ABSTRACT
Hoxb8 mutant mice show compulsive behavior similar to trichotillomania, a human obsessive-compulsive-spectrum disorder. The only Hoxb8 lineage-labeled cells in the brains of mice are microglia, suggesting that defective Hoxb8 microglia caused the disorder. What is the source of the Hoxb8 microglia? It has been posited that all microglia progenitors arise at embryonic day (E) 7.5 during yolk sac hematopoiesis, and colonize the brain at E9.5. In contrast, we show the presence of two microglia subpopulations: canonical, non-Hoxb8 microglia and Hoxb8 microglia. Unlike non-Hoxb8 microglia, Hoxb8 microglia progenitors appear to be generated during the second wave of yolk sac hematopoiesis, then detected in the aorto-gonad-mesonephros (AGM) and fetal liver, where they are greatly expanded, prior to infiltrating the E12.5 brain. Further, we demonstrate that Hoxb8 hematopoietic progenitor cells taken from fetal liver are competent to give rise to microglia in vivo. Although the two microglial subpopulations are very similar molecularly, and in their response to brain injury and participation in synaptic pruning, they show distinct brain distributions which might contribute to pathological specificity. Non-Hoxb8 microglia significantly outnumber Hoxb8 microglia, but they cannot compensate for the loss of Hoxb8 function in Hoxb8 microglia, suggesting further crucial differences between the two subpopulations.
KEY WORDS: Tmem119, Microglia, Hoxb8 microglia, Non-Hoxb8 microglia, Microglia ontogeny, Yolk sac, AGM and fetal liver hematopoiesis, Fetal HSCs, OCD, Obsessive-compulsive-spectrum disorders, Trichotillomania
Summary: Defective Hoxb8 microglia, a proposed cause of trichotillomania-like behavior in mice, have a distinct ontogeny relative to canonical microglia, providing the brain with greater microglial functional diversity.
INTRODUCTION
Microglia are recognized as crucial players in the shaping and fine-tuning of newly formed brain circuits. Failure of proper microglia function can lead to behavioral pathologies (Kettenmann et al., 2013; Li et al., 2012; Nimmerjahn et al., 2005; Prinz and Priller, 2014; Schafer et al., 2013; Tremblay et al., 2011; Zhan et al., 2014). The loss of Hoxb8 function in a subpopulation of microglia appears causative for compulsive grooming and a hair removal pathology that resembles the obsessive-compulsive-spectrum disorder (OCD), trichotillomania (Chen et al., 2010). Such new behavioral roles for microglia greatly extend our vision of how these brain sentinels contribute to maintenance of brain homeostasis, beyond their accepted roles as guardians of order through their phagocytic activity.
Based on temporal lineage tracing analysis, it has been reported that all microglia progenitors originate from the first wave of yolk sac hematopoiesis at embryonic day (E) 7.5, and then directly populate the developing brain at E9.5 (Ashwood et al., 2006; Ginhoux et al., 2010; Gomez-Perdiguero et al., 2015; Hoeffel et al., 2015; Kierdorf et al., 2013; Schulz et al., 2012; Sheng et al., 2015). The nature of the progenitors generated during the second wave of yolk sac hematopoiesis is currently under debate. Data presented in Gomez-Perdiguero et al. (2015) and McGrath et al. (2015) support the generation of yolk sac erythromyeloid progenitors (EMPs) at E8.5 that subsequently give rise to a broad range of self-sustaining, tissue-resident macrophages. On the other hand, Sheng et al. (2015), using c-KitMerCreMer cell fate mapping, suggest that the second wave of yolk sac hematopoiesis is characterized predominantly by a stem cell antigen-1 (Sca-1; Ly6a – Mouse Genome Informatics)-expressing progenitor cell population in the AGM and fetal liver, designated fetal hematopoietic stem cells (f-HSCs), which give rise to tissue-resident macrophages, except for microglia. After the formation of the blood brain barrier (BBB), resident microglia have little exchange with the peripheral immune system, and microglial homeostasis is maintained through a balance of self-renewal and apoptosis (Ginhoux et al., 2010; Mildner et al., 2007).
In the course of characterizing a pathological grooming behavior in Hoxb8 mutant mice, we identified a new subset of microglia in the brain that is uniquely labeled by a Hoxb8 lineage reporter (Chen et al., 2010). The developmental source of this subpopulation was not determined. Henceforth, we will designate the population of microglia labeled by the Hoxb8 cell lineage marker as Hoxb8 microglia, and the canonical microglia population as non-Hoxb8 microglia. Because grooming behavior is controlled by the central nervous system, and the only cells in the brain labeled by the Hoxb8 lineage reporter appeared to be microglia, we suggested that defective Hoxb8 microglia caused the associated pathology (Chen et al., 2010). Consistent with this interpretation, the pathological overgrooming behavioral deficit could be rescued by transplantation of normal bone marrow into lethally irradiated Hoxb8 mutant mice. Further, the pathology is recapitulated by conditionally restricting the Hoxb8 mutation to the hematopoietic lineage, but not, for example, by restricting the mutation to Hoxb8-expressing interneurons (Chen et al., 2010).
Herein, we show that the Hoxb8 microglia progenitors appear to be generated during the second wave of yolk sac hematopoiesis, and are not detected in the developing brain until E12.5, long after the arrival of non-Hoxb8 microglia. Either a subpopulation of non-Hoxb8 microglia turn on Hoxb8 expression in the developing brain at E12.5, or Hoxb8 microglia progenitors enter the brain later because they have transited, for example, through other hematopoietic tissues, such as the para-aortic-splanchnopleural (P-Sp)/aorto-gonad-mesonephros (AGM) region and/or the fetal liver. No Hoxb8 RNA transcripts are ever detected in either the developing or adult mouse brain using either quantitative reverse transcription polymerase chain reaction (qRT-PCR) or deep sequencing to identify such transcripts, refuting the first hypothesis. Thus, our data strongly support the second hypothesis that Hoxb8 microglia enter the brain at E12.5 from a hematopoietic source within the embryo proper. Further, cell transplantation experiments involving the injection of genetically marked Hoxb8 microglia progenitor cells, derived from E12.5 fetal livers, into neonatal mouse brains, clearly demonstrate that the purified Hoxb8 microglia progenitor cells are capable of generating mature parenchymal microglia in vivo, as defined by their morphology and expression of the microglia-specific gene, Tmem119 (Bennett et al., 2016).
Direct comparison of RNA transcripts by deep sequencing shows that adult Hoxb8 and non-Hoxb8 microglia subpopulations are very similar molecularly, with only 21 genes showing significant differential expression. Moreover, two recently described markers that distinguish parenchymal microglia from other macrophages in the periphery or brain, Tmem119 and Sall1 (Bennett et al., 2016; Buttgereit et al., 2016), are similarly expressed in the two microglial subpopulations. Both subpopulations also share similarities in their response to brain injury and participation in synaptic pruning. These two populations of microglia, however, exhibit distinct stereotypic spatial distributions in the adult mouse brain. Interestingly, Hoxb8 microglia, unlike non-Hoxb8 microglia, show their highest concentration in the brain in regions previously defined by functional magnetic resonance imaging (fMRI) in humans as the ‘OCD circuit’ (Graybiel and Rauch, 2000). The juxtaposition of Hoxb8 microglia within the ‘OCD neural circuit’ could provide a mechanism for the specificity of the behavioral disorder associated with the Hoxb8 mutation. In summary, our data define Hoxb8 microglia as a bona fide microglia subpopulation. Although open to other interpretations, the expression of Hoxb8 in the early hematopoietic progenitor population, as well as the kinetics of amplification and distribution of the Hoxb8 hematopoietic progenitors within hematopoietic tissues, suggest that this population of microglia progenitor cells appears to be born during the second wave of yolk sac hematopoiesis and then transits through the AGM and fetal liver, where these cells are greatly amplified prior to their infiltration into the brain at 12.5 days of gestation. This alternative developmental pathway and route of entry into the developing brain could provide Hoxb8 microglia with opportunities to acquire functional diversity relative to non-Hoxb8 microglia.
RESULTS
Hoxb8 microglia progenitors are born during the second wave of yolk sac hematopoiesis and expanded in the AGM and fetal liver prior to colonizing the E12.5 brain
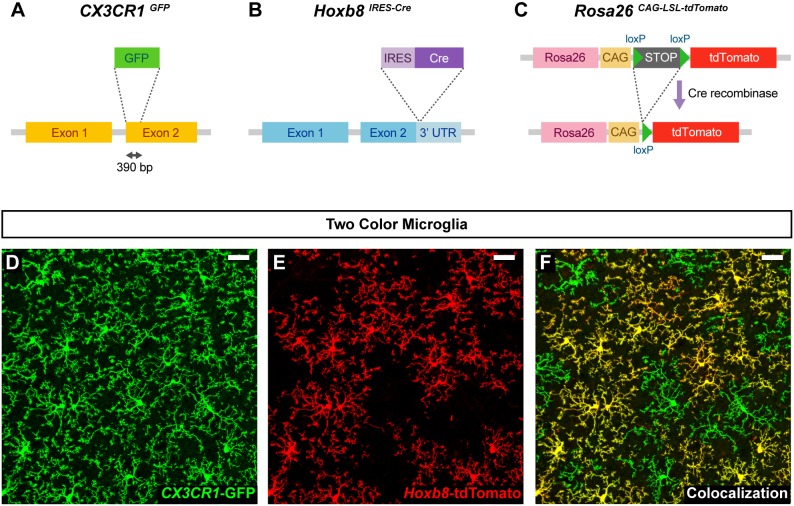
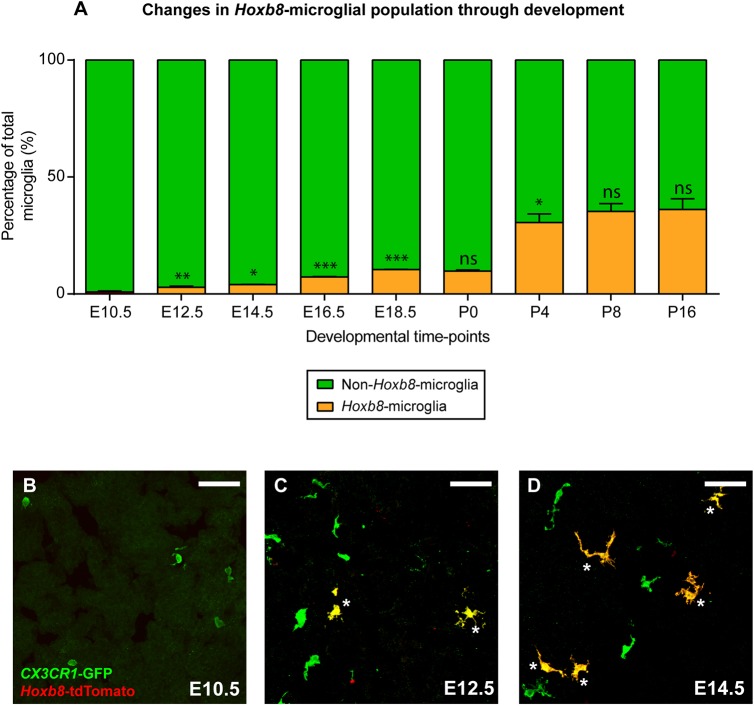
The Hoxb8 lineage marker labels a subpopulation of parenchymal microglia (Chen et al., 2010); however, their developmental origin has not been previously defined. To determine the origin of Hoxb8 microglia, we generated Cx3cr1GFP/+; Hoxb8IRES-Cre/+; ROSA26CAG-LSL-tdTomato/+ mice. In these mice, Cre is expressed in all cells that express Hoxb8, leading to the constitutive expression of the red, tdTomato fluorescent protein (Fig. 1B,C). This marker is maintained in the cells that expressed Hoxb8 and their progeny, irrespective of continued Hoxb8 expression. In these triple-transgenic mice, all parenchymal microglia are labeled with green fluorescent protein (GFP) due to Cx3cr1GFP expression (Fig. 1A), whereas only Hoxb8 microglia are co-labeled with tdTomato (Fig. 1D-F). Initially, we monitored the appearance of GFP+ only and GFP+ tdTomato+ microglia in the brain, as a function of developmental time. At early time points (between E9.5 and E11.5), only GFP+ non-Hoxb8 microglia were observed in the embryonic brain by confocal microscopy (Fig. 2A,B). GFP+ tdTomato+ double-positive microglia, were absent at these early time points, but are consistently detected in the brain at E12.5, although such cells are rare at this stage (Fig. 2A,C). From E14.5 onward, GFP+ tdTomato+ double-positive microglia are readily apparent in the brain (Fig. 2A,D). Hoxb8 microglia comprise ∼25% of the total microglia population in the adult brain cortex (Fig. 2A). The delayed appearance of Hoxb8 microglia could be explained by either of two hypotheses: (1) a subpopulation of non-Hoxb8 microglia activates the expression of Hoxb8 in the developing brain starting around E12.5, or (2) a separate population of microglial progenitors, marked by expression of the Hoxb8-tdTomato reporter, starts to infiltrate the brain at E12.5. To address these hypotheses, we sought to quantify the spatial-temporal expression pattern of Hoxb8 using qRT-PCR in Hoxb8 microglia progenitors residing in the distinct hematopoietic tissues.
