Abstract
Cities and adjacent regions represent foci of intense human activity and provide unique opportunities for studying human-mediated dispersal and gene flow. We examined the effect of landscape features on gene flow in the invasive grass Brachypodium sylvaticum across an urban–rural interface at the edge of its expanding range. We used genome-wide single-nucleotide polymorphism surveys of individuals from 22 locations. Resistance surfaces were created for each landscape feature, using ResistanceGA to optimize resistance parameters. Our Structure analysis identified three distinct clusters, and diversity analyses support the existence of at least three local introductions. Multiple regression on distance matrices showed no evidence that development, roads, canopy cover or agriculture had a significant influence on genetic distance in B. sylvaticum. Geographical distance was a mediocre predictor of genetic distance and reflected geographical clustering. The model of rivers acting as a conduit explained a large portion of variation in genetic distance, but the lack of evidence of directional gene flow eliminated hydrochory as a dispersal mechanism. These results and observations of the distribution of populations in disturbed sites indicate that the influence of rivers on patterns of dispersal of B. sylvaticum probably reflects seed dispersal due to human recreational activity.
Keywords: biological invasions, directed dispersal, human-mediated dispersal, landscape genetics, isolation by resistance
1. Introduction
Urbanization generates a novel set of problems for species undergoing range expansion. While the creation of vast tracts of developed land can act as barriers to traditional modes of dispersal, the movement of large numbers of humans through these spaces may allow weedy and invasive species to increase their dispersal through these landscapes. The potential for human-mediated dispersal of introduced plants is particularly high in cities and adjacent rural areas where human activity tends to be concentrated. While there has been much speculation about the role of humans in the spread of weedy plants [1], there is a general lack of information on the importance of human-mediated seed dispersal for the spread of invasive plants (e.g. [2,3]). While several studies have provided evidence that humans are responsible for long-distance dispersal and introduction to novel regions [4,5], there is much less information available on the role of humans for post-introduction range expansion and success of invasive plants in the introduced range.
Range expansion of invasive species provides natural experiments for studying the impact of landscape features as they affect the behaviour of vectors to influence patterns of gene flow and dispersal. Landscape genetic analysis of invading species also provides opportunities for the study of patterns of colonization and ongoing gene flow and range expansion, and to evaluate the relative importance of different dispersal vectors. In most native species, historical range expansion, contraction and ongoing meta-population processes can complicate interpretation of genetic relatedness among populations [6]. By contrast, population differentiation in newly invasive species undergoing range expansion primarily reflects diversity of the initial colonization events [7]. However, recurrent gene flow will generate patterns of genetic similarity even among recently colonized sites [8], allowing identification of landscape features influencing dispersal pathways. While all species have expanded their ranges at some point in the past, this expansion is more evident in newly invasive species. Species that are invading new ranges exhibit genetic signatures of range expansion and present an opportunity to evaluate colonization processes characterizing patterns of range expansion and dispersal.
Invasive plants are particularly tractable for studying the impact of landscape features on patterns of gene flow, as their stationary nature simplifies population delineation and geographical sampling. Previous studies indicate that long-distance dispersal is necessary to account for the rate of historical range expansions and is disproportionately important for the establishment of new populations [9–11]. However, long-distance dispersal often results in severe genetic bottlenecks because newly established populations are isolated and established by few propagules [12]. For self-compatible species, just one individual is required to establish a new population (Baker's law [1]), creating an extreme bottleneck. After establishment, subsequent gene flow among populations can boost genetic diversity, alleviating initial fitness reductions from low genetic diversity [13–15].
Human activities often facilitate the spread of invasive species, particularly by increasing their dispersal frequency [16]. Movement of invasive propagules by humans directs dispersal towards human-disturbed areas, where heightened propagule pressure allows invasive species to become established [17–20]. If humans are moving plant propagules, we may also expect long-distance dispersal events to be more common, while also directing dispersal towards areas dominated by human activity. Directed dispersal of plant propagules to disturbed areas should reinforce the effects of human-mediated dispersal even further [21,22].
If humans and other vectors moved propagules without respect to landscape features, then we would expect to see a direct correlation between increasing geographical distance and increasing genetic distance, termed isolation by distance (IBD) [23]. IBD has been found in a wide range of plant species [24,25]; however, landscape features including human modifications such as roads, agriculture and urban developments can influence the movement of dispersal vectors of pollen and seeds, resulting in a more complex relationship between genetic distance and geographical distance. The effects of landscape features can be accounted using estimates of isolation by resistance (IBR), which uses geographical information system (GIS) mapping to assign relative resistances and estimates the overall landscape connectivity using cost-weighted movement [26,27]. Commute distance uses an algorithm similar to Circuitscape to calculate resistance values across a landscape for all pairs of populations [28]. These pairwise landscape resistance values can be compared to pairwise genetic distances to identify which landscape features best predict the patterns of genetic diversity seen in the species. Patterns of genetic distance can then be used to determine which dispersal agents cause a landscape feature to act as a conduit or a barrier to gene flow among populations.
We used the recently introduced invasive grass Brachypodium sylvaticum (Huds.) Beauv. (Poaceae; slender false brome) to study potential impacts of land use and landscape features on gene flow at the edge of an expanding range. Brachypodium sylvaticum is a diploid, self-compatible perennial bunchgrass that is wind pollinated and is native throughout Europe, North Africa and Asia [29]. Brachypodium sylvaticum has been naturalized in the wild in North America for approximately 80 years and is actively expanding its range from introduction points in Oregon to neighbouring regions of the Pacific Northwest of North America [30,31]. In addition, smaller invasions that may represent independent introductions from Europe have been found in California, Virginia and New York [32]. Long-distance spread of B. sylvaticum in its invasive range in Oregon is suspected to be linked to movement of logging equipment [33]. As its range has expanded, B. sylvaticum has encountered increasingly urbanized areas, which may have influenced its rates of range expansion and patterns of dispersal. Brachypodium sylvaticum seeds possess a barbed awn which may predispose the species to animal-mediated seed dispersal [34]. While B. sylvaticum is dispersed primarily by large ungulates in its native range [35], its occurrence along roadsides and waterways has brought into question whether its dispersal in the invasive range is linked to human use of these features [36–38].
