcNAc (Busse-Wicher et al., 2014). The GT64 domain in EXT is responsible for the transfer of GlcNAc from UDP-GlcNAc to GlcA. Recently, it was shown that some Arabidopsis tissues contain GIPCs that have an IPC core linked to a GlcA (GlcA-IPC) linked to a GlcN(Ac) [GlcN(Ac)-GlcA-IPC; Fig. 1; Tellier et al., 2014; Luttgeharm et al., 2015]. Therefore, we considered AtGINT1 a good candidate for the GT responsible for the addition of GlcNAc to GlcA-IPC in these tissues.
Tissue-Specific Expression in Arabidopsis
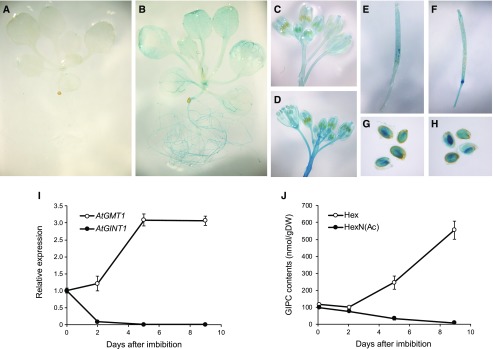
GlcN(Ac)-containing GIPCs are most abundant in Arabidopsis seeds and undetectable in vegetative tissue (Tellier et al., 2014), and therefore we predicted that AtGINT1 gene expression should reflect this. Both publicly available microarray data (Supplemental Fig. S2, B and C; Winter et al., 2007) and expression of a reporter construct containing Escherichia coli β-GLUCURONIDASE driven by the AtGINT1 promoter (GINT1pro:GUS; Fig. 2; Supplemental Fig. S2, D and E) showed that AtGINT1 is specifically and highly expressed in the developing embryo and mature seed. Almost no signal was observed in vegetative tissues, but weak GUS staining was detected in stigma and pollen (Fig. 2), consistent with the presence of GlcN(Ac)-containing GIPCs in flowers (Fig. 3) and pollen (Luttgeharm et al., 2015). In contrast, AtGMT1, the predominant GIPC mannosyl transferase (Fang et al., 2016), was expressed in all tissues, in accordance with the ubiquitous distribution of Hex-GlcA-IPCs in Arabidopsis. Reverse transcription quantitative PCR (RT-qPCR) and sphingolipidomic analysis also demonstrated the correlation of transcriptional levels of the two putative sugar transferases and the composition of the GIPC sugar headgroup in seed and germinating seedlings. AtGINT1 was highly expressed in dry seed but drastically downregulated after seed imbibition in parallel with a rapid decline of HexN(Ac)-containing GIPC content, whereas the levels of AtGMT1 transcript and Hex-GIPCs were synchronously elevated during seed germination (Fig. 2).
Figure 2.
Tissue-specific expression of AtGINT1 and AtGMT1 and comparison of GIPC subclasses. A to H, Promoter-GUS staining indicating expression patterns of AtGINT1 (A, C, E, and G) and AtGMT1 (B, D, F, and H) in young seedlings (A and B), flowers (C and D), developing siliques (E and F), and mature seeds (G and H). I and J, GT expression analysis (I) and quantification of GIPC contents (J) during seed germination. Data are means of three or four biological replicates ± sd.
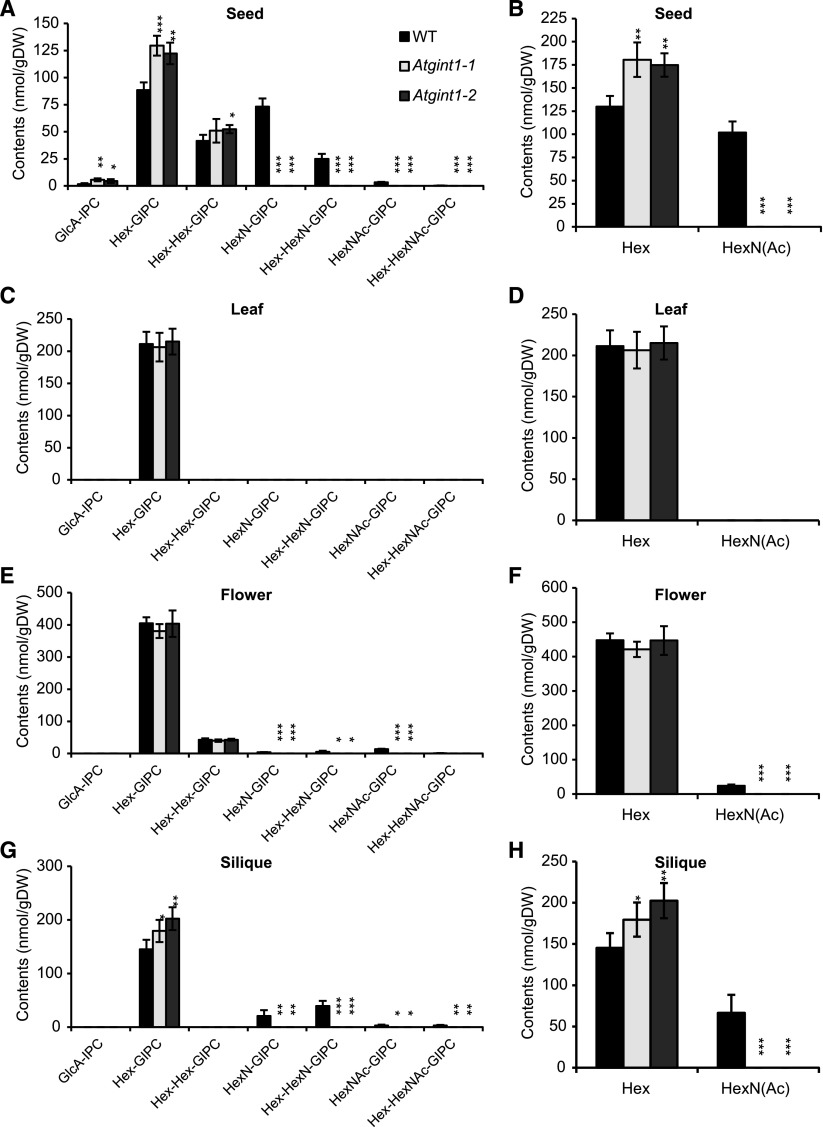
Figure 3.
GIPC composition of Atgint1 plants. A to H, LC-MS/MS was used to determine the GIPC composition of dry seeds (A and B), leaves of 3-week-old plants (C and D), flowers (E and F), and developing green siliques (G and H). A, C, E, and G, Accumulation profiles of all the major GIPC classes. B, D, F, and H, Overall amounts of either Hexn-only GIPCs (Hex) or HexN(Ac)-containing GIPCs [HexN(Ac)]. Data are means of three biological replicates ± sd. Asterisks indicate significant difference compared to that in the wild type (Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001).
Isolation of Arabidopsis T-DNA Lines
We next obtained two allelic T-DNA insertion mutants, Atgint1-1 (SALK_002825) and Atgint1-2 (GK_834G06), both in the Columbia-0 (Col-0) ecotype background (Supplemental Fig. S3A). We isolated plants homozygous for the T-DNA insertion, confirmed the location of the insertions by sequencing, and showed that both alleles lacked detectable AtGINT1 transcript by RT-PCR (Supplemental Fig. S3B). Both mutants grew normally on agar and soil under standard conditions and showed no gross morphological phenotype compared to that of wild type.
Analysis of GIPCs in Atgint1 Plants
Next, we analyzed GIPC composition by sphingolipidomics using multiple reaction monitoring liquid chromatography-tandem mass spectrometry (LC-MS/MS; Fang et al., 2016; Ishikawa et al., 2016), which is necessary to survey the extensive variation in both the ceramide moiety and the type and degree of glycosylation attached to the GlcA-IPC core. GIPCs from Arabidopsis leaves are almost exclusively composed of Hex-GlcA-IPCs, but those from flowers, siliques, and seeds also contain HexN(Ac)-GlcA-IPCs in wild-type plants (Fig. 3; Supplemental Data Set 1). In Atgint1 plants, HexN(Ac)-GlcA-IPCs were undetectable in these tissues, whereas Hex-GlcA-IPCs accumulated in siliques and seeds at significantly higher levels than in wild-type plants. No significant differences in the ceramide and glucosylceramide fractions were observed between Atgint1 and wild type (Supplemental Fig. S4). It should be noted that AtGMT1 expression levels did not change in Atgint1 (Supplemental Fig. S5) and that GlcA-IPC did not accumulate. These data suggest that AtGINT1 is a HexNAc-specific GIPC sugar transferase.