Fig. 1.
Two-color microglia mouse model. (A-F) All microglia in the brain are labeled with the fractalkine receptor (Cx3cr1) disrupted by the GFP gene (A,D) (Jung et al., 2000), and Hoxb8 microglia are also labeled by the Hoxb8 cell lineage marker Hoxb8IRES-Cre, ROSA26CAG-LSL-tdTomato (B,C,E) (Madisen et al., 2010). (F) Note that only a subpopulation of microglia is co-labeled with the Hoxb8 cell lineage marker. The above confocal micrographs depict an image from the frontal cortex of an adult mouse. Scale bars: 30 µm.
Fig. 2.
Hoxb8 microglia enter the brain significantly later than non-Hoxb8 microglia. (A) Graph showing the Hoxb8 microglial population (orange) as a percentage of total microglia in the developing mouse brain cortex (E10.5-P16) as determined by confocal microscopy. P-values were determined from comparisons with the previous time point. (B-D) Hoxb8 (yellow, white asterisks) and non-Hoxb8 (green) microglia in E10.5 (B), E12.5 (C) and E14.5 (D) brain parenchyma. Scale bars: 50 µm. n=3 biological replicates per time point, ns, non-significant; *P<0.05, **P<0.005, ***P<0.0001; data are mean±s.e.m. See also Figs S3 and S4.
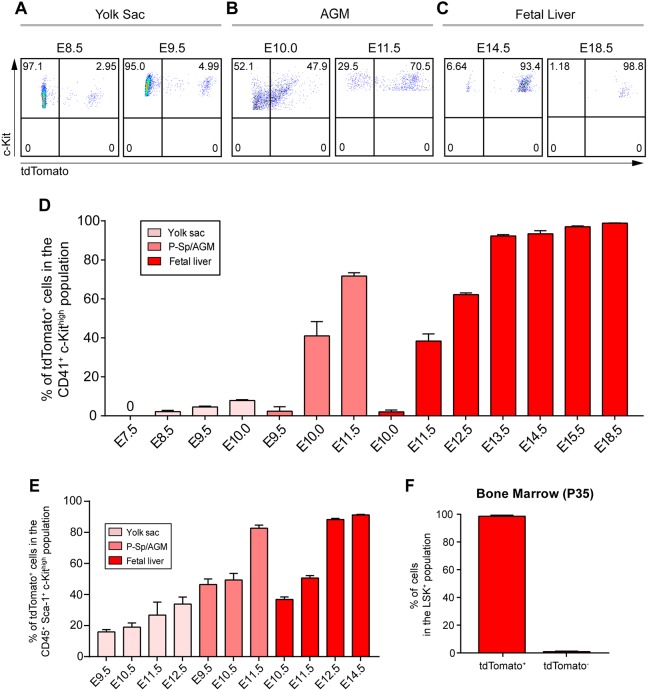
The emergence of Hoxb8 microglia progenitors during embryogenesis (appearance of tdTomato-labeled CD41+ c-Kithigh- or CD45+ c-Kithigh-expressing cells) was determined by fluorescence-activated cell sorting (FACS) analysis of yolk sac tissue from E7.5 to E10.0, in the P-Sp/AGM region from E9.5 to E11.5 and fetal liver from E10.0 to E18.5. CD41 (Itga2b) is commonly used as a marker for early, less mature, hematopoietic progenitors, whereas CD45 (Ptprc) marks more mature hematopoietic progenitors (Ferkowicz et al., 2003; Mikkola et al., 2003). Hoxb8 microglial progenitors (CD41+ c-Kithigh tdTomato+) were not detectable during the early stages of yolk sac hematopoiesis at E7.5 (Fig. 3D). Only low levels of tdTomato-positive progenitors (∼5.0%) were apparent in the yolk sac between E8.5 and E10.0 (Fig. 3A,D). In the AGM, a significant fraction (>40%) of CD41+ c-Kithigh progenitor cells was labeled with the Hoxb8 cell lineage marker tdTomato by E10.0 during the early stages of P-Sp/AGM hematopoiesis (Fig. 3B,D). It is evident from P-Sp/AGM cell-counting experiments, that the number of tdTomato+ cells is greatly and selectively amplified, relative to those present in the yolk sac (see below). By E11.5, greater than 75% of these progenitor cells were tdTomato+ (Fig. 3D). On the other hand, at E10.0 in the fetal liver, we could barely detect CD41+ c-Kithigh tdTomato+ progenitor cells, but by E11.5, greater than 38% of these progenitor cells were tdTomato+ (Fig. 3D). Again, by comparing numbers of tdTomato+ cells in fetal liver to yolk sac or AGM, the number of Hoxb8 progenitor cells in fetal liver relative to even AGM was further amplified. By E18.5, almost 100% of the CD41+ hematopoietic progenitor population in the fetal liver was tdTomato+ (Fig. 3C,D). Because many progenitor cells in the fetal liver begin to express CD45, we also quantified the CD45+ c-Kithigh tdTomato+ population. These results were comparable with those from cell sorting for CD41+ c-Kithigh and tdTomato+ except that CD45 marks a larger pool of hematopoietic progenitor cells in the later stages of fetal liver development (Fig. S1). The temporal and quantitative analysis of the Hoxb8 lineage-labeled progenitors in the yolk sac, P-Sp/AGM region and fetal liver suggests that this progenitor cell population follows a developmental pathway that originates during the second wave of yolk sac hematopoiesis. Expansion of this population begins in the AGM, followed by further selective expansion in the fetal liver prior to colonization of the brain. When we compared hematopoietic progenitor cell expansion rates between the yolk sac, AGM and fetal liver during their respective optimal periods of progenitor production, we observed a 16.6-fold increase in AGM, and a 281-fold increase in fetal liver, relative to yolk sac. This represents approximately four and eight cell doublings, respectively.
Fig. 3.
Hoxb8 microglia progenitors are selectively expanded during AGM and fetal liver hematopoiesis. (A) FACS profiles of CD41+ cells gated for c-Kit and tdTomato from the yolk sac of E8.5 and E9.5 Cx3cr1GFP/+; Hoxb8IRES-Cre/+; ROSA26CAG-LSL-tdTomato/+ embryos. (B) FACS profile of CD41+ cells gated for c-Kit and tdTomato isolated from the AGM of E10.0 and E11.5 embryos. (C) FACS profiles of CD45+ cells gated for c-Kit and tdTomato from fetal livers of E14.5 and E18.5 Cx3cr1GFP/+; Hoxb8IRES-Cre/+; ROSA26CAG-LSL-tdTomato/+ embryos. (D) Graph showing the percentage of Hoxb8 hematopoietic progenitor cells (CD41+ c-Kithigh tdTomato+) through embryonic development in different tissues. n=2-4 replicates or pooled replicates per time point; data are mean±s.e.m. See also Fig. S1. (E) Graph showing the percentage of Hoxb8 f-HSCs (CD45+ Sca-1+ c-Kithigh tdTomato+) through development in different tissue. n=2-6 replicates or pooled replicates per time point; data are mean±s.e.m. (F) Graph depicting the percentage of Hoxb8 HSCs (Lin− Sca-1+ c-Kithigh tdTomato+) in adult bone marrow. n=5 replicates; data are mean±s.e.m. See also Figs S1 and S2.
Sca-1 is a marker commonly used to identify hematopoietic stem cells (HSCs) and is expressed in multipotent cells derived from E8.5 yolk sacs (Inlay et al., 2014). To test whether Hoxb8 progenitors express Sca-1, we used FACS to count progenitor cells derived from the yolk sac, P-Sp/AGM region and fetal liver during embryonic development, by gating for CD45+ Sca-1+ c-Kithigh tdTomato+ (Fig. 3E). tdTomato+ labels a significant fraction of the Sca-1+ f-HSC population in the yolk sac, P-Sp/AGM region and fetal liver. By E12.5, a time when Hoxb8 microglia first appear in the brain, the percentage of tdTomato+ cells in the CD45+ Sca-1+ c-Kithigh population in the fetal liver approaches 90% (Fig. 3E).
The Runx1 transcription factor has been shown to label microglia progenitors during early mouse embryonic development (Ginhoux et al., 2010). To determine the extent of overlap between the Hoxb8 lineage-labeled cells and the progenitor population labeled by Runx1, we generated Runx1IRES-GFP/+; Hoxb8IRES-Cre/+; ROSA26CAG-LSL-tdTomato/+ compound mice and analyzed the yolk sac at E9.5 for CD41+ c-Kithigh GFP+ tdTomato− and CD41+ c-Kithigh GFP+ tdTomato+ progenitor cells (Fig. S2). We found that 100% of all CD41+ c-Kithigh cells in the yolk sac at E9.5 expressed GFP, confirming results published by Ginhoux et al. (2010). The percentage of the GFP+ hematopoietic progenitor cells also labeled by tdTomato was ∼8%.
Because the Hoxb8 lineage contributes to the hematopoietic progenitor cell population in the yolk sac, P-Sp/AGM region and fetal liver, we next asked whether adult HSCs are also derived from the Hoxb8 lineage. To address this issue, bone marrow was isolated from Hoxb8IRES-Cre/+; ROSA26CAG-LSL-tdTomato/+ adult mice and FACS analyzed for HSCs by gating for Lin− Sca-1+ c-Kithigh (LSK) tdTomato+ cells. Virtually all LSK+ cells were tdTomato+ (Fig. 3F). These sorted cells were further analyzed for the presence of Hoxb8 transcripts by qRT-PCR and none were detectable. As adult bone marrow LSK+ cells do not express Hoxb8, the Hoxb8 lineage marker, tdTomato, must have been activated at an earlier hematopoietic progenitor state (i.e. in f-HSCs).
