Abstract
Sexually dimorphic behaviors are a feature common to species across the animal kingdom, however how such behaviors are generated from mostly sex-shared nervous systems is not well understood. Building on our previous work which described the sexually dimorphic expression of a neuroendocrine ligand, DAF-7, and its role in behavioral decision-making in C. elegans (Hilbert and Kim, 2017), we show here that sex-specific expression of daf-7 is regulated by another neuroendocrine ligand, Pigment Dispersing Factor (PDF-1), which has previously been implicated in regulating male-specific behavior (Barrios et al., 2012). Our analysis revealed that PDF-1 signaling acts sex- and cell-specifically in the ASJ neurons to regulate the expression of daf-7, and we show that differences in PDFR-1 receptor activity account for the sex-specific effects of this pathway. Our data suggest that modulation of the sex-shared nervous system by a cascade of neuroendocrine signals can shape sexually dimorphic behaviors.
Research organism: C. elegans
Introduction
Behavioral differences between the sexes of animal species can make major contributions to the reproductive fitness of the organism. While sex-specific behaviors can be readily observed, the mechanistic basis of such behavioral differences is less well understood. Morphological differences, including the existence of sex-specific neurons, have been documented in the nervous systems of many species, but differences in sex-shared neurons have also been implicated in generating sex-specific behaviors. In particular, how sex-specific behavioral circuits are generated within the features of the nervous system common to both sexes has been the focus of recent studies in diverse organisms. Studies of the mouse vomeronasal organ (VNO) has suggested that the functional circuits for both male- and female-specific behaviors such as courtship and aggression are intact in the brains of both sexes and are modulated by VNO activity in response to pheromone cues (Kimchi et al., 2007; Stowers et al., 2002). In a similar vein, the Drosophila male pheromone 11-cis Vaccenyl acetate (cVa) has been shown to be sensed by the same neurons in the two sexes but stimulates distinct sex-specific behavioral responses (Datta et al., 2008; Kohl et al., 2013; Kurtovic et al., 2007; Ruta et al., 2010). These examples and others have provided some insight into the sexual dimorphisms present in the nervous system and their contributions to behavior, although many open questions remain (Dulac and Kimchi, 2007; Stowers and Logan, 2010; Yang and Shah, 2014).
In the nematode Caenorhabditis elegans, behavioral differences between the two sexes—hermaphrodites and males—range from behaviors exclusively performed by one sex, such as egg laying by hermaphrodites and the mating program of males (Liu and Sternberg, 1995), to those in which the two sexes differ in their responses to the same stimuli, including differing responses to pheromone (Fagan et al., 2018; Jang et al., 2012; Srinivasan et al., 2008), food-related cues (Ryan et al., 2014), and conditioning to aversive stimuli (Sakai et al., 2013; Sammut et al., 2015). While sex-specific neurons regulate corresponding behaviors in C. elegans, the 294 neurons that are common to the nervous systems of both hermaphrodites and males have emerged as major contributors to a number of different sexually dimorphic behaviors (Barr et al., 2018; Barrios et al., 2012, 2008; Fagan et al., 2018; Lee and Portman, 2007; Mowrey et al., 2014; Sakai et al., 2013). In particular, recent work has uncovered sexually dimorphic differences in axonic and dendritic morphology and synaptic connectivity within the sex shared nervous system, which can modulate neuronal circuits and behavior (Hart and Hobert, 2018; Oren-Suissa et al., 2016; Serrano-Saiz et al., 2017a; Weinberg et al., 2018). In addition, studies of sexually dimorphic gene expression (Hilbert and Kim, 2017; Ryan et al., 2014; Serrano-Saiz et al., 2017a) and neurotransmitter identity (Gendrel et al., 2016; Pereira et al., 2015; Serrano-Saiz et al., 2017a, 2017b) have suggested that sexual differentiation of neurons within the sex-shared nervous system of C. elegans is critical for the establishment of sexually dimorphic behaviors.
We have previously demonstrated that daf-7, which encodes a TGFβ family neuroendocrine ligand that regulates diverse aspects of C. elegans behavior and physiology (Chang et al., 2006; Fletcher and Kim, 2017; Gallagher et al., 2013; Greer et al., 2008; Milward et al., 2011; Ren et al., 1996; Shaw et al., 2007; White and Jorgensen, 2012; You et al., 2008), is expressed in a sex-specific and context-dependent manner in the sex-shared ASJ chemosensory neurons and functions to promote exploratory behaviors (Hilbert and Kim, 2017; Meisel et al., 2014). Regulation of daf-7 expression in the ASJ neurons requires the integration of sensory and internal state information including the sex and age of the animal, its nutritional state, and the type of bacterial species it encounters in its environment (Hilbert and Kim, 2017). These stimuli feed into the regulation of daf-7 expression in the two ASJ neurons in a hierarchical manner, which enables the animal to make behavioral decisions taking into account past experiences as well as its current environment.
Here, we report the identification of a second neuroendocrine signaling pathway, the Pigment Dispersing Factor (PDF-1) pathway, which functions to regulate the expression of daf-7 and its effects on behavior in a sex-specific manner. We show that PDF-1 pathway signaling, which has previously been shown to be essential for male mate-searching behavior (Barrios et al., 2012), functions sex-specifically in the ASJ neurons themselves to regulate daf-7 expression. Further, we demonstrate that the sex-specificity of PDF-1 regulation of daf-7 derives from differences in the activation of PDF-1 signaling downstream of the PDF-1 receptor gene, pdfr-1, in the ASJ neurons. Our data suggest that the gating of neuronal responses to neuropeptide modulators through sex-specific restriction of receptor activity is a mechanism by which sex-specific behaviors can be generated from the largely sex-shared nervous system of C. elegans.
Results and discussion
PDF-1 neuropeptide signaling regulates the sex-specific expression of daf-7 in the ASJ chemosensory neurons
To explore the molecular and genetic mechanisms that underlie the sex-specificity of daf-7 expression, we identified a number of candidate genes that had previously been shown to be involved in the regulation of mate-searching behavior or other aspects of male physiology and tested mutants of these genes for effects on daf-7 expression in the male ASJ neurons. Through this approach, we identified the PDF-1 neuropeptide signaling pathway as a regulator of daf-7 expression in the ASJ neurons (Figure 1). The PDF neuropeptide signaling pathway is conserved among insects, crustaceans and nematodes. In Drosophila melanogaster, PDF signaling has been well studied for its critical role in the regulation of circadian rhythmicity (Helfrich-Förster, 1995; Park and Hall, 1998; Park et al., 2000; Renn et al., 1999), but it has also been shown to modulate geotaxis (Mertens et al., 2005), pheromone production and mating behaviors (Fujii and Amrein, 2010; Kim et al., 2013; Krupp et al., 2013). In C. elegans, PDF-1 signaling has been established as an important regulator of locomotion, roaming behaviors, quiescence, and notably, male mate-searching behavior (Figure 1A; Barrios et al., 2012; Choi et al., 2013; Flavell et al., 2013; Janssen et al., 2008, 2009; Meelkop et al., 2012).