Complementation of Atgint1 by GINT1 Expression
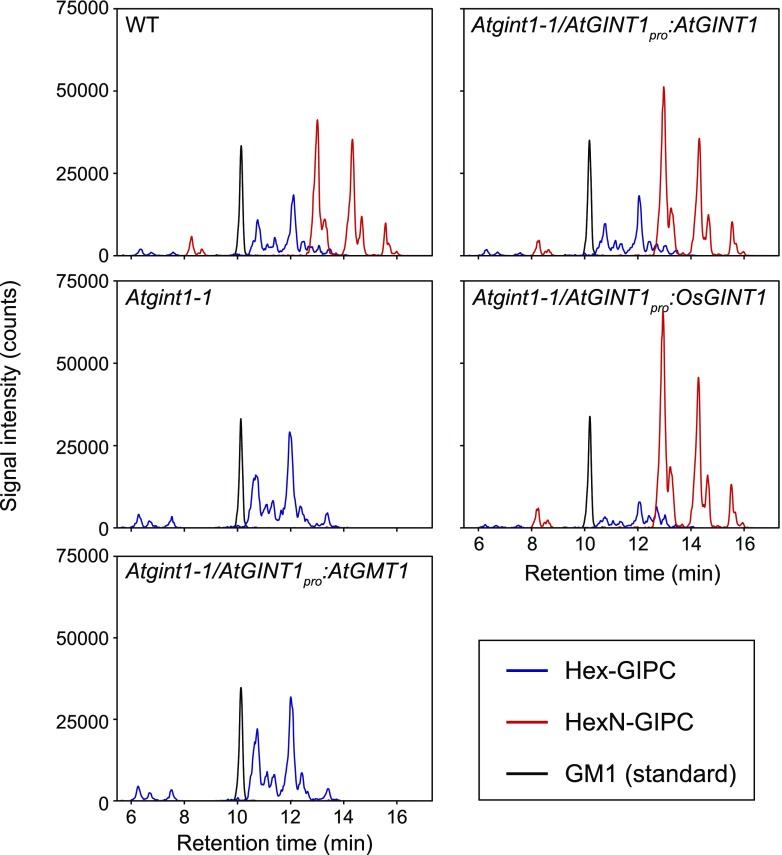
To evaluate the hypothesized difference in substrate specificity between AtGINT1 and AtGMT1, we prepared heterologous complementation lines by expressing AtGINT1pro:AtGINT1 and AtGINT1pro:GMT1 in Atgint1-1. The loss of HexN(Ac)-GIPCs in dry seeds was completely recovered in genomic AtGINT1-complemented lines. The rice AtGINT1 homolog, OsGINT1, also rescued the deficiency, whereas expression of AtGMT1 did not rescue the phenotype in any of the independent transgenic lines tested (Fig. 4).
Figure 4.
Complementation of the HexN-GIPC deficiency in Atgint1. LC-MS/MS chromatograms of the dry seed sphingolipidome in Atgint1-1, Atgint1-1 expressing AtGINT1pro:AtGINT1, AtGINT1pro:OsGINT1, and AtGINT1pro:AtGMT1. Total ion counts corresponding to Hex-GIPCs (blue line), HexN-GIPCs (red line), and Ganglioside GM1 (internal standard, black line) are shown.
Since HexN(Ac)-containing GIPCs are undetectable in Arabidopsis vegetative tissue, we expressed AtGINT1 and OsGINT1 in wild-type Arabidopsis plants, driven by the cauliflower mosaic virus (CaMV) 35S constitutive promoter (35Spro) (Fig. 5). In both cases, we could now successfully detect HexN(Ac)-GIPCs in leaf tissue by LC-MS/MS (Fig. 5A). These transgenic plants did not show any visible phenotypes, probably because the comparatively low levels of HexN(Ac)-GIPC (∼4 mol% for 35Spro:AtGINT1 and ∼20 mol% for 35Spro:OsGINT1), and the unchanged Hex-GIPC levels (Fig. 5B).
Figure 5.
De novo synthesis of HexN-GlcA-IPCs in Arabidopsis leaves following constitutive expression of AtGINT1 or OsGINT1. A, LC-MS/MS chromatograms of Hex-GIPC (t18:1-h24:1, left) and HexN-GIPC (t18:1-h24:1, right) are shown. B, Total quantity of HexN- and HexN(Ac)-GIPC in leaves of Arabidopsis expressing 35Spro:AtGINT1 or 35Spro:OsGINT1.
Phenotype of Atgint1 Seeds
Since GlcN(Ac)-GlcA-IPCs are highly enriched in Arabidopsis seeds and AtGINT1 is specifically expressed in developing embryos and seeds, we investigated how the loss of AtGINT1 might affect seed morphology and germination.
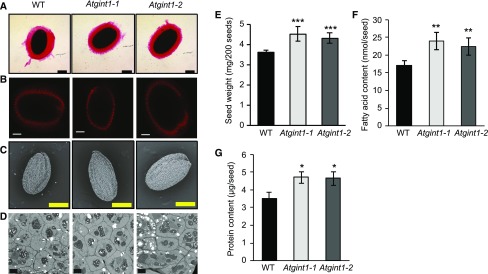
First, we investigated mucilage production. During imbibition, Arabidopsis seed coat cells produce a large quantity of mucilage, which can be visualized using ruthenium red staining (Fig. 6A). This was unaffected in the Atgint1 seeds. We then used scanning electron microscopy to investigate the seed coat in more detail. The seed coat of Atgint1 seeds appeared normal (Fig. 6C), although the seeds were slightly larger. Indeed, the Atgint1 seeds are significantly heavier (Fig. 6E) and have a significant increase in seed storage lipid (Fig. 6F) and protein content (Fig. 6G). Transmission electron microscopy (TEM) revealed that Atgint1 seed coat cells are slightly larger than those of the wild type (Fig. 6D) but had a normal distribution of oil and starch bodies. Examination of developing siliques did not reveal abnormal embryo development (Supplemental Table S1).
Figure 6.
Microscopy analysis and cell wall composition of wild-type and Atgint1 seeds. A, Ruthenium red staining of imbibed seeds indicating mucilage layer. Bar = 200 µm. B, Scarlet4B staining of imbibed seeds indicating cellulose rays in mucilage. Bar = 100 µm. C, Scanning electron microscopy. Bar = 200 µm. D, TEM. Bar = 4 µm. E, Seed weight. F, Seed fatty acid content. G, Seed protein content. A to D, representative images are shown. E to G, Data are means of 3 or 4 biological replicates ± sd. Asterisks indicate significant difference compared to the wild type (Student’s t test, *P < 0.05, **P < 0.01, ***P < 0.001).
