Abstract
Polybrominated diphenyl ethers (PBDEs) are persistent environmental contaminants with well characterized toxicities in host organs. Gut microbiome is increasingly recognized as an important regulator of xenobiotic biotransformation; however, little is known about its interactions with PBDEs. Primary bile acids (BAs) are metabolized by the gut microbiome into more lipophilic secondary BAs that may be absorbed and interact with certain host receptors. The goal of this study was to test our hypothesis that PBDEs cause dysbiosis and aberrant regulation of BA homeostasis. Nine-week-old male C57BL/6 conventional (CV) and germ-free (GF) mice were orally gavaged with corn oil (10 mg/kg), BDE-47 (100 μmol/kg), or BDE-99 (100 μmol/kg) once daily for 4 days (n = 3–5/group). Gut microbiome was characterized using 16S rRNA sequencing of the large intestinal content in CV mice. Both BDE-47 and BDE-99 profoundly decreased the alpha diversity of gut microbiome and differentially regulated 45 bacterial species. Both PBDE congeners increased Akkermansia muciniphila and Erysipelotrichaceae Allobaculum spp., which have been reported to have anti-inflammatory and antiobesity functions. Targeted metabolomics of 56 BAs was conducted in serum, liver, and small and large intestinal content of CV and GF mice. BDE-99 increased many unconjugated BAs in multiple biocompartments in a gut microbiota-dependent manner. This correlated with an increase in microbial 7α-dehydroxylation enzymes for secondary BA synthesis and increased expression of host intestinal transporters for BA absorption. Targeted proteomics showed that PBDEs downregulated host BA-synthesizing enzymes and transporters in livers of CV but not GF mice. In conclusion, there is a novel interaction between PBDEs and the endogenous BA-signaling through modification of the “gut-liver axis”.
Introduction
Polybrominated diphenyl ethers (PBDEs) are flame retardants commonly used in a variety of consumer products, such as plastics, rubbers, textiles, furniture, and electronic devices. Health concerns about exposure to PBDEs have increased significantly in recent years as their presence has been detected in environmental samples and in human tissues (Frederiksen et al., 2009). PBDEs have been shown to cause developmental neurotoxicity, reproductive toxicity, thyroid hormone disruption, liver toxicity, and potential cancer development in rodent studies (Gascon et al., 2011; Linares et al., 2015). Among the 209 PBDE congeners, 2,2′,4,4′-tetrabromodiphenyl ether (BDE-47) and 2,2′,4,4′,5-pentabromodiphenyl ether (BDE-99) are the most predominant congeners detected in the environment and in human samples (Erratico et al., 2011). BDE-47 and BDE-99 activate the major xenobiotic-sensing nuclear receptors pregnane X receptor (PXR/Nr1i2) and constitutive androstane receptor (CAR/Nr1i3) in both rodents and human hepatocytes, leading to induced expression of cytochrome P450s (P450s), which may cause adverse drug reactions or drug-drug interactions (Pacyniak et al., 2007; Sueyoshi et al., 2014). Using RNA-Seq, we recently demonstrated that BDE-47 and BDE-99 differentially regulated many other phase-I drug-metabolizing enzymes (i.e., enzymes involved in oxidation, reduction, and hydrolysis), phase-II enzymes (i.e., enzymes involved in conjugation), as well as transporters in mouse liver. We also demonstrated that the lack of gut microbiota in GF mice altered the oxidative metabolites of BDE-47 and BDE-99 and profoundly modified the PBDE-mediated transcriptomic responses in liver (Li et al., 2017). This provided the first evidence that there is a novel interaction between gut microbiota and PBDEs that influences the host hepatic xenobiotic biotransformation. However, very little is known regarding the interactions between PBDEs and gut microbiota on intermediary metabolism.
Gut microbiome communicates with the host liver to modify hepatic xenobiotic biotransformation and nutrient homeostasis (Shreiner et al., 2015; Fu and Cui, 2017). Disruptions in the composition of gut microbial communities and altered microbiota-host interactions have been linked to several diseases, such as cancer and metabolic disorders (Jumpertz et al., 2011). Bile acids (BAs), a group of steroids produced in liver from cholesterol, are important signaling molecules for intermediary metabolism within the gut-liver axis and in extrahepatic organs, such as brown adipose tissue and muscle (Broeders et al., 2015). The primary BAs are secreted across the canalicular membrane of hepatocytes via the bile salt export pump (Bsep) into bile for release into the lumen of the duodenum. In intestine, gut microbiota converts primary BAs to secondary BAs via dehydroxylation, deconjugation, and epimerization reactions. The intestinal microbiota contains enzymes that metabolize BAs, such as bile salt hydrolases (Bsh), which remove glycine or taurine from conjugated BAs. The bacteria Clostridia contain hydroxysteroid dehydrogenases and 7-dehydratases that produce BA intermediates and secondary BAs (Ridlon et al., 2006). Ninety-five percent of BAs undergo reabsorption from the intestinal lumen via active transport and recirculate to liver by the portal blood (Chiang, 2003). Upon reaching the liver, BAs are taken up across the sinusoidal membrane of hepatocytes via sodium taurocholate cotransporting polypeptide (Ntcp/Slc10a1) and organic anion-transporting polypeptide 1b2 (Oatp1b2/Slco1b2) transporters (Russell, 2003; Klaassen and Cui, 2015). Accumulation of BAs in hepatocytes or the biliary tract may lead to oxidative stress, inflammation, and cholestatic liver injury (Perez and Briz, 2009).
Clinically, many therapeutic drugs have been shown to produce cholestatic liver injury by elevating hepatic BA levels (Padda et al., 2011). In laboratory animals, activation of the xenobiotic sensor CAR by phenobarbital decreased BAs in mouse liver but increased fecal excretion of muricholic acids (Sberna et al., 2011; Lickteig et al., 2016). Lack of gut microbiota in GF mice resulted in BA concentrations in serum, liver, bile, and ileum higher than in conventional (CV) mice with a microbiome (Selwyn et al., 2015b). To investigate to what extent PBDEs modulate host BA homeostasis and the potential involvement of the “gut-liver axis,” the present study used a multiomics approach to test our working hypothesis that the gut microbiome serves as a critical interface in modulating the crosstalk between PBDEs and BA synthesis and metabolism pathways (Fig. 1).
Fig. 1.
A diagram illustrating the experimental design and dosing regimen of mice. Briefly, 9-week-old C57BL/6 CV and GF mice were exposed to vehicle (corn oil, 10 ml/kg, oral gavage), BDE-47 (100 μmol/kg, oral gavage), or BDE-99 (100 μmol/kg, oral gavage) once daily for 4 consecutive days. Tissues were collected 24 hours after the final dose (n = 3–5 per group). The expression of genes involved in BA synthesis, conjugation, and transport was determined by RNA-Seq and targeted proteomics using LC-MS/MS. The gut microbiota from LIC of CV mice was determined by 16S rRNA sequencing. BAs in serum, liver, SIC, and LIC were determined by targeted metabolomics. A total of 56 BAs were quantified (as listed in Supplemental Table 1), including primary and secondary BAs in either conjugated or unconjugated form.
Materials and Methods
Chemicals.
BDE-47 was purchased from Chem Service, Inc. (part no.: N-10522-10MG, CAS: 5436-43-1; West Chester, PA) (PubChem CID: 95170; link to chemical structure: https://pubchem.ncbi.nlm.nih.gov/compound/95170#section=Top), and BDE-99 (PubChem CID: 36159; link to chemical structure: https://pubchem.ncbi.nlm.nih.gov/compound/36159) was purchased from AccuStandard, Inc. (cat. no.: FF-BDE-099N-80MG; New Haven, CT). Phosphate-buffered saline (PBS, 10×, pH 7.4, PubChem CID: 24978514), liquid chromatography-mass spectrometry (LC-MS)-grade methanol, water, and acetonitrile were purchased from Thermo Fisher Scientific (Grand Island, NY). Bile acid standards and deuterated internal standards (IS) lithocholic acid-2,2,4,4-D4 (LCA-D4) (PubChem CID of LCA: 9903), deoxycholic acid-2,2,4,4-D4 (DCA-D4) (PubChem CID of DCA: 222528), cholic acid-2,2,4,4-D4 (CA-D4) (PubChem CID of CA: 221493), glycochenodeoxycholic acid-2,2,4,4-D4 (GCDCA-D4) (PubChem CID of GCDCA: 12544), and glycocholic acid-2,2,4,4-D4 (GCA-D4) (PubChem CID of GCA: 10140) were purchased from Steraloids, Inc. (Newport, RI). All other chemicals and reagents, unless indicated otherwise, were purchased from MilliporeSigma (St. Louis, MO).
Animals and Procedures.
Eight-week-old male C57BL/6 CV mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and were acclimated to the animal facility at University of Washington for 1 week prior to experiments. The initial breeding colony of GF mice in the C57BL/6 background was established with mice purchased from the National Gnotobiotic Rodent Resource Center (University of North Carolina, Chapel Hill). All mice were housed according to the Association for Assessment and Accreditation of Laboratory Animal Care International guidelines, and the animal studies were approved by the Institutional Animal Care and Use Committee at the University of Washington. The CV and GF mice were exposed to the same diet (laboratory autoclaved rodent diet no. 5010; LabDiet, St. Louis, MO), water (nonacidified autoclaved water), and bedding (autoclaved Enrich-N’Pure; Andersons Lab Bedding, Maumee, OH). All chemical solutions were sterilized using the Steriflip Vacuum-driven Filtration System with a 0.22-μM Millipore Express Plus Membrane (EMD Millipore, Temecula, CA). All gavage needles and syringes were sterilized by autoclave. As described in Fig. 1, at 9 weeks of age, male CV and GF mice were randomly allocated for exposure to vehicle (corn oil, 10 ml/kg), BDE-47 (100 μmol/kg), or BDE-99 (100 μmol/kg) via oral gavage once daily for 4 consecutive days between 8:00 AM and noon (n = 3–5 per group). On the 5th day (24 hours after the final dose), various tissues were collected. Blood was collected via cardiac puncture and centrifuged at 1500g for 10 minutes at 4°C to obtain serum. Livers were removed and immediately frozen in liquid nitrogen. Small and large intestinal contents (SIC and LIC) were flushed using PBS containing 10 mM dithiothreitol (MilliporeSigma) and centrifuged at 20,000g for 30 minutes at 4°C to isolate the solid intestinal content pellet. The intestinal tissues were separated into duodenum, jejunum, ileum, and large intestine (colon and cecum). All samples were stored at −80°C until further analysis.
Quantification of Bacterial DNA and 16S rRNA Gene Sequencing.
Total DNA was extracted from the LIC of CV mice using E.Z.N.A. DNA Stool Kit (Omega Bio-tek, Inc., Norcross, GA) according to the manufacturer’s instructions. The concentration of DNA was determined using a Qubit 2.0 Fluorometer (Life Technologies/Thermo Fisher Scientific, Grand Island, NY), and the integrity and quality of DNA samples were confirmed using an Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA). The V4 region of 16S rRNA gene was amplified and sequenced using a HiSEq. 2500 platform (250-bp paired-end) (BGI Americas, Cambridge, MA) (n = 3 per group). The paired-end sequence reads were merged, demultiplexed, and chimera-filtered using QIIME version 1.9.1 (Quantitative Insights Into Microbial Ecology (Caporaso et al., 2010). Operational-taxonomy-unit clustering and taxonomy classification were performed using open-reference operational taxonomic unit picking method (UCLUST version 1. 2. 22q; The University of Chicago Research Computing Center) against the 99% representative databases for Greengenes (version 13.8; The Greengenes Database Consortium) (DeSantis et al., 2006; Wang et al., 2007; Edgar, 2010). Functional profiles of microbial communities were predicted using PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States; Langille et al., 2013). Differentially enriched pathways were plotted as two-way hierarchical clustering dendrograms using JMP Genomics (SAS Institute, Inc., Cary, NC). Selected differentially regulated bacteria were validated by quantitative polymerase chain reaction (qPCR) using Bio-Rad CFX384 Real-Time PCR Detection System (Hercules, CA). The 16S rRNA primers for the detection of Akkermansia muciniphila, Erysipelotrichaceae Allobaculum spp., and C. scindens were designed on the basis of the 16S rRNA sequences of these bacteria (Supplemental Table 1). The primers recognizing the universal bacterial 16S rRNA sequences were provided by the National Gnotobiotic Rodent Resource Center core facilities, University of North Carolina (Supplemental Table 1) (Packey et al., 2013). All primers were synthesized by Integrated DNA Technologies (Coralville, IA). The abundances of the genomic DNA were expressed as mean delta-delta cycle value (ddCq) of the quantitative PCR compared with the universal bacteria.