The relatively short history of B. sylvaticum in its invasive range coupled with its active expansion presents the opportunity to study interactions between newly established populations and landscape features in human-dominated areas. Here, we investigate B. sylvaticum gene flow to understand how both man-made and natural landscape features influence and potentially facilitate dispersal at the range edge in the Clackamas Watershed within the Portland Metro Region (Portland, OR, USA). We ask (i) what is the invasion history in this watershed, (ii) what landscape features have influenced gene flow among invasive populations, and (iii) if there is evidence for human involvement in this movement. We expect that populations in this region will display characteristics of recent range expansion including populations differing in genetic diversity that reflects their age since colonization [38]. Our analyses provide an assessment of the primary vectors responsible and the importance of urbanization for patterns of range expansion and gene flow in this newly invasive grass.
2. Material and methods
(a). Sampling
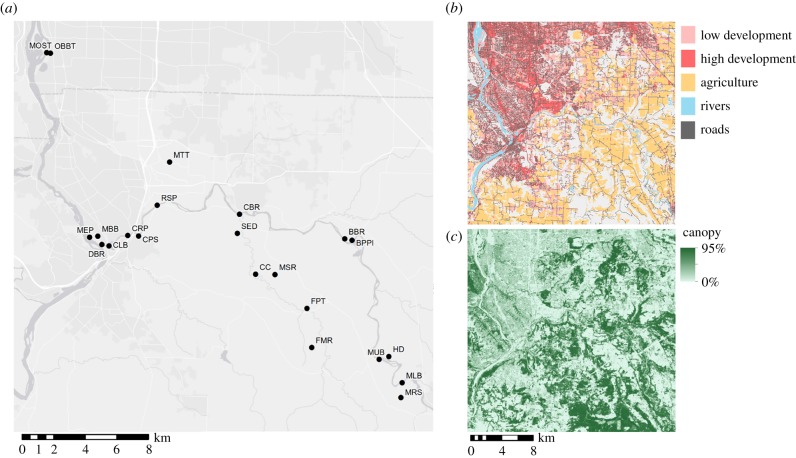
A total of 22 B. sylvaticum locations were sampled to span the diversity of landscape features represented in the Clackamas Watershed in the Portland, Oregon, metropolitan region (figure 1). The Clackamas Watershed is at the leading edge of the expanding range of B. sylvaticum and encompasses a diversity of land use types, varying from heavily developed urban areas to natural secondary growth evergreen forests. At each sampling location, we collected approximately 3 cm of leaf tissue from 3 to 12 individuals for a total of 176 individuals (an average of eight per population; electronic supplementary material, table S1). Individual plants were sampled at a minimum of 1 m apart to prevent double sampling of the same genet, as it was often difficult to delineate individuals growing in dense monocultures. This distance was considered adequate because B. sylvaticum individuals form dense clumps of tillers that are rarely larger than 25 cm in diameter.
Figure 1.
Geographical distribution of (a) B. sylvaticum sampling locations, (b) land use and (c) canopy cover. Sampling locations were chosen to span an urban-to-rural (northwest to southeast) gradient in the Clackamas watershed.
Leaf tissue was stored at −4°C for up to 24 h prior to extraction of total genomic DNA using DNeasy Plant 96 Kits (Qiagen, Leusden, The Netherlands). We used genotyping-by-sequencing (GBS) to concentrate sequencing around restriction enzyme cut sites, allowing genome-wide discovery of single-nucleotide polymorphisms (SNPs) while maintaining sufficient read depth and consistency across samples for genotyping [39]. GBS libraries were constructed using the ApeKI (GCWGC) restriction enzyme by the Cornell University Institute of Biotechnology Genomics Facility. Samples were sequenced on Illumina HiSeq 2500, yielding 100 bp single-end reads.
(b). Bioinformatics
Sequence data were filtered for quality and analysed for accurate genotyping using a reduced-representation mock reference to identify reliable variants. The use of a mock reference was necessary due to a lack of a full genome reference for B. sylvaticum. We used the GBS–SNP–CROP pipeline for quality control, generation of the reference and variant calling [40]. Illumina Truseq 3 SE adapters were removed using IlluminaClip in Trimmomatic 0.35 [41] with a clip threshold of 10 and allowing up to 2 bp mismatches between the read and the adapter. Reads were trimmed when their average quality score dropped below 30 in a 4 bp sliding window; leading and trailing bases were trimmed if their quality score fell below 30. Any reads below 32 bp in length following trimming were considered too short for analysis and were discarded. A reduced-representation mock reference of consensus sequences for regions directly surrounding GBS cut sites was generated using USEARCH with a nucleotide identity cut-off of 93% [42]. Trimmed reads from all samples were aligned to the mock reference using BWA [43]. Unmapped reads and reads with poor (less than 30) alignment quality were removed using SAMtools view [44]. Variant SNPs were then called using SAMtools mpileup with a minimum mapping quality of 30, the coefficient for downgrading the quality score of reads with excessive mismatches set to 50 and with probabilistic realignment of base alignment quality disabled [44].
Variants were filtered following recommendations for diploid data in the GBS–SNP–CROP pipeline [40]. SNPs were retained if found in at least 75% of individuals with an average depth of 10–200 reads, and with a minimum of nine independent reads in at least three individuals supporting existence of the alternate allele. Non-biallelic variants were excluded from analysis by removing SNPs with a proportion of alternate reads to other non-primary below 0.92. An individual was called homozygous at a locus when there were at least five supporting reads if there were no reads showing the alternate allele; when there was one alternate read, a minimum of 20 primary allele reads were required to call the individual homozygous. A minimum of five alternate reads were required to call an individual heterozygous, with a minimum ratio of primary reads to alternate reads of 0.3. SNPs with a proportion of heterozygous individuals greater than 0.5 or a minor allele frequency below 0.05 were removed from analysis, as these probably resulted from misalignment or collapse of homologous regions in the mock reference and do not represent a true variant. A total of 2178 SNPs remained following filtering and were used in subsequent analysis.
(c). Population structure
Population statistics including the observed heterozygosity (Ho), expected heterozygosity (Hs; a measure of genetic diversity) and the inbreeding coefficient (Fis; calculated as the average difference between the observed and expected heterozygosities) were calculated using Hierfstat in R [45]. The number of alleles exhibiting significant interallelic linkage disequilibrium (%LD) was found using Adegenet in R. We conducted a Structure analysis to characterize the number of subpopulations (K) in the watershed and to assess the amount of recent admixture among sampling locations [46]. Structure v. 2.3.4 was run with a burn-in of 50 000 and a run of 100 000 for values of K ranging from 1 to 10; 10 replicates were run for each value of K and the optimal K was found using the Evanno method [47].