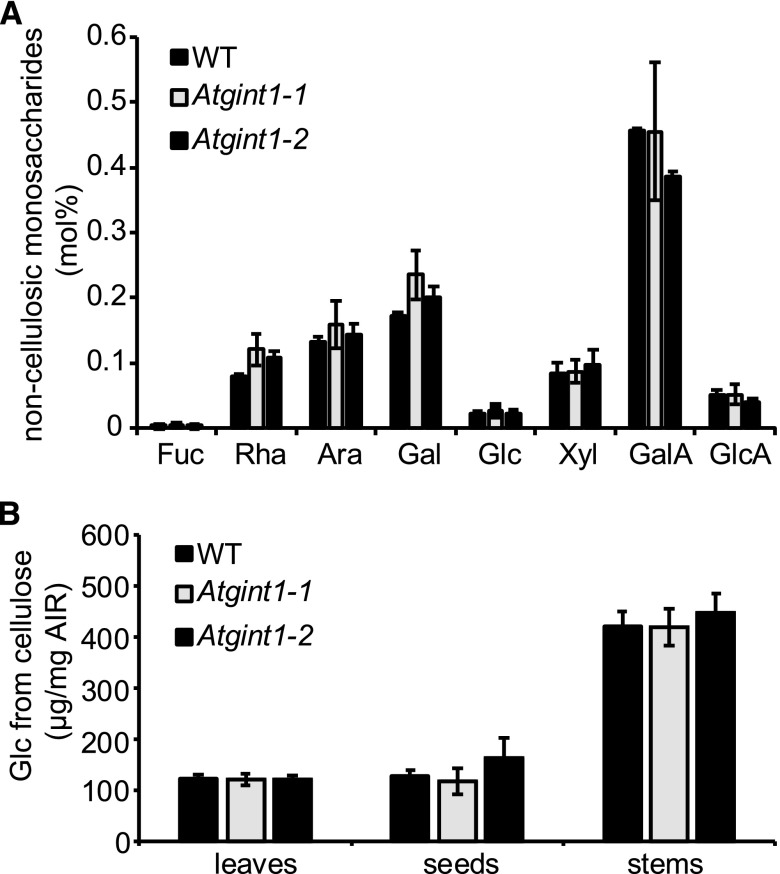
Both Cellulosic and Noncellulosic Cell Wall Polysaccharides Are Unaffected in Atgint1
Previously, we showed that Atgmt1 had reduced crystalline cellulose, but an otherwise unchanged cell wall composition (Fang et al., 2016). To see if the loss of AtGINT1 impacted the seed cell wall, we prepared destarched alcohol insoluble residue (AIR) from dry seeds. We then performed trifluoroacetic acid (TFA) hydrolysis to release the noncellulosic monosaccharides, which were quantified by high performance anion-exchange chromatography-pulsed amperometric detection (Fig. 7; Supplemental Data Set 2). No significant difference was detected between Atgint1 and the wild type (Fig. 7A). The TFA-insoluble residue was then further hydrolyzed using sulfuric acid to release the Glc from cellulose. No significant difference from the wild type was detected for mutant lines carrying either Atgint1 allele (Fig. 7B). Since cell-specific variation in polysaccharide composition may be masked when using these bulk analysis techniques, we also used Scarlet4B staining to investigate the production of cellulose rays in the mucilage of the germinating seeds (Fig. 6B). Again, these were normal in the Atgint1 seeds.
Figure 7.
Cell wall composition of Atgint1. A, Monosaccharide composition of seed AIR following TFA hydrolysis. B, Glc released from TFA-insoluble AIR from leaves, stems, and seeds following Saeman hydrolysis. Data are means of three biological replicates ± sd. No statistically significant difference (Student’s t test, P < 0.05) was observed between wild-type and Atgint1 plants.
Although AtGINT1 is expressed mainly in the developing seed (Supplemental Fig. S2), we also tested the cell wall composition of vegetative tissues. However, there were no significant changes in the cell wall composition of Atgint1 mature stems and young leaves compared to that of the wild type (Supplemental Fig. S6; Fig. 7B).
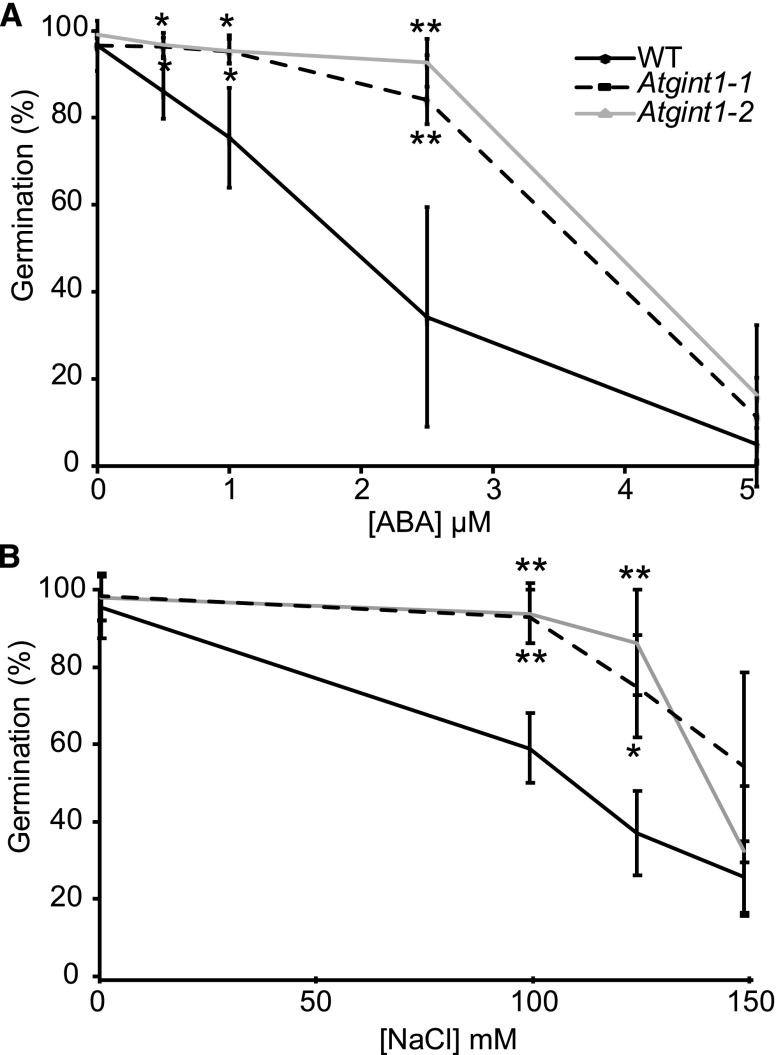
Germination of Atgint1 Seeds Is Resistant to Abscisic Acid and Salt
To further explore the possible functional role for GlcN(Ac)-GIPCs in Arabidopsis seeds, we explored Atgint1 germination rates under various growth conditions. Under standard conditions, there was no significant difference between wild-type and Atgint1 germination (Supplemental Fig. S7; Fig. 8). Next, we tested whether mimicking ageing, via storage of seeds in high humidity and temperature, differentially affected germination (Supplemental Fig. S7). Again, no significant difference was seen.
Figure 8.
Effect of ABA and NaCl on wild-type and Atgint1 seed germination. A, Seed germination rates in the presence of ABA; seeds were scored for radicle emergence after 3 d. B, Seed germination rates in the presence of NaCl, with seeds scored for radicle emergence after 2 d. Data are means of three biological replicates (>50 seeds per replicate) ± sd. An asterisk indicates statistically significant difference from the wild type (Student’s t test, *P < 0.05, **P < 0.01).
The plant hormone abscisic acid (ABA) inhibits radicle growth emergence in imbibed seeds. Previous work has shown that Arabidopsis plants with a partial loss of SPHINGOSINE KINASE1 were less sensitive to application of exogenous ABA (Worrall et al., 2008). The Atgint1 seeds showed a similar phenotype with significantly higher germination than wild-type seeds when germinated on 0.5× Murashige and Skoog media containing either 0.5, 1, or 2.5 µm ABA (Fig. 8A). Since ABA-insensitive mutants can have reduced sensitivity to salinity during germination, we also tested whether this was the case with Atgint1-1 and Atgint1-2. Indeed, both mutant lines had significantly higher germination rates than the wild type in the presence of 100 and 125 mm NaCl, although this difference was lost at the highest NaCl concentration tested (Fig. 8B).
OsGINT1 Is Responsible for HexN(Ac)-GIPC Synthesis and Normal Growth in Rice
GINT1 expression and HexN(Ac)-GIPC production are tissue-specific in Arabidopsis, in contrast to the ubiquitous presence of Hex-GIPCs produced by AtGMT1. In rice, on the other hand, OsGINT1 is highly expressed in almost all tissues, whereas OsGMT1 expression is barely detectable (Supplemental Fig. S8), which corresponds with the distribution of the respective GIPC head group in rice, i.e. HexN(Ac)-GIPCs are ubiquitous but Hex-GIPCs are undetectable in most tissues (Ishikawa et al., 2016).