Targeted Metabolomics of Bile Acids Using Liquid Chromatography–Tandem Mass Spectrometry.
A standard stock solution of each BA and deuterated IS were prepared at a concentration of 2 mg/ml in methanol. IS (10 μM of LCA-D4, DCA-D4, CA-D4, GCDCA-D4, and GCA-D4) were added to the samples, and BAs were extracted from the serum, liver, SIC, and LIC using methods reported previously (Zhang and Klaassen, 2010). Fifty-six BAs were quantified using liquid chromatography–tandem mass spectrometry (LC-MS/MS) as listed in Supplemental Table 2. Among them, tauro-α muricholic acid (T-αMCA) and T-βMCA were summed together and expressed as T-α/βMCA. The concentrations of individual BAs were analyzed relative to their corresponding internal standards.
For serum BA extraction, 50-μl serum samples were spiked with 25 μl of the mixed internal standard solution. The samples were vortexed and put on ice for 5 minutes. Cold methanol (500 μl) was added for protein precipitation, and samples were vortexed 3 × 10 seconds and maintained on ice for 10 minutes. After centrifugation at 12,000g for 10 minutes at 4°C, supernatants were transferred to clean tubes. The residues were reconstituted in 500 μl methanol, vortexed for 10 minutes, and centrifuged at 12,000g for 10 minutes at 4°C. The two extraction supernatants were pooled and dried under vacuum. The residue was then reconstituted by adding 100 μl methanol solution (methanol/water = 50:50, v/v), and centrifuged at 20,000g for 10 minutes at 4°C. The supernatants were transferred to individual glass vials for MS analysis.
For liver BA extraction, approximately 60 mg frozen liver was accurately weighed and homogenized in 5 volumes of water (300 μl for 60 mg). For each 300-μl of homogenate (50 mg of liver in 250 μl of water), 1.5 ml of acetonitrile (ACN) with 5% ammonia hydroxide (NH4OH) and 25 μl of IS solution was added. Tubes were vortexed and shaken for 1 hour at room temperature. The mixtures were centrifuged at 12,000g for 15 minutes at 4°C, and supernatants were transferred to clean tubes. A second BA extraction was performed with 750 μl methanol, shaken for 20 minutes, sonicated for 5–10 minutes, and centrifuged at 15,000g for 20 minutes. Finally, the two extraction supernatants were pooled and dried under vacuum. The residue was then reconstituted by addition of 100 μl 50% MeOH, vortexed, transferred onto the 0.22-μm Costar Spin-X centrifuge tube filter (MilliporeSigma), and centrifuged at 20,000g for 10 minutes before injection.
For BA extraction from SIC and LIC, approximately 60-mg frozen pellets of the flushed intestinal contents isolated by centrifugation at 20,000g for 30 minutes at 4°C were accurately weighed and homogenized in 5 volumes of water (300 μl). For each 280-μl of homogenate (46.667 mg of liver in 233.333 μl of water), 1.5 ml ACN with 5% NH4OH and 20 μl IS were added. The remaining procedures were same as the liver BA extraction.
For BA measurements, 2 μl of each prepared sample was injected into a LC-MS/MS system (Waters Acquity I-Class UPLC TQS-micro MS; Waters Technologies Corporation, Milford, MA) for analysis using negative ionization mode. The LC-MS/MS system was equipped with an electrospray ionization source. Chromatographic separation was achieved on a Waters XSelect HSS T3 column (2.5 μm, 2.1 × 150 mm). The mobile phase A was 5 mM ammonium acetate in H2O with 0.1% acetic acid, and the mobile phase B was acetonitrile with 0.1% acetic acid. After 1 minute of isocratic elution of 75% of solvent A, the mobile phase composition changed to 5% A at t = 15 minutes. It was then maintained at 5% A for 10 minutes, followed by a rapid increase at t = 25 minutes to 75% A to prepare for the next injection. The total experimental time for each injection was 40 minutes. The flow rate was 0.3 ml/min, the autosampler temperature was 4°C, and column compartment temperature was 40°C. The targeted LC-MS/MS data were acquired using multiple-reaction-monitoring (MRM) mode. The experimental parameters for all 56 BAs and five ISs, such as MRM transitions and retention times, were validated by spiking mixtures of standard compounds into a pooled study sample. TargetLynx software (Waters) was used to integrate extracted MRM peaks. To determine absolute BA concentrations, calibration curves were first constructed for all the BA standards in reference to their corresponding internal standard (Supplementary Table 2). Concentrations of the BAs in the study samples were then calculated from the peak areas and the slopes of the calibration curves. Intersubject differences were normalized by the volume of serum (50 μl) and mass of liver (50 mg) or frozen intestinal pellet (46.667 mg).
Total RNA Isolation.
Total RNA was isolated from liver and intestinal tissue sections using RNA-Bee reagent (Tel-Test Inc., Friendswood, TX). The concentration of total RNA was quantified using NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA) at 260 nm. The quality of RNA was evaluated by formaldehyde-agarose gel electrophoresis with visualization of 18S and 28S rRNA bands under UV light and was confirmed using an Agilent 2100 Bioanalyzer (Agilent Technologies Inc.). Liver samples with RNA integrity number (RIN) above 8.0 were used for RNA sequencing.
Messenger RNA Quantification.
The mRNA of Cyp7a1 (Fig. 9A) as well as the mRNAs of other BA-processing genes in liver were retrieved from our previously published RNA-Seq dataset (Li et al., 2017) and are accessible though the NCBI Gene Expression Omnibus Series accession number GSE101650. Intestinal mRNAs of various BA-processing genes were determined using reverse-transcription qPCR. Briefly, total RNA from intestinal tissue sections was transcribed to cDNA using a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). The cDNAs were amplified by PCR using SsoAdvanced Universal SYBR Green Supermix in a BioRad CFX384 Real-Time PCR Detection System (Bio-Rad). The PCR primers were synthesized by Integrated DNA Technologies and the primer sequences are listed in Supplemental Table 1. The delta-delta cycle values were calculated for each target gene and were normalized to the expression of the housekeeping gene β-actin.
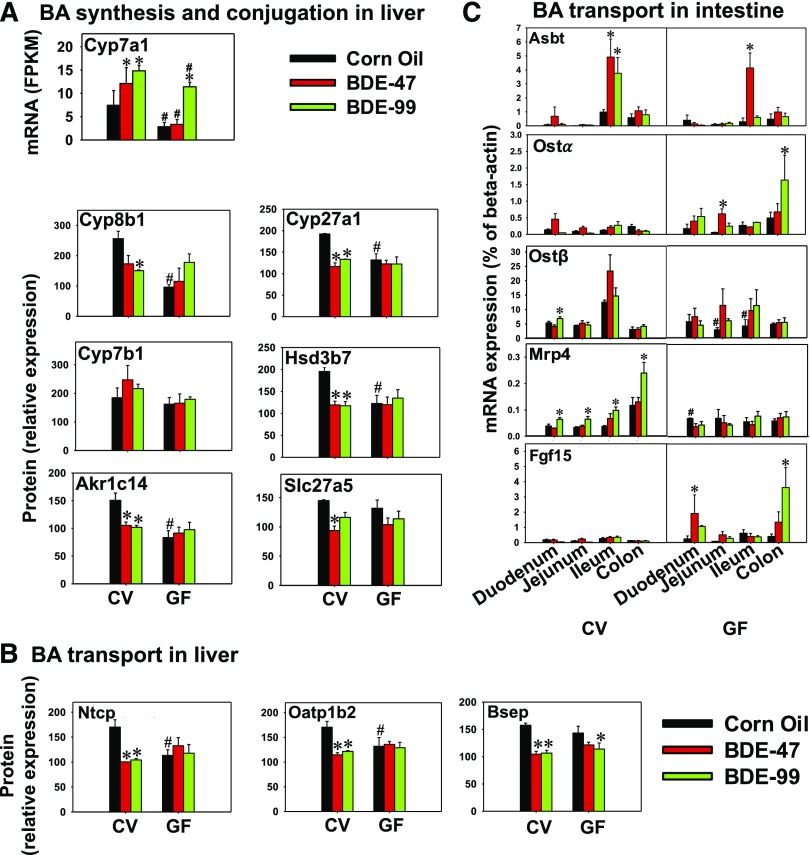
Fig. 9.
Messenger RNA and protein expression of various BA-processing genes in liver and intestine of CV and GF mice exposed to corn oil, BDE-47 (100 μmol/kg), or BDE-99 (100 μmol/kg). (A) Hepatic mRNA of Cyp7a1 and protein expression of other genes involved in BA synthesis (Cyp8b1, Cyp27a1, Cyp7b1, Hsd3b7, and Akr1c14) and conjugation (Slc27a5). Cyp7a1 mRNA expression was substituted for protein expression because the proteomics method cannot accurately detect proteins with an FPKM less than 20. (B) Hepatic protein expression of uptake transporters Ntcp and Oatp1b2, as well as the efflux transporter Bsep. (C) Messenger RNA expression of Asbt, Ostα, Ostβ, Mrp4, and Fgf15 in intestine of CV and GF mice. Asterisks represent statistically significant differences between corn oil- and PBDE-exposed groups within CV or GF mouse colonies. Hash mark (#) represents statistically significant differences between CV and GF mice under the same exposure (P < 0.05).
Targeted Proteomics by LC-MS/MS.
The procedure is similar to a previous publication with a few modifications (Selwyn et al., 2015a; Li et al., 2017). Briefly, the membrane proteins of the mouse livers were isolated using a Pierce Mem-PER Plus Membrane Protein Extraction Kit (Thermo Fisher Scientific). Eighty microliters of the tissue extract (2 mg/ml total protein) was mixed with 10 μl of dithiothreitol (250 mM), 20 μl of 0.2 mg/ml bovine serum albumin, 10 μl of 10 mg/ml human serum albumin, and 40 μl of ammonium bicarbonate buffer (100 mM, pH 7.8), and incubated at 95°C for 5 minutes (denaturation and reduction). Subsequently, 20 μl of iodoacetamide (500 mM) was added and the sample was incubated for 30 minutes at ambient temperature in the dark (alkylation). Ice-cold methanol (0.5 ml), chloroform (0.1 ml), and water (0.4 ml) were added to each sample. After centrifugation at 16,000g for 5 minutes at 4°C, the upper and lower layer were removed, and the pellet was washed with ice-cold methanol (0.5 ml) and centrifuged at 8000g for 5 minutes at 4°C. The pellet was resuspended with 60 μl of 50 mM ammonium bicarbonate buffer. Trypsin (20 μl) was added at a 1:80 trypsin/protein ratio (w/w), and samples were incubated for 16 hours at 37°C. Trypsin digestion was stopped by adding 10 μl of chilled quenching solvent (80% acetonitrile with 0.5% formic acid) followed by 20 μl of heavy peptide internal standard. Samples were centrifuged for 5 minutes at 4000g under 4°C, and supernatant was collected into a LC-MS vial. The stable isotope-labeled heavy peptides were used as internal standards (Thermo Fisher Scientific). LC-MS/MS consisted of an Acquity LC (Waters Technologies Corporation) coupled to an AB Sciex Triple Quadrupole 6500 MS system (Framingham, MA). One to three surrogate peptides per protein were designed for the quantification of selected proteins (Supplemental Table 3) according to a previously published protocol (Prasad and Unadkat, 2014). The peptide separation was achieved on an Acquity UPLC column (HSS T3 1.8 μm, 2.1 × 100 mm; Waters Limited, Hertfordshire, UK). Mobile phase A (water with 0.1% formic acid; v/v) and mobile phase B (ACN with 0.1% formic acid; v/v) were used with a flow rate of 0.3 ml/min in a gradient manner. Peak integration and quantification were performed using Analyst (Version 1.6, Mass Spectrometry Toolkit v3.3; AB Sciex). A robust strategy was used to ensure optimal reproducibility when these proteins were quantified (Bhatt and Prasad, 2017). For example, ion suppression was addressed by using heavy peptide. Bovine serum albumin (BSA) was used as an exogenous internal standard added to each sample to correct for protein loss during processing and for digestion efficiency. In total, three-step data normalization was used: first, average peak areas of light peptides were divided by corresponding average heavy peptide peak areas. This ratio was then divided by BSA light/heavy peak area ratio. Finally, the data were normalized to average quality control values (pooled representative sample). Specific peptides targeting these proteins are listed in Supplemental Table 3. Owing to instrument sensitivity, bile acid synthesis genes with an FPKM (fragments per kilobase of transcript per million mapped reads) for mRNA expression greater than 20 were selected for targeted proteomics.
