Abstract
MADS-box transcription factors are important for plant growth and development, and hundreds of MADS-box genes have been functionally characterized in plants. However, less is known about the functions of these genes in the economically important allopolyploid oil crop, Brassica napus. We identified 307 potential MADS-box genes (BnMADSs) in the B. napus genome and categorized them into type I (Mα, Mβ, and Mγ) and type II (MADS DNA-binding domain, intervening domain, keratin-like domain, and C-terminal domain [MIKC]c and MIKC*) based on phylogeny, protein motif structure, and exon-intron organization. We identified one conserved intron pattern in the MADS-box domain and seven conserved intron patterns in the K-box domain of the MIKCc genes that were previously ignored and may be associated with function. Chromosome distribution and synteny analysis revealed that hybridization between Brassica rapa and Brassica oleracea, segmental duplication, and homologous exchange (HE) in B. napus were the main BnMADSs expansion mechanisms. Promoter cis-element analyses indicated that BnMADSs may respond to various stressors (drought, heat, hormones) and light. Expression analyses showed that homologous genes in a given subfamily or sister pair are highly conserved, indicating widespread functional conservation and redundancy. Analyses of BnMADSs provide a basis for understanding their functional roles in plant development.
Introduction
The MADS-box gene family includes important transcription factors that are relevant across Eukarya, including fungi, animals, and plants [1]. This gene family commonly possesses a highly conserved N-terminal DNA binding domain (MADS-box), a moderately conserved coiled-coil domain for protein reciprocity (K-box), a lightly conserved domain (I domain) between the MADS-box and K-box, and a non-conserved C-terminal. The MADS-box is a DNA binding domain of approximately 58 amino acids that binds DNA at consensus recognition sequences known as CArG boxes [CC(A/T)6GG][2, 3]. The I terminal is less conserved and contributes to the specification of dimerization. The K domain is characterized by a coiled-coil structure that facilitates the dimerization of MADS-box proteins [4, 5]. Evolution of motifs in different C-terminals results in different functions for MADS-box proteins [4, 6].
The MADS-box gene family is divided into two main lineages, type-I and type-II, that originated from a duplication of ancestor genes during the divergence of plants and animals [7]. Type-I genes were further divided into an animal serum-response factor (SRF) type and plant type-I, while type-II genes were divided into fungi myocyte enhancer factor 2 (MEF2) type and plant type-II[7]. A major difference between type-I and type-II genes is that type-II genes possess the K-box domain. Genes in the type-II lineage are also termed MADS DNA-binding domain, intervening domain, keratin-like domain, and C-terminal domain (MIKC) MADS-box genes, attributable to their four domains, including the MADS-box domain, I domain, K-box domain, and C terminal [8, 9]. MIKC-type genes can be further divided into MIKCc and MIKC* clades, both of which were identified in a common ancestor of moss and vascular plants, suggesting they are ancestral gene types [10].
The MADS-box gene family is known to contain flower organ identity genes and participate in homologous transformation during floral ontogeny. Floral organ identity genes are subdivided into A, B, C, D, and E classes (ABCDE model), providing different homeotic functions with different combinations, including A sepal, A+B petal, B+C stamen, C carpel, D ovary formation and ovule development, E formation of floral organ in four rounds, and B+C+E male flowers[11–15]. Moreover, the MADS-box gene family also contains flowering-time genes and fruit dehiscence-related genes [16–18]. In addition, MADSs play regulatory roles during the vegetative stage, including embryo, root, and leaf development [16, 19–22]. MADSs also have been identified in gymnosperms and mosses without flowers or fruits, proving they are not limited to flower and fruit development [10, 23–27].
To date, studies regarding the MADS-box gene family have been performed at the genome level in many plant species, including Arabidopsis and Brassica rapa [28, 29]. Brassica napus is one of the most important oil crops and ornamental plants in China; B. napus fluoresces, which is considered decorative, while its seeds can be used to produce edible oils. Thus, identification and functional analyses of the MADS-box gene family in B. napus would contribute to our understanding of the developmental mechanisms associated with flowering, as well as improving floral organ features. For example, silique dehiscence is a serious problem that results in a reduction in seed yield in B. napus [30], and the molecular mechanisms underlying silique dehiscence are closely related to the ABCDE model. Thus, corresponding analyses in B. napus could be an alternative way to solve this problem.
In this study, we performed a complete analysis of the MADS-box family of genes in B. napus (BnMADSs) by identifying putative genes, evaluating the division of different subfamilies based on phylogeny, and exploring the protein structure and exon-intron characters. We proved that the evolutionary mechanisms underlying intron patterns remain consistent with those of functional diversification. Chromosome localization combined with gene synteny analyses revealed the expansion of BnMADSs in B. napus. In addition, a cis-acting regulatory element analysis of promoters indicated possible roles for BnMADSs in plant development and stress response. Moreover, an expression profile analysis revealed abundant roles for BnMADSs in B. napus.
Material and methods
Sequence retrieval
A preliminary search for B. napus MADS-box proteins in Genoscope (http://www.genoscope.cns.fr/brassicanapus/) was performed using the basic local alignment search tool-protein (BLASTP) with at least one representative sequence of the MADS-box domain for each MADS-box subfamily. We discarded redundant sequences with the same chromosome locus to ensure the candidate genes mapped to unique loci in their respective genomes. We then confirmed the putative non-redundant sequences to ensure that the putative proteins contained MADS-box domains using ExPASy (http://expasy.org/prosite/) [31] and MEGA 7.0 [32] software. Finally, all candidate genes were named according to the chromosome locus.
Phylogenetic tree construction
Multiple sequence alignment was performed using MADS-box protein sequences of Arabidopsis and B. napus via the multiple alignment fast Fourier transform (MAFFT) software (http://mafft.cbrc.jp/alignment/server/) [33]. The phylogenetic tree was constructed based on alignment of the MADS-box domains using MEGA7.0 [32] and the neighbor-joining (NJ) method, with 1000 iterations for the bootstrap values, p-distance model, and pairwise deletion for gap treatment. Tree files were viewed and edited using FigTree v1.3.1 (http://tree.bio.ed.ac.uk/software/figtree/).
Chromosome localization
Information regarding chromosome length and gene locations was obtained from the Brassica Database (http://brassicadb.org/brad/index.php) and B. napus Genome Browser (http://jacob.cea.fr/drf/ifrancoisjacob/Pages/Departements/Genoscope.aspx), respectively. Mapchart software was used to draw a chromosome map of the BnMADSs. Locations of the MADS-box genes in Arabidopsis, B. rapa, and Brassica oleracea were also determined using the same method.
Intron/Exon structure analysis
To find the intron and exon distribution and splicing phase in the candidate BnMADSs, we compared and viewed the coding DNA sequences (CDS) and DNA sequences of BnMADSs using GSDS software (http://gsds.cbi.pku.edu.cn/) [34] and then manually located the intron insertion sites of protein sequences. Information about the MADSs intron insertion sites in other species included in this study were gathered from Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html#).
Cis-element analysis of promoters
Cis-acting regulatory elements as binding sites of transcription factors connect gene structure and function [35] and play important roles in gene expression. The cis-elements in BnMADSs promoters were predicted using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Identification of conserved motifs
Full-length protein sequences of candidate BnMADSs were analysed using MEME software (http://meme-suite.org/tools/meme) [36], with the following parameters: optimum motif width ≥ 6 and ≤ 150 and maximum number of motifs to identify = 15. In addition, we used the PFAM tool (http://pfam.xfam.org/) to identify whether any remaining motifs matched well-known motifs [37].
Gene synteny analysis
We used CoGe software (https://genomevolution.org/coge/) to conduct a gene synteny analysis of MADS-box genes in Arabidopsis, B. napus, B. rapa, and B. oleracea.