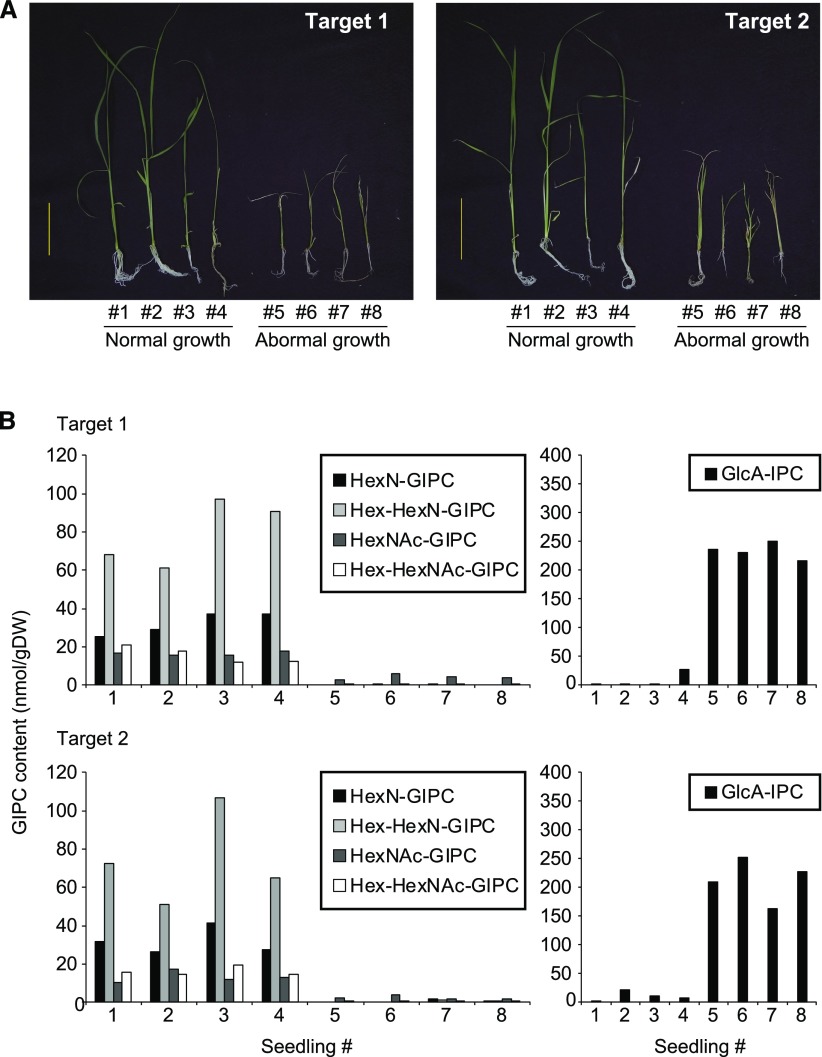
To test the function of HexN(Ac)-GIPCs in rice, we used the CRISPR/Cas9 gene-editing tool to disrupt OsGINT1 using two different target sequences (Supplemental Fig. S9). Most transgenic calli grew as well as the wild type, but upon transfer of calli to regeneration medium followed by transfer to rooting medium, some shoots became stunted and eventually died (Fig. 9A). Sphingolipidomic analysis of the regenerated shoots revealed that wild-type shoots and transgenic shoots with a normal growth phenotype had wild-type levels of HexN(Ac)-GIPCs present (Fig. 9B). However, regenerated shoots with abnormal growth displayed a drastic reduction in HexN(Ac)-GIPCs, as well as an accumulation of GlcA-IPCs, the proposed substrate of GINT1/GMT1 (Fig. 9B).
Figure 9.
Seedling phenotype and GIPC content of OsGINT1-targeted CRISPR/Cas9 rice. A, Regenerated T0 shoots harboring T-DNA with two independent CRISPR/Cas9 OsGINT1 target sequences were further cultured on 0.5× Murashige and Skoog media for 2–3 weeks. Some of the regenerated lines showed severely retarded growth (#5–8), which later proved to be seedling lethal. B, The GIPC content for each shoot shown in A as determined by LC-MS/MS. Hex-containing GIPCs were not detected.
This corresponds to the observations from Atgmt1, which also accumulates GlcA-IPCs (Fang et al., 2016). It should be noted that Atgmt1 also grows well on callus-inducing medium (Fang et al., 2016) but fails to regenerate on shoot-inducing medium (Supplemental Fig. S10). These results indicate indispensable roles for the predominant GIPC head groups, specifically Hex in Arabidopsis and HexN(Ac) in rice.
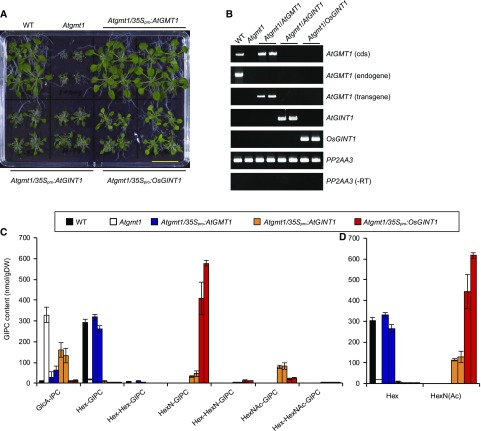
Complementation of Atgmt1 by GINT1 Expression
Our observations demonstrate that both Hex- and HexN(Ac)-GIPCs are crucial for plant development in a plant species- and tissue-specific manner. This raises the question as to whether the two headgroup sugars have common or distinct functions. To evaluate this, the Atgmt1 mutant was complemented by expression of either AtGMT1, AtGINT1, or OsGINT1 driven by 35Spro. Since homozygous Atgmt1 plants have severely compromised growth and fertility, heterozygous plants were transformed with the constructs, and lines homozygous for the loss of a functional copy of AtGMT1 were selected in the T1 and T2 generation (Fig. 10). As expected, both the growth (Fig. 10A) and the GIPC profile (Fig. 10C) of Atgmt1 were fully rescued by expression of 35Spro:AtGMT1. This contrasted with the 35Spro:AtGINT1 and 35Spro:OsGINT1 lines, which only partially restored the Atgmt1 growth phenotype (Fig. 10A). The 35SproOsGINT1 lines exhibited slightly increased growth compared to the 35Spro:AtGINT1 lines, which seems parallel to the metabolic complementation rate of HexN(Ac)-GIPC (Fig. 10C). It should also be noted though that the ratio of HexN-GIPC and HexN(Ac)-GIPC was different between the AtGINT1 and OsGINT1 overexpression lines. Taken together, these data indicate that the two different monosaccharides can only partially substitute for each other’s function.
Figure 10.
Heterologous complementation of Atgmt1 with GINT1. A, Heterozygous Atgmt1 plants were transformed with CaMV 35Spro:AtGMT1, 35Spro:AtGINT1, or 35Spro:OsGINT1. Bar = 2 cm. T2 seedlings (two independently transformed lines per construct) were analyzed by RT-PCR (B). Sphingolipidomic analyses using LC-MS/MS (C and D). C, Profiles of all the major classes of GIPCs detected in the tissues tested, and D, amounts of either Hexn-only GIPCs (Hex) or HexN(Ac)-containing GIPCs [HexN(Ac)]. PP2AA3, PROTEIN PHOSPHATASE 2A SUBUNIT A3; −RT, reaction lacking reverse transcriptase enzyme. Data are means of three biological replicates (>50 seeds per replicate) ± sd.
DISCUSSION
GIPCs are critical for plant growth and development (Wang et al., 2008; Mortimer et al., 2013; Rennie et al., 2014; Fang et al., 2016; Tartaglio et al., 2017). However, their synthesis and function are poorly understood, particularly of the glycan headgroup. Identification of the enzymes responsible for glycosylation of these lipids will provide some of the tools necessary to investigate GIPC function.