Statistical Analysis.
Data are presented as mean ± S.E. Statistically significant differences were determined by analysis of variance (ANOVA) followed by Duncan post-hoc test (P < 0.05). Asterisks (*) represent statistically significant differences between corn oil- and PBDE-exposed groups within CV or GF mouse colonies. Hash mark (#) represent significant differences between CV and GF mice under the same exposure. A hierarchical clustering dendrogram (Ward minimum variance method, distance scale) of the differentially enriched pathways was generated by JMP Genomics software (SAS Institute, Inc.). Pearson correlations between BAs and bacterial species were performed with R (corrplot; Wei T and Simko V, 2017, R package “corrplot”: Visualization of a correlation matrix (version 0.84), https://github.com/taiyun/corrplot).
Results
Effect of PBDEs on Gut Microbiome Using 16S rRNA Sequencing.
To determine the effect of oral exposure to PBDEs on gut microbiome, 16S rRNA sequencing targeting V4 region was performed in the LIC of CV mice exposed to corn oil, BDE-47 (100 μmol/kg), or BDE-99 (100 μmol/kg; n = 3 per group). The sequence analysis yielded a total of 104,282 ± 3574 reads per sample (Supplemental Table 4). Rarefaction analysis (QIIME) was performed from 0 to 80,000 reads with an increment of 10,000 reads. The alpha diversity (Chao 1 index), which describes species richness, demonstrated that both BDE-47 and BDE-99 profoundly decreased microbial richness compared with corn oil-exposed control group in the LIC of CV mice (Fig. 2A). The beta diversity, which describes the species differences among different samples, was calculated using weighted UniFrac distance matrix and visualized with EMPeror by principle coordinates analysis (PCoA) (Vazquez-Baeza et al., 2013). As shown in Fig. 2B, there was a clear separation between PBDE-exposed samples and control samples. The BDE-47 exposed samples were clustered separately from BDE-99 exposed samples at PC2 direction in the PCoA plot (Fig. 2B).
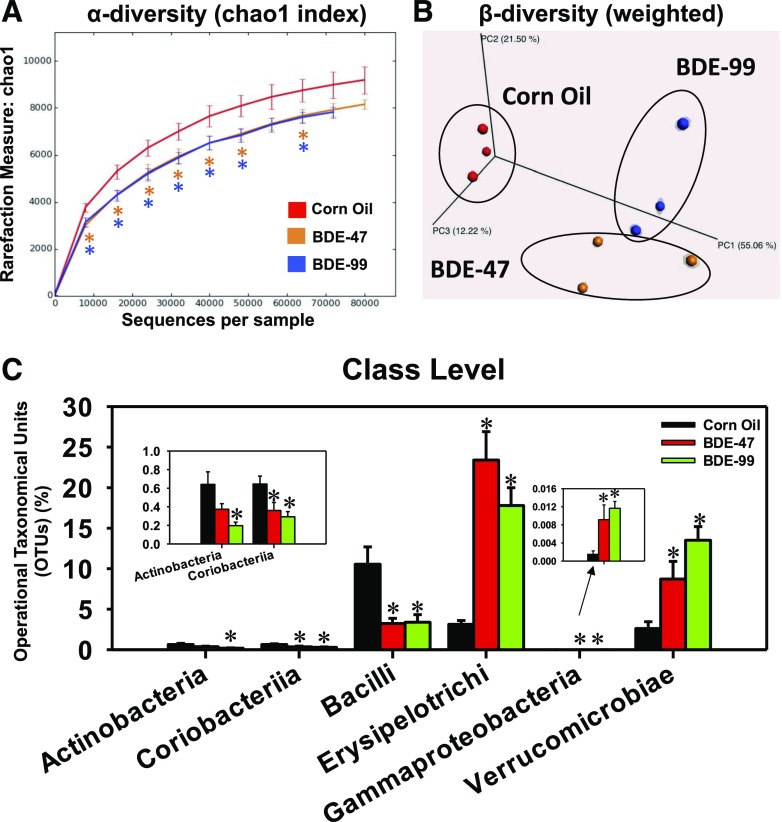
Fig. 2.
Alpha diversity (A) and beta diversity (B) of gut microbiota in LIC of CV mice exposed to corn oil, BDE-47 (100 μmol/kg, oral gavage), or BDE-99 (100 μmol/kg, oral gavage; n = 3 per group). Data were analyzed using QIIME as described in Materials and Methods. Asterisks represent statistically significant differences compared with corn oil-treated group (t test, P < 0.05). (C) The differentially regulated taxa at class level by BDE-47 and BDE-99 compared with corn oil-exposed group. Data were presented as mean percentage of operational taxonomical units (OTUs) ± S.E. Asterisks represent statistically significant differences compared with corn oil-exposed group (ANOVA, Duncan post-hoc test, P < 0.05).
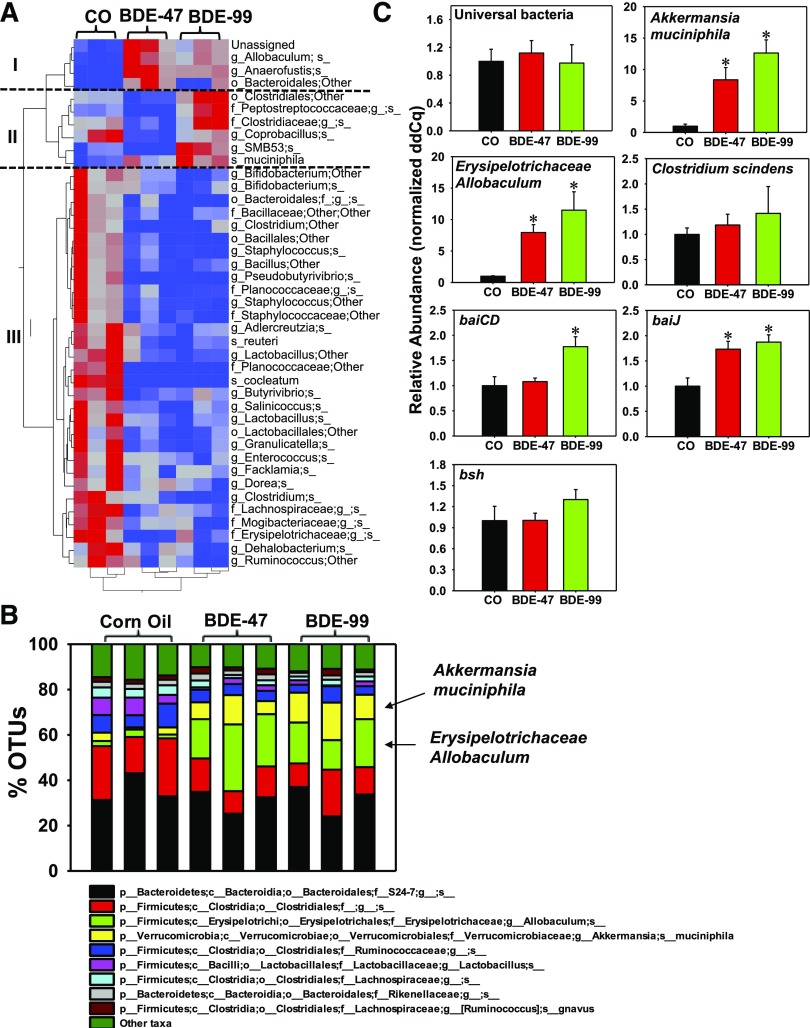
The majority of operational taxonomic units were assigned to the phyla Firmicutes, Bacteroidetes, and Verrucomicrobia (Supplemental Fig. 1); Actinobacteria, Tenericutes, Fusobacteria, Proteobacteria, Acidobacteria, Deferribacteres, Chlorobi, Planctomycetes, Crenarchaeota, Fibrobacteres, Nitrospirae, and Chloroflexi were detected at levels <1%. In addition, 2.5 ± 0.4% of sequences were not assigned to any bacterial phylum (Supplemental Fig. 1). At class level (Fig. 2C), BDE-99 decreased the abundance of Actinobacteria in the Actinobacteria phylum. Coriobacteriia in Actinobacteria phylum and Bacilli in Firmicutes phylum were decreased by both BDE-47 and BDE-99. Conversely, Erysipelotrichia in Firmicutes phylum, Gammaproteobacteria in Proteobacteria phylum, and Verrucomicrobiae in Verrucomicrobia phylum were profoundly increased by both BDE-47 and BDE-99 exposure (Fig. 2C). Analysis conducted at the species level identified 154 bacterial taxa, of which 41 were differentially regulated by PBDEs compared with corn oil-exposed control group (Fig. 3A). These bacterial species partitioned into three distinct patterns: Pattern I includes four taxa that were increased by both BDE-47 and BDE-99; pattern II included six taxa that were decreased by BDE-47 but increased by BDE-99; pattern III included 31 taxa that were consistently decreased by both BDE-47 and BDE-99. The relative abundance of bacterial species was determined across all exposure groups with Fig. 3B showing the top nine most abundant bacterial species and all other summed and labeled as “other taxa” in the 10th category. On average, across all exposure groups, the predominant species were S24-7 in Bacteroidetes phylum and Clostridiales in Firmicutes phylum. Most notably, Akkermansia muciniphila in Verrucomicrobia phylum and Erysipelotrichaceae Allobaculum spp. in Firmicutes phylum were minimally present in corn oil-exposed control group but were markedly increased after PBDE exposure (Fig. 3B). The elevated levels of Akkermansia muciniphila and Erysipelotrichaceae Allobaculum spp. by PBDEs were further confirmed by qPCR (Fig. 3C).
Fig. 3.
Differentially regulated taxa at the species level by PBDEs in LIC of CV mice. (A) A two-way hierarchical clustering dendrogram of the differentially regulated taxa at the species level (41 in total). Data were analyzed using JMP Genomics as described in Materials and Methods. Red represents relatively high abundance and blue represents relatively low abundance. (B) The top 10 most abundant bacterial species that were differentially regulated by PBDEs in LIC of CV mice. The taxa that were lower than the top nine were summed and presented as “other taxa” as the 10th category. (C) qPCR quantification of Akkermansia muciniphila, Erysipelotrichaceae Allobaculum spp., and C. scindens, as well as microbial enzymes involved in secondary BA synthesis (baiJ, baiCD, and bsh) in LIC of CV mice (n = 3 per group). Asterisks represent statistically significant differences as compared with corn oil-exposed group (ANOVA, Duncan post-hoc test, P < 0.05).
To further examine the effect of PBDEs on BA-metabolizing bacteria and microbial enzymes, qPCR was performed on C. scindens, which is a bacterium with known 7α dehydroxylation functions (Ridlon et al., 2006); the BA-inducible operons baiCD and baiJ, which are microbial enzymes involved in BA dehydroxylation (i.e., secondary BA synthesis); as well as bsh, which performs BA deconjugation. Interestingly, although PBDEs did not alter the abundance of C. scindens, baiCD was increased by BDE-99 and baiJ was increased by both BDE-47 and BDE-99. Bsh appeared to be increased by BDE-99 although a statistical significance was not achieved (Fig. 3C). Together these data indicate that bacteria other than C. scindens, which are probably from pattern I and pattern II may be responsible for the upregulation of microbial BA-metabolizing enzymes.