(d). Landscape features
We compiled GIS layers representing landscape features likely to influence the behaviour of dispersal vectors and converted them into hypothetical landscapes in which features resisted or conducted gene flow. Roads were represented by the National Transportation Dataset (NTD) [48]. Polylines in the NTD were converted to rasters using ArcMap's Polygon to Raster tool (ESRI, Redlands, CA, USA). The National Land Cover Dataset (NCLD) [49] represented land use and major water features in the study area (figure 1b). Cells in NCLD identified as pasture, hay and cultivated crops in NCLD were combined into one class representing agricultural activity. Developed areas were classified based on the degree of impervious surface cover, with greater than 50% impervious surfaces classified as highly developed (e.g. apartment complexes, commercial/industrial areas and dense single family homes) and less than 50% classified as low development (golf courses, large-lot single family homes and recreational areas). Finally, we used the NCLD Tree Cover Analytical raster to represent percentage canopy cover in 30 m2 areas (figure 1c). All layers were represented at a 30 m resolution and clipped to a 2 km buffer surrounding the study locations to avoid edge effects in resistance modelling.
(e). Statistics
Data were analysed to evaluate the relationship between our hypothetical resistance landscapes and genetic distance. We calculated Nei's genetic distance using Adegenet in R [50,51]. Optimal resistance values for landscape layers were found using ResistanceGA in R [26]. ResistanceGA iteratively varies the resistance of features and finds the overall resistance between populations using commuteDistance in the gdistance package [28]. ResistanceGA then evaluates the feature's performance using a regression against genetic distance, choosing the resistance that maximizes the Akaike information criterion. Results from optimization with commute distance have been found to be equivalent to Circuitscape analysis, yet ResistanceGA optimization with commute distance is approximately two times faster than with Circuitscape [52]. Resistance surfaces with categorical identification of landscape features (rivers, roads, agriculture, natural areas, and high and low development) were prepared for input into ResistanceGA by setting cells representing the landscape feature of interest to 10 and all other cells to 1. Canopy was quantified on a gradient and was input directly into ResistanceGA without modification of cell values. ResistanceGA was run with 10 000 steps and a convergence p-value of 0.05 for each landscape feature.
As landscape features were optimized separately, substantial collinearity in resistance distances due to spatial autocorrelation between different landscape models was expected. To control for this collinearity, we evaluated model significance using the multiple regression on distance matrices (MRM) function in the Ecodist package in R [53]. This allowed us to partition variation in genetic distance among the different models and to remove non-significant features from analysis using a backwards selection model until only significant landscape models remained. Variance inflation (VIF) was calculated using fmsb in R for each MRM model to determine the degree of collinearity between features in the model [54]. Our initial model included all landscape resistance models, and we then used backwards model selection to remove non-significant features.
3. Results
(a). Invasion history
Patterns of diversity in range-edge populations of B. sylvaticum were consistent with expectations in a recently colonized area. Individual populations varied from very low genetic diversity (CPS, HS = 0.046) to very high diversity to the point of heterozygosity excess (CC, HS = 0.311; electronic supplementary material, table S1). A previously published study on B. sylvaticum populations established ranges of genetic diversity associated with different stages of invasion, inferred from population genetic parameters such as genetic diversity (Hs) [38]. Overall genetic diversity in our study area (Hs = 0.182) fell within the moderate age range described previously for B. sylvaticum (Hs 0.1–0.25). Five sampling locations fell within the highest age rank described by Ramakrishnan et al. [38] (Hs > 0.25), indicating that these are probably the oldest populations sampled as they have had the opportunity to accumulate genetic diversity, and potentially represent points of local introduction. Three of these high-diversity locations (CBR, CC and RSP) are clustered in the centre of the study area near Clear Creek, one is located at the northwest edge (MOST), while one (MLB) is at the southeast edge in McIver State Park.
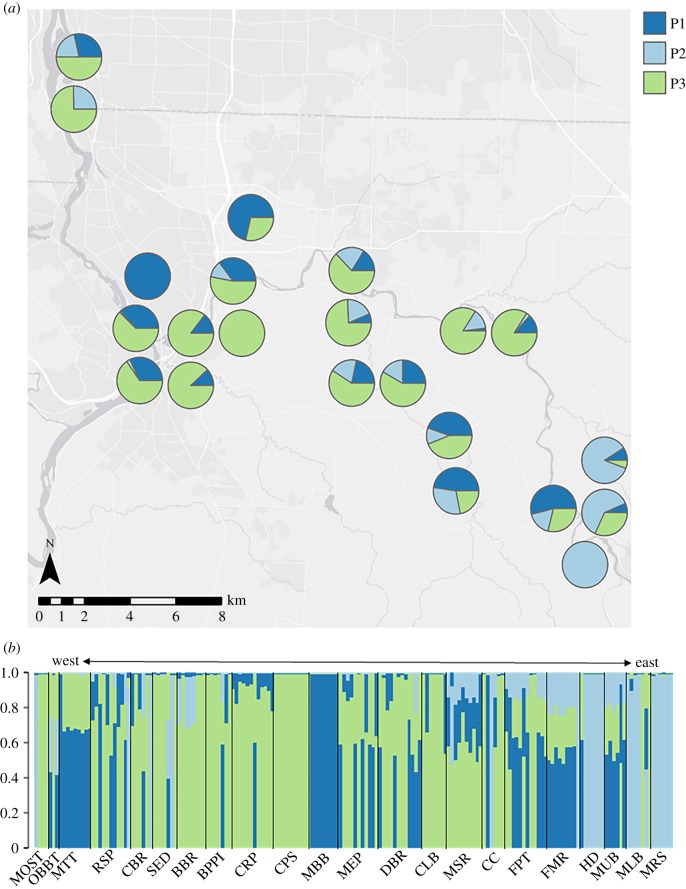
Structure analysis identified K = 3 as the optimum number of clusters in the study area (electronic supplementary material, figure S2). A large number of individuals were assigned to one cluster by Structure; however, the majority of sampling locations were composed of individuals with mixed assignment probabilities (figure 2b). Only three locations were assigned entirely to one cluster (CPS, MBB and MRS; figure 2b); all three had very low genetic diversity and low interallelic genetic disequilibria (Hs < 0.1, %LD < 0.05; electronic supplementary material, table S1). Low genetic diversity and disequilibrium indicate that these locations were recently established from a single source [38]. Sampling locations at the southeast edge of the watershed were assigned primarily to cluster 1 in Structure analysis (figure 2a), which is largely absent from the rest of the area, while the three central and one northwestern sampling locations identified as potential introduction points are primarily composed of Structure clusters 1 and 3 (CBR, CC, RSP and MOST; figure 2b). From this and our population genetic analysis, we can infer multiple introductions into the Clackamas watershed, at least one in the centre of the study area (sampling locations CBR, CC, MOST and RSP) and a second in McIver State Park (location MLB).