Here, we have identified and characterized a GT, GINT1, which we propose is responsible for the addition of GlcNAc to the glycan headgroup of GIPCs in Arabidopsis and rice. GINT1 is a member of CAZy family GT64, and a homolog of GMT1, which we previously identified as the GIPC mannosyl transferase. Production of HexN(Ac)-GIPCs in imbibing Arabidopsis seeds follows the expression of AtGINT1, and Arabidopsis plants lacking a functional copy of GINT1 do not produce detectable HexN(Ac)-GIPCs. The dominant GIPCs in Arabidopsis (Hex-GIPCs) were unaffected. Both AtGINT1 and OsGINT1 restored the loss of HexN(Ac)-GIPCs in Atgint1 when expressed under the AtGINT1 promoter, unlike AtGMT1. Whereas in this report we did not show the in vitro activity for AtGINT1 or OsGINT1, constitutive expression of GINT1 in Arabidopsis results in the de novo production of HexN(Ac)-GIPCs in leaves. Tobacco, like rice, lacks Hex-GIPCs (Carter et al., 1958; Kaul and Lester, 1975; Hsieh et al., 1981; Buré et al., 2011). Previously, we have also overexpressed AtGINT1 in tobacco BY2 cells, and again, we did not see the production of Hex-GIPCs, whereas overexpression of AtGMT1 produced significant quantities (Fang et al., 2016). Together, these data show that, in planta, GINT1 and AtGMT1 have different substrate specificities but can use the same acceptors (GlcA-IPCs).
Considering the tissue-specific distribution of the HexN(Ac)-GIPCs in Arabidopsis, we explored whether the loss of this entire class of GIPCs in Atgint1 affected seed development and germination. Unexpectedly, Atgint1 seeds developed and germinated normally, although they were slightly, but significantly, larger than those of the wild type. However, the germination rate of Atgint1 seeds was significantly less sensitive to the presence of exogenous ABA or salt. Previously, it has been reported that plants with disrupted sphingolipid biosynthesis have altered responses to ABA, including reduced sensitivity to ABA-dependent stomatal closure and suppression of germination (Ng et al., 2001; Coursol et al., 2003; Worrall et al., 2008; Wu et al., 2015). Therefore, analogous to their role in mammalian systems, sphingolipid molecules such as sphingosine-1-phosphate and ceramide may act as signaling molecules. Our data suggest that the structure of the glycan headgroup of GIPCs is also involved in ABA responses, at least in the seed. How this could be mediated is unclear, but detergent-resistant membrane fractions are highly enriched in GIPCs (Borner et al., 2005; Cacas et al., 2016). It has been proposed that the extended length of the acyl chain in GIPCs is critical both for plasma membrane microdomain stability and for defining a specific endomembrane trafficking pathway (Markham et al., 2011; Cacas et al., 2016). Altered GIPC headgroup glycosylation results in a constitutive defense response mediated by increased salicylic acid biosynthesis (Mortimer et al., 2013; Fang et al., 2016; Tartaglio et al., 2017), as well as a decrease in cellulose content (cellulose biosynthesis also occurs at the plasma membrane). Therefore, we conclude that there is evidence that the mis-glycosylation of GIPCs impacts plasma-membrane-regulated physiological processes, particularly those, such as signaling, which are often associated with lipid microdomains.
Rice vegetative tissue does not contain detectable Hex-GIPCs (Ishikawa et al., 2016) despite the presence of a GMT1 ortholog; therefore, we hypothesized that HexN(Ac)-GIPCs may have a critical role in all tissues, in much the same way as Hex-GIPCs have in Arabidopsis. Inactivation of the rice AtGINT1 ortholog, OsGINT1, resulted in abnormal growth phenotypes in seedlings regenerated from calli, which were eventually seedling lethal. These seedlings almost completely lacked HexN(Ac)-GIPCs and accumulated the substrate of OsGINT1, GlcA-IPC, to very high levels. This reinforces the data showing that OsGINT1 can complement Atgint1, and strongly supports our conclusion that these enzymes have the same biochemical function in planta.
Since the distribution of HexN(Ac)-containing GIPCs is very different in rice and Arabidopsis, the characterization of GINT1 provided an opportunity to test the effects of de novo synthesis of atypical GIPC glycans on plant growth and development. Our data show that replacing a Man with a glucosamine can only partially rescue the Atgmt1 phenotype. This implies that the identity of the sugar, as well as the sugar chain length, is important for GIPC function, depending on the tissue and species context. Alternatively, the Man-GlcA-IPC may be an important substrate for some further glycosylation steps, and these unusual longer headgroups could have specialized functions.
In line with previous studies on Arabidopsis (Mortimer et al., 2013; Fang et al., 2016), rice cells with altered GIPC headgroup structures grew normally as calli on callus-induction medium (solid and liquid), but when seedlings were regenerated or grown from seed, a severe or lethal phenotype was observed. This suggests a possible role in cell adhesion or differentiation for the GIPC headgroup, perhaps via interaction with cell wall polysaccharides, as has been proposed previously (Voxeur and Fry, 2014). Indeed, the Atgmt1 mutant was initially identified as a cell adhesion mutant (Singh et al., 2005), and a Volvox carteri mutant lacking a GONST1 homolog is unable to undergo inversion at the end of embryogenesis (Ueki and Nishii, 2009). In mammalian systems, carbohydrate-carbohydrate interactions, such as Lewisx-Lewisx (where Lewisx is the terminal Gal-Fuc-GlcNAc trisaccharide decorating many surface glycoproteins and glycolipids) have been shown to play a role in cell adhesion (Zhang et al., 2016). The plants developed in this publication will be used to further explore this hypothesis.
Studies using tobacco and bean have reported that the HexN(Ac) is GlcN(Ac) and that the hexose is Gal and/or Man (Carter et al., 1958; Carter and Koob, 1969). We have previously reported that the first hexose on Arabidopsis Hex-GIPCs is Man (Mortimer et al., 2013; Fang et al., 2016). However, in this study, due to the use of MS, we have referred to the GIPCs as HexN(Ac)-GIPCs and Hex-HexN-GIPCs, as we have not identified the sugars beyond their masses. Nevertheless, GlcN(Ac) is the only amino sugar known to be present in plants, and previous reports have identified GlcNAc in GIPCs from rice vegetative tissue (Kojima et al., 1991) and Arabidopsis seeds (Luttgeharm et al., 2015) is GlcN(Ac). The identity of the Hex is unknown. We predict that GINT1 transfers GlcNAc rather than GlcN, since the donor UDP-GlcNAc is abundant in plants but UDP-GlcN has not yet been detected (Ito et al., 2014). Thus, the different ratio of HexN- and HexNAc-GIPC between AtGINT1- and OsGINT1-expressing gmt1 leaves (Fig. 10) was possibly due to a deacetylation reaction, for which the responsible enzyme remains unidentified.
To conclude, we propose that GINT1 is a GIPC α-GlcNAc transferase that likely uses UDP-GlcNAc as substrate. GlcN(Ac)-containing GIPCs are essential for rice but have a minor role in seed development and germination in Arabidopsis. We also showed that the identity of the monosaccharides on the GIPC headgroup and not just the presence of a certain number of sugar units, are important for function. This implies that there is some recognition of these structures by the plant and a potential signaling role for glycosylated GIPCs.
MATERIALS AND METHODS
Bioinformatics
Plant GT64 family genes were searched by BLAST in the Phytozome database (https://phytozome.jgi.doe.gov) for Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa), moss (Physcomitrella patens), liverwort (Marchantia polymorpha), and green alga (Chlamydomonas reinhardtii), and the NCBI database (http://www.ncbi.nlm.nih.gov/BLAST/) for charophyte (Klebsormidium flaccidum). The amino acid sequences were aligned by ClustalW (http://www.genome.jp/tools-bin/clustalw), and the phylogenetic tree was constructed using NJPLOT software (Perrière and Gouy, 1996).
Sample Collection
For all experiments, unless explicitly stated, one biological replicate represents a pool of plants grown at the same time, alongside their respective control.
Plant Growth Conditions
Arabidopsis seeds were surface sterilized and sown on solid medium containing 0.5× Murashige and Skoog salts including vitamins and 1% (w/v) Suc. Following stratification (48 h, 4°C, in the dark), plates were transferred to a growth room (22°C, 100–200 µmol m−2 s−1, 14 h light/10 h dark, 60% humidity). After 2 to 3 weeks, plants were transferred to soil.