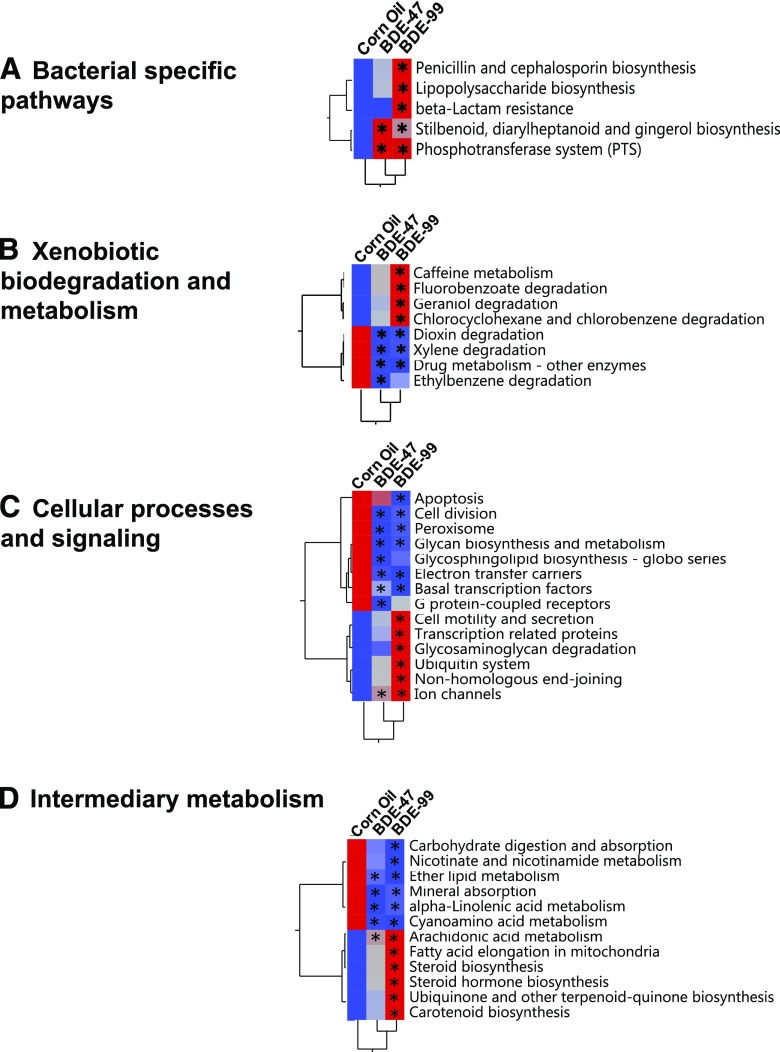
Predicted Functional Composition of Gut Microbiome with PICRUSt.
The metagenomic functional content of gut microbiota was predicted by PICRUSt (Langille et al., 2013) across all three exposure groups. A total of 39 differentially enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways by PBDEs were identified and plotted using the mean value for each exposure (Fig. 4). These pathways were grouped into four categories: bacteria-specific processes (Fig. 4A), xenobiotic biodegradation and metabolism (Fig. 4B), basal cellular processes (Fig. 4C), and intermediary metabolism (Fig. 4D). For bacteria-specific processes, BDE-99 increased the abundance of five KEGG pathways, namely penicillin and cephalosporin biosynthesis; lipopolysaccharide biosynthesis; β-lactam resistance; stilbenoid, diarylheptanoid, and gingerol biosynthesis; and phosphotransferase system (PTS, a bacterial-specific pathway for carbohydrate metabolism). BDE-47 also increased the stilbenoid, diarylheptanoid, and gingerol biosynthesis and the PTS pathways (Fig. 4A). These data indicate that there may be a compensatory mechanism to upregulate these microbial processes to fight against PBDE-induced dysbiosis.
Fig. 4.
Hierarchical clustering dendrograms of differentially regulated KEGG pathways predicted by PICRUSt, as described in Materials and Methods. (A) Bacteria-specific processes. (B) Xenobiotic biodegradation and metabolism. (C) Basal cellular processes. (D) Intermediary metabolism. Data were analyzed using JMP Genomics as described in Materials and Methods. Red represents relatively high expression, and blue represents relatively low expression. Asterisks represent statistically significant differences as compared with corn oil-exposed group (ANOVA, Duncan post-hoc test, P < 0.05).
For xenobiotic biodegradation and metabolism (Fig. 4B), BDE-99 upregulated four KEGG pathways: caffeine metabolism, fluorobenzoate degradation, geraniol degradation, and chlorocyclohexane and chlorobenzene degradation. Conversely, both BDE-47 and BDE99 decreased the KEGG pathways related to dioxin degradation, xylene degradation, drug metabolism, and ethylbenzene degradation. These data suggest that the microbial biotransformation of PBDEs may interact and influence the metabolism of other xenobiotics.
Regarding basal cellular functions (Fig. 4C), BDE-47 increased the KEGG pathway of ion channels but decreased the KEGG pathways involved in cell division, glycan biosynthesis and metabolism, glycosphingolipid biosynthesis, electron transfer carriers, basal transcription factors, and G protein-coupled receptors. BDE-99 increased the KEGG pathways involved in cell motility and secretion, transcription-related proteins, glycosaminoglycan degradation, ubiquitin system, nonhomologous end-joining, and ion channels but decreased the KEGG pathways involved in apoptosis, cell division, glycan biosynthesis and metabolism, electron transfer carriers, and basal transcription factors. These data further suggest that there is a compensatory mechanism of bacterial functions to fight against the PBDE-induced insult on the basal functions of the gut microbiota.
Within intermediary metabolism pathways (Fig. 4D), BDE-47 decreased the KEGG pathways involved in ether lipid metabolism, mineral absorption, α-linolenic acid metabolism, and cyanoamino acid metabolism but increased arachidonic acid metabolism. BDE-99 showed a similar trend in regulating these pathways. In addition, BDE-99 decreased the KEGG pathways involved in carbohydrate digestion and absorption, nicotinate and nicotinamide metabolism but increased arachidonic acid metabolism, fatty acid elongation in mitochondria, steroid and steroid hormone biosynthesis, ubiquinone and other terpenoid-quinone biosynthesis, and carotenoid biosynthesis. Because of the decrease in the anti-inflammatory α-linolenic metabolism pathway and an increase in the proinflammatory arachidonic acid metabolism pathway, these data suggest that PBDEs may lead to a pro-inflammatory stage in the gastrointestinal (GI) tract and perturb the absorption and biotransformation of essential micronutrients.
BAs in Serum, Liver, SIC, and LIC of CV Mice.
To determine how oral exposure to PBDEs affects BA homeostasis, targeted metabolomic analysis of BAs was performed in serum, liver, SIC, and LIC of CV mice exposed to corn oil, BDE-47 (100 μmol/kg), or BDE-99 (100 μmol/kg) (n = 3 per group). Abbreviations for all BAs are in Supplemental Table 2.
We estimated relative S.D. (RSD) values from the quality control samples that were injected multiple times during the experiment runs. For serum samples, the detected BAs had a median RSD of 7.3%, ranging from 0.2% to 16.6% (0.2% for CA and 16.6% for T-UDCA). For liver tissue, the median RSD was 5.9%, ranging from 0.1% to 28.0% (0.1% for MCA and 28.0% for ωMCA). For SIC and LIC samples, the median RSD was 8.7%, ranging from 2.5% to 35.7% (2.5% for G-CDCA and 35.7% for 9(11),(5β)-cholenic acid-3α-ol-12-one). We obtained acceptable recovery rates between 80.0% and 112.1% (80.0% for T-DCA and 112.1% for alloLCA) with a median of 98.1% for detected BAs in all samples. The limit of detection (LOD) is different for each BA. The median LOD of each BA is 4.5 × 10−4 nmol, ranging from 1.1 × 10−4 to 3.2 × 10−3 nmol (1.1 × 10−4 nmol for T-UDCA and 3.2 × 10−3 nmol for HDCA).
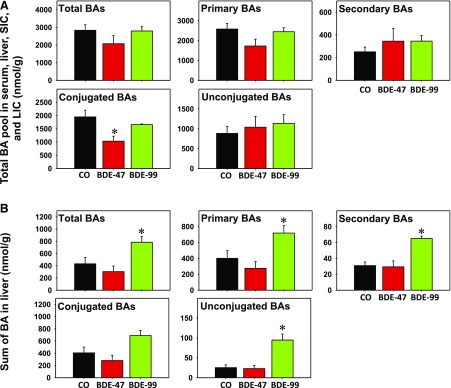
To evaluate the overall changes in the regulation of BA pool, the sum of all BAs for each compartment (serum, liver, SIC, and LIC) were summed and categorized by group (Fig. 5A). Overall, total BAs, primary BAs, secondary BAs, and unconjugated BAs did not change owing to BDE-99 or BDE-47. Conjugated BAs were decreased by BDE-47 exposure but remained unchanged by BDE-99 exposure. Conversely, in the liver, BDE-99 exposure increased total BAs, primary BAs, secondary BAs, and unconjugated BAs but conjugated BAs were not altered by BDE-99 or BDE-47 (Fig. 5B).
Fig. 5.
Evaluation of the overall changes by BDE-99 or BDE-47 in the regulation of the BA pool. Sum of all BAs for serum, liver, SIC, and LIC (A) and liver (B).
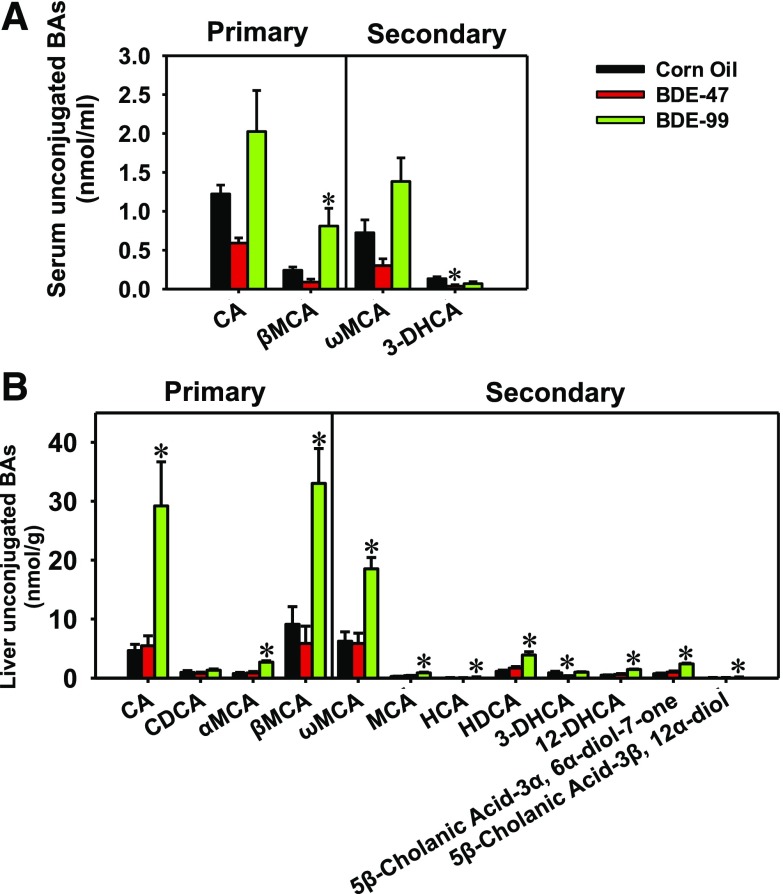
In serum, 12 BAs were detectable in CV mice, including four unconjugated BAs (Fig. 6A) and eight taurine (T)- or glycine (G)-conjugated BAs (Supplemental Fig. 2A). In general, PBDEs did not markedly alter the primary or secondary conjugated BAs in serum, except for a moderate decrease in G-CA by BDE-99 and an apparent decrease in T-α/βMCA by BDE-47 (Supplemental Fig. 2A). Regarding the unconjugated BAs in serum, BDE-99 increased the primary BA βMCA and also tended to increase the primary BA CA and the secondary BA ωMCA; conversely, BDE-47 decreased the secondary BA 3-DHCA and tended to decrease the primary BAs, CA and βMCA, and the secondary BA, ωMCA.
Fig. 6.
Concentrations of unconjugated primary and secondary BAs in serum (A) and liver (B) of CV mice exposed to corn oil, BDE-47 (100 μmol/kg), or BDE-99 (100 μmol/kg). BAs were quantified using LC-MS/MS as described in Materials and Methods. Data were presented as mean ± S.E. (n = 3 per group). Asterisks represent statistically significant differences compared with corn oil-exposed group by ANOVA (Duncan post-hoc test, P < 0.05).
In the liver, 23 BAs were detected in CV mice, including 12 unconjugated BAs (Fig. 6B) and 11 T- or G-conjugated BAs (Supplemental Fig. 2B). In general, PBDEs had less effect on the conjugated BA profiles, except for an increase in the primary BA G-CA and the secondary BA T-DCA by BDE-99. In contrast, BDE-99 markedly increased most unconjugated primary BAs, including CA, αMCA, and βMCA, as well as most unconjugated secondary BAs, including ωMCA, MCA, HDCA, 12-DHCA, 5β-cholanic acid-3α, 6α-diol-7-one, and 5 β-cholanic acid-3β, 12α-diol (Fig. 6B).