Expression analysis of BnMADSs
The temporal and spatial expression patterns of candidate BnMADSs were further analysed using the RNA-seq data of 50 different tissues (including roots, stems, leaves, flowers, seeds, and siliques) from the B. napus cultivar Zhongshuang 11 (ZS11) at different developmental stages (germination, seedling, budding, initial flowering, and full-bloom stages). We recently created and deposited ZS11 in the BioProject (ID PRJNA358784). BnMADSs expression was analysed using Cluster 3.0 software based on fragments per kilobase of exon per million reads mapped (FPKM) with default parameters, and the heatmap was created using Cluster 3.0 [38] and Java Treeview software [39].
Results
Identification of MADS-box genes in B. napus
With the aim of defining the B. napus MADS-box gene family, we applied BLASTP for genes encoding MADS-box proteins in Genoscope using representative DNA-binding domain sequences of typical MADS-box proteins as queries (S1 Table). After removing redundant sequences, 312 primary putative MADS-box proteins were obtained in B. napus for this study.
To verify the reliability, we performed a PROSITE profile analysis to ensure all the putative MADS-box protein sequences contained the typical MADS-box domain. Consequently, BnaA08g31660D and BnaA09g05900D were deleted, as they had poor homology with atypical MADS-box proteins; BnaA09g30870D, BnaA02g31770D, and BnaC05g45970D were deleted because of mass deletions, especially at the N-terminal; while BnaC01g34010D, BnaC02g43730D, BnaAnng04820D, and BnaA05g35580D were reserved because the deletion occurred at the C-terminal, which did not impact the subsequent phylogenetic analyses.
Finally, a total of 307 BnMADSs with relatively complete open reading frame (ORF) regions were identified in this study, constituting the largest known plant MADS-box gene family to date. The candidate genes were then named according to the chromosomal distribution order (S1 Table).
Sequence characteristics of BnMADS proteins
To investigate sequence features, we performed a multiple alignment analysis of type-I and type-II BnMADS proteins, respectively. The MADS-box domain of type-I proteins is relatively less conserved and studied than those of type-II proteins; therefore, we manually identified type-I proteins using type-I MADS-box proteins in Arabidopsis as a reference [28].
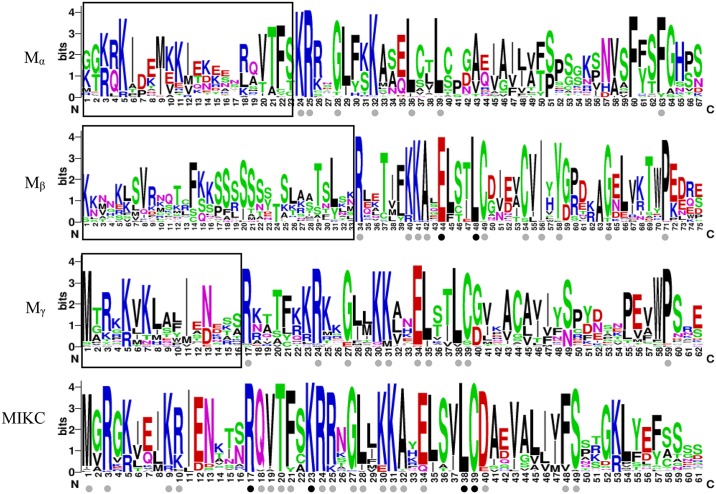
Our results show that MADS-box domains of type-I proteins are quite divergent in length and sequence characteristics (Fig 1). The length of the MADS-box domains in Mα, Mβ, and Mγ were 67aa, 75aa, and 62aa, respectively. In type-I, the N-terminal of the MADS-box domain was relatively more divergent than the C-terminal. In contrast, the MADS-box domains of type-II proteins were highly conserved, and the basic region consisted of 61 basic residues with rare deletions or insertions (approximately 1.6%). The residues Arg-17, Lys-23, Leu-38, and Cys-39 were completely conserved in the type-II MADS-box domains, and another 18 residues were also highly conserved (>90%), including Met-1, Arg-3, Lys-9, Ile-11, Gln-18, Val-19, Thr-20, Phe-21, Arg-24, Arg-25, Gly-27, Leu-28, Lys-30, Lys-31, Ala-32, Glu-34, Asp-40, Phe-48, and Ser-49. In addition, we found that the Leu-38 was completely conserved in Mβ MADS-box domains. Cys-39 was also highly conserved in type-I MADS-box domains, except in Mα. As with its counterparts in Arabidopsis, the lightly conserved Cys-39 residue in Mα was partially replaced by Ser. Similarly, Lys-24, Arg-25, and Lys-32 residues were relative highly conserved (>90%) in both type-I and type-II proteins.
Fig 1. MADS-box domain of MADSs-box genes in the Brassica napus (BnMADSs) genome.
A multiple alignment analysis was performed using the MAFFT program. The sequence logos are based on the alignments of all type I (Mα, Mβ, and Mγ) and type II (MIKC) B. napus MADS-box domains. Bit scores indicate the information content for each position in the sequence. Black and grey dots indicate 100%- and 90%-conserved residues, respectively.
Phylogenetic analysis of BnMADS proteins
To determine the evolutionary relationship of BnMADSs between B. napus and Arabidopsis, a neighbor-joining (NJ) phylogenetic tree was constructed based on the alignment of 307 and 107 MADS-box domains in B. napus and Arabidopsis, respectively, using bootstrap values (1000 iterations).
The 414 MADS-box proteins were divided into two obvious lineages; specifically, type-I and II, with 127 and 180 genes, respectively. Six BnMADSs homologous for AGL33 were assigned to the MIKC* subgroup based on the findings of research on the Arabidopsis MADS-box gene family [28]. To better examine the phylogenetic relationships between these two individual lineages, we re-constructed the phylogenetic trees of type I and type II proteins (Fig 2 and S1 Fig). Type-I genes made up a smaller percentage (37.5%) in B. napus than in Arabidopsis (64%), with significantly more type-II genes than type-I genes. This result is consistent with the trend observed in other species [28, 29] and may result from differences in gene-loss bias between these two species.
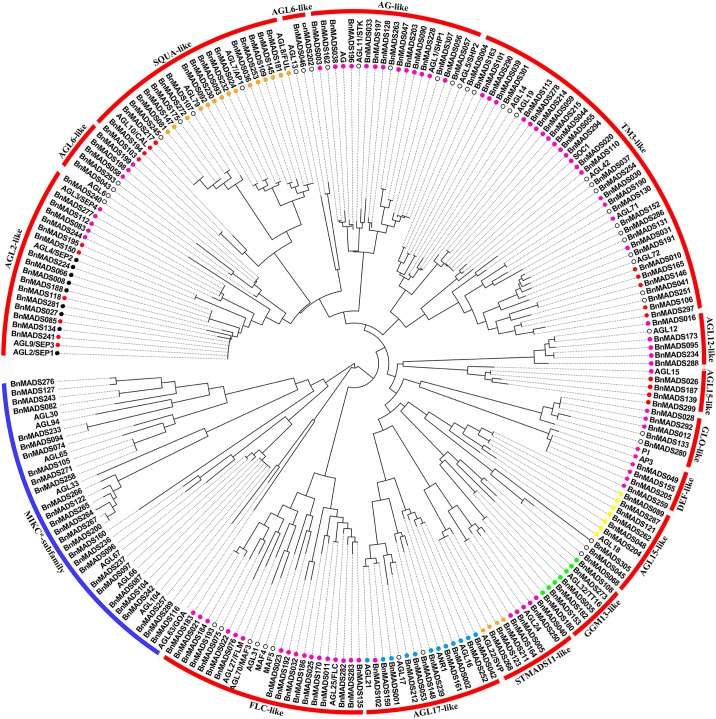
Fig 2. Phylogenetic relationships of type II BnMADS proteins investigated in this study.