Figure 2.
(a) Sampling location cluster composition in relation to geography of the study area and (b) individual assignment probabilities from Structure arranged by sampling location.
(b). Isolation by resistance
Most landscape features were assigned lower resistance values than the surrounding areas (matrix resistance greater than 1) by ResistanceGA, suggesting that these features are potentially acting as conduits to gene flow (feature resistance = 1; table 1). Conduit features include rivers, both high and low levels of development, and roads. Agriculture was assigned a feature resistance value higher than surrounding areas (matrix resistance = 1), indicating that it may act as a barrier to gene flow (table 1). Original values for per cent canopy cover were found by ResistanceGA to have an optimal relationship with genetic distance after undergoing an inverse Ricker transformation to their final resistance values, resulting in a high level of resistance in areas of high and low levels of canopy cover (electronic supplementary material, figure S2).
Table 1.
Backwards selection results for IBR models optimized using ResistanceGA. All feature layers were analysed with the natural log of geographical distance as a covariable in MRM to correct for IBD. Most features acted as conduits for dispersal (feature resistance = 1) and had lower resistance values for the surrounding landscape (matrix resistance), while agriculture had a high feature resistance, indicating that it acted as a dispersal barrier. Significant features in italics.
| landscape feature | feature resistance | matrix resistance | initial model |
final model |
||||
|---|---|---|---|---|---|---|---|---|
| coefficient | probability | VIF | coefficient | probability | VIF | |||
| geographical distance | n.a. | n.a. | −0.058 | 0.030 | 4.77 | −0.035 | 0.075 | 2.376 |
| rivers | 1 | 72.05 | 3.65 × 10−5 | 0.019 | 4.97 | 3.88 × 10−05 | <0.001 | 2.376 |
| development (high) | 1 | 97.40 | 1.63 × 10−5 | 0.067 | 11.90 | — | — | — |
| roads | 1 | 48.73 | −8.87 × 10−6 | 0.195 | 2.44 | — | — | — |
| agriculture | 271.15 | 1 | −1.32 × 10−5 | 0.185 | 6.40 | — | — | — |
| development (low) | 1 | 57.48 | −1.38 × 10−7 | 0.972 | 6.88 | — | — | — |
| canopy covera | n.a. | n.a. | −5.90 × 10−8 | 0.987 | 5.49 | — | — | — |
aCanopy cover was a continuous surface with an inverse Ricker relationship to optimize resistance cell values (see electronic supplementary material, figure S2).
Optimized landscape models representing the impact of development, roads, canopy cover and agriculture all were non-significant in our initial model (table 1). Only geographical distance and rivers acting as a conduit were significant and carried forward into the final model. Rivers as a conduit continued to have a significant relationship with genetic distance in the final model (coefficient = 3.88 × 10−5, p < 0.001), while geographical distance dropped to marginal significance (p = 0.075; table 1). While it would be possible to run an additional MRM demonstrating the impact of rivers alone, keeping geographical distance in the model allows us to control for the impact of spatial autocorrelation on these results. The optimization of resistance values with ResistanceGA on this landscape assigned rivers a resistance value approximately 60 times lower than surrounding areas and the final model explained a large portion of variation in genetic distance (table 1; R2 = 0.373, p < 0.001).
To determine if gene flow along rivers occurs primarily in the direction of water flow or bidirectionally, we regressed the genetic diversity (Hs) of sampling locations within 1 km of the Clackamas River against their distance along the river from farthest upstream location. If gene flow was moving in the direction of river flow, then we would expect to see a gradient of decreasing genetic diversity in populations close to the water moving down river. There was no significant relationship between distance along the river and genetic diversity (R2 < 0.001, p = 0.912; electronic supplementary material, figure S3), indicating that gene flow along rivers is bidirectional. We conducted MRM with distance along the river as a predictor of genetic distance and found a significant relationship (R2 = 0.155, p = 0.005), indicating the presence of IBD along rivers in addition to IBR with rivers acting as a conduit.
4. Discussion
Our investigation of the history of invasion of B. sylvaticum in the Portland metropolitan area revealed a history of multiple introductions into the Clackamas watershed. We also found evidence of extensive ongoing gene flow, represented in a large number of individuals with mixed assignment probabilities across the watershed, as well as sites that appear to have been recently colonized. While analysis of landscape features identified rivers as the single most important conduit for dispersal, it is not clear that seed movement along the rivers is due to water dispersal of seeds. Evidence for bidirectional gene flow along rivers indicates that river-mediated dispersal may be more likely to be due to the association between the rivers and human recreation activities, and possibly movement of other animals along riparian corridors.
(a). Invasion history
Five areas were identified as potential local introduction points for B. sylvaticum in the Clackamas watershed: three in the area surrounding Clear Creek (CC, CBR and RSP), one in the Northwest (MOST) and one in Milo McIver State Park (MRS). The genetic cluster represented in populations near McIver State Park suggests a separate introduction, but the presence of an additional two distinct genetic clusters throughout the study area suggests that more than one introduction occurred elsewhere in the watershed. These introductions and the one at McIver State Park probably originated from the original introduction near Corvallis, Oregon, which came from multiple source locations in Europe [31]. It is possible that independent dispersal events from the relatively high level of genetic diversity present in populations associated with the original introduction near Corvallis are responsible for multiple introductions in the Clackamas watershed at Clear Creek and McIver State Park. Moreover, these sites display evidence of historical logging activity, which has been associated with long-distance dispersal of B. sylvaticum from Oregon State University's McDonald-Dunn Experimental Forest near Corvallis to locations across Oregon [38]. While we cannot infer the exact history of introduction into the Clackamas watershed, we can state that the significant genetic structure found in the watershed is very unlikely to have arisen since local colonization, and therefore there were probably at least three independent colonization events in this region.