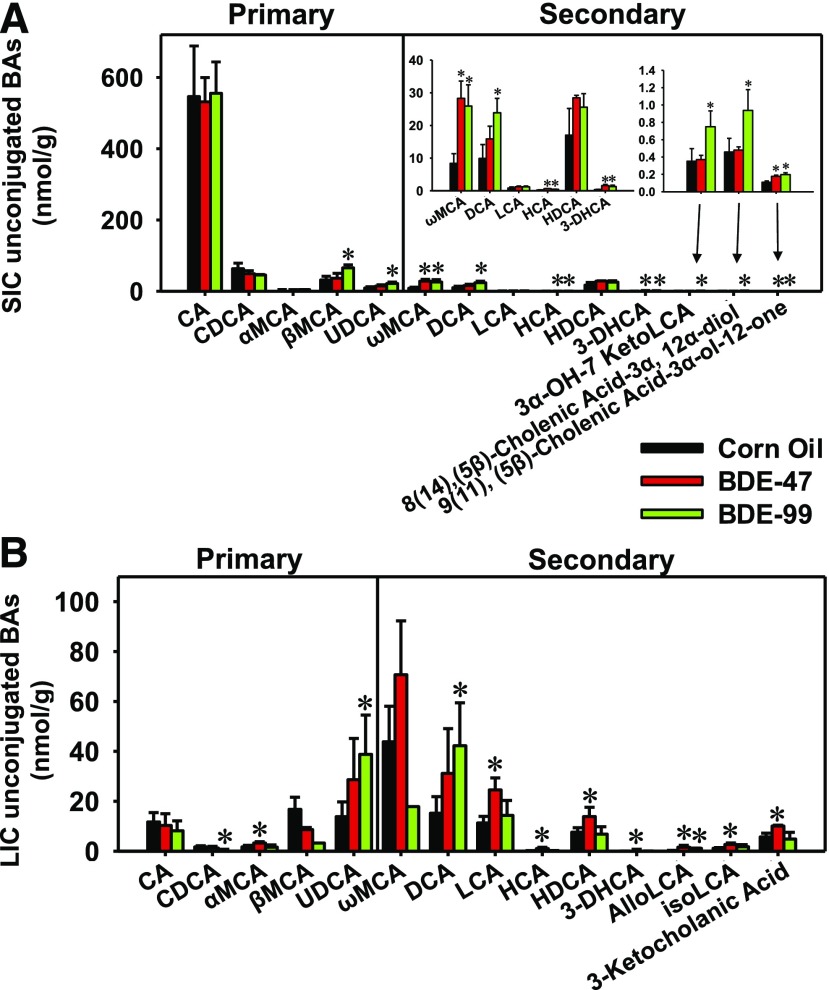
In the SIC, 24 BAs were detected in CV mice, including 14 unconjugated BAs (Fig. 7A) and 10 T- or G-conjugated BAs (Supplemental Fig. 3A). For conjugated BAs, PBDEs in general decreased many primary and secondary BAs in SIC: BDE-47 and BDE-99 decreased T-UDCA, G-CA, T-LCA, and T-HDCA. BDE-47 also decreased T-α/βMCA, whereas BDE-99 decreased T-HCA. Conversely, BDE-99 increased unconjugated primary BAs, such as the primary BAs βMCA and UDCA, and most unconjugated secondary BAs, including ωMCA, DCA, HCA, 3-DHCA, 3α-OH-12 ketolithocholic acid, 8(14), (5β)-cholenic acid-3α, 12α-diol, and 9(11), (5β)-cholenic acid-3α-ol-12-one. BDE-47 also increased several unconjugated secondary BAs, including ωMCA, HCA, 3 DHCA, and 9(11), (5β)-cholenic acid-3α-ol-12-one (Fig. 7A).
Fig. 7.
Concentrations of unconjugated primary and secondary BAs in SIC (A) and LIC (B) of CV mice exposed to corn oil, BDE-47(100 μmol/kg), or BDE-99 (100 μmol/kg). BAs were quantified using LC-MS/MS as described in Materials and Methods. Data were presented as mean ± S.E. (n = 3 per group). Asterisks represent statistically significant differences compared with corn oil-exposed group by ANOVA (Duncan post-hoc test, P < 0.05).
In the LIC, 25 BAs were detected in CV mice, including 14 unconjugated BAs (Fig. 7B) and 11 T- or G-conjugated BAs (Supplemental Fig. 3B). For conjugated BAs, BDE-47 increased the primary BA T-α/βMCA and the secondary BA T-DCA, whereas BDE-99 increased the primary BA G-CDCA and the secondary BA T-HDCA (Supplemental Fig. 3B). Among unconjugated BAs, BDE-47 increased the primary BA αMCA, as well as the secondary BAs LCA, HCA, HDCA, 3-DHCA, alloLCA, isoLCA, and 3-ketocholanic acid. BDE-99 decreased the primary BA CDCA, but increased UDCA, as well as increased the secondary BAs DCA and alloLCA (Fig. 7B).
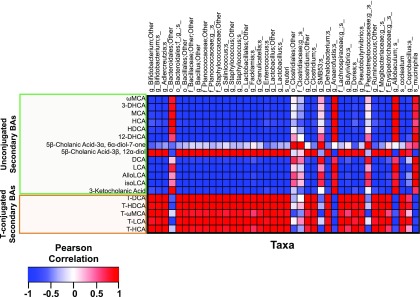
Pearson Correlation between Secondary BAs and Bacterial Species.
Because secondary BAs were exclusively produced by gut microbiota, to determine which bacterial species may contribute to altered total secondary BA pool in CV mice, individual secondary BAs from all four compartments of CV mice were summed and then correlated with 41 bacterial species that were differentially regulated by PBDEs (Fig. 8). Both unconjugated secondary BAs (which were exclusively produced by gut microbiota) and conjugated secondary BAs (which were produced by bacteria and then conjugated in liver) were considered. A Pearson correlation analysis revealed that 32 out of 41 bacterial species were positively correlated with taurine-conjugated secondary BAs (including T-ωMCA, T-HCA, T-HDCA, and T-LCA) but negatively correlated with many unconjugated secondary BAs. Conversely, four taxa, namely Bacteroidales other, Anaerofustis, Allobaculum spp., and A. muciniphila, were in general negatively correlated with conjugated secondary BAs but in general positively correlated with unconjugated secondary BAs, suggesting that these taxa may directly or indirectly contribute to BA deconjugation.
Fig. 8.
Pearson correlation between secondary BAs (including both taurine-conjugated and unconjugated BAs) and bacterial species that were differentially regulated by PBDE exposure (n = 3). Individual secondary BAs from all four compartments of CV mice were summed and used as an input (unit: nmol/ml). Each column represents one bacterial species (unit: operational taxonomical unit). Red represents positively correlated, and blue represents negatively correlated.
Comparison of PBDE-Mediated Changes in BA Compositions in Serum, Liver, SIC, and LIC between CV and GF Mice.
To determine the BA compositional changes caused by PBDEs and the necessity of gut microbiota in modulating the basal and PBDE-regulated BA profiles, the proportions of individual BAs in serum, liver, SIC, and LIC of CV and GF mice were analyzed across all three exposure groups (Supplemental Figs. 4–7).
In serum (Supplemental Fig. 4), compared with corn oil-exposed CV (CV CO) mice with more diversified BA profiles, the predominant BAs in corn oil-exposed GF (GF CO) mice were the primary BAs T-α/βMCA and T-CA, whereas other BAs were minimally detected. Following PBDE exposure, BDE-99 had a more prominent effect on BA compositional changes than BDE-47 in CV mice, namely, an apparent increase in the proportion of major primary conjugated BA T-α/βMCA. In PBDE-exposed GF mice, T-α/βMCA and T-CA were still predominant in serum BA profiles. In particular, BDE-47 exposure led to an apparent increase in the proportion of T-α/βMCA, whereas BDE-99 exposure led to an apparent increase in the proportions of T-HDCA and T-UDCA.
In liver (Supplemental Fig. 5), under CV control conditions, T-α/βMCA and T-CA were the predominant BAs detected, whereas lack of gut microbiota increased the proportion of T-α/βMCA but decreased the proportion of T-CA; in addition, there was an increase in the proportion of hepatic βMCA in control GF mice. Following PBDE exposure in CV mice, both BDE-47 and BDE-99 increased the proportions of T-CA and doubled the proportions of the major secondary BAs T-DCA and ωMCA. BDE-99 also doubled the proportions of the primary unconjugated BAs βMCA and CA in livers of CV mice. In livers of GF mice, PBDEs had less effect, except for an apparent decrease in the proportions of T-α/βMCA by BDE-99.
In SIC (Supplemental Fig. 6), in general, BA profiles were more diverse in CV mice than in GF mice. Under CV control conditions, CA in its unconjugated form became a major BA in this compartment, in addition to other major BAs, including T-α/βMCA and T-CA, suggesting extensive deconjugation reactions converting T-CA to CA in the presence of gut microbiota. In contrast, in control GF mice, the predominant BAs were T-α/βMCA and T-CA, whereas other BAs were minimal. Following PBDE exposure, the proportions of BAs remained relatively constant in GF mice. Conversely, in CV mice, there was a marked increase in the proportion of CA (from 25% in controls to 42% by BDE-47 and to 35% by BDE-99), and correspondingly, the proportion of the primary BA T-CA was reduced by BDE-99 in CV mice. In addition, the proportion of T-α/βMCA was reduced by both BDE-47 and BDE-99. These data suggested that PBDEs further enhanced the BA deconjugation reactions in a microbiota-dependent manner.
In LIC (Supplemental Fig. 7), under CV control conditions, the BA profiles were more diverse than those in GF mice, and the most predominant BAs were T-α/βMCA and ωMCA. In GF mice, the predominant LIC BAs were T-α/βMCA followed by T-CA, with all other BA levels minimal. Following PBDE exposure, BAs in GF mice remained relatively constant; in contrast, in CV mice, both PBDEs increased the proportion of T-α/βMCA but decreased the proportions of ωMCA and βMCA. BDE-99 exposure also led to an apparent increase in the proportions of the secondary BAs DCA and T-HDCA, as well as the primary BA UDCA.
Host BA-Processing Gene Expression in Livers of CV and GF Mice.
To determine to what extent BA-metabolizing enzymes and transporters are affected by PBDEs and gut microbiota interactions, the expression of major BA-processing genes involved in BA biosynthesis (Cyp8b1, Cyp27a1, Cyp7b1, Hsd3b7, Akr1c14), conjugation [Slc27a5 (also known as bile acid ligase)], and transport (Ntcp, Oatp1b2, Bsep) were quantified using targeted proteomics; Cyp7a1 was below the detection limit, and therefore the mRNA level is shown.
Cyp7a1 and Cyp8b1 are involved in the classic pathway of BA synthesis. Under control conditions, the mRNA of the rate-limiting enzyme Cyp7a1 was higher in CV mice than GF mice (Fig. 9A). Following PBDE exposure, Cyp7a1 mRNA was increased by BDE-47 and BDE-99 in CV mice. However, in GF mice, Cyp7a1 mRNA was not altered by BDE-47 but increased 4.2-fold by BDE-99, suggesting that the BDE-47-mediated increase in Cyp7a1 mRNA was dependent on gut microbiota. Cyp8b1 protein was lower in GF mice than CV mice under control conditions, whereas BDE-99 decreased Cyp8b1 protein expression in CV mice, and the PBDE-mediated decrease in Cyp8b1 was completely abolished in livers of GF mice. Cyp27a1 and Cyp7b1 are key enzymes in the alternative pathway of BA synthesis. The Cyp27a1 protein was lower in control GF mice compared with control CV mice, whereas both BDE-47 and BDE-99 decreased Cyp27a1 protein in livers of CV mice but not in GF mice, suggesting that the downregulation of Cyp27a1 protein by PBDEs is gut microbiota-dependent. The Cyp7b1 protein was not altered by either PBDEs or gut microbiota. The proteins of the downstream BA-synthetic enzymes, namely Hsd3b7, which catalyzes the inversion of 3β-hydroxyl group of cholesterol to the 3α-hydroxyl group of bile acids, and Akr1c14 (aldo-keto reductase) were also determined. Both enzymes were lower in control GF mice compared with control CV mice and were decreased by BDE-47 and BDE-99 in CV mice. This downregulation in their protein expression was absent in livers of GF mice. Regarding BA conjugation, Slc27a5 is a bile acid-CoA ligase that mediates the conjugation of primary BAs with glycine or taurine before excretion into bile canaliculi. The Slc27a5 protein was decreased by BDE-47 in CV mice but not in GF mice.