A neighbor-joining tree representing relationships among 187 BnMADS proteins translated from B. napus and 45 from Arabidopsis are shown. The proteins are clustered into 16 subfamilies. Coloured dots indicate the corresponding intron distribution patterns, as shown in Fig 3.
Based on our results, we grouped the 120 type-I BnMADSs into Mα, Mβ, and Mγ clades containing 57, 30, and 33 genes, respectively. The remaining 187 MIKC genes (type II) were classed into MIKC* and MIKCc subfamilies, containing 25 and 161 genes, respectively. Among the 161 MIKCc genes, BnMADS116 and BnMADS289, along with the homologous AGL63, could not be assigned to any MIKCc subfamily, possibly attributable to high sequence divergence in the conserved regions, which is consistent with the situation observed in Arabidopsis [28]. The MIKCc subfamily was further divided into 13 clades with at least 57% bootstrap support, including AGL2-like, AGL6-like, SQUA-like, AG-like, TM3-like, AGL12-like, AGL15-like, GLO-like, DEF-like, GGM13-like, STMADS11-like, AGL17-like, and FLC-like with 17, 7 19, 15, 33, 5, 13, 5, 4, 6, 7, 11, and 17 members, respectively. Our results indicate that B. napus and Arabidopsis MADS-box proteins were not equal within a given clade, and we commonly observed two or more putative orthologous BnMADSs of a single Arabidopsis gene. Genes from the AGL6-like and AGL15-like clades were separated into two clades in our phylogenetic tree, indicating that a possible functional divergence appeared between them.
Intron patterns of BnMADSs
Intron and exon structures are important clues to understand the evolutionary gene relationship and functional diversification within a gene family [40]. In the present study, the intron and exon patterns were determined by comparing the full length CDS and DNA sequences of candidate BnMADSs using the GSDS web server.
We observed a very striking bimodal distribution of introns between types I and II in the BnMADSs family (S1 Table). Type-I genes had fewer intron insertions, mostly varying from 0 to 2, except for BnMADS077 and BnMADS156 which had three and BnMADS300 which had six introns. In contrast, type-II genes had more introns, ranging from 0 to 15, with an average of 5.95. The BnMADS005 gene had 15 introns, which is the maximum number. Type-II genes with few introns usually did not have Arabidopsis orthologs. Candidate MADS-box genes identified from other species such as Arabidopsis and B. rapa also showed the same bimodal distribution of introns [28, 29, 41–43], indicating that this trend is evolutionarily conserved. Moreover, the entire MADS-box domain was located in the first exon, accompanied by an intron insertion with phase 2 in type II genes (S2 Fig).
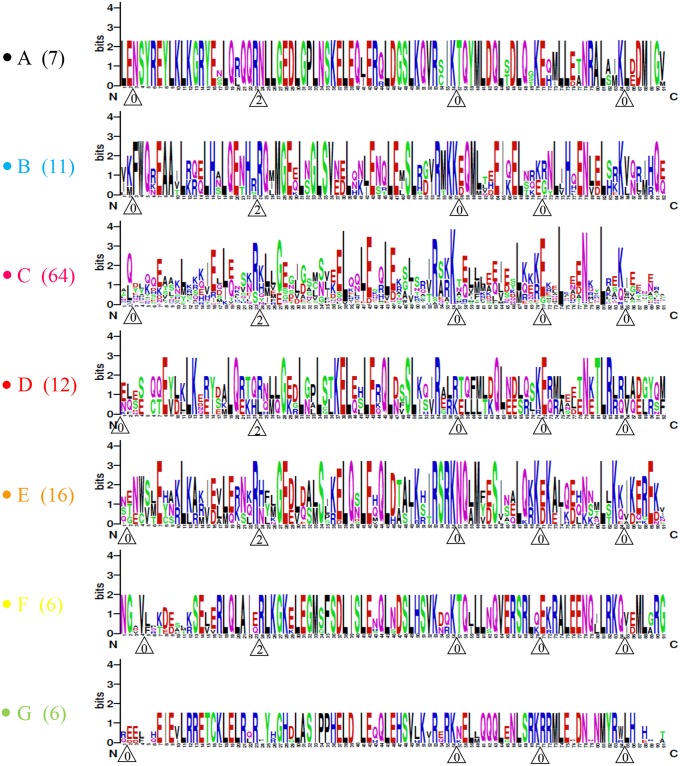
The K-box was the second conserved domain of the MIKCc genes, with 4–5 intron insertion sites generally conserved. A total of 7 highly conserved intron patterns (A–G) were first identified in terms of conserved intron insertion sites and phases in this study. Among these conserved intron patterns, C was the main type with the largest number, accounting for 52.5% of type II genes; pattern A accounted for 5.7%, B for 9.0%, D for 9.8%, E for 13.1%, F for 4.9%, and G for 4.9% (Fig 3). Intron insertion sites were conserved in each type, varying from 4 to 5 in different types. Except for the first intron insertion site, the other 4 insertion sites were conserved in all types, with the type A and G lacking the second and fifth intron, respectively. Moreover, amino acids near the intron insertion site were somewhat conserved (Fig 3). Interestingly, in the phylogenetic tree, genes in the same clade generally shared the same intron patterns (Fig 2). This finding constitutes an independent criterion for testing the reliability of our phylogenetic analysis.
Fig 3. Schematic diagram of intron distribution patterns within the K-box of proteins translated from type II BnMADSs.
Alignment of the K-box domains is representative of 7 intron patterns, designated A to G. Intron locations are indicated by white triangles, and the number within each triangle indicates the splicing phases: 0 refers to phase 0; 1 to phase 1; and 2 to phase 2. The number of BnMADSs within each pattern is presented on the left. The correlation between intron distribution patterns and phylogenetic subfamilies is provided in Fig 2.
Common motifs identified in the non-conserved domain supports subgroup designation
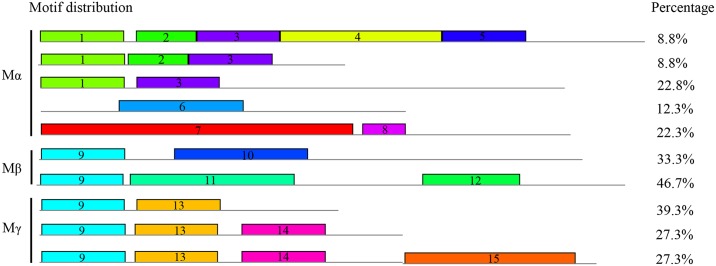
To discover motifs shared among related proteins within subfamilies, the MEME tool was used to analyse the relatively non-conserved regions of BnMADSs. Since regions following MADS-box domains in type-II BnMADS proteins are well known as the I-domain, K-box, and C-terminal, we only included type-I BnMADS proteins in the MEME analysis.
In total, 15 conserved motifs of variable length (15–108 amino acids) were detected in the C-terminals of type I proteins (S2 Table). We found that members of the same clade generally shared one or more motifs (Fig 4). For example, 8 motifs (1–8) were identified in Mα, which encompassed 40.4%, 17.6%, 40.4%, 8.8%, 8.8%, 12.3%, 22.3%, and 22.3% respectively; Mβ protein members contained motifs 9–12, with up to 90% contained motif 9; members of Mγ contained motif 9 and 14–15. In general, most of these 15 motifs were clade-specific rather than subfamily-specific, except motif 9, which was shared by Mβ and Mγ, supporting the evolutionary relationship of each subfamily. However, the function of these motifs remains unknown.
Fig 4. Architecture of conserved protein motifs translated from type I BnMADSs.
The percentage of BnMADS proteins with each pattern is presented on the right. Numbers in the colored bar indicate different motifs.
Chromosomal distribution and duplication events of BnMADSs
To investigate the relationship between genetic divergence and gene duplication within the B. napus MADS-box gene family, we analysed the chromosomal locations of BnMADSs based on information from the Genoscope genomic database.