The Hoxb8IRES-Cre and ROSA26CAG-LSL-tdTomato alleles provide a convenient and sensitive marking system for the Hoxb8 microglia lineage, but they do not reflect real-time Hoxb8 gene expression. Once the cell lineage marker is expressed, it stays on in those cells and all of their progeny. Further, because the system is dependent on the production and function of Cre protein, visualization of the reporter gene takes ∼24 h (i.e. for the synthesis of Cre protein, translocation to the nucleus, excision of the lox-stop-lox cassette, as well as production of the reporter gene product).
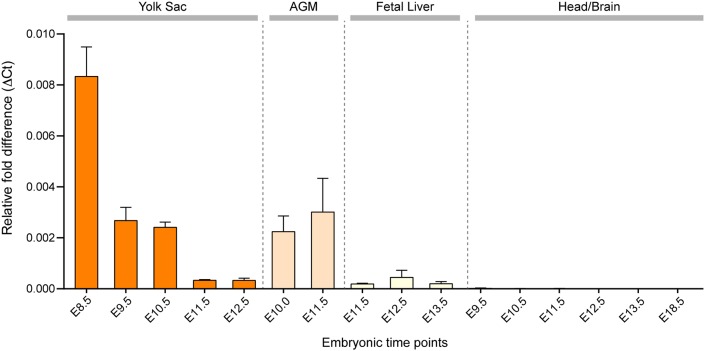
In order to determine more accurately when and where Hoxb8 transcripts are produced, we used qRT-PCR to analyze the yolk sac, AGM, fetal liver and brain in developing mouse embryos. Interestingly, Hoxb8 transcripts were most abundant in the yolk sac at E8.5 (Fig. 4). Although expression of Hoxb8 was detectable in the AGM and fetal liver, this expression was significantly less than in the yolk sac at E8.5. In stark contrast, we did not detect any Hoxb8 transcripts at any time point analyzed in the head region of developing embryos from E9.5 to birth (Fig. 4). In addition, by RNA deep sequencing, Hoxb8 transcripts could not be detected in mature Hoxb8 microglia present in the adult brain parenchyma (see below). Thus, Hoxb8 is predominantly expressed in a subpopulation of yolk sac hematopoietic progenitor cells during a very narrow window of Hoxb8 microglia progenitor development, long before their infiltration into the brain. Taken together, (1) the gene expression pattern of Hoxb8 in the yolk sac, AGM and fetal liver, (2) the lack of expression of Hoxb8 in the developing and mature brain, (3) the distribution pattern of tdTomato-expressing progenitors from the yolk sac, to AGM and fetal liver, and (4) the time of entry of Hoxb8-labeled progenitor cells into the developing brain, we concluded that Hoxb8 microglia have a significantly different embryonic history, relative to non-Hoxb8 microglia.
Fig. 4.
Hoxb8 expression through embryonic development. Hoxb8 expression is predominantly detected in the yolk sac during primitive hematopoiesis as determined by qRT-PCR in the respective tissues as a function of developmental time. n=3-5 replicates per time point; data are mean±s.e.m. Data were normalized to GAPDH expression.
In summary, Hoxb8 microglia progenitors appear to be born during the second wave of yolk sac hematopoiesis. Sheng et al. (2015) has suggested that the progenitor population coincident with the second wave of hematopoiesis is characterized by expression of Sca-1. Consistent with this interpretation, most Hoxb8 hematopoietic progenitor cells express Sca-1, which is not known to be expressed during the first wave of yolk sac hematopoiesis. The Hoxb8 progenitor population then appears in the AGM and fetal liver, where Hoxb8 lineage cells extensively and selectively expand in number (which is also characteristic of the second wave of yolk sac hematopoiesis), prior to their infiltration into the developing brain beginning at E12.5. The added exposure of Hoxb8 microglia progenitor cells to the AGM and fetal liver prior to their entry into the brain provides opportunities for signaling inputs from these hematopoietic organs, and thereby potential acquisition of additional genetic and epigenetic complexity that might be translated into a new repertoire of functions.
Early postnatal dynamics of Hoxb8 microglia
Having shown that Hoxb8 microglia have a distinct ontogeny compared to canonical microglia, we next set out to determine whether the distinct embryonic history affected Hoxb8 microglia biology. Hoxb8 microglia cell distribution and morphology change considerably during early postnatal development (Fig. S3), consistent with previously reported observations on early postnatal development of canonical microglia (Dalmau et al., 2003).
The proportion of Hoxb8 microglia compared to non-Hoxb8 microglia increases several fold during the first postnatal week and remains relatively constant at ∼20-25% thereafter (Fig. 2A). Following BBB closure, infiltration of microglia should be severely limited and the proportion of the two microglia populations should remain constant throughout development. We therefore sought to determine whether the relative enrichment of postnatal Hoxb8 microglia was due to higher proliferation rates. Using incorporation of 5-ethynyl-2´-deoxyuridine (EdU) into dividing cells, we found that the overall proliferation in cortical microglia was high during the first postnatal week but decreased to barely measurable values by postnatal day (P) 16 (Fig. S4A). Hoxb8 microglia show a higher proliferation rate just after birth (Fig. S4A). Beyond P4, proliferation in the two subpopulations did not appear to differ significantly. To complement the proliferation assays with a measurement of cell death, we evaluated microglia for the presence of activated caspase-3, a primary effector of apoptosis. Microglial apoptotic rates were observed to be relatively high at P0 (∼1.5%) (Fig. S4B), but decreased dramatically by P4 and remained at nearly undetectable levels thereafter. No significant differences in cell death were detected between the two subpopulations of microglia.
Gene expression profile of Hoxb8 microglia
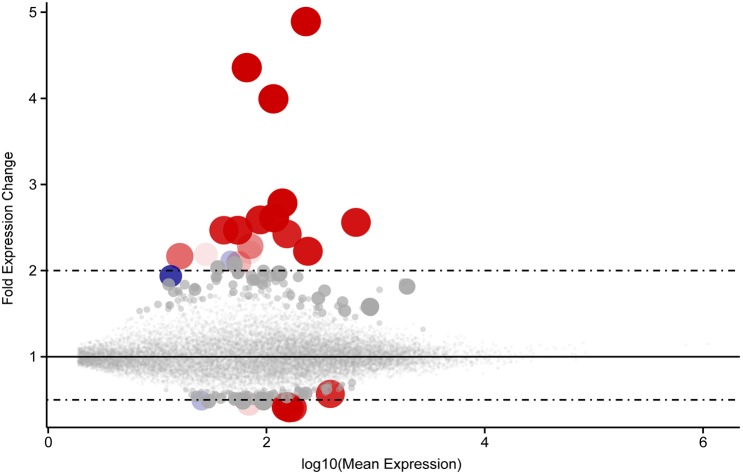
Next, we compared the gene expression profiles of non-Hoxb8 microglia and Hoxb8 microglia. Microglia were isolated from 12-week-old animals carrying the Cx3cr1GFP/+; Hoxb8IRES-Cre/+; ROSA26CAG-LSL-tdTomato/+ alleles. To reduce potential contamination from peripheral monocytes, the brains were perfused and the meninges removed. Additionally, microglia were isolated from forebrain and midbrain tissue to eliminate potential contamination resulting from tdTomato-labeled interneuron projections emanating from the spinal cord (Fig. 8D). Microglia were sorted, gating on tdTomato and GFP. The barcoded complementary DNA (cDNA) libraries were sequenced on Illumina HiSeq platforms, resulting in 25-50 million sequences per biological sample. The data sets were very similar with only 21 genes showing differential expression levels between the non-Hoxb8 microglia and Hoxb8 microglia subpopulations (Table S1). Fig. 5 provides a summary of the RNA sequence data, illustrating the fold difference in gene expression (relative to expression in non-Hoxb8 microglia) for all genes with a quantifiable expression level. Each point on the plot represents a single gene as identified in the GRCm38v3 mouse genomic assembly. The size of the point is proportional to the statistical significance of the observed fold change values for each gene. The largest symbols denote genes with the lowest adjusted P-values from our DEseq2 analysis. The color of the dot denotes relative ranking in this set (see Fig. 5 legend).
Fig. 8.
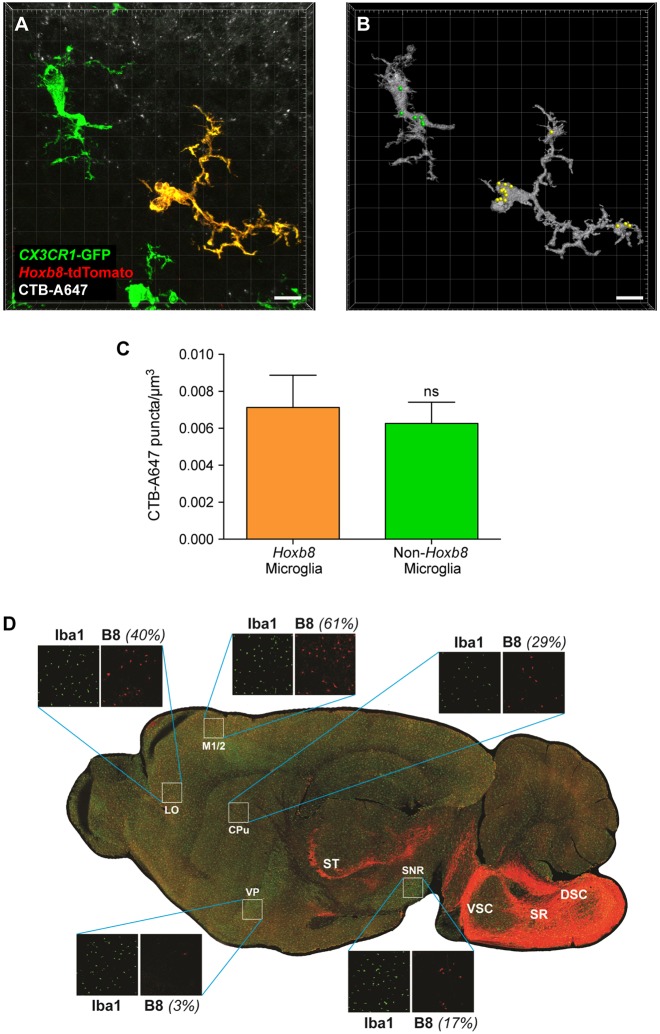
Synaptic pruning behavior of Hoxb8 and non-Hoxb8 microglia is similar, whereas the distributions of the two microglial populations in the adult brain are significantly different. (A) Confocal image of Hoxb8 and non-Hoxb8 microglia, situated close to the CTB-A647-labeled neurons in the dLGN. (B) 3D rendering of the microglial cells shown in A. The yellow and green spots indicate the phagocytosed tracer puncta detected inside the Hoxb8 and non-Hoxb8 microglia, respectively. Scale bars: 10 μm. (C) Histogram showing the average number of neuronal pieces, phagocytosed detected per unit volume of a microglial cell. n=3 mice; ns, non-significant; **P<0.005; data are mean±s.e.m. (D) Spatial distribution of Hoxb8 microglia. Comparative distribution patterns of non-Hoxb8 microglia (green) and Hoxb8 microglia (yellow) in the adult mouse brain. LO, lateral orbital cortex; CPu, caudate putamen; DSC and VSC, dorsal and ventral spinocerebellar tracts; M1/2, primary and secondary motor cortex; VP, ventral pallidum; SNR, substantia nigra; SR, spinoreticular tracts; ST, spinothalamic track. The strong fluorescence in the posterior region of the brain reflects Hoxb8-expressing interneuron projections emanating from the spinal cord.