For analyses of gene expression and GIPC profiles during seed germination, dry seeds (day 0) were imbibed for 2 d in the dark at 4°C (day 2) and incubated on Murashige and Skoog medium for 3 and 7 d (days 5 and 9) under continuous light at 22°C. Whole-plant tissues including seed coats and germinating seedlings were collected at each time point and used for RT-qPCR and GIPC analyses as follows.
GUS Staining
The 5′ flanking region with a part of N-terminal coding sequence (10 amino acids) of AtGINT1 (−2,016 to +30) and AtGMT1 (−1,052 to +30) was amplified by genomic PCR and cloned into pDONR207 by BP reaction (Thermo Fisher Scientific). The fragments were transferred to pMDC164 (Curtis and Grossniklaus, 2003) and used for Agrobacterium tumefaciens-mediated transformation of Arabidopsis. GUS staining was performed using tissues of homozygous T3 plants from at least five independent lines as described by Kawai-Yamada et al. (2009).
RT-qPCR
Total RNA was extracted from dry seeds and germinating seedlings as described by (Meng and Feldman, 2010). In brief, plant materials were ground in liquid nitrogen with a mortar and pestle and homogenized in extraction buffer (100 mm Tris-HCl, pH 9.5, 150 mm NaCl, 1% [w/v] sarkosyl, 1% [v/v] 2-mercaptoethanol). Insoluble materials were removed by centrifugation (15,000g, 5 min), and the supernatant was washed with 0.5 volumes each of chloroform and phenol. After centrifugation, the upper layer was collected and mixed with 0.1 volumes of 3 m sodium acetate (pH 5.2) and 0.75 volumes of 2-propanol. After incubation (10 min) and centrifugation, the supernatant was removed, and the pellet was washed with 70% (v/v) ethanol and briefly air dried. The pellet was resuspended in TRIzol reagent (Life Technologies), and total RNA was prepared according to the manufacturer’s protocol. cDNA was prepared using High Capacity cDNA reverse transcription kit (Applied Biosystems), and qPCR was conducted with Power SYBR Green PCR Master Mix and a 7300 real-time PCR system (Applied Biosystems). PP2AA3 was used as a reference. Primer sequences are shown in Supplemental Table S2.
GIPC Analysis
Lyophilized plant tissues (5–10 mg) were homogenized in 450 µL of methanol/1-butanol (1:2, v/v). After heat-denaturation of enzymes at 80°C for 10 min, 300 µL of 1 n KOH was added. The mixture was further incubated at 60°C for 30 min to eliminate glycerolipids. The extract was then acidified with 1.5 mL of 0.4 n HCl and extracted with additional 1 mL 1-butanol. After vigorous shaking and centrifugation, the upper 1-butanol layer was collected and evaporated. The residue was dissolved in 150 µL of THF/methanol/water (2:1:2, v/v/v) containing 0.1% formic acid and GIPC composition was analyzed by LC-MS/MS according to previous reports (Fang et al., 2016; Ishikawa et al., 2016).
Mutant Identification
Seeds of AT5g04500 T-DNA mutants were obtained from the Arabidopsis Biological Resource Center (gint1-1, SALK_002825C and gint1-2, GK_834G06), which are both ecotype Col-0. Plants homozygous for the T-DNA insertion were identified by PCR (Phire Plant Direct PCR kit; Finnzymes), and the site of insertion was confirmed by sequencing. RT-PCR was used to test whether the alleles were transcriptionally null as follows: Total RNA was extracted from green siliques using the RNeasy Plant Mini Kit (Qiagen). RNA was first treated with DNase I (Qiagen) and first-strand cDNA synthesis was performed using the SuperScript II RT (Life Technologies). ACTIN7 was used as an internal control. All primers used in this study are listed in Supplemental Table S2.
Promoter Swap and Complementation of gint1
Genomic fragment of AtGINT1 including 5′ upstream (2,016 bp) and 3′ downstream (710 bp) of the coding sequence was amplified by PCR. For the promoter exchange, the 5′ and 3′ sequences of AtGINT1 were separately amplified and joined to the coding sequence of AtGMT1 or OsGINT1 by overlap extension PCR to prepare the chimeric complementation construct. These fragments were cloned into pDONR207 and transferred into pMDC99 (Curtis and Grossniklaus, 2003) by the Gateway system and used for transformation of gint1. Seeds obtained from homozygous T2 plants were used for GIPC analysis.
Seed Weight, Storage Lipid, and Protein Content
Seed weight was measured using around 200 seeds. Storage lipids were extracted by the Bligh-Dyer method (Bligh and Dyer, 1959) and subjected to methanolysis using 5% (w/v) HCl in methanol (100°C, 2 h). Margaric acid (17:0) was added as an internal standard prior to methanolysis. Fatty acid methyl esters were quantified by gas chromatography-mass spectrometry (GCMS2010; Shimadzu) using a TC-70 column (30 m × 0.25 µm × 0.25 µm; GL Science) held at 120°C for 3 min, increased to 240°C by 6°C/min and held for 5 min. Temperature of injector, interface, and ion source were kept at 250°C. Carrier gas was He at 30 cm/s. MS scan range was m/z 50 to 400 monitored between 7 and 25 min. Seed storage proteins were extracted by homogenizing seeds in Tris-buffered saline (50 mm Tris-HCl, pH 7.4, and 150 mm NaCl) and quantified with Bio-Rad protein assay kit using bovine serum albumin as a standard. Seed grain numbers used for lipid and protein analyses were counted and the contents were expressed as amount of lipids or proteins per seed grain.
Ruthenium Red Staining
Seed coat mucilage was visualized by ruthenium red staining as described by Western (2011). Dry seeds were hydrated in 50 mm EDTA for 2 h with vigorous shaking and stained with 0.01% (w/v) ruthenium red for 1 h. Seeds were washed with water and observed under a microscope.
Scanning Electron Microscopy
Scanning electron microscopy of seed surface of gint1 mutants was performed with a TM-1000 Miniscope (Hitachi).
TEM
Arabidopsis seeds were sterilized with 10% (v/v) household bleach and imbibed overnight in distilled water at 4°C. The following day, a slit was carefully cut in the seed coat to help aid infiltration, followed by fixation in 0.1 m cacodylate buffer (pH 7.2) containing 2% (v/v) EM-grade glutaraldehyde, with gentle pulling under vacuum for 30 s. Seeds remained in fixation buffer overnight at 4°C. Seeds were rinsed three times with 0.1 m cacodylate buffer (pH 7.2), stained for 1 h with 1% (w/v) osmium tetraoxide, rinsed three times with buffer, followed by an additional three rinses with distilled water. Seeds were then dehydrated in an acetone gradient (35/50/70/80/95/100/100% v/v) and infiltrated in acetone:epon resin at 2:1, 1:1, and 1:2 dilution ratios for 1 h each, followed by pure epon resin that was freshly changed after 1 h and allowed to infiltrate overnight. The following day, seeds were infiltrated with pure resin + accelerator for 2 h and embedded in Pelco molds containing fresh resin + accelerator, which were left in a 65°C oven to polymerize for 2 d. Ultra-thin 70-nm-thick sections were collected on to grids using a diamond knife and Reichert microtome. Grids were stained with 2% (v/v) aqueous uranyl acetate for 5 min, rinsed with distilled water five times, stained with lead citrate for 5 min, and rinsed another five times with distilled water using a Pelco Grid Staining System. Grids were imaged using a Technai 1200 electron microscope. For consistency, we analyzed two grids per seed, using a total of three seeds per genotype.
AIR Cell Wall Preparation
Plant tissues (lower half of the inflorescence stems [8-week-old plants], young leaves [15-d-old plants], and dry seeds) were harvested and incubated in 96% (v/v) ethanol for 30 min at 70°C to inactivate cell wall degrading enzymes. The tissue was homogenized using a Retsch mixer mill and centrifuged at 4,000g for 15 min. The pellet was washed with 100% ethanol and twice with chloroform:methanol (2:1), followed by three successive washes with 65% (v/v), 80% (v/v), and 100% ethanol. The pellet was air-dried overnight. The starch in the samples was degraded with α-amylase, amyloglucosidase, and pullulanase (Megazyme) as described previously (Harholt et al., 2006). The destarched residue is referred to as AIR.