Results show that BnMADSs were distributed across all 19 chromosomes, and the number of genes within each sub-genome appeared to be uneven (Fig 5). The An-subgenome had an average of 12.1 BnMADS on its 10 chromosomes; A08 had a minimum number of 7, while A03 and A09 had a maximum of 18 genes. The average number of BnMADSs in the Cn-subgenome was 13.3, with the C05 containing a minimum of 9 genes and the C04 having as many as 17 genes. Thus, there is not a biased tendency between these two subgenomes. Moreover, the distribution of candidates on each chromosome was not quite balanced, except for A07 and A09; the genes tended to distribute on the terminal parts. This pattern was similar to the MADS-box genes observed in B. rapa and Arabidopsis [28].
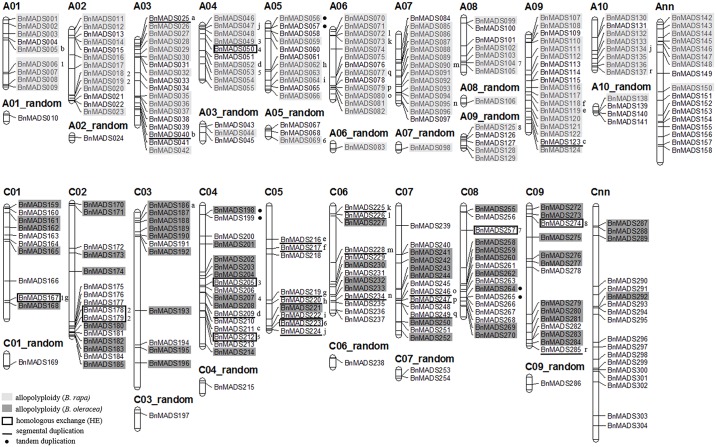
Fig 5. Chromosomal locations and duplications of BnMADSs.
The chromosome number is indicated above each chromosome. The scale is in megabases (Mb). Genes inherited from B. rapa or B. oleracea genomes are under grey and black backgrounds respectively; genes originating from segmental duplication are underlined by black lines accompanied with the same letter (e.g., a, b, and c); genes arising from homologous exchange (HE) are indicated using black boxes accompanied with the same number (e.g., 1, 2, and 3); and genes originating from tandem duplication are marked with black dots.
A total of 201 BnMADSs were identified to have syntenic relationships, of which 174 BnMADSs were inherited from B. rapa or B. oleracea genomes (S3 Table). In contrast, only 18 genes from 18 pairs originated from segmental duplication in the B. napus genome, and 3 genes from 3 pairs were from tandem duplication. Among those genes from segmental duplications, 11 of 18 genes belonged to the type-I subfamily, while the remaining 7 genes belonged to type-II, confirming a higher frequency of segmental gene duplication in the type-I subfamily [44]. Moreover, according to the distribution on chromosome and sequence similarity, BnMADS056/057, BnMADS198/199, and BnMADS264/265 were identified as tandem duplication gene pairs.
Together, genes obtained from tandem duplication (3) and segmental duplication (18) took up 0.98% and 5.86% of the BnMADS gene family, respectively, providing an outstanding example of genome-wide allopolyploidization between B. rapa and B. oleracea that mainly contributed to the large BnMADSs expansion in B. napus (56.68%).
Cis-acting regulatory elements in the BnMADSs promoters
Cis-elements in the promoter of a gene can regulate the initiation and efficiency of gene transcription by binding with transcription factors. The cis-acting regulatory elements of BnMADS promoters were predicted using PlantCARE.
In all, three cis-element groups in BnMADS promoters were classified (A–C) depending on their functional annotations (Fig 6 and S4 Table). The A group contained elements responding to light, the B group consisted of elements responding to biotic and/or abiotic stress induction, and the C group contained elements responding to hormones.
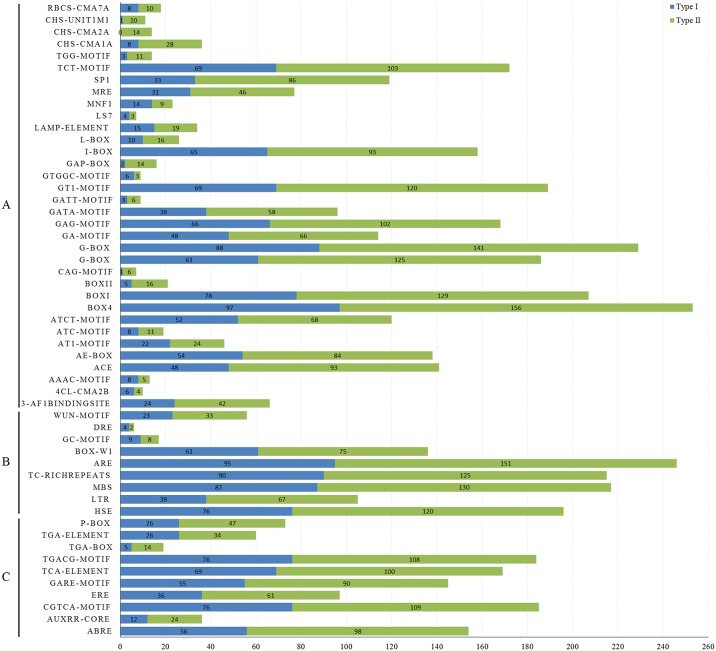
Fig 6. Classification of the cis-elements in BnMADS promoters.
106 Cis-elements were identified in the promoters of all 307 BnMADSs and were classified into three main groups (A–C). The number of type I and type II genes with the same type of cis-element is marked in different colours.
In the A group, 34 elements were related to the light response, implying that BnMADSs expression is induced by light, which is consistent with the function of many MADS-box genes in plants that use a fluorescent process widely regulated by light. Elements in the B group consisted of many stress response elements, such as MYB transcription factor binding site (MBS, drought stress response element), heat shock element (HSE, heat stress response), and TC-rich repeats (stress reaction), indicating that BnMADSs may respond to abiotic stress responses. There are some cis-elements in the C group, including the salicylic acid (TCA) response element, gibberellin response element (GARE-motif), CGTCA-motif/TGACG-motif (methyl jasmonate [MeJA] response), and abscisic acid response (ABRE), accounting for over 50% of both type-I and type-II genes and indicating possible roles for candidate genes in hormone response processes.
Genome-wide expression of BnMADSs in B. napus by RNA-seq
Gene expression patterns have been examined in relation to gene function [45], therefore, we investigated the expression of all 307 BnMADSs in 50 different tissues at different stages.
As shown in Fig 7, most BnMADSs were differentially expressed in different tissues in both vegetative and reproductive organs at different stages, where the type II genes had much higher and broader expression patterns. In contrast, the majority of type I genes had no or very low expression levels, with a few exceptions, indicating that type II genes may have abundant functions while type I genes have limited functions or are only expressed under special conditions in B. napus, such as stress and induction of hormones.
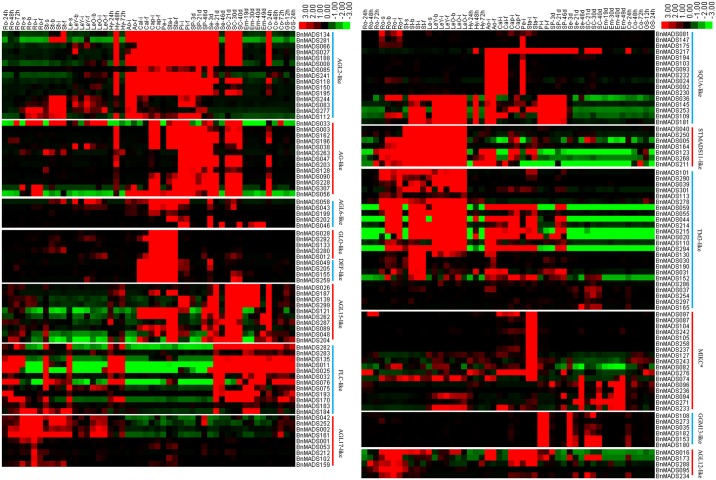
Fig 7. Expression profiles of type II BnMADSs across different developmental stages and organs.