Fig. 5.
Few genes are differentially expressed between Hoxb8 and non-Hoxb8 microglia. A representation of differentially expressed genes with quantifiable expression levels plotted, comparing Hoxb8 microglia with non-Hoxb8 microglia. Note the linear scale on the y-axis (fold change expression). Each point on the plot represents a single gene as identified in the GRCm38v3 mouse genomic assembly. Plot symbol size is proportional to the statistical significance of the observed fold change values for each gene. The largest symbols denote genes with the lowest adjusted P-values (padj) from our DEseq2 analysis. Genes considered statistically significant according to the default DESeq2 guidelines (padj <0.1) are coded red; the color saturation indicates their relative ranking in this set (padj <0.1). Thus, the most significantly differentially regulated gene is displayed as a fully saturated red circle, while the gene with the highest padj value still less than 0.1 is displayed as a faint red circle. Similarly, genes within padj values 0.1-0.2 are displayed as blue circles, while genes with padj between 0.2-1.0 are displayed as gray circles. For actual fold differences of the top differentially expressed genes, see Table S1 and padj values for all genes provided in Table S2.
The list of significantly differentially expressed genes is remarkably small (Table S1). It is an interesting list of genes, noted for their expression in myeloid cells representing a broad range of activities: receptors participating in cell signaling pathways, ligands such as cytokines and chemokines, activators of cell mobility, protein kinases, inhibitors of tyrosine kinase signaling, and proteins associated with inflammatory responses. Although this is a provocative list, what is strikingly clear is that these two microglia subpopulations share highly similar expression profiles.
When we looked at the level of Hoxb8 expression, the number of Hoxb8 transcripts detected in adult Hoxb8 microglia is well below one transcript per cell. This observation, coupled with the qRT-PCR data (Fig. 4), shows that Hoxb8 expression in embryonic and adult brains is not detectable, indicating that the Hoxb8 cell lineage marker (tdTomato) is activated in Hoxb8 microglia progenitor cells well before their infiltration into the brain.
Recently, the Barres laboratory described a marker expressed in mature microglia, Tmem119, which distinguishes parenchymal microglia from other macrophages in the brain and periphery (Bennett et al., 2016). In our comparative mouse transcript analysis of non-Hoxb8 and Hoxb8 microglia, we found that Tmem119 was significantly and similarly expressed in both microglial subpopulations (10,940 and 11,765 reads, respectively; Table S2). The presence of the Tmem119 marker in both subpopulations further supports our classification of Hoxb8 microglia as parenchymal microglia.
Sall1 has also been described as a marker whose expression is largely restricted to microglia and has not been detected in other peripheral or brain macrophages (Buttgereit et al., 2016). Notably, this marker is also expressed at equivalent levels in the two microglial subpopulations (9500 versus 9116 reads in non-Hoxb8 and Hoxb8 microglia, respectively; Table S2).
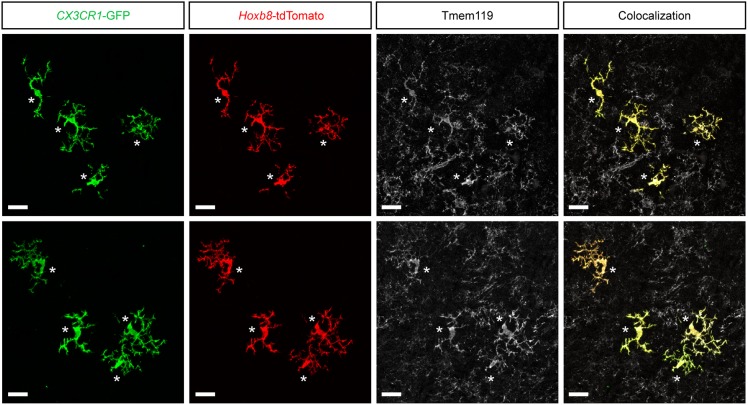
The Hoxb8 lineage reporter system, Hoxb8IRES-Cre; ROSA26CAG-LSL-tdTomato also labels peripheral macrophages. The question then arises: how many of the tdTomato+ cells in the brain are legitimate parenchymal microglia as opposed to other myeloid cells present in the normal brain? To answer this question we used an anti-Tmem119 antibody to quantify the percentage of tdTomato+ cells that are specifically microglia. Examination of brain sections derived from adult mice (n=3) counterstained with the anti-Tmem119 antibody (Fig. S5) and quantified for co-labeled cells revealed that greater than 98% of the tdTomato+ cells in these sections were parenchymal Hoxb8 microglia and greater than 99% of the GFP only cells were parenchymal non-Hoxb8 microglia.
Hoxb8 hematopoietic progenitor cells derived from E12.5 fetal liver give rise to mature parenchymal microglia when introduced into neonatal mouse brains
Having shown that Hoxb8 microglia in adult mice are labeled with the microglia-specific marker, Tmem119, we were positioned to directly determine whether fetal liver derived Hoxb8-labeled c-Kithigh, hematopoietic progenitor cells, are capable of giving rise to mature parenchymal microglia when transplanted into neonatal mouse brains. Freshly sorted hematopoietic progenitor cells (tdTomato+ c-Kithigh) derived from E12.5 fetal livers of Cx3cr1GFP/+; Hoxb8IRES-Cre/+; ROSA26CAG-LSL-tdTomato/+ mice were injected into the brains of C57Bl6/J pups at P4, which were allowed to develop to P14-P17 to provide time for the expression of the mature microglial-specific marker, Tmem119. Brain sections of these mice were stained with anti-tdTomato, anti-GFP and anti-Tmem119 antibodies, and scanned by confocal microscopy. Representative images are shown in Fig. 6. Transplantation of 25,000 c-kithigh tdTomato+ hematopoietic progenitor cells into C57Bl6/J pups resulted in low engraftment of GFP+ tdTomato+ cells (∼50 cells/brain), all of which exhibited the morphology of mature microglia and co-labeled with Tmem119. Doubling the number of transplanted progenitor cells resulted in higher engraftment (∼350 cells/brain). However, some of these cells appeared morphologically less mature and 83% of these cells co-labeled with Tmem119. Thus, overabundance of transplanted cells appears to slow down the maturation of the engrafted cells. As expected, under both experimental conditions, all tdTomato+ cells also expressed GFP resulting from the presence of the Cx3cr1GFP allele in the donor cells. Finally, the engraftment efficiency of the fetal liver-derived Hoxb8 progenitor cells was enormously increased by using Csfr1−/− mutant pups as recipients in order to ablate the resident microglia population in the recipients prior to transplantation (Fig. S6) (Erblich et al., 2011). The recipient Csf1r−/− brain was engrafted with thousands of tdTomato+ GFP+ Hoxb8 microglia cells. The latter experiments suggest that most of the newly introduced Hoxb8 progenitor cells in the previous experiments were eliminated from the recipient brains by the resident brain microglia in the recipient wild-type brain. Because the only source of tdTomato+ GFP+ cells in these pups were donor-derived, purified hematopoietic progenitor cells obtained from the fetal livers of E12.5 embryo donors, we conclude that these progenitor cells are indeed capable of giving rise to mature parenchymal microglia when introduced into wild-type and Csf1r−/− mutant mouse brains.
Fig. 6.
E12.5 Hoxb8 fetal liver-derived hematopoietic progenitors give rise to parenchymal microglia in vivo. Mature GFP+ tdTomato+ Tmem119+ microglia, generated from transplanted Hoxb8 lineage progenitor cells, are marked by white asterisks. Tmem119 immunohistochemistry identifies both donor-derived GFP+ tdTomato+ microglia and recipient resident microglia (no fluorescence). Scale bars: 20 µm. See also Fig. S6.
Physiological properties of Hoxb8 microglia
The activation response of microglia following an insult to brain tissue is one of the most well-characterized microglial functions. We analyzed acute and chronic injury responses of the two microglial subpopulations in the cortex of the adult mouse brain to compare their effectiveness as brain macrophages.
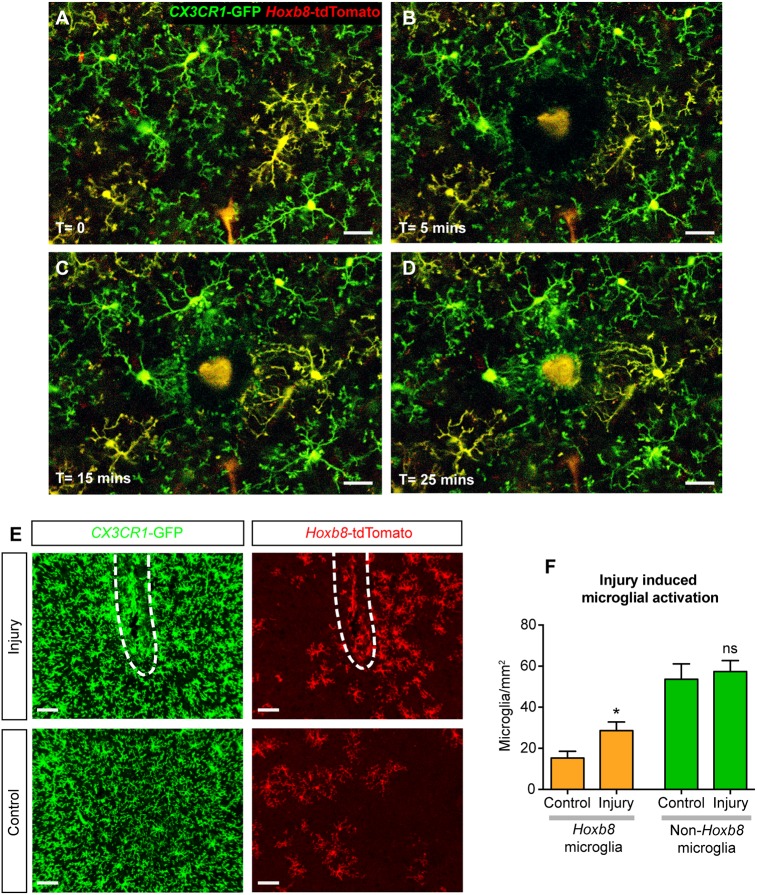
To study the acute response to injury, we employed multiphoton microscopy for live imaging in the mouse brain cortex. For this purpose, the Cx3cr1GFP/+; Hoxb8IRES-Cre/+; ROSA26CAG-LSL-tdTomato/+ triple-transgenic mouse line was used to visualize both subpopulations of microglia simultaneously (Fig. 7A-D). The response of activated microglia to focal laser ablation in the brain cortex was observed in anesthetized mice (Movie 1). Immediately following laser-induced ablation, the microglial processes started to advance toward the damage, and within 20-25 min they reached the site of injury (Fig. 7A-D, Movie 1). Both classes of microglia responded with similar speed (Hoxb8 microglia, 1.34±0.19 µm/min; non-Hoxb8 microglia, 1.49±0.4 µm/min), showing that both subpopulations have indistinguishable acute responses.
Fig. 7.