Following introduction, additional dispersal into the watershed and subsequent gene flow generated patterns of local genetic similarity. This pattern of multiple introductions via long-distance dispersal at the edge of the range is consistent with results in other invasive species [55–59]. While founder events lower genetic diversity at the range edge, multiple introductions increase diversity and evolutionary potential on the edges of the range [12,60]. Gene flow between the multiple invasive lineages present in the Clackamas watershed has probably alleviated loss of genetic diversity from founder events, potentially providing the diversity needed to continue expanding the range.
(b). Landscape influences on gene flow
While we have found that rivers are significant corridors for gene flow, it is not clear that seed movement along the rivers is due to water dispersal of seeds. There are multiple possible explanations for rivers acting as a conduit for gene flow in invasive B. sylvaticum. First, seeds could be dispersed by the flow of water. Second, it is possible that the gaps created in canopies by rivers act as conduits for pollen dispersal. Finally, recreationalists or deer could be transporting seeds among river access points.
There is no evidence of directional gene flow along the river, as would be expected if seeds were being moved by water [61]. There is also no evidence of decreasing genetic diversity (HS) at sites further downriver, as would expected when seeds are dispersed by water [62]. While this does not completely rule out B. sylvaticum seed movement by water, we can conclude that movement of seeds by river flow is not likely to be the primary dispersal mechanism. Similarly, we see no effect of canopy cover on genetic distance independent of rivers, as would be expected if pollen dispersal by wind in the river's canopy opening over were the primary cause of rivers acting as conduits [63,64]. While pollen movement is very likely to be contributing to the overall high amount of gene flow in the study area, it is unlikely that pollen dispersal above rivers is the cause of rivers acting as a conduit for gene flow.
River-mediated dispersal is more likely to be related to movement along rivers by humans and other animals. Brachypodium sylvaticum seeds possess a barbed awn and are known to disperse via large animals in the native range [35]. White-tailed deer are present in large numbers in the Pacific Northwest and are known to disperse a wide range of both native and invasive plant species [65,66]. Deer have been shown to move preferentially in riparian corridors [67], which could cause deer-mediated dispersal to create the pattern of genetic diversity seen in our landscape analysis. In addition to dispersal, disturbance caused by deer could facilitate the success of B. sylvaticum [68,69]. However, as deer movement is known to respond to suburban land use and other habitat features [70,71], we would expect to see an effect of other land use features on genetic distance if deer were the primary dispersal vectors.
The lack of a clear gradient of genetic diversity could likewise reflect the bidirectional movement of people and their dogs among access points along the river. The primary water feature in the study area is frequented by recreational fishermen, boaters and rafters. Brachypodium sylvaticum is common at river access points throughout the area, often lining boat ramps and informal paths leading to the water. There is ample opportunity for recreationalists to come into contact with and incidentally disperse B. sylvaticum, and dispersal of seeds in shoes and clothing has been demonstrated in other weedy grasses [72,73]. When we consider dogs frequently accompany humans visiting these recreational areas (M.B.C. 2018, personal observation), the opportunity for seed transport becomes very high. Dogs have been shown to be effective dispersers of seeds in forest habitats [74] and are particularly adept at dispersing seeds with morphological features associated with epizoochory grown on tall inflorescences such as B. sylvaticum [75]. The prevalence of B. sylvaticum in recreational access points and its relatively low abundance in other areas probably implicates movement of seeds in recreational gear and dog fur as a primary cause of gene flow along rivers.
All known populations of this invasive grass in the Portland metropolitan region are associated with human activity—they are most commonly distributed along roads in pullouts, along hiking trails and at river access points (M.B.C. 2018, personal observation). Given that humans are likely to be contributing to movement of B. sylvaticum propagules, we should expect to see some movement of seeds along roads in addition to along the river. Dispersal of invasive plant seeds by vehicles is known to occur [76–78]. However, a significant effect of roads was not seen in our analysis. An assumption of resistance modelling is that genetic distance should increase proportionally with distance through the landscape, with shorter dispersal distances occurring more frequently than long distances. However, if seeds are transported on clothing and pet fur, then we can expect long-distance movement to be more common than short-distance dispersal. It is possible that our analysis is unable to detect seed movement from long-distance dispersal events resulting from movement of people by cars at this scale. A previous study of gene flow in B. sylvaticum found evidence of IBD along one road where plants were distributed continuously along a residential road, but an association with geographical distance was lacking along a highway where populations were mostly limited to roadside parking areas [38]. This pattern is consistent with the distribution of populations in the Clackamas watershed, where populations are mostly limited to roadside areas that offer vehicle access, as well as parks and other locations that have intense human activity.
Newly invasive plants are excellent models for understanding processes affecting range expansion. Our landscape genetic analyses of dispersal in the Clackamas watershed provide evidence that humans both create habitat for B. sylvaticum through disturbance and disperse seeds directly to these disturbed locations. Once introduced into an area, facilitation of B. sylvaticum dispersal by recreation creates a network of invasion foci and creates gene flow among invaded locations. Human involvement in dispersal can drastically increase and direct gene flow of invasive species to suitable habitat, and may have a major influence on the abundance and distribution of alien plant species. We suggest that the dispersal of B. sylvaticum by humans both to new areas and between existing populations will aid the continued range expansion and success of this aggressively invasive species.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
The authors gratefully acknowledge Pamela Thompson for her valuable and constructive criticism during preparation of this manuscript. We also thank Jessica Persinger, Avery Pheil and Cammille Mitchell for their assistance in conducting field and laboratory work.
Data accessibility
Data and scripts for ResistanceGA are archived in Dryad (http://dx.doi.org/10.5061/dryad.99j7793) [79]. Sequencing reads are deposited in NCBI under BioProject number PRJNA476867.
Authors' contributions
T.M.A. conducted molecular laboratory work, bioinformatics, data analysis and drafted the manuscript. G.L.M. conceived of the study, helped conduct molecular laboratory work and conducted fieldwork. M.B.C. oversaw the study and helped draft the manuscript. All authors gave final approval for publication.
Competing interests
We declare we have no competing interests.
Funding
Funding provided by the Portland State Faculty Enhancement Grant and the Forbes-Lea Endowed Fund for Student Research.