The genes and their corresponding clade are on the right. The tissues used for expression analysis are indicated at the top of each column. GS, germinate seed; Hy, hypocotyl; Ao, anthocaulus; Ro, root; St, stem; Le, leaf; Cal, calyx; Cap, capillament; Pe, petal; Sta, stamen; Pi, pistil; SP, silique; Se, seed; SC, seed coat; Em, embryo; Co, cotyledon. s, seedling stage; b, bud stage; i, initial flowering stage; and f, full-bloom stage. The time after seed germination is indicated as 24, 48, and 72 h. The number of days after pollination (DAP) is indicated as 3, 19, 21, 30, 40, and 46 d. The colour bar represents log2 expression values (FPKM). The genes with weak or no expression are supplied in S1 Table.
In the type II subfamily, 20 of 187 (10.69%) genes were not expressed in any tissues, 18 (9.63%) genes had very weak expression levels (S1 Table), and the expression patterns of the remaining 149 (79.68%) were clustered into three main blocks, with some genes (38, 25.5%) having higher expression levels in the root, stem, and leaf during the seedling, budding, and flowering stages (Fig 7). Furthermore, some genes (83, 55.7%) had relatively higher expression levels in the flower and/or early developmental seed tissues, while some (28, 18.79%) had higher expression levels in the seed tissues 19 days after pollination (DAP). It is obvious that the main function of BnMADSs may involve reproductive organs (flower and/or seed tissues). Moreover, the expression patterns of BnMADSs in different tissues of the same organ, such as the flower and seeds, were generally different. For example, highly expressed genes belonging to the GGM13-like clade were primarily in female-reproductive related organs such as the pistil, and highly expressed members of the MIKC* subfamily primarily occurred in male-reproductive organs such as the stamen. However, homologous genes in the same subfamily usually had a conserved expression profile, suggesting functional redundancy. For instance, most members of the AGL2-like clade were highly expressed in flowers and seed tissues during the early development stage, and members of the STMADS11-like clade were highly expressed in the root and/or stem and leaves.
In the type-I subfamily, 17 out of 120 (14.17%) genes were not expressed in any tissues, 84 (70%) had very low expression levels (S1 Table), and 19 (15.83%) showed relatively high expression levels in B. napus (S3 Fig). Moreover, expression patterns of the genes with high expression levels were broad in terms of reproductive and vegetable organs and were not conserved between genes. In addition, the entire Mβ subfamily of genes showed very low expression levels.
Taken together, the type II genes had much wider and higher expression levels than type I genes in B. napus, and the expression levels were generally conserved in each subfamily but were different across distinct subfamilies.
Discussion
A good example of gene expansion and loss of the MADS-box gene family in land plants
In the present study, we identified 307 MADS-box members in the B. napus genome (BnMADSs), which is the largest number reported in plants to date [28, 29, 41–43]. Among the identified genes, type-I genes made up 37.5% of the BnMADSs gene family, while in Arabidopsis the percentage is 64% [28]. In the case of B. rapa and B. oleracea, 40% [29] and 34% of MADS-box genes are type I, respectively. Similarly, in higher plants such as rice, maize, soybean, and sorghum, the percentage of type II genes is much higher than type I genes [41, 42, 46]. In lower land plants such as Selaginella, this trend is similar to that observed in Arabidopsis [47]. Overall, it is obvious that the percentage of type-I genes generally diminishes during the evolution to higher plants. The functions of type-I genes have been minimally reported to date; however, we speculated that the presence of many non-functional type-I genes (i.e., pseudogenes) indicates inactivity. In contrast, the number of type II genes in higher plants obviously increased, and type-II genes appear to have differentiated functionally in a relatively short time, likely attributable to type II genes being maintained as functional genes in the genome to perform more complex functions during flower and organ development [44].
Type-I and II genes differentiated in duplication patterns. According to synteny analysis, 201 out of 307 BnMADSs had syntenic relationships with MADS-box genes from B. rapa or B. oleracea, respectively, proving that the majority of BnMADSs were obtained from inheritance and genome-wide allopolyploidy of B. rapa and B. oleracea genomes. Among these, 57 type-I genes (47.5%) come from allopolyploidy and 11 genes (9.17%) are segmental duplications in B. napus. In contrast, 117 type-II genes come from allopolyploidy (62.57%), while 7 genes (3.74%) and 3 genes (1.6%) are segmental and tandem duplications, respectively.
In addition, there were 109 and 90 syntenic genes in the An- and Cn-subgenomes, respectively. After allopolyploidy, the gene retainment ratio was mainly balanced between these two subgenomes, with the An-subgenome slightly advantaged. Furthermore, homologous exchange (HE), meaning replacement of a chromosomal region with a duplicated copy from the corresponding homologous subgenome, was found to be frequent between B. napus subgenomes [48]. Accordingly, we identified 18 genes from 8 pairs located in known HE blocks [48], and the number of type I genes (6, 5%) was higher than the number of type II genes (3, 1.6%), indicating HE was an important source underlying the evolution of BnMADSs after allopolyploidization, especially for type I genes. Meanwhile, among HE events, more genes from the Cn-genome were replaced by genes from the An-genome, indicating that genes from the latter were more retained.
Overall, genome-wide allopolyploidy, segmental duplication and HE were the main expansion mechanisms of BnMADSs, and subsequent duplications were preferred in the type-I genes over the type-II genes in the B. napus genome.
An ignored evolutionary story of intron insertion patterns in the K-box domain
Many studies have revealed the conservation of intron insertion sites in relatively conserved DNA binding domains of many transcription factor gene families [49, 50]. Similarly, nearly all studies of the MADS-box gene family focus on the MADS-box domain. In this study, we found that all MADS-box domains of the BnMADS gene family are in the first exon, with no intron insertions observed in this domain.
Interestingly, we found that the intron insertion patterns in the K-box domain are highly conserved in type-II genes, and type-II genes could be further classified into 7 conserved intron patterns in terms of absolute conserved intron insertion positions and phases (A–G types) (Fig 3). The gene structure and sequence characteristics of these 7 types were highly similar, indicating a relatively close relationship. There were 4–5 conserved introns inserted into the K domain in each type. While the location of the first intron was variable across different gene types, the remaining were highly conserved. Genes in the MIKCc clade were excluded in the analysis, attributable to an incomplete K-box domain. Interestingly, as discussed below, we found that genes in the MIKCc clade were not expressed.
To verify the intron patterns in different species of land plants, we extended this analysis to 19 other species, including chlorophytes (Chlamydomonas reinhardtii and Volvox carteri), embryophytes (Marchantia polymorpha, Physcomitrella patens, and Sphagnum fallax), tracheophytes (Selaginella moellendorffii), basic angiosperm (Amborella trichopoda and Zostera marina), monocots (Brachypodium distachyon, Oryza sativa, Sorghum bicolor, and Zea mays), and eudicots (Sorghum bicolor, Vitis vinifera, Populus trichocarpa, Citrus sinensis, Glycine max, and Malus domestica) (S5 Table). We found that the C type was distributed in all species investigated except chlorophytes, while the A and B types existed in some embryophyte species, indicating that the C type is older. The E type was distributed in basic angiosperms (Amborella trichopoda and Zostera marina), indicating it is relatively older than the last three types (D, F, and G). The D and G types were found in all flowering plants, including monocots and dicots, suggesting they may predate divergence of these two lineages. F types were only distributed in the Brassica species investigated in this study, indicating a new origin in this lineage. These results proved that the intron patterns in the K-box are highly conserved across land plants, thus should be a typical feature for the MADS-box gene family.