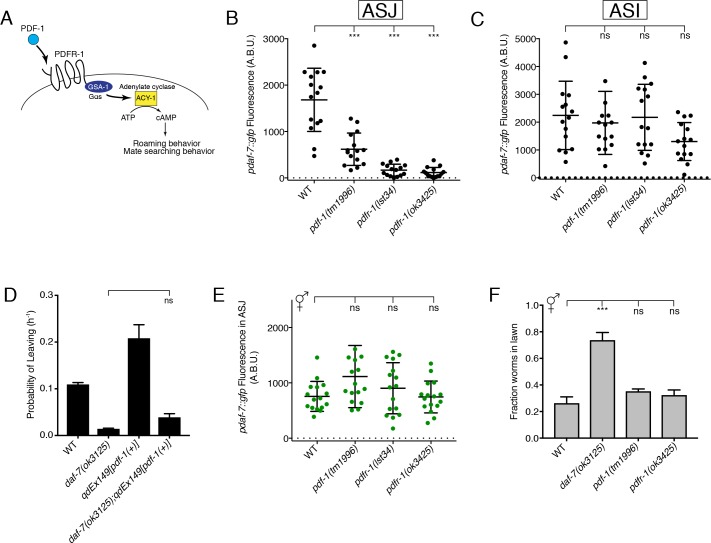
Figure 1. The PDF-1 pathway is required for the male-specific expression of daf-7 in the ASJ neurons and its effects on male mate-searching behavior.
(A) PDF-1 signaling activates cAMP production and regulates both roaming behavior and male mate-searching behavior in C. elegans. (B–C) Maximum fluorescence values of pdaf-7::gfp in the ASJ (B) and ASI (C) neurons of adult male animals. ***p<0.001 as determined by ordinary one-way ANOVA followed by Dunnett’s multiple comparisons test. Error bars represent standard deviation (SD). ns, not significant. n = 15 animals for all genotypes. (D) Probability of leaving values for epistasis experiment between daf-7(ok3125) and a PDF-1 overexpressing line. Values plotted are the mean +SEM for three independent experiments. Significance determined by unpaired t-test with Welch’s correction. ns, not significant. n = 60 total animals for all genotypes except the daf-7(ok3125); qdEx149 strain where n = 48. (E) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of hermaphrodites after 16 hr on P. aeruginosa. Significance determined by ordinary one-way ANOVA followed by Dunnett’s multiple comparisons test. Error bars represent SD. ns, not significant. n = 15 animals for all genotypes. (F) Lawn occupancy of animals on P. aeruginosa after 16 hr. ***p<0.001 as determined by ordinary one-way ANOVA followed by Dunnett’s multiple comparisons test. Values plotted indicate the mean + SD for three replicates. Number of animals assayed are as follows: WT (n = 89), daf-7 (n = 66), pdf-1 (n = 117), pdfr-1 (n = 105).
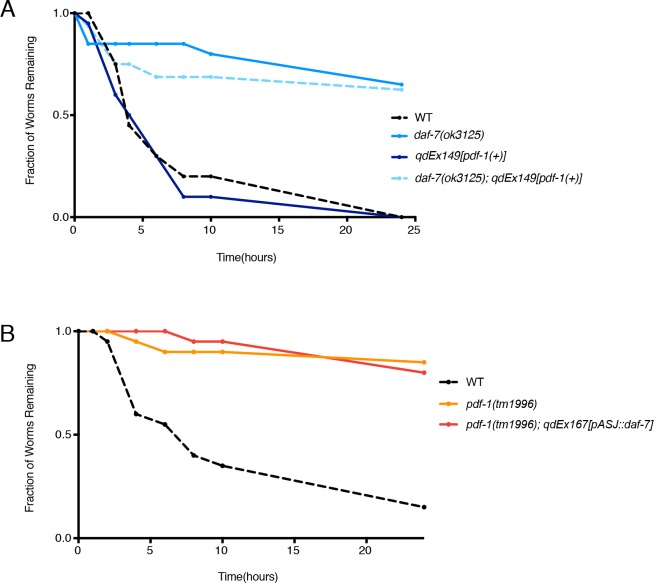
Figure 1—figure supplement 1. The PDF-1 pathway regulates mate-searching behavior via male-specific modulation of daf-7 expression in ASJ and through parallel mechanisms.
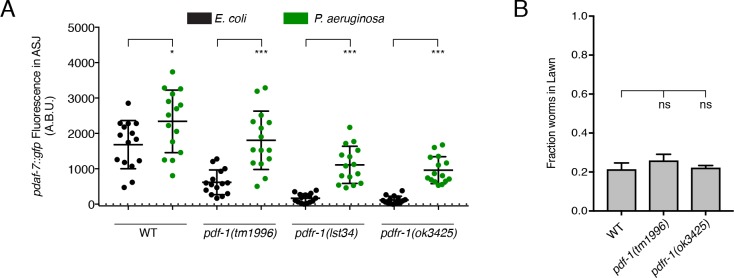
Figure 1—figure supplement 2. PDF-1 pathway mutant males can upregulate daf-7 expression in ASJ in response to P. aeruginosa exposure and show intact pathogen avoidance behavior.
We observed that males with mutation of either the PDF-1 neuropeptide ligand or its receptor, PDFR-1, have markedly attenuated expression of daf-7 in the ASJ neuron pair (Figure 1B). The ASI chemosensory neurons are established sites of daf-7 expression in both male and hermaphrodite animals (Ren et al., 1996; Schackwitz et al., 1996), so we asked if the PDF-1 signaling pathway also regulates daf-7 expression in these neurons. In the PDF-1 pathway mutant males, we observe no difference in daf-7 levels in the ASI neurons when compared to WT (Figure 1C), suggesting that the PDF-1 pathway specifically affects the regulation of daf-7 in the ASJ neuron pair.
Expression of daf-7 in the ASJ neuron pair of males is required for the male-specific mate-searching behavioral program (Hilbert and Kim, 2017), while the PDF-1 pathway has similarly been implicated as a regulator of this same behavior (Barrios et al., 2012). Given the role that this PDF-1 pathway plays in regulating the expression of daf-7, we set out to determine if the effects of the PDF-1 pathway on mate-searching behavior are the result of PDF-1 and DAF-7 functioning through a single pathway or through separate parallel pathways. Overexpression of the pdf-1 genomic sequence confers increased mate-searching behavior in male animals (Figure 1D; Barrios et al., 2012). We introduced a daf-7 mutation into these transgenic PDF-1 overexpressing lines and observed that the effect of PDF-1 overexpression on mate-searching behavior was suppressed by loss of daf-7 function (Figure 1D and Figure 1—figure supplement 1A). However, we observed that overexpression of daf-7 in the ASJ neurons of pdf-1(tm1996) mutant males could not rescue the mate-searching defects of these animals (Figure 1—figure supplement 1B). Taken together, the results of this epistasis analysis suggest that PDF-1 regulates mate-searching behavior in males through the regulation of daf-7 expression in the ASJ neurons and through additional mechanisms functioning in parallel to DAF-7 signaling.
We have previously reported that daf-7 expression serves a dual role in the ASJ neurons, functioning in males to promote food-leaving behaviors (Hilbert and Kim, 2017), but also being induced by the presence of Pseudomonas aeruginosa in both sexes to promote pathogen avoidance behaviors (Meisel et al., 2014). Given this and our interest in identifying male-specific regulators of daf-7 expression, we asked if the PDF-1 pathway is required for the upregulation of daf-7 expression in response to P. aeruginosa. We did not observe a requirement for PDF-1 signaling in the induction of daf-7 expression in the ASJ neurons after 16 hr on P. aeruginosa; both pdf-1 and pdfr-1 mutant hermaphrodites had equivalent levels of daf-7 expression when compared to control animals (Figure 1E). Similarly, males that are mutant for either the PDF-1 ligand or receptor (and show little to no daf-7 expression in their ASJ neurons on E. coli, see Figure 1B) were capable of upregulating daf-7 expression in ASJ upon exposure to P. aeruginosa (Figure 1—figure supplement 2A). Given the previously established function of daf-7 expression in the ASJ neurons of hermaphrodites in promoting pathogen avoidance behavior (Meisel et al., 2014), these results predict that mutants in the PDF-1 pathway should have no defects in their ability to avoid a lawn of pathogenic P. aeruginosa. Consistent with this expectation, we observed that while daf-7 mutant hermaphrodites fail to avoid a lawn of pathogenic bacteria, the pdf-1 and pdfr-1 mutant hermaphrodites appear wild-type for their ability to perform this behavior (Figure 1F). Similarly, pdf-1 and pdfr-1 mutant males also displayed robust pathogen avoidance despite their defects in the male-specific mate searching behavior (Figure 1—figure supplement 2B; Barrios et al., 2012). These data suggest that the PDF-1 signaling pathway acts sex-specifically to regulate daf-7 expression in the ASJ neurons and its effects on downstream sexually dimorphic behavioral programs.