Hoxb8 microglia have similar response to damage as non-Hoxb8 microglia. (A-D) Stills from a time-lapse movie of microglial response to damage induced by focused laser ablation in brain cortex, using in vivo multiphoton imaging. Scale bars: 30 μm. (A) Hoxb8 (yellow) and non-Hoxb8 (green) microglia in their resting state before injury. A video of the acute response is provided (Movie 1). (B-D) Successive time points as the microglial processes converge towards the site of injury. (E) Microglial activation induced by needle-poke injury (white dashed lines) in the mouse brain cortex observed 7 days postinjury. Scale bars: 50 μm. (F) Hoxb8 microglia show a 1.9-fold increase in numbers around the injury site compared with a 1.1-fold increase in non-Hoxb8 microglia. n=5 mice, ns, non-significant; *P<0.05; data are mean±s.e.m. See also Fig. S7 and Movie 1.
To analyze the response to chronic injury, adult mouse brains were subjected to a small, localized needle poke via intracranial injection, followed by a recovery period of 7 days. This recovery period provides time for microglia surrounding the injection site to be activated and migrate to the site of injury. Hoxb8 microglia demonstrated a greater tendency to accumulate around the site of injection (Fig. 7E,F). These cells exhibited a 1.9-fold increase in cell density around the injury site compared with a 1.1-fold increase in the density of non-Hoxb8 microglia (Fig. 7E,F). Because this mechanical injury created some damage to the surrounding vasculature, it might also have led to the infiltration of tdTomato-expressing Hoxb8 lineage macrophages present in peripheral blood. To distinguish the resident Hoxb8 microglia from peripheral macrophages, we stained the cells with the microglia-specific 4D4 antibody (Dr Oleg Butovsky, Harvard Medical School, Boston, MA, USA) (Fig. S7). The 4D4 antibody labels resident brain microglia specifically in both the resting and activated states. This analysis showed that the majority of Hoxb8 lineage cells (>93%) around the site of injury were resident brain Hoxb8 microglia with a small contribution from peripheral cells. These results suggest that Hoxb8 microglia are more responsive than non-Hoxb8 microglia to chronic injury in the brain.
Synaptic pruning
Early in the normal development of the brain, far more synaptic connections are formed than are maintained. Activity-dependent synaptic pruning is used to eliminate weaker connections and thereby maintain strengthened synapses (Hua and Smith, 2004; Katz and Shatz, 1996; Sanes and Lichtman, 1999). Microglia have been shown to actively participate in this synaptic remodeling program (Paolicelli et al., 2011; Schafer et al., 2012, 2013; Stephan et al., 2012). A particularly attractive system to assess the role of microglia in activity-dependent synaptic pruning is the reduction of excess synapses in the mouse retinogeniculate nucleus (Schafer et al., 2012; Stephan et al., 2012). These neurons can be labeled through ocular injection of anterograde tracers, such as the cholera toxin β subunit conjugated to Alexa Fluor 647 (CTB-A647) (Schafer et al., 2012). By carrying out these experiments in our triple-transgenic mice, we directly compared the efficiency of non-Hoxb8 microglia and Hoxb8 microglia to phagocytose labeled neurons. The two microglia populations were equally competent in mediating activity-dependent synaptic pruning in the retinogeniculate nucleus during its peak activity in P5 mice (Fig. 8A-C).
Distribution of Hoxb8 microglia in the adult mouse brain
During embryogenesis and early postnatal development, we observed higher concentrations of Hoxb8 microglia in the cortex than in other neighboring regions of the brain. To determine whether this bias persisted into adulthood, we examined the distribution of the two microglial subpopulations in the adult brain. In Fig. 8D, the strong tdTomato signal in the posterior brain (right side) is associated with ascending fibers emanating from the spinal cord, such as the dorsal and ventral spinocerebellar tracts (DSC and VSC), and the spinoreticular tracts (SR). These fibers strongly express tdTomato because Hoxb8 is strongly expressed in the dorsal spinal cord interneurons that receive inputs from peripheral sensory neurons (Holstege et al., 2008). Although these interneurons express high levels of Hoxb8, selective disruption of Hoxb8 in these interneurons using conditional mutagenesis did not recapitulate the compulsive pathological overgrooming behavior characteristic of Hoxb8 mutant mice (Chen et al., 2010), ruling out any contribution of these interneurons to the pathological overgrooming phenotype.
In the adult brain parenchyma, the concentration of Hoxb8 microglia relative to non-Hoxb8 microglia was noticeably higher in the cortex, specifically in the frontal association cortex, the primary and secondary motor cortex (M1/2), and the lateral orbital cortex (LO). The dorsal striatum (caudate putamen, CPu) and the olfactory bulb also display relatively higher concentrations of Hoxb8 microglia (Fig. 8D). In contrast, other regions such as the ventral pallidum (VP) and substantia nigra (SNR) show much lower densities of Hoxb8 microglia. Interestingly, the regions of high Hoxb8 microglia concentrations coincided with brain regions previously designated as the ‘OCD circuit’, based on fMRI studies of OCD and control human populations (Graybiel and Rauch, 2000). The relative distribution patterns of non-Hoxb8 and Hoxb8 microglia in the adult mouse brain were reproducible from mouse to mouse. The distinct spatial distribution of Hoxb8 microglia in the normal brain might contribute to the specificity of behavioral phenotype observed in Hoxb8 mutant mice.
DISCUSSION
A hallmark of the immune system is its diversity and complexity. Thus, it should not be surprising that diversity and complexity are also characteristics of microglia, a cell type that is derived from the immune system and is noted for performing multiple biological functions in the brain. Herein, we show that, in contrast to previously published results, mammalian microglia have more than one developmental route to colonize the embryonic brain. The canonical microglia progenitors, generated during the first wave of yolk sac hematopoiesis at E7.5, transit directly from extraembryonic hematopoietic tissue (i.e. yolk sac) to the developing brain. Although open to alternative interpretations, parsimony and consistency with the data derived from both Hoxb8 expression profiling as well as the progressive locations of the Hoxb8 hematopoietic progenitor population during development as shown by FACS analysis, suggest that Hoxb8 microglia progenitors are generated during the second wave of yolk sac hematopoiesis then seed the AGM and fetal liver, where they are greatly amplified prior to their infiltration into the brain at E12.5. Consistent with the above hypothesis, Hoxb8 microglia progenitors enter the brain significantly later (E12.5) than non-Hoxb8 microglia (E9.5) (Ginhoux et al., 2010; Hoeffel et al., 2015; Kierdorf et al., 2013; Schulz et al., 2012; Sheng et al., 2015). Furthermore, purified Hoxb8 hematopoietic progenitor cells, derived from E12.5 fetal livers, injected into P4 mouse brains, were capable of giving rise to mature parenchymal microglia expressing the microglia-specific marker, Tmem119. Interestingly, two separate embryonic sources of microglia have also been recently shown to exist in zebrafish (Xu et al., 2015).
Hoxb8 gene expression is highest during yolk sac hematopoiesis. Both the later timing of the appearance of the Hoxb8 lineage-labeled progenitors in the yolk sac, and the expression of Sca-1, support the hypothesis that the Hoxb8 microglia progenitors are generated during the second wave of yolk sac hematopoiesis. Sheng et al. (2015) have suggested that a prominent hematopoietic population present in the yolk sac during the second wave of hematopoiesis is composed of f-HSCs expressing Sca-1. The Hoxb8 lineage marker also labels virtually all bone marrow-derived HSCs. Because Hoxb8 is not expressed in adult hematopoietic stem cells (HSCs), this suggests that the Hoxb8 cell lineage marker is activated in a progenitor population that gives rise to bone marrow HSCs (i.e. f-HSCs). Our observations are consistent with the hypothesis posited by Sheng et al. (2015), that a principal hematopoietic progenitor cell population is f-HSCs. As reported by Sheng et al. (2015) for f-HSCs, the Hoxb8-labeled hematopoietic progenitors also appear in the P-Sp/AGM region and fetal liver, where they are significantly expanded in number. That Hoxb8 expression is undetectable in the brain during embryonic development or in adult microglia further supports the hypothesis that the Hoxb8 lineage in the brain infiltrates from the embryonic periphery, rather than arising from a subpopulation of non-Hoxb8 microglia that subsequently express Hoxb8 in the developing brain.
The passage of Hoxb8 microglia progenitors through the P-Sp/AGM region and fetal liver, prior to infiltration into the developing brain, provides added opportunities for these microglia to acquire additional genetic and epigenetic complexity and thereby potential for a greater repertoire of functions. The second wave of microglia infiltration into the brain occurs prior to maturation of the BBB and uptake of these progenitor cells would likely terminate once the BBB is closed. As with non-Hoxb8 microglia, Hoxb8 microglia are therefore within a closed environment and maintained at steady-state concentrations in the brain through a balance of self-renewal and apoptosis.
Comparison of the RNA transcriptome of Hoxb8 microglia with non-Hoxb8 microglia shows that the two populations have extremely similar expression profiles. Also, two molecular markers recently characterized as specific to parenchymal microglia, Tmem119 and Sall1 (Bennett et al., 2016; Buttgereit et al., 2016), are equally and prominently expressed in both microglial subpopulations. Not only are these two populations of microglia molecularly very similar, but they also show similar responses to injury and synaptic pruning. Two types of injury responses were examined: a fast response, within 30 min of a focal laser-induced injury, and a response to more extensive damage induced by a needle poke. The Hoxb8 microglia response to the needle poke injury appeared to be more extensive than observed for the non-Hoxb8 microglia. Both populations of microglia responded aggressively and similarly to acute injury by immediate extensions of their projections. Finally, activity-dependent postnatal pruning in the retinogeniculate nucleus was indistinguishable between the two microglial subpopulations.
Although these populations of microglia have much in common molecularly and functionally, they were also distinguishable. Interestingly, the relative concentration of Hoxb8 microglia was high in regions of the brain previously characterized by fMRI as belonging to the ‘OCD circuit’ (Graybiel and Rauch, 2000). The distinct distribution of Hoxb8 microglia in the adult brain might be an important contributor to the specific dysfunctional behavioral output observed in Hoxb8 mutant mice. The mechanisms responsible for generating the distinct distributions of non-Hoxb8 microglia and Hoxb8 microglia, whether passive or active, has not been determined.
Additional functional differences between the two subpopulations of microglia could emerge. It appears that the effects of the Hoxb8 mutation on Hoxb8 microglia, which comprise ∼25% of total brain microglia, cannot be compensated for by the presence of non-Hoxb8 microglia (∼75% of total microglia) or the mouse would not exhibit an aberrant behavioral pathology associated with this mutation.
An association of a broad spectrum of neuropsychiatric disorders with immune dysfunctions is well documented in the literature (Ashwood et al., 2006; da Rocha et al., 2008; Hounie et al., 2008; Kronfol and Remick, 2000; Lang et al., 2007; Leonard and Myint, 2009; Strous and Shoenfeld, 2006). Further, among genes revealed by genome-wide association studies of multiple neuropsychiatric disorders are a common class of genes known for their prominent roles in immunology (International Schizophrenia et al., 2009; Shi et al., 2009; Stefansson et al., 2009). What was unclear from the above reports was which sets of genes are causative. In our case, we have argued for the hypothesis that defective Hoxb8 microglia appear causative for a specific behavioral dysfunction. Aside from Hoxb8 microglia being in the appropriate regions of the adult brain, why is the behavioral output in Hoxb8 mutant mice so specific? In a separate study, we have shown that Hoxb8 mutant mice also exhibit high levels of anxiety, and these mutant mice responded favorably to serotonin uptake blockers such as fluoxetine (Nagarajan et al., 2017). High levels of anxiety have also been associated with multiple neuropsychiatric disorders, particularly obsessive-compulsive-spectrum disorders. Patients with OCD, during periods of high anxiety, often participate in their pathological compulsive activity, whereas when they are not anxious, they can control their compulsive behavior (Markarian et al., 2010). A reasonable argument can be made that particularly among social animals like humans and mice, one of the multiple roles for microglia is to maintain brain homeostasis, including protection from excessive anxiety. Failure of such maintenance may lead to behavioral disorders, with the particular disorder being dependent upon the constellation of genetic and environmental insults experienced by the individual patient. Regardless of the targets of microglia for maintaining homeostasis, what is clear is that we are in the midst of a glia revolution with the recognition that glia have much broader roles in brain function, brain homeostasis and brain pathology than we ever imagined.