Cell Wall Monosaccharide Composition
AIR (5 mg) was hydrolyzed with fresh 2 m TFA at 121°C for 1 h. The supernatant was retained, dried under vacuum, and resuspended in 1 mL water. To release the Glc from the crystalline cellulose fraction, the TFA-insoluble material was washed with water and further hydrolyzed with 72% (v/v) sulfuric acid containing 10 µg myo-inositol for 1 h at room temperature. The sulfuric acid was then diluted to 1 m with water, and following incubation at 100°C for 3 h, neutralized with BaCO3. All samples were filtered through a 96-well 0.45 µm filter plate (Millipore) and analyzed by high-performance anion-exchange chromatography on an ICS-5000 instrument (Thermo Fisher Scientific) equipped with a CarboPac PA20 (3 mm × 150 mm; Thermo Fisher Scientific) analytical anion-exchange column, PA20 guard column (3 mm × 30 mm), borate trap, and a pulsed amperometric detector using the elution profile described (Fang et al., 2016).
Germination Rate
Surface sterilized seeds were sowed on solid 0.5× Murashige and Skoog salt media containing either ABA (mixed isomers; Sigma-Aldrich A1049) or NaCl. Following stratification for 48 h (dark, 4°C), plates were moved to the light, and seeds were scored for radicle emergence daily. Each genotype was sown in triplicate (80–100 seeds from an individual plant per petri dish).
Embryo Characterization
For phenotypic analysis of Atgint1 embryos, mature green siliques were dissected onto a slide and examined using a light microscope. The ratio of green (healthy) to white (arrested development) embryos was tallied.
Seed Aging
An accelerated seed aging test was performed as described by Sattler et al. (2004). Dry seeds were treated with 100% relative humidity in a tight-closed container at 40°C for 3 d and germinated on Murashige and Skoog media under continuous light at 22°C. Nontreated dry seeds were used as control. Germination rate was counted daily for 7 d.
CRISPR/Cas9-Mediated Mutagenesis of OsGINT1
Two target sequences for CRISPR/Cas9-mediated mutagenesis of OsGINT1 were designed using the CRISPR-P tool (Lei et al., 2014; Supplemental Fig. S9). Each fragment was ligated to the BbsI site of the pU6gRNA plasmid. The OsU6pro::gRNA::polyT cassette was digested by AscI and PacI and introduced into pZH_gYSA_MMCas9 (Mikami et al., 2015). The binary plasmid was used for A. tumefaciens-mediated transformation of rice calli (Toki et al., 2006). Hygromycin-resistant calli were subcultured three to four times and transferred to regeneration medium (Murashige and Skoog salts containing vitamins, 3% [w/v] Suc, 3% [w/v] sorbitol, 0.4% [w/v] casamino acid, 2 µg/L 1-naphthaleneacetic acid, 2 mg/L kinetin, 30 mg/L hygromycin). After 2 to 3 weeks on regeneration media, emerging shoots (1–3 cm) were transferred to hormone-free rooting medium (0.5× Murashige and Skoog salts containing vitamins, 1% [w/v] Suc, 30 mg/L hygromycin) and further cultured for 2 to 3 weeks (28°C, 300 µmol m−2 s−1, 12 h light/12 h dark). Some of the regenerated shoots showed retarded growth and finally died, although the most were grown well without dead tissues. Four independent shoots showing the typical growth phenotype were used for GIPC analysis.
Heterologous Complementation of Atgmt1
Full coding sequences of AtGMT1, AtGINT1, and OsGINT1 were introduced into pH2GW7 (Karimi et al., 2002) for CaMV35Spro-driven constitutive expression (Supplemental Fig. S3). AtGMT1/Atgmt1 heterozygotes were transformed with the constructs and hygromycin-resistant T1 plants were selected. Homozygous Atgmt1 mutants complemented with the transgene were screened in the T1 and T2 generations by genomic PCR and used for RT-PCR and GIPC analyses.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers NM_120532.3 and AP014961.1.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenetic tree of the plant GT64 family.
Supplemental Figure S2. Expression profile of AtGINT1.
Supplemental Figure S3. Summary of Atgint1 alleles and constructs used in this study.
Supplemental Figure S4. GlcCer and Cer content in Atgint1 tissues.
Supplemental Figure S5. AtGMT1 expression in the dry seeds of Atgint1 mutants.
Supplemental Figure S6. Noncellulosic monosaccharide composition of Atgint1 stem and young leaf AIR.
Supplemental Figure S7. Seed longevity assay.
Supplemental Figure S8. Expression profile of OsGINT1, OsGINT2, and OsGMT1 across development.
Supplemental Figure S9. Target sequences of CRISPR/Cas9-mediated mutagenesis of OsGINT1.
Supplemental Figure S10. Shoot regeneration phenotype of Atgmt1 mutant.
Supplemental Table S1. Embryo lethality rate in Atgint1 green siliques.
Supplemental Table S2. Primers used in this study.
Supplemental Data Set 1. Sphingolipidomic data underlying Figures 2, 3, 9, and 10.
Supplemental Data Set 2. Monosaccharide composition data underlying Figure 7 and Supplemental Figure S6.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Dr. Masaki Endo, Dr. Seiichi Toki (National Food Research Institute, NARO), and Mr. Masafumi Mikami (Yokohama City University) for providing CRISPR/Cas9 vectors. We thank Dr. Mi-Yeon Lee for assistance with maintaining the plant growth facilities and Dr. Jonathan Markham for advice on Arabidopsis seed GIPC analysis.
Footnotes
This work was part of the Department of Energy Joint BioEnergy Institute supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy. E.A.R. was supported by a Life Sciences Research Foundation fellowship from the Gordon and Betty Moore Foundation. This work was also supported by the Japan Society for the Promotion of Science (KAKENHI grants 17K15411 and 26292190 to T.I. and M.K.-Y.) and by the National Science Foundation (MCB-1158500 to E.B.C.).
Articles can be viewed without a subscription.