PDF-1 signaling acts cell-autonomously in the ASJ neurons to promote daf-7 expression
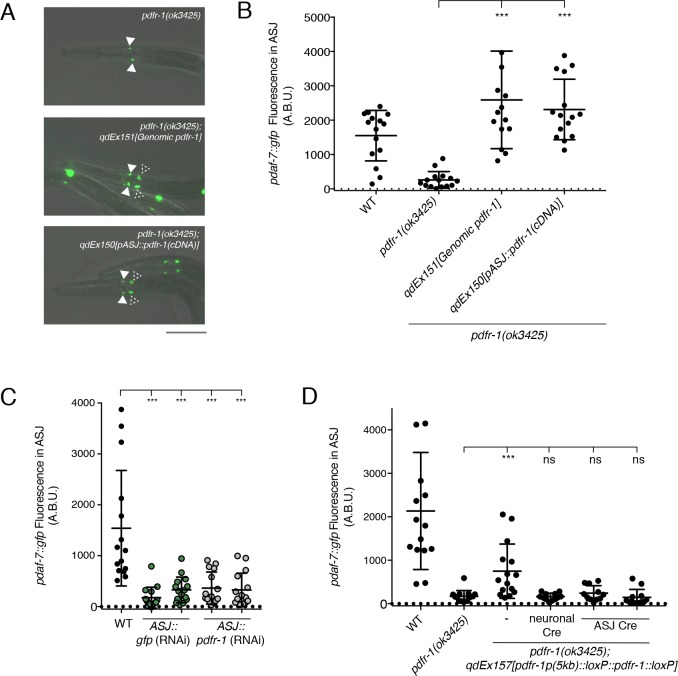
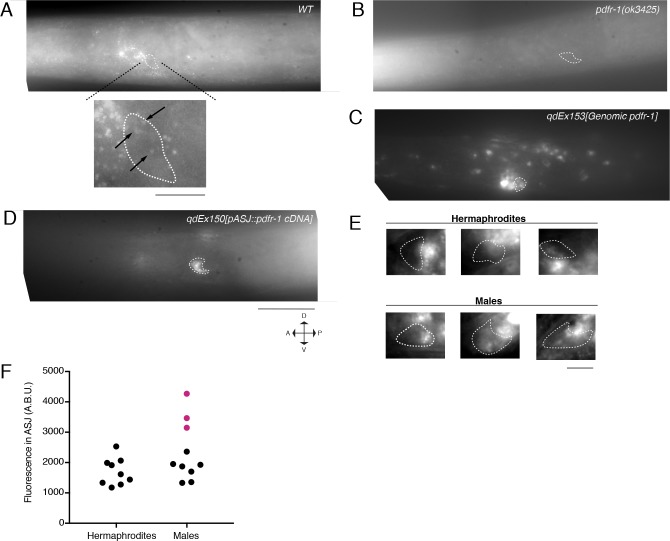
The PDF-1 neuropeptide ligand is secreted from multiple neurons in the head region of the animal where a similarly large number of neurons express the PDFR-1 receptor (Barrios et al., 2012; Janssen et al., 2009; Meelkop et al., 2012). To identify the relevant site of action for this pathway in the regulation of daf-7 expression in males, we used the pdfr-1(ok3425) mutant animals and introduced pdfr-1 cDNA transgenes into specific neurons using heterologous cell-specific promoters. We observed that while the pdfr-1 mutant males lack daf-7 expression in the ASJ neurons, introduction of a genomic DNA fragment carrying the pdfr-1 locus fully rescued this phenotype and restored daf-7 expression in the ASJ neurons (Figure 2A and B). Furthermore, we observed that expression of pdfr-1 under the control of the ASJ-specific trx-1 promoter was sufficient to rescue daf-7 expression in the ASJ neurons of the mutant male animals, suggesting that PDF-1 signals to its receptor, PDFR-1, in the ASJ neurons to influence daf-7 expression specifically in the male (Figure 2A and B).
Figure 2. PDF-1 signaling is necessary and sufficient in the ASJ neurons for the regulation of daf-7 expression in male C. elegans.
(A) pdaf-7::gfp expression in pdfr-1(ok3425) mutant (top), genomic rescue (middle), and ASJ-specific rescue (bottom) male animals. Filled arrowheads indicate the ASI neurons; dashed arrowheads indicate the ASJ neurons. Scale bar indicates 50 μm. (B) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of pdfr-1 rescue males. ***p<0.001 as determined by ordinary one-way ANOVA followed by Dunnett’s multiple comparisons test. Error bars represent SD. n = 15 animals for all genotypes. (C) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of WT males (black) and animals with ASJ-specific RNAi of either GFP (green) or pdfr-1(gray). ***p<0.001 as determined by ordinary one-way ANOVA followed by Dunnett’s multiple comparisons test. Error bars indicate SD. n = 15 animals for all conditions. (D) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of WT, pdfr-1(ok3425) mutants, and animals with floxed pdfr-1 rescued under the control of a 5 kb distal reporter as reported in Flavell et al. (2013) (left three columns). pdfr-1 function was removed either in all neurons or specifically in ASJ using cell-specific expression of Cre recombinase (right three columns). ***p<0.001 as determined by ordinary one-way ANOVA followed by Dunnett’s multiple comparisons test. ns, not significant. Error bars indicate SD. n = 12–15 animals for each condition.
To assess the necessity of pdfr-1 function in ASJ for the regulation of daf-7 expression in males, we knocked down pdfr-1 expression in the ASJ neurons via cell-specific RNAi (Esposito et al., 2007). We observed that animals with RNAi targeting pdfr-1 in the ASJ neurons exhibited reduced daf-7 expression in ASJ comparable to what we observed with ASJ-specific RNAi of GFP, our positive control (Figure 2C). Additionally, we generated transgenic animals carrying a floxed copy of the pdfr-1 cDNA under the control of a 5 kb region of the endogenous pdfr-1 promoter, which has been previously reported to rescue roaming behaviors in the C. elegans hermaphrodite (Flavell et al., 2013). We observed that this construct partially rescued daf-7 expression in the ASJ neurons of pdfr-1(ok3425) mutant males, and ASJ-specific expression of the Cre recombinase suppressed the rescuing effects of this transgene (Figure 2D), strongly suggestive that PDFR-1 activity in the ASJ neurons is required for daf-7 expression. These results indicate that the PDF-1 signaling pathway functions cell-autonomously in the ASJ neuron pair to regulate the sexually dimorphic expression of daf-7.