References
- 1.Baker HG, Stebbins GL. 1965. The genetics of colonizing species. New York, NY: Academic Press. [Google Scholar]
- 2.Auffret AG, Cousins SAO. 2013. Humans as long-distance dispersers of rural plant communities. PLoS ONE 8, e62763 ( 10.1371/journal.pone.0062763) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taylor K, Brummer T, Taper ML, Wing A, Rew LJ. 2012. Human-mediated long-distance dispersal: an empirical evaluation of seed dispersal by vehicles. Divers. Distrib. 18, 942–951. ( 10.1111/j.1472-4642.2012.00926.x) [DOI] [Google Scholar]
- 4.Von der Lippe M, Kowarik I. 2012. Interactions between propagule pressure and seed traits shape human-mediated seed dispersal along roads. Perspect. Plant Ecol. Evol. Syst. 14, 123–130. ( 10.1016/j.ppees.2011.09.006) [DOI] [Google Scholar]
- 5.Mack RN, Lonsdale WM. 2001. Humans as global plant dispersers: getting more than we bargained for. Bioscience 51, 95 ( 10.1641/0006-3568(2001)051%5B0095:HAGPDG%5D2.0.CO;2) [DOI] [Google Scholar]
- 6.Richardson JL, Brady SP, Wang IJ, Spear SF. 2016. Navigating the pitfalls and promise of landscape genetics. Mol. Ecol. 25, 849–863. ( 10.1111/mec.13527) [DOI] [PubMed] [Google Scholar]
- 7.Lee CE. 2002. Evolutionary genetics of invasive species. Trends Ecol. Evol. 17, 386–391. ( 10.1016/S0169-5347(02)02554-5) [DOI] [Google Scholar]
- 8.Dlugosch KM, Parker IM. 2008. Founding events in species invasions: genetic variation, adaptive evolution, and the role of multiple introductions. Mol. Ecol. 17, 431–449. ( 10.1111/j.1365-294X.2007.03538.x) [DOI] [PubMed] [Google Scholar]
- 9.Levin SA, Muller-Landau HC, Nathan R, Chave J. 2003. The ecology and evolution of seed dispersal: a theoretical perspective. Annu. Rev. Ecol. Evol. Syst. 34, 575–604. ( 10.1146/annurev.ecolsys.34.011802.132428) [DOI] [Google Scholar]
- 10.Clark JS, et al. 1998. Reid's paradox of plant rapid migration. BioScience 48, 13–24. [Google Scholar]
- 11.Hewitt GM. 1999. Postglacial re-colonisation of European biota. Biol. J. Linn. Soc. 68, 87–112. ( 10.1111/j.1095-8312.1999.tb01160.x) [DOI] [Google Scholar]
- 12.Bialozyt R, Ziegenhagen B, Petit RJ. 2006. Contrasting effects of long distance seed dispersal on genetic diversity during range expansion. J. Evol. Biol. 19, 12–20. ( 10.1111/j.1420-9101.2005.00995.x) [DOI] [PubMed] [Google Scholar]
- 13.Courchamp F, Clutton-Brock T, Grenfell B. 1999. Inverse density dependence and the Allee effect. Trends Ecol. Evol. 14, 405–410. ( 10.1016/S0169-5347(99)01683-3) [DOI] [PubMed] [Google Scholar]
- 14.Marchini GL, Sherlock NC, Ramakrishnan AP, Rosenthal DM, Cruzan MB. 2016. Rapid purging of genetic load in a metapopulation and consequences for range expansion in an invasive plant. Biol. Invasions 18, 183–196. ( 10.1007/s10530-015-1001-5) [DOI] [Google Scholar]
- 15.Frankham R. 2015. Genetic rescue of small inbred populations: meta-analysis reveals large and consistent benefits of gene flow. Mol. Ecol. 24, 2610–2618. ( 10.1111/mec.13139) [DOI] [PubMed] [Google Scholar]
- 16.Mack RN, Simberloff D, Lonsdale WM, Evans H, Clout M, Bazzaz FA. 2000. Biotic invasions: causes, epidemiology, global consequences, and control. Ecol. Appl. 10, 689–710. ( 10.1890/1051-0761(2000)010%5B0689:BICEGC%5D2.0.CO;2) [DOI] [Google Scholar]
- 17.Lockwood JL, Cassey P, Blackburn T. 2005. The role of propagule pressure in explaining species invasions. Trends Ecol. Evol. 20, 223–228. ( 10.1016/j.tree.2005.02.004) [DOI] [PubMed] [Google Scholar]
- 18.Colautti RI, Grigorovich IA, MacIsaac HJ. 2006. Propagule pressure: a null model for biological invasions. Biol. Invasions 8, 1023–1037. ( 10.1007/s10530-005-3735-y) [DOI] [Google Scholar]
- 19.Simberloff D. 2009. The role of propagule pressure in biological invasions. Annu. Rev. Ecol. Evol. Syst. 40, 81–102. ( 10.1146/annurev.ecolsys.110308.120304) [DOI] [Google Scholar]
- 20.Taylor LAV, Cruzan MB. 2015. Propagule pressure and disturbance drive the invasion of perennial false-brome (Brachypodium sylvaticum). Invasive Plant Sci. Manag. 8, 169–180. ( 10.1614/IPSM-D-14-00042.1) [DOI] [Google Scholar]
- 21.Howe HE, Smallwood J. 1982. Ecology of seed dispersal. Annu. Rev. Ecol. Syst. 13, 201–228. ( 10.1146/annurev.es.13.110182.001221) [DOI] [Google Scholar]
- 22.Wenny DG. 2001. Advantages of seed dispersal: a re-evaluation of directed dispersal. Evol. Ecol. Res. 3, 51–74. [Google Scholar]
- 23.Wright S. 1943. Isolation by distance. Genetics 28, 114–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jenkins DG, et al. 2010. A meta-analysis of isolation by distance: relic or reference standard for landscape genetics? Ecography 33, 315–320. ( 10.1111/j/1600-0587.2010.06285.x) [DOI] [Google Scholar]
- 25.Sexton JP, Hangartner SB, Hoffmann AA. 2014. Genetic isolation by environment or distance: which pattern of gene flow is most common? Evolution 68, 1–15. ( 10.1111/evo.12258) [DOI] [PubMed] [Google Scholar]
- 26.Peterman WE. 2018. ResistanceGA: an R package for the optimization of resistance surfaces using genetic algorithms. Methods Ecol. Evol. 9, 1638–1647. ( 10.1111/2041-210X.12984) [DOI] [Google Scholar]