MATERIALS AND METHODS
Animals
Gt(ROSA)26Sortm14(CAT-tdTomato)Hze (Ai14, #007908), Cx3cr1GFP mice (#005582) and Csf1rtm1.2Jwp (#021212) mice were obtained from the Jackson Laboratory. Hoxb8IRES-Cre mice were generated in our laboratory and reported in Chen et al. (2010). Runx1IRES-GFP mice were a kind gift from Dr James Downing, Department of Pathology, St. Jude Children's Research Hospital, Memphis, TN, USA.
Embryo isolation
Timed matings were set up between male and female mice of the required genotypes. The uterine horns were removed and placed on ice-cold 1× PBS. The embryos were isolated from the uterus with the help of sharp forceps, washed in ice-cold 1× PBS and incubated in 4% (wt/vol) paraformaldehyde (PFA; EMS 15713) in 1× PBS overnight. Embryonic tissues/brains were then processed for immunohistochemistry as described below.
Immunohistochemistry
For adult brain isolation, mice were overdosed with avertin and transcardially perfused with 4% PFA. The brains were isolated and incubated in 4% PFA for 2 h on a bench-top shaker at room temperature. Embryonic and adult samples were kept in 10% sucrose overnight at 4°C on a shaker, followed by another overnight incubation at 4°C in 30% sucrose until brain tissues sink to the bottom. These samples were then embedded in Tissue-Tek OCT (Sakura 4583), frozen on liquid nitrogen and stored at −80°C. For immunohistochemistry, brain samples were sectioned at 20-25 μm using a Leica CM1900 cryostat and mounted on positively charged microscope slides (Fisherbrand Tissue Path SuperFrost Plus Gold slides, 22-035813, Fisher Scientific). Sections were washed with 1× PBS and permeabilized with 0.2% Triton X-100, 1% sodium deoxycholate solution, then incubated overnight with primary antibody mixture at 4°C. The following day the sections were washed and incubated with secondary antibodies for 2 h at room temperature. Finally, the sections were stained with 4′,6-diamidino-2-phenylindole (DAPI; D1306, Molecular Probes) and mounted with ProLong Gold antifade reagent (P36934, Molecular Probes) and microscope cover glass (Fisherbrand 22-266882). Images were acquired on Leica TCS SP5 confocal microscope and processed and analyzed using Imaris 7.7 (Bitplane), as described below.
Antibodies for immunohistochemistry
Primary antibodies
Primary antibodies were as follows: chicken anti-GFP (1:500, GFP-1020, Aves Labs), rabbit anti-RFP (1:500, 600-401-379, Rockland), rat anti-RFP (5F8) (1:250, 5f8-100, Chromotek), guinea pig anti tdTomato (1:250, AB 2631185, Frontier Institute), rabbit anti-cleaved caspase-3 (Asp175) (1:500, 9661, Cell Signaling Technology), rat 4D4 antibody (1:200, provided by Dr Oleg Butovsky, Harvard Medical School), rabbit anti-Tmem119 (28-3) (1:500, 209064, Abcam).
Secondary antibodies
Secondary antibodies were as follows: goat anti-chicken Alexa Fluor 488 (1:500, A-11039, Thermo Fisher Scientific), goat anti-rabbit Alexa Fluor 555 (1:500, A-21428, Thermo Fisher Scientific), goat anti-rabbit Alexa Fluor 647 (1:500, 111-605-144, Jackson ImmunoResearch), goat anti-rat Alexa Fluor 555/647 (1:250, A-21434/ A-21247, Thermo Fisher Scientific) and goat anti-guinea pig Alexa Fluor 555 (1:250, A21435, Thermo Fisher Scientific).
Both the primary and secondary antibodies used have been monitored for specificity and cross-reactivity.
EdU labeling
To label proliferating microglia at different time points (P0, P4, P8, P16 and P32, n=3 per time point), Cx3cr1GFP/+; Hoxb8IRES-Cre/+; ROSA26CAG-LSL-tdTomato/+ mice were anesthetized with isoflurane and injected (dosage 10 µl/gm body weight) with 10 mM EdU (A10044, Molecular Probes), twice every 1.5 h. After the final incubation period the brains were isolated and processed for immunohistochemistry and confocal imaging as described above. The EdU-labeled cells were identified using the Click-iT EdU Alexa Fluor 647 Imaging Kit (C10340, Molecular Probes).
Embryonic tissue processing for FACS
Embryos were removed from the uterus and placed on ice in 5% fetal bovine serum (FBS, Atlanta Biologicals) in 1× Hanks' balanced salt solution (HBSS, Gibco). Dissections of the following tissues were performed at their respective time points: yolk sac (E7.5, E8.5, E9.5, E10.0, E10.5, E11.5, E12.5), P-Sp/AGM region (E9.5, E10.0, E10.5, E11.5), fetal liver (E10.0, E10.5, E11.5, E12.5, E13.5, E14.5, E15.5, E18.5) and bone marrow (P35). Dissected tissues before E12.5 were pooled together to obtain enough cells at these time points for flow cytometry. The tissue was broken up into single cells using a 26G needle. The dissociated tissues were spun down at 1500 g for 5 min at 4°C. Cells were washed three times with 1× HBSS and spun down at 1500 g for 5 min. To obtain a single-cell suspension, the cells were passed through an 80 µm mesh. The suspension was incubated in 20 µl of red blood cell lysis buffer (0.15 M ammonium chloride, 1 mM KHCO3) for 5 min, then ∼13 ml of HBSS was added and incubation continued for 5 min to lyse red blood cells, followed by another 5 min spin at 200 g. The anti-mouse antibodies used consisted of the following: TER-119 PerCP-Cy5.5 (1:50, 116228, BioLegend), CD41 APC-eFluor 780 (1:100, 47-0411-82, eBioscience), CD45 APC (1:100, 103112, BioLegend), c-Kit PE-Cy7 (1:100, 105814, BioLegend), Sca-1 APC (1:100, 108112, BioLegend), Sca-1 APC/FIRE 750 (1:100, 108146, BioLegend), Lin-1 Alexa Fluor 700 (1:100, 133313, BioLegend) in 1× HBSS. Cells were incubated in the dark with their respective antibody cocktail for 30 min on ice. Cells were washed with 1× HBSS and spun down at 1500 g for 5 min, then resuspended in 1× HBSS with DAPI (1:500). Flow cytometry data were obtained using the BD Bioscience FACSCanto II flow cytometry sorter. All FACS data were analyzed with FlowJo 10.0.7 (Celeza).
FACS gating strategy for embryonic and adult tissue populations
For all embryonic tissue populations, live cells were gated with side scatter (SSC) and forward scatter (FSC) parameters. Viability and singlets were gated with FSC and DAPI parameters. For Cx3cr1GFP/+; Hoxb8IRES-Cre/+; ROSA26CAG-LSL-tdTomato/+ triple-transgenic mice, the following gates were applied depending on the developmental stage of the embryo: time points for yolk sac and P-Sp/AGM region (E7.5, E8.5, 9.5, E10.0, E10.5, E11.5) included CD41 and TER119 (Ly76) parameters in order to exclude platelets, red blood cells and erythrocytes, and select CD41+ or CD45+ hematopoietic cells. These cells were further analyzed for tdTomato and c-Kit or Sca-1 to examine the percentage of tdTomato+ cells in the hematopoietic progenitor or f-HSC compartment. Time points for fetal liver (E10.0, E10.5, E11.5, E12.5, E13.5, E14.5, E15.5, E18.5) were gated in a similar fashion except that CD45 was used to gate CD45+ hematopoietic cells. For bone marrow, viable cells (DAPI−) were gated for Lin-1−, c-Kithigh and Sca-1+ to identify the HSC cell population to examine the percentage of tdTomato+ cells in the HSC compartment. For Runx1IRES-GFP+/−; Hoxb8IRES-Cre+/−; ROSA26CAG-LSL-tdTomato/+ compound transgenic mice: TER119− CD41+ cells were selected and gated with c-Kit to identify hematopoietic progenitors. Hematopoietic progenitors were further analyzed to examine the percentage of tdTomato+ cells that were GFP+.
Postnatal brain injections
Hematopoietic progenitor cells were isolated (described above) from E12.5 fetal livers of Cx3cr1GFP/+; Hoxb8IRES-Cre/+; ROSA26CAG-LSL-tdTomato/+ embryos and sorted by flow cytometry using the BD Bioscience FACS ARIA flow cytometry sorter. Sorted Hoxb8 lineage progenitor cells consisted of the following immunophenotype: DAPI− TER119− c-Kithigh tdTomato+. Freshly sorted cells were directly injected bilaterally into the motor cortex (12.5-25K cells/hemisphere) of wild-type (C57Bl6/J) or Csf1r−/− newborns (P0-P4). These brains were harvested, sectioned and stained with antibodies against tdTomato, GFP and Tmem119, from P14 to P17 to examine whether fetal liver-derived Hoxb8 lineage hematopoietic progenitor cells injected into P0-P4 pups can colonize the postnatal brain and become Hoxb8 microglia. Microglia were observed in seven of nine brains injected.
qRT-PCR
Total RNA was isolated from either yolk sac (E8.5, E9.5, E10.5, E11.5, E12.5), P-Sp/AGM region (E10.0, E11.5), fetal liver (E11.5, E12.5, E13.5) or head/brain (E9.5, E10.5, E11.5, E12.5, E13.5, E18.5), and purified using the RNeasy Kit (Qiagen). The concentration and quality of total RNA was determined by using a NanoDrop2000c spectrophotometer (Thermo Fisher Scientific), and 500 µg of total RNA was used for reverse transcription into cDNA using the SuperScript III First-Strand Synthesis SuperMix (Invitrogen). For qRT-PCR, an Applied Biosystems 7900HT instrument was used to amplify 50 ng Hoxb8-specific cDNA and 50 ng Gapdh-specific cDNA (an endogenous housekeeping control) using a Hoxb8 probe (Thermo Fisher Scientific) and a Gapdh probe (Thermo Fisher Scientific), respectively. Each tissue sample was run in triplicate. Hoxb8 gene expression was quantified using the comparative CT method (ΔCT) by normalizing to Gapdh expression in age-matched tissues.