References
- Bligh EG, Dyer WJ (1959) A rapid method of total lipid extraction and purification. Can J Biochem Physiol 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Borner GH, Sherrier DJ, Weimar T, Michaelson LV, Hawkins ND, Macaskill A, Napier JA, Beale MH, Lilley KS, Dupree P (2005) Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol 137: 104–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buré C, Cacas JL, Wang F, Gaudin K, Domergue F, Mongrand S, Schmitter JM (2011) Fast screening of highly glycosylated plant sphingolipids by tandem mass spectrometry. Rapid Commun Mass Spectrom 25: 3131–3145 [DOI] [PubMed] [Google Scholar]
- Busse-Wicher M, Wicher KB, Kusche-Gullberg M (2014) The exostosin family: proteins with many functions. Matrix Biol 35: 25–33 [DOI] [PubMed] [Google Scholar]
- Cacas J-L, Buré C, Grosjean K, Gerbeau-Pissot P, Lherminier J, Rombouts Y, Maes E, Bossard C, Gronnier J, Furt F, et al. (2016) Revisiting plant plasma membrane lipids in tobacco: A focus on sphingolipids. Plant Physiol 170: 367–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter HE, Koob JL (1969) Sphingolipids in bean leaves (Phaseolus vulgaris). J Lipid Res 10: 363–369 [PubMed] [Google Scholar]
- Carter HE, Gigg RH, Law JH, Nakayama T, Weber E (1958) Biochemistry of the sphingolipides. XI. Structure of phytoglycolipide. J Biol Chem 233: 1309–1314 [PubMed] [Google Scholar]
- Coursol S, Fan LM, Le Stunff H, Spiegel S, Gilroy S, Assmann SM (2003) Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature 423: 651–654 [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn TM, Lynch DV, Michaelson LV, Napier JA (2004) A post-genomic approach to understanding sphingolipid metabolism in Arabidopsis thaliana. Ann Bot 93: 483–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Ishikawa T, Rennie EA, Murawska GM, Lao J, Yan J, Tsai AY, Baidoo EE, Xu J, Keasling JD, et al. (2016) Loss of inositol phosphorylceramide sphingolipid mannosylation induces plant immune responses and reduces cellulose content in Arabidopsis. Plant Cell 28: 2991–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, Chang HY, Dosztányi Z, El-Gebali S, Fraser M, et al. (2017) InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res 45: D190–D199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harholt J, Jensen JK, Sørensen SO, Orfila C, Pauly M, Scheller HV (2006) ARABINAN DEFICIENT 1 is a putative arabinosyltransferase involved in biosynthesis of pectic arabinan in Arabidopsis. Plant Physiol 140: 49–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TC, Lester RL, Laine RA (1981) Glycophosphoceramides from plants. Purification and characterization of a novel tetrasaccharide derived from tobacco leaf glycolipids. J Biol Chem 256: 7747–7755 [PubMed] [Google Scholar]
- Ishikawa T, Ito Y, Kawai-Yamada M (2016) Molecular characterization and targeted quantitative profiling of the sphingolipidome in rice. Plant J 88: 681–693 [DOI] [PubMed] [Google Scholar]
- Ito J, Herter T, Baidoo EE, Lao J, Vega-Sánchez ME, Michelle Smith-Moritz A, Adams PD, Keasling JD, Usadel B, Petzold CJ, et al. (2014) Analysis of plant nucleotide sugars by hydrophilic interaction liquid chromatography and tandem mass spectrometry. Anal Biochem 448: 14–22 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kaul K, Lester RL (1975) Characterization of inositol-containing phosphosphingolipids from tobacco leaves: Isolation and identification of two novel, major lipids: N-acetylglucosamidoglucuronidoinositol phosphorylceramide and glucosamidoglucuronidoinositol phosphorylceramide. Plant Physiol 55: 120–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai-Yamada M, Hori Z, Ogawa T, Ihara-Ohori Y, Tamura K, Nagano M, Ishikawa T, Uchimiya H (2009) Loss of calmodulin binding to Bax inhibitor-1 affects Pseudomonas-mediated hypersensitive response-associated cell death in Arabidopsis thaliana. J Biol Chem 284: 27998–28003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima M, Ohta K, Ohnishi M, Ito S (1991) Chemical characterization of sphingophosphoglycolipids in rice leafy stems. Res. Bull. Obihiro. Univ. I 17: 143–148 [Google Scholar]
- Lei Y, Lu L, Liu HY, Li S, Xing F, Chen LL (2014) CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant 7: 1494–1496 [DOI] [PubMed] [Google Scholar]
- Lenarčič T, Albert I, Böhm H, Hodnik V, Pirc K, Zavec AB, Podobnik M, Pahovnik D, Žagar E, Pruitt R, et al. (2017) Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins. Science 358: 1431–1434 [DOI] [PubMed] [Google Scholar]
- Luttgeharm KD, Kimberlin AN, Cahoon RE, Cerny RL, Napier JA, Markham JE, Cahoon EB (2015) Sphingolipid metabolism is strikingly different between pollen and leaf in Arabidopsis as revealed by compositional and gene expression profiling. Phytochemistry 115: 121–129 [DOI] [PubMed] [Google Scholar]
- Markham JE, Jaworski JG (2007) Rapid measurement of sphingolipids from Arabidopsis thaliana by reversed-phase high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry. Rapid Commun Mass Spectrom 21: 1304–1314 [DOI] [PubMed] [Google Scholar]
- Markham JE, Li J, Cahoon EB, Jaworski JG (2006) Separation and identification of major plant sphingolipid classes from leaves. J Biol Chem 281: 22684–22694 [DOI] [PubMed] [Google Scholar]
- Markham JE, Molino D, Gissot L, Bellec Y, Hématy K, Marion J, Belcram K, Palauqui JC, Satiat-Jeunemaître B, Faure JD (2011) Sphingolipids containing very-long-chain fatty acids define a secretory pathway for specific polar plasma membrane protein targeting in Arabidopsis. Plant Cell 23: 2362–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markham JE, Lynch DV, Napier JA, Dunn TM, Cahoon EB (2013) Plant sphingolipids: function follows form. Curr Opin Plant Biol 16: 350–357 [DOI] [PubMed] [Google Scholar]
- Meng L, Feldman L (2010) A rapid TRIzol-based two-step method for DNA-free RNA extraction from Arabidopsis siliques and dry seeds. Biotechnol J 5: 183–186 10.1002/biot.200900211 [DOI] [PubMed] [Google Scholar]
- Mikami M, Toki S, Endo M (2015) Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol Biol 88: 561–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer JC, Yu X, Albrecht S, Sicilia F, Huichalaf M, Ampuero D, Michaelson LV, Murphy AM, Matsunaga T, Kurz S, et al. (2013) Abnormal glycosphingolipid mannosylation triggers salicylic acid-mediated responses in Arabidopsis. Plant Cell 25: 1881–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng CK, Carr K, McAinsh MR, Powell B, Hetherington AM (2001) Drought-induced guard cell signal transduction involves sphingosine-1-phosphate. Nature 410: 596–599 [DOI] [PubMed] [Google Scholar]
- Perrière G, Gouy M (1996) WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78: 364–369 [DOI] [PubMed] [Google Scholar]
- Rennie EA, Ebert B, Miles GP, Cahoon RE, Christiansen KM, Stonebloom S, Khatab H, Twell D, Petzold CJ, Adams PD, et al. (2014) Identification of a sphingolipid α-glucuronosyltransferase that is essential for pollen function in Arabidopsis. Plant Cell 26: 3314–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler SE, Gilliland LU, Magallanes-Lundback M, Pollard M, DellaPenna D (2004) Vitamin E is essential for seed longevity and for preventing lipid peroxidation during germination. Plant Cell 16: 1419–1432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Eland C, Harholt J, Scheller HV, Marchant A (2005) Cell adhesion in Arabidopsis thaliana is mediated by ECTOPICALLY PARTING CELLS 1—a glycosyltransferase (GT64) related to the animal exostosins. Plant J 43: 384–397 [DOI] [PubMed] [Google Scholar]
- Tartaglio V, Rennie EA, Cahoon R, Wang G, Baidoo E, Mortimer JC, Cahoon EB, Scheller HV (2017) Glycosylation of inositol phosphorylceramide sphingolipids is required for normal growth and reproduction in Arabidopsis. Plant J 89: 278–290 [DOI] [PubMed] [Google Scholar]
- Tellier F, Maia-Grondard A, Schmitz-Afonso I, Faure JD (2014) Comparative plant sphingolipidomic reveals specific lipids in seeds and oil. Phytochemistry 103: 50–58 [DOI] [PubMed] [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47: 969–976 [DOI] [PubMed] [Google Scholar]
- Ueki N, Nishii I (2009) Controlled enlargement of the glycoprotein vesicle surrounding a volvox embryo requires the InvB nucleotide-sugar transporter and is required for normal morphogenesis. Plant Cell 21: 1166–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voxeur A, Fry SC (2014) Glycosylinositol phosphorylceramides from Rosa cell cultures are boron-bridged in the plasma membrane and form complexes with rhamnogalacturonan II. Plant J 79: 139–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yang X, Tangchaiburana S, Ndeh R, Markham JE, Tsegaye Y, Dunn TM, Wang GL, Bellizzi M, Parsons JF, et al. (2008) An inositolphosphorylceramide synthase is involved in regulation of plant programmed cell death associated with defense in Arabidopsis. Plant Cell 20: 3163–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Western TL. (2011) The sticky tale of seed coat mucilages: production, genetics, and role in seed germination and dispersal. Seed Sci Res 22: 1–25 [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrall D, Liang Y-K, Alvarez S, Holroyd GH, Spiegel S, Panagopulos M, Gray JE, Hetherington AM (2008) Involvement of sphingosine kinase in plant cell signalling. Plant J 56: 64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J-X, Li J, Liu Z, Yin J, Chang Z-Y, Rong C, Wu J-L, Bi F-C, Yao N (2015) The Arabidopsis ceramidase AtACER functions in disease resistance and salt tolerance. Plant J 81: 767–780 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lu D, Sollogoub M, Zhang Y (2016) Carbohydrate-carbohydrate interaction: from hypothesis to confirmation. In Rauter AR, Lindhorst TK, Queneau Y, eds, Carbohydrate Chemistry, Vol 41 The Royal Society of Chemistry, Cambridge, pp 238–254 [Google Scholar]