In the phylogenetic tree, the intron patterns of BnMADSs were generally conserved within most of the clades, except for genes lacking a full ORF, suggesting a common origin of members in each clade. For example, members of the AGL17-like clade shared the B intron pattern, while members of the AGL15-like clade shared the F type (Fig 2). However, this pattern was not absolute, as the intron patterns of the AGL2-like clade were mixed, containing both the A and D types. Given that type A exists in lower land plants and members having similar functions to those of pattern D in flower development, we speculated that type D likely developed from type A. Interestingly, it has been reported that AGL18 (type F) and AGL15 (type C) are functionally redundant in floral repress [51] and could be classified into one clade [28]. However, in our phylogenetic tree, the homologous nature of these two groups included different intron patterns; therefore, we divided them into two clades (AGL15-like and AGL18-like clades). This result proves that intron patterns support classification of the phylogenetic tree. Type F existed only in Cruciferae plants, while pattern C was widely distributed in all species investigated; these results indicated that type F originated from type C in the Brassica lineage.
Furthermore, the function of the A, C, D, and E types relates to the flower and fruit [51–59]; the B type relates to root development [60] and seeds [61]; and the G type is important for the colour of the seed epidermis [62], which likely explains the closer relationship between the A, C, D, and E types while the B and G types were more independent. It is also interesting to note that there are 65 C-type BnMADSs, which is higher than the number of other types in B. napus (Fig 3). Similar results were also observed in all the flowering plants investigated in this study (S5 Table). Further, the B pattern, which is related to root development, took up the highest percentage in Selaginella. These phenomena support our hypothesis that the intron insertion pattern is related to a certain function in land plants.
Functional conservation and redundancy of BnMADSs based on expression analysis
Our results show that the expression of BnMADSs is mainly conserved in each family and is very similar to that of the homologous genes in Arabidopsis. These results suggest functional conservation of homologous genes across different plants. For example, the genetic and molecular basis of floral development have revealed that numerous MADS-box genes are involved in specifying floral organ identity in Arabidopsis [11, 58, 62–64]. Accordingly, the Arabidopsis SEP1/2/3 genes in the AGL2-like clade were identified as class E genes and function as organ-identity genes in development of the petal, stamen, and carpel. Similarly, BnMADSs in the AGL2-like clade are primarily highly expressed in the flower (calyx, capillament, petal, stamen, and pistil) and seed tissues during the early development stages (seed and seed coat) (Fig 7). The short vegetative phase (SVP) gene in the STMADS11-like clade was identified as a floral repressor and plays an essential role in determining the length of vegetative growth; BnMADSs in the same clade are all highly expressed in vegetative organs at different stages. The transparent Testa16 (TT16) gene in the GGM13-like clade is involved in the determination of seed coat colour; BnMADSs in this clade are primarily highly expressed in the female-reproductive organs (pistil) and seed tissues during the early stages. Trends in the other subfamilies were the same.
Moreover, 65 pairs of 142 type II paralogous genes were identified in this study (S6 Table), including 56 pairs with two genes (six genes had some deletions in the ORF regions), six pairs with three genes, and three pairs with four genes. While six genes had some deletions in the ORF region, the last of the 50 pairs contained two genes and shared a very high degree of sequence identity or similarity in the binding domain and full protein length, with an average identity of approximately 99% in the binding domain and an average identity of 85% in the full-length proteins. In addition, the DNA binding domains of 42 out of the 65 pairs were the same (Fig 7). Accordingly, most of the paralogous BnMADSs shared a similar or the same expression patterns, implying a functional redundancy. For example, BnMADS103/194, BnMADS030/190, BnMADS045/068, BnMADS035/182, and BnMADS153/180 showed the same expression pattern. Furthermore, the expressions of sister pair genes showed no bias, as candidates from the An- and/or Cn-subgenome were quite similar. For instance, BnMADS092/230 contained two genes from the An- and Cn-subgenome and shared a similar expression pattern, while BnMADS045/068 contained two genes from the An-subgenome and had nearly the same expression patterns. In addition, some sister pairs may represent neo-functionalization or sub-functionalization, as they showed similar expression patterns with slight diversification. For instance, BnMADS150/195 were both highly expressed in the calyx, capillament, and petal; however, BnMADS150 was highly expressed in the pistil, whereas BnMADS195 was not (Fig 7). The expression of several pairs was obviously different, which may indicate functional divergence; BnMADS032 from the BnMADS032/192 gene pair was highly expressed in vegetative organs and seeds, while BnMADS192 was not expressed. As mentioned above, the 20 type II BnMADSs that were not expressed in any tissues investigated were generally the homologous genes in a given clade; five were duplications and/or sister pair genes (S3 and S6 Tables). These data suggested functional redundancy of highly homologous genes.
Furthermore, sequence analysis showed that the promoter regions of sister pairs were also highly homologous, with the average sequence identity ranging from 48% to 97% (S6 Table). In general, expression patterns and promoter sequence identity are closely related. Expression of 55 of the 65 pairs was similar or the same, indicating functional redundancy; the last generally had more sequence identity in the ORF regions, but less in the promoter regions, suggesting that functional divergence may first occur in the promoter region of sister pairs during evolution.
Thus, our data show that expression patterns of homologous genes in a given subfamily or sister pair are generally conserved, revealing a major functional conservation as well as redundancy of BnMADSs during evolution.
Conclusions
Results of the present work confirm many features described in other species. However, some novel characteristics were found in the B. napus MADS-box gene family, including intron patterns in the K-box and expansion mechanisms after allopolyploidy between B. rapa and B. oleracea. Further, our phylogenetic analyses provide a useful reference for postulating functional hypotheses for uncharacterized BnMADSs. In this study, we evaluated the roles and evolution of uncharacterized MADS-box genes in B. napus using a comparative phylogenetic analysis of B. napus, B. rapa, B. oleracea, and Arabidopsis, as well as information from previous research.
Supporting information
A neighbor-joining tree representing relationships among 120 type I MADS-box proteins from B. napus and 63 from Arabidopsis is shown. The proteins are clustered into 3 clades, including Mα, Mβ, and Mγ.
(PDF)
Exon(s) are indicated by yellow boxes, MADS-box domain(s) by orange boxes, untranslated region(s) by grey boxes, and spaces between the coloured boxes correspond to introns. Exon and intron size can be estimated using the horizontal scale bar. (A): the structures of type I BnMADSs; (B): the structures of type II BnMADSs.
(PDF)
The tissues used for expression analysis are indicated at the top of each column. GS, germinate seed; Hy, hypocotyl; Ao, anthocaulus; Ro, root; St, stem; Le, leaf; Cal, calyx; Cap, capillament; Pe, petal; Sta, stamen; Pi, pistil; SP, silique; Se, seed; SC, seed coat; Em, embryo; Co, cotyledon. s, seedling stage; b, bud stage; i, initial flowering stage; and f, full-bloom stage. The time after seed germination is indicated as 24, 48, and 72 h. Days after pollination (DAP) are indicated as 3, 19, 21, 30, 40, and 46 d. The colour bar represents log2 expression values (FPKM). Genes with weak or no expression are supplied in S1 Table.
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the “111” Project of China (B12006), National Natural Science Foundation of China (31671727 and 31471528), the National Basic Research Program of China (973 Program; 2015CB150201).