Microglia isolation
Cortical microglia were isolated from four independent ∼12-week-old two-color animals (Cx3Cr1GFP/+; Hoxb8IRES-Cre/+; ROSA26CAG-LSL-tdTomato/+). Animals were humanely euthanized by isoflurane, cortical tissue dissected and meninges removed, minced and trypsin digested (1% trypsin, 37°C, 30 min in HBSS). Cortical single-cell suspensions were generated by mechanical trituration and successive filtration (70 and 30 µm mesh) and glia enriched via capture with anti-Cd11b (Itgam) magnetic beads (Miltenyi Biotec). Bead-associated cortical microglia were then sorted on a BD FACSAriaII gating on tdTomato and GFP with collection into RNAlater solution, flash frozen and stored at −80°C.
RNA sequencing isolation and analyses
Total RNA was isolated and cloned by the University of Utah High-throughput Genomics Core using a Clontech UltraLow RNA isolation kit, and barcoded libraries sequenced on an Illumina HiSeq platform (50 bp nonstranded single-end reads), resulting in ∼25-30 million sequences per biological replicate. Sequence quality was confirmed by FastQC. Raw fastq reads were mapped using STAR (2.4.1c) (Dobin et al., 2013). Indices for STAR were built based upon the Gencode GRCm38v3 release M5 of the mouse genome. The index for STAR was constructed based upon the primary assembly and included splice junction information from the GTF file. The cDNA sequences corresponding to Cre, tdTomato and eGFP were appended to the fasta files prior to index generation. Mapping was accomplished using default parameters for STAR necessary to the utilize splice junction information built into the corresponding index. STAR read counts at the gene level were generated using feature counts from the SubRead package.
Deseq2 analyses were conducted independently on all count data sets to compare relative transcript levels between the green (non-Hoxb8, control) and yellow (Hoxb8, experimental) microglia data sets. Deseq2 was run using default parameters and differentially expressed genes determined based upon a padj cutoff value <0.1. The list of differentially expressed genes was generated (Table S1).
Microglial activation using focused laser ablation
Live brain imaging was performed using a Prairie Technologies Ultima Multiphoton Microscopy System. For live imaging in the cerebral cortex, an open skull window was prepared on 1- to 2-month-old Cx3cr1GFP/+; Hoxb8IRES-Cre/+; ROSA26CAG-LSL-tdTomato/+ mice. The open skull window prep was performed according to the previously described protocol by Pozner et al. (2015). The mice were mounted on a custom-built head frame and imaged under 1.5% isofluorane anesthesia. Imaging was performed with a Chameleon Ti:Sapphire laser at 950 nm and a 16× water-immersion objective (0.8 NA, Nikon) with 3× optical zoom. The signals were acquired using GaSP detectors with 490-560 nm bandpass filter for GFP and 570-620 nm bandpass filter for tdTomato. Microglial cells were imaged 50-80 μm below the pial surface at 1024×1024 pixel resolution and a sampling rate of 0.05 frames/s for 30-35 min. Focused laser injury was induced at a small area (8×8 pixels) within the region of interest that was scanned at a frequency of 1 Hz for 2 min with the laser power increased by 75%. Image acquisition was achieved using the Prairie View 5.2 software and images were analyzed using Imaris (Bitplane).
Microglial activation using intracranial injections
To assess microglial activation response, we induced focal injury in the motor cortex of 3- to 4-month-old Cx3cr1GFP/+; Hoxb8IRES-Cre/+; ROSA26CAG-LSL-tdTomato/+ mice. Microglial activation was induced by a needle poke injury through stereotaxic intracranial injections. Mice were anesthetized with isoflurane (4% for induction, 1.5% for surgery) and their heads were mounted on a stereotaxic frame (Kopf). Anesthetized mice were placed on a heating pad (FHC) to regulate and maintain their body temperature at 37°C, and the depth of anesthesia was tested and monitored by toe-pinch reflex and respiration rate. A subcutaneous injection of 50-60 µl of 0.5% marcaine (Hospira) was applied as local anesthesia at the incision site. An incision of ∼7 mm was made along the anterioposterior axis to expose the skull. The skull was cleaned with ferric chloride solution and saline, and dried with sterile cotton tipped applicators. Using a high-speed dental drill, a burr hole was created at the stereotaxic position anteroposterior (AP) −1.5 mm; mediolateral (ML) −1.9 mm; dorsoventral (DV) −1 mm, using Bregma as reference. Tissue damage was induced by intracranial injection of 0.1 μl of sterile 1× PBS, delivered at a rate of 20 nl/min using a Quintessential Stereotaxic Injector (QSI) (Stoelting). Following the injection, saline was applied to the exposed skull and skin, then the incision was sutured. Seven days following the injection, the mice were humanely euthanized by isoflurane, and the brains were isolated and analyzed with immunohistochemistry and confocal imaging.
Synaptic pruning assay
To study the pruning behavior of each microglial population, P4 pups (Cx3cr1GFP/+; Hoxb8IRES-Cre/+; ROSA26CAG-LSL-tdTomato/+) were administered bilateral intravitreal injections of 1 mg/ml Alexa Fluor 647-conjugated cholera toxin B subunit (CTB-A647, C34778, Molecular Probes). After 24 h, the mice were humanely euthanized by isoflurane, their brains were harvested, then fixed in 4% PFA, passed through the sucrose gradient, embedded in OCT, and stored at −80°C. Cryosections (25 µm) were collected for immunohistochemistry to fluorescently label the Hoxb8 and non-Hoxb8 microglia, as described above. Brains chosen for analysis were those that exhibited sufficient dye fills in the dorsal lateral geniculate nucleus (dLGN).
Confocal imaging parameters
Images were acquired on Leica TCS SP5 confocal system. The imaging parameters used for these different types of analyses are described below.
Microglia counting
For density analysis, the brain sections were imaged with a 10× objective (0.4 NA, Leica) and 1.5× digital zoom, acquired at 512×512 resolution and 400-600 Hz scan speed, using a 5.0 µm z-depth through the tissue.
Synaptic pruning analysis
Confocal images of the dLGN regions of the thalamus were acquired with a 63× oil immersion objective (1.40-0.60 NA, Leica), at 1024×1024 resolution, 0.5 µm z-depth.
Imaris image analysis
The images acquired with the confocal and two photon microscopy were processed using Imaris Image Analysis Software ×64 (v 7.7.2, Bitplane). The parameters used for each type of analysis are mentioned below.
Microglia counting
To count the number of microglia per unit area (mm2), the ‘Spots’ function was used. To further identify subsets of cells that co-label with another marker (proliferating cells, apoptotic cells, etc.), the spots were filtered using ‘mean intensities’ of the fluorescence of the marker. To quantify the area of the region of analysis, ‘Surface’ function was used.
Movement of microglial processes
To assess the speed of the microglial processes moving towards the site of focused laser injury (see experiment above), at first, the site of injury was identified with ‘Surfaces’ to set it as the point of reference. This was followed by performing the following Imaris Xtension functions: ‘Time subtract average’ to exclude the stationary site of injury from the subsequent movement/tracking calculations, and ‘Drift correction’ to correct for any artifacts caused by the movement of the test subject (owing to breathing, etc.) during image acquisition. To follow the motion of the microglial processes, the microglia were identified with ‘Spots’ (the bulbous ends of the processes). Xtension ‘Autoregressive motion’ identified the movements (tracks) of these chosen processes through the entire time period. Following that, running the Xtension ‘Distance transformation’ on the site of injury surface finally produced the numerical values of the different parameters, such as speed and distance, that were used for the analysis of the movement of the microglial processes.
Synaptic pruning analysis
First, each microglial cell surface/cellular volume was isolated using ‘Surface’ function, and the CTB-A647 puncta were identified using ‘Spots’ function. The numbers of CTB-A647 spots that co-localize with the microglia were quantified using the Imaris XTension ‘Split into surface objects’.
Statistical analysis
Unless otherwise stated, all results are reported as mean±s.e.m. and statistical tests were deemed significant when P<0.05. Statistical calculations (Student’s t-test) were performed with GraphPad Prism 6.0 (GraphPad Software).
Supplementary Material
Acknowledgements
We thank our tissue culture team, S. Barnett and C. Lenz; animal husbandry support team, K. Lustig, J. Wangerin, R. Beglarian, R. Focht, K. Prettyman, J. Hayes, M. Rudd and K. Smith-Fry; K. Gilleese for the preparation of the manuscript; and all members of the Capecchi laboratory. Sequencing was performed at the High Throughput Genomics Core Facility, University of Utah. We thank M. Vetter, Department of Neurobiology and Anatomy, University of Utah, for providing Cx3cr1GFP and Csf1r−/− mice; B. Barres, Stanford School of Medicine, for providing the anti-Tmem119 primary antibody; and O. Butovsky, Center of Neurologic Diseases, Harvard Medical School, for providing the 4D4 primary antibody.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.D., D.V.D., A.B., S.T., M.R.C.; Methodology: S.D., D.V.D., E.P., A.B., S.T., M.R.C.; Software: S.D., D.V.D., M.H.; Validation: S.D., D.V.D., E.P., M.H.; Formal analysis: S.D., D.V.D., M.H.; Investigation: S.D., D.V.D., E.P.; Resources: S.D., D.V.D., E.P.; Data curation: S.D., M.H.; Writing - original draft: S.D., D.V.D., M.H., M.R.C.; Writing - review & editing: S.D., D.V.D., A.B., S.T., M.R.C.; Visualization: S.D., M.R.C.; Supervision: A.B., S.T., M.R.C.; Project administration: S.T., M.R.C.; Funding acquisition: M.R.C.