- 27.McRae BH. 2006. Isolation by resistance. Evolution 60, 1551–1561. ( 10.1111/j.0014-3820.2006.tb00500.x) [DOI] [PubMed] [Google Scholar]
- 28.van Etten J. 2017. R package gdistance: distances and routes on geographical grids. J. Stat. Softw. 76, 1–21. ( 10.18637/jss.v076.i13) [DOI] [Google Scholar]
- 29.Steinwand MA, Young HA, Bragg JN, Tobias CM, Vogel JP. 2013. Brachypodium sylvaticum, a model for perennial grasses: transformation and inbred line development. PLoS ONE 8, 1–11. ( 10.1371/journal.pone.0075180) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller BM, Aitken RJ, Oldham MJ, Reznicek AA. 2012. Slender false brome (Brachypodium sylvaticum, Poaceae), an invasive grass new to Ontario, Canada. Can. Field Nat. 125, 235–240. ( 10.22621/cfn.v125i3.1226) [DOI] [Google Scholar]
- 31.Rosenthal DM, Ramakrishnan AP, Cruzan MB. 2008. Evidence for multiple sources of invasion and intraspecific hybridization in Brachypodium sylvaticum (Hudson) Beauv. in North America. Mol. Ecol. 17, 4657–4669. ( 10.1111/j.1365-294X.2008.03844.x) [DOI] [PubMed] [Google Scholar]
- 32.Daniel S, Werier D. 2010. Slender false brome (Brachypodium Sylvaticum ssp. Sylvaticum): a new invasive plant in New York. N. Y. Flora Assoc. Q. Newsl. 21, 1–5. [Google Scholar]
- 33.Kaye TN, Blakeley-Smith M. 2006. False-brome (Brachypodium sylvaticum). In Invasive species in the Pacific Northwest (eds Boersma PD, Reichard SE, van Buren AN), pp. 80–81. Seattle, WA: University of Washington Press. [Google Scholar]
- 34.Kulic IM, Mani M, Mohrbach H, Thaokar R, Mahadevan L. 2009. Botanical ratchets. Proc. R. Soc. B 276, 2243–2247. ( 10.1098/rspb.2008.1685) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heinken T, Raudnitschka D. 2002. Do wild ungulates contribute to the dispersal of vascular plants in central European forests by epizoochory? A case study in NE Germany. Forstwissenschaftliches Cent. 121, 179–194. ( 10.1046/j.1439-0337.2002.02029.x) [DOI] [Google Scholar]
- 36.Kaye T.2003. Invasive plant alert: false-brome (Brachypodium sylvaticum). Corvallis, OR: False-Brome Working Group.
- 37.Holmes SE, Roy BA, Reed JP, Johnson BR. 2010. Context-dependent pattern and process: the distribution and competitive dynamics of an invasive grass, Brachypodium sylvaticum. Biol. Invasions 12, 2303–2318. ( 10.1007/s10530-009-9645-7) [DOI] [Google Scholar]
- 38.Ramakrishnan AP, Musial T, Cruzan MB. 2010. Shifting dispersal modes at an expanding species' range margin. Mol. Ecol. 19, 1134–1146. ( 10.1111/j.1365-294X.2010.04543.x) [DOI] [PubMed] [Google Scholar]
- 39.Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE. 2011. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6, 1–10. ( 10.1371/journal.pone.0019379) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Melo ATO, Bartaula R, Hale I. 2016. GBS-SNP-CROP: a reference-optional pipeline for SNP discovery and plant germplasm characterization using variable length, paired-end genotyping-by-sequencing data. BMC Bioinform. 17, 29 ( 10.1186/s12859-016-0879-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. ( 10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. ( 10.1093/bioinformatics/btq461) [DOI] [PubMed] [Google Scholar]
- 43.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. ( 10.1093/bioinformatics/btp324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. ( 10.1093/bioinformatics/btp352) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goudet J. 2005. HIERFSTAT, a package for R to compute and test hierarchical F-statistics. Mol. Ecol. Notes 5, 184–186. ( 10.1111/j.1471-8278.2004.00828.x) [DOI] [Google Scholar]
- 46.Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155, 945–959. ( 10.1111/j.1471-8286.2007.01758.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14, 2611–2620. ( 10.1111/j.1365-294X.2005.02553.x) [DOI] [PubMed] [Google Scholar]
- 48.Witt EC. 2015. Geospatial resources for the geology community: the USGS national map. J. Geol. 123, 283–294. ( 10.1086/682008) [DOI] [Google Scholar]
- 49.Homer CG, et al. 2015. Completion of the 2011 national land cover database for the conterminous United States-representing a decade of land cover change information. Photogramm. Eng. Remote Sens. 81, 345–354. ( 10.14358/PERS.81.5.345) [DOI] [Google Scholar]
- 50.Nei M. 1972. Genetic distance between populations. Am. Nat. 106, 283–292. ( 10.2307/2678832) [DOI] [Google Scholar]
- 51.Jombart T. 2008. Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405. ( 10.1093/bioinformatics/btn129) [DOI] [PubMed] [Google Scholar]
- 52.Marrotte RR, Bowman J. 2017. The relationship between least-cost and resistance distance. PLoS ONE 12, 1–19. ( 10.1371/journal.pone.0174212) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lichstein JW. 2007. Multiple regression on distance matrices: a multivariate spatial analysis tool. Plant Ecol. 188, 117–131. ( 10.1007/s11258-006-9126-3) [DOI] [Google Scholar]
- 54.Nakazawa M. 2015. fmsb: Functions for medical statistics book with some demographic data. R package version 0.6.0.