References
- 1.Shore P, Sharrocks AD. The MADS-box family of transcription factors. Eur J Biochem. 1995; 229(1):1–13. 10.1111/j.1432-1033.1995.0001l.x . [DOI] [PubMed] [Google Scholar]
- 2.Hayes TE, Sengupta P, Cochran BH. The human c-fos serum response factor and the yeast factors GRM/PRTF have related DNA-binding specificities. Genes Dev. 1988; 2(12B):1713–1722. 10.1101/gad.2.12b.1713 . [DOI] [PubMed] [Google Scholar]
- 3.Riechmann JL, Wang M, Meyerowitz EM. DNA-Binding Properties of Arabidopsis MADS Domain Homeotic Proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res. 1996; 24(16):3134–3141. 10.1093/nar/24.16.3134 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davies B, Egea-Cortines M, Silva EA, Saedler H, Sommer H. Multiple interactions amongst floral homeotic MADS box proteins. EMBO J. 1996; 15(16):4330–4343. [PMC free article] [PubMed] [Google Scholar]
- 5.Fan HY, Hu Y, Tudor M, Ma H. Specific interactions between the K domains of AG and AGLs, members of the MADS domain family of DNA binding proteins. Plant J. 1997; 12(5):999–1010. 10.1046/j.1365-313X.1997.12050999.x . [DOI] [PubMed] [Google Scholar]
- 6.Honma T, Goto K. Complexes of MADS-BOX proteins are sufficient to convert leaves into floral organs. Nature. 2001; 409(6819):525–529. 10.1038/35054083 . [DOI] [PubMed] [Google Scholar]
- 7.Alvarezbuylla ER, Pelaz S, Liljegren SJ, Gold SE, Burgeff C, Ditta GS, et al. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc Natl Acad Sci U S A. 2000; 97(10):5328–5333. 10.1073/pnas.97.10.5328 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Theissen G, Kim JT, Saedler H. Classification and phylogeny of the MADS-box multigene family suggest defined roles of MADS-box gene subfamilies in the morphological evolution of eukaryotes. J Mol Evol. 1996; 43(5):484–516. 10.1007/BF02337521 . [DOI] [PubMed] [Google Scholar]
- 9.Kaufmann K, Melzer R, Theissen G. MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene. 2005; 347(2):183–198. 10.1016/j.gene.2004.12.014 . [DOI] [PubMed] [Google Scholar]
- 10.Henschel K, Kofuji R, Hasebe M, Saedler H, Münster T, Theissen G. Two ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens. Mol Biol Evol. 2002; 19(6):801–814. [DOI] [PubMed] [Google Scholar]
- 11.Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF. B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature. 2000; 405(6783):200–203. 10.1038/35012103 . [DOI] [PubMed] [Google Scholar]
- 12.Angenent GC, Colombo L. Molecular control of ovule development. Trends Plant Sci. 1996; 1(7):228–232. 10.1016/1360-1385(96)86900-0. [DOI] [Google Scholar]
- 13.Theissen G. Development of floral organ identity: stories from the MADS house. Curr Opin Plant Biol. 2001; 4(1):75–85. 10.1016/S1369-5266(00)00139-4 . [DOI] [PubMed] [Google Scholar]
- 14.Theissen G, Saedler H. Plant biology. Floral quartets. Nature, 2011; 409(6819), 469–471. 10.1038/35054172 . [DOI] [PubMed] [Google Scholar]
- 15.Weigel D, Meyerowitz EM. The ABCs of floral homeotic genes. Cell. 1994; 78(2):203–209. 10.1016/0092-8674(94)90291-7 . [DOI] [PubMed] [Google Scholar]
- 16.Rounsley SD, Ditta GS, Yanofsky MF. Diverse roles for MADS box genes in Arabidopsis development. Plant Cell. 1995; 7(8):1259–1269. 10.1105/tpc.7.8.1259 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan MR, Khan IU, Ali GM. MPF2-like MADS-box genes affecting expression of SOC1 and MAF1 are recruited to control flowering time. Mol Biotechnol. 2013; 54(1):25–36. 10.1007/s12033-012-9540-9 . [DOI] [PubMed] [Google Scholar]
- 18.Moore S, Vrebalov J, Payton P, Giovannoni J. Use of genomics tools to isolate key ripening genes and analyse fruit maturation in tomato. J Exp Bot. 2002; 53(377):2023–2030. [DOI] [PubMed] [Google Scholar]
- 19.Alvarez-Buylla ER, Liljegren SJ, Pelaz S, Gold SE, Burgeff C, Ditta GS, et al. MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. Plant J. 2000; 24(4):457–466. 10.1111/j.1365-313X.2000.00891.x . [DOI] [PubMed] [Google Scholar]
- 20.Huang H, Tudor M, Weiss CA, Hu Y, Ma H. The Arabidopsis MADS-box gene AGL3 is widely expressed and encodes a sequence-specific DNA-binding protein. Plant Mol Biol. 1995; 28(3):549–567. 10.1007/BF00020401 . [DOI] [PubMed] [Google Scholar]
- 21.Ma H, Yanofsky MF, Meyerowitz EM. AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev. 1991; 5(3):484–495. 10.1101/gad.5.3.484 . [DOI] [PubMed] [Google Scholar]
- 22.Theissen G. Plant biology. Shattering developments. Nature. 2000; 404 (6779):711–713. 10.1038/35008171 . [DOI] [PubMed] [Google Scholar]
- 23.Krogan NT, Ashton NW. Ancestry of plant MADS-box genes revealed by bryophyte (Physcomitrella patens) homologues. New Phytol. 2000; 147(3):505–517. 10.1046/j.1469-8137.2000.00728.x [DOI] [PubMed] [Google Scholar]
- 24.Münster T, Pahnke J, Rosa AD, Kim JT, Martin W, Saedler H, et al. Floral homeotic genes were recruited from homologous MADS-box genes preexisting in the common ancestor of ferns and seed plants. Proc Natl Acad Sci U S A. 1997; 94(6):2415–2420. 10.1073/pnas.94.6.2415 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nilsson L, Carlsbecker A, Sundås-Larsson A, Vahala T. APETALA2 like genes from Picea abies show functional similarities to their Arabidopsis homologues. Planta. 2007. February;225(3):589–602. 10.1007/s00425-006-0374-1 . [DOI] [PubMed] [Google Scholar]
- 26.Tandre K, Albert VA, Sundås A, Engström P. Conifer homologues to genes that control floral development in angiosperms. Plant Mol Biol. 1995; 27(1):69–78. 10.1007/BF00019179 [DOI] [PubMed] [Google Scholar]
- 27.Winter KU, Becker A, Münster T, Kim JT, Saedler H, Theissen G. MADS-box genes reveal that gnetophytes are more closely related to conifers than to flowering plants. Proc Natl Acad Sci U S A. 1999; 96(13):7342–7347. 10.1073/pnas.96.13.7342 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parenicová L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell. 2003; 15(7):1538–51. 10.1105/tpc.011544 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saha G, Park JI, Jung HJ, Ahmed NU, Kayum MA, Chung MY, et al. Genome-wide identification and characterization of MADS-box family genes related to organ development and stress resistance in Brassica rapa. BMC Genomics. 2015; 16:178 10.1186/s12864-015-1349-z . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaradat MR, Ruegger M, Bowling A, Butler H, Cutler AJ. A comprehensive transcriptome analysis of silique development and dehiscence in Arabidopsis and Brassica integrating genotypic, interspecies and developmental comparisons. GM Crops Food. 2014; 5(4):302–320. 10.4161/21645698.2014.947827 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012; 40(Web Server issue):W597–603. 10.1093/nar/gks400 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016; 33(7):1870–1874. 10.1093/molbev/msw054 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013; 30(4):772–780. 10.1093/molbev/mst010 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu B, Jin J, Guo A, Zhang H, Luo J, Gao G. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015; 31(8):1296–1297. 10.1093/bioinformatics/btu817 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rani V. Computational methods to dissect cis-regulatory transcriptional networks. J Biosci. 2007; 32(3):1325–1330. . [DOI] [PubMed] [Google Scholar]