Funding
This work was supported by the Foundation for the National Institutes of Health [R01 MH093595 to M.R.C.], the National Cancer Institute [5P30CA042014-24], the National Center for Research Resources [1S10RR026802-01] and the University of Utah Flow Cytometry Facility. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.152306.supplemental
References
- Ashwood P., Wills S. and Van de Water J. (2006). The immune response in autism: a new frontier for autism research. J. Leukoc. Biol. 80, 1-15. 10.1189/jlb.1205707 [DOI] [PubMed] [Google Scholar]
- Bennett M. L., Bennett F. C., Liddelow S. A., Ajami B., Zamanian J. L., Fernhoff N. B., Mulinyawe S. B., Bohlen C. J., Adil A., Tucker A. et al. (2016). New tools for studying microglia in the mouse and human CNS. Proc. Natl. Acad. Sci. USA 113, E1738-E1746. 10.1073/pnas.1525528113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttgereit A., Lelios I., Yu X., Vrohlings M., Krakoski N. R., Gautier E. L., Nishinakamura R., Becher B. and Greter M. (2016). Sall1 is a transcriptional regulator defining microglia identity and function. Nat. Immunol. 17, 1397-1406. 10.1038/ni.3585 [DOI] [PubMed] [Google Scholar]
- Chen S.-K., Tvrdik P., Peden E., Cho S., Wu S., Spangrude G. and Capecchi M. R. (2010). Hematopoietic origin of pathological grooming in Hoxb8 mutant mice. Cell 141, 775-785. 10.1016/j.cell.2010.03.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalmau I., Vela J. M., González B., Finsen B. and Castellano B. (2003). Dynamics of microglia in the developing rat brain. J. Comp. Neurol. 458, 144-157. 10.1002/cne.10572 [DOI] [PubMed] [Google Scholar]
- da Rocha F. F., Correa H. and Teixeira A. L. (2008). Obsessive-compulsive disorder and immunology: a review. Prog. Neuropsychopharmacol. Biol. Psychiatry 32, 1139-1146. 10.1016/j.pnpbp.2007.12.026 [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M. and Gingeras T. R. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15-21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erblich B., Zhu L., Etgen A. M., Dobrenis K. and Pollard J. W. (2011). Absence of colony stimulation factor-1 receptor results in loss of microglia, disrupted brain development and olfactory deficits. PLoS ONE 6, e26317 10.1371/journal.pone.0026317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferkowicz M. J., Starr M., Xie X., Li W., Johnson S. A., Shelley W. C., Morrison P. R. and Yoder M. C. (2003). CD41 expression defines the onset of primitive and definitive hematopoiesis in the murine embryo. Development 130, 4393-4403. 10.1242/dev.00632 [DOI] [PubMed] [Google Scholar]
- Ginhoux F., Greter M., Leboeuf M., Nandi S., See P., Gokhan S., Mehler M. F., Conway S. J., Ng L. G., Stanley E. R. et al. (2010). Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science 330, 841-845. 10.1126/science.1194637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Perdiguero E., Klapproth K., Schulz C., Busch K., Azzoni E., Crozet L., Garner H., Trouillet C., de Bruijn M. F., Geissmann F. et al. (2015). Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518, 547-551. 10.1038/nature13989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybiel A. M. and Rauch S. L. (2000). Toward a neurobiology of obsessive-compulsive disorder. Neuron 28, 343-347. 10.1016/S0896-6273(00)00113-6 [DOI] [PubMed] [Google Scholar]
- Hoeffel G., Chen J., Lavin Y., Low D., Almeida F. F., See P., Beaudin A. E., Lum J., Low I., Forsberg E. C. et al. (2015). C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 42, 665-678. 10.1016/j.immuni.2015.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege J. C., de Graaff W., Hossaini M., Cardona Cano S., Jaarsma D., van den Akker E. and Deschamps J. (2008). Loss of Hoxb8 alters spinal dorsal laminae and sensory responses in mice. Proc. Natl. Acad. Sci. USA 105, 6338-6343. 10.1073/pnas.0802176105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hounie A. G., Cappi C., Cordeiro Q., Sampaio A. S., Moraes I., Rosário M. C., Palácios S. A., Goldberg A. C., Vallada H. P., Machado-Lima A. et al. (2008). TNF-alpha polymorphisms are associated with obsessive-compulsive disorder. Neurosci. Lett. 442, 86-90. 10.1016/j.neulet.2008.07.022 [DOI] [PubMed] [Google Scholar]
- Hua J. Y. and Smith S. J. (2004). Neural activity and the dynamics of central nervous system development. Nat. Neurosci. 7, 327-332. 10.1038/nn1218 [DOI] [PubMed] [Google Scholar]
- Inlay M. A., Serwold T., Mosley A., Fathman J. W., Dimov I. K., Seita J. and Weismann I. L. (2014). Identification of multipotent progenitors that emerge prior to hematopoietic stem cells in embryonic development. Stem Cell Reports 2, 457-472. 10.1016/j.stemcr.2014.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Schizophrenia Consortium, Purcell S. M., Wray N. R., Stone J. L., Visscher P. M., O'Donovan M. C., Sullivan P. F. and Sklar P. (2009). Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S., Aliberti J., Graemmel P., Sunshine M. J., Kreutzberg G. W., Sher A. and Littman D. R. (2000). Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell. Biol. 20, 4106-4114. 10.1128/MCB.20.11.4106-4114.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L. C. and Shatz C. J. (1996). Synaptic activity and the construction of cortical circuits. Science 274, 1133-1138. 10.1126/science.274.5290.1133 [DOI] [PubMed] [Google Scholar]
- Kettenmann H., Kirchhoff F. and Verkhratsky A. (2013). Microglia: new roles for the synaptic stripper. Neuron 77, 10-18. 10.1016/j.neuron.2012.12.023 [DOI] [PubMed] [Google Scholar]
- Kierdorf K., Erny D., Goldmann T., Sander V., Schulz C., Perdiguero E. G., Wieghofer P., Heinrich A., Riemke P., Hölscher C. et al. (2013). Microglia emerge from erythromyeloid precursors via Pu.1- and Irf8-dependent pathways. Nat. Neurosci. 16, 273-280. 10.1038/nn.3318 [DOI] [PubMed] [Google Scholar]
- Kronfol Z. and Remick D. G. (2000). Cytokines and the brain: implications for clinical psychiatry. Am. J. Psychiatry 157, 683-694. 10.1176/appi.ajp.157.5.683 [DOI] [PubMed] [Google Scholar]
- Lang U. E., Puls I., Müller D. J., Strutz-Seebohm N. and Gallinat J. (2007). Molecular mechanisms of schizophrenia. Cell. Physiol. Biochem. 20, 687-702. 10.1159/000110430 [DOI] [PubMed] [Google Scholar]
- Leonard B. E. and Myint A. (2009). The psychoneuroimmunology of depression. Hum. Psychopharmacol. 24, 165-175. 10.1002/hup.1011 [DOI] [PubMed] [Google Scholar]
- Li Y., Du X.-F., Liu C.-S., Wen Z.-L. and Du J.-L. (2012). Reciprocal regulation between resting microglial dynamics and neuronal activity in vivo. Dev. Cell 23, 1189-1202. 10.1016/j.devcel.2012.10.027 [DOI] [PubMed] [Google Scholar]
- Madisen L., Zwingman T. A., Sunkin S. M., Oh S. W., Zariwala H. A., Gu H., Ng L. L., Palmiter R. D., Hawrylycz M. J., Jones A. R. et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 13, 133-140. 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markarian Y., Larson M. J., Aldea M. A., Baldwin S. A., Good D., Berkeljon A., Murphy T. K., Storch E. A. and McKay D. (2010). Multiple pathways to functional impairment in obsessive-compulsive disorder. Clin. Psychol. Rev. 30, 78-88. 10.1016/j.cpr.2009.09.005 [DOI] [PubMed] [Google Scholar]
- McGrath K. E., Frame J. M., Fegan K. H., Bowen J. R., Conway S. J., Catherman S. C., Kingsley P. D., Koniski A. D. and Palis J. (2015). Distinct sources of hematopoietic progenitors emerge before HSCs and provide functional blood cells in the mammalian embryo. Cell Rep. 11, 1892-1904. 10.1016/j.celrep.2015.05.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkola H. K. A., Fujiwara Y., Schlaeger T. M., Traver D. and Orkin S. H. (2003). Expression of CD41 marks the initiation of definitive hematopoiesis in the mouse embryo. Blood 101, 508-516. 10.1182/blood-2002-06-1699 [DOI] [PubMed] [Google Scholar]
- Mildner A., Schmidt H., Nitsche M., Merkler D., Hanisch U.-K., Mack M., Heikenwalder M., Brück W., Priller J. and Prinz M. (2007). Microglia in the adult brain arise from Ly-6ChiCCR2+ monocytes only under defined host conditions. Nat. Neurosci. 10, 1544-1553. 10.1038/nn2015 [DOI] [PubMed] [Google Scholar]
- Nagarajan N., Jones B. W., West P. J., Marc R. E. and Capecchi M. R. (2017). Corticostriatal circuit defects in Hoxb8 mutant mice. Mol. Psychiatry, 10.1038/mp.2017.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn A., Kirchhoff F. and Helmchen F. (2005). Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science 308, 1314-1318. 10.1126/science.1110647 [DOI] [PubMed] [Google Scholar]
- Paolicelli R. C., Bolasco G., Pagani F., Maggi L., Scianni M., Panzanelli P., Giustetto M., Ferreira T. A., Guiducci E., Dumas L. et al. (2011). Synaptic pruning by microglia is necessary for normal brain development. Science 333, 1456-1458. 10.1126/science.1202529 [DOI] [PubMed] [Google Scholar]
- Pozner A., Xu B., Palumbos S., Gee J. M., Tvrdik P. and Capecchi M. R. (2015). Intracellular calcium dynamics in cortical microglia responding to focal laser injury in the PC::G5-tdT reporter mouse. Front. Mol. Neurosci. 8, 12 10.3389/fnmol.2015.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz M. and Priller J. (2014). Microglia and brain macrophages in the molecular age: from origin to neuropsychiatric disease. Nat. Rev. Neurosci. 15, 300-312. 10.1038/nrn3722 [DOI] [PubMed] [Google Scholar]
- Sanes J. R. and Lichtman J. W. (1999). Development of the vertebrate neuromuscular junction. Annu. Rev. Neurosci. 22, 389-442. 10.1146/annurev.neuro.22.1.389 [DOI] [PubMed] [Google Scholar]
- Schafer D. P., Lehrman E. K., Kautzman A. G., Koyama R., Mardinly A. R., Yamasaki R., Ransohoff R. M., Greenberg M. E., Barres B. A. and Stevens B. (2012). Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691-705. 10.1016/j.neuron.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer D. P., Lehrman E. K. and Stevens B. (2013). The “quad-partite” synapse: microglia-synapse interactions in the developing and mature CNS. Glia 61, 24-36. 10.1002/glia.22389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz C., Gomez Perdiguero E., Chorro L., Szabo-Rogers H., Cagnard N., Kierdorf K., Prinz M., Wu B., Jacobsen S. E., Pollard J. W. et al. (2012). A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science 336, 86-90. 10.1126/science.1219179 [DOI] [PubMed] [Google Scholar]
- Sheng J., Ruedl C. and Karjalainen K. (2015). Most tissue-resident macrophages except microglia are derived from fetal hematopoietic stem cells. Immunity 43, 382-393. 10.1016/j.immuni.2015.07.016 [DOI] [PubMed] [Google Scholar]
- Shi J., Levinson D. F., Duan J., Sanders A. R., Zheng Y., Pe'er I., Dudbridge F., Holmans P. A., Whittemore A. S., Mowry B. J. et al. (2009). Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature 460, 753-757. 10.1038/nature08192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H., Ophoff R. A., Steinberg S., Andreassen O. A., Cichon S., Rujescu D., Werge T., Pietilainen O. P., Mors O., Mortensen P. B. et al. (2009). Common variants conferring risk of schizophrenia. Nature 460, 744-747. 10.1038/nature08186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan A. H., Barres B. A. and Stevens B. (2012). The complement system: an unexpected role in synaptic pruning during development and disease. Annu. Rev. Neurosci. 35, 369-389. 10.1146/annurev-neuro-061010-113810 [DOI] [PubMed] [Google Scholar]
- Strous R. D. and Shoenfeld Y. (2006). Schizophrenia, autoimmunity and immune system dysregulation: a comprehensive model updated and revisited. J. Autoimmun. 27, 71-80. 10.1016/j.jaut.2006.07.006 [DOI] [PubMed] [Google Scholar]
- Tremblay M.-E., Stevens B., Sierra A., Wake H., Bessis A. and Nimmerjahn A. (2011). The role of microglia in the healthy brain. J. Neurosci. 31, 16064-16069. 10.1523/JNEUROSCI.4158-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhu L., He S., Wu Y., Jin W., Yu T., Qu J. Y. and Wen Z. (2015). Temporal-spatial resolution fate mapping reveals distinct origins for embryonic and adult microglia in zebrafish. Dev. Cell 34, 632-641. 10.1016/j.devcel.2015.08.018 [DOI] [PubMed] [Google Scholar]
- Zhan Y., Paolicelli R. C., Sforazzini F., Weinhard L., Bolasco G., Pagani F., Vyssotski A. L., Bifone A., Gozzi A., Ragozzino D. et al. (2014). Deficient neuron-microglia signaling results in impaired functional brain connectivity and social behavior. Nat. Neurosci. 17, 400-406. 10.1038/nn.3641 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.