Regarding BA transporters, the proteins of the major basolateral uptake transporter for conjugated BAs, namely sodium taurocholate cotransporting polypeptide (Ntcp/Slc10a1), as well as the major basolateral uptake transporter for unconjugated BAs, namely organic anion-transporting polypeptide (Oatp/Slco) 1b2, were both lower in control GF mice than those in control CV mice. Both BDE-47 and BDE-99 decreased the proteins of Ntcp and Oatp1b2 in a gut microbiota-dependent manner. The protein of the rate-limiting canalicular BA efflux transporter Bsep was similar between livers of CV and GF mice under control conditions, whereas both BDE-47 and BDE-99 decreased its expression in livers of CV mice. This downregulation was dampened by the lack of gut microbiota, since there was no change in Bsep protein expression by BDE-47 and decrease by BDE-99 in livers of GF mice (Fig. 9B).
The mRNA expression of several other genes involved in BA homeostasis was also quantified in livers of CV and GF mice, including the BA-synthetic enzymes Cyp39a1 and Akr1d1, the BA-conjugation enzyme Baat (bile acid CoA/amino acid N-acyltransferase), the basolateral BA-efflux transporters Ostα (Slc51a) and Ostβ (Slc51b), the cholesterol efflux transporter Abca1, the lipid uptake transporter Atp8b1, the BA-efflux transporter Asbt (Slc10a2), as well as various transcription factors: Nr1h4 (or FXR), Nr0b2 (or Shp), Mafg (musculoaponeurotic fibrosarcoma oncogene homolog G), Crip2 (cysteine-rich intestinal protein 2), Zfp385a (Zinc-finger protein 385a), Nr1h2 (or LXRβ), and Nr1h3 (or LXRα) (Supplemental Fig. 4). The mRNAs of Abcb4, Abcg5, and Abcg8 were not altered by lack of gut microbiota or PBDEs (data not shown). Under control conditions, lack of gut microbiome moderately decreased Cyp39a1 mRNA. Following PBDE exposure, many of the mRNAs in CV mice were not affected. However, in GF mice, BDE-47 increased the mRNAs of Cyp39a1 and Slc51b, and BDE-99 increased Akr1d1, Slc51b, Abca1, Atp8b1, Slc10a2, and Mafg. Conversely, the mRNAs of the majority of transcription factors were decreased by BDE-99 in GF mice, including Nr0b2, Crip2, Zfp385a, Nr1h2, and Nr1h3 (Supplemental Fig. 8).
BA-Transporters and Fgf15 Gene Expression in Various Parts of the Intestine.
The mRNA expression of major intestinal transporters involved in enterohepatic circulation of BAs was determined in four intestinal sections of CV and GF mice, including the uptake transporter Asbt and efflux transporters Ostα, Ostβ, and Mrp4. The intestinal hormone Fgf15, which regulates the hepatic BA-synthesis, was also determined (Fig. 9C).
Apical sodium-bile acid transporter (Asbt) brings BAs from the intestinal lumen into enterocytes. In ileum, where the peak basal expression of Asbt was observed, both BDE-47 and BDE-99 markedly increased Asbt mRNA in CV mice; however, in GF mice, only BDE-47 increased Asbt mRNA. Asbt mRNA was not affected by either lack of gut microbiota or PBDE exposure in other sections of intestine. The basolateral BA efflux transporters Ostα and Ostβ transport the reabsorbed BAs from enterocytes to portal blood. The mRNA expression of Ostα was not affected by lack of gut microbiota under control conditions, and PBDE exposure did not affect Ostα in CV mice. In GF mice, BDE-47 increased Ostα mRNA in jejunum, and BDE-99 increased Ostα mRNA in colon. The basal mRNA expression of Ostβ was lower in jejunum and ileum of GF mice. In CV mice, BDE-99 moderately increased Ostβ in duodenum. For the basolateral efflux transporter Mrp4, its basal expression in duodenum was higher in GF mice, and interestingly Mrp4 mRNA was consistently upregulated by BDE-99 in all four sections of intestine of CV mice in a gut microbiota-dependent manner. The mRNA of the intestinal hormone Fgf15 was not altered by lack of gut microbiota under control conditions and remained unchanged following PBDE exposure in CV mice. However, in GF mice, BDE-47 increased Fgf15 mRNA in duodenum and BDE-99 increased Fgf15 mRNA in colon.
Discussion
The present study provides the first evidence of the effect of PBDEs on gut microbiota and BA homeostasis. Specifically, PBDEs decreased the bacterial richness and differentially regulated various bacterial species in the large intestine of mice. Most notably, A. muciniphila and Erysipelotrichaceae Allobaculum spp., which have been shown to have antiobesogenic effects, were overwhelmingly increased by PBDEs, suggesting that there may be a novel interaction between environmental PBDEs and nutrition. Regarding BA profiles, PBDEs, in particular BDE-99, increased the unconjugated BAs in serum, liver, SIC, and LIC of CV mice but not in GF mice. The lack of gut microbiome was associated with a shift in BA composition toward an increase in conjugated muricholic acids in all four compartments of GF mice.
As the human intestinal microbiota is dominated strictly by anaerobic bacteria, there is limited information regarding how GI bacteria respond to PBDE exposure. The present study provided the first evidence of the effect of PBDEs on bacterial community in the GI tract. At class level, we found oral exposure to BDE-47 or BDE-99 decreased Actinobacteria, Coriobacteriia, and Bacilli, but increased Erysipelotrichia, Gammaproteobacteria, and Verrucomicrobiae in LIC of CV mice. The elevated abundances of Erysipelotrichia and Gammaproteobacteria in the present study is similar to previous findings from sediment samples. Using an in vitro culture approach, previous studies have reported positive correlation of five bacterial classes (Clostridia, Bacilli, Betaproteobacteria, Alphaproteobacteria, and Gammaproteobacteria) with anaerobic degradation of BDE-209 and BDE-28 in the sediment (Robrock et al., 2008; Yang et al., 2017). Another study also demonstrated a positive relationship between bacteria (Actinobacteria and Erysipelotrichia) and biodegradation of lesser-brominated PBDE congeners, tri-BDE, and tetra-BDE (Xu et al., 2012). Therefore, the increased Erysipelotrichia and Gammaproteobacteria following PBDE exposure suggests that these two bacterial classes may be involved in debromination of BDE-47 and BDE-99 in vivo. Although Bacilli and Actinobacteria have been reported to be positively correlated with degradation of PBDEs (BDE-209 and BDE-28) in sediment, we observed decreased abundances of Bacilli, Actinobacteria, and Coriobacteriia (which is also from Actinobacteria phylum) by PBDEs. It is possible that these bacterial classes are more sensitive and susceptible to BDE-47 and BDE-99-mediated insults, since tetra- and penta-BDEs are considered to be more toxic and bioaccumulative than higher brominated congeners such as BDE-209 (Siddiqi et al., 2003).
The present study provides the first evidence linking A. muciniphila and Erysipelotrichaceae Allobaculum spp. with exposure to PBDEs. A. muciniphila is a mucin-degrading bacteria that resides in the mucus layer of the human intestinal tract and helps to maintain the gut barrier and can be found in the colon of healthy human subjects at 1%–4% of the bacterial population (van Passel et al., 2011; Zhao et al., 2017). Studies have shown that A. muciniphila may have anti-inflammatory effects in humans, and inverse relationships have been observed between A. muciniphila colonization and inflammatory conditions, such as irritable bowel syndrome (Png et al., 2010). Reduced levels of A. muciniphila have also been found in patients with metabolic disorders such as diabetes and obesity, suggesting A. muciniphila may also have antiobesogenic properties (Derrien et al., 2017). Likewise, Allobaculum spp. have also been shown to have antiobesogenic effects. Lower abundances of Allobaculum spp. were found in mice fed with high-fat diet compared with mice fed with control diet, and Allobaculum spp. were enriched in weight-reduced mice (Ravussin et al., 2012). Moreover, both A. muciniphila and Allobaculum spp. were negatively correlated with circulating leptin concentrations in obese mice (Ravussin et al., 2012). The mechanisms underlying the effects of A. muciniphila and Allobaculum spp. in inflammation and metabolic disorders are not understood, and no published studies have linked these two bacterial species with environmental chemical exposure. The involvement of PBDEs in modulating the nutrition response by altering these two bacteria is worth investigating in future studies.
In the present study, the bacterial species that were differentially regulated by PBDEs have been linked with multiple pathways: bacteria-specific processes, basal cellular processes, xenobiotic metabolism, and intermediary metabolism (Fig. 4). The increased bacteria-specific processes by BDE-47 and BDE-99 suggest that there may be a compensatory mechanism to upregulate these microbial processes to fight against PBDE-induced dysbiosis. Conversely, many xenobiotic metabolism-related pathways were decreased by both BDE-47 and BDE99, suggesting that the microbial biotransformation of PBDEs may affect the metabolism of other xenobiotics. In addition, predictive analysis by PICRUSt showed that BDE-99 may increase cell motility and secretion, transcription-related proteins, glycosaminoglycan degradation, ubiquitin system, nonhomologous end-joining, and ion channels. These data further suggested that there is a compensatory mechanism of bacterial functions to fight against the PBDE-induced insult on the basal functions of the gut microbiota. Regarding intermediary metabolism, both BDE-47 and BDE-99 decreased ether lipid metabolism, mineral absorption, α-linolenic acid metabolism (which is anti-inflammatory), and cyanoamino acid metabolism; whereas BDE-99 markedly increased arachidonic acid metabolism, fatty acid elongation in mitochondria, steroid and steroid hormone biosynthesis, ubiquinone and other terpenoid-quinone biosynthesis, and carotenoid biosynthesis. These data suggest that PBDEs may lead to a proinflammatory stage in the GI tract and perturb the absorption and biotransformation of essential micronutrients. The BDE-99-mediated increase in steroid/steroid hormone synthesis also coincides with increased microbial biotransformation of BAs (Fig. 3C), which are part of steroid hormone signaling.
The increased unconjugated secondary BAs by PBDEs correlated with the increased abundance of bacteria (Clostridiales and Clostridiaceae) and the increased expression of microbial enzymes (baiCD and baiJ) that are involved in BA metabolism (Fig. 3, A and C). The microbial enzymes baiCD and baiJ convert primary BAs to secondary BAs via dihydroxylation and Bsh removes glycine or taurine conjugates via deconjugation. Bsh activity is widely present in bacteria in the large and small intestines, including Clostridium, Enterococcus, Bifidobacterium, Lactobacillus, and Bacteroides (Ridlon et al., 2006). In the present study, we found a slight increase of Bsh following BDE-99 exposure, although a statistical significance was not achieved. In addition to the bacterial species that are mentioned above, there are also many other bacterial species involved in BA metabolism, including genera Bacteroides, Clostridium, Lactobacillus, Bifidobacterium, Enbacterium, and Escherichia (Ridlon et al., 2006). Therefore, future studies discovering novel bacteria and microbial enzymes involved in BA deconjugation and dehydroxylation will help us further understand the connection between the gut microbiome and BA homeostasis.
The BA profiles of GF mice differ markedly from those of CV mice under both control and PBDE exposed conditions. As expected, GF mice have increased conjugated BAs and decreased secondary BAs. Under control condition, the proportion of T-α/βMCA is much higher in GF mice than in CV mice in all four compartments (Supplemental Figs. 4–7). These findings are consistent with previous studies (Sayin et al., 2013; Hu et al., 2014; Miyata et al., 2015; Selwyn et al., 2015b). T-CA was increased in all compartments, except liver. In contrast, the unconjugated BAs were decreased in all four compartments of GF mice compared with those of CV mice.
Many enzymes and transporters that are involved in BA synthesis, conjugation, and transport were decreased by PBDEs, in a gut microbiome-dependent manner. It is well known that BAs can regulate their own synthesis by activating farnesoid X receptor (FXR) in liver and in intestine. In the liver, activation of FXR induces the expression of Shp, which subsequently decreases the BA-processing genes for BA synthesis. However, the hepatic mRNA expression of Shp was not induced by PBDEs in the present study. In the ileum, activation of FXR by BAs induces the secretion of Fgf15, which could also decrease the transcription of BA-synthesis genes such as Cyp7a1 (Chiang, 2004). However, we did not observe increased Fgf15 in the ileum of CV mice, suggesting that mechanisms other than FXR signaling are responsible for the reduced expression of BA-processing genes in liver. Since BDE-47 and BDE-99 are known activators of human and mouse PXR, and activation of human PXR by its prototypical ligand rifampicin has been shown to reduce hepatic BA in cholestatic patients (Geenes et al., 2015), it is possible that PBDE exposure may affect PXR signaling to modulate the BA pathway in CV mice with a minor effect from FXR activation.