- 55.Suarez AV, Holway DA, Case TJ. 2001. Patterns of spread in biological invasions dominated by long-distance jump dispersal: insights from Argentine ants. Proc. Natl Acad. Sci. USA 98, 1095–1100. ( 10.1073/pnas.98.3.1095) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kirk H, Paul J, Straka J, Freeland JR. 2011. Long-distance dispersal and high genetic diversity are implicated in the invasive spread of the common reed, Phragmites australis (Poaceae), in northeastern North America. Am. J. Bot. 98, 1180–1190. ( 10.3732/ajb.1000278) [DOI] [PubMed] [Google Scholar]
- 57.Parisod C, Bonvin G. 2008. Fine-scale genetic structure and marginal processes in an expanding population of Biscutella laevigata L. (Brassicaceae). Heredity 101, 536–542. ( 10.1038/hdy.2008.95) [DOI] [PubMed] [Google Scholar]
- 58.Genton BJ, Shykoff JA, Giraud T. 2005. High genetic diversity in French invasive populations of common ragweed, Ambrosia artemisiifolia, as a result of multiple sources of introduction. Mol. Ecol. 14, 4275–4285. ( 10.1111/j.1365-294X.2005.02750.x) [DOI] [PubMed] [Google Scholar]
- 59.Petit RJ, Pineau E, Demesure B, Bacilieri R, Ducousso A, Kremer A. 1997. Chloroplast DNA footprints of postglacial recolonization by oaks. Proc. Natl Acad. Sci. USA 94, 9996–10 001. ( 10.1073/pnas.94.18.9996) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berthouly-Salazar C, Hui C, Blackburn TM, Gaboriaud C, Van Rensburg BJ, Van Vuuren BJ, Le Roux JJ. 2013. Long-distance dispersal maximizes evolutionary potential during rapid geographic range expansion. Mol. Ecol. 22, 5793–5804. ( 10.1111/mec.12538) [DOI] [PubMed] [Google Scholar]
- 61.Miller NP, Matlack GR. 2010. Population expansion in an invasive grass, Microstegium vimineum: a test of the channelled diffusion model. Divers. Distrib. 16, 816–826. ( 10.1111/j.1472-4642.2010.00690.x) [DOI] [Google Scholar]
- 62.Nilsson C, Brown RL, Jansson R, Merritt DM. 2010. The role of hydrochory in structuring riparian and Wetland vegetation. Biol. Rev. 85, 837–858. ( 10.1111/j.1469-185X.2010.00129.x) [DOI] [PubMed] [Google Scholar]
- 63.Young A, Boyle T, Brown T. 1996. The population genetic consequences of habitat fragmentation for plants. Trends Ecol. Evol. 11, 413–418. ( 10.1016/0169-5347(96)10045-8) [DOI] [PubMed] [Google Scholar]
- 64.Imbert E, Lefèvre F. 2003. Dispersal and gene flow of Populus nigra (Salicaceae) along a dynamic river system. J. Ecol. 91, 447–456. ( 10.1046/j.1365-2745.2003.00772.x) [DOI] [Google Scholar]
- 65.Myers JA, Vellend M, Gardescu S, Marks PL. 2004. Seed dispersal by white-tailed deer: implications for long-distance dispersal, invasion, and migration of plants in eastern North America. Oecologia 139, 35–44. ( 10.1007/s00442-003-1474-2) [DOI] [PubMed] [Google Scholar]
- 66.Vellend M, Myers JA, Gardescu S, Marks PL. 2003. Dispersal of Trillium seeds by deer: implications for long-distance migration of forest herbs. Ecology 84, 1067–1072. ( 10.1890/0012-9658(2003)084%5B1067:DOTSBD%5D2.0.CO;2) [DOI] [Google Scholar]
- 67.Finder RA, Roseberry JL, Woolf A. 1999. Site and landscape conditions at white-tailed deer/vehicle collision locations in Illinois. Landsc. Urban Plan. 44, 77–85. ( 10.1016/S0169-2046(99)00006-7) [DOI] [Google Scholar]
- 68.Knight TM, Dunn JL, Smith LA, Davis J, Kalisz S. 2009. Deer facilitate invasive plant success in a Pennsylvania forest understory. Nat. Areas J. 29, 110–116. ( 10.3375/043.029.0202) [DOI] [Google Scholar]
- 69.Taylor LAV, Hasenkopf EA, Cruzan MB. 2015. Barriers to invasive infilling by Brachypodium sylvaticum in Pacific Northwest forests. Biol. Invasions 17, 2247–2260. ( 10.1007/s10530-015-0871-x) [DOI] [Google Scholar]
- 70.Kilpatrick H, Spohr S. 2000. Spatial and temporal use of a suburban landscape by female white-tailed deer. Wildl. Soc. Bull. 28, 1023–1029. [Google Scholar]
- 71.Felix AB, Walsh DP, Hughey BD, Campa H, Winsterstein SR. 2007. Applying landscape-scale habitat-potential models to understand deer spatial structure and movement patterns. J. Wildl. Manage. 71, 804–810. ( 10.2193/2006-366) [DOI] [Google Scholar]
- 72.Ansong M, Pickering C. 2014. Weed seeds on clothing: a global review. J. Environ. Manage. 144, 203–211. ( 10.1016/j.jenvman.2014.05.026) [DOI] [PubMed] [Google Scholar]
- 73.Vibrans H. 1999. Epianthropochory in Mexican weed communities. Am. J. Bot. 86, 476–481. ( 10.2307/2656808) [DOI] [PubMed] [Google Scholar]
- 74.Heinken T. 2000. Diepersal of plants by a dog in a deciduous forest. Bot. Jahrb. Syst. 122, 449–467. [Google Scholar]
- 75.Graae BJ. 2002. The role of epizoochorous seed dispersal of forest plant species in a fragmented landscape. Seed Sci. Res. 12, 113–120. ( 10.1079/SSR2002103) [DOI] [Google Scholar]
- 76.von der Lippe M, Kowarik I. 2006. Do cities export biodiversity? Traffic as dispersal vector across urban-rural gradients. Divers. Distrib. 14, 18–25. ( 10.1111/j.1472-4642.2007.00401.x?2) [DOI] [Google Scholar]
- 77.Zwaenepoel A, Roovers P, Hermy M. 2006. Motor vehicles as vectors of plant species from road verges in a suburban environment. Basic Appl. Ecol. 7, 83–93. ( 10.1016/j.baae.2005.04.003) [DOI] [Google Scholar]
- 78.Veldman JW, Putz FE. 2010. Long-distance dispersal of invasive grasses by logging vehicles in a tropical dry forest. Biotropica 42, 697–703. ( 10.1111/j.1744-7429.2010.00647.x) [DOI] [Google Scholar]
- 79.Arredondo TM, Marchini GL, Cruzan MB. Date from: Evidence for human-mediated range expansion and gene flow in an invasive grass Dryad Digital Repository. ( 10.5061/dryad.99j7793) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Arredondo TM, Marchini GL, Cruzan MB. Date from: Evidence for human-mediated range expansion and gene flow in an invasive grass Dryad Digital Repository. ( 10.5061/dryad.99j7793) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Data and scripts for ResistanceGA are archived in Dryad (http://dx.doi.org/10.5061/dryad.99j7793) [79]. Sequencing reads are deposited in NCBI under BioProject number PRJNA476867.