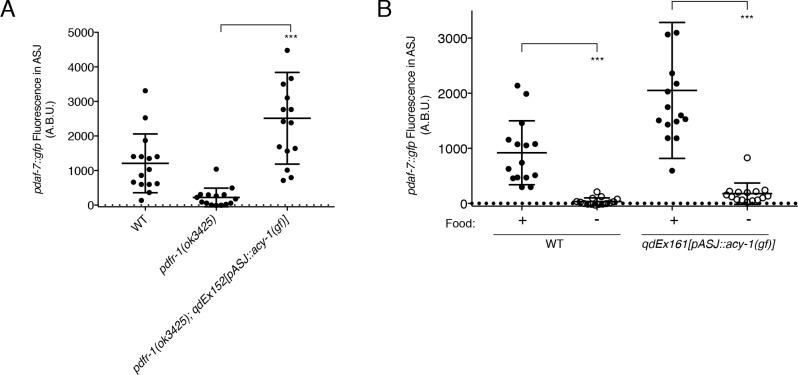
The PDFR-1 receptor is a secretin-family G-protein coupled receptor (GPCR), which has been shown to stimulate Gαs signaling and upregulation of cAMP production in transfected cells as well as in both Drosophila melanogaster and C. elegans neurons (Figure 1A; Flavell et al., 2013; Hyun et al., 2005; Janssen et al., 2008; Lear et al., 2005; Mertens et al., 2005; Shafer et al., 2008). Using a gain-of-function variant of the adenylate cyclase, ACY-1 (Flavell et al., 2013; Saifee et al., 2011; Schade et al., 2005), we asked if activation of the pathway downstream of PDFR-1 specifically in ASJ was sufficient to rescue the defects in daf-7 expression that we observe in the pdfr-1 mutant males. We observed that in pdfr-1 mutant males with transgenic expression of the acy-1(gf) cDNA only in the ASJ neurons, daf-7 expression was fully rescued (Figure 3A). This ability to bypass PDFR-1 by activation of cAMP production specifically in the ASJ neuron pair further suggest that the PDF-1 signaling pathway acts directly on the ASJ neurons in order to regulate daf-7 expression in male animals. Given the hierarchical nature of the regulation of daf-7 expression in the male ASJ neurons (Hilbert and Kim, 2017), we asked if acy-1 function may serve a broader role coordinating the many inputs of this hierarchy into changes in daf-7 expression. We previously reported that starvation of adult male animals efficiently suppresses daf-7 expression in the ASJ neurons in wild-type animals (Hilbert and Kim, 2017; Figure 3B). Notably, ASJ-specific expression of the acy-1(gf) variant in starved males did not suppress the effects of starvation on daf-7 expression in ASJ (Figure 3B), suggestive that the effects of starvation act downstream of or in parallel to PDFR-1-ACY-1 signaling.
Figure 3. ACY-1 acts downstream of PDFR-1 to regulate daf-7 expression in male ASJ neurons.
(A) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of males expressing the gain-of-function ACY-1(P260S) cDNA specifically in ASJ. ***p<0.001 as determined by unpaired t-test with Welch’s correction. Error bars represent SD. n = 15 animals for all genotypes. (B) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of WT and ASJ-specific acy-1(gf) expressing fed (filled circles) and starved (open circles) males. ***p<0.001 as determined by unpaired t-test with Welch’s correction. Error bars indicate SD. n = 15 animals for all conditions.
Sex differences in PDF-1 receptor activity underlie the sex-specific regulation of daf-7 transcription in ASJ
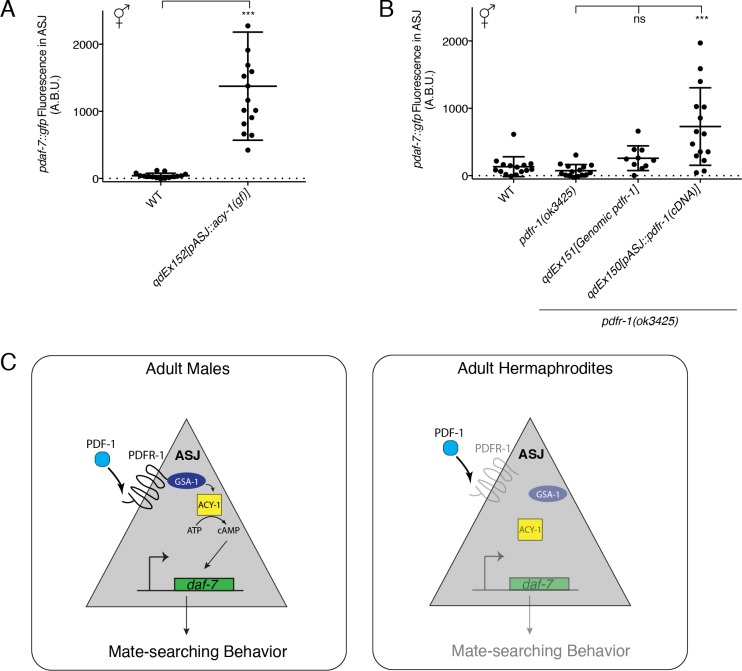
The sex-specificity of the effects of the PDF-1 pathway on daf-7 regulation and mate searching behavior is intriguing given very little evidence of differences in the expression or function of this neuropeptide pathway between the two C. elegans sexes (Barrios et al., 2012; Janssen et al., 2008, 2009). It was recently shown that pdf-1 is produced by the newly identified male-specific MCM neurons and is required for the regulation of sex-specific learning in males, but interestingly, ablation of these neurons has no effect on mate-searching behavior (Sammut et al., 2015). Nevertheless, we wondered if there might be unidentified sex differences in the signaling or expression of this PDF-1 neuropeptide pathway in neurons such as ASJ, which would confer its sex-specific effects on the regulation of daf-7 gene expression. To this end, we asked if activation of the PDF-1 signaling pathway in the ASJ neurons of hermaphrodites might be sufficient to drive daf-7 expression inappropriately in these animals. We first looked at hermaphrodite animals carrying the same ASJ-expressed acy-1(gf) transgene and observed significant upregulation of daf-7 expression in the ASJ neurons of these hermaphrodites (Figure 4A). We next asked whether we could observe daf-7 expression in the ASJ neurons of hermaphrodite animals with heterologous expression of pdfr-1 in only the ASJ neurons. Strikingly, we found that in hermaphrodites with overexpression of pdfr-1 cDNA in the ASJ neurons, daf-7 expression was also upregulated similar to what we observed in the acy-1(gf) transgenic strains (Figure 4B). We also quantified daf-7 expression in the ASJ neurons of hermaphrodites carrying the genomic pdfr-1 fragment with all of the endogenous regulatory sequence and observed no upregulation of expression in those animals. This control suggests that daf-7 expression in ASJ cannot be triggered simply as the result of overexpression of pdfr-1 (Figure 4B), rather, these results suggest that expression of PDFR-1 specifically in the hermaphrodite ASJ neurons is sufficient to allow daf-7 expression in these neurons.
Figure 4. Heterologous activation of the PDF-1 pathway in ASJ is sufficient to activate daf-7 transcription in adult hermaphrodites.
(A) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of WT hermaphrodites and hermaphrodites where ACY-1 has been activated (via the gain of function P260S mutant) specifically in ASJ. ***p<0.001 as determined by unpaired t-test with Welch’s correction. Error bars represent SD. n = 15 animals for both genotypes. (B) Maximum fluorescence values of pdaf-7::gfp in the ASJ neurons of hermaphrodites overexpressing pdfr-1 from either a genomic fragment or under the control of a heterologous ASJ-specific promoter. ***p<0.001 as determined by ordinary one-way ANOVA followed by Dunnett’s multiple comparisons test. Error bars represent SD. ns, not significant. n = 15 animals for all genotypes except pdfr-1(ok3425); qdEx151 where n = 10 animals. (C) Model for the sex-specific regulation of daf-7 expression in the ASJ chemosensory neurons by the PDF-1 signaling pathway.
Figure 4—figure supplement 1. FISH imaging of endogenous pdfr-1 mRNA transcripts.
We note that establishing the neuronal expression pattern of pdfr-1 has been challenging because of the apparent complexity of defining putative regulatory regions of the gene (Barrios et al., 2012; Flavell et al., 2013; Janssen et al., 2008). We sought to examine the transcription of pdfr-1 in male and hermaphrodite animals using fluorescence in situ hybridization (FISH). We generated fluorescent probes for a region of the pdfr-1 coding sequence that is shared among all isoforms and verified the specificity of these probes for the pdfr-1 coding sequence by examining expression in the pdfr-1(ok3425) deletion mutant, where we observed no fluorescent signal (Figure 4—figure supplement 1B), and in our ASJ-specific rescue lines, where we could only observe fluorescence in the ASJ neurons (Figure 4—figure supplement 1D). Imaging of pdfr-1 transcription in WT animals revealed a diffuse expression pattern with fluorescent signal observable in muscle tissue as well as in neurons, but with few cells having strong signal and many cells with only scattered fluorescent spots, including the ASJ neurons (Figure 4—figure supplement 1A). To corroborate and confirm these observations, we also imaged pdfr-1 transcripts in animals carrying our genomic rescuing fragment, which amplified probe fluorescence throughout the nervous system and muscle (Figure 4—figure supplement 1C). We expect that because of the intact endogenous regulatory sequence on this genomic fragment, the mRNA localization we observe in this strain should still be representative of the wild-type expression pattern of pdfr-1. While we observe qualitative differences in the abundance of pdfr-1 mRNA in the ASJ neurons between males and hermaphrodites, we did not detect pdfr-1 mRNA in all male animals examined. We observed puncta of pdfr-1 mRNA in the ASJ neurons of about 20% of adult male animals (Figure 4—figure supplement 1E and F), whereas similarly aged hermaphrodites did not have a corresponding subpopulation of animals with puncta of pdfr-1 mRNA. Further analysis of the expression pattern of pdfr-1 will be required to definitively identify any sexual dimorphism in the expression of this gene. Nevertheless, our functional studies demonstrating that PDFR-1 expression in the ASJ neurons is both necessary and sufficient for daf-7 expression suggest that the expression or activity of the PDFR-1 receptor may be regulated in a sexually dimorphic manner in these neurons.
The PDF-1-DAF-7 neuroendocrine signaling cascade regulates sex-specific behavior through the sex-shared ASJ neurons
Building on our previous work on the sexually dimorphic regulation of the neuroendocrine gene daf-7 and its role in promoting male decision-making behaviors (Hilbert and Kim, 2017), we have presented here a set of experiments which implicate the PDF-1 neuropeptide signaling pathway as a critical male-specific regulator of daf-7 expression in the ASJ neurons. Our data suggest that sexually dimorphic regulation of the PDF-1 receptor, PDFR-1, may serve as a gating mechanism, allowing the ASJ neurons of adult male C. elegans to respond to the PDF-1 ligand. We suggest that this ligand-receptor interaction activates a downstream signaling cascade in ASJ terminating in the transcriptional activation of daf-7, which in turn promotes male-specific decision-making behaviors (Figure 4C, left). We hypothesize that the relative lack of expression or activity of pdfr-1 in the hermaphrodite ASJ neurons prevents the activation of this pathway and consequently daf-7 expression is not induced under normal growth conditions in adult hermaphrodites (Figure 4C, right). Strikingly, heterologous expression of the PDF-1 receptor in the hermaphrodite ASJ neurons was sufficient to drive daf-7 expression in an inappropriate physiological context (the hermaphrodite nervous system). All together, our data suggest that the PDF-1 pathway plays an integral role in facilitating sex-specific differences in gene expression and behavior.
While recent work has revealed sexual dimorphisms at the level of gene expression, neuronal connectivity and neurotransmitter release in the sex-shared nervous system of C. elegans (Hart and Hobert, 2018; Hilbert and Kim, 2017; Oren-Suissa et al., 2016; Pereira et al., 2015; Ryan et al., 2014; Serrano-Saiz et al., 2017a, 2017b; Weinberg et al., 2018), the role of neuromodulators and other neuroendocrine signals in facilitating sex-specific responses of neurons in the shared neuronal circuitry has been relatively unexplored. Here, we propose a model in which two pathways, the PDF-1 and DAF-7/TGFβ pathways, act in concert as a neuroendocrine signaling cascade to regulate sex-specific behavior within the context of the sex-shared ASJ neurons (Figure 4C). Our data suggest that the PDF-1 pathway functions in tuning the response of the ASJ neurons to this endogenous neuromodulator in a sex-specific manner. Interestingly, recent work in mice has uncovered a similar phenomenon wherein the neuromodulator oxytocin facilitates sex-specific social preference in male mice by modulating the ability of subsets of neurons to respond to social cues (Yao et al., 2017). The parallels between this work and ours underscore the role of neuroendocrine signaling through sex-shared nervous system components in shaping sexually dimorphic neuronal activity and behavior in evolutionarily diverse animals.
Materials and methods
Key resources table.
| Reagent type (species) or resource |
Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Caenorhabditis elegans) |
pdf-1 | NA | WBGene00020317 | |
| Gene (C. elegans) | pdfr-1 | NA | WBGene00015735 | |
| Gene (C. elegans) | daf-7 | NA | WBGene00000903 | |
| Gene (C. elegans) | acy-1 | NA | WBGene00000068 | |
| Genetic reagent (C. elegans) |
ksIs2 | PMID: 11677050 | WBTransgene00000788 | pdaf-7::gfp |
| Recombinant DNA reagent |
Moerman Fosmid Library |
Source Bioscience | WRM0629dH07, WRM0627cG01, WRM0641dA07, WRM068aD11 |
|
| Recombinant DNA reagent |
pPD95.75 | Fire Lab C. elegans
Vector Kit |
Addgene plasmid # 1494 | |
| Recombinant DNA reagent |
pCFJ90 | PMID:18953339 | Addgene plasmid # 19327 |
pmyo-2::mCherry, used as co-injection marker |
| Recombinant DNA reagent |
pZH42 | this paper | pdf-1 genomic DNA in pUC19 | |
| Recombinant DNA reagent |
pZH48 | this paper |
ptrx-1::pdfr-1(cDNA, b isoform,
no STOP codon)::F2A::mCherry:: unc-54 3'UTR |
|
| Recombinant DNA reagent |
pZH53 | this paper | ptrx-1::ACY-1(P260S)::unc-54 3'UTR | |
| Recombinant DNA reagent |
pZH58 | this paper |
pdfr-1p(distal, 5 kb)::loxP::pdfr-1
cDNA(B isoform)::loxP::unc-54 3'UTR |
|
| Recombinant DNA reagent |
pZH59 | this paper | ptrx-1::nCre | |
| Recombinant DNA reagent |
pJDM30 | PMID: 25303524 | ptrx-1::daf-7 | |
| Recombinant DNA reagent |
pSF11 | PMID: 23972393 |
ptag-168::nCre, gift of C. Bargmann and S. Flavell |
|
| Commercial assay or kit |
NEBuilder HiFi DNA Assembly Master Mix |
NEB | E2621 | |
| Software, algorithm | GraphPad Prism | GraphPad | RRID:SCR_002798 | |
| Other | Alexa Fluor 647 DHS ester |
Invitrogen/Thermo Fisher Scientific |
A20006 | dye used for conjugation of FISH probes |
C. elegans strains
C. elegans strains were cultured as previously described (Brenner, 1974; Hilbert and Kim, 2017). For a complete list of strains used in this study please see Supplementary file 1.
Cloning and transgenic strain generation
For the pdf-1 overexpression transgene, a 6.5 kb region of sequence containing the pdf-1 promoter, coding sequence and 3’UTR were amplified from the fosmid WRM0641dA07 from the Moerman fosmid library. This fragment was cloned into the pUC19 vector backbone by Gibson assembly (Gibson et al., 2009) to generate plasmid pZH42. ASJ-specific overexpression of daf-7 transgenes in the pdf-1(tm1996) mutant background were established by reinjection of the plasmid pJDM30 which contains daf-7 cDNA under the control of the trx-1 promoter (Meisel et al., 2014).
For the ASJ-specific pdfr-1 rescue construct, the B isoform of the pdfr-1 cDNA with no stop codon was amplified from cDNA generated with an Ambion RetroScript kit using primers based on previously described annotation of the isoform (Barrios et al., 2012). The trx-1 ASJ-specific promoter was amplified as previously described (Hilbert and Kim, 2017). An F2A::mCherry fragment was amplified off a plasmid that was a gift from C. Pender and H.R. Horvitz. All fragments were cloned into the pPD95.75 backbone with an intact unc-54 3’UTR by Gibson assembly to generate plasmid pZH48. Genomic rescue of pdfr-1 was done by injection of the WRM0629dH07 fosmid from the Moerman fosmid library.
Cloning for cell-specific RNAi experiments was carried out similarly to the method previously described (Esposito et al., 2007). A 1.2 kb fragment of the ASJ-specific trx-1 promoter was amplified from the fosmid WRM0627cG01 from the Moerman fosmid library with overlap to either pdfr-1 or GFP in either the sense or antisense direction. A 1.8 kb exon rich region of the pdfr-1 coding sequence was amplified from the fosmid WRM0629dH07. A 1 kb fragment containing part of the GFP coding sequence was amplified from the plasmid, pPD95.75. The GFP and pdfr-1 fragments were cloned in the sense and anti-sense directions with the trx-1 promoter into the pUC19 plasmid backbone using Gibson Assembly. PCR with nested primers was then used to amplify only the promoter and gene sequence off the plasmid backbone and these PCR products were purified and used for injections. Both sense and anti-sense PCR products were injected at a concentration of 20 ng/μL along with pCFJ90 (Frøkjaer-Jensen et al., 2008) at 2.5 ng/μL and 1 kb ladder as carrier DNA.
For the floxed pdfr-1 rescue strain, the pdfr-1 cDNA was amplified with primers carrying loxP sequences on either side. The 5 kb pdfr-1 promoter was amplified from the fosmid WRM068aD11. These fragments were cloned into a pPD95.75 backbone with an intact unc-54 3’UTR by Gibson Assembly to generate plasmid pZH58. ASJ-specific Cre lines were generated by swapping the trx-1 promoter into the plasmid pSF11 (gift of S. Flavell and C. Bargmann) in place of the tag-168 pan-neuronal promoter to generate plasmid pZH59. Pan-neuronal Cre lines were generated by re-injection of pSF11 at a concentration of 20 ng/μL. For all Cre lines, pCFJ90 was used as a co-injection marker at a concentration of 2.5 ng/μL.
For the ACY-1(gf) construct, the 3.8 kb acy-1(P260S) fragment was amplified from genomic DNA extracted from the strain CX15050 (gift from S. Flavell and C. Bargmann) which carries a transgenic array with the acy-1(P260S) cDNA under the control of a different promoter. This fragment was cloned into a plasmid backbone carrying the trx-1 promoter and unc-54 3’UTR to generate pZH53. All fosmids and plasmids were verified by sequencing and injected at a concentration of 50 ng/μL along with a plasmid carrying pofm-1::gfp at 50 ng/μL as a co-injection marker unless otherwise noted. At least three independent transgenic lines were obtained and analyzed for each construct and one or two representative lines are shown. For a list of all primers used in this paper, please see Supplementary file 2.
Measurement of gene expression in ASI and ASJ neurons
Quantification of daf-7 expression was performed as described in (Hilbert and Kim, 2017) using the ksIs2(pdaf-7::gfp) transgene (Murakami et al., 2001). All adult quantifications were done on animals 72 hr after egg lay. Quantification of animals on P. aeruginosa were performed as before.
Starvation assays
Starvation assays and measurement of pdaf-7::gfp fluorescence in the ASJ neurons of starved males was performed as previously described (Hilbert and Kim, 2017).
Mate-Searching assays
Mate-searching assays were performed as previously described (Hilbert and Kim, 2017; Lipton et al., 2004).
P. aeruginosa lawn avoidance assays
P. aeruginosa plates were prepared as described in (Hilbert and Kim, 2017). Animals were synchronized by treatment with bleach and allowed to hatch and arrest as L1 larvae before being dropped onto E. coli plates. L4 animals were transferred to the center of the P. aeruginosa lawn, incubated at 25˚C and then scored for avoidance after 16 hr.
For male lawn avoidance assays shown in Figure 1—figure supplement 2B, plates and animals were prepared using the same method as for hermaphrodites, but males were placed individually onto plates seeded with P. aeruginosa as L4s. Plates were incubated at 25˚C and then scored for avoidance after 16 hr. These experiments were repeated three times, with 30 individual animals per genotype in each replicate.
Fluorescence In Situ Hybridization
FISH was performed as previously described (Hilbert and Kim, 2017; Raj et al., 2008). The pdfr-1 probe was constructed by pooling together 36 unique 20 nucleotide oligos that tile across base-pairs 580–1540 in the pdfr-1 B-isoform cDNA. This sequence is contained in all isoforms of pdfr-1 so should anneal to any endogenous pdfr-1 mRNA. After pooling, oligos were coupled to Alexa Fluor 647 NHS ester (Invitrogen/Thermo Fisher Scientific) and then purified by HPLC.
Statistical analysis
All statistical analysis was performed using the Graphpad Prism software. Statistical tests used for each experiment are listed in the figure legend.
Acknowledgements
We thank C Bargmann, S Flavell, HR Horvitz, J Kaplan, and the Caenorhabditis Genetics Center (which is funded by the NIH Office of Research Infrastructure Programs P40 OD010440) for strains and reagents. We thank S Flavell and members of the Kim and Horvitz labs for helpful discussions in the development of the manuscript.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Contributor Information
Dennis H Kim, Email: dhkim@mit.edu.
Oliver Hobert, Howard Hughes Medical Institute, Columbia University, United States.
Funding Information
This paper was supported by the following grants:
National Institutes of Health GM084477 to Dennis H Kim.
National Institutes of Health T32GM007287 to Zoë A Hilbert.
Additional information
Competing interests
No competing interests declared.
Author contributions
Conceptualization, Formal analysis, Investigation, Visualization, Methodology, Writing—original draft, Writing—review and editing.
Conceptualization, Supervision, Funding acquisition, Visualization, Writing—review and editing.
Additional files
A comprehensive list of the strains used in this study. Strain source (this study or others) is indicated.
A comprehensive list of the oligos created for this study. All sequences are listed in the 5’ to 3’ direction.
Data availability
All data generated or analysed during this study are included in the manuscript and supporting files.
References
- Barr MM, García LR, Portman DS. Sexual dimorphism and sex differences in Caenorhabditis elegans neuronal development and behavior. Genetics. 2018;208:909–935. doi: 10.1534/genetics.117.300294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrios A, Ghosh R, Fang C, Emmons SW, Barr MM. PDF-1 neuropeptide signaling modulates a neural circuit for mate-searching behavior in C. elegans. Nature Neuroscience. 2012;15:1675–1682. doi: 10.1038/nn.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrios A, Nurrish S, Emmons SW. Sensory regulation of C. elegans male mate-searching behavior. Current Biology. 2008;18:1865–1871. doi: 10.1016/j.cub.2008.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AJ, Chronis N, Karow DS, Marletta MA, Bargmann CI. A distributed chemosensory circuit for oxygen preference in C. elegans. PLoS Biology. 2006;4:e274. doi: 10.1371/journal.pbio.0040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S, Chatzigeorgiou M, Taylor KP, Schafer WR, Kaplan JM. Analysis of NPR-1 reveals a circuit mechanism for behavioral quiescence in C. elegans. Neuron. 2013;78:869–880. doi: 10.1016/j.neuron.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta SR, Vasconcelos ML, Ruta V, Luo S, Wong A, Demir E, Flores J, Balonze K, Dickson BJ, Axel R. The Drosophila pheromone cVA activates a sexually dimorphic neural circuit. Nature. 2008;452:473–477. doi: 10.1038/nature06808. [DOI] [PubMed] [Google Scholar]
- Dulac C, Kimchi T. Neural mechanisms underlying sex-specific behaviors in vertebrates. Current Opinion in Neurobiology. 2007;17:675–683. doi: 10.1016/j.conb.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G, Di Schiavi E, Bergamasco C, Bazzicalupo P. Efficient and cell specific knock-down of gene function in targeted C. elegans neurons. Gene. 2007;395:170–176. doi: 10.1016/j.gene.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Fagan KA, Luo J, Lagoy RC, Schroeder FC, Albrecht DR, Portman DS. A Single-Neuron chemosensory switch determines the valence of a sexually dimorphic sensory behavior. Current Biology. 2018;28:902–914. doi: 10.1016/j.cub.2018.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Pokala N, Macosko EZ, Albrecht DR, Larsch J, Bargmann CI. Serotonin and the neuropeptide PDF initiate and extend opposing behavioral states in C. elegans. Cell. 2013;154:1023–1035. doi: 10.1016/j.cell.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher M, Kim DH. Age-Dependent neuroendocrine signaling from sensory neurons modulates the effect of dietary restriction on longevity of Caenorhabditis elegans. PLOS Genetics. 2017;13:e1006544. doi: 10.1371/journal.pgen.1006544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nature Genetics. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii S, Amrein H. Ventral lateral and DN1 clock neurons mediate distinct properties of male sex drive rhythm in Drosophila. PNAS. 2010;107:10590–10595. doi: 10.1073/pnas.0912457107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher T, Kim J, Oldenbroek M, Kerr R, You YJ. ASI regulates satiety quiescence in C. elegans. Journal of Neuroscience. 2013;33:9716–9724. doi: 10.1523/JNEUROSCI.4493-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendrel M, Atlas EG, Hobert O. A cellular and regulatory map of the GABAergic nervous system of C. elegans. eLife. 2016;5:e17686. doi: 10.7554/eLife.17686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Greer ER, Pérez CL, Van Gilst MR, Lee BH, Ashrafi K. Neural and molecular dissection of a C. elegans sensory circuit that regulates fat and feeding. Cell Metabolism. 2008;8:118–131. doi: 10.1016/j.cmet.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart MP, Hobert O. Neurexin controls plasticity of a mature, sexually dimorphic neuron. Nature. 2018;553:165–170. doi: 10.1038/nature25192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfrich-Förster C. The period clock gene is expressed in central nervous system neurons which also produce a neuropeptide that reveals the projections of circadian pacemaker cells within the brain of Drosophila melanogaster. PNAS. 1995;92:612–616. doi: 10.1073/pnas.92.2.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilbert ZA, Kim DH. Sexually dimorphic control of gene expression in sensory neurons regulates decision-making behavior in C. elegans. eLife. 2017;6:e21166. doi: 10.7554/eLife.21166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyun S, Lee Y, Hong ST, Bang S, Paik D, Kang J, Shin J, Lee J, Jeon K, Hwang S, Bae E, Kim J. Drosophila GPCR han is a receptor for the circadian clock neuropeptide PDF. Neuron. 2005;48:267–278. doi: 10.1016/j.neuron.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Jang H, Kim K, Neal SJ, Macosko E, Kim D, Butcher RA, Zeiger DM, Bargmann CI, Sengupta P. Neuromodulatory state and sex specify alternative behaviors through antagonistic synaptic pathways in C. elegans. Neuron. 2012;75:585–592. doi: 10.1016/j.neuron.2012.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen T, Husson SJ, Lindemans M, Mertens I, Rademakers S, Ver Donck K, Geysen J, Jansen G, Schoofs L. Functional characterization of three G protein-coupled receptors for pigment dispersing factors in Caenorhabditis elegans. Journal of Biological Chemistry. 2008;283:15241–15249. doi: 10.1074/jbc.M709060200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen T, Husson SJ, Meelkop E, Temmerman L, Lindemans M, Verstraelen K, Rademakers S, Mertens I, Nitabach M, Jansen G, Schoofs L. Discovery and characterization of a conserved pigment dispersing factor-like neuropeptide pathway in Caenorhabditis elegans. Journal of Neurochemistry. 2009;111:228–241. doi: 10.1111/j.1471-4159.2009.06323.x. [DOI] [PubMed] [Google Scholar]
- Kim WJ, Jan LY, Jan YN. A PDF/NPF neuropeptide signaling circuitry of male Drosophila melanogaster controls rival-induced prolonged mating. Neuron. 2013;80:1190–1205. doi: 10.1016/j.neuron.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimchi T, Xu J, Dulac C. A functional circuit underlying male sexual behaviour in the female mouse brain. Nature. 2007;448:1009–1014. doi: 10.1038/nature06089. [DOI] [PubMed] [Google Scholar]
- Kohl J, Ostrovsky AD, Frechter S, Jefferis GS. A bidirectional circuit switch reroutes pheromone signals in male and female brains. Cell. 2013;155:1610–1623. doi: 10.1016/j.cell.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp JJ, Billeter JC, Wong A, Choi C, Nitabach MN, Levine JD. Pigment-dispersing factor modulates pheromone production in clock cells that influence mating in Drosophila. Neuron. 2013;79:54–68. doi: 10.1016/j.neuron.2013.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtovic A, Widmer A, Dickson BJ. A single class of olfactory neurons mediates behavioural responses to a Drosophila sex pheromone. Nature. 2007;446:542–546. doi: 10.1038/nature05672. [DOI] [PubMed] [Google Scholar]
- Lear BC, Merrill CE, Lin JM, Schroeder A, Zhang L, Allada R. A G protein-coupled receptor, groom-of-PDF, is required for PDF neuron action in circadian behavior. Neuron. 2005;48:221–227. doi: 10.1016/j.neuron.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Lee K, Portman DS. Neural sex modifies the function of a C. elegans sensory circuit. Current Biology. 2007;17:1858–1863. doi: 10.1016/j.cub.2007.10.015. [DOI] [PubMed] [Google Scholar]
- Lipton J, Kleemann G, Ghosh R, Lints R, Emmons SW. Mate searching in Caenorhabditis elegans: a genetic model for sex drive in a simple invertebrate. Journal of Neuroscience. 2004;24:7427–7434. doi: 10.1523/JNEUROSCI.1746-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu KS, Sternberg PW. Sensory regulation of male mating behavior in Caenorhabditis elegans. Neuron. 1995;14:79–89. doi: 10.1016/0896-6273(95)90242-2. [DOI] [PubMed] [Google Scholar]
- Meelkop E, Temmerman L, Janssen T, Suetens N, Beets I, Van Rompay L, Shanmugam N, Husson SJ, Schoofs L. PDF receptor signaling in Caenorhabditis elegans modulates locomotion and egg-laying. Molecular and Cellular Endocrinology. 2012;361:232–240. doi: 10.1016/j.mce.2012.05.001. [DOI] [PubMed] [Google Scholar]
- Meisel JD, Panda O, Mahanti P, Schroeder FC, Kim DH. Chemosensation of bacterial secondary metabolites modulates neuroendocrine signaling and behavior of C. elegans. Cell. 2014;159:267–280. doi: 10.1016/j.cell.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens I, Vandingenen A, Johnson EC, Shafer OT, Li W, Trigg JS, De Loof A, Schoofs L, Taghert PH. PDF receptor signaling in Drosophila contributes to both circadian and geotactic behaviors. Neuron. 2005;48:213–219. doi: 10.1016/j.neuron.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Milward K, Busch KE, Murphy RJ, de Bono M, Olofsson B. Neuronal and molecular substrates for optimal foraging in Caenorhabditis elegans. PNAS. 2011;108:20672–20677. doi: 10.1073/pnas.1106134109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowrey WR, Bennett JR, Portman DS. Distributed effects of biological sex define sex-typical motor behavior in Caenorhabditis elegans. Journal of Neuroscience. 2014;34:1579–1591. doi: 10.1523/JNEUROSCI.4352-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Koga M, Ohshima Y. DAF-7/TGF-beta expression required for the normal larval development in C. elegans is controlled by a presumed guanylyl cyclase DAF-11. Mechanisms of Development. 2001;109:27–35. doi: 10.1016/S0925-4773(01)00507-X. [DOI] [PubMed] [Google Scholar]
- Oren-Suissa M, Bayer EA, Hobert O. Sex-specific pruning of neuronal synapses in Caenorhabditis elegans. Nature. 2016;533:206–211. doi: 10.1038/nature17977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Hall JC. Isolation and chronobiological analysis of a neuropeptide pigment-dispersing factor gene in Drosophila melanogaster. Journal of Biological Rhythms. 1998;13:219–228. doi: 10.1177/074873098129000066. [DOI] [PubMed] [Google Scholar]
- Park JH, Helfrich-Förster C, Lee G, Liu L, Rosbash M, Hall JC. Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. PNAS. 2000;97:3608–3613. doi: 10.1073/pnas.97.7.3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira L, Kratsios P, Serrano-Saiz E, Sheftel H, Mayo AE, Hall DH, White JG, LeBoeuf B, Garcia LR, Alon U, Hobert O. A cellular and regulatory map of the cholinergic nervous system of C. elegans. eLife. 2015;4:e12432. doi: 10.7554/eLife.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nature Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren P, Lim CS, Johnsen R, Albert PS, Pilgrim D, Riddle DL. Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science. 1996;274:1389–1391. doi: 10.1126/science.274.5291.1389. [DOI] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell. 1999;99:791–802. doi: 10.1016/S0092-8674(00)81676-1. [DOI] [PubMed] [Google Scholar]
- Ruta V, Datta SR, Vasconcelos ML, Freeland J, Looger LL, Axel R. A dimorphic pheromone circuit in Drosophila from sensory input to descending output. Nature. 2010;468:686–690. doi: 10.1038/nature09554. [DOI] [PubMed] [Google Scholar]
- Ryan DA, Miller RM, Lee K, Neal SJ, Fagan KA, Sengupta P, Portman DS. Sex, age, and hunger regulate behavioral prioritization through dynamic modulation of chemoreceptor expression. Current Biology. 2014;24:2509–2517. doi: 10.1016/j.cub.2014.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saifee O, Metz LB, Nonet ML, Crowder CM. A gain-of-function mutation in adenylate cyclase confers isoflurane resistance in Caenorhabditis elegans. Anesthesiology. 2011;115:1162–1171. doi: 10.1097/ALN.0b013e318239355d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai N, Iwata R, Yokoi S, Butcher RA, Clardy J, Tomioka M, Iino Y. A sexually conditioned switch of chemosensory behavior in C. elegans. PLoS One. 2013;8:e68676. doi: 10.1371/journal.pone.0068676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sammut M, Cook SJ, Nguyen KCQ, Felton T, Hall DH, Emmons SW, Poole RJ, Barrios A. Glia-derived neurons are required for sex-specific learning in C. elegans. Nature. 2015;526:385–390. doi: 10.1038/nature15700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schackwitz WS, Inoue T, Thomas JH. Chemosensory neurons function in parallel to mediate a pheromone response in C. elegans. Neuron. 1996;17:719–728. doi: 10.1016/S0896-6273(00)80203-2. [DOI] [PubMed] [Google Scholar]
- Schade MA, Reynolds NK, Dollins CM, Miller KG. Mutations that rescue the paralysis of Caenorhabditis elegans ric-8 (synembryn) mutants activate the G alpha(s) pathway and define a third major branch of the synaptic signaling network. Genetics. 2005;169:631–649. doi: 10.1534/genetics.104.032334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Saiz E, Oren-Suissa M, Bayer EA, Hobert O. Sexually dimorphic differentiation of a C. elegans Hub neuron is cell autonomously controlled by a conserved transcription factor. Current Biology. 2017a;27:199–209. doi: 10.1016/j.cub.2016.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Saiz E, Pereira L, Gendrel M, Aghayeva U, Battacharya A, Howell K, Garcia LR, Hobert O. A neurotransmitter atlas of the Caenorhabditis elegans male nervous system reveals sexually dimorphic neurotransmitter usage. Genetics. 2017b;206:1251–1269. doi: 10.1534/genetics.117.202127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Kim DJ, Dunbar-Yaffe R, Nikolaev VO, Lohse MJ, Taghert PH. Widespread receptivity to neuropeptide PDF throughout the neuronal circadian clock network of Drosophila revealed by real-time cyclic AMP imaging. Neuron. 2008;58:223–237. doi: 10.1016/j.neuron.2008.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw WM, Luo S, Landis J, Ashraf J, Murphy CT. The C. elegans TGF-beta Dauer pathway regulates longevity via insulin signaling. Current Biology. 2007;17:1635–1645. doi: 10.1016/j.cub.2007.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan J, Kaplan F, Ajredini R, Zachariah C, Alborn HT, Teal PE, Malik RU, Edison AS, Sternberg PW, Schroeder FC. A blend of small molecules regulates both mating and development in Caenorhabditis elegans. Nature. 2008;454:1115–1118. doi: 10.1038/nature07168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowers L, Holy TE, Meister M, Dulac C, Koentges G. Loss of sex discrimination and male-male aggression in mice deficient for TRP2. Science. 2002;295:1493–1500. doi: 10.1126/science.1069259. [DOI] [PubMed] [Google Scholar]
- Stowers L, Logan DW. Sexual dimorphism in olfactory signaling. Current Opinion in Neurobiology. 2010;20:770–775. doi: 10.1016/j.conb.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg P, Berkseth M, Zarkower D, Hobert O. Sexually dimorphic unc-6/Netrin expression controls Sex-Specific maintenance of synaptic connectivity. Current Biology. 2018;28:623–629. doi: 10.1016/j.cub.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JQ, Jorgensen EM. Sensation in a single neuron pair represses male behavior in hermaphrodites. Neuron. 2012;75:593–600. doi: 10.1016/j.neuron.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CF, Shah NM. Representing sex in the brain, one module at a time. Neuron. 2014;82:261–278. doi: 10.1016/j.neuron.2014.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Bergan J, Lanjuin A, Dulac C. Oxytocin signaling in the medial amygdala is required for sex discrimination of social cues. eLife. 2017;6:e31373. doi: 10.7554/eLife.31373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You YJ, Kim J, Raizen DM, Avery L. Insulin, cGMP, and TGF-beta signals regulate food intake and quiescence in C. elegans: a model for satiety. Cell Metabolism. 2008;7:249–257. doi: 10.1016/j.cmet.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]