In the present study, we found that the mRNA of Asbt was upregulated by both BDE-47 and BDE-99 in the ileum. The ileal BA uptake transporter Asbt is responsible for the reabsorption of most of the BAs from the intestine. Combined with the decreased total BAs in SIC and increased total BAs in serum and liver following BDE-99 exposure (Supplemental Table 5), these results suggest that there is an increase in the reabsorption of BAs. In addition, the mRNA of basolateral efflux transporter Mrp4, which actively transports BAs from enterocytes to portal circulation, was also increased by BDE-99 in all four sections of intestine in CV mice (Fig. 9C). This also correlated with the increased total BAs in serum and liver and suggests more BAs are retrieved back from the gut.
In the present study, we dosed CV and GF mice with 100 μmol/kg of PBDEs (which is 48.5 mg/kg body weight for BDE-47 and 56.5 mg/kg body weight for BDE-99) for four consecutive days in an exposure scheme similar that used to induce P450s in adult mice (Pacyniak et al., 2007). Another study showed that a single oral exposure of BDE-99 at 20 mg/kg resulted in serum concentrations of 3.6 pg/g serum 2 days later (Lundgren et al., 2013). These acute doses are higher than environmental exposure to humans. In the United States, human serum concentrations for BDE-47 ranged from 3.9 to 1000 pg/g serum, and the concentration range for BDE-99 was 1.3–290 pg/g serum (Makey et al., 2016). Therefore, the circulating levels of PBDEs in this study were at concentrations similar to that in humans.
The vehicle used in the present study was corn oil, which contains a variety of saturated, monounsaturated, and polyunsaturated fatty acids, and is especially high in linoleic acid (42%–56%). This was consistent with the observation that linoleic acid metabolism pathway was high in differentially regulated microbial taxa from large intestinal content of vehicle-exposed CV mice (Fig. 4D), indicating that this was probably owing to vehicle-effect. For the present study, we did not quantify the bacterial profiles between vehicle and naïve (unexposed) mice because the goal of the study was to determine the effect of PBDEs, which were dissolved in corn oil, and the effect of corn oil itself was expected to be similar between the vehicle and PBDE-exposed groups. In the literature, it has been demonstrated that certain fatty acids are associated with dysbiosis (Alcock and Lin, 2015), and certain fatty acids themselves can also regulate bile acid metabolism by altering the serum cholesterol levels (Liu et al., 2017). Therefore, results from the present study should be interpreted as the effects from a combination of PBDEs and corn oil.
Prior to our work, there were no studies in the literature investigating the association between PBDE exposure and perturbation of the gut microbiome. Our study is the first showing that PBDEs can cause dysbiosis in the large intestinal content of mice, and this also leads to disrupted BA metabolism. Previous studies on PBDEs have focused primarily on the host effect, such as thyroid toxicity, neurodevelopmental disorders, and oxidative stress. However, considering that the gut microbiome conducts the majority of the reduction reactions of the body, and also contributes to deconjugation reactions (such as reversing the glucuronidation of toxic chemicals including PBDEs), it is critical to unveil such novel interactions between PBDE exposure and gut microbiome, so as to take a first step in further understanding idiosyncratic adverse reactions to chemicals in humans. One particular example is that we identified that PBDEs preferably increased unconjugated BAs in multiple biologic compartments in mice (Figs. 6 and 7). Especially in small and large intestinal contents, there were increases in multiple unconjugated secondary BAs (Fig. 7), which are positively associated with colon cancer (Imray et al., 1992). Because secondary BAs can be carcinogens (Ajouz et al., 2014), it is possible that people at a higher risk of PBDE exposure, in particular BDE-47 exposure (Fig. 7B), may also have increased risks of developing colon cancer. Indeed, brominated compounds tend to induce colon cancer in rodents (Dunnick et al., 1997), and transcript patterns following PBDE exposure in rodents is similar to human colon cancer gene expression profiles (Dunnick et al., 2012). More studies are needed to understand the potential PBDE-induced colon cancer mediated by unconjugated BAs. In addition, it has been shown in mice that unconjugated BAs are substrates for the major basolateral xenobiotic uptake transporter Oatp1b2, which is the mouse ortholog to human OATP1B1/1B3 (Csanaky et al., 2011). Genetic polymorphisms of OATP1B1/1B3 are implicated in altered pharmacokinetics of many clinical drugs such as statins and the subsequent adverse drug reactions (Akaike et al., 1991). Because PBDEs increased the endogenous Oatp1b2/OATP1B1/1B3 substrates (i.e., the unconjugated BAs), it is possible that PBDEs may interfere with the hepatic uptake of many drugs that are OATP substrates. Further studies are necessary to determine the potential inhibitory effects of PBDE-induced increases in OATP-substrate BAs that may alter the disposition and efficacy of drugs. In summary, in addition to their well characterized toxicities, PBDEs may produce additional adverse effects on human health by interfering with gut microbiome and bile acid metabolism, as well as competing with hepatic uptake of other chemicals.
In general, unconjugated BAs are thought to be more toxic than their conjugated counterparts at excessive concentrations, evidenced by impaired liver functions and cholestasis (Fischer et al., 1996; Perez and Briz, 2009). In this study, unconjugated BAs were increased by PBDE exposure (Figs. 6 and 7), suggesting that increased exposure to PBDEs could be a risk factor for liver injury. In particular, unconjugated BAs were increased almost 3-fold in liver (Fig. 5B). This fold increase is similar to cholestatic liver injury observed in male adult mice exposed to oleanolic acid (Liu et al., 2013). The bacterial deconjugation enzyme Bsh had an increasing trend following BDE-99 exposure, although a statistical significance was not achieved; furthermore, among the 41 differentially regulated taxa, five were positively correlated with unconjugated secondary BAs, indicating that deconjugation of BAs by gut microbiota may not be the only mechanism for increased unconjugated BAs. A small number of BAs are conjugated to glucuronic acid in the liver by uridine 5′-diphospho-glucuronosyltransferases (UGTs) to decrease the toxicity and urinary excretion of unconjugated BAs (Barbier et al., 2009; Perreault et al., 2013; Chatterjee et al., 2014). Using human liver microsomes, it was shown that hydroxylated PBDEs can be glucuronidated by hepatic UGTs to increase urinary excretion (Erratico et al., 2015). However, in rat liver microsomes, glucuronidation of PBDEs inhibited the glucuronidation of 17β-estradiol (Lai et al., 2012). Therefore, the increased concentration of unconjugated BAs may be attributable to competitive inhibition by the glucuronidation of hydroxylated PBDEs.
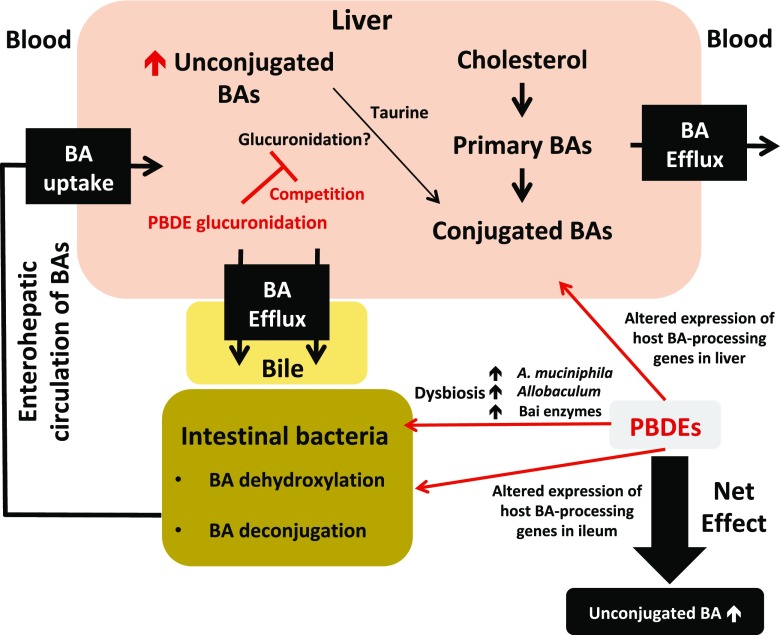
In conclusion, the present study using multiomics approaches is among the first to show that oral exposure to PBDEs altered the gut microbiome and BA homeostasis (Fig. 10). The key findings are: 1) PBDEs decreased bacterial diversity, but increased the taxa A. muciniphila and Allobaculum spp.; 2) PBDEs, and especially BDE-99, increased unconjugated BAs in the serum, liver, SIC, and LIC of CV mice; 3) the lack of gut microbiome mediated a shift in BA composition toward an increase in T-α/βMCA in GF mice; and 4) PBDEs downregulated enzymes and transporters involved in BA metabolism, in a gut microbiome-dependent manner. The present study focused on the correlative rather than causal relationship between PBDE exposure and gut microbiome. It remains to be determined which specific bacteria are responsible for intestinal degradation of PBDEs. In addition, further research should be carried out to investigate the mechanism of A. muciniphila and Allobaculum spp. in host metabolism.
Fig. 10.
A diagram illustrating the major findings of the present study. Briefly, 1) PBDEs decreased bacterial diversity, but increased the taxa A. muciniphila and Allobaculum spp., 2) PBDEs increased unconjugated BAs in the serum, liver, SIC, and LIC of CV mice, 3) the lack of gut microbiome led to a shift in BA composition toward an increase in T-α/βMCA in GF mice, and 4) PBDEs down-regulated host enzymes and transporters involved in BA metabolism in a gut microbiota-dependent manner.
Acknowledgments
The authors thank the GNAC technical support for the germ-free mouse experiment, Debbie Nickerson’s laboratory at UW Genome Sciences for sequencer access and support for the A-Seq experiments, Matt Karasu from UW School of Pharmacy for the proteomics data analysis, and other members of Cui laboratory for help in tissue collection and manuscript revision.
Abbreviations
- ACN
acetonitrile
- Akr1c14
aldo-keto reductase
- ANOVA
analysis of variance
- bai
BA-inducible operons
- BAs
bile acids
- BDE-47
2,2′,4,4′-tetrabromodiphenyl ether
- BDE-99
2,2′,4,4′, 5-pentabromodiphenyl ether
- BSA
bovine serum albumin
- Bsep
bile salt export pump
- bsh
bile salt hydrolase
- CA-D4
cholic acid-2,2,4,4-D4
- CV
conventional
- DCA-D4
deoxycholic acid-2,2,4,4-D4
- FXR
farnesoid X receptor
- GCA-D4
glycocholic acid-2,2,4,4-D4
- GCDCA-D4
glycochenodeoxycholic acid-2,2,4,4-D4
- GF
germ-free
- IS
internal standard
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LCA-D4
lithocholic acid-2,2,4,4-D4
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- LIC
large intestinal content
- MCA
muricholic acid
- Ntcp
sodium taurocholate cotransporting polypeptide
- Oatp
organic anion-transporting polypeptide
- P450
cytochrome P450
- PBDEs
polybrominated diphenyl ethers
- PICRUSt
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States
- PXR
pregnane X receptor
- QIIME
Quantitative Insights Into Microbial Ecology
- qPCR
quantitative polymerase chain reaction
- SIC
small intestinal content
- T-α/βMCA
tauro-α/β muricholic acid
Authorship Contributions
Participated in research design: Li, Gu, Cui.
Conducted experiments: Li, Wang, Lee, Dempsey, Weigel, Fei, Bhatt, Prasad, Gu, Cui.
Performed data analysis: Li, Wang, Lee, Dempsey, Weigel, Bhatt, Fei, Prasad, Gu, Cui.
Wrote or contributed to the editing of the manuscript: Li, Dempsey, Weigel, Raftery, Prasad, Gu, Cui.
Footnotes
This work was supported by National Institutes of Health (NIH) Grants [GM111381, ES019487, ES025708, T32 ES007032], the University of Washington Center for Exposures, Diseases, Genomics, and Environment [P30 ES0007033], and the Murphy Endowment.
 This article has supplemental material available at dmd.aspetjournals.org.
This article has supplemental material available at dmd.aspetjournals.org.