- 36.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006; 34(Web Server issue):W369–373. 10.1093/nar/gkl198 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonnhammer EL, Eddy SR. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins. 1997; 28(3):405–420. . [DOI] [PubMed] [Google Scholar]
- 38.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20(9):1453–1454 10.1093/bioinformatics/bth078 . [DOI] [PubMed] [Google Scholar]
- 39.Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004; 20(17):3246–3248. 10.1093/bioinformatics/bth349 . [DOI] [PubMed] [Google Scholar]
- 40.Wei B, Zhang RZ, Guo JJ, Liu DM, Li AL, Fan RC, et al. Genome-wide analysis of the MADS-box gene family in Brachypodium distachyon. PLoS One. 2014; 9(1):e84781 10.1371/journal.pone.0084781 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, et al. MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductie development and stress. BMC genomics. 2007; 8:242 10.1186/1471-2164-8-242 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fan CM, Xu W, Wang YW, Hu RB, Zhang XM, Chen JX, et al. Genome-Wide Expression Analysis of Soybean MADS Genes Showing Potential Function in the Seed Development. Plos One. 2013; 8(4):e62288 10.1371/journal.pone.0062288 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thangavel G, Nayar S. A Survey of MIKC Type MADS-Box Genes in Non-seed Plants: Algae, Bryophytes, Lycophytes and Ferns. Front Plant Sci. 2018; 9:510 10.3389/fpls.2018.00510 eCollection . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nam J, Kim J, Lee S, An G, Ma H, Nei M. Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc Natl Acad Sci U S A. 2004; 101(7):1910–1915. 10.1073/pnas.0308430100 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kong H, Landherr LL, Frohlich MW, Leebens-Mack J, Ma H, Depamphilis CW. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J. 2010; 50(5):873–885. 10.1111/j.1365-313x.2007.03097.x . [DOI] [PubMed] [Google Scholar]
- 46.Zhang Z, Li H, Zhang D, Liu Y, Fu J, Shi Y. et al. Characterization and expression analysis of six MADS-box genes in maize (Zea mays L.). J Plant Physiol. 2012; 169(8):797–806. 10.1016/j.jplph.2011.12.020 . [DOI] [PubMed] [Google Scholar]
- 47.Gramzow L, Barker E, Schulz C, Ambrose B, Ashton N, Theißen G, et al. Selaginella Genome Analysis—Entering the “Homoplasy Heaven” of the MADS World. Front Plant Sci. 2012; 3:214 10.3389/fpls.2012.00214 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chalhoub B, Denoeud F, Liu S, Parkin IA, Tang H, Wang X, et al. Plant genetics. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014; 345(6199):950–953. 10.1126/science.1253435 . [DOI] [PubMed] [Google Scholar]
- 49.Du H, Liang Z, Zhao S, Nan MG, Tran LS, Lu K, et al. The Evolutionary History of R2R3-MYB Proteins Across 50 Eukaryotes: New Insights Into Subfamily Classification and Expansion. Sci Rep. 2015; 5:11037 10.1038/srep11037 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan J, Ma Z, Xu X, Guo AY. Evolution, functional divergence and conserved exon–intron structure of bHLH/PAS gene family. Mol Genet Genomics. 2014; 289(1):25–36. 10.1007/s00438-013-0786-0 . [DOI] [PubMed] [Google Scholar]
- 51.Fernandez DE, Wang CT, Zheng Y, Adamczyk B, Singhal R, Hall PK, et al. The MADS-domain factors AGL15 and AGL18, along with SVP and AGL24, are necessary to block floral gene expression during the vegetative phase. Plant Physiol. 2014; 165(4):1591–1603. 10.1104/pp.114.242990 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yanofsky MF, Ma H, Bowman JL, Drews GN, Feldmann KA, Meyerowitz EM. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature. 1990; 346(6279):35–39. 10.1038/346035a0 . [DOI] [PubMed] [Google Scholar]
- 53.Scortecci KC, Michaels SD, Amasino RM. Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 2001; 26(2):229–236. 10.1046/j.1365-313x.2001.01024.x . [DOI] [PubMed] [Google Scholar]
- 54.Goto K, Meyerowitz EM. Function and regulation of the Arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 1994; 8(13):1548–1560. 10.1101/gad.8.13.1548 . [DOI] [PubMed] [Google Scholar]
- 55.Moon J, Suh SS, Lee H, Choi KR, Hong CB, Paek NC, et al. The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 2003; 35(5):613–623. 10.1046/j.1365-313X.2003.01833.x . [DOI] [PubMed] [Google Scholar]
- 56.Michaels SD, Ditta G, Gustafson-Brown C, Pelaz S, Yanofsky M, Amasino RM. AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J. 2003; 33(5):867–874. 10.1046/j.1365-313X.2003.01671.x . [DOI] [PubMed] [Google Scholar]
- 57.Michaels SD, Amasino RM. FLOWERING LOCUS C Encodes a Novel MADS Domain Protein That Acts as a Repressor of Flowering. Plant Cell. 1999; 11(5):949–956. 10.1105/tpc.11.5.949 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liljegren SJ, Ditta GS, Eshed Y, Savidge B, Bowman JL, Yanofsky MF. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature. 2000; 404(6779):766–770. 10.1038/35008089 . [DOI] [PubMed] [Google Scholar]
- 59.Gu Q, Ferrándiz C, Yanofsky MF, Martienssen R. The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development. 1998; 125(8):1509–1517. . [DOI] [PubMed] [Google Scholar]
- 60.Zhang H, Forde BG. An Arabidopsis MADS box gene that controls nutrient-induced changes in root architecture. Science. 1998; 279(5349):407–409. 10.1126/science.279.5349.407 . [DOI] [PubMed] [Google Scholar]
- 61.Yu LH, Wu J, Zhang ZS, Miao ZQ, Zhao PX, Wang Z, et al. Arabidopsis MADS-Box Transcription Factor AGL21 Acts as Environmental Surveillance of Seed Germination by Regulating ABI5 Expression. Mol Plant. 2017; 10(6):834–845. 10.1016/j.molp.2017.04.004 . [DOI] [PubMed] [Google Scholar]
- 62.Nesi N, Debeaujon I, Jond C, Stewart AJ, Jenkins GI, Caboche M, et al. The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat. Plant cell. 2002; 14(10):2463–2479. 10.1105/tpc.004127 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kater MM, Dreni L, Colombo L. Functional conservation of MADS-box factors controlling floral organ identity in rice and Arabidopsis. J Exp Bot. 2006; 57(13):3433–3444. 10.1093/jxb/erl097 . [DOI] [PubMed] [Google Scholar]
- 64.Hartmann U, Höhmann S, Nettesheim K, Wisman E, Saedler H, Huijser P. Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J. 2010; 21(4):351–360. 10.1046/j.1365-313x.2000.00682.x . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A neighbor-joining tree representing relationships among 120 type I MADS-box proteins from B. napus and 63 from Arabidopsis is shown. The proteins are clustered into 3 clades, including Mα, Mβ, and Mγ.
(PDF)
Exon(s) are indicated by yellow boxes, MADS-box domain(s) by orange boxes, untranslated region(s) by grey boxes, and spaces between the coloured boxes correspond to introns. Exon and intron size can be estimated using the horizontal scale bar. (A): the structures of type I BnMADSs; (B): the structures of type II BnMADSs.
(PDF)
The tissues used for expression analysis are indicated at the top of each column. GS, germinate seed; Hy, hypocotyl; Ao, anthocaulus; Ro, root; St, stem; Le, leaf; Cal, calyx; Cap, capillament; Pe, petal; Sta, stamen; Pi, pistil; SP, silique; Se, seed; SC, seed coat; Em, embryo; Co, cotyledon. s, seedling stage; b, bud stage; i, initial flowering stage; and f, full-bloom stage. The time after seed germination is indicated as 24, 48, and 72 h. Days after pollination (DAP) are indicated as 3, 19, 21, 30, 40, and 46 d. The colour bar represents log2 expression values (FPKM). Genes with weak or no expression are supplied in S1 Table.
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.