References
- Ajouz H, Mukherji D, Shamseddine A. (2014) Secondary bile acids: an underrecognized cause of colon cancer. World J Surg Oncol 12:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akaike T, Kobayashi A, Tobe S, Takei Y, Goto M, Kobayashi K. (1991) [A preview of the practical application of hybrid artificial liver]. Nippon Geka Gakkai Zasshi 92:1272–1275. [PubMed] [Google Scholar]
- Alcock J, Lin HC. (2015) Fatty acids from diet and microbiota regulate energy metabolism. F1000 Res 4 (F1000 Faculty Rev):738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbier O, Trottier J, Kaeding J, Caron P, Verreault M. (2009) Lipid-activated transcription factors control bile acid glucuronidation. Mol Cell Biochem 326:3–8. [DOI] [PubMed] [Google Scholar]
- Bhatt DK, Prasad B. (2017) Critical issues and optimized practices in quantification of protein abundance level to determine interindividual variability in DMET proteins by LC-MS/MS proteomics. Clin Pharmacol Ther 103:619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broeders EP, Nascimento EB, Havekes B, Brans B, Roumans KH, Tailleux A, Schaart G, Kouach M, Charton J, Deprez B, et al. (2015) The bile acid chenodeoxycholic acid increases human brown adipose tissue activity. Cell Metab 22:418–426. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, et al. (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Bijsmans IT, van Mil SW, Augustijns P, Annaert P. (2014) Toxicity and intracellular accumulation of bile acids in sandwich-cultured rat hepatocytes: role of glycine conjugates. Toxicol In Vitro 28:218–230. [DOI] [PubMed] [Google Scholar]
- Chiang JY. (2003) Bile acid regulation of hepatic physiology: III. Bile acids and nuclear receptors. Am J Physiol Gastrointest Liver Physiol 284:G349–G356. [DOI] [PubMed] [Google Scholar]
- Chiang JY. (2004) Regulation of bile acid synthesis: pathways, nuclear receptors, and mechanisms. J Hepatol 40:539–551. [DOI] [PubMed] [Google Scholar]
- Csanaky IL, Lu H, Zhang Y, Ogura K, Choudhuri S, Klaassen CD. (2011) Organic anion-transporting polypeptide 1b2 (Oatp1b2) is important for the hepatic uptake of unconjugated bile acids: studies in Oatp1b2-null mice. Hepatology 53:272–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M, Belzer C, de Vos WM. (2017) Akkermansia muciniphila and its role in regulating host functions. Microb Pathog 106:171–181. [DOI] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, Huber T, Dalevi D, Hu P, Andersen GL. (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnick JK, Brix A, Cunny H, Vallant M, Shockley KR. (2012) Characterization of polybrominated diphenyl ether toxicity in Wistar Han rats and use of liver microarray data for predicting disease susceptibilities. Toxicol Pathol 40:93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnick JK, Heath JE, Farnell DR, Prejean JD, Haseman JK, Elwell MR. (1997) Carcinogenic activity of the flame retardant, 2,2-bis(bromomethyl)-1,3-propanediol in rodents, and comparison with the carcinogenicity of other NTP brominated chemicals. Toxicol Pathol 25:541–548. [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. [DOI] [PubMed] [Google Scholar]
- Erratico CA, Moffatt SC, Bandiera SM. (2011) Comparative oxidative metabolism of BDE-47 and BDE-99 by rat hepatic microsomes. Toxicol Sci 123:37–47. [DOI] [PubMed] [Google Scholar]
- Erratico C, Zheng X, Ryden A, Marsh G, Maho W, Covaci A. (2015) Human hydroxylated metabolites of BDE-47 and BDE-99 are glucuronidated and sulfated in vitro. Toxicol Lett 236:98–109. [DOI] [PubMed] [Google Scholar]
- Fischer S, Beuers U, Spengler U, Zwiebel FM, Koebe HG. (1996) Hepatic levels of bile acids in end-stage chronic cholestatic liver disease. Clin Chim Acta 251:173–186. [DOI] [PubMed] [Google Scholar]
- Frederiksen M, Vorkamp K, Thomsen M, Knudsen LE. (2009) Human internal and external exposure to PBDEs--a review of levels and sources. Int J Hyg Environ Health 212:109–134. [DOI] [PubMed] [Google Scholar]
- Fu ZD, Cui JY. (2017) Remote sensing between liver and intestine: importance of microbial metabolites. Curr Pharmacol Rep 3:101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascon M, Vrijheid M, Martínez D, Forns J, Grimalt JO, Torrent M, Sunyer J. (2011) Effects of pre and postnatal exposure to low levels of polybromodiphenyl ethers on neurodevelopment and thyroid hormone levels at 4 years of age. Environ Int 37:605–611. [DOI] [PubMed] [Google Scholar]
- Geenes V, Chambers J, Khurana R, Shemer EW, Sia W, Mandair D, Elias E, Marschall HU, Hague W, Williamson C. (2015) Rifampicin in the treatment of severe intrahepatic cholestasis of pregnancy. Eur J Obstet Gynecol Reprod Biol 189:59–63. [DOI] [PubMed] [Google Scholar]
- Hu X, Bonde Y, Eggertsen G, Rudling M. (2014) Muricholic bile acids are potent regulators of bile acid synthesis via a positive feedback mechanism. J Intern Med 275:27–38. [DOI] [PubMed] [Google Scholar]
- Imray CH, Radley S, Davis A, Barker G, Hendrickse CW, Donovan IA, Lawson AM, Baker PR, Neoptolemos JP. (1992) Faecal unconjugated bile acids in patients with colorectal cancer or polyps. Gut 33:1239–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumpertz R, Le DS, Turnbaugh PJ, Trinidad C, Bogardus C, Gordon JI, Krakoff J. (2011) Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr 94:58–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen CD, Cui JY. (2015) Review: mechanisms of how the intestinal microbiota alters the effects of drugs and bile acids. Drug Metab Dispos 43:1505–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Y, Lu M, Lin S, Cai Z. (2012) Glucuronidation of hydroxylated polybrominated diphenyl ethers and their modulation of estrogen UDP-glucuronosyltransferases. Chemosphere 86:727–734. [DOI] [PubMed] [Google Scholar]
- Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, et al. (2013) Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CY, Lee S, Cade S, Kuo LJ, Schultz IR, Bhatt DK, Prasad B, Bammler TK, Cui JY. (2017) Novel interactions between gut microbiome and host drug-processing genes modify the hepatic metabolism of the environmental chemicals PBDEs. Drug Metab Dispos 45:1197–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickteig AJ, Csanaky IL, Pratt-Hyatt M, Klaassen CD. (2016) Activation of constitutive androstane receptor (CAR) in mice results in maintained biliary excretion of bile acids despite a marked decrease of bile acids in liver. Toxicol Sci 151:403–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares V, Bellés M, Domingo JL. (2015) Human exposure to PBDE and critical evaluation of health hazards. Arch Toxicol 89:335–356. [DOI] [PubMed] [Google Scholar]
- Liu J, Lu YF, Zhang Y, Wu KC, Fan F, Klaassen CD. (2013) Oleanolic acid alters bile acid metabolism and produces cholestatic liver injury in mice. Toxicol Appl Pharmacol 272:816–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang Y, Zhang X, Xu Q, Yang X, Xue C. (2017) Medium-chain fatty acids reduce serum cholesterol by regulating the metabolism of bile acid in C57BL/6J mice. Food Funct 8:291–298. [DOI] [PubMed] [Google Scholar]
- Lundgren M, Darnerud PO, Ilbäck NG. (2013) The flame-retardant BDE-99 dose-dependently affects viral replication in CVB3-infected mice. Chemosphere 91:1434–1438. [DOI] [PubMed] [Google Scholar]
- Makey CM, McClean MD, Braverman LE, Pearce EN, He XM, Sjödin A, Weinberg JM, Webster TF. (2016) Polybrominated diphenyl ether exposure and thyroid function tests in North American adults. Environ Health Perspect 124:420–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata M, Hayashi K, Yamakawa H, Yamazoe Y, Yoshinari K. (2015) Antibacterial drug treatment increases intestinal bile acid absorption via elevated levels of ileal apical sodium-dependent bile acid transporter but not organic solute transporter α protein. Biol Pharm Bull 38:493–496. [DOI] [PubMed] [Google Scholar]
- Packey CD, Shanahan MT, Manick S, Bower MA, Ellermann M, Tonkonogy SL, Carroll IM, Sartor RB. (2013) Molecular detection of bacterial contamination in gnotobiotic rodent units. Gut Microbes 4:361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacyniak EK, Cheng X, Cunningham ML, Crofton K, Klaassen CD, Guo GL. (2007) The flame retardants, polybrominated diphenyl ethers, are pregnane X receptor activators. Toxicol Sci 97:94–102. [DOI] [PubMed] [Google Scholar]
- Padda MS, Sanchez M, Akhtar AJ, Boyer JL. (2011) Drug-induced cholestasis. Hepatology 53:1377–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez MJ, Briz O. (2009) Bile-acid-induced cell injury and protection. World J Gastroenterol 15:1677–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perreault M, Białek A, Trottier J, Verreault M, Caron P, Milkiewicz P, Barbier O. (2013) Role of glucuronidation for hepatic detoxification and urinary elimination of toxic bile acids during biliary obstruction. PLoS One 8:e80994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Png CW, Lindén SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin TH. (2010) Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol 105:2420–2428. [DOI] [PubMed] [Google Scholar]
- Prasad B, Unadkat JD. (2014) Optimized approaches for quantification of drug transporters in tissues and cells by MRM proteomics. AAPS J 16:634–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravussin Y, Koren O, Spor A, LeDuc C, Gutman R, Stombaugh J, Knight R, Ley RE, Leibel RL. (2012) Responses of gut microbiota to diet composition and weight loss in lean and obese mice. Obesity (Silver Spring) 20:738–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridlon JM, Kang DJ, Hylemon PB. (2006) Bile salt biotransformations by human intestinal bacteria. J Lipid Res 47:241–259. [DOI] [PubMed] [Google Scholar]
- Robrock KR, Korytár P, Alvarez-Cohen L. (2008) Pathways for the anaerobic microbial debromination of polybrominated diphenyl ethers. Environ Sci Technol 42:2845–2852. [DOI] [PubMed] [Google Scholar]
- Russell DW. (2003) The enzymes, regulation, and genetics of bile acid synthesis. Annu Rev Biochem 72:137–174. [DOI] [PubMed] [Google Scholar]
- Sayin SI, Wahlström A, Felin J, Jäntti S, Marschall HU, Bamberg K, Angelin B, Hyötyläinen T, Orešič M, Bäckhed F. (2013) Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab 17:225–235. [DOI] [PubMed] [Google Scholar]
- Sberna AL, Assem M, Gautier T, Grober J, Guiu B, Jeannin A, Pais de Barros JP, Athias A, Lagrost L, Masson D. (2011) Constitutive androstane receptor activation stimulates faecal bile acid excretion and reverse cholesterol transport in mice. J Hepatol 55:154–161. [DOI] [PubMed] [Google Scholar]
- Selwyn FP, Cheng SL, Bammler TK, Prasad B, Vrana M, Klaassen C, Cui JY. (2015a) Developmental regulation of drug-processing genes in livers of germ-free mice. Toxicol Sci 147:84–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selwyn FP, Csanaky IL, Zhang Y, Klaassen CD. (2015b) Importance of large intestine in regulating bile acids and glucagon-like peptide-1 in germ-free mice. Drug Metab Dispos 43:1544–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shreiner AB, Kao JY, Young VB. (2015) The gut microbiome in health and in disease. Curr Opin Gastroenterol 31:69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqi MA, Laessig RH, Reed KD. (2003) Polybrominated diphenyl ethers (PBDEs): new pollutants-old diseases. Clin Med Res 1:281–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueyoshi T, Li L, Wang H, Moore R, Kodavanti PR, Lehmler HJ, Negishi M, Birnbaum LS. (2014) Flame retardant BDE-47 effectively activates nuclear receptor CAR in human primary hepatocytes. Toxicol Sci 137:292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Passel MW, Kant R, Zoetendal EG, Plugge CM, Derrien M, Malfatti SA, Chain PS, Woyke T, Palva A, de Vos WM, et al. (2011) The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS One 6:e16876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. (2013) EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience 2:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Chen X, Qiu M, Zeng X, Xu J, Deng D, Sun G, Li X, Guo J. (2012) Bar-coded pyrosequencing reveals the responses of PBDE-degrading microbial communities to electron donor amendments. PLoS One 7:e30439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CW, Huang HW, Chang BV. (2017) Microbial communities associated with anaerobic degradation of polybrominated diphenyl ethers in river sediment. J Microbiol Immunol Infect 50:32–39. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Klaassen CD. (2010) Effects of feeding bile acids and a bile acid sequestrant on hepatic bile acid composition in mice. J Lipid Res 51:3230–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Liu W, Wang J, Shi J, Sun Y, Wang W, Ning G, Liu R, Hong J. (2017) Akkermansia muciniphila improves metabolic profiles by reducing inflammation in chow diet-fed mice. J Mol Endocrinol 58:1–14. [DOI] [PubMed] [Google